- 1Key Laboratory of Integrated Pest Management on Crops in Northwestern Oasis, Ministry of Agriculture and Rural Affairs, Institute of Plant Protection, Xinjiang Academy of Agricultural Sciences, Urumqi, China
- 2State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 3Guangdong Laboratory for Lingnan Modern Agriculture (Shenzhen Branch), Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
The bark beetle, Scolytus schevyrewi (S. schevyrewi), is an economically important pest in China that causes serious damage to the fruit industry, particularly, in Xinjiang Province. Chemical signals play an important role in the behavior of most insects, accordingly, ecofriendly traps can be used to monitor and control the target pests in agriculture. In order to lay a foundation for future research on chemical communication mechanisms at the molecular level, we generate antennal transcriptome databases for male and female S. schevyrewi using RNA sequencing (RNA-seq) analysis. By assembling and analyzing the adult male and female antennal transcriptomes, we identified 47 odorant receptors (ORs), 22 ionotropic receptors (IRs), 22 odorant-binding proteins (OBPs), and 11 chemosensory proteins (CSPs). Furthermore, expression levels of all the candidate OBPs and CSPs were validated in different tissues of male and female adults by semiquantitative reverse transcription PCR (RT-PCR). ScosOBP2 and ScosOBP18 were highly expressed in female antennae. ScosCSP2, ScosCSP3, and ScosCSP5 were specifically expressed in the antennae of both males and females. These results provide new potential molecular targets to inform and improve future management strategies of S. schevyrewi.
Introduction
Olfaction serves to detect environmental chemical information necessary for insect behavior such as finding food sources, mates, and oviposition sites (Hanson, 1999; Clyne et al., 2000). Insects have a sophisticated olfactory system that begins with the reception of odorants at the peripheral chemosensory system. Insect olfaction is dependent on olfactory receptor neurons (ORNs) in sensilla (Leal, 2013) distributed mainly in antennae and also in maxillary palps or labial palps (Stoker et al., 1990). The research of molecular mechanisms of olfactory reception in insects has predominantly been in the model organism Drosophila melanogaster. These studies have shown diverse olfactory genes encoding proteins, such as odorant receptors (ORs), ionotropic receptors (IRs), odorant-binding proteins (OBPs), and chemosensory proteins (CSPs), involved in different chemical signal transduction processes (Benton et al., 2009; Wilson, 2013; Xiao et al., 2019).
Odorant receptors play a critical role in recognizing thousands of odorant molecules in the insect olfactory system. Insect ORs were first identified in Drosophila which has the characteristic feature of a seven-transmembrane domain (TMD) structure that is unrelated to the ORs in vertebrates (Clyne et al., 1999; Benton et al., 2006). Every ORN can express a single or two OR genes (Vosshall and Hanson, 2011). Specificity of OR relies on the ligand-banding ORs (Dobritsa et al., 2003; Elmore et al., 2003; Hallem et al., 2004), while Orco functions as an obligatory chaperon for the Orco-OR complex (Larsson et al., 2004; Benton et al., 2006; Stengl, 2017).
Evolved from the ionotropic glutamate receptor superfamily, IRs have been shown to be involved in odor reception. They are expressed in the sensory neurons that respond to many distinct odors, such as acids, amines, and other chemicals that cannot be recognized by ORs (Benton et al., 2006). Aside from olfaction, IRs serve various functions, such as cool sensation (Ni et al., 2015), hygrosensation (Knecht et al., 2016), circadian clock (Chen et al., 2015), and detection of carbon dioxide (CO2) (Breugel et al., 2018).
In addition to ORs and IRs, other multigene families encode proteins that also play critical roles in olfaction. OBPs are small soluble proteins secreted in the sensillar lymph. They are characterized by an N-terminal signal peptide sequence and a set of six conserved cysteine residues that form three disulfide bridges (Pelosi et al., 2005, 2006). Studies of defective mutants and wild-type counterparts of OBP76a (also known as LUSH) in Drosophila have shown that this protein has a key role in the perception of alcohol and 11-cis vaccenyl acetate (Kim et al., 1998; Xu et al., 2005; Gomez-Diaz et al., 2013). OBPs have also been reported as a pheromone-binding protein in Lepidoptera (Jing et al., 2019). Some OBPs operate similar to LUSH in response to pheromones. In vivo studies have shown that OBPs significantly affect pheromone perception in moths. Knocking out these OBPs significantly reduced electrophysiological responses to pheromones in several species, such as Helicoverpa armigera (Ye et al., 2017), Spodoptera litura (Liu et al., 2012; Zhu et al., 2016a), and Chilio suppressalis (Dong et al., 2017).
Chemosensory proteins are also small soluble proteins but are shorter in amino acid sequence length than that of OBPs, and CSPs share the same structure of having four conserved cysteines forming two disulfide bridges (Pelosi et al., 2005, 2006; Honson et al., 2015). As semiochemical carriers, some CSPs are involved in chemodetection (Pelosi et al., 2018; Li et al., 2021) because CSPs are abundant in the lymph of chemosensory hairs (Angeli et al., 1999; Jacquin-Joly et al., 2001; Monteforti et al., 2002; Sun et al., 2014). Some of them already have been reported to function such as OBPs, e.g., CSP3 of the honeybee, which specially binds some components of brood pheromone (Briand et al., 2002).
Bark beetles (Coleoptera; Curculionidae; Scolytinae) feed on woods and several of them pose serious threats to forestry, e.g., Ips typographus (Wermelinger, 2004), Dendroctonus ponderosae (Andersson et al., 2013). Since their host-finding relies on chemical communication, e.g., aggregation behavior based on male-produced pheromone (Schlyter et al., 1987), pheromone-based technique could be used for the detection and control of this pest. In order to develop this technique efficiently, one way is to exploit olfactory genes that are critical for successful mate and host finding. Transcriptomic and genomics studies have been performed for searching olfactory genes in bark beetles (Andersson et al., 2013, 2019; Mitchell et al., 2019), and functional studies were limited to only seven ORs (Hou et al., 2021; Yuvaraj et al., 2021). Scolytus schevyrewi (S. schevyrewi) (Cleoptera: Scolylidae) is one of the most destructive insect pests of fruit trees in China. It has a wide host range and has been reported to attack several families of trees in Xinjiang province (Li et al., 1995). Several studies have focused on the identification and field bioassay of chemical attractants in the bark beetle (Fan et al., 2014). In order to provide a molecular basis for gene targets for putative chemical lures of this pest, we performed Illumina Hiseq 2000 sequencing of the transcriptome of adult male and female antennae samples.
Materials and Methods
Insect Rearing and Tissue Collection
Scolytus schevyrewi larvae were reared on the branches of their host plants (Armeniaca vulgaris) collected from Baren County, Xinjiang province, China (39.0°N, 75.8°E) and maintained in the lab under the following conditions of 26.5°C, a cycle of 14-h light:10-h dark, and 65% relative humidity. Pupae were placed on a branch and the emerged adults were collected every day. Two-day-old adults were used to collect male and female antennae, heads (without antennae), thorax, abdomen, legs, and wings using the fine-tip forceps, immediately frozen in liquid nitrogen and stored at−80°C until RNA isolation.
RNA Extraction
Total RNA from different tissues of S. schevyrewi was obtained using TRIzol reagent (Invitrogen, Carlsbad, California, USA) following the instruction of manufacturer. The total RNA from each pair of antennae, legs, and wings were separately obtained from each adult, totaling 300 males and 300 females. Heads (without antennae), thoraxes, and abdomens were separately collected from 20 to 30 adult males and 20 to 30 females. Total RNA was dissolved in RNase-free water, and RNA integrity was verified by gel electrophoresis. RNA concentration and purity were determined on the Nanodrop ND-2000 spectrophotometer (NanoDrop products, Wilmington, DE, USA).
cDNA Library Construction and Sequencing
A total of 1 μg of total RNA of each sample of male and female antennae were used to construct two separate cDNA libraries, one for each sex. Paired-end reads of 100 bp were sequenced using the Illumina HiSeq 2000 platform to obtain library-sequencing information at Beijing Genome Institute (Shenzhen, China). The detailed protocols for cDNA library construction and sequencing applied have been described in the previous studies (Cao et al., 2014; Zhang et al., 2015). The raw data were uploaded to the NCBI SRA database (Accession: PRJNA732801, https://www.ncbi.nlm.nih.gov/sra).
Assembly
Low-quality reads were filtered out, low-quality nucleotides at each end were trimmed, and 3′ adaptors and poly-A/T were removed from the raw reads to generate the clean reads. Subsequently, the clean reads were used to form a de novo assembly using the Trinity platform (v2.1.0) with default parameters (Grabherr et al., 2011). The Trinity outputs were then clustered by TIGR gene indices clustering tools (TGICL) to generate the final unigene dataset (Pertea et al., 2003).
Identification of Olfactory Genes
Unigenes were annotated using blastx against NCBI nonredundant (nr) sequences with e < 1e−5. The blast results were then imported into the Blast2Go (version 3.1) with default parameters (Conesa et al., 2005). OR, IR, OBP, and CSP genes of the candidates were selected according to the nr sequence annotation results in the remote server from the lab. All candidate olfactory genes were manually checked using the blastx program against the nr sequence database. The open-reading frames (ORFs) of the putative olfactory genes were predicted using the ExPASy (Expert Protein Analysis System) translate tool (https://web.expasy.org/translate/). The TMDs of ORs and IRs were predicted using TMHMM server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM). Putative N-terminal signal peptides of OBPs and CSPs were predicted using the SignalP 4.0 server (http://www.cbs.dtu.dk/services/SignalP-4.0/) with default parameters.
Phylogenetic Analysis
Olfactory genes from S. schevyrewi, Ips typographus, Dendroctonus ponderosae (Andersson et al., 2013), and Holotrichia parallela (Yi et al., 2018) were selected for the phylogenetic analysis. Sequence information was listed in Supplementary Table 2. Amino acid sequences were aligned by MAFFT (https://www.ebi.ac.uk/Tools/msa/mafft/). Phylogenetic trees of olfactory genes were constructed using RAxML version 8 with the Jones-Taylor-Thornton amino acid substitution model. Node support was assessed using a bootstrap method based on 1,000 replicates. The trees were visualized, and color-coded in FigTree 1.4.3. For ORs, the tree was rooted in the Orco lineage.
Expression Analysis of the Candidate OBPs and CSPs by Semiquantitative Reverse Transcription PCR (RT-PCR)
Reverse transcription-PCR was performed to verify the expression patterns of OBPs and CSPs of S. schevyrewi. Total RNA from male and female antennae, heads (without antennae), thoraxes, legs, abdomens, and wings were used to synthesize cDNA by RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). Gene-specific primers were designed using Primer 5 and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) (Supplementary Table 1). PCR was performed with the Veriti Thermal Cycler (Applied Biosystems, Carlsbad, CA, USA) under the following conditions: 95°C for 3 min, 25 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, and 72°C for 10 min. PCR amplification products were run on a 2% agarose gel. Because it is difficult to acquire massive amounts of RNA from antennae samples of S. schevyrewi, only two technical repeats were performed for each gene. Uncropped gel images were uploaded as supplements (Supplementary Figure 2).
Results
Transcriptome Assembly
The transcriptomes of male and female S. schevyrewi antennae were separately sequenced by the Illumina HiSeq 2000 platform. Then after filtering, 26,804,894 and 29,176,485 clean reads with 98.60 and 98.55% Q20 scores were generated for male and female samples, respectively. The clean reads were assembled subsequently and generated 40,666 and 36,216 unigenes, respectively. After merging and clustering, a final transcript dataset was revealed with 34,098 unigenes consisting of 14,071 clusters and 20,027 distinct singletons. The dataset was 46.7~57.4 Mb in size and with unigenes having a mean length of 1,684 bp and N50 of 3,179 bp.
Gene Identification and Functional Annotation
The functional annotations of the unigenes were performed mainly based on the blastx results against the nr sequence database. We matched 22,815 (66.9%) unigenes to known proteins by blastx. Among those annotated genes, 16,725 (73.3%) unigenes showed strong homology (e-values lower than 1e−45), while 6,090 (26.7%) unigenes showed poor matches with e-values between 1e−15 and 1e−5. The similarity analysis showed that 11,514 (50.5%) unigenes had more than 60% similarity with known proteins. Most of the annotated unigenes were matched to Tribolium castaneum (67.3%), followed by D. ponderosae (13.7%) and others species (19.0%).
Gene ontology (GO) annotations of the entire set of unigenes were performed using the Blast2GO pipeline based on the blastx searches against nr sequences. A total of 12,720 unigenes were assigned various GO terms. In the molecular function category, genes involved in the binding activity and catalytic activity were most abundant. In the cellular component category, genes involved in cell, cell part, macromolecular, membrane, organelle, and organelle part were enriched. In the biological process category, genes involved in the cellular process, metabolic process and single-organism process were the most represented.
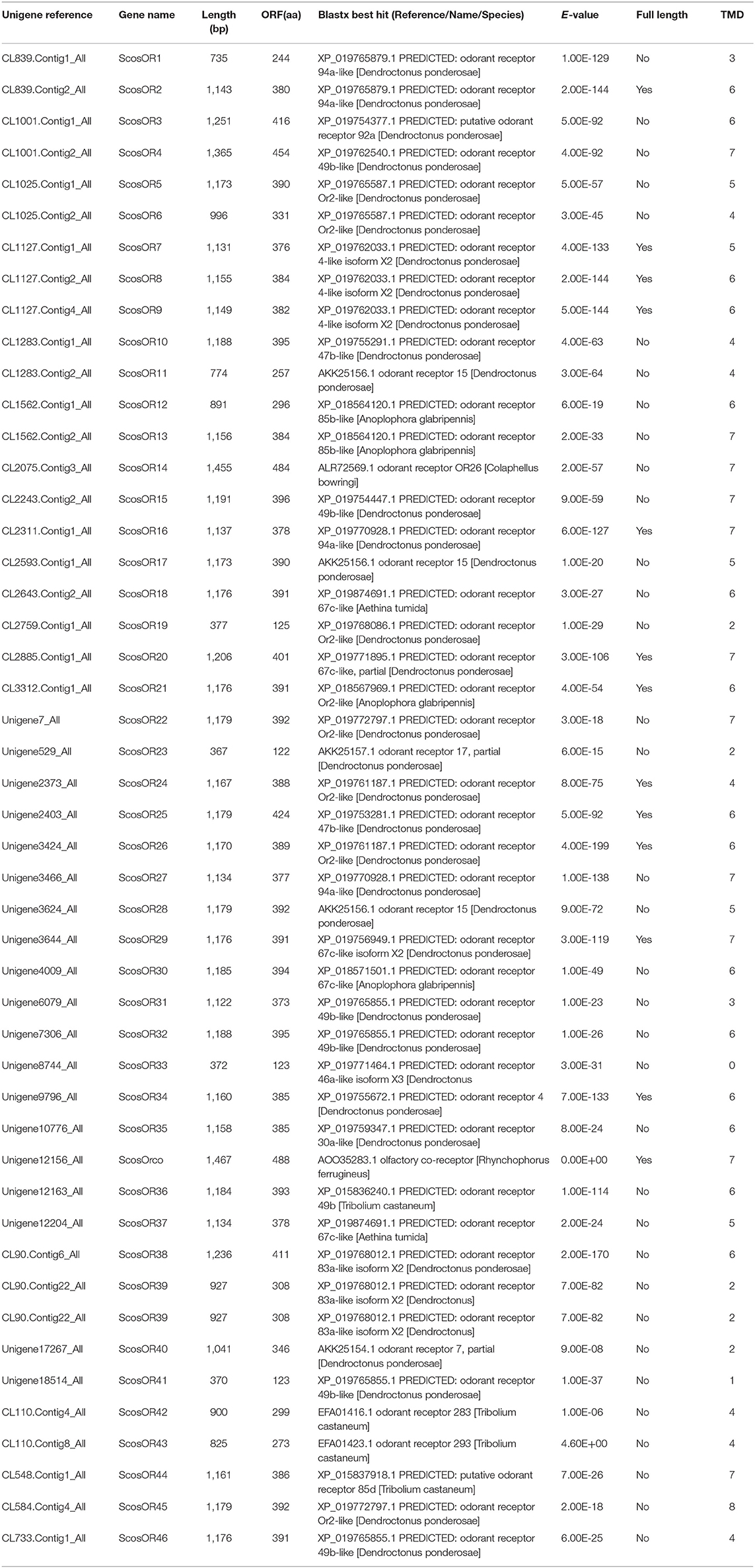
Identification of Candidate Odorant Receptors
The candidate ORs were identified by keyword search of the blastx annotations. We identified 47 putative OR genes. Thirteen of them were full-length putative OR genes ranging from 1,100 to 1,400 bp with complete ORFs and 5 to 7 TMDs, which are characteristics of typical insect ORs. This includes the full-length ScosOrco gene encoding 488 amino acids. Seven of the predicted incomplete ORs were shorter in length and contained a deduced protein longer than 300 amino acids. Four of the predicted incomplete ORs were even shorter than 200 amino acids.
The blastx results indicated that the identities of the most predicted ORs shared with known insect ORs were very low, ranging from 24 to 49%. Nine predicted ORs (ScosOR1, ScosOR27, ScosOR7, ScosOR38, ScosOR39, ScosOR2, ScosOR8, ScosOR9, and ScosOR34) had greater identity (52–62%) with the OR from D. ponderosae. ScosOrco had 88% identity with the Orco from Rhynchophorus ferrugineus. Phylogenetic analysis was performed with ORs from D. ponderosae, I. typographus, H. parallela, and S. schevyrewi (Figure 1). A branch for Orco was identified in the phylogenetic tree. Two expanded branches in this species relative to others in the comparison were also identified. One branch consisted of ScosOR5, ScosOR6, ScosOR10, ScosOR11, ScosOR25, and ScosOR28 and the other consisted of ScosOR17, ScosOR18, Scos22, Scos31, Scos32, Scos37, Scos40, and Scos45. Most of the branches in the tree were supported by high-local support values and few branches were not reliable.
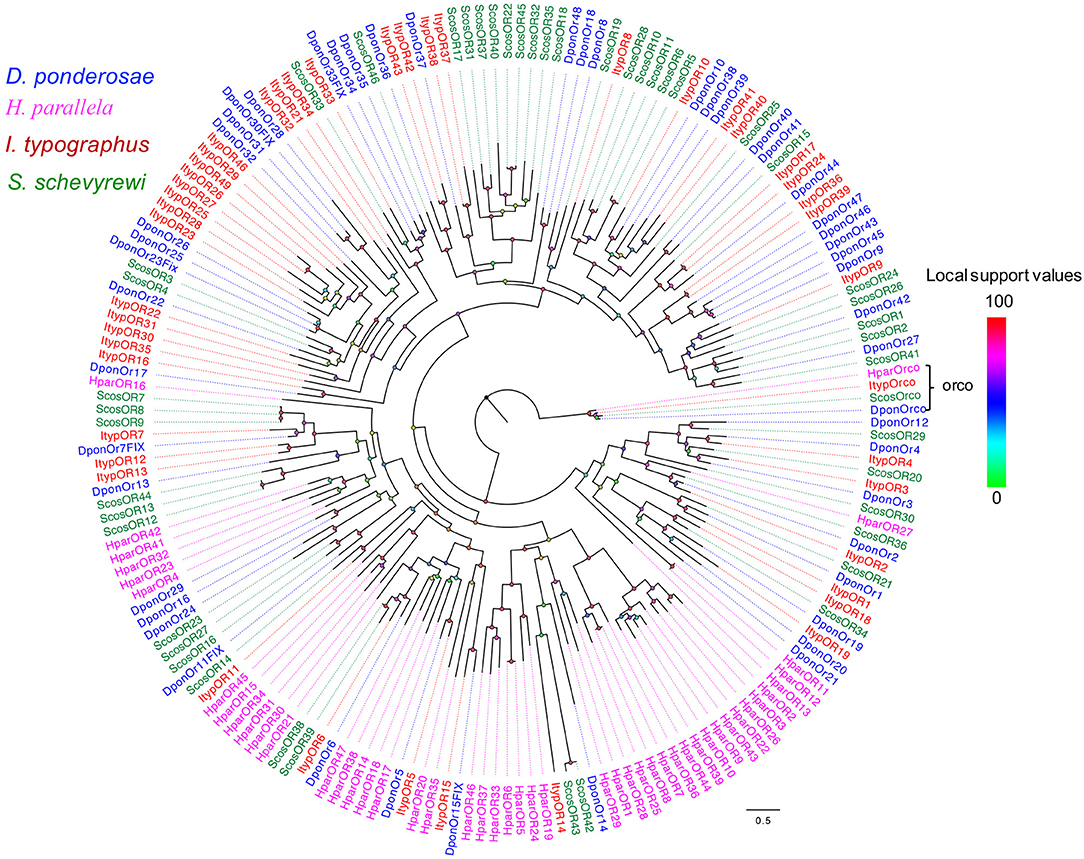
Figure 1. Phylogenetic tree of candidate ScosORs with known Coleoptera odorant receptors sequences. Dpon, D. ponderosae; Hpar, H. parallela; Ityp, I. typographus; Scos, S. schevyrewi.
Information on unigene reference, length, and best blastx hit of all 47 ORs are listed in Table 1.
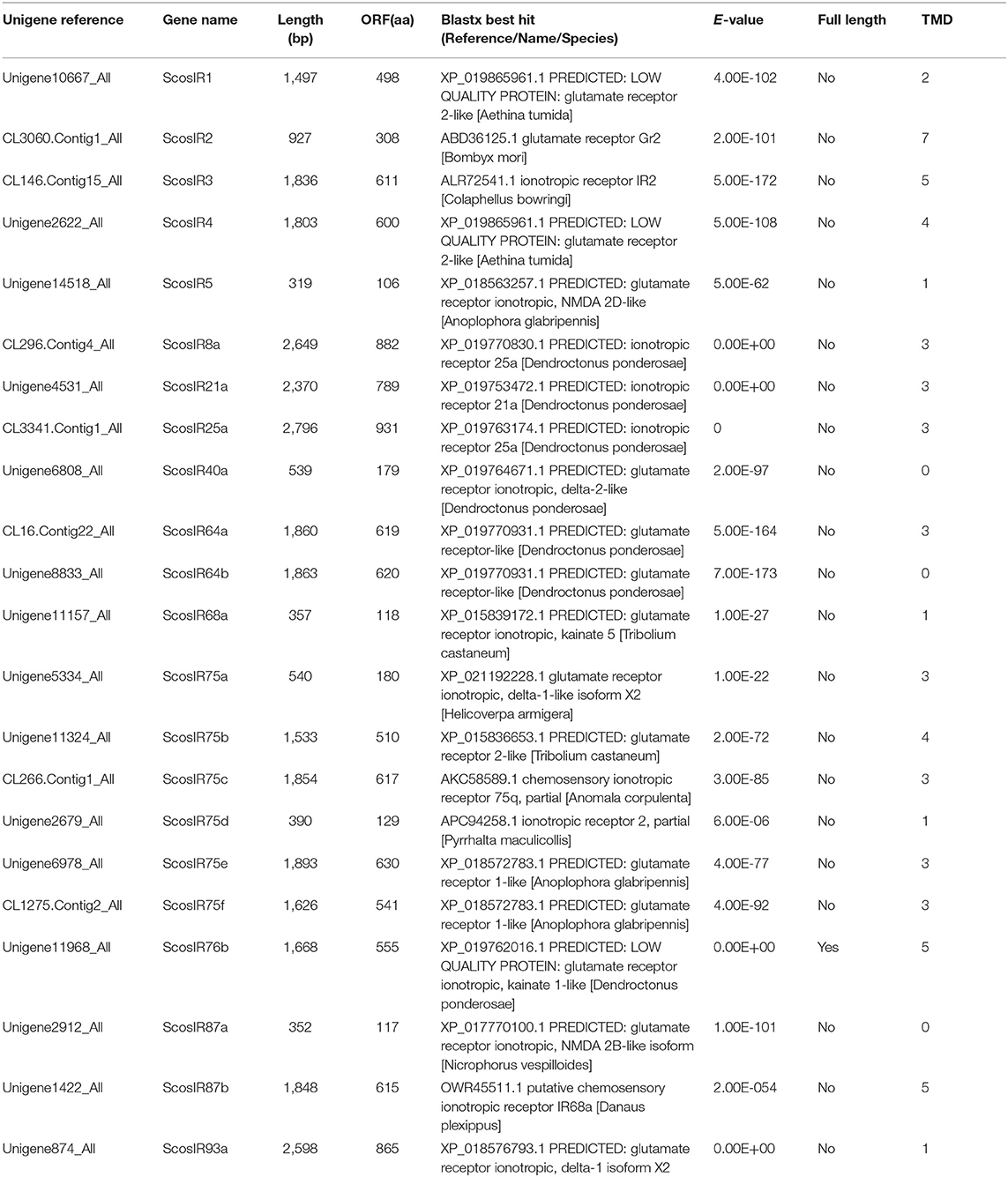
Identification of Candidate Ionotropic Receptors
Bioinformatics analysis identified 22 putative IRs in the S. schevyrewi transcriptome. Only ScosIR76b was a full-length sequence with 555 amino acids and five TMDs; the other IRs were incomplete due to the lack of the 5′ or 3′ terminus.
The blastx results showed that more than half of the predicted IRs (ScosIR1, ScosIR75a, ScosIR75b, ScosIR75c, ScosIR75d, ScosIR75e, ScosIR75f, ScosIR3, ScosIR4, ScosIR64a, ScosIR64b, and ScosIR87b) shared low identity (24–49%) with known insect IRs. A total of 10 predicted IRs (ScosIR68a, ScosIR93a, ScosIR76b, ScosIR2, ScosIR21a, ScosIR8a, ScosIR40a, ScosIR87a, ScosIR25a, and ScosIR5) had greater identity (54–95%) with known insect IRs, most of which were from D. ponderosae. Candidate genes with high identity (87%) to DponIR25a were deemed IR 25a homologs. A phylogenetic tree was constructed based on the IR sequences from D. ponderosae, I. typographus, H. parallela, and S. schevyrewi (Figure 2). ScosIR8a and ScosIR25a were identified as putative IR8a and IR25a homlogs due to the IR8a/IR25a branch.
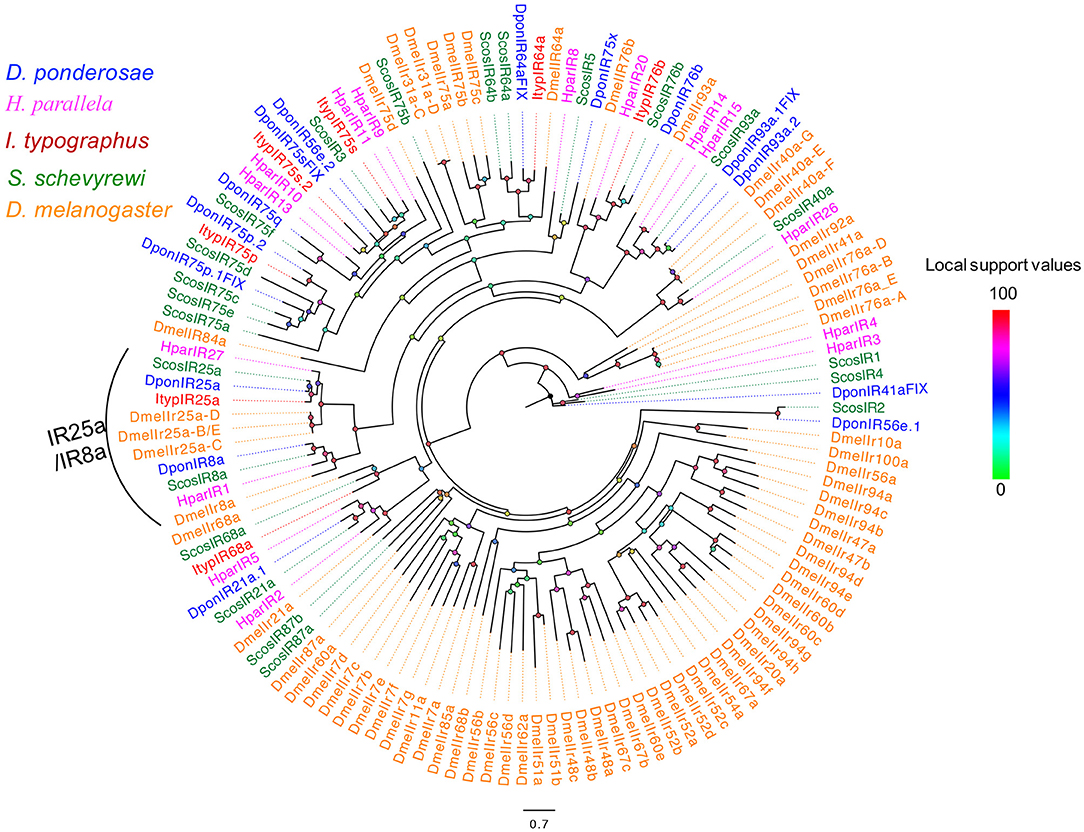
Figure 2. Phylogenetic tree of candidate ScosIRs with ionotropic receptor sequences from other insects. Tcas, T. castaneum; Dpon, D. ponderosae; Hpar, H. parallela; Ityp, I. typographus; Dmel, D. melanogaster; Scos, S. schevyrewi.
Information on unigene reference, length, and best blastx hit of all 22 IRs are listed in Table 2.
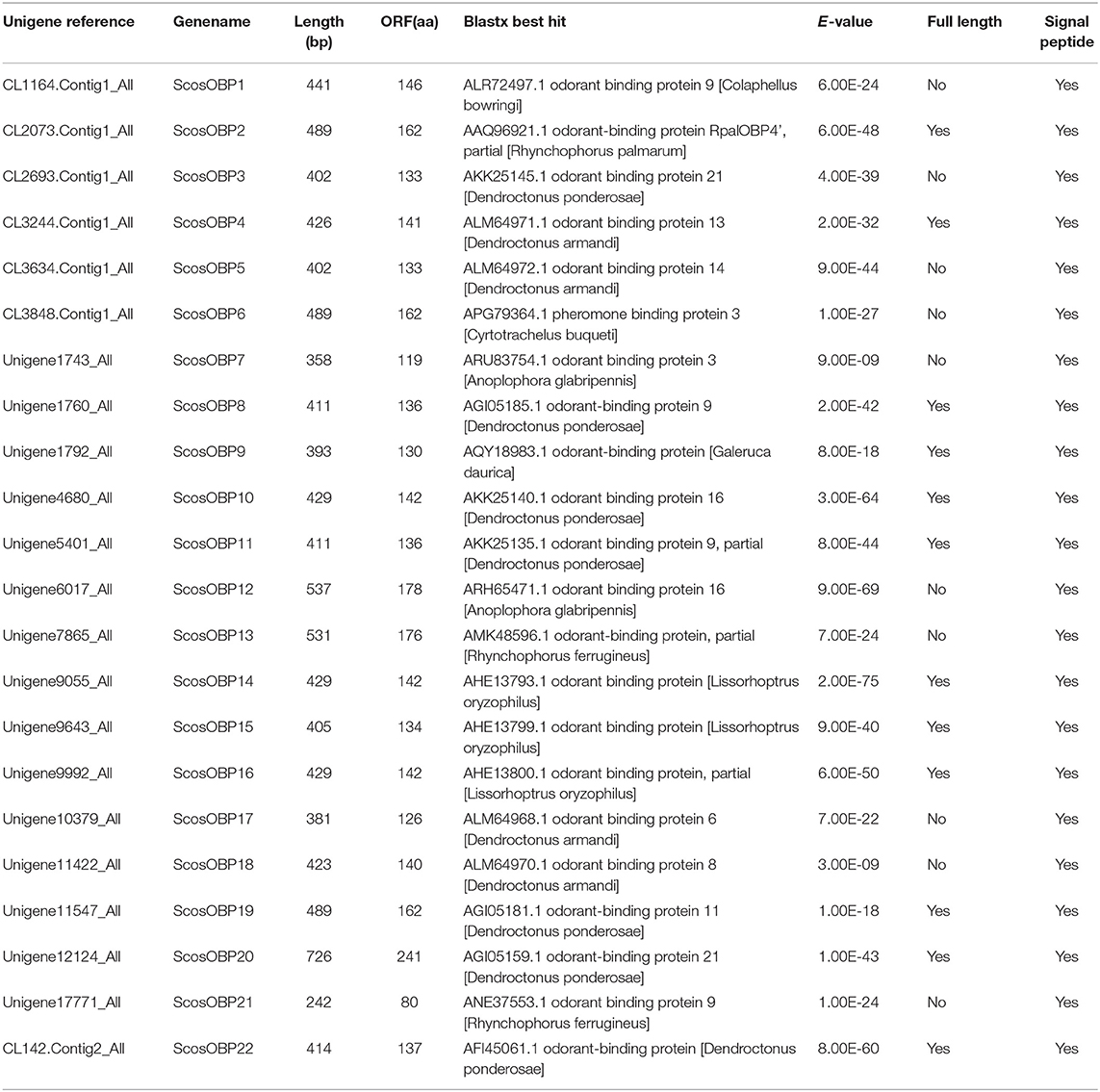
Identification of Odorant-Binding Proteins
In the S. schevyrewi antennal transcriptome, 22 different sequences encoding putative OBPs were identified. More than half of them (ScosOBP2, ScosOBP4, ScosOBP8, ScosOBP9, ScosOBP10, ScosOBP11, ScosOBP14, ScosOBP15, ScosOBP16, ScosOBP19, ScosOBP20, and ScosOBP22) were identified as full-length sequences. The lengths of all full-length ScosOPBs ranged from 130 to 241 amino acids.
Nearly, half of the predicted OBPs (ScosOBP7, ScosOBP18, ScosOBP9, ScosOBP1, ScosOBP20, ScosOBP19, ScosOBP17, ScosOBP4, ScosOBP4, ScosOBP6, and ScosOBP8) shared relatively low identity with known insect OBPs (31–49%). A total of 12 predicted OBPs (ScosOBP13, ScosOBP15, ScosOBP11, ScosOBP16, ScosOBP21, ScosOBP5, ScosOBP2, ScosOBP3, ScosOBP22, ScosOBP12, ScosOBP10, and ScosOBP14) had greater identity (51–83%) with known OBPs, a majority of which were D. ponderosae OBPs. Sequence alignment showed that 10 OBPs (ScosOBP1, 2, 3, 10, 12, 14, 16, 17, 18, and 22) matched in amino acid sequence to the sequence structure of classic OBPs, and eight OBPs (ScosOBP4, 5, 8, 9, 11, 13, 15, and 19) matched the sequence structure of Minus-C OBPs (Figure 3). Other OBPs were not analyzed by sequence alignment due to their limited sequence lengths. In the phylogenetic analysis of OBPs of different coleopterans, ScosOBPs were found across various branches and generally formed small subgroups together with OBPs from other coleopterans (Figure 4). No species-specific branch was discovered.
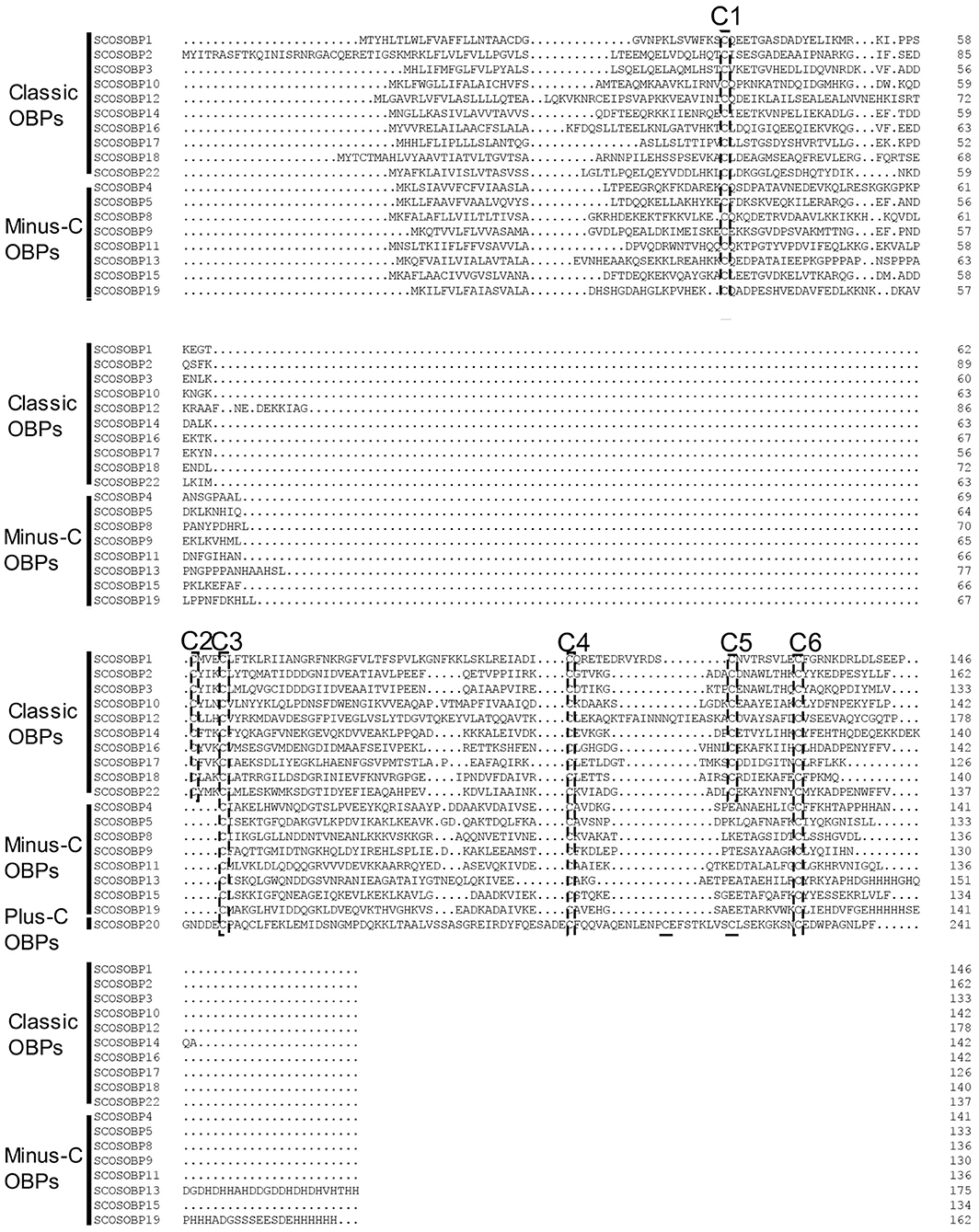
Figure 3. Sequence alignment of candidate ScosOBPs. The six conserved cysteine residues are marked as C1–C6.
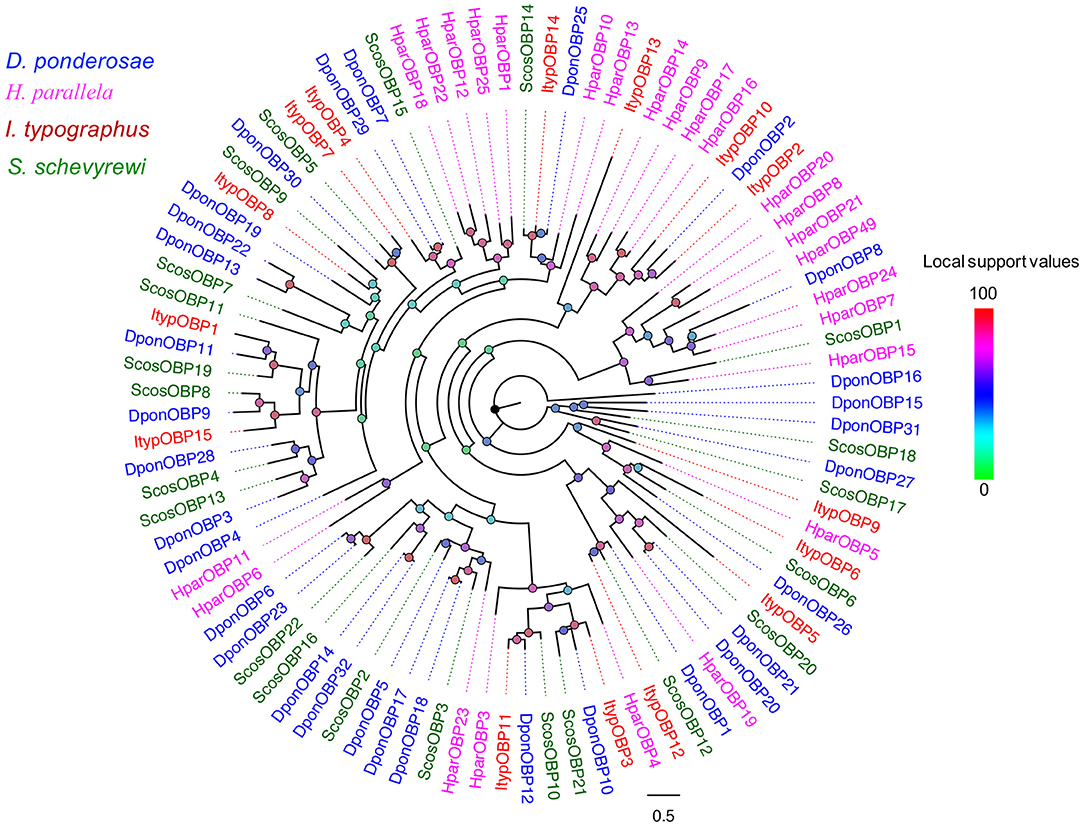
Figure 4. Phylogenetic tree of candidate ScosOBPs with known Coleoptera odorant-binding proteins sequences. Tcas, T. castaneum; Dpon, D. ponderosae; Hpar, H. parallela; Ityp, I. typographus; Scos, S. schevyrewi.
Information on unigene reference, length, and best blastx hit of all 22 OBPs are listed in Table 3.
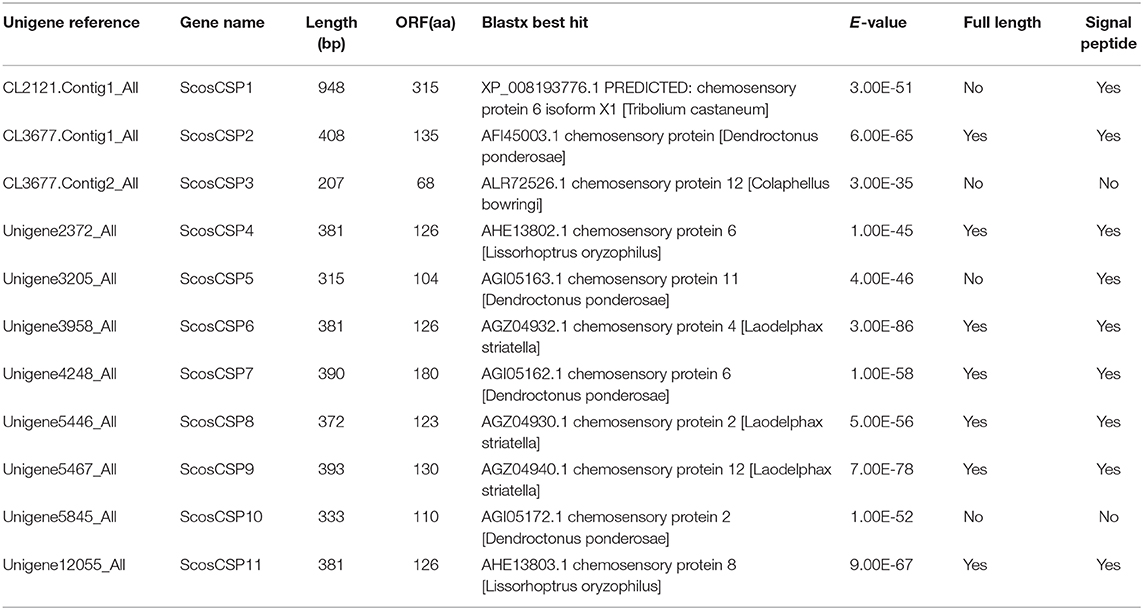
Identification of Putative Chemosensory-Binding Proteins
A total of 11 putative CSPs were identified from the S. schevyrewi antennal transcriptome. Seven of them had full-length ORFs and nine of them had the predicted signal peptide. All of them shared the typical structure of a CSP except ScosCSP3 and ScosCSP10 because these two lacked the signal peptide.
All of the predicted CSPs shared relatively high identity (57–100%) with known insect CSPs. The phylogenetic analysis of the CSPs in different beetles showed that most of the ScosCSPs were clustered with orthologs of D. ponderosae, I. typographus, and H. parallela in a separate clade (Figure 5). Only ScosCSP2 and ScosCSP3 formed a small subgroup.
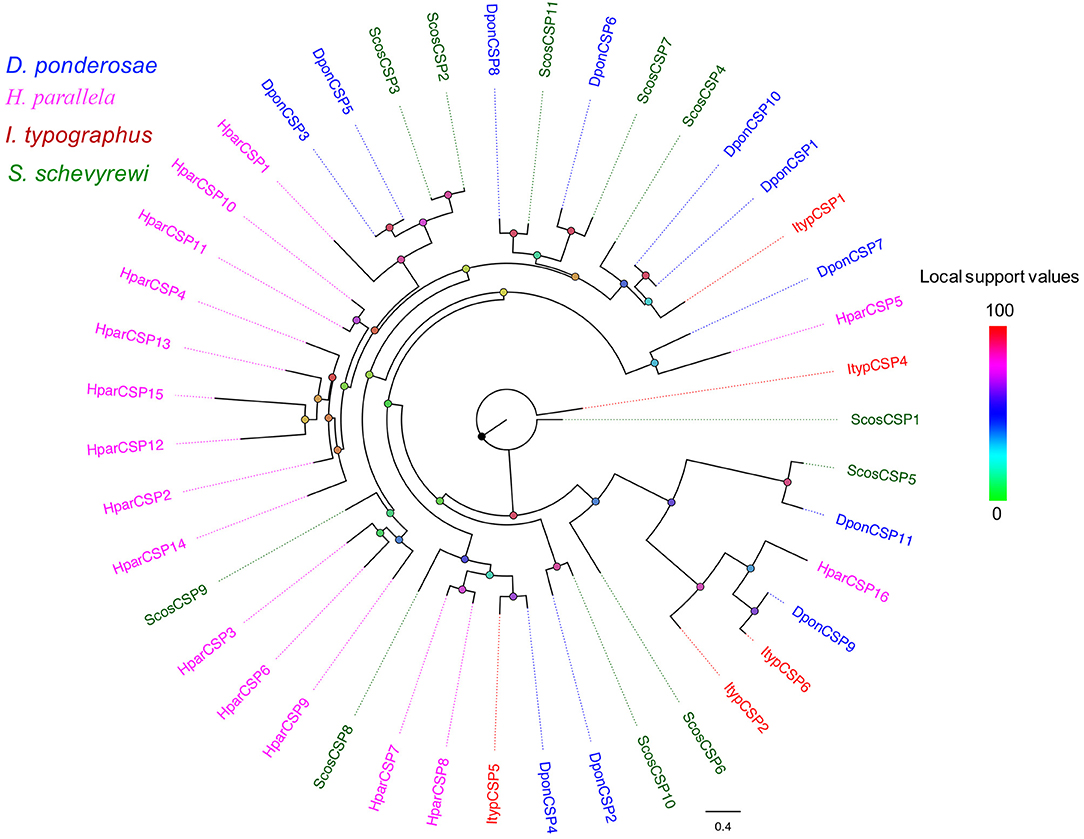
Figure 5. Phylogenetic tree of candidate ScosCSPs with known Coleoptera chemosensory proteins sequences. Tcas, T. castaneum; Dpon, D. ponderosae; Hpar, H. parallela; Ityp, I. typographus; Scos, S. schevyrewi.
Information on sequence alignment, unigene reference, length, and best blastx hit of all 11 CSPs are shown in Figure 6 and Table 4.

Figure 6. Sequence alignment of candidate ScosCSPs. The four conserved cysteine residues are marked as C1–C4. Additional residues that conserved in this species were also marked.
Tissue- and Sex-Specific Expression of Candidate ScosOBP and ScosCSP Genes
The expression patterns of ScosOBPs and ScosCSPs were analyzed by RT-PCR and are shown in Figures 7, 8. ScosOBP1, 2, 3, 7, 9, 10, 16, 17, 18, 20, and 22 were highly expressed or specifically expressed in the antennae and head tissues. Among them, ScosOBP2 and OBP18 expressed at higher levels in female antennae than in male antennae. ScosOBP4, 5, 6, 11, 12, 13, 15, 16, and 19 were generally expressed in multiple tissues. Among them, ScosOBP12 and ScosOBP19 expressions were stronger in the female than in the male antennae. ScosOBP8 and ScosOBP21 were not detected by RT-PCR possibly because their expression levels were too low to detect.
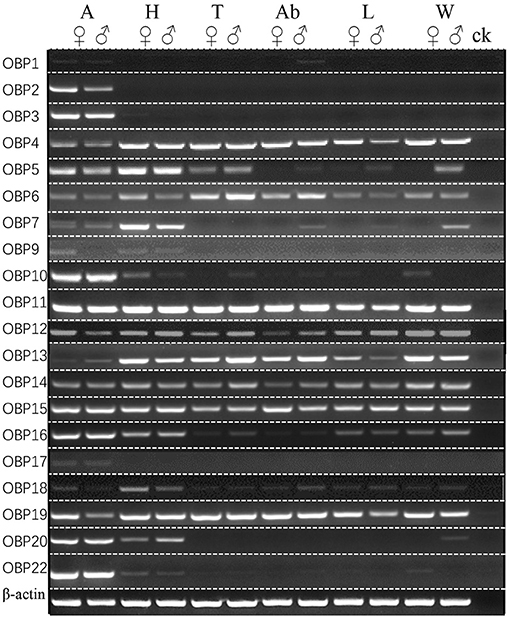
Figure 7. Sex- and tissue-specific expression of candidate ScosOBPs. A, antennae; H, head; T, thorax; Ab, abdomen; L, leg; W, wing; ck, control [ultra-pure water (germ-free)].
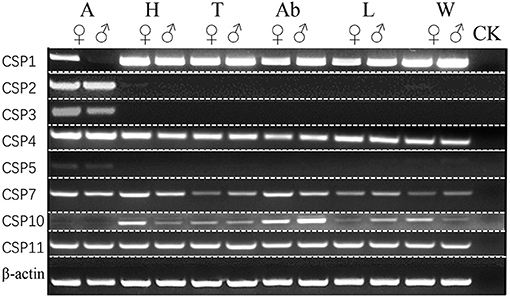
Figure 8. Sex- and tissue-specific expression of candidate ScosCSPs. A, antennae; H, head; T, thorax; Ab, abdomen; L, leg; W, wing; ck, control [ultra-pure water (germ-free)].
ScosCSP2, ScosCSP3, and ScosCSP5 were specifically expressed in the male and female antennae. Other ScosCSPs were expressed in multiple tissues. Among them, ScosCSP1 was not detected in male antennae and ScosCSP10 was not detected in the antennae of both the sexes. Potentially due to undetectable expression levels, ScosCSP6, ScosCSP8, and ScosCSP9 were not detected by RT-PCR.
Discussion
The genes reported in our study provide additional knowledge on the pool of identified olfactory genes in coleopterans. Compared with a large number of studies on Lepidopteran species, the current understanding of olfactory genes in Coleoptera is mainly sourced from a few reported studies on T. castaneum (Engsontia et al., 2008), Megacyllene caryae (Mitchell et al., 2012), I. typographus, and D. ponderosae (Andersson et al., 2013), Leptinotarsa decemlineata (Liu et al., 2015), H. parallela (Yi et al., 2018), Rhynchophorus ferrugineus (Antony et al., 2016; Gonzalez et al., 2021), etc. S. schevyrewi belongs to the genus of bark beetles and shares similar biology with the related species that are destructive forest pests, such as I. typographus and D. ponderosae. Aggregation behaviors are critical for bark beetle survival and rely on chemical communication (Byers, 1989). The genes we identified might contribute to aggregation behavior and provide molecular targets for novel pest management techniques.
We identified a total of 47 OR genes in the S. schevyrewi antennae transcriptome. In another coleopteran, 265 candidate OR genes were annotated in the T. castaneum genome (Richards, 2008), which is much more than the known number of OR genes reported by other beetles. The numbers of ORs in M. caryae (Mitchell et al., 2012), I. typographus (Andersson et al., 2013), D. ponderosae (Andersson et al., 2013), and H. parallela (Yi et al., 2018) range from 43 to 57. The number of ScosORs identified in this study is consistent with that identified in these reports. Most of the predicted ORs in S. schevyrewi share greater identity with ORs of D. ponderosae, another bark beetle, indicating that these two species may be able to share the same ecological environments and detect similar semiochemicals. Functional studies in OR from bark beetles were relatively limited to only seven ORs (Hou et al., 2021; Yuvaraj et al., 2021). ItypOR46 and ItypOR49 were responsive to single enantiomers of the common bark beetle pheromone compounds ipsenol and ipsdienol, respectively (Yuvaraj et al., 2021). The other five ItypORs were responsive to monoterpenoids of different ecological origins (Hou et al., 2021). Future studies should be focused on deorphinized ScosORs with similar functions to provide potential molecular targets for detection and control methods.
We identified, in total, 22 IR genes in the S. schevyrewi antennae transcriptome. ScosIR8a and ScosIR25a were identified as coreceptors. The numbers of IR genes in I. typographus, D. ponderosae, and H. parallela (Yi et al., 2018) are 7, 15, and 27, respectively (Andersson et al., 2013; Yi et al., 2018). The number of ScosIRs identified in this study is considerable compared with the numbers reported in the previous studies. More than half of the predicted IRs shared relatively low identity with other coleopteran IRs. These IRs with low identity were probably not conserved in Coleoptera, and they might serve diverse functions in S. schevyrewi.
Within the S. schevyrewi antennae transcriptome, a total of 22 OBPs were predicted. Genome annotation indicated that 46 OBPs were identified in T. castaneum (Richards, 2008). Fewer OBPs were found in other coleopteran antennae transcriptomes. In D. ponderosae, I. typographus, and H. parallela, respectively, 31, 15, and 25 OBPs were annotated (Andersson et al., 2013; Ju et al., 2014). Our number of ScosOBPs is consistent with the numbers stated in these reports.
A total of 11 CSPs were identified in the S. schevyrewi antennae transcriptome. In the T. castaneum genome, a total of 40 CSPs were identified (Richards, 2008). Other coleopterans have fewer CSPs in their antennae transcriptomes; in D. ponderosae, I. typographus, and H. parallela, 11, 6, and 16 were annotated, respectively (Andersson et al., 2013; Ju et al., 2014). Our recorded number of ScosCSPs is comparable with these reports. The high level of sequence conservation (57–100%) indicates the function of CSPs is likely conserved among coleopterans.
All ScosOBPs and ScosCSPs contain a typical OBP and CSP motif, respectively. ScosOBP1, 2, 3, 10, 12, 14, 16, 17, 18, and 22 generally possessed the “classic” OBP motif C1-X22−44-C2-X3-C3-X21−42-C4-X8−12-C5-X8-C6 of coleopteran insects (Xu et al., 2009). ScosOBP4, 5, 8, 9, 11, 13, 15, and 19 generally contained the “minus-C” OBP motif C1-X30−32-C2-X38-C3-X16−18-C4 (Ju et al., 2014). The cysteine-spacing pattern of ScosOBP20 followed the “plus-C” OBP motif C1-X24−28-C2-X3-C3-X43-C4-X13−15-C4a-X9-C5-X8-C6p-X9-C6a from D. melanogaster and Anopheles gambiae (Zhou et al., 2008). All the CSPs were conserved in having the motif C1-X6-C2-X18-C3-X2-C4 (Xu et al., 2009).
Scolytinae beetles respond to volatiles that emanate from both the host and non-host plants (Zhang and Schlyter, 2004; Erbilgin et al., 2007; Andersson et al., 2010). However, most individuals locate target trees by relying on an important signal called an aggregation pheromone released by beetles that have already attacked a tree (Andersson et al., 2013). Thus, olfactory signals and proteins serve critical roles in insect behavior. In this study, ScosOBP1, 2, 3, 7, 9, 10, 16, 17, 18, 20, and 22 might be important in odor perception because they were only expressed in the antennae and head, especially, ScosOBP2 and ScosOBP18. These two may be the key proteins in female olfactory behavior based on the specificity of protein expression we observed. ScosCSP2, ScosCSP3, and ScosCSP5 might also be important in olfaction due to their antennae-specific expression. Other ScosOBPs and ScosCSPs might not be involved in odor reception. Studies have shown a multitude of other roles that insect OBPs and CSPs have in Pelosi et al. (2018) releasing semiochemicals in pheromone glands (Benton, 2007), regeneration and development (Cheng et al., 2015), anti-inflammatory action (Isawa et al., 2002), nutrition (Zhu et al., 2016b), carrying visual pigments (Wang et al., 2007), and insecticide resistance (Bautista et al., 2015).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA732801.
Author Contributions
XZ and WL designed the research, analyzed the data, and wrote the paper. YL gave a lot of adivces and help to revise the paper. AK, BS, and HC provided biological samples. XZ, BX, and ZQ performed the experiment. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (Grant No. 31660518) and the earmarked fund of the Xinjiang apricot Industrial technology (Grant No. XJCYTX-03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Sai Zhang and Yilu Feng for rearing the experimental larvae for the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.717698/full#supplementary-material
Supplementary Figure 1. GO classification analysis of unigenes in all-unigene. GO functions are shown in the X-axis. The right Y-axis shows the number of genes that have the GO function, and the left Y-axis shows the percentage.
Supplementary Figure 2. Uncropped gel images of candidate ScosOBPs and candidate ScosCSPs. A, antennae; H, head; T, thorax; Ab, abdomen; L, leg; W, wing; ck, control [ultra-pure water (germ free)].
Supplementary Figure 3. Uncropped gel images for candidate ScosOBPs and ScosCSPs.
Supplementary Table 1. Forward and reverse primer sequence used in RT-PCR.
Supplementary Table 2. Sequence information.
References
Andersson, M. N., Grosse-Wilde, E., Keeling, C. I., Bengtsson, J. M., Yuen, M., Li, M., et al. (2013). Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genom. 14:198. doi: 10.1186/1471-2164-14-198
Andersson, M. N., Keeling, C., and Mitchell, R. F. (2019). Genomic content of chemosensory genes correlates with host range in wood-boring beetles (Dendroctonus ponderosae, Agrilus planipennis and Anoplophora glabripennis). BMC Genom. 20:690. doi: 10.1186/s12864-019-6054-x
Andersson, M. N., Larsson, M. C., BlaŽenec, M., Jakuš, R., Zhang, Q. H., and Schlyter, F. (2010). Peripheral modulation of pheromone response by inhibitory host compound in a beetle. J. Exp. Biol. 213, 3332–3339. doi: 10.1242/jeb.044396
Angeli, S., Ceron, F., Scaloni, A., Monti, M., Monteforti, G., Minnocci, A., et al. (1999). Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistoceraca gregaria. Eur. J. Biochem. 262, 745–754. doi: 10.1046/j.1432-1327.1999.00438.x
Antony, B., Soffan, A., Jakse, J., Mahmoud, M. M., Aldosari, S. A., and Aldawood, A. S. (2016). Identification of the genes involved in odorant reception and detection in the palm weevil Rhynchophorus ferrugineus, an important quarantine pest, by antennal transcriptome analysis. BMC Genom. 17:69. doi: 10.1186/s12864-016-2362-6
Bautista, M. A., Bhandary, B., Wijeratne, A. J., Michel, A. P., Hoy, C. W., and Mittapalli, O. (2015). Evidence for trade-offs in detoxification and chemosensation gene signatures in Plutella xylostella. Pest. Manag. Sci. 71, 423–432. doi: 10.1002/ps.3822
Benton, R. (2007). Sensitivity and specificity in Drosophila pheromone perception. Trends.Neurosci. 30, 512–519. doi: 10.1016/j.tins.2007.07.004
Benton, R., Sachse, S., Michnick, S. W., and Vosshall, L. B. (2006). A typical memobrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS. Biol. 4:e20. doi: 10.1371/journal.pbio.0040020
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Breugel, F. V., Huda, A., and Dickinson, M. H. (2018). Distinct activity-gated pathways mediate attraction and aversion to CO2 in Drosophila. Nature 564, 420–424. doi: 10.1038/s41586-018-0732-8
Briand, L., Swasdipan, N., Nespoulous, C., Bézirard, V., Blon, F., Huet, J. C., et al. (2002). Characterization of a chemosensory protein (ASP3c) from honeybee (Apis mellifera L.) as a brood pheromone carrier. Eur. J. Biochem. 269, 4586–4596. doi: 10.1046/j.1432-1033.2002.03156.x
Cao, D., Liu, Y., Wei, J., Liao, X., Walker, W. B., Li, J., et al. (2014). Identification of Candidate olfactory genes in Chilo suppressalis by antennal transcriptome analysis. Int. J. Biol. Sci. 10, 846–860. doi: 10.7150/ijbs.9297
Chen, C. H., Buhl, E., Xu, M., Croset, V., Rees, J. S., Lilley, K. S., et al. (2015). Drosophila ionotropic receptor 25a mediates circadian clock resetting by temperature. Nature 527, 516–520. doi: 10.1038/nature16148
Cheng, D. F., Lu, Y. Y., Zeng, L., Liang, G. W., and He, X. F. (2015). Si-CSP9 regulates the integument and moulting process of larvae in the red imported fire ant, Solenopsis invicta. Sci. Rep. 5:9245. doi: 10.1038/srep09245
Clyne, P. J., Warr, C. G., and Carlson, J. R. (2000). Candidate taste receptors in Drosophila. Science 287, 1830–1834. doi: 10.1126/science.287.5459.1830
Clyne, P. J., Warr, C. G., Marc, R. F., Lessing, D., Kim, J. Y., and Carlson, J. R. (1999). A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22, 327–338. doi: 10.1016/S0896-6273(00)81093-4
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21:3674–3676. doi: 10.1093/bioinformatics/bti610
Dobritsa, A. A., van der Goes van Naters, W., Warr, C. G., Steinbrecht, R. A., and Carlson, J. R. (2003). Integrating the molecular and cellular basis of odor codling in the Drosophila antenna. Neuron 37, 827–841. doi: 10.1016/S0896-6273(03)00094-1
Dong, X. T., Liao, H., Zhu, G. H., Khuhro, S. A., Ye, Z. F., Yan, Q., et al. (2017). CRISPR/Cas9-mediated PBP1 and PBP3 mutagenesis induced significant reduction in electrophysiological response to sex pheromones in male chilo suppressalis. Insect. Sci. 26, 388–399. doi: 10.1111/1744-7917.12544
Elmore, T., Ignell, R., Carlson, J., and Smith, D. (2003). Targeted mutation of a Drosophila odor receptor defines receptor requirement in a novel class of sensillum. J. Neurosci. 23, 9906–9912. doi: 10.1523/JNEUROSCI.23-30-09906.2003
Engsontia, P., Sanderson, A. P., Matthew, C., Walden, K., Robertson, H. M., and Browna, S. (2008). The red flour beetle's large nose: An expanded odorant receptor gene family in Tribolium castaneum. Insect. Biochem. Mol. Biol. 38, 387–397. doi: 10.1016/j.ibmb.2007.10.005
Erbilgin, N., Mori, R. I., Sun, J. H., Stein, J. D., Owen, D. R., and Merrill, L. D. (2007). Response to host volatiles by native and introduced populations of Dendroctonus valens (Coleoptera: Curculionidae, Scolytinae) in North America and China. J. Chem. Ecol. 33, 131–146. doi: 10.1007/s10886-006-9200-2
Fan, L. H., Niu, H. L., Zhang, J. T., Liu, J. L., Yang, M. H., and Zong, S. X. (2014). Extraction and identification of aggregation pheromone components of Scolytus schevyrewi Semenov(Coleoptera: Scolytidae) and trapping test. Acta Ecol. Sinica 35, 892–899. doi: 10.5846/stxb201311032656
Gomez-Diaz, C., Reina, J. H., Cambillau, C., and Benton, R. (2013). Ligands for pheromone-sensing neurons are not conformatinally activated odorant binding proteins. PLoS Biology. 11:e1001546. doi: 10.1371/journal.pbio.1001546
Gonzalez, F., Johny, J., Walker, I. I. I. W. B., Guan, Q. T., Mfarrej, S., and Jakse, J. (2021). Antennal transcriptome sequencing and identification of candidate chemoreceptor proteins from an invasive pest, the American plam weevil, Rhynchophorus palmarus. Sci. Rep. 11:8334. doi: 10.1038/s41598-021-87348-y
Grabherr, M. C., Haas, B. J., Yassour, M., Levin, J. Z., Tompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biothchnol. 29, 644–652. doi: 10.1038/nbt.1883
Hallem, E. A., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979. doi: 10.1016/j.cell.2004.05.012
Honson, N., Gong, Y., and Plettner, E. (2015). “Structure and function of insect odorant and pheromone-binding proteins (OBPs and PBPs) and chemosensory-specific proteins (CSPs),” in Chemical Ecology and Phytochemistry of Forest Ecosystems, ed J. T. Romeo (Oxford: Elsevier Ltd.), 227–268. doi: 10.1016/S0079-9920(05)80010-3
Hou, X. Q., Yuvaraj, K. J., Roberts, R. E., Zhang, D. D., Uneilius, C. R., Löfstedt, C., et al. (2021). Functional evolution of a bark beetle odorant receptor clade detecting monoterpenoids of different ecological origins. Mol. Biol. Evol. msab218. doi: 10.1093/molbev/msab218
Isawa, H., Yuda, M., Orito, Y., and Chinzei, Y. (2002). A mosquito salivary protein inhibits activation of the plasma contact system by binding to factor XII and high molecular weight kininogen. J. Biol. Chem. 277, 27651–27658. doi: 10.1074/jbc.m203505200
Jacquin-Joly, E., Vogt, R. G., François, M. C., and Nagnan-Le Meillour, P. (2001). Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem. Senses 26, 833–844. doi: 10.1093/chemse/26.7.833
Jing, D., Zhang, T., Bai, S., Prabu, S., He, K., Dewer, Y., et al. (2019). GOBP1 plays a key role in sex pheromones and plant volatiles recognition in yellow peach moth, conogethes punctiferalis (Lepidoptera: Crambidae). Insects 10:302. doi: 10.3390/insects10090302
Ju, Q., Li, X., Jiang, J. X., Qu, M. J., Guo, X. Q., Han, Z. J., et al. (2014). Transcriptome ranscriptome and tissue-specific expression analysis of obp and csp genes in the dark black chafer. Arch. Insect. Biochem. 87, 177–200. doi: 10.1002/arch.21188
Kim, M. S., Repp, A., and Smith, D. P. (1998). LUSH odorant binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics 150, 711–721. doi: 10.1093/genetics/150.2.711
Knecht, Z. A., Silbering, A. F., Ni, L. N., Klein, M., Budelli, G., Bell, R., et al. (2016). Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. Elife 5:e17879. doi: 10.1101/056267
Larsson, M. C., Domingos, A. I., Jones, W. D., Chiappe, M. E., Amrein, H., and Vosshall, L. B. (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Li, F., Dewer, Y., Li, D., Qu, C., and Luo, C. (2021). Functional and evolutionary characterization of chemosensory protein CSP2 in the whitefly, Bemisia tabaci. Pest. Manag. Sci. 77, 378–388. doi: 10.1002/ps.6027
Li, J. L., Zhang, T., Li, X. T., and Ma, S. K. (1995). Studies on the bionomics of Scolytus seulensis Murayama and its control. Plant Protection 21, 8–10. doi: 10.1016/j.aspen.2008.04.001
Liu, S. J., Liu, N. Y., He, P., Li, Z. Q., Dong, S. L., and Mu, L. F. (2012). Molecular characterization, expression patterns, and ligand-binding properties of two OBP genes from Orthaga achatina(Butler) (Lepidoptera: Pyralidae). Arch. Insect. Biochem. 80, 123–139. doi: 10.1002/arch.21036
Liu, Y., Sun, L. J., Cao, D. P., Walker, W. B., Zhang, Y. Q., and Wang, G. R. (2015). Identification of candidate olfactory genes in Leptinotarsa decemlineata by antennal transcriptome analysis. Font. Ecol. Evol. 19:60. doi: 10.3389/fevo.2015.00060
Mitchell, R. F., Hughes, D. T., Luetje, C. W., Millar, J. G., Soriano-Agatónc, F., Hanks, L. M., et al. (2012). Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae. Insect Biochem. Mol. Biol. 42, 499–505. doi: 10.1016/j.ibmb.2012.03.007
Mitchell, R. F., Schneider, T. M., Schwartz, A. M., Andersson, M. N., and McKenna, D. D. (2019). The diversity and evolution of odorant receptors in beetles (Coleoptera). Insect Mol. Biol. 29, 77–91. doi: 10.1111/imb.12611
Monteforti, G., Angeli, S., Petacchi, R., and Minnocci, A. (2002). Ultrastructural characterization of antennal sensilla and immunocytochemical localization of a chemosensory protein in Carausius morosus Brünner (Phasmida: Phasmatidae). Arthropod Struct. Dev. 30, 195–205. doi: 10.1016/S1467-8039(01)00036-6
Ni, L., Klein, M., Svec, K., Budelli, G., Chang, E. C., Ferrer, A. J., et al. (2015). The Ionotropic Receptors IR21a and IR25a mediate cool sensing in Drosophila. Elife. 5. doi: 10.7554/eLife.13254
Pelosi, P., Calvello, M., and Ban, L. P. (2005). Diversity of odorant binding protein and chemosensory proteins in insects. Chem. Senses 30, 1291–1292. doi: 10.1093/chemse/bjh229
Pelosi, P., Iovinella, I., Zhu, J., Wang, G. R., and Dani, F. R. (2018). Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Bio. Rev. 93, 184–200. doi: 10.1111/brv.12339
Pelosi, P., Zhou, J. J., Ban, L. P., and Calvello, M. (2006). Solube proteins in insect chemical communication. Cell. Mol. Life. Sci. 63, 1658–1676. doi: 10.1007/s00018-005-5607-0
Pertea, G., Huang, X. Q., Liang, F., Antonescu, V., Razvan, S. R., Svetlana, K. S., et al. (2003). TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. doi: 10.1093/bioinformatics/btg034
Richards, S. (2008). The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955. doi: 10.5167/uzh-2931
Schlyter, F., Birgersson, G., Byers, J. A., Löfqvist, J., and Bergström, G. (1987). Field response of spruce bark beetle, Ips typographus, to aggregation pheromo candidates. J. Chem. Ecol. 13:701–716.
Stengl, M. (2017). Chemosensory transduction in arthropods,” in Oxford Handbooks Online. The Oxford Handbook of Invertebrate Neurobiology, ed J. H. Byrne (Oxford University Press), 1–42. doi: 10.1093/oxfordhb/9780190456757.013.15
Stoker, R. F., Lienhard, M. C., Borst, A, and Fischbach, K. F. (1990). Neuronal architecture of the antennal lobe in Drosophila melanogaster. Cell. Tissue Res. 262, 9–34.
Sun, H. Y., Guan, L., Feng, H. L., Yin, J., Cao, Y. Z., Xi, J. H., et al. (2014). Functional characterization of chemosensory proteins in the scarab beetle, holotrichia oblita faldermann (Coleoptera: Scarabaeida). PLoS ONE 9:e107059. doi: 10.1371/journal.pone.0107059
Vosshall, L. B., and Hanson, B. S. (2011). A unified nomenclature system for the insect olfactory coreceptor. Chem. Senses 36, 497–498. doi: 10.1093/chemse/bjr022
Wang, T., Jiao, Y., and Montell, C. (2007). Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J. Cell. Biol. 177, 305–316. doi: 10.1083/jcb.200610081
Wermelinger, B. (2004). Ecology and management of the spruce bark beetle Ips typographus-a review of recent research. For. Ecol. Manag. 202, 67–82. doi: 10.1016/j.foreco.2004.07.018
Wilson, R. I. (2013). Early olfactory processing in Drosophila: mechanisms and principles. Annu. Rev. Neurosci. 36, 217–241. doi: 10.1146/annurev-neuro-062111-150533
Xiao, S., Sun, J. S., and Carlson, J. R. (2019). Robust olfactory responses in the absence of odorant binding proteins. Elife: e51040. doi: 10.7554/eLife.51040
Xu, P. X., Atkinson, R., Jones, D. N. M., and Smith, D. P. (2005). Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45, 192–200. doi: 10.1016/j.neuron.2004.12.031
Xu, Y. L., He, P., Zhang, L., Fang, S. Q., Dong, S. L., Zhang, Y. J., et al. (2009). Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC. Genom. 10:632. doi: 10.1186/1471-2164-10-632
Ye, Z. F., Liu, X. L., Han, Q., Liao, H., Dong, X. T., Zhu, G. H., et al. (2017). Functional characterization of PBP1 gene in Helicoverpa armigera (Lepidoptera: Noctuidae). Sci. Rep. 7:8769. doi: 10.1038/s41598-017-08769-2
Yi, J. K., Yang, S., Wang, S., Wang, J., Zhang, X. X., and Liu, Y. (2018). Identification of candidate chemosensory receptors in the antennal transcriptome of the large black chafer Holotrichia parallela Motschulsky (Coleoptera: Scarabaeidae). Comp. Biochem. Phys. D 28, 63–71. doi: 10.1016/j.cbd.2018.06.005
Yuvaraj, J. K., Roberts, R. E., Sonntag, Y., Hou, X. Q., Grosse-Wilde, E., Machara, A., et al. (2021). Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. BMC Biol. 19:16. doi: 10.1186/s12915-020-00946-6
Zhang, J., Wang, B., Dong, S. L., Cao, D. P., Dong, J. F., Walker, W. B., et al. (2015). Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, helicoverpa armigera and H. assulta. PloS ONE 10. doi: 10.1371/journal.pone.0117054
Zhang, Q. H., and Schlyter, F. (2004). Olfactory recognition and behavioural avoidance of angiosperm nonhost volatiles by conifer-inhabiting bark beetles. Agr. Forest. Entomol. 6, 1–19. doi: 10.1111/j.1461-9555.2004.00202.x
Zhou, J. J., He, X. L., Pickett, J. A., and Field, L. M. (2008). Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect Mol. Biol. 17, 147–163. doi: 10.1111/j.1365-2583.2007.00789.x
Zhu, G. H., Xu, J., Cui, Z., Dong, X. T., Ye, Z. F., Niu, D. J., et al. (2016a). Functional characterization of SlitPBP3 in Spodoptera litura by CRISPR/Cas9 mediated genome editing. Insect Biochem. Mol. Biol. 75, 1–9. doi: 10.1016/j.ibmb.2016.05.006
Keywords: Scolytus schevyrewi, transcriptome, olfactory genes, expression analysis, antennae
Citation: Zhu X, Xu B, Qin Z, Kader A, Song B, Chen H, Liu Y and Liu W (2021) Identification of Candidate Olfactory Genes in Scolytus schevyrewi Based on Transcriptomic Analysis. Front. Physiol. 12:717698. doi: 10.3389/fphys.2021.717698
Received: 31 May 2021; Accepted: 30 August 2021;
Published: 04 October 2021.
Edited by:
Kayvan Etebari, The University of Queensland, AustraliaReviewed by:
Youssef Dewer, Agricultural Research Center, EgyptMatan Shelomi, National Taiwan University, Taiwan
William Benjamin Walker III, United States Department of Agriculture (USDA-ARS), United States
Daniele Silva De Oliveira, Federal University of Rio de Janeiro, Brazil
Copyright © 2021 Zhu, Xu, Qin, Kader, Song, Chen, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Liu, bGl1d2VpMTFAY2Fhcy5jbg==
 Xiaofeng Zhu1
Xiaofeng Zhu1 Zhenjie Qin
Zhenjie Qin Yang Liu
Yang Liu Wei Liu
Wei Liu