
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 23 December 2021
Sec. Aquatic Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.809529
Despite substantial research on neuronal circuits in nudipleuran gastropods, few peptides have been implicated in nudipleuran behavior. In this study, we expanded the understanding of peptides in this clade, using three species with well-studied nervous systems, Hermissenda crassicornis, Melibe leonina, and Pleurobranchaea californica. For each species, we performed sequence homology analysis of de novo transcriptome predictions to identify homologs to 34 of 36 prohormones previously characterized in the gastropods Aplysia californica and Lymnaea stagnalis. We then used single-cell mass spectrometry to characterize peptide profiles in homologous feeding interneurons: the multifunctional ventral white cell (VWC) in P. californica and the small cardioactive peptide B large buccal (SLB) cells in H. crassicornis and M. leonina. The neurons produced overlapping, but not identical, peptide profiles. The H. crassicornis SLB cells expressed peptides from homologs to the FMRFamide (FMRFa), small cardioactive peptide (SCP), LFRFamide (LFRFa), and feeding circuit activating peptides prohormones. The M. leonina SLB cells expressed peptides from homologs to the FMRFa, SCP, LFRFa, and MIP-related peptides prohormones. The VWC, previously shown to express peptides from the FMRFa and QNFLa (a homolog of A. californica pedal peptide 4) prohormones, was shown to also contain SCP peptides. Thus, each neuron expressed peptides from the FMRFa and SCP families, the H. crassicornis and M. leonina SLB cells expressed peptides from the LFRFa family, and each neuron contained peptides from a prohormone not found in the others. These data suggest each neuron performs complex co-transmission, which potentially facilitates a multifunctional role in feeding. Additionally, the unique feeding characteristics of each species may relate, in part, to differences in the peptide profiles of these neurons. These data add chemical insight to enhance our understanding of the neuronal basis of behavior in nudipleurans and other gastropods.
Due to their relatively simple nervous systems and individually identifiable neurons, nudipleuran gastropods have yielded considerable insight into the neuronal basis of behavior (Katz and Quinlan, 2019). The neuronal circuits governing certain behaviors are well described (Elliott and Susswein, 2002; Crow, 2004; Gillette and Brown, 2015; Sakurai and Katz, 2015), yet there is relatively little information on the peptides and hormones regulating these circuits. Only one nudipleuran, Tritonia diomedea, has been the subject of a broad scale peptidomic study (Senatore et al., 2015) and physiological studies have largely focused on only three peptides: pedal peptide (Beck et al., 2000; Baltzley et al., 2011), FMRFamide (FMRFa; Lillvis et al., 2012; Webber et al., 2017), and one of the small cardioactive peptides (SCP; Watson and Willows, 1992; Lillvis et al., 2012; Watson et al., 2020). Because 100s of peptides and hormones are present in many species (Ma et al., 2009; Cafe-Mendes et al., 2014; Gan et al., 2015; Christie and Pascual, 2016; Van Camp et al., 2017), and peptidergic signaling is diverse and essential to animal behavior (Liu et al., 2008), the limited peptidomic insight constitutes a considerable gap in our understanding of nudipleuran physiology. Research on other gastropods further highlights this gap. Numerous peptides and peptide prohormones have been identified in the gastropods Aplysia californica and Lymnaea stagnalis (Hummon et al., 2003b; Di Cosmo and Di Cristo, 2006; Feng et al., 2009; Cropper et al., 2018; Wood et al., 2021), and recent large-scale transcriptomic studies have examined peptides in the snails Theba pisana (Adamson et al., 2015), Deroceras reticulatum (Ahn et al., 2017), and Charonia tritonis (Bose et al., 2017). Identifying more peptides in nudipleurans will help to rectify this deficit.
Nudipleurans also permit analysis of individual, homologous neurons, that is, neurons with conserved neuroanatomy and function (Bulloch and Ridgway, 1995; Sakurai and Katz, 2019). By comparing homologous neurons one can examine the evolution of behavior at the level of a single neuron (Croll, 1987), and moreover, enables examination of traits at the level of a clade rather than a single species, revealing trends in specific traits (Jourjine and Hoekstra, 2021). In most species, it is difficult to reproducibly identify individual neurons, but nudipleurans, with their large neuronal cell bodies, simple behaviors, and relatively few neurons (less than 10,0000; Boyle et al., 1983), allow for such identification (Katz and Quinlan, 2019). Additionally, the large neuronal sizes are well suited for single-cell peptidomic analysis, performed using matrix-assisted laser desorption/ionization (MALDI)-time-of-flight (TOF) mass spectrometry (MS; Garden et al., 1996; Li et al., 2000a). Several homologous neurons have been characterized across nudipleurans (Baltzley et al., 2011; Lillvis et al., 2012; Newcomb et al., 2012), and detailed analyses have uncovered the subtle differences in their circuit-level roles (Katz, 2016; Sakurai and Katz, 2019). Characterizing the peptide profiles of homologous neurons is a logical next step to this research.
This study characterized peptides in both homologous neurons and central nervous system (CNS) transcriptomes for three nudipleuran sea slugs, Hermissenda crassicornis, Melibe leonina, and Pleurobranchaea californica. Each species has been the subject of extensive neurophysiological research (Crow and Tian, 2006; Sakurai et al., 2014; Gillette and Brown, 2015), has a publicly deposited CNS transcriptome, and an easily identified, homologous feeding interneuron in its buccal ganglion. In each species, this neuron drives fictive feeding rhythms and extends axons to extensively innervate the esophagus. In P. californica, this cell, the ventral white cell (VWC), has been shown to be multifunctional (Gillette et al., 1980; Gillette and Gillette, 1983), playing both a command and a motor role in feeding behavior. In H. crassicornis and M. leonina, these are the SCPB large buccal (SLB) cells, which stain for SCPB (Watson and Willows, 1992). Furthermore, each species has unique feeding characteristics that can be correlated with peptide identities. M. leonina lacks a buccal mass or radula for food breakdown, is a filter feeder, engages in hours-long feeding bouts, and has only approximately 40 neurons in its buccal ganglion (Watson and Trimarchi, 1992; Lee and Watson, 2016). H. crassicornis feeds primarily on polyps and jellyfish (Hoover et al., 2012), and P. californica is an active, generalist predator that indulges in cannibalism (Noboa and Gillette, 2013). These species thus allow for a meaningful comparison of homologous interneurons, using existing resources for rapid annotation of their transcriptomes.
For prohormone annotation, we obtained the CNS de novo transcriptome assemblies because no genomic assemblies for H. crassicornis, M. leonina, and P. californica are available. Using 36 known A. californica and L. stagnalis prohormones, we identified homologous prohormones within each species, and from these compiled a library of putative encoded peptides for each species. We then performed single-cell MS on individual VWCs and SLB cells and used the peptide library to perform peptide mass fingerprinting (Thiede et al., 2005) on the resulting spectra. In H. crassicornis we detected peptides from homologs to the FMRFa, SCP, LFRFamide (LFRFa), and feeding circuit activating peptides (FCAP) prohormones, and in M. leonina, peptides from homologs to the FMRFa, SCP, LFRFa, and myoinhibitory peptide (MIP)-related prohormones. The P. californica VWC has previously been shown to have peptides from the FMRFa and QNFLa [a homolog of the A. californica pedal peptide 4; (Green et al., 2018)] prohormones, and we found that it also contains peptides from the SCP prohormone. Thus, each species’ neuron expressed peptides from both the FMRFa and SCP prohormones, and both H. crassicornis and M. leonina expressed peptides from the LFRFa prohormone; however, each neuron also expressed peptides not seen in the others. This work provides an untargeted peptidomic characterization of single homologous neurons and a large-scale prohormone annotation of multiple nudipleuran sea slugs.
P. californica and H. crassicornis were trapped by the Monterey Abalone Company (Monterey Bay, CA) and shipped overnight to the University of Illinois Urbana-Champaign. There they were housed individually in artificial seawater at 12°C. M. leonina were collected at the San Juan Islands, either off the docks of Friday Harbor Labs (San Juan, WA), or via snorkel/scuba diving at Park’s Bay (Shaw Island, WA), and maintained at Friday Harbor Labs in sea tables with flow-through sea water.
For each species, prohormone annotations were performed on publicly available de novo RNA transcriptome assemblies from the NCBI Sequence Read Archive (Kodama et al., 2012; Christie, 2017; Southey et al., 2020). Species-specific information is as follows: H. crassicornis – SRR1719366 (Goodheart et al., 2017), M. leonina – SRR1950947 and SRR3738852 (Goodheart et al., 2017), and P. californica – SRR026692, SRR026693, SRR026694, SRR026695, SRR1505130, and SRR3928990 (Zapata et al., 2014). For each experimental data set, de novo assemblies were created without any preprocessing of reads using MEGAHIT (Li et al., 2015), SOAPdenovo (Luo et al., 2012), and Trinity (Grabherr et al., 2011; Haas et al., 2013) with default settings. De novo assemblies from the same species were combined into a single BLAST (Altschul et al., 1997) database.
For each species, A TBLASTN search was performed on a database of de novo assemblies for 34 A. californica and two L. stagnalis neuropeptide prohormones obtained from the UniProt database (Apweiler et al., 2004), yielding both RNA and protein matches. RNA matches were translated using the ExPaSy “Translate” tool (Gasteiger et al., 2003), and the longest predicted protein sequence from the matched region was selected for further analysis. SignalP 5.0 (Armenteros et al., 2019) and Phobius (Kall et al., 2007) were then used to analyze both translated proteins and direct protein sequences for the presence of a signal sequence, which is required for targeting into the secretory pathway (Rusch and Kendall, 1995). Finally, potential neuropeptides from each matching protein, whether complete (i.e., possessing a signal sequence) or not, were predicted using NeuroPred (Southey et al., 2006a,b, 2008) with the Mollusc model (Hummon et al., 2003a) and common PTMs selected. The resulting predicted peptides were compiled to form a putative peptide library for each species.
We followed prior approaches (Li et al., 2000a) for single-cell isolation and MALDI MS characterization. Subjects were pinned out in dissecting trays, and buccal ganglia were surgically removed. Ganglia were then incubated for 6 min in 1% type 14 protease prepared in saline (460 mm NaCl, 10 mm KCl, 10 mm CaCl2, 25 mm MgCl2, 25 mm MgSO4−, 10 mm HEPES, pH = 7.6), which loosened the surrounding connective sheath. Moria scissors were then used to cut through the connective sheath (one layer in M. leonina and H. crassicornis, two layers in P. californica) to expose the neurons. Neurons were identified visually by their distinct morphology, color, and landmark location within the ganglion (Gillette et al., 1980; Watson and Willows, 1992), and then carefully teased away from the rest of the ganglion using either pulled glass capillaries or tungsten needles. Isolated neurons were then aspirated into a custom-made transfer pipette and spotted onto a ground steel MALDI sample plate (Bruker Corp., Billerica, MA), and 0.5 μl of matrix solution (dihydroxybenzoic acid, 20 mg/ml in deionized water) was applied to each neuron. Following drying and matrix crystallization, samples were analyzed by MALDI-TOF MS using an ultrafleXtreme mass spectrometer (Bruker Corp.) in positive reflectron mode, with a surveyed mass range of 530–5,000 m/z and external calibration. Once the spectra had been collected, detected masses were matched to those in the peptide libraries by peptide mass fingerprinting (Thiede et al., 2005) with an allowed mass match error of 200 ppm.
De novo transcriptomes of the three nudipleurans were queried against 34 A. californica and two L. stagnalis neuropeptide prohormones to identify 35 transcripts in H. crassicornis and M. leonina, and 34 P. californica (Table 1; Supplementary Tables S1–S3). This included two SCP prohormones in H. crassicornis and M. leonina, and two M. leonina temptin proteins. Two proteins, A. californica attractin and egg-laying hormone, were searched but did not yield matches in any of the three species. The majority (27 in H. crassicornis, 27 in M. leonina, and 26 in P. californica) contained a predicted signal sequence. However, only 19, 17, and 12 transcripts of H. crassicornis, M. leonina, and P. californica, respectively, had sequence lengths of at least 95% of A. californica and L. stagnalis neuropeptide prohormone sequence lengths. Each prohormone encoded peptides homologous to those found in the A. californica/L. stagnalis versions of the prohormone.
Further analysis of the transcriptomes found two protein isoforms for the SCP prohormone in H. crassicornis and four in M. leonina (Figure 1). The M. leonina isoforms all expressed identical signal sequences and both SCPA and SCPB neuropeptides, and three of the four shared the same 94 C-terminal amino acids. Two of the isoforms also expressed a novel peptide, SCPC, which differed from the other SCPs in that it has a serine at the C-terminus, lacked amidation, and is 10 amino acids long rather than nine. However, it retains the YXXFPRM motif seen in all other SCPs, including those found in A. californica (P09892), L. stagnalis (O97374), the snail T. pisana (A0A0S1RSH0), and the snail D. reticulatum (A0A1X9WEF6; Figure 1). Although SCPC has not been observed in any of the above species, it was identified in one of two SCP protein isoforms in T. diomedea.

Figure 1. Multiple sequence alignment of SCP prohormones across species and predicted protein isoforms. Melibe leonina yielded four SCP prohormone isoforms, and Hermissenda crassicornis and Tritonia diomedea each yielded two. Each prohormone encoded SCPB (orange highlight) and SCPA (green highlight), and one T. diomedea and two M. leonina isoforms encoded SCPC (blue highlight). Conserved cysteine (red font) and predicted or known cleavage sites (purple font) are also indicated.
Mass spectrometric analysis of individual SLB cells found that the peptides SCPA and SCPB, encoded by species-specific homologues of the A. californica SCP prohormone (UniProt accession number: P09892), were present (Figures 2, 3). The M. leonina SLB cells contained two other peptides encoded by the SCP prohormone (GGCA01092244.1), including SCPC. In addition, peptides encoded by several other neuropeptide genes co-localized in the SLB cells, although combinations differed by species (Table 2). The H. crassicornis SLB cells contained FMRFa and peptides from homologs to the L. stagnalis LFRFa prohormone (Q5U900) (Supplementary Figure S1) and A. californica FCAP prohormone (Q8ISH7) (Supplementary Figure S2). The H. crassicornis LFRFa prohormone encodes five different peptides with a conserved LFRFa motif and amidated C-terminus (Supplementary Figure S1), and each was present within the H. crassicornis SLB cells. Finally, the H. crassicornis FCAP prohormone encoded three structurally similar peptides, and each was confirmed by MS in the neuron.
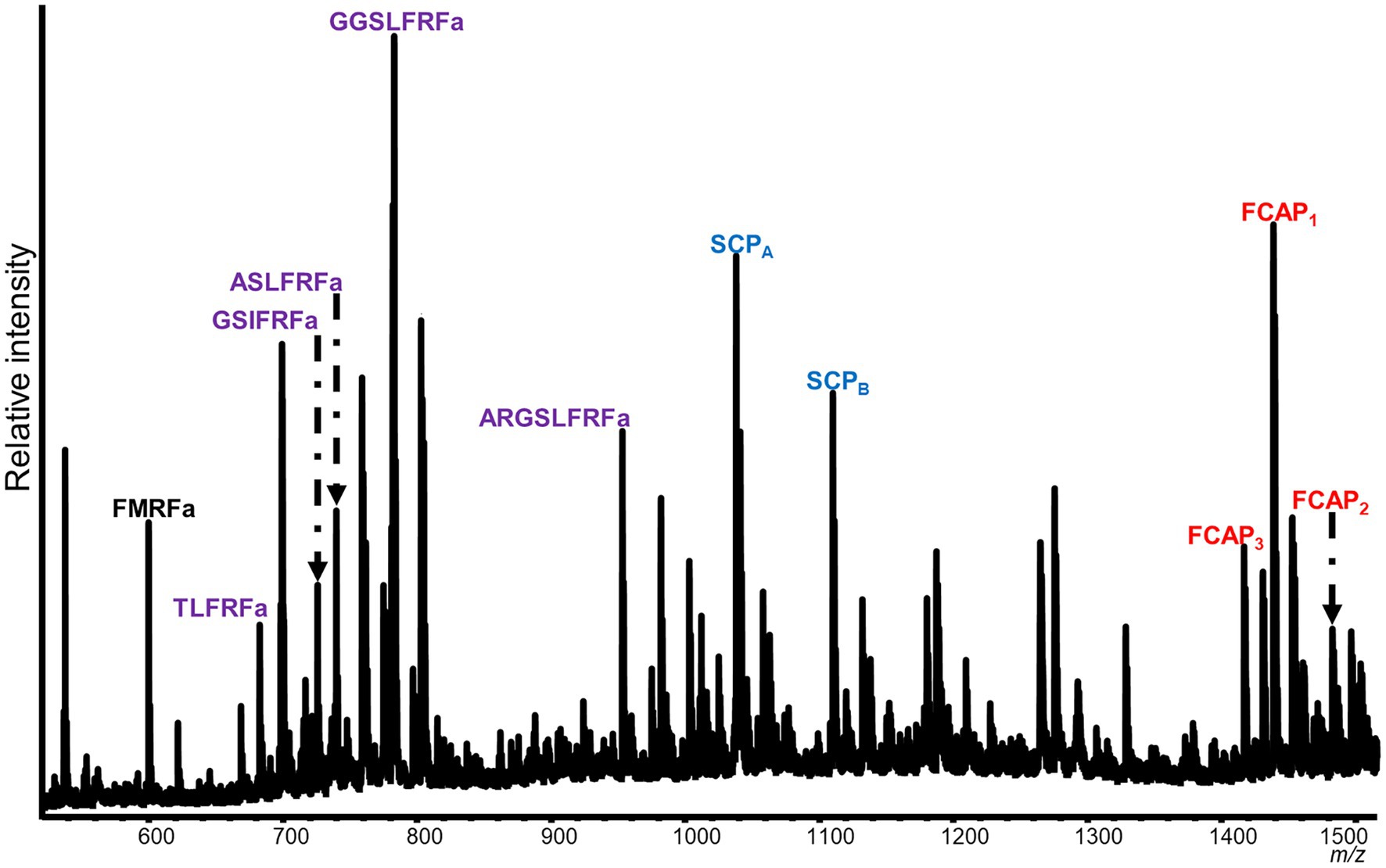
Figure 2. Representative spectrum from a H. crassicornis SLB cell. Six neurons were analyzed, and peptides from the FMRFa (black), SCPB (blue), LFRFa (purple), and FCAP (red) prohormones were present. The LFRF prohormone is predicted to produce five peptides with a C-terminal LFRFa motif, the FCAP prohormone, three versions of the FCAP peptide, and the SCP prohormone, both SCPA and SCPB; all of these were present in the cell. The FMRFa prohormone is predicted to produce multiple tetrapeptides, but only FMRFa was present in the cell.
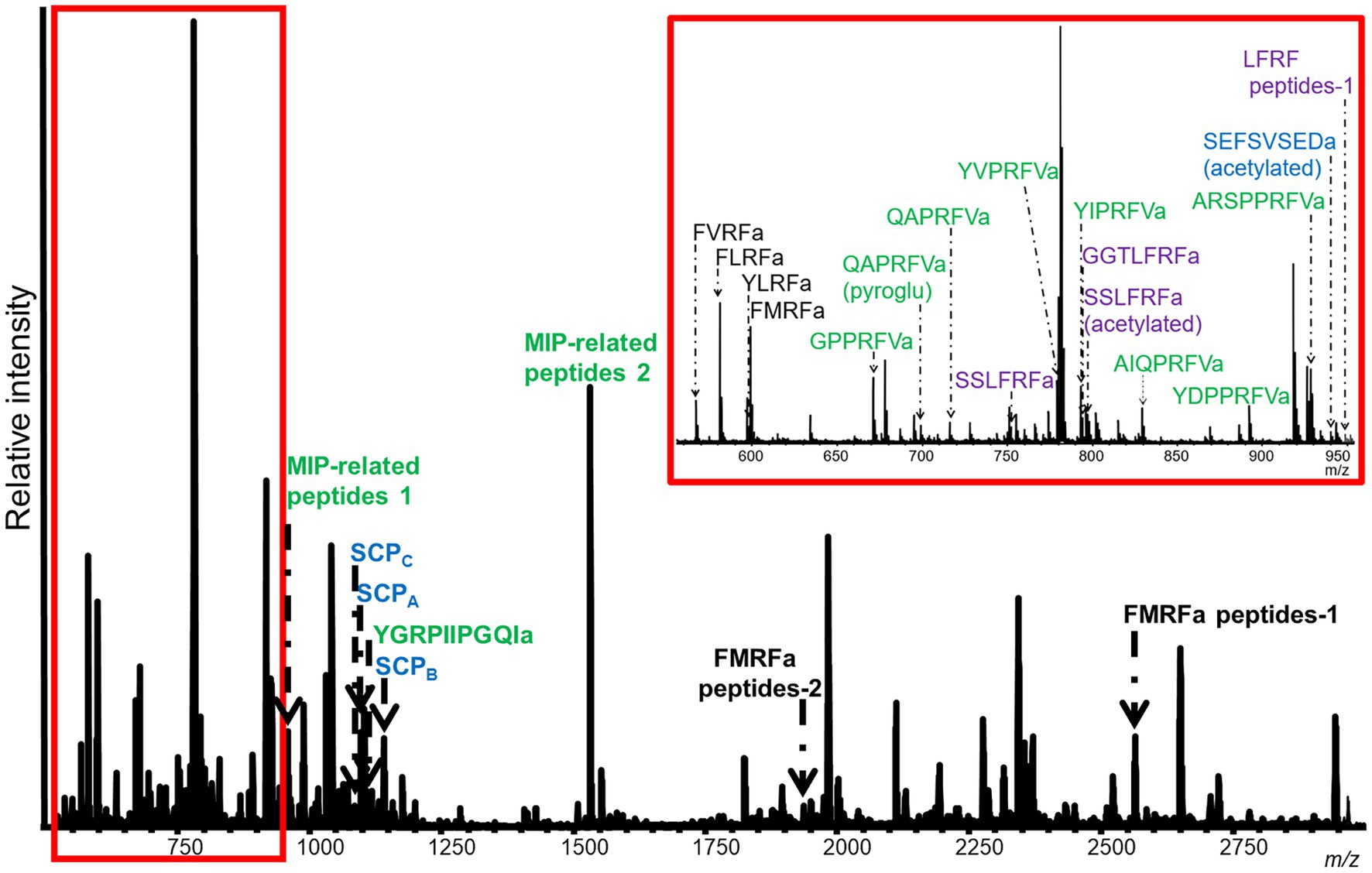
Figure 3. Representative MALDI-TOF MS spectrum from a M. leonina SLB cell. Four neurons were analyzed. This cell expressed 11 peptides from the MIP-related prohormone (green), six peptides from the FMRFa prohormone (black), and four peptides from the SCP (blue) and LFRFa (purple) prohormones. Peptides from the MIP-related prohormone included eight, with either a PRFVa or PTFVa C-terminal motif. The peptides from the SCP prohormone included SCPA, SCPB, and SCPC. Four of the six peptides from the FMRFa were tetrapeptides, including FMRFa itself. Lastly, the peptides from the LFRFa prohormone included both an acetylated and unacetylated version of SSLFRFa.
In addition to peptides from the SCP prohormone, the M. leonina SLB cells contained peptides matching three other prohormones (Figure 3): a homolog to the A. californica MIP-related prohormone (Q9NDE8) (Supplementary Figure S3), a homolog to the A. californica FMRFa prohormone (P08021) (Supplementary Figure S4), and a homolog to the L. stagnalis LFRFa prohormone (Supplementary Figure S1). Eleven peptides derived from the MIP-related prohormone, including eight with a C-terminal amidation and a PRFV or PTFV motif, similar to the PRFX motif found in the A. californica MIP-related prohormone. Six peptides came from the FMRFa prohormone, including FMRFa and three other -RFa tetrapeptides, and four peptides from the LFRFa prohormone (Table 2). Two of these four had the same amino acid sequence (GGTLFRF), differing only in the post-translational addition of an acetyl group, and a third peptide also shared the LFRFa motif. Interestingly, two other peptides with an LFRFa motif were putatively encoded on the M. leonina LFRFa prohormone, yet were not detected in the SLB cells by MS. Finally, the P. californica VWC also contained both SCPA and SCPB (Figure 4; Table 2).
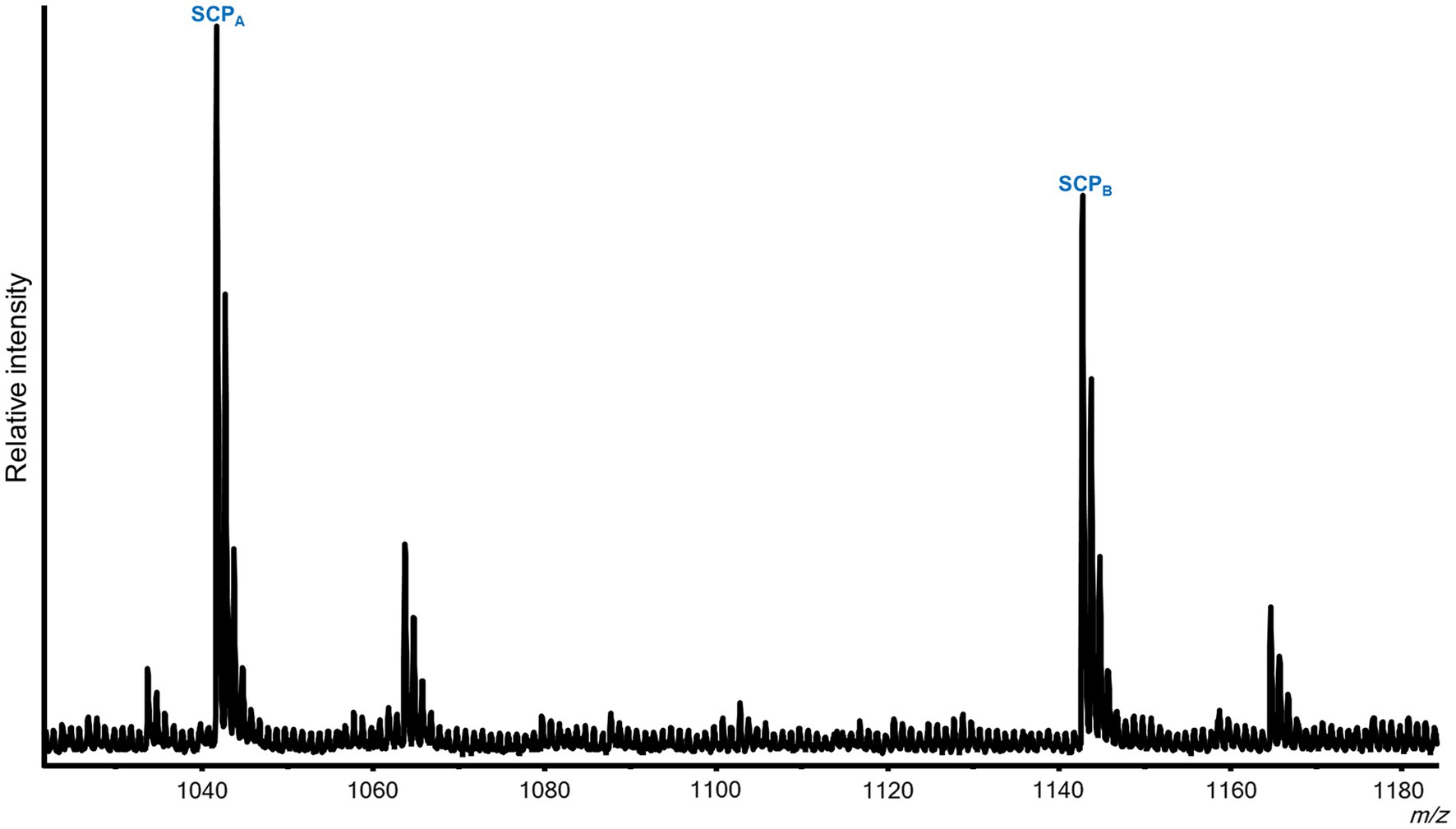
Figure 4. Representative MALDI-TOF MS spectrum from a Pleurobranchaea californica VWC. Five neurons were analyzed. In addition to the previously described peptides from the FMRFa and QNFLa prohormones (Green et al., 2018), the VWC expressed both SCPA and SCPB.
Peptides in nudipleurans remain understudied and yet have been extensively studied in the gastropods A. californica and L. stagnalis. Exploring peptides in nudipleurans can enhance the existing understanding of their neuronal circuits while also allowing for comparison with other species, an essential task for the study of brain evolution (Webber et al., 2017; Moroz, 2018). Using de novo transcriptome assemblies, we predicted putative peptide prohormones for H. crassicornis, M. leonina, and P. californica, and identified several homologs to prohormones previously characterized in A. californica and L. stagnalis. Additionally, in M. leonina we identified alternatively spliced SCP transcripts that encoded a novel peptide. Although we recovered homologs to almost every searched transcript, in some instances we could not recover the full protein sequence, and thus it is possible that we missed certain peptides. It is also possible that the transcripts not found here are indeed present in these species, but simply not expressed in the tissues used to generate the transcriptome assemblies.
We found that the neuropeptide complements of the VWC and SLB cells overlapped but were not identical (Figure 5), which may reflect adaptation to the species’ different feeding habits. SCP peptides were present in every cell, consistent with earlier immunological work (Watson and Willows, 1992) and suggesting a conserved role for these peptides in feeding. In P. californica, VWC firing drives esophageal dilation (Gillette and Gillette, 1983), and in M. leonina, SCPB application causes esophageal contractions (Watson et al., 2020), so the data suggest that these neurons use SCPB to regulate esophageal movement.
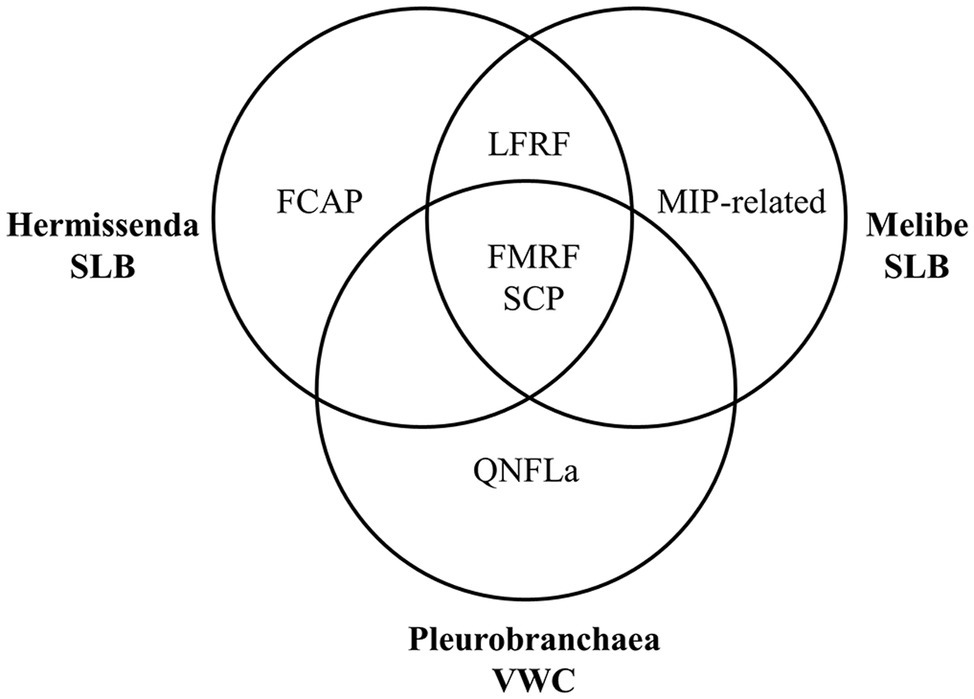
Figure 5. Summary of the prohormone families present in each species’ neuron. Each species has peptides from the FMRFa and SCP prohormone families, both nudibranchs have peptides from the LFRFa prohormone family, and each species has a prohormone family not detected in the other species’ neurons.
Meanwhile, SCPs are extensively involved in the control of feeding motor programs in A. californica (Lloyd, 1986; Lloyd et al., 1987) and L. stagnalis (Santama et al., 1994; Perry et al., 1999). The A. californica B1 and B2 and the L. stagnalis B2 neurons contain SCPA and SCPB and project axons to the esophageal nerve (Lloyd et al., 1988; Santama et al., 1994; Perry et al., 1998, 1999), and additional A. californica SCP-immunoreactive neurons innervate buccal musculature (Lloyd, 1988; Church et al., 1991). In both species, the SCPs co-localize with each other (Perry et al., 1998; Perry et al., 1999; Li et al., 2000b), and act as co-transmitters with both classical neurotransmitters (Weiss et al., 1992; Perry et al., 1999) and other neuropeptides (Santama et al., 1994). SCP also drives rhythmic bursting in the buccal ganglion of the snail Helisoma trivolvis (Murphy et al., 1985) and has even been implicated in feeding in Octopus vulgaris, as it drives contraction of the radula protractor muscle, and is transcribed in the buccal ganglion (Kanda and Minakata, 2006). The studies discussed here are insufficient to determine if A. californica and L. stagnalis have homologs to the VWC/SLB cells, but a clear conserved role for SCP can be seen in feeding-related movements.
FMRFa was previously found in the VWCs (Green et al., 2018) and was also found here within the SLB cells, which was surprising given that it inhibits feeding in other gastropods. In A. californica, the FMRFa peptide partially shifts feeding rhythms from ingestive to egestive and is released from sensory neurons to reduce accessory radula closer (ARC) muscle contractions (Vilim et al., 2010). Meanwhile, in L. stagnalis (Kyriakides and McCrohan, 1989) and H. trivolvis (Murphy et al., 1985), FMRFa perfusion inhibits the feeding rhythm, although in L. stagnalis it appears to be released from a pleural interneuron involved in defensive responses (Alania et al., 2004), rather than from an element of the feeding neural network. The L. stagnalis buccal mass is immunopositive for FMRFa and the buccal ganglion contains a single, bilateral neuron pair with immunoreactivity to the related peptide SEQPDVDDYLRDWLQSEEPLY (Santama et al., 1994), but FMRFa itself has not been detected in the L. stagnalis buccal ganglion by MS. Meanwhile, numerous sensory and motor neurons express FMRFa in the A. californica buccal ganglion (Vilim et al., 2010). Does the FMRFa released from the VWC/SLB cells in some way attenuate feeding, or does the presence of FMRFa in these cells reflect divergence from its role in A. californica and L. stagnalis? Our finding opens the door for future functional studies to address this question.
LFRFa peptides were observed in the H. crassicornis and M. leonina SLB cells but not the P. californica VWC, a pattern that perhaps reflects the three species’ phylogeny. The nudipleuran clade separates into nudibranchia and pleurobranchomorpha; H. crassicornis and M. leonina are nudibranchs whereas P. californica is a pleurobranch. In A. californica, LFRFa peptides have a similar effect as FMRFa, modulating contraction of the ARC muscle and weakening ingestive feeding rhythms (Cropper et al., 1994; Vilim et al., 2010). In L. stagnalis, MS analysis of the buccal ganglion found the presence of the six peptides encoded by the LFRFa prohormone, which inhibit neurons that regulate metabolism (Hoek et al., 2005). However, immunostaining and single-cell analysis have not been carried out thus far, nor is it known if these peptides have a role in L. stagnalis feeding circuitry. Thus, it will be of interest to determine the roles of LFRFa peptides in feeding in other species. Similarly, it will be of interest will be to determine if other cells within the P. californica buccal ganglion express LFRFa.
Finally, each species’ neuron expressed peptides from a prohormone not detected by MS in the others. First, the H. crassicornis SLB cells contained three peptides from the homolog to the A. californica FCAP prohormone and may contain more, as our annotation of the H. crassicornis FCAP prohormone returned an incomplete protein. FCAP drives feeding rhythms in A. californica, and interestingly is co-expressed with SCPB in a mechanosensory neuron (Sweedler et al., 2002), but its effects on feeding appear to come via the cerebral ganglion neuron CBI-2 (Friedman et al., 2015). FCAP has not been implicated in feeding in any other species. The M. leonina SLB cells express peptides from the MIP-related prohormone, which is found in one bilaterally paired set of buccal neurons in A. californica (Fujisawa et al., 1999), and many small buccal neurons in L. stagnalis and Helix pomatia (Elekes et al., 2000). In each species, application of MIP-related peptides drives contractions of the gut. Finally, the P. californica VWC contains peptides from the QNFLa prohormone that is a homolog to the A. californica pedal peptide 4 prohormone (Green et al., 2018). Pedal peptide 4 has not been investigated physiologically, but in Biomphalaria glabrata was observed to be less abundant 12 days post-infection with the parasite Schistosoma (Wang et al., 2017).
What are the implications of peptide co-localization in these neurons? Co-localization suggests co-transmission, which can increase the flexibility of post-synaptic control. Co-transmitters, particularly those released from different prohormones, can confer numerous possible abilities onto a single neuron, notably, the modulation of a different neurotransmitter’s effects (Kiss, 2011), more refined control of a single target (Brezina et al., 1995; Vilim et al., 2010), or the differential control of multiple targets (Svensson et al., 2019). This final mechanism seems especially possible in the VWC/SLB cells, which affect both feeding circuitry and the gut. Interestingly, SCP and FMRF co-localize in a cerebral interneuron in five different nudipleurans, including H. crassicornis, M. leonina, and P. californica (Lillvis et al., 2012). We cannot say definitively what each peptide does in these three cells, but it seems possible that SCP is released to drive esophageal contractions, and the other peptides to regulate feeding circuits.
Additionally, what are the functional consequences of the unique aspects of each neurons’ peptide profiles? Differences in in the intrinsic properties and synaptic wiring of homologous neurons can lead to subtle differences in behavior (Newcomb et al., 2012; Ding et al., 2019), and it may be that these chemical differences are another mechanism of this change. M. leonina differs markedly from the other species in this study in the lack of a buccal mass, and differs further in its prey capture apparatus, feeding mechanics, feeding bout duration, and prey. P. californica and H. crassicornis differ from each other in prey choice and the relative size of their feeding apparatuses. Additionally, the M. leonina buccal ganglion is considerably smaller than that of the others, consisting of only 30 to 40 neurons (Trimarchi and Watson, 1992). The differences in peptide profiles may relate in part to these anatomical and behavioral differences.
Finally, in prior studies, neuron homology has been inferred based on synaptic wiring, neuroanatomical position, function, and overlap in immunohistochemical staining (Faulkes, 2008; Lillvis et al., 2012; Sakurai and Katz, 2019). The data in this study do not address the first three characteristics, but do suggest that limits should be placed on interpretations made based on immunohistochemical staining. Our data suggest that at least some of the peptides expressed in homologous neurons will not overlap, and thus if staining is performed for a peptide found in only some of the neurons, it may lead to incorrect conclusions regarding homology.
Characterization of the neuropeptides present in a variety of animals is essential to our understanding of neurotransmission. Combining de novo transcriptomics and peptidomics allows us to examine the functional consequences of different peptide profiles without requiring a genomic assembly. The usage of different species in this work helped reveal what is “typical” of neuropeptide signaling, which is essential to the translatability of comparative research. Examining neuropeptides in nudipleuran sea slugs furthers this goal, and moreover, does so in a clade that has provided great insight into neuronal circuits.
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
CL – conceptualization, methodology, data collection, and writing. ER – conceptualization, methodology, and writing. BS – methodology, data collection, and writing. RG and JS – conceptualization and writing. All authors contributed to the article and approved the submitted version.
The project described was supported by Award Number P30DA018310 from the National Institute on Drug Abuse (NIDA) and the Friday Harbor Labs Research Fellowship Endowment. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Trevor Fay, Amanda Fay, Art Seavey, and Reyn Yoshioka for collecting specimens for us.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys. 2021.809529/full#supplementary-material
ARC, accessory radula closer; CNS, central nervous system; FMRFa, FMRFamide; LFRFa, LFRFamide; MALDI, matrix-assisted laser desorption/ionization; MIP, myoinhibitory peptide; MS, mass spectrometry; SCP, small cardioactive peptide; SLB, small cardioactive peptide B large buccal; VWC, ventral white cell; TOF, time-of-flight.
Adamson, K. J., Wang, T. F., Zhao, M., Bell, F., Kuballa, A. V., Storey, K. B., et al. (2015). Molecular insights into land snail neuropeptides through transcriptome and comparative gene analysis. BMC Genomics 16:308. doi: 10.1186/s12864-015-1510-8
Ahn, S. J., Martin, R., Rao, S., and Choi, M. Y. (2017). Neuropeptides predicted from the transcriptome analysis of the gray garden slug Deroceras reticulatum. Peptides 93, 51–65. doi: 10.1016/j.peptides.2017.05.005
Alania, M., Sakharov, D. A., and Elliott, C. J. H. (2004). Multilevel inhibition of feeding by a peptidergic pleural interneuron in the mollusc Lymnaea stagnalis. J. Comp. Physiol. A 190, 379–390. doi: 10.1007/s00359-004-0503-x
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., et al. (2004). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 32, 115D–1119D. doi: 10.1093/nar/gkh131
Armenteros, J. J. A., Tsirigos, K. D., Sonderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Baltzley, M. J., Sherman, A., Cain, S. D., and Lohmann, K. J. (2011). Conservation of a Tritonia pedal peptides network in gastropods. Invertebr. Biol. 130, 313–324. doi: 10.1111/j.1744-7410.2011.00242.x
Beck, J. C., Cooper, M. S., and Willows, A. O. D. (2000). Immunocytochemical localization of pedal peptide in the central nervous system of the gastropod mollusc Tritonia diomedea. J. Comp. Neurol. 425, 1–9. doi: 10.1002/1096-9861(20000911)425:1<1::aid-cne1>3.0.co;2-y
Bose, U., Suwansa-ard, S., Maikaeo, L., Motti, C. A., Hall, M. R., and Cummins, S. F. (2017). Neuropeptides encoded within a neural transcriptome of the giant triton snail Charonia tritonis, a crown-of-thorns starfish predator. Peptides 98, 3–14. doi: 10.1016/j.peptides.2017.01.004
Boyle, M. B., Cohen, L. B., Macagno, E. R., and Orbach, H. (1983). The number and size of neurons in the CNS of gastropod mollusks and their suitability for optical-recording of activity. Brain Res. 266, 305–317. doi: 10.1016/0006-8993(83)90662-5
Brezina, V., Bank, B., Cropper, E. C., Rosen, S., Vilim, F. S., Kupfermann, I., et al. (1995). Nine members of the myomodulin family of peptide cotransmitters at the B16-ARC neuromuscular junction of Aplysia. J. Neurophysiol. 74, 54–72. doi: 10.1152/jn.1995.74.1.54
Bulloch, A. G. M., and Ridgway, R. L. (1995). “Comparative aspects of gastropod neurobiology,” in The Nervous Systems of Invertebrates: An Evolutionary and Comparative Approach: With a Coda Written by T.H. Bullock. O. Breidbach and W. Kutsch (Eds.) (Basel: Birkhäuser Basel), 89–113.
Cafe-Mendes, C. C., Ferro, E. S., Britto, L. R. G., and Martins-de-Souza, D. (2014). Using mass spectrometry-based Peptidomics to understand the brain and disorders such as Parkinson’s disease and schizophrenia. Curr. Top. Med. Chem. 14, 369–381. doi: 10.2174/1568026613666131204120747
Christie, A. E. (2017). Neuropeptide discovery in Proasellus cavaticus: prediction of the first large-scale peptidome for a member of the isopoda using a publicly accessible transcriptome. Peptides 97, 29–45. doi: 10.1016/j.peptides.2017.09.003
Christie, A. E., and Pascual, M. G. (2016). Peptidergic signaling in the crab Cancer borealis: tapping the power of transcriptomics for neuropeptidome expansion. Gen. Comp. Endocrinol. 237, 53–67. doi: 10.1016/j.ygcen.2016.08.002
Church, P. J., Cohen, K. P., Scott, M. L., and Kirk, M. D. (1991). Peptidergic motoneurons in the buccal ganglia of Aplysia californica: immunocytochemical, morphological, and physiological characterizations. J. Comp. Physiol. A 168, 323–336. doi: 10.1007/BF00198352
Croll, R. P. (1987). “Identified neurons and cellular homologies,” in Nervous Systems in Invertebrates. M. A. Ali (Ed.) (Boston, MA: Springer US), 41–59.
Cropper, E. C., Brezina, V., Vilim, F. S., Harish, O., Price, D. A., Rosen, S., et al. (1994). FRF peptides in the ARC neuromuscular system of Aplysia: purification and physiological actions. J. Neurophysiol. 72, 2181–2195. doi: 10.1152/jn.1994.72.5.2181
Cropper, E. C., Jing, J., Vilim, F. S., Barry, M. A., and Weiss, K. R. (2018). Multifaceted expression of Peptidergic modulation in the feeding system of Aplysia. ACS Chem. Neurosci. 9, 1917–1927. doi: 10.1021/acschemneuro.7b00447
Crow, T. (2004). Pavlovian conditioning of Hermissenda: current cellular, molecular, and circuit perspectives. Learn. Memory 11, 229–238. doi: 10.1101/lm.70704
Crow, T., and Tian, L. M. (2006). Pavlovian conditioning in Hermissenda: A circuit analysis. Biol. Bull. 210, 289–297. doi: 10.2307/4134565
Di Cosmo, A., and Di Cristo, C. (2006). “Molluscan peptides and reproduction,” in Handbook of Biologically Active Peptides ed. A. J. Kastin (USA: Academic Press), 241–246.
Ding, Y., Lilivis, J. L., Cande, J., Berman, G. J., Arthur, B. J., Long, X., et al. (2019). Neural evolution of context-dependent Fly song. Curr. Biol. 29, 1089–1099.e7. doi: 10.1016/j.cub.2019.02.019
Elekes, K., Kiss, T., Fujisawa, Y., Hernadi, L., Erdelyi, L., and Muneoka, Y. (2000). Mytilus inhibitory peptides (MIP) in the central and peripheral nervous system of the pulmonate gastropods, Lymnaea stagnalis and Helix pomatia: distribution and physiological actions. Cell Tissue Res. 302, 115–134. doi: 10.1007/s004410000252
Elliott, C. J. H., and Susswein, A. J. (2002). Comparative neuroethology of feeding control in molluscs. J. Exp. Biol. 205, 877–896. doi: 10.1242/jeb.205.7.877
Faulkes, Z. (2008). Turning loss into opportunity: the key deletion of an escape circuit in decapod crustaceans. Brain Behav. Evol. 72, 251–261. doi: 10.1159/000171488
Feng, Z. P., Zhang, Z., van Kesteren, R. E., Straub, V. A., van Nierop, P., Jin, K., et al. (2009). Transcriptome analysis of the central nervous system of the mollusc Lymnaea stagnalis. BMC Genomics 10:451. doi: 10.1186/1471-2164-10-451
Friedman, A. K., Weiss, K. R., and Cropper, E. C. (2015). Specificity of repetition priming: The role of chemical coding. J. Neurosci. 35, 6326–6334. doi: 10.1523/jneurosci.4562-14.2015
Fujisawa, Y., Furukawa, Y., Ohta, S., Ellis, T. A., Dembrow, N. C., Li, L., et al. (1999). The Aplysia Mytilus inhibitory peptide-related peptides: identification, cloning, processing, distribution, and action. J. Neurosci. 19, 9618–9634. doi: 10.1523/JNEUROSCI.19-21-09618.1999
Gan, L., Xie, L. W., Zuo, F. Y., Xiang, Z. H., and He, N. J. (2015). Transcriptomic analysis of Rongchang pig brains and livers. Gene 560, 96–106. doi: 10.1016/j.gene.2015.01.051
Garden, R. W., Moroz, L. L., Moroz, T. P., Shippy, S. A., and Sweedler, J. V. (1996). Excess salt removal with matrix rinsing: direct peptide profiling of neurons from marine invertebrates using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Mass Spectrom. 31, 1126–1130. doi: 10.1002/(SICI)1096-9888(199610)31:10<1126::AID-JMS403>3.0.CO;2-7
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gillette, R., and Brown, J. W. (2015). The sea slug, Pleurobranchaea californica: A signpost species in the evolution of complex nervous systems and behavior. Integr. Comp. Biol. 55, 1058–1069. doi: 10.1093/icb/icv081
Gillette, M. U., and Gillette, R. (1983). Bursting neurons command consummatory feeding behavior and coordinated visceral receptivity in the predatory mollusk Pleurobranchaea. J. Neurosci. 3, 1791–1806. doi: 10.1523/jneurosci.03-09-01791.1983
Gillette, R., Gillette, M. U., and Davis, W. J. (1980). Action-potential broadening and endogenously sustained bursting are substrates of command ability in a feeding neuron of Pleurobranchaea. J. Neurophysiol. 43, 669–685. doi: 10.1152/jn.1980.43.3.669
Goodheart, J. A., Bazinet, A. L., Valdes, A., Collins, A. G., and Cummings, M. P. (2017). Prey preference follows phylogeny: evolutionary dietary patterns within the marine gastropod group Cladobranchia (Gastropoda: Heterobranchia: Nudibranchia). BMC Evol. Biol. 17:221. doi: 10.1186/s12862-017-1066-0
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Green, D. J., Huang, R. C., Sudlow, L., Hatcher, N., Potgieter, K., McCrohan, C., et al. (2018). cAMP, Ca2+, pH(i), and NO regulate H-like Cation channels That underlie feeding and locomotion in the Predatory Sea slug Pleurobranchaea californica. ACS Chem. Neurosci. 9, 1986–1993. doi: 10.1021/acschemneuro.8b00187
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hoek, R. M., Li, K. W., van Minnen, J., Lodder, J. C., de Jong-Brink, M., Smit, A. B., et al. (2005). LFRFamides: a novel family of parasitation-induced -RFamide neuropeptides that inhibit the activity of neuroendocrine cells in Lymnaea stagnalis. J. Neurochem. 92, 1073–1080. doi: 10.1111/j.1471-4159.2004.02927.x
Hoover, R. A., Armour, R., Dow, I., and Purcell, J. E. (2012). Nudibranch predation and dietary preference for the polyps of Aurelia labiata (Cnidaria: Scyphozoa). Hydrobiologia 690, 199–213. doi: 10.1007/s10750-012-1044-x
Hummon, A. B., Hummon, N. P., Corbin, R. W., Li, L., Vilim, F. S., Weiss, K. R., et al. (2003a). From precursor to final peptides: a statistical sequence-based approach to predicting prohormone processing. J. Proteome Res. 2, 650–656. doi: 10.1021/pr034046d
Hummon, A. B., Sweedler, J. V., and Corbin, R. W. (2003b). Discovering new neuropeptides using single-cell mass spectrometry. Trac. Trends Anal. Chem. 22, 515–521. doi: 10.1016/s0165-9936(03)00901-4
Jourjine, N., and Hoekstra, H. E. (2021). Expanding evolutionary neuroscience: insights from comparing variation in behavior. Neuron 109, 1084–1099. doi: 10.1016/j.neuron.2021.02.002
Kall, L., Krogh, A., and Sonnhammer, E. L. L. (2007). Advantages of combining transmembrane topology and signal peptide prediction - the Phobius web serve. Nucleic Acids Res. 35, W429–W432. doi: 10.1093/nar/gkm256
Kanda, A., and Minakata, H. (2006). Isolation and characterization of a novel small cardioactive peptide-related peptide from the brain of Octopus vulgaris. Peptides 27, 1755–1761. doi: 10.1016/j.peptides.2005.12.006
Katz, P. S. (2016). Evolution of central pattern generators and rhythmic behaviours. Philos. Trans. R. Soc. B 371:20150057. doi: 10.1098/rstb.2015.0057
Katz, P. S., and Quinlan, P. D. (2019). The importance of identified neurons in gastropod molluscs to neuroscience. Curr. Opin. Neurobiol. 56, 1–7. doi: 10.1016/j.conb.2018.10.009
Kiss, T. (2011). Diversity and abundance: the basic properties of neuropeptide action in molluscs. Gen. Comp. Endocrinol. 172, 10–14. doi: 10.1016/j.ygcen.2011.02.016
Kodama, Y., Shumway, M., and Leinonen, R. (2012). The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 40, D54–D56. doi: 10.1093/nar/gkr854
Kyriakides, M. A., and McCrohan, C. R. (1989). Effect of putative neuromodulators on rhythmic buccal motor output in Lymnaea stagnalis. J. Neurobiol. 20, 635–650. doi: 10.1002/neu.480200704
Lee, C. A., and Watson, W. H. (2016). The influence of stomach distention on feeding in the nudibranch molluskMelibe leonina. Mar. Freshwater Behav. Physiol. 49, 277–290. doi: 10.1080/10236244.2016.1192305
Li, L. J., Garden, R. W., and Sweedler, J. V. (2000a). Single-cell MALDI: a new tool for direct peptide profiling. Trends Biotechnol. 18, 151–160. doi: 10.1016/s0167-7799(00)01427-x
Li, D., Liu, C. M., Luo, R., Sadakane, K., and Lam, T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Li, L. J., Romanova, E. V., Rubakhin, S. S., Alexeeva, V., Weiss, K. R., Vilim, F. S., et al. (2000b). Peptide profiling of cells with multiple gene products: combining immunochemistry and MALDI mass spectrometry with on-plate microextraction. Anal. Chem. 72, 3867–3874. doi: 10.1021/ac000260z
Lillvis, J. L., Gunaratne, C. A., and Katz, P. S. (2012). Neurochemical and neuroanatomical identification of central pattern generator neuron homologues in Nudipleura Molluscs. PLoS One 7:e31737. doi: 10.1371/journal.pone.0031737
Liu, F., Baggerman, G., Schoofs, L., and Wets, G. (2008). The construction of a bioactive peptide database in Metazoa. J. Proteome Res. 7, 4119–4131. doi: 10.1021/pr800037n
Lloyd, P. E. (1986). The small cardioactive peptides: a class of modulatory neuropeptides in Aplysia. Trends Neurosci. 9, 428–432. doi: 10.1016/0166-2236(86)90138-4
Lloyd, P. E. (1988). Fast axonal transport of modulatory neuropeptides from central ganglia to components of the feeding system in Aplysia. J. Neurosci. 8, 3507–3514. doi: 10.1523/JNEUROSCI.08-09-03507.1988
Lloyd, P. E., Frankfurt, M., Stevens, P., Kupfermann, I., and Weiss, K. R. (1987). Biochemical and immunocytological localization of the neuropeptides FMRFamide, SCPA, SCPB, to neurons involved in the regulation of feeding in Aplysia. J. Neurosci. 7, 1123–1132. doi: 10.1523/JNEUROSCI.07-04-01123.1987
Lloyd, P. E., Kupfermann, I., and Weiss, K. R. (1988). Central peptidergic neurons regulate gut motility in Aplysia. J. Neurophysiol. 59, 1613–1626. doi: 10.1152/jn.1988.59.5.1613
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18
Ma, M. M., Wang, J. H., Chen, R. B., and Li, L. J. (2009). Expanding the crustacean Neuropeptidome using a multifaceted mass spectrometric approach. J. Proteome Res. 8, 2426–2437. doi: 10.1021/pr801047v
Moroz, L. L. (2018). NeuroSystematics and periodic system of neurons: model vs reference species at single-cell resolution. ACS Chem. Neurosci. 9, 1884–1903. doi: 10.1021/acschemneuro.8b00100
Murphy, A. D., Lukowiak, K., and Stell, W. K. (1985). Peptidergic modulation of patterned motor activity in identified neurons of Helisoma. Proc. Natl. Acad. Sci. U. S. A. 82, 7140–7144. doi: 10.1073/pnas.82.20.7140
Newcomb, J. M., Sakurai, A., Lillvis, J. L., Gunaratne, C. A., and Katz, P. S. (2012). Homology and homoplasy of swimming behaviors and neural circuits in the Nudipleura (Mollusca, Gastropoda, Opisthobranchia). Proc. Natl. Acad. Sci. U. S. A. 109, 10669–10676. doi: 10.1073/pnas.1201877109
Noboa, V., and Gillette, R. (2013). Selective prey avoidance learning in the predatory sea slug Pleurobranchaea californica. J. Exp. Biol. 216, 3231–3236. doi: 10.1242/jeb.079384
Perry, S. J., Dobbins, A. C., Schofield, M. G., Piper, M. R., and Benjamin, P. R. (1999). Small cardioactive peptide gene: structure, expression and mass spectrometric analysis reveals a complex pattern of co-transmitters in a snail feeding neuron. Eur. J. Neurosci. 11, 655–662. doi: 10.1046/j.1460-9568.1999.00472.x
Perry, S. J., Straub, V. A., Kemenes, G., Santama, N., Worster, B. M., Burke, J. F., et al. (1998). Neural modulation of gut motility by myomodulin peptides and acetylcholine in the snail Lymnaea. J. Neurophysiol. 79, 2460–2474. doi: 10.1152/jn.1998.79.5.2460
Rusch, S. L., and Kendall, D. A. (1995). Protein transport via amino-terminal targeting sequences: common themes in diverse systems. Mol. Membr. Biol. 12, 295–307. doi: 10.3109/09687689509072431
Sakurai, A., Gunaratne, C. A., and Katz, P. S. (2014). Two interconnected kernels of reciprocally inhibitory interneurons underlie alternating left-right swim motor pattern generation in the mollusk Melibe leonina. J. Neurophysiol. 112, 1317–1328. doi: 10.1152/jn.00261.2014
Sakurai, A., and Katz, P. S. (2015). Phylogenetic and individual variation in gastropod central pattern generators. J. Comp. Physiol. A 201, 829–839. doi: 10.1007/s00359-015-1007-6
Sakurai, A., and Katz, P. S. (2019). Command or obey? Homologous neurons differ in hierarchical position for the generation of homologous behaviors. J. Neurosci. 39, 6460–6471. doi: 10.1523/jneurosci.3229-18.2019
Santama, N., Brierley, M., Burke, J. F., and Benjamin, P. R. (1994). Neural network controlling feeding in Lymnaea stagnalis: immunocytochemical localization of myomodulin, small cardioactive peptide, buccalin, and FMRFamide-related peptides. J. Comp. Neurol. 342, 352–365. doi: 10.1002/cne.903420304
Senatore, A., Edirisinghe, N., and Katz, P. S. (2015). Deep mRNA sequencing of the Tritonia diomedea brain transcriptome provides access to gene homologues for neuronal excitability, synaptic transmission and peptidergic signalling. PLoS One 10:e0118321. doi: 10.1371/journal.pone.0118321
Southey, B. R., Amare, A., Zimmerman, T. A., Rodriguez-Zas, S. L., and Sweedler, J. V. (2006a). NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 34, W267–W272. doi: 10.1093/nar/gkl161
Southey, B. R., Rodriguez-Zas, S. L., Rhodes, J. S., and Sweedler, J. V. (2020). Characterization of the prohormone complement in Amphiprion and related fish species integrating genome and transcriptome assemblies. PLoS One 15:e0228562. doi: 10.1371/journal.pone.0228562
Southey, B. R., Rodriguez-Zas, S. L., and Sweedler, J. V. (2006b). Prediction of neuropeptide prohormone cleavages with application to RFamides. Peptides 27, 1087–1098. doi: 10.1016/j.peptides.2005.07.026
Southey, B. R., Sweedler, J. V., and Rodriguez-Zas, S. L. (2008). A python analytical pipeline to identify prohormone precursors and predict prohormone cleavage sites. Front. Neuroinform. 2:7. doi: 10.3389/neuro.11.007.2008
Svensson, E., Apergis-Schoute, J., Burnstock, G., Nusbaum, M. P., Parker, D., and Schioth, H. B. (2019). General principles of neuronal co-transmission: insights From multiple model systems. Front. Neural Circuits 12:117. doi: 10.3389/fncir.2018.00117
Sweedler, J. V., Li, L., Rubakhin, S. S., Alexeeva, V., Dembrow, N. C., Dowling, O., et al. (2002). Identification and characterization of the feeding circuit-activating peptides, a novel neuropeptide family of Aplysia. J. Neurosci. 22, 7797–7808. doi: 10.1523/JNEUROSCI.22-17-07797.2002
Thiede, B., Hohenwarter, W., Krah, A., Mattow, J., Schmid, M., Schmidt, F., et al. (2005). Peptide mass fingerprinting. Methods 35, 237–247. doi: 10.1016/j.ymeth.2004.08.015
Trimarchi, J., and Watson, W. H. (1992). The role of the Melibe buccal ganglia in feeding behavior. Mar. Behav. Physiol. 19, 195–209. doi: 10.1080/10236249209378808
Van Camp, K. A., Baggerman, G., Blust, R., and Husson, S. J. (2017). Peptidomics of the zebrafish Danio rerio: In search for neuropeptides. J. Proteome 150, 290–296. doi: 10.1016/j.jprot.2016.09.015
Vilim, F. S., Sasaki, K., Rybak, J., Alexeeva, V., Cropper, E. C., Jing, J., et al. (2010). Distinct mechanisms produce functionally complementary actions of neuropeptides That are structurally related But derived from different precursors. J. Neurosci. 30, 131–147. doi: 10.1523/jneurosci.3282-09.2010
Wang, T. F., Zhao, M., Liang, D., Bose, U., Kaur, S., McManus, D. P., et al. (2017). Changes in the neuropeptide content of Biomphalaria ganglia nervous system following Schistosoma infection. Parasites Vectors 10:275. doi: 10.1186/s13071-017-2218-1
Watson, W. H. III, and Willows, A. O. (1992). Evidence for homologous peptidergic neurons in the buccal ganglia of diverse nudibranch mollusks. J. Neurobiol. 23, 173–186. doi: 10.1002/neu.480230208
Watson, W. H., Nash, A., Lee, C., Patz, M. D., and Newcomb, J. M. (2020). The distribution and possible roles of small Cardioactive peptide in the Nudibranch Melibe leonina. Int. Org. Biol. 2:obaa016. doi: 10.1093/iob/obaa016
Watson, W. H., and Trimarchi, J. (1992). A quantitative description of Melibe feeding behavior and its modification by prey density. Mar. Behav. Physiol. 19, 183–194. doi: 10.1080/10236249209378807
Webber, M. P., Thomson, J. W. S., Buckland-Nicks, J., Croll, R. P., and Wyeth, R. C. (2017). GABA-, histamine-, and FMRFamide-immunoreactivity in the visual, vestibular and central nervous systems of Hermissenda crassicornis. J. Comp. Neurol. 525, 3514–3528. doi: 10.1002/cne.24286
Weiss, K. R., Březina, V., Cropper, E. C., Hooper, S. L., Miller, M. W., Probst, W. C., et al. (1992). Peptidergic co-transmission in Aplysia: functional implications for rhythmic behaviors. Experientia 48, 456–463. doi: 10.1007/bf01928164
Wood, E. A., Stopka, S. A., Zhang, L. W., Mattson, S., Maasz, G., Pirger, Z., et al. (2021). Neuropeptide localization in Lymnaea stagnalis: From the central nervous system to subcellular compartments. Front. Mol. Neurosci. 14:670303. doi: 10.3389/fnmol.2021.670303
Keywords: mass spectrometry, bioinformatics, peptidomics, neuroethology, mollusk, invertebrate, evolution
Citation: Lee CA, Romanova EV, Southey BR, Gillette R and Sweedler JV (2021) Comparative Analysis of Neuropeptides in Homologous Interneurons and Prohormone Annotation in Nudipleuran Sea Slugs. Front. Physiol. 12:809529. doi: 10.3389/fphys.2021.809529
Received: 05 November 2021; Accepted: 02 December 2021;
Published: 23 December 2021.
Edited by:
Andrew Gracey, University of Southern California, United StatesReviewed by:
Akira Sakurai, Georgia State University, United StatesCopyright © 2021 Lee, Romanova, Southey, Gillette and Sweedler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan V. Sweedler, anN3ZWVkbGVAaWxsaW5vaXMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.