
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 14 December 2022
Sec. Chronobiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.1062632
 Nirav Thakkar1,2
Nirav Thakkar1,2 Astrid Giesecke3
Astrid Giesecke3 Olga Bazalova1
Olga Bazalova1 Jan Martinek1
Jan Martinek1 Vlastimil Smykal1
Vlastimil Smykal1 Ralf Stanewsky3*
Ralf Stanewsky3* David Dolezel1,2*
David Dolezel1,2*Circadian clocks are timing devices that rhythmically adjust organism’s behavior, physiology, and metabolism to the 24-h day-night cycle. Eukaryotic circadian clocks rely on several interlocked transcription-translation feedback loops, where protein stability is the key part of the delay between transcription and the appearance of the mature proteins within the feedback loops. In bilaterian animals, including mammals and insects, the circadian clock depends on a homologous set of proteins. Despite mostly conserved clock components among the fruit fly Drosophila and mammals, several lineage-specific differences exist. Here we have systematically explored the evolution and sequence variability of insect DBT proteins and their vertebrate homologs casein kinase 1 delta (CKIδ) and epsilon (CKIε), dated the origin and separation of CKIδ from CKIε, and identified at least three additional independent duplications of the CKIδ/ε gene in Petromyzon, Danio, and Xenopus. We determined conserved regions in DBT specific to Diptera, and functionally tested a subset of those in D. melanogaster. Replacement of Lysine K224 with acidic residues strongly impacts the free-running period even in heterozygous flies, whereas homozygous mutants are not viable. K224D mutants have a temperature compensation defect with longer free-running periods at higher temperatures, which is exactly the opposite trend of what was reported for corresponding mammalian mutants. All DBTs of dipteran insects contain the NKRQK motif at positions 220–224. The occurrence of this motif perfectly correlates with the presence of BRIDE OF DOUBLETIME, BDBT, in Diptera. BDBT is a non-canonical FK506-binding protein that physically interacts with Drosophila DBT. The phylogeny of FK506-binding proteins suggests that BDBT is either absent or highly modified in non-dipteran insects. In addition to in silico analysis of DBT/CKIδ/ε evolution and diversity, we have identified four novel casein kinase 1 genes specific to the Drosophila genus.
To cope with and anticipate daily environmental changes, organisms have evolved circadian clocks. These genetically determined time-measuring devices “tick” with a free-running period (τ) close to 24 h (Dunlap, 1999). The circadian clock runs with almost the same τ within the physiological temperature range; this phenomenon, known as temperature compensation, seemingly contradicts the basic principles of biochemical reactions (Arrhenius, 1889). At the molecular level, circadian clocks in eukaryotes comprise interlocked negative transcription-translation feedback loops (TTFL; Dunlap, 1999). The positive regulators are transcription factors driving the expression of mRNAs encoding the negative regulators. Once the negative regulators are present in the nucleus, they inhibit their own expression by suppressing the activity of the positive regulator(s). Importantly, the mere transcription-translation process would be much faster than the required 24-h cycle, thus, additional steps delaying the entire process must be involved. Firstly, the negative regulator proteins are initially destabilized, which delays their accumulation. In addition, the translocation of the negative regulators to the cell nucleus might require dimerization with a partner protein, and often larger complexes are formed (Aryal et al., 2017). At a biochemical level, both positive and negative regulators undergo various posttranslational modifications, of which protein phosphorylation is the most prominent. In the end, well-timed depletion of the negative regulators is key for the start of the next cycle and contributes to the resulting τ.
The circadian clock of mammals and the fruit fly Drosophila melanogaster relies on homologous components. The positive regulators CLOCK and BMAL/CYCLE belong to the basic helix-loop-helix (bHLH) PER-ARNT-SIM (PAS) transcription factors (King et al., 1997; Darlington et al., 1998; Hogenesch et al., 1998; Rutila et al., 1998). PERIOD (PER), which also belongs to the PAS protein family, is a negative regulator shared among vertebrates and insects (Hardin et al., 1990; Zylka et al., 1998).
In D. melanogaster, PER interacts with Drosophila-type TIMELESS protein (dTIM) (despite the general conservation of the molecular mechanisms and genetic components among various vertebrates and insects, some important differences exist. Furthermore, the gene/PROTEIN names vary in the literature as they were historically evolving. Here, we use the prefix m-for the so-called mammalian type and the prefix d-for the Drosophila-type proteins. See the supplementary text for more detailed notes on circadian clock gene terminology).
PER:dTIM dimerization in the cytoplasm is necessary for subsequent nuclear localization of PER and dTIM (Saez and Young, 1996; Meyer et al., 2006). dTIM is an essential component of the fruit fly circadian clock, because tim null mutations result in complete arrhythmicity (Sehgal et al., 1994), whereas missense mutations affect τ (Rothenfluh et al., 2000; Wulbeck et al., 2005), and certain d-tim mutations affect the temperature compensation of the circadian clock (Matsumoto et al., 1999; Singh et al., 2019). Furthermore, dTIM is a key component of the light-mediated synchronization in Drosophila (Hunter-Ensor et al., 1996; Myers et al., 1996; Zeng et al., 1996), which involves light-dependent interaction with Drosophila-type CRYPTOCHROME (dCRY) (Ceriani et al., 1999; Peschel et al., 2009). dCRY serves as a deep brain circadian photoreceptor (Emery et al., 2000) with no impact on the behavioral rhythmicity in constant-dark conditions (DD) at ambient temperature (Stanewsky et al., 1998), although d-cry depletion reduced rhythmicity at 18°C (Dolezelova et al., 2007). Interestingly, d-cry mutations abolish transcriptional oscillations in peripheral clocks, which allowed the identification of this mutant in a luciferase reporter-based screen (Stanewsky et al., 1998). In mice, mammalian-type CRYPTOCHROME (mCRY) is present as two paralogous and closely related proteins, that are essential for (light-independent) clock function, while dimerizing with one of the three mammalian PER proteins (Zylka et al., 1998; Kume et al., 1999; Putker et al., 2021).
An important feature of the negative TTFL is the temporal regulation of subcellular localization of participating proteins. In addition to nuclear localization signals (NLS), some circadian clock proteins also contain nuclear export signals (NES) (Saez and Young, 1996; Vielhaber et al., 2001; Ashmore et al., 2003; Yildiz et al., 2005; Hara et al., 2011; Saez et al., 2011; Jang et al., 2015; Singh et al., 2019; Giesecke et al., 2021). Thus, the resulting nuclear import/export strongly affects the suppression potential of the negative feedback loop and thereby τ. The stability and subcellular localization of the negative complex, such as PER and dTIM, is regulated by posttranslational modifications (Li et al., 2019; Crosby and Partch, 2020), including phosphorylation and dephosphorylation by several kinases and phosphatases (Sathyanarayanan et al., 2004; Leloup 2009; Reischl and Kramer, 2011; Agrawal and Hardin, 2016; Narasimamurthy and Virshup., 2021). One of the most explored circadian clock kinases is DBT which was first identified as a clock component in a Drosophila screen when the short- (DBTS) and long- (DBTL) free-running period mutants were identified (Price et al., 1998; Kloss et al., 1998, Figure 8). However, as it turned out later, dbt is also known as discs overgrown, a gene which had been discovered for its role during development (Jursnich et al., 1990; Zilian et al., 1999). Mammalian homologs of DBT are CKIδ/ε, which were shown to be essential for the clock in the hamster, human, and mice (Lowrey et al., 2000; Xu et al., 2005; Meng et al., 2008). The interaction between this kinase and PER is remarkably stable (Kloss et al., 2001; Lee et al., 2001; Aryal et al., 2017). Overexpression of either DBTL or DBTS variants in Drosophila resulted in the same τ as was produced by the corresponding alleles of the endogenous gene (Muskus et al., 2007), whereas in vitro studies using non-physiological substrates implied, surprisingly, that both mutants have reduced kinase activity (Kivimaë et al., 2008; Venkatesan et al., 2019). Furthermore, unlike most enzymes, CKIδ/ε activity is temperature insensitive (Isojima et al., 2009), but paradoxically, the hamster CK1εtau mutant is a temperature compensation mutant (Tosini and Menaker, 1998). The conundrum started to unravel in the context of the PER phosphorylation pattern elicited upon the action of multi-kinase hierarchical activities identified in several model organisms (Xu et al., 2007; Ko et al., 2010; Chiu et al., 2011; Lam et al., 2018). The current phosphoswitch model involves two competing phosphorylation sites on mouse (Mus musculus) PER2, the phosphodegron and the FASP (familial advanced sleep phase, Toh et al., 2001) sites, which regulate PER2 stability in opposing ways (Zhou et al., 2015; Masuda et al., 2020). Thus, the temperature-sensitive phosphoswitch slows down PER2 degradation at higher temperatures, resulting in a global temperature-compensated system. Somewhat similar, multiple phospho-clusters are detected on Drosophila PER, which cumulatively contribute to PER stability and transcriptional repressor activity (Chiu et al., 2008; Kivimaë et al., 2008; Garbe et al., 2013; Top et al., 2018). Therefore, the phosphoswitch mechanism might be conserved across species, even though the details differ, as a phosphodegron with functionally heterogeneous sites was recently reported for Drosophila (Joshi et al., 2022).
Both in mammals and Drosophila, the PER phosphorylation pattern is defined by the synergistic action of multiple kinases (see the text above), phosphatases (Sathyanarayanan et al., 2004; Fang et al., 2007; Reischl and Kramer, 2011), and some additional post-translational modifications, such as O-GlcNAcylation and acetylation (Kaasik et al., 2013; Li et al., 2019). The PER phosphorylation dynamics is regulated by yet another level of complexity, as is indicated by the distinct capacity of CKIδ splice isoforms. CKIδ1 and CKIε (both similar in the last 16 amino acids of their carboxy-terminal tails, here abbreviated as “C-terminal tails”) are more active in priming kinase activity at the FASP site, whereas CKIδ2 is more potent in priming the degron site (Fustin et al., 2018; Narasimamurthy et al., 2018). The CKIδ/ε C-terminal tail autophosphorylation inhibits its kinase activity (Graves and Roach, 1995; for review see Narasimamurthy and Virshup, 2021). As was shown for Drosophila DBT, the C-terminal tail stabilizes interactions between the kinase and the substrate, while the C-terminal tail autophosphorylation inhibits substrate binding (Dahlberg et al., 2009; Fan et al., 2015). Furthermore, two residues on the DBT kinase domain influence its affinity to PER (Dahlberg et al., 2009). However, no splicing isoforms of DBT exist in Drosophila as dbt is an intronless gene in this species.
The temperature-independent activity of CK1δ/ε was connected to sequence motifs close to the active site of the kinase, where Lysine 224 was identified as key for the temperature-compensated primed phosphorylation (Shinohara et al., 2017). Importantly, the K224D mutation in CK1δ shortens τ and affects temperature compensation in the mammalian system in vitro. Notably, the corresponding region of Drosophila DBT was systematically explored by Venkatesan et al. (2019) who identified a second NLS in positions 220–224. This region in DBT is further important for its interaction with BRIDE of DOUBLETIME (BDBT) (Venkatesan et al., 2015), a non-canonical FK506-binding protein with tetratricopeptide repeats that might promote the assembly of larger protein complexes (Fan et al., 2013).
Although the circadian clock is in general conserved among bilaterian species, some notable variations in the PER/dTIM/dCRY/mCRY feedback exist with some functional implications (Kotwica-Rolinska et al., 2022a). Therefore, we decided to explore and define the variability in insect DBT proteins. As a reference, we analyzed deuterostomian homologs of DBT and dated the origin and separation of CKIδ from CKIε. Furthermore, we have identified four novel casein kinase I genes specific to the Drosophila genus. We identified conserved regions in DBT specific to Diptera, functionally tested some of them in D. melanogaster, and analyzed their impact on temperature compensation of the circadian clock.
Recent progress in genome and transcriptome sequencing (Misof et al., 2014; Johnson et al., 2018; Kawahara et al., 2019; McKenna et al., 2019; Wipfler et al., 2019) allowed us to systematically explore casein kinases and FK506-binding proteins across all major insect orders. In essence, we applied an approach similar to that of Smykal et al. (2020), when multiple rounds of Basic Local Alignment Search Tool (BLAST) searches followed by fast phylogenetic analyses were conducted to retrieve evolutionary informative sequences from the genomes and transcriptomes of all major insect lineages. Although a reasonable collection of sequences could be retrieved from the protein database using BLASTP algorithm, more detailed and taxon-focused TBLASTN searches (search in translated nucleotide databases using a protein query) were used to explore transcriptome shotgun assemblies (TSAs). Multiple query sequences were tested in all searches described above (fruit fly Drosophila melanogaster DBT, firebrat Thermobia domestica DBT, and house mouse Mus musculus CKIε/CKIδ). For well-annotated genomes (zebrafish Danio rerio, African clawed frog Xenopus laevis, M. musculus, human Homo sapiens, etc.), all protein variants were retrieved directly from gene models. To retrieve non-DBT/CKIε/CKIδ kinases, multiple rounds of BLASTP and TBLASTN were performed. To test whether Drosophila-specific CKI genes (CG9962, CG2577, CKIalpha-like I, and CKIalpha-like II) could be identified outside of Drosophila, TBLASTN was performed in TSA of all insects with the exclusion of the Drosophila genus (NCBI:txid7215). In addition, reciprocal BLAST searches were performed when the identified sequence served as a query in the next rounds of BLASTs. Additional dbt sequences were obtained by PCR and 3′RACE from the housefly Musca domestica (Bazalova and Dolezel, 2017) and Chymomyza costata (Kobelkova et al., 2010), with support from Illumina-based transcriptome (Poupardin et al., 2015). See Supplementary Tables S1, S2 for accession numbers.
To reconstruct the evolution of BDBT, all FK506-binding protein homologs were retrieved from D. melanogaster, the monarch butterfly Danaus plexippus, the red flour beetle Tribolium castaneum, the brown marmorated stink bug Halyomorpha halys, and M. musculus. Then, multiple rounds of order- and species-specific searches in insects were employed and fast phylogenetic analyses performed. First, proteins were aligned using the algorithm MAFFT E-INS-i in Geneious 11 (Biomatters). Then, a FAST tree algorithm in Geneious 11 (Biomatters) was used to infer preliminary trees and identify duplicates. For detailed analyses, protein sequences were aligned using MAFFT algorithm with the E-INS-i multiple alignment method and the BLOSUM80 scoring matrix, and the trees were inferred using RAxML maximum likelihood GAMMA-based model and the bootstrap values calculated from 100 replicates (both as a package of Geneious 11 software, Biomatters). The datasets consisted of 239 sequences used for CKI evolution in Figure 2, whereas 31 sequences were used for vertebrate-specific duplication analyses (Figure 4), and 280 sequences were used for BDBT/FK506-binding proteins (Figure 5).
Details of Oxford Nanopore Technology (ONT) transcriptome sequencing will be described elsewhere. Briefly, P. apterus brains and other tissues were dissected and poly A+ mRNA was isolated using Dynabeads mRNA DIRECT Kit (Life Technologies) according to the manufacturer’s instructions and 100 ng of the polyA+ mRNA was then reverse-transcribed, turned to double-stranded DNA, and the sequencing adaptors were added using PCR-free Direct cDNA Sequencing kit (SQK-DCS109; Oxford Nanopore Technology) according to the manufacturer’s instructions. The library was immediately sequenced on a MinION device (Oxford Nanopore Technology). Base calling was performed after the run using Guppy 3.6.0 at a high-accuracy setting. Obtained tissue-specific transcriptomes were used in exhaustive searches using P. apterus dbt mRNA sequence as a query. All dbt transcripts were retrieved, manually inspected, and mapped to the in-house P. apterus genome (hybrid assembly of Illumina and ONT data, which will also be published separately), and a dbt gene model was built. All dbt transcripts were sequentially mapped to four defined individual dbt isoforms and only reads unequivocally distinguishing specific dbt isoform (protein)-coding sequences were counted.
The putative phosphorylation sites were predicted in silico using NetPhos 3.1 server at http://www.cbs.dtu.dk/services/NetPhos/ and scores higher than 0.5 were plotted in alignments.
The target site was designed to induce a double-strand strand brake in the C-terminal tail coding part of dbt gene. Two gRNA sequences (PAM, which is not part of the gRNA, is shown in square brackets) targeting GCGATGCTGGGCGGCAATGG[AGG] and GTCGGCCTTCGATACGGATG[CGG], respectively, were prepared from custom-synthesized oligonucleotides and cloned into pBFv-U6.2 (Kondo and Ueda, 2013) obtained from fly stocks of National Institute of Genetics, Japan (NIG-FLY). Plasmids were injected into y1 v1 P{nos-phiC31\int.NLS}X; attP40 (II) (NIG-FLY#: TBX-0002) flies with docking site on the second chromosome, transformants identified by eye color rescue, and balanced by y2 cho2 v1/Yhs-hid; Sp/CyO (NIG-FLY#: TBX-0008).
Flies expressing Cas9 specifically in germ cells (nos-Cas9) from the second chromosome insertion (NIG-FLY#: CAS-0001; y2 cho2 v1; attP40{nos-Cas9}/CyO) were crossed with U6gRNA-encoded transgenic strains (also located on the second chromosome). Resulting F1 offspring thus expressed both gRNA and CAS9 on second chromosomes, which potentially targeted the dbt gene located on the third chromosome and induce insertions and deletions as a result of the non-homologous-end-joining (NHEJ) mechanism. The resulting F1 offspring were crossed to y2 cho2 v1; Pr Dr/TM6C, Sb Tb (NIG-FLY#: TBX-0010) to balance the modified third chromosomes with TM6C. Individual F1 flies were used in heteroduplex mobility shift assay (Kotwica-Rolinska et al., 2019) to identify flies with the highest degree of mosaicism in the targeted dbt locus, thus, the crosses with the highest frequency of NHEJ-induced mutants were identified. From these selected crosses, F2 males and females with third chromosome balancer were individually crossed back to y2 cho2 v1; Pr Dr/TM6C, Sb Tb flies (NIG-FLY#: TBX-0010) to establish lines with identically modified third chromosomes. Mutated region was identified by polymerase chain reaction (PCR) and sequencing.
Target gRNA sites were selected so that Cas9-mediated cleavage was directed to a target locus of 100 bp upstream and downstream of the dbt K244 site. To avoid off-target cleavage optimal target sites were identified using CRISPR target finder (http://flycrispr.molbio.wisc.edu/tools). One gRNA target was chosen that was close to the target locus. Complementary target site oligos also contained a 5′ guanine for transcription from the U6 promoter and a 3 bp overhang compatible with BbsI sites. Oligos were annealed using standard primer annealing reactions and cloned into BbsI linearized pCFD3 plasmid (Port et al., 2014) via T4 DNA ligation.
Donor plasmids that contain the desired dbt mutations and all elements necessary for homologous recombination were constructed in 3 subsequent cloning steps. In each round of cloning the 1.5 kb 5′ homology arm and the 1.5 kb 3′ homology arm were individually PCR amplified from nos-Cas9 flies (Port et al., 2014) using outside primers dbtBMHRF and dbtBMHRR in combination with respective internal primers. Outside primers dbtBMHRF and dbtBMHRR contain a 15 bp overhang for In-Fusion cloning that is homologous to linearized vector ends. Inside primers have 5′ 15–20 bp extensions that are complementary to each other in addition to one defined mutation for each round of cloning. In the initial round of cloning a silent SalI site was introduced that can be used to screen for transformants. The two fragments (5′ homology arm and 3′ homology arm) were assembled into plasmid pBS-KS-attB1-2-PT-SA-SD-0-2xTY1-V5 (Addgene) that was linearized with XbaI and HindIII using In-Fusion cloning. In a second round of cloning the homology arms were amplified again using the pBS donor plasmid from the previous round as a template. Outside primers were as described above while the inside primers introduced either the K224D or the K224E mutation, respectively. In-Fusion cloning was used to assemble the fragments as described above. The resulting plasmid was then used in a final round of PCR to introduce PAM site mutations to avoid unwanted Cas9 cleavage within the donor plasmid. See Supplementary table S3 for a detailed list of all primers.
Donor plasmids containing the desired mutation along with gRNA plasmids were verified by sequence analysis and scaled up for injections using Qiagen plasmid midiprep. 6 µg of each plasmid were precipitated and eluted in injection buffer. gRNA construct and donor plasmids were mixed prior to injection and the mix was injected into freshly laid embryos of nos-Cas9 flies (Port et al., 2014). Surviving adults were backcrossed in batch crosses to y w; +; Dr/TM3 flies to balance 3rd chromosome modifications. Individual male and female flies from this cross were crossed again to y w; +; Dr/TM3. After letting the females lay eggs for 3–5 days, adult transformant flies were used for molecular screening.
In general, a total of 95 flies for each mutation were screened using PCR in combination with restriction digests. A ∼800 bp target locus was amplified by PCR using genomic DNA from individual flies. 20 units of SalI were then added to half of the PCR reaction and incubated for 2 h at 37°C. The resulting products were analyzed on agarose gels. The remaining PCR product of samples that showed digested products of the correct size were then used for sequencing to verify the presence of the desired mutations.
No homozygous flies could be obtained for both K224 dbt mutations and both stocks are balanced over TM3. Thus, for behavior experiments, flies harboring the dbt mutations were crossed against y w controls and only flies without TM3 were tested. For dbt C-terminal tail mutants, homozygous flies were tested.
Two to four-day old males were loaded into glass tubes containing 5% sucrose in 2% agar and loaded into the DAM2 TriKinetics system (Waltham, MA) and locomotor activity was recorded as previously described (Pfeiffenberger et al., 2010). K224 mutant flies were exposed to 12 h Light: 12 h Dark regime (LD) for 3 days, followed by 5–7 days in constant darkness (DD) to assess their free-running periods at constant temperatures of 18°C, 25°C, or 29°C. Period length and their significance (RS values) were determined using autocorrelation and Chi-square periodogram analysis functions of the fly toolbox implemented in MATLAB (MathWorks) (Levine et al., 2002). Period values with associated RS values ≥1.5 were considered rhythmic (Levine et al., 2002).
Two to four-day old C-terminal tail mutant males were loaded into the DAM2 TriKinetics system as described above, exposed to LD for 5 days, followed by 10 days in DD to assess their free-running periods at constant temperatures of 17°C, 20°C, 25°C, or 28°C. To determine τ during the first 10 days in DD, Lombe-Scargle periodogram analysis was performed using ActogramJ (Schmid et al., 2011) and double-plotted actograms were eye inspected in parallel.
Although mammalian CKIε, CKIδ, and Drosophila DBT are conserved components of the circadian clock, the mouse CKIε sequence did not rescue either the lethality or the rhythmicity of dbt-deficient Drosophila (Sekine et al., 2008). Therefore, to identify the key differences, we compared the protein sequences of mouse CKIε and CKIδ, Drosophila melanogaster DBT, and DBT of the most basal insect, the firebrat Thermobia domestica (Figure 1). All four proteins consist of the conserved casein kinase domain with a substantial C-terminal tail, whereas the N-terminal extension is minimal. However, a detailed inspection revealed two major differences. Within the kinase domain, the region 210–244 of D. melanogaster DBT differs from all three sequences. Notably, D. melanogaster DBT contains Asparagine (N) instead of Threonine (T) in position 220, which contrasts with DBT homologs from Thermobia, mouse, (Figure 1A), and more distant kinases. A recent study indicates that autophosphorylation of T220 influences substrate specificity (Cullati et al., 2022), thus the N at 220 prevents this posttranslational regulatory modification. Remarkable sequence divergence is observed in the C-terminal tail. Although the tail contains positions conserved even among both mouse CKI sequences and T. domestica DBT, surprisingly, the tail sequence is quite different in Drosophila with major insertions and deletions compared to the mammalian and firebrat sequences. These data indicate that during insect evolution DBT acquired substantial changes present in recent D. melanogaster DBT.
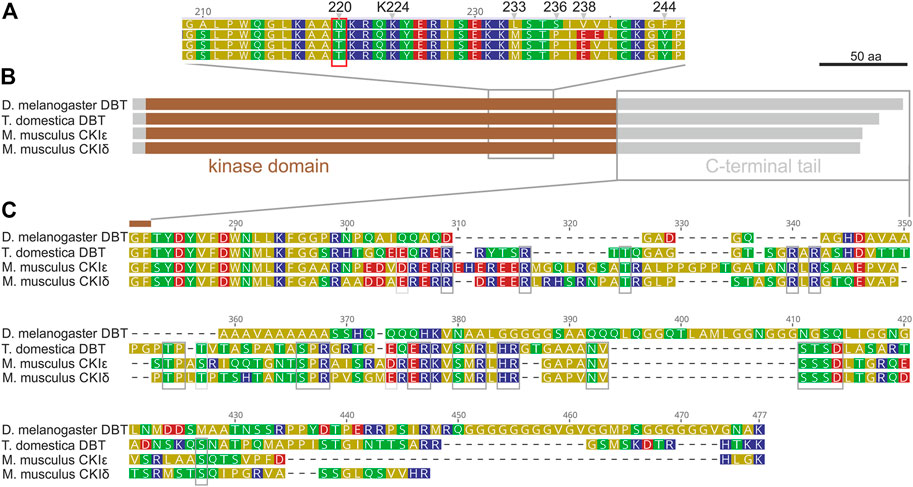
FIGURE 1. Drosophila melanogaster DBT differs from DBT of ancestral insect Thermobia domestica and both mouse (M. musculus) homologs, CKIε and CKIδ. (A) A detail of the kinase domain with highlighted conserved Lysine K224 and Drosophila-specific differences: N220, L233, S236, V238, and F244. (B) A schematic depiction of proteins with highlighted kinase domain, N- and C-terminal tails, and positions of detailed alignments shown in panels (A,C). (C) Detail of the C-terminal tail, where grey boxes indicate residues conserved among CKIε, CKIδ, and T. domestica DBT.
To be able to perform a comprehensive analysis of DBT/CKIε/CKIδ evolution, we first explored the phylogeny of casein kinases I in insects and used representative deuterostomian species as a reference. Tau-tubulin kinase served as an outgroup. CKI formed five distinct clusters (Figure 2A and Supplementary Figure S1): DBT/CKIε/CKIδ, CKIα, CKIγ, and two additional clusters not assigned to a specific CKI-type. These two clusters, provisionally labeled as CG9962 and CG2577, seem to be specific to the Drosophila genus, as no representative was found even in the dipteran genera Musca or Ceratitis. Similarly, two Drosophila-specific clusters are branching at the base of CKIα; thus, we use the provisional terms CKIα-like I and CKIα-like II (see Supplementary Table S1 for all non-DBT acc. numbers and Supplementary Table S2 for DBT, CKIε, and CKIδ acc. numbers). In line with the observed phylogenetic clustering of CKI, differences were identified in the N- and C-terminal tail lengths (Figure 2B) and in the sequence motifs within the kinase domain, including the activation loop (Philpott et al., 2020) and residues N275 and R279, which are responsible for enhanced substrate-specific binding to DBT (Dahlberg et al., 2009) (Figure 2C).
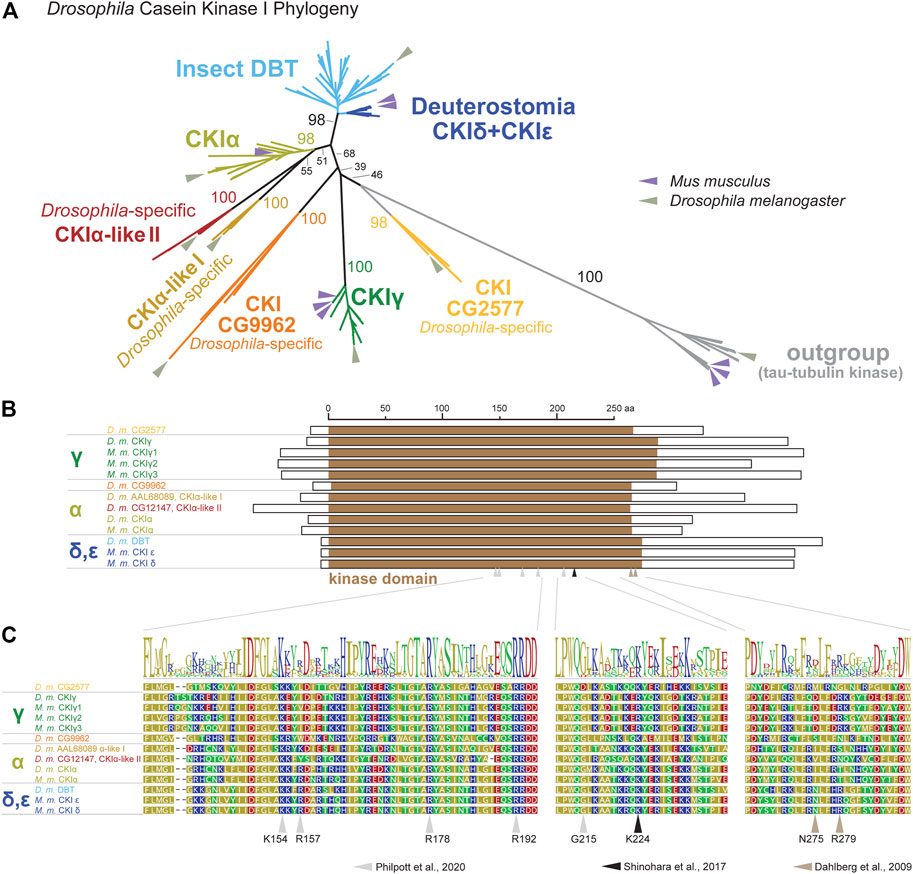
FIGURE 2. Phylogeny of bilaterian Casein kinase I (CKI) reveals seven clearly separated CKI-coding genes in fruit flies (Drosophila genus). (A) A tree illustrating relatedness among CKI proteins (values above branches indicate bootstrap support), in which well-separated clusters are color-coded. Insect DBTs (cerulean) branch together with deuterostomian sequences (cobalt blue) including CKIε and CKIδ from fish, amphibia, reptiles, and mammals. Besides well-established isoforms CKIα and CKIγ (the latter encoded by gilgamesh in Drosophila), four Drosophila genus-specific clusters were detected. Two of them branch at the base of CKIα and are therefore labeled as CKIα-like I (peanut) and CKIα-like II (cinnamon brown). Two additional clusters are separated from established CKI isoforms and are labeled according to the D. melanogaster nomenclature as CG9962 (orange) and CG2577 (apricot). Tau-tubulin kinases (a.k.a. asator in D. melanogaster) serve as an outgroup. Positions of the fruit fly D. melanogaster and the mouse Mus musculus proteins are highlighted by arrows in sage green and lavender, respectively. In established CKI terminology, the term isoform (α, γ, δ, ε) is used to refer to kinases encoded by distinct genes, although some of these genes might also encode different splice variants (for clarity, we use the term “splicing isoforms”). Multiple arrows in the mouse refer to gene multiplications, not to splicing isoforms. The phylogenetic analysis strongly supports the existence of the new groups of CKI genes in Drosophila and confirms already established groups. However, the relationship among CKI groups is sometimes poorly supported and the tree should not be interpreted as a focused analysis of CKI history. The tree was inferred using RAxML maximum likelihood of 239 protein sequences (final GAMMA-based score of the best tree -72993.763796) using Geneious 11 software (Biometters). Bootstrap support was calculated from 100 replications. See Supplementary Tables S1, S2 for accession numbers of analyzed sequences. (B) Schematic illustration of CKI proteins with highlighted kinase domain (brown), N- and C-terminal tails are shown as empty rectangles. (C) Details of protein alignment with highlighted residues that are important for the function of CKIδ (Shinohara et al., 2017; Philpott et al., 2020) and DBT (Dahlberg et al., 2009).
Having unambiguously identified CKI types, we performed a systematic audit of the DBT sequences across insects with three goals: explore possible patterns in the C-terminal tail variability, determine when the NKRQK motif arose, and identify whether we may correlate these changes in DBT with additional changes in the circadian clock setup.
In our comprehensive analysis, we identified and further analyzed DBT sequences from 55 species representing 20 insect orders and 9 deuterostomian classes (Figure 3, Supplementary Figures S2–S7, and Supplementary Table S2). Whereas only one dbt gene was found in all analyzed insects, up to as many as three dbt paralogs were identified in the zebrafish Danio and four paralogs in the clawed frog Xenopus. In mammals, reptiles, and birds, two paralogous genes, CKIε and CKIδ, are known. Similarly, in the sea lamprey Petromyzon marinus, two dbt-like genes were discovered; however, a detailed sequence comparison indicates that these dbt-like genes result from lamprey-specific gene duplication (Figure 4). The CKIε/CKIδ separation is observed in sharks, rays, and fishes, and thus seems to be a result of gene duplication specific to Gnathostomata. The second duplication of CKIδ led to two CKIδ genes present in the zebrafish Danio rerio and probably a similar but independent (Xenopus-specific) duplication happened in the ancestor of the clawed frog Xenopus laevis. In addition to gene duplications and quadruplications, a various number of protein isoforms can be produced in some organisms from individual genes as a result of alternative splicing (see the C-terminal tail analysis below).
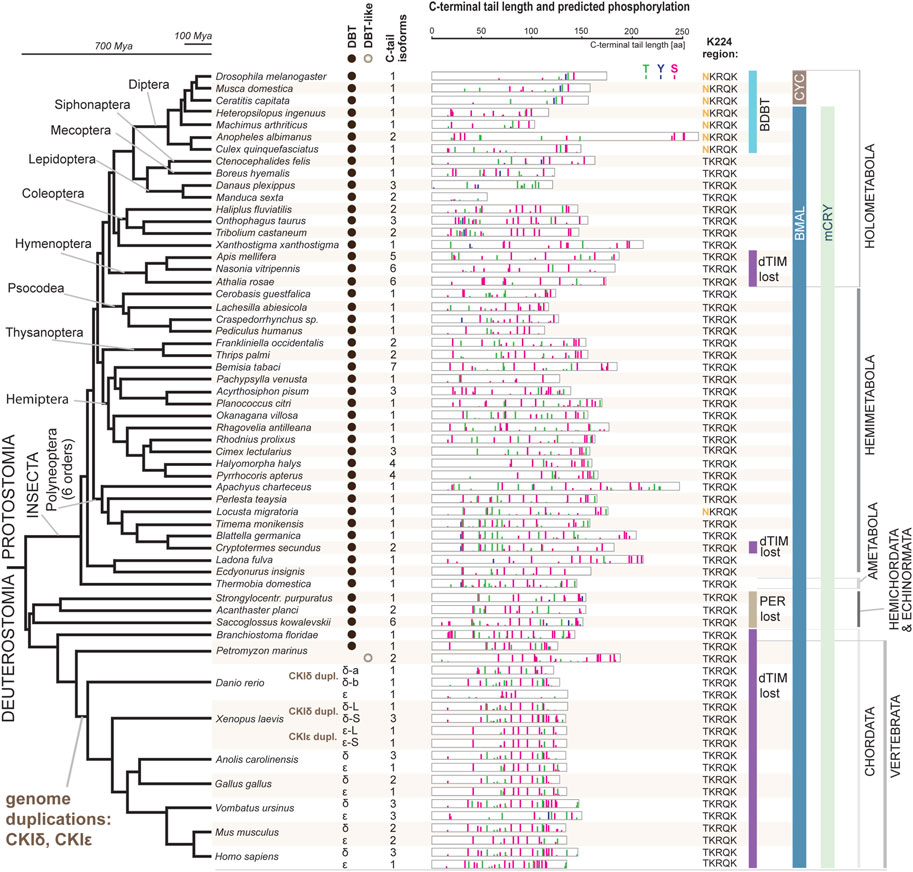
FIGURE 3. DBT and CKIδ/ε genes and proteins mapped on insect and deuterostomian phylogeny. The phylogenetic tree corresponds to a consensus of recent phylogenomic studies (Misof et al., 2014; Johnson et al., 2018; Kawahara et al., 2019; McKenna et al., 2019; Wipfler et al., 2019). Representative species are shown at the terminal nodes. The first column indicates the presence of DBT-coding genes (note a Petromyzon-specific gene duplication). Greek letters refer to the presence of CKIδ and CKIε; note two CKIδ paralogs in Danio and Xenopus and two CKIε paralogs in Xenopus (for details on CKIδ and CKIε phylogeny, see Figure 4). The second column indicates how many splicing isoforms affecting the C-terminal tail protein sequence were identified. The C-terminal tail of the longest isoform is depicted for each gene in each species (see Supplementary Figures S2–S7 for all isoforms and additional species). The color bars indicate in silico predicted phosphorylation patterns of threonine (T, green), tyrosine (Y, blue), and serine residues (S, pink). The bar’s height refers to the predicted score between 0.5–1.0. K224 region indicates whether NKRQK or TKRQK motif was found in the region corresponding to the 220–224 position within the catalytic domain of D. melanogaster DBT. Major changes in the circadian clock setup are depicted: presence of CRY mammalian (mCRY) type, loss of TIMELESS-drosophila type (dTIM), loss of PERIOD (PER) (Kotwica-Rolinska et al., 2022b), and transition of BMAL (activation domain is present) to CYC (activation domain lost) (Supplementary Figure S8). DBT-interacting protein paralogs BRIDE of DBT (BDBT) was found only in Diptera (for details and BDBT phylogeny see Figure 5).
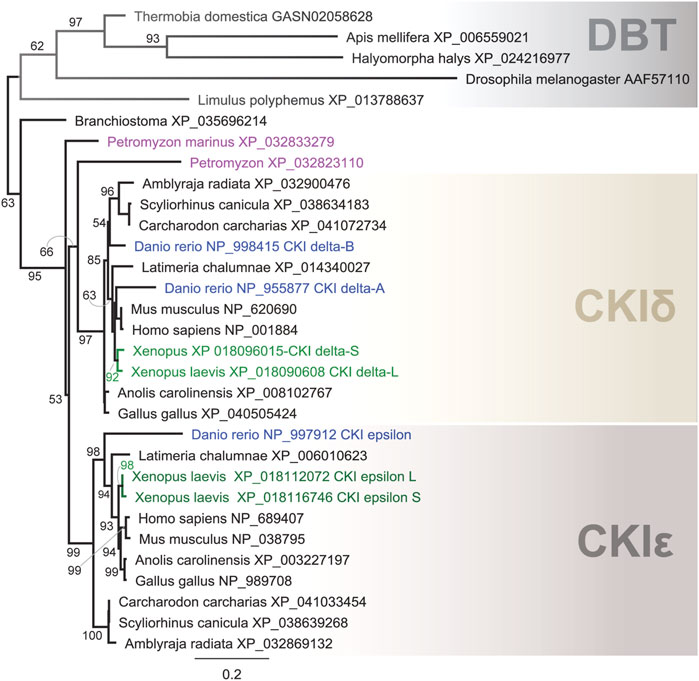
FIGURE 4. Phylogeny of DBT, CKIδ, and CKIε indicates several gene duplications in vertebrates. In the lancelet Branchiostoma, only one CKI gene was found to precede the δ- and ε-isoforms. The major duplication that gave rise to the δ and ε isoforms dates back to the ancestor of Gnatostomata (vertebrates with jaws), while two CKI genes in the sea lamprey Petromyzon marinus resulted from Petromyzon-specific gene duplication. CKIδ was duplicated in the zebrafish Danio rerio, resulting in the so-called CKIδ-A and CKIδ-B. In the African clawed frog Xenopus laevis, a large genome duplication resulted in two CKIδ (L and S) and CKIε (L and S) genes. DBT sequences from protostomian representatives (the firebrat Thermobia domestica, the honey bee Apis mellifera, the marmorated sting bud Halyomorpha halys, the fruit fly Drosophila melanogaster, and the horseshoe crab Limulus polyphemus) were used as outgroups. The tree was inferred using RAxML maximum likelihood of 31 protein sequences using Geneious 11 software (Biomatters). Bootstrap support was calculated using 100 replications.
The NKRQK motif (positions 220–224 in Drosophila DBT) within the catalytic domain distinguishing D. melanogaster DBT from the mouse homologs (Figure 1) was identified in all analyzed dipteran insects (Figure 3). Apart from the desert locust Locusta migratoria (the sequence was confirmed by Sanger sequencing and only one dbt gene was identified in the genome), all non-dipteran species contain the TKRQK motif. Therefore, we compared the presence of NKRQK to known changes in the circadian clock setup, such as the presence of mCRY, loss of dTIM (Kotwica-Rolinska et al., 2022a), and transformation of BMAL to CYC (Meireles-Filho et al., 2006). The transition of BMAL, a transcription factor with a transactivation domain, to CYC, a transcription factor which lacks the transactivation domain, was identified exclusively in Cyclorrhapha, a subset of Diptera (Supplementary Figure S8). However, this change perfectly corresponds to the loss of mCRY and does not agree with the presence of NKRQK (Figure 3).
Since none of the known changes in the clock setup correlated with the presence of the NKRQK motif in DBT, we analyzed the evolution of BDBT, a non-canonical FK506-binding protein interacting with DBT in Drosophila (Fan et al., 2013). First, we performed a phylogenetic analysis of available known FK506-binding proteins. The unrooted phylogenetic tree in Figure 5 (and the full tree version in Supplementary Figure S9) represents how various FK506-binding proteins evolved over time. Notably, dipteran BDBTs form a clear cluster that is separated from all remaining proteins. Furthermore, FK-506 binding proteins from Mecoptera (Scorpionflies) and Siphonaptera (Fleas), the closest relatives of Diptera, do not cluster with BDBT. Therefore, BDBT has been so heavily modified in Diptera that we cannot reliably identify the corresponding BDBT in any non-dipteran insect. Interestingly, in Diptera, the rise of the BDBT gene correlates with the transition from TKRQK to NKRQK motifs. The only other occurrence of the NKRQK motif was observed in Locusta (Orthoptera), whereas the sister polyneopteran lineages (termites, roaches, phasmids, Mantophasmatodea) contain TKRQK. However, FK-506 binding proteins of all polyneopteran lineages (including Orthoptera) branch together independently of TKRQK to NKRQK motifs.
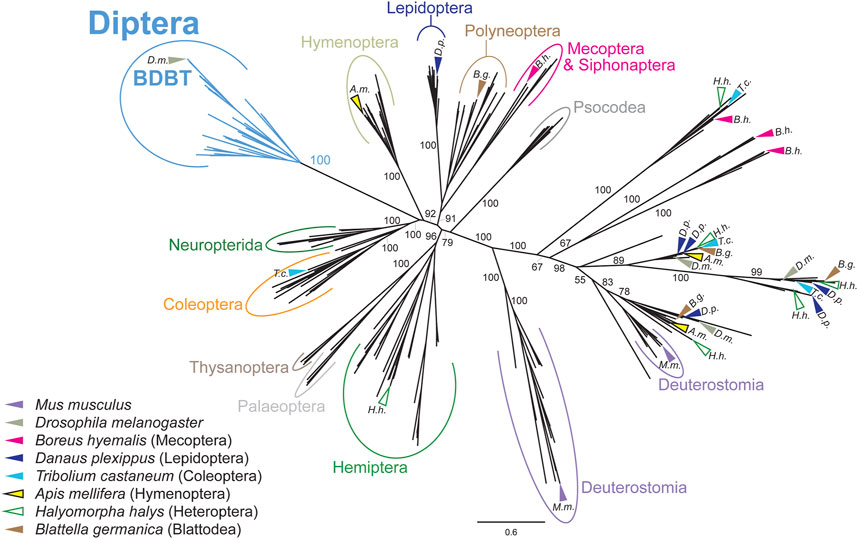
FIGURE 5. Phylogeny of FK506-binding proteins revealed a clear separation of dipteran BRIDE of DBT (BDBT) from all remaining clusters. Major insect orders are highlighted. Arrows indicate the position of sequences from representative species. FK506-binding proteins from Mecoptera and Siphonaptera do branch far away from Diptera. The tree was inferred using RAxML maximum likelihood of 280 protein sequences (final GAMMA-based score of the best tree −104683.7791) using Geneious 11 software (Biomatters). Bootstrap support was calculated using 100 replications.
The well-established impact of the C-terminal tail on the priming capacity of CKIδ splice isoforms in mice (Narasimamurthy and Virshup, 2021) prompted us to explore the C-terminal tail variability in the identified DBT/CKI dataset. The in silico predicted phosphorylation pattern was depicted for probabilities >0.5 (Figure 3 and Supplementary Figures S2–S7). When compared to the kinase domain, the C-terminal tails are the most variable parts of the proteins. Somewhat similar phosphorylation patterns and comparable lengths are found among C-terminal tails in vertebrates. In insects, however, the length and sequence of the C-terminal tails are remarkably variable. The shortest tails were identified in Lepidoptera (in several species only around 50 amino acids), whereas the longest tail in the Anopheles mosquito exceeded 250 amino acids (Figure 3). Putative phosphorylation was more prevalent in the C part of the C-terminal tail; however, the predicted phosphorylation patterns were quite variable in insects.
Alternative splicing of mouse CKIδ transcripts affects the biochemical properties of resulting CKIδ protein isoforms. In all analyzed vertebrate species, CKIδ was alternatively spliced with impact on the predicted phosphorylation pattern in the terminal part of the C-terminal tail. In contrast, CKIε was alternatively spliced only in a few vertebrate species (Supplementary Figure S7). Alternative splicing was detected in dbt of many insects, including the mosquito Anopheles albimanus (Diptera), all analyzed beetles (Coleoptera), all analyzed butterflies/moths (Lepidoptera), all analyzed hymenopteran species (the honey bee Apis mellifera, the jewel wasp Nasonia vitripennis, and the turnip sawfly Athalia rosae), the pea aphid (Acyrthosiphon pisum), both analyzed species of Thysanoptera (the western flower thrips Frankliniella occidentalis and the melon thrips Thrips palmi), the silverleaf whitefly Bemisia tabaci, true bugs (Heteroptera: the water strider Rhagovelia antilleana, the kissing bug Rhodnius prolixus, the common bed bug Cimex lectularius, the brown marmorated stink bug Halyomorpha halys, and the linden bug Pyrrhocoris apterus), and the drywood termite Cryptotermes secundus (Supplementary Figures S3–S5). As a representative of the true bugs, having access to the linden bug P. apterus brain transcriptome obtained by Oxford Nanopore Technology, we analyzed the presence and abundance of all four identified dbt isoforms (Figure 6). The three most abundant isoforms comprising 99% of dbt transcripts encode proteins with predicted altered phosphorylation patterns. Thus, alternative splicing of dbt might serve as a regulatory step influencing and modulating the properties of DBT in some insects. In flies including Drosophila, however, dbt is an intronless gene. A similar intronless gene organization might exist in other insect species. However, the identification of only one transcript variant should be interpreted with caution. The reliable decision whether dbt gene contains introns requires a good non-fragmented genome assembly, ideally with well-annotated gene models. On the other hand, the identification of only one dbt isoform in the transcriptome of a particular species may reflect only shallow sequencing or might be affected by transcript assembly and post-sequencing processing.
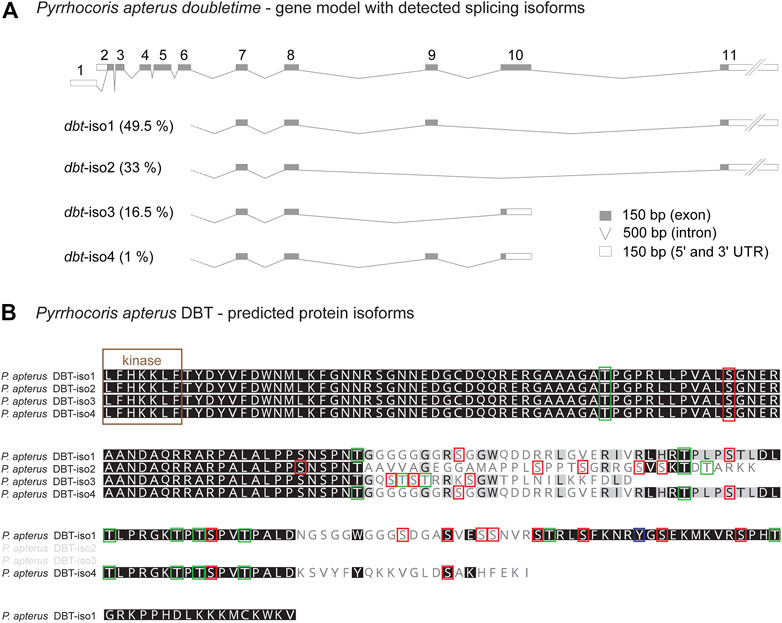
FIGURE 6. Alternative splicing of doubletime (dbt) gene in the linden bug Pyrrhocoris apterus. (A) A gene model illustrating the length of all dbt exons and introns. Two alternative transcription starts and splicing at the 5’end (exon 1 or 2, respectively) do not influence the predicted protein coding sequence. Alternative skipping of exons 9 and/or 10 results in four isoforms (dbt-iso1, 2, 3, 4). The expression level of each dbt isoform mRNA in the brain was detected from the Oxford nanopore transcriptome and depicted as % contribution to all dbt molecules (100%—all isoforms). (B) Protein alignment of the C-terminal tail illustrating the isoform-specific and variable regions (the more conserved residue the darker the color). Color rectangles refer to the theoretical prediction of phosphorylation pattern (>0.5; NetPhos-3.1) for serine (S), threonine (T), and tyrosine (Y).
The absence of introns in dbt genes of flies and the remarkable diversity of the C-terminal tail among flies and mosquitoes motivated a detailed comparison of this part of DBT in Diptera. Protein alignment of 15 species representing major dipteran lineages revealed a conserved region in the C-terminal tail, where a short 12 amino acid motif is identified in all dipteran species, and an even longer motif is shared among Cyclorrhapha (Figure 7). This conserved motif contains residues with high scores predicting their phosphorylation.
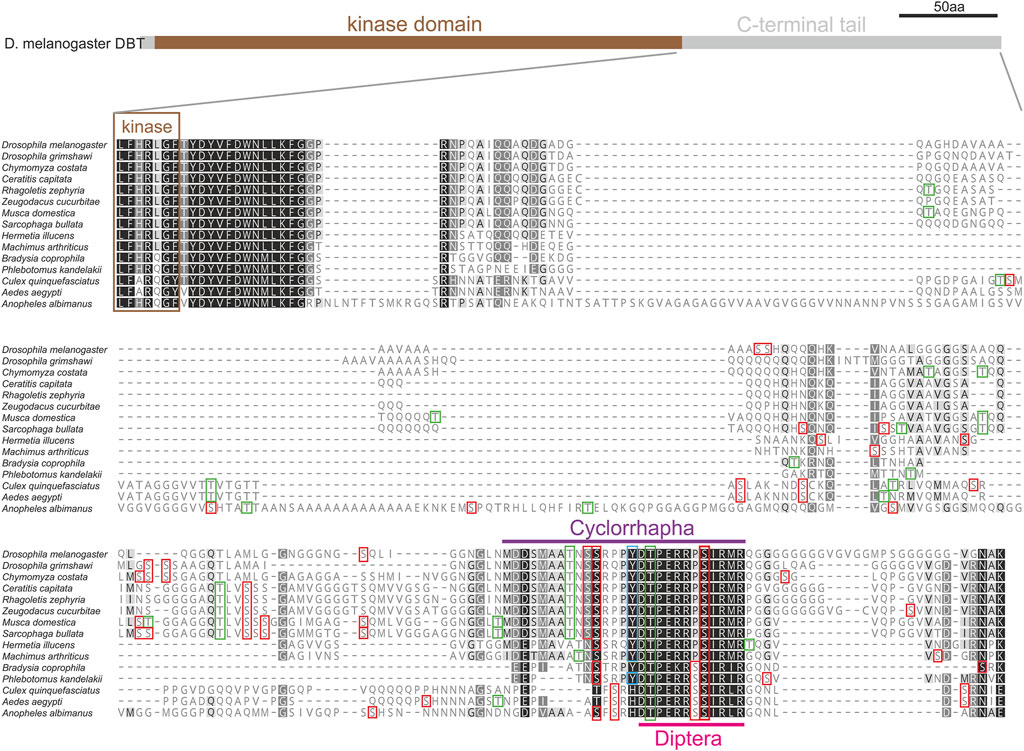
FIGURE 7. C-terminal tail of DBT in 15 representative species of Diptera. Protein alignment points to variable and conserved regions in the protein sequence (the more conserved the darker the color; a hyphen corresponds to a gap in the alignment). Color rectangles refer to the theoretical prediction of phosphorylation pattern (>0.5, NetPhos-3.1) for serin (S), threonine (T), and tyrosine (Y). The C-terminal tail contains a motif conserved in Diptera (light purple), which can be further extended in Cyclorhapha (Deep Purple).
To test whether the identified sequence motifs in DBT have an impact on the function of the circadian clock in vivo, we applied reverse genetic tools to the fruit fly D. melanogaster and focused on two regions: 1) residue K224, which was established as important for temperature compensation in mammalian CKIδ (Shinohara et al., 2017), and which is also part of NKRQK motif (Figure 3), and 2) the C-terminal tail (Figure 7).
Three different C-terminal tail mutants were created, encompassing or bordering the conserved C-terminal tail domain (Figures 8A, B). All of them are homozygous viable and displayed only very mild, yet significant circadian phenotypes. The deletion of amino acids downstream of position 370, that is the part comprising conserved cyclorrhaphan and dipteran motifs, slightly extended τ when compared to controls (Figures 8C, D and Supplementary Table S4). Deletions of the very end of the C-terminal tail and frameshift (∆411–440) and a 15-bp in-frame deletion upstream of conserved motifs (∆366–370) mildly shortened τ (Figures 8E, F and Supplementary Table S4).
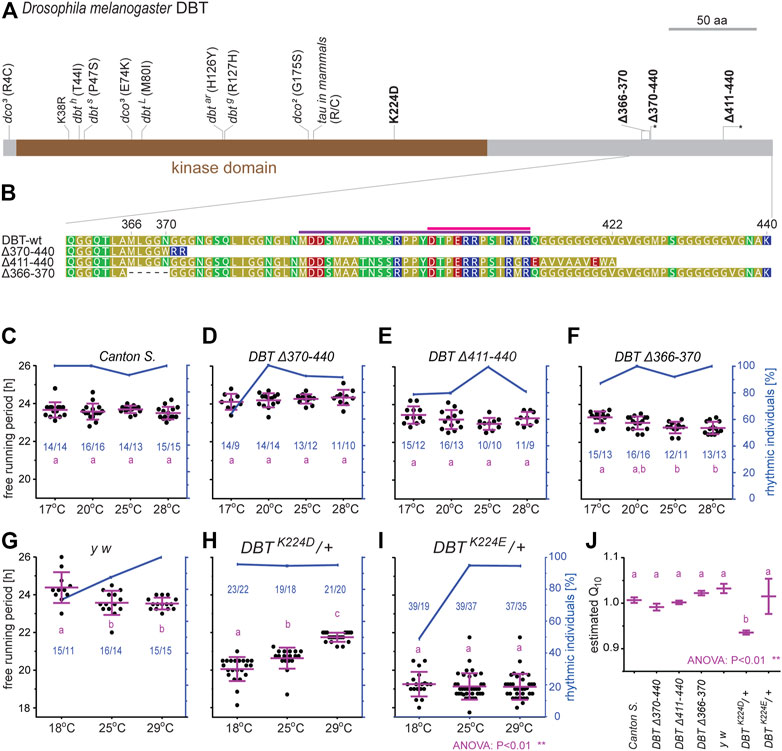
FIGURE 8. Functional analysis of new dbt mutants created in D. melanogaster. (A) A schematic depiction of DBT protein with highlighted mutations; here-created mutants are in bold, the asterisk in the scheme refers to a premature protein termination due to a frameshift. (B) Detail of the C-terminal tail in the wild type (wt) and three mutant lines. The dipteran and cyclorrhaphan motifs are highlighted by light and deep purple, respectively. (C–I) Circadian clock phenotype [each dot in panels (C–I) represents the free-running period tau, τ, of individual male flies] was recorded for 10 days in DD at the specified temperature. Blue dots connected with a blue line indicate the percentage of rhythmic individuals at a particular temperature. A scale indicating the percentage rhythmicity is on the right y-axis, the blue numbers in the chart represented ntotal/nrhythmic. (J) The temperature compensation depicted as Q10 was calculated from data presented in (C–I). Magenta bars represent means ± SEM.
The basic lysine residue K224 was replaced by the acidic residues aspartic acid (D) or glutamic acid (E), respectively. In both cases, homozygous mutants were not viable, thus, heterozygous flies were analyzed. In both mutants, τ was significantly shorter compared to control flies. Moreover, these two mutants differ in their temperature compensation (Figures 8G–J). The K224D heterozygotes displayed a τ of ∼20 h at 18°C and lengthening of τ up to 22 h at higher temperatures (Q10 = 0.94) (Figures 8H, J). The K224E heterozygotes displayed an approximately 5-h faster clock compared to the wild type (τ ∼ 19) at all three tested temperatures and showed reduced rhythmicity at 18°C (Figures 8I, J). The shorter τ corresponded to earlier evening activity peaks in LD regimes in both mutant lines, K224D (Supplementary Figure S11) and K224E (Supplementary Figure S12), when compared to control y w flies (Supplementary Figure S10).
Recent remarkable progress in the genome and transcriptome sequencing allowed us to retrieve dbt/CKI genes from all major insect orders and from representative vertebrate lineages, and perform their sequence comparison. The available data point to several mutually independent CKI duplications observed in the deuterostomian lineage. Our analysis suggests that two dbt-homologous genes identified in P. marinus are the result of a Petromyzon-/lamprey-specific duplication, whereas duplication leading to the rise of epsilon and delta isoforms of CKI is dated to the common ancestor of Gnatohostomata. The complexity of the circadian clock setup in Danio and Xenopus has been shaped by independent genome duplications. In Danio, this gene duplication reflects complexity identified in many teleost species (Garcia-Concejo and Larhammar, 2021), however, in Xenopus, we see the outcome of a very recent species-limited genome duplication (Uno et al., 2013).
In addition to gene duplications, further diversity of CKIδ/ε is achieved via alternative splicing. Notably, CKIδ is alternatively spliced in all tetrapods (amphibia, reptiles, birds, and mammals), where the splicing influences the C-terminal tail of the protein including its putative phosphorylation pattern. In Danio, only one splice variant was identified for each CKIδ paralogue, thus, the protein diversity of CKIδ might be achieved by independent genes in this species.
The diversity of DBT sequences in insects is remarkable, and we suggest that the role of alternative splicing will most likely differ among various insect lineages. As we show in the linden bug P. apterus, not only are multiple splicing isoforms encoded by the dbt gene but three of them are expressed in the brain. Since dbt silencing results in a remarkable extension of τ in P. apterus (Kotwica-Rolinska et al., 2022a), functional tests of each splicing isoform would be an interesting research direction. Unfortunately, the isoform-specific exon 9 is only 159 nt long. Therefore, we are reaching technical limitations of ds RNA mediated interference, even though a 288 bp long dsRNA was successfully used to knock down isoform-specific transcripts (Bajgar et al., 2013). An alternative approach could utilize gene editing, a method established and used for circadian research in P. apterus (Kotwica-Rolinska et al., 2019; Kotwica-Rolinska et al., 2022b).
In some insects, however, no alternative splicing was detected and in certain lineages, such as flies, the dbt gene is intronless. Interestingly, our comprehensive analysis revealed three new Drosophila genus-specific CKI genes. To our knowledge, the role and function of these casein kinases are not established. Given the participation of both DBT and CKIα in the fruit fly circadian clock (Lam et al., 2018), one cannot rule out the involvement of here-identified CKI genes in the circadian clock. Although this is entirely speculative, the combination of multiple kinases encoded by independent genes would provide an alternative source to the isoform repertoire produced by alternative splicing from an individual gene in some other species.
Functional analysis of the C-terminal tail in D. melanogaster revealed negligible effects on rhythmicity or changes in τ. However, only three simple deletion mutants were created here, thus, a full evaluation of the role for the C-terminal tail in DBT is rather premature. The second set of mutants, K224 modifications, resulted in homozygous lethality, similar to known strong dbt/dco alleles disrupting the developmental function of CK1 during the pupal stage (Price et al., 1998; Zilian et al., 1999). The equivalent mammalian mutations are not homozygous lethal, presumably because CK1δ and CK1ε are able to complement each other. Heterozygous mutant K224D/+ and K224E/+ flies produced profound shortenings of τ, almost identical to homozygous CK1δ K224D mutant mice (Shinohara et al., 2017, Figures 8G–I). Acidic K224 substitutions presumably bypass the phospho relay embedded in the perS serine cluster (FASP serine cluster in mPER2), and immediately phosphorylate the respective serine in the PER phosphodegron. This is because in mammalian CK1, K224 (together with R178) forms an anion-binding site required for phosphorylation of primed substrates (Narasimamurthy and Virshup, 2021). In flies and mammals, phosphorylation of the perS/FASP serine cluster delays PER degradation by preventing premature phosphorylation at the phosphodegron site, which would lead to rapid PER turnover (phosphoswitch). However, phosphorylation of multiple serine residues within the perS/FASP cluster by DBT/CK1 requires a priming phosphorylation at a particular serine within each cluster. Binding of DBT/CK1 to this primed substrate requires the basic anion binding pocket formed by R178 and K224, which most likely can not form in acidic K224D and K224E mutants. Therefore, the short periods observed in K224D, K224E, and R178 (= hamster tau mutant: Ralph and Menaker, 1988; Lowrey et al., 2000) are presumably the consequence of impaired perS/FASP region phosphorylation and the resulting acceleration of phosphorylation at the phosphodegron site. Moreover, the K224D mutation affects temperature compensation, both in flies and mouse organ cultures. However, the temperature-dependent lengthening of τ associated with the K224D mutation in flies is opposite to what was detected for mPer2-luc expression in suprachiasmatic nucleus (SCN) slices of homozygous K224D mice, which showed a shortening of τ with increasing temperature (Shinohara et al., 2017). The opposite temperature compensation phenotypes of K224D in flies and mice, as well as the lack of temperature-dependent period lengthening in the very similar K224E mutants, suggest that K224D overcompensation in flies is not simply caused by potential thermal instability of the K224D protein. Moreover, temperature overcompensation is not generally linked to decreased protein stability at higher temperatures (Giesecke et al., 2021). The phenotypic differences between the fly K224D and K224E mutants are surprising (temperature overcompensation in K224D and reduced rhythmicity at 18°C in K224E, Figure 8). These differences demonstrate that the two replacements have different consequences (apart from both shortening the free-running period), even though both introduce an acidic residue. Although very similar in structure, glutamic acid is slightly larger compared to aspartic acid, which may influence substrate binding in a temperature-dependent manner. In addition, the presence of an Asparagine (N) at position 220 in fly DBT, as opposed to a Threonine (T) at this position in CKIδ, might contribute to the opposite temperature compensation phenotypes of K224D mutants in flies and mice. Replacing the fly Asparagine (N220) with a Threonine would be interesting, not only in the light of temperature compensation differences, but also in the context of recently published autophosphorylation of Threonine, which is the preferred amino acid in corresponding positions in the majority of homologous kinases (Cullati et al., 2022). Secondly, the possible interaction with BDBT in the fruitfly might be an interesting and experimentally testable explanation for the altered temperature compensation in K224D. On the other hand, the NKRQK motif arose independently in Locusta and other orthopteran species and, given the diversity of FK-506 binding proteins, can hardly be interpreted as BDBT-dependent modification. Moreover, given the broad range of targets phosphorylated by DBT and even its non-catalytic role in the circadian clock (Yu et al., 2009), the mechanistic model might be quite complex.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
DD and RS designed the study. NT, AG, OB, and JM performed Drosophila experiments. DD run all phylogenetic analyses with help of VS, who also established the P. apterus dbt gene model. DD wrote the manuscript with input from NT and RS.
The work, DD, VS, and NT were supported by the Czech Science Foundation (GACR) grant number 22-10088S. AG and RS were supported by the Deutsche Forschungsgemeinschaft grant STA 421/7-1. ONT sequencing experiments were supported from European Research Council (ERC) under the European Union’s Horizon 2020 Program Grant Agreement 726049. We acknowledge support from the Open Access Publishing Fund of University of Münster.
We thank Dr. Samarjeet Singh for advice on transgenesis and CRISPR/CAS9 mutagenesis, and Bulah C. Wu for technical assistance with ONT genome and transcriptome sequencing and assembly. For ONT processing we thank Computational resources which were supplied by the project “e-Infrastruktura CZ” (e-INFRA CZ LM2018140) supported by the Ministry of Education, Youth and Sports of the Czech Republic.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.1062632/full#supplementary-material
Agrawal P., Hardin P. E. (2016). An RNAi screen to identify protein phosphatases that function within the Drosophila circadian clock. G3 6, 4227–4238. doi:10.1534/g3.116.035345
Arrhenius S. (1889). Über die Reaktionsgeschwindigkeit bei der Inversion von Rohrzucker durch Säuren. Zeitschrit fuer Phys. Chem. 4, 226–248. doi:10.1515/zpch-1889-0416
Aryal R. P., Kwak P. B., Tamayo A. G., Gebert M., Chiu P. L., Walz T., et al. (2017). Macromolecular assemblies of the mammalian circadian clock. Mol. Cell 67, 770–782. doi:10.1016/j.molcel.2017.07.017
Ashmore L. J., Sathyanarayanan S., Silvestre D. W., Emerson M. M., Schotland P., Sehgal A. (2003). Novel insights into the regulation of the timeless protein. J. Neurosci. 23, 7810–7819. doi:10.1523/jneurosci.23-21-07810.2003
Bajgar A., Jindra M., Dolezel D. (2013). Autonomous regulation of the insect gut by circadian genes acting downstream of juvenile hormone signaling. Proc. Natl. Acad. Sci. U. S. A. 110, 4416–4421. doi:10.1073/pnas.1217060110
Bazalova O., Dolezel D. (2017). Daily activity of the housefly, Musca domestica, is influenced by temperature independent of 3’ UTR period gene splicing. G3 7, 2637–2649. doi:10.1534/g3.117.042374
Ceriani M. F., Darlington T. K., Staknis D., Mas P., Petti A. A., Weitz C. J., et al. (1999). Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285, 553–556. doi:10.1126/science.285.5427.553
Chiu J. C., Ko H. W., Edery I. (2011). NEMO/NLK Phosphorylates PERIOD to initiate a time delay phosphorylation circuit that sets circadian clock speed. Cell 145, 357–370. doi:10.1016/j.cell.2011.04.002
Chiu J. C., Vanselow J. T., Kramer A., Edery I. (2008). The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 22, 1758–1772. doi:10.1101/gad.1682708
Crosby P., Partch C. L. (2020). New insights into non-transcriptional regulation of mammalian core clock proteins. J. Cell Sci. 133, jcs241174. doi:10.1242/jcs.241174
Cullati S. N., Chaikuad A., Chen J. S., Gebel J., Tesmer L., Zhubi R., et al. (2022). Kinase domain autophosphorylation rewires the activity and substrate specificity of CK1 enzymes. Mol. Cell 82, 2006–2020.e8. doi:10.1016/j.molcel.2022.03.005
Dahlberg C. L., Nguyen E. Z., Goodlett D., Kimelman D. (2009). Interactions between Casein kinase Iepsilon (CKIepsilon) and two substrates from disparate signaling pathways reveal mechanisms for substrate-kinase specificity. PLoS One 4 (3), e4766. doi:10.1371/journal.pone.0004766
Darlington T. K., Wager-Smith K., Ceriani M. F., Staknis D., Gekakis N., Steeves T. D. L., et al. (1998). Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280, 1599–1603. doi:10.1126/science.280.5369.1599
Dolezelova E., Dolezel D., Hall J. C. (2007). Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics 177, 329–345. doi:10.1534/genetics.107.076513
Dunlap J. C. (1999). Molecular bases for circadian clocks. Cell 96, 271–290. doi:10.1016/s0092-8674(00)80566-8
Emery P., Stanewsky R., Helfrich-Forster C., Emery-Le M., Hall J. C., Rosbash M. (2000). Drosophila CRY is a deep brain circadian photoreceptor. Neuron 26, 493–504. doi:10.1016/S0896-6273(00)81181-2
Fan J. Y., Agyekum B., Venkatesan A., Hall D. R., Keightley A., Bjes E. S., et al. (2013). Noncanonical FK506-binding protein BDBT binds DBT to enhance its circadian function and forms foci at night. Neuron 80, 984–996. doi:10.1016/j.neuron.2013.08.004
Fan J. Y., Means J. C., Bjes E. S., Price J. L. (2015). Drosophila DBT autophosphorylation of its C-terminal domain antagonized by SPAG and involved in UV-induced apoptosis. Mol. Cell. Biol. 35, 2414–2424. doi:10.1128/MCB.00390-15
Fang Y., Sathyanarayanan S., Sehgal A. (2007). Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1). Genes Dev. 21, 1506–1518. doi:10.1101/gad.1541607
Fustin J. M., Kojima R., Itoh K., Chang H. Y., Shiqi Y., Zhuang B., et al. (2018). Two Ck1δ transcripts regulated by m6A methylation code for two antagonistic kinases in the control of the circadian clock. Proc. Natl. Acad. Sci. U. S. A. 115, 5980–5985. doi:10.1073/pnas.1721371115
Garbe D. S., Fang Y., Zheng X. Z., Sowcik M., Anjum R., Gygi S. P., et al. (2013). Cooperative interaction between phosphorylation sites on PERIOD maintains circadian period in Drosophila. PLoS Genet. 9, e1003749. doi:10.1371/journal.pgen.1003749
Garcia-Concejo A., Larhammar D. (2021). Protein kinase C family evolution in jawed vertebrates. Dev. Biol. 479, 77–90. doi:10.1016/j.ydbio.2021.07.013
Giesecke A., Johnstone P. S., Lamaze A., Landskron J., Atay E., Chen K.-F., et al. (2021). Nuclear export of Drosophila PERIOD contributes to temperature compensation of the circadian clock. Cold Spring Harbor, NY. bioRxiv. doi:10.1101/2021.10.25.465663
Graves P. R., Roach P. J. (1995). Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J. Biol. Chem. 270, 21689–21694. doi:10.1074/jbc.270.37.21689
Hara T., Koh K., Combs D. J., Sehgal A. (2011). Post-translational regulation and nuclear entry of TIMELESS and PERIOD are affected in new timeless mutant. J. Neurosci. 31, 9982–9990. doi:10.1523/JNEUROSCI.0993-11.2011
Hardin P. E., Hall J. C., Rosbash M. (1990). Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–540. doi:10.1038/343536a0
Hogenesch J. B., Gu Y. Z., Jain S., Bradfield C. A. (1998). The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. U. S. A. 95, 5474–5479. doi:10.1073/pnas.95.10.5474
Hunter-Ensor M., Ousley A., Sehgal A. (1996). Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 84 (5), 677–685. doi:10.1016/s0092-8674(00)81046-6
Isojima Y., Nakajima M., Ukai H., Fujishima H., Yamada R. G., Masumoto K. H., et al. (2009). CKI epsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc. Natl. Acad. Sci. U. S. A. 106, 15744–15749. doi:10.1073/pnas.0908733106
Jang A. R., Moravcevic K., Saez L., Young M. W., Sehgal A. (2015). Drosophila TIM binds importin α1, and acts as an adapter to transport PER to the nucleus. PLoS Genet. 11, e1004974. doi:10.1371/journal.pgen.1004974
Johnson K. P., Dietrich C. H., Friedrich F., Beutel R. G., Wipfler B., Peters R. S., et al. (2018). Phylogenomics and the evolution of hemipteroid insects. Proc. Natl. Acad. Sci. U. S. A. 115, 12775–12780. doi:10.1073/pnas.1815820115
Joshi R., Cai Y. D., Xia Y., Chiu J. C., Emery P. (2022). PERIOD phosphoclusters control temperature compensation of the Drosophila circadian clock. Front. Physiol. 981, 888262. doi:10.3389/fphys.2022.888262
Jursnich V. A., Fraser S. E., Held L. I., Ryerse J., Bryant P. J. (1990). Defective gap-junctional communication associated with imaginal disc overgrowth and degeneration caused by mutations of the dco gene in Drosophila. Dev. Biol. 140, 413–429. doi:10.1016/0012-1606(90)90090-6
Kaasik K., Kivimae S., Allen J. J., Chalkley R. J., Huang Y., Baer K., et al. (2013). Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17, 291–302. doi:10.1016/j.cmet.2012.12.017
Kawahara A. Y., Plotkin D., Espeland M., Meusemann K., Toussaint E. F. A., Donath A., et al. (2019). Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc. Natl. Acad. Sci. U. S. A. 116, 22657–22663. doi:10.1073/pnas.1907847116
King D. P., Zhao Y. L., Sangoram A. M., Wilsbacher L. D., Tanaka M., Antoch M. P., et al. (1997). Positional cloning of the mouse circadian Clock gene. Cell 89, 641–653. doi:10.1016/S0092-8674(00)80245-7
Kivimaë S., Saez L., Young M. W. (2008). Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 6, e183–e1583. doi:10.1371/journal.pbio.0060183
Kloss B., Price J. L., Saez L., Blau J., Rothenfluh A., Wesley C. S., et al. (1998). The Drosophila clock gene double-time encodes a protein closely related to human casein kinase I epsilon. Cell 94, 97–107. doi:10.1016/S0092-8674(00)81225-8
Kloss B., Rothenfluh A., Young M. W., Saez L. (2001). Phosphorylation of PERIOD is influenced by cycling physical associations of DOUBLE-TIME, PERIOD, and TIMELESS in the Drosophila clock. Neuron 30, 699–706. doi:10.1016/S0896-6273(01)00320-8
Ko H. W., Kim E. Y., Chiu J., Vanselow J. T., Kramer A., Edery I. (2010). A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3 beta/SGG in circadian clocks. J. Neurosci. 30, 12664–12675. doi:10.1523/Jneurosci.1586-10.2010
Kobelkova A., Bajgar A., Dolezel D. (2010). Functional molecular analysis of a circadian clock Gene timeless promoter from the drosophilid fly Chymomyza costata. J. Biol. Rhythms 25, 399–409. doi:10.1177/0748730410385283
Kondo S., Ueda R. (2013). Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721. doi:10.1534/genetics.113.156737
Kotwica-Rolinska J., Damulewicz M., Chodáková L., Krištofová L., Dolezel D. (2022b). Pigment Dispersing Factor is a circadian clock output and regulates photoperiodic response in the linden bug, Pyrrhocoris apterus. Front. Physiol. 13, 884909. doi:10.3389/fphys.2022.884909
Kotwica-Rolinska J., Chodakova L., Chvalova D., Kristofova L., Fenclova I., Provaznik J., et al. (2019) CRISPR/Cas9 genome editing introduction and optimization in the non-model insect Pyrrhocoris apterus. Front. Physiology. 10: 891. doi:10.3389/fphys.2019.00891
Kotwica-Rolinska J., Chodakova L., Smykal V., Damulewicz M., Provaznik J., Wu B. C. H., et al. (2022a). Loss of timeless Underlies an evolutionary transition within the circadian clock. Mol. Biol. Evol. 39, msab346. doi:10.1093/molbev/msab346
Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X. W., et al. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205. doi:10.1016/S0092-8674(00)81014-4
Lam V. H., Li Y. H., Liu X., Murphy K. A., Diehl J. S., Kwok R. S., et al. (2018). CK1α collaborates with DOUBLETIME to regulate PERIOD function in the Drosophila circadian clock. J. Neurosci. 38, 10631–10643. doi:10.1523/JNEUROSCI.0871-18.2018
Lee C., Etchegaray J. P., Cagampang F. R. A., Loudon A. S. I., Reppert S. M. (2001). Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867. doi:10.1016/S0092-8674(01)00610-9
Leloup J. C. (2009). Circadian clocks and phosphorylation: Insights from computational modeling. Open Life Sci. 4, 290–303. doi:10.2478/s11535-009-0025-1
Levine J. D., Funes P., Dowse H. B., Hall J. C. (2002). Signal analysis of behavioral and molecular cycles. BMC Neurosci. 3, 1. doi:10.1186/1471-2202-3-1
Li Y. H., Liu X., Vanselow J. T., Zheng H., Schlosser A., Chiu J. C. (2019). O-GlcNAcylation of PERIOD regulates its interaction with CLOCK and timing of circadian transcriptional repression. PLoS Genet. 15 (1), e1007953. doi:10.1371/journal.pgen.1007953
Lowrey P. L., Shimomura K., Antoch M. P., Yamazaki S., Zemenides P. D., Ralph M. R., et al. (2000). Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288, 483–492. doi:10.1126/science.288.5465.483
Masuda S., Narasimamurthy R., Yoshitane H., Kim J. K., Fukada Y., Virshup D. M. (2020). Mutation of a PER2 phosphodegron perturbs the circadian phosphoswitch. Proc. Natl. Acad. Sci. U. S. A. 117, 10888–10896. doi:10.1073/pnas.2000266117
Matsumoto A., Tomioka K., Chiba Y., Tanimura T. (1999). Timrit lengthens circadian period in a temperature-dependent manner through suppression of PERIOD protein cycling and nuclear localization. Mol. Cell. Biol. 19, 4343–4354. doi:10.1128/mcb.19.6.4343
McKenna D. D., Shin S., Ahrens D., Balke M., Beza-Beza C., Clarke D. J., et al. (2019). The evolution and genomic basis of beetle diversity. Proc. Natl. Acad. Sci. U. S. A. 116, 24729–24737. doi:10.1073/pnas.1909655116
Meireles-Filho A. C., Amoretty P. R., Souza N. A., Kyriacou C. P., Peixoto A. A. (2006). Rhythmic expression of the cycle gene in a hematophagous insect vector. BMC Mol. Biol. 7, 38. doi:10.1186/1471-2199-7-38
Meng Q. J., Logunova L., Maywood E. S., Gallego M., Lebiecki J., Brown T. M., et al. (2008). Setting clock speed in mammals: The CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58, 78–88. doi:10.1016/j.neuron.2008.01.019
Meyer P., Saez L., Young M. W. (2006). PER-TIM interactions in living Drosophila cells: An interval timer for the circadian clock. Science 311, 226–229. doi:10.1126/science.1118126
Misof B., Liu S., Meusemann K., Peters R. S., Donath A., Mayer C., et al. (2014). Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. doi:10.1126/science.1257570
Muskus M. J., Preuss F., Fan J. Y., Bjes E. S., Price J. L. (2007). Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol. Cell. Biol. 27, 8049–8064. doi:10.1128/Mcb.00680-07
Myers M. P., Wager Smith K., Rothenfluh Hilfiker A., Young M. W., Rothenfluh-Hilfiker A. (1996). Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science 271, 1736–1740. doi:10.1126/science.271.5256.1736
Narasimamurthy R., Hunt S. R., Lu Y., Fustin J. M., Okamura H., Partch C. L., et al. (2018). CK1δ/ε protein kinase primes the PER2 circadian phosphoswitch. Proc. Natl. Acad. Sci. U. S. A. 115, 5986–5991. doi:10.1073/pnas.1721076115
Narasimamurthy R., Virshup D. M. (2021). The phosphorylation switch that regulates ticking of the circadian clock. Mol. Cell 81, 1133–1146. doi:10.1016/j.molcel.2021.01.006
Peschel N., Chen K. F., Szabo G., Stanewsky R. (2009). Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, Jetlag, and timeless. Curr. Biol. 19 (3), 241–247. doi:10.1016/j.cub.2008.12.042
Pfeiffenberger C., Lear B. C., Keegan K. P., Allada R. (2010). Locomotor activity level monitoring using the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb. Protoc. 2010 (11), prot5518. doi:10.1101/pdb.prot5518
Philpott J. M., Narasimamurthy R., Ricci C. G., Freeberg A. M., Hunt S. R., Yee L. E., et al. (2020). Casein kinase 1 dynamics underlie substrate selectivity and the PER2 circadian phosphoswitch. Elife 9, e52343. doi:10.7554/eLife.52343
Port F., Chen H. M., Lee T., Bullock S. L. (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 111, E2967–E2976. doi:10.1073/pnas.1405500111
Poupardin R., Schottner K., Korbelova J., Provaznik J., Dolezel D., Pavlinic D., et al. (2015). Early transcriptional events linked to induction of diapause revealed by RNAseq in larvae of drosophilid fly, Chymomyza costata. BMC Genomics 16, 720. doi:10.1186/s12864-015-1907-4
Price J. L., Blau J., Rothenfluh A., Abodeely M., Kloss B., Young M. W. (1998). Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94, 83–95. doi:10.1016/S0092-8674(00)81224-6
Putker M., Wong D. C., Seinkmane E., Rzechorzek N. M., Zeng A., Hoyle N. P., et al. (2021). CRYPTOCHROMES confer robustness, not rhythmicity, to circadian timekeeping. EMBO J. 40 (7), e106745. doi:10.15252/embj.2020106745
Ralph M. R., Menaker M. A. (1988). A mutation of the circadian system in golden hamsters. Science 2241 (4870), 1225–1227. doi:10.1126/science.3413487
Reischl S., Kramer A. (2011). Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 10, 1393–1399. doi:10.1016/j.febslet.2011.02.038
Rothenfluh A., Abodeely M., Price J. L., Young M. W. (2000). Isolation and analysis of six timeless alleles that cause short- or long-period circadian rhythms in Drosophila. Genetics 156, 665–675. doi:10.1093/genetics/156.2.665
Rutila J. E., Suri V., Le M., So W. V., Rosbash M., Hall J. C. (1998). CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93, 805–814. doi:10.1016/S0092-8674(00)81441-5
Saez L., Derasmo M., Meyer P., Stieglitz J., Young M. W. (2011). A key temporal delay in the circadian cycle of Drosophila is mediated by a nuclear localization signal in the timeless protein. Genetics 188, 591–600. doi:10.1534/genetics.111.127225
Saez L., Young M. W. (1996). Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron 17, 911–920. doi:10.1016/S08966273(00)80222-6
Sathyanarayanan S., Zheng X., Xiao R., Sehgal A. (2004). Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 116, 603–615. doi:10.1016/s0092-8674(04)00128-x
Schmid B., Helfrich-Forster C., Yoshii T. (2011). A new ImageJ plug-in “ActogramJ” for chronobiological analyses. J. Biol. Rhythms 26, 464–467. doi:10.1177/0748730411414264
Sehgal A., Price J. L., Man B., Young M. W. (1994). Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263, 1603–1606. doi:10.1126/science.8128246
Sekine T., Yamaguchi T., Hamano K., Young M. W., Shimoda M., Saez L. (2008). Casein kinase I epsilon does not rescue double-time function in Drosophila despite evolutionarily conserved roles in the circadian clock. J. Biol. Rhythms 23, 3–15. doi:10.1177/0748730407311652
Shinohara Y., Koyama Y. M., Ukai-Tadenuma M., Hirokawa T., Kikuchi M., Yamada R. G., et al. (2017). Temperature-sensitive substrate and product binding underlie temperature-compensated phosphorylation in the clock. Mol. Cell 67, 783–798. doi:10.1016/j.molcel.2017.08.009
Singh S., Giesecke A., Damulewicz M., Fexova S., Mazzotta G. M., Stanewsky R., et al. (2019). New Drosophila circadian clock mutants affecting temperature compensation induced by targeted mutagenesis of timeless. Front. Physiol. 10, 1442. doi:10.3389/fphys.2019.01442
Smykal V., Pivarci M., Provaznik J., Bazalova O., Jedlicka P., Luksan O., et al. (2020). Complex evolution of insect insulin receptors and homologous decoy receptors, and functional significance of their multiplicity. Mol. Biol. Evol. 37, 1775–1789. doi:10.1093/molbev/msaa048
Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K., Kay S. A., et al. (1998). The cry(b) mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692. doi:10.1016/S0092-8674(00)81638-4
Toh K. L., Jones C. R., He Y., Eide E. J., Hinz W. A., Virshup D. M., et al. (2001). An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040–1043. doi:10.1126/science.1057499
Top D., O'Neil J. L., Merz G. E., Dusad K., Crane B. R., Young M. W. (2018). CK1/Doubletime activity delays transcription activation in the circadian clock. Elife 7, e32679. doi:10.7554/eLife.32679
Tosini G., Menaker M. (1998). The tau mutation affects temperature compensation of hamster retinal circadian oscillators. Neuroreport 9, 1001–1005. doi:10.1097/00001756-199804200-00009
Uno Y., Nishida C., Takagi C., Ueno N., Matsuda Y. (2013). Homoeologous chromosomes of Xenopus laevis are highly conserved after whole-genome duplication. Hered. (Edinb) 111 (5), 430–436. doi:10.1038/hdy.2013.65
Venkatesan A., Fan J. Y., Bouyain S., Price J. L. (2019). The circadian tau mutation in casein kinase 1 is part of a larger domain that can Be mutated to shorten circadian period. Int. J. Mol. Sci. 20, 813. doi:10.3390/ijms20040813
Venkatesan A., Fan J. Y., Nauman C., Price J. L. (2015). A doubletime nuclear localization signal mediates an interaction with bride of doubletime to promote circadian function. J. Biol. Rhythms 30, 302–317. doi:10.1177/0748730415588189
Vielhaber E. L., Duricka D., Ullman K. S., Virshup D. M. (2001). Nuclear export of mammalian PERIOD proteins. J. Biol. Chem. 276 (49), 45921–45927. doi:10.1074/jbc.M107726200
Wipfler B., Letsch H., Frandsen P. B., Kapli P., Mayer C., Bartel D., et al. (2019). Evolutionary history of Polyneoptera and its implications for our understanding of early winged insects. Proc. Natl. Acad. Sci. U. S. A. 116 (8), 3024–3029. doi:10.1073/pnas.1817794116
Wulbeck C., Szabo G., Shafer O. T., Helfrich-Forster C., Stanewsky R. (2005). The novel Drosophila tim(blind) mutation affects behavioral rhythms but not periodic eclosion. Genetics 169 (2), 751–766. doi:10.1534/genetics.104.036244
Xu Y., Padiath Q. S., Shapiro R. E., Jones C. R., Wu S. C., Saigoh N., et al. (2005). Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 434, 640–644. doi:10.1038/nature03453
Xu Y., Toh K. L., Jones C. R., Shin J. Y., Fu Y. H., Ptacek L. J. (2007). Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell 128, 59–70. doi:10.1016/j.cell.2006.11.043
Yildiz O., Doi M., Yujnovsky I., Cardone L., Berndt A., Hennig S., et al. (2005). Crystal structure and interactions of the PAS repeat region of the Drosophila clock protein PERIOD. Mol. Cell 17 (1), 69–82. doi:10.1016/j.molcel.2004.11.022
Yu W. J., Zheng H., Price J. L., Hardin P. E. (2009). DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol. Cell. Biol. 29 (6), 1452–1458. doi:10.1128/Mcb.01777-08
Zeng H. K., Qian Z. W., Myers M. P., Rosbash M. (1996). A light-entrainment mechanism for the Drosophila circadian clock. Nature 380 (6570), 129–135. doi:10.1038/380129a0
Zhou M., Kim J. K., Eng G. W., Forger D. B., Virshup D. M. (2015). A Period2 phosphoswitch regulates and temperature compensates circadian period. Mol. Cell 60, 77–88. doi:10.1016/j.molcel.2015.08.022
Zilian O., Frei E., Burke R., Brentrup D., Gutjahr T., Bryant P. J., et al. (1999). double-time is identical to discs overgrown, which is required for cell survival, proliferation and growth arrest in Drosophila imaginal discs. Development 126 (23), 5409–5420. doi:10.1242/dev.126.23.5409
Keywords: casein kinase 1, doubletime, evolution, circadian clock, temperature compensation, bride of doubletime
Citation: Thakkar N, Giesecke A, Bazalova O, Martinek J, Smykal V, Stanewsky R and Dolezel D (2022) Evolution of casein kinase 1 and functional analysis of new doubletime mutants in Drosophila. Front. Physiol. 13:1062632. doi: 10.3389/fphys.2022.1062632
Received: 06 October 2022; Accepted: 28 November 2022;
Published: 14 December 2022.
Edited by:
Ueli Schibler, Université de Genève, SwitzerlandReviewed by:
Charalambos P. Kyriacou, University of Leicester, United KingdomCopyright © 2022 Thakkar, Giesecke, Bazalova, Martinek, Smykal, Stanewsky and Dolezel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralf Stanewsky, c3RhbmV3c2t5QHd3dS5kZQ==; David Dolezel, ZG9sZXplbEBlbnR1LmNhcy5jeg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.