
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 02 June 2022
Sec. Chronobiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.888262
Ambient temperature varies constantly. However, the period of circadian pacemakers is remarkably stable over a wide-range of ecologically- and physiologically-relevant temperatures, even though the kinetics of most biochemical reactions accelerates as temperature rises. This thermal buffering phenomenon, called temperature compensation, is a critical feature of circadian rhythms, but how it is achieved remains elusive. Here, we uncovered the important role played by the Drosophila PERIOD (PER) phosphodegron in temperature compensation. This phosphorylation hotspot is crucial for PER proteasomal degradation and is the functional homolog of mammalian PER2 S478 phosphodegron, which also impacts temperature compensation. Using CRISPR-Cas9, we introduced a series of mutations that altered three Serines of the PER phosphodegron. While all three Serine to Alanine substitutions lengthened period at all temperatures tested, temperature compensation was differentially affected. S44A and S45A substitutions caused undercompensation, while S47A resulted in overcompensation. These results thus reveal unexpected functional heterogeneity of phosphodegron residues in thermal compensation. Furthermore, mutations impairing phosphorylation of the pers phosphocluster showed undercompensation, consistent with its inhibitory role on S47 phosphorylation. We observed that S47A substitution caused increased accumulation of hyper-phosphorylated PER at warmer temperatures. This finding was corroborated by cell culture assays in which S47A slowed down phosphorylation-dependent PER degradation at high temperatures, causing PER degradation to be excessively temperature-compensated. Thus, our results point to a novel role of the PER phosphodegron in temperature compensation through temperature-dependent modulation of the abundance of hyper-phosphorylated PER. Our work reveals interesting mechanistic convergences and differences between mammalian and Drosophila temperature compensation of the circadian clock.
Circadian clocks are present across almost all life forms and allow them to adapt their activities with daily changes in their surrounding environment. In eukaryotes, the molecular circadian pacemaker is a self-sustained transcriptional feedback loop that repeats itself every ∼24 h, thus producing molecular, physiological and behavioral rhythms that closely match the duration of the Earth’s revolution around its own axis (Dunlap, 1999). In Drosophila, the circadian clock comprises four core components (Hardin, 2011). The transcription factors CLOCK (CLK) and CYCLE (CYC) form a heterodimer that binds to E-box motifs upstream of the period (per) and timeless (tim) genes to promote their transcription. PER and TIM heterodimerize in the cytoplasm before entering the nucleus. Once inside the nucleus, the PER/TIM heterodimer binds to CLK/CYC and thus inhibits per and tim transcription. PER, TIM, and CLK undergo various post-translational modifications that affect the period of the clock, the most notable being the progressive phosphorylation driven by kinases such as DOUBLETIME (DBT) (Kloss et al., 1998; Price et al., 1998), SHAGGY (Martinek et al., 2001), Proline-directed kinases including p38 MAP Kinase (Ko et al., 2010; Dusik et al., 2014), CASEIN KINASE 1 alpha (CK1α) (Lam et al., 2018), CASEIN KINASE 2 (CK2) (Lin et al., 2002; Lin et al., 2005; Akten et al., 2003; Szabó et al., 2013), AMPK (Cho et al., 2019) and NEMO (NMO) (Chiu et al., 2011; Yu et al., 2011).
Circadian clocks exhibit three key properties (Pittendrigh, 1960). First, these clocks free-run with a period of ∼24 h in the absence of external temporal cues (referred to as Zeitgebers) such as the daily light and temperature cycles. Second, these Zeitgebers can reset the clocks, thus anchoring the clock’s phase to the day/night cycle. Third, the period of circadian clocks is temperature-compensated. As ambient temperature increases, most enzymatic reactions speed up. However, under constant conditions, circadian clocks exhibit ∼24 h periodicities over a wide range of physiological temperatures, even in ectotherms. This was first observed by C. Pittendrigh while studying eclosion rhythms of Drosophila pseudoobscura (Pittendrigh, 1954).
The mechanism of temperature compensation is one of the long lasting mysteries of circadian rhythms. Different models have been proposed. For example, the network model proposes that multiple temperature-sensitive reactions cancel each other with changes in temperature. Early studies on Dinoflagellates indeed suggested that two temperature-sensitive reactions with opposite effects would help achieving constant period (Hastings and Sweeney, 1957). At the biochemical level, phosphorylation mediated by CK1 and CK2 has received the most attention across species. Mutations in the β1 and α subunits of CK2 in Neurospora alter temperature compensation (Mehra et al., 2009). Temperature compensation in Arabidopsis is modulated by the inhibitory effect of CK2 phosphorylation on CIRCADIAN ASSOCIATED CLOCK A1 (CCA1) binding. CCA1 binding to its target promoters increases with temperature, which is kept in check by the opposing effect of CK2 phosphorylation (Portolé and Má, 2010). In mammals, phosphorylation of a PER2 peptide by CK1δ is temperature-insensitive, owing to opposite temperature sensitivity of substrate and product binding by this kinase (Isojima et al., 2009; Shinohara et al., 2017). Interestingly, the CK1εTau mutant shows an undercompensation phenotype (Tosini and Menaker, 1998; Zhou et al., 2015). Furthermore, CK1δ/ε activity on PER2 results in a temperature-dependent phosphoswitch involving two phosphorylation sites (Eide et al., 2005; Zhou et al., 2015; Masuda et al., 2020). One of them is Serine (S)-478, also called phosphodegron. S478 phosphorylation causes βTrCP (β-Transducing repeat Containing Protein)-dependent ubiquitination and degradation of mPER2 (Eide et al., 2005; Reischl et al., 2007; Masuda et al., 2020). The second is S659, the mouse homolog to the site of Familial Advanced Sleep Phase Syndrome (FASPS) mutations (Toh et al., 2001). S659 phosphorylation inhibits phosphorylation of the phosphodegron, and thus PER2 degradation (Shanware et al., 2011; Zhou et al., 2015). S478 is preferentially phosphorylated at colder temperatures while FASPS domain phosphorylation increases at warmer temperatures, consequently balancing out PER stability (Zhou et al., 2015; Narasimamurthy et al., 2018). Along with phosphorylation, other processes such as sumoylation in plants (Hansen et al., 2017), nuclear cytoplasmic ratio of PER (Giesecke et al., 2021) and TIM (Singh et al., 2019), PER intermolecular and PER-TIM interactions (Gekakis et al., 1995; Huang et al., 1993, Huang et al., 1995) have been proposed to modulate temperature compensation of circadian rhythms in flies.
Over the past several decades, circadian mutants have fuelled our understanding of the molecular clockwork. Therefore, to gain better understanding into the mechanism of temperature compensation in Drosophila we turned towards Drosophila mutants known to have a temperature compensation defect. Interestingly, of the numerous mutations impacting circadian rhythm period, only a subset of mutants show a defect in temperature compensation (Bao et al., 2001; Ewer et al., 1990; Hamblen et al., 1998; Konopka et al., 1994; Matsumoto et al., 1999; Peixoto et al., 1998; Rothenfluh et al., 2000; Sawyer et al., 1997). We were intrigued by the mutant perSLIH (Some Like It Hot, Figure 1A) due to its striking undercompensation phenotype: circadian period length decreases as temperature increases in this mutant instead of remaining approximately the same. (Hamblen et al., 1998). The perSLIH mutation causes a Serine (S) to Tyrosine (Y) substitution at the 45th residue, which is a part of Drosophila PER’s phosphodegron (S44, 45, and 47) (Chiu et al., 2008). The PER phosphodegron is regulated in a similar manner as its mammalian homolog (Chiu et al., 2011): it is phosphorylated by the CK1δ/ε homolog DBT (Kloss et al., 1998; Price et al., 1998; Chiu et al., 2008), and its phosphorylation is inhibited by the pershort (pers) phosphocluster, which is equivalent to the mammalian FASPS domain (Zhou et al., 2015). Phosphorylation events at the PER phosphodegron, S47 in particular, are critical for Supernumerary limbs (SLIMB)-mediated PER degradation and thus period length (Grima et al., 2002; Ko et al., 2002; Chiu et al., 2008). SLIMB is the fly homolog of βTrCP. Here, we show that, unexpectedly, the residues of the Drosophila phosphodegron are functionally heterogeneous and differentially modulate temperature compensation, even though they all lengthen circadian period. In addition, we found that the pers phosphocluster, which negatively regulates S47 phosphorylation, also regulates temperature compensation. Furthermore, we show that substituting PER’s S47 residue to an Alanine (A) results in an overcompensated circadian period, i.e., circadian period lengthens when ambient temperature increases. This overcompensation phenotype correlates with increased hyper-phosphorylated PER levels in vivo at warm temperature and overcompensated PER degradation in a well-established cell culture model. These results converge to support an important role of the phosphodegron in temperature compensation in Drosophila.
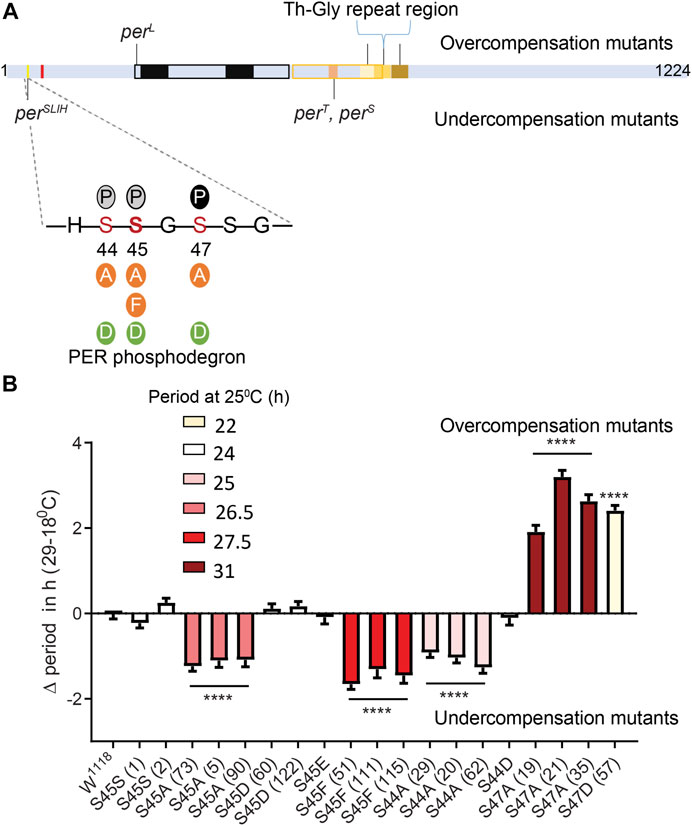
FIGURE 1. Differential modulation of temperature compensation by three Serine residues in the PER phosphodegron. (A) Schematic of PER protein depicting the relative positions of amino acids modified in various mutants affecting temperature compensation in Drosophila. Mutants above the PER schematic are overcompensated while mutants depicted below are undercompensated mutants. The magnified region of PER shows residues of the dPER phosphodegron and the various substitutions (Serine to Alanine, Phenylalanine, Aspartate or Glutamate) we generated using CRISPR/CAS9. Bold S at the 45th residue indicates the position of perSLIH mutation. Solid P within black circle, shows that S47 position undergoes phosphorylation. Grey circles indicate potential additional phosphorylation sites at S44 and S45 (Chiu et al., 2008). Red mark indicate a putative NLS close to the phosphodegron (Chang and Reppert, 2003). (B) Graphical representation of the difference in period values of various mutants of dPER phosphodegron at 29 and 18°C, relative to controls. ΔPeriod values for each genotype were offset by the average difference in period observed at 29 and 18°C in S45SI and S45SII control lines, so that overcompensation or undercompensation compared to control is clearly represented. The color scheme denotes period of each mutant line at 25°C. Interestingly, S44A, S45A, and S47A all produce long period phenotype at 25°C. However, the directionality of their temperature compensation phenotypes is opposite between S44A/S45A and S47A. S44A, and S45A produce significant undercompensation while S47A and S47D shows overcompensation. Statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparison test.
The unique location of the perSLIH mutation (Figure 1A) prompted us to evaluate the role of residues of the PER phosphodegron in temperature compensation. DBT, a Serine/Threonine kinase, phosphorylates S47, with S44 and S45 being additional potential targets (Chiu et al., 2008). To explore the possibility that phosphorylation at these residues modulates temperature compensation, we generated mutant per alleles encoding phosphomimics [S to D (Aspartate) or E (Glutamate)] and phosphoinhibitors [S to Alanine (A)] at PER S44, S45, and S47 residues using CRISPR-CAS9 (Figure 1A). Moreover, we generated a mutant per allele creating a S45F (S to Phenylalanine) substitution to mimic contribution of a bulky hydrophobic amino acid similar to Tyrosine (Y) to validate previous observations on perSLIH. We used S45S as a control, which contains only the silent substitutions introduced at the protospacer adjacent motifs (PAM site) during CRISPR mutagenesis. In our hands white1118 (w1118) and S45S control strains showed a decrease in period of ∼0.9 h (Table 1) over the range of 18–29°C. To accurately represent the effect of mutations on temperature compensation, values plotted on Figure 1B are relative to the average of the two S45S control strains, which was set to 0. Note that an undercompensation phenotype is represented with a minus value, since period shortens as temperature increases, while overcompensation is represented by a positive value.
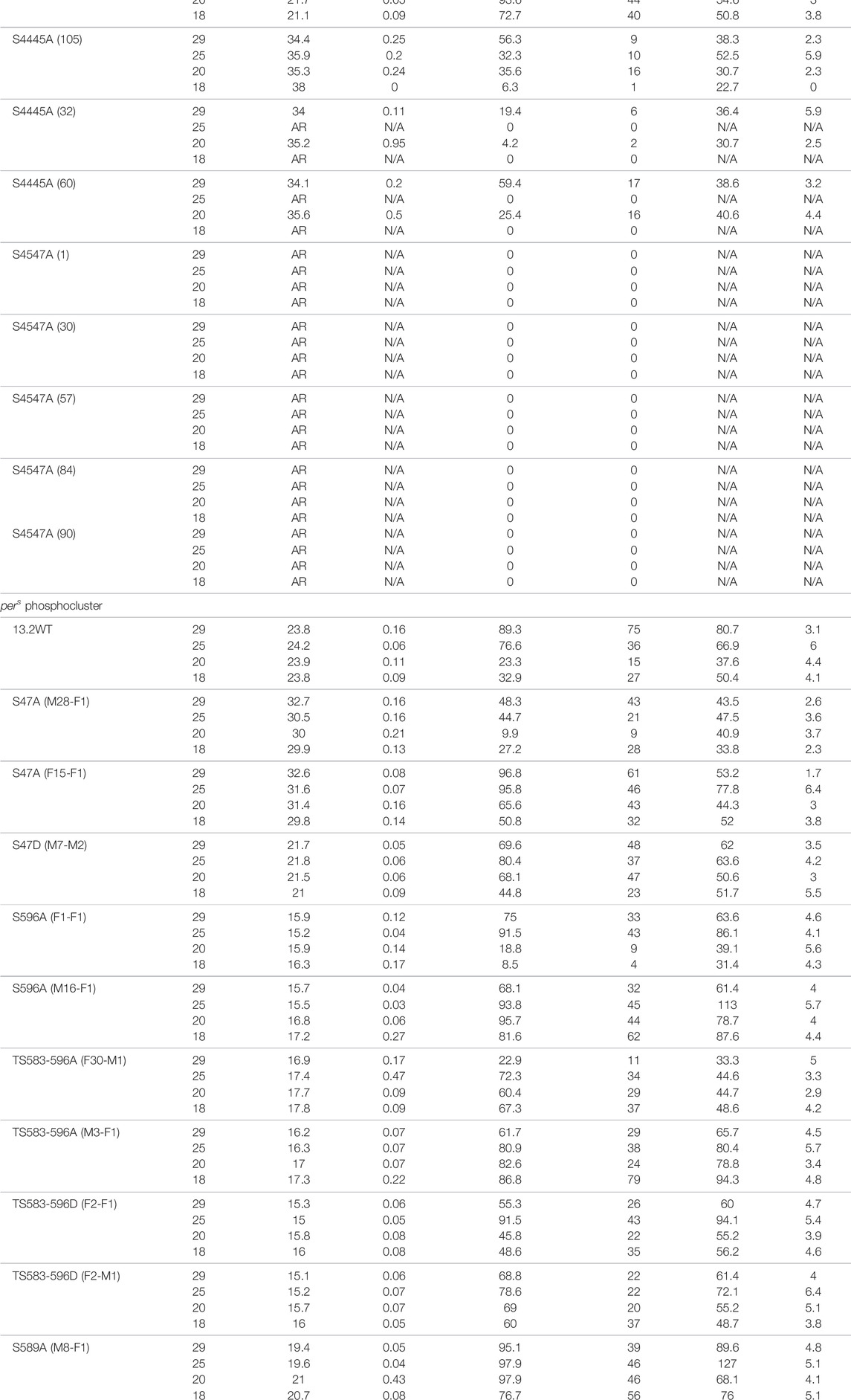
TABLE 1. Circadian behavior of control and per mutant flies under different temperatures. Mutations in the phosphoclusters were generated by CRISPR/Cas9-mediated genome editing. Amino acid substitutions are indicated in the left column. w1118 is a control strain. For the pers phosphocluster, per0 flies were rescued with various transgenes. 13.2WT carries a wild-type per genomic region, while the other transgenes carry the indicated substitutions. AR = arrhythmic ND = Not done, N/A = Not applicable.
Consistent with previous observations (Chiu et al., 2008), we observed that the phosphoinhibitory substitution S47A produced lengthened period. In addition, we found that S44A, S45A, and S45F also resulted in longer periods, albeit to a lesser extent at 25°C (Figure 1B; Table 1). S44A lengthened the period by ∼1.5 h, S45A by 2.5 h, and S47A by 6.5 h at 25°C. Interestingly, the effect of these mutations on temperature compensation did not correlate with period length. S44A and S45 A/F were undercompensated by ∼1–2 h, compared to controls. However, S47A showed an opposite phenotype: it caused a strong overcompensation of more than 2 h (Figure 1B).
Since it is possible that mutations at a residue affect phosphorylation events at the neighboring residues, we generated the double mutants S44-45A and S45-47A. S44-45A double mutants exhibited a very long period (>34 h), but the effect on temperature compensation was difficult to interpret due to poor rhythmicity, particularly at 18°C (Table 1). However, the synergistic effects on period and rhythmicity indicate that S44 and 45 work together to control circadian rhythms. S45–47A mutants were completely arrhythmic at both temperatures tested (Table 1).
We also tested phosphomimic substitutions (S to D/E). S45D and S45E did not show any substantial effect on either period length or temperature compensation (Figure 1; Table 1). S44D lengthened the period to 26.6 h at 25°C, but had no effect on temperature compensation. The behavior of S47D was the most peculiar. It showed a short rhythm period phenotype (>21 h), as previously reported by Chiu et al. (2008) with per genomic rescue constructs in an amorphic per0 background (Konopka and Benzer, 1971). However, period was overcompensated by more than 2 h. This could either be because phosphomimics do not correctly or fully reproduce the structural changes caused by phosphorylation, or because phosphomimics are neither temporally controlled nor reversible and could thus have unexpected impact on temperature compensation.
Thus, taken together our results show that the PER phosphodegron modulates temperature compensation, and shows functional temperature compensation heterogeneity within its residues.
To further strengthen evidence for a role of the PER phosphodegron in temperature compensation, we analyzed the impact of mutations in the pers domain (T/S583-596). Indeed, this region also undergoes phosphorylation by DBT and NMO, and phosphorylation in the pers domain temporally precedes and inhibits phosphorylation at S47 (Chiu et al., 2011). Therefore we tested point mutations in the pers domain for their role in temperature compensation.
We used previously described transgenic lines (Chiu et al., 2011), in which the per0 mutation is rescued with per transgenes carrying various mutations in the pers domain. In addition, we included transgenic flies carrying S47A and S47D substitutions (Chiu et al., 2008). Strikingly, the period length and overcompensation phenotypes observed with transgenes encoding PER S47A were similar to those observed with the corresponding mutants generated with CRISPR-CAS9 (Figures 1, 2), confirming the critical role played by the S47 residue in temperature compensation. For S47D, effect on temperature compensation was weaker with a single mutant transgene than with the CRISPR-Cas9 generated mutant line, and was just above the significance cut-off. Mutants in the pers domain exhibited shortened period length at 25°C [as described previously (Chiu et al., 2011)]. They also showed consistently an undercompensation phenotype of about 1–1.5 h, except for one S596A line (Figure 2B). This could be because of an insertional effect of P-elements, since per transgenes were randomly inserted in the fly genome. Positional effect of transgene insertions might also explain the weaker phenotype observed with the S47D transgene. It should be noted that the perT (G593D substitution) and pers (S589N substitution) mutations have also been reported to be undercompensated (Konopka et al., 1994; Rothenfluh et al., 2000; Bao et al., 2001), and that in a recent study similar results as ours have been obtained with the same transgenic lines (Giesecke et al., 2021). We would also like to point out that TS583-596A and TS583-596D mutations both showed an undercompensation phenotype in line with their short period phenotype. Short period length of TS583-596D mutants was also observed previously (Chiu et al., 2011). As discussed above, the inability of Aspartate to fully mimic a dynamic phosphorylation event might explain the similar phenotypes observed with S to A and S to D substitutions.
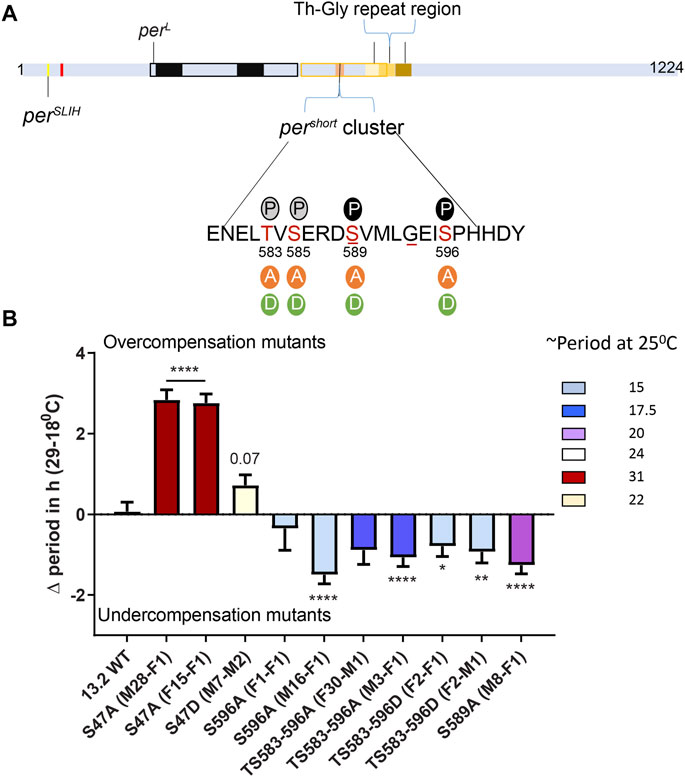
FIGURE 2. The pers phosphocluster impacts temperature compensation. (A) PER schematic depicting the relative position of various mutants affecting temperature compensation, as shown in Figure 1A. The magnified region shows key Serine residues of the pers phosphocluster and the various substitutions that were tested. Solid P within black circle at 589 and 596 shows sites of DBT and NMO mediated phosphorylation, respectively. Grey circles show predicted phosphorylation sites at S585 and S583. Underlines at S589 and G593 denote positions of pers and perT mutant, respectively. (B) Graphical representation of the difference in period of various mutants of the pers domain at 29 and 18°C. The color scheme denotes period of each mutant line at 25°C. All mutants in the pers domain produce short period phenotype at 25°C. Most show significant undercompensation. Transgenic lines with S47A recapitulated our observations with CRISPR-Cas9 genome editing (Figure 1). Transgenic S47D showed a weaker phenotype than genome edited S47D, just short of statistical significance (p = 0.07). Statistical significance was calculated using two-way ANOVA with Sidak’s multiple comparison test.
Taken together, our results and the aforementioned previous and recent reports show that the pers domain is important for temperature compensation. Since S to A/D substitutions in this domain have opposite effects than S47A, and since mutagenesis studies revealed that phosphorylation of the pers domain blocks S47 phosphorylation (Chiu et al., 2011), it seems likely that the pers domain controls temperature compensation through S47 phosphorylation.
The results just described provide strong evidence for a key role of the PER phosphodegron in temperature compensation. To gain further mechanistic insights, we examined PER protein and phosphorylation cycling in two phosphodegron mutants that showed opposite temperature-compensation phenotypes: S45A and S47A. We examined PER rhythms in these mutants at 18°C and 29°C in head extracts. Note that for phosphorylation quantifications, we divided PER signal in three categories, as previously done for CLK (Kim and Edery, 2006; Yu et al., 2006): hypo-phosphorylated (hypo-PER), intermediate (inter-PER) and hyperphosphorylated (hyper-PER) (Figures 3A,D,I). Overall PER phosphorylation rhythms are shown in Figures 3B,E,J, where phosphorylated PER (P-PER) include both inter- and hyper-PER isoforms. We also quantified and compared hyper-PER and hypo-PER signals in Figures 3G,H to determine whether the PER S47A substitution had a specific effect on hyper-phosphorylated PER degradation, given the key role played by the phosphodegron in degrading hyperphosphorylated PER (Chiu et al., 2008), and the functional relevance of hyper-PER on transcriptional repression (Nawathean and Rosbash, 2004; Kivimäe et al., 2008).
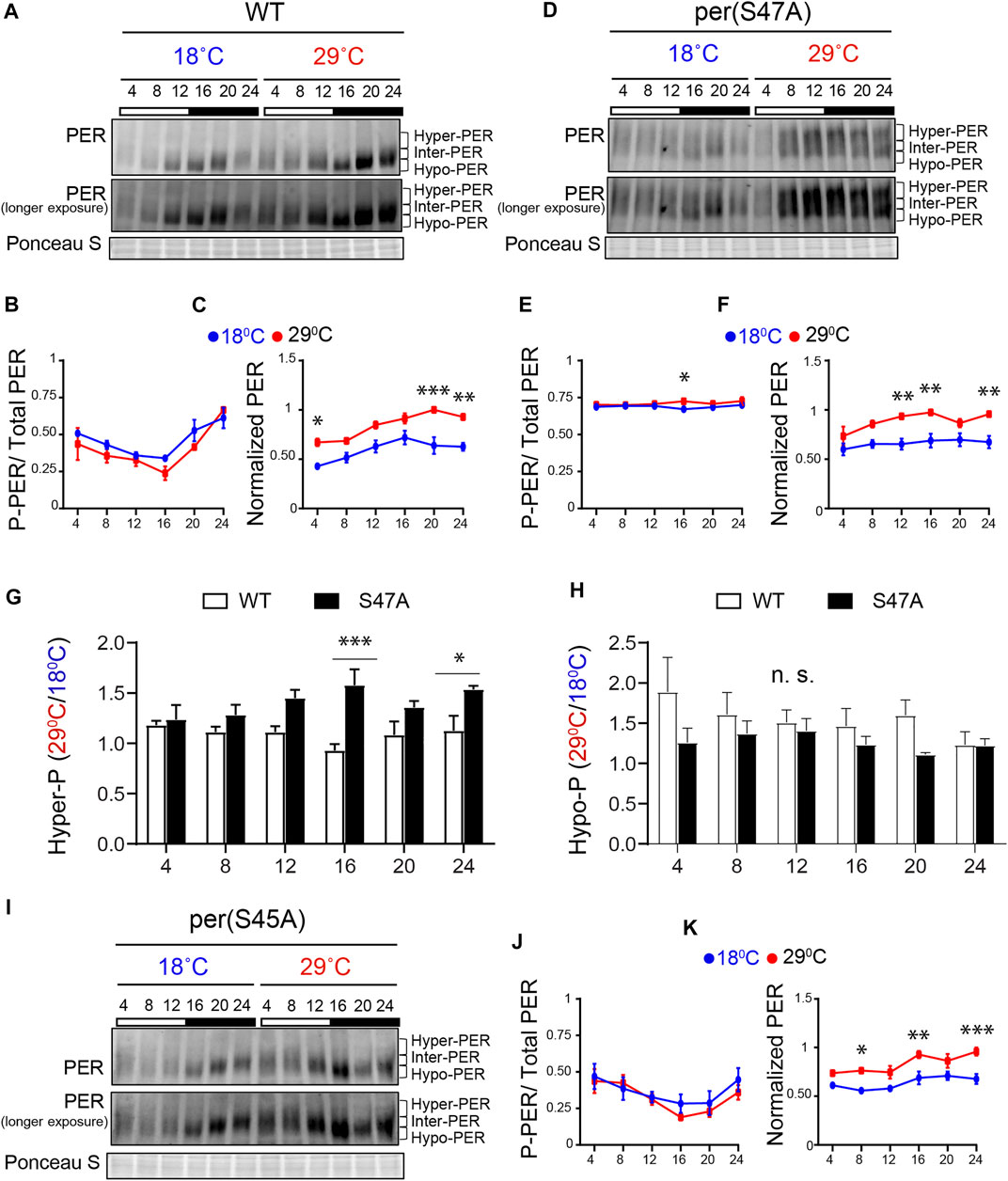
FIGURE 3. S47A mutation causes hyper-phosphorylated PER to accumulate preferentially at high temperature. (A,D,I) Representative western blots probing head extracts from flies entrained to LD at 18 and 29°C. PER abundance was monitored throughout the day at the indicated time points. Respective genotypes are represented at the top of each gel. Brackets indicate hyper-, intermediate- (inter-) and hypo-phosphorylated PER isoforms. Two different exposures of the same blot are shown. Ponceau S was used as a loading control and for normalization. (B,F) and (J,K) Graphs show PER abundance and phosphorylation (inter + hyper) quantifications. In control flies, both PER phosphorylation (B) and abundance (C) were scored as statistically rhythmic at 29 and 18°C, using the JTK_CYCLE. For phosphorylation: p < 0.05, phase = 2 at 18°C and p < 0.001, phase = 2 at 29°C. For abundance: p < 0.05, phase = 18 at 18°C and p < 0.001, phase = 20 at 29°C. For S47A, neither phosphorylation (E) nor abundance (F) were scored as statistically rhythmic, although PER levels were reproducibly lower at ZT4. For S45A (J,K), only PER abundance at 18°C scored as statistically rhythmic. p < 0.05, phase = 18. Red lines represent data at 29°C and blue lines represent data at 18°C. *p < 0.05, **p < 0.01, ***p < 0.001, two-way ANOVA followed by Sidak’s multiple comparison to test for time and temperature-dependent differences (n = 3 biological replicates). (G,H) Ratio of PER levels measured at 29°C vs. 18°C. G shows the ratio for hyper-phosphorylated PER, and H for hypo-phosphorylated PER. S47A accumulates more hyper-phosphorylated PER during the night than control flies at warm temperatures.
Under LD, control (w1118) flies showed rhythmic PER abundance and phosphorylation regardless of the temperature, albeit with significantly higher abundance at 29°C (Figures 3A–C). This observation is in line with increased TIM abundance at warm temperature (Kidd et al., 2015). S47A also showed increased PER abundance at 29°C compared to 18°C, but PER abundance and phosphorylation rhythms were perturbed at both temperature (Figures 3D–F). Importantly, S47A mutants showed exaggerated temperature-sensitive accumulation of hyper-phosphorylated PER compared to controls, which appeared most pronounced at night (see ZT16 and ZT24 in Figure 3G). This was not the case for hypo-phosphorylated PER, which tend to be lower (Figure 3H). Increased accumulation of hyper-phosphorylated PER is likely to result in longer CLK/CYC repression phase, thus lengthening circadian period (Kivimäe et al., 2008) at warmer temperatures. These results therefore reveal a temperature-dependent effect on PER phosphorylation and/or degradation of the S47A substitution, which could underlie the observed overcompensation phenotype.
Though S45A showed a long period and an undercompensation behavior, the effect of S45A mutation on PER cycling was milder than S47A, with patterns of phosphorylation similar to that of WT (Figures 3I–K). However, PER cycling amplitude at 29°C was reduced and did not clear statistical significance. Phosphorylation rhythm amplitude of S45A was reduced at both temperatures as well.
The increase in the accumulation of hyper-phosphorylated PER, observed at high temperature with the S47A substitution, could be the result of abnormal protein stability, possibly due to disrupted phosphorylation. We therefore turned to S2 cell culture assays (Ko et al., 2002) to determine the effect of temperature on PER phosphorylation and degradation kinetics. In these assays, recombinant DBT is expressed under the inducible metallothionine promoter, while PER is expressed constitutively. Since PER is phosphorylated on multiple sites by DBT and thus produces several phosphorylated isoforms with various mobility that are subjected to SLIMB-mediated proteasomal degradation, we quantified the disappearance of the hypo-phosphorylated PER after DBT induction to determine PER phosphorylation kinetics (Figure 4). Interestingly, phosphorylation kinetics of PER S47A was significantly slowed down at low temperature 6 h after DBT induction (Figures 4A,C), while no significant effect was observed with either wild-type PER or S45A at this time point (Figures 4A,B,D). However, it seems unlikely that a slower phosphorylation kinetic at cold temperature would result in the shorter circadian behavior period observed in S47A mutants at 18°C compared to 29°C.
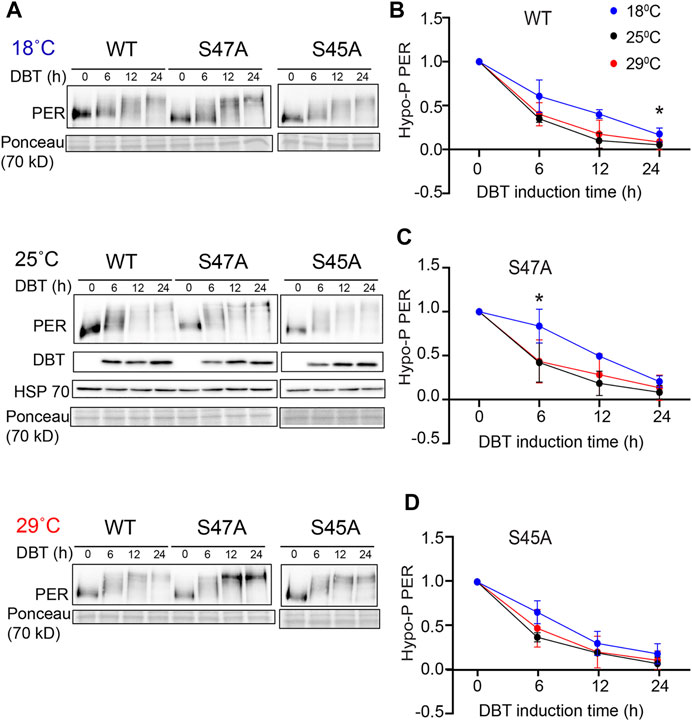
FIGURE 4. S47A phosphorylation kinetics is slowed down at cold temperature. (A) Representative western blots probing cell extracts from S2 cells expressing DBT and either wild-type (WT) PER, S47A or S45A. Cells were incubated at the indicated temperatures, and collected at the indicated time points after DBT induction. Top panel shows PER immunoblotting, bottom panel shows Ponceau S staining, used as loading control and for normalization. (B–D) Western Blot quantifications. PER signal shown at time point 0 is used as a reference to classify PER isoforms as hypo-phosphorylated. Graphs representing the relative amount of hypo-phosphorylated PER at 18, 25, and 29°C for the respective genotypes at different time points after DBT induction. S47A shows increased hypo-phosphorylated PER 6 h after DBT induction.*p < 0.05, 18 compared to 29°C, two-way ANOVA followed by Sidak’s multiple comparison to test for time and temperature-dependent differences (n = 3 biological replicates).
Since S47A caused increased hyper-phosphorylated PER accumulation at 29°C, we also determined S47A’s effects on PER stability. We again induced DBT and after 6 h added cycloheximide to block protein synthesis. To quantify the amount of PER protein more accurately, we deliberately condensed different phosphorylated PER isoforms in this assay by running a higher percentage SDS-PAGE gel. Wild-type PER was more stable than S47A at 18°C, while at 29°C there was no obvious difference (Figures 5B,C). Interestingly, S47A’s degradation kinetics was slowed down at higher temperature, while for WT, it was accelerated (Figures 5A,D,E). Thus, S47A results in a slower degradation of PER at high temperatures, which we propose would contribute to the excessive hyper-phosphorylated PER visible in flies expressing S47A, and thus would cause overcompensation of circadian period (Figure 6).
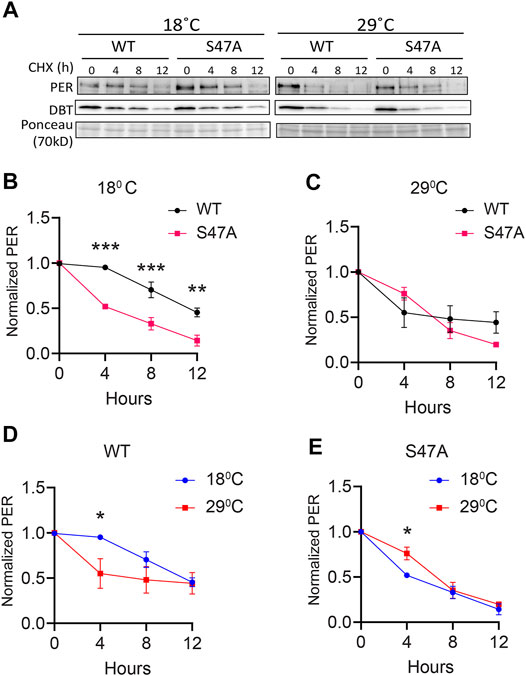
FIGURE 5. S47A degradation kinetics is excessively temperature-compensated. (A) Representative western blots probing cell extracts from S2 cells expressing DBT and either wild-type (WT) PER, S47A or S45A. Cells were collected at the indicated time points after cycloheximide addition at the indicated temperatures. Top panel shows PER immunoblotting, bottom panel shows Ponceau S staining, used as loading control and for normalization. (B–E) Western Blot quantifications. All experiments were performed with three independent replicates. *p < 0.05, **p < 0.01, ***p < 0.001, two-way ANOVA followed by Sidak’s multiple comparison to test for time and temperature-dependent differences. In (B,C), WT and S47A mutants are compared to each other at the indicated temperature. (D,E) show the same data, but degradation kinetics of WT and S47A are compared between high and low temperatures.
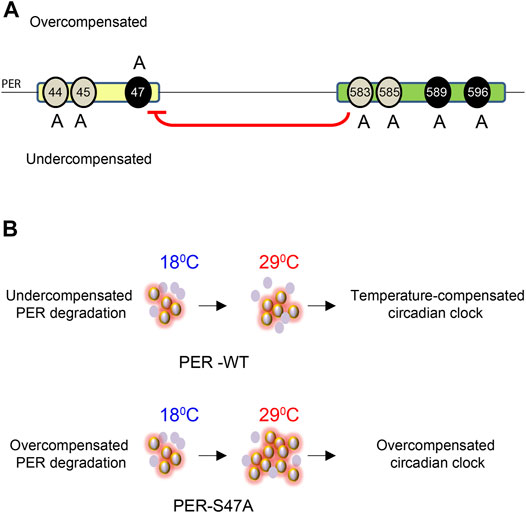
FIGURE 6. Model for the role of PER phosphoclusters in temperature compensation. (A) The PER phosphodegron (yellow) and pers phosphocluster (green) modulate temperature compensation. Mutations of different residues of the PER phosphodegron have opposite effects on temperature compensation. S44A and S45A cause undercompensation while S47A results in overcompensation of circadian period. Mutations in the pers phosphocluster cause undercompensation, consistent with its inhibitory role on S47 phosphorylation (red arrow). Black ovals around 47, 589, and 596 show DBT and NMO mediated phosphorylation sites on PER. Grey ovals around 44, 45, 583, and 585 depict putative additional phosphorylation sites. (B) S47 action on temperature compensation. In wild-type flies, PER abundance and rate of PER degradation increase with increase in temperature. Hyper- and hypo-phosphorylated isoforms remain in balance and the clock is properly temperature-compensated, presumably because of S47 phosphorylation. In S47A mutant flies, PER degradation is overcompensated and thus slows down at high temperature, leading to excessive accumulation of hyperphosphorylated isoforms. This is predicted to prolong repression of CLK/CYC-transcription and thus lengthened period (Kivimäe et al., 2008). The clock is then overcompensated. Violet ovals represent PER protein, Halo around violet ovals show hyper-phosphorylated PER.
Phosphorylation events play an important role in the temperature compensation of the circadian clock across phyla (Tosini and Menaker, 1998; Mehra et al., 2009; Portolé and Má, 2010; Zhou et al., 2015; Masuda et al., 2020). In Drosophila, PER phosphorylation has been studied in detail, and interestingly three per mutations affecting two PER domains undergoing DBT phosphorylation are undercompensated. perSLIH causes the most robust undercompensation phenotype (Hamblen et al., 1998), while pers and perT have milder effects (Ewer et al., 1990; Konopka et al., 1994; Rothenfluh et al., 2000; Bao et al., 2001). We therefore systematically tested phosphorylated residues in both the PER phosphodegron and the pers domain to understand their impact on temperature compensation. Overall, our results point at a central role for residue S47 through temperature-dependent modulation of PER degradation, and a differential, yet to be elucidated, role of S44 and S45. In addition we found that phosphorylated residues in the pers phosphorylation cluster also play a role in temperature compensation, as expected from their ability to inhibit S47 phosphorylation (Chiu et al., 2011).
Indeed, the S47A substitution resulted in a striking overcompensation. This suggests that phosphorylation at this residue is important for the compensation. Actually, we found that PER levels were exaggeratedly temperature-dependent in vivo, with an increase in hyper-phosphorylated PER levels at high temperature. Moreover, PER degradation induced by DBT phosphorylation was slowed down at high temperature by the S47A substitution, as opposed to WT PER in a Drosophila cell culture assay. These results thus suggest that the S47 residue impacts the kinetics of PER degradation in a temperature-dependent manner, protecting degradation from being overcompensated. It should be noted that our molecular observations were made with whole-head extracts (and S2 cells), but rhythmic locomotor behavior is controlled by a small number of circadian pacemaker neurons, the small ventral lateral neurons (sLNvs), that contribute minimally to whole-head protein extracts (Zeng et al., 1994; Renn et al., 1999; Stoleru et al., 2005). Differences have been noted between the molecular clock of these neurons and other circadian neurons and tissues (Lin et al., 2002; Lim et al., 2011; Top et al., 2016; Reviewed in; Top and Young, 2018). We therefore cannot exclude that S47A impacts differently PER rhythms in sLNvs than in other tissues, but we note importantly that DBT-mediated PER phosphorylation and degradation is mechanistically conserved in clock neurons and peripheral tissues, and closely recapitulated in S2 cells (Kloss et al., 1998, 2001; Price et al., 1998; Ko et al., 2002; Chiu et al., 2008, 2011; Garbe et al., 2013).
Surprisingly, our results from cycloheximide chase assays suggest that S47A is less stable than WT at 18°C, while at 29°C we observed a mild increase in stability of S47A compared to WT. These results appear to differ from those reported in Chiu et al. (2008) that indicated a robust stabilization of S47A at 25°C. These discrepancies could be the result of slightly different growth conditions or other changes in the state of our S2 cell cultures, which could affect expression of endogenous proteins important for PER degradation. Nevertheless, since the increase in stability of the S47A protein at 29°C correlates well with the overcompensation of circadian period in S47A flies, our observations in S2 cells provide a coherent mechanistic explanation for S47A’s temperature-compensation phenotype. However, that circadian behavior period is lengthened at all temperatures in S47A mutant flies seems at odd with the kinetics of S47A degradation observed in cell culture, which is accelerated at 18°C or similar to wild-type PER at 29°C (Figures 5B,C). The cycloheximide chase assay we used models DBT-dependent PER degradation in the absence of any feedback such as PER repression of its own transcription. The S47A mutation could thus have a lengthening effect on period independent of its impact on temperature compensation, for example by modulating the ability of PER to function as a transcriptional repressor in vivo. A different mechanism for period determination and temperature compensation by the phosphodegron is in agreement with our observation that S to A substitutions at residues 44, 45 or 47 all cause period lengthening, while the valence of the temperature compensation phenotype is dependent on the specific residue being substituted.
Mutations in the pers phosphocluster provide further support to the key role played by phosphorylation at S47 in temperature compensation. Indeed, mutations in the pers phosphocluster, which inhibit S47 phosphorylation, have opposite effects on temperature compensation than S47A: flies carrying these mutations are undercompensated. Unexpectedly though, the phosphomimetic substitution S47D did not produce an undercompensation phenotype. Rather, an overcompensation phenotype was observed. This could be explained by the substitution to Aspartate not fully mimicking the structure of a phosphorylated Serine, or that constantly mimicking S47 phosphorylation impacts thermal compensation of the clock through a different mechanism than the S47A substitution. Notably it is not uncommon that phosphomimetic mutations do not produce opposite phenotype than phosphoinhibitory mutations, including in circadian proteins (Lin et al., 2005; Chiu et al., 2011; Top et al., 2018; Cai et al., 2021). Thus, S47D could lead to overcompensation via yet unknown, different mechanism. Given the opposite effect of S47A and S47D on SLIMB binding (Chiu et al., 2008) and consequently period length, PER-SLIMB interaction and thus PER degradation is unlikely to be the mechanism mediating S47D temperature overcompensation.
Another unexpected observation is that Serine to Alanine substitution at residues 44 and 45 have opposite effects on temperature compensation than S47A, even though all these substitutions lengthen circadian period. Residues of the phosphodegron are thus functionally heterogeneous when it comes to temperature compensation. Indeed, unlike S47A, S45A did not cause obvious change in temperature-dependent PER stability in vivo or in cell culture. It should be noted that unlike S47, S44 or S45 phosphorylation has not been unambiguously demonstrated (Chiu et al., 2008). It remains possible that these residues are not phosphorylated and play a structural role in the phosphodegron, and this could explain functional diversity in temperature compensation between S44/45 and S47. It is also possible that S44/45’s mechanism of action on period length and/or temperature compensation is independent of S47. Clearly, further studies are needed to elucidate S44 and S45’s exact function in thermal compensation. Of note, the S45F substitution, which introduces a more bulky amino acid than S45A, caused a more severe phenotype that better mimicked the perSLIH phenotype resulting from a S45Y substitution [Figure 1B; Table 1 (Hamblen et al., 1998)]. This could indicate an important structural role for this amino acid, perhaps controlling specific protein-protein interactions not implicated in PER degradation. We note that a putative Nuclear Localization Signal (NLS) is located close to the phosphodegron (Figure 1A, red mark) and perhaps a bulky amino acid at S45 can interfere in a temperature dependent manner with nuclear translocation. However, at least in S2 cells, this NLS has little impact on nuclear localization (Chang and Reppert, 2003).
Interestingly, our work, combined with previous work in mammals, converge to support the model that CKIδ/ε-mediated phosphorylation of PER proteins and their proteasomal degradation are critical for temperature compensation of the circadian clock. Surprisingly, however, there appear to be important mechanistic differences. Indeed, in Drosophila, mutation of the key Serine residue (S47) in the phosphodegron increases temperature compensation, while in mammals mutation of the homologous S478 residue has the opposite effect [Figures 1B, 2B; Table 1 (Zhou et al., 2015; Masuda et al., 2020)]. Intriguingly, in mammalian cell culture assays, phosphodegron mutation (S478A in mammals) causes degradation kinetics to become essentially insensitive to temperature (Zhou et al., 2015), while S47A degradation in flies is mildly overcompensated. However, in mammals, wild-type mPER2 degradation kinetics accelerates when temperature drops, while in flies we observed the opposite effect, perhaps because the phosphodegrons, though conserved, are located in different location in the PER and mPER2 proteins. This explains why mutations that both renders PER degradation less sensitive to temperature have opposite effect on temperature compensation.
Finally, it is important to point out that so far, any mutation or manipulation of phosphorylation of key circadian protein in any system only partially disrupts temperature compensation, or even increases it. It is therefore clear that multiple mechanisms are implicated, and recent results by Giesecke and collaborators (Giesecke et al., 2021) in flies for example point to an important role for nuclear export. It is therefore clear that temperature compensation is proving to be a complex, systemic process, as first proposed by Hastings and Sweeny in their seminal work on Dinoflagellates (Hastings and Sweeney, 1957).
Fly stocks were maintained on standard cornmeal agar at 25°C under 12:12 h light: dark (LD). The following available strains were used w1118, yw, FM7a, w[1118]; PBac[y(+mDint2) = vas-Cas9]VK00027 (BL51324), y[1] M(Act5C-Cas9.P.RFP-)ZH-2A w[1118] DNAlig4[169] (BL58492). Transgenic flies used in Figure 2 were described previously (Chiu et al., 2008, Chiu et al., 2011).
Single stranded oligos (ssODNs, Table 2) were injected into either BL51324 or BL58492 from Bloomington. The injected embryos, upon eclosion, were crossed with FM7a balancer flies. F1 progeny was screened with PCR and sequencing for the presence of the desired modifications. All the resultant lines were backcrossed into the w1118 genetic background. For lines generated from BL58492 (S45S# I, S45S# II, S45D #122, #67, S45A 98, S4445A# 105, S45E, S45F #51, S44D, and S47D), we determined whether the Cas9 transgene located on the X chromosome had been removed. S45E, S44D, and S47D fly lines unexpectedly retained Cas9 even after multiple attempts to remove it. It seems however highly unlikely that Cas9 could somehow interfere with temperature compensation, particularly since in both S45E and S44D temperature compensation was unaffected (Figure 1B). Primers used for molecular screenings are listed in Table 3.
A conveniently located PAM site (TGG) was identified using CRISPR fly design. The guide RNA sequence chosen was—5′AGCCACTGCTGCCGGAGGAGTGG. The guide RNAs were cloned into plasmid pCFD3 (Addgene, Cat no: 49410). Silent modifications (AGC-AGT) were introduced into the seed sequence to prevent re-cutting and for subsequent molecular screening.
2–5 days old male flies were loaded into behavior tubes. Flies were entrained at the defined temperature for at least 3 days under 12:12 LD cycle and released into constant darkness (DD). Activity was recorded using the DAM (Drosophila Activity Monitoring) system (Trikinetics, Waltham, MA, United States) in 136-LL incubators (Percival). Behavior was analyzed and plotted using FaasX software (courtesy of F. Rouyer, Centre National de la Recherche Scientifique, Gif-sur- Yvette, France). Rhythmicity was defined by following the criteria: power >20, width >1.5 h, using the c2 periodogram analysis. 5 days of DD data were used to determine period. Period and power of all individual flies under various conditions can be found in supplemental file Supplementary Figures S1, S2; Supplementary Table S1 —datasets. xls.
Whether difference in period at 29 and 18°C were statistically significant was determined by using a two-way ANOVA followed by Sidak’s multiple comparison tests. Since in Figure 1 control flies showed a significant period shortening at warm temperatures compared to cold, we did factor this drift prior to run statistical test by subtracting the average period lengthening observed in S45SI, and S45SII (1.03 h) from the period observed at 18°C for each genotypes.
For graphical representation in Figures 1, 2, mean difference between 29 and 18°C were plotted on the Y axis and Standard Error (SE) of difference were used for error bars. Mean difference and SE of difference were obtained from the Two-way Anova statistics details is Prism. Western blots were analyzed using Two-way ANOVA with Tukey’s multiple comparison test. Rhythms in PER phosphorylation and abundance in Figure 3 were analyzed by JTK-CYCLE as in (Hughes et al., 2010).
Drosophila S2 cells were maintained at 22°C in Schneider’s Drosophila medium [Life Technologies (Carlsbad, CA, United States)] supplemented with 10% Fetal Bovine Serum (FBS) (VWR, Radnor, PA). Plasmids expressing pAc-per(WT)-V5, pAc-per(S45A)-V5, pAc-per(S47A)-V5, and pMT-dbt-V5 were described previously (Ko et al., 2002; Chiu et al., 2008). For all cell culture experiments, S2 cells were seeded at 1 × 106 cells/ml in a 6-well plate and transfected using Effectene (Qiagen, Germantown, MD, United States). For mobility shift assay (Figure 4), S2 cells were co-transfected with 0.2 μg of pMT-dbt-V5 and 0.8 μg of pAc-per(X)-V5, where X is either WT, S45A or S47A. dbt expression were induced with 500 μM CuSO4 36 h after transfection and cells were moved into incubators with indicated temperature and cells were harvested at 0, 6, 12, 24 h after induction. Proteins were analyzed by Western Blotting. For cycloheximide (CHX) chase experiment (Figure 5), S2 cells were co-transfected with 0.2 μg of pMT-dbt-V5 and 0.8 μg pAc-per(X)-V5, where X is either WT or S47A. dbt expression were induced with 500 μM CuSO4 at 36 h after transfection. 6 h after DBT induction, CHX (Sigma) was added at a final concentration of 10 μg/ml. Cells were harvested and lysed with EB2 (20 mM HEPES pH 7.5, 100 mM KCl, 5% glycerol, 5 mM EDTA, 1 mM DTT, 0.1% Triton X-100, 10 μg/ml Aprotinin, 5 μg/ml Leupeptin, 1 μg/ml Pepstatin A, 0.5 mM PMSF, 25 mM NaF) at the indicated times. Proteins were analyzed by Western Blotting.
Protein extractions from Drosophila S2 cells and adult fly heads, western blotting, and image analysis was performed as previously described (Chiu et al., 2008; Cai et al., 2021). 2–5 day old flies were entrained to 12:12 light-dark cycle for 3 days at indicated temperature. On the 4th day of LD, flies were collected at the indicated time points. ∼40 fly heads for each time point were extracted with RBS buffer (20 mM HEPES pH7.5, 50mM KCl, 10% glycerol, 2 mM EDTA, 1 mM DTT, 1% Triton X-100, 0.4% NP-40, 10 μg/ml Aprotinin, 5 μg/ml Leupeptin, 1 μg/ml Pepstatin, 0.5 mM PMSF, 25 mM NaF). Protein concentration was measured using Pierce Coomassie Plus Assay Reagents (Thermo Fisher Scientific). 2X SDS sample buffer was added and the mixture boiled at 95°C for 5 min. Equal amounts of proteins were resolved by polyacrylamide-SDS gel electrophoresis (PAGE) and transferred to nitrocellulose membrane (Bio-Rad, Hercules, CA, United States) using Semi-Dry Transfer Cell (Bio-Rad). Membranes were stained with Ponceau S solution (0.1% Ponceau S, 5% acetic acid) to stain total proteins as loading control (Chen et al., 2011). Membranes were then incubated in 5% Blocking Buffer (Bio-Rad) for 40 min, incubated with primary antibodies for 16–20 h. Blots were then washed with 1X TBST for 1 h, incubated with secondary antibodies for 1 h, washed again prior to treatment of chemiluminescence Clarity ECL reagent (Bio-Rad). The following percentage of polyacrylamide-SDS gel were used: 6% for PER (Figures 3, 4); 10% for DBT and HSP70.10% gel was used for PER in order to condense the bands for quantification of total PER abundance in CHX chase experiment (Figure 5). For Figures 3B,E,J phospho-PER (intermediate and hyperphosphorylated moieties were quantified as showed in the brackets in Figures 3A,D,I. For Figures 3G,H, only hyper-phosphorylated form of PER was used for analysis. WT, ZT 16 was used to define hypo-phosphorylated form of PER.
Primary antibodies: α-V5 (Thermo Fisher Scientific) at 1:3000 for PER-V5 and DBT-V5, α-PER (GP5620; RRID:AB_2747405) at 1:2000 for PER, α-HSP70 at 1:10000 for HSP70. Secondary antibodies conjugated with HRP were added as follows: α-mouse IgG (Sigma) at 1:2000 for α-V5 detection, 1:10000 for α-HSP70 detection, α-guinea pig IgG (Sigma) at 1:1000 for α-PER detection. All quantifications are included in supplemental file Supplementary Figures S3-S5 - datasets.xlsx.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
RJ, YC, JC, and PE contributed to conception and design of the study. RJ, YC, and YX performed experiments. RJ, YC, JC, and PE analyzed the data. RJ wrote the first draft of the manuscript. RJ, YC, JC, and PE wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by MIRA award 1R35GM118087 from the National Institute of General Medicine Sciences (NIGMS) to PE, and R01 DK124068 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) to JC.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank VInh Phan for technical support, and members of the Emery, Weaver and Anaclet labs for helpful discussions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.888262/full#supplementary-material
Akten B., Jauch E., Genova G. K., Kim E. Y., Edery I., Raabe T., et al. (2003). A Role for CK2 in the Drosophila Circadian Oscillator. Nat. Neurosci. 6, 251–257. doi:10.1038/nn1007
Bao S., Rihel J., Bjes E., Fan J.-Y., Price J. L. (2001). TheDrosophila Double-timeSMutation Delays the Nuclear Accumulation ofperiodProtein and Affects the Feedback Regulation ofperiodmRNA. J. Neurosci. 21, 7117–7126. doi:10.1523/jneurosci.21-18-07117.2001
Cai Y. D., Xue Y., Truong C. C., Del Carmen-Li J., Ochoa C., Vanselow J. T., et al. (2021). CK2 Inhibits TIMELESS Nuclear Export and Modulates CLOCK Transcriptional Activity to Regulate Circadian Rhythms. Curr. Biol. 31, 502–514. doi:10.1016/J.CUB.2020.10.061
Chang D. C., Reppert S. M. (2003). A Novel C-Terminal Domain of Drosophila PERIOD Inhibits dCLOCK:CYCLE-Mediated Transcription. Curr. Biol. 13, 758–762. doi:10.1016/S0960-9822(03)00286-0
Chen K. F., Peschel N., Zavodska R., Sehadova H., Stanewsky R. (2011). QUASIMODO, a Novel GPI-Anchored Zona Pellucida Protein Involved in Light Input to the Drosophila Circadian Clock. Curr. Biol. 21, 719–729. doi:10.1016/J.CUB.2011.03.049
Chiu J. C., Ko H. W., Edery I. (2011). NEMO/NLK Phosphorylates PERIOD to Initiate a Time-Delay Phosphorylation Circuit that Sets Circadian Clock Speed. Cell. 145, 357–370. doi:10.1016/j.cell.2011.04.002
Chiu J. C., Vanselow J. T., Kramer A., Edery I. (2008). The Phospho-Occupancy of an Atypical SLIMB-Binding Site on PERIOD that Is Phosphorylated by DOUBLETIME Controls the Pace of the Clock. Genes. Dev. 22, 1758–1772. doi:10.1101/gad.1682708
Cho E., Kwon M., Jung J., Hyun Kang D., Jin S., Choi S.-E., et al. (2019). AMP-activated Protein Kinase Regulates Circadian Rhythm by Affecting CLOCK in Drosophila. J. Neurosci. 39, 3537–3550. doi:10.1523/JNEUROSCI.2344-18.2019
Dunlap J. C. (1999). Molecular Bases for Circadian Clocks. Cell. 96, 271–290. doi:10.1016/S0092-8674(00)80566-8
Dusik V., Senthilan P. R., Mentzel B., Hartlieb H., Wülbeck C., Yoshii T., et al. (2014). The MAP Kinase P38 Is Part of Drosophila melanogaster's Circadian Clock. PLOS Genet. 10, e1004565. doi:10.1371/JOURNAL.PGEN.1004565
Eide E. J., Woolf M. F., Kang H., Woolf P., Hurst W., Camacho F., et al. (2005). Control of Mammalian Circadian Rhythm by CKIε-Regulated Proteasome-Mediated PER2 Degradation. Mol. Cell. Biol. 25, 2795–2807. doi:10.1128/MCB.25.7.2795-2807.2005
Ewer J., Hamblen-Coyle M., Rosbash M., Hall J. C. (1990). Requirement forPeriodGene Expression in the Adult and Not during Development for Locomotor Activity Rhythms of ImaginalDrosophila Melanogaster. J. Neurogenetics 7, 31–73. doi:10.3109/01677069009084151
Garbe D. S., Fang Y., Zheng X., Sowcik M., Anjum R., Gygi S. P., et al. (2013). Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in Drosophila. PLOS Genet. 9, e1003749. doi:10.1371/JOURNAL.PGEN.1003749
Gekakis N., Saez L., Delahaye-Brown A.-M., Myers M. P., Sehgal A., Young M. W., et al. (1995). Isolation of Timeless by PER Protein Interaction: Defective Interaction between Timeless Protein and Long-Period Mutant PER L. Science 270, 811–815. doi:10.1126/science.270.5237.811
Giesecke A., Johnstone P. S., Lamaze A., Landskron J., Atay E., Chen K.-F., et al. (2021). Nuclear Export of Drosophila PERIOD Contributes to Temperature Compensation of the Circadian Clock. bioRxiv. doi:10.1101/2021.10.25.465663
Grima B., Lamouroux A., Chélot E., Papin C., Limbourg-Bouchon B., Rouyer F. (2002). The F-Box Protein Slimb Controls the Levels of Clock Proteins Period and Timeless. Nature 420, 178–182. doi:10.1038/nature01122
Hamblen M. J., White N. E., Emery P. T. J., Kaiser K., Hall J. C. (1998). Molecular and Behavioral Analysis of Four Period Mutants in Drosophila melanogaster Encompassing Extreme Short, Novel Long, and Unorthodox Arrhythmic Types. Genetics 149, 165–178. doi:10.1093/genetics/149.1.165
Hansen L. L., van den Burg H. A., van Ooijen G. (2017). Sumoylation Contributes to Timekeeping and Temperature Compensation of the Plant Circadian Clock. J. Biol. Rhythms 32, 560–569. doi:10.1177/0748730417737633
Hardin P. E. (2011). Molecular Genetic Analysis of Circadian Timekeeping in Drosophila. Adv. Genet. 74, 141–173. doi:10.1016/B978-0-12-387690-4.00005-2
Hastings J. W., Sweeney B. M. (1957). On the Mechanism of Temperature Independence in a Biological Clock. Proc. Natl. Acad. Sci. U.S.A. 43, 804–811. doi:10.1073/pnas.43.9.804
Huang Z. J., Curtin K. D., Rosbash M. (1995). PER Protein Interactions and Temperature Compensation of a Circadian Clock in Drosophila. Science 267, 1169–1172. doi:10.1126/science.7855598
Huang Z. J., Edery I., Rosbash M. (1993). PAS Is a Dimerization Domain Common to Drosophila Period and Several Transcription Factors. Nature 364, 259–262. doi:10.1038/364259a0
Hughes M. E., Hogenesch J. B., Kornacker K. (2010). JTK_CYCLE: an Efficient Nonparametric Algorithm for Detecting Rhythmic Components in Genome-Scale Data Sets. J. Biol. Rhythms 25, 372–380. doi:10.1177/0748730410379711
Isojima Y., Nakajima M., Ukai H., Fujishima H., Yamada R. G., Masumoto K.-h., et al. (2009). Ckiε/δ-dependent Phosphorylation Is a Temperature-Insensitive, Period-Determining Process in the Mammalian Circadian Clock. Proc. Natl. Acad. Sci. U.S.A. 106, 15744–15749. doi:10.1073/pnas.0908733106
Kidd P. B., Young M. W., Siggia E. D. (2015). Temperature Compensation and Temperature Sensation in the Circadian Clock. Proc. Natl. Acad. Sci. U.S.A. 112, E6284–E6292. doi:10.1073/pnas.1511215112
Kim E. Y., Edery I. (2006). Balance between DBT/CKIε Kinase and Protein Phosphatase Activities Regulate Phosphorylation and Stability of Drosophila CLOCK Protein. Proc. Natl. Acad. Sci. U.S.A. 103, 6178–6183. doi:10.1073/PNAS.0511215103
Kivimäe S., Saez L., Young M. W. (2008). Activating PER Repressor through a DBT-Directed Phosphorylation Switch. PLoS Biol. 6, e183–1583. doi:10.1371/journal.pbio.0060183
Kloss B., Price J. L., Saez L., Blau J., Rothenfluh A., Wesley C. S., et al. (1998). The Drosophila Clock Gene Double-Time Encodes a Protein Closely Related to Human Casein Kinase Iε. Cell. 94, 97–107. doi:10.1016/S0092-8674(00)81225-8
Kloss B., Rothenfluh A., Young M. W., Saez L. (2001). Phosphorylation of PERIOD Is Influenced by Cycling Physical Associations of DOUBLE-TIME, PERIOD, and TIMELESS in the Drosophila Clock. Neuron 30, 699–706. doi:10.1016/S0896-6273(01)00320-8
Ko H. W., Jiang J., Edery I. (2002). Role for Slimb in the Degradation of Drosophila Period Protein Phosphorylated by Doubletime. Nature 420, 673–678. doi:10.1038/nature01272
Ko H. W., Kim E. Y., Chiu J., Vanselow J. T., Kramer A., Edery I. (2010). A Hierarchical Phosphorylation Cascade that Regulates the Timing of PERIOD Nuclear Entry Reveals Novel Roles for Proline-Directed Kinases and GSK-3/SGG in Circadian Clocks. J. Neurosci. 30, 12664–12675. doi:10.1523/JNEUROSCI.1586-10.2010
Konopka R. J., Benzer S. (1971). Clock Mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116. doi:10.1073/pnas.68.9.2112
Konopka R. J., Hamblen-Coyle M. J., Jamison C. F., Hall J. C. (1994). An Ultrashort Clock Mutation at the Period Locus of Drosophila melanogaster that Reveals Some New Features of the Fly's Circadian System. J. Biol. Rhythms 9, 189–216. doi:10.1177/074873049400900303
Lam V. H., Li Y. H., Liu X., Murphy K. A., Diehl J. S., Kwok R. S., et al. (2018). CK1α Collaborates with DOUBLETIME to Regulate PERIOD Function in the Drosophila Circadian Clock. J. Neurosci. 38, 10631–10643. doi:10.1523/JNEUROSCI.0871-18.2018
Lim C., Lee J., Choi C., Kilman V. L., Kim J., Park S. M., et al. (2011). The Novel Gene Twenty-Four Defines a Critical Translational Step in the Drosophila Clock. Nature 470, 399–403. doi:10.1038/nature09728
Lin J.-M., Kilman V. L., Keegan K., Paddock B., Emery-Le M., Rosbash M., et al. (2002). A Role for Casein Kinase 2α in the Drosophila Circadian Clock. Nature 420, 816–820. doi:10.1038/nature01235
Lin J.-M., Schroeder A., Allada R. (2005). In Vivo Circadian Function of Casein Kinase 2 Phosphorylation Sites in Drosophila PERIOD. J. Neurosci. 25, 11175–11183. doi:10.1523/JNEUROSCI.2159-05.2005
Martinek S., Inonog S., Manoukian A. S., Young M. W. (2001). A Role for the Segment Polarity Gene shaggy/GSK-3 in the Drosophila Circadian Clock. Cell. 105, 769–779. doi:10.1016/S0092-8674(01)00383-X
Masuda S., Narasimamurthy R., Yoshitane H., Kim J. K., Fukada Y., Virshup D. M. (2020). Mutation of a PER2 Phosphodegron Perturbs the Circadian Phosphoswitch. Proc. Natl. Acad. Sci. U.S.A. 117, 10888–10896. doi:10.1073/PNAS.2000266117
Matsumoto A., Tomioka K., Chiba Y., Tanimura T. (1999). Tim Rit Lengthens Circadian Period in a Temperature-dependent Manner through Suppression of PERIOD Protein Cycling and Nuclear Localization. Mol. Cell. Biol. 19, 4343–4354. doi:10.1128/mcb.19.6.4343
Mehra A., Shi M., Baker C. L., Colot H. V., Loros J. J., Dunlap J. C. (2009). A Role for Casein Kinase 2 in the Mechanism Underlying Circadian Temperature Compensation. Cell. 137, 749–760. doi:10.1016/j.cell.2009.03.019
Narasimamurthy R., Hunt S. R., Lu Y., Fustin J.-M., Okamura H., Partch C. L., et al. (2018). CK1δ/ε Protein Kinase Primes the PER2 Circadian Phosphoswitch. Proc. Natl. Acad. Sci. U.S.A. 115, 5986–5991. doi:10.1073/pnas.1721076115
Nawathean P., Rosbash M. (2004). The Doubletime and CKII Kinases Collaborate to Potentiate Drosophila PER Transcriptional Repressor Activity. Mol. Cell. 13, 213–223. doi:10.1016/S1097-2765(03)00503-3
Peixoto A. A., Hennessy J. M., Townson I., Hasan G., Rosbash M., Costa R., et al. (1998). Molecular Coevolution within a Drosophila Clock Gene. Proc. Natl. Acad. Sci. U.S.A. 95, 4475–4480. doi:10.1073/pnas.95.8.4475
Pittendrigh C. S. (1960). Circadian Rhythms and the Circadian Organization of Living Systems. Cold Spring Harb. Symposia Quantitative Biol. 25, 159–184. doi:10.1101/SQB.1960.025.01.015
Pittendrigh C. S. (1954). On Temperature Independence in the Clock System Controlling Emergence Time in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 40, 1018–1029. doi:10.1073/pnas.40.10.1018
Portolés S., Más P. (2010). The Functional Interplay between Protein Kinase CK2 and CCA1 Transcriptional Activity Is Essential for Clock Temperature Compensation in Arabidopsis. Plos Genet. 6, e1001201. doi:10.1371/journal.pgen.1001201
Price J. L., Blau J., Rothenfluh A., Abodeely M., Kloss B., Young M. W. (1998). Double-time Is a Novel Drosophila Clock Gene that Regulates PERIOD Protein Accumulation. Cell. 94, 83–95. doi:10.1016/S0092-8674(00)81224-6
Reischl S., Vanselow K., Westermark P. O., Thierfelder N., Maier B., Herzel H., et al. (2007). β-TrCP1-Mediated Degradation of PERIOD2 Is Essential for Circadian Dynamics. J. Biol. Rhythms 22, 375–386. doi:10.1177/0748730407303926
Renn S. C. P., Park J. H., Rosbash M., Hall J. C., Taghert P. H. (1999). A Pdf Neuropeptide Gene Mutation and Ablation of PDF Neurons Each Cause Severe Abnormalities of Behavioral Circadian Rhythms in Drosophila. Cell. 99, 791–802. doi:10.1016/S0092-8674(00)81676-1
Rothenfluh A., Abodeely M., Price J. L., Young M. W. (2000). Isolation and Analysis of Six Timeless Alleles that Cause Short- or Long-Period Circadian Rhythms in Drosophila. Genetics 156, 665–675. doi:10.1093/genetics/156.2.665
Sawyer L. A., Hennessy J. M., Peixoto A. A., Rosato E., Parkinson H., Costa R., et al. (1997). Natural Variation in a Drosophila Clock Gene and Temperature Compensation. Science 278, 2117–2120. doi:10.1126/science.278.5346.2117
Shanware N. P., Hutchinson J. A., Kim S. H., Zhan L., Bowler M. J., Tibbetts R. S. (2011). Casein Kinase 1-dependent Phosphorylation of Familial Advanced Sleep Phase Syndrome-Associated Residues Controls PERIOD 2 Stability. J. Biol. Chem. 286, 12766–12774. doi:10.1074/jbc.M111.224014
Shinohara Y., Koyama Y. M., Ukai-Tadenuma M., Hirokawa T., Kikuchi M., Yamada R. G., et al. (2017). Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock. Mol. Cell. 67, 783–798. doi:10.1016/J.MOLCEL.2017.08.009
Singh S., Giesecke A., Damulewicz M., Fexova S., Mazzotta G. M., Stanewsky R., et al. (2019). New Drosophila Circadian Clock Mutants Affecting Temperature Compensation Induced by Targeted Mutagenesis of Timeless. Front. Physiol. 10, 1442. doi:10.3389/FPHYS.2019.01442/FULL
Stoleru D., Peng Y., Nawathean P., Rosbash M. (2005). A Resetting Signal between Drosophila Pacemakers Synchronizes Morning and Evening Activity. Nature 438, 238–242. doi:10.1038/NATURE04192
Szabó Á., Papin C., Zorn D., Ponien P., Weber F., Raabe T., et al. (2013). The CK2 Kinase Stabilizes CLOCK and Represses its Activity in the Drosophila Circadian Oscillator. PLOS Biol. 11, 1001645. doi:10.1371/JOURNAL.PBIO.1001645
Toh K. L., Jones C. R., He Y., Eide E. J., Hinz W. A., Virshup D. M., et al. (2001). An H Per2 Phosphorylation Site Mutation in Familial Advanced Sleep Phase Syndrome. Science 291, 1040–1043. doi:10.1126/science.1057499
Top D., Harms E., Syed S., Adams E. L., Saez L. (2016). GSK-3 and CK2 Kinases Converge on Timeless to Regulate the Master Clock. Cell. Rep. 16, 357–367. doi:10.1016/J.CELREP.2016.06.005
Top D., O'Neil J. L., Merz G. E., Dusad K., Crane B. R., Young M. W. (2018). CK1/doubletime Activity Delays Transcription Activation in the Circadian Clock. Elife 7, 1–21. doi:10.7554/eLife.32679
Top D., Young M. W. (2018). Coordination between Differentially Regulated Circadian Clocks Generates Rhythmic Behavior. Cold Spring Harb. Perspect. Biol. 10, a033589. doi:10.1101/CSHPERSPECT.A033589
Tosini G., Menaker M. (1998). The Tau Mutation Affects Temperature Compensation of Hamster Retinal Circadian Oscillators. Neuroreport 9, 1001–1005. doi:10.1097/00001756-199804200-00009
Yu W., Houl J. H., Hardin P. E. (2011). NEMO Kinase Contributes to Core Period Determination by Slowing the Pace of the Drosophila Circadian Oscillator. Curr. Biol. 21, 756–761. doi:10.1016/J.CUB.2011.02.037
Yu W., Zheng H., Houl J. H., Dauwalder B., Hardin P. E. (2006). PER-dependent Rhythms in CLK Phosphorylation and E-Box Binding Regulate Circadian Transcription. Genes. Dev. 20, 723–733. doi:10.1101/GAD.1404406
Zeng H., Hardin P. E., Rosbash M. (1994). Constitutive Overexpression of the Drosophila Period Protein Inhibits Period mRNA Cycling. EMBO J. 13, 3590–3598. doi:10.1002/J.1460-2075.1994.TB06666.X
Keywords: circadian rhythms, temperature compensation, period, phophorylated amino acids, drosophila
Citation: Joshi R, Cai YD, Xia Y, Chiu JC and Emery P (2022) PERIOD Phosphoclusters Control Temperature Compensation of the Drosophila Circadian Clock. Front. Physiol. 13:888262. doi: 10.3389/fphys.2022.888262
Received: 02 March 2022; Accepted: 29 April 2022;
Published: 02 June 2022.
Edited by:
Elzbieta M. Pyza, Jagiellonian University, PolandReviewed by:
Charlotte Helfrich-Förster, Julius Maximilian University of Würzburg, GermanyCopyright © 2022 Joshi, Cai, Xia, Chiu and Emery. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick Emery, cGF0cmljay5lbWVyeUB1bWFzc21lZC5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.