- 1School of Agriculture, Ludong University, Yantai, China
- 2College of Life Sciences, Yantai University, Yantai, China
- 3Shandong Fisheries Development and Resources Conservation Center, Yantai, China
- 4Yantai Haiyu Marine Science and Technology Co. Ltd., Yantai, China
The metabolic processes of organisms are very complex. Each process is crucial and affects the growth, development, and reproduction of organisms. Metabolism-related mechanisms in Octopus ocellatus behaviors have not been widely studied. Brood-care is a common behavior in most organisms, which can improve the survival rate and constitution of larvae. Octopus ocellatus carried out this behavior, but it was rarely noticed by researchers before. In our study, 3,486 differentially expressed genes (DEGs) were identified based on transcriptome analysis of O. ocellatus. We identify metabolism-related DEGs using GO and KEGG enrichment analyses. Then, we construct protein–protein interaction networks to search the functional relationships between metabolism-related DEGs. Finally, we identified 10 hub genes related to multiple gene functions or involved in multiple signal pathways and verified them using quantitative real-time polymerase chain reaction (qRT-PCR). Protein–protein interaction networks were first used to study the effects of brood-care behavior on metabolism in the process of growing of O. ocellatus larvae, and the results provide us valuable genetic resources for understanding the metabolic processes of invertebrate larvae. The data lay a foundation for further study the brood-care behavior and metabolic mechanisms of invertebrates.
Introduction
Cephalopods are marine mollusks that are widely distributed throughout the world’s oceans. The artificial breeding of cephalopods has gradually appeared all over the world in the last few years because of the difficult catching (Arkhipkin, 1985; Budelmann, 1994; Schnell et al., 2020). Previous studies have shown that the reproductive cycle of cephalopods is generally around 2 months (Zheng et al., 2009; Wang et al., 2010). They lay eggs about a month after mating, and the eggs hatch after 25–40 days. It’s worth noting that water temperature significantly affects the incubation time, suitable water temperature will greatly reduce the incubation time (Wang et al., 2010; Zheng et al., 2011). Normally, the incubation rates of cephalopod larvae are 70–80%, and the survival rate is more than 90% (Zheng et al., 2009, 2011; Wang et al., 2010). Octopus ocellatus has become a sought-after breed for its high nutritional value, short growth cycle, rapid growth, strong adaptability to the breeding environment, and ease of reproduction (Boletzky, 1975; Navarro and Villanueva, 2003; Rosas et al., 2013; Wei et al., 2015). In China, the research on O. ocellatus larvae has mainly focused on survival, feeding, and palatability factors, with relatively less research on the breeding methods of O. ocellatus during the hatching period (Boletzky, 1989; Li et al., 2019).
There are many factors that affect the growth and development of O. ocellatus larvae, including water quality, dissolved oxygen, light, temperature, and brood-care (Boletzky, 1989; Quiroga and Garcia, 2003; Huffard, 2013; Vidal et al., 2014; Seibel, 2016; Chatterjee et al., 2018). The brood-care behavior of female O. ocellatus can reduce the occurrence of hatching diseases and improve the hatching rate (Boletzky, 1989; Baeza and Fernández, 2002). Similarly, in marine vertebrates such as catfish, perch, and other fish, parents may display brood-care behavior, and this has an impact on the birth rate and bodily functions of fish larvae (Bunn et al., 2000; Baldridge and Lodge, 2013; Foster et al., 2016). The brood-care behavior affects the growth of O. ocellatus larvae and determines the immune and metabolic functions of the larvae to a certain extent (Zheng et al., 2009; Wang et al., 2010).
Metabolism is the synthesis and biochemical processes of cells and tissues (Harper and Patti, 2020). The metabolic network of organisms is very complex, and multiple metabolic processes share the metabolic functions together. Changes in one metabolic process often lead to changes in other processes. A good understanding of metabolic functions is conducive to the study of the physiological and biochemical functions of organisms (Hosseini et al., 2016; Matsumoto et al., 2018; Xu and Zheng, 2020). In studies of invertebrate metabolism, energy accounts for a large proportion; for example, glucose metabolism in marine mollusks or the comparison of energy metabolism between Bivalvia and Brachiopoda (Martínez-Quintana and Yepiz-Plascencia, 2012; Bruning et al., 2013; Payne et al., 2014). Studies of cephalopod metabolic impact factors mainly focus on external environmental factors, such as water depth and habitat temperature, while other studies consider metabolic mechanisms through starvation, individual size, and other aspects (Seibel et al., 1997; Pimentel et al., 2012; Ikeda, 2016; Speers-Roesch et al., 2016). The relationships between metabolism and brood-care behavior have not been reported up to now.
We divide O. ocellatus eggs laid by the same parent into Pro group (brood-care group) and Unp (brood-careless group) in our research. We performed transcriptome sequencing and bioinformatics analyses using these larvae in two groups at 0 h, 4 h, 12 h, and 24 h after hatching, including gene function annotation, differentially expressed genes analysis, GO enrichment analysis, KEGG functional enrichment analysis, and metabolism-related protein–protein interaction networks (PPI network) analysis. Among these analyses, 3,486 DEGs were identified, which may be related to metabolic processes. The results of a heatmap indicate that brood-care has a great impact on the metabolism of larvae. Ten metabolism-related KEGG signaling pathways were enriched in our study. They are closely related to larval metabolism and maintain the metabolic functions. We then construct a protein–protein interaction network using 49 genes enriched in these pathways and find multiple key genes most related to larval metabolism. In the end, quantitative RT-PCR (qRT-PCR) was used to identify and verify 10 hub genes. These results supply us with novel insights for further comprehending the relationships between O. ocellatus brood-care behavior and larval metabolic mechanisms. This conclusion lays a foundation for the artificial breeding technology of O. ocellatus.
Materials and Methods
Ethics Statement
Octopus ocellatus samples were obtained from a commercial hatchery. This research was conducted in accordance with the protocols of the Institutional Animal Care and Use Committee of the Ludong University (protocol number LDU-IRB20210308NXY) and the China Government Principles for the Utilization and Care of Invertebrate Animals Used in Testing, Research, and Training (State Science and Technology Commission of the People’s Republic of China for No. 2, October 31, 19881).
Sample Collection and RNA Preparation
We caught wild O. ocellatus parents in Rizhao sea area to prepare for this experiment, the latitude and longitude of animals sampling site are 34.91005177008973 and 119.63820016113283. The eggs spawned by parents were collected after temporary feeding, and these were divided into two groups (Pro and Unp). Among them, the eggs in Pro group continued to be brooded by their parents, and the eggs in Unp group were hatched in flowing seawater without parent brood. Then, the primary incubation larvae were temporarily cultured in floating seawater for 24 h. The temperature of seawater was stable at the optimum temperature of eggs hatching (19–20.8°C) in 29 days of hatching. The larvae at 0h, 4h, 12h, and 24h after hatching were collected and stored in liquid nitrogen container until RNA extraction using TRIzol method.
We randomly selected nine primary incubation larvae at each time point in two groups for RNA extraction: brood-care larvae grow for 0h (Pro-C), brood-care larvae grow for 4h (Pro-4h), brood-care larvae grow for 12h (Pro-12h), and brood-care larvae grow for 24h (Pro-24h); brood-careless larvae grow for 0h (Unp-C), brood-careless larvae grow for 4h (Unp-4h), brood-careless larvae grow for 12h (Unp-12h), and brood-careless larvae grow for 24h (Unp-24h). Three of the nine larvae in a group at each time point were randomly selected, and their equal molar mass of RNA was concentrated in a replicate as a template for constructing the transcriptome library; the same method was used to pooled equal molar mass RNA from the three larvae into the second replicate, and the equal molar mass RNA of the remaining larvae was concentrated in the third replicate. We subsequent qRT-PCR verification using remaining RNA that has been preserved.
Library Construction and Illumina Sequencing
The method of Li et al. (2017) description was used to construct the library. And Illumina Hiseq 4000 platform was used to sequence samples.
The Expression and Function Annotation of Genes
We splice Trinity with RSEM to obtain reference sequences and map the clean reads to these sequences. Then, a great quantity of read counts of each sample mapped to each gene was obtained. To understand the expression and abundance of genes, we transform read counts into FPKM. The result showed that there was a positive correlation between FPKM and the expression level of samples. The unigenes were annotated by searching the sequences against the NR, NT, GO, SwissProt, and KOG databases using BLASTX with a cut-off of Evalue ≤ 1e-5. Meanwhile, we annotated unigenes into Pfam database with Hmmer 3.0 package (Evalue ≤ 0.01).
The Screening and Analysis of Differentially Expressed Genes
Differentially expressed genes (DEGs) were screened out using the DESeq2 package for R, and the filter parameters were | log2 Fold Change| ≥ 4 and q-value ≤ 0.01. We identified DEGs distribution and multiple GO terms by analyzing DEGs with GO functional enrichment analysis and conduct the DEGs statistical analyses. We used KEGG enrichment analysis to further study the specific functions of DEGs, and the metabolism-related pathways involved in DEGs were annotated. Finally, the signaling pathways with significant enrichment of DEGs were selected, and we count the species and number of DEGs after selecting.
The Construction of Functional Protein Association Networks
STRING v11.0 with default parameters was used to construct PPI networks to study relationships between functions of metabolism-related genes (Damian et al., 2018).
The Validation of Quantitative Real-Time Polymerase Chain Reaction
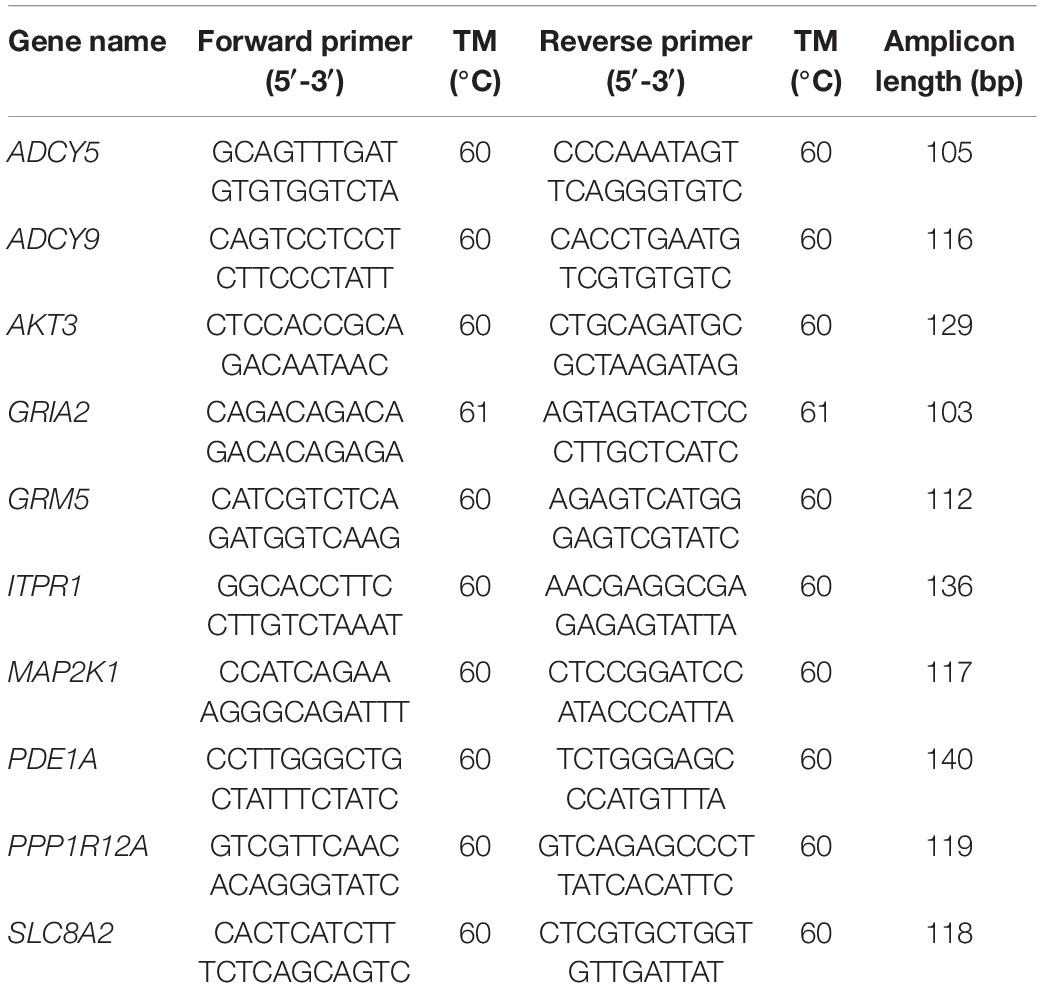
We verified the accuracy of our RNA-Seq results using quantitative real-time polymerase chain reaction (qRT-PCR) to validate 10 selected genes. In this experiment, each group contained three biological replicates, and Primer Premier 5.0 software was used to project gene-specific primers. We screened 10 genes for verification, and their names and primer sequences are listed in Table 1. The stability of three genes including 18S, β-actin, and GAPDH were evaluated in different tissues and embryo development stages of O. ocellatus. By comparison, we used O. ocellatusβ-actin as an endogenous control based on its expression level which tended to be stable. This experiment is based on Li et al.’s (2019) description method for qRT-PCR.
Results
Apparent Results of Octopus ocellatus Larvae
Brood-care behavior affects the hatching rate and survival rate of O. ocellatus larvae. The survival rate of brood-careless eggs was significantly lower than that of normal hatching larvae, and the hatching rate was reduced by 30–40%. However, there were no significant differences in morphology and development of larvae between Pro and Unp.
The Results and Quality Assessment of Sequencing
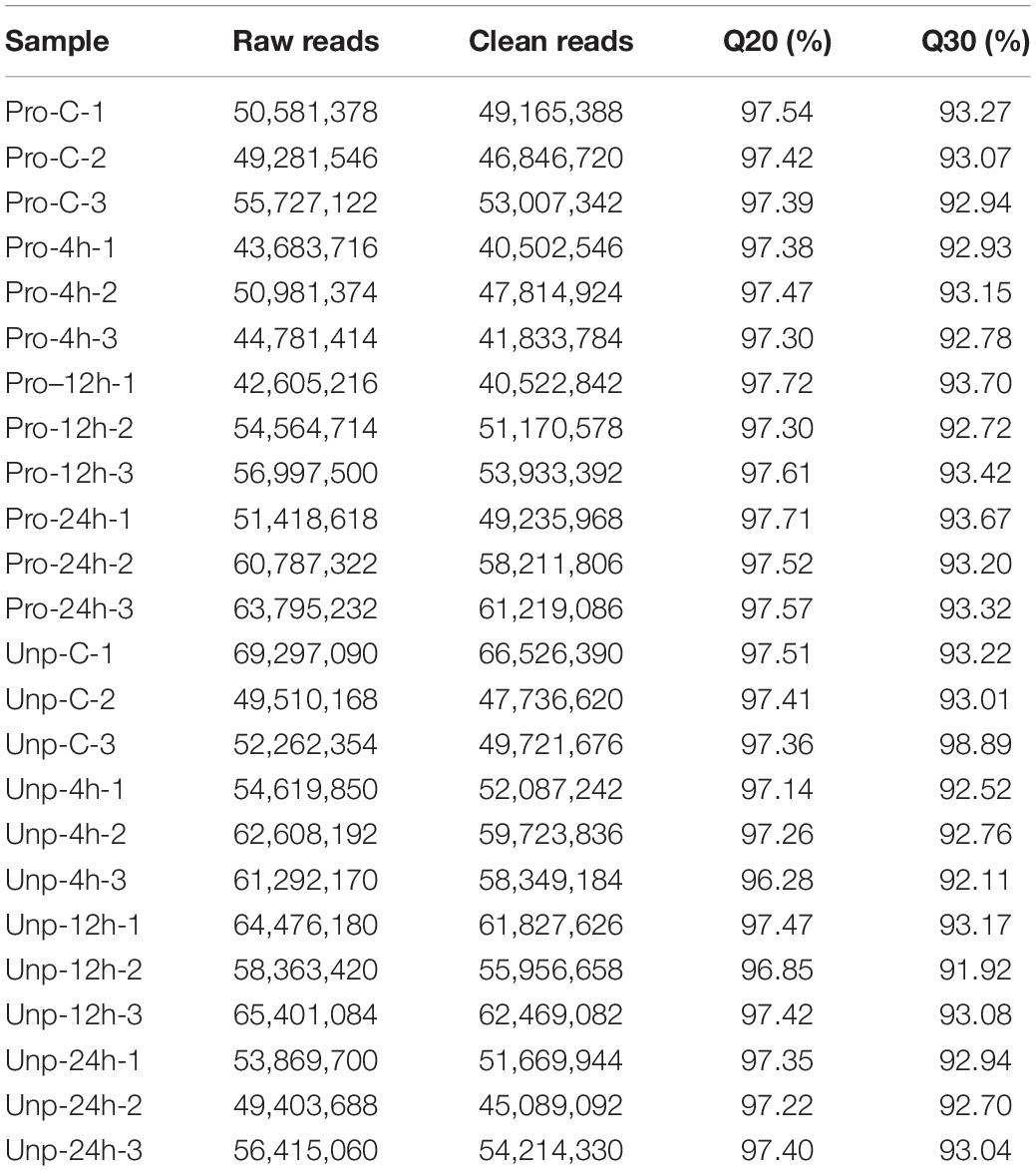
Samples at four time points in two groups were sequenced by RNA-Seq. Table 2 shows the detailed results of sequencing. Raw sequencing reads were submitted to Sequence Read Archive in NCBI; the SRA accession numbers were SRR15204591, SRR15204592, SRR15204593, SRR15204594, SRR15204595, SRR15204596, SRR15204597, SRR15204598, SRR15204599, SRR15204600, SRR15204601, SRR15204602, SRR15927331, SRR15927332, SRR15927333, SRR15927334, SRR15927336, SRR15927337, SRR15927338, SRR15927339, SRR15927340, SRR15927341, SRR15927342, and SRR159273432.
Differential Expression Analysis
Through differential expression analysis, we found 229, 583, 659, and 2,328 DEGs in Pro group at 0h, 4h, 12h, and 24h after hatching compared with Unp group. Among these, 101 DEGs were up-regulated, and 128 DEGs were down-regulated at 0h after hatching; 298 DEGs were up-regulated, and 285 DEGs were down-regulated at 4h after hatching; 323 DEGs were up-regulated, and 336 DEGs were down-regulated at 12 h after hatching; 1,657 DEGs were up-regulated, and 671 DEGs were down-regulated at 12 h after hatching (Figure 1). Venn diagram helps us find the union of DEGs at three time points (Figure 2). The clustering distribution of DEGs is shown in Figure 3 intuitively.
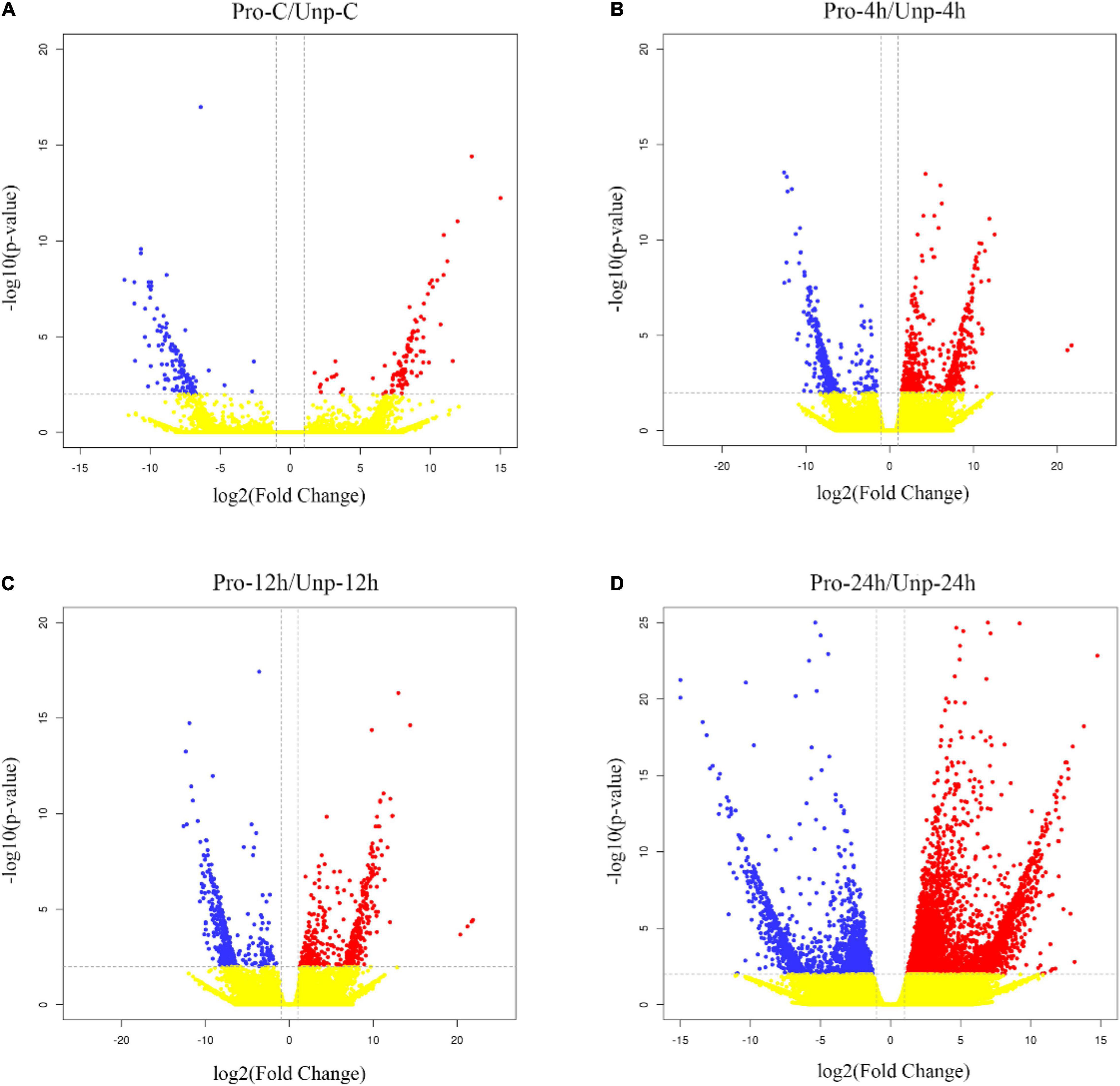
Figure 1. (A) Volcano plot of DEGs distribution trends between Pro-C and Unp-C. Each dot stands for a gene. Red dots indicate up-regulated DEGs; blue dots are down-regulated DEGs; and yellow dots stand for the genes with no difference. (B) Volcano Plot of DEGs distribution trends between Pro-4h and Unp-4h. (C) Volcano Plot of DEGs distribution trends between Pro-12h and Unp-12h. (D) Volcano Plot of DEGs distribution trends between Pro-24h and Unp-24h.
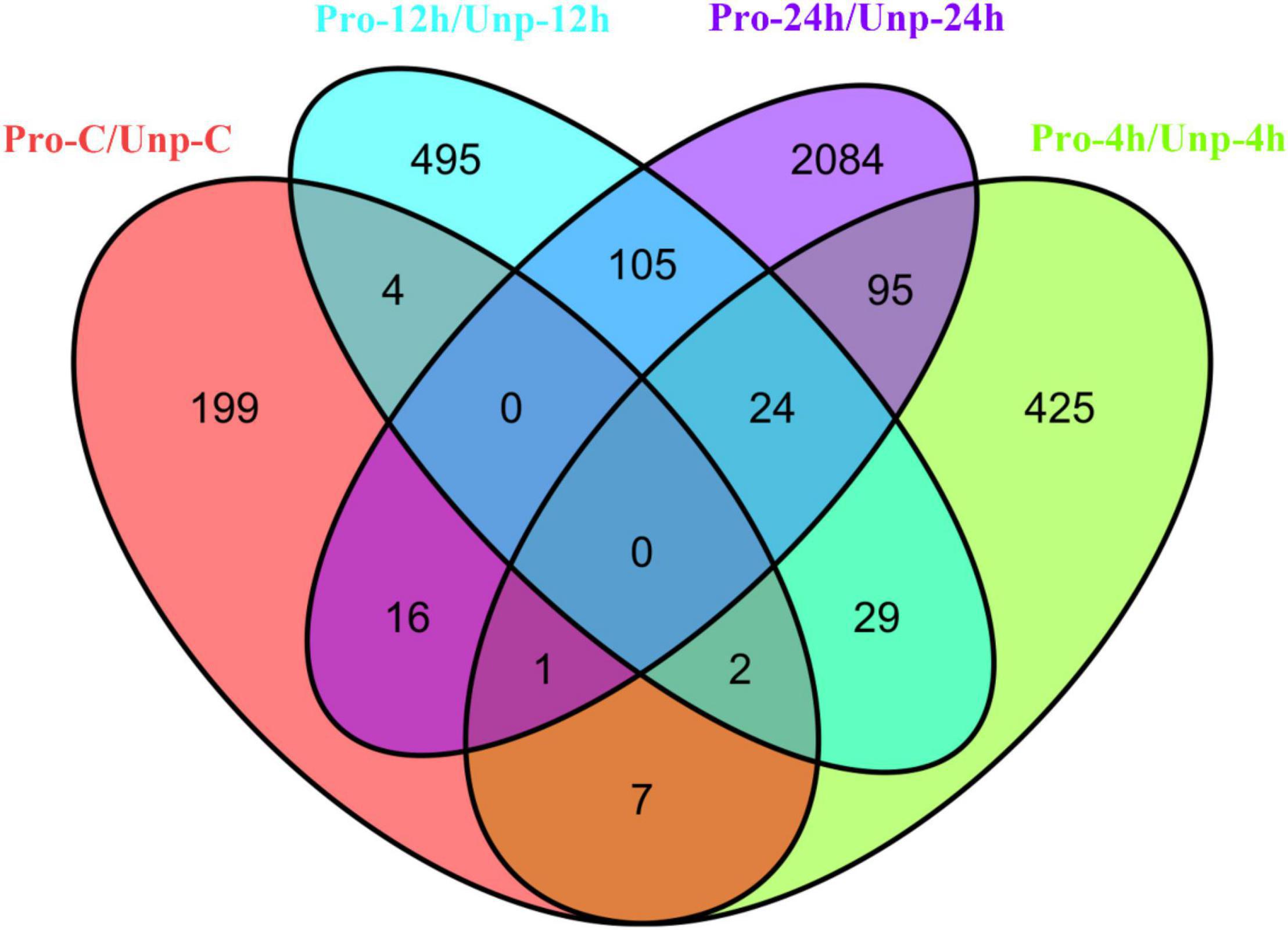
Figure 2. The Venn diagram shows the overlap of the DEGs at 0h (red), 4h (green), 12h (blue), and 24h (purple) after hatching.
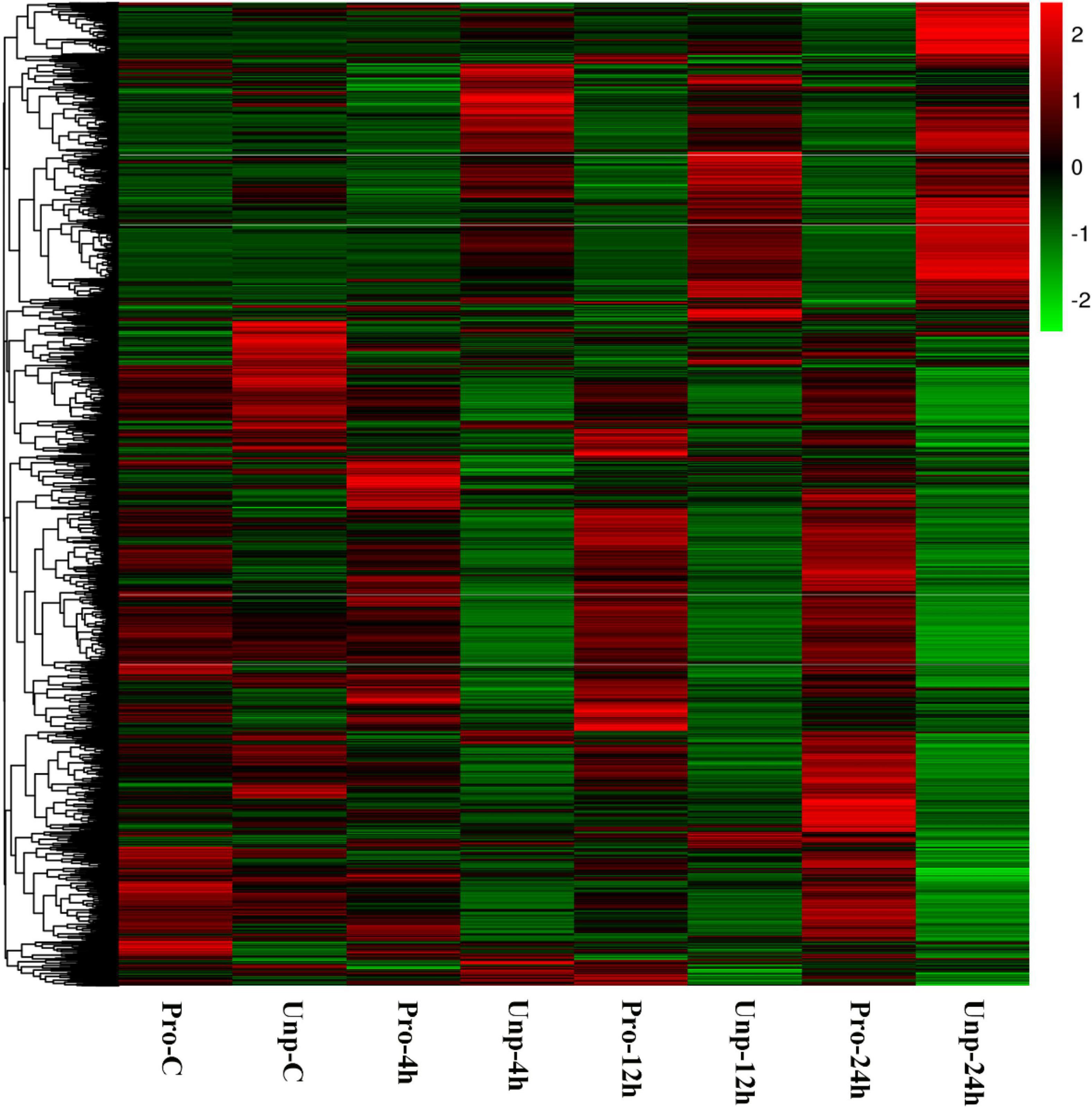
Figure 3. Heatmap analysis of hierarchical clustering of DEGs at four time points in two groups. Each row represents one gene, and each column stands for a time point. The colors range from green to red, indicating the level of expression from low to high.
GO and KEGG Enrichment Analyses of Differentially Expressed Genes
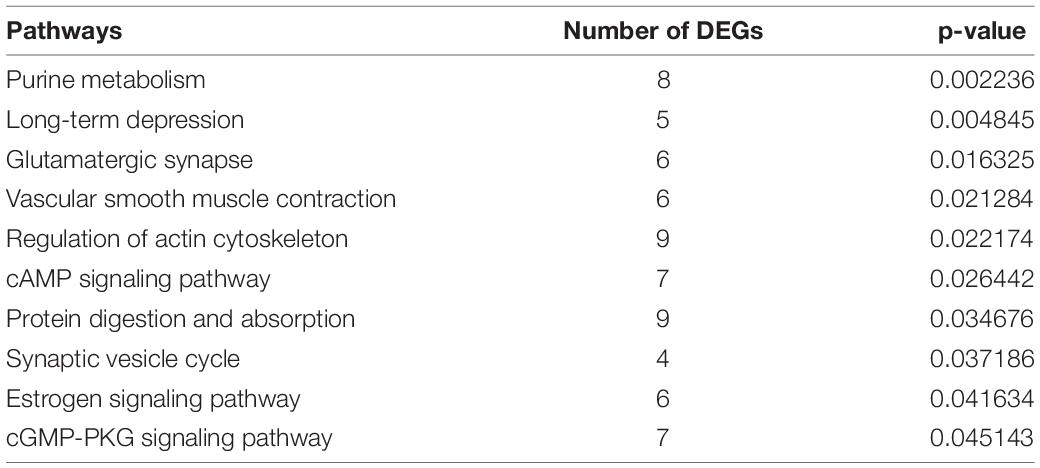
We identified 269 level-3 terms based on GO enrichment analysis including 151 biological process subclasses, 72 molecular function subclasses, and 46 cellular component subclasses. And the top 10 level-3 terms were shown in Figure 4. Meanwhile, The KEGG signaling pathways enriched with DEGs was analyzed to help us further understand the gene functions. We enriched 179 level-2 KEGG classes pathways containing 1,777 selected DEGs (Figure 5), and 10 significantly enriched metabolism-related pathways (Table 3) were identified in our study.
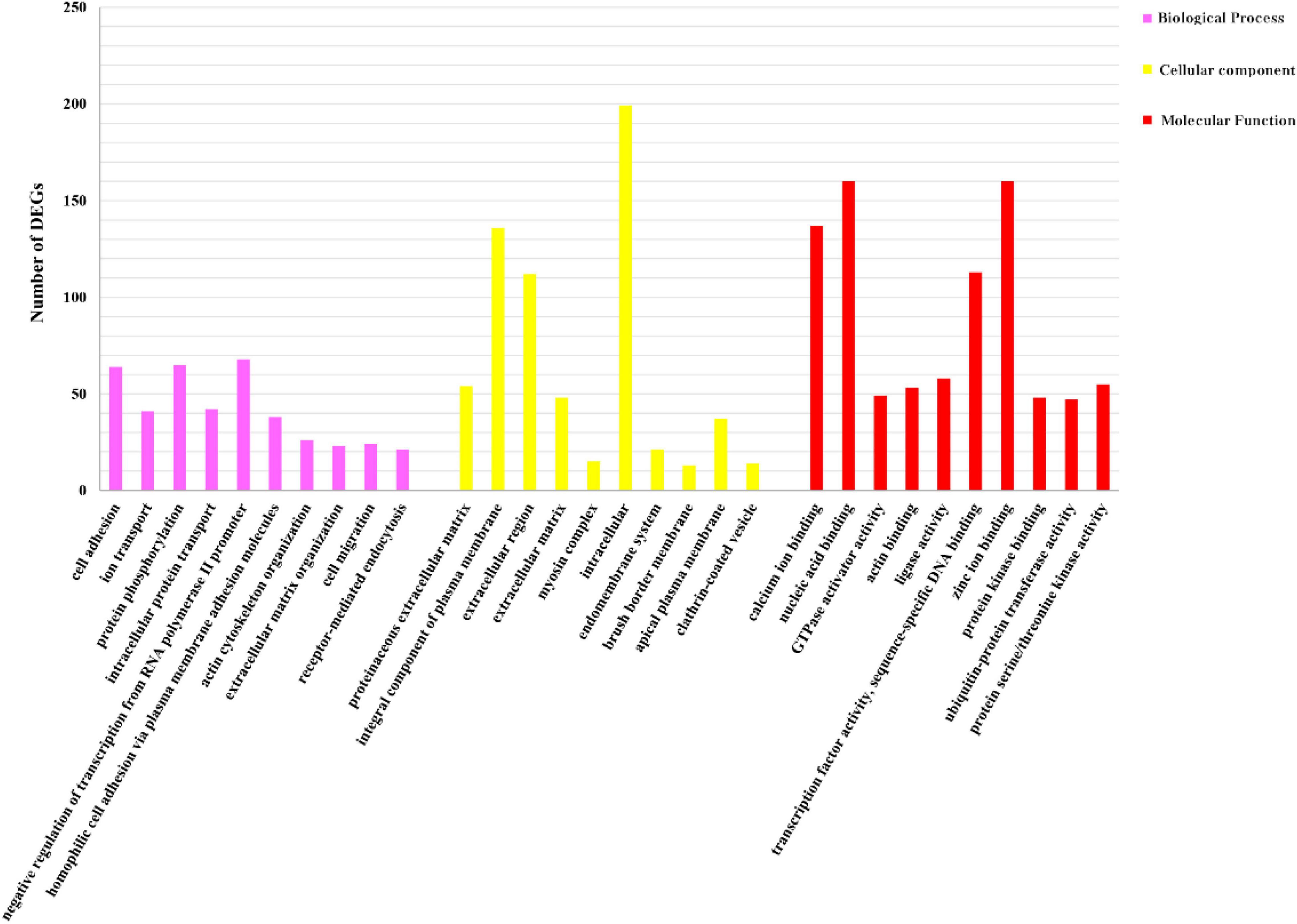
Figure 4. GO analysis of DEGs. Distribution of level-3 GO annotation in three categories. The y-axis represents the corresponding number of DEGs; the x-axis stands for the gene functional classification based on GO.
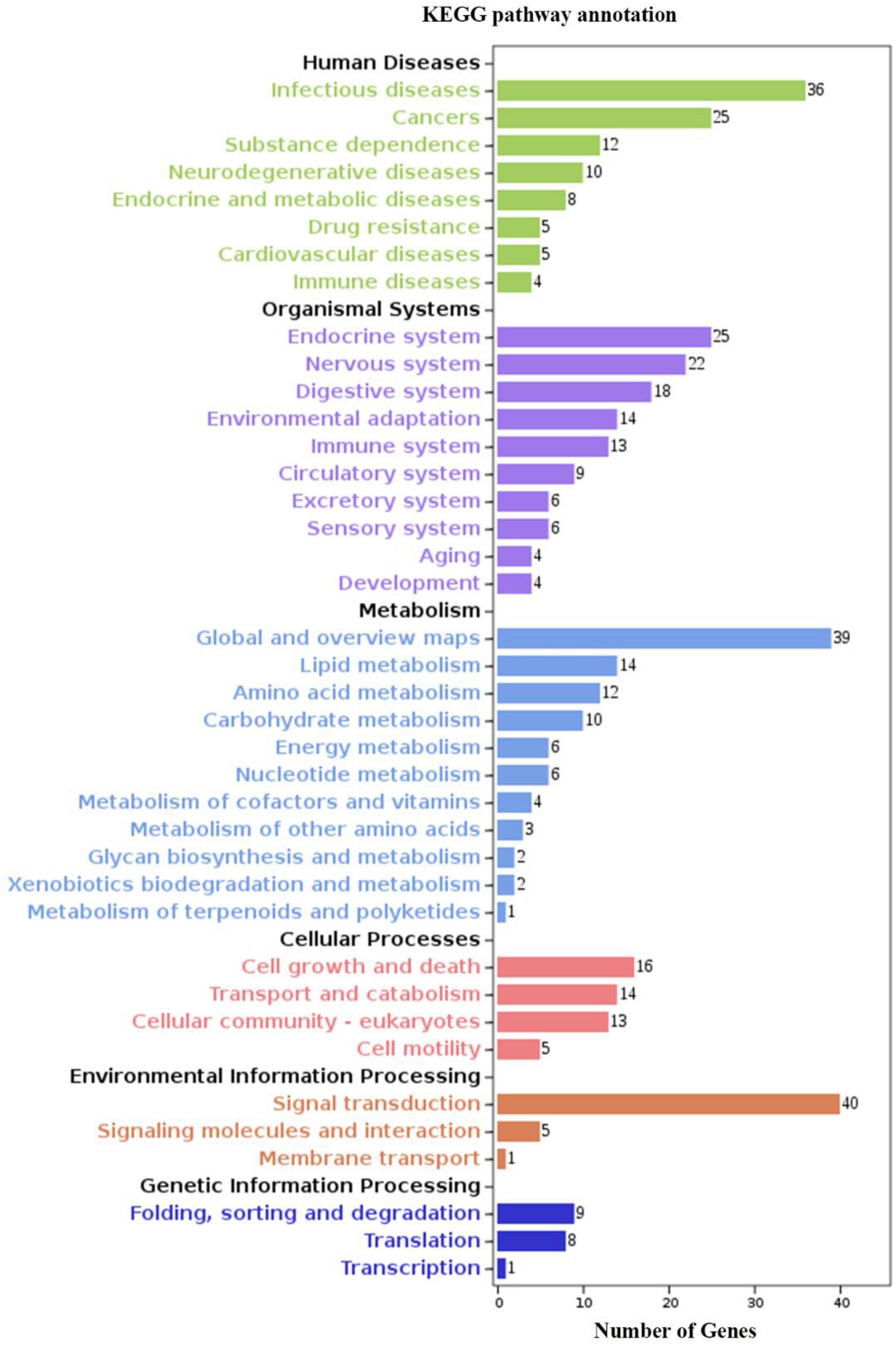
Figure 5. KEGG analysis of DEGs. The y-axis indicates level-2 KEGG classes; the x-axis represents the corresponding number of DEGs.
Construction of Metabolism-Related Protein–Protein Interaction Networks
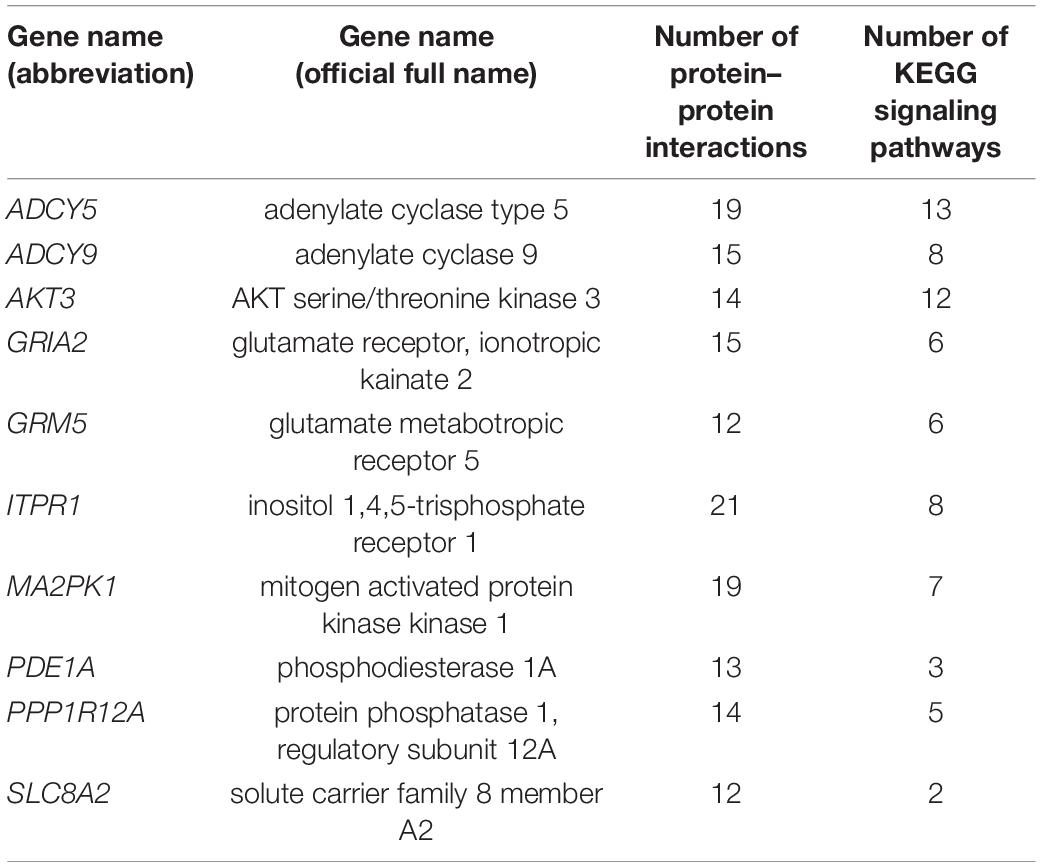
Proteins exist in all organisms as the main components of cells and tissues. All significant parts of the organism need protein participation, which is the main undertaker of life activities. We can identify key genes in the metabolic gene population by constructing protein–protein interaction networks. In our research, we used 49 gene protein sequences in the signaling pathways in Table 3 to construct the protein–protein interaction networks. The networks were shown in Figure 6. We found that the ITPR1 gene had the highest number of protein-protein interactions, and other genes were interacted with more than 10 proteins. Relevant parameter information of them was listed in Table 4. There were 44 nodes and 197 edges in the network, and each node was interacted with an average of 8.95 nodes. The clustering coefficient was 0.5, and the p-value of this network was ≤ 1.0e-16.
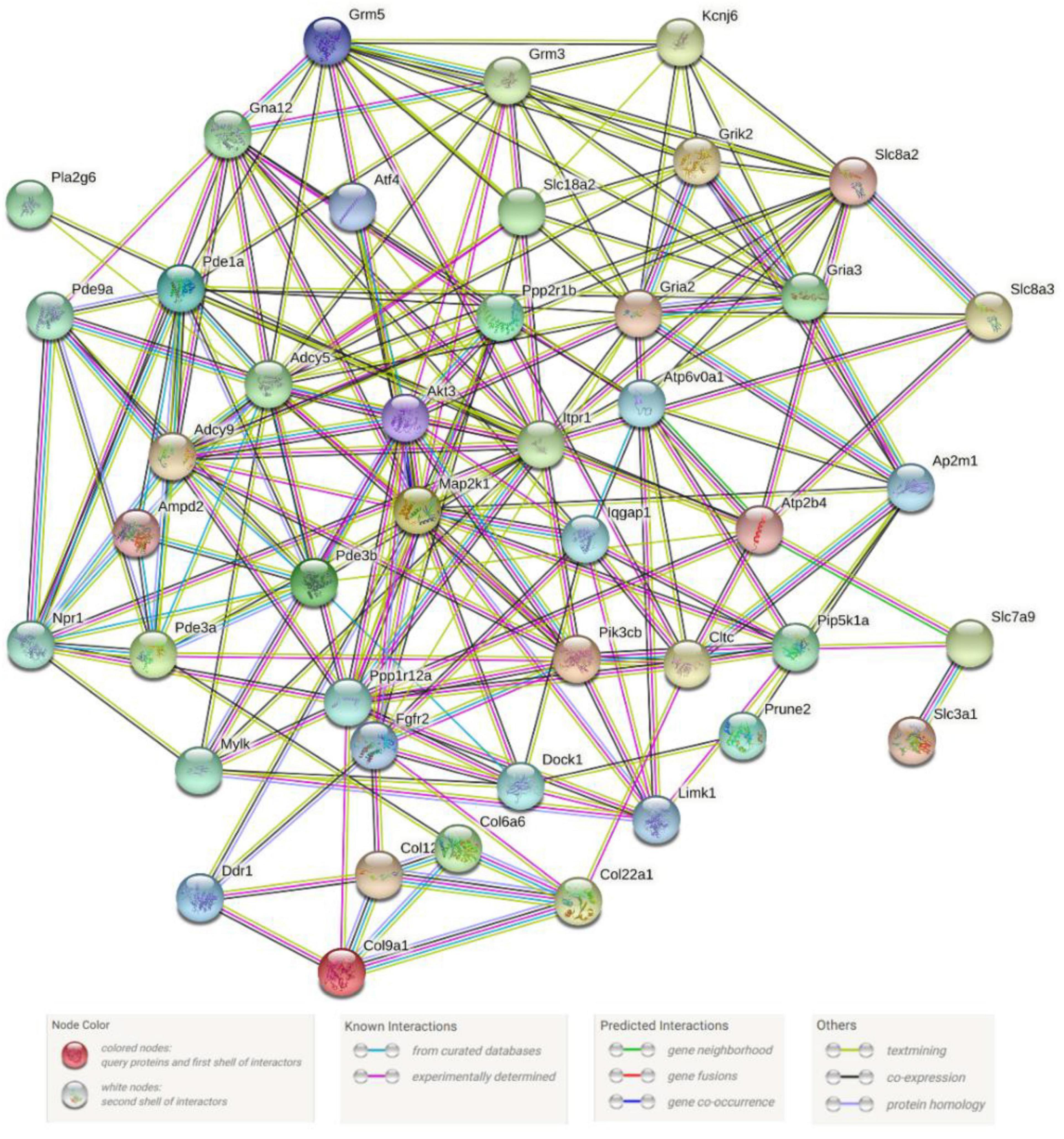
Figure 6. Metabolism-related protein–protein interaction networks. Network nodes stand for proteins. The legend represents the relationships between nodes.
Analysis of Key Metabolism-Related Differentially Expressed Genes
The interaction relationships between key metabolism-related DEGs were studied in our research. We identified 10 genes (Table 5) with high protein interactions or involved in multiple signaling pathways based on KEGG enrichment and PPI networks analyses, and then explored their interaction relationships. We divided these 10 key DEGs into five categories: solute carrier family, purine metabolism signaling pathway, cAMP signaling pathway, protein digestion and absorption signaling pathway, and other significant metabolism-related DEGs. The family and signaling pathways were gone hand in hand to metabolic processes, analyzing them will promote us to further understand the metabolic mechanisms of differences of larval growth between Pro group and Unp group.
Differentially Expressed Genes Verification by Quantitative Real-Time Polymerase Chain Reaction
We detected the expression of 10 metabolism-related DEGs at each time point in the two groups using qRT-PCR, and verified the consistency of qRT-PCR and RNA-Seq expression. The results of qRT-PCR suggested that all DEGs measured were single products. We compared the gene expression profiles between qRT-PCR results and RNA-Seq results (Figure 7). And the result suggested that their results were significantly correlated, and showed the same trend pattern.
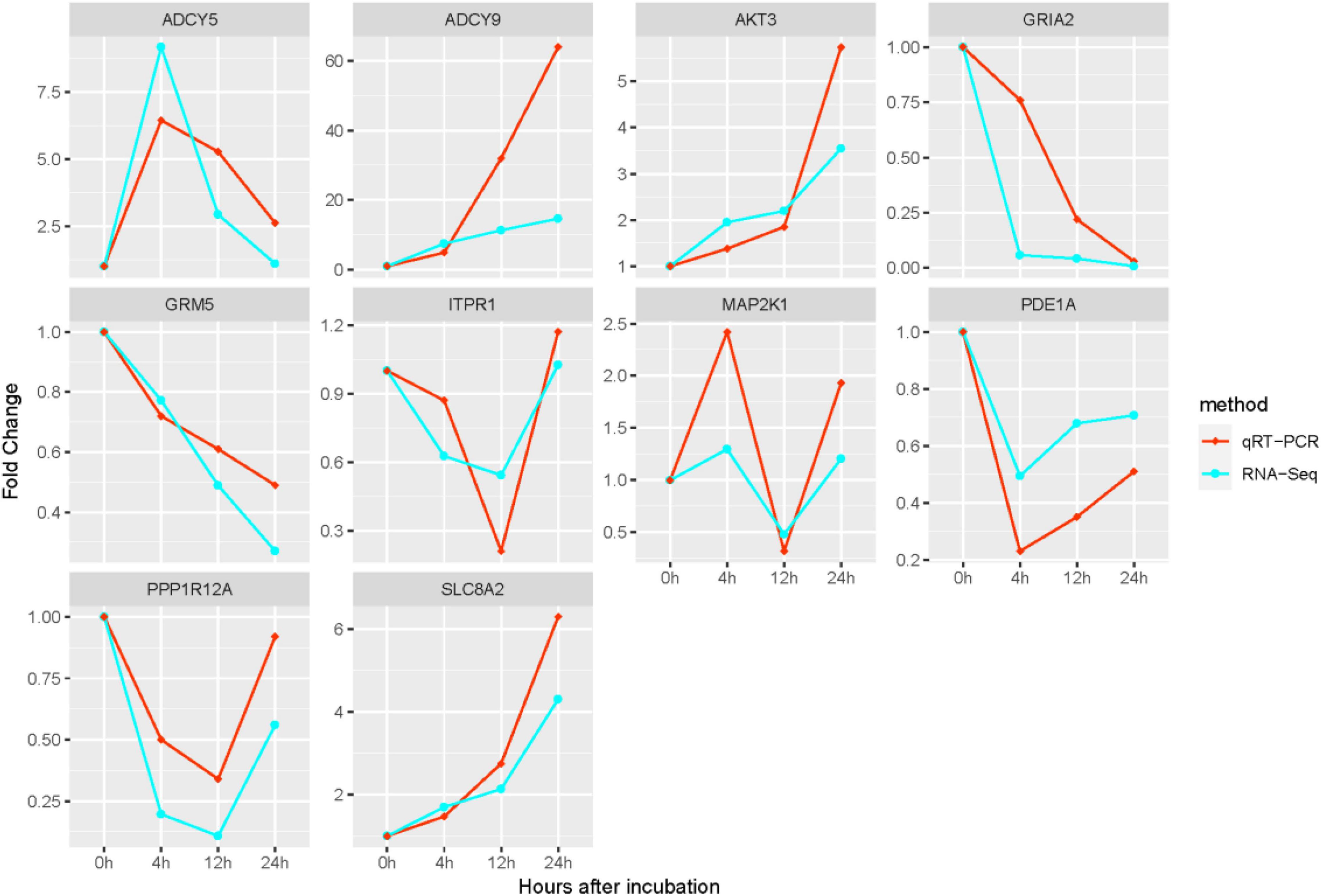
Figure 7. Comparison of expression of 10 hub genes between qRT-PCR and RNA-Seq results. The transcript expression levels of the selected DEGs were each normalized to that of the β-actin gene. The x-axis represents the growth time after hatching; the y-axis stands for fold change.
Discussion
The Purpose and Significance of This Study
Due to the rich nutrition and excellent taste, O. ocellatus is gradually favored by people (Boletzky, 1975; Wei et al., 2018). As an economically important species employed in mariculture, the artificial breeding of O. ocellatus is an issue of concern. We understand that multiple octopuses have the brood-care behavior based on previous studies, and the behavior directly affects the growth and development of their larvae. Thus, understanding the effects of brood-care behavior is vital to O. ocellatus larval growth and artificial breeding. Our research screened 3,486 DEGs, and these genes are considered to have significant relationships with O. ocellatus larval metabolic processes. A heatmap showed that the genes in two groups have significant differences of the same point in time. It is further indicated that there are quite metabolic differences between the brood-care larvae and the brood-careless larvae. Then, we identified 10 metabolism-related KEGG signaling pathways, they interact with each other and share the metabolic function of organisms. Finally, we construct protein-protein interaction networks using 49 DEGs in these signaling pathways to further explore O. ocellatus larval metabolic mechanisms.
Enrichment Analyses of Metabolism-Related GO Terms and KEGG Pathways
Many metabolism-related terms and pathways were yielded through GO and KEGG enrichment analyses, including cell adhesion term, protein phosphorylation term, cAMP signaling pathway, purine metabolism signaling pathway, and other key pathways and terms. These pathways and terms are directly related to metabolic processes, suggesting that there were several metabolism-related DEGs between larvae in two groups, and thus resulted in complex metabolic mechanisms of larvae. We comprehensively analyzed these terms and signaling pathways. The results will help us to understand the metabolic processes and molecular mechanisms of O. ocellatus under different incubation conditions within 24h of incubation, so as to lay a foundation for the artificial breeding of O. ocellatus.
Speculation of Hub Genes
Protein is an essential component of an organism and is closely related to metabolic processes. Studying their interaction relationships helps us to enrich our understanding of biological metabolic mechanisms We construct protein–protein interaction networks based on 49 key genes contained in metabolism-related signaling pathways. These proteins interacted significantly better than randomly selected groups of proteins of similar size. This conclusion suggests that the above proteins may function as a group. We thus speculated that proteins interact with more other proteins in the network were hub proteins in metabolism. The genes corresponding to hub proteins are further studied.
Functional Analyses of Metabolism-Related Hub Genes and Signaling Pathways
In our research, we analyze the differences in metabolic mechanisms between larvae under different hatching conditions using transcriptome profiling. The results help us to further understand the effect of O. ocellatus brood-care behavior on metabolic mechanisms. Ten hub genes were speculated and screened involved in multiple protein interactions or metabolism-related signaling pathways, and these hub genes, pathways, and the larval metabolic differences between two groups at the same time point were investigated.
Analysis of Three Metabolic Pathways
Purine metabolism is one of the key ways for aquatic animals to detoxify and obtain energy, which plays a major part in maintaining the metabolic stability of organisms. In the liver of marine organisms, purines are converted by degrading enzymes into urea and glyoxylate, and then excreted by other metabolic pathways (Noguchi et al., 1982). In recent studies, we found that there is a urease that can decompose urea into NH3 in marine invertebrates. NH3 is very soluble in water, organisms can excrete urea in the form of NH3 and CO2 through purine metabolism to prevent poisoning caused by excessive urea in the body (Yeldandi et al., 1996). In addition to detoxification, purine metabolism will release ATP, ADP, and other derivatives, providing the energy necessary for biological life activities (Furchgott, 1966; Sakuraba et al., 1996). In our research, key metabolism-related genes such as NPR1 and AMPD2 were enriched in this pathway. Compared with the Unp group, NPR1 was continuously up-regulated with the growth of O. ocellatus larvae, AMPD2 was down-regulated slightly in the first 4 h of growth, and up-regulated significantly in the last 20 h. These results indicate that brood-care larvae have higher detox and energy production abilities than brood-careless larvae, and that they have higher metabolic capacity; cAMP is an essential substance in biological cells. It regulates many biological functions, such as proliferation, differentiation, and apoptosis of cells. Meanwhile, cAMP also plays a role in regulating the balance of energy and nutrition. cAMP is widely distributed in many organelles, such as nucleus and cytoplasm. In mitochondria, cAMP can stabilize the function of mitochondria, and promote the growth of O. ocellatus larvae by regulating the stress response of cells and mitochondrial activity (Zhang et al., 2016). cAMP signaling pathway acts a key part in regulating the metabolism of saccharides and fat. It maintains the metabolic homeostasis by regulating the content of various saccharides. At the same time, it can regulate the formation of fat and the distribution of lipid in adipose tissue, control the differentiation of adipocytes, and maintain the fat content in vivo (Ravnskjaer et al., 2015). The maladjustment of cAMP signaling pathway may lead to cell metabolic disorders and the occurrence of various diseases (Tang et al., 2021). In our research, a large number of DEGs were significantly enriched in this pathway. And most DEGs were up-regulated in Pro group compared with Unp group, suggesting that the metabolic mechanisms of the brood-care larvae were more complex, and the ability to regulate this metabolic process was stronger. This phenomenon explains that the O. ocellatus larvae in Pro group had a stronger metabolic capacity and higher survival rate compared with those in Unp group; Protein digestion and absorption signaling pathway regulates the decomposition of proteins by digestive organs to produce favorable carbohydrates and energy (Freeman and Kim, 1978). This pathway was significantly enriched in our research, indicating that the digestive organs of O. ocellatus larvae were active, and metabolized rapidly. Moreover, we found multiple metabolism-related genes enriched in this pathway were significantly up-regulated compared with brood-careless larvae, including COL22A1, COL12A1, and other key genes. However, COL9A1 were down-regulated in our research, the specific reasons need to additional explore. All of these pathways are important pathways for the regulation of biological metabolism, and they are significantly enriched. Compared with the Unp group, most of the genes that are beneficial to biological metabolism enriched in above three important pathways were up-regulated, which suggests that the metabolic efficiency of O. ocellatus larvae in the Pro group was higher and more stable. And it was beneficial to the growth and development of O. ocellatus larvae.
Solute Carrier Family
Solute carrier families are the main factors affecting solute transport in organisms. There are many subunits in the family, and they act roles in almost all organs. In this study, we significantly enriched five family members, which play a significant regulatory part in substance transport and metabolism. SLC8A2, also known as NCX2, is a plasma membrane transporter that regulates intracellular calcium concentration and plays a major part in the regulation of Ca2+ in stomach, intestine, vascular smooth muscle, and other organs (Azuma et al., 2012). Similarly, as another subtype of the solid carrier family, SLC8A3 is highly expressed in the brain, and can regulate the concentration of Ca2+ in the excitatory cells. It is worth mentioning that SLC8A3 also acts a key role in maintaining blood transport in the brain (Sokolow et al., 2004; Yuan Z. et al., 2016). Ca2+ is an essential part of biological metabolism processes, which can regulate the synthesis and release of neurotransmitters and hormones. Above two genes regulate the concentration of Ca2+, indicating that they are important genes regulating the metabolic process (Wu et al., 2020). SLC3A1 and SLC7A9 are cystine transport factors, they regulate the metabolic processes of organisms by regulating the concentration of cysteine and the synthesis of glutathione (Alter et al., 2016). As an ammonia transport factor, SLC18A2 is active in a variety of organisms. Lack of SLC18A2 will result in obstruction of the motor system, which will affect the larvae from normal activities (Jin et al., 2014). The five genes of solute carrier family enriched in our study were all metabolism-related, and their high expression was helpful to O. ocellatus larval growth and development. Beside SLC3A1, the other four genes were significantly up-regulated in brood-care larvae compared with brood-careless larvae, indicating that larvae in the Pro group had more complex metabolic processes and higher metabolic capacity.
Three Significant Hub Genes Analysis
Among all hub genes, ITPR1, MAP2K1, and ADCY5 were enriched in more than seven metabolism-related signaling pathways, and they possessed higher numbers of protein–protein interactions. ITPR1, also known as IP3R1, is a phosphate receptor that regulates the concentration of Ca2 +, and promotes the metabolic activity of organisms by increasing the concentration of Ca2+ (Wakai and Fissore, 2019). In somatic cells, Ca2 + is transported to mitochondria to stimulate mitochondrial metabolism. And in oocytes, Ca2 + can increases cell activity and ATP content (Valladares et al., 2018; Wakai and Fissore, 2019). Moreover, ITPR1 plays a significant part in regulating vasodilation and maintaining normal blood pressure (Yuan Q. et al., 2016). MAP2K1 is a protein kinase, which exists in most cells, and can be activated by many extracellular signals to regulate metabolism processes of organisms (Crews et al., 1992). MAP2K1 can regulate cell growth, proliferation, and differentiation, and maintain the stability of biological metabolism (Chung et al., 2011). ADCY5 is an important subtype of adenyl cycles family, which transforms ATP into cAMP in vivo. As a protein kinase activator, cAMP is widely used in various organisms. It regulates the physiological function and material metabolism to control cell activity and hormone application, which plays an important part in carbohydrate, fat metabolism, nucleic acid, and protein synthesis (Richter-Landsberg and Jastorff, 1986; Frezza et al., 2018). The above three genes are significantly enriched, and have the highest protein interaction number, which are closely related to metabolism. Compared with the Unp group, the expression of ADCY5 gene was up-regulated at 4h after hatching, and down-regulated from 4 to 24 h in the Pro group; the expression of ITPR1 was down-regulated in the first 12 h, and significantly up-regulated in the second 12 h; the expression of MAP2K1 was down-regulated from 4h to 12h, and significantly up-regulated within 0 to 4 h and 12 to 24 h, indicating that the brood-care eggs had more complex metabolic mechanisms. These results explain that the O. ocellatus larvae in Pro group had a stronger metabolic capacity compared with those in Unp group. However, the specific functions of these genes in larval metabolism need to be further explored.
Other Signaling Pathways and Hub Genes
In addition to the above signaling pathways and hub genes, other metabolism-related key genes and signaling pathways involved in the metabolism of O. ocellatus larvae were identified such as long-term depression signaling pathway, glutamatergic synapse signaling pathway, and two key genes AKT3 and ADCY9. In previous studies, these two genes and signaling pathways have been reported to be involved in biological metabolic processes and act significant roles (Marty and Llano, 1995; Rodríguez and Ortega, 2017; Teixeira et al., 2017; Chen et al., 2018; DuBois et al., 2019). The effects of genes and pathways on metabolism of O. ocellatus under different brood-care behaviors need to be further explored from now on.
Conclusion
We performed transcriptome profiling of gene expression in larval growth from brood-care and brood-careless eggs and constructed a PPI network. Ten hub genes with numerous protein–protein interaction relationships or that involved in multiple KEGG signaling pathways were identified. In our research, we first study the effects of brood-care behavior on metabolic mechanisms of O. ocellatus larval growth using PPI networks. Our results provide valuable gene resources for understanding the metabolism of invertebrate larvae. Meanwhile, the data lays a foundation for further understanding invertebrate brood-care behaviors.
Data Availability Statement
The original contributions presented in the study are publicly available. These data can be found here: SRR15204591–SRR15204602, SRR15927331–SRR15927334, and SRR15927336–SRR15927343.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of the Ludong University (protocol number LDU-IRB20210308NXY) and the China Government Principles for the Utilization and Care of Invertebrate Animals Used in Testing, Research, and Training (State Science and Technology Commission of the People’s Republic of China for No. 2, October 31, 1988; http://www.gov.cn/gongbao/content/2011/content_1860757.htm).
Author Contributions
ZL and JY designed and supervised the study. XB, XL, BY, YL, MC, and BL prepared the samples. XB, XL, YF, GS, WW, and XX analyzed all sequencing data. XB and ZL wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This research was supported by the China Agriculture Research System of MOF and MARA, the Natural Science Foundation of Shandong Province (No. ZR2019BC052), and the National Natural Science Foundation of China (No. 42006077).
Conflict of Interest
BL was employed by the company Yantai Haiyu Marine Science and Technology Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ http://www.gov.cn/gongbao/content/2011/content_1860757.htm
- ^ https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP329195&o=acc_s%3Ad
References
Alter, S. P., Stout, K. A., Lohr, K. M., Taylor, T. N., Shepherd, K. R., Wang, M., et al. (2016). Reduced vesicular monoamine transport disrupts serotonin signaling but does not cause serotonergic degeneration. Exp. Neurol. 275(Pt 1), 17–24. doi: 10.1016/j.expneurol.2015.09.016
Arkhipkin, A. I. (1985). Reproductive system structure, development and function in cephalopods with a new general scale for maturity stages. J. Northw. Atl. Fish. Sci. 12, 63–74.
Azuma, Y. T., Nishiyama, K., Kita, S., Komuro, I., Nakajima, H., Iwamoto, T., et al. (2012). Na+/Ca2+ exchanger 2-heterozygote knockout mice display decreased acetylcholine release and altered colonic motility in vivo. Neurogastroenterol. Motil. 24:e600-10. doi: 10.1111/nmo.12029
Baeza, J. A., and Fernández, M. (2002). Active brood care in Cancer setosus (Crustacea: Decapoda): the relationship between female behaviour, embryo oxygen consumption and the cost of brooding. Funct. Ecol. 16, 241–251.
Baldridge, A. K., and Lodge, D. M. (2013). Intraguild predation between spawning smallmouth bass (Micropterus dolomieu) and nest-raiding crayfish (Orconectes rusticus): implications for bass nesting success. Freshw. Biol. 58, 2355–2365.
Boletzky, S. V. (1975). A contribution to the study of yolk absorption in the Cephalopoda. Zoomorphology 80, 229–246.
Boletzky, S. V. (1989). Recent studies on spawning, embryonic development, and hatching in the Cephalopoda. Adv. Mar. Biol. 25, 85–115. doi: 10.1016/s0065-2881(03)44003-0
Bruning, A., González, A., Gaitán-Espitia, J. D., Bartheld, J. L., Toader-Williams, A., Mondaca, F., et al. (2013). Energy metabolism, heart rate and physiological differentiation in the pulmonate gastropod Cornu Aspersum. J. Molluscan Stud. 79, 257–262. doi: 10.1093/mollus/eyt021
Budelmann, B. U. (1994). Cephalopod sense organs, nerves and the brain: adaptations for high performance and life style. Mar. Freshw. Behav. Physiol. 25, 13–33.
Bunn, N. A., Fox, C. J., and Webb, T. (2000). A Literature Review of Studies on Fish Egg Mortality: Implications for the Estimation of Spawning Stock Biomass by the Annual Egg Production Method (Funded Under MAFF Project Code MF0426: Fish Egg Development and Mortality Studies in the Irish Sea). Science Series Technical Report, Vol. 111. Lowestoft: CEFAS, 1565–1575.
Chatterjee, A., Norton-Baker, B., Bagge, L. E., Patel, P., and Gorodetsky, A. A. (2018). An introduction to color-changing systems from the Cephalopod protein reflectin. Bioinspir. Biomim. 13:045001. doi: 10.1088/1748-3190/aab804
Chen, T. J., Kula, B., Nagy, B., Barzan, R., Gall, A., Ehrlich, I., et al. (2018). In vivo regulation of oligodendrocyte precursor cell proliferation and differentiation by the AMPA-receptor subunit GluA2. Cell Rep. 25, 852–861.e7. doi: 10.1016/j.celrep.2018.09.066
Chung, E., Hsu, C. L., and Kondo, M. (2011). Constitutive MAP kinase activation in hematopoietic stem cells induces a myeloproliferative disorder. PLoS One 6:e28350. doi: 10.1371/journal.pone.0028350
Crews, C. M., Alessandrini, A., and Erikson, R. L. (1992). The primary structure of MEK, a protein kinase that phosphorylates the ERK gene product. Science 258, 478–480. doi: 10.1126/science.1411546
Damian, S., Gable, A. L., David, L., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2018). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
DuBois, J. C., Ray, A. K., Gruber, R. C., Zhang, Y., Aflakpui, R., and Macian-Juan, F. (2019). Akt3-mediated protection against inflammatory demyelinating disease. Front. Immunol. 10:1738. doi: 10.3389/fimmu.2019.01738
Foster, J. G., Algera, D. A., Brownscombe, J. W., Zolderdo, A. J., and Cooke, S. J. (2016). Consequences of different types of littoral zone light pollution on the parental care behaviour of a freshwater teleost fish. Water Air Soil Pollut. 227:404.
Freeman, H. J., and Kim, Y. S. (1978). Digestion and absorption of protein. Annu. Rev. Med. 29, 99–116.
Frezza, E., Martin, J., and Lavery, R. (2018). A molecular dynamics study of adenylyl cyclase: the impact of ATP and G-protein binding. PLoS One 13:e0196207. doi: 10.1371/journal.pone.0196207
Furchgott, R. F. (1966). Metabolic factors that influence contractility of vascular smooth muscle. Bull. N. Y. Acad. Med. 42, 996–1006.
Harper, M. E., and Patti, M. E. (2020). Metabolic terminology: what’s in a name? Nat. Metab. 2, 476–477. doi: 10.1038/s42255-020-0216-7
Hosseini, S. R., Martin, O. C., and Wagner, A. (2016). Phenotypic innovation through recombination in genome-scale metabolic networks. Proc. R. Soc. B Biol. Sci. 283:20161536. doi: 10.1098/rspb.2016.1536
Huffard, C. L. (2013). Cephalopod neurobiology: an introduction for biologists working in other model systems. Invertebr. Neurosci. 13, 11–18. doi: 10.1007/s10158-013-0147-z
Ikeda, T. (2016). Routine metabolic rates of pelagic marine fishes and cephalopods as a function of body mass, habitat temperature and habitat depth. J. Exp. Mar. Biol. Ecol. 480, 74–86.
Jin, J., Lao, A. J., Katsura, M., Caputo, A., Schweizer, F. E., and Sokolow, S. (2014). Involvement of the sodium-calcium exchanger 3 (NCX3) in ziram-induced calcium dysregulation and toxicity. Neurotoxicology 45, 56–66. doi: 10.1016/j.neuro.2014.09.004
Li, Z., Fan, T., Liu, X., Liu, X., Wang, W., Wang, Q., et al. (2019). Characterization and functional study on Octopus ocellatus interleukin-17. J. Ocean Univ. China 18, 1443–1450. doi: 10.1007/s11802-019-4116-y
Li, Z., Liu, X., Liu, J., Zhang, K., Yu, H., He, Y., et al. (2017). Transcriptome profiling based on protein–protein interaction networks provides a core set of genes for understanding blood immune response mechanisms against Edwardsiella tarda infection in Japanese flounder (Paralichthys olivaceus). Dev. Comp. Immunol. 78, 100–113. doi: 10.1016/j.dci.2017.09.013
Martínez-Quintana, J. A., and Yepiz-Plascencia, G. (2012). Glucose and other hexoses transporters in marine invertebrates: a mini review. Electron. J. Biotechnol. 15:16.
Marty, A., and Llano, I. (1995). Modulation of inhibitory synapses in the mammalian brain. Curr. Opin. Neurobiol. 5, 335–341. doi: 10.1016/0959-4388(95)80046-8
Matsumoto, T., Tanaka, T., and Kondo, A. (2018). Sortase A-assisted metabolic enzyme ligation in Escherichia coli for enhancing metabolic flux. Synth. Biol. 1772, 125–136. doi: 10.1007/978-1-4939-7795-6_6
Navarro, J. C., and Villanueva, R. (2003). The fatty acid composition of Octopus vulgaris paralarvae reared with live and inert food: deviation from their natural fatty acid profile. Aquaculture 219, 613–631. doi: 10.1016/s0044-8486(02)00311-3
Noguchi, T., Fujiwara, S., Takada, Y., Mori, T., and Nagano, M. (1982). Metabolism of urea and glyoxylate, degradative products of purines in marine animals. J. Biochem. 92, 525–529. doi: 10.1093/oxfordjournals.jbchem.a133960
Payne, J. L., Heim, N. A., Knope, M. L., and McClain, C. R. (2014). Metabolic dominance of bivalves predates brachiopod diversity decline by more than 150 million years. Proc. R. Soc. B Biol. Sci. 281:20133122. doi: 10.1098/rspb.2013.3122
Pimentel, M. S., Truebenbach, K., Faleiro, F., Boavida-Portugal, J., Repolho, T., and Rosa, R. (2012). Impact of ocean warming on the early ontogeny of cephalopods: a metabolic approach. Mar. Biol. 159, 2051–2059. doi: 10.1007/s00227-012-1991-9
Quiroga, R. Q., and Garcia, H. (2003). Single-trial event-related potentials with wavelet denoising. Clin. Neurophysiol. 114, 376–390. doi: 10.1016/s1388-2457(02)00365-6
Ravnskjaer, K., Madiraju, A., and Montminy, M. (2015). “Role of the cAMP pathway in glucose and lipid metabolism,” in Handbook of Experimental Pharmacology, ed. S. Herzig (Cham: Springer), 29–49. doi: 10.1007/164_2015_32
Richter-Landsberg, C., and Jastorff, B. (1986). The role of cAMP in nerve growth factor-promoted neurite outgrowth in PC12 cells. J. Cell Biol. 102, 821–829. doi: 10.1083/jcb.102.3.821
Rodríguez, A., and Ortega, A. (2017). Glutamine/glutamate transporters in glial cells: much more than participants of a metabolic shuttle. Adv. Neurobiol. 16, 169–183. doi: 10.1007/978-3-319-55769-4_8
Rosas, C., Valero, A., Caamal-Monsreal, C., Uriarte, I., Farias, A., Gallardo, P., et al. (2013). Effects of dietary protein sources on growth, survival and digestive capacity of Octopus maya juveniles (Mollusca: Cephalopoda). Aquac. Res. 44, 1029–1044. doi: 10.1111/j.1365-2109.2012.03107.x
Sakuraba, H., Fujiwara, S., and Noguchi, T. (1996). Metabolism of glyoxylate, the end product of purin degradation, in liver peroxisomes of fresh water fish. Biochem. Biophys. Res. Commun. 229, 603–606. doi: 10.1006/bbrc.1996.1850
Schnell, A. K., Amodio, P., Boeckle, M., and Clayton, N. S. (2020). How intelligent is a cephalopod? Lessons from comparative cognition. Biol. Rev. 96, 162–178. doi: 10.1111/brv.12651
Seibel, B. A. (2016). Cephalopod susceptibility to asphyxiation via ocean incalescence, deoxygenation, and acidification. Physiology 31, 418–429. doi: 10.1152/physiol.00061.2015
Seibel, B. A., Thuesen, E. V., and Childress, J. J. (1997). Decline in pelagic cephalopod metabolism with habitat depth reflects differences in locomotory efficiency. Biol. Bull. 192, 262–278. doi: 10.2307/1542720
Sokolow, S., Manto, M., Gailly, P., Molgó, J., Vandebrouck, C., Vanderwinden, J. M., et al. (2004). Impaired neuromuscular transmission and skeletal muscle fiber necrosis in mice lacking Na/Ca exchanger 3. J. Clin. Invest. 113, 265–273. doi: 10.1172/JCI18688
Speers-Roesch, B., Callaghan, N. I., MacCormack, T. J., Lamarre, S. G., Sykes, A. V., and Driedzic, W. R. (2016). Enzymatic capacities of metabolic fuel use in cuttlefish (Sepia officinalis) and responses to food deprivation: insight into the metabolic organization and starvation survival strategy of cephalopods. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 186, 711–725. doi: 10.1007/s00360-016-0991-3
Tang, C., Liu, D., Fan, Y., Yu, J., Li, C., Su, J., et al. (2021). Visualization and bibliometric analysis of cAMP signaling system research trends and hotspots in cancer. J. Cancer 12, 358–370. doi: 10.7150/jca.47158
Teixeira, H. M., Alcantara-Neves, N. M., Barreto, M., Figueiredo, C. A., and Costa, R. S. (2017). Adenylyl cyclase type 9 gene polymorphisms are associated with asthma and allergy in Brazilian children. Mol. Immunol. 82, 137–145. doi: 10.1016/j.molimm.2017.01.001
Valladares, D., Utreras-Mendoza, Y., Campos, C., Morales, C., Diaz-Vegas, A., Contreras-Ferrat, A., et al. (2018). IP3 receptor blockade restores autophagy and mitochondrial function in skeletal muscle fibers of dystrophic mice. Biochim. Biophys. Acta 1864, 3685–3695. doi: 10.1016/j.bbadis.2018.08.042
Vidal, E. A., Villanueva, R., Andrade, J. P., Gleadall, I. G., Iglesias, J., Koueta, N., et al. (2014). Cephalopod culture: current status of main biological models and research priorities. Adv. Mar. Biol. 67, 1–98. doi: 10.1016/B978-0-12-800287-2.00001-9
Wakai, T., and Fissore, R. A. (2019). Constitutive IP3R1-mediated Ca2+ release reduces Ca2+ store content and stimulates mitochondrial metabolism in mouse GV oocytes. J. Cell Sci. 132:jcs225441. doi: 10.1242/jcs.225441
Wang, W., Yang, J., Zhou, Q., Zheng, X., Zhang, Y., Sun, G., et al. (2010). Reproductive behavior and process of embryonic development of Octopus ocellatus. J. Fish. Sci. China 17, 1157–1165.
Wei, X., Xu, J., Yang, J., Liu, X., Zhang, R., Wang, W., et al. (2015). Involvement of a Serpin serine protease inhibitor (OoSerpin) from mollusc Octopus ocellatus in antibacterial response. Fish Shellfish Immunol. 42, 79–87. doi: 10.1016/j.fsi.2014.10.028
Wei, X., Zhao, T., Ai, K., Li, H., Jiang, X., Li, C., et al. (2018). Role of scavenger receptor from Octopus ocellatus as a co-receptor of toll-like receptor in initiation of TLR-NF-κB signaling during anti-bacterial response. Dev. Comp. Immunol. 84, 14–27. doi: 10.1016/j.dci.2018.01.023
Wu, K. C., Reisman, S. A., and Klaassen, C. D. (2020). Tissue distribution, hormonal regulation, ontogeny, diurnal expression, and induction of mouse cystine transporters Slc3a1 and Slc7a9. Free Radic. Res. 54, 525–534. doi: 10.1080/10715762.2020.1812597
Xu, R., and Zheng, X. (2020). Hemocytes transcriptomes reveal metabolism changes and detoxification mechanisms in response to ammonia stress in Octopus minor. Ecotoxicology 29, 1441–1452. doi: 10.1007/s10646-020-02279-0
Yeldandi, A. V., Chu, R., Pan, J., Zhu, Y., and Usuda, N. (1996). Peroxisomal purine metabolism. Ann. N. Y. Acad. Sci. 804, 165–175.
Yuan, Q., Yang, J., Santulli, G., Reiken, S. R., Wronska, A., Kim, M. M., et al. (2016). Maintenance of normal blood pressure is dependent on IP3R1-mediated regulation of eNOS. Proc. Natl. Acad. Sci. U.S.A. 113, 8532–8537. doi: 10.1073/pnas.1608859113
Yuan, Z., Liu, C., Tian, Y., Zhang, X., Ye, H., Jin, L., et al. (2016). Higher levels of magnesium and lower levels of calcium in whole blood are positively correlated with the metabolic syndrome in a Chinese population: a case-control study. Ann. Nutr. Metab. 69, 125–134. doi: 10.1159/000450761
Zhang, F., Zhang, L., Qi, Y., and Xu, H. (2016). Mitochondrial cAMP signaling. Cell. Mol. Life Sci. 73, 4577–4590. doi: 10.1007/s00018-016-2282-2
Zheng, X., Han, S., Lin, X., and Li, Q. (2009). Research progress in Cephalopod reproductive behaviors. J. Fish. Sci. China 16, 459–465.
Keywords: Octopus ocellatus, transcriptome, protein–protein interaction networks, metabolism, brood-care behavior
Citation: Bao X, Liu X, Yu B, Li Y, Cui M, Wang W, Feng Y, Xu X, Sun G, Li B, Li Z and Yang J (2022) Transcriptome Profiling Based on Larvae at Different Time Points After Hatching Provides a Core Set of Gene Resource for Understanding the Metabolic Mechanisms of the Brood-Care Behavior in Octopus ocellatus. Front. Physiol. 12:762681. doi: 10.3389/fphys.2021.762681
Received: 22 August 2021; Accepted: 29 October 2021;
Published: 07 January 2022.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Neeraj Kumar, National Institute of Abiotic Stress Management (ICAR), IndiaXiaoshang Ru, Institute of Oceanology, Chinese Academy of Sciences (CAS), China
Copyright © 2022 Bao, Liu, Yu, Li, Cui, Wang, Feng, Xu, Sun, Li, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zan Li, bGl6YW5seG1AMTYzLmNvbQ==; Jianmin Yang, bGFkZGVydXBAMTI2LmNvbQ==
 Xiaokai Bao1
Xiaokai Bao1 Weijun Wang
Weijun Wang Xiaohui Xu
Xiaohui Xu Zan Li
Zan Li Jianmin Yang
Jianmin Yang