
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol. , 11 November 2022
Sec. Cell Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.1004330
Acute leukemia is a common hematologic tumor with highly genetic heterogeneity, and many factors are involved in the pathogenesis and drug-resistance mechanism. Emerging evidence proves that E3 ubiquitin ligases participate in the acute leukemic signaling pathways via regulating substrates. This review summarized the E3 ligases which can affect the leukemic signal. It is worth noting that the abnormal signal is often caused by a deficiency or a mutation of the E3 ligases. In view of this phenomenon, we envisioned perspectives associated with targeted agonists of E3 ligases and proteolysis-targeting chimera technology. Moreover, we emphasized the significance of research into the upstream factors regulating the expression of E3 ubiquitin ligases. It is expected that the understanding of the mechanism of leukemic signaling pathways with which that E3 ligases are involved will be beneficial to accelerating the process of therapeutic strategy improvement for acute leukemia.
Acute leukemia is a hematopoietic stem cell malignancy that is highly heterogeneous. Most abnormally proliferating cells are primitive and naïve cells due to the stagnation of leukocyte differentiation in the early stage. According to the type of cells involved, acute leukemia can generally be divided into two major groups: acute myeloid leukemia (AML), characterized by an uncontrolled clonal proliferation of abnormal myeloid stem/progenitor cells, and acute lymphoblastic leukemia (ALL), characterized by abnormal proliferation and aggregation of immature lymphocytes in the bone marrow and extramedullary tissue. The common clinical manifestation of acute leukemia presented with anemia, hemorrhage, fever, hepatomegaly, splenomegaly, or enlarged lymph nodes. Formal diagnosis has progressed to dimensional of MICM. This means the combination of morphology, immunology, cytogenetics, and molecular biology (Chang et al., 2021; Newell and Cook, 2021). Leukemogenesis is the result of the interaction of various complex mechanisms, among which chromosomal aberrations and gene mutations play an important role. Identifying the molecular abnormalities contributes to formulating risk stratification and improving treatment strategies. At present, some breakthroughs have been achieved, such as all-trans retinoic acid targeted PML-RARα fusion gene and Imatinib targeted BCR-ABL fusion gene; Chimeric Antigen Receptor T-Cell Immunotherapy (CAR-T) targeted CD19; and Gemtuzumab ozogamicin targeted CD33-positive AML (Baron and Wang, 2018; Zhu and Gao, 2019; Levin et al., 2021; Yilmaz et al., 2021). Despite this, the enhancement of overall survival in patients with leukemia remains challenging, and a good knowledge of the acute leukemic signal transduction involved would bring a significant improvement in treatment regimens.
Protein translation modifications are one of the most important regulatory mechanisms for intracellular proteins, enabling cells to respond to changes in the internal and external environment through rapid and reversible modifications to the structure, function, and location of a specific protein. More than two hundred protein translation modifications have been discovered, including phosphorylation, methylation, acetylation, ubiquitination, to name a few, which play an important role in cell growth, metabolism, differentiation, apoptosis, and other processes (Dunphy et al., 2021). Ubiquitination refers to the covalent binding of ubiquitin to a target protein (Figure 1), a process that involves the synergistic action of three ubiquitin enzymes: E1 ubiquitin activating enzyme, E2 ubiquitin conjugating enzyme, and E3 ubiquitin ligase (Nandi et al., 2006). Ubiquitin is a 76-residue small protein in which the C-terminal cysteine is activated by E1 ubiquitin activating enzyme and then transferred onto the activate site of an E2 ubiquitin conjugating enzyme through trans-thioesterification (Mansour, 2018; Wijk et al., 2019). Subsequently, E2-ubiquitin intermediate is linked to the target protein by E3 ubiquitin ligases-mediated isopeptide bond formation between the C-terminal glycine of ubiquitin and the substrate lysine residue. In this series of enzymatic cascade reactions, E3 plays a unique role in recognizing target proteins and regulating ubiquitination system activity. E3 ubiquitin ligases fall into three main types: the really interesting new gene/U (UFD2)-box (RING/U-box), the homologous to E6AP carboxyl-terminus (HECT), and the RING-between-RING (RBR) families (Berndsen and Wolberger, 2014; Wang et al., 2017). These E3 ligases can target a variety of substrates, then trigger ubiquitination and proteasome degradation. When this protein degradation process is out of balance due to changes in the E3 ligases, a variety of diseases can be caused or promoted, such as neurodegenerative disorders, cardiovascular disease, and cancer.
With the development of proteolysis-targeting chimeras (PROTAC), E3 ligases have become the key target. This technique can remove unwanted or damaged proteins by forming a stable target protein/PROTAC/E3 ternary complex (Figure 2), solving the problem of undruggable cases to some extent (Sakamoto et al., 2001; Moon and Lee, 2018). In addition to PROTAC, there are targeted inhibitors of E3 ligases, targeted agonists of E3 ligases, and molecular glues, thereby arousing our interest in the mechanism of E3 ubiquitin ligases in the acute leukemic signaling pathways. Combined with these techniques, E3 ligases can be excellent regulatory targets affecting anti-leukemia potential. In this review, we summarized E3 ligases that their changes are involved in the abnormal activated signaling pathway, ultimately promoting the occurrence or progression of leukemia.
RING family is the most common E3 ligases; the RING domain of RING E3s harbors two zinc ions, providing Zn-coordination in a cross-braced configuration for domain folding (Das et al., 2021). RING E3s exerted their E3 activity with a highly diverse quaternary architecture and different modes of assembly: monomer, homodimer, heterodimer, and compositions of multiple subunits (also known as higher order oligomers) (Buetow and Huang, 2016). Additionally, the U-box domain is the same as the RING fold, but without zinc.
The tumor necrosis factor receptor-associated factor (TRAF) family consists of five members: TRAFs 1, 2, 3, 5, and 6, participating in cell proliferation, differentiation, survival, apoptosis, immune and inflammatory responses. Among these members, TRAF2 and TRAF6 have a typical TRAF structure, including an N-terminal RING domain, four or five zinc finger motifs that provide structural support for RING domain activity, and a C-terminal domain that consists of a coiled coil domain and a TRAFC domain (also known as the MATH domain) (Table 1) (Lamothe et al., 2008; Arkee and Bishop, 2020). Besides, the common functional domains of the Casitas B-cell lymphoma (CBL) family (mainly c-Cbl and Cbl-b) include the tyrosine kinase binding domain consisting of four helix bundles, an EF hand, and an Src homology 2 (SH2) domain, which is responsible for recognition of the substrate; a highly conserved helical linker; and one RING finger domain that can work together with the helical linker to form the structural platform for binding to an E2 ubiquitin conjugating enzyme (Table 1) (Meng et al., 1999; Zheng et al., 2000). Cbl-b is a larger protein than c-Cbl and contains an additional 69 amino acids at the C-terminus (Lyle et al., 2019). Specific substrates of CBL are epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor (PDGFR), c-Kit, FLT3, ZAP70 and SYK (Thien and Langdon, 2005; Mohapatra et al., 2013). In addition, the tripartite motif containing (TRIM) proteins contain a large family of RING type E3 ligases. There are three common structural features of TRIMs, containing the N-terminal catalytic RING domain, one or two B-boxes, and a coiled-coil domain (Table 1) (Meroni and Diez-Roux, 2005). Functionally, it has been identified that TRIM56 and TRIM65 can stimulate type I interferon expression combined with nucleic acid-sensing receptors, such as STING, RIG-I, and MDA5 (Tsuchida et al., 2010; Versteeg et al., 2013; Kamanova et al., 2016). Furthermore, TRIAD1 (Two RING fingers and DRIL) is a cysteine-rich domain of around 200 amino acids, consisting of a DRIL motif located between two RING fingers (Reijden et al., 1999). The N-terminal is an acid domain while the C-terminal region contains a helical ring domain (Table 1). TRIAD1 is a proapoptotic protein that promotes p53 activation that plays a critical role in cell differentiation and apoptosis and is ubiquitinated by Mouse double minute 2 (MDM2) (Marteijn et al., 2005; Bae et al., 2012). MDM2 is a p53-specific E3 ubiquitin ligase, acting to limit the p53 growth-suppressive function in unstressed cells (Moll and Petrenko, 2003). MDM2 N-terminal domain is responsible for recognizing and binding to substrates; the C-terminal domain catalysis the transfer of ubiquitin to substrates (Table 1) (Rotin and Kumar, 2009). MDM2 expression is up-regulated in numerous cancers, resulting in a loss of p53-dependent activities, such as apoptosis and cell-cycle arrest (Oliner et al., 2016).
F-box protein functions as the substrate receptor in Cullin-RING E3 ubiquitin ligases (CRLs), which consists of three categories: FBXWs, FBXLs, and FBXOs. Among the RING-type E3 ligases, the Cullin is the largest family. CRLs belong to RING-type E3s with a characterized Cullin-RING heterodimeric complex, containing seven Cullin (CUL) proteins, CUL1, CUL2, CUL3, CUL4A, CUL4B, CUL5, and CUL7. The core structure of CRL consists of four parts: the CUL scaffold, a substrate receptor, adaptor proteins that connect the substrate receptor to CUL, and a RING finger protein binding to E2 ubiquitin conjugating enzyme. The C-terminal domain of CUL protein forms a core ligase complex in combination with a RING finger protein, either RBX1 or RBX2, while the N-terminal domain interchangeably assembles with CUL-specific substrate receptors through adaptors. For instance, CRL1, also named SKP1–CUL1–F-box (SCF), uses SKP1 as its adaptor, and F-box as substrate receptor (Skowyra et al., 1997; Nakagawa et al., 2020).
S-phase kinase-associated protein 2 (SKP2), which belongs to F-box protein is a class of proteins containing a sequence homologous to cyclin F and can catalyze the substrate by binding to SKP1 and CUL1 (Bai et al., 1994; Jin et al., 2004; Nguyen and Busino, 2020). The CRL1SKP2 consists of four components: CUL1, SKP1, the F-box domain of SKP2 (F-boxSKP2), and RBX1 (Table 1). Functionally, SKP2 targets various cyclin-dependent kinases inhibitors (CKI) for degradation, such as p21Cip1, p27Kip1, and p57Kip2, controlling cell cycle regulatory proteins (Frescas and Pagano, 2008). Therefore, the dysfunction of SKP2 causes cell cycle entry or arrest. F-box and WD-40 repeat domain-containing protein 7 (FBW7), a component of an SCF ubiquitin ligase complex, belongs to the F-box protein family. The structure of FBW7 is organized in three domains: the D domain, F-box domain, and seven tandem WD40-repeat domains. The D domain is mainly responsible for FBW7 dimerization, regulating substrate binding and ubiquitylation; the F-box domain is essential for FBW7 interaction with SKP1 whereas the WD40-repeat domain plays a role in the recognition of phosphorylated substrates (Table 1) (Hao et al., 2007; Tang et al., 2007). The classical substrates of FBW7 include c-Myc (Yada et al., 2004) and cyclin-E (Koepp et al., 2001), although other target proteins have been identified such as neurogenic locus Notch homolog protein 1 (Notch 1) (Fryer et al., 2004), NF-κB (Fukushima et al., 2012), MCL-1 (Wertz et al., 2011), c-Jun (Wei et al., 2005), granulocyte colony stimulating factor receptor (Lochab et al., 2013), heat shock transcription factor 1 (Kourtis et al., 2015), CCAAT/enhancer-binding protein-alpha (C/EBPα) (Bengoechea-Alonso and Ericsson, 2010), and glucocorticoid receptor α (Malyukova et al., 2013). Additionally, the expression of FBW7 is regulated by a series of genes, including p53 (Kimura et al., 2003), miR-223 (Mansour et al., 2013), miR-25 (Xiang et al., 2015), miR-182 (Li et al., 2014), miR-503 (Li et al., 2014), miR-92a (Zhou et al., 2015), RBP-J-interaction, tubulin-associated (RITA) protein (Wang et al., 2014), NF-κB1 (Huang et al., 2014).
Different from the scaffolding role (combining E2 closely with substrates) of RING-type E3 ligases, HECT-type E3 ligases play a catalytic role. For the HECT E3 ligase, it consists of an N-terminal substrate-binding domain and a C-terminal HECT domain. The C-terminal HECT domain was first discovered in human papillomavirus E6-associated protein (E6AP) 5, containing almost 350 amino acids (Rotin and Kumar, 2009; Singh and Sivaraman, 2020); there are two lobes in the conserved HECT domain that are connected by a flexible hinge loop, with the N-terminal lobe (N-lobe) binding to E2∼ubiquitin and the C-terminal lobe (C-lobe) having the catalytic cysteine residue (Huang et al., 1999). Based on the different N-terminal domains, HECT E3s can be divided into NEDD4 family, HERC family, and HECTs with other protein-protein interaction domains (Singh et al., 2021). Ubiquitin ligase for ARF (ULF) was identified in nucleophosmin (NPM) protein complexes. It includes an N-terminal ARM domain and a C-terminal HECT domain, with a centrally located WWE motif (Table 1) (Chen et al., 2010a). The main function of ULF is to mediate the polyubiquitination and proteasomal degradation of alternative reading frame (ARF) protein (Chen et al., 2010a). As a member of the NEDD4 family, WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) composes a C2 domain in N-terminal and a catalytic HECT domain in C-terminal, with four WW domains in its central part (Hu et al., 2021). Functionally, WWP1 interacts with a variety of substrates, such as TβR1 (W1Komuro et al., 2004), Smad2 (Chen et al., 2007), ErbB4/HER4 (Feng et al., 2009), RNF11 (Chen et al., 2008), SPG20 (Edwards et al., 2009), RUNX2 (Shen et al., 2006), p63 (Li et al., 2008), and p27 (Cao et al., 2011). These interactions regulate numerous physiology processes, including TGF signaling, osteoblast differentiation, differentiation and apoptosis of cancer cells, senescence.
FBW7 is deemed to be a p53-dependent tumor suppressor (Kimura et al., 2003; Mao et al., 2004). In the incidence of T-ALL, FBW7 deletion without other tumor-promoting factors is one of the causes (Matsuoka et al., 2008). Evidence showed that 59% FBW7 deletion mice developed T-ALL, along with Notch1 and c-Myc proteins accumulated in leukemic cells while p53 protein level was decreased. Notch1 is a transmembrane protein that serves as a ligand-activated transcription factor. The Notch1/c-Myc signaling pathway plays a pivotal role in T-ALL (Weng et al., 2004; Weng et al., 2006). FBW7 mutations, which are mainly located in codons that encode for arginine residues R479Q, R505C, and R465H, have been identified in T-ALL, accounting for 31% of all T-ALL cases (O’Neil et al., 2007; Akhoondi et al., 2007). It is worth noting that spontaneously developed leukemia was not seen in mice that carry Cre-inducible FBW7 heterozygote mutants (King et al., 2013). Notch1 mutation, as an important oncogenic factor in ALL, is also associated with FBW7 mutation. Notch1 mutations mainly occur in the PEST domain and the heterodimeric domain. The reason for the name PEST is that this mutation truncates the C-terminal domain of protein, among which the proline, glutamine, serine, and threonine have a high frequency of truncation and are known as PEST domains (Chiang et al., 2016). Mechanistically, disruption or deletion of the PEST domain can be found in 15% T-ALL and has a relationship with FBW7 mutation, causing increased intracellular Notch1 (ICN1) protein stabilization; heterodimeric domain point mutation can be found in 25% T-ALL and has a relationship with PEST or FBW7 mutations, contributing to ligand-independent activation of the receptor (Weng et al., 2004). ICN1 splits from the cell membrane and migrates into the nucleus. The extended half-life of ICN1 causes sustained activation of Notch1 signaling pathway, ultimately leading to blocking of apoptosis in normal cells and the abnormal proliferation of non-functional T cells. Adult T cell leukemia/lymphoma (ATL), an aggressive lymphoproliferative disease with poor prognosis, is caused by the human T-cell leukemia virus type 1 (HTLV-1). In HTLV-1-transformed ATL cells, FBW7 acts as a tumor suppressor as well. According to the study of O’Neil et al. (2007), the FBW7 mutation, which is located in the beta-propeller domain adjacent to the pocket for substrate binding, activates the Notch1 signaling pathway. To be more specific, FBW7 mutants T416A and W406R retain the ability to interact and degrade ICN1, while the majority of FBW7 mutants lose that ability. In contrast to T416A and W406R mutants, the FBW7 mutants D510E can normally interact with ICN1, but lose its degradation ability, leading to the continued activation of the Notch1 signaling pathway (Figure 3). Interestingly, FBW7 D510E can target c-Myc, cyclin E, and MCL-1. The reason why FBW7 D510E selectively degrades substrates remains unknown. In addition, hyperphosphorylation of cyclin E, which is related to the FBW7 mutation, is highly correlated with polyploidy and aneuploidy (Table 2). The latter is one of the characteristics of acute ATL (Rajagopalan et al., 2004).
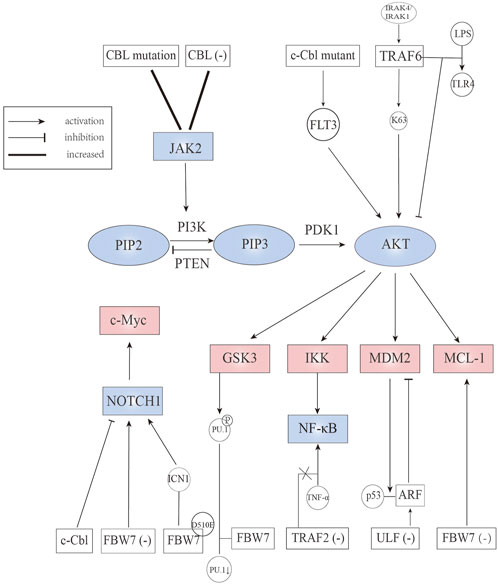
FIGURE 3. E3 ubiquitin ligases involved in JAK2, Notch, PI3K/AKT, and downstream signaling pathways.
In T-ALL, ICN1 is the potential therapeutic target, an aspect that is described in detail in the section covering FBW7. However, although γ-secretase inhibitors (GSI) can prevent the release of ICN1, they also lead to off-target effects. As for c-Cbl (a member of the CBL family), such mutation is frequent in myeloid leukemia but not in T-ALL, making it a possible drug target. Recent studies have shown that flavone (2-phenyl-4H-1-benzopyran-4-one), the core structure of flavonoids, can induce ICN1 degradation through up-regulation of the level of c-Cbl, inhibiting cell proliferation (Zhu et al., 2020). In addition to Notch1, Notch3 was identified as another member of the Notch family that plays a critical role in the development of T-ALL. In the absence of pTα, Notch3 can be degraded in a c-Cbl-dependent manner (Checquolo et al., 2010). Furthermore, SKP2 was reported to play a crucial role in T-ALL. Recent research advances demonstrated that SKP2 genetic ablation can delay T-ALL progression in vivo, while the pharmacological blockade of SKP2 can inhibit the proliferation of human T-ALL cells (Rodriguez et al., 2020). This arises because SKP2 can degrade the cyclin-dependent kinase inhibitor p27Kip1, inducing T cell progression into the cell cycle and coordinating cell proliferation and cell differentiation. Moreover, it has been reported that Notch activation promotes the role of SKP2 in T-ALL, forming a Notch/SKP2/p27Kip1 forward feedback loop (Table 2) (Zhang et al., 2000; Barata et al., 2001; Dohda et al., 2007; Rodriguez et al., 2020).
Cytokine dependent Janus kinase 2 (JAK2) signaling is crucial to hematopoietic stem/progenitor cells. Uncontrolled activation of JAK2 signal triggers the occurrence of hematological malignancy. A study assessed whether CBL influence JAK2 protein levels showed that CBL knock-down can prolong JAK2 activation (Lv et al., 2017). Similarly, an elevated level and extended half-life of JAK2 was observed in CBL mutant AML cells. However, independent studies have shown that compared to CBL wild-type AML cells, CBL mutant AML cells are more sensitive to quizartinib (a FLT3 inhibitor) than ruxolitinib (a JAK2 inhibitor) (Lv et al., 2017). As c-Cbl mutant protein can stimulate FLT3 signaling, we envisioned the existence of a c-Cbl, JAK2, and FLT3 signaling axis, regulating the development of AML (Figure 3) (Table 2) (Rathinam et al., 2010; Taylor et al., 2015).
Phosphatidylinositol 3-kinase (PI3K)/Protein Kinase B (PKB, also named AKT) is known to be associated with cell metabolism, proliferation, differentiation, and apoptosis. The interaction between PI3K and AKT is related to two metabolites and two coding genes: the small molecules PI (4,5) P2 and PI (3,4,5) P3 and the proteins phosphatase and tensin homologue (PTEN) and 3-phosphoinositide dependent protein kinase-1 (PDK1). In normal cells, PI (3,4,5) P3 is rapidly metabolized and dephosphorylated by lipid phosphatases (such as PTEN) to terminate the PI3K signal (Vara et al., 2004). Large deposits of PI(3,4,5)P3 or the loss-of-function of PTEN are often found in cancer cells, suggesting that abnormal regulatory genes play an important role in cancer. In addition, an increasing number of studies show that PI3K/AKT regulates the occurrence and development of leukemia and drug resistance (Nepstad et al., 2020; Yao et al., 2021).
Currently, the link between TRAF6 and AML is widely studied. TRAF6 is considered to play an essential role in signal transduction of the Toll-like-receptor (TLR) superfamily (classified as immune receptors). TRAF6 inhibits the activation of AKT after stimulation of TLR-4 with lipopolysaccharide in FLT3-ITD AML cells (Schnetzke et al., 2013a). Down-regulation of TRAF6 results in enhanced constitutive AKT activation through phosphorylating residues Thr308 and Ser473 in MV4-11 cells (FLT3-ITD AML cells). Interestingly, Chan et al. (2012) suggested that the ubiquitination and activation of AKT were affected by diverse growth factors utilizing distinct E3 ligases. For example, the SKP2-SCF complex was required for EGFR induced AKT activation; SKP2 underwent tyrosine and serine/threonine phosphorylation after EGF stimulation. TRAF6 not only acts in AML, but also in ALL. In primary T cell, TRAF6 was activated through MyD88/IL-1 receptor-associated kinase (IRAK) 4/IRAK 1 signaling, then catalyzing K63 polyubiquitination (which is known to activate AKT directly), and ultimately stabilizing antiapoptotic protein MCL-1 (Figure 3) (Li et al., 2015). The study also showed that inhibition of IRAK reduced the stability of MCL-1 and enhanced the sensitivity to chemotherapy (Table 2) (Li et al., 2015).
Deletion and loss-of-function mutation of c-Cbl can be found mainly in myeloproliferative neoplasms (MPNs) but also occured in other hematopoietic malignancies such as AML (Dunbar et al., 2008). AML patients with CBL mutation always need an intense chemotherapy regimen or accept hemopoietic stem cell transplantation, showing highly malignant characteristics (Makishima et al., 2009). Studies in animal models have shown that single copy mutant c-CblA/− mice developed aggressive myeloid leukemia with significantly increased white blood cell counts. The stimulation of FLT3 signaling caused by c-Cbl mutant protein results in the constitutive activation of the AKT pathway (Figure 3) (Table 2), suggesting that FLT3 kinase is a potential therapeutic target for the treatment of c-Cbl mediated leukemia (Rathinam et al., 2010; Taylor et al., 2015). In this regard, it is worth mentioning a recent study showing that Cbl-b was implicated in FLT3 kinase inhibitor-resistant AML. According to Park et al. (2017), the mutant FLT3-ITD AML cells could inactivate p53, resisting the effects of FLT3 inhibitors.
NF-κB, a generic name of a family of transcription factors, regulates a series of genes involved in cell survival, proliferation, differentiation, and immune and inflammatory responses. Mounting evidence shows that constitutive activation of NF-κB is frequently associated with hematological malignancies. NF-κB selective inhibitory drugs have been developed for targeting tumor cells (Breccia and Alimena, 2010). At present, NF-κB inhibitors such as bortezomib and carfilzomib are some of the potent therapeutically useful drugs for multiple myeloma. Activation of NF-κB signaling can be caused by stimulation of lipopolysaccharide-mediated TLR-4 activation and tumor necrosis factor-α (TNF-α) (Lomaga et al., 1999; Verstrepen et al., 2008; Schnetzke et al., 2013a). Previous studies have shown that TRAF6 is involved in the NF-κB signaling pathway (Min et al., 2018). However, more studies are warranted to ascertain whether TRAF6 activates NF-κB. Besides TRAF6, TRAF2 is of great importance in NF-κB activation by TNF-α stimulation (Chung et al., 2007). The anti-apoptosis effect of NF-κB is part of the reason for TNF-α mediated avoidance of programmed cell death (Ren et al., 2007). In the heterozygous FLT3-ITD positive MOLM-13 cell line, cells presented a higher susceptibility to TNF-α after TRAF2 knock-down compared with control cells, and induction of apoptosis and impaired proliferation after TNF-α exposure were observed (Table 2) (Schnetzke et al., 2013b). This result proves the potential antiapoptotic role of TRAF2.
FBW7 promotes ubiquitylation and proteasomal degradation of glucocorticoid receptor α through a conserved, GSK3β phosphorylated degron (Malyukova et al., 2013). Glucocorticoid is an essential part of ALL chemotherapy regimens and glucocorticoid sensitivity can be restored through overexpression of glucocorticoid receptor (Schmidt et al., 2006). Malyukova et al. (2013) confirmed that FBW7 inactivation leads to up-regulation of glucocorticoid receptor and enhanced glucocorticoid sensitivity in glucocorticoid-resistant cells. Additionally, FBW7 targets MCL-1 for ubiquitination and degradation in a GSK3-dependent manner (Wertz et al., 2011). FBW7 deletion can up-regulate the expression of MCL-1 in T-ALL cell line, which is an anti-apoptotic protein of the BCL-2 family, causing increased sensitivity to sorafenib (a tyrosine kinase inhibitor) but decreased sensitivity to ABT-737 (a pan-inhibitor of the Bcl-2 family of anti-apoptotic proteins) (Yang-Yen, 2006; Inuzuka et al., 2011). Furthermore, FBW7 mutation is partly the reason for resistance to GSI, which is effective against T-ALL cell lines harboring wild-type PTEN deletion on chromosome ten (Weng et al., 2004; Hales et al., 2014). GSK3β, a multifunctional serine/threonine kinase that is overexpressed and hyperactivated in AML, is considered a tumor promoter in mixed lineage leukemia (MLL) (Wang et al., 2008). Several studies showed that GSK3β inhibition contributes to differentiation and apoptosis of leukemic cells (Zhou et al., 2011; Gupta et al., 2012; Gupta et al., 2016). Mechanically, in AML cell lines THP1 and U937, both GSK3β inhibition and proteasome inhibition can enhance the expression of Purine Rich Box-1 (PU.1), which is a central regulator of the differentiation of all hematopoietic cell lineages (Dakic et al., 2005; Mishra et al., 2021). Suppression of PU.1 leads to leukemic transformation of myeloid cells, indicating that PU.1 plays a tumor suppressor role in myeloid cells (Metcalf et al., 2006). Further experiments indicated that PU.1 is phosphorylated by GSK3β and then recognized by FBW7, resulting in ubiquitination and degradation (Figure 3) (Table 2) (Mishra et al., 2021). Taken together, these studies suggest that targeting the GSK3-FBW7 signaling axis provides a possibility of inhibiting AML growth and induce myeloid differentiation.
As a tumor suppressor, ARF can work through p53 pathway activation but also in a p53-independent form. For ARF-p53 axis, the activation of p53 involves multiple mechanisms. On the one hand, a nucleolar form of ARF can bind to MDM2 to sequester it in the nucleolus, and nucleoplasmic forms of ARF can suppress the ubiquitin ligase activity of MDM2, contributing to p53 activation and stabilization (Weber et al., 1999; Llanos et al., 2001). MDM2, a RING-type E3 ubiquitin ligase, is the main regulator of p53 (Vousden and Prives, 2005). On the other hand, ARF can inhibit the enzyme activity of ARF-BP1 (a HECT type E3 ligase), causing p53 stabilization (Chen et al., 2005; Chen et al., 2006). The ubiquitination and degradation of ARF are mediated by ULF, which can directly interact with the C-terminus of NPM to disrupt the protection role for ARF (Chen et al., 2010b). NPM1, one of the most common genetic mutations in AML, can elevate autophagy activity, which contributes to cell survival in NPM1 mutated AML. NPM mutation is found in 35% of AML cells, leading to cytoplasmic dislocation of nucleophosmin (NPM-c) that cannot retain ARF in the nucleoli (Quentmeier et al., 2005). In NPM-c AML cells, ARF is unstable and can be degraded rapidly, whereas ULF knock-down can stabilize ARF and activate the ARF-p53 axis (Figure 3) (Table 2), making ULF an effective target in AML cells (Chen et al., 2010b).
Wnt/β-catenin signaling is critical for leukemic stem cell maintenance, with dishevelled2 (DVL2) functioning as the core component. DVL2 can disassemble the APC/Axin/CK1α/GSK3β degradation complex under the binding between Wnt ligands and Fzd receptors, leading to β-catenin stabilization; while absence of Wnt ligands made β-catenin degrade in the manner of ubiquitin-proteasome (Nusse and Clevers, 2017). The degradation of β-catenin correlates with the leukemic stem cell, contributing to the promotion of AML (Soares-Lima et al., 2020). Yan et al. (2021) studied the mechanism of nuclear paraspeckle assembly transcript 1 (NEAT1) (which belongs to long non-coding RNAs) in leukemogenesis and leukemic stem cell function, finding that loss of NEAT1_1 promotes murine leukemogenesis. Besides, they confirmed that NEAT1 can enhance the degradation of DVL2 by TRIM56, then inactivating the Wnt signaling pathway. Collectively, these finding indicate that E3 ubiquitin ligase TRIM56 combined with DVL2 participate in the inhibition of the Wnt signaling (Figure 4) (Table 2), in a process that is mediated by NEAT1_1.
Receptor tyrosine kinases (RTKs), which consists of about twenty subfamilies, are implicated in regulating pathways for cell growth, differentiation, adhesion, and cell death. The RTKs commonly referred to as type III, which include c-Kit, CSF1R, FLT3, and PDGFR, have a major impact on leukemogenesis and transformation into AML (Carter et al., 2020). The genetic characteristic of MLL is the chromosome 11q23 abnormality, and this type of leukemia is associated with adverse prognosis. Emergency granulopoiesis is known to accelerate leukemogenesis in MLL1-rearranged AML and this occurs in an RTK-dependent manner (Wang et al., 2020). Moreover, it has been reported that in MLL1-ELL AML mice, TRIAD1 expression is decreased, contributing to sustained RTK signaling and failing to terminate emergency granulopoiesis (Figure 4) (Wang et al., 2018; Wang et al., 2020). Using TRIAD1-substrate RTKs inhibitors can terminate emergency granulopoiesis, delay leukemogenesis during emergency granulopoiesis, and normalize innate immune responses when combined with chemotherapy (Table 2) (Wang et al., 2020). In addition, as a RTK inhibitor, nintedanib can reverse activation (phosphorylation) of AKT in MLL1-ELL+ LIN− cells that are associated with inhibited GSK3β phosphorylation and β-catenin destabilization (Wang et al., 2020).
Autophagy is an intracellular evolutionarily conserved catabolic degradation process, mediated by lysosome sustains (Yu et al., 2018). The process of autophagy is divided into four steps: initiation, nucleation, maturation, and degradation (Feng et al., 2014). The first two steps promote the formation of the autophagic vesicle membrane. During the maturation step, protein conjugation events are necessary for autophagosome formation, involving a series of proteins from the autophagy related gene (ATG) family (Figure 5) (Onorati et al., 2018). LC3-I is formed through the cleavage of LC3 (ATG8) by ATG4. It can conjugate to phosphatidylethanolamine of the autophagosome by ATG3 and ATG7. The level of LC3-II (lipid form of LC3) is proportional to the degree of autophagy (Onorati et al., 2018). According to Sanarico et al. (2018), WWP1 depletion transformed the cytosolic LC3-I to the lipid-bound LC3-II form accompanied by the accumulation of autophagy-associated protein ATG7. Meanwhile, the level of SQSTM1/p62 was decreased, which is a cargo receptor that recruits cargo destined for autophagic degradation to LC3-II on forming autophagasomes (Figure 5) (Sanarico et al., 2018). Overall, WWP1 depletion promotes the autophagic degradation of oncogenic proteins, such as PML-RARα and FLT3-ITD, inducing differentiation of AML cells.
In conclusion, a line of evidence has highlighted the role of E3 ubiquitin ligases in the abnormal activation of leukemic signaling pathways. An interesting observation is that the absence of the E3 ligases is often the key to causing the abnormal signal. For this situation, targeted agonists of E3 ligases such as CC-90009 represent an advance in AML treatment. CC-90009 can co-opt the CUL4-DDB1-CRBN-RBX1 (CRL4CRBN) E3 ubiquitin ligase complex to target G1 to S phase transition 1 (GSPT1) selectively for ubiquitination and proteasomal degradation, inducing AML apoptosis (Hansen et al., 2021). This provides a promising approach to other agonists of E3 ligases. In this regard, it is crucial to clarify what types of factors regulate the expression of E3 ubiquitin ligases because the understanding of upstream mechanisms is beneficial to developing new therapeutic regimens. Additionally, PROTAC can remove unwanted or damaged proteins without the need for a specific target. Whether it is possible to link a normal E3 ligases to a mutant E3 ligase so that the aberrant one can be ubiquitination and proteasomal degradation?
QZ and HZ wrote the manuscript. BW draw the table. QZ drafted the figures. NZ and LZ revised the paper. All authors approved the final vision of the manuscript and the submission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akhoondi S., Sun D., Lehr N., Apostolidou S., Klotz K., Maljukova A., et al. (2007). FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 67 (19), 9006–9012. doi:10.1158/0008-5472.CAN-07-1320
Arkee T., Bishop G. A. (2020). TRAF family molecules in T cells: Multiple receptors and functions. J. Leukoc. Biol. 107 (6), 907–915. doi:10.1002/JLB.2MR1119-397R
Bae S., Jung J. H., An I., Kim O. Y., Lee M. J., Lee J. H., et al. (2012). TRIAD1 is negatively regulated by the MDM2 E3 ligase. Oncol. Rep. 28 (5), 1924–1928. doi:10.3892/or.2012.2005
Bai C., Richman R., Elledge S. J. (1994). Human cyclin F. EMBO J. 13 (24), 6087–6098. doi:10.1002/j.1460-2075.1994.tb06955.x
Barata J. T., Cardoso A. A., Nadler L. M., Boussiotis V. A. (2001). Interleukin-7 promotes survival and cell cycle progression of T-cell acute lymphoblastic leukemia cells by down-regulating the cyclin-dependent kinase inhibitor p27(kip1). Blood 98 (5), 1524–1531. doi:10.1182/blood.v98.5.1524
Baron J., Wang E. S. (2018). Gemtuzumab ozogamicin for the treatment of acute myeloid leukemia. Expert Rev. Clin. Pharmacol. 11 (6), 549–559. doi:10.1080/17512433.2018.1478725
Bengoechea-Alonso M. T., Ericsson J. (2010). The ubiquitin ligase Fbxw7 controls adipocyte differentiation by targeting C/EBPalpha for degradation. Proc. Natl. Acad. Sci. U. S. A. 107 (26), 11817–11822. doi:10.1073/pnas.0913367107
Berndsen C. E., Wolberger C. (2014). New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21 (4), 301–307. doi:10.1038/nsmb.2780
Breccia M., Alimena G. (2010). NF-κB as a potential therapeutic target in myelodysplastic syndromes and acute myeloid leukemia. Expert Opin. Ther. Targets 14 (11), 1157–1176. doi:10.1517/14728222.2010.522570
Buetow L., Huang D. T. (2016). Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 17 (10), 626–642. doi:10.1038/nrm.2016.91
Cao X., Xue L., Han L., Ma L., Chen T., Tong T. (2011). WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) delays cellular senescence by promoting p27(Kip1) degradation in human diploid fibroblasts. J. Biol. Chem. 286 (38), 33447–33456. doi:10.1074/jbc.M111.225565
Carter J. L., Hege K., Yang J., Kalpage H. A., Su Y., Edwards H., et al. (2020). Targeting multiple signaling pathways: The new approach to acute myeloid leukemia therapy. Signal Transduct. Target. Ther. 5 (1), 288. doi:10.1038/s41392-020-00361-x
Chan C., Li C., Yang W., Gao Y., Lee S. W., Feng Z., et al. (2012). The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 149 (5), 1098–1111. doi:10.1016/j.cell.2012.02.065
Chang J. H., Poppe M. M., Hua C., Marcus K. J., Esiashvili N. (2021). Acute lymphoblastic leukemia. Pediatr. Blood Cancer 68 (2), e28371. doi:10.1002/pbc.28371
Checquolo S., Palermo R., Cialfi S., Ferrara G., Oliviero C., Talora C., et al. (2010). Differential subcellular localization regulates c-Cbl E3 ligase activity upon Notch3 protein in T-cell leukemia. Oncogene 29 (10), 1463–1474. doi:10.1038/onc.2009.446
Chen C., Sun X., Guo P., Dong X. Y., Sethi P., Zhou W., et al. (2007). Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene 26 (16), 2386–2394. doi:10.1038/sj.onc.1210021
Chen C., Zhou Z., Liu R., Li Y., Azmi P. B., Seth A. K. (2008). The WW domain containing E3 ubiquitin protein ligase 1 upregulates ErbB2 and EGFR through RING finger protein 11. Oncogene 27 (54), 6845–6855. doi:10.1038/onc.2008.288
Chen D., Brooks C. L., Gu W. (2006). ARF-BP1 as a potential therapeutic target. Br. J. Cancer 94 (11), 1555–1558. doi:10.1038/sj.bjc.6603119
Chen D., Kon N., Li M., Zhang W., Qin J., Gu W. (2005). ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 121 (7), 1071–1083. doi:10.1016/j.cell.2005.03.037
Chen D., Shan J., Zhu W., Qin J., Gu W. (2010). Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature 464 (7288), 624–627. doi:10.1038/nature08820
Chen D., Yoon J., Gu W. (2010). Reactivating the ARF-p53 axis in AML cells by targeting ULF. Cell. Cycle 9 (15), 2946–2951. doi:10.4161/cc.9.15.12355
Chiang M. Y., Radojcic V., Maillard I. (2016). Oncogenic Notch signaling in T-cell and B-cell lymphoproliferative disorders. Curr. Opin. Hematol. 23 (4), 362–370. doi:10.1097/MOH.0000000000000254
Chung J. Y., Lu M., Yin Q., Wu H. (2007). Structural revelations of TRAF2 function in TNF receptor signaling pathway. Adv. Exp. Med. Biol. 597, 93–113. doi:10.1007/978-0-387-70630-6_8
Dakic A., Metcalf D., Rago L. D., Mifsud S., Wu L., Nutt S. L., et al. (2005). PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J. Exp. Med. 201 (9), 1487–1502. doi:10.1084/jem.20050075
Das A., Middleton A. J., Padala P., Ledgerwood E. C., Mace P. D., Day C. L. (2021). The structure and ubiquitin binding properties of TRAF RING heterodimers. J. Mol. Biol. 433 (8), 166844. doi:10.1016/j.jmb.2021.166844
Dohda T., Maljukova A., Liu L., Heyman M., Grander D., Brodin D., et al. (2007). Notch signaling induces SKP2 expression and promotes reduction of p27Kip1 in T-cell acute lymphoblastic leukemia cell lines. Exp. Cell. Res. 313 (14), 3141–3152. doi:10.1016/j.yexcr.2007.04.027
Dunbar A. J., Gondek L. P., O'Keefe C. L., Makishima H., Rataul M. S., Szpurka H., et al. (2008). 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 68 (24), 10349–10357. doi:10.1158/0008-5472.CAN-08-2754
Dunphy K., Dowling P., Bazou D., O'Gorman P. (2021). Current methods of post-translational modification analysis and their applications in blood cancers. Cancers (Basel) 13 (8), 1930. doi:10.3390/cancers13081930
Edwards T. L., Clowes V. E., Tsang H. T., Connell J. W., Sanderson C. M., Luzio J. P., et al. (2009). Endogenous spartin (SPG20) is recruited to endosomes and lipid droplets and interacts with the ubiquitin E3 ligases AIP4 and AIP5. Biochem. J. 423 (1), 31–39. doi:10.1042/BJ20082398
Feng S. M., Muraoka-Cook R. S., Hunter D., Sandahl M. A., Caskey L. S., Miyazawa K., et al. (2009). The E3 ubiquitin ligase WWP1 selectively targets HER4 and its proteolytically derived signaling isoforms for degradation. Mol. Cell.. Biol. 29 (3), 892–906. doi:10.1128/MCB.00595-08
Feng Y., He D., Yao Z., Klionsky D. J. (2014). The machinery of macroautophagy. Cell. Res. 24 (1), 24–41. doi:10.1038/cr.2013.168
Frescas D., Pagano M. (2008). Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the scales of cancer. Nat. Rev. Cancer 8 (6), 438–449. doi:10.1038/nrc2396
Fryer C. J., White J. B., Jones K. A. (2004). Mastermind recruits CycC: CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 16 (4), 509–520. doi:10.1016/j.molcel.2004.10.014
Fukushima H., Matsumoto A., Inuzuka H., Zhai B., Lau A. W., Wan L., et al. (2012). SCF(Fbw7) modulates the NFkB signaling pathway by targeting NFkB2 for ubiquitination and destruction. Cell. Rep. 1, 434–443. doi:10.1016/j.celrep.2012.04.002
Gupta K., Gulen F., Sun L., AguileRa R., ChAkrAbArti A., Kiselar J., et al. (2012). GSK3 is a regulator of RAR-mediated differentiation. Leukemia 26 (6), 1277–1285. doi:10.1038/leu.2012.2
Gupta K., Stefan T., Ignatz-Hoover J., Moreton S., Parizher G., Saunthararajah Y., et al. (2016). GSK-3 inhibition sensitizes acute myeloid leukemia cells to 1, 25d-mediated differentiation. Cancer Res. 76 (9), 2743–2753. doi:10.1158/0008-5472.CAN-15-2290
Hales E. C., Taub J. W., Matherly L. H. (2014). New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: Targeted therapy of γ-secretase inhibitor resistant T-cell acute lymphoblastic leukemia. Cell. Signal. 26 (1), 149–161. doi:10.1016/j.cellsig.2013.09.021
Hansen J. D., Correa M., Alexander M., Nagy M., Huang D., Sapienza J., et al. (2021). CC-90009: A cereblon E3 ligase modulating drug that promotes selective degradation of GSPT1 for the treatment of acute myeloid leukemia. J. Med. Chem. 64 (4), 1835–1843. doi:10.1021/acs.jmedchem.0c01489
Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P. (2007). Structure of a fbw7-skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell. 26 (1), 131–143. doi:10.1016/j.molcel.2007.02.022
Hu X., Yu J., Lin Z., Feng R., Wang Z. W., Chen G. (2021). The emerging role of WWP1 in cancer development and progression. Cell. Death Discov. 7 (1), 163. doi:10.1038/s41420-021-00532-x
Huang H., Ma L., Li J., Yu Y., Zhang D., Wei J., et al. (2014). NF-κB1 inhibits c-Myc protein degradation through suppression of FBW7 expression. Oncotarget 5 (2), 493–505. doi:10.18632/oncotarget.1643
Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., et al. (1999). Structure of an e6ap-UbcH7 complex: Insights into ubiquitination by the E2-E3 enzyme cascade. Science 286 (5443), 1321–1326. doi:10.1126/science.286.5443.1321
Inuzuka H., Shaik S., Onoyama I., Gao D., Tseng A., Maser R. S., et al. (2011). SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471 (7336), 104–109. doi:10.1038/nature09732
Jin J., Cardozo T., Lovering R. C., Elledge S. J., Pagano M., Harper J. W. (2004). Systematic analysis and nomenclature of mammalian F-box proteins. Genes. Dev. 18 (21), 2573–2580. doi:10.1101/gad.1255304
Kamanova J., Sun H., Lara-Tejero M., Galán J. E. (2016). The Salmonella effector protein SopA modulates innate immune responses by targeting TRIM E3 ligase family members. PLoS Pathog. 12 (4), e1005552. doi:10.1371/journal.ppat.1005552
Kimura T., Gotoh M., Nakamura Y., Arakawa H. (2003). hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Sci. 94 (5), 431–436. doi:10.1111/j.1349-7006.2003.tb01460.x
King B., Trimarchi T., Reavie L., Xu L., Mullenders J., Ntziachristos P., et al. (2013). The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 153 (7), 1552–1566. doi:10.1016/j.cell.2013.05.041
Koepp D. M., Schaefer L. K., Ye X., Keyomarsi K., Chu C., Harper J. W., et al. (2001). Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294, 173–177. doi:10.1126/science.1065203
Kourtis N., Moubarak R. S., Aranda-Orgilles B., Lui K., Aydin I. T., Trimarchi T., et al. (2015). FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat. Cell. Biol. 17 (3), 322–332. doi:10.1038/ncb3121
Lamothe B., Campos A. D., Webster W. K., Gopinathan A., Hur L., Darnay B. G. (2008). The RING domain and first zinc finger of TRAF6 coordinate signaling by interleukin-1, lipopolysaccharide, and RANKL. J. Biol. Chem. 283 (36), 24871–24880. doi:10.1074/jbc.M802749200
Levin A. G., Rivière I., Eshhar Z., Sadelain M. (2021). CAR T cells: Building on the CD19 paradigm. Eur. J. Immunol. 51 (9), 2151–2163. doi:10.1002/eji.202049064
Li L., Sarver A. L., Khatri R., Hajeri P. B., Kamenev I., French A. J., et al. (2014). Sequential expression of miR-182 and miR-503 cooperatively targets FBXW7, contributing to the malignant transformation of colon adenoma to adenocarcinoma. J. Pathol. 234 (4), 488–501. doi:10.1002/path.4407
Li Y., Zhou Z., Chen C. (2008). WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell. Death Differ. 15 (12), 1941–1951. doi:10.1038/cdd.2008.134
Li Z., Younger K., Gartenhaus R., Joseph A. M., Hu F., Baer M. R., et al. (2015). Inhibition of IRAK1/4 sensitizes T cell acute lymphoblastic leukemia to chemotherapies. J. Clin. Investig.. 125 (3), 1081–1097. doi:10.1172/JCI75821
Llanos S., Clark P. A., Rowe J., Peters G. (2001). Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat. Cell. Biol. 3 (5), 445–452. doi:10.1038/35074506
Lochab S., Pal P., Kapoor I., Kanaujiya J. K., Sanyal S., Behre G., et al. (2013). E3 ubiquitin ligase Fbw7 negatively regulates granulocytic differentiation by targeting G-CSFR for degradation. Biochim. Biophys. Acta 1833 (12), 2639–2652. doi:10.1016/j.bbamcr.2013.06.018
Lomaga M. A., Yeh W. C., Sarosi I., Duncan G. S., Furlonger C., Ho A., et al. (1999). TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes. Dev. 13 (8), 1015–1024. doi:10.1101/gad.13.8.1015
Lv K., Jiang J., Donaghy R., Riling C. R., Cheng Y., Chandra V., et al. (2017). CBL family E3 ubiquitin ligases control JAK2 ubiquitination and stability in hematopoietic stem cells and myeloid malignancies. Genes. Dev. 31 (10), 1007–1023. doi:10.1101/gad.297135.117
Lyle C. L., Belghasem M., Chitaliac-Cbl V. C. (2019). c-Cbl: An important regulator and a target in angiogenesis and tumorigenesis. Cells 8 (5), 498. doi:10.3390/cells8050498
Makishima H., Cazzolli H., Szpurka H., Dunbar A., Tiu R., Huh J., et al. (2009). Mutations of e3 ubiquitin ligase cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J. Clin. Oncol. 27 (36), 6109–6116. doi:10.1200/JCO.2009.23.7503
Malyukova A., Brown S., Papa R., O'Brien R., Giles J., Trahair T. N., et al. (2013). FBXW7 regulates glucocorticoid response in T-cell acute lymphoblastic leukaemia by targeting the glucocorticoid receptor for degradation. Leukemia 27 (5), 1053–1062. doi:10.1038/leu.2012.361
Mansour M. A. (2018). Ubiquitination: Friend and foe in cancer. Int. J. Biochem. Cell. Biol. 101, 80–93. doi:10.1016/j.biocel.2018.06.001
Mansour M. R., Sanda T., Lawton L. N., Li X., Kreslavsky T., Novina C. D., et al. (2013). The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J. Exp. Med. 210 (8), 1545–1557. doi:10.1084/jem.20122516
Mao J., Perez-Losada J., Wu D., Delrosario R., Tsunematsu R., Nakayama K. I., et al. (2004). Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 432 (7018), 775–779. doi:10.1038/nature03155
Marteijn J. A. F., Emst L., Erpelinck-Verschueren C. A. J., Nikoloski G., Menke A., de Witte T., et al. (2005). The E3 ubiquitin-protein ligase Triad1 inhibits clonogenic growth of primary myeloid progenitor cells. Blood 106 (13), 4114–4123. doi:10.1182/blood-2005-04-1450
Matsuoka S., Oike Y., Onoyama I., Iwama A., Arai F., Takubo K., et al. (2008). Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes. Dev. 22 (8), 986–991. doi:10.1101/gad.1621808
Meng W., Sawasdikosol S., Burakoff S. J., Eck M. J. (1999). Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature 398 (6722), 84–90. doi:10.1038/18050
Meroni G., Diez-Roux G. (2005). TRIM/RBCC, a novel class of 'single protein RING finger' E3 ubiquitin ligases. Bioessays 27 (11), 1147–1157. doi:10.1002/bies.20304
Metcalf D., Dakic A., Mifsud S., Di Rago L., Wu L., Nutt S. (2006). Inactivation of PU.1 in adult mice leads to the development of myeloid leukemia. Proc. Natl. Acad. Sci. U. S. A. 103 (5), 1486–1491. doi:10.1073/pnas.0510616103
Min Y., Kim M. J., Lee S., Chun E., Lee K. Y. (2018). Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy 14 (8), 1347–1358. doi:10.1080/15548627.2018.1474995
Mishra M., Thacker G., Sharma A., Singh A. K., Upadhyay V., Sanyal S., et al. (2021). FBW7 inhibits myeloid differentiation in acute myeloid leukemia via GSK3-dependent ubiquitination of PU.1. Mol. Cancer Res. 19 (2), 261–273. doi:10.1158/1541-7786.MCR-20-0268
Mohapatra B., Ahmad G., Nadeau S., Zutshi N., An W., Scheffe S., et al. (2013). Protein tyrosine kinase regulation by ubiquitination: Critical roles of cbl-family ubiquitin ligases. Biochim. Biophys. Acta 1833 (1), 122–139. doi:10.1016/j.bbamcr.2012.10.010
Moon S., Lee B. (2018). Chemically induced cellular proteolysis: An emerging therapeutic strategy for undruggable targets. Mol. Cells 41 (11), 933–942. doi:10.14348/molcells.2018.0372
Nakagawa T., Nakayama K., Nakayama K. I. (2020). Knockout Mouse models provide insight into the biological functions of CRL1 components. Adv. Exp. Med. Biol. 1217, 147–171. doi:10.1007/978-981-15-1025-0_10
Nandi D., Tahiliani P., Kumar A., Chandu D. (2006). The ubiquitin-proteasome system. J. Biosci. 31 (1), 137–155. doi:10.1007/BF02705243
Nepstad I., Hatfield K. J., Grønningsæter I. S., Reikvam H. (2020). The PI3K-Akt-mTOR signaling pathway in human acute myeloid leukemia (AML) cells. Int. J. Mol. Sci. 21 (8), 2907. doi:10.3390/ijms21082907
Newell L. F., Cook R. J. (2021). Advances in acute myeloid leukemia. BMJ 375, n2026. doi:10.1136/bmj.n2026
Nguyen K. M., Busino L. (2020). The biology of F-box proteins: The SCF family of E3 ubiquitin ligases. Adv. Exp. Med. Biol. 1217, 111–122. doi:10.1007/978-981-15-1025-0_8
Nusse R., Clevers H. (2017). Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell. 169 (6), 985–999. doi:10.1016/j.cell.2017.05.016
O'Neil J., Grim J., Strack P., Rao S., Tibbitts D., Winter C., et al. (2007). FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 204 (8), 1813–1824. doi:10.1084/jem.20070876
Oliner J. D., Saiki A. Y., Caenepeel S. (2016). The role of MDM2 amplification and overexpression in tumorigenesis. Cold Spring Harb. Perspect. Med. 6 (6), a026336. doi:10.1101/cshperspect.a026336
Onorati A. V., Dyczynski M., Ojha R., Amaravadi R. K. (2018). Targeting autophagy in cancer. Cancer 124 (16), 3307–3318. doi:10.1002/cncr.31335
Park I., Blum W., Baker S. D., Caligiuri M. A. (2017). E3 ubiquitin ligase Cbl-b activates the p53 pathway by targeting Siva1, a negative regulator of ARF, in FLT3 inhibitor-resistant acute myeloid leukemia. Leukemia 31, 502. doi:10.1038/leu.2016.293
Quentmeier H., Martelli M. P., Dirks W. G., BolliN. , Liso A., Macleod R. A. F., et al. (2005). Cell line OCI/AML3 bears exon-12 NPM gene mutation-A and cytoplasmic expression of nucleophosmin. Leukemia 19 (10), 1760–1767. doi:10.1038/sj.leu.2403899
Rajagopalan H., Jallepalli P. V., Rago C., Velculescu V. E., Kinzler K. W., Vogelstein B., et al. (2004). Inactivation of hCDC4 can cause chromosomal instability. Nature 428 (6978), 77–81. doi:10.1038/nature02313
Rathinam C., Thien C. B. F., Flavell R. A., Langdon W. Y. (2010). Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell. 18 (4), 341–352. doi:10.1016/j.ccr.2010.09.008
Reijden B. A., Erpelinck-Verschueren V. A., Löwenberg B., Jansen J. H. (1999). TRIADs: A new class of proteins with a novel cysteine-rich signature. Protein Sci. 8 (7), 1557–1561. doi:10.1110/ps.8.7.1557
Ren X., Xu Z., Myers J. N., Wu X. (2007). Bypass NFkappaB-mediated survival pathways by TRAIL and Smac. Cancer Biol. Ther. 6 (7), 1031–1035. doi:10.4161/cbt.6.7.4206
Rodriguez S., Abundis C., Boccalatte F., Mehrotra P., Chiang M. Y., Yui M. A., et al. (2020). Therapeutic targeting of the E3 ubiquitin ligase SKP2 in T-ALL. Leukemia 34 (5), 1241–1252. doi:10.1038/s41375-019-0653-z
Rotin D., Kumar S. (2009). Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 10 (6), 398–409. doi:10.1038/nrm2690
Sakamoto K. M., Kim K. B., Kumagai A., Crews C. M., Deshaies R. J. (2001). Protacs: Chimeric molecules that target proteins to the skp1-cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. U. S. A. 98 (15), 8554–8559. doi:10.1073/pnas.141230798
Sanarico A. G., Ronchini C., Croce A., Memmi E. M., Cammarata U. A., De Antoni A., et al. (2018). The E3 ubiquitin ligase WWP1 sustains the growth of acute myeloid leukaemia. Leukemia 32 (4), 911–919. doi:10.1038/leu.2017.342
Schmidt S., Irving J. A. E., Minto L., Matheson E., Nicholson L., Ploner A., et al. (2006). Glucocorticoid resistance in two key models of acute lymphoblastic leukemia occurs at the level of the glucocorticoid receptor. FASEB J. 20 (14), 2600–2602. doi:10.1096/fj.06-6214fje
Schnetzke U., Fischer M., Kuhn A., Spies-Weisshart B., Zirm E., Hochhaus A., et al. (2013). The E3 ubiquitin ligase TRAF6 inhibits LPS-induced AKT activation in FLT3-ITD-positive MV4-11 AML cells. J. Cancer Res. Clin. Oncol. 139 (4), 605–615. doi:10.1007/s00432-012-1362-4
Schnetzke U., Fischer M., Spies-Weisshart B., Zirm E., Hochhaus A., Muller J. P., et al. (2013). The E3 ubiquitin ligase TRAF2 can contribute to TNF-α resistance in FLT3-ITD-positive AML cells. Leuk. Res. 37 (11), 1557–1564. doi:10.1016/j.leukres.2013.08.004
Shen R., Chen M., Wang Y., Kaneki H., Xing L., O'keefe R. J., et al. (2006). Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. J. Biol. Chem. 281 (6), 3569–3576. doi:10.1074/jbc.M506761200
Singh S., Ng J., Sivaraman J. (2021). Exploring the "Other" subfamily of HECT E3-ligases for therapeutic intervention. Pharmacol. Ther. 224, 107809. doi:10.1016/j.pharmthera.2021.107809
Singh S., Sivaraman J. (2020). Crystal structure of HECT domain of UBE3C E3 ligase and its ubiquitination activity. Biochem. J. 477 (5), 905–923. doi:10.1042/BCJ20200027
Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 91 (2), 209–219. doi:10.1016/s0092-8674(00)80403-1
Soares-Lima S. C., Pombo-de-Oliveira M. S., Carneiro F. R. G. (2020). The multiple ways Wnt signaling contributes to acute leukemia pathogenesis. J. Leukoc. Biol. 108 (4), 1081–1099. doi:10.1002/JLB.2MR0420-707R
Tang X., Orlicky S., Lin Z., Willems A., Neculai D., Ceccarelli D., et al. (2007). Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 129 (6), 1165–1176. doi:10.1016/j.cell.2007.04.042
Taylor S. J., Thien C. B. F., Dagger S. A., Duyvestyn J. M., Grove C. S., Lee B. H., et al. (2015). Loss of c-Cbl E3 ubiquitin ligase activity enhances the development of myeloid leukemia in FLT3-ITD mutant mice. Exp. Hematol. 43 (3), 191–206. doi:10.1016/j.exphem.2014.11.009
Thien C. B. F., Langdon W. Y. (2005). Negative regulation of PTK signalling by Cbl proteins. Growth factors. 23 (2), 161–167. doi:10.1080/08977190500153763
Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y., et al. (2010). The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33 (5), 765–776. doi:10.1016/j.immuni.2010.10.013
Vara J. A. F., Casado E., Castro J., Cejas P., Belda-Iniesta C., Gonzalez-Baron M. (2004). PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 30 (2), 193–204. doi:10.1016/j.ctrv.2003.07.007
Versteeg G. A., Rajsbaum R., Sánchez-Aparicio M. T., Maestre A. M., Valdiviezo J., Shi M., et al. (2013). The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 38 (2), 384–398. doi:10.1016/j.immuni.2012.11.013
Verstrepen L., Bekaert T., Chau T., Tavernier J., ChAriot A., BeyaeRt R. (2008). TLR-4, IL-1R and TNF-R signaling to NF-kappaB: Variations on a common theme. Cell.. Mol. Life Sci. 65 (19), 2964–2978. doi:10.1007/s00018-008-8064-8
Vousden K. H., Prives C. (2005). P53 and prognosis: New insights and further complexity. Cell. 120 (1), 7–10. doi:10.1016/j.cell.2004.12.027
W1Komuro A., Imamura T., Saitoh M., Yoshida Y., Yamori T., Miyazono K., et al. (2004). Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1). Oncogene 23 (41), 6914–6923. doi:10.1038/sj.onc.1207885
Wang D., Ma L., Wang B., Liu J., Wei W. (2017). E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 36 (4), 683–702. doi:10.1007/s10555-017-9703-z
Wang H., Bei L., Shah C. A., Huang W., Platanias L. C., Eklund E. A. (2018). The E3 ubiquitin ligase TRIAD1 influences development of Mll-Ell-induced acute myeloid leukemia. Oncogene 37 (19), 2532–2544. doi:10.1038/s41388-018-0131-5
Wang H., Shah C. A., Hu L., Huang W., Platanias L. C., Eklund E. A. (2020). An aberrantly sustained emergency granulopoiesis response accelerates postchemotherapy relapse in MLL1-rearranged acute myeloid leukemia in mice. J. Biol. Chem. 295 (28), 9663–9675. doi:10.1074/jbc.RA120.013206
Wang H., Yang Z., Liu C., Huang S., Wang H., Chen Y., et al. (2014). RBP-J-interacting and tubulin-associated protein induces apoptosis and cell cycle arrest in human hepatocellular carcinoma by activating the p53-Fbxw7 pathway. Biochem. Biophys. Res. Commun. 454 (1), 71–77. doi:10.1016/j.bbrc.2014.10.023
Wang Z., Smith K. S., Murphy M., Piloto O., Somervaille T. C. P., Cleary M. L. (2008). Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature 455 (7217), 1205–1209. doi:10.1038/nature07284
Weber J. D., Taylor L. J., Roussel M. F., Sherr C. J., Bar-Sagi D. (1999). Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell. Biol. 1 (1), 20–26. doi:10.1038/8991
Wei W., Jin J., Schlisio S., Harper J. W., Kaelin W. G. (2005). The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 8 (1), 25–33. doi:10.1016/j.ccr.2005.06.005
Weng A. P., Ferrando A. A., Lee W., Morris J. P., Silverman L. B., Sanchez-Irizarry C., et al. (2004). Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306 (5694), 269–271. doi:10.1126/science.1102160
Weng A. P., Millholland J. M., Yashiro-Ohtani Y., Arcangeli M. L., Lau A., Wai C., et al. (2006). c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes. Dev. 20 (15), 2096–2109. doi:10.1101/gad.1450406
Wertz I. E., Kusam S., Lam C., Okamoto T., Sandoval W., Anderson D. J., et al. (2011). Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471 (7336), 110–114. doi:10.1038/nature09779
Wijk S. J., Fulda S., Dikic I., Heilemann M. (2019). Visualizing ubiquitination in mammalian cells. EMBO Rep. 20 (2), e46520. doi:10.15252/embr.201846520
Xiang J., Hang J. B., Che J. M., Li H. C. (2015). MiR-25 is up-regulated in non-small cell lung cancer and promotes cell proliferation and motility by targeting FBXW7. Int. J. Clin. Exp. Pathol. 8 (8), 9147–9153. eCollection 2015.
Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., et al. (2004). Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO 23, 2116–2125. doi:10.1038/sj.emboj.7600217
Yan H., Wang Z., Sun Y., Hu L., Bu P. (2021). Cytoplasmic NEAT1 suppresses AML stem cell self-renewal and leukemogenesis through inactivation of Wnt signaling. Adv. Sci. 8 (22), e2100914. doi:10.1002/advs.202100914
Yang-Yen H. (2006). Mcl-1: A highly regulated cell death and survival controller. J. Biomed. Sci. 13 (2), 201–204. doi:10.1007/s11373-005-9064-4
Yao Q., Zhang L., Wang Y., Liu J., Yang L. (2021). LncRNA UCA1 elevates the resistance of human leukemia cells to daunorubicin by the PI3K/AKT pathway via sponging miR-613. Biosci. Rep. 41 (6), BSR20201389. doi:10.1042/BSR20201389
Yilmaz M., Kantarjian H., Ravandi F. (2021). Acute promyelocytic leukemia current treatment algorithms. Blood Cancer J. 11 (6), 123. doi:10.1038/s41408-021-00514-3
Yu L., Chen Y., Tooze S. A. (2018). Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14 (2), 207–215. doi:10.1080/15548627.2017.1378838
Zhang S., Lawless V. A., Kaplan M. H. (2000). Cytokine-stimulated T lymphocyte proliferation is regulated by p27Kip1. J. Immunol. 165 (11), 6270–6277. doi:10.4049/jimmunol.165.11.6270
Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000). Structure of a c-cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 102 (4), 533–539. doi:10.1016/s0092-8674(00)00057-x
Zhou C., Shen L., Mao L., Wang B., Li Y., Yu H. (2015). miR-92a is upregulated in cervical cancer and promotes cell proliferation and invasion by targeting FBXW7. Biochem. Biophys. Res. Commun. 458 (1), 63–69. doi:10.1016/j.bbrc.2015.01.066
Zhou F., Zhang L., Laar T., van Dam H., Ten Dijke P. (2011). GSK3β inactivation induces apoptosis of leukemia cells by repressing the function of c-Myb. Mol. Biol. Cell. 22 (18), 3533–3540. doi:10.1091/mbc.E11-06-0483
Zhu H., Gao F. (2019). Regulatory molecules and corresponding processes of BCR-ABL protein degradation. J. Cancer 10 (11), 2488–2500. doi:10.7150/jca.29528
Keywords: E3 ubiquitin ligases, acute leukemia, Notch, JAK2, NF-κB, PI3K/AKT, Wnt/β-catenin
Citation: Zhan Q, Zhang H, Wu B, Zhang N and Zhang L (2022) E3 ubiquitin ligases in the acute leukemic signaling pathways. Front. Physiol. 13:1004330. doi: 10.3389/fphys.2022.1004330
Received: 27 July 2022; Accepted: 28 October 2022;
Published: 11 November 2022.
Edited by:
Victor M. Bolanos-Garcia, Oxford Brookes University, United KingdomReviewed by:
Kevin Rouault-Pierre, Queen Mary University of London, United KingdomCopyright © 2022 Zhan, Zhang, Wu, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijun Zhang, bHpoYW5nMjAyMDAzQDE2My5jb20=; Naijin Zhang, bmp6aGFuZ0BjbXUuZWR1LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.