
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Physiol., 06 January 2022
Sec. Mitochondrial Research
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.806426
This article is part of the Research TopicTargeting Signals in Protein Trafficking and TransportView all 15 articles
The vast majority of mitochondrial proteins are encoded in the nuclear genome and synthesized on cytosolic ribosomes as precursor proteins with specific mitochondrial targeting signals. Mitochondrial targeting signals are very diverse, however, about 70% of mitochondrial proteins carry cleavable, N-terminal extensions called presequences. These amphipathic helices with one positively charged and one hydrophobic surface target proteins to the mitochondrial matrix with the help of the TOM and TIM23 complexes in the outer and inner membranes, respectively. Translocation of proteins across the two mitochondrial membranes does not take place independently of each other. Rather, in the intermembrane space, where the two complexes meet, components of the TOM and TIM23 complexes form an intricate network of protein–protein interactions that mediates initially transfer of presequences and then of the entire precursor proteins from the outer to the inner mitochondrial membrane. In this Mini Review, we summarize our current understanding of how the TOM and TIM23 complexes cooperate with each other and highlight some of the future challenges and unresolved questions in the field.
Eukaryotic cells are defined by the presence of different membrane-enclosed compartments, cell organelles, that contain specific sets of proteins and provide specific chemical milieus. The obvious advantage of the subcellular compartmentalization is that a wide variety of cellular processes can take place simultaneously under vastly different conditions. The obvious disadvantage, however, is that eukaryotic cells needed to develop very precise mechanisms that would ensure that each protein is correctly sorted to the specific organelle where it can perform its function. In general, intracellular protein sorting relies on the presence of specific targeting signals within the proteins and on the respective receptors, usually localized on the surface of the organelle, that recognize these signals (Blobel, 2000). Upon recognition of targeting signals, proteins are translocated across or inserted into the organelle membrane, usually through some form of the translocation channel and with the input of energy. Though this basic concept also applies to protein translocation into mitochondria, the complex structure of this organelle brings many additional challenges. One of them is the need to translocate the majority of its proteins across two membranes in a coordinated manner. In this Mini Review, we briefly summarize and discuss our current understanding of this process.
Mitochondria are double-membrane-bounded organelles with four subcompartments: two membranes, the outer membrane (OM) and the inner membrane (IM), that define two aqueous subcompartments, the intermembrane space (IMS) and the innermost matrix. Though mitochondria possess their own genome, the mitochondrial DNA (mtDNA), and a complete apparatus for its expression, out of ca. 1,000–1,500 mitochondrial proteins, only 8 in yeast Saccharomyces cerevisiae and 13 in humans are encoded in the mtDNA. The vast majority of mitochondrial proteins are encoded by nuclear genes and translated on cytosolic ribosomes as precursor proteins with specific mitochondrial targeting signals. Upon initial recognition by cytosol-exposed receptors in the OM of mitochondria, precursor proteins are imported into their final place of function with the help of highly specific protein translocases present in all mitochondrial subcompartments (Neupert, 2015; Wiedemann and Pfanner, 2017; Hansen and Herrmann, 2019). The mitochondrial targeting signals are very diverse, mirroring the complex structure of the organelle. Still, about 70% of the mitochondrial precursor proteins carry at their N-termini typically 15–55 amino acids long, cleavable extensions called presequences (Vögtle et al., 2009). Presequences are characterized by the ability to form an amphipathic helix with a net positive charge (typically +3 to +6) on one side and a hydrophobic surface on the opposite side. By default, presequences target precursor proteins to the mitochondrial matrix, however, in combination with additional targeting signals, presequence-containing precursor proteins can also be targeted to the IM, IMS, and even OM. Translocation of presequence-containing precursor proteins across the two mitochondrial membranes is mediated by the TOM and TIM23 complexes in the outer and inner membranes, respectively (Figure 1). This import pathway is also termed the “presequence pathway.” As the main entry gate of mitochondria, the TOM complex is not only involved in the presequence pathway but also in import of essentially all nuclear-encoded mitochondrial proteins along other mitochondrial protein import pathways involving TOB/SAM, MIM, MIA and TIM22 complexes (Neupert, 2015; Schulz et al., 2015; Wiedemann and Pfanner, 2017; Hansen and Herrmann, 2019; Pfanner et al., 2019).
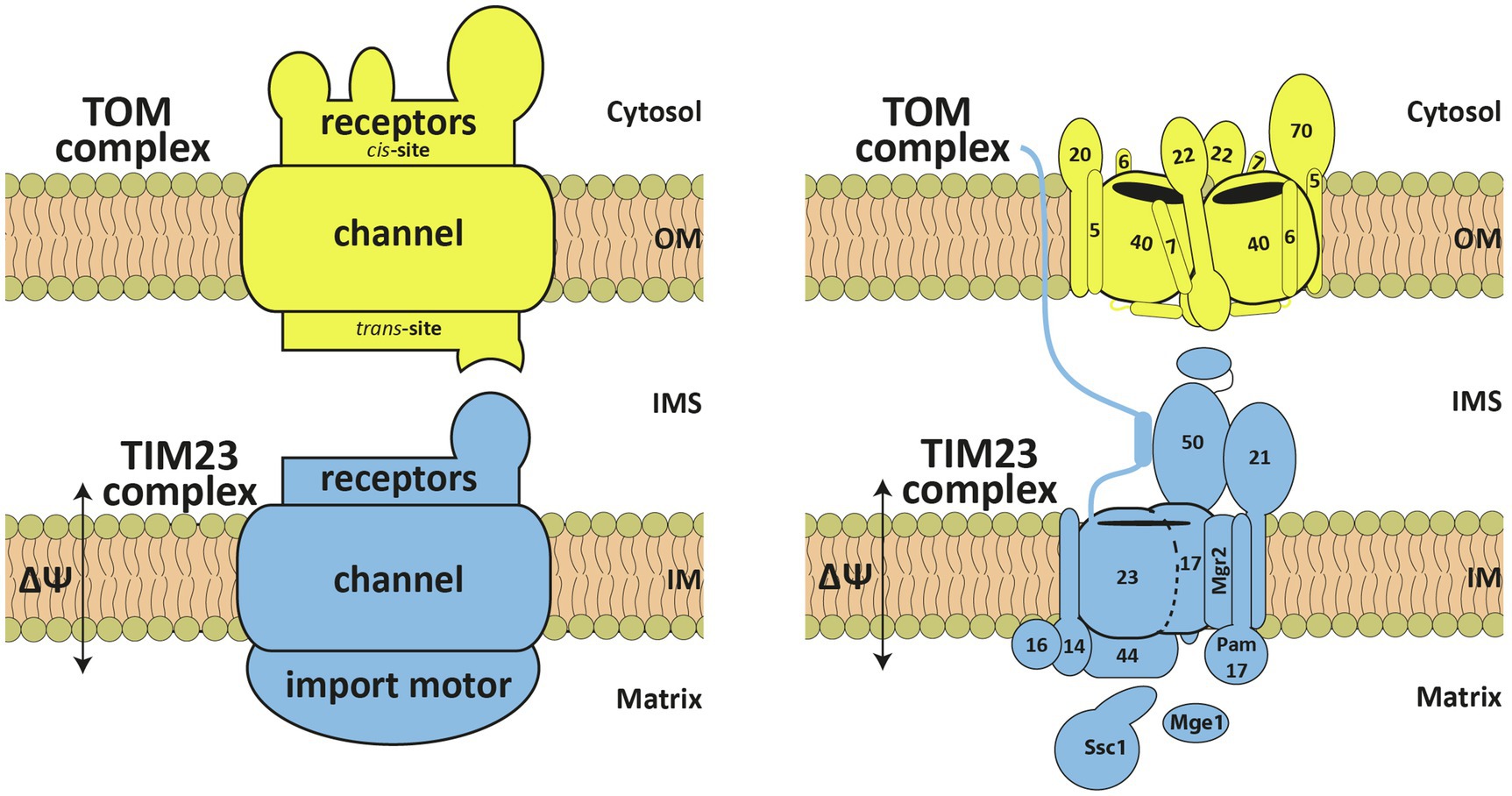
Figure 1. Schematic representation of the TOM and TIM23 complexes. The TOM complex consists of the receptors, Tom20, Tom22, and Tom70, and a channel unit, formed by Tom40, associated with three small proteins Tom5, Tom6 and Tom7. It possesses cis and trans presequence-binding sites. The TIM23 complex can be functionally divided into receptors, translocation channel and import motor – Tim23, Tim17, Tim50, Tim44, Tim14, Tim16, mtHsp70 (Ssc1), Mge1, Tim21, Mgr2, and Pam17. See text for details. OM, outer membrane; IMS, intermembrane space; IM, inner membrane.
Newly synthesized mitochondrial precursor proteins are bound to cytosolic chaperones that keep them in a largely unfolded, import-competent state (Becker et al., 2019; Avendaño-Monsalve et al., 2020; Bykov et al., 2020). Presequences are recognized on the cytosolic surface of the OM, the so-called cis site, by the receptors of the TOM complex, Tom20 and Tom22 (Figure 1). The TOM complex has an additional receptor, Tom70. Though initial data suggested that Tom70 is specifically involved in recognition of internal targeting signals within mitochondrial proteins, recent work shows that its predominant function is in tethering cytosolic chaperones to the surface of mitochondria (Backes et al., 2021), suggesting a more general role for Tom70 in protein translocation into mitochondria. After initial recognition by Tom20 and Tom22, presequences are transferred to the translocation channel of the TOM complex formed by the β-barrel protein Tom40. Transmembrane segments of Tom22 and of three small Tom proteins, Tom5, Tom6, and Tom7, are bound on the outer surface of the Tom40 barrel. On the IMS face of the channel, presequences bind to the so-called trans site of the TOM complex formed by the IMS-exposed segments of Tom22, Tom40, and Tom7. Already at this stage, presequences are recognized by the IMS-exposed receptors of the TIM23 complex, Tim50 and Tim23 (Figure 1). In a membrane-potential dependent step, presequences are subsequently inserted into the still mysterious translocation channel of the TIM23 complex, formed by the membrane-embedded segments of Tim23 and Tim17. Once in the matrix, presequences are proteolytically removed by the mitochondrial processing peptidase. Translocation of the complete polypeptide chain into the matrix requires the ATP-dependent action of the import motor (Craig, 2018; Mokranjac, 2020). The peripheral membrane protein Tim44 recruits mtHsp70 (Ssc1), the ATP-consuming subunit of the motor, and its cochaperones Tim14 (Pam18), Tim16 (Pam16), and Mge1, to the translocation channel in the inner membrane. If the presequence is the only targeting signal present, precursor proteins will be completely translocated into the matrix. However, if an additional hydrophobic sorting signal (“stop-transfer signal”) is present downstream of the presequence, translocation into the matrix will be arrested and the hydrophobic segment will be inserted laterally into the IM. The TIM23 complex contains three nonessential subunits, Pam17, Tim21, and Mgr2, that appear to play a role in the differential sorting of proteins into the matrix and the IM.
It is likely that all major players of the presequence pathway are identified by now. However, molecular understanding of how presequences are recognized and handed over along this pathway is still very rudimentary – the only high-resolution structure of the receptor bound to the presequence peptide is that of the cytosolic domain of Tom20 (Abe et al., 2000; Saitoh et al., 2007). Below we present and discuss our current knowledge of how TOM and TIM23 cooperate during transfer of presequences between outer and inner mitochondrial membranes.
Upon solubilization of mitochondria, the TOM complex does not interact in a stable manner with the TIM23 complex, or with any other of the downstream translocases. Studies with isolated OM vesicles and purified and reconstituted TOM complex showed that the TOM complex is able to recognize presequence-containing precursor proteins and initiate their translocation across the OM; however, they also showed that the TOM complex on its own is not able to translocate proteins completely across the membrane (Mayer et al., 1995; Künkele et al., 1998). On the other hand, experiments performed with mitoplasts, mitochondria in which the OM was artificially removed, showed that the TIM23 complex is, on its own, able to recognize and import precursor proteins across the IM (Hwang et al., 1989). In intact mitochondria, however, N-terminal presequences can be proteolytically removed in the matrix while the C-terminal part of the protein is still in the cytosol. Also, the TOM-TIM23 supercomplex can be stabilized with precursor proteins arrested as the TOM-TIM23 spanning intermediates (Chacinska et al., 2003; Popov-Celeketić et al., 2008). These experiments show that the TOM and TIM23 complexes do not operate as isolated units but rather mediate import of presequence-containing precursor proteins in a tightly controlled and coordinated manner. The cooperation of the TOM and TIM23 complexes is likely to take place in the IMS where the two complexes meet. The subunits implicated in TOM and TIM23 cooperation are Tom22, Tom7, and Tom40, from the TOM side, and Tim50, Tim23, and Tim21, from the TIM23 side (Figure 2A).
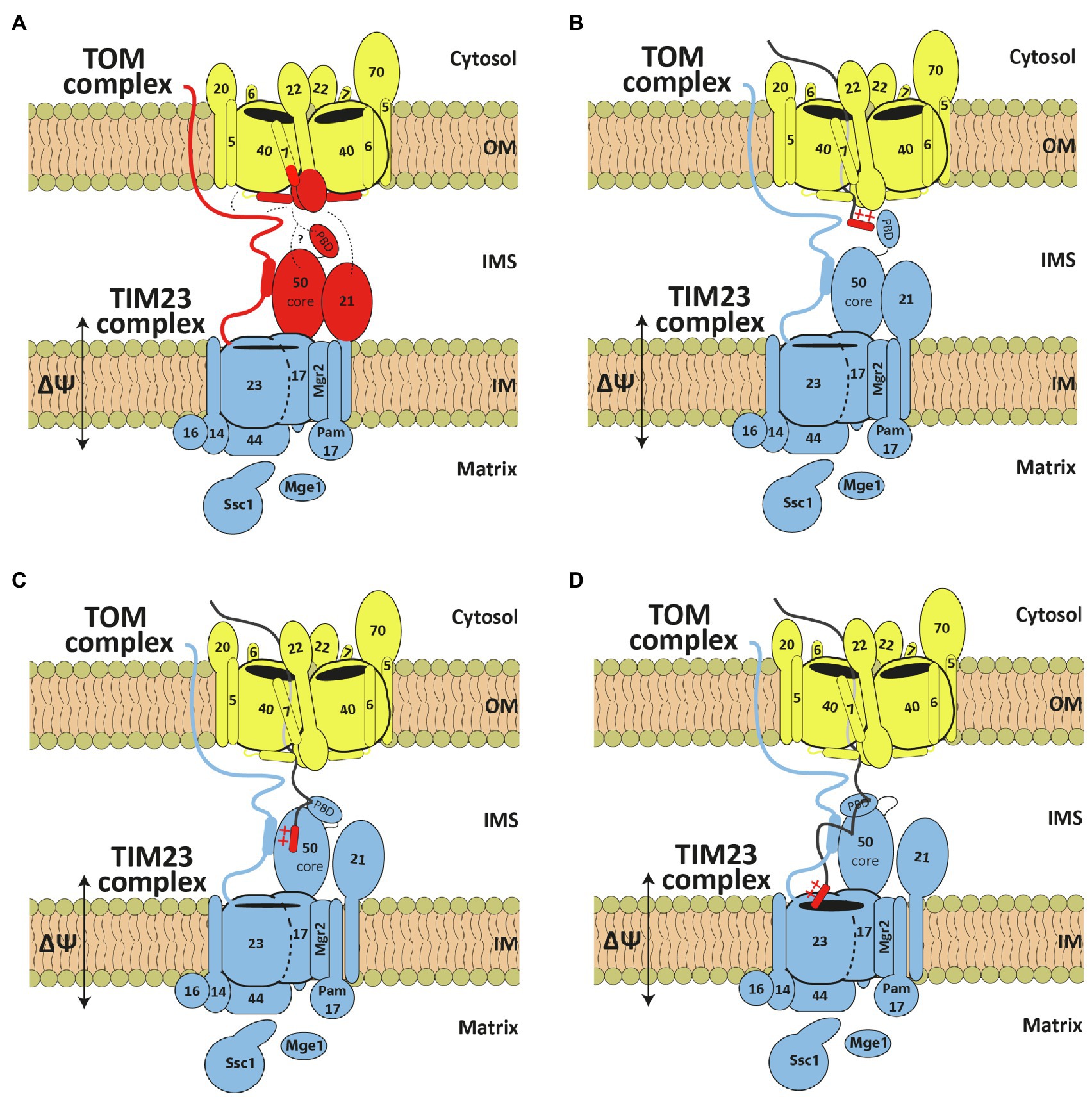
Figure 2. TOM and TIM23 cooperation during precursor translocation across two mitochondrial membranes. (A) Subunits implicated in TOM and TIM23 cooperation are highlighted in red. Dashed lines represent identified interaction points. (B–D) Current working model for transfer of precursors from the TOM to the TIM23 complex. See text for details. OM, outer membrane; IMS, intermembrane space; IM, inner membrane.
Biochemical and genetic experiments suggested that the trans site of the TOM complex is formed by the IMS-facing segments of Tom22, Tom7, and Tom40. All three proteins can be crosslinked to precursor proteins arrested in the TOM complex (Kanamori et al., 1999; Esaki et al., 2004). Simultaneous deletion of Tom7 and the IMS segment of Tom22 leads to the accumulation of precursor forms of mitochondrial proteins in the cytosol, indicative of impaired import. These cells are not able to grow on a fermentable carbon source at higher temperatures and not at all on nonfermentable carbon sources (Esaki et al., 2004). The recently determined cryo-EM structure of the TOM complex indeed suggests that they are all found close to each other at the Tom40 dimer interface (Araiso et al., 2019; Tucker and Park, 2019). Unfortunately, the actual IMS-exposed segments of Tom40 and Tom22, where the presequences most likely bind, were not resolved in the structure. However, biochemical evidence has been presented that the presequence-containing precursor proteins exit the channel in the middle of the dimer where the trans site of the TOM complex is expected to be (Araiso et al., 2019).
Experiments performed in intact mitochondria and with isolated recombinant proteins showed that the TIM23 complex interacts with the trans site of the TOM complex even in the absence of protein translocation (Figure 2A). Using chemical and/or site-specific UV crosslinking, Tim23 and Tim50 were crosslinked to the IMS-exposed segments of Tom22 and Tom40 (Tamura et al., 2009; Shiota et al., 2011; Waegemann et al., 2015; Araiso et al., 2019; Günsel et al., 2020). Recombinantly expressed and purified IMS segments of Tim23 and Tim21 bound to the IMS segment of Tom22 in vitro (Chacinska et al., 2005; Mokranjac et al., 2005; Albrecht et al., 2006; Bajaj et al., 2014). A direct interaction of any of these TIM23 subunits with Tom7 has not been demonstrated yet, however, considering the recently shown proximity of Tom7 to the IMS-exposed segments of Tom22 and Tom40 (Araiso et al., 2019; Tucker and Park, 2019), it is likely that Tom7 is present in the vicinity of and/or interacts with at least one of the three TIM23 subunits. Indeed, Tom7 genetically interacts with the N-terminal segment of Tim23, as does the IMS segment of Tom22 (Waegemann et al., 2015). The unique feature of the N-terminal segment of Tim23 is that it is accessible to externally added proteases in intact mitochondria (Donzeau et al., 2000). Though still controversial, the exposure of Tim23 on the mitochondrial surface depends on the interaction between Tim23 and Tim50 in the IMS (Yamamoto et al., 2002; Gevorkyan-Airapetov et al., 2009; Tamura et al., 2009), the dynamics of the TOM complex (Waegemann et al., 2015), the energetic state of the inner membrane (Günsel et al., 2020) and the translocation activity of the TIM23 complex (Popov-Celeketić et al., 2008). Whether Tim23 reaches the cytosol through the lipid bilayer, through the TOM channel or by some other, still unknown mechanism remains unclear. Even though this segment of Tim23 is not essential for cell viability (Chacinska et al., 2003; Waegemann et al., 2015), it is only logical that, by crossing two mitochondrial membranes, Tim23 would bring them closer, facilitating transfer of proteins between TOM and TIM23 complexes.
The IMS-exposed segments of the TIM23 subunits not only interact with the trans site of the TOM complex but also with each other. The high-resolution structural information on these interactions is unfortunately still missing. Still, biochemical experiments showed that Tim21 binds to Tim23 and to Tim50, as judged by both in organello crosslinking and interactions between recombinantly expressed and purified proteins (Tamura et al., 2009; Lytovchenko et al., 2013; Bajaj et al., 2014). The interaction between Tim23 and Tim50 has been extensively analyzed in vivo, in organello, and in vitro, and residues in both proteins have been identified that contribute to their interaction (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003, 2009; Meinecke et al., 2006; Alder et al., 2008; Gevorkyan-Airapetov et al., 2009; Tamura et al., 2009; Qian et al., 2011; Schulz et al., 2011; Lytovchenko et al., 2013; Günsel et al., 2020; Gomkale et al., 2021). Interestingly, Tim23 seems to bind to two distinct patches on the surface of Tim50 (Tamura et al., 2009; Qian et al., 2011; Dayan et al., 2019). Also, lipids seem to play an important role in the interaction between the two proteins (Malhotra et al., 2017). Despite many efforts, the picture of Tim23-Tim50 interaction and particularly its dynamics remains blurry, likely due to the intrinsically disordered character of the Tim23 segment involved in the interaction (Gevorkyan-Airapetov et al., 2009; de La Cruz et al., 2010; Günsel et al., 2020).
Presequences are recognized by several components of the presequence pathway and precursor proteins influence not only the interactions between TOM and TIM23 complexes but also the interactions among the TIM23 subunits. Precursor proteins arrested at the trans site of the TOM complex can already be crosslinked to Tim50 (Yamamoto et al., 2002; Mokranjac et al., 2003, 2009), making it the first subunit of the TIM23 complex to recognize and bind presequences. The actual binding of presequences to the IMS segment of Tim50 was subsequently confirmed in a reconstituted system (Marom et al., 2011). Details of how translocating precursor proteins are transferred from the trans site of the TOM complex to Tim50 remain unclear. Precursor proteins increased the efficiency of chemical crosslinking between Tim50 and Tom22 (Waegemann et al., 2015), but they decreased a site-specific crosslink between the two proteins (Shiota et al., 2011). The situation got complicated even further when it was shown that the IMS segment of Tim50 consists of two domains: the highly evolutionary conserved core domain and the fungi-specific presequence-binding domain (PBD; Schulz et al., 2011). Though PBD was initially suggested to be solely responsible for recognizing and binding presequences, subsequent experiments showed that the core domain also binds presequences and even with similar affinity as PBD (Schulz et al., 2011; Lytovchenko et al., 2013). Recent NMR experiments indicate that the two domains of Tim50 bind to each other and that their interaction is modulated by presequences (Rahman et al., 2014). The receptor function of Tim50 depends on its interaction with Tim23 (Mokranjac et al., 2009; Tamura et al., 2009). Tim23 on its own also binds to presequences, however, with far lower affinity than Tim50 (Bauer et al., 1996; de La Cruz et al., 2010; Marom et al., 2011; Lytovchenko et al., 2013). Presequences dissociated the interaction of Tim50 with Tim21, indicating that Tim21 modulates the dynamic interplay of the TOM and TIM23 subunits in the IMS with presequences (Lytovchenko et al., 2013). Recent purification of the TOM-TIM23 supercomplex followed by crosslinking and mass spectrometry identified many new potential TOM-TIM23 interactions (Gomkale et al., 2021). Particularly interesting are the multiple contacts between Tim21 and Tom22 as well as the ones between Tim23 and Tom5 and Tom40. Unfortunately, these new contacts were not yet analyzed in intact mitochondria. It is also surprising that the TOM-TIM23 crosslinks previously identified in intact mitochondria were not recapitulated in this work.
Based on the available data, the current working model of how presequences are transferred from the trans site of the TOM complex to the translocation channel in the inner membrane would suggest the following scenario. Precursor proteins exit the TOM channel at the trans site where presequences are recognized by Tim50 (Figure 2B). The changes in multiple interactions between TOM and TIM23 complexes, induced by the recognition of presequences, would allow the presequences to be released from the trans site of the TOM complex, likely, to the PBD of Tim50. Binding of presequences to the PBD would then induce structural rearrangements within Tim50 so that presequences are further transferred to the core domain of Tim50 and the IMS-exposed segment of Tim23 (Figure 2C). In a membrane-potential dependent step, presequences are finally inserted into the TIM23 channel for translocation across the IM (Figure 2D).
Even ca. 30 years after identification of its first components, the presequence pathway still withholds many of its secrets. The major players involved in cooperation between the TOM and TIM23 complexes are probably all identified; however, we are only beginning to understand their multiple and dynamic interactions that underlie transfer of precursor proteins between two mitochondrial membranes and many open questions remain. Many of the TOM-TIM23 contacts have only been identified and their dynamics during translocation of proteins have not yet been analyzed. The different steps during transfer of presequences from the TOM to TIM23 complex remain speculative and unclear even on the level of components involved at different proposed steps. Understanding of presequence recognition in the IMS is still very limited, both on the level of the TOM and TIM23 complexes. Do the two domains of Tim50 bind presequences individually, do they together form a presequence-binding site or are they maybe involved in recognition of presequences during different stages of protein translocation? How does Tim23 contribute to presequence recognition in the IMS? Which domain of Tim50 is involved in which of the identified interactions of Tim50 and how do they change during translocation of proteins into mitochondria? If PBD of Tim50 is indeed only fungi-specific, how do higher eukaryotes deal with the lack of this domain? On a more general note, it will be interesting to know whether newly synthesized precursor proteins in the cytosol already know which TOM complexes are associated with TIM23 complexes or if the coordination of the two complexes predominantly happens after the presequences have reached the trans site of the TOM complex. If former, what distinguishes TOM complexes bound to TIM23 from the rest?
The successful use of the recent developments in the cryo-EM to solve the high-resolution structure of the TOM complex (Araiso et al., 2019; Tucker and Park, 2019; Wang et al., 2020) represents a milestone toward understanding the molecular mechanisms of protein import through the TOM complex and its coordination with other protein translocases. The structure of at least part of the TIM23 complex will hopefully become available soon (Sim et al., 2021). The ability to generate and purify the TOM-TIM23 supercomplex gives hope that the same developments can also be used to solve the structure of the supercomplex. The structures will certainly help in putting all the already available data in the structural context but also in raising novel hypotheses that can then be tested in careful biochemical experiments. The secrets of the presequence pathway seem less out of reach now than they were just few years ago.
MG and DM wrote the manuscript together and authors approved the submitted version. MG made the figures with the input from DM.
This work was supported by the Deutsche Forschungs gemeinschaft (MO1944/3-1 to DM).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the present and past members of the Mokranjac group and the Mito Club for the stimulating and inspiring discussions on mitochondrial biogenesis.
Abe, Y., Shodai, T., Muto, T., Mihara, K., Torii, H., Nishikawa, S., et al. (2000). Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100, 551–560. doi: 10.1016/s0092-8674(00)80691-1
Albrecht, R., Rehling, P., Chacinska, A., Brix, J., Cadamuro, S. A., Volkmer, R., et al. (2006). The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep. 7, 1233–1238. doi: 10.1038/sj.embor.7400828
Alder, N. N., Sutherland, J., Buhring, A. I., Jensen, R. E., and Johnson, A. E. (2008). Quaternary structure of the mitochondrial TIM23 complex reveals dynamic association between Tim23p and other subunits. MBoC 19, 159–170. doi: 10.1091/mbc.e07-07-0669
Araiso, Y., Tsutsumi, A., Qiu, J., Imai, K., Shiota, T., Song, J., et al. (2019). Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature 575, 395–401. doi: 10.1038/s41586-019-1680-7
Avendaño-Monsalve, M. C., Ponce-Rojas, J. C., and Funes, S. (2020). From cytosol to mitochondria: the beginning of a protein journey. Biol. Chem. 401, 645–661. doi: 10.1515/hsz-2020-0110
Backes, S., Bykov, Y. S., Flohr, T., Räschle, M., Zhou, J., Lenhard, S., et al. (2021). The chaperone-binding activity of the mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress. Cell Rep. 35:108936. doi: 10.1016/j.celrep.2021.108936
Bajaj, R., Jaremko, Ł., Jaremko, M., Becker, S., and Zweckstetter, M. (2014). Molecular basis of the dynamic structure of the TIM23 complex in the mitochondrial intermembrane space. Structure 22, 1501–1511. doi: 10.1016/j.str.2014.07.015
Bauer, M. F., Sirrenberg, C., Neupert, W., and Brunner, M. (1996). Role of Tim23 as voltage sensor and Presequence receptor in protein import into mitochondria. Cell 87, 33–41. doi: 10.1016/S0092-8674(00)81320-3
Becker, T., Song, J., and Pfanner, N. (2019). Versatility of Preprotein transfer from the cytosol to mitochondria. Trends Cell Biol. 29, 534–548. doi: 10.1016/j.tcb.2019.03.007
Blobel, G. (2000). Protein targeting (Nobel lecture). Chembiochem 1, 86–102. doi: 10.1002/1439-7633(20000818)1:2<86::AID-CBIC86>3.0.CO;2-A
Bykov, Y. S., Rapaport, D., Herrmann, J. M., and Schuldiner, M. (2020). Cytosolic events in the biogenesis of mitochondrial proteins. Trends Biochem. Sci. 45, 650–667. doi: 10.1016/j.tibs.2020.04.001
Chacinska, A., Lind, M., Frazier, A. E., Dudek, J., Meisinger, C., Geissler, A., et al. (2005). Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120, 817–829. doi: 10.1016/j.cell.2005.01.011
Chacinska, A., Rehling, P., Guiard, B., Frazier, A. E., Schulze-Specking, A., Pfanner, N., et al. (2003). Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 22, 5370–5381. doi: 10.1093/emboj/cdg532
Craig, E. A. (2018). Hsp70 at the membrane: driving protein translocation. BMC Biol. 16:11. doi: 10.1186/s12915-017-0474-3
Dayan, D., Bandel, M., Günsel, U., Nussbaum, I., Prag, G., Mokranjac, D., et al. (2019). A mutagenesis analysis of Tim50, the major receptor of the TIM23 complex, identifies regions that affect its interaction with Tim23. Sci. Rep. 9:2012. doi: 10.1038/s41598-018-38353-1
de La Cruz, L., Bajaj, R., Becker, S., and Zweckstetter, M. (2010). The intermembrane space domain of Tim23 is intrinsically disordered with a distinct binding region for presequences. Protein Sci. 19, 2045–2054. doi: 10.1002/pro.482
Donzeau, M., Káldi, K., Adam, A., Paschen, S., Wanner, G., Guiard, B., et al. (2000). Tim23 links the inner and outer mitochondrial membranes. Cell 101, 401–412. doi: 10.1016/S0092-8674(00)80850-8
Esaki, M., Shimizu, H., Ono, T., Yamamoto, H., Kanamori, T., Nishikawa, S., et al. (2004). Mitochondrial protein import. Requirement of presequence elements and tom components for precursor binding to the TOM complex. J. Biol. Chem. 279, 45701–45707. doi: 10.1074/jbc.M404591200
Geissler, A., Chacinska, A., Truscott, K. N., Wiedemann, N., Brandner, K., Sickmann, A., et al. (2002). The mitochondrial presequence translocase. Cell 111, 507–518. doi: 10.1016/S0092-8674(02)01073-5
Gevorkyan-Airapetov, L., Zohary, K., Popov-Celeketic, D., Mapa, K., Hell, K., Neupert, W., et al. (2009). Interaction of Tim23 with Tim50 is essential for protein translocation by the mitochondrial TIM23 complex. J. Biol. Chem. 284, 4865–4872. doi: 10.1074/jbc.M807041200
Gomkale, R., Linden, A., Neumann, P., Schendzielorz, A. B., Stoldt, S., Dybkov, O., et al. (2021). Mapping protein interactions in the active TOM-TIM23 supercomplex. Nat. Commun. 12:5715. doi: 10.1038/s41467-021-26016-1
Günsel, U., Paz, E., Gupta, R., Mathes, I., Azem, A., and Mokranjac, D. (2020). In vivo dissection of the intrinsically disordered receptor domain of Tim23. J. Mol. Biol. 432, 3326–3337. doi: 10.1016/j.jmb.2020.03.031
Hansen, K. G., and Herrmann, J. M. (2019). Transport of proteins into mitochondria. Protein J. 38, 330–342. doi: 10.1007/s10930-019-09819-6
Hwang, S., Jascur, T., Vestweber, D., Pon, L., and Schatz, G. (1989). Disrupted yeast mitochondria can import precursor proteins directly through their inner membrane. J. Cell Biol. 109, 487–493. doi: 10.1083/jcb.109.2.487
Kanamori, T., Nishikawa, S., Nakai, M., Shin, I., Schultz, P. G., and Endo, T. (1999). Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc. Natl. Acad. Sci. 96, 3634–3639. doi: 10.1073/pnas.96.7.3634
Künkele, K.-P., Heins, S., Dembowski, M., Nargang, F. E., Benz, R., Thieffry, M., et al. (1998). The preprotein translocation channel of the outer membrane of mitochondria. Cell 93, 1009–1019. doi: 10.1016/S0092-8674(00)81206-4
Lytovchenko, O., Melin, J., Schulz, C., Kilisch, M., Hutu, D. P., and Rehling, P. (2013). Signal recognition initiates reorganization of the presequence translocase during protein import. EMBO J. 32, 886–898. doi: 10.1038/emboj.2013.23
Malhotra, K., Modak, A., Nangia, S., Daman, T. H., Gunsel, U., Robinson, V. L., et al. (2017). Cardiolipin mediates membrane and channel interactions of the mitochondrial TIM23 protein import complex receptor Tim50. Sci. Adv. 3:e1700532. doi: 10.1126/sciadv.1700532
Marom, M., Dayan, D., Demishtein-Zohary, K., Mokranjac, D., Neupert, W., and Azem, A. (2011). Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 protein complex. J. Biol. Chem. 286, 43809–43815. doi: 10.1074/jbc.M111.261040
Mayer, A., Neupert, W., and Lill, R. (1995). Mitochondrial protein import: reversible binding of the presequence at the trans side of the outer membrane drives partial translocation and unfolding. Cell 80, 127–137. doi: 10.1016/0092-8674(95)90457-3
Meinecke, M., Wagner, R., Kovermann, P., Guiard, B., Mick, D. U., Hutu, D. P., et al. (2006). Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312, 1523–1526. doi: 10.1126/science.1127628
Mokranjac, D. (2020). How to get to the other side of the mitochondrial inner membrane – the protein import motor. Biol. Chem. 401, 723–736. doi: 10.1515/hsz-2020-0106
Mokranjac, D., Paschen, S. A., Kozany, C., Prokisch, H., Hoppins, S. C., Nargang, F. E., et al. (2003). Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 22, 816–825. doi: 10.1093/emboj/cdg090
Mokranjac, D., Popov-Celeketić, D., Hell, K., and Neupert, W. (2005). Role of Tim21 in mitochondrial translocation contact sites. J. Biol. Chem. 280, 23437–23440. doi: 10.1074/jbc.C500135200
Mokranjac, D., Sichting, M., Popov-Celeketić, D., Mapa, K., Gevorkyan-Airapetov, L., Zohary, K., et al. (2009). Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. MBoC 20, 1400–1407. doi: 10.1091/mbc.e08-09-0934
Neupert, W. (2015). A perspective on transport of proteins into mitochondria: a myriad of open questions. J. Mol. Biol. 427, 1135–1158. doi: 10.1016/j.jmb.2015.02.001
Pfanner, N., Warscheid, B., and Wiedemann, N. (2019). Mitochondrial proteins: from biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 20, 267–284. doi: 10.1038/s41580-018-0092-0
Popov-Celeketić, D., Mapa, K., Neupert, W., and Mokranjac, D. (2008). Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 27, 1469–1480. doi: 10.1038/emboj.2008.79
Qian, X., Gebert, M., Höpker, J., Yan, M., Li, J., Wiedemann, N., et al. (2011). Structural basis for the function of Tim50 in the mitochondrial presequence translocase. J. Mol. Biol. 411, 513–519. doi: 10.1016/j.jmb.2011.06.020
Rahman, B., Kawano, S., Yunoki-Esaki, K., Anzai, T., and Endo, T. (2014). NMR analyses on the interactions of the yeast Tim50 C-terminal region with the presequence and Tim50 core domain. FEBS Lett. 588, 678–684. doi: 10.1016/j.febslet.2013.12.037
Saitoh, T., Igura, M., Obita, T., Ose, T., Kojima, R., Maenaka, K., et al. (2007). Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 26, 4777–4787. doi: 10.1038/sj.emboj.7601888
Schulz, C., Lytovchenko, O., Melin, J., Chacinska, A., Guiard, B., Neumann, P., et al. (2011). Tim50’s presequence receptor domain is essential for signal driven transport across the TIM23 complex. J. Cell Biol. 195, 643–656. doi: 10.1083/jcb.201105098
Schulz, C., Schendzielorz, A., and Rehling, P. (2015). Unlocking the presequence import pathway. Trends Cell Biol. 25, 265–275. doi: 10.1016/j.tcb.2014.12.001
Shiota, T., Mabuchi, H., Tanaka-Yamano, S., Yamano, K., and Endo, T. (2011). In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc. Natl. Acad. Sci. U. S. A. 108, 15179–15183. doi: 10.1073/pnas.1105921108
Sim, S. I., Chen, Y., and Park, E. (2021). Structural basis of mitochondrial protein import by the TIM complex. BioRxiv. [ahead of preprint]. doi: 10.1101/2021.10.10.463828
Tamura, Y., Harada, Y., Shiota, T., Yamano, K., Watanabe, K., Yokota, M., et al. (2009). Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184, 129–141. doi: 10.1083/jcb.200808068
Tucker, K., and Park, E. (2019). Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat. Struct. Mol. Biol. 26, 1158–1166. doi: 10.1038/s41594-019-0339-2
Vögtle, F.-N., Wortelkamp, S., Zahedi, R. P., Becker, D., Leidhold, C., Gevaert, K., et al. (2009). Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439. doi: 10.1016/j.cell.2009.07.045
Waegemann, K., Popov-Čeleketić, D., Neupert, W., Azem, A., and Mokranjac, D. (2015). Cooperation of TOM and TIM23 complexes during translocation of proteins into mitochondria. J. Mol. Biol. 427, 1075–1084. doi: 10.1016/j.jmb.2014.07.015
Wang, W., Chen, X., Zhang, L., Yi, J., Ma, Q., Yin, J., et al. (2020). Atomic structure of human TOM core complex. Cell Discov. 6:67. doi: 10.1038/s41421-020-00198-2
Wiedemann, N., and Pfanner, N. (2017). Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 86, 685–714. doi: 10.1146/annurev-biochem-060815-014352
Keywords: mitochondria, protein translocation, presequence pathway, TOM-TIM23 contacts, precursor transfer, intermembrane space, TOM complex, TIM23 complex
Citation: Genge MG and Mokranjac D (2022) Coordinated Translocation of Presequence-Containing Precursor Proteins Across Two Mitochondrial Membranes: Knowns and Unknowns of How TOM and TIM23 Complexes Cooperate With Each Other. Front. Physiol. 12:806426. doi: 10.3389/fphys.2021.806426
Received: 31 October 2021; Accepted: 03 December 2021;
Published: 06 January 2022.
Edited by:
Nathan Alder, University of Connecticut, United StatesReviewed by:
Johannes M. Herrmann, University of Kaiserslautern, GermanyCopyright © 2022 Genge and Mokranjac. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dejana Mokranjac, bW9rcmFuamFjQGJpby5sbXUuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.