- 1Institute of Plant Protection, Jiangxi Academy of Agricultural Sciences, Nanchang, China
- 2Institute of Insect Sciences/Ministry of Agriculture Key Laboratory of Molecular Biology of Crop Pathogens and Insect Pests, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 3Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, Jiangsu Key Laboratory for Food and Safety-State Key Laboratory Cultivation Base of Ministry of Science and Technology, Nanjing, China
- 4Institute of Animal Husbandry and Veterinary Medicine, Jiangxi Academy of Agricultural Sciences, Nanchang City, China
- 5Institute of Soil Fertilizer and Environmental Resource, Jiangxi Academy of Agricultural Sciences, Nanchang, China
- 6Institute of Red Soil and Germplasm Resources in Jiangxi, Nanchang, China
Long non-coding RNAs, referred to as lncRNAs, perform essential functions in some biological processes, including reproduction, metamorphosis, and other critical life functions. Yet, lncRNAs are poorly understood in pesticide resistance, and no reports to date have characterized which lncRNAs are associated with chlorantraniliprole resistance in Chilo suppressalis. Here, RNA-seq was performed on two strains of C. suppressalis exposed to chlorantraniliprole: one is a susceptible strain (S), and the other is a resistant strain (R). In total, 3,470 lncRNAs were identified from 40,573 merged transcripts in six libraries, including 1,879 lincRNAs, 245 intronic lncRNAs, 853 sense lncRNAs, and 493 antisense lncRNAs. Moreover, differential expression analysis revealed 297 and 335 lncRNAs upregulated in S and R strains, respectively. Differentially expressed (DE) lncRNAs are usually assumed to be involved in the chlorantraniliprole resistance in C. suppressalis. As potential targets, adjacent protein-coding genes (within <1000 kb range upstream or downstream of DE lncRNAs), especially detoxification enzyme genes (cytochrome P450s, carboxyl/cholinesterases/esterases, and ATP-binding cassette transporter), were analyzed. Furthermore, the strand-specific RT-PCR was conducted to confirm the transcript orientation of randomly selected 20 DE lincRNAs, and qRT-PCR was carried out to verify the expression status of 8 out of them. MSTRG.25315.3, MSTRG.25315.6, and MSTRG.7482.1 were upregulated in the R strain. Lastly, RNA interference and bioassay analyses indicated overexpressed lincRNA MSTRG.7482.1 was involved in chlorantraniliprole resistance. In conclusion, we represent, for the first time, the genome-wide identification of chlorantraniliprole-resistance-related lncRNAs in C. suppressalis. It elaborates the views underlying the mechanism conferring chlorantraniliprole resistance in lncRNAs.
Introduction
The striped stem borer (termed SSB), Chilo suppressalis Walker (Lepidoptera: Pyralidae), is one of the most destructive insect pests of rice in China, causing significant yield losses. The SSB larvae feed within plant stems, and damage leads to death of sheaths and cores, resulting in white tassels in rice (Sheng et al., 2003; He et al., 2013). Management of this insect pest mainly relies on insecticide application. Monosultap, triazophos, abamectin, and chlorantraniliprole are currently the most widely used insecticides in reducing the population of SSB (Shuijin et al., 2017). Chlorantraniliprole is the most effective of these options; it is effective against SSB and stem borers and is utilized most heavily in areas known to consistently have high densities of the pest (Sun et al., 2021; Dai et al., 2022). Chlorantraniliprole is an anthranilic diamide insecticide that functions via activation of the insect ryanodine receptors located on the sarcoplasmic reticulum in muscle cells, causing sustained release of calcium levels within the cytosol which leads to muscle contraction, paralysis, and eventual death of the organism (Lahm et al., 2005; Ebbinghaus-Kintscher et al., 2006). However, SSB have developed a low-level and medium-level (Su et al., 2014) resistance to chlorantraniliprole in a short time, eventually developing high-level resistance (Yao et al., 2017), followed very soon by extremely-high-level resistance (Sun et al., 2018).
Understanding of resistance mechanism is helpful in formulating more reasonable resistance management strategies. Target resistance can be determined by the interaction pattern between chlorantraniliprole and RyR (Guo et al., 2014). In SSB, four RyR mutants (I4758M, G4910E, I4891F, and Y4667D) were reported, and they were recently functionally confirmed (Yao et al., 2017; Sun et al., 2018; Huang et al., 2020). The metabolic detoxification was also tested already in chlorantraniliprole. Over-expression of cytochrome P450 genes, CYP321F3, CYP6CV5, CYP32412, and CYP9A68, caused chlorantraniliprole insensitivity (Xu et al., 2019). Increased esterase activity possibly improved chlorantraniliprole resistance as well (Sun et al., 2018). A genome-wide study discovered dozens of ATP-binding cassettes (ABCs) and found ABCA1, -D2, and -H2 are upregulated in all three resistant SSB strains (Peng et al., 2021). However, these previous studies mainly focus on the analyses of protein-coding genes, whereas non-coding RNA genes associated with chlorantraniliprole resistance in SSB have not been characterized.
LncRNAs are an important component of >200 nT non-coding RNA (Quinn and Chang, 2016) and were previously regarded as “transcriptional noise” (Struhl, 2007). More recently, evidence has shown that they perform essential regulatory functions in various biological processes or events, like imprinting genomic loci, X-chromosome silencing, shaping chromosome conformation, and carcinogenesis (Angrand et al., 2015; Chen et al., 2016; Diederichs et al., 2016; Quinn and Chang, 2016). Because the genome is difficult to obtain, these in-depth mechanism studies are limited to humans, mice, and the other model species (Li et al., 2019). For insects, the recent progress in sequencing technology, genome-wide investigation, and preliminary functional analysis was the main focus in lncRNA studies (Zhu et al., 2017; Chang et al., 2020; Yang et al., 2021; Wu et al., 2022). They identified 1309, 4516, 6171, and 11978 lncRNAs in Plutella xylostella (Zhu et al., 2017), Tribolium castaneum (Yang et al., 2021), Bactrocera dorsalis (Meng et al., 2021), and Spodoptera litura (Shi et al., 2022), respectively. In addition, the functions of insect lncRNAs could be involved in RNAi pathways, wing development, response to heat stress, plant–insect interactions, and pesticide resistance (Bernabo et al., 2020; Chen et al., 2020; Guan et al., 2020; Shang et al., 2021; Zhu et al., 2021).
Non-coding RNAs, such as lncRNAs, are hypothesized to play much more important roles in multiple biological processes, and uncovering their function in pesticide resistance will lead to novel insights and expanded avenues for integrated pest management (IPM). In this study, lncRNAs of SSB in susceptible (S) and resistant (R) strains were obtained via next-generation sequencing (NGS), and the differentially expressed (DE) lncRNAs in both S and R strains were investigated and located in the genome. Subsequently, adjacent protein-coding genes within 1000 kb apart from DE lncRNAs were mapped in the genome, including P450s, carboxyl/cholinesterases/esterases, and ABC genes. Also, the detoxification enzyme gene family was further analyzed to study their possible association with chlorantraniliprole resistance. Moreover, RT-PCR and qRT-PCR were conducted to validate the selected DE lncRNAs. Lastly, RNAi and chlorantraniliprole bioassay were performed to explore the lncRNA functions in chlorantraniliprole resistance. These findings report the first characterization of lncRNAs in SSB and elaborate the knowledge on the existing mechanism of development of chlorantraniliprole resistance in non-coding RNAs.
Materials and methods
Insect strains
A susceptible strain (S) of SSB was sourced from the Institute of Plant Protection, Chinese Academy of Agricultural Sciences (Haidian District, Beijing), provided by Dr. Lanzhi Han and reared on an artificial diet (Shuijin et al., 2017) under constant laboratory conditions (temperature: 27°C ± 1°C, relative humidity: 70%–80%, and photoperiod: 16:8) without insecticides for over 50 generations. The S strain was screened under chlorantraniliprole treatment for over 30 generations, and the resistance ratio (RR) of the R strain was 110.4 compared to the S strain, as previously reported (Sun et al., 2018). The SSB of both S and R strains were reared on an artificial diet with or without chlorantraniliprole treatment.
RNA extraction, library preparation, and sequencing
Total RNA was extracted from three to four 4th instar SSB larvae using the TRIzol reagent kit (Life Technologies, United States), following the manufacturer’s instructions. RNA quality was first checked by 1% (w/v) agarose gel electrophoresis, followed by a NanoDrop spectrophotometer (RNA model, California, United States). Lastly, Qubit 2.0 accurately quantified the RNA concentration. We finished the library construction and RNA-seq (Novogene, China). The total RNA of six samples (three independent biological replicates for S and R strains, respectively) was purified and qualified first, followed by the removal of rRNA using the Ribo-Zero kit (Epicentre, United States), the random disturbance of RNA in fragmentation buffer (NEB, United States), and the cDNA synthesis with 6-bp random hexamers. The purified double-stranded cDNA was repaired and adapter-ligated. Then, the cDNA library was enriched by PCR after being purified. Sequencing was performed on the NovaSeq 6000 platform.
lncRNA identification, gene structural features, and differential expression analysis
To isolate SSB lncRNAs from the sequencing dataset, a computational pipeline was constructed, which was referred to report with minor modifications (Liu et al., 2017). At the very beginning, Trimmomatic software was used to filter low-quality reads (Bolger et al., 2014). The raw reads from the six libraries were then mapped to the SSB genome via TopHat (Trapnell et al., 2009). In detail, the reads from each library were first mapped to the scaffolds, and the junction outputs from every RNA-seq dataset were combined together to generate a “pooled junction set,” which was further used to map all of the reads from different RNA-seq datasets to the scaffolds via TopHat. This step supplied a junction set for Cufflinks (Trapnell et al., 2012). Then, the six datasets were combined into a whole transcriptome by Cuffcompare, according to the annotated genomic information. The transcripts that are longer than 200 nT and have more than two exons were reserved, and 24,267 transcripts were obtained in this step. At this point, the candidate protein-coding genes were excluded after blasting to the NR database (e < 001). Next, transcripts >300 nT ORF were removed by getorf software (http://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html). Then, the protein-coding transcripts were predicted by Coding Potential Assessment Tool (CPAT), and other transcripts were used as the template to search in the Pfam database with HMMER software (Finn et al., 2015). The transcripts that have no conserved domain/motif potential were reserved, while other non-coding RNAs except lncRNAs, the known tRNAs, snoRNAs, snRNAs, and rRNAs were all removed by the Infernal and BLASTN search (Zhao et al., 2016), generating the final lncRNA sets.
Aligning lncRNAs with the SSB genome (Ma et al., 2020), we analyzed the lncRNA exon–intron structure by Geneious (Kearse et al., 2012) and also checked its distribution among the scaffolds.
The transcript enrichment of the identified lncRNAs was examined by counting reads and normalized by Cuffdiff software, using the t-test to display the significance of DE. The lncRNA expression level was detected with FPKM (fragments per kilobase of transcript per million fragments mapped) as an indicator. Q-value is the FDR-adjusted p-value. A lncRNA which meets these criteria will be defined as specifically expressed: 1) the value of |log2 fold-change|≥1; 2) p-value <.01; and 3) q-value<.01. The fold change refers to the ratio of expression amount between two strains. The lncRNAs obtained by the differential expression analysis are displayed in the form of a volcano plot.
Strand-specific RT-PCR
Sample preparation, RNA extraction, and quality control are as described previously. Three reactions during the cDNA synthesis are forward (F) primer with reverse transcriptase (RT), reverse (R) primer with RT, and F + R primers without RT. The primers for RT-PCR (Supplementary Table S1) were designed using http://www.idtdna.com/Scitools/and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China).
RNA isolation, cDNA synthesis, and quantitative real-time PCR
The extraction and purification of total RNA were described previously. cDNA was obtained by using the PrimeScript®RT Master Mix kit (TaKaRa, Japan) with 500 ng of total RNA as the template. qRT-PCR was carried out with a SYBR Premix Ex Taq kit (TaKaRa, Japan) on an ABI 7300 real-time PCR system (Thermo Fisher Scientific, United States) as described as follows: 30 s denaturation at 95°C, 40 cycles of 5 s at 95°C, and 31 s at 60°C, 15 s at 95°C, 15 s at 60°C, and 15 s at 95°C again. Actin A1 was used as an internal reference. Data were analyzed by the 2−ΔΔCt method (Pfaffl 2001). The primers used are shown in Supplementary Table S2.
Expression profile across life-time and RNAi assay of the lincRNA MSTRG.7482.1
Samples of different development stages were collected including the egg, 1st, 2nd, 3rd, 4th, 5th, and 6th stages, pupae, and male and female moths. For each stage, 3–4 individuals were collected as one biological replicate, and four biological replicates were performed. These samples were then used to conduct RNA extraction, cDNA collection, and qRT-PCR for the lincRNA MSTRG.7482.1.
The chemically synthesized small interference RNA (siRNA) came from GenePharma Co., Ltd. (Shanghai, China) (see Supplementary Table S2 to find its sequence).The HPLC-purified double-stranded siRNAs were dissolved in diethylpyrocarbonate-treated water (Milli-Q-grade) to make a 4 mg/ml solution. Then, 1 μl (4 μg) of siRNA was injected into fourth-instar larvae with a micro-needle, pulling the needles from glass capillaries which have 1.0-mm outer diameter and 50-mm inner diameter by using a micropipette puller (Model P-87, Sutter Instruments Co., CA), keeping needles still for 30 s at the injection point to avoid siRNA leakage. The shuffled siRNAs were taken as negative control (Supplementary Table S2). About 20 to 30 specimens were examined for each treatment, with triplicate experiments. The fourth larvae were collected 24 and 48 h post-micro-injection to determine the efficiency of RNAi.
For RNAi assay, the larvae were removed onto an artificial diet without any pesticide treatment for 24 h post-injection. The larvae that died or whose movement was blocked due to mechanical injury were discarded. The remaining specimens were removed onto an artificial diet with chlorantraniliprole treatment, according to the LC50 dose in S or R strains for another 96 h, respectively. Lastly, these specimens were removed onto an artificial diet without treatment. The mortality was calculated day by day from the fourth day post-treatment with chlorantraniliprole.
Data analysis
For the location of lncRNA and protein-coding genes on the genome of SSB, information on the chromosome length, lncRNA/gene density, and location of the lncRNA/gene to be located was collected. The R package RIdeogram was used to display the aforementioned information. Regarding the qRT-PCR result, Tukey’s test and one-way ANOVA statistical analysis were performed to compare the different gene expression levels between S and R strains.
Results
Identification and characterization of lncRNAs in SSB
Strand-specific RNA-seq was finished via NovaSeq 6000, Illumina. Raw data on RNA-seq were submitted to the public database: National Genomics Data Center (https://ngdc.cncb.ac.cn/). The assigned accession of the submission is CRA009124. Then, 43,490,487 to 55,131,982 clean reads were obtained from a total of 94.73 G clean data (Supplementary Table S3). Mapping these reads to an updated version genome of SSB (Ma et al., 2020), the highest alignment rate of 91.85% was yielded. An average of 121,998 transcripts was assembled in each sample from aligned reads. Finally, a total of 40,573 merged transcripts were determined (Supplementary Table S3). Twenty-two transcripts shorter than 200 nT were first removed from 40,573 merged transcripts (Figure 1A). Then, 16,284 transcripts which contained single exons were also discarded. Up to 24,267 transcripts with multiple exons were filtered by CPAT, Swiss-Prot, and Pfam databases to exclude the possibility of protein-coding genes. Finally, a total of 3470 lncRNAs were discovered and characterized in SSB after removing small non-coding RNAs via NONCODE and Rfam databases. These lncRNAs were further divided into four categories, namely, intronic lncRNAs, lincRNAs, sense lncRNAs, and antisense lncRNAs, according to their relative positions and orientations on the genome (Figure 1B). Most lncRNAs (1879, 54.12%) were located in intergenic regions, the second-ranked sense lncRNAs (853, 24.57%) shared an overlap with the exon region of protein-coding genes, and 14.23% and 7.09% lncRNAs were antisense lncRNAs (493) and intronic lncRNAs (245) (Figure 1B). Among 3470 SSB lncRNAs, 2454 of which contained two exons, accounting for 70.68% (Figure 2A), and only 2.13% of lncRNAs contained seven or more exons. In contrast, only 13.27% protein-coding genes harbored two exons. In addition, the protein-coding genes with three exons accounted for the largest proportion (17.16%). The exons whose length was shorter than 200 bp made up for more than 70% in protein-coding genes, but there was less than 40% in exons shorter than 200 bp in lncRNAs (Figure 2B). As shown in Figure 2C, the average length of the SSB lncRNA transcript and the protein-coding genes were 1049 and 199 bp, respectively.
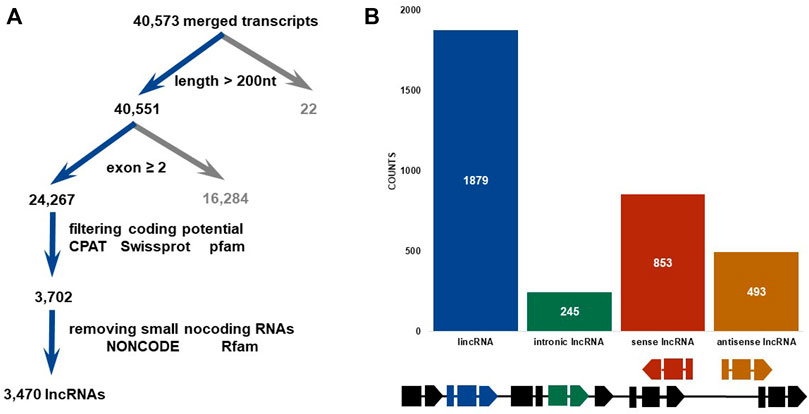
FIGURE 1. Identification and classification of lnc RNAs in SSB. (A) Brief computational pipeline for identifying lncRNAs. (B) Histogram of lncRNAs categorized as lincRNA (long intergenic non-coding RNA), intronic lncRNA, sense exon lncRNA, and antisense exon lncRNA. The schematic diagram at the bottom shows the differences of the gene structure of different types of lncRNAs in the genome.
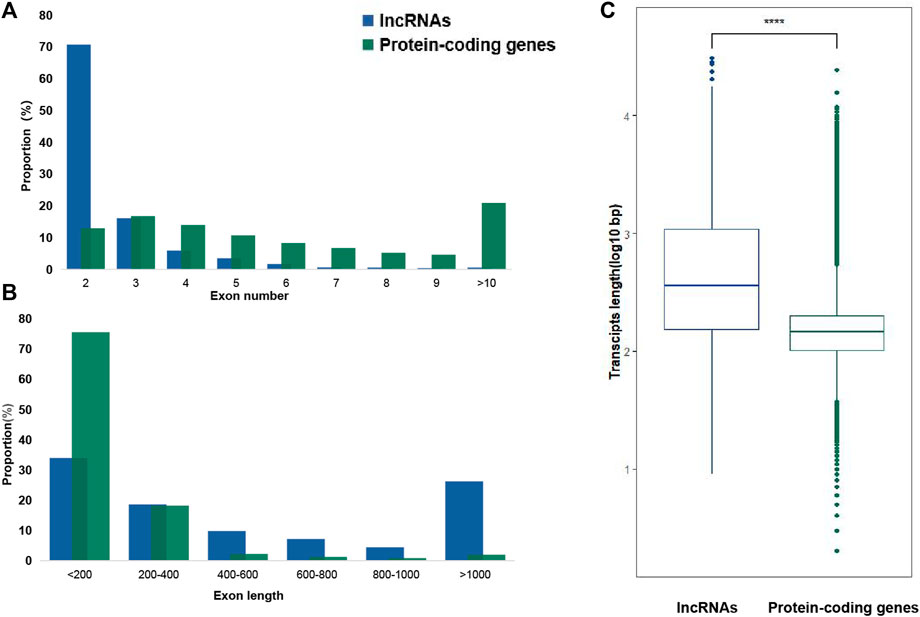
FIGURE 2. LncRNA gene structural features of SSB. (A) Exon numbers of lncRNAs and protein-coding genes. (B) Distribution of exon sizes of lncRNA and protein-coding genes. (C) Transcript length comparison of lncRNAs and protein-coding genes. On average, lncRNAs have longer transcripts than protein-coding genes.
Function analysis of differentially expressed lncRNA and adjacent protein genes
Up to 3356 lncRNAs were gathered in S or R strains after removing 116 lncRNAs with FPKM=0 in all six libraries. There were 1343 lncRNAs dysregulated with |log2 fold-change|>1. Also, there were 798 and 545 lncRNAs highly expressed in R and S strains, respectively (Figure 3A). Moreover, a total of 632 differentially expressed (DE) lncRNAs met strict screening criteria, that is, p-value < 01, p-adj < 01, and |log2 fold-change|>1 (Figure 3B).There were 297 and 335 lncRNAs upregulated in S and R strains. Among these lncRNAs, 365, 31, 141, and 95 belonged to lincRNAs, intronic lncRNAs, sense lncRNAs, and antisense lncRNAs, respectively.
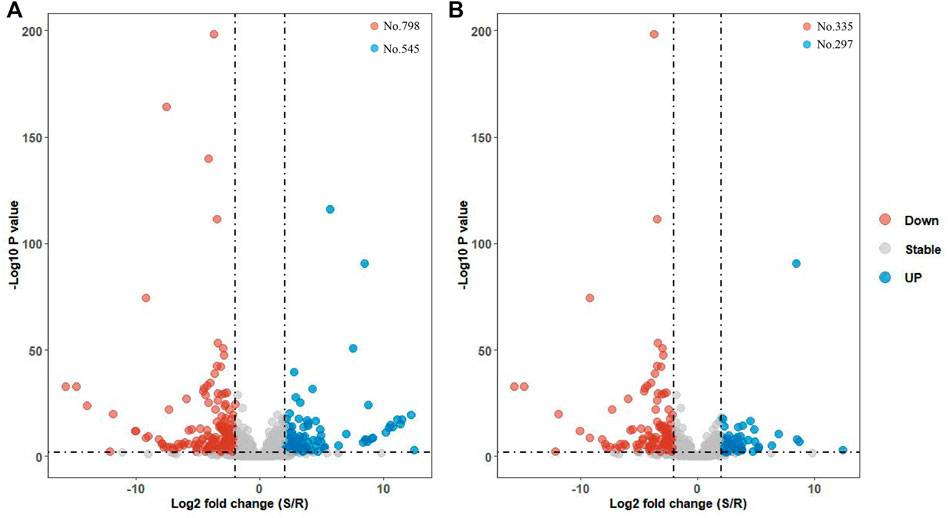
FIGURE 3. Volcano map of differentially expressed lncRNA genes. (A) Expression profile of all 1,343 dysregulated lncRNAs in S and R strains were plotted via the volcano map. (B) Expression profile of 632 DE lncRNAs in S and R strains. They met a strict screening criteria, that is, |log2 fold-change|>1, p-value < 01, and p-adj < 01.
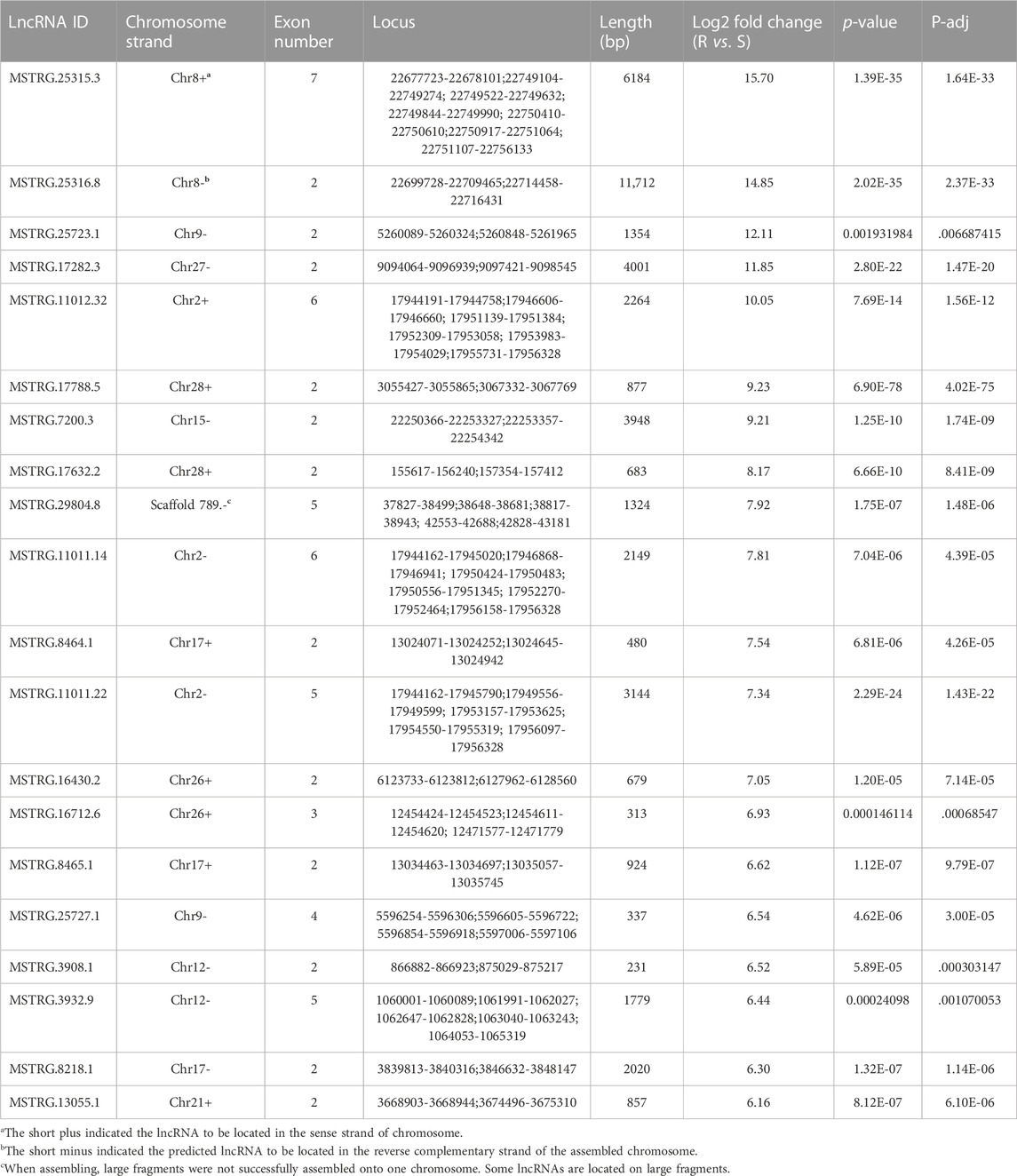
In the detoxification enzyme gene family, P450s, carboxyl/cholinesterases/esterases, and ABC genes were related to the chlorantraniliprole resistance in SSB (Wang et al., 2015; Xu et al., 2019; Peng et al., 2021). Also, a 1000-kb range between the center of an lncRNA gene and a neighboring protein-coding gene transcription start site (TSS) was defined as a regulatory zone by the Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al., 2010). To study the potential function of lncRNAs in chlorantraniliprole resistance, both the detoxification enzyme genes and DE lncRNAs were located in the genome of SSB. Sixty-six P450 genes were located on 20 different chromosomes (Figure 4A; Supplementary Table S4). Among the 38 lncRNAs adjacent to these P450 genes, the lincRNA, sense lncRNA, antisense lncRNA, and intronic lncRNA were 24, 7, 5, and 2, respectively. On chromosome10, the lincRNA MSTRG.2776.2 was upregulated in the S strain as 8.45 log2 fold change (log2FC) as in the R strain, and it was 944, 528 bp apart from CYP9A12. For esterases, there were 67 carboxyl/choline esterases located on 16 chromosomes (Figure 4B; Supplementary Table S5), and a total of 27 lncRNAs were located nearby. On chromosome 9, the lincRNA MSTRG.25727.1 was upregulated in the R strain as 6.54 log2FC as in the S strain, and CsuEst35 antennal esterase was 887, 454 bp apart from it. Forty-three of the 47 ABC transporter genes previously reported were successfully located in the genome of SSB. They are distributed on 16 different chromosomes (Figure 4C; Supplementary Table S6). At the same time, 51 lncRNAs were adjacent to them, including 31 lincRNAs, 12 sense lncRNAs, 6 antisense lncRNAs, and 2 intronic lncRNAs. ABCB4, D3, and G12 were located on chromosome 8, and the expression of D3 was upregulated and that of G12 was downregulated in the R strain (Peng et al., 2021). A total of 18 lncRNAs were dysregulated in the R strain (Figure 4C; Supplementary Table S6) as lincRNAs MSTRG.25315.3 and MSTRG.25316.8 were upregulated.
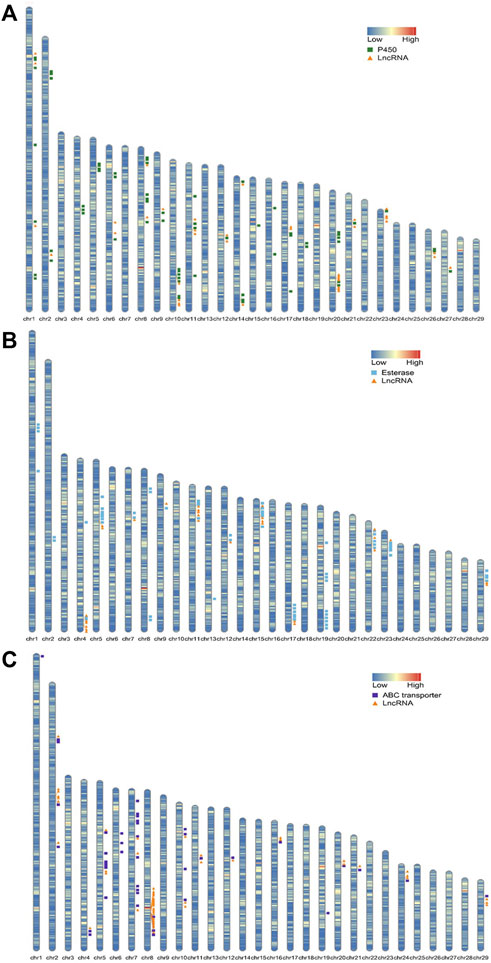
FIGURE 4. Chromosome maps showing the location of genes related to detoxification metabolic enzymes and dysregulated lncRNAs which were adjacent within 1000 kb. (A) P450 genes and dysregulated lncRNAs. (B) Esterase genes and dysregulated lncRNAs. (C) ABC genes and dysregulated lncRNAs.
Differentially expressed long intergenic non-coding RNAs
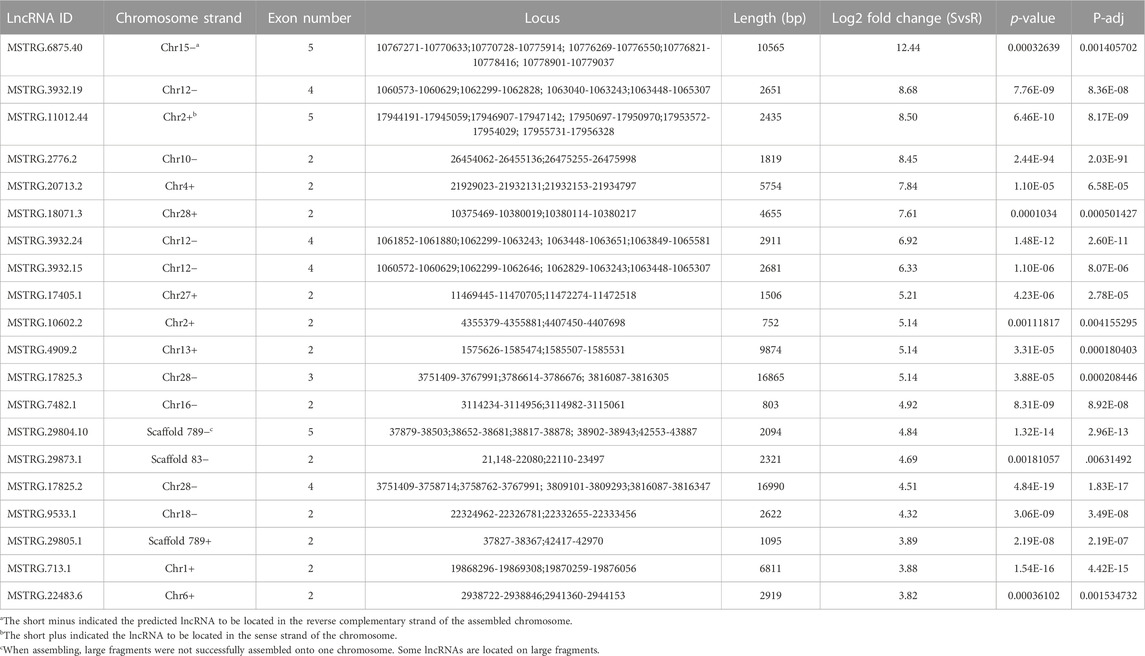
Considering lincRNAs, also known as long intergenic non-coding RNAs, accounted for 54.10% of the number of lncRNAs (Figure 1B, 1879 out of 3470), our focused analyses revealed there were 167 and 198 lincRNAs differentially highly expressed in S and R strains, respectively. Those lincRNAs whose value of |log2FC| ranked in the top 20 were chosen in the following analysis (Tables 1, 2). The log2FC of lincRNA MSTRG.6875.40 was 12.44, ranking it the highest in the top in the S strain (Table 1), whereas MSTRG.25315.3 was 15.70, ranked first in the R strain (Table 2). The log2FC values of differentially expressed MSTRG.22483.6 and MSTRG.13055.1 were 3.82 and 6.16, as they were listed last. These 40 lincRNAs were distributed on 17 different chromosomes (Chr1, 2, 4, 6, 8, 9, 10, 12, 13, 15, 16, 17, 18, 21, 26, 27, and 28) or scaffolds (scaffold 789 and scaffold 83). The length of them varied widely, ranging from 231 to 11,712 bp. The exon numbers varied from 2 to 7. For example, there were seven, six, and five exons in MSTRG.25315.3, MSTRG.11012.32 (Table 2), and MSTRG.6875.40 (Table 1), respectively.
Protein-coding genes within 1000 kb adjacent to lincRNA were collected. MSTRG.25315.3 was the top differentially upregulated gene in the R strain (Table 2, 15.70 of log2FC) with 83 coding genes located. Heat shock protein 70–2 (GenBank accession: AGR84224.1) was only 1,770 bp close to MSTRG.25315.3 (Figure 5). The lincRNA MSTRG.25727.1 was 337 bp long, but it harbored four exons and was elevated to a log2FC value of 6.54 in the R strain compared to the S strain. Among 20 mRNA genes nearby, antennal esterase was 887, 454 bp adjacent to MSTRG.25727.1. In addition, sodium channel protein was 429, 858 bp adjacent to MSTRG.7200.3 (Table 2).
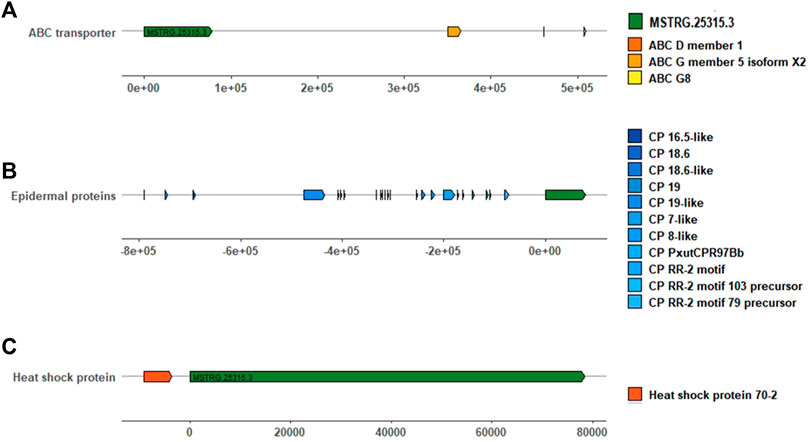
FIGURE 5. LincRNA MSTRG.25315.3 and its adjacent protein-coding genes involved in pesticide resistance within 1000 kb. (A) Green square represents MSTRG.25315.3, and orange, brownish red, and yellow squares represent ABC genes. (B) Gradient blue squares represent different cuticle protein genes. (C) Red square represents the heat shock protein gene. The long solid line and the number marked above represent the distance (bp) between them, MSTRG.25315.3 and adjacent genes.
With regards to the top 20 highly expressed lincRNAs in the S strain, MSTRG.2776.2 was 961, 794 bp adjacent to cytochrome 9A20 (annotated from Bombyx mori, GenBank accession: BAI47532.1). Sixty-four protein-coding genes were close to MSTRG.6875.40 within 1000 kb, and UDP-glucuronosyl transferase 2B31-like was 990, 115 bp apart from it. MSTRG.20713.2 was upregulated in the S strain (Table 1, 7.84 of log2FC), and it was 494, 273 bp away from 4A1-like gene, which is a family member of solute carrier organic anion transporters, encoding membrane proteins that are involved in solute (charged and uncharged organic molecules, and inorganic ions) transportation. They are likely to be involved in pesticide metabolism and transport.
Validation of DE lincRNAs in two strains by RT-PCR and qRT-PCR
We randomly selected 20 lincRNA candidates shown in Tables 1, 2 to be validated by strand-specific RT-PCR. Results showed 13 lincRNAs were strand-specific, that is, five came out forward stranded (MSTRG.25723.1, 3932.19, 3932.24, 7482.1, and 6875.40) and eight were reverse stranded (MSTRG.8464.1, 25727.1, 11012.32, 29804.8, 29804.10, 22483.6, 25315.3, and 25316.8) (Figure 6). Of the 13 strand-specific lincRNAs, only four were in the same direction as the originally labeled strand (Tables 1, 2, MSTRG.25316.8, 29804.8, 25727.1, and 29804.10). The other seven lincRNAs can be amplified by both primers (Figure 6).
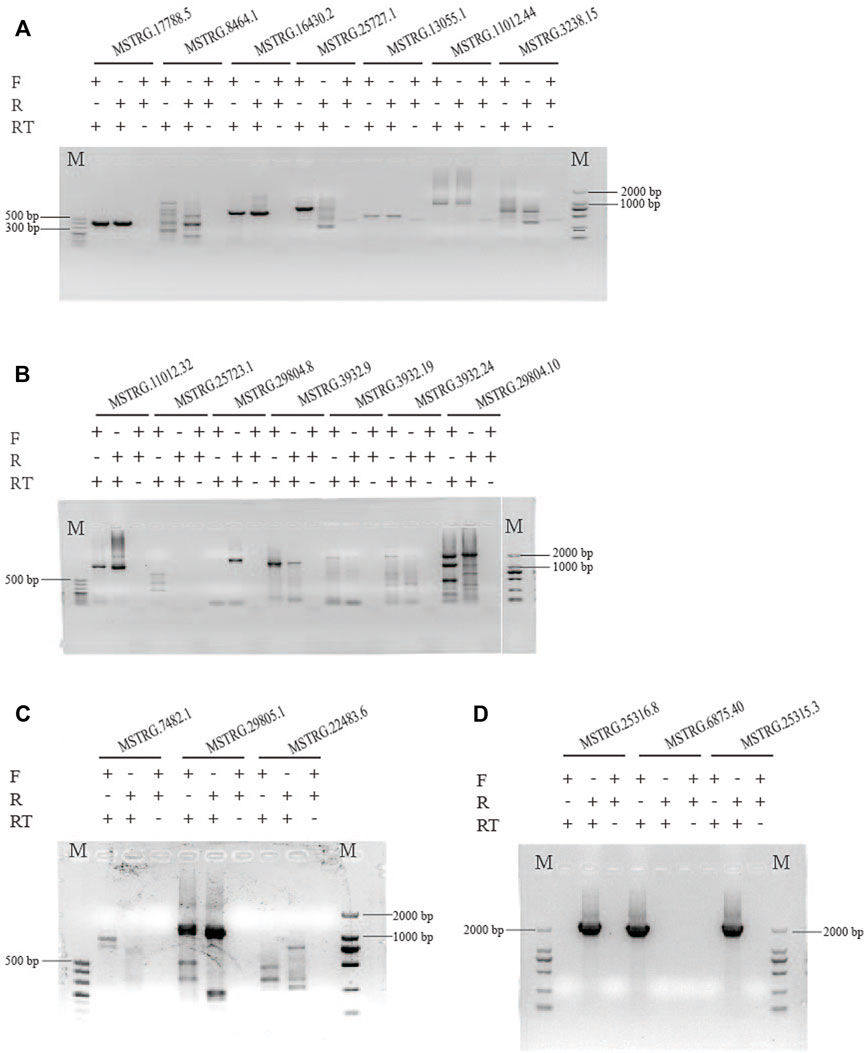
FIGURE 6. Strand-specific PCR of 20 randomly selected lincRNAs to determine transcription orientation. The different electrophoretic bands of lncRNAs with similar PCR product size were shown in (A–D). The results indicated that 13 lincRNAs are strand-specific, that is, five came out as forward-stranded (MSTRG.25723.1, 3932.19, 3932.24, 7482.1, and 6875.40) and eight were reverse-stranded (MSTRG.8464.1, 25727.1, 11012.32, 29804.8, 29804.10, 22483.6, 25315.3, and 25316.8. The other seven lincRNAs can be amplified by both primers.
To verify the RNA-seq results, the relative expression levels of three randomly selected DE lincRNAs in the R strain (MSTRG.25315.3, MSTRG.25316.8, and MSTRG.17788.5) and five lincRNAs in the S strain (MSTRG.3932.19, MSTRG.3932.15, MSTRG.7482.1, MSTRG.29805.1, and MSTRG.22483.6) were examined by qRT-PCR (Figure 7). Consistent results were observed between the qRT-PCR results of most of the selected strains and the sequencing data except for MSTRG.17788.5 and MSTRG.7482.1. The two lincRNAs displayed the opposite trend. The expression of MSTRG.17788.5 was 9.23 times the log2FC value in the R strain as that in the S strain (Table 2), but it turned out 0.69 times in the R strain as that in the S strain. On the contrary, the expression of MSTRG.7482.1 was first 4.92 of log2FC in the S strain as that in the R strain (Table 1), but it turned out 35.86 times in the R strain confirmed by qRT-PCR.
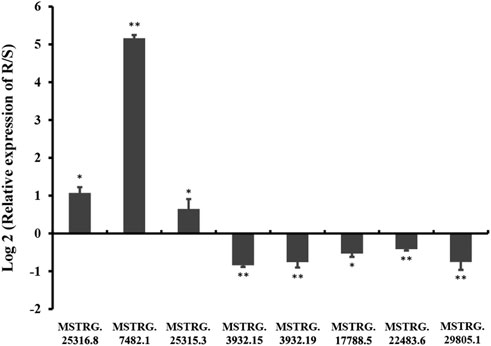
FIGURE 7. Relative expression levels of lincRNAs in chlorantraniliprole-resistant strains (R) and susceptible strains (S).The column represents the value of log2 (relative expression levels of lincRNAs in the R strain compared to the S strain). Each column represents the mean of three to four biological samples. Error bars represent the standard deviation from the mean. The change in the expression level was calculated using the 2−ΔΔCt method. Data were normalized to the expression of housekeeping genes (Actin A1). Asterisks on the error bars show significant differences (p < 05 or p < 01).
Functional analysis of the lincRNA MSTRG.7482.1
MSTRG.7482.1 had the highest differential expression folder change among the three lincRNAs, which is highly expressed in the R strain (Figure 7). We first used qRT-PCR to examine the expression of MSTRG.7482.1 in different stages of development, like eggs, each instar larvae, pupae, and male and female moths (Supplementary Figure S1). Altogether, MSTRG.7482.1 expressed highly from eggs until the sixth-instar larvae. Then, it declined sharply in the pupal stage and was expressed to the lowest in the female moth. The expression profile suggested MSTRG.7482.1 functions mainly in larvae, which is, the feeding stage. The siRNA was designed from position 231 based on the sequence of MSTRG.7482.1, and we successfully knocked down MSTRG.7482.1 gene by injecting siRNA in the fourth-instar larvae (Figure 8). The expression of MSTRG.7482.1 was knocked to about 40% compared to control in both 24 and 48 h after injection.
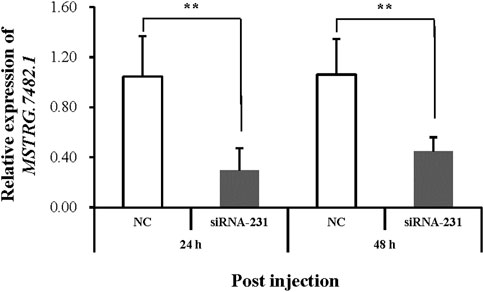
FIGURE 8. Relative expression of MSTRG.7482.1 after injection with siRNA. Asterisks on the error bars show significant differences (p < 01).
To detect the effect of the downregulated MSTRG.7482.1 gene on chlorantraniliprole sensitivity, the fourth-instar larvae injected with siRNA-231 were removed onto an artificial diet with chlorantraniliprole treatment, according to the LC50 dose in the S or R strain for 96 h, respectively. In the S strain, the RNAi group showed 70.09% average mortality, which is significantly higher than that of control (mortality: 38.62%) four days after pesticide treatment (Figure 9A). On day 8 post-chlorantraniliprole treatment, both mortality of siRNA-231 and the control group declined and it turned out closer (39.36% vs. 26.75%) (Figure 9B). In the R strain, the average mortality of the siRNA-231 group was higher than that of the control at 4 and 8 days after pesticide treatment; there was no statistically significant difference (Figures 9C, D).
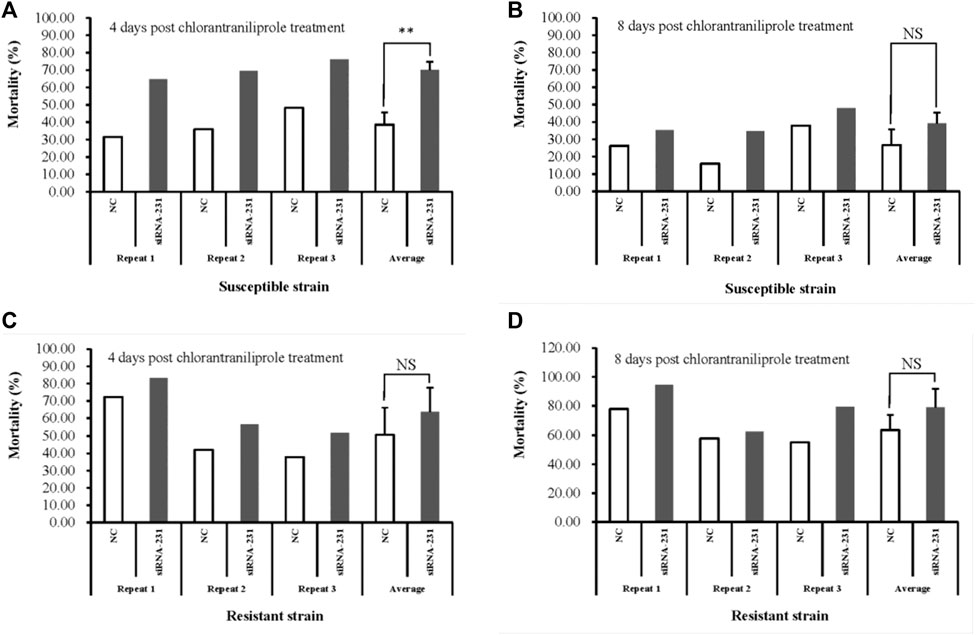
FIGURE 9. Susceptibility to chlorantraniliprole after silencing MSTRG.7482.1 with siRNA. The toxicity assay was conducted in both the S strain (A,B) and R strain (C,D). Mortality was calculated 96 h after chlorantraniliprole exposure. All experiments were conducted in triplicate. Asterisks on the error bars show significant differences (p < 01). NS on the error bars, however, show no significant differences.
Discussion
LncRNA has attracted much attention due to its unique features and molecular mechanisms involved in variety of biological functions (Wang and Chang 2011; Quinn and Chang 2016). In Drosophila, lncRNAs are responsible for the regulation of development (such as neuromuscular junction, embryo, and organ), behavior (sheep, mating, courtship, and locomotion), sex control, and dosage compensation. In addition, lncRNAs also help fruit flies against some stressors, including heat, bacterial infection, and wasp attacks (Li et al., 2019). As the sequencing technology advances, much more lncRNAs have been discovered in other non-model insects. A total of 2,914 lncRNAs were identified in Aphis citricidus, and down-regulation of Ac_lnc54106.1 resulted in malformed wings, which showed lncRNAs mediated the wing plasticity (Shang et al., 2021). 20-Hydroxyecdysone (20E) was injected into the hemolymph of silkworms to study autophagy. Predictive 6,493 cis pairs and 42,797 trans pairs of lncRNA–mRNA were found, and functional analysis of LNC_000560 suggested it potentially regulated Atg4B and participated in the 20E-induced autophagy of the fat body (Qiao et al., 2021). In addition, lncRNAs might have key roles in conferring insecticide resistance and regulating the metamorphosis development in P. xylostella (Liu et al., 2017). Unfortunately, the molecular mechanism of how lncRNAs regulate pesticide resistance of SSB is still unclear.
SSB has developed severe resistance to chlorantraniliprole (Sun et al., 2018; Huang et al., 2020). Here, a total of 3,470 lncRNAs were identified at last from six strand-specific libraries in both S and R strains. The total number of lncRNAs was smaller than 6,171 reported in B. dorsalis (Meng et al., 2021) and 11,978 lncRNAs in S. litura (Shi et al., 2022), but it was greater than that reported in P. xylostella (Zhu et al., 2017). Interestingly, the number of lncRNAs was different even in the same species. In P. xylostella, 3,324 lncRNAs were found from 13 RNA-seq datasets, 1,309 lncRNAs were identified in 9 libraries, and 3,844 lincRNAs were discovered based on 7 RNA sequencing libraries (Etebari et al., 2015; Liu et al., 2017; Zhu et al., 2017). The difference in quantity may come from many factors, such as different methods of library construction, different tissues obtained from samples, and different insecticide exposure. There were 10 kinds of classification for lncRNAs, according to different criteria in the early stage (St Laurent et al., 2015). For example, lncRNA was divided into six types, that is, convergent, divergent, overlapping, intergenic, enhancer, host of miRNA, and intronic in Drosophila (Li et al., 2019). At present, much research refers to the location of lncRNAs to classify them into four categories, including intronic lncRNAs, lincRNAs, sense lncRNAs, and antisense lncRNAs (Zhu et al., 2017; Meng et al., 2021; Yang et al., 2021). Although the composition proportion of intronic lncRNA, sense, and antisense lncRNA is different, the proportion of lincRNA is the highest. This is also reflected in SSB, where lincRNA accounts for 54.10% of lncRNA (1879 in 3470). Up to 70.68% SSB lncRNAs harbored two exons, which was similar to that in N. lugens (77.9%) (Xiao et al., 2015), P. xylostella (74.49%) (Liu et al., 2017), and B. dorsalis (84.36%) (Meng et al., 2021). Usually, the average transcript length of lncRNAs is shorter than protein-coding genes. The average lengths of the P. xylostella lncRNA transcript and protein-coding genes were 912 bp and 1,385 bp, respectively (Liu et al., 2017). However, the average transcript length of SSB lncRNAs (1049 bp) was much longer than that of protein-coding genes (199 bp). This may be a result of species specificity.
The association between lncRNAs and adjacent genes is one of many methods to study the regulatory function of lncRNAs. However, the distance between lncRNA and its neighbor genes varied. In N. lugens, adjacent protein-coding genes less than 5 kb apart from lncRNAs were statistically significant than randomly selected coding genes (Xiao et al., 2015). Zhu et al. predicted cis regulation of many protein-coding genes, which were found within a 10-kb range upstream or downstream from the target lncRNAs (Zhu et al., 2017). In B. dorsalis, 793 target genes were predicted via searching 100 kb upstream and downstream (Meng et al., 2021).To more comprehensively understand the association between lncRNA and potential target genes, we set the distance between the protein-coding gene and lncRNA within 1000 kb (McLean et al., 2010; Mitchell et al., 2017). It is well known that RyR is the target of chlorantraniliprole, and a previous report attempted to study the relation of chlorantraniliprole resistance with lncRNA and RyR (Zhu et al., 2017). Two co-expressed lncRNAs with RyR, TCONS_00013329 and TCONS_00056155, were found in P. xylostella. In SSB, there was no RyR found within 1000 kb of differentially expressed lincRNAs in this region. This might be related to the dataset used in the analysis that we focused on lincRNAs rather than all the four types of lncRNAs. Other three types of lncRNAs will be investigated in the future research and is beyond the scope of this study.
Fortunately, there were meaningful findings in eight lincRNAs that had been verified by qRT-PCR. MSTRG.25316.8, MSTRG.25315.3, and MSTRG.7482.1 were highly expressed in the R strain. Among 83 adjacent genes near to MSTRG.25315.3, hsp70-2 (GenBank accession: AGR84224.1) was only 1,770 bp nearby. Exposed to carbaryl, three small-molecule heat shock (smhsp) genes (hsp20. 3, hsp19. 1, and hsp17. 0) were upregulated in Lymantria dispar to varying degrees (Peng et al., 2017). These results suggested hsp may be involved in responding to the pesticide stress. In addition, three ATP-binding cassette sub-family genes G5, D3, and G8 were located in 310, 421, and 468 kb. In a previous study, CsABCG5 and G8 were significantly highly expressed in the resistant strain of SSB against chlorantraniliprole (Peng et al., 2021).
The expression of MSTRG.7482.1 in the R strain was 35.86 times as that in the S strain (Figure 7), and RNAi assay showed that knock-down of MSTRG.7482.1 resulted in an increase of average mortality by 31.47% (Figure 9A). However, such a statistic significant phenomenon was only observed at four days post-pesticide treatment in the S strain. In the test of the R strain, the mortality of the treatment group in all groups was higher than that of the control group to varying degrees, but it was not statistically significant (p-value >.05). On the whole, although not all repeated experimental results were statistically significant, the mortality of the RNAi group is indeed higher than that of the control. In other words, RNAi of MSTRG.7482.1 increased the sensitivity to chlorantraniliprole. In S. litura, RNAi of LNC_004867 or LNC_006576 increased the mortality from 14.68% to 34.69% against indoxacarb (Shi et al., 2022). Because LNC_006576 and LNC_004867 were significantly expressed in the developmental stages of 1-6-instar larvae, it suggested the two lncRNAs may play important roles in the metabolism of insecticides or plant chemicals in S. litura. In SSB, the lincRNA MSTRG.7482.1 was also expressed highly across the feeding larval stage (Fig. S1), and it might be associated with xenobiotic detoxification. A total of 35 adjacent protein-coding genes within 1000 kb apart from lincRNA MSTRG.7482.1 were searched (Supplementary Table S7). There were 14 undefined protein residues. Among the other 21 genes, indole-3-acetaldehyde oxidase-like isoform X1 (Bombyx mandarina) was the nearest which is 85, 966bp to MSTRG.7482.1. Alpha-(1, 6)-fucosyltransferase (Galleria mellonella) plays an important role in cell recognition, proliferation, and metabolic activities. At a distance of 433, 455 bp, there was cuticle secretory protein (Ostrinia furnacalis). However, the function of these adjacent genes and their association with resistance mechanisms are unknown. In future, more precise target prediction and carefully functional studies are needed to elucidate the relation to the mechanism of chlorantraniliprole resistance.
Conclusion
In this study, a total of 3,470 lncRNAs were identified from six RNA-seq libraries of SSB, including 1,879 intergenic lncRNAs, 245 intronic lncRNAs, 853 sense lncRNAs, and 493 antisense lncRNAs. In addition, 632 DE lncRNAs were discovered. Adjacent protein-coding genes of each of the 20 top DE lincRNAs in both S and R stains were analyzed. Strand-specific RT-PCR was conducted to determine the transcript orientation of 20 randomly selected lincRNAs. qRT-PCR was performed to verify the expression level of eight lincRNAs, including MSTRG.25315.3 and MSTRG.7482.1. RNAi and bioassay analyses indicated that MSTRG.7482.1 was involved in chlorantraniliprole resistance. All the results provide the basis for better understanding about the roles of lncRNAs in regulating the resistance of chlorantraniliprole and other insecticides in SSB.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/, CRA009124.
Ethics statement
Ethical review and approval was not required for the animal study because the test object is rice pests, and the sample is raised indoors.
Author contributions
YS, SJH, FL, and HL conceived and designed research. DJ, GHL, TW, YH, and WJQ conducted experiments. LX, YYH, and KH analyzed data. YS and HL wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was financed by the National Natural Science Foundation of China (NSFC) (31860501 and 31972309) and the Natural Science Foundation of Zhejiang Province (LY22C140005), with additional finance provided by the Earmarked Fund for China Agriculture Research System (CARS-01-40) and the Jiangxi postdoctoral scientific research project (2019RC16 and 2020KY22).
Acknowledgments
The authors would like to thank the reviewers for their constructive suggestions. They thank DBM editing for improving the English language expression of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YD declared a past co-authorship with the author LX to the handling editor.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.1091232/full#supplementary-material
Supplementary Figure S1 | Expression profile of MSTRG.7482.1 in different stages across life time in the S strain. Each column represents the mean of three biological samples. Error bars represent the standard deviation from the mean. The change in the expression level was calculated using the 2-ΔΔCt method. Data were normalized to the expression of housekeeping genes (Actin A1). Different letters on the error bars show significant differences (p < .05).
References
Angrand P. O., Vennin C., Le Bourhis X., Adriaenssens E. (2015). The role of long non-coding RNAs in genome formatting and expression. Front. Genet. 165, 165. doi:10.3389/fgene.2015.00165
Bernabo P., Viero G., Lencioni V. (2020). A long non-coding RNA acts as a post-transcriptional regulator of heat shock protein (HSP70) synthesis in the cold hardy Diamesa tonsa under heat shock. PLoS One 15 (4), e0227172. doi:10.1371/journal.pone.0227172
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 15, 2114–2120. doi:10.1093/bioinformatics/btu170
Chang Z. X., Ajayi O. E., Guo D. Y., Wu Q. F. (2020). Genome-wide characterization and developmental expression profiling of long non-coding RNAs in Sogatella furcifera. Insect Sci. 27 (5), 987–997. doi:10.1111/1744-7917.12707
Chen C. K., Blanco M., Jackson C., Aznauryan E., Ollikainen N., Surka C., et al. (2016). Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354 (6311), 468–472. doi:10.1126/science.aae0047
Chen Y., Singh A., Kaithakottil G. G., Mathers T. C., Gravino M., Mugford S. T., et al. (2020). An aphid RNA transcript migrates systemically within plants and is a virulence factor. Proc. Natl. Acad. Sci. U. S. 117 (23), 12763–12771. doi:10.1073/pnas.1918410117
Dai C., Li H., Wei Q., Hu Y. (2022). Toxicity and control effect of five insecticides on different instar larvae of Chilo suppressalis. Hybrid. Rice 37 (3), 25–28. doi:10.16267/j.cnki.1005-3956.20210531.201
Diederichs S., Bartsch L., Berkmann J. C., Frose K., Heitmann J., Hoppe C., et al. (2016). The dark matter of the cancer genome: Aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol. 8 (5), 442–457. doi:10.15252/emmm.201506055
Ebbinghaus-Kintscher U., Luemmen P., Lobitz N., Schulte T., Funke C., Fischer R., et al. (2006). Phthalic acid diamides activate ryanodine-sensitive Ca2+ release channels in insects. Cell Calcium 39, 21–33. doi:10.1016/j.ceca.2005.09.002
Etebari K., Furlong M. J., Asgari S. (2015). Genome wide discovery of long intergenic non-coding RNAs in Diamondback moth (Plutella xylostella) and their expression in insecticide resistant strains. Sci. Rep. 5, 14642. doi:10.1038/srep14642
Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., et al. (2015). HMMER web server: 2015 update. Nucleic Acids Res. 43 (W1), W30–W38. doi:10.1093/nar/gkv397
Guan R., Li H., Zhang H., An S. (2020). Comparative analysis of dsRNA-induced lncRNAs in three kinds of insect species. Arch. Insect Biochem. Physiol. 103 (1), e21640. doi:10.1002/arch.21640
Guo L., Liang P., Zhou X., Gao X. (2014). Novel mutations and mutation combinations of ryanodine receptor in a chlorantraniliprole resistant population of Plutella xylostella (L.). Sci. Rep. 4, 6924. doi:10.1038/srep06924
He Y., Zhang J., Gao C., Su J., Chen J., Shen J. (2013). Regression analysis of dynamics of insecticide resistance in field populations of Chilo suppressalis (Lepidoptera: Crambidae) during 2002-2011 in China. J. Econ. Entomol. 106 (4), 1832–1837. doi:10.1603/ec12469
Huang J. M., Rao C., Wang S., He L. F., Zhao S. Q., Zhou L. Q., et al. (2020). Multiple target-site mutations occurring in lepidopterans confer resistance to diamide insecticides. Insect Biochem. Mol. 121, 103367. doi:10.1016/j.ibmb.2020.103367
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28 (12), 1647–1649. doi:10.1093/bioinformatics/bts199
Lahm G. P., Selby T. P., Freudenberger J. H., Stevenson T. M., Myers B. J., Seburyamo G., et al. (2005). Insecticidal anthranilic diamides: A new class of potent ryanodine receptor activators. Bioorg Med. Chem. Lett. 15 (22), 4898–4906. doi:10.1016/j.bmcl.2005.08.034
Li K., Tian Y., Yuan Y., Fan X., Yang M., He Z., et al. (2019). Insights into the functions of LncRNAs in Drosophila. Int. J. Mol. Sci. 20 (18), 4646. doi:10.3390/ijms20184646
Liu F., Guo D., Yuan Z., Chen C., Xiao H. (2017). Genome-wide identification of long non-coding RNA genes and their association with insecticide resistance and metamorphosis in diamondback moth, Plutella xylostella. Sci. Rep. 7 (1), 15870. doi:10.1038/s41598-017-16057-2
Ma W., Zhao X., Yin C., Jiang F., Du X., Chen T., et al. (2020). A chromosome-level genome assembly reveals the genetic basis of cold tolerance in a notorious rice insect pest, Chilo suppressalis. Mol. Ecol. Resour. 20 (1), 268–282. doi:10.1111/1755-0998.13078
McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., et al. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28 (5), 495–501. doi:10.1038/nbt.1630
Meng L. W., Yuan G. R., Chen M. L., Dou W., Jing T. X., Zheng L. S., et al. (2021). Genome-wide identification of long non-coding RNAs (lncRNAs) associated with malathion resistance in Bactrocera dorsalis. Pest Manag. Sci. 77 (5), 2292–2301. doi:10.1002/ps.6256
Mitchell R. D., Wallace A. D., Hodgson E., Roe R. M. (2017). Differential expression profile of lncRNAs from primary human hepatocytes following DEET and fipronil exposure. Int. J. Mol. Sci. 18 (10), 2104. doi:10.3390/ijms18102104
Peng L., Lili S., qihui Z., chuanwang C. (2017). Analysis of small molecule heat shock protein gene of Lymantria dispar and its response to carbaryl stress. J. Beijing For. Univ. 39 (1), 7. doi:10.13332/j.1000-1522.20160179
Peng Y., Zhao J., Sun Y., Wan P., Hu Y., Luo G., et al. (2021). Insights into chlorantraniliprole resistance of Chilo suppressalis: Expression profiles of ATP-binding cassette transporter genes in strains ranging from low- to high-level resistance. J. Asia-Pacific Entomology24 24 (2), 224–231. doi:10.1016/j.aspen.2021.02.006
Pfaffl M. W. (2001). A new mathematical model for relative quantification in realtime RT-PCR. Nucleic Acids Research29 6, e45. doi:10.1093/nar/29.9.e45
Qiao H., Wang J., Wang Y., Yang J., Wei B., Li M., et al. (2021). Transcriptome analysis reveals potential function of long non-coding RNAs in 20-hydroxyecdysone regulated autophagy in Bombyx mori. BMC Genomics 22 (1), 374. doi:10.1186/s12864-021-07692-1
Quinn J. J., Chang H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17 (1), 47–62. doi:10.1038/nrg.2015.10
Shang F., Ding B. Y., Zhang Y. T., Wu J. J., Pan S. T., Wang J. J. (2021). Genome-wide analysis of long non-coding RNAs and their association with wing development in Aphis citricidus (Hemiptera: Aphididae). Insect Biochem. Mol. 139, 103666. doi:10.1016/j.ibmb.2021.103666
Sheng C., Wang H., Sheng S., Gao L., Xuan W. (2003). Pest status and loss assessment of crop damage caused by the rice borers, Chilo suppressalis and Tryporyza incertulas in China. Chin. Bull. Entomology 40 (04), 289–294.
Shi L., Li W. L., Zeng H. X., Shi Y., Liao X. L. (2022). Systematic identification and functional analysis of long non-coding RNAs involved in indoxacarb resistance in Spodoptera litura. Insect Sci. 29, 1721–1736. doi:10.1111/1744-7917.13015
Shuijin H., Qiong C., Wenjing Q., Yang S., Houguo Q. (2017). Resistance monitoring of four insecticides and a description of an artificial diet incorporation method for chilo suppressalis (Lepidoptera: Crambidae). J. Econ. Entomol. 110 (6), 2554–2561. doi:10.1093/jee/tox266
St Laurent G., Wahlestedt C., Kapranov P. (2015). The Landscape of long non-coding RNA classification. Trends Genet. 31 (5), 239–251. doi:10.1016/j.tig.2015.03.007
Struhl K. (2007). Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. 14 (2), 103–105. doi:10.1038/nsmb0207-103
Su J., Zhang Z., Wu M., Gao C. (2014). Geographic susceptibility of chilo suppressalis walker (Lepidoptera: Crambidae), to chlorantraniliprole in China. Pest Manag. Sci. 70 (6), 989–995. doi:10.1002/ps.3640
Sun Y., Hu Y., Qin W., Wan P., Zou F., Huang S. (2021). Resistance of asiatic rice borer Chilo suppressalis (Lepidoptera: Crambidae) population to four insecticides in Jiangxi Province from 2017 to 2019. J. Plant Prot. 48 (2), 396–406. doi:10.13802/j.cnki.zwbhxb.2021.2020049
Sun Y., Xu L., Chen Q., Qin W., Huang S., Jiang Y., et al. (2018). Chlorantraniliprole resistance and its biochemical and new molecular target mechanisms in laboratory and field strains of Chilo suppressalis (Walker). Pest Manag. Sci. 74 (6), 1416–1423. doi:10.1002/ps.4824
Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25 (9), 1105–1111. doi:10.1093/bioinformatics/btp120
Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7 (3), 562–578. doi:10.1038/nprot.2012.016
Wang B., Wang Y., Zhang Y., Han P., Li F., Han Z. (2015). Genome-wide analysis of esterase-like genes in the striped rice stem borer, Chilo suppressalis. Genome 58 (6), 323–331. doi:10.1139/gen-2014-0082
Wang K. C., Chang H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43 (6), 904–914. doi:10.1016/j.molcel.2011.08.018
Wu H., Yue S., Huang Y., Zhao X., Cao H., Liao M. (2022). Genome-wide identification of the long noncoding RNAs of Tribolium castaneum in response to terpinen-4-ol fumigation. Insects 13, 283. (3). doi:10.3390/insects13030283
Xiao H., Yuan Z., Guo D., Hou B., Yin C., Zhang W., et al. (2015). Genome-wide identification of long noncoding RNA genes and their potential association with fecundity and virulence in rice Brown planthopper, Nilaparvata lugens. BMC Genomics 749, 749. doi:10.1186/s12864-015-1953-y
Xu L., Zhao J., Sun Y., Xu D., Xu G., Xu X., et al. (2019). Constitutive overexpression of cytochrome P450 monooxygenase genes contributes to chlorantraniliprole resistance in Chilo suppressalis (Walker). Pest Manag. Sci. 75 (3), 718–725. doi:10.1002/ps.5171
Yang L., Wang Y. W., Lu Y. Y., Li B., Chen K. P., Li C. J. (2021). Genome-wide identification and characterization of long non-coding RNAs in Tribolium castaneum. Insect Sci. 28 (5), 1262–1276. doi:10.1111/1744-7917.12867
Yao R., Zhao D. D., Zhang S., Zhou L. Q., Wang X., Gao C. F., et al. (2017). Monitoring and mechanisms of insecticide resistance in Chilo suppressalis (Lepidoptera: Crambidae), with special reference to diamides. Pest Manag. Sci. 73, 1169–1178. doi:10.1002/ps.4439
Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., et al. (2016). Noncode 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 44 (D1), D203–D208. doi:10.1093/nar/gkv1252
Zhu B., Li L., Wei R., Liang P., Gao X. (2021). Regulation of GSTu1-mediated insecticide resistance in Plutella xylostella by miRNA and lncRNA. PLoS Genet. 17, e1009888. doi:10.1371/journal.pgen.1009888
Keywords: chlorantraniliprole resistance, long non-coding RNAs, adjacent protein-coding genes, genome-wide identification, Chilo suppressalis
Citation: Huang S, Jing D, Xu L, Luo G, Hu Y, Wu T, Hu Y, Li F, He K, Qin W, Sun Y and Liu H (2023) Genome-wide identification and functional analysis of long non-coding RNAs in Chilo suppressalis reveal their potential roles in chlorantraniliprole resistance. Front. Physiol. 13:1091232. doi: 10.3389/fphys.2022.1091232
Received: 06 November 2022; Accepted: 16 December 2022;
Published: 09 January 2023.
Edited by:
Peng He, Guizhou University, ChinaReviewed by:
Hu Wan, Huazhong Agricultural University, ChinaYoussef Dewer, Agricultural Research Center, Egypt
Xingliang Wang, Nanjing Agricultural University, China
Copyright © 2023 Huang, Jing, Xu, Luo, Hu, Wu, Hu, Li, He, Qin, Sun and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Sun, c3VuMjAwN3lhbmdAMTI2LmNvbQ==; Hui Liu, MTMxMDk3MzU1MEBxcS5jb20=
†These authors have contributed equally to this work and share first authorship
 Shuijin Huang1†
Shuijin Huang1† Lu Xu
Lu Xu Guanghua Luo
Guanghua Luo Fei Li
Fei Li Kang He
Kang He Yang Sun
Yang Sun