- 1Department of Orthopedics, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, Shanghai, China
- 2Department of Physiology, School of Basic Medical Sciences, Cheeloo College of Medicine, Shandong University, Jinan, China
Purpose: This study aimed to identify the biological functions of small intestine intestinal epithelial cell derived exosomes (IEC-Exos) and further distinguished the difference proteins in IEC-Exos between ileum and jejunum related to function of the digestive system and occurrence of several diseases.
Materials and Methods: IECs of Male C57BL/6J mice were isolated. IEC-Exos were extracted from jejunum and ileum epithelial cell culture fluid by ultracentrifugation. In addition, isobaric tags for relative and absolute quantitation (iTRAQ) combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) were used to detect IEC-Exo proteins and conduct biological information analysis.
Results: The results showed that compared with jejunum IEC-Exos from ileum IEC-Exos, there were 393 up-regulated proteins and 346 down-regulated proteins. IECs-Exos, especially derived from jejunum, were rich in angiotensin-converting enzyme 2 (ACE2). The highly expressed proteins from ileum IEC-Exos were mostly enriched in genetic information processing pathways, which mainly mediate the processes of bile acid transport, protein synthesis and processing modification. In contrast, the highly expressed proteins from jejunum IEC-Exos were mainly enriched in metabolic pathways involved in sugar, fatty acid, amino acid, drug, and bone metabolism, etc. The differentially expressed proteins between ileum and jejunum IEC-Exos were not only related to the function of the digestive system but also closely related to the occurrence of infectious diseases, endocrine diseases and osteoarthritis, etc.
Conclusion: IEC-Exos there were many differentially expressed proteins between ileum and jejunum, which played different roles in regulating intestinal biological functions. ACE2, the main host cell receptor of SARS-CoV-2, was highly expressed in IEC-Exos, which indicated that IEC-Exos may be a potential route of SARS-CoV-2 infection.
Introduction
Intestinal epithelial cells (IECs), which have the characteristics of continuous migration, differentiation, and renewal, play an important role in maintaining intestinal function. As an organ involved in nutrient digestion and absorption, microbial defense, and endocrine functions, the intestine has important physiological functions, such as nutrient absorption, secretion, and transport. In addition, the intestine is also involved in drug metabolism, energy metabolism, endocrine responses, oxidative stress, and immune defense mechanisms (Peterson and Artis, 2014; Kiela and Ghishan, 2016; Satsu, 2017). The occurrence and development of clinical diabetes, atherosclerosis, intestinal inflammation, neurodegenerative diseases, and osteoarthritis are all correlated with intestinal epithelial dysfunction (de Wit et al., 2008; Gribble and Reimann, 2019; Tajik et al., 2020). Angiotensin-converting enzyme 2 (ACE2), which is highly expressed on IECs, is a receptor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected cells and has an important effect on the occurrence and development of coronavirus disease 2019 (COVID-19) (Lamers et al., 2020; Ziegler et al., 2020). Recent studies confirmed that the exosomes are important intercellular communication vehicles, which play key role in cell biology. However, Intestinal epithelial cell derived exosomes (IEC-Exos) have not been studied in detail. Studies have shown that the biological function of IECs is closely related to their paracrine exosomes. Exosomes selectively enrich biologically active components derived from parent cells, including nucleic acids, proteins, lipids, amino acids, and metabolites, which are the basis for information and material exchange between cells. Once ingested by target cells, exosomes can regulate a variety of biological behaviors, such as gene expression and survival, proliferation, and migration of cells (Kalluri and LeBleu, 2020). IEC-Exos play pivotal roles in regulating intestinal antigen presentation, intestinal inflammation, intestinal homeostasis and systemic immune responses (van Niel and Heyman, 2002; Zhang H. et al., 2019). High-fat-induced IEC-Exos are involved in the occurrence and development of obesity, diabetes, and vascular complications (Xia et al., 2019). Many studies have demonstrated that exosomes play pivotal roles in the transmission of viruses and aggravation of adverse reactions and have potential application value in preventing viral infections (Hassanpour et al., 2020). Nevertheless, there are no reports on IEC-Exos in SARS-CoV-2 infection. At the same time, many significant biological functions, and mechanisms of IEC-Exos have not been fully elucidated.
The jejunum and ileum are the main components of the small intestine. The functional differences of exosomes derived from the jejunum and ileum are poorly understood. As a high-throughput screening method, proteomics has attracted increasing attention in the study of the protein components and biological effects of exosomes. In this study, extracted mouse jejunum and ileum IEC-Exos were identified by transmission electron microscopy, western blotting, and nanoparticle tracking analysis. For the first time, differentially expressed proteins of IEC-Exos were detected and compared through isobaric tags for relative and absolute quantitation (iTRAQ) combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS). Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein–protein interaction (PPI) bioinformatics analyses were used to explore the function, distribution and related signaling pathways of differentially expressed proteins from ileum and jejunum IEC-Exos. Our study provides a basis for better understanding the function of IECs and for performing further research on the diagnosis and treatment of related diseases.
Materials and Methods
Cell Culture
Twenty-four male C57BL/6J mice (six mice per group) were sacrificed via cervical dislocation. The abdominal cavity of mice was opened, and the jejunum and ileum were gently cut out. The tissue was placed in a culture dish filled with cold normal saline. Then, the surplus mesenteric adipose tissue was gently removed, and the intestine was rinsed with cold saline. Next, the small intestine was cut longitudinally, and the stool was gently scraped off with a coverslip. The tissue was cut into small pieces (approximately 2 mm) and placed in a 50 mL centrifuge tube. DTT (30 ml, 10 mmol/L) was added to the tube and shaken for 5 min to remove mucus. After removing mucus, the tissue was transferred to a tube containing 30 mL 8 mmol/L EDTA and shaken for 30 min at 4°C. The EDTA was discarded, and 30 ml DPBS was added to a 50 mL centrifuge tube. The mixture was blown evenly with a 10 mL pipette, and the supernatant was collected. The above steps were repeated approximately six times until the supernatant became clear. The supernatant was mixed and centrifuged at 300 g for 5 min at 4°C. The supernatant was discarded, and the precipitate was collected. The precipitate was mainly composed of IECs. IECs of the jejunum and ileum were cultured with PBS, placed in a cell incubator at 37°C and 5% CO2, and cultured for 15 min. The culture fluid was collected to extract exosomes.
Exosome Purification
Exosomes were isolated by differential centrifugation. In short, three-step centrifugation at 300 × g for 10 min, 2000 × g for 10 min, and 10,000 × g for 10 min were used to separate living cells, possible apoptotic bodies and large cell fragments from the culture fluid. Then, it was centrifuged at 100,000 × g for 70 min to collect EVs-containing pellets. After washing with phosphate buffered saline (PBS), ultracentrifugation was centrifuged at 100,000 × g for 70 min with a Beckman 32Ti rotor. The pellet was resuspended in PBS to obtain an IEC-Exo suspension of the jejunum and ileum. The size and particle number of EVs were characterized by electron microscope and nanoparticle tracking analysis (NTA).
Nanoparticle Tracking Analysis
The exosome samples were first diluted to a final dilution of 1:5000 in filtered sterile PBS. The particle size and concentration distribution of the IEC-Exos were measured by NTA (ZetaVIEWS/N 17-310, PARTICLE METRIX, Meerbusch, Germany) according to the manufacturer’s instructions.
Transmission Electron Microscopy
Twenty microliters of the exosome suspension was added to 2 nm copper grids, placed at room temperature for 3 min, and stained with a 3% phosphotungstic acid solution. The samples were then visualized using a transmission electron microscope (Tecnai G2 spirit Biotwin, FEI, Hillsboro, OR, United States).
Western Blotting Analysis
The collected IEC-Exos were fully lysed by adding RIPI and a protease inhibitor (100:1). The lysis solution was centrifuged at 12,000 rpm for 20 min at 4°C. The supernatant was collected and mixed with 5x loading buffer. The mixture was placed in a heater for 10 min at 100°C to fully denature the protein. The expression of TSG 101 (Santa Cruz, sc-7964, dilution rate 1:500) and CD 81 (Santa Cruz, sc-166029, dilution rate 1:500) was detected by western blot analysis.
Protein Extraction and Digestion
Frozen IEC-Exo samples from the ileum group and jejunum group were removed (four samples for each group, six mice per sample). The samples were homogenized with a hand-held homogenizer in the presence of lysate buffer on ice and then ultrasonically processed. The resulting solutions were centrifuged to remove any pellets and precipitates. A bicinchoninic acid assay (BCA) was performed to determine the protein concentration of every sample. Twenty micrograms of each sample of the same group were mixed together. Then, six volumes of acetone were added to each obtained group supernatant, and the mixed solutions were placed at –20°C overnight. After centrifugation, the supernatants were removed. The precipitated proteins were washed with acetone and resuspended in 500 mM triethylammonium bicarbonate/6 M guanidine hydrochloride (TEAB). BCA was performed to determine the protein concentration. A filter-aided sample preparation (FASP) strategy described in a previous report (Wisniewski et al., 2009) was performed for subsequent sample preparation. Briefly, 200 μg of protein from each group was reduced, alkylated and digested in centrifugal units. After digestion at 37°C overnight, the peptide solutions were centrifuged, and the filtrates were collected. Sequentially, the digested peptides were dried by vacuum centrifugation and stored at –80°C until further use.
Isobaric Tags for Relative and Absolute Quantitation Labeling
One hundred micrograms samples from each group were labeled using a Reagent-8-plex Multiplex Kit according to the manufacturer’s instructions (AB Sciex, Darmstadt, Germany). The four ileum samples were labeled with reagents 113, 114, 117, 118 and the four jejunum samples were labeled with reagents 115, 116, 119 and 121. After resuspension in dissolution buffer, the digested peptides from each sample were incubated with a specific iTRAQ tag for 3 h at room temperature. The labeled samples were equally mixed and dried under a SpeedVac. After resuspension in 30 μL of 20 mM ammonium formate, 20 μL of the pooled sample was prefractionated by high-pH reverse-phase liquid chromatography using an ACQUITY UPLC H-Class Bio HPLC system (Thermo Scientific, Waltham, MA, United States), and finally, 10 consolidated fractions were acquired. Labeled peptides in each fraction were dried and redissolved in 30 μL 2% acetonitrile/0.1% formic acid for LC-MS/MS analysis.
Liquid Chromatography-Tandem Mass Spectrometry
The labeled peptide mixtures were separated using an EASY-nLC 1200 system (Thermo Scientific, United States). The peptides were separated on a PepMap100 RSLC C18, 2 μm, 75 μm × 15 cm analytic column using a 110-min mobile phase gradient from 5 to 30%. Mass spectra were recorded on an Orbitrap Exploris 480 mass spectrometer conFigured with a Nano-ESI source (Thermo Scientific, United States). Full scan MS spectra were acquired in the m/z range of 350–1600 at a resolution of 70,000. The top 10 precursors were selected for high-energy collision-induced dissociation (HCD) with a collision energy of 35%, and the product ions were detected at a resolution of 17,500 in data-dependent acquisition mode.
Isobaric Tags for Relative and Absolute Quantitation Data Analysis
The raw data were searched against the UniProt database using MASCOT2.3.2 (Thermo Scientific, United States; Matrix Science, London, United Kingdom), a search engine built in the Proteome Discoverer 1.4 software suite. Protein level changes were compared between paired ileum IEC-Exos and jejunum IEC-Exos, and differentially expressed proteins (fold change ≥ 1.3 or ≤0.769 and P < 0.05) were defined. The data were analyzed by Bohao Biotechnologies Co., Ltd. (Shanghai, China).
Bioinformatic Analysis
Gene Ontology, KEGG and PPI bioinformatics analyses were performed on the differentially expressed proteins between the ileum and jejunum IEC-Exos. Enrichment analysis of the GO terms, including molecular function (MF), biological process (BP) and cellular component (CC), and KEGG pathway analysis of the dysregulated proteins were performed using the DAVID web server. Fisher’s exact test was used to test the significance of the overlap between various gene sets, and P values < 0.05 were considered significant. The smallest 15 P-values or the 30 items with the smallest P-values between the up- or down-regulated differentially expressed proteins were selected to draw bar plots or dotplots. The PPI network was analyzed with the String database1 and visualized with Cytoscape software.
Results
Intestinal Epithelial Cell Derived Exosomes Identification
Under transmission electron microscopy, both ileum and jejunum IEC-Exos presented as round or elliptical vesicles with clear double-layer membrane structures (Figure 1A). The results of nanoparticle tracking analysis showed that the concentration of ileum IEC-Exos was 1.0E + 8 particles/mL, and the diameter of IEC-Exos was mainly concentrated at 30–130 nm. The concentration of jejunum IEC-Exos was 1.2E + 8 particles/mL with a diameter of 30–120 nm (Figure 1B). In addition, the exosomal intracellular proteins CD81 and TSG101 were also detected by western blotting (Figure 1C).
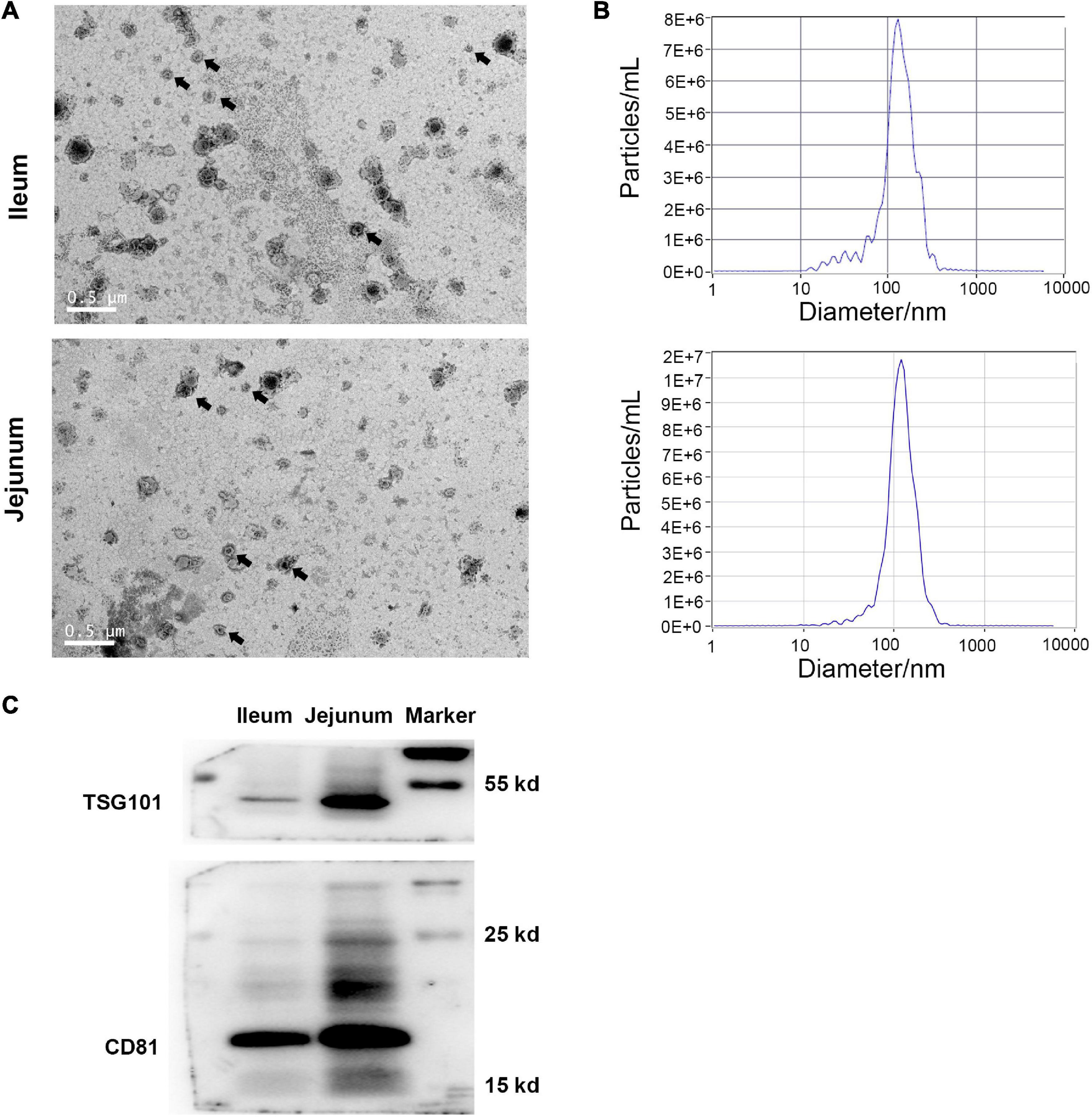
Figure 1. Identification of IEC-Exos from ileum and jejunum. (A) Transmission electron microscopic observation (scale = 0.5 μm), arrows indicated the typical exosomes. (B) Nanoparticle tracking analyzer Size Distribution. (C) Immunoblot of CD81 and TSG101 in IEC-Exos.
Intestinal Epithelial Cell Derived Exosomes Proteins Quantification
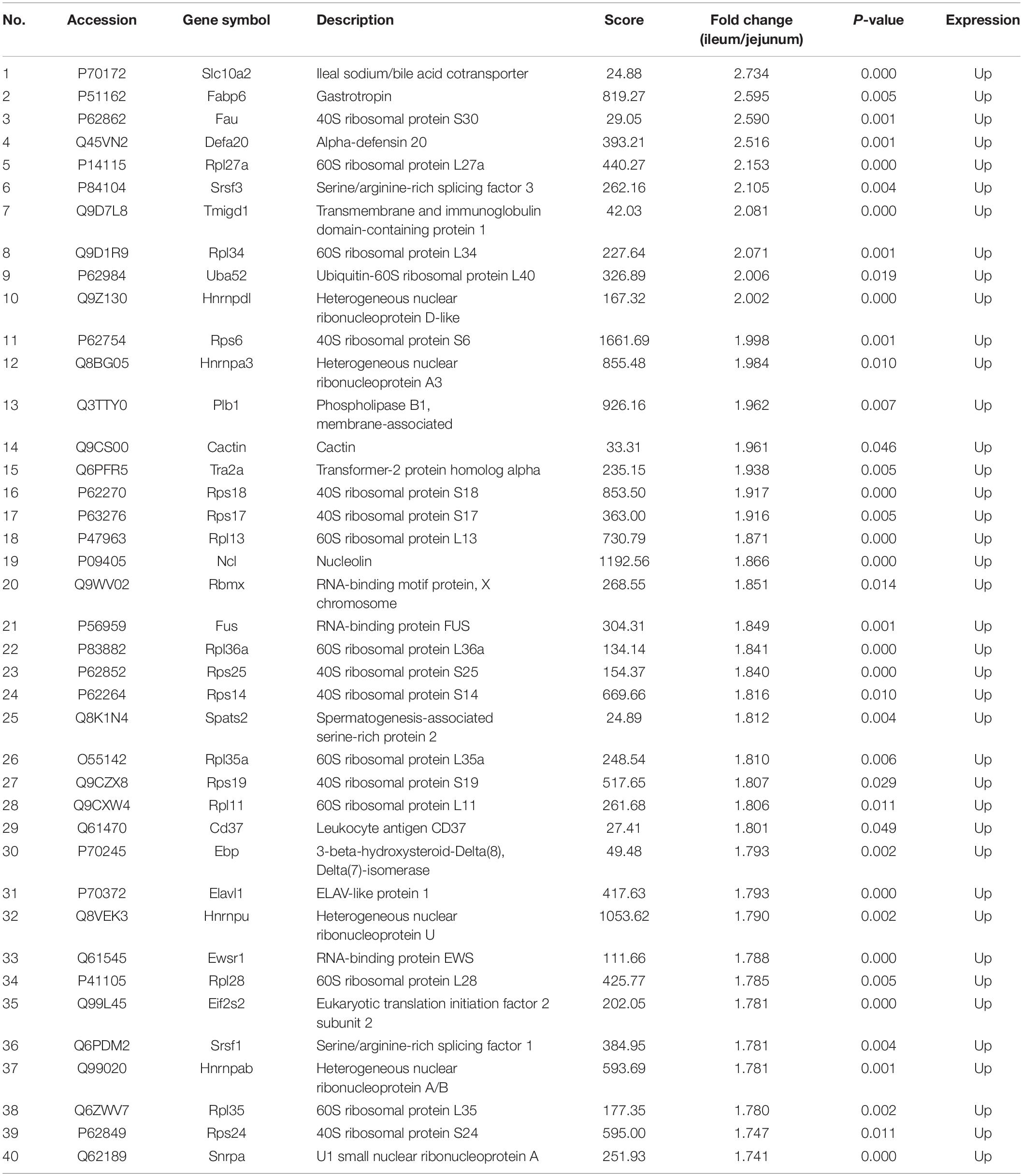
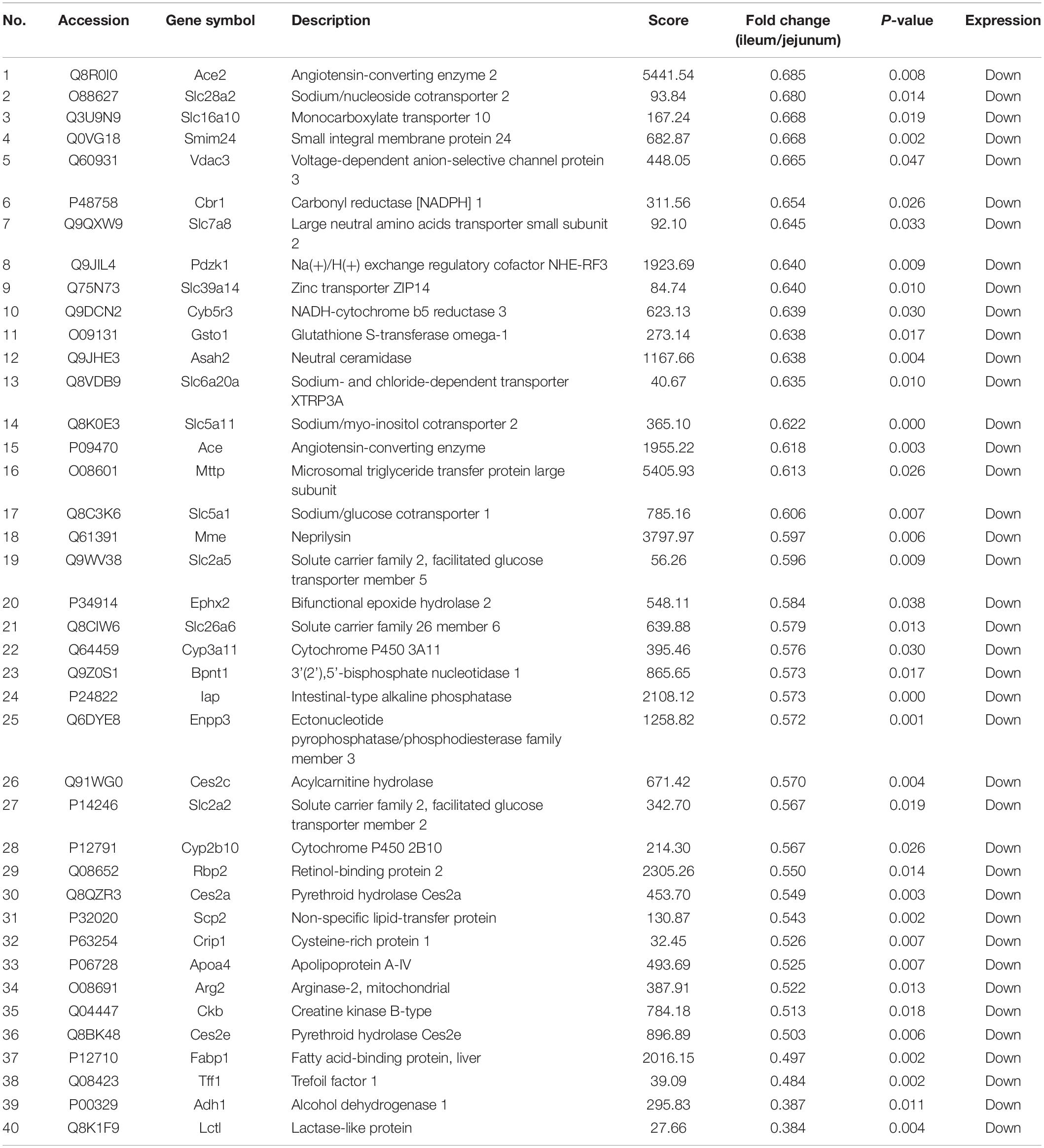
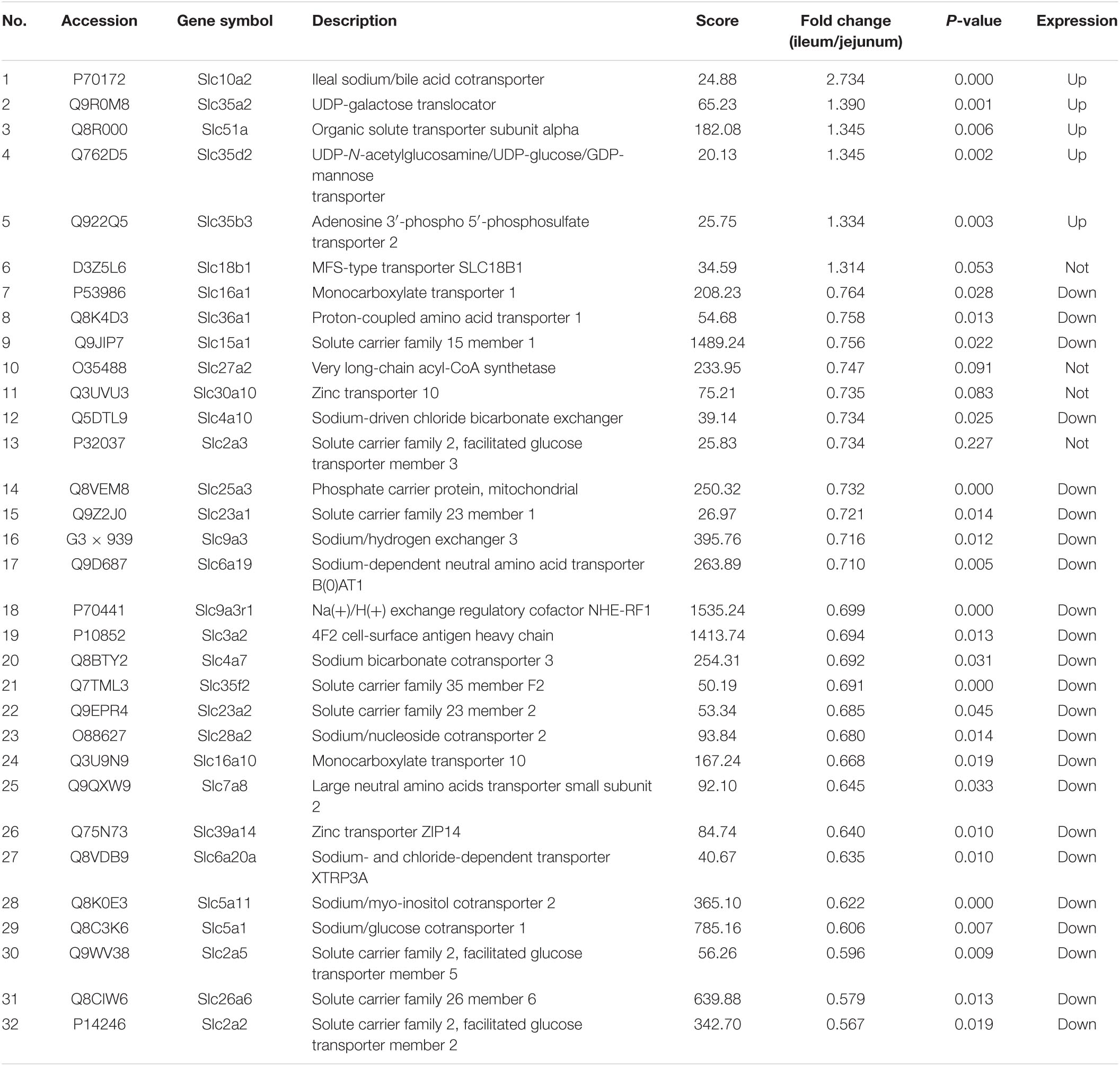
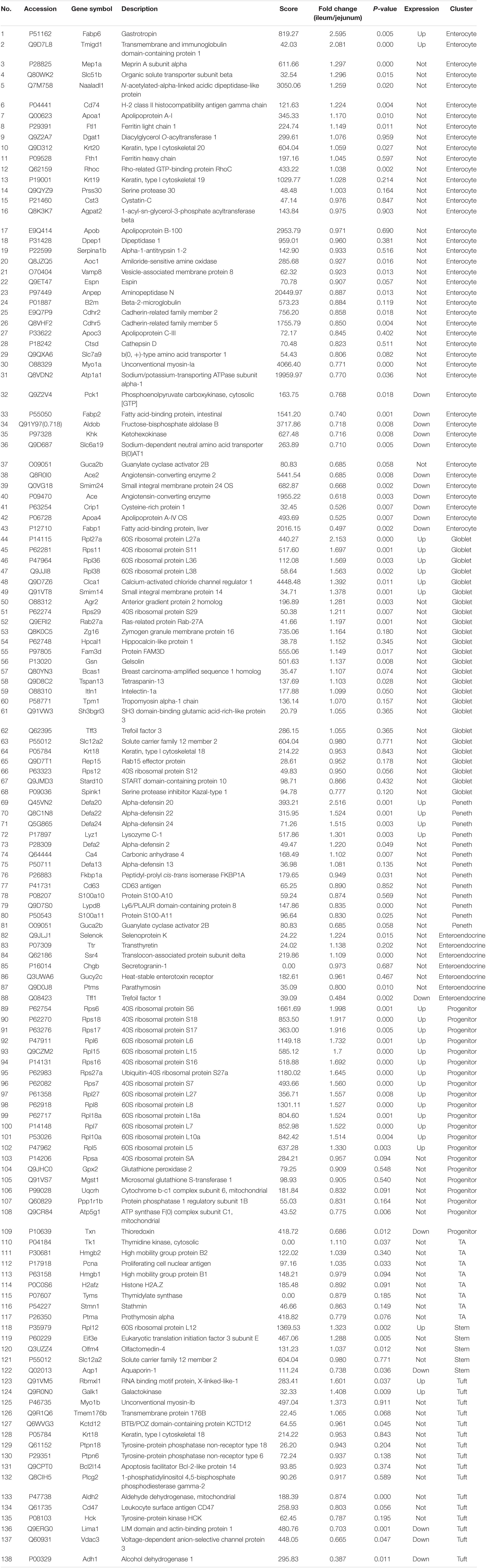
The protein quantification results showed that 4140 proteins were identified in IEC-Exos, and compared with jejunum IEC-Exos, there were 739 differentially expressed proteins(fold change ≥ 1.3 or ≤0.769 and P < 0.05) from ileum IEC-Exos, including 393 up-regulated proteins and 346 down-regulated proteins. Moreover, there was no difference in the expression of 3401 proteins between the two types of IEC-Exos (Figure 2). Ribosomal proteins were the majority of the 40 up-regulated proteins (Table 1). Solute carrier (SLC) transporters carbohydrates, amino acids, nucleic acids, ions and drugs were the majority of the down-regulated proteins (Table 2 and Schedule 1). However, Slc10a2 (solute carrier family 10 member 2), which is related to the uptake of intestinal bile acids in the distal ileum, was significantly up-regulated in IEC-Exos (Table 1 and Schedule 1). Based on previously reported cell type–specific markers and intestinal single-cell sequencing results(Haber et al., 2017; Wang Y. et al., 2020), we confirmed that the IEC-Exos we isolated were mainly derived from eight known intestinal epithelial cells, including enterocyte cells, goblet cells, Paneth cells, enteroendocrine cells, tuft cells, progenitor cells, transit amplifying cells (TACs) and stem cells (Schedule 2).
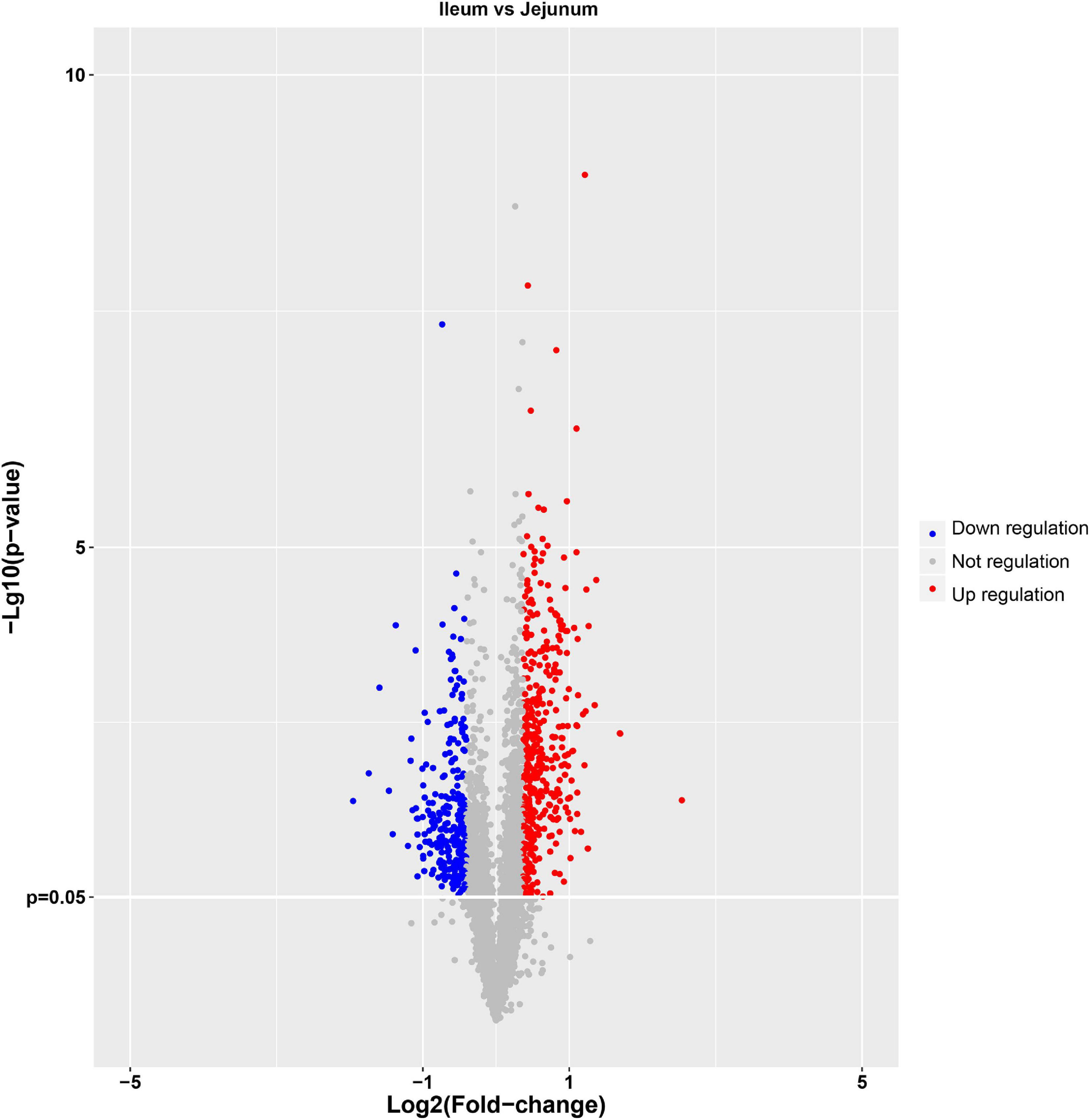
Figure 2. Volcano map of differentially expressed proteins. The abscissa represents the fold change (log2 value) of differentially expressed proteins, and the ordinate represents the P-value (–log10). Gray points represent proteins with insignificant differences, red points represent up-regulated proteins, and the blue points represent down-regulated proteins.
Bioinformatics Analysis
Gene Ontology Analysis of Intestinal Epithelial Cell Derived Exosomes Proteins From the Ileum and Jejunum
Differentially expressed proteins between ileum and jejunum IEC-Exos were mainly involved in the molecular functions of binding and hydrolase activity, suggesting that these proteins were mainly related to biological functions through target cell fusion (Figure 3A). CC was mainly distributed in bound organelles, the cytosol, the organelle lumen, the nucleus, the ribonucleoprotein complex, and the endomembrane system, reflecting the formation of IEC-Exo proteins (Figure 3B). BP was mainly involved in gene expression, metabolic processes, biosynthetic processes, regulation processes and transport (Figure 3C).
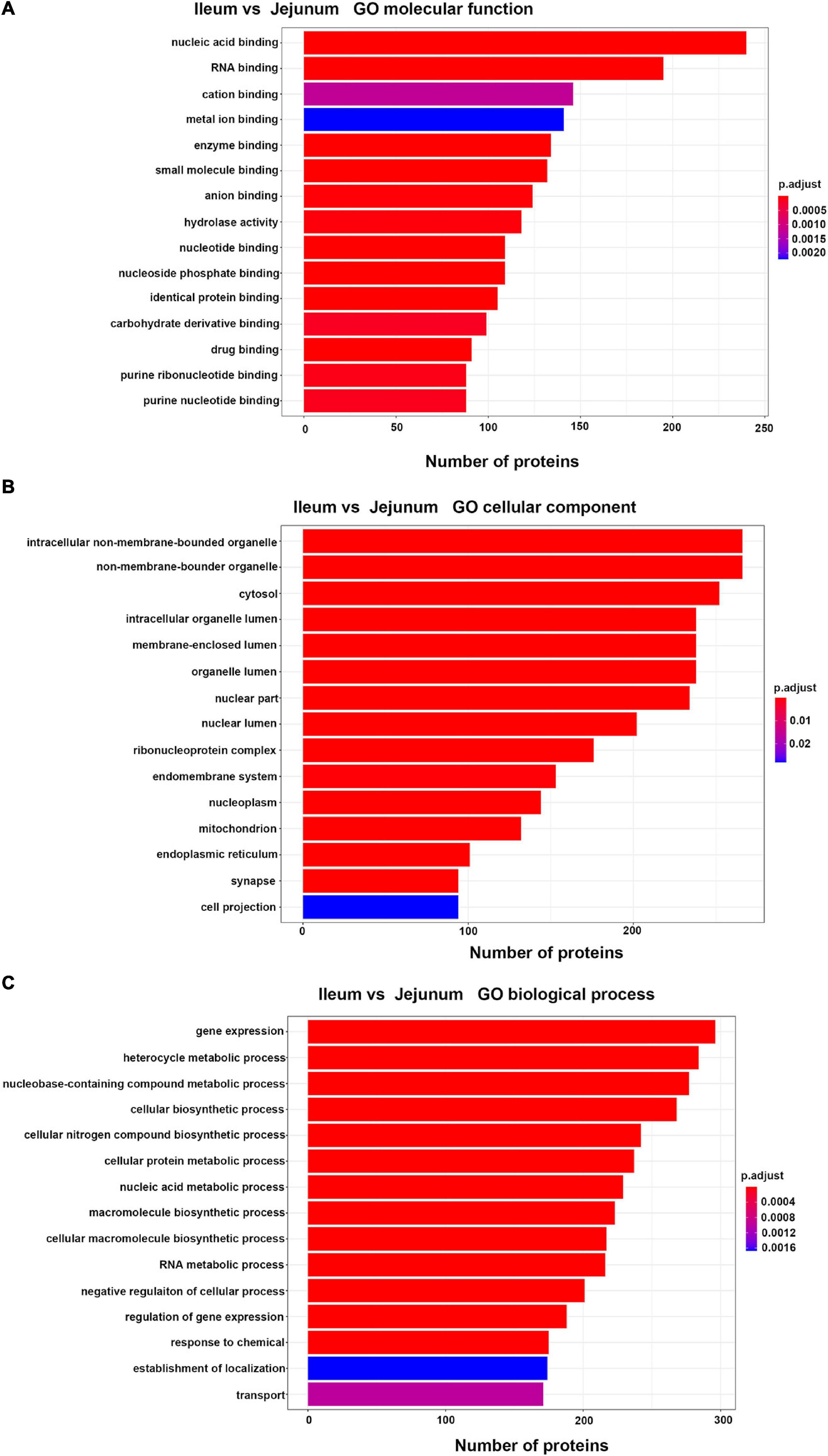
Figure 3. Top 15 of GO enrichment analysis in differentially expressed proteins between ileum and jejunum IEC-Exos. (A) GO molecular function analysis. (B) GO biological process analysis. (C) GO cellular component analysis.
Gene Ontology Analysis of Up-Regulated Proteins
Up-regulated proteins were involved in poly (A) binding, proteasome-activating ATPase activity, mRNA5′-UTR binding, RS domain binding and other complex molecular functions. CC was mainly distributed in cytosolic ribosomes, the large ribosomal subunit and the small ribosomal subunit. BP was mainly involved in cotranslational protein targeting to the membrane, the endoplasmic reticulum, the establishment of protein localization, and nuclear-transcribed mRNA catabolic processes (Figure 4A). It was suggested that the differentially expressed proteins with high expression in IEC-Exos from the ileum were mainly involved in protein biosynthesis and processing modifications.
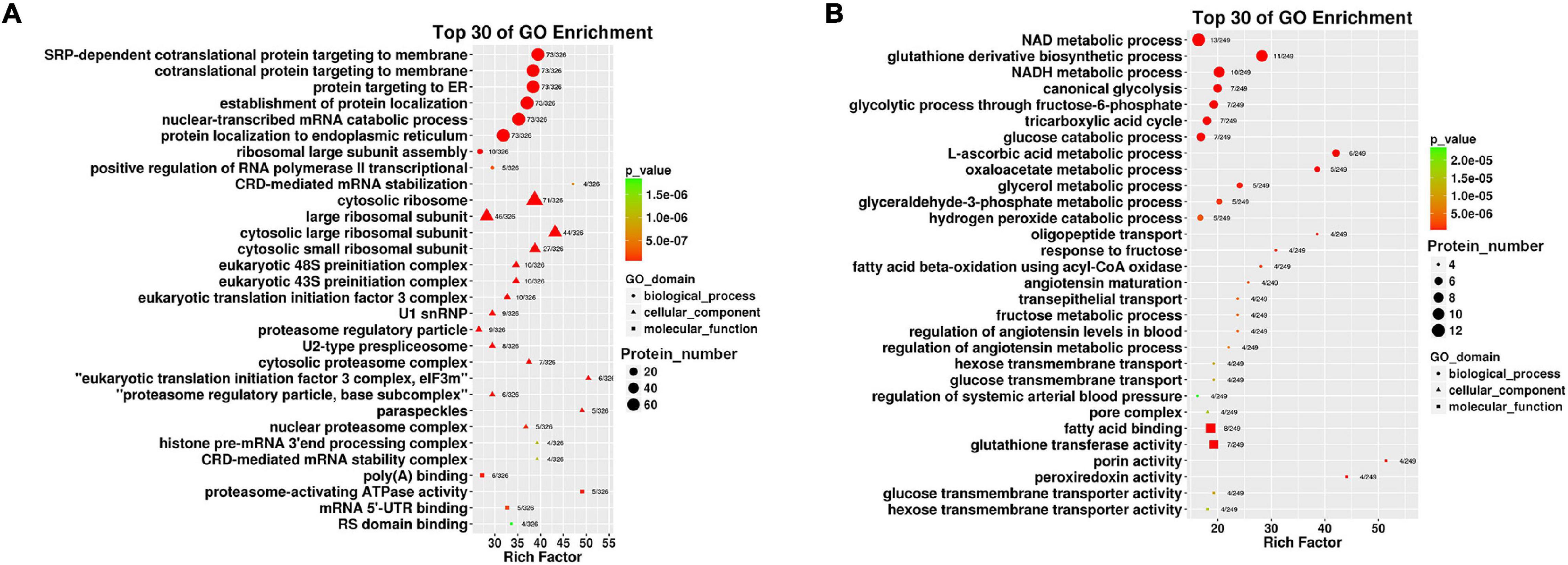
Figure 4. Top 30 of GO enrichment in up/down regulated proteins in IEC-Exos. (A) GO enrichment analysis of up-regulated protein. (B) GO enrichment analysis of down-regulated protein.
Gene Ontology Analysis of Down-Regulated Proteins
Down-regulated proteins were involved in fatty acid binding, glutathione transferase activity, porin activity, peroxiredoxin activity, the glucose transmembrane transporter, the hexose transmembrane transporter and other complex molecular functions. CC was mainly distributed in the pore complex. BP was mainly involved in NAD metabolic processes, NADH metabolic processes, glutathione derivative biosynthetic processes, glucose metabolism, the tricarboxylic acid cycle and glycerol metabolic processes (Figure 4B). It was suggested that the differentially expressed proteins with high expression in IEC-Exos from the jejunum were mainly involved in the metabolism of the three major nutrients, sugar, fat and protein, and the redox reaction (antioxidant effect, integrated detoxification effect).
Kyoto Encyclopedia of Genes and Genomes Pathway Analysis
Kyoto Encyclopedia of Genes and Genomes Pathway Analysis of IEC-Exo Proteins From the Ileum and Jejunum
Differentially expressed proteins between ileum and jejunum IEC-Exos mediated metabolic pathways, ribosomes, spliceosomes, RNA transport and drug metabolism, etc. (Figure 5A).
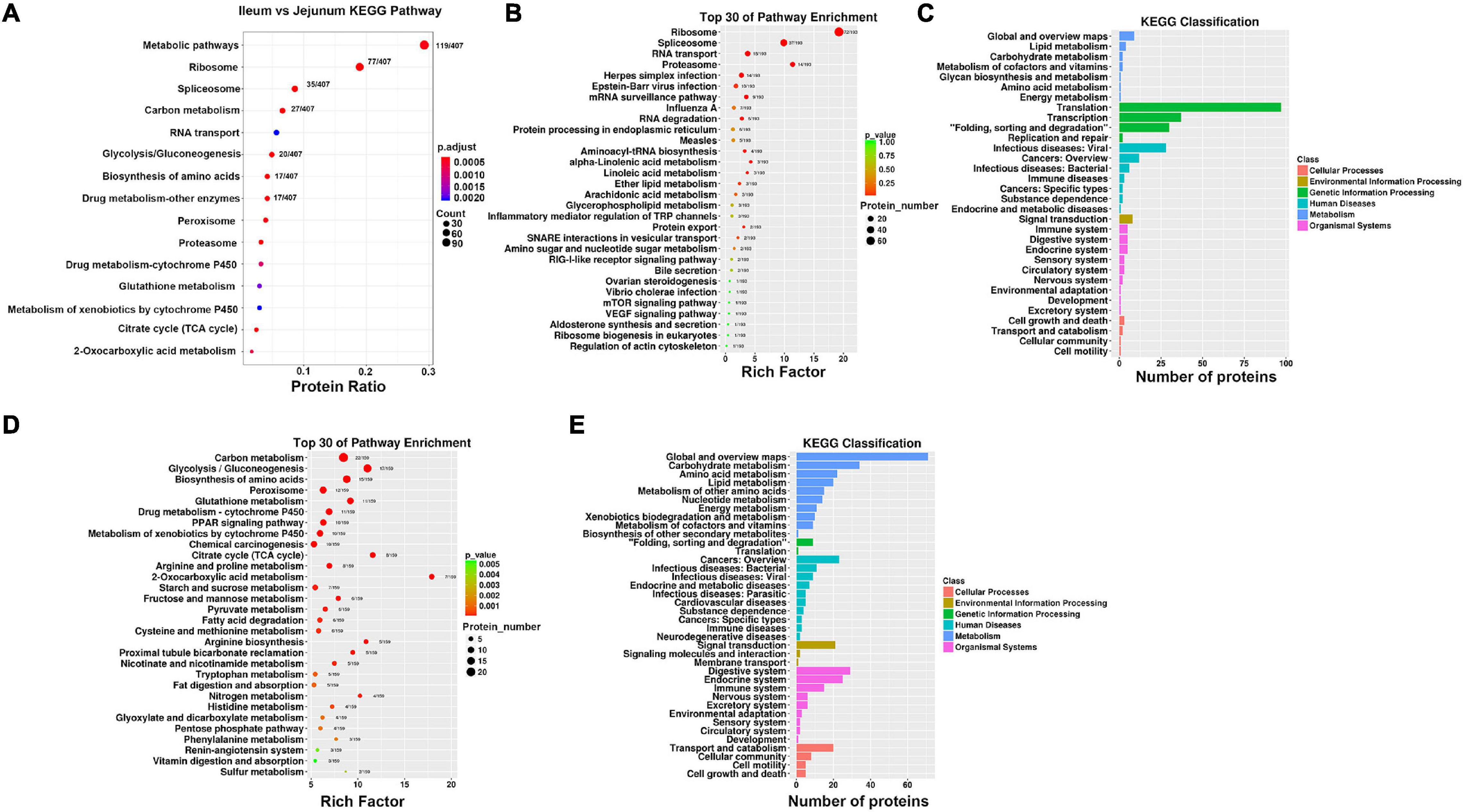
Figure 5. Enrichment analysis of KEGG pathway. (A) KEGG pathway analysis of differentially expressed proteins between ileum and jejunum IEC-Exos. (B) KEGG pathway enrichment analysis of up-regulated protein. (C) KEGG pathway classification of up-regulated protein. (D) KEGG pathway enrichment analysis of down-regulated protein. (E) KEGG pathway classification of down-regulated protein.
Kyoto Encyclopedia of Genes and Genomes Pathway Analysis of Up-Regulated Proteins
The differentially expressed proteins with high expression in IEC-Exos from the ileum were mainly enriched in signaling pathways such as ribosome, spliceosome, RNA transport, and proteasome pathways (Figure 5B). The KEGG analysis results showed that these proteins were mainly involved in genetic information processing, including translation, transcription, folding, sorting, and degradation. These proteins mainly affected biological processes such as cell growth and death, lipid metabolism and signal transduction. In addition to being related to the function of the digestive system, these proteins were also related to the functions of the immune system, endocrine system, circulatory system and nervous system. In addition, these proteins were closely related to infectious diseases (viral or bacterial), cancers and immune diseases (Figure 5C).
Kyoto Encyclopedia of Genes and Genomes Pathway Analysis of Down-Regulated Proteins
The differentially expressed proteins with high expression in IEC-Exos from the jejunum were mainly enriched in signaling pathways such as metabolism, peroxisome, drug metabolism, and the renin angiotensin system (Figure 5D). The KEGG analysis results showed that these proteins were mainly involved in metabolic pathways, including carbohydrate metabolism, amino acid metabolism, lipid metabolism and other amino acid metabolism. These proteins mainly affected processes such as cell transport and catabolism, cellular immunity and signal transduction. In addition to being closely related to the function of the digestive system, these proteins were also related to the functions of the endocrine system, immune system, nervous system, excretory system, circulatory system, etc. Furthermore, these proteins were related to infectious diseases (bacterial, viral or parasitic), endocrine and metabolic diseases, cardiovascular diseases, cancers, immune diseases, neurodegenerative diseases and other diseases (Figure 5E).
Protein–Protein Interaction Network
The hub of the differentially expressed proteins between ileum and jejunum IEC-Exos included ribosomal proteins (Figure 6A). The PPI network showed that the up-regulated proteins in ileum IEC-Exos were FABP6, Slc10a2, and RPL27a (Figure 6B), which are mainly involved in bile acid transport, fatty acid metabolism and protein synthesis pathways. However, the up-regulated proteins in jejunum IEC-Exos mainly included ACE2, ACE, Pdzk1, Asah2, ADH1, ARG2, Slc2a2, and Rbp2 (Figure 6C), which mediate sugar metabolism, fatty acid metabolism, amino acid metabolism, drug metabolism, bone metabolism, vitamin absorption, the renin-angiotensin system (RAS), NO, etc.
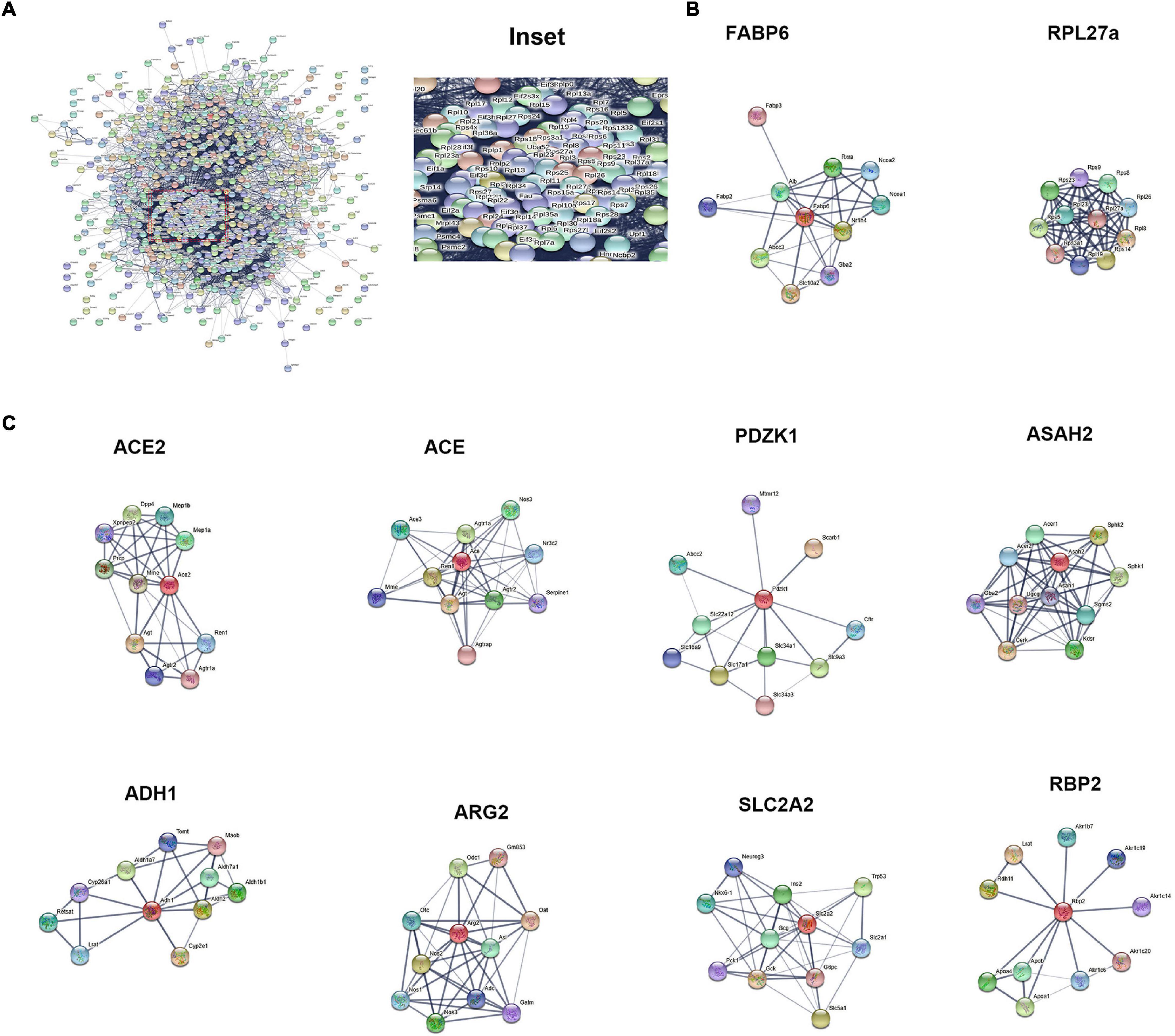
Figure 6. Protein–protein interaction network. (A) PPI network analysis of the hub of the differentially expressed proteins between ileum and jejunum IEC-Exos. (B) PPI network analysis of up-regulated proteins. (C) PPI network analysis of up-regulated proteins.
Discussion
In this study, ileum and jejunum IEC-Exos were extracted by ultracentrifugation. Under transmission electron microscopy, both ileum and jejunum IEC-Exos presented as round or elliptical vesicles with a clear double-layered membrane structure, which conformed to the characteristics of exosomes. Subsequently, we detected the expression of the exosomal-specific marker proteins CD81 and TSG101 by western blotting. The results of nanoparticle tracking analysis showed that the diameter of ileum IEC-Exos was mainly concentrated at 30–130 nm, and the diameter of jejunum IEC-Exos was mainly concentrated at 30–120 nm, both of which were in the range of the diameter of exosomes.
Exosomes are rich in active substances, such as proteins, nucleic acids, and lipids. The proteins in exosomes play important roles in the processes of cellular material transport, signal transduction and antigen presentation. Exosomes derived from different cells have differences in the compositions and functions of their contents (Mathivanan et al., 2010; Pegtel and Gould, 2019). Proteomics studies on exosomes are of great significance for revealing the pathogenesis of related diseases, identifying biomarkers for disease diagnosis and prognosis, and screening disease treatment targets (Xiao et al., 2009; Kalluri and LeBleu, 2020).
In this study, for the first time, the iTRAQ quantitative proteomic approach combined with LC-MS/MS technology was used to detect IEC-Exo proteins, which was helpful for identifying exosomal proteins. The results showed that compared with jejunum IEC-Exos from ileum IEC-Exos, there were 393 up-regulated proteins and 346 down-regulated proteins, which indicated that differentially expressed proteins between ileum and jejunum IEC-Exos might perform different biological functions. Through analysis of biological information, we studied and compared the biological functions and related signaling pathways of the differential expressed proteins between ileum and jejunum IEC-Exos, which improved our understanding of the functions of the ileum and jejunum. Abundant exosomal proteins may have important significance in revealing the different biological functions between the ileum and jejunum.
The mammalian intestine is covered by a single layer of epithelial cells that is renewed every 4–5 days and performs the main functions of digestion and water and nutrient absorption. In addition, epithelial cells form a barrier against pathogens in the cavity. Due to the technological advancement of large-scale single-cell transcriptome profiling, more precise and comprehensive descriptions of cell types have been obtained from a multitude of organs.
Based on the expression of known marker genes in humans (Wang Y. et al., 2020) and mice (Haber et al., 2017), eight different cell types were identified, including enterocyte cells, goblet cells, Paneth cells, enteroendocrine cells, tuft cells, progenitor cells, transit amplifying (TA) cells and stem cells. In our study, the expression of IEC-Exo proteins was consistent with the abovementioned mRNA expression, indicating that various small intestinal epithelial cells can secrete exosomes. Furthermore, IEC-Exos contained a variety of cell-derived proteins and played an important role in regulating the biological functions of the intestine.
The small intestine is the main organ involved in nutrient digestion and absorption. Different sections of the small intestine have different digestion and absorption capacities for nutrient molecules (Borgstrom et al., 1957; Silk et al., 1974). Most of the digestion products of carbohydrates, proteins and lipids are absorbed in the duodenum and jejunum. When the nutrients in food reach the ileum, they are mostly digested and absorbed, so the ileum is the reserve part for absorption. However, the ileum has a unique role of actively absorbing bile salts and vitamin B12 and plays an important role in the digestion and absorption of fatty acids. Moreover, the small intestine is the main organ involved in oral drug absorption and drug metabolism (Billat et al., 2017). Studies have shown that small intestinal epithelial cells contain a large number of bioconverting enzymes. The longitudinal distributions of small intestinal cytochrome P450, glutathione S-transferase and bilirubin-uridine 5′-diphosphate (UDP)-glucuronyltransferase have a downward trend from the duodenum to the ileum (Tahir et al., 1988; Peters et al., 1989), suggesting that the antioxidant and detoxification effects of the proximal small intestine are stronger than that of the distal small intestine.
Intestinal epithelial cell derived exosomes selectively enrich protein bioactive components derived from small intestinal epithelial cells, which can regulate multiple functions and biological processes of cells after being taken up by target cells. Our study explored the MF, CC, and BP of differentially expressed proteins from ileum and jejunum IEC-Exos based on GO analysis. These proteins were mainly involved in gene expression, metabolic processes, biosynthetic processes, regulatory processes, and transport. The highly expressed differentially proteins – in ileum IEC-Exos were closely related to the transport of bile acids and the digestion and absorption of fatty acids. The level of participation of jejunum IEC-Exo proteins in of sugar, fat, protein, and drug metabolism and redox reactions was higher than that of ileum IEC-Exos, suggesting that the highly expressed differentially proteins in ileum and jejunum IEC-Exos played different biological roles, which was consistent with the aforementioned functions of the small intestine.
Kyoto encyclopedia of genes and genomes biological information analysis showed that the differentially expressed proteins between ileum and jejunum IEC-Exos were mainly involved in metabolic processes, the ribosome, the spliceosome, RNA transport and drug metabolism-other enzymes. The highly expressed differentially expressed proteins in ileum IEC-Exos were mainly enriched in genetic information processing pathways, which mainly affected processes such as cell growth and death, lipid metabolism and signal transduction. The highly expressed differentially expressed proteins in jejunum IEC-Exos were mainly enriched in metabolic pathways, which mainly affected cell transport and catabolism, cellular immunity, and signal transduction. These results indicated that the differentially expressed between the ileum and jejunum IEC-Exos participated in different signaling pathways and played important roles in regulating intestinal biological functions.
Ribosomal proteins and RNA form the ribosome, which is the organelle involved in protein synthesis. In addition to participating in protein synthesis, ribosomal proteins also have a wide range of in vitro functions in ribosomes, such as regulation of gene transcription, mRNA translation, cell proliferation, differentiation, and apoptosis (Warner and McIntosh, 2009; Zhou et al., 2015). In our study, PPI analysis showed that the core differentially expressed proteins between ileum and jejunum IEC-Exos included ribosomal proteins, which had the characteristics of active proliferation, strong self-renewing ability, and extensive biological functions of small intestinal epithelial cells. In particular, ileum IEC-Exos contained a variety of ribosomal proteins, suggesting that ileum epithelial cells participate in more physiological functions and pathophysiological processes than jejunum epithelial cells and have a stronger renewal speed and damage repair potential. Partial small bowel resection experiments performed in rats found (Ziegler et al., 1998) that the ileum exhibited a stronger adaptive growth ability than the jejunum, which was consistent with our results.
The core proteins FABP6 and SLC10A2, which were highly expressed in ileum IEC-Exos, are important bile acid transporters that are regulated by bile acids, cholesterol and hormones (Xiao and Pan, 2017). Dysfunctions of FABP6 and SLC10A2 are mainly involved in hepatobiliary diseases, inflammatory bowel disease, metabolic diseases and intestinal tumors. Thus, FABP6 and SLC10A2 are expected to become new targets for the treatment of related diseases and drug discovery (Zhang Y. et al., 2019; Yang et al., 2020). RPL27a is an important ribosomal protein that interacts with other ribosomal proteins to participate in protein synthesis. Clinical studies have found that RPL27a-related genes and pathways are closely related to the occurrence and development of intestinal tumors (Yajima et al., 2007; Yu et al., 2019). FABP6, SLC10A2, and RPL27a were highly expressed in ileum IEC-Exos, indicating that ileum IEC-Exos played key roles in bile acid transport, fatty acid digestion and absorption, and protein synthesis, which are associated with the occurrence and development of ileum functions and related diseases.
The SLC superfamily is one of the most important membrane transporter families in the cell membrane. The SLC superfamily is involved in essential physiological functions, such as intercellular substance transport, energy transfer, nutrition and metabolism, and signal transduction (Schumann et al., 2020). SLC transporters were found in jejunum IEC-Exos, indicating that they play an important role in the occurrence and development of material metabolism and related diseases, and are expected to become new therapeutic targets for the treatment of metabolic diseases.
Protein–protein interaction analysis further showed that ACE2, ACE, Pdzk1, Asah2, ADH1, ARG2, Slc2a2, and Rbp2 were enriched in jejunum IEC-Exos and participated in many signaling pathways. ACE2 and angiotensin-converting enzyme (ACE) are the key regulators of the RAS. The ACE2-Ang(1-7)-Mas receptor axis and ACE-Ang II-AT1 receptor axis are antagonistic to each other and interact with Mme, which plays an important role in regulating cardiovascular function, respiratory function, water and electrolyte balance, intestinal homeostasis, bone metabolism and nervous system function (Imai et al., 2010).
A novel coronavirus, SARS-CoV-2, has caused a global pandemic of COVID-19. Viral infection with SARS-CoV-2 causes a series of respiratory illnesses, including severe respiratory syndrome, indicating that the virus most likely infects respiratory epithelial cells and mainly spreads via the respiratory tract from human to human. However, gastrointestinal symptoms have been found in a substantial proportion of patients with COVID-19. Viral RNA has been detected in respiratory and stool specimens of patients, suggesting that SARS-CoV-2 may cause an enteric infection, in addition to a respiratory infection (Galanopoulos et al., 2020; Jin et al., 2021). Research has confirmed robust SARS-CoV-2 replication in human intestinal organoids, suggesting that the human intestinal tract may be a transmission route of SARS-CoV-2 (Zhou et al., 2020). It has been reported that ACE2 is the main host cell receptor of SARS-CoV-2 and that it plays a crucial role in the entry of virus into the cell to cause infection (Ziegler et al., 2020; Bickler et al., 2021). Furthermore, ACE2 expression has mainly been observed in human enterocytes, renal tubules, gallbladder, cardiomyocytes, vasculature and the lung (Hikmet et al., 2020). The distribution of viral receptors in different cell types of diverse tissues may indicate viral tropism and potential transmission routes, and SARS-CoV-2 productively infects human gut enterocytes (Lamers et al., 2020). IEC-Exos contained ACE2, which is new evidence that SARS-CoV-2 can infect intestinal cells.
Proteins, genomic molecules and receptors from infected cells make healthy cells more susceptible to infection. Exosome-mediated transfer of viruses may participate in viral infection (Pocsfalvi et al., 2020; Todd and Tripp, 2020) but has not yet been fully elucidated for coronaviruses. IEC-Exos may be a potential route of SARS-CoV-2 infection and may provide new ideas for further in-depth exploration of the mechanism of multiple organ damage in COVID-19. Jejunum IEC-Exos contained a higher level of ACE2 than ileum IEC-Exos, suggesting that the jejunum might be more susceptible to SARS-CoV-2. These findings provide a rich resource for future investigations of COVID-19 and its pathogenesis.
The small intestine is not only the main site of oral drug absorption but is also an important site of drug metabolism. The main site of intestinal absorption and metabolism of most oral drug formulations is the proximal small intestine because the proximal small intestine contains a large number of transporters and drug metabolism-related enzymes. The PDZ kinase 1 (PDZK1) protein is a member of a family of transporter adaptor proteins containing PDZ domains. The PDZK1 protein mainly binds to drug transporters and regulates their location, expression and function, which mediate the transmembrane transport of a variety of nutrients, endogenous substances and drugs (Sugiura et al., 2008; DeGorter et al., 2012). Jejunum IEC-Exos enriched a higher level of the PDZK1 protein than ileum IEC-Exos, providing further evidence that the jejunum plays a major role in drug absorption and metabolism.
The neutral ceramidase N-acylsphingosine amide hydrolase 2 (ASAH2) is a key enzyme in ceramide metabolism. ASAH2, which is expressed in the intestine, plays a major role in ceramide metabolism in the gut (Kono et al., 2006; Ohlsson et al., 2007), and ceramide mainly affects metabolic and disease states. An accumulation of ceramide has been observed in type 2 diabetes mellitus (T2DM), non-alcoholic fatty liver disease (NAFLD), inflammatory bowel disease (IBD), Alzheimer’s disease (AD), and cancer. Clarifying the role of ASAH2 in human diseases and determining its potential for use in the treatment of metabolic disorders and neurodegenerative diseases are critical (Parveen et al., 2019).
Traditionally, alcohol dehydrogenase 1 (ADH1) and aldehyde dehydrogenase 2 (ALDH2) in the liver are key enzymes in the ethanol metabolism pathway. Studies have shown that both ADH1 and ALDH2 are expressed in epithelial cells in the human digestive tract, especially in the proximal small intestine, which is the main site of ethanol metabolism (Chiang et al., 2012). ALDH1 (the ADH1 gene cluster: ADH1A, ADH1B, and ADH1C) plays a key role in the metabolic pathways of substances such as alcohol and retinol. Abnormal or unregulated expression of ALDH1 can cause liver disease, tumors, alcohol addiction and vitamin A absorption disorder (Gaviria-Calle et al., 2018; Wang S.C. et al., 2020; Sun et al., 2021). Jejunum IEC-Exos contained higher levels of ADH1, indicating that it plays an important role in ethanol metabolism and vitamin A absorption.
Arginase-2 (Arg-2) is expressed in the small intestine, kidney, brain, lactating mammary glands, monocytes, and macrophages (Kohler et al., 2008; Choi et al., 2012). Arg-2 and nitric oxide synthase (NOS) participate in the intracellular arginine metabolism pathway, regulating the intracellular concentration of arginine and the synthesis of NO, proline and polyamines in organisms. Furthermore, Arg-2 and NOS play important roles in regulating cardiovascular function, the inflammatory response, oxidative stress, immune function, and tumor occurrence and development. Inhibition of arginase is proposed as a method to improve antitumor immune responses (via activation and proliferation of T and NK cells) (Borek et al., 2020). Clinical studies have shown that L-Arginine/Nitric Oxide pathway was closely related to Crohn’s disease (CD) (Krzystek-Korpacka et al., 2020), CD can occur anywhere in the entire digestive tract, but is more common in the distal ileum and right colon. Compared with normal tissues, Arg-2 was down-regulated in the inflamed small intestine with CD. We found that IEC-Exos enriched Arg-2, especially ileum IEC-Exos contained lower levels of Arg-2, indicating that IEC-Exos may be involved in the pathogenesis of CD.
Slc2a2 (GLUT2) is expressed in small intestinal epithelial cells and mediates the glucose metabolism pathway, glucose absorption, and gluconeogenesis. In addition, GLUT2 participates in the glucagon and insulin signaling pathways, which are closely related to the occurrence of diabetes (O’Brien et al., 2018; Holman, 2020).
Retinol-binding protein 2 (Rbp2) is highly expressed in proximal small intestinal epithelial cells. The interactions between RBP2 and apolipoprotein are mainly involved in the binding, absorption, transport and metabolism of vitamin A and lipids (Blaner et al., 2020). Jejunum IEC-Exos contained high levels of RBP2, which could participate in the proliferation, migration, metastasis, and drug resistance of cancer cells. Thus, RBP2 may be a target for tumor treatment (Qi et al., 2014).
In addition to its important digestion and absorption functions, the intestine is also a very important immune organ and endocrine organ of the human body. Small intestinal epithelial cells synthesize and secrete a variety of cytokines to participate in the digestion and absorption of nutrients, microbial defense and immune responses, and endocrine functions. In our study, it was observed that the differentially expressed proteins enriched in ileum and jejunum IEC-Exos were not only closely related to the function of the digestive system but also related to infectious diseases, endocrine and metabolic diseases, cardiovascular diseases, cancers, immune diseases, neurodegenerative diseases and osteoarthritis. This study provides a new experimental basis for further in-depth study of the digestive system and disease occurrence.
Conclusion
In summary, IEC-Exos contained a variety of cytokines secreted by intestinal epithelial cells, and there were many differentially expressed proteins between ileum and jejunum IEC-Exos, which played different roles in regulating intestinal biological functions. The highly expressed differentially expressed proteins in ileum IEC-Exos mainly mediated the functions of bile acid transport, fatty acid metabolism, protein synthesis and processing modifications. Moreover, the proteins in jejunum IEC-Exos mainly mediated sugar metabolism, fatty acid metabolism, amino acid metabolism, drug metabolism, bone metabolism, vitamin absorption, the RAS and the NO signaling system. IEC-Exos, especially in the jejunum, contained high levels of ACE2, which provided further evidence that SARS-CoV-2 infection could occur in the intestine. This study provided an important basis for further in-depth study of the function of small intestinal epithelial cells and related diseases. However, these results were approached in the manner of bioinformatics analysis; therefore, further verification is required.
Data Availability Statement
The data presented in the study are deposited in the ProteomeXchange repository (http://www.proteomexchange.org/), accession number PXD030945.
Ethics Statement
The animal study was reviewed and approved by 2020-0215.
Author Contributions
ZD and CZ designed the research, performed the collection of exosome samples, and contributed to writing scripts. ZD, CZ, and BZ performed the proteome analysis and data analysis. QL supervised and guided the project. All the authors contributed to the article and approved the submitted version.
Funding
Supported by grants from the National Natural Science Foundation of China (81770542).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Bickler, S. W., Cauvi, D. M., Fisch, K. M., Prieto, J. M., Sykes, A. G., Thangarajah, H., et al. (2021). Extremes of age are associated with differences in the expression of selected pattern recognition receptor genes and ACE2, the receptor for SARS-CoV-2: implications for the epidemiology of COVID-19 disease. BMC Med. Genomics 14:138. doi: 10.1186/s12920-021-00970-7
Billat, P. A., Roger, E., Faure, S., and Lagarce, F. (2017). Models for drug absorption from the small intestine: where are we and where are we going? Drug Discov. Today 22, 761–775. doi: 10.1016/j.drudis.2017.01.007
Blaner, W. S., Brun, P. J., Calderon, R. M., and Golczak, M. (2020). Retinol-binding protein 2 (RBP2): biology and pathobiology. Crit. Rev. Biochem. Mol. Biol. 55, 197–218. doi: 10.1080/10409238.2020.1768207
Borek, B., Gajda, T., Golebiowski, A., and Blaszczyk, R. (2020). Boronic acid-based arginase inhibitors in cancer immunotherapy. Bioorg. Med. Chem. 28:115658. doi: 10.1016/j.bmc.2020.115658
Borgstrom, B., Dahlqvist, A., Lundh, G., and Sjovall, J. (1957). Studies of intestinal digestion and absorption in the human. J. Clin. Invest. 36, 1521–1536.
Chiang, C. P., Wu, C. W., Lee, S. P., Ho, J. L., Lee, S. L., Nieh, S., et al. (2012). Expression pattern, ethanol-metabolizing activities, and cellular localization of alcohol and aldehyde dehydrogenases in human small intestine. Alcohol Clin. Exp. Res. 36, 2047–2058. doi: 10.1111/j.1530-0277.2012.01836.x
Choi, S., Park, C., Ahn, M., Lee, J. H., and Shin, T. (2012). Immunohistochemical study of arginase 1 and 2 in various tissues of rats. Acta Histochem. 114, 487–494. doi: 10.1016/j.acthis.2011.09.002
de Wit, N. J., Bosch-Vermeulen, H., de Groot, P. J., Hooiveld, G. J., Bromhaar, M. M., Jansen, J., et al. (2008). The role of the small intestine in the development of dietary fat-induced obesity and insulin resistance in C57BL/6J mice. BMC Med. Genomics 1:14. doi: 10.1186/1755-8794-1-14
DeGorter, M. K., Xia, C. Q., Yang, J. J., and Kim, R. B. (2012). Drug transporters in drug efficacy and toxicity. Annu. Rev. Pharmacol. Toxicol. 52, 249–273.
Galanopoulos, M., Gkeros, F., Doukatas, A., Karianakis, G., Pontas, C., Tsoukalas, N., et al. (2020). COVID-19 pandemic: pathophysiology and manifestations from the gastrointestinal tract. World J. Gastroenterol. 26, 4579–4588. doi: 10.3748/wjg.v26.i31.4579
Gaviria-Calle, M., Duque-Jaramillo, A., Aranzazu, M., Di Filippo, D., Montoya, M., Roldan, I., et al. (2018). Polymorphisms in alcohol dehydrogenase (ADH1) and cytochrome p450 2E1 (CYP2E1) genes in patients with cirrhosis and/or hepatocellular carcinoma. Biomedica 38, 555–568. doi: 10.7705/biomedica.v38i4.3897
Gribble, F. M., and Reimann, F. (2019). Function and mechanisms of enteroendocrine cells and gut hormones in metabolism. Nat. Rev. Endocrinol. 15, 226–237. doi: 10.1038/s41574-019-0168-8
Haber, A. L., Biton, M., Rogel, N., Herbst, R. H., Shekhar, K., Smillie, C., et al. (2017). A single-cell survey of the small intestinal epithelium. Nature 551, 333–339. doi: 10.1038/nature24489
Hassanpour, M., Rezaie, J., Nouri, M., and Panahi, Y. (2020). The role of extracellular vesicles in COVID-19 virus infection. Infect. Genet. Evol. 85:104422. doi: 10.1016/j.meegid.2020.104422
Hikmet, F., Mear, L., Edvinsson, A., Micke, P., Uhlen, M., and Lindskog, C. (2020). The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 16:e9610. doi: 10.15252/msb.20209610
Holman, G. D. (2020). Structure, function and regulation of mammalian glucose transporters of the SLC2 family. Pflugers Arch. 472, 1155–1175. doi: 10.1007/s00424-020-02411-3
Imai, Y., Kuba, K., Ohto-Nakanishi, T., and Penninger, J. M. (2010). Angiotensin-converting enzyme 2 (ACE2) in disease pathogenesis. Circ. J. 74, 405–410. doi: 10.1253/circj.cj-10-0045
Jin, B., Singh, R., Ha, S. E., Zogg, H., Park, P. J., and Ro, S. (2021). Pathophysiological mechanisms underlying gastrointestinal symptoms in patients with COVID-19. World J. Gastroenterol. 27, 2341–2352. doi: 10.3748/wjg.v27.i19.2341
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science 367:eaau6977. doi: 10.1126/science.aau6977
Kiela, P. R., and Ghishan, F. K. (2016). Physiology of intestinal absorption and secretion. Best Pract. Res. Clin. Gastroenterol. 30, 145–159.
Kohler, E. S., Sankaranarayanan, S., van Ginneken, C. J., van Dijk, P., Vermeulen, J. L., Ruijter, J. M., et al. (2008). The human neonatal small intestine has the potential for arginine synthesis; developmental changes in the expression of arginine-synthesizing and -catabolizing enzymes. BMC Dev. Biol. 8:107. doi: 10.1186/1471-213X-8-107
Kono, M., Dreier, J. L., Ellis, J. M., Allende, M. L., Kalkofen, D. N., Sanders, K. M., et al. (2006). Neutral ceramidase encoded by the Asah2 gene is essential for the intestinal degradation of sphingolipids. J. Biol. Chem. 281, 7324–7331. doi: 10.1074/jbc.M508382200
Krzystek-Korpacka, M., Fleszar, M. G., Bednarz-Misa, I., Lewandowski, Ł, Szczuka, I., Kempiński, R., et al. (2020). Transcriptional and metabolomic analysis of L-arginine/nitric oxide pathway in inflammatory bowel disease and its association with local inflammatory and angiogenic response: preliminary findings. Int. J. Mol. Sci. 21:1641. doi: 10.3390/ijms21051641
Lamers, M. M., Beumer, J., van der Vaart, J., Knoops, K., Puschhof, J., Breugem, T. I., et al. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54. doi: 10.1126/science.abc1669
Mathivanan, S., Ji, H., and Simpson, R. J. (2010). Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 73, 1907–1920. doi: 10.1016/j.jprot.2010.06.006
O’Brien, P., Hewett, R., and Corpe, C. (2018). Sugar sensor genes in the murine gastrointestinal tract display a cephalocaudal axis of expression and a diurnal rhythm. Physiol. Genomics 50, 448–458. doi: 10.1152/physiolgenomics.00139.2017
Ohlsson, L., Palmberg, C., Duan, R. D., Olsson, M., Bergman, T., and Nilsson, A. (2007). Purification and characterization of human intestinal neutral ceramidase. Biochimie 89, 950–960. doi: 10.1016/j.biochi.2007.03.009
Parveen, F., Bender, D., Law, S. H., Mishra, V. K., Chen, C. C., and Ke, L. Y. (2019). Role of ceramidases in sphingolipid metabolism and human diseases. Cells 8:1573. doi: 10.3390/cells8121573
Pegtel, D. M., and Gould, S. J. (2019). Exosomes. Annu. Rev. Biochem. 88, 487–514. doi: 10.1016/b978-0-12-816053-4.00021-3
Peters, W. H., Nagengast, F. M., and van Tongeren, J. H. (1989). Glutathione S-transferase, cytochrome P450, and uridine 5’-diphosphate-glucuronosyltransferase in human small intestine and liver. Gastroenterology 96, 783–789.
Peterson, L. W., and Artis, D. (2014). Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14, 141–153. doi: 10.1038/nri3608
Pocsfalvi, G., Mammadova, R., Ramos Juarez, A. P., Bokka, R., Trepiccione, F., and Capasso, G. (2020). COVID-19 and extracellular vesicles: an intriguing interplay. Kidney Blood Press. Res. 45, 661–670. doi: 10.1159/000511402
Qi, L., Zhu, F., Li, S. H., Si, L. B., Hu, L. K., and Tian, H. (2014). Retinoblastoma binding protein 2 (RBP2) promotes HIF-1alpha-VEGF-induced angiogenesis of non-small cell lung cancer via the Akt pathway. PLoS One 9:e106032. doi: 10.1371/journal.pone.0106032
Satsu, H. (2017). Molecular and cellular studies on the absorption, function, and safety of food components in intestinal epithelial cells. Biosci. Biotechnol. Biochem. 81, 419–425. doi: 10.1080/09168451.2016.1259552
Schumann, T., Konig, J., Henke, C., Willmes, D. M., Bornstein, S. R., Jordan, J., et al. (2020). Solute carrier transporters as potential targets for the treatment of metabolic disease. Pharmacol. Rev. 72, 343–379. doi: 10.1124/pr.118.015735
Silk, D. B., Webb, J. P., Lane, A. E., Clark, M. L., and Dawson, A. M. (1974). Functional differentiation of human jejunum and ileum: a comparison of the handling of glucose, peptides, and amino acids. Gut 15, 444–449. doi: 10.1136/gut.15.6.444
Sugiura, T., Kato, Y., Wakayama, T., Silver, D. L., Kubo, Y., Iseki, S., et al. (2008). PDZK1 regulates two intestinal solute carriers (Slc15a1 and Slc22a5) in mice. Drug Metab. Dispos. 36, 1181–1188. doi: 10.1124/dmd.107.020321
Sun, H. W., Chen, J., Wu, W. C., Yang, Y. Y., Xu, Y. T., Yu, X. J., et al. (2021). Retinoic acid synthesis deficiency fosters the generation of polymorphonuclear myeloid-derived suppressor cells in colorectal cancer. Cancer Immunol. Res. 9, 20–33. doi: 10.1158/2326-6066.CIR-20-0389
Tahir, M. K., Ozer, N., and Mannervik, B. (1988). Isoenzymes of glutathione transferase in rat small intestine. Biochem. J. 253, 759–764. doi: 10.1042/bj2530759
Tajik, N., Frech, M., Schulz, O., Schalter, F., Lucas, S., Azizov, V., et al. (2020). Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat. Commun. 11, 1–14. doi: 10.1038/s41467-020-15831-7
Todd, K. V., and Tripp, R. A. (2020). Exosome-mediated human norovirus infection. PLoS One 15:e0237044. doi: 10.1371/journal.pone.0237044
van Niel, G., and Heyman, M. (2002). The epithelial cell cytoskeleton and intracellular trafficking. II. Intestinal epithelial cell exosomes: perspectives on their structure and function. Am. J. Physiol. Gastrointest. Liver Physiol. 283, G251–G255. doi: 10.1152/ajpgi.00102.2002
Wang, S. C., Chen, Y. C., Chen, S. J., Lee, C. H., and Cheng, C. M. (2020). Alcohol addiction, gut microbiota, and alcoholism treatment: a review. Int. J. Mol. Sci. 21:6413. doi: 10.3390/ijms21176413
Wang, Y., Song, W., Wang, J., Wang, T., Xiong, X., Qi, Z., et al. (2020). Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med. 217:e20191130. doi: 10.1084/jem.20191130
Warner, J. R., and McIntosh, K. B. (2009). How common are extraribosomal functions of ribosomal proteins? Mol. Cell. 34, 3–11. doi: 10.1016/j.molcel.2009.03.006
Wisniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Xia, F., Ding, F., Lv, Y., Di, W., Sheng, Y., and Ding, G. (2019). A high efficient method to isolate exosomes from small intestinal epithelium. Mol. Biotechnol. 61, 325–331. doi: 10.1007/s12033-019-00163-9
Xiao, L., and Pan, G. (2017). An important intestinal transporter that regulates the enterohepatic circulation of bile acids and cholesterol homeostasis: the apical sodium-dependent bile acid transporter (SLC10A2/ASBT). Clin. Res. Hepatol. Gastroenterol. 41, 509–515. doi: 10.1016/j.clinre.2017.02.001
Xiao, Z., Blonder, J., Zhou, M., and Veenstra, T. D. (2009). Proteomic analysis of extracellular matrix and vesicles. J. Proteomics 72, 34–45. doi: 10.1016/j.jprot.2008.11.011
Yajima, S., Ishii, M., Matsushita, H., Aoyagi, K., Yoshimatsu, K., Kaneko, H., et al. (2007). Expression profiling of fecal colonocytes for RNA-based screening of colorectal cancer. Int. J. Oncol. 31, 1029–1037.
Yang, N., Dong, Y. Q., Jia, G. X., Fan, S. M., Li, S. Z., Yang, S. S., et al. (2020). ASBT(SLC10A2): a promising target for treatment of diseases and drug discovery. Biomed. Pharmacother. 132:110835. doi: 10.1016/j.biopha.2020.110835
Yu, C., Hong, H., Zhang, S., Zong, Y., Ma, J., Lu, A., et al. (2019). Identification of key genes and pathways involved in microsatellite instability in colorectal cancer. Mol. Med. Rep. 19, 2065–2076. doi: 10.3892/mmr.2019.9849
Zhang, H., Wang, L., Li, C., Yu, Y., Yi, Y., Wang, J., et al. (2019). Exosome-induced regulation in inflammatory bowel disease. Front. Immunol. 10:1464. doi: 10.3389/fimmu.2019.01464
Zhang, Y., Zhao, X., Deng, L., Li, X., Wang, G., Li, Y., et al. (2019). High expression of FABP4 and FABP6 in patients with colorectal cancer. World J. Surg. Oncol. 17:171. doi: 10.1186/s12957-019-1714-5
Zhou, J., Li, C., Liu, X., Chiu, M. C., Zhao, X., Wang, D., et al. (2020). Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 26, 1077–1083. doi: 10.1038/s41591-020-0912-6
Zhou, X., Liao, W. J., Liao, J. M., Liao, P., and Lu, H. (2015). Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 7, 92–104.
Ziegler, C. G. K., Allon, S. J., Nyquist, S. K., Mbano, I. M., Miao, V. N., Tzouanas, C. N., et al. (2020). SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016.e9–1035.e9. doi: 10.1016/j.cell.2020.04.035
Keywords: exosomes, intestinal epithelial cells, proteomics, exosome proteins, ACE2
Citation: Ding Z, Zhang C, Zhang B and Li Q (2022) Unraveling the Proteomic Landscape of Intestinal Epithelial Cell-Derived Exosomes in Mice. Front. Physiol. 13:773671. doi: 10.3389/fphys.2022.773671
Received: 10 September 2021; Accepted: 14 January 2022;
Published: 23 February 2022.
Edited by:
Stephen J. Pandol, Cedars-Sinai Medical Center, United StatesReviewed by:
Nur Izzah Ismail, The Chinese University of Hong Kong, ChinaRaquel Arifa, Minas Gerais State University, Brazil
Copyright © 2022 Ding, Zhang, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenyu Ding, ZHpoZW55dUBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Zhenyu Ding
Zhenyu Ding Cuiyu Zhang
Cuiyu Zhang Baokun Zhang1
Baokun Zhang1