- 1Department of Biological Sciences, Vanderbilt University, Nashville, TN, United States
- 2Department of Chemical and Biomolecular Engineering, Vanderbilt University, Nashville, TN, United States
The study of circadian rhythms in bacteria was transformed by studies of the cyanobacterium Synechococcus elongatus. However, in a number of respects S. elongatus is atypical, and while those unusual characteristics were helpful for rapid progress in the past, another commonly used cyanobacterial species, Synechocystis sp. PCC 6803, may be more representative and therefore more productive for future insights into bacterial clock mechanisms. In the past, circadian studies of Synechocystis have suffered from not having an excellent reporter of circadian gene expression, but we introduce here a new luminescence reporter that rivals the reporters that have been used so successfully in S. elongatus. Using this new system, we generate for the first time in Synechocystis circadian period mutants resulting from point mutations. The temperature compensation and dark-pulse resetting that mediates entrainment to the environment is characterized. Moreover, we analyse the complex organization of clock genes in Synechocystis and identify which genes are essential for circadian rhythmicity and adaptive fitness for entrainment and optimal phase alignment to environmental cycles (and which genes are not). These developments will provide impetus for new approaches towards understanding daily timekeeping mechanisms in bacteria.
Introduction
Before the mid-1980s, chronobiologists thought that endogenous circadian rhythms were an exclusive property of eukaryotic organisms, and it became a dogma that prokaryotic organisms were either too “simple” or grew too rapidly to have evolved a bona fide circadian timekeeper (Johnson et al., 1996). That dogma cracked in 1986 with reports that the diazotrophic cyanobacterium Synechococcus RF-1 displayed daily rhythms of nitrogen fixation in LD cycles that persist in constant light (LL) (Grobbelaar et al., 1986; Huang et al., 1990; Chen et al., 1991). The salient properties of circadian rhythms–persistence, entrainment, and temperature compensation (Johnson et al., 2017) were established in these early studies, but progress on the genetics and mechanism of the cyanobacterial clockwork was hindered because no genetic tools were available for Synechococcus RF-1 (e.g., transformation, homologous recombination, luminescence/fluorescence reporters, etc.).
Several years after the reports of circadian phenomena in Synechococcus RF-1, we and our collaborators began the analysis of circadian rhythmicity in the cyanobacterium Synechococcus elongatus PCC 7942 (hereafter S. elongatus), which was a species for which genetic tools were available, and for which a luciferase reporter strain (PpsbAI::luxAB) had already been generated for the analysis of light intensity regulation of gene expression (Kondo et al., 1993; Johnson & Xu, 2009). The availability of genetic tools and a robust luminescence reporter of rhythmic gene expression enabled spectacular progress on understanding the mechanism and adaptive significance of circadian rhythmicity in cyanobacteria, transforming S. elongatus into one of the best understood circadian model organisms (Johnson et al., 2017; Chavan et al., 2021; Ito-Miwa et al., 2021; Johnson & Rust, 2021).
On the other hand, the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis) has long been a model organism for photosynthesis research and biotechnological applications (Nixon et al., 1992; Pakrasi & Vermaas, 1992; Shen et al., 1993; Ikeuchi & Tabata, 2001; Jagannathan & Golbeck, 2009; Santos-Merino et al., 2019; Wang et al., 2021b) partially due to the early discovery of its natural competence for genetic transformation (Grigorieva & Shestakov, 1982). For example, Synechocystis has served as a model platform for the structural and functional characterization of photosynthetic mechanisms, especially of photosystem II (Nixon et al., 1992; Pakrasi & Vermaas, 1992; Larom et al., 2010). When the complete genome sequence of Synechocystis became available in 1996, it became among the first of all phototrophic organisms to have that wealth of genetic information (Kaneko et al., 1996; Nakamura et al., 1998). As a result of this genomic data, research productivity involving this strain dramatically accelerated. In particular, Synechocystis-derived strains were explored as microbial cell factories to produce a variety of fuels and chemicals (Wang, et al., 2021b), including precursors to (biodegradable) plastics, such as poly-3-hydroxybutyrate (Wu et al., 2001; Agarwal et al., 2022), 3-hydroxybutyrate (Wang et al., 2018b), 3-hydroxypropionate (Wang Y. et al., 2016), lactate (Varman et al., 2013; Angermayr et al., 2014), and ethylene (Ungerer et al., 2012; Zhu et al., 2015; Wang et al., 2021a).
However, in comparison with S. elongatus, Synechocystis has been understudied in terms of its circadian properties (Aoki & Onai, 2009; Schmelling et al., 2021). The analysis of circadian phenomena in Synechocystis may have been hampered by the fact that the PdnaK::luxAB reporter developed for Synechocystis (Aoki et al., 1995) was never as robust as the PpsbAI::luxAB reporter that revolutionized circadian analyses in S. elongatus (Kondo et al., 1993; Kondo et al., 1994; Johnson & Xu, 2009; Johnson et al., 2017). Nevertheless, understanding and manipulating the circadian system of Synechocystis specifically would be beneficial for multiple reasons. For example, some strains of Synechocystis can grow photoheterotrophically by using glucose in the medium (Anderson & McIntosh, 1991), whereas S. elongatus is an obligate photoautotroph and must have light to grow. Therefore, photoheterotropic characteristics of Synechocystis could be useful in the analysis of circadian behavior in constant darkness using a luciferase reporter (Aoki et al., 1997), which is a capability that is not possible in the obligate photoautotroph S. elongatus. Moreover, in terms of biotechnological applications, we previously showed in S. elongatus that manipulating the circadian system can be used to enhance foreign gene expression (Xu et al., 2013b), and similar tools could be applied to the versatile biotech platform Synechocystis.
To overcome the deficit of an inadequate circadian assay, we have developed a new luminescence reporter for Synechocystis that exhibits many of the advantageous properties of S. elongatus’ PpsbAI::luxAB reporter in terms of brightness and excellent peak-to-trough amplitude. Surprisingly, our new reporter is based on using a promoter that is not endogenous to Synechocystis, but is derived from a chloroplast gene from a higher plant. We show here that the PpsbAAh::luxAB reporter as applied to Synechocystis identifies circadian properties that are equivalent to those that have been extensively characterized in S. elongatus and enabled the isolation of period mutants as well as rigorous tests of adaptive significance in Synechocystis.
Results
A strong promoter system was developed independently for biotechnological purposes using Synechocystis that derived a hybrid promoter from the chloroplast psbA gene of the higher plant Amaranthus hybridus coupled to an optimized ribosome binding site (RBS) (Salis et al., 2009; Xiong et al., 2015). We tested whether this hybrid promoter would display circadian rhythms in Synechocystis by designing a bacterial luciferase reporter and recombining it into the genome. This hybrid promoter/reporter (PpsbAAh::luxAB) displays excellent circadian rhythms of luminescence (Figure 1). In comparison with the previously used PdnaK::luxAB reporter (Aoki et al., 1995), luminescence signals are 5–10 times brighter with the PpsbAAh::luxAB reporter and display robust circadian rhythms with a period τ) slightly longer than 24 h in constant light (LL) at 30°C (Figure 1B, Supplementary Figure S1). As in the case of S. elongatus, we take this luminescence rhythm to be a reporter of pervasively rhythmic gene expression in Synechocystis, which has also been assessed by microarray studies (Liu et al., 1995; Woelfle et al., 2007; Ito et al., 2009; Vijayan et al., 2009). An expanded scale is shown for the data of the PdnaK::luxAB reporter in Figure 1C, and this is among the most robustly rhythmic traces we have observed for this reporter, whereas the trace for the data of the PpsbAAh::luxAB reporter in Figure 1B is of average robustness. Clearly the quality of rhythms is excellent with the PpsbAAh::luxAB reporter, and its brightness is also an advantage because it is less demanding of the sensitivity of the monitoring instruments. In addition, the PpsbAAh::luxAB reporter exhibited robust rhythms over a broad range of temperatures, whereas the rhythms of PdnaK::luxAB reporter became poor or arhythmic at lower temperatures (see below).
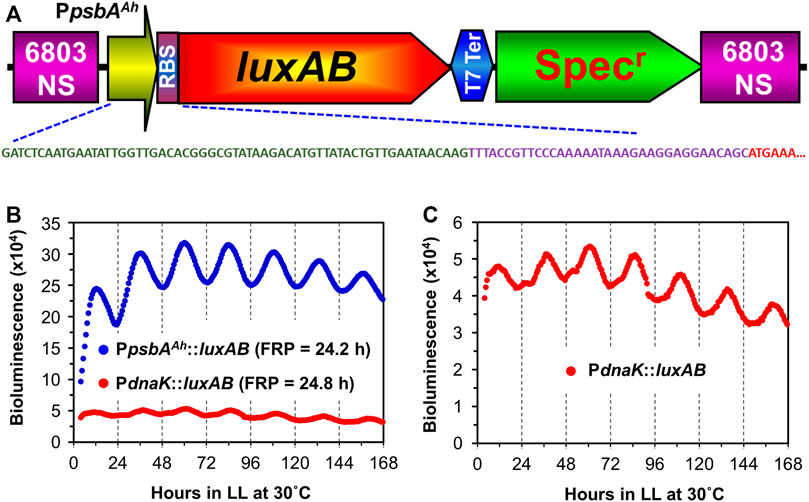
FIGURE 1. Comparison of rhythmic luxAB expression between the plant psbA promoter and the native dnaK promoter from colonies of Synechocystis sp. PCC 6803 on agar. (A). Diagram of the PpsbAAh::luxAB expression construct with a spectinomycin resistance marker inserted into the genome of PCC 6803. The DNA sequence shows the nucleotide sequences of the psbA promoter from the chloroplast of Amaranthus hybridus (green), an artificial ribosomal binding site (purple), and the partial N-terminal coding region of luxA (red). The physical map and full nucleotide sequence of the PpsbAAh::luxAB expression plasmid is presented in Supplementary Figure S1. (B). Representative luminescence rhythms and calculated free-running periods (FRPs) of the PpsbAAh::luxAB and PdnaK::luxAB reporters monitored in constant light at 30°C with the Kondotron. (C). The same luminescence rhythm for the PdnaK::luxAB reporter shown in panel B but on an expanded scale.
We therefore applied the PpsbAAh::luxAB reporter to address a previously unresolved question, namely what is the role of the various kaiC genes in circadian rhythmicity in Synechocystis? In S. elongatus, there is a single kaiABC clock gene cluster which encodes the central core clock proteins mediating the clockwork (Ishiura et al., 1998; Johnson et al., 2017), but in Synechocystis, there are three kaiC homologs organized with and without kaiA and kaiB homologs (Figure 2A) (Aoki & Onai, 2009). When the presence of kaiC homologs among bacterial species is assessed globally, it is not unusual to find species with two or more kaiC homologs (Aoki & Onai, 2009; Schmelling et al., 2021). Since S. elongatus is able to elaborate a precise clockwork with only one kaiC homolog, what is the function of multiple kaiC genes in those species that harbor more than one copy? Synechocystis is an excellent test case, so we undertook to make null strains in which each kai cluster (and each kai gene individually) was knocked out to determine if its presence was necessary to enable robust rhythmicity of gene expression as reported by PpsbAAh::luxAB (see Supplementary Figure S2 for the genotyping of the knock-out strains). Figure 2 illustrates the results of the gene knockouts upon the luminescence rhythm, where Figure 2B shows the data for three representative wild-type (WT) colonies. As was found for the case of the kaiABC cluster in S. elongatus (Ishiura et al., 1998), knocking out the kaiAB1C1 cluster or the kaiB1 or kaiC1 genes individually led to immediate arhythmicity (Figure 2C). Of the kai homologs in Synechocystis, the kaiB1 and kaiC1 genes are the most similar to kaiB and kaiC from S. elongatus (GroupA kai homologs (Aoki & Onai, 2009).
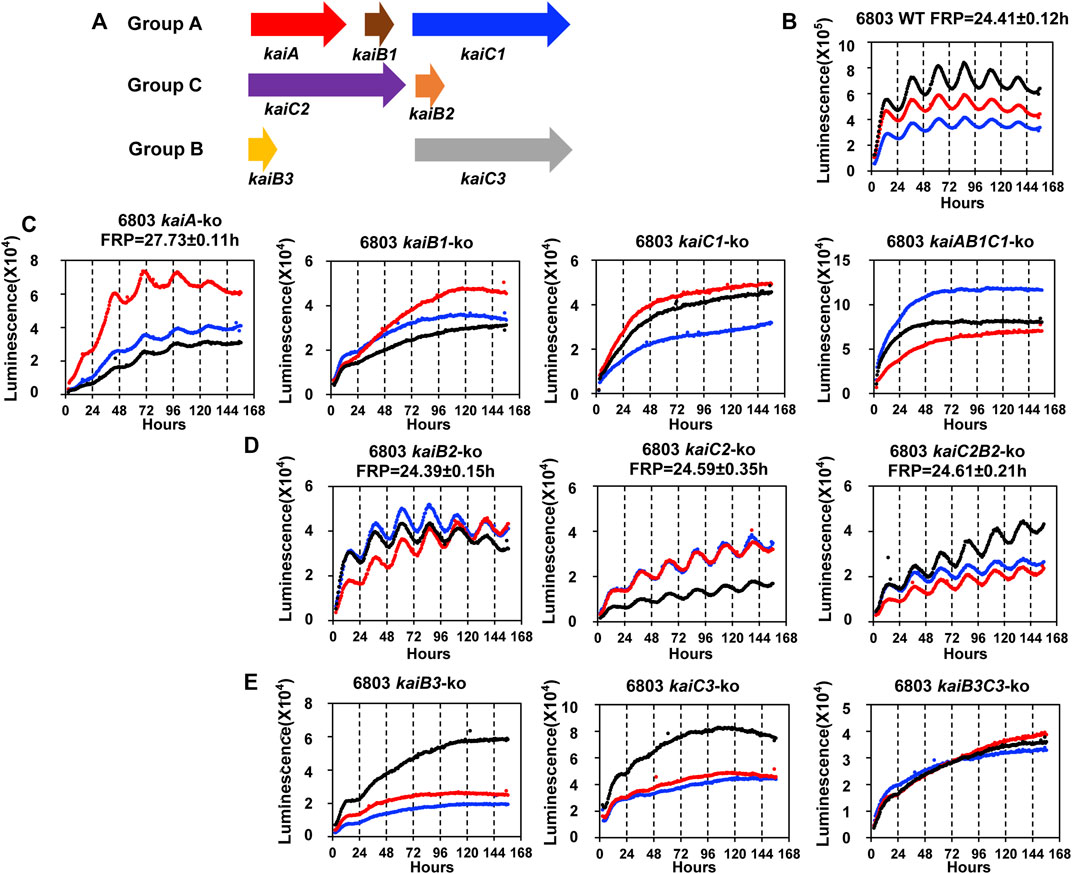
FIGURE 2. The rhythmicity of Synechocystis sp. PCC 6803 WT and kai null mutants on agar medium harboring the PpsbAAh::luxAB reporter in LL. (A). Schematic diagram of the three sets of kai genes in Synechocystis. The kaiA, kaiB1 and kaiC1 genes are localized together on the chromosome as a clock gene cluster, and kaiC1 belongs to GroupA that includes only kaiCs from cyanobacteria (Aoki & Onai, 2009). The kaiC2 and kaiB2 genes also localize together as a separate cluster, and kaiC2 belongs to GroupC which includes kaiC homologs from Archaea and Proteobacteria. The kaiB3 and kaiC3 localize separately on the chromosome, and kaiC3 belongs to GroupB which includes kaiC homologs from cyanobacteria, Proteobacteria and Chloroflexi (Aoki & Onai, 2009). (B). The rhythmicity of WT Synechocystis colonies in LL as monitored with the Kondotron. The free-running period (FRP) of WT in this experiment was 24.41 ± .12 h (n = 3). Three representative colonies are shown. (C). GroupA null mutants: expression patterns of kaiA-ko, kaiB1-ko, kaiC1-ko, kaiAB1C1-ko strains. All these null mutants were arhythmic except kaiA-ko, which expressed a reduced amplitude and long free-running period. (D). GroupC null mutants: all three null mutants (kaiB2-ko, kaiC2-ko and kaiC2B2-ko) were rhythmic with the free-running periods indicated on the panels. (E). GroupB null mutants: All three null mutants (kaiB3-ko, kaiC3-ko and kaiB3C3-ko) were arhythmic. Three representative traces are shown in each panel. FRP = free-running period.
Surprisingly–and unlike the case for S. elongatus–knocking out the kaiA gene did not cause total arhythmicity (although the amplitude of the rhythm was reduced and the free-running period (FRP) was lengthened). In non-cyanobacterial species, there is precedence for the hypothesis that KaiB and KaiC can generate a daily timekeeping process in the absence of KaiA (Ma et al., 2016). Indeed, even in S. elongatus, the deletion of KaiA has been reported to allow the expression of a damped circadian oscillation under some conditions (Kawamoto et al., 2020). Another interpretation of the data in Figure 2C based on the known mechanism for the cyanobacterial clockwork of S. elongatus is that there might be another protein in Synechocystis that exhibits KaiA-like functionality. Based on the operon structure of kaiABC in S. elongatus (Ishiura et al., 1998), it is possible that the knockout of kaiA has disrupted the promoter region of kaiB1C1, but this is not likely because 1) the kaiA-ko strain is still rhythmic whereas the kaiB1-ko, kaiC1-ko, and kaiAB1C1-ko strains are all arhythmic, and 2) computational prediction of the kaiB1C1 promoter is most likely to occur in the 126bp intergenic region between kaiA and kaiB1 (Supplementary Figure S3).
Another surprise was that knocking out the GroupC kaiCB cluster had essentially no effect on the circadian rhythm (Figure 2D), despite the suggestion of circadian-like functions mediated by GroupC kaiB and kaiC in the purple bacterium Rhodopseudomonas (Ma et al., 2016). The final unexpected result was that knocking out kaiB3 and/or kaiC3 also abolished (or at least dramatically reduced the amplitude of) the rhythm even though kaiC2 is more similar to kaiC1 (and kaiC from S. elongatus) than is kaiC3 (Figure 2E). Therefore, in terms of the circadian gene expression rhythm reported by PpsbAAh::luxAB, kaiAB1C1 and kaiB3/kaiC3 appear to be important for rhythmicity, whereas kaiC2B2 is not. The adaptive fitness value of each of these kai clusters was assessed by competition experiments described below.
In addition to persisting free-running rhythmicity under constant conditions, the other two defining properties of circadian rhythms are “temperature compensation” of the FRP and entrainment by environmental cycles (usually light/dark signals; (Pittendrigh, 1960; Johnson et al., 2017), both of which were established for the S. elongatus rhythms (Kondo et al., 1993). Consistent with a previous report (Aoki et al., 1995), temperature compensation of the FRP for Synechocystis is clearly demonstrated by the near-independence of FRP over a temperature gradient of 25°C–35°C (Figure 3A). Over this temperature range, the FRP alters by only ∼2 h, leading to a Q10 value of 1.08 (1.00 would be perfectly temperature independent), which is in the typical range for circadian clocks (Sweeney & Hastings, 1960). Panels B & C of Figure 3 underscore the value of using the PpsbAAh::luxAB reporter at 25°C and 35°C, where the amplitude of the PpsbAAh::luxAB rhythms are robust, whereas the amplitudes reported by PdnaK::luxAB are poor (and even somewhat bimodal) at the lower and higher temperatures. Entrainment to environmental cycles is another defining property, which can often be assessed by measuring the phase-dependent resetting by light or dark pulses that can be graphed as a Phase Response Curve (PRC) (Johnson, 1990). Because we are measuring Synechocystis rhythms in LL, we used dark pulses to elicit phase resetting. As expected, phase resetting to 6-h dark pulses exhibit a PRC with significant phase advances in the subjective day (CT 0-12) and phase delays in the subjective night (CT 12-24, see Figure 4), which is a commonly observed pattern for dark pulse PRCs from microorganisms to mammals (Boulos & Rusak, 1982; Johnson et al., 1989), and is appropriately in antiphase with the previously reported light-pulse PRC for Synechocystis (Aoki et al., 1997). Thus, all three defining properties of circadian pacemakers are satisfied by the rhythms reported by PpsbAAh::luxAB in Synechocystis.
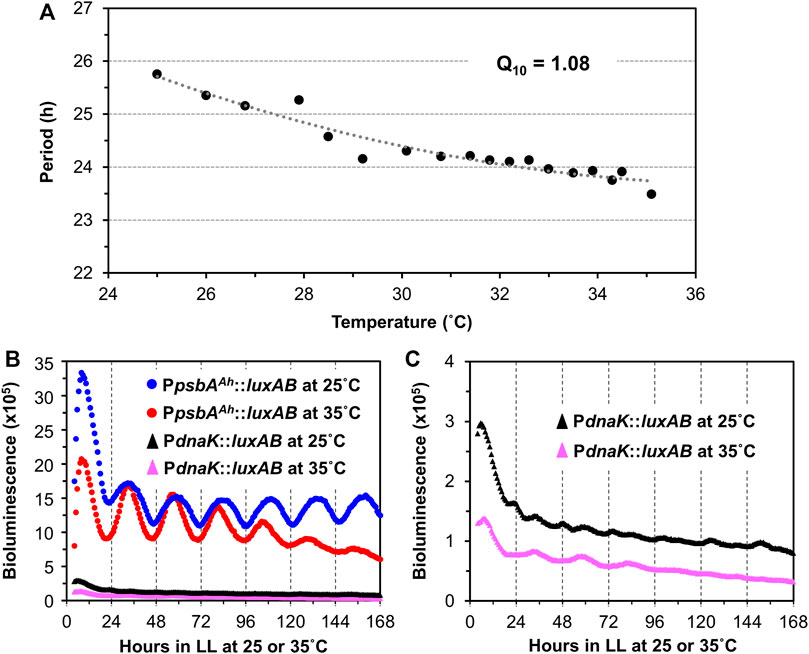
FIGURE 3. Temperature compensation of the PpsbAAh::luxAB-driving luminescence rhythms in Synechocystis PCC 6803. (A). Free-running periods (FRPs) and Q10 value of the PpsbAAh::luxAB reporter measured from WT Synechocystis colonies on Petri dishes containing agar medium at various temperatures in LL by the Taylortron. Q10 was calculated as described in the Materials and Methods. (B). Comparison of representative luminescence rhythms of the PpsbAAh::luxAB and PdnaK::luxAB reporters monitored in constant light at 25°C and 35°C with the Taylortron. (C). The same luminescence rhythms for the PdnaK::luxAB reporter shown in panel B but on an expanded scale.
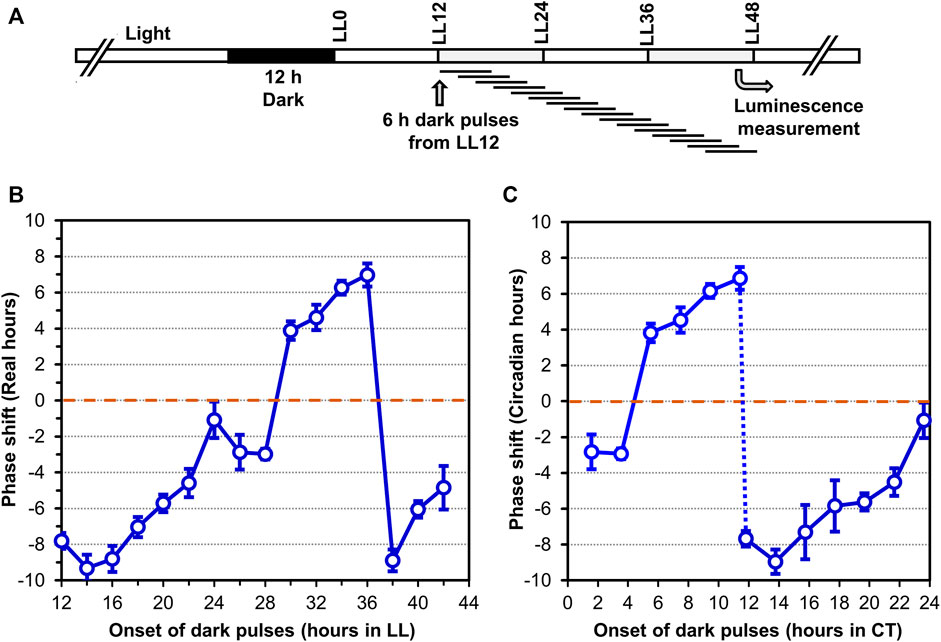
FIGURE 4. Phase Response Curve (PRC) to 6-h dark pulses of the luminescence rhythms reported by PpsbAAh::luxAB in Synechocystis PCC 6803. (A). Protocol of the experiment where 6-h dark pulses were applied at different time points in constant light (LL) beginning 12 h after the synchronizing 12-h dark exposure. After all dark pulses were completed (at LL48), the cultures were placed in the Kondotron turntable luminescence monitoring apparatus and the bioluminescence rhythms were monitored for the next 7 days. (B,C). Phase shifts caused by the 6-h dark pulses are plotted as a function of progressive time in constant light (Panel B) or as Circadian Time (=CT; Panel (C). Advance versus delay phase shifts are determined on the basis of whether the shifted peak is 12 h or less earlier than the control (= Advance phase shifts, plotted as + values), versus 12 h or less later than the control (= Delay phase shifts, plotted as–values). Data are averages and standard deviations from six replicates.
In S. elongatus, the PpsbAI::luxAB reporter enabled a mutant screening that identified a stable of FRP mutants that ultimately led to the identification of the kaiABC gene cluster (Kondo et al., 1994; Ishiura et al., 1998). The brightness and robustness of the PpsbAAh::luxAB reporter should make possible such a mutant screen in Synechocystis. However, given 1) the large quantity and variety of FRP mutants in S. elongatus for which the altered sequence is known and 2) the excellent sequence conservation between kaiABC in S. elongatus with that of kaiAB1C1 in Synechocystis, it might be possible to create the first FRP mutants in Synechocystis on the basis of the known mutations in S. elongatus. We set out to test that prediction by directed mutagenesis (Figure 5A), and indeed this prediction was upheld. Mutations of Synechocystis’ kaiA gene based on the work of Nishimura and coworkers with S. elongatus (Nishimura et al., 2002) led to the generation of three mutant strains with FRPs from ∼28 h to ∼23 h (kaiAD119E, kaiAE103K, kaiAF224S, Figures 5C–E). Moreover, based on the fantastic range of FRPs generated by mutations to a single residue (Tyrosine402) of S. elongatus’ kaiC that was reported from the Kondo lab (Ito-Miwa et al., 2020), we revealed three mutations in Y402 of Synechocystis’ kaiC1 that ranged from ∼23 h to ∼27 h (kaiC1Y402F, kaiC1Y402M, kaiC1Y402W, Figures 5F–H). The similarity of the effect of these mutations in Synechocystis with those in S. elongatus confirms that the kaiAB1C1 gene is the master clock locus in Synechocystis. However, the results in Figure 2E implies an essential role for kaiC3 that could create a kaiC1-kaiC3 redundancy that might mask mutations in kaiC1, but at least so far, this has not been observed. It would probably be worthwhile to test the effects of the mutations shown in Figure 5 in the kaiB3/kaiC3 background, as well as testing point mutations in kaiC3.
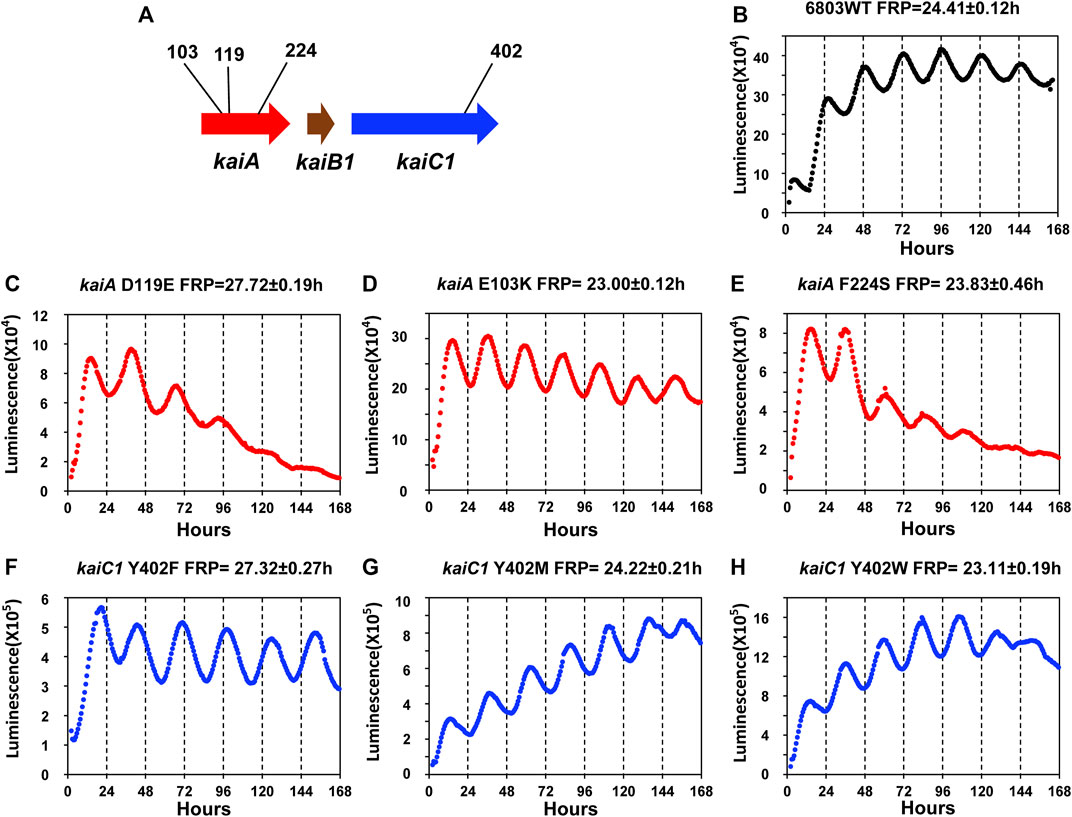
FIGURE 5. Luminescence phenotypes of period mutants in Synechocystis generated by point mutations that were informed by mutations identified in S. elongatus. (A). Schematic diagram of the kaiA and kaiC1 loci identifying the positions of the point mutations. Panels (B–H): rhythmicity of representative colonies of the indicated strain/mutant of Synechocystis in LL monitored by the Kondotron. (B). WT Synechocystis (FRP = 24.41 ± .12 h). (C). kaiAD119E (FRP = 27.72 ± .19 h). (D). kaiAE103K (FRP = 23.00 ± .12 h). (E). kaiAF224S (FRP = 23.83 ± .46 h). (F). kaiCY402F (FRP = 27.32 ± .27 h). (G). kaiCY402M (FRP = 24.22 ± .21 h). (H). kaiCY402W (FRP = 23.11 ± .19 h). All FRP data are means and S.D. from three replicates.
To test if the various kai clusters in Synechocystis influence the fitness of the cells in LL or LD, we performed mixed-strain competitions modeled upon those we pioneered for assessing the adaptive significance of circadian periodicity in S. elongatus (Figure 6; (Woelfle et al., 2004). First, we assessed the single-strain growth rates of the WT, kaiAB1C1-ko, kaiC2B2-ko, and kaiB3/kaiC3-ko strains individually in LL, which is the non-selective environmental condition (Figure 6B). In LL, only the kaiB3/kaiC3-ko strain appears to grow significantly slower than the other three strains. In mixed cultures, WT outcompetes kaiAB1C1-ko even though there was not a significant difference in their growth rates in pure cultures (Figure 6C). The rhythm of the kaiB3/kaiC3-ko strain was severely disrupted (Figure 2E), and its growth was slower which apparently leads to out-competition by WT in mixed cultures (Figures 6B,C). In the clock-selective condition of LD 12:12, the kaiAB1C1-ko has a significantly lower growth rate. And both of the strains whose rhythms are disrupted (Figures 2C,E), namely kaiAB1C1-ko and kaiB3/kaiC3-ko, are out-competed by WT in LD (Figure 6E). Interestingly, rhythms in kaiC2B2-ko seem to be largely unaffected (Figure 2D), and the growth rate and competitive ability of this strain appears to be relatively equivalent to WT in both LL and LD (Figure 6).
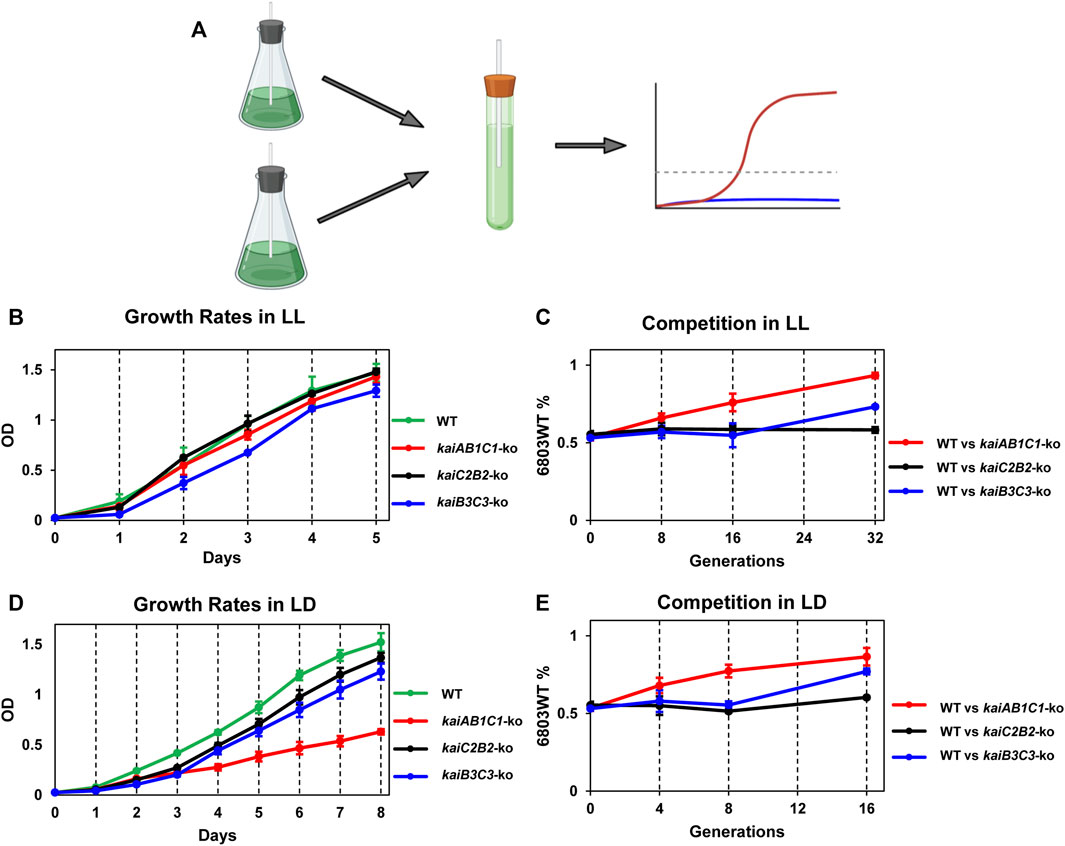
FIGURE 6. Adaptive fitness of the various kai gene clusters assessed by growth rates of monocultures and competition assays. The Synechocystis strains that were tested include wild-type (WT) and knockouts of the three different kai gene clusters (kaiAB1C1-ko, kaiC2B2-ko and kaiB3/kaiC3-ko). (A). Mixed-strain cultures were grown in competition and changes in population structure quantified by QPCR of strain-specific sequences. (B). Growth curves of the four strains in LL as assessed by light scattering of the cell culture (measured at OD730) as a function of time. (C). Competition of WT against the kaiAB1C1-ko, kaiC2B2-ko or kaiB3C3-ko strains over generations/time in LL. The ordinate plots the percentage of the WT strain over time under competition. (D). Growth curves of the four strains in LD 12:12 as assessed by light scattering of the cell culture (measured at OD730) as a function of time. (E). Competition of WT against the kaiAB1C1-ko, kaiC2B2-ko or kaiB3C3-ko strains over generations/time in LD 12:12. The ordinate plots the percentage of the WT strain over time under competition. Data are plotted as means ± S.D. from three biological replicates.
Discussion
Clearly, circadian rhythmicity in Synechocystis is robust (Figures 1–5) and adaptive (Figure 2; Figure 6). In hindsight, it was fortuitous that the first cyanobacterial species to be analyzed genetically for circadian rhythmicity was S. elongatus rather than Synechocystis (Kondo et al., 1993; Kondo et al., 1994; Ishiura et al., 1998). First, S. elongatus has a single kaiABC cluster (Ishiura et al., 1998), whereas Synechocystis and many other cyanobacterial species have multiple copies/clusters of kai genes that might have complicated the initial analysis of the genetic basis of the circadian clockwork (Aoki & Onai, 2009; Schmelling et al., 2021). Second, the first luciferase reporter created in S. elongatus for other experimental goals, the PpsbAI::luxAB reporter, turned out to exhibit excellent high-amplitude rhythms (Kondo et al., 1993) that were optimal for enlistment to the high-throughput screen for clock mutations (Kondo et al., 1994) that ultimately led to the discovery of the kaiABC clock gene cluster (Ishiura et al., 1998). However, PpsbAI::luxAB is not uniquely high amplitude in S. elongatus–in fact, in contrast to Synechocystis, most promoters from S. elongatus exhibit high amplitude rhythms (Liu et al., 1995). Therefore, serendipitously S. elongatus turned out to be an optimal species to begin circadian analyses in cyanobacteria.
At first, it might seem surprising that a promoter from a very different organism exhibits a better rhythm than the endogenous promoters that have been tested in Synechocystis. The example of S. elongatus, however, again exemplifies this principle; the conII promoter from E. coli expresses a beautiful rhythm in S. elongatus when coupled to a luxAB reporter (Xu, et al., 2013b). In S. elongatus, this result has been interpreted to mean that the entire chromosome is undergoing circadian cycles of supercoiling/compaction that confer circadian rhythmicity on all promoters, even heterologous promoters (Woelfle et al., 2007; Xu et al., 2013b). The PpsbAAh promoter used for driving the expression of luxAB in this study is among the strongest constitutive promoters characterized in cyanobacteria so far. It is a hybrid of the core promoter region of a chloroplast psbA gene (from Amaranthus hybridus) and an artificial RBS generated using the RBS Calculator (Salis et al., 2009; Xiong et al., 2015). This promoter-RBS hybrid, sometimes with subtle modifications, exhibited superior performance in expressing a variety of genes, such as the ethylene-forming enzyme (Efe), catalase (KatE), and two synthetic hydrogenase operons, rendering up to 12% of the total soluble protein as the target enzyme in Synechocystis (Xiong et al., 2015; Wang et al., 2018a). In addition, the same promoter was also able to drive strong expression of Efe and a limonene synthase in S. elongatus, and this capability was used to eliminate pathway bottlenecks (Wang X. et al., 2016; Wang et al., 2021a). Investigating the behaviors of biotechnology-relevant strong promoters, such as the herein studied PpsbAAh promoter, in response to the circadian rhythm regulation in Synechocystis could provide important information for guiding the synthetic biology and metabolic engineering efforts in Synechocystis in the future.
As we previously found for S. elongatus (Woelfle et al., 2004), the rhythmic WT strain of Synechocystis was able to outcompete arhythmic strains in LD (Figure 6). This experimental result clearly indicates the adaptiveness of appropriately aligning an internal oscillator with the external environmental cycle, and therefore that “desynchrony” of these two rhythms hampers competitiveness. Interestingly, arhythmic strains are able to compete successfully with WT in the non-selective LL conditions for S. elongatus (Woelfle et al., 2004), whereas WT Synechocystis is also dominant in LL (Figure 6), implying a fitness advantage for temporal organization in Synechocystis even when the environment is not rhythmic. On the basis of these knockout studies, the kaiC2B2 cluster appears to not contribute to rhythmicity nor to fitness (Figure 2; Figure 6). This result is unexpected, given that GroupC kai genes do contribute to fitness in rhythmic environments in Rhodopseudomonas (Ma et al., 2016). Interestingly, we found that knocking out kaiB3 and/or kaiC3 drastically attenuated the circadian rhythm in Synechocystis, a phenotype that was unexpected when compared to the findings from a previous report in which ΔkaiC3 mutant did not exhibit any apparent growth defect relative to the WT Synechocystis strain under photoautotrophic and photomixotrophic conditions during spot assays (Dörrich et al., 2014). In the current study, we not only found that growth of the ΔkaiB3C3 mutant was significantly impaired (Figure 6), but also that the circadian rhythmicity of Synechocystis cells was eliminated when either kaiB3 or kaiC3 was knocked out (Figure 2). Our results hence provided sufficient evidence that Group B kai genes, i.e., kaiB3 and kaiC3, are essential to the circadian clock of Synechocystis.
In this study, we developed a new methodology for measuring the proportions of two strains in mixed cultures based on QPCR rather than on our previous method of differential antibiotic sensitivity and colony-forming unit (CFU) counting (Woelfle et al., 2004). An advantage of our new methodology is that both strains in a mixed culture can harbor the same antibiotic resistance, thereby avoiding the complication of whether having different antibiotic resistance genes in the two strains are having a fitness effect (we always undertake our competition assays in the total absence of antibiotics, but the resistance genes themselves might have an impact on intracellular metabolism). A potential disadvantage of the new QPCR method is that we are measuring chromosome copy numbers rather than cell numbers or CFUs. WT Synechocystis cells have an average number of chromosome copies of three to six per cell (Pope et al., 2020). However, if the number of chromosome copies has been changed in any of the kai knockout strains, then the absolute proportion of the cells might be biased. We consider this to be a minor liability because even if the two tested strains have different numbers of chromosomes/cell, the relative proportions and their change over time will still be valid. And for the competition assay, the most important factor to be measured is the change over time of the proportion of one strain to the other. Therefore, we believe that the advantages of the new method outweigh its potential disadvantages.
As stated above, S. elongatus has functioned as an excellent system that provided superb insights into circadian mechanisms and adaptiveness, but its “simple” kaiABC genetic background makes it impossible to study the orchestration of Kai proteins from different Kai Gene Groups. On the other hand, the more complicated kai genetics of Synechocystis may provide a more realistic view of redundant genetics in a prokaryote. Moreover, the mechanism of the GroupB and GroupC Kai proteins may be most easily addressed in Synechocystis and subsequently those insights can be applied to the analysis of other species of bacteria that harbor those other versions of the kai genes. In that sense, Synechocystis may be a more productive “springboard” towards understanding circadian rhythmicity in non-cyanobacterial species than has been S. elongatus. Therefore, Synechocystis may be the model system that enlarges the scope of our comprehensive understanding and appreciation of daily timekeeping mechanisms in bacteria.
Materials and methods
Strains and growth conditions
The cyanobacterium Synechocystis sp. PCC 6803 wild-type (WT) strain was the basis of these studies. The cyanobacteria were grown on modified BG11 medium on agar plates or in liquid (Rippka et al., 1979; Bustos & Golden, 1991) with appropriate antibiotics (Xue et al., 2014; Taton et al., 2020). Cells were grown at 30 °C under constant cool-white fluorescence light (LL, 40–50 µE/m2s), or given one or two 12-h light/12-h dark cycles (e.g., LD 12:12) before release to LL to synchronize the cells in the population. NEB® 5-alpha Competent E. coli (New England Biolabs) were used for the plasmid construction and gene cloning with appropriate antibiotics. E. coli cells were grown on LB medium (Bertani, 1951, 2004), on agar plates or in liquid media at 37 °C.
Generation of PpsbAAh::luxAB reporter
The core promoter sequence from the chloroplast psbA gene of the plant Amaranthus hybridus was coupled to an engineered ribosome binding site (Xiong et al., 2015; Wang et al., 2018a) and assembled with the coding sequences of the Vibrio harveyi luciferase structure gene luxAB to generate a 2.18 kb fragment of PpsbAAh::luxAB (Figure 1A). A 1.24 kb BbvC I/Hind III fragment on pJU158 (Xiong et al., 2015) was replaced with the PpsbAAh::luxAB fragment to produce the PpsbAAh-driving luminescence expression construct with a spectinomycin selection marker and the sir0168-flanking sequences (Xiong et al., 2015) of Synechocystis genome (Supplemental Figure S1). The transformed PpsbAAh::luxAB reporter was confirmed by luminescence expression and colony PCR with a 2.2 kb DNA product using a pair of primers 5-ATCTCAATGAATATTGGTTGA-3 and 5-ACGAGTGGTATTTGACGATGTTG-3.
Construction of null and period mutant strains
All the kai null mutants as well as the period mutants were constructed based on the PpsbAAh::luxAB reporter strain. The knockout plasmids were constructed in 5-alpha Competent E. coli first, and thereafter transformed into Synechocystis sp. PCC 6803 wild type strain harboring the PpsbAAh::luxAB reporter. Briefly, approximately 1 Kb upstream and 1 Kb downstream fragments of the target genes were amplified separately through PCR, then linked together with an antibiotic resistance gene. This larger DNA fragment was cloned into the linearized pMiniT™ 2.0 cloning vector (NEB® PCR Cloning Kit, New England Biolabs) to prepare for the transformation into cyanobacteria. To obtain the kaiA knockout plasmid (kaiA-ko-Em), 1 kb upstream and 1 kb downstream regions of the kaiA orf were amplified through PCR, then linked by the Erythromycin (Em) resistance gene, and the resulting fragment was inserted into the linearized pMiniT™ 2.0 cloning vector (NEB® PCR Cloning Kit, New England Biolabs). Equivalent methods were used to generate the kaiB1-ko-Em and kaiB3-ko-Em knockout plasmids with Erythromycin resistance. Similarly, we used a Kanamycin (Km) resistance knockout plasmid to generate knockouts of the kaiC1, kaiAB1C1, and kaiB2 genes (kaiC1-ko-Km, kaiAB1C1-ko-Km, and kaiB2-ko-Km). We used a Chloramphenicol (Cm) resistant knockout plasmid to generate knockouts of the kaiC2 and kaiC2B2 genes (kaiC2-ko-Cm and kaiC2B2-ko-Cm). Finally, we used a Gentamycin (Gm) resistant knockout plasmid to generate a knockout of the kaiC3 gene (kaiC3-ko-Gm). See Supplemental Figure S2 for the genotyping data that confirm these knockouts, Supplemental Figure S4 for the physical maps & DNA sequences of all the plasmids for generating null mutants, and Supplemental Table S1 for the primer sequences used.
The period mutants were constructed by site-directed mutagenesis of the Synechocystis genome to produce mutations based on circadian mutants discovered in S. elongatus (Ishiura et al., 1998; Nishimura et al., 2002; Ito-Miwa et al., 2020). These mutations were introduced into the kaiAB1C1 locus. The plasmid contained the wild type Promoter-kaiAB1C1 DNA sequence was first assembled into the NEB cloning vector (NEB® PCR Cloning Kit, New England Biolabs), then point mutations were introduced into either the kaiA or the kaiC1 gene with QuikChange II Site-Directed Mutagenesis Kit (Agilent). Briefly, a pair of primers containing the desired mutations were used to amplify the backbone plasmid through PCR with PfuUltra HF DNA polymerase. DpnI was used to digest the methylated template DNA, then the purified DNA was transformed to E. coli competent cells. The correct mutations were confirmed with Sanger Sequencing. The resulting plasmid which contained the chloramphenicol resistance cassette was then transformed into the kaiAB1C1 null mutant strain to integrate the mutated version of kaiAB1C1 into the endogenous site by homologous recombination, thereby reconstituting the kaiAB1C1 locus with the mutation, and the resulting period mutants were resistant to both Spectinomycin and Chloramphenicol. See Supplementary Figure S4 for the physical maps & DNA sequences of all the plasmids for generating period mutants, and Supplementary Table S1for the primer sequences used.
Homologous recombination and natural transformation were used to obtain all the null and period mutants (Eaton-Rye, 2004; Clerico et al., 2007). Approximately 1 µg of plasmid DNA as well as 4–5 ml of a cyanobacterial liquid culture in log phase growth (OD730 ∼.4–.6) were mixed together in an Eppendorf tube. The mixture was left in the dark for 3–4 h, then spread on a BG11 agar plate without antibiotics and left in the light for 12–16 h. Top agar with appropriate antibiotics was poured on top of the plate. The recombinant colonies appeared in ∼10 days. Fully segregated mutants were obtained after streaking 4-5 times on the agar plates with antibiotics. The concentrations of the antibiotics supplemented in the media for the selection of Synechocystis mutants were: Spectinomycin 30 μg/ml, Kanamycin 10 μg/ml, Chloramphenicol 7.5 μg/ml, Erythromycin 10 μg/ml, and Gentamycin 5 µg/ml.
Bioluminescence monitoring
Synechocystis luminescence reporter strains were grown in modified BG11 medium on agar plates that was supplemented with appropriate antibiotics at 30°C under continuous cool-white illumination (LL; 40–50 µE/m2s). Circadian rhythms of luminescence from Synechocystis are generally more robust from colonies on agar plates as compared with liquid cultures. Fresh agar cultures grown in LL for two or 3 days were tooth-picked onto freshly made agar plates and after further growth in LL for one or days, a 12 h dark exposure was given to synchronize the clocks in the populations. Bioluminescence measurements of the Synechocystis colonies on agar media in LL were carried out using automated luminescence measuring systems (Kondo et al., 1993; Kondo et al., 1994; Johnson & Xu, 2009). A custom computer controlled turntable/CCD camera apparatus called the “Kondotron” was used to monitor the in vivo luminescence of single colonies on agar plates (Kondo et al., 1994). For measurement of luminescence rhythms over a broad range of temperatures, a custom apparatus called the “Taylortron” was used (Johnson & Xu, 2009). The Taylortron comprises a hollow metal bar with 30 positions, each of which can hold a 20-ml scintillation vial with a cyanobacterial culture on liquid or agar media. A temperature-controlled heater blows hot air into one end of the metal bar, thereby generating a temperature gradient along the bar that can be adjusted by appropriately altering the heater’s setting relative to the setpoint of the temperature-controlled room that houses the Taylortron. In the Taylortron, a computer-controlled cart with a photomultiplier tube moves from position to position and automatically monitors the intensity of the luminescence emission from the 20 ml vials containing 3–5 ml of liquid or agar medium. For the temperature gradient assay in this study, 12 colonies from fresh agar cultures of the WT reporter strain were tooth-picked onto the surface of the agar inside each vial, and then an open Eppendorf tube containing 0.1 ml oil with 1% decanal was also put inside the vial. After a 12 h dark synchronization, the luminescence rhythms were measured at a gradient of temperatures in the Taylortron in LL at an intensity of ∼50 µE/m2s. The Taylortron was used to monitor the rhythms in the experiments of Figure 3, whereas the turntable “Kondotron” (Johnson & Xu, 2009) was used to monitor the rhythms in the experiments of Figures 1–5.
Q10 measurement and calculations
The different periods of luminescence rhythms of the wild type Synechocystis sp. PCC 6803 over the temperature ranges from 25°C to 35°C were analyzed with ChronoAnalysis II, version 10.1 (courtesy of T. Roenneberg) (Xu et al., 2013a), and the temperature coefficient Q10 for the evaluation of Temperature Compensation was estimated by fitting Eq 1 to a set of the data points (i.e., circadian periods that were experimentally obtained at various temperatures).
where the dependent variable
Dark-pulse phase response curve
Colonies of wild type Synechocystis on agar plates were given a 12-h light/12-h dark cycle (LD 12:12) to synchronize all the cells, then the colonies were released into LL. Starting from 12 h in the light (LL12), the agar plates were sequentially given a 6-h dark treatment followed by the next plate 2 h later (from LL12 to LL42). A control plate remained in LL without any dark-pulse treatment. Phase shifts that resulted from these dark pulses were calculated with ChronoAnalysis II, version 10.1 (courtesy of Dr. T. Roenneberg); the values at each timepoint were calculated from eight replicate colonies (Xu et al., 2000).
Intra-species competition
Single-strain monocultures of Synechocystis WT, kaiAB1C1-ko, kaiC2B2-ko and kaiB3C3-ko were grown under 30°C and constant cool-white fluorescence light (LL, 40–50 µE/m2s) to OD730 .4-.6. Then WT was mixed with kaiAB1C1-ko, kaiC2B2-ko and kaiB3C3-ko respectively in 30 ml liquid BG11 to an OD730 ratio of 1:1. The starting OD730 of the mixed-strain culture was .2. The competition of the mixed-strain cultures (3 biological replicates for each combination) was conducted in either LL or LD 12:12 with the cultures bubbled with air to provide aeration (especially CO2). Cultures were diluted every 8 d, and samples were collected for quantification on Days 0, 4, 8 and 16. Quantitative QPCR was used to quantify the copy numbers of the integrated antibiotic resistance genes as a proxy for chromosome number in the WT strain versus the other strain in each competition combination.
Genome DNA extraction and absolute quantitative QPCR calculations
For the measurement of the relative percentages of Synechocystis chromosome copy numbers of WT and kai mutants in the competition experiments, absolute quantitative QPCR was applied. A pair of primers targeting the Spectinomycin resistant gene was used to quantify both strains in each combination (PpsbAAh::luxAB was inserted into the genome with the Spectinomycin resistance gene cartridge), whereas primers targeting the Kanamycin, Chloramphenicol, and Erythromycin resistance genes were used to quantify the kaiAB1C1, kaiC2B2 and kaiB3C3 null mutants respectively. We first amplified the DNA fragments targeting the regions on Spectinomycin, Kanamycin, Chloramphenicol and Erythromycin resistance genes, purified them with the Monarch® DNA Gel Extraction Kit (New England Biolabs) and measured their concentrations. Then we did serial dilutions of each specific DNA fragment to known concentrations (e.g., to 10–13 mol/µL, 10–14 mol/µL, 10–15 mol/µL, 10–16 mol/µL, 10–17 mol/µL and 10–18 mol/µL). Henceforth, those DNA fragments were used as the standard samples to calculate DNA concentrations in the actual samples.
Ct (cycle time) values from the wells that contained the standard samples (with known concentrations) were used to draw standard curves to quantify the DNA concentrations and the Ct values, as shown inSupplementary Figure S5. From the wells that contained the actual samples (genome DNA extracted with Favorgen plant genomic DNA extraction Mini Kit FAPGK001-2), we used the Ct numbers obtained in QPCR to quantify the DNA concentrations of each strain separately, which were used to estimate chromosome copy number with the standard curves; in Synechocystis, the average number of chromosome copies is three to six per cell (Pope et al., 2020). By these comparisons, we calculated the relative percentages of each strain in the mixed cultures under competition.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
CZ, YX, and CJ designed the experiments, CZ and YX performed the experiments and analyzed the data, and BW provided essential reagents, materials, and information. All authors contributed to the writing and editing of the manuscript. CJ was responsible for overall supervision of the study and generated the funding for the research. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funding from the United States National Institutes of Health (GM107434 and GM067152) to CJ.
Acknowledgments
We thank Jacob Gussert for his assistance in the early phases of this project, and we are grateful to Dr. Ken-ichi Kucho of Kagoshima University (Japan) for his gift of the pTS1PdnaK::luxAB plasmid that was used to construct the PdnaK::luxAB reporter. The program for analysis of circadian data, ChronoAnalysis II version 10.1, was provided courtesy of Dr. Till Roenneberg. We are also grateful to Dr. Jianping Yu of National Renewable Energy Laboratory for sharing the pJU158 plasmid that includes the PpsbA promoter from Amaranthus hybridus and RBSv4.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.1085959/full#supplementary-material
References
Agarwal P., Soni R., Kaur P., Madan A., Mishra R., Pandey J., et al. (2022). Cyanobacteria as a promising alternative for sustainable environment: Synthesis of biofuel and biodegradable plastics. Front. Microbiol. 13, 939347. doi:10.3389/fmicb.2022.939347
Anderson S. L., Mcintosh L. (1991). Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: A blue-light-requiring process. J. Bacteriol. 173, 2761–2767. doi:10.1128/jb.173.9.2761-2767.1991
Angermayr S. A., Van Der Woude A. D., Correddu D., Vreugdenhil A., Verrone V., Hellingwerf K. J. (2014). Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803. Biotechnol. Biofuels 7, 99–15. doi:10.1186/1754-6834-7-99
Aoki S., Kondo T., Ishiura M. (1995). Circadian expression of the dnaK gene in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 177, 5606–5611. doi:10.1128/jb.177.19.5606-5611.1995
Aoki S., Kondo T., Wada H., Ishiura M. (1997). Circadian rhythm of the cyanobacterium Synechocystis sp. strain PCC 6803 in the dark. J. Bacteriol. 179, 5751–5755. doi:10.1128/jb.179.18.5751-5755.1997
Aoki S., Onai K. (2009). “Circadian clocks of Synechocystis sp. strain PCC 6803, Thermosynechococcus elongatus, Prochlorococcus spp., Trichodesmium spp. and other species,” in Bacterial circadian programs. Editors J. L. Ditty, S. R. Mackey, and C. H. Johnson (Berlin, Heidelberg: Springer), 259–282.
Bertani G. (2004). Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186, 595–600. doi:10.1128/JB.186.3.595-600.2004
Bertani G. (1951). Studies on lysogenesis I: The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300. doi:10.1128/jb.62.3.293-300.1951
Boulos Z., Rusak B. (1982). Circadian phase response curves for dark pulses in the hamster. J. Comp. Physiol. 146, 411–417. doi:10.1007/BF00609437
Bustos S. A., Golden S. S. (1991). Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J. Bacteriol. 173, 7525–7533. doi:10.1128/jb.173.23.7525-7533.1991
Chavan A. G., Swan J. A., Heisler J., Sancar C., Ernst D. C., Fang M., et al. (2021). Reconstitution of an intact clock reveals mechanisms of circadian timekeeping. Science 374, eabd4453. doi:10.1126/science.abd4453
Chen T.-H., Chen T.-L., Hung L.-M., Huang T.-C. (1991). Circadian rhythm in amino acid uptake by Synechococcus RF-1. Plant Physiol. 97, 55–59. doi:10.1104/pp.97.1.55
Clerico E. M., Ditty J. L., Golden S. S. (2007). “Specialized techniques for site-directed mutagenesis in cyanobacteria,” in Circadian rhythms. Editor E. Rosato (Totowa, NJ: Humana Press), 155–171.
Dörrich A. K., Mitschke J., Siadat O., Wilde A. (2014). Deletion of the Synechocystis sp. PCC 6803 kaiAB1C1 gene cluster causes impaired cell growth under light-dark conditions. Microbiology 160, 2538–2550. doi:10.1099/mic.0.081695-0
Eaton-Rye J. J. (2004). “The construction of gene knockouts in the cyanobacterium Synechocystis sp. PCC 6803,” in Photosynthesis research protocols. Editor R. Carpentier (Totowa, NJ: Humana Press), 309–324.
Grigorieva G., Shestakov S. (1982). Transformation in the cyanobacterium Synechocystis sp. 6803. FEMS Microbiol. Lett. 13, 367–370. doi:10.1111/j.1574-6968.1982.tb08289.x
Grobbelaar N., Huang T., Lin H., Chow T. (1986). Dinitrogen-fixing endogenous rhythm in Synechococcus RF-1. FEMS Microbiol. Lett. 37, 173–177. doi:10.1111/j.1574-6968.1986.tb01788.x
Huang T.-C., Tu J., Chow T.-J., Chen T.-H. (1990). Circadian rhythm of the prokaryote Synechococcus sp. RF-1. Plant Physiol. 92, 531–533. doi:10.1104/pp.92.2.531
Ikeuchi M., Tabata S. (2001). Synechocystis sp. PCC 6803—A useful tool in the study of the genetics of cyanobacteria. Photosynth. Res. 70, 73–83. doi:10.1023/A:1013887908680
Ishiura M., Kutsuna S., Aoki S., Iwasaki H., Andersson C. R., Tanabe A., et al. (1998). Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523. doi:10.1126/science.281.5382.1519
Ito H., Mutsuda M., Murayama Y., Tomita J., Hosokawa N., Terauchi K., et al. (2009). Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. U. S. A. 106, 14168–14173. doi:10.1073/pnas.0902587106
Ito-Miwa K., Furuike Y., Akiyama S., Kondo T. (2020). Tuning the circadian period of cyanobacteria up to 6.6 days by the single amino acid substitutions in KaiC. Proc. Natl. Acad. Sci. U. S. A. 117, 20926–20931. doi:10.1073/pnas.2005496117
Ito-Miwa K., Terauchi K., Kondo T. (2021). “Mechanism of the cyanobacterial circadian clock protein KaiC to measure 24 hours,” in Circadian rhythms in bacteria and microbiomes. Editors C. H. Johnson, and M. J. Rust (Cham, Switzerland: Springer), 79–91.
Jagannathan B., Golbeck J. (2009). “Photosynthesis: Microbial,” in Encyclopedia of microbiology. Editor M. Schaechter (Amsterdam: Elsevier), 325–341.
Johnson C. H. (1990). An atlas of phase responses curves for circadian and circatidal rhythm. Available at: https://as.vanderbilt.edu/johnsonlab/prcatlas/[Accessed December 3, 2022].
Johnson C. H., Golden S. S., Ishiura M., Kondo T. (1996). Circadian clocks in prokaryotes. Mol. Microbiol. 21, 5–11. doi:10.1046/j.1365-2958.1996.00613.x
Johnson C. H., Miwa I., Kondo T., Hastings J. W. (1989). Circadian rhythm of photoaccumulation in Paramecium bursaria. J. Biol. Rhythms 4, 405–415. doi:10.1177/074873048900400402
Johnson C. H., Rust M. J. (2021). Circadian rhythms in bacteria and microbiomes. Cham, Switzerland: Springer, 415.
Johnson C. H., Xu Y. (2009). “The Decade of Discovery: How Synechococcus elongatus became a model circadian system 1990–2000,” in Bacterial circadian programs. Editors J. L. Ditty, S. R. Mackey, and C. H. Johnson (Berlin, Heidelberg: Springer), 63–86.
Johnson C. H., Zhao C., Xu Y., Mori T. (2017). Timing the day: What makes bacterial clocks tick? Nat. Rev. Microbiol. 15, 232–242. doi:10.1038/nrmicro.2016.196
Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136. doi:10.1093/dnares/3.3.109
Kawamoto N., Ito H., Tokuda I. T., Iwasaki H. (2020). Damped circadian oscillation in the absence of KaiA in Synechococcus. Nat. Commun. 11, 2242. doi:10.1038/s41467-020-16087-x
Kondo T., Strayer C. A., Kulkarni R. D., Taylor W., Ishiura M., Golden S. S., et al. (1993). Circadian rhythms in prokaryotes: Luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 90, 5672–5676. doi:10.1073/pnas.90.12.5672
Kondo T., Tsinoremas N. F., Golden S. S., Johnson C. H., Kutsuna S., Ishiura M. (1994). Circadian clock mutants of cyanobacteria. Science 266, 1233–1236. doi:10.1126/science.7973706
Larom S., Salama F., Schuster G., Adir N. (2010). Engineering of an alternative electron transfer path in photosystem II. Proc. Natl. Acad. Sci. U. S. A. 107, 9650–9655. doi:10.1073/pnas.1000187107
Liu Y., Tsinoremas N. F., Johnson C. H., Lebedeva N. V., Golden S. S., Ishiura M., et al. (1995). Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 9, 1469–1478. doi:10.1101/gad.9.12.1469
Ma P., Mori T., Zhao C., Thiel T., Johnson C. H. (2016). Evolution of KaiC-dependent timekeepers: A proto-circadian timing mechanism confers adaptive fitness in the purple bacterium Rhodopseudomonas palustris. PLoS Genet. 12, e1005922. doi:10.1371/journal.pgen.1005922
Nakamura Y., Kaneko T., Hirosawa M., Miyajima N., Tabata S. (1998). CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803. Nucleic Acids Res. 26, 63–67. doi:10.1093/nar/26.1.63
Nishimura H., Nakahira Y., Imai K., Tsuruhara A., Kondo H., Hayashi H., et al. (2002). Mutations in KaiA, a clock protein, extend the period of circadian rhythm in the cyanobacterium Synechococcus elongatus PCC 7942. Microbiology 148, 2903–2909. doi:10.1099/00221287-148-9-2903
Nixon P. J., Chisholm D. A., Diner B. A. (1992). “Isolation and functional analysis of random and site-directed mutants of photosystem II,” in Plant protein engineering. Editors P. R. Shewry, and S. Gutteridge (Cambridge: Cambridge University Press), 93–141.
Pakrasi H. B., Vermaas W. J. (1992). “Protein engineering of photosystem II,” in The photosystems: Structure, function and molecular biology. Editor J. Barber (Amsterdam: Elsevier), 231–257.
Pittendrigh C. S. (1960). Circadian rhythms and the circadian organization of living systems, 25. New York: Cold Spring Harbor Symposia on Quantitative Biology, 159–184.
Pope M. A., Hodge J. A., Nixon P. J. (2020). An improved natural transformation protocol for the cyanobacterium Synechocystis sp. PCC 6803. Front. Plant Sci. 11, 372. doi:10.3389/fpls.2020.00372
Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111, 1–61. doi:10.1099/00221287-111-1-1
Salis H. M., Mirsky E. A., Voigt C. A. (2009). Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27, 946–950. doi:10.1038/nbt.1568
Santos-Merino M., Singh A. K., Ducat D. C. (2019). New applications of synthetic biology tools for cyanobacterial metabolic engineering. Front. Bioeng. Biotechnol. 7, 33. doi:10.3389/fbioe.2019.00033
Schmelling N. M., Scheurer N., Köbler C., Wilde A., Axmann I. M. (2021). in Diversity of timing systems in cyanobacteria and BeyondCircadian rhythms in bacteria and microbiomes. Editors C. H. Johnson, and M. J. Rust (Cham, Switzerland: Springer), 179–202.
Shen G., Boussiba S., Vermaas W. (1993). Synechocystis sp PCC 6803 strains lacking photosystem I and phycobilisome function. Plant Cell 5, 1853–1863. doi:10.1105/tpc.5.12.1853
Sweeney B. M., Hastings J. W. (1960). Effects of temperature upon diurnal rhythms, 25. New York: Cold Spring Harbor Symposia on Quantitative Biology, 87–104.
Taton A., Erikson C., Yang Y., Rubin B. E., Rifkin S. A., Golden J. W., et al. (2020). The circadian clock and darkness control natural competence in cyanobacteria. Nat. Commun. 11, 1688. doi:10.1038/s41467-020-15384-9
Ungerer J., Tao L., Davis M., Ghirardi M., Maness P.-C., Yu J. (2012). Sustained photosynthetic conversion of CO2 to ethylene in recombinant cyanobacterium Synechocystis 6803. Energy Environ. Sci. 5, 8998–9006. doi:10.1039/C2EE22555G
Varman A. M., Yu Y., You L., Tang Y. J. (2013). Photoautotrophic production of D-lactic acid in an engineered cyanobacterium. Microb. Cell Factories 12, 117–118. doi:10.1186/1475-2859-12-117
Vijayan V., Zuzow R., O'shea E. K. (2009). Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 106, 22564–22568. doi:10.1073/pnas.0912673106
Wang B., Eckert C., Maness P.-C., Yu J. (2018a). A genetic toolbox for modulating the expression of heterologous genes in the cyanobacterium Synechocystis sp. PCC 6803. ACS Synth. Biol. 7, 276–286. doi:10.1021/acssynbio.7b00297
Wang B., Xiong W., Yu J., Maness P.-C., Meldrum D. R. (2018b). Unlocking the photobiological conversion of CO2 to (R)-3-hydroxybutyrate in cyanobacteria. Green Chem. 20, 3772–3782. doi:10.1039/C8GC01208C
Wang B., Xu Y., Wang X., Yuan J. S., Johnson C. H., Young J. D., et al. (2021a). A guanidine-degrading enzyme controls genomic stability of ethylene-producing cyanobacteria. Nat. Commun. 12, 5150. doi:10.1038/s41467-021-25369-x
Wang B., Young J. D., Xu Y. (2021b). “Reprogramming metabolic networks and manipulating circadian clocks for biotechnological applications,” in Circadian rhythms in bacteria and microbiomes. Editors C. H. Johnson, and M. J. Rust (Cham, Switzerland: Springer), 259–296.
Wang X., Liu W., Xin C., Zheng Y., Cheng Y., Sun S., et al. (2016a). Enhanced limonene production in cyanobacteria reveals photosynthesis limitations. Proc. Natl. Acad. Sci. U. S. A. 113, 14225–14230. doi:10.1073/pnas.1613340113
Wang Y., Sun T., Gao X., Shi M., Wu L., Chen L., et al. (2016b). Biosynthesis of platform chemical 3-hydroxypropionic acid (3-HP) directly from CO2 in cyanobacterium Synechocystis sp. PCC 6803. Metab. Eng. 34, 60–70. doi:10.1016/j.ymben.2015.10.008
Woelfle M. A., Ouyang Y., Phanvijhitsiri K., Johnson C. H. (2004). The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr. Biol. 14, 1481–1486. doi:10.1016/j.cub.2004.08.023
Woelfle M. A., Xu Y., Qin X., Johnson C. H. (2007). Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 104, 18819–18824. doi:10.1073/pnas.0706069104
Wu G., Wu Q., Shen Z. (2001). Accumulation of poly-β-hydroxybutyrate in cyanobacterium Synechocystis sp. PCC6803. Bioresour. Technol. 76, 85–90. doi:10.1016/s0960-8524(00)00099-7
Xiong W., Morgan J. A., Ungerer J., Wang B., Maness P.-C., Yu J. (2015). The plasticity of cyanobacterial metabolism supports direct CO2 conversion to ethylene. Nat. Plants 1, 15053–15057. doi:10.1038/nplants.2015.53
Xu Y., Ma P., Shah P., Rokas A., Liu Y., Johnson C. H. (2013a). Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495, 116–120. doi:10.1038/nature11942
Xu Y., Mori T., Johnson C. H. (2000). Circadian clock-protein expression in cyanobacteria: Rhythms and phase setting. EMBO J. 19, 3349–3357. doi:10.1093/emboj/19.13.3349
Xu Y., Weyman P. D., Umetani M., Xiong J., Qin X., Xu Q., et al. (2013b). Circadian yin-yang regulation and its manipulation to globally reprogram gene expression. Curr. Biol. 23, 2365–2374. doi:10.1016/j.cub.2013.10.011
Xue Y., Zhang Y., Cheng D., Daddy S., He Q. (2014). Genetically engineering Synechocystis sp. Pasteur Culture Collection 6803 for the sustainable production of the plant secondary metabolite p-coumaric acid. Proc. Natl. Acad. Sci. U. S. A. 111, 9449–9454. doi:10.1073/pnas.1323725111
Keywords: circadian, cyanobacteria, synechocystis, synechococcus elongatus, biological clocks
Citation: Zhao C, Xu Y, Wang B and Johnson CH (2023) Synechocystis: A model system for expanding the study of cyanobacterial circadian rhythms. Front. Physiol. 13:1085959. doi: 10.3389/fphys.2022.1085959
Received: 31 October 2022; Accepted: 12 December 2022;
Published: 04 January 2023.
Edited by:
Julie S. Pendergast, University of Kentucky, United StatesReviewed by:
Vincent M. Cassone, University of Kentucky, United StatesHideo Iwasaki, Waseda University, Japan
Copyright © 2023 Zhao, Xu, Wang and Johnson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carl Hirschie Johnson, Y2FybC5oLmpvaG5zb25AdmFuZGVyYmlsdC5lZHU=
 Chi Zhao1
Chi Zhao1 Yao Xu
Yao Xu Bo Wang
Bo Wang Carl Hirschie Johnson
Carl Hirschie Johnson