
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 27 October 2021
Sec. Invertebrate Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.752768
This article is part of the Research TopicMethods and Applications in Invertebrate PhysiologyView all 12 articles
 Gothandapani Sellamuthu1
Gothandapani Sellamuthu1 Shan Amin1,2
Shan Amin1,2 Jan Bílý1
Jan Bílý1 Jirí Synek1
Jirí Synek1 Roman Modlinger1
Roman Modlinger1 Madhab Kumar Sen3
Madhab Kumar Sen3 Amrita Chakraborty4
Amrita Chakraborty4 Amit Roy1,4*
Amit Roy1,4*Ips sexdentatus (Coleoptera: Curculionidae: Scolytinae) is one of the most destructive and economically important forest pests. A better understanding of molecular mechanisms underlying its adaptation to toxic host compounds may unleash the potential for future management of this pest. Gene expression studies could be considered as one of the key experimental approaches for such purposes. A suitable reference gene selection is fundamental for quantitative gene expression analysis and functional genomics studies in I. sexdentatus. Twelve commonly used reference genes in Coleopterans were screened under different experimental conditions to obtain accurate and reliable normalization of gene expression data. The majority of the 12 reference genes showed a relatively stable expression pattern among developmental stages, tissue-specific, and sex-specific stages; however, some variabilities were observed during varied temperature incubation. Under developmental conditions, the Tubulin beta-1 chain (β-Tubulin) was the most stable reference gene, followed by translation elongation factor (eEF2) and ribosomal protein S3 (RPS3). In sex-specific conditions, RPS3, β-Tubulin, and eEF2 were the most stable reference genes. In contrast, different sets of genes were shown higher stability in terms of expression under tissue-specific conditions, i.e., RPS3 and eEF2 in head tissue, V-ATPase-A and eEF2 in the fat body, V-ATPase-A and eEF2 in the gut. Under varied temperatures, β-Tubulin and V-ATPase-A were most stable, whereas ubiquitin (UbiQ) and V-ATPase-A displayed the highest expression stability after Juvenile Hormone III treatment. The findings were validated further using real-time quantitative reverse transcription PCR (RT-qPCR)-based target gene expression analysis. Nevertheless, the present study delivers a catalog of reference genes under varied experimental conditions for the coleopteran forest pest I. sexdentatus and paves the way for future gene expression and functional genomic studies on this species.
Differential gene expression (DGE) studies are fundamental to evaluate the effects of physiological responses on biological variation in insect populations at the molecular level. Real-time quantitative reverse transcription PCR (RT-qPCR) is a reliable technique and is widely used to analyze the expression of target genes due to its high sensitivity, accuracy, specificity, reproducibility, and speed (Heid et al., 1996; Bustin et al., 2005; VanGuilder et al., 2008; Wang et al., 2020). In addition, RT-qPCR allows simultaneous measurement of gene expression in many different samples for a limited number of target genes and is particularly suitable when only a limited number of samples are available (Higuchi et al., 1993; Vandesompele et al., 2002). Nevertheless, expression results vary due to initial sample size, RNA integrity (quantity and quality), reverse transcription, messenger RNA (mRNA) recovery, PCR efficiency, and primer design (Gao et al., 2020). Therefore, internal control genes (alternatively called reference genes) are commonly used to normalize mRNA levels for more accurate gene expression quantification as their expression should not vary under different biological or experimental conditions (Nicot et al., 2005; Lu et al., 2018). Furthermore, it is strongly recommended to use multiple reference genes for target gene expression normalization for more authenticity (García-Reina et al., 2018; Shakeel et al., 2018; Wang et al., 2020). There are several studies on gene function analysis in insects using reference genes such as β-actin (Actin), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), elongation factor 1α (EF1A), β-Tubulin (β-Tubulin), ribosomal proteins (RPs), ubiquitin (UbiQ), superoxide dismutase (SOD), heat shock protein 90 (HSP90), and vacuolar-type H+-ATPase subunit B (V-ATPase-B) for gene expression normalization (Lu et al., 2018; Qu et al., 2018; Gao et al., 2020; Wang et al., 2020). This suggests that reference genes can be differentially expressed in insects under varied experimental conditions. Alternatively, no universal reference gene is available, which is stably expressed under different experimental conditions (Lu et al., 2018). Therefore, identification and validation of reference genes under different experimental conditions, life stages, and tissue-specific stages are essential on a case-by-case basis for accurate quantification of gene expression in any insects, including I. sexdentatus (Pfaffl et al., 2004; Rodrigues et al., 2014; Qu et al., 2018; Basu et al., 2019; Gao et al., 2020; Gurusamy et al., 2021; Xie et al., 2021).
The wood-boring insect I. sexdentatus (Coleoptera: Curculionidae: Scolytinae; hereby referred to as ISx), also known as the six-toothed bark beetle, is one of the most destructive and economically important insects, causing severe damage to coniferous species throughout Europe and Asia (Etxebeste and Pajares, 2011; Seidl et al., 2017; Douglas et al., 2019). ISx is an opportunistically aggressive species of bark beetles (Wermelinger et al., 2008; Chakraborty et al., 2020a). It usually attacks old scots pine trees and can occupy young trees up to a diameter of 10 cm in dbh (diameter at breast height). It was primarily a sparse specimen (Pfeffer, 1955; Postner, 1974) in the past, but now ISx is considered a significant pest in some European countries (Gregoire and Evans, 2004). Several drought periods during the last decade highly favored its current upliftment on the pest status, and we are registering higher population densities of ISx in most of the Czech forests. At least from 2018, the south Moravian forests suffered from the severe outbreak of this species. The volume of the harvested pine due to bark beetles in the most affected region was 12 times higher than in the previous years (Lubojacký and Knížek, 2020).
Bark beetles preferentially colonize weakened, wilted, or recently dead trees. However, favorable conditions such as drought can lead to an outbreak (epidemic phase). Beetles start attacking the healthy standing trees due to weakened defenses caused by warmer temperatures (Bouhot et al., 1988; Marini et al., 2017; Pettit et al., 2020) and larger windthrows (Kausrud et al., 2012; Biedermann et al., 2019; Sommerfeld et al., 2020). Furthermore, conifers have self-defense mechanisms to ward off bark beetle infestations with secondary metabolites such as terpenes (Ferrenberg et al., 2014; Denham et al., 2019). These defense mechanisms can also be overcome during mass insect attacks by detoxifying plant-derived secondary metabolites with the help of symbiotic microbes (Chakraborty et al., 2020b; Huang et al., 2020). To elucidate molecular underpinnings shaping environment-conifer-bark beetle (i.e., I. sexdentatus) interactions, gene expression studies need to be conducted in the future. Such studies advocate the requirements for stable reference genes under different experimental conditions.
Hence, we extensively evaluated 12 commonly known reference genes in Colepterans and other insects (Supplementary Figure 1 and Supplementary Table 1) for the expression stability in ISx under varied experimental conditions in the present study. Using available in-house transcriptome data of ISx, we have obtained gene sequences of β-actin (Actin), translation EF 2 (eEF2), Tubulin beta-1 chain (β-Tubulin), myosin regulatory light chain 2 (Myosin L), V-type proton ATPase catalytic subunit A (V-ATPase-A), NADH dehydrogenase subunit 1 (NADH), ubiquitin C variant (UbiQ), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), arginine kinase isoform X1(ArgK), RPS3, RPL17, and HSP83 for expression stability evaluation. The candidate reference genes were screened using different parameters, such as developmental stages (i.e., larvae, pupae, and adult stages), tissue- and sex-specific [head, gut, fat body, and whole body (WB) except the head, gut, fat body], and different treatments [such as temperature; Juvenile hormone III (JHIII); wild-collected vs. long-term laboratory-reared; and different host feeding] to obtain accurate reference genes for future genomic and functional studies. Our study delivers a catalog of genes that should be used for DGE studies on ISx under varied experimental conditions and commences the possibility for aggressive management of bark beetles using molecular approaches such as RNA interference (RNAi) (Joga et al., 2021).
Ips sexdentatus (ISx)-infested Pinus sylvestris were collected from Kostelec nad Cernými lesy (50°00'07.2”N 14°50'56.3”E, under School Forest Enterprise) maintained insect rearing chambers at Faculty of Forestry and Wood Sciences, Czech University of Life Sciences, Prague. Beetles were maintained with fresh pine logs at 27 ± 1°C under 70 ± 5% humidity and a 16:8-h light/dark (L:D) photoperiod. ISx is the largest beetle of the genus Ips, at 6-8 mm in length. The life stages include three larval stages, pupae, callow (just emerged), and fed adult stage (flying for new host colonization). Both sexes have six spines at each side of the elytral declivity. The fourth spine is the largest and is capitate. The wild population was supplied with fresh pine logs for the next generation and maintained for 40–45 days for one life cycle. Similarly, the wild population was continuously maintained for three generations to study the difference between wild and lab-reared beetle conditions.
Samples were collected from different life stages of ISx: three larval stages (L1, L2, and L3), pupae, (P), callow (newly emerged) male (CM) and female (CF), and fed adult (mature adults move toward trunks of pines to lay eggs for next-generation) male (AMF) and female (AFF). For sufficient nucleic acid extraction for downstream processing, each biological replicate was prepared using pooled individuals from various developmental stages, such as five first instar larvae (L1)/replicate, three second and third instar larvae (L2 and L3)/ replicate, three pupae/ replicate, and two adults/replicate. Four biological replicates were used in all experiments.
Tissues such as head, fat body, gut, and WB minus head, fat body, and gut (hereby referred to as WB) were dissected from the callow and fed adult stage of both male and female ISx producing experimental samples, such as CMH, callow male beetle head; CMFB, callow male beetle fat body; CMMG, callow male beetle midgut; CMWB, callow male beetle WB; CFH, callow female beetle head; CFFB, callow female beetle fat body; CFMG, callow female beetle midgut; CFWB, callow female beetle WB; and AMFH, fed adult male head; AMFFB, fed adult male fat body; AMFMG, fed adult male midgut; AMFWB, fed adult male WB; AFFH, fed adult female head; AFFFB, fed adult female fat body; AFFMG, fed adult female midgut; and AFFWB, fed adult female WB. Four independent biological replicates were collected and stored at −80°C. Each replicate was derived by pooling tissues from 10 individual beetles.
For the temperature treatment, the freshly emerged adults were placed in small glass tubes and exposed to a range of temperatures (4, 27, and 37°C) for 72 h in a temperature-controlled chamber. After exposure, survivors were frozen in liquid nitrogen and stored at −80°C. Adults maintained at 27°C were used as control. At least four beetles were randomly collected per replication, and four independent biological replications were used from each temperature treatment.
For JHIII treatment, emerged beetles were sorted based on gender and kept at 4°C on moist paper towels. Beetles were treated topically on the ventral surface of the abdomen with 10 μg JHIII (dissolved in acetone) (Sigma-Aldrich, St. Louis, MO, USA) and only acetone as control (Aw et al., 2010; Sun et al., 2021). The beetles were incubated at 27 ± 1°C under 70 ± 5% humidity and a 16:8-h light/dark (L:D) photoperiod in groups of 20 in 60 ml plastic containers for 72 h. After incubation, beetles were immediately shock frozen in liquid nitrogen and stored at −80°C for downstream processing. Four biological replicates were used for JHIII treatment and control.
Under different host feeding treatments, ISx was placed into the phloem tissue of freshly cut lodgepole pine and spruce. Beetles were placed under the bark in randomly chosen male-female pairs and maintained under standard rearing conditions. ISx was allowed to feed under the bark for one generation (40–45 days). We removed ISx adults from galleries showing the excavation of frass (an indication of feeding). The beetles were collected and immersed in insect ringer solution; the gut tissues were excised. Gut tissues were gently purged of their contents (i.e., malpighian tubules, fat body) and then frozen in liquid nitrogen and stored at −80°C.
Total RNA from bark beetle tissue samples was extracted using TRIzol® (Invitrogen, Carlsbad, CA) following the protocol of the manufacturer. Isolated RNA was further treated with DNases using a TURBO DNAase Kit (Ambion, USA) to remove any DNA contamination. RNA quantity and quality were evaluated using 1.5% agarose gel and quantified by NanoDrop 2000/2000c from Thermo Fisher Scientific® (Waltham, MA, USA) and stored at −80°C. Complementary DNA (cDNA) (first-strand) was synthesized from 1 μg of total RNA in triplicate using the high-capacity cDNA reverse transcription kits (Applied Biosystems-Life Technologies) following the recommendations of the manufacturer and stored at −20°C.
This study selected 12 potential reference genes (Supplementary Table 2) that function as reference genes in other Coleoptera species (Supplementary Figure 1 and Supplementary Table 1). These genes were obtained from our functionally annotated in-house transcriptome of ISx (manuscript in preparation). The primers for those selected genes were designed via Integrated DNA Technologies, Inc. (IDT) (Table 1).
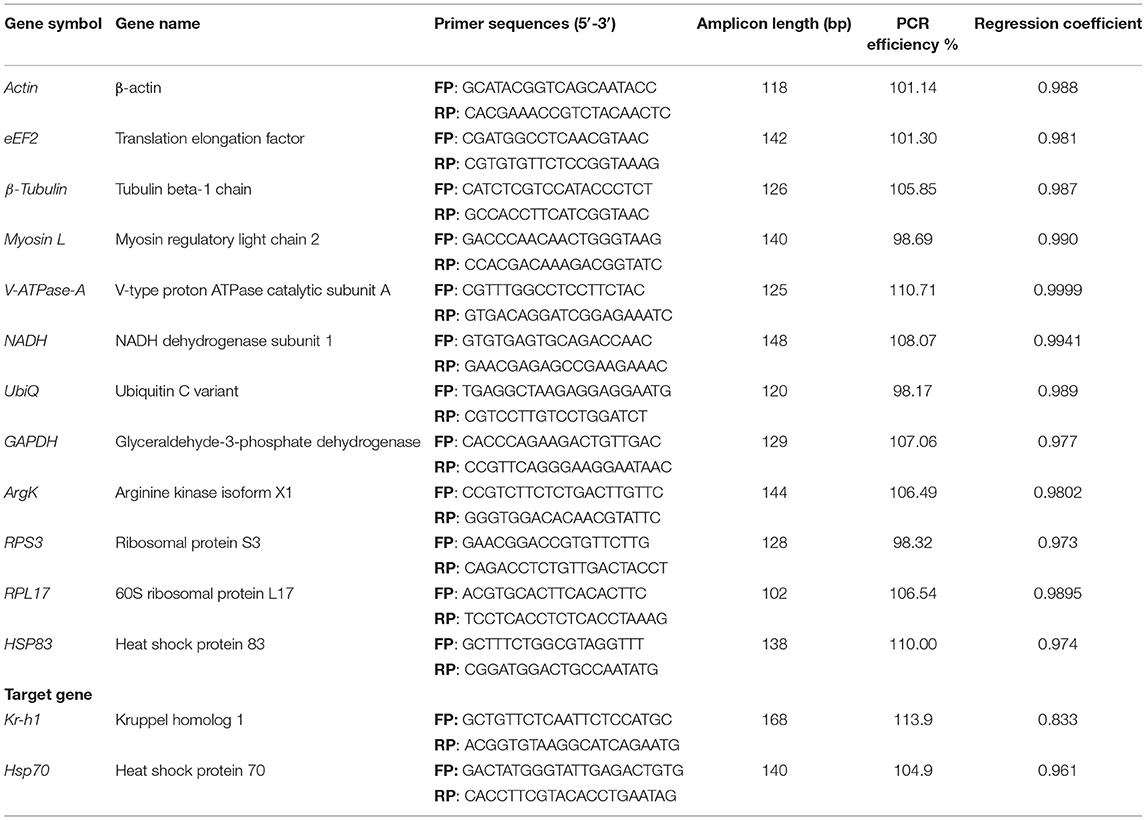
Table 1. Details of primer sequence, amplicon length, and RT-qPCR analysis of candidate reference genes and target genes.
cDNA samples were diluted 1:20 before being used in RT-qPCR. Four independent biological replicates from each treatment, developmental stage, and tissue-type were included in each RT-qPCR run. RT-qPCR run was performed using the Applied Biosystems™ StepOne™ Real-Time PCR System (Applied Biosystems) with a reaction mix containing 5.0 μL of SYBR® Green PCR Master Mix (Applied Biosystems), 1.0 μL of cDNA, 1.0 μL optimized concentrations of primers (Table 1), and RNase-free water (Invitrogen) to a total volume of 10.0 μL. Amplification conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. In order to confirm the primer specificity, we performed melt curve analysis using default parameters by a steady increase in temperature from 60 to 95°C. All RT-qPCR assays were carried out in four biological replicates, including two or three technical replicates.
The raw cycle threshold (Ct) values were obtained with 7500 software v2.0.5 (Applied Biosystems®). Gene expression and stability were analyzed using four different commonly used tools and algorithms such as geNorm (Vandesompele et al., 2002), BestKeeper (Pfaffl et al., 2004), NormFinder (Andersen et al., 2004), and ΔCt method (Silver et al., 2006).
Precisely, geNorm assesses the expression stability value (M) as the average pairwise variation of one of the genes against all control genes present in the experiment. The program estimates the mean pairwise variation between genes across all samples, and the gene with the lowest M value is considered most stable (Vandesompele et al., 2002). NormFinder calculates the SD for each target gene and juxtaposes it with other gene expressions. The gene displaying the lowest variation between intra- and inter-group comparisons is considered most stable (Andersen et al., 2004). On the contrary, BestKeeper is a data processing method based on crossing points that compares all genes across all samples and provides a stability index for each reference gene (Pfaffl et al., 2004). The comparative delta-Ct method juxtaposes Ct values and the relative expression of “gene pairs” within each sample (Silver et al., 2006). The sample-specific mean Ct values of each reference gene from each experiment are given as input data and subsequently processed using the web-based tool RefFinder (https://www.heartcure.com.au/reffinder/).
Furthermore, pairwise variation (V), estimated by geNorm, was used to decide the optimal number of reference genes for accurate RT-qPCR normalization. The Vn/Vn+1 value reflected the pairwise variation between two sequential normalization factors. A cutoff threshold, i.e., Vn/Vn+1 = 0.15, was set for a valid normalization (Vandesompele et al., 2002).
We analyzed the relative expression levels of Kr-h1 and Hsp70 genes after JHIII treatment and different temperature exposure to evaluate the selected reference genes. To compare the impact of different normalization strategies, the expression of target genes was normalized using both selected reference genes individually and in combinations using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Target genes expression were analyzed using one-way ANOVA using GraphPad Prism software. P-value < 0.05 was considered to identify significant differences between samples.
In the current study, 12 genes, namely Actin, eEF2, β-Tubulin, Myosin L, V-ATPase-A, NADH, UbiQ, GAPDH, ArgK, RPS3, RPL17, and HSP83, were screened for identifying suitable reference gene or gene combination from ISx. RT-qPCR products generated with each primer set (forward+ reverse) against target genes were evaluated by the occurrence of a single peak in melting curve analyses (Supplementary Figure 2) and specific bands of the expected size in agarose gel electrophoresis (Supplementary Figure 3). The amplification efficiency for each primer pair ranged from 98.17 to 107.06%, and the correlation coefficients (R2) were greater than 0.98 (Table 1). The Ct values of the 12 candidate reference genes ranged from 19.87 to 34.78 and covered all experimental conditions (Figure 1A). While most Ct values ranged from 19 to 27, Actin, eEF2, β-Tubulin, and RPS3 were the most abundant transcripts under almost all experimental conditions. The least frequently expressed reference genes were NADH, RPL17, and HSP83. The five remaining reference genes were expressed at moderate levels.
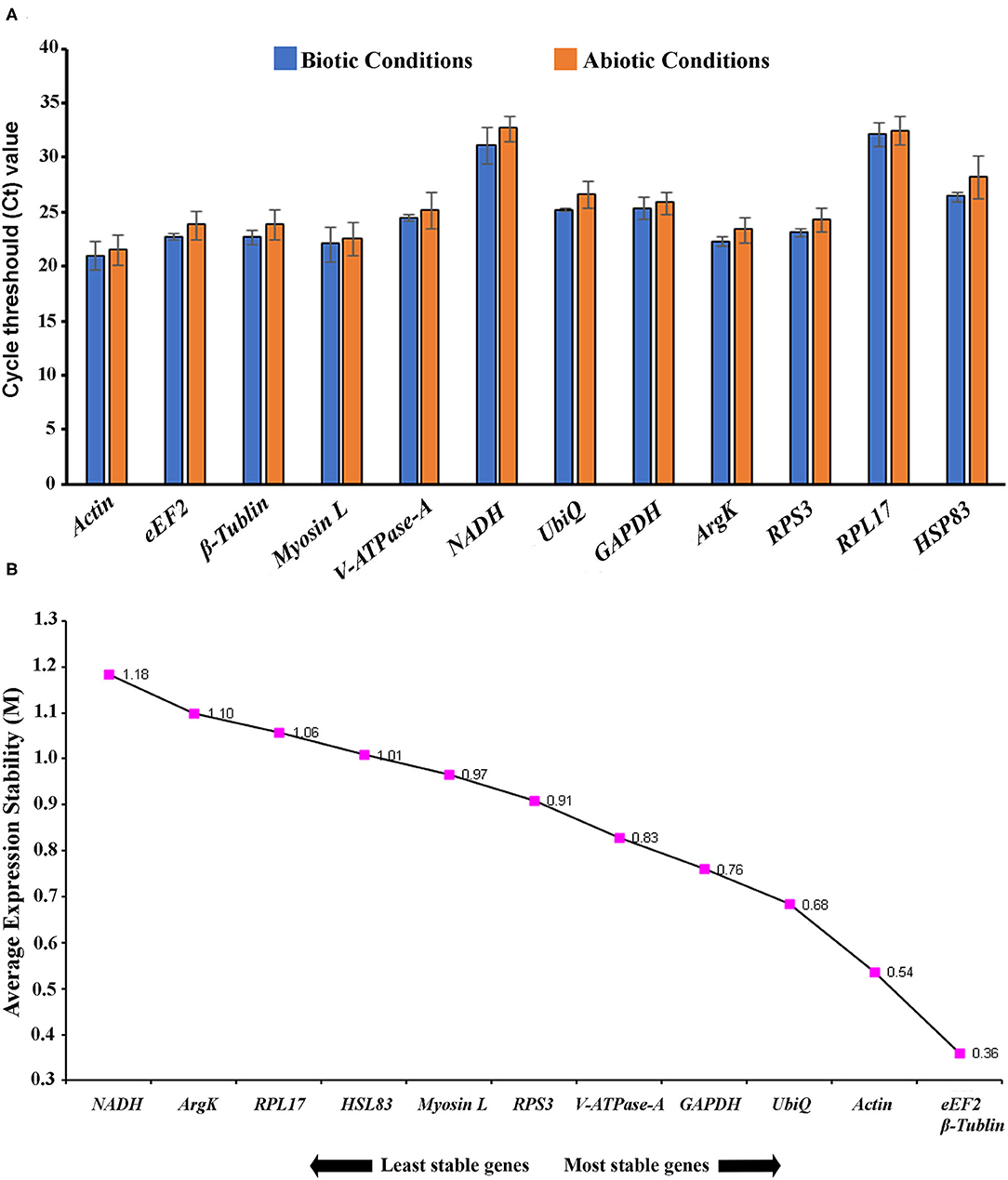
Figure 1. (A) Expression range of cycle threshold (Ct) values of candidate reference genes under different experimental conditions in I. sexdentatus (ISx). The different conditions included in biotic conditions: developmental stages (i.e., larvae, pupae, and adult stages), sex-specific (male and female), tissue types [head, midgut, fat body, and whole body (WB) except head, gut, fat body (Abdomen)] and abiotic conditions: different treatments (such as temperature; Juvenile hormone III; wild-collected vs. long-term laboratory-reared; and different host feeding). (B) The average expression stability values (M) of 12 reference genes in different developmental stages (larvae, pupae, and adult stages) were plotted from the least stable (left) to the most stable (right).
For identifying stable reference genes, four different algorithms (geNorm, NormFinder, BestKeeper, and delta-Ct) were deployed to evaluate the stability of candidate reference genes under different experimental conditions [(i.e., developmental stages, tissue-specific, sex-specific) and different treatments (JHIII; wild-collected vs. long-term laboratory-reared; and different host feeding)] by using the RefFinder web-based tool that ranks reference genes.
For the developmental stages, the order of stability of the first four most stable genes, namely β-Tubulin, eEF2, RPS3, and GADPH, identified by four programs, was inconsistent (Table 2). The least stable genes nominated by four programs were Actin, RPL17, and HSP83. The most stable genes were β-Tubulin, eEF2, and RPS3 determined based on Normfnder rankings of 0.343, 0.347, and 0.379, respectively (Table 2). As per RefFinder, the stability ranking of the reference genes from most stable to least stable across different developmental stages were as follows: eEF2, β-Tubulin, Actin, UbiQ, GAPDH, V-ATPase-A, RPS3, Myosin L, HSP83, RPL17, ArgK, and NADH (Figure 1B). RefFinder identified the top three candidates, eEF2, β-Tubulin, and Actin, across developmental stages by integrating the results from all four programs.
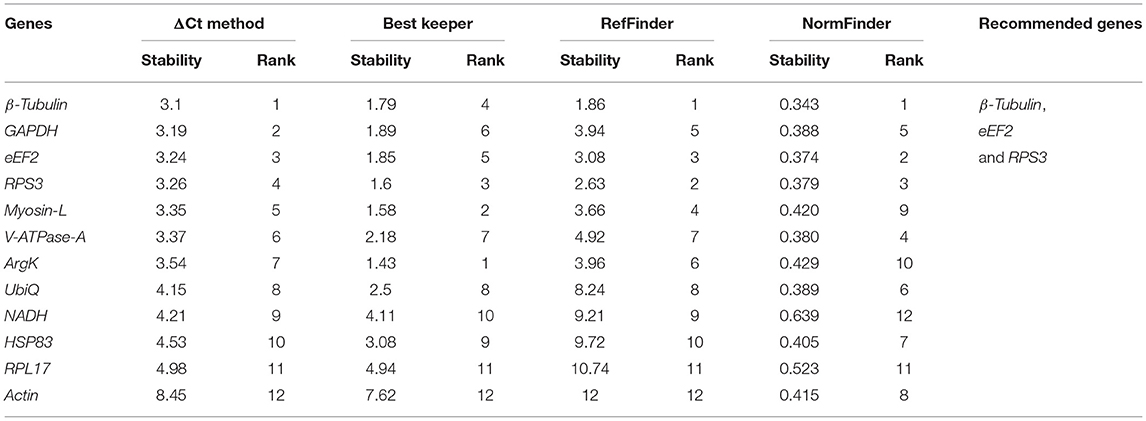
Table 2. Ranking of the candidate reference genes based on stability values performed by Delta Ct, BestKeeper, RefFinder, and NormFinder, in different developmental stages [i.e., larval stages (L1, L2, and L3), pupae (P), callow (newly emerged) male (CM), and female (CF)] and fed adult (mature adults move toward trunks of pines to lay eggs for next-generation) male (AMF) and female (AFF).
Sex-specific reference gene expressions were calculated separately for male (callow and fed adult) and female (callow and fed adult) insect tissues (i.e., head, fat body, gut, WB except for the head, fat body, and gut collected from male and female, or in short, abdomen). In male tissues, the order of stability of the first four most stable genes (RPS3, GAPDH, eEF2, and β-Tubulin) determined by four programs was inconsistent (Table 3). The least stable genes recommended by four programs were Actin, RPL17, and Myosin L. The stability ranking of the reference genes of the most stable and the top three candidates, β-Tubulin, GAPDH, and RPS3, was constant according to RefFinder (Figure 2A). In contrast, the female genes eEF2, RPS3, β-Tubulin, and UbiQ were the most stable genes (Table 3). The least stable gene was ArgK. The top three most stable reference genes, namely eEF2, β-Tubulin, and RPS3, were constant for females in the RefFinder analysis (Figure 2B).
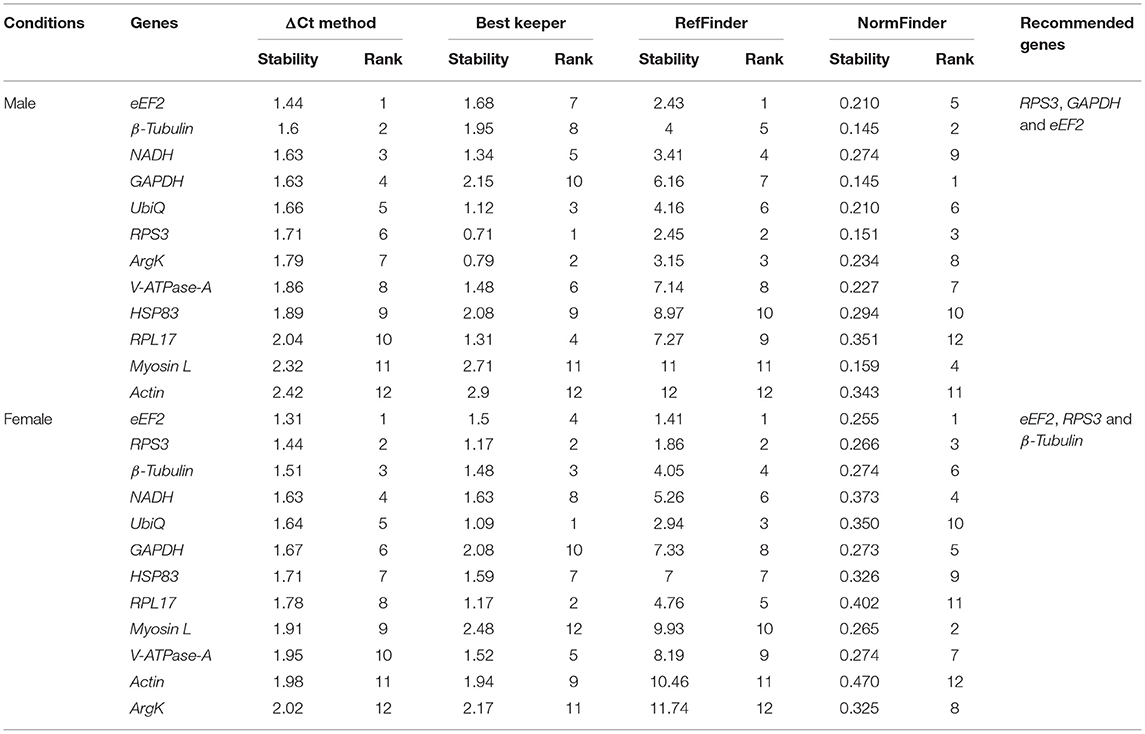
Table 3. Ranking of the candidate reference genes based on stability values performed by Delta Ct, BestKeeper, RefFinder, and NormFinder, in sex-specific conditions (male and female).
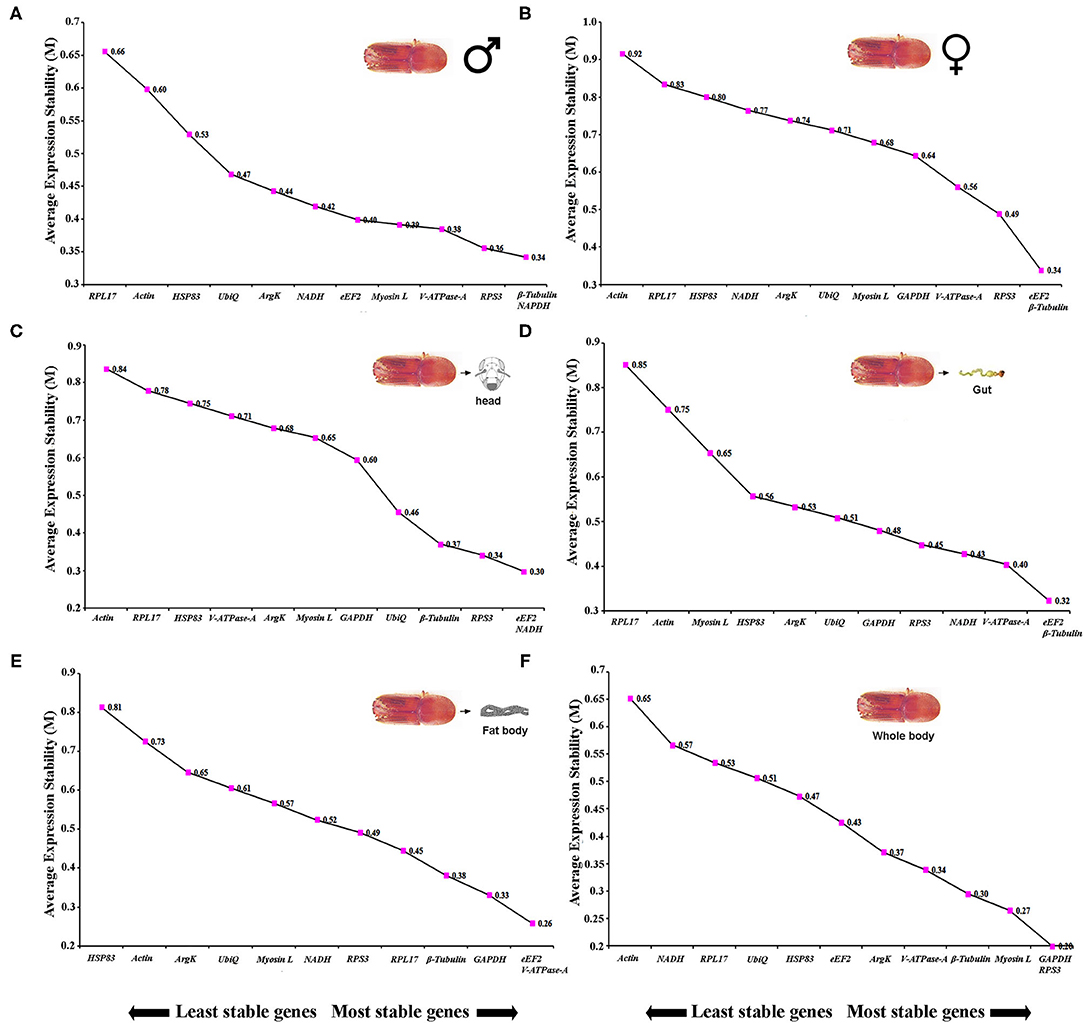
Figure 2. The average expression stability values (M) of 12 reference genes under different conditions (sex-specific and tissue types) were calculated by geNorm from the least stable (left) to the most stable (right). (A) male, (B) female, (C) head, (D) gut, (E) fat body, and (F) whole body (WB).
Tissue sections were obtained to evaluate the stability of candidate reference genes among different tissue (i.e., head, fat body, gut, and WB). In head tissues, the order of stability of the first four most stable genes (i.e., RPS3, eEF2, ArgK, and V-ATPase-A) recognized by four programs was inconsistent (Table 4). The least stable genes determined by four programs were GAPDH, RPL17, and HSP83. According to RefFinder, the stability ranking of reference genes from the most stable and the top three candidates were eEF2, NADH, and RPS3 within head tissues (Figure 2C). Whereas among fat body tissues, V-ATPase-A, eEF2, NADH, and β-Tubulin genes were the most consistently expressed (Table 4). The least stable genes were RPL17, Myosin L, and Actin. The stability ranking (top three) of the reference genes was eEF2, β-Tubulin, and V-ATPase-A (Figure 2D) in the RefFinder analysis for the fat body. However, V-ATPase-A, eEF2, RPL17, and NADH were the most stable genes in the gut tissue (Table 4). The least stable genes in the gut were HSP83, Myosin L, and Actin. The final top three stable reference genes after RefFinder analysis were eEF2, V-ATPase-A, and GAPDH in gut tissues (Figure 2E). Similarly, WB tissue showed that RPS3, β-Tubulin, HSP83, and eEF2 genes were highly expressed and the most stable genes within these tissues (Table 4), while RPL17, UbiQ, and ArgK genes were found to be least stably expressed (Figure 2F). The best reference gene combination and stability ranking for WB tissues were GAPDH, RPS3, and Myosin L. The most stable reference gene among all four tissue stages was eEF2 after assessing all expression results. The optimal reference genes for all tissue stages combined (head, fat body, gut, and WB) conditions were β-Tubulin, eEF2, and RPS3 based on all algorithms (Supplementary Table 3).
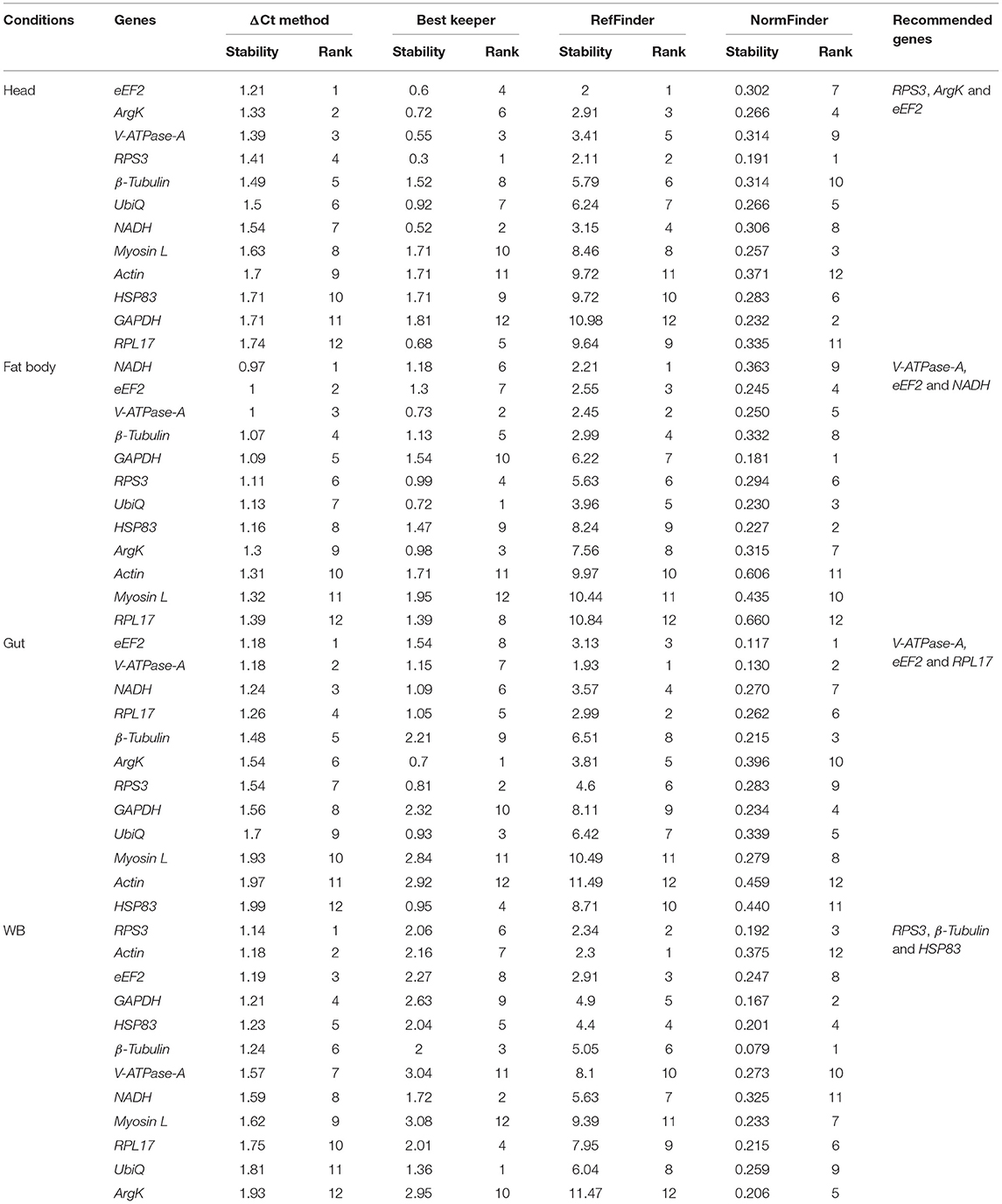
Table 4. Ranking of the candidate reference genes based on stability values performed by Delta Ct, BestKeeper, RefFinder, and NormFinder in various tissue types [head, midgut, fat body, and whole body (WB) except head, gut, and fat body (Abdomen)].
The same four algorithms were used to identify the most appropriate reference genes under four different abiotic conditions such as temperature, JHIII treatment, laboratory-reared vs. wild beetles, and pine-fed vs. spruce-fed beetles. For the temperature treatment, β-Tubulin, V-ATPase-A, ArgK, and GAPDH genes were shown to have the most stable expression using the four algorithms (Table 5). RefFinder confirmed that the three most stable gene expressions were V-ATPase-A, ArgK, and β-Tubulin (Figure 3A). After JHIII treatment, the stability of the first four most stable genes, such as UbiQ, V-ATPase-A, RPS3, and Myosin L determined by four programs was inconsistent (Table 5). The least stable genes were found to be Actin, RPL17, and NADH. According to RefFinder, the stability ranking of the reference genes of the most stable and the top three candidates were RPS3, HSP83, and UbiQ (Figure 3B). However, in beetles from wild and laboratory rearing conditions, most stable expressions were observed for Actin, ArgK, Myosin L, and GAPDH via ΔCt method (Table 5). The top three most stable reference genes via RefFinder were Actin, β-Tubulin, and V-ATPase-A (Figure 3C). Similarly, pine- and spruce-fed beetle gut showed stable expression for V-ATPase-A, UbiQ, ArgK, and β-Tubulin in RefFinder (Table 5 and Figure 3D). The overall stability ranking and combination of reference genes among various tested abiotic conditions were UbiQ, GAPDH, and β-Tubulin.
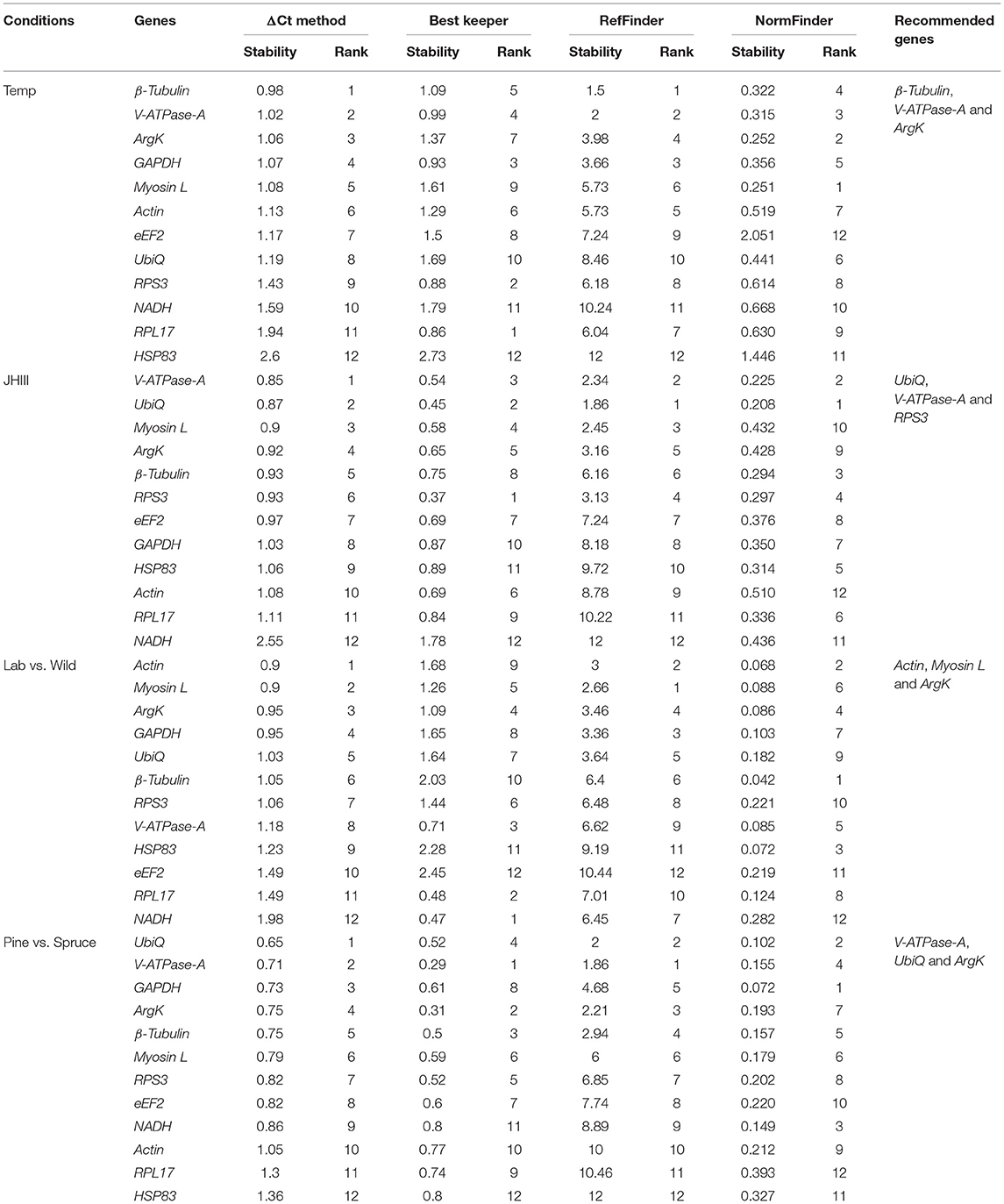
Table 5. Ranking of the candidate reference genes based on stability values performed by Delta Ct, BestKeeper, RefFinder, and NormFinder under the influence of various abiotic factors such as temperature (Temp), Juvenile hormone III (JHIII), laboratory-reared vs. wild beetles, and pine-fed vs. spruce-fed beetles.
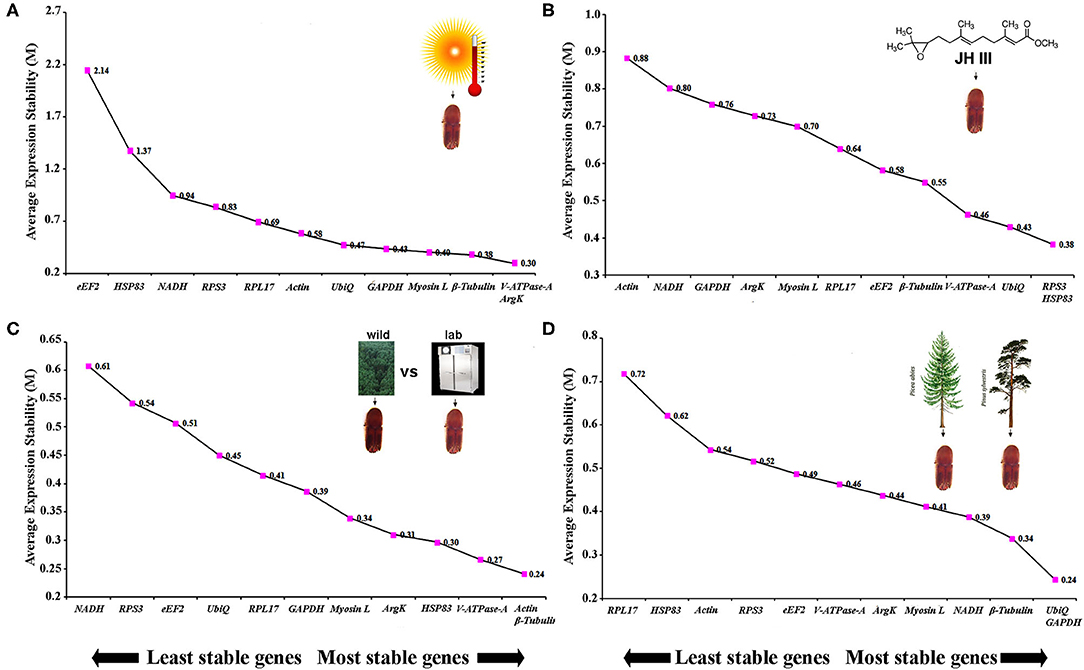
Figure 3. The average expression stability values (M) of 12 reference genes under diverse conditions were calculated by geNorm from the least stable (left) to the most stable (right). (A) Temperature, (B) juvenile hormone III, (C) laboratory-reared vs. wild beetles, and (D) pine-fed vs. spruce-fed beetles.
To generate more accurate and reliable gene expression results, often more than one reference gene is recommended. According to Vandesompele et al. (2002), a Vn/Vn+1 value under 0.15 means adding the n+1 reference gene is unnecessary. Alternatively, the first reference gene is sufficient to normalize the target gene expression in those cases. We also calculated the optimal reference gene number based on geNorm algorithm analysis for each condition. We found that at least two reference genes were required for the head, female tissues, and temperature conditions based on the pairwise values (Figures 4A,B).
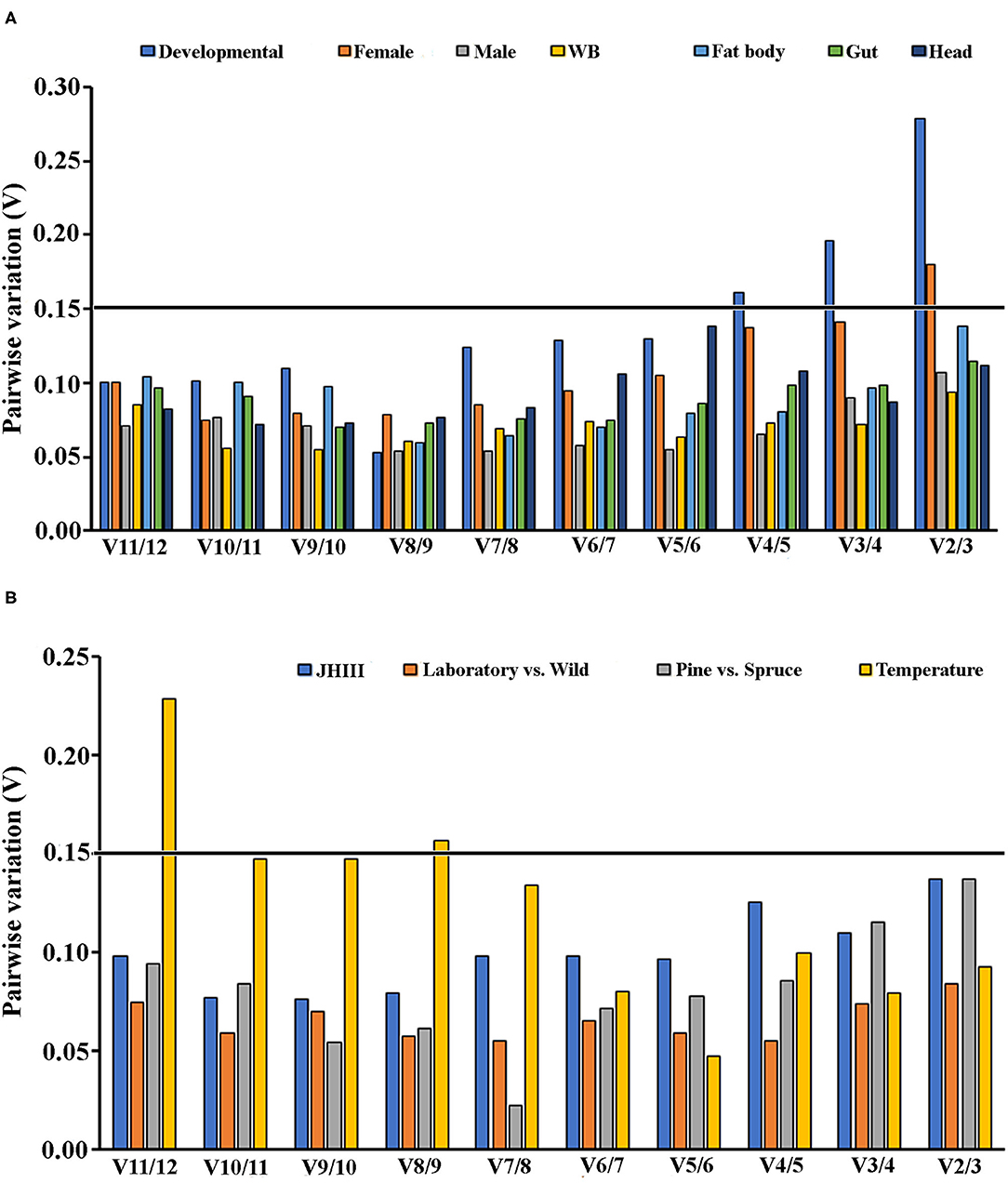
Figure 4. An optimal number of reference genes for the normalization of ISx under selected extrinsic experimental conditions. Based on geNorm analysis, average pairwise variations were calculated between the normalization factors NFn and NFn + 1. Values <0.15 indicate that n + 1 genes were not required for the normalization of gene expression. (A) Biotic conditions: developmental stages (i.e., larvae, pupae, and adult stages), tissue types [head, midgut, fat body, and whole body (WB) except head, gut, fat body] and (B) Abiotic conditions: different treatments (temperature; Juvenile hormone III; wild-collected vs. long-term laboratory-reared; and different host feeding). Values <0.15 indicate that additional genes are not necessary for normalization.
Krüppel-homolog 1 (Kr-h1) encodes a key transcription factor and plays a critical role in regulating insect metamorphosis within the juvenile hormone signaling pathway (Li et al., 2018a; Roy and Palli, 2018). The relative expression of Kr-h1 in response to the JHIII treatment was normalized with single reference genes or gene combinations recommended by geNorm (Figures 4A,B). The two most stable reference genes, individually and in combination, and the least stable gene were used in this experiment. The results showed that the Kr-h1 gene was expressed in male and female beetles (Figure 5A). The expression levels of Kr-h1 normalized with NADH (least stable) in males reduced from 2.1 to 0.6 fold, and females were increased from 1.8 to 3.8 fold higher than those of Kr-h1 normalized with stable reference gene or gene combinations, respectively. Similarly, HSP70 is a key protein closely related to the molecular mechanism underlying insect resistance to the environment (Štětina et al., 2015). The relative expression of Hsp70 showed a stable expression difference after normalizing with the single and most stable reference gene combination during temperature exposure. On the contrary, the Hsp70 gene expression level after normalizing with the least stable reference gene showed higher variation after different temperature incubation (Figure 5B). Henceforth, the most stable genes used for normalization, either individually or in combination, resulted in more consistent and trustworthy target gene expression patterns.
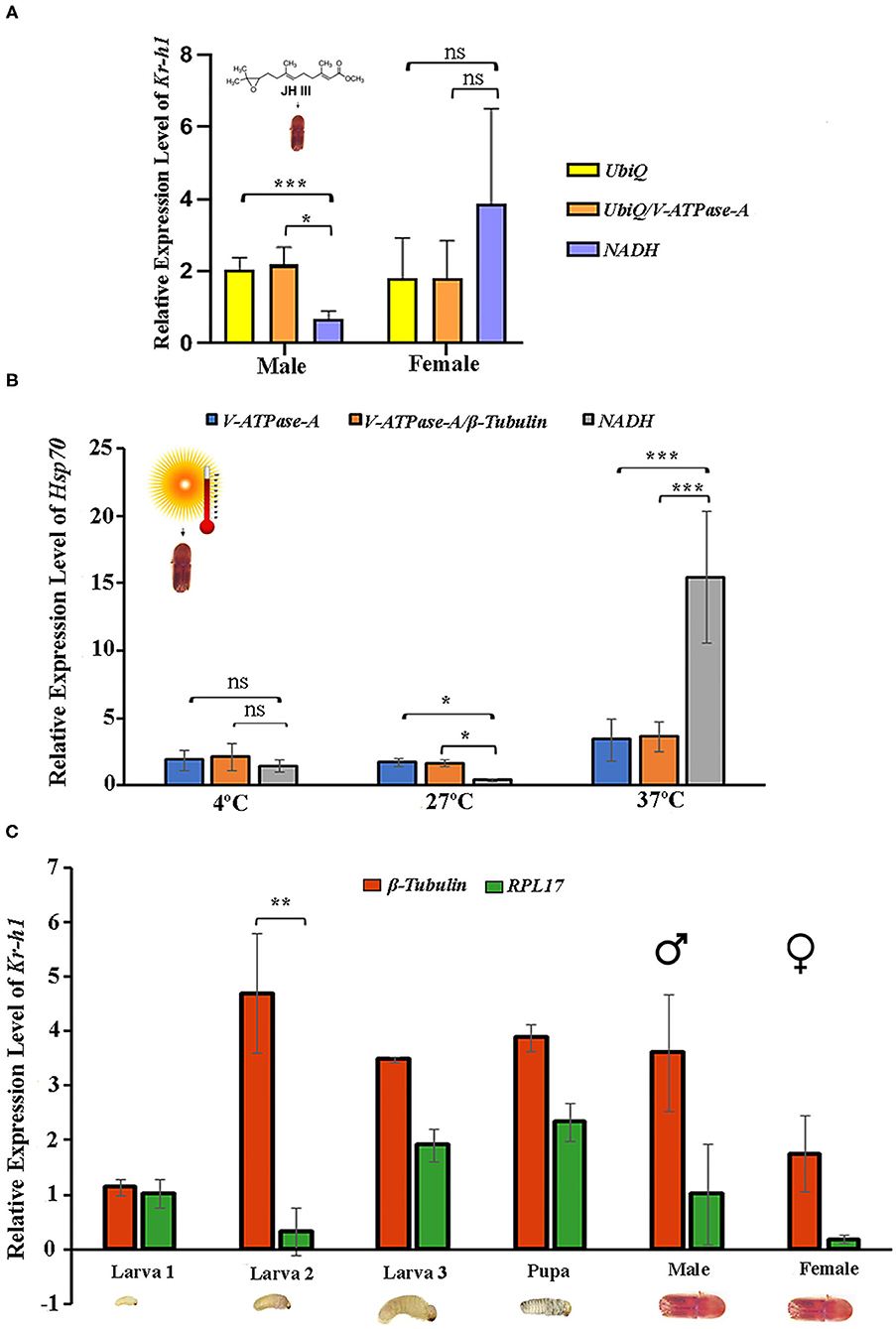
Figure 5. Validation of the recommended reference genes. The relative expression levels of the target genes Kr-h1 and Hsp70 were studied under Juvenile hormone III treatment and temperature treatment by normalizing the selected reference gene. The most stable reference gene is UbiQ and V-ATPase-A for juvenile hormone III; V-ATPase-A and β-Tubulin for temperature, the least stable reference gene for both treatments NADH. The respective combination of stable reference genes UbiQ+V-ATPase-A and V-ATPase-A+β-Tubulin. (A) Juvenile hormone III and (B) Temperature. (C) The relative expression levels of the target gene Kr-h1 were calculated under developmental stages (larvae, pupae, and adult stages), normalized with the most stable reference gene β-Tubulin and the least stable reference gene RPL17. Data represent mean values ± SD of four biological replicates. Asterisks indicate significant differences in the expression of the target gene normalized separately by different reference genes (***P < 0.001, **P < 0.01, *P < 0.05, ns indicate no significant difference).
In addition, Kr-h1 gene expression was evaluated for the various developmental stages to validate the reference gene findings taking the most and least stable gene for expression normalization. Results showed that the expression level of Kr-h1 in the second instar larvae was almost 4-fold higher than the first instar larvae when normalization was performed based on the most stable reference gene (β-Tubulin) (Figure 5C). In contrast, when the least stable reference gene (RPL17) was used for normalization, the Kr-h1 expression was considerably low. Furthermore, normalization with β-Tubulin resulted in lower expression in all stages except second instar larvae signifying the importance of having optimal reference genes for expression normalization.
The wood-boring coleopteran pest, ISx, is one of the most destructive forest pests, causing severe damage to coniferous species throughout Europe and Asia (Jeger et al., 2017). Environmental stress significantly affects host colonization and can provoke transitions from endemic to epidemic bark beetle development (Kausrud et al., 2012). However, to understand the molecular mechanisms of beetle-host interactions, the advent of high-throughput sequencing technologies must advance to increase genetic information. Reference genes with high expression stability under different environmental conditions will be needed to study further a particular gene expression (Fu and Meyer-Rochow, 2021). However, there is no universal reference gene for all samples and tissue types with diverse conditions to date. Therefore, evaluating the stable reference gene under various treatments is essential before aiming DGE study (Lu et al., 2018). The present study examined 12 reference genes commonly applied to Coleoptera using four algorithms (geNorm, NormFinder, BestKeeper, and the ΔCt method) under various biotic and abiotic conditions. There are copious studies regarding the validation of reference genes in other insects (Lu et al., 2018), but no information has previously been reported in any Ips species. Hence, this is the first report for suitable reference genes in any Ips bark beetles.
Accurate normalization using a stable reference gene is necessary to conduct gene expression studies under specific experimental conditions and to avoid erroneous differences in target gene expression (Andersen et al., 2004; Bustin et al., 2005; Ferguson et al., 2010; Cheng et al., 2013; García-Reina et al., 2018; Xie et al., 2021). The results obtained in this study indicate that the stability of reference genes in ISx can differ under various experimental conditions, including developmental stage, sex, and tissue-specific conditions, and exposure to abiotic conditions (Tables 2–5 and Figures 1B–4) as observed in other reference gene finding studies in insects (Lu et al., 2018). Among the 12 reference genes studied in this study, we found that β-Tubulin, eEF2, RPS3, and V-ATPase-A were the most stable in the developmental stage, sex-specific, and tissue-specific conditions from all four algorithms (Tables 2–4); and geNorm (Figures 1B, 2A–F). In addition, both β-Tubulin and eEF2 were more stable than V-ATPase-A and RPS3 at various developmental stages. Teng et al. (2012) reported EF (eEF2) as the most stably expressed gene in different developmental stages of Plutella xylostella. The higher stability of β-Tubulin and eEF2 in two biotic factors (developmental stages and tissues) was also documented in reference gene analyses for Agrilus planipennis (Rajarapu et al., 2012); Sogatella furcifera (An et al., 2016); Mythimna separata (Li et al., 2018b); Chilo partellus (Adeyinka et al., 2019); and Hippodamia variegate (Xie et al., 2021). Alternatively, many studies have reported these genes as unsuitable for normalization because of expression variability in different experimental conditions (developmental stages, tissue stages) as in Drosophila melanogaster (Ponton et al., 2011). However, our results confirmed that tissue-specific expression of reference gene eEF2 was highly stable. It was consistently the top-ranking gene for the developmental, head, fat body, and gut except for WB tissue in ISx.
Similarly, RPs have been evaluated and showed highly stable expression in different insects (Lu et al., 2018). Earlier research findings documented that RP-encoding genes are among the most stably expressed reference genes and have been widely used to normalize gene expression levels in insect molecular investigations during the past 10 years (Lu et al., 2018). For instance, RPS13 and RPS7 exhibited the most stable expression under larval-crowding conditions in Mythimna separata (Li et al., 2018b). RPS3 also exhibited high stability under larval tissues in Lucilia sericata (Baumann et al., 2015). Similarly, RPL9 and RPL10 genes showed higher expression stability in different developmental stages and tissues of Sogatella furcifera (An et al., 2016), whereas RPS26 and RPL32 genes showed the same in Thermobia domestica (Bai et al., 2021). Furthermore, RPS18 and RPL13 genes showed the highest expression stability in Rhopalosiphum padi tissues (Li et al., 2021). Wang et al. (2014) reported RPL22e as the most stable reference gene comparing male and female Mylabris cichorii. Similarly, our results suggested that the RP, RPS3, was the most stable gene among the 12 candidates in sex-specific and tissue-specific (except fat body and gut) conditions tested in ISx (Tables 3, 4 and Figure 2).
Vacuolar-type ATPase (V-ATPase-A) is a proton translocating pump responsible for ATP hydrolysis, one of the most highly conserved eukaryotic proteins. The V-ATPase-A gene had been commonly used as a reference gene in Amrasca biguttula for experiments involving starvation stress and different life stages (Singh et al., 2018); for sex-specific experiments in Cicindela campestris (García-Reina et al., 2018). On the contrary, V-ATPase-A gene expression was highly unstable in the developmental and tissue stages of Mythimna separata (Li et al., 2018c). Nevertheless, our results demonstrated stable expression of V-ATPase-A in the fat body and gut tissues of ISx (Table 4 and Figures 2D,E).
Experiments on ISx involving alteration of abiotic conditions revealed a varied set of genes as references, such as V-ATPase-A, ArgK, and β-Tubulin for temperature incubation; UbiQ, V-ATPase-A, and RPS3 for JHIII treatment; Actin, Myosin L, and β-Tubulin in between laboratory vs. wild beetles, and V-ATPase-A, UbiQ, and ArgK among pine vs. spruce-fed beetle gut tissues (Table 5 and Figure 3). The reference gene V-ATPase-A was used previously for normalization of Mythimna separata gene expression after temperature treatment (Li et al., 2018c), whereas β-Tubulin was applied for a similar purpose for Bemisia tabaci (Dai et al., 2017); Phenacoccus solenopsis (Arya et al., 2017); Amphitetranychus viennensis (Yang et al., 2019); and Hippodamia variegate (Xie et al., 2021).
In the present study, host plants among all treatments caused the highest expression variations of the reference genes. In the dataset of gut samples collected from the adults fed on different plants (i.e., pine and spruce), the stability ranking of tested reference genes was different according to five algorithms. The most stable reference gene was UbiQ and GAPDH according to the geNorm algorithm (Figure 3D). However, V-ATPase-A, UbiQ, and ArgK were the most stable reference genes evaluated by the other four algorithms (delta Ct, BestKeeper, RefFinder, and Normfinder, respectively) (Table 5). Interestingly, similar studies in other arthropods have not found such dramatic variations so far (Arya et al., 2017). Furthermore, in laboratory-reared and wild-collected beetle gut tissues, Actin, Myosin L, and ArgK were the most stable reference genes after delta Ct, BestKeeper, RefFinder, and Normfinder analysis (Table 5), which differed from the geNorm algorithm analysis (Figure 3C). In our opinion, plant diet causes drastic changes at the gene expression level in ISx, hence higher variation in the reference gene expression.
Further, the expression of Kr-h1 and Hsp70 was evaluated in various developmental stages and respective treatments (JHIII and temperature) to corroborate the suitability of the identified reference genes. Although, the results demonstrated that the expression trends in different conditions were accordant using various reference gene or gene combinations. The Kr-h1 gene expression was induced by JHIII, as expected from similar treatments on other beetles (Roy et al., 2017; Roy and Palli, 2018; Xu et al., 2018). However, our results displayed that using less stable reference genes may generate erroneous interpretation, whereas stable reference gene combinations can reduce bias during normalization (Figure 5A). Alternatively, there was no significant expression difference of Hsp70 at 4°C temperature, but considerable expression differences were observed after 27 and 37°C incubation using the most stable reference gene (β-Tubulin) separately and together (β-Tubulin/ V-ATPase-A), and least reference gene (NADH) as normalizer. It is worth mentioning that the expression change was more extensive at 37°C for Hsp70 when normalized with NADH, the least stable gene. Recently, elimination of such erroneous findings was achieved by normalization of gene expression with combinations of stable reference genes in different experimental conditions in Helicoverpa armigera (Zhang et al., 2015), Aphis gossypii (Ma et al., 2016), and Cydia pomonella (Wei et al., 2020). Additionally, with the selected single reference genes, such as β-Tubulin (most stable) and RPL17 (least stable), we performed RT-qPCR to study Kr-h1 expression patterns in several developmental stages of ISx (Figure 5C). More convincing results were obtained when two genes were used for expression normalization (Figures 5A,B). Our results corroborate the current notion of using two or three genes for target gene expression normalization to enhance accuracy (Arya et al., 2017; Li et al., 2018c; Wei et al., 2020; Bai et al., 2021; Fu and Meyer-Rochow, 2021).
Similar to other published studies in the field, our analyses also documented different results under diverse experimental conditions for all five algorithms; however, the results were comparable among some treatments. Interestingly, the most conclusive observation from the current assessments showed that β-Tubulin and eEF2 were the most stable reference genes across all developmental stages, sex, and tissue-specific conditions investigated under the present study (Tables 2–4 and Figures 1A, 2). Our findings suggested that the recommended number of reference genes should be two for many comparisons in the study, based on the pairwise values (>0.15) obtained with geNorm (Figures 4A,B). Furthermore, these results imply that a single reference gene is not optimal to normalize the target gene expression in different experimental conditions. Hence, we endorsed optimal reference genes for the specific experimental conditions in ISx (Table 6).
In conclusion, temperate and boreal forests have recently undergone unprecedented pressure from bark beetle outbreaks, reducing forest biodiversity and their role in global carbon sequestration. It also affects their economic value and endangered wildlife habitat. Current management approaches are proven progressively deficient against outbreaking bark beetle populations, urging investigations into novel mitigation strategies using state-of-the-art molecular methodologies. Future gene expression and functional genomics studies are crucial to alleviate the ongoing bark beetle-mediated forest depletion. Hence, dedicated reference gene selection studies are of utmost importance on economically important bark beetles. The present work represents the first reference gene validation study in I. sexdentatus, an ecologically important wood-boring beetle. It provides valuable information on reference genes (Table 6) for future molecular studies (i.e., DGE studies) on host-beetle interactions and functional genomic studies (i.e., RNAi) on this bark beetle and similar Ips beetles (Coleoptera: Curculionidae: Scolytinae).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s. The reference gene sequences used in the study were submitted under the NCBI accession numbers from OK143445 to OK143455.
GS, AC, and AR conceived and designed the research and prepared the final manuscript. SA, RM, and JS collected, sorted, and dissected beetles. JB and SA prepared samples for in-house transcriptome. GS conducted real-time experiments and wrote the first draft. GS and MKS analyzed the data. All authors have read and approved the final manuscript.
The project was funded by the Internal Grant Agency (IGA) from the Faculty of Forestry and Wood Sciences, Czech University of Life sciences. Infrastructural support and salary for GS, JB, and AR are obtained from grant EXTEMIT-K, No. CZ.02.1.01/0.0/0.0/15_003/0000433 financed by OP RDE. The financial support for AC was provided by grant EVA 4.0, No. CZ.02.1.01/0.0 /0.0/16_019 /0000803 financed by OP RDE.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.752768/full#supplementary-material
Adeyinka, O. S., Tabassum, B., Nasir, I. A., Yousaf, I., and Husnain, T. (2019). Identification and validation of potential reference gene for effective dsRNA knockdown analysis in Chilo partellus. Sci. Rep. 9:13629. doi: 10.1038/s41598-019-49810-w
An, X. K., Hou, M. L., and Liu, Y. D. (2016). Reference gene selection and evaluation for gene expression studies using qRT-PCR in the white-backed Planthopper, Sogatella furcifera (Hemiptera: Delphacidae). J. Econ. Entomol. 109:879. doi: 10.1093/jee/tov333
Andersen, C. L., Jensen, J. L., and Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Arya, S. K., Jain, G., Upadhyay, S. K., Sarita, S. H., Dixit, S., and Verma, P. C. (2017). Reference genes validation in Phenacoccus solenopsis under various biotic and abiotic stress conditions. Sci. Rep. 7:13520. doi: 10.1038/s41598-017-13925-9
Aw, T., Schlauch, K., Keeling, C. I., Young, S., Bearfield, J. C., Blomquist, G. J., et al. (2010). Functional genomics of mountain pine beetle (Dendroctonus ponderosae) midguts and fat bodies. BMC Genom. 11:215. doi: 10.1186/1471-2164-11-215
Bai, Y., Lv, Y. N., Zeng, M., Jia, P. Y., Lu, H. N., Zhu, Y. B., et al. (2021). Selection of reference genes for normalization of gene expression in Thermobia domestica (Insecta: Zygentoma: Lepismatidae). Genes 12:21. doi: 10.3390/genes12010021
Basu, S., Pereira, A. E., Pinheiro, D. H., Wang, H., Valencia-Jiménez, A., Siegfried, B. D., et al. (2019). Evaluation of reference genes for real-time quantitative PCR analysis in southern corn rootworm, Diabrotica undecimpunctata howardi (Barber). Sci. Rep. 9:10703. doi: 10.1038/s41598-019-47020-y
Baumann, A., Lehmann, R., Beckert, A., Vilcinskas, A., and Franta, Z. (2015). Selection and evaluation of tissue specific reference genes in Lucilia sericata during an immune challenge. PLoS ONE. 10:e0135093. doi: 10.1371/journal.pone.0135093
Biedermann, P. H. W., Muller, J., Gregoire, J. C., Gruppe, A., Hagge, J., Hammerbacher, A., et al. (2019). Bark beetle population dynamics in the anthropocene: challenges and solutions. Trends Ecol. Evol. 34, 914–924. doi: 10.1016/j.tree.2019.06.002
Bouhot, L., Lieutier, F., and Debouzie, D. (1988). Spatial and temporal distribution of attacks by Tomicus piniperda L. and Ips sexdentatus Boern. (Col., Scolytidae) on Pinus sylvestris. J. Appl. Entomol. 106, 356–371. doi: 10.1111/j.1439-0418.1988.tb00604.x
Bustin, S. A., Benes, V., Nolan, T., and Pfaffl, M. W. (2005). Quantitative real-time RT-PCR–a perspective. J. Mol. Endocrinol. 34, 597–601. doi: 10.1677/jme.1.01755
Chakraborty, A., Ashraf, M. Z., Modlinger, R., Synek, J., Schlyter, F., and Roy, A. (2020b). Unravelling the gut bacteriome of Ips (Coleoptera: Curculionidae: Scolytinae): identifying core bacterial assemblage and their ecological relevance. Sci. Rep. 10:18572. doi: 10.1038/s41598-020-75203-5
Chakraborty, A., Modlinger, R., Ashraf, M. Z., Synek, J., Schlyter, F., and Roy, A. (2020a). Core mycobiome and their ecological relevance in the gut of five Ips bark beetles (Coleoptera: Curculionidae: Scolytinae). Front. Microbiol. 11:568853. doi: 10.3389/fmicb.2020.568853
Cheng, D., Zhang, Z., He, X., and Liang, G. (2013). Validation of reference genes in Solenopsis invicta in different developmental stages, castes and tissues. PLoS ONE 8:e57718. doi: 10.1371/journal.pone.0057718
Dai, T. M., Lü, Z. C., Liu, W. X., and Wan, F. H. (2017). Selection and validation of reference genes for qRT-PCR analysis during biological invasions: the thermal adaptability of Bemisia tabaci MED. PLoS ONE 12:e0173821. doi: 10.1371/journal.pone.0173821
Denham, S. O., Coyle, D. R., Oishi, A. C., Bullock, B. P., Heliövaara, H., and Novick, K. A. (2019). Tree resin flow dynamics during an experimentally induced attack by Ips avulsus, I. calligraphus, and I. Grandicollis. Can. J. For. Res. 49:1. doi: 10.1139/cjfr-2018-0024
Douglas, H. B., Cognato, A. I., Grebennikov, V., and Savard, K. (2019). Dichotomous and matrix-based keys to the Ips bark beetles of the World (Coleoptera: Curculionidae: Scolytinae). Can. J. Arthropod. Identifi. 38:234. doi: 10.3752/cjai.2019.38
Etxebeste, I., and Pajares, J. A. (2011). Verbenone protects pine trees from colonization by the six-toothed pine bark beetle, Ips f sexdentatus Boern. (Col.: Scolytinae). J. Appl. Entomol. 135, 258–268. doi: 10.1111/j.1439-0418.2010.01531.x
Ferguson, B. S., Nam, H., Hopkins, R. G., and Morrison, R. F. (2010). Impact of reference gene selection for target gene normalization on experimental outcome using real-time qRT-PCR in adipocytes. PLoS ONE 5:e15208. doi: 10.1371/journal.pone.0015208
Ferrenberg, S., Kane, J. M., and Mitton, J. B. (2014). Resin duct characteristics associated with tree resistance to bark beetles across lodgepole and limber pines. Oecologia 174, 1283–1292. doi: 10.1007/s00442-013-2841-2
Fu, X., and Meyer-Rochow, V. B. (2021). Selection and validation of suitable reference genes for RT-qPCR analysis in the rare aquatic firefly Aquatica leii (Coleoptera: Lampyridae). Insects 12:359. doi: 10.3390/insects12040359
Gao, P., Wang, J., and Wen, J. (2020). Selection of reference genes for tissue/organ samples of adults of Eucryptorrhynchus scrobiculatus. PLoS ONE 15:e0228308. doi: 10.1371/journal.pone.0228308
García-Reina, A., Rodríguez-García, M. J., and Galián, J. (2018). Validation of reference genes for quantitative real-time PCR in tiger beetles across sexes, body parts, sexual maturity and immune challenge. Sci. Rep. 8:10743. doi: 10.1038/s41598-018-28978-7
Gregoire, J. C., and Evans, H. F. (2004). “Damage and control of BAWBILT organisms an overview,” in Bark And Wood Boring Insects in Living Trees in Europe, A Synthesis, eds F. Lieutier, K. R. Day, A. Battisti, J.-C. Grégoire, H. F. Evans (Dordrecht: Springer), 19–37. doi: 10.1007/978-1-4020-2241-8_4
Gurusamy, D., Howell, J. L., Chereddy, S. C. R. R., Mogilicherla, K., and Palli, S. R. (2021). Improving RNA interference in the southern green stink bug, Nezara viridula. J. Pest Sci. 4, 1461–1472. doi: 10.1007/s10340-021-01358-3
Heid, C. A., Stevens, J., Livak, K. J., and Williams, P. M. (1996). Real time quantitative PCR. Genome Res. 6, 986–994. doi: 10.1101/gr.6.10.986
Higuchi, R., Fockler, C., Dollinger, G., and Watson, R. (1993). Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology. 11, 1026–1030. doi: 10.1038/nbt0993-1026
Huang, J., Kautz, M., Trowbridge, A. M., Hammerbacher, A., Raffa, K. F., Adams, H. D., et al. (2020). Tree defence and bark beetles in a drying world: carbon partitioning, functioning and modelling. New Phytol. 225, 26–36. doi: 10.1111/nph.16173
Jeger, M., Bragard, C., Caffier, D., Candresse, T., Chatzivassiliou, E., Dehnen-Schmutz, K., et al. (2017). Scientific Opinion on the pest categorisation of Ips sexdentatus. EFSA J. 15:4999. doi: 10.2903/j.efsa.2017.4999
Joga, M. R., Mogilicherla, K., Smagghe, G., and Roy, A. (2021). RNA interference-based forest protection products (FPPs) against wood-boring coleopterans: hope or hype? Front. Plant Sci. 12:733608. doi: 10.3389/fpls.2021.733608
Kausrud, K., Økland, B., Skarpaas, O., Grégoire, J. C., Erbilgin, N., and Stenseth, N. C. (2012). Population dynamics in changing environments: the case of an eruptive forest pest species. Biol. Rev. 87, 34–51. doi: 10.1111/j.1469-185X.2011.00183.x
Li, H. B., Dai, C. G., Zhang, C. R., He, Y. F., Ran, H. Y., and Chen, S. H. (2018b). Screening potential reference genes for quantitative real-time PCR analysis in the oriental armyworm, Mythimna separata. PLoS ONE 13:e0195096. doi: 10.1371/journal.pone.0195096
Li, K., Xu, N., Yang, Y. J., Zhang, J. H., and Yin, H. (2018c). Identification and validation of reference genes for RT-qPCR normalization in Mythimna separata (Lepidoptera: Noctuidae). Biomed. Res. Int. 14:1828253. doi: 10.1155/2018/1828253
Li, K. L., Yuan, S. Y., Nanda, S., Wang, W. X., Lai, F. X., Fu, Q., et al. (2018a). The roles of E93 and Kr-h1 in metamorphosis of Nilaparvata lugens. Front. Physiol. 9:1677. doi: 10.3389/fphys.2018.01677
Li, M., Li, X., Wang, C., Li, Q., Zhu, S., Zhang, Y., et al. (2021). Selection and validation of reference genes for qRT-PCR analysis of Rhopalosiphum padi (Hemiptera: Aphididae). Front. Physiol. 12:663338. doi: 10.3389/fphys.2021.663338
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta C(T)] method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, J., Yang, C., Zhang, Y., and Pan, H. (2018). Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: a systematic review. Front. Physiol. 9:1560. doi: 10.3389/fphys.2018.01560
Lubojacký, J., and Knížek, M. (2020). “Podkorní Hmyz,” in Výskyt lesních škodlivých činitelu v roce 2019 a jejich očekávaný stav v roce 2020. Strnady, eds M. Knížek and J. Liška (VULHM), 22–35.
Ma, K. S., Li, F., Liang, P. Z., Chen, X. W., Liu, Y., and Gao, X. W. (2016). Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae). J. Insect Sci. 16:17. doi: 10.1093/jisesa/iew003
Marini, L., Okland, B., Jonsson, A. M., Bentz, B., Carroll, A., Forster, B., et al. (2017). Climate drivers of bark beetle outbreak dynamics in Norway spruce forests. Ecography 40, 1426–1435. doi: 10.1111/ecog.02769
Nicot, N., Hausman, J. F., Hoffmann, L., and Evers, D. (2005). Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2914. doi: 10.1093/jxb/eri285
Pettit, J., Voelker, S., DeRose, R. J., and Burton, J. (2020). Spruce beetle outbreak was not driven by drought stress: evidence from a tree-ring iso-demographic approach indicate temperatures were more important. Glob. Chang. Biol. 2020, 1–15. doi: 10.1111/gcb.15274
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pairwise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Pfeffer, A. (1955). Fauna of Czechoslovakia: Bark beetles – Scolytoidea, Fauna CSR., svazek 6. Kurovci–Scolytoidea. 1. vyd. Praha: Czechoslovak Academy of Sciences-Czechoslovak Academy of Sciences, 324.
Ponton, F., Chapuis, M. P., Pernice, M., Sword, G. A., and Simpson, S. J. (2011). Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57, 840–850. doi: 10.1016/j.jinsphys.2011.03.014
Postner, M. (1974). “Scolytidae (=ipidae), borkenkaäfer,” in Die forstschaädlinge Europas, II Käfer, vol 2, ed W. Schwenke (Hamburg: Verlag Paul Parey), 334–482.
Qu, C., Wang, R., Che, W., Zhu, X., Li, F., and Luo, C. (2018). Selection and evaluation of reference genes for expression analysis using quantitative real-time PCR in the Asian Ladybird Harmonia axyridis (Coleoptera: Coccinellidae). PLoS ONE 13:e0192521. doi: 10.1371/journal.pone.0192521
Rajarapu, S. P., Mamidala, P., and Mittapalli, O. (2012). Validation of reference genes for gene expression studies in the emerald ash borer (Agrilus planipennis). Insect Sci. 19, 41–46. doi: 10.1111/j.1744-7917.2011.01447.x
Rodrigues, T. B., Khajuria, C., Wang, H., Matz, N., Cunha, C. D., Valicente, F. H., et al. (2014). Validation of reference housekeeping genes for gene expression studies in western corn rootworm (Diabrotica virgifera virgifera). PLoS ONE 9:e109825. doi: 10.1371/journal.pone.0109825
Roy, A., George, S., and Palli, S. R. (2017). Multiple functions of CREB-binding protein during postembryonic development: identification of target genes. BMC Genom. 18:996. doi: 10.1186/s12864-017-4373-3
Roy, A., and Palli, S. R. (2018). Epigenetic modifications acetylation and deacetylation play important roles in juvenile hormone action. BMC Genom. 19:934. doi: 10.1186/s12864-018-5323-4
Seidl, R., Thom, D., Kautz, M., Martin-Benito, D., Peltoniemi, M., Vacchiano, G., et al. (2017). Forest disturbances under climate change. Nat. Clim. Change. 7, 395–402. doi: 10.1038/nclimate3303
Shakeel, M., Rodriguez, A., Tahir, U. B., and Jin, F. (2018). Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol. Lett. 40, 227–236. doi: 10.1007/s10529-017-2465-4
Silver, N., Best, S., Jiang, J., and Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7:33. doi: 10.1186/1471-2199-7-33
Singh, S., Gupta, M., Pandher, S., Kaur, G., Rathore, P., and Palli, S. R. (2018). Selection of housekeeping genes and demonstration of RNAi in cotton leafhopper, Amrasca biguttula biguttula (Ishida). PLoS ONE 13:e0191116. doi: 10.1371/journal.pone.0191116
Sommerfeld, A., Rammer, W., Heurich, M., Hilmers, T., Müller, J., and Seidl, R. (2020). Do bark beetle outbreaks amplify or dampen future bark beetle disturbances in Central Europe? J. Ecol. 2020, 1–13. doi: 10.1111/1365-2745.13502
Štětina, T., Koštál, V., and Korbelová, J. (2015). The role of inducible Hsp70, and other heat shock proteins, in adaptive complex of cold tolerance of the fruit fly (Drosophila melanogaster). PLoS ONE 10:e0128976. doi: 10.1371/journal.pone.0128976
Sun, Y., Fu, F., Kang, X., Liu, B., Ning, H., and Chen, H. (2021). Function of mevalonate pathway genes in the synthesis of frontalin in Chinese white pine beetle, Dendroctonus armandi (curculionidae: Scolytinae). Arch. Insect Biochem. Physiol. 107, 1–13. doi: 10.1002/arch.21828
Teng, X., Zhang, Z., He, G., Yang, L., and Li, F. (2012). Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Four Lepidopteran insects. J. Insect Sci. 12, 1–17. doi: 10.1673/031.012.6001
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Roy, N. V., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 1–11. doi: 10.1186/gb-2002-3-7-research0034
VanGuilder, H. D., Vrana, K. E., and Freeman, W. M. (2008). Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44, 619–626. doi: 10.2144/000112776
Wang, Y., Wang, Z. K., Huang, Y., Liao, Y. F., and Yin, Y. P. (2014). Identification of suitable reference genes for gene expression studies by qRT-PCR in the blister beetle Mylabris cichorii. J. Insect Sci. 14:94. doi: 10.1673/031.014.94
Wang, Z., Meng, Q., Zhu, X., Sun, S., Liu, A., Gao, S., et al. (2020). Identification and evaluation of reference genes for normalization of gene expression in developmental stages, sexes, and tissues of Diaphania caesalis (Lepidoptera, Pyralidae). J. Insect Sci. 20:1. doi: 10.1093/jisesa/iez130
Wei, Z., Liu, M., Hu, C., and Yang, X. (2020). Overexpression of glutathione S-transferase genes in field λ-cyhalothrin-resistant population of Cydia pomonella: reference gene selection and expression analysis. J. Agr. Food Chem. 68, 5825–5834. 10.1021/acs.jafc.0c01367 doi: 10.1021/acs.jafc.0c01367
Wermelinger, B., Rigling, D., Schneider, M. D., and Dobbertin, M. (2008). Assessing the role of bark- and wood-boring insects in the decline of Scots pine (Pinus sylvestris) in the Swiss Rhone valley. Ecol. Entomol. 33, 239–249. doi: 10.1111/j.1365-2311.2007.00960.x
Xie, J., Liu, T., Khashaveh, A., Yi, C., Liu, X., and Zhang, Y. (2021). Identification and evaluation of suitable reference genes for RT-qPCR analysis in Hippodamia variegata (Coleoptera: Coccinellidae) under different biotic and abiotic conditions. Front. Physiol. 12:669510. doi: 10.3389/fphys.2021.669510
Xu, J., Roy, A., and Palli, S. R. (2018). CREB-binding protein plays key roles in juvenile hormone action in the red flour beetle, Tribolium Castaneum. Sci. Rep. 8:1426. doi: 10.1038/s41598-018-30083-8
Yang, J., Gao, Y., Liu, Z., Lu, J., Zhang, Y., Zhang, P., et al. (2019). Selection of reference genes for RT-qPCR analysis under intrinsic conditions in the hawthorn spider mite, Amphitetranychus viennensis (Acarina: Tetranychidae). Front. Physiol. 10:1427. doi: 10.3389/fphys.2019.01427
Keywords: Ips sexdentatus, reference gene, RT-qPCR, differential gene expression, housekeeping genes, bark beetles, Scolytinae
Citation: Sellamuthu G, Amin S, Bílý J, Synek J, Modlinger R, Sen MK, Chakraborty A and Roy A (2021) Reference Gene Selection for Normalizing Gene Expression in Ips Sexdentatus (Coleoptera: Curculionidae: Scolytinae) Under Different Experimental Conditions. Front. Physiol. 12:752768. doi: 10.3389/fphys.2021.752768
Received: 03 August 2021; Accepted: 24 September 2021;
Published: 27 October 2021.
Edited by:
Fernando Ariel Genta, Oswaldo Cruz Foundation (FIOCRUZ), BrazilReviewed by:
Xue-Qing Yang, Shenyang Agricultural University, ChinaCopyright © 2021 Sellamuthu, Amin, Bílý, Synek, Modlinger, Sen, Chakraborty and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit Roy, cm95QGZsZC5jenUuY3o=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.