
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Physiol. , 08 April 2021
Sec. Aquatic Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fphys.2021.666842
This article is part of the Research Topic Molecular Physiology in Molluscs, Volume II View all 26 articles
 Kaidi Jin1†
Kaidi Jin1† Qianqian Jin1†
Qianqian Jin1† Zhongqiang Cai2
Zhongqiang Cai2 Baoyu Huang1
Baoyu Huang1 Lei Wei1
Lei Wei1 Meiwei Zhang1
Meiwei Zhang1 Wen Guo3
Wen Guo3 Yaqiong Liu1*
Yaqiong Liu1* Xiaotong Wang1*
Xiaotong Wang1*Retinoic acid (RA) signaling pathways mediated by RA receptors (RARs) are essential for many physiological processes such as organ development, regeneration, and differentiation in animals. Recent studies reveal that RARs identified in several mollusks, including Pacific oyster Crassostrea gigas, have a different function mechanism compared with that in chordates. In this report, we identified the molecular characteristics of CgRAR to further explore the mechanism of RAR in mollusks. RT-qPCR analysis shows that CgRAR has a higher expression level in the hemocytes and gonads, indicating that CgRAR may play roles in the processes of development and metabolism. The mRNA expression level of both CgRAR and CgRXR was analyzed by RT-qPCR after injection with RA. The elevated expression of CgRAR and CgRXR was detected upon all-trans-RA (ATRA) exposure. Finally, according to the results of Yeast Two-Hybrid assay and co-immunoprecipitation analysis, CgRAR and CgRXR can interact with each other through the C-terminal region. Taken together, our results suggest that CgRAR shows a higher expression level in gonads and hemocytes. ATRA exposure up-regulates the expression of CgRAR and CgRXR. Besides, CgRAR can interact with CgRXR to form a heterodimer complex.
Retinoic acid (RA), an important hormone derived from vitamin A, plays crucial roles in regulating many development and differentiation processes, such as axial patterning, tissue formation, nervous system development, and regeneration (Das et al., 2014; Ghyselinck and Duester, 2019; Pawlikowski et al., 2019). It has been reported that RA-activated responses are mediated by the RA receptors (RARs) and the retinoid X receptors (RXRs) in vertebrates (Balmer and Blomhoff, 2002; Gutierrez-Mazariegos et al., 2014; Ghyselinck and Duester, 2019). Both RARs and RXRs belong to the steroid hormone/thyroid hormone nuclear receptor superfamily, containing a well-defined DNA-binding domain (DBD) and a C-terminal ligand-binding domain (LBD) (Bourguet et al., 2000; Le Maire and Bourguet, 2014).
Many studies have revealed that the RAR gene undergoes duplications through evolution. As a result, there are three paralogous RAR genes that exist in most vertebrates, including RARα, RARβ, and RARγ (Bastien and Rochette-Egly, 2004; Escriva et al., 2006). In vertebrates, RARs interact with RXRs to form different combinations of a heterodimer. There are several RA isomers, including all-trans-RA (ATRA), 9-cis-RA (9cRA), and 13-cis-RA. ATRA is the primary ligand sensed by RAR–RXR heterodimers during the development process (Mic et al., 2003; Cunningham and Duester, 2015). After being combined with ATRA, RAR–RXR heterodimers regulate transcription by combining with RA response elements (RAREs) in the regulatory regions of target genes (Kastner et al., 1995; Bastien and Rochette-Egly, 2004). In addition, RARs identified in urochordates Polyandrocarpa misakiensis and cephalochordate Branchiostoma lanceolatum can form a heterodimer with RXRs and combine with RA to activate transcription of target genes, with a similar function to that in vertebrates (Kamimura et al., 2000; Escriva et al., 2006).
It was once thought that RA signaling was unique to chordate animals for a long time until the signaling pathway was found in non-chordate animals, including ambulacrarians and lophotrochozoans, over the past one or two decades (Bertrand et al., 2004; Gutierrez-Mazariegos et al., 2014). Many studies focused on RA signaling pathway and the function of RARs in mollusks over the years. In Lymnaea stagnalis, RA modifies invertebrate electrical synapses of central neurons and function in the formation and modulation of invertebrate central synapses (Rothwell et al., 2017). Disruption of RA signaling in Lymnaea embryos using RAR antagonists resulted in abnormal eye and shell development (Carter et al., 2015). In addition, RA reduces intracellular calcium levels rapidly and affects calcium signaling in adult molluscan neurons of L. stagnalis (Vesprini et al., 2015). In Thais clavigera and Nucella lapillus, RXR is involved in the organotin-induced development of imposex (Nishikawa et al., 2004; Castro et al., 2007). Moreover, RXR identified from Biomphalaria glabrata can combine with 9cRA and activate the transcription of targets genes (Bouton et al., 2005). TcRAR and NlRAR, identified from T. clavigera and N. lapillus, respectively, can interact with RXR to form heterodimers but appear not to be activated by RA when detecting the transcription activity in mammalian cells (Urushitani et al., 2013; Juliana et al., 2014). In Crassostrea gigas, both in silico analysis and molecular experiments indicated that CgRXR shows high potential to combine with RA, while CgRAR loses the ability to interact with natural or synthetic RA ligands. In addition, RA can activate the transcriptional activity of CgRXR but not CgRAR (Vogeler et al., 2017; André et al., 2019; Huang et al., 2020). Thus, effects of RA on mollusks might be RAR independent. RAR may have different functions in these mollusks, including C. gigas.
Pacific oyster C. gigas is an important marine shellfish in the world, with great ecological and economic significance. It is of great significance to explore the RA signaling mechanism and RAR function in C. gigas. In the current report, we investigate the molecular characteristics of CgRAR. Tissue expression pattern and mRNA expression of CgRAR upon ATRA exposure were analyzed in C. gigas first. Then the subcellular localization of CgRAR was detected in HEK293T cells. To get a better understanding of the possible functions of CgRAR, the interaction between CgRAR and CgRXR was detected, and the results show that CgRAR interacts with CgRXR through the C-terminal region. Besides, both CgRAR and CgRXR can interact with themselves to form homodimers in yeast and mammalian cells.
Oysters used in this study were collected from a local culture zone (Yantai, China), with an average shell length of 65 cm. All the oysters were acclimated in aerated seawater at 15–22°C for at least 1 week before experiment. For tissue expression analysis, mantle (Man), gill (Gil), adductor muscle (Amu), hemolymph (Hae), digestive gland (Dgl), gonad (Gon), and labial palps (Lpa) were collected from nine wild oyster individuals. For RA exposure analysis, 48 oysters were divided into two groups randomly. ATRA was dissolved in dimethylsulfoxide (DMSO); oysters in two groups were injected with DMSO and 2 μg/μl of ATRA and given a supplementary injection every 2 days for a total of five injections.
Total RNA was isolated from different oyster tissues or RA-treated oysters using an RNA extraction kit (Tiangen). Briefly, tissue blocks were ground into a homogenate in liquid nitrogen, the cracking buffer was added to the homogenate, the supernatant was obtained by centrifugation, and subsequent operations were performed according to the kit instructions. cDNA synthesis using PrimeScriptTM RT Master Mix (TaKaRa) was performed according to the instructions. RT-qPCR analysis was performed using SYBR Premix Ex Taq II (TaKaRa) and a Bio-Rad CFX Connect PCR instrument. RS18 was used as reference gene for normalization of gene expression. 2–ΔΔCt method was used to calculate the relative expression level. Primers utilized for RT-qPCR are listed in Supplementary Table 1.
Full-length coding sequence (CDS) of CgRAR was cloned into pEGFP-N1 plasmid (Clontech) and then transferred into HEK293T cells with Lipofectamine 3000 (Invitrogen) when the confluence of cells reached 60%. Transfected cells were fixed with 4% paraformaldehyde and then incubated with DAPI for 5 min to stain the nuclei 24 h after transfection. Laser-Scanning Confocal Microscopy System FluoView FV1000 (Olympus, Japan) was used to observe fluorescent signal. Primers used for fusion vector construction are listed in Supplementary Table 1.
The yeast strain Yeast Two-Hybrid (Y2H) Gold (Clontech) was used to assess protein–protein interactions in this study. The CDS fragments of CgRAR, CgRXR, C-terminal of CgRAR, and C-terminal of CgRXR were cloned into pGAD T7 and pGBK T7 to generate CgRAR-AD/BD, CgRXR-AD/BD, CgRARC-AD/BD, and CgRXRC-AD/BD fusion plasmids; fusion plasmids were co-transferred into Y2H Gold yeast strain according to the manufacturer’s instructions (Clontech). Primers used for fusion plasmids construction are listed in Supplementary Table 1. Yeast transformants were grown on SD/-Leu/-Trp double drop out (DDO) medium for 3–5 days and then screened on selective SD/-Leu/-Trp/-His/-Ade quadruple drop-out (QDO) medium with aureobasidin A (AbA) and X-α-gal.
The full-length CDSs of CgRAR and CgRXR were constructed into pCMV-Myc and pCMS-flag to generate the fusion vectors. Primers used for fusion vectors construction are listed in Supplementary Table 1. The fusion vectors were introduced into the HEK293T cells by Lipofectamine 3000 (Invitrogen). Thirty-six hours after transfection, proteins were extracted from co-transferred HEK293T cells with lysis buffer (Beyotime, China). Anti-flag magnetic beads (Sigma, United States) were used for co-immunoprecipitation (co-IP) as indicated. After the IP, beads were washed at least three times in wash buffer. Input samples were separated from the cell lysate without anti-flag magnetic beads. Samples were boiled for 5 min in 2× protein sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (TAKAEA) and subjected to western blot (WB) analysis using anti-flag and anti-myc (Sigma, United States).
Transcriptional expression pattern of CgRAR was detected in different tissues isolated from healthy adult oysters under normal growing conditions, including the mantle, adductor muscle, gill, digestive gland, gonad, labial palp, and hemocytes. As a result, the CgRAR gene is widely expressed in all the detected tissues, with a higher expression in hemocytes and gonads (Figure 1A), suggesting that CgRAR may function in these tissues.
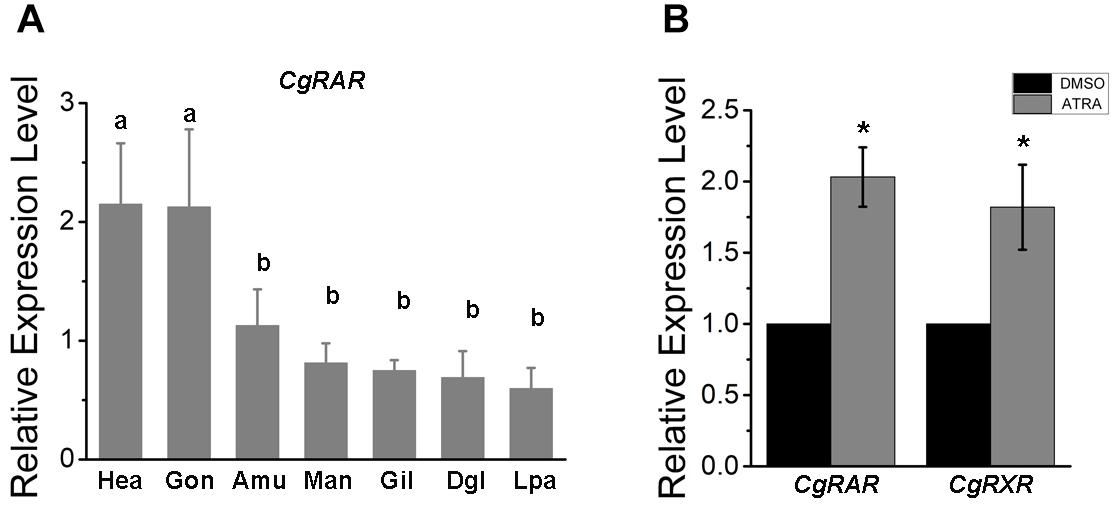
Figure 1. Relative expression of CgRAR in different tissues (A) and in response to retinoic acid (RA) treatment (B). Relative mRNA expression levels of CgRAR and CgRXR was normalized to that of CgRS18. Bars represent means of three replicates ± SD (standard deviation). One-way ANOVA and Student’s t-test (*P < 0.05) were used for significance analysis in (A,B) respectively. Hae, hemolymph; Gon, gonad; Amu, adductor muscles; Man, mantle; Gil, gill; Dgl, digestive gland; Lpa, labial palps.
To explore the relationship between CgRAR and RA-induced responses, we performed RT-qPCR analysis to detect the relative expression level of CgRAR and CgRXR after ATRA treatment. As shown in Figure 1B, the transcript level of both CgRAR and CgRXR was up-regulated after ATRA injection. This result indicates that mRNA expression of CgRAR and CgRXR can be up-regulated by ATRA exposure.
To reveal the subcellular localization of CgRAR protein, a green fluorescent protein (GFP)-tagged CgRAR was transferred into HEK293T cells. GFP plasmid was also transferred into HEK293T as control. As show in Figure 2, fluorescent single was observed on cytoplasm in GFP-transfected cells; in the cells transferred with CgRAR–GFP, fluorescent single was detected in the nucleus. This result suggests that CgRAR protein localized in the nucleus in HEK293T cells.
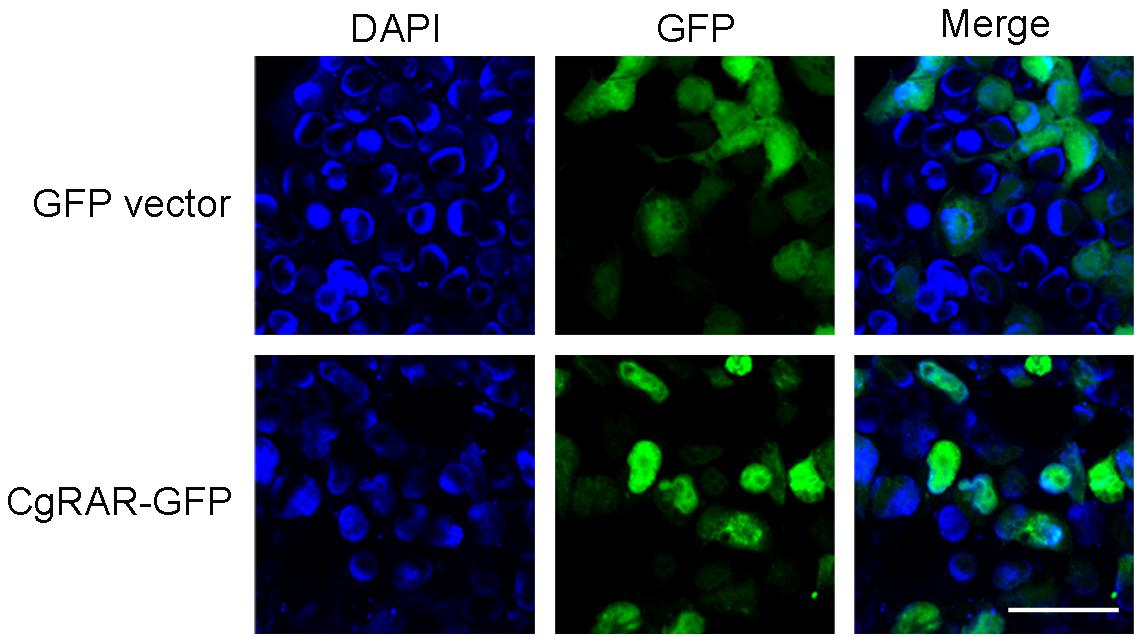
Figure 2. Subcellular localization of CgRAR in HEK293T cells. Confocal microscopy images of green fluorescent protein (GFP) (top) and CgRAR–GFP (bottom). HEK293T cells were transfected with pEGFP-N1 and CgRAR–GFP respectively. Bar, 50 μm.
To verify the interaction between CgRAR and CgRXR, a Y2H assay was carried out using the full-length CDSs of CgRAR and CgRXR first. Both CgRAR and CgRXR fusion vectors exhibit strong self-activation (Figure 3B). Thus, the DBD domain of CgRAR and CgRXR was deleted to generate CgRARC-AD/BD and CgRXRC-AD/BD fusion vectors, respectively (Figure 3A). Yeast strain co-expressing CgRARC and CgRXRC can be grown on QDO selection medium without self-activation (Figure 3C), indicating that CgRAR interacts with CgRXR through the C-terminal region. Co-IP analysis was also taken to confirm the interaction between CgRAR and CgRXR (Figure 3D). As a result, CgRAR was co-precipitated with CgRXR in HEK293T cells. In addition, both CgRAR and CgRXR can interact with themselves to form homodimers (Figures 3C,D). Taken together, these results demonstrate that CgRAR can either form a heterodimer by binding CgRXR or form a homodimer with itself in yeast and in HEK293T cells.
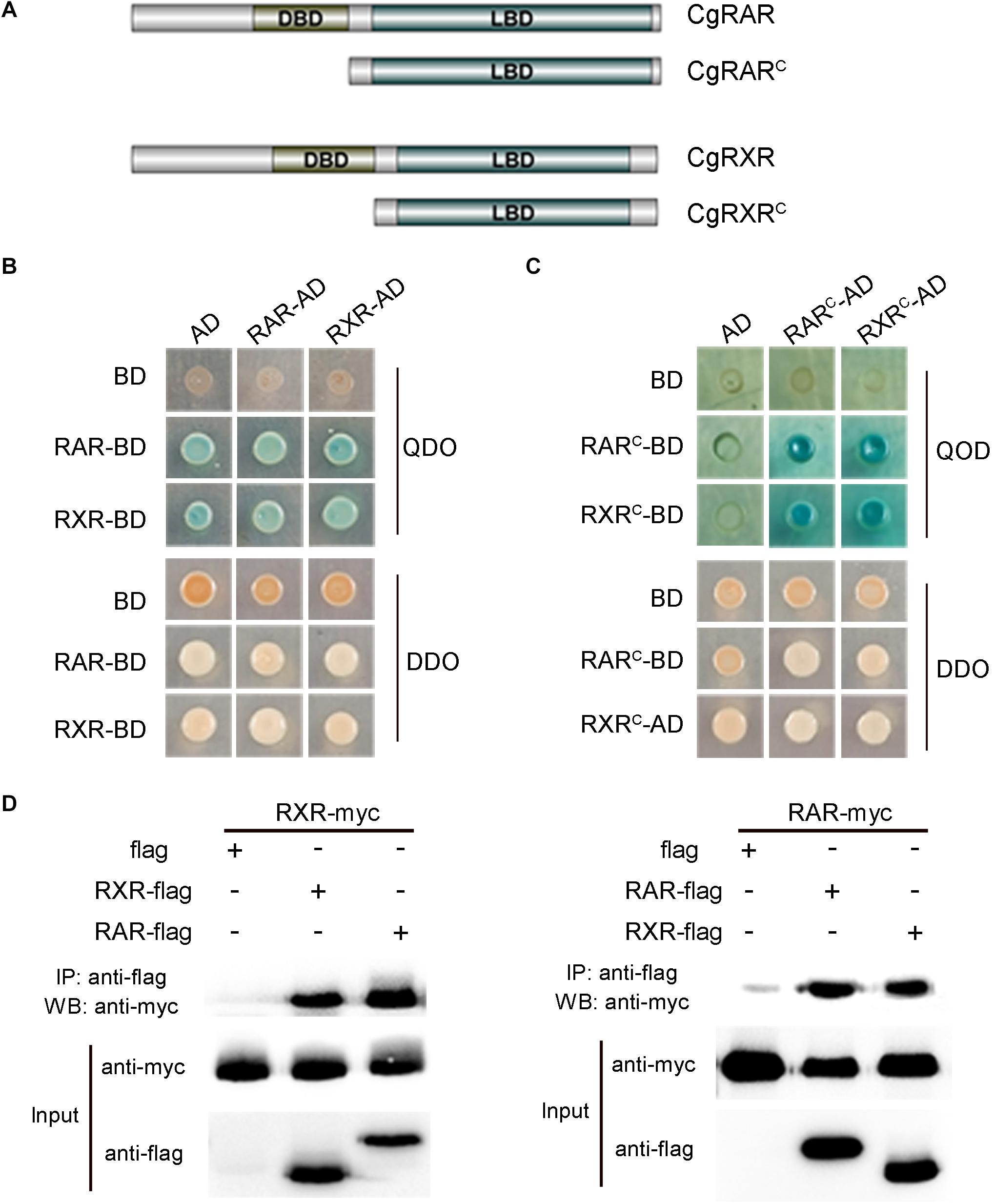
Figure 3. CgRAR physically interacts with CgRXR. (A) Representation of truncated CgRAR and CgRXR proteins. (B) Yeast two-hybrid assay for interactions between CgRAR and CgRXR protein. (C) Yeast two-hybrid assay for interactions between truncated CgRAR and CgRXR protein. AD and BD empty vectors were co-transformed as negative control. QDO/X/A, quadruple drop-out medium to test interaction; DDO, double drop-out as control. QDO, SD/-Trp/-Leu/-His/-Ade; DDO, SD-Leu-Trp; X, X-α-gal; A, aureobasidin A (AbA). (D) Co-immunoprecipitation assays verified the interaction between CgRAR and CgRXR protein. WB analysis using anti-myc and anti-flag on total protein extracts (“Input”) and on eluted proteins after immunoprecipitation (IP) with anti-flag magnetic beads.
Retinoic acid is an important hormone playing critical roles in the processes of organogenesis, neuronal differentiation, and embryonic development in vertebrates (Duester, 2008; Kam et al., 2012; Janesick et al., 2015). The important roles of RA signaling were also found in physiological process in mollusks, such as maturation of neurons, imposex, and formation of central synapses (Nishikawa et al., 2004; Vesprini et al., 2015; Rothwell et al., 2017). RARs are the primary receptors that sense RA ligand and regulate target gene transcription in chordates. However, the RA signaling mechanism in non-chordate animals, especially in some mollusks, seems different from that in chordates (André et al., 2019).
To help predict the possible function of CgRAR, tissue expression pattern was first examined in adult oysters under normal indoor culture environment in this study. A universal distribution of CgRAR gene was detected in different tissues of oyster, with a higher expression in gonads and hemocytes (Figure 1A). Since both gonads and hemocytes are tissues closely related to the processes of reproduction, differentiation, and development, we speculated that the elevated expression of CgRAR in these two tissues suggested that CgRAR might be involved in the development process of C. gigas. In chordates, RARs and RXRs form a heterodimer to regulate the expression of target genes in the perception of ATRA (Niederreither and Dolle, 2008; Gutierrez-Mazariegos et al., 2014). A recent study revealed that CgRXR is widely expressed in all tissues examined in Pacific oyster, with the highest relative expression in the mantle and lowest in gonads (Huang et al., 2020). Thus, the main function of CgRAR and CgRXR may not be exactly the same.
All-trans-RA exposure activates the mRNA expression of both CgRAR and CgRAR in Pacific oyster (Figure 1B). This result indicates that transcriptions of both CgRAR and CgRXR can be respond to ATRA exposure. It is worth noting that activation at the transcriptional level does not mean that CgRAR and CgRXR proteins can sense and bind to ATRA ligands. Whether CgRAR and CgRXR function in this process and what role they play need to be further studied.
Both RARs and RXRs belong to the nuclear receptor family and can regulate target gene transcription in the nucleus in vertebrates (Bastien and Rochette-Egly, 2004). A recent study demonstrated that CgRXR localized in the nucleus in the human cell line (Huang et al., 2020). CgRAR–GFP fusion plasmid was constructed and transferred into HEK293T cells to explore the subcellular location of CgRAR. The results showed that CgRAR was also localized in the nucleus in the human cell line (Figure 2). Therefore, we speculate that CgRAR’s function in the nucleus may be to regulate the transcription of target genes as a transcription factor.
In vertebrates, RA–RAR–RXR complex targets RARE of downstream genes, activating or repressing gene transcription (Kastner et al., 1995; Cunningham and Duester, 2015). Several studies have shown that RAR gene identified in some mollusks, including T. clavigera, Patella vulgata, N. lapillus, and C. gigas, lose the ability to bind ligand RA. In addition, the transcriptional activity of these RARs cannot be activated by RA (Urushitani et al., 2013; Juliana et al., 2014; Vogeler et al., 2017; André et al., 2019). It is reported that the RAR–RXR heterodimer still has the ability to bind RAREs and recruit co-repressors to negatively regulate transcription in the absence of ligand (Niederreither and Dolle, 2008). In this study, the physical interaction between CgRAR and CgRXR was confirmed by Y2H assay and Co-IP analysis (Figure 3). Whether CgRAR or CgRAR–CgRXR heterodimer regulate target gene transcription in the nucleus remains to be further studied. Taken together, this study indicates that CgRAR localized in the nuclear and can interact with CgRXR to form a heterodimer complex. The transcription of CgRAR can respond to ATRA, and CgRAR may function during the development process. This work may help to better understand the possible functions of CgRAR and to provide data reference for the further research.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethical review and approval was not required for the animal study because Crassostrea gigas is not an endangered or protected species and is not a vertebrate.
XW and YL designed the experiment. KJ, QJ, BH, LW, MZ, and YL carried out the experiments. ZC and WG contributed reagents, materials, and analysis tools. YL wrote the manuscript. XW supervised the study and revised the manuscript. All authors reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Key R&D Program of China (No. 2018YFD0901400), the National Natural Science Foundation of China (Nos. 42076088, 41876193, and 41906088), the Special Funds for Taishan Scholars Project of Shandong Province, China (No. tsqn201812094), the Shandong Provincial Natural Science Foundation, China (No. ZR2019MC002), the Fine Agricultural Breeds Project of Shandong Province, China (2019LZGC020), the Modern Agricultural Industry Technology System of Shandong Province, China (SDAIT-14-03), and the Plan of Excellent Youth Innovation Team of Colleges and universities in Shandong Province, China (2019KJF004).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.666842/full#supplementary-material
Supplementary Table 1 | Primers used for RT-qPCR analysis and fusion vectors construction.
André, A., Ruivo, R., Fonseca, E., Froufe, E., Castro, L. F. C., and Santos, M. M. (2019). The retinoic acid receptor (RAR) in molluscs: function, evolution and endocrine disruption insights. Aquat. Toxicol. 208, 80–89.
Balmer, J. E., and Blomhoff, R. (2002). Gene expression regulation by retinoic acid. J. Lipid Res. 43, 1773–1808. doi: 10.1194/jlr.r100015-jlr200
Bastien, J., and Rochette-Egly, C. (2004). Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328, 1–16. doi: 10.1016/j.gene.2003.12.005
Bertrand, S., Brunet, F. G., Escriva, H., Parmentier, G., Laudet, V., and Robinson-Rechavi, M. (2004). Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol. Biol. Evol. 21, 1923–1937. doi: 10.1093/molbev/msh200
Bourguet, W., Vivat, V., Wurtz, J. M., Chambon, P., Gronemeyer, H., and Moras, D. (2000). Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell 5, 289–298. doi: 10.1016/s1097-2765(00)80424-4
Bouton, D., Escriva, H., De Mendonça, R. L., Glineur, C., Bertin, B., Noël, C., et al. (2005). A conserved retinoid X receptor (RXR) from the mollusk Biomphalaria glabrata transactivates transcription in the presence of retinoids. J. Mol. Endocrinol. 34, 567–582. doi: 10.1677/jme.1.01766
Carter, C. J., Rand, C., Mohammad, I., Lepp, A., Vesprini, N., Wiebe, O., et al. (2015). Expression of a retinoic acid receptor (RAR)-like protein in the embryonic and adult nervous system of a protostome species. J. Exp. Zool. B Mol. Dev. Evol. 324, 51–67. doi: 10.1002/jez.b.22604
Castro, L. F., Lima, D., Machado, A., Melo, C., Hiromori, Y., Nishikawa, J., et al. (2007). Imposex induction is mediated through the retinoid X receptor signalling pathway in the neogastropod Nucella lapillus. Aquat. Toxicol. 85, 57–66. doi: 10.1016/j.aquatox.2007.07.016
Cunningham, T. J., and Duester, G. (2015). Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 16, 110–123. doi: 10.1038/nrm3932
Das, B. C., Thapa, P., Karki, R., Das, S., Mahapatra, S., Liu, T. C., et al. (2014). Retinoic acid signaling pathways in development and diseases. Bioorg. Med. Chem. 22, 673–683.
Duester, G. (2008). Retinoic acid synthesis and signaling during early organogenesis. Cell 134, 921–931. doi: 10.1016/j.cell.2008.09.002
Escriva, H., Bertrand, S., Germain, P., Robinson-Rechavi, M., Umbhauer, M., Cartry, J., et al. (2006). Neofunctionalization in vertebrates: the example of retinoic acid receptors. PLoS Genet. 2:e102. doi: 10.1371/journal.pgen.0020102
Ghyselinck, N. B., and Duester, G. (2019). Retinoic acid signaling pathways. Development 146:dev167502. doi: 10.1242/dev.167502
Gutierrez-Mazariegos, J., Schubert, M., and Laudet, V. (2014). Evolution of retinoic acid receptors and retinoic acid signaling. Subcell. Biochem. 70, 55–73. doi: 10.1007/978-94-017-9050-5_4
Huang, W., Wu, Q., Xu, F., Li, L., Li, J., Que, H., et al. (2020). Functional characterization of retinoid X receptor with an emphasis on the mediation of organotin poisoning in the Pacific oyster (Crassostrea gigas). Gene 753:144780. doi: 10.1016/j.gene.2020.144780
Janesick, A., Wu, S. C., and Blumberg, B. (2015). Retinoic acid signaling and neuronal differentiation. Cell. Mol. Life Sci. 72, 1559–1576.
Juliana, G. M., Kumar, N. E., Daniela, L., Keely, P., Jones, J. W., Maureen, K., et al. (2014). A mollusk retinoic acid receptor (RAR) ortholog sheds light on the evolution of ligand binding. Endocrinology 155, 4275–4286. doi: 10.1210/en.2014-1181
Kam, R. K., Deng, Y., Chen, Y., and Zhao, H. (2012). Retinoic acid synthesis and functions in early embryonic development. Cell Biosci. 2:11. doi: 10.1186/2045-3701-2-11
Kamimura, M., Fujiwara, S., Kawamura, K., and Yubisui, T. (2000). Functional retinoid receptors in budding ascidians. Dev. Growth Differ. 42, 1–8. doi: 10.1046/j.1440-169x.2000.00478.x
Kastner, P., Mark, M., and Chambon, P. (1995). Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell 83, 859–869. doi: 10.1016/0092-8674(95)90202-3
Le Maire, A., and Bourguet, W. (2014). Retinoic acid receptors: structural basis for coregulator interaction and exchange. Subcell. Biochem. 70, 37–54. doi: 10.1007/978-94-017-9050-5_3
Mic, F. A., Molotkov, A., Benbrook, D. M., and Duester, G. (2003). Retinoid activation of retinoic acid receptor but not retinoid X receptor is sufficient to rescue lethal defect in retinoic acid synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 7135–7140. doi: 10.1073/pnas.1231422100
Niederreither, K., and Dolle, P. (2008). Retinoic acid in development: towards an integrated view. Nat. Rev. Genet. 9, 541–553. doi: 10.1038/nrg2340
Nishikawa, J., Mamiya, S., Kanayama, T., Nishikawa, T., Shiraishi, F., and Horiguchi, T. (2004). Involvement of the retinoid X receptor in the development of imposex caused by organotins in gastropods. Environ. Sci. Technol. 38, 6271–6276. doi: 10.1021/es049593u
Pawlikowski, B., Wragge, J., and Siegenthaler, J. A. (2019). Retinoic acid signaling in vascular development. Genesis 57:e23287. doi: 10.1002/dvg.23287
Rothwell, C. M., De Hoog, E., and Spencer, G. E. (2017). The role of retinoic acid in the formation and modulation of invertebrate central synapses. J. Neurophysiol. 117, 692–704. doi: 10.1152/jn.00737.2016
Urushitani, H., Katsu, Y., Ohta, Y., Shiraishi, H., Iguchi, T., and Horiguchi, T. (2013). Cloning and characterization of the retinoic acid receptor-like protein in the rock shell, Thais clavigera. Aquat. Toxicol. 142-143, 403–413. doi: 10.1016/j.aquatox.2013.09.008
Vesprini, N. D., Dawson, T. F., Yuan, Y., Bruce, D., and Spencer, G. E. (2015). Retinoic acid affects calcium signaling in adult molluscan neurons. J. Neurophysiol. 113, 172–181. doi: 10.1152/jn.00458.2014
Keywords: Crassostrea gigas, retinoic acid receptor, retinoid X receptor, molecular characterization, retinoic acid
Citation: Jin K, Jin Q, Cai Z, Huang B, Wei L, Zhang M, Guo W, Liu Y and Wang X (2021) Molecular Characterization of Retinoic Acid Receptor CgRAR in Pacific Oyster (Crassostrea gigas). Front. Physiol. 12:666842. doi: 10.3389/fphys.2021.666842
Received: 11 February 2021; Accepted: 01 March 2021;
Published: 08 April 2021.
Edited by:
Jianmin Zhao, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Qingheng Wang, Guangdong Ocean University, ChinaCopyright © 2021 Jin, Jin, Cai, Huang, Wei, Zhang, Guo, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaqiong Liu, bHlxcGthQDE2My5jb20=; Xiaotong Wang, d2FuZ3hpYW90b25nOTk5QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.