- 1Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, China
- 2Inflammation and Immune Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, School of Pharmacy, Anhui Medical University, Hefei, China
- 3School of Chinese Medicine, The University of Hong Kong, Hong Kong, China
- 4Department of Chinese Medicine, The University of Hong Kong-Shenzhen Hospital, Shenzhen, China
Diabetic nephropathy (DN) leads to high morbidity and disability. Inflammation plays a critical role in the pathogenesis of DN, which involves renal cells and immune cells, the microenvironment, as well as extrinsic factors, such as hyperglycemia, chemokines, cytokines, and growth factors. Epigenetic modifications usually regulate gene expression via DNA methylation, histone modification, and non-coding RNAs without altering the DNA sequence. During the past years, numerous studies have been published to reveal the mechanisms of epigenetic modifications that regulate inflammation in DN. This review aimed to summarize the latest evidence on the interplay of epigenetics and inflammation in DN, and highlight the potential targets for treatment and diagnosis of DN.
Introduction
The latest Diabetes Atlas by the International Diabetes Federation indicates that the current number of patients with diabetes mellitus (DM) is 463 million in 2019, which is estimated to increase to 578 million by 2030 and to 700 million by 2045 (International Diabetes Federation, 2019). DM and its complications seriously affect patients’ quality of life and result in tremendous socioeconomic burdens (GBD, 2017 Disease and injury incidence and prevalence collaborators, 2018; Lin et al., 2020b). Diabetic nephropathy (DN), one of the most common microvascular complications of DM, is the major contributor to chronic kidney disease (CKD) and end-stage renal disease (Ruiz-Ortega et al., 2020). Approximately 30–40% of DM patients gradually develop DN (Lim, 2014). Current therapies, including intensive glucose control and the treatment of hypertension through renin-angiotensin-aldosterone system (RAAS) blockers, only slow down the progression of DN and fail to reverse or stop it (Sanz et al., 2019; Ruiz-Ortega et al., 2020). Therefore, early diagnosis and novel treatment for DN are of great significance while recognizing its etiology remains urgent.
The biologist Conrad Waddington firstly introduced ‘epigenetics’ which describes a phenomenon of inheritance that is independent of DNA sequence (Russo et al., 1996; Goldberg et al., 2007). This concept has become one of the frontiers of genetic research over the years. Epigenetic modifications modulate gene expressions through DNA methylation, histone modification, and non-coding RNAs involving in the pathogenesis of DN (Keating and El-Osta, 2013; Reddy et al., 2015). Studies have also shown that the modifications are reversible indicating potential therapeutic value for DN (Hotamisligil, 2017; Kato and Natarajan, 2019). Low grade chronic inflammation is a major characteristic in the pathogenesis of DN, but the pathophysiological relevance between epigenetics and inflammation has not been fully summarized. In this review, we highlighted recent epigenetic modifications relevant to inflammation and its signaling pathways in DN. The prespecified search strategies were shown in the Supplementary Material 1.
Inflammation in the Progression of DN
The Role of Renal Resident and Immune Cells in the Inflammatory Response
Hyperglycemia and glucose metabolites such as advanced glycation end products (AGEs) have long been regarded as initial factors of DN which promote the loss of podocytes, the hyperfiltration of endothelial cells, the expansion of mesangial cells and the thickening of glomerular basement membrane, and finally result in the deposition of extracellular matrix in the glomerulus (Schena and Gesualdo, 2005; Grabias and Konstantopoulos, 2014). The injured resident cells in kidney release chemokines and cytokines to attract the infiltration of immune cells (e.g., monocytes, macrophages, dendritic cells, and lymphocytes) (Tang and Yiu, 2020). Macrophages/monocytes are found to be the most predominant immune cells through both clinical and experimental studies. Previous study shows that macrophages are positively associated with pathological lesions in DN (Chow et al., 2004). A recent study of single cell RNA sequencing (scRNA-seq) indicates proportions of endothelial cells and immune cells are significantly increased while mesangial cells and podocytes are decreased in the glomerular cells in diabetic mouse kidney (Fu et al., 2019a). Among of immune cells in this study, macrophages are predominant, particularly M1 phenotype macrophages (Fu et al., 2019a). It has been also demonstrated that infiltration of macrophages in the glomeruli and tubulointerstitial tissues was increased in renal biopsies of patients (Klessens et al., 2017). In addition, the depletion of macrophages significantly reduces proteinuria and glomerular pathological changes in diabetic mice (You et al., 2013). The scRNA-seq analysis of kidney cortex from diabetic (n = 3) and non-diabetic patients (n = 3) shows patients in early diabetic nephropathy have 78 folds of leukocytes, including T cells, B cells, monocytes and plasma cells, compared to non-diabetic patients (Wilson et al., 2019). Few macrophages are observed in early diabetic kidneys (Wilson et al., 2019). Proportions of kidney cells and immune cells, and their roles at different stages of DN needed to be further studies. Epigenetic modifications in diabetic kidneys are shown in Figure 1.
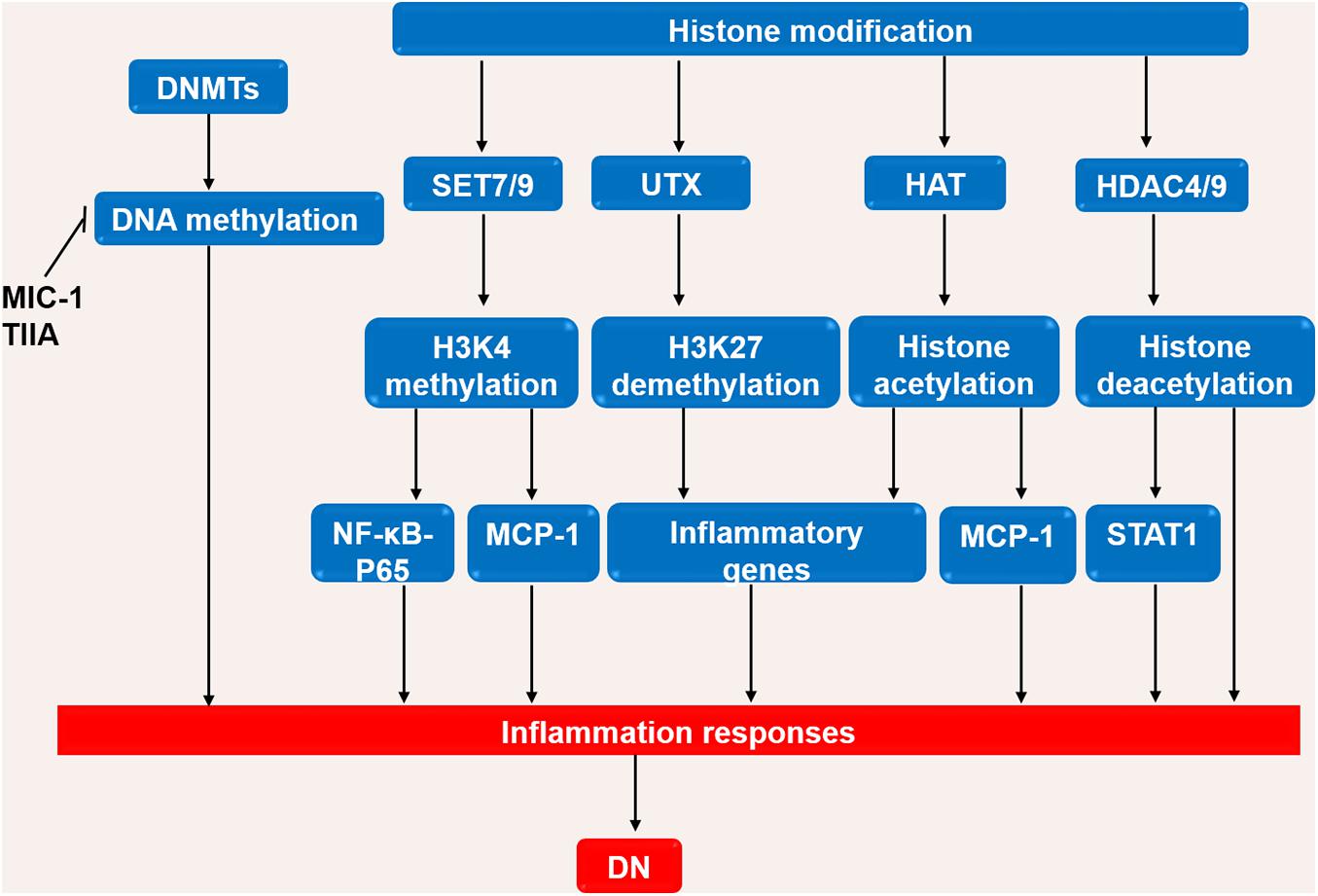
Figure 1. Interaction of immune cells, kidney intrinsic cells and epigenetic modifications. DNA methylation, histone modifications, and non-coding RNA modifications activate inflammatory pathways by interactions of immune cells and kidney intrinsic cells. OS, oxidative stress; ROS, reactive oxygen species; HG, high glucose; Ang II, angiotensin II; AGE-RAGE, advanced glycation end-products-receptor for advanced glycation end products; IL, interleukin; TNF, tumor necrosis factor; MCP-1, monocyte chemoattractant protein 1; NF-κB, nuclear factor-κB; JAK-STAT, Janus kinase/signal transducer and activator of transcription; NRF2, Nuclear Factor-2 Erythroid Related Factor; NLRP, NOD-like receptor pyrin domain-containing protein.
The infiltration of macrophages is promoted by chemokines and adhesion molecules which are released from resident cells under the stimulation of high glucose and AGEs (Hickey and Martin, 2013). Notably, MCP-1 is an important mediator in the infiltration of macrophages and the progression of inflammation (Chow et al., 2006). The deletion of MCP-1 in mice and inhibition of MCP-1 in type 2 diabetic patients have been shown to improve renal function (Chow et al., 2006). Previous studies have shown that an increase in M1 macrophages is negatively associated with renal function (Wang et al., 2017), while the induction of M2 macrophages has been shown to attenuate renal damage in DN mouse model (Sun et al., 2015). High glucose and AGEs promote macrophages to M1 polarization and the release of inflammatory cytokines, such as tumor necrosis factor (TNF), contributing to pathogenesis in the early stage of diabetes (Webster et al., 1997). Additionally, macrophages can also act as myofibroblasts through the process of macrophage-myofibroblast transition (MMT) to deteriorate renal fibrosis, replace parenchyma tissue with (Tang et al., 2020b) extracellular matrix (ECM), and also contribute to the production of reactive oxygen species (ROS) and proteases (Meng et al., 2014; Torres et al., 2020). Orchestrated by TGF-β/Smad signaling pathway, MMT is a newly known fibrosis process which has been rarely found neither in acute inflammation, nor in normal kidney, indicating that chronic inflammation was the principle contributor to fibrosis (Meng et al., 2016; Tang et al., 2019). A recent study has found that brain-specific transcription factor POU4F1 is the only transcription factor taking part in the TGF-β1/Smad3-driven MMT and thus could be a new therapeutic target in chronic inflammation induced MMT fibrosis (Tang et al., 2020b). The proto-oncogene tyrosine protein kinase SRC presents as a direct SMAD3 target gene and is also essential for MMT in macrophages (Tang et al., 2018b). In general, the accumulation of macrophage are not only related to the degree of inflammation and kidney function, but also correlated to glomerulosclersis and the degree of interstitial fibrosis (Tang et al., 2019). Studies have shown that aberrant intrarenal infiltration and activation of T cells are involved in the pathogenesis of DN in both clinical samples and streptozotocin (STZ)-induced diabetes mice (Moon et al., 2012). Clinical findings show that T cell immunity and TNF-α signaling pathway are activated during the early development of DN in patients (Moon et al., 2012; Lampropoulou et al., 2020). The proportions of T helper cells (Th1, Th2, Th17 and regulatory T (Treg) cells) in DN are altered with the increased levels of Th1 and Th17, and the decreased level of Treg (Zhang et al., 2014). Adoptive transfer of CD4 + Foxp3 + Treg cells in mice have been found to ameliorate diabetic kidney injuries and insulin resistance by inhibiting inflammation (Eller et al., 2011).
The Role of Inflammatory Mediators and Signaling Pathways in DN
Several signaling pathways contribute to the inflammation and the release of inflammatory cytokines (Figure 2; Newton and Dixit, 2012). Interleukins (ILs) play critical roles in the regulation of the immune system. Studies have shown that the circulating level of IL-6 is positively correlated with the progression of DN in patients (Saraheimo et al., 2003), and IL-1β, IL-18, and IL-17A are associated with the occurrence and development of DN (Cortvrindt et al., 2017; Lemos et al., 2018; Lin et al., 2020a). TNF-α is involved in the development of various diseases, such as psoriasis, rheumatoid arthritis, and CKD (Elliott et al., 1994; Pina et al., 2016). Studies have demonstrated that macrophages are the main source of renal TNF-α (Awad et al., 2015). In diabetic mice, the inhibition of TNF-α leads to decreased urinary albumin excretion, and in a clinical trial where DN patients were treated with pentoxifylline, a methylxanthine derivative with anti-inflammatory function, the reduction in urinary TNF-α concentration was directly correlated with the change in albuminuria, suggesting the role of TNF-α in the pathogenesis of DN (Moriwaki et al., 2007; Navarro-González et al., 2015).
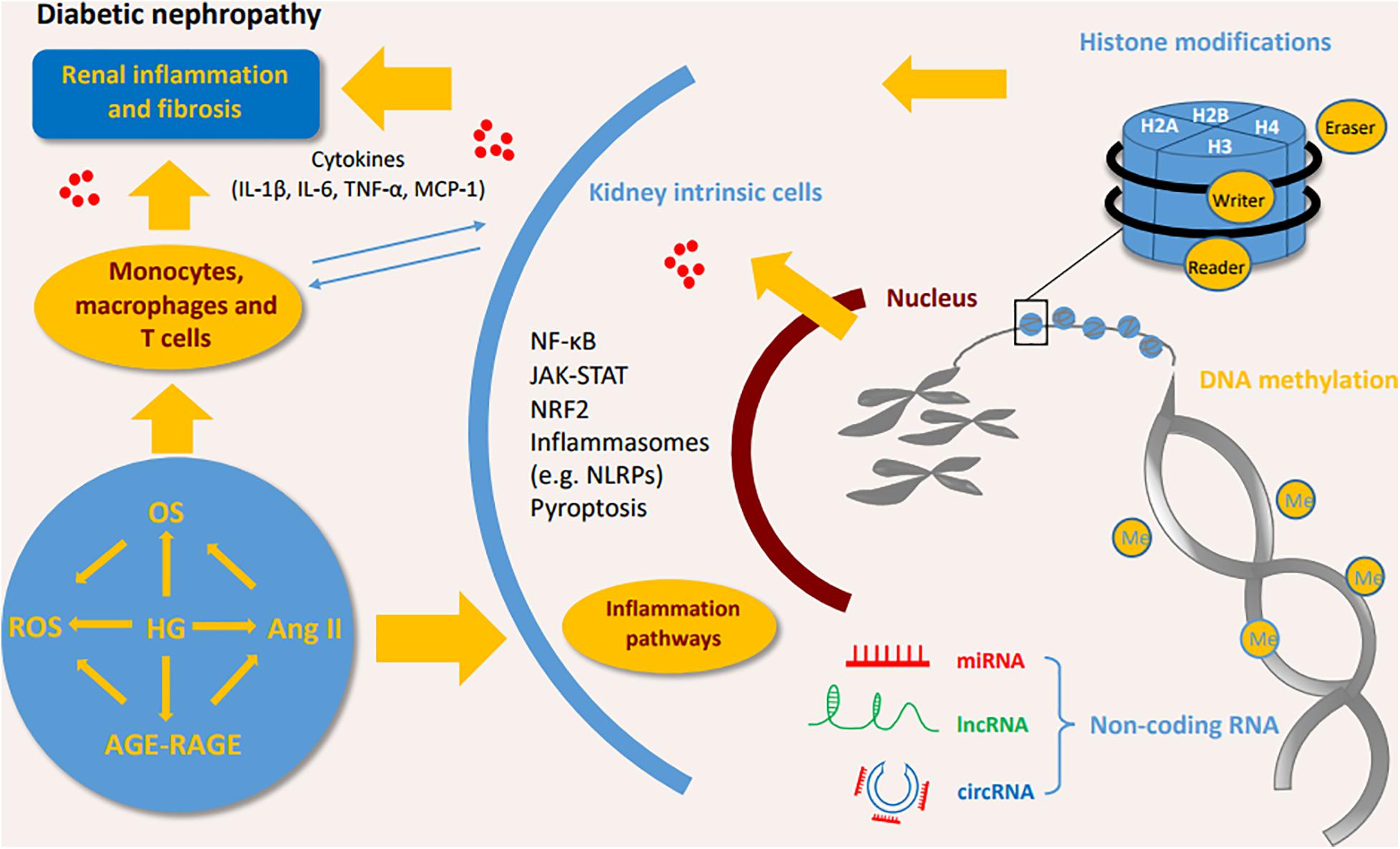
Figure 2. The inflammatory pathways involved in the DN process. High glucose or stimulating factors cause the activation of multiple pathways, including P38/MAPK, PI3K/AKT, TLR4/NLRP3 and the novel CRP/DPP4/CD32b, which all further activate NF-κB pathway, and transcriptionally promote the expression of multiple inflammatory cytokines to enhance inflammation. SMAD7 can inhibit NF-κB activity. NRF2 has anti-inflammatory and antioxidant functions in DN. In addition, JAK/STAT pathway induces IL-6 to promote the inflammatory response in DN. DN, diabetic nephropathy; IL, interleukin; TNF, tumor necrosis factor; NF-κB, Nuclear factor-κB; MCP-1, monocyte chemoattractant protein-1; STAT, Signal transducer and activator of transcription; NRF2, Nuclear factor-2 erythroid related factor; NLRP3, NOD-like receptor protein 3; ROS, reactive oxygen species; DPP4, dipeptidyl peptidase-4; MAPK, mitogen-activated protein kinase; JAK2, Janus kinase; TLR4, Toll-like receptors 4; CRP, C-reactive protein; AGE, advanced glycation end product; RAGE, Receptor for advanced glycosylation end products; Ang II, angiotensin II; TGF, Transforming growth factor.
Nuclear factor-κB (NF-κB) is the basic transcription factor that plays a pivotal role in inflammation in DN patients. Activated by upstream signals such as AGEs, angiotensin II, and oxidative stress (OS), NF-κB dissociates from its inhibitor IκB proteins and is transferred into the nucleus to regulate the expression of inflammatory gene including cytokines, chemokines, and adhesion molecules such as IL-6, TNF-α, and MCP-1 (Wada and Makino, 2016; Mikuda et al., 2018). One of the upstream signal pathways stimulated by AGEs is called the p38 mitogen-activated protein kinase (MAPK) pathway (Wu et al., 2002). The p38 MAPK pathway induces the activation of NF-κB in the infiltrating macrophages of DN (Adhikary et al., 2004). In turn, in renal parenchymal cells, elevated IL-1 and TNF-α have been shown to promote the phosphorylation of p38 MAPK, demonstrating their inflammatory roles in DN (Adhikary et al., 2004). Similarly, PI3K/AKT/mTOR is a widely studied signaling pathway that mediates the phenotype and injury of podocytes in DN. Stimulated by AGEs, PI3K/AKT can also promote NF-κB and aggravate inflammation (Ahmad et al., 2013; Hong et al., 2017). Recently, C-reactive protein (CRP) has been found to trigger a novel NF-κB-involved signaling pathway in the progression of DN, more narrowly, in human CRP transfected-db/db mice and cultured renal tubular epithelial cells, CRP is proved to promote inflammation via the evoking and dimerization of dipeptidyl peptidase-4 (DPP4) through DPP4/CD32b/NF-kB signaling circuit. The blockage of the circult by the DPP4 inhibitor, linagliptin, attenuates DN, suggesting the potential therapeutic effect for DN (Tang et al., 2021).
TGF-β/SMAD signaling pathway plays a criticial role in diabetic kidney injuries (Chen et al., 2011, 2014a; Liu et al., 2011; Lan, 2012; Zhong et al., 2013; Li et al., 2014; Zhang et al., 2019b; Xu et al., 2020a; Yang et al., 2020). In the diabetic kidney, high glucose and AGEs enhance the phosphorylation of SMAD3 and decrease the phosphorylation SMAD7. SMAD3 deficiency prevents renal inflammation and fibrosis in SMAD3-db/db mice via regulations of lncRNA Erbb4-IR.transcription and miR-29b (Xu et al., 2020a). SMAD3 deficiency protects against diabetes-associated beta cell dysfunction and loss in DN mice (Sheng et al., 2021). SMAD3 also promotes autophagy dysregulation and kidney injury (Yang et al., 2020). SMAD7 inhibits IκBα, an NF-κB inhibitor, suppressing the activation of NF-κB pathway (Chung et al., 2009). The deletion of SMAD7 significantly aggravates renal inflammation as evidenced by the upregulation of IL-1β, TNF-α, and MCP-1 in diabetic mice by crosstalk with NF-κB pathway, and the addition of SMAD7 attenuates the kidney injuries (Chen et al., 2011). Thus, TGF-β/SMAD and NF-κB crosstalk pathway may act as a novel prevention and therapeutic targets for diabetic nephropathy.
Activation of OS signaling pathways contributes to renal inflammation in DN. Nuclear factor-2 erythroid related factor (NRF2) is a protein that has the ability to alleviate inflammation and act as an antioxidant mediator in the process of OS. NRF2 reduces the infiltration of macrophages and proinflammatory cytokines by ameliorating oxidative overload. Clinical studies have demonstrated that the NRF2 activator, such as bardoxolone methyl, improves kidney function in diabetic patients (Pergola et al., 2011).
The nucleotide-binding oligomerization domain (NOD) family and NOD-like receptor pyrin domain-containing protein (NLRP) family are involved in DN. NOD2 promotes the endothelial-to-mesenchymal transition in DN (Shang et al., 2017). NLRP3 promotes the generation of IL-1β and IL-18 by activating the NLRP3-Caspase-1-IL-1β pathway in diabetic kidneys (Chi et al., 2020; Wang et al., 2020). NLRP3 has interactions with Toll-like receptors, ROS and NF-κB pathway to promote inflammation in DN (Shahzad et al., 2015; Wang et al., 2020).
The Janus kinase/signal transducer and activator of transcription (JAK-STAT) pathway is involved in processing extracellular signals (cytokines and chemokines) to the cell nucleus, resulting in gene expression (O’shea et al., 2013). The clinical findings show that JAK2 is increased in the podocytes of patients with early DN (Berthier et al., 2009). Podocyte-specific JAK2 overexpression in diabetic mice aggravates glomerulopathy while the inhibition of JAK1/2 attenuates the phenotypic changes of diabetic kidney (Zhang et al., 2017).
DN and Epigenetic Modifications Involved in Inflammation
DNA Methylation Involved in Inflammation of DN
DNA methylation promotes inflammatory activation of immune cells in diabetic kidney disease and demethylating agents prevent the progressive kidney disease (Larkin et al., 2018; Chen et al., 2019b). DNA methylation is a process in which the methyl group of S-adenosylmethionine is transferred to the cytosine of DNA under the catalysis of DNA methyltransferases (DNMTs), resulting in down-regulation of the gene expression (Yagi et al., 2012). DNMTs mainly include DNMT1, DNMT3a, and DNMT3b. DNMT1 has been found to contribute to the maintenance of methylation and the other DNMTs are related to de novo methylation (Hsieh, 1999). DNA methylation occurs specifically at the 5’ site of the CpG dinucleotide cytosine residue, hindering the binding of transcription factors and promoters, subsequently inhibiting transcription (Yagi et al., 2012). The genome-wide DNA methylation analysis shows that DNA methylation is associated the kidney injuries and kidney inflammation in DN patients (VanderJagt et al., 2015; Park et al., 2019). In vivo study also indicates high-glucose induced high levels of methylation in kidney cells. It is found that there are 173 differentially methylated regions (DMRs) in high glucose (HG)-treated mesangial cells compared to the low-glucose (LG) treatment (Li et al., 2020d). Suppression of methylation by bioactive constituent extracted from plants, e.g., moringa isothiocyanate (MIC-1), potentially down-regulates expression of TGF-β1, and changes the Nrf2, Col4a2, Tceal3, Ret, and Agt expressions (Li et al., 2020d; Cheng et al., 2019).
Aberrant cytosine methylation of the upstream regulators of the mammalian target of rapamycin (mTOR) promotes inflammation by the upregulation of DNMT1 in DN (Chen et al., 2019b). Notably, DNA methylation is dynamic and can be altered by environmental factors. Studies have found that hyperglycemia in T2DM patients triggers a self-regulatory mechanism leading to the reduction of 5mC levels in the peripheral blood, which indicates that the DNA might undergo demethylation via the upregulation of ten-eleven-translocation 2 (TET2), a DNA demethylation enzyme (Yuan et al., 2019).
Histone Modifications Involved in Inflammation of DN
The nucleosome is the basic unit of chromatin consisting of DNA and wrapped histone proteins. The post-translational modifications (PTMs) on chromatin histone include acetylation, ubiquitination, phosphorylation, and methylation. Recently, the genome-wide analysis of chromatin binding proteins and histone modifications has been conducted through chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) or by microarrays. The modifications are mainly mediated by three types of enzymes: writer, eraser, and reader. Writers/erasers carry on the modifications by adding/removing methyl or acetyl groups at amino acid residues in histone, such as histone acetyltransferase, histone methyltransferase (HMT), histone deacetylase (HDAC) and histone demethylase (HDM) (Bhatt et al.). Readers are the effectors that can identify and interpret post-translational modifications. Histone acetylation promotes gene transcription, while histone methylation promotes or inhibits gene transcription (Kouzarides, 2007). Specifically, the methylation of histone mostly happens on the residues of lysine and arginine. There are three types of methylation in lysine, namely monomethylation, dimethylation and trimethylation, and all three types of methylation of H3 at lysine 4 (H3K4me1, H3K4me2 and H3K4me3, respectively) exert an active effect (Kato and Natarajan, 2019). Similarly, H3K36me2 and H3K36me3 are enriched at transcriptional activation genome regions (Kato and Natarajan, 2019). Conversely, the methylation of H3K9me3, H3K27me3 and H4K20me3 are associated with gene repression (Kato and Natarajan, 2019). These modifications usually happen at promoters, insulators, enhancers, and other cis-regulatory regions, and finally lead to aberrant gene expression (Barski et al., 2007; Heintzman et al., 2009; Pradeepa et al., 2016).
Histone PTMs are involved in the pathogenesis of DN (Kato and Natarajan, 2019). HG and other danger signals increase the expression of pro-inflammatory genes by histone PTMs (Kato and Natarajan, 2019). TXNIP, pro-inflammatory gene, has been demonstrated to play an important role in the development of DN (Chen et al., 2008). In hyperglycemia-induced DN mice, HG-induced Txnip expression is associated with the enrichment of activated histone marks H3K9ac, H3K4me3, H3K4me1, and the repressive histone mark H3K27me3 at the promoter region of the gene, which has also been proved in human mesanginal cells (De Marinis et al., 2016). Furthermore, histone methylation take part in the process of inflammation via the secretion of inflammatory cytokines in diabetes. Specifically, H3K4 methylation could be mediated by HMT SET7 (Cheng et al., 2005). It is reported that transient HG causes the recruitment of HMT SET7 and increases H3K4 methylation at the NF-κB -P65 promoter, which promotes the expression of P65, MCP-1 and VCAM-1 in endothelial cells (El-Osta et al., 2008). Meanwhile, in endoplasmic reticulum (ER) stress induced kidney model of db/db mice, the increased expression of Mcp-1 is associated with the enrichment of H3K4me1 at Mcp-1 promoters, and could be significantly attenuated by the methyltransferase SET7/9 gene silencing (Chen et al., 2014c). The other study indicates that SET7/9 modifies chromatin histone lysine at promoters of MCP-1and TNF-α which promotes the inflammation in THP-1 monocytes (Li et al., 2008). In contrast, UTX (ubiquitously transcribed tetratricopeptide repeat, X chromosome) is a histone demethylase that can remove di- and tri-methyl groups from H3K27 (Choi et al., 2015). Studies have reported that the expression of UTX is upregulated in podocytes, tubular and mesangial cells of DN patients in vitro and in vivo (Majumder et al., 2018). Moreover, the knockout of UTX or the treatment of UTX inhibitor, GSK-J4, can reduce palmitic acid-induced increase of inflammation and DNA damage (Chen et al., 2019c). Furthermore, one study demonstrated that the inhibition of UTX could inhibit hypertrophy, a key event in glomerular dysfunction (Jia et al., 2019). In parallel, TGF-β down-regulates Enhancer of Zeste homolog 2 (EZH2), a H3K27me3 methyltransferase, by inducing miR-101b, which targets the 3’-untranslated region (3’-UTR) of EZH2. Meanwhile, TGF-β up-regulates UTX, a key role for H3K27me3 demethylases in renal mesangial cells. TGF-β-induced the inhibition of H3K27me3 augments pathological genes via dysregulation of associated histone-modifying enzymes and miR-101b in DN (Jia et al., 2019). Another H3K27me3 demethylase JMJD3 regulates inflammatory genes in macrophages (De Santa et al., 2007). To conclude, these studies suggest that the inhibition of H3K27me3 augments the expression of inflammation genes and the progression of DN.
Similarly, acetylation and deacetylation of histones via histone acetyltransferases (HATs) and histone deacetylase (HDACs) contribute to the pathogenesis of DN. Hyperglycemia promotes chromatin histone acetylation at inflammatory genes promoter regions and enhances inflammatory gene expressions in vivo (Miao et al., 2004). Levels of H3K9ac, H3K9ac/S10p, H3K18ac, H3K23ac and H3K56ac are increased in the kidneys of db/db mice (Huang et al., 2015). Furthermore, the expression of Sirt6 (a histone deacetylase) is reduced in podocytes of STZ-induced mice which results high levels of H3K9ac at promoters of Notch1 and Notch4, and exacerbates the inflammation in kidney (Liu et al., 2017). The silencing of HDAC9 attenuates renal injuries as demonstrated by the decrease in glomerulosclerosis, inflammatory cytokines, and alteration of podocyte apoptosis (Liu et al., 2016). The elevated HDAC4 in diabetic kidney exacerbates inflammation via suppressing STAT1 signaling and the silencing of HDAC4 is associated with the decreases of cytokines (TNF-α, TGF-β, IL-8, MCP-1) (Wang et al., 2014). The roles of DNA methylation and histone modification in the DN process are briefly shown in Figure 3.
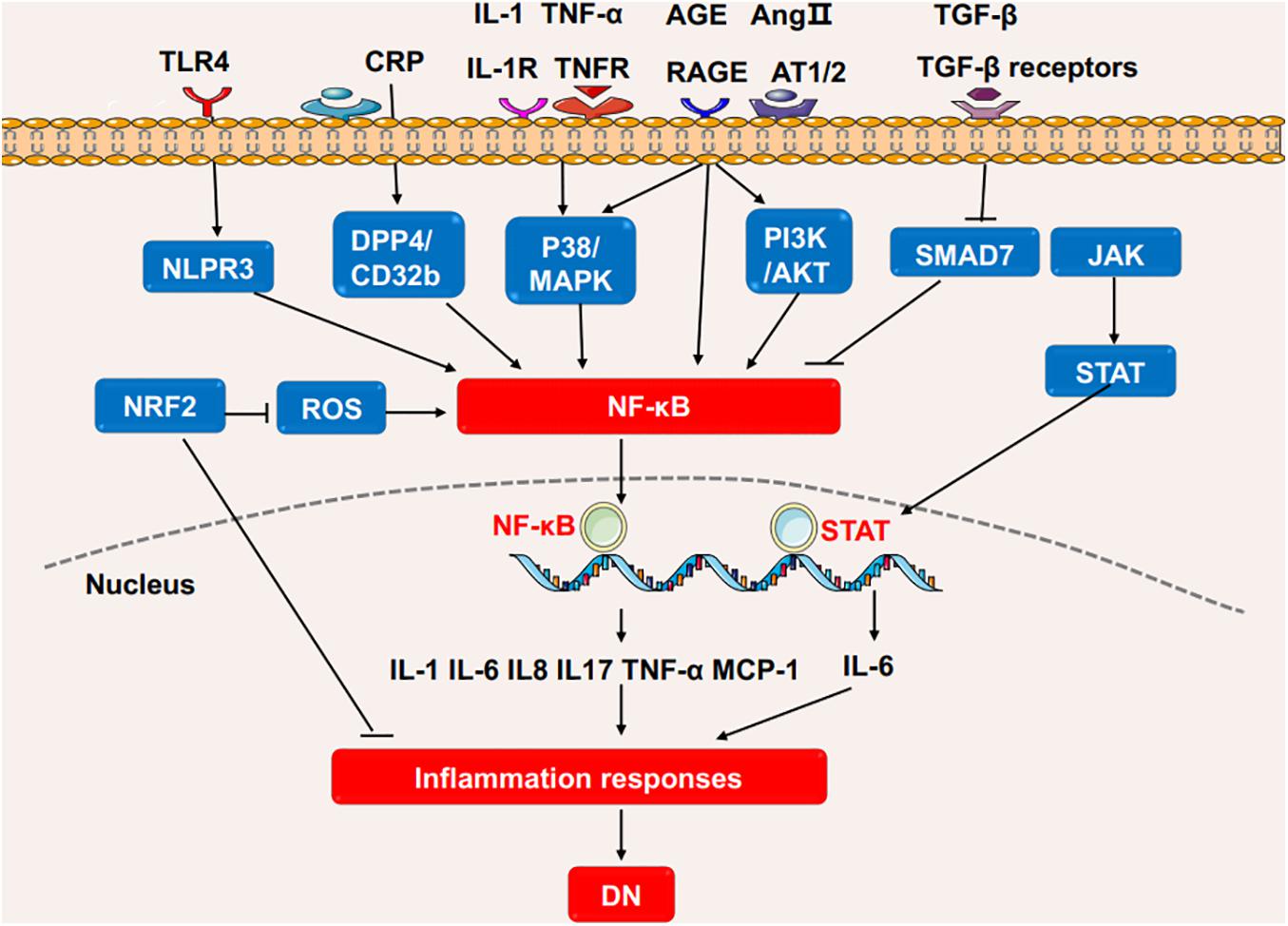
Figure 3. The roles of DNA methylation and histone modification in the DN process. High glucose or stimulating factors cause DNA methylation and histone modification. DNA methylation is mainly regulated by DNMTs. MIC-1 and TIIA inhibits DNA methylation and reduces inflammation in DN. In the processes of histone modification, SET7/9 regulates H3K4 methylation, UTX regulates H3K27 demethylation, HAT promotes histone acetylation, and HDAC4/9 promotes histone deacetylation. The above processes regulate inflammatory genes, in turn affects the inflammatory response in DN. DN, diabetic nephropathy; DNMTs, DNA methyltransferases; UTX, ubiquitously transcribed tetratricopeptide repeat, X chromosome; HAT, histone acetyltransferases; HDAC, histone deacetylase; NF-κB, Nuclear factor-κB; MCP-1, monocyte chemoattractant protein-1; STAT1, Signal transducer and activator of transcription 1.
Non-coding RNAs Involved in Inflammation of DN
Non-coding RNAs (ncRNAs) commonly include transfer RNA, ribosomal RNA, long ncRNA (lncRNA), small ncRNA (e.g., microRNA, piRNAs, snoRNA, snRNA, exRNA) and circular RNA (circRNA) (Storz, 2002; Yang, 2015). Roles of microRNA (miRNA), lncRNA and circRNA in DN have been recently studied (Loganathan et al., 2020; Zhou et al., 2021). MiRNA is the best characterized non-coding RNA for transcriptional gene regulation by targeting the 3′-UTR of a specific mRNA. Typically, miRNAs exert their inhibitory actions on the gene via RNA silencing and translational repression (Wilczynska and Bushell, 2015).
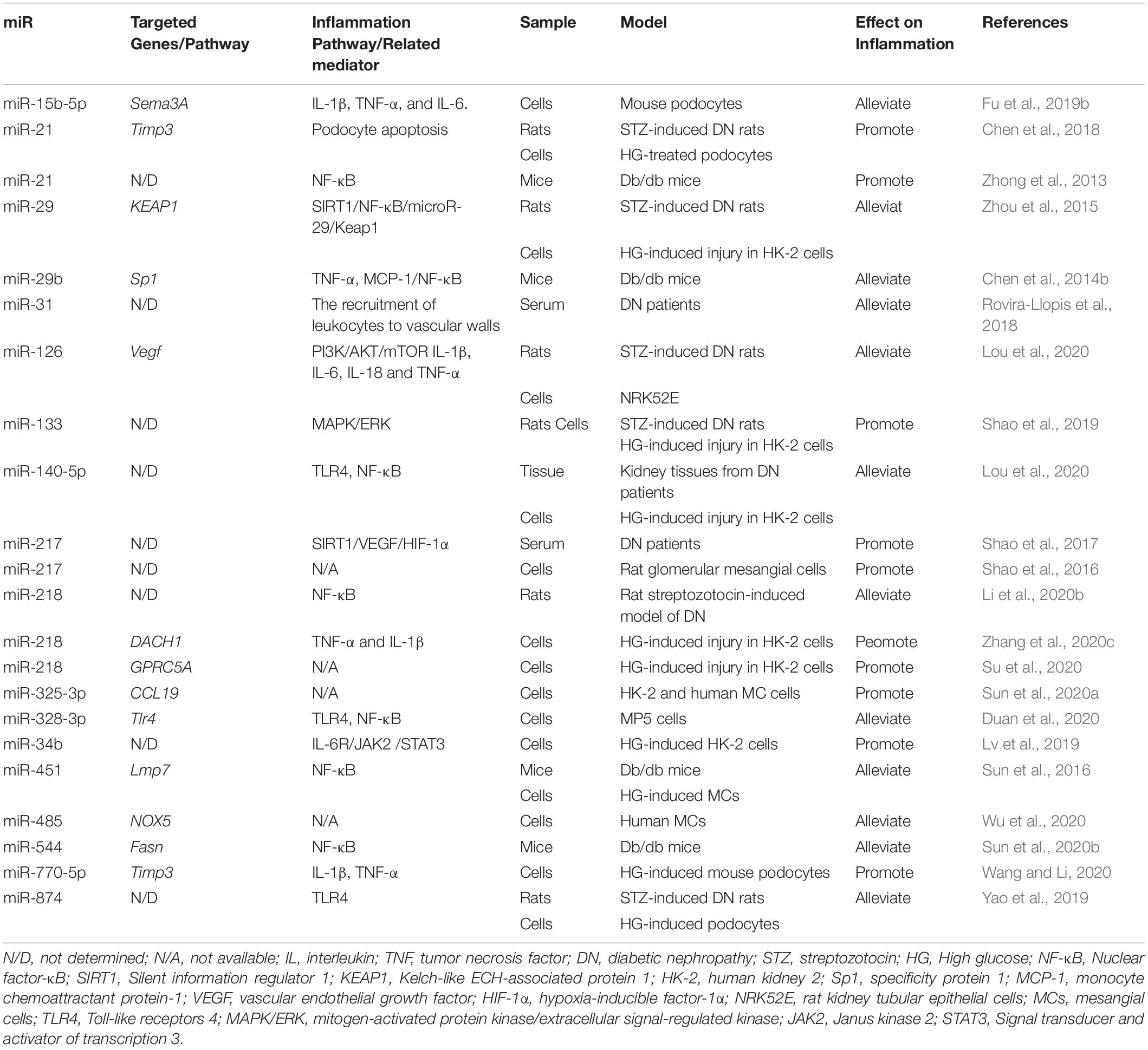
MiRNAs play significant roles in regulating inflammation in DN (Zhou et al., 2021). Recent studies involving models of DN podocytes have found that downregulation of the miR-17∼92 cluster ameliorates inflammation and podocyte injury by targeting ABCA1 (ATP-binding cassette transporter A1) (Fan et al., 2020). Similarly, the inhibition of miR-21-5p in a macrophage-derived extracellular vesicle model could also exert podocyte protective effect by the restraint of inflammasome activation (Ding et al., 2020). Moreover, miRNAs are also found to regulate inflammation in renal tubular epithelial cells. The overexpression of miR-199a-3p improves the injury in high glucose induced HK-2 cell damage model, following with decreased IL-1, IL-6 and TNF-α level, which is also consistent with the clinical finding that miR-199a-3p is negatively correlated with the progression of DN (Zhang et al., 2020b). The protective effects of miR-199a-3p is via suppressing miR-199a-3p mediated IKKβ/NF-κB pathway (Zhang et al., 2020b). In vitro experiments, the overexpression of miR-26a-5p significantly inhibits the bovine serum albumin (BSA)-induced IL-6 and TNF-α expression in HK2 cells while the inhibition of miR-26a-5p promotes the expression of inflammatory cytokines (Li et al., 2020c). MiR-26a-5p is also found to activate NF-κB pathway by targeting on CHAC1 and TLR4 genes (Zhong et al., 2018; Li et al., 2020c). MiR-155 and miR-146a have also been found to be correlated with renal damage, possibly due to the increased expression of TNF-α, TGF-β1, and NF-κB, and their roles in inflammation-mediated glomerular endothelial damage (Huang et al., 2014). Moreover, miRNAs regulate inflammation by modulating macrophage polarization. As mentioned before, macrophage M1 polarization act as an inflammation driver. In miR-146a deficiency diabetic mice, the expression of M1 markers is increased while the M2 response is diminished which is in accordance with the upregulated pro-inflammatory cytokines, suggesting the anti-inflammatory proporities of miR-146a (Bhatt et al., 2016). M2 macrophages ameliorate podocyte injury is related to miR-25-3p (Huang et al., 2020). It is found that autophagy deficiency in diabetic mice increases macrophage infiltration in proximal tubules (Ma et al., 2020), and the induction of miR-214 enhances the autophagy impairment, thus aggravating renal inflammation (Li et al., 2011). MiR-214 in monocytes is upregulated by AGEs, which in turn impairs the expression of the phosphatase and tensin homolog (PTEN) and delays spontaneous apoptosis of monocytes (Li et al., 2011). Additionally, miR-27a is downregulated by an adipokine, omentin-1, which alleviates inflammation and OS by directly targeting the 3′-UTR of Nrf2 (Song et al., 2018). MiR-29b attenuates podocyte injury by targeting the 3′-UTR of HADC4 in DN (Gondaliya et al., 2020). MiR-125b has been found to inhibit the chromatin histone H3K9 methyltransferase to regulate inflammatory genes in diabetic mice (Villeneuve et al., 2010). Hyperglycemia induces miR-101b, which targets the EZH2, leading to mesangial dysfunction in DN (Jia et al., 2019).
Moreover, accumulating evidence shows that a lot of miRNAs are involved in the regulation of inflammation in DN as shown in Table 1.
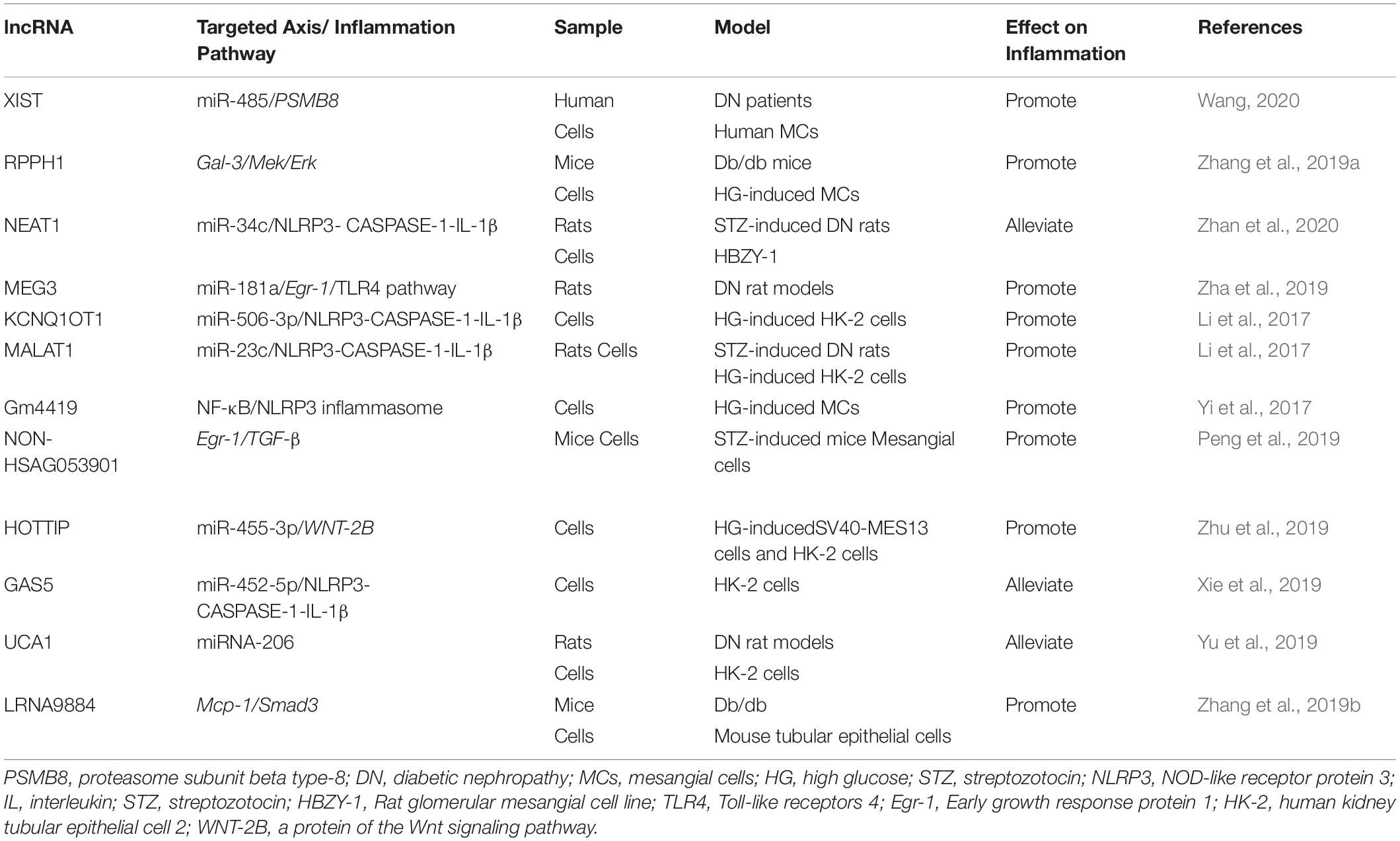
LncRNAs also contribute to the development and progression of DN. LncRNA myocardial infarction associated transcript (MIAT) promotes hyperglycemia-induced podocyte inflammation by sponging miR-130a-3p and the regulation of TLR4 (Zhang et al., 2020a). LncRNA 4930556M19Rik has been found to protect against HG-induced podocyte damage by downregulation miR-27a-3p (Fan and Zhang, 2020). Macrophage-specific lncRNA_7949 mediates macrophage-induced kidney inflammation by the controlling of MCP-1 transcription through TLR4/NF-κB pathway (Lv et al., 2015). TGF-β/Smad3 transits the miRNA profile and promotes renal diseases via regulating transcriptional levels of non-coding RNAs. SMAD3-dependent lncRNAs have been recently uncovered in kidney diseases (Tang et al., 2018a, 2020a). LncRNA Erbb4-IR is responsible for TGF-β/Smad3-regulated renal fibrosis by inhibiting SMAD7 (Feng et al., 2018). It has been reported that lncRNA Erbb4-IR enhances diabetic kidney injury by mediating miR-29b in db/db Mice. Deletion of SMAD3 could down-regulate the lncRNA Erbb4-IR transcription, and therefore protect against renal injury in db/db mice (Sun et al., 2018). LRNA9884, a novel SMAD3-dependent lncRNA, is not only involved into NF-κB-mediated inflammatory responses by activation of macrophage migration inhibitory factor (MIF) in AKI, but also enhances diabetic renal injury via promoting MCP-1-dependent renal inflammation in db/db mice (Zhang et al., 2019b, 2020d; Xu et al., 2020a). The lncRNAs involved in the inflammation of DN are shown in Table 2.
CircRNAs regulate gene expressions by acting as sponges of miRNA (Kristensen et al., 2019), and play an important role in renal diseases (Jin et al., 2020). As a sponge of miR-135a, circRNA_010383 is markedly decreased in the kidney of db/db mice and HG-induced kidney resident cells, and overexpression of circRNA_010383 in kidney protects kidney from proteinuria and fibrosis in DN (Peng et al., 2021). CircLRP6, as a sponge of miR-205, activates TLR4/NF-κB pathway and induces inflammation in high glucose treated mesangial cells (Chen et al., 2019a). CircACTR2 induces inflammation and pyroptosis in high glucose treated renal tubular cells (Wen et al., 2020). Circ_0003928 attenuates the high glucose-induced inflammation in HK-2 cells by targeting miR-151-3p/Anxa2 (An et al., 2020). CircWBSCR17 aggravates inflammation and fibrosis in high glucose-induced HK-2 cells via miR-185-5p/SOX6 axis (Li et al., 2020a). Circ0000285 enhances inflammation via sponging miR-654-3p in high glucose treated podocytes and diabetic mouse kidney (Yao et al., 2020).
Discussion
The current evidence reveals epigenetics (methylation, acetylation, and non-coding RNA modification) modulate inflammation via intrinsic cells, immune cells, and numerous inflammatory pathways in the development of DN. Persistent inflammation in DN promotes the renal fibrosis, thus resulting in CKD and even end-stage renal disease (Tang et al., 2020a). Anti-inflammatory therapy has long been considered to have enormous benefits for either the alleviation or the prevention of DN (Barutta et al., 2015). In this review, we summarized the evidence linking epigenetic modifications and inflammation in DN. Thus, it may be an effective approach to target these modifications for DN treatment. As for histone modification, the inhibition of HATs/HDACs provides as a class of new agents or therapeutic targets for the treatment of DN. Most of agents are non-selective inhibitors hindering the clinical application (Wang et al., 2014). Valproic acid is a specific HDAC1 inhibitor, which attenuates proteinuria, fibrosis, and inflammatory effects and even acute pancreatitis (Van Beneden et al., 2011; Jain et al., 2019). However, effects of specific HDAC inhibitors for DN remain largely unexplored.
LncRNAs have been considered the novel markers as well as the potential therapeutic targets, and novel drug delivery vehicles (e.g., exosome-ncRNAs). Metformin has been found to protect against inflammation and ECM accumulation in mesangial cells via the H19/miR-143-3p/TGF-β1 axis, suggesting that the H19/miR-143-3p/TGF-β1 axis could be a potential therapeutic target for the management of DN (Xu et al., 2020b). The competing endogenous RNA (ceRNA) network analysis on human miRNA indicates that RP11-363E7.4/TTN-AS1/HOTAIRM1-hsa-miR-106b-5p-PTGER3 and LINC00960-hsa-miR-1237-3p-MMP-2 interaction pairs are significant in diabetic kidney (Yu et al., 2021). Drugs such as iloprost, treprostinil, and captopril that target PTGER3 and MMP-2 might benefit patients with DN (Yu et al., 2021).
Intriguingly, several studies show miRNA-192 is upregulated in diabetic patients with microalbuminuria, but downregulated in macroalbumnuria compared to normalbuminuria (Krupa et al., 2010; Jia et al., 2016). However, another study shows that miR-192 is increased in DN patients with over proteinuria (ACT >300 mg/g) compared to microalbuminuria (Chien et al., 2016). These studies indicate miRNA-mediated epigenetic modifications may have various roles in different stages of a disease.
Besides DNA methylation, histone modification and non-coding RNA, RNA methylation plays an important role in the mRNA post-translational modification. For example, N6-methyladenosine (m6A) methylation is the most chemically modified form of eukaryotic messenger RNA (mRNA) which modifies the adenosine at the 3’-UTR and the stop codon of a mRNA (Fu et al., 2014; Roundtree et al., 2017). Roles of epigenetic modifications are not fully elucidated. Recently, single nucleus ATCT-seq integrated with snRNA-seq has been used to detect the cell-type-specific chromatin accessibility which enable to deep understanding of cell heterogeneity in kidney (Bansal et al., 2020; Muto et al., 2021). It may provide a new approach to understand the epigentic modifications in DN.
Collectively, further studies are warranted to reveal the precise regulatory mechanisms in the different stages of DN as well as potential therapeutic targets and diagnostic biomarkers for DN.
Author Contributions
H-YC and X-MM conceived, revised and edited the manuscript. B-YS and S-FZ collected studies and drafted the manuscript. H-DL assisted in data extraction and revised the manuscript. All authors approved the final version of the manuscript for publication.
Funding
The study was supported by the General Research Fund (17113416, 17109019), Seed Fund for Basic Research (201910159215), and Shenzhen Science & Innovation Fund (JCYJ20180306173745092).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.649587/full#supplementary-material
References
Adhikary, L., Chow, F., Nikolic-Paterson, D. J., Stambe, C., Dowling, J., Atkins, R. C., et al. (2004). Abnormal p38 mitogen-activated protein kinase signalling in human and experimental diabetic nephropathy. Diabetologia 47, 1210–1222. doi: 10.1007/s00125-004-1437-0
Ahmad, A., Biersack, B., Li, Y., Kong, D., Bao, B., Schobert, R., et al. (2013). Targeted regulation of PI3K/Akt/mTOR/NF-κB signaling by indole compounds and their derivatives: mechanistic details and biological implications for cancer therapy. Anti Cancer Agents Med. Chem. 13, 1002–1013. doi: 10.2174/18715206113139990078
An, L., Ji, D., Hu, W., Wang, J., Jin, X., Qu, Y., et al. (2020). Interference of Hsa_circ_0003928 alleviates high glucose-induced cell apoptosis and inflammation in HK-2 cells via miR-151-3p/Anxa2. Diabetes Metab Syndr Obes 13, 3157–3168. doi: 10.2147/dmso.s265543
Awad, A. S., You, H., Gao, T., Cooper, T. K., Nedospasov, S. A., Vacher, J., et al. (2015). Macrophage-derived tumor necrosis factor-α mediates diabetic renal injury. Kidney Int. 88, 722–733. doi: 10.1038/ki.2015.162
Bansal, A., Balasubramanian, S., Dhawan, S., Leung, A., Chen, Z., and Natarajan, R. (2020). Integrative omics analyses reveal epigenetic memory in diabetic renal cells regulating genes associated with kidney dysfunction. Diabetes 69, 2490–2502. doi: 10.2337/db20-0382
Barski, A., Cuddapah, S., Cui, K., Roh, T.-Y., Schones, D. E., Wang, Z., et al. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. doi: 10.1016/j.cell.2007.05.009
Barutta, F., Bruno, G., Grimaldi, S., and Gruden, G. (2015). Inflammation in diabetic nephropathy: moving toward clinical biomarkers and targets for treatment. Endocrine 48, 730–742. doi: 10.1007/s12020-014-0437-1
Berthier, C. C., Zhang, H., Schin, M., Henger, A., Nelson, R. G., Yee, B., et al. (2009). Enhanced expression of a janus kinase–signal transducer and activator of transcription pathway members in human diabetic nephropathy. Diabetes 58, 469–477. doi: 10.2337/db08-1328
Bhatt, K., Lanting, L. L., Jia, Y., Yadav, S., Reddy, M. A., Magilnick, N., et al. (2016). Anti-inflammatory role of microRNA-146a in the pathogenesis of diabetic nephropathy. J. Am. Soc. Nephrol. 27, 2277–2288. doi: 10.1681/asn.2015010111
Chen, B., Li, Y., Liu, Y., and Xu, Z. (2019a). circLRP6 regulates high glucose-induced proliferation, oxidative stress, ECM accumulation, and inflammation in mesangial cells. J. Cell. Physiol. 234, 21249–21259. doi: 10.1002/jcp.28730
Chen, G., Chen, H., Ren, S., Xia, M., Zhu, J., Liu, Y., et al. (2019b). Aberrant DNA methylation of mTOR pathway genes promotes inflammatory activation of immune cells in diabetic kidney disease. Kidney Int. 96, 409–420. doi: 10.1016/j.kint.2019.02.020
Chen, H., Huang, Y., Zhu, X., Liu, C., Yuan, Y., Su, H., et al. (2019c). Histone demethylase UTX is a therapeutic target for diabetic kidney disease. J. Physiol. 597, 1643–1660. doi: 10.1113/jp277367
Chen, H., Zhong, X., Huang, X. R., Meng, X. M., You, Y., Chung, A. C., et al. (2014a). MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol. Ther. 22, 842–853. doi: 10.1038/mt.2013.235
Chen, H. Y., Huang, X. R., Wang, W., Li, J. H., Heuchel, R. L., Chung, A. C., et al. (2011). The protective role of Smad7 in diabetic kidney disease: mechanism and therapeutic potential. Diabetes 60, 590–601. doi: 10.2337/db10-0403
Chen, H. Y., Zhong, X., Huang, X. R., Meng, X. M., You, Y., Chung, A. C., et al. (2014b). MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol. Ther. 22, 842–853.
Chen, J., Guo, Y., Zeng, W., Huang, L., Pang, Q., Nie, L., et al. (2014c). ER stress triggers MCP-1 expression through SET7/9-induced histone methylation in the kidneys of db/db mice. Am. J. Physiol. R. Physiol. 306, F916–F925.
Chen, J., Saxena, G., Mungrue, I. N., Lusis, A. J., and Shalev, A. (2008). Thioredoxin-interacting protein: a critical link between glucose toxicity and β-cell apoptosis. Diabetes 57, 938–944. doi: 10.2337/db07-0715
Chen, X., Zhao, L., Xing, Y., and Lin, B. (2018). Down-regulation of microRNA-21 reduces inflammation and podocyte apoptosis in diabetic nephropathy by relieving the repression of TIMP3 expression. Biomed. Pharmacother. 108, 7–14. doi: 10.1016/j.biopha.2018.09.007
Cheng, D., Gao, L., Su, S., Sargsyan, D., Wu, R., Raskin, I., et al. (2019). Moringa isothiocyanate activates Nrf2: potential role in diabetic nephropathy. AAPS J. 21:31.
Cheng, X., Collins, R. E., and Zhang, X. (2005). Structural and sequence motifs of protein (histone) methylation enzymes. Annu. Rev. Biophys. Biomol. Struct. 34, 267–294. doi: 10.1146/annurev.biophys.34.040204.144452
Chi, K., Geng, X., Liu, C., Cai, G., and Hong, Q. (2020). Research progress on the role of inflammasomes in kidney disease. Med. Inf. 2020:8032797.
Chien, H. Y., Chen, C. Y., Chiu, Y. H., Lin, Y. C., and Li, W. C. (2016). Differential microRNA profiles predict diabetic nephropathy progression in taiwan. Int. J. Med. Sci. 13, 457–465. doi: 10.7150/ijms.15548
Choi, H. J., Park, J. H., Park, M., Won, H. Y., Joo, H. S., Lee, C. H., et al. (2015). UTX inhibits EMT-induced breast CSC properties by epigenetic repression of EMT genes in cooperation with LSD 1 and HDAC 1. EMBO Rep. 16, 1288–1298. doi: 10.15252/embr.201540244
Chow, F., Nikolic-Paterson, D. J., Ozols, E., Atkins, R. C., Rollin, B., and Tesch, G. H. (2006). Monocyte chemoattractant protein-1 promotes the development of diabetic renal injury in streptozotocin-treated mice. Kidney Int. 69, 73–80. doi: 10.1038/sj.ki.5000014
Chow, F., Ozols, E., Nikolic-Paterson, D. J., Atkins, R. C., and Tesch, G. H. (2004). Macrophages in mouse type 2 diabetic nephropathy: correlation with diabetic state and progressive renal injury. Kidney Int. 65, 116–128. doi: 10.1111/j.1523-1755.2004.00367.x
Chung, A. C., Huang, X. R., Zhou, L., Heuchel, R., Lai, K. N., and Lan, H. Y. (2009). Disruption of the Smad7 gene promotes renal fibrosis and inflammation in unilateral ureteral obstruction (UUO) in mice. Nephrol. Dialysis Transplant. 24, 1443–1454. doi: 10.1093/ndt/gfn699
Cortvrindt, C., Speeckaert, R., Moerman, A., Delanghe, J. R., and Speeckaert, M. M. (2017). The role of interleukin-17A in the pathogenesis of kidney diseases. Pathology 49, 247–258. doi: 10.1016/j.pathol.2017.01.003
De Marinis, Y., Cai, M., Bompada, P., Atac, D., Kotova, O., Johansson, M. E., et al. (2016). Epigenetic regulation of the thioredoxin-interacting protein (TXNIP) gene by hyperglycemia in kidney. Kidney Int. 89, 342–353. doi: 10.1016/j.kint.2015.12.018
De Santa, F., Totaro, M. G., Prosperini, E., Notarbartolo, S., Testa, G., and Natoli, G. (2007). The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094. doi: 10.1016/j.cell.2007.08.019
Ding, X., Jing, N., Shen, A., Guo, F., Song, Y., Pan, M., et al. (2020). MiR-21-5p in macrophage-derived extracellular vesicles affects podocyte pyroptosis in diabetic nephropathy by regulating A20. J. Endocrinol. Invest. Epub ahead of print,
Duan, Y., Luo, Q., Wang, Y., Ma, Y., Chen, F., Zhu, X., et al. (2020). Adipose mesenchymal stem cell-derived extracellular vesicles containing microRNA-26a-5p target TLR4 and protect against diabetic nephropathy. J. Biol. Chem. 295, 12868–12884. doi: 10.1074/jbc.ra120.012522
Eller, K., Kirsch, A., Wolf, A. M., Sopper, S., Tagwerker, A., Stanzl, U., et al. (2011). Potential role of regulatory T cells in reversing obesity-linked insulin resistance and diabetic nephropathy. Diabetes 60, 2954–2962. doi: 10.2337/db11-0358
Elliott, M. J., Maini, R. N., Feldmann, M., Kalden, J. R., Antoni, C., Smolen, J. S., et al. (1994). Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor α (cA2) versus placebo in rheumatoid arthritis. Lancet 344, 1105–1110. doi: 10.1016/s0140-6736(94)90628-9
El-Osta, A., Brasacchio, D., Yao, D., Pocai, A., Jones, P. L., Roeder, R. G., et al. (2008). Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J. Exp. Med. 205, 2409–2417. doi: 10.1084/jem.20081188
Fan, H., and Zhang, W. (2020). Overexpression of Linc 4930556M19Rik Suppresses High Glucose-Triggered Podocyte Apoptosis, Fibrosis and Inflammation via the miR-27a-3p/Metalloproteinase 3 (TIMP3) Axis in Diabetic Nephropathy. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 26:e925361.
Fan, X., Hao, Z., Li, Z., Wang, X., and Wang, J. (2020). Inhibition of miR-17~92 cluster ameliorates high glucose-induced podocyte damage. Med. Inf. 2020:6126490.
Feng, M., Tang, P. M.-K., Huang, X.-R., Sun, S.-F., You, Y.-K., Xiao, J., et al. (2018). TGF-β mediates renal fibrosis via the Smad3-Erbb4-IR long noncoding RNA axis. Mol. Ther. 26, 148–161. doi: 10.1016/j.ymthe.2017.09.024
Fu, J., Akat, K. M., Sun, Z. G., Zhang, W. J., Schlondorff, D., Liu, Z. H., et al. (2019a). Single-cell RNA profiling of glomerular cells shows dynamic changes in experimental diabetic kidney disease. J. Am. Soc. Nephrol. 30, 533–545. doi: 10.1681/asn.2018090896
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene expression regulation mediated through reversible m 6 A RNA methylation. Nat. Rev. Gene. 15, 293–306. doi: 10.1038/nrg3724
Fu, Y., Wang, C., Zhang, D., Chu, X., Zhang, Y., and Li, J. (2019b). miR-15b-5p ameliorated high glucose-induced podocyte injury through repressing apoptosis, oxidative stress, and inflammatory responses by targeting Sema3A. J. Cell. Physiol. 234, 20869–20878. doi: 10.1002/jcp.28691
GBD. (2018). global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet 392, 1789–1858.
Goldberg, A. D., Allis, C. D., and Bernstein, E. (2007). Epigenetics: a landscape takes shape. Cell 128, 635–638. doi: 10.1016/j.cell.2007.02.006
Gondaliya, P. A., Jash, K., Tekade, R. K., Srivastava, A., and Kalia, K. (2020). miR-29b attenuates histone deacetylase-4 mediated podocyte dysfunction and renal fibrosis in diabetic nephropathy. J. Diabetes Metab Dis. 19, 13–27. doi: 10.1007/s40200-019-00469-0
Grabias, B. M., and Konstantopoulos, K. (2014). The physical basis of renal fibrosis: effects of altered hydrodynamic forces on kidney homeostasis. Am. J. Physiol. R. Physiol. 306, F473–F485.
Heintzman, N. D., Hon, G. C., Hawkins, R. D., Kheradpour, P., Stark, A., Harp, L. F., et al. (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112. doi: 10.1038/nature07829
Hickey, F. B., and Martin, F. (2013). Diabetic kidney disease and immune modulation. Curr. Opin. Pharmacol. 13, 602–612. doi: 10.1016/j.coph.2013.05.002
Hong, J. N., Li, W. W., Wang, L. L., Guo, H., Jiang, Y., Gao, Y. J., et al. (2017). Jiangtang decoction ameliorate diabetic nephropathy through the regulation of PI3K/Akt-mediated NF-κB pathways in KK-Ay mice. Chin. Med. 12, 1–16.
Hotamisligil, G. S. (2017). Inflammation, metaflammation and immunometabolic disorders. Nature 542, 177–185. doi: 10.1038/nature21363
Hsieh, C. L. (1999). In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol. 19, 8211–8218. doi: 10.1128/mcb.19.12.8211
Huang, H., Liu, H., Tang, J., Xu, W., Gan, H., Fan, Q., et al. (2020). M2 macrophage-derived exosomal miR-25-3p improves high glucose-induced podocytes injury through activation autophagy via inhibiting DUSP1 expression. IUBMB Life 72, 2651–2662. doi: 10.1002/iub.2393
Huang, J., Wan, D., Li, J., Chen, H., Huang, K., and Zheng, L. (2015). Histone acetyltransferase PCAF regulates inflammatory molecules in the development of renal injury. Epigenetics 10, 62–72. doi: 10.4161/15592294.2014.990780
Huang, Y., Liu, Y., Li, L., Su, B., Yang, L., Fan, W., et al. (2014). Involvement of inflammation-related miR-155 and miR-146a in diabetic nephropathy: implications for glomerular endothelial injury. BMC Nephrol. 15:142.
International Diabetes Federation (2019). IDF DIABETES ATLAS, 9th Edn. Available online at: https://diabetesatlas.org/en/ (accessed October 10, 2020).
Jain, A., Haque, I., Tayal, V., and Roy, V. (2019). Valproic acid-induced acute pancreatitis. Indian J. Psychiatry 61, 421–422. doi: 10.4103/psychiatry.indianjpsychiatry_383_18
Jia, Y., Guan, M., Zheng, Z., Zhang, Q., Tang, C., Xu, W., et al. (2016). miRNAs in urine extracellular vesicles as predictors of early-stage diabetic nephropathy. J. Diabetes Res. 2016:7932765. doi: 10.1155/2016/7932765
Jia, Y., Reddy, M. A., Das, S., Oh, H. J., Abdollahi, M., Yuan, H., et al. (2019). Dysregulation of histone H3 lysine 27 trimethylation in transforming growth factor-β1–induced gene expression in mesangial cells and diabetic kidney. J. Biol. Chem. 294, 12695–12707. doi: 10.1074/jbc.ra119.007575
Jin, J., Sun, H., Shi, C., Yang, H., Wu, Y., Li, W., et al. (2020). Circular RNA in renal diseases. J. Cell. Mol. Med. 24, 6523–6533.
Kato, M., and Natarajan, R. (2019). Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat. Rev. Nephrol. 15, 327–345. doi: 10.1038/s41581-019-0135-6
Keating, S. T., and El-Osta, A. (2013). Glycemic memories and the epigenetic component of diabetic nephropathy. Curr. Diabetes Rep. 13, 574–581. doi: 10.1007/s11892-013-0383-y
Klessens, C. Q., Zandbergen, M., Wolterbeek, R., Bruijn, J. A., Rabelink, T. J., Bajema, I. M., et al. (2017). Macrophages in diabetic nephropathy in patients with type 2 diabetes. Nephrol. Dialysis Trans. 32, 1322–1329.
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Gene. 20, 675–691.
Krupa, A., Jenkins, R., Luo, D. D., Lewis, A., Phillips, A., and Fraser, D. (2010). Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J. Am. Soc. Nephrol. 21, 438–447. doi: 10.1681/asn.2009050530
Lampropoulou, I. T., Stangou, M., Sarafidis, P., Gouliovaki, A., Giamalis, P., Tsouchnikas, I., et al. (2020). TNF-α pathway and T-cell immunity are activated early during the development of diabetic nephropathy in type II diabetes mellitus. Clin. Immunol. 215:108423. doi: 10.1016/j.clim.2020.108423
Lan, H. Y. (2012). Transforming growth factor-beta/smad signalling in diabetic nephropathy. Clin. Exp. Pharmacol. Physiol. 39, 731–738. doi: 10.1111/j.1440-1681.2011.05663.x
Larkin, B. P., Glastras, S. J., Chen, H., Pollock, C. A., and Saad, S. (2018). DNA methylation and the potential role of demethylating agents in prevention of progressive chronic kidney disease. FASEB J. 32, 5215–5226. doi: 10.1096/fj.201800205r
Lemos, D. R., Mcmurdo, M., Karaca, G., Wilflingseder, J., Leaf, I. A., Gupta, N., et al. (2018). Interleukin-1β activates a MYC-dependent metabolic switch in kidney stromal cells necessary for progressive tubulointerstitial fibrosis. J. Am. Soc. Nephrol. 29, 1690–1705. doi: 10.1681/asn.2017121283
Li, G., Qin, Y., Qin, S., Zhou, X., Zhao, W., and Zhang, D. (2020a). Circ_WBSCR17 aggravates inflammatory responses and fibrosis by targeting miR-185-5p/SOX6 regulatory axis in high glucose-induced human kidney tubular cells. Life Sci. 259:118269. doi: 10.1016/j.lfs.2020.118269
Li, L. M., Hou, D. X., Guo, Y. L., Yang, J. W., Liu, Y., Zhang, C. Y., et al. (2011). Role of microRNA-214–targeting phosphatase and tensin homolog in advanced glycation end product-induced apoptosis delay in monocytes. J. Immunol. 186, 2552–2560. doi: 10.4049/jimmunol.1001633
Li, M., Guo, Q., Cai, H., Wang, H., Ma, Z., and Zhang, X. (2020b). miR-218 regulates diabetic nephropathy via targeting IKK−β and modulating NK−κB-mediated inflammation. J. Cell. Physiol. 235, 3362–3371. doi: 10.1002/jcp.29224
Li, R., Chung, A. C., Yu, X., and Lan, H. Y. (2014). MicroRNAs in diabetic kidney disease. Int. J. Endocrinol. 2014:593956.
Li, S., Jia, Y., Xue, M., Hu, F., Zheng, Z., Zhang, S., et al. (2020c). Inhibiting Rab27a in renal tubular epithelial cells attenuates the inflammation of diabetic kidney disease through the miR-26a-5p/CHAC1/NF-kB pathway. Life Sci. 261:118347. doi: 10.1016/j.lfs.2020.118347
Li, S., Li, W., Wu, R., Yin, R., Sargsyan, D., Raskin, I., et al. (2020d). Epigenome and transcriptome study of moringa isothiocyanate in mouse kidney mesangial cells induced by high glucose, a potential model for diabetic-induced nephropathy. AAPS J. 22:8.
Li, X., Zeng, L., Cao, C., Lu, C., Lian, W., Han, J., et al. (2017). Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp. Cell Res. 350, 327–335. doi: 10.1016/j.yexcr.2016.12.006
Li, Y., Reddy, M. A., Miao, F., Shanmugam, N., Yee, J.-K., Hawkins, D., et al. (2008). Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-κB-dependent inflammatory genes: relevance to diabetes and inflammation. J. Biol. Chem. 283, 26771–26781. doi: 10.1074/jbc.m802800200
Lim, A. K. (2014). Diabetic nephropathy–complications and treatment. Int. J. Nephrol. Renovascular Dis. 7:361. doi: 10.2147/ijnrd.s40172
Lin, J., Cheng, A., Cheng, K., Deng, Q., Zhang, S., Lan, Z., et al. (2020a). New insights into the mechanisms of pyroptosis and implications for diabetic kidney disease. Int. J. Mol. Sci. 21:7057. doi: 10.3390/ijms21197057
Lin, X., Xu, Y., Pan, X., Xu, J., Ding, Y., Sun, X., et al. (2020b). Global, regional, and national burden and trend of diabetes in 195 countries and territories: an analysis from 1990 to 2025. Sci. Rep. 10:14790.
Liu, F., Chen, H. Y., Huang, X. R., Chung, A. C., Zhou, L., Fu, P., et al. (2011). C-reactive protein promotes diabetic kidney disease in a mouse model of type 1 diabetes. Diabetologia 54, 2713–2723. doi: 10.1007/s00125-011-2237-y
Liu, F., Zong, M., Wen, X., Li, X., Wang, J., Wang, Y., et al. (2016). Silencing of histone deacetylase 9 expression in podocytes attenuates kidney injury in diabetic nephropathy. Sci. Rep. 6:33676.
Liu, M., Liang, K., Zhen, J., Zhou, M., Wang, X., Wang, Z., et al. (2017). Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nat. Commun. 8, 1–15. doi: 10.1016/j.biochi.2014.12.015
Loganathan, T. S., Sulaiman, S. A., Abdul Murad, N. A., Shah, S. A., Abdul Gafor, A. H., Jamal, R., et al. (2020). Interactions among non-coding RNAs in diabetic nephropathy. Front. Pharmacol. 11:191. doi: 10.3389/fphar.2020.0019
Lou, Z., Li, Q., Wang, C., and Li, Y. (2020). The effects of microRNA-126 reduced inflammation and apoptosis of diabetic nephropathy through PI3K/AKT signalling pathway by VEGF. Arch. Physiol. Biochem. 25, 1–10. doi: 10.1080/13813455.2020.1767146
Lv, L., Tang, P., You, Y. K., Huang, X., Liu, B.-C., and Lan, H.-Y. (2015). Long noncoding RNA-7949 regulates macrophage activation in renal inflammation via the TLR4/NF-KB pathway. Hong Kong J. Nephrol. 2:S76.
Lv, N., Li, C., Liu, X., Qi, C., and Wang, Z. (2019). miR-34b alleviates high glucose-induced inflammation and apoptosis in human HK-2 cells via IL-6R/JAK2/STAT3 signaling pathway. Med. Sci. Monitor Int. Med. J. Exp. Clin. Res. 25:8142. doi: 10.12659/msm.917128
Ma, Z., Li, L., Livingston, M. J., Zhang, D., Mi, Q., Zhang, M., et al. (2020). p53/microRNA-214/ULK1 axis impairs renal tubular autophagy in diabetic kidney disease. J. Clin. Invest. 130, 5011–5026. doi: 10.1172/jci135536
Majumder, S., Thieme, K., Batchu, S. N., Alghamdi, T. A., Bowskill, B. B., Kabir, M. G., et al. (2018). Shifts in podocyte histone H3K27me3 regulate mouse and human glomerular disease. J. Clin. Invest. 128, 483–499. doi: 10.1172/jci95946
Meng, X. M., Nikolic-Paterson, D. J., and Lan, H. Y. (2014). Inflammatory processes in renal fibrosis. Nat. Rev. Nephrol. 10, 493–503. doi: 10.1038/nrneph.2014.114
Meng, X. M., Wang, S., Huang, X. R., Yang, C., Xiao, J., Zhang, Y., et al. (2016). Inflammatory macrophages can transdifferentiate into myofibroblasts during renal fibrosis. Cell Death Dis. 7:e2495. doi: 10.1038/cddis.2016.402
Miao, F., Gonzalo, I. G., Lanting, L., and Natarajan, R. (2004). In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J. Biol. Chem. 279, 18091–18097. doi: 10.1074/jbc.m311786200
Mikuda, N., Kolesnichenko, M., Beaudette, P., Popp, O., Uyar, B., Sun, W., et al. (2018). The IκB kinase complex is a regulator of mRNA stability. EMBO J. 37:e98658.
Moon, J. Y., Jeong, K. H., Lee, T. W., Ihm, C. G., Lim, S. J., and Lee, S. H. (2012). Aberrant recruitment and activation of T cells in diabetic nephropathy. Am. J. Nephrol. 35, 164–174. doi: 10.1159/000334928
Moriwaki, Y., Inokuchi, T., Yamamoto, A., Ka, T., Tsutsumi, Z., Takahashi, S., et al. (2007). Effect of TNF-α inhibition on urinary albumin excretion in experimental diabetic rats. Acta Diabetologica 44, 215–218. doi: 10.1007/s00592-007-0007-6
Muto, Y., Wilson, P., Wu, H., Waikar, S., and Humphreys, B. (2021). Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat. Commun. 12:2190. doi: 10.1038/s41467-021-22368-w
Navarro-González, J. F., Mora-Fernández, C., De Fuentes, M. M., Chahin, J., Méndez, M. L., Gallego, E., et al. (2015). Effect of pentoxifylline on renal function and urinary albumin excretion in patients with diabetic kidney disease: the PREDIAN trial. J. Am. Soc. Nephrol. 26, 220–229. doi: 10.1681/asn.2014010012
Newton, K., and Dixit, V. M. (2012). Signaling in innate immunity and inflammation. Cold Spring Harbor Perspect. Biol. 4:a006049.
O’shea, J. J., Kontzias, A., Yamaoka, K., Tanaka, Y., and Laurence, A. (2013). Janus kinase inhibitors in autoimmune diseases. Ann. Rheumatic Dis. 72:ii111. doi: 10.1136/annrheumdis-2012-202576
Park, J., Guan, Y., Sheng, X., Gluck, C., Seasock, M. J., Hakimi, A. A., et al. (2019). Functional methylome analysis of human diabetic kidney disease. JCI Insight 4:e128886.
Peng, F., Gong, W., Li, S., Yin, B., Zhao, C., Liu, W., et al. (2021). circRNA_010383 acts as a sponge for miR-135a, and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes 70, 603–615. doi: 10.2337/db20-0203
Peng, W. F., Huang, S., Shen, L. S., Tang, Y. B., Li, H. H., and Shi, Y. Q. (2019). Long noncoding RNA NONHSAG053901 promotes diabetic nephropathy via stimulating Egr-1/TGF-beta-mediated renal inflammation. J. Cell. Physiol. 234, 18492–18503. doi: 10.1002/jcp.28485
Pergola, P. E., Raskin, P., Toto, R. D., Meyer, C. J., Huff, J. W., Grossman, E. B., et al. (2011). Bardoxolone methyl and kidney function in CKD with type 2 diabetes. New Engl. J. Med. 365, 327–336.
Pina, T., Corrales, A., Lopez-Mejias, R., Armesto, S., Gonzalez-Lopez, M. A., Gómez-Acebo, I., et al. (2016). Anti-tumor necrosis factor-alpha therapy improves endothelial function and arterial stiffness in patients with moderate to severe psoriasis: a 6-month prospective study. J. Dermatol. 43, 1267–1272. doi: 10.1111/1346-8138.13398
Pradeepa, M. M., Grimes, G. R., Kumar, Y., Olley, G., Taylor, G. C., Schneider, R., et al. (2016). Histone H3 globular domain acetylation identifies a new class of enhancers. Nat. Genet. 48:681. doi: 10.1038/ng.3550
Reddy, M. A., Zhang, E., and Natarajan, R. (2015). Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia 58, 443–455. doi: 10.1007/s00125-014-3462-y
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Rovira-Llopis, S., Escribano-Lopez, I., Diaz-Morales, N., Iannantuoni, F., Lopez-Domenech, S., Andújar, I., et al. (2018). Downregulation of miR-31 in diabetic nephropathy and its relationship with inflammation. Cell. Physiol. Biochem. 50, 1005–1014. doi: 10.1159/000494485
Ruiz-Ortega, M., Rayego-Mateos, S., Lamas, S., Ortiz, A., and Rodrigues-Diez, R. R. (2020). Targeting the progression of chronic kidney disease. Nat. Rev. Nephrol. 16, 269–288.
Russo, V. E., Martienssen, R. A., and Riggs, A. D. (1996). Epigenetic Mechanisms of Gene Regulation. New York, NY: Cold Spring Harbor Laboratory Press.
Sanz, A. B., Ramos, A. M., Soler, M. J., Sanchez-Niño, M. D., Fernandez-Fernandez, B., Perez-Gomez, M. V., et al. (2019). Advances in understanding the role of angiotensin-regulated proteins in kidney diseases. Exp. Rev. Proteomics 16, 77–92. doi: 10.1080/14789450.2018.1545577
Saraheimo, M., Teppo, A.-M., Forsblom, C., Fagerudd, J., Groop, P.-H., and Group, F. S. (2003). Diabetic nephropathy is associated with low-grade inflammation in type 1 diabetic patients. Diabetologia 46, 1402–1407. doi: 10.1007/s00125-003-1194-5
Schena, F. P., and Gesualdo, L. (2005). Pathogenetic mechanisms of diabetic nephropathy. J. Am. Soc. Nephrol. 16, S30–S33.
Shahzad, K., Bock, F., Dong, W., Wang, H., Kopf, S., Kohli, S., et al. (2015). Nlrp3-inflammasome activation in non-myeloid-derived cells aggravates diabetic nephropathy. Kidney Int. 87, 74–84. doi: 10.1038/ki.2014.271
Shang, J., Zhang, Y., Jiang, Y., Li, Z., Duan, Y., Wang, L., et al. (2017). NOD2 promotes endothelial-to-mesenchymal transition of glomerular endothelial cells via MEK/ERK signaling pathway in diabetic nephropathy. Biochem. Biophys. Res. Commun. 484, 435–441. doi: 10.1016/j.bbrc.2017.01.155
Shao, X., Kong, W. X., and Li, Y. T. (2019). MiR-133 inhibits kidney injury in rats with diabetic nephropathy via MAPK/ERK pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 10957–10963.
Shao, Y., Lv, C., Wu, C., Zhou, Y., and Wang, Q. (2016). Mir-217 promotes inflammation and fibrosis in high glucose cultured rat glomerular mesangial cells via Sirt1/HIF-1α signaling pathway. Diabetes Metab. Res. Rev. 32, 534–543. doi: 10.1002/dmrr.2788
Shao, Y., Ren, H., Lv, C., Ma, X., Wu, C., and Wang, Q. (2017). Changes of serum Mir-217 and the correlation with the severity in type 2 diabetes patients with different stages of diabetic kidney disease. Endocrine 55, 130–138. doi: 10.1007/s12020-016-1069-4
Sheng, J., Wang, L., Tang, P. M., Wang, H. L., Li, J. C., Xu, B. H., et al. (2021). Smad3 deficiency promotes beta cell proliferation and function in db/db mice via restoring Pax6 expression. Theranostics 11, 2845–2859. doi: 10.7150/thno.51857
Song, J., Zhang, H., Sun, Y., Guo, R., Zhong, D., Xu, R., et al. (2018). Omentin-1 protects renal function of mice with type 2 diabetic nephropathy via regulating miR-27a-Nrf2/Keap1 axis. Biomed. Pharmacother. 107, 440–446. doi: 10.1016/j.biopha.2018.08.002
Storz, G. (2002). An expanding universe of noncoding RNAs. Science 296, 1260–1263. doi: 10.1126/science.1072249
Su, S.-S., Li, B.-P., Li, C.-L., Xiu, F.-R., Wang, D.-Y., and Zhang, F.-R. (2020). Downregulation of MiR-218 can alleviate high-glucose-induced renal proximal tubule injury by targeting GPRC5A. Biosci. Biotechnol. Biochem. 84, 1123–1130. doi: 10.1080/09168451.2020.1717330
Sun, H., Tian, J., Xian, W., Xie, T., and Yang, X. (2015). Pentraxin-3 attenuates renal damage in diabetic nephropathy by promoting M2 macrophage differentiation. Inflammation 38, 1739–1747. doi: 10.1007/s10753-015-0151-z
Sun, J., Wang, J., Lu, W., Xie, L., Lv, J., Li, H., et al. (2020a). MiR-325-3p inhibits renal inflammation and fibrosis by targeting CCL19 in diabetic nephropathy. Clin. Exp. Pharmacol. Physiol. 47, 1850–1860.
Sun, S. F., Tang, P. M., Feng, M., Xiao, J., Huang, X. R., Li, P., et al. (2018). Novel lncRNA Erbb4-IR promotes diabetic kidney injury in db/db mice by targeting miR-29b. Diabetes 67, 731–744. doi: 10.2337/db17-0816
Sun, T., Liu, Y., Liu, L., and Ma, F. (2020b). MicroRNA-544 attenuates diabetic renal injury via suppressing glomerulosclerosis and inflammation by targeting FASN. Gene 723:143986. doi: 10.1016/j.gene.2019.143986
Sun, Y., Peng, R., Peng, H., Liu, H., Wen, L., Wu, T., et al. (2016). miR-451 suppresses the NF-kappaB-mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. Mol. Cell. Endocrinol. 433, 75–86. doi: 10.1016/j.mce.2016.06.004
Tang, P. C.-T., Zhang, Y.-Y., Chan, M. K.-K., Lam, W. W.-Y., Chung, J. Y.-F., Kang, W., et al. (2020a). The emerging role of innate immunity in chronic kidney diseases. Int. J. Mol. Sci. 21:4018. doi: 10.3390/ijms21114018
Tang, P. M. K., Nikolic-Paterson, D. J., and Lan, H. Y. (2019). Macrophages: versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 15, 144–158. doi: 10.1038/s41581-019-0110-2
Tang, P. M. K., Zhang, Y. Y., Hung, J. S. C., Chung, J. Y. F., Huang, X. R., To, K. F., et al. (2021). DPP4/CD32b/NF-κB Circuit: a novel druggable target for inhibiting CRP-driven diabetic nephropathy. Mol. Ther. 29, 365–375. doi: 10.1016/j.ymthe.2020.08.017
Tang, P. M. K., Zhang, Y. Y., Mak, T. S. K., Tang, P. C. T., Huang, X. R., and Lan, H. Y. (2018a). Transforming growth factor−β signalling in renal fibrosis: from Smads to non-coding RNAs. J. Physiol. 596, 3493–3503. doi: 10.1113/jp274492
Tang, P. M. K., Zhang, Y. Y., Xiao, J., Tang, P. C. T., Chung, J. Y. F., Li, J., et al. (2020b). Neural transcription factor Pou4f1 promotes renal fibrosis via macrophage–myofibroblast transition. Proc. Natl. Acad. Sci. 117, 20741–20752. doi: 10.1073/pnas.1917663117
Tang, P. M. K., Zhou, S., Li, C. J., Liao, J., Xiao, J., Wang, Q. M., et al. (2018b). The proto-oncogene tyrosine protein kinase Src is essential for macrophage-myofibroblast transition during renal scarring. Kidney Int. 93, 173–187. doi: 10.1016/j.kint.2017.07.026
Tang, S. C. W., and Yiu, W. H. (2020). Innate immunity in diabetic kidney disease. Nat. Rev. Nephrol. 16, 206–222. doi: 10.1038/s41581-019-0234-4
Torres, Á, Muñoz, K., Nahuelpán, Y., R Saez, A.-P., Mendoza, P., Jara, C., et al. (2020). Intraglomerular monocyte/macrophage infiltration and macrophage–myofibroblast transition during diabetic nephropathy is regulated by the A2B adenosine receptor. Cells 9:1051. doi: 10.3390/cells9041051
Van Beneden, K., Geers, C., Pauwels, M., Mannaerts, I., Verbeelen, D., Van Grunsven, L. A., et al. (2011). Valproic acid attenuates proteinuria and kidney injury. J. Am. Soc. Nephrol. 22, 1863–1875. doi: 10.1681/asn.2010111196
VanderJagt, T. A., Neugebauer, M. H., Morgan, M., Bowden, D. W., and Shah, V. O. (2015). Epigenetic profiles of pre-diabetes transitioning to type 2 diabetes and nephropathy. World J. Diabetes 6:1113. doi: 10.4239/wjd.v6.i9.1113
Villeneuve, L. M., Kato, M., Reddy, M. A., Wang, M., Lanting, L., and Natarajan, R. (2010). Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes 59, 2904–2915. doi: 10.2337/db10-0208
Wada, J., and Makino, H. (2016). Innate immunity in diabetes and diabetic nephropathy. Nat. Rev. Nephrol. 12:13. doi: 10.1038/nrneph.2015.175
Wang, J., Shen, X., Liu, J., Chen, W., Wu, F., Wu, W., et al. (2020). High glucose mediates NLRP3 inflammasome activation via upregulation of ELF3 expression. Cell Death Dis. 11, 1–14.
Wang, L., and Li, H. (2020). MiR-770-5p facilitates podocyte apoptosis and inflammation in diabetic nephropathy by targeting TIMP3. Biosci. Rep. 40:BSR20193653.
Wang, Q. (2020). XIST silencing alleviated inflammation and mesangial cells proliferation in diabetic nephropathy by sponging miR-485. Arch. Physiol. Biochem. 15, 1–7. doi: 10.1080/13813455.2020.1789880
Wang, X., Liu, J., Zhen, J., Zhang, C., Wan, Q., Liu, G., et al. (2014). Histone deacetylase 4 selectively contributes to podocyte injury in diabetic nephropathy. Kidney Int. 86, 712–725. doi: 10.1038/ki.2014.111
Wang, X., Yao, B., Wang, Y., Fan, X., Wang, S., Niu, A., et al. (2017). Macrophage cyclooxygenase-2 protects against development of diabetic nephropathy. Diabetes 66, 494–504. doi: 10.2337/db16-0773
Webster, L., Abordo, E. A., Thornalley, P. J., and Limb, G. A. (1997). Induction of TNFα and IL-1β mRNA in monocytes by methylglyoxal-and advanced glycated endproduct-modified human serum albumin. Biochem. Soc. Trans. 25:250S. doi: 10.1042/bst025250s
Wen, S., Li, S., Li, L., and Fan, Q. (2020). circACTR2: a novel mechanism regulating high glucose-induced fibrosis in renal tubular cells via pyroptosis. Biol. Pharm. Bull. 43, 558–564. doi: 10.1248/bpb.b19-00901
Wilczynska, A., and Bushell, M. (2015). The complexity of miRNA-mediated repression. Cell Death Differ. 22, 22–33. doi: 10.1038/cdd.2014.112
Wilson, P. C., Wu, H. J., Kirita, Y., Uchimura, K., Ledru, N., Rennke, H. G., et al. (2019). The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc. Natl. Acad. Sci. U.S.A. 116, 19619–19625. doi: 10.1073/pnas.1908706116
Wu, C. H., Huang, C. M., Lin, C. H., Ho, Y. S., Chen, C. M., and Lee, H. M. (2002). Advanced glycosylation end products induce NF-κB dependent iNOS expression in RAW 264.7 cells. Mol. Cell. Endocrinol. 194, 9–17. doi: 10.1016/s0303-7207(02)00212-5
Wu, J., Lu, K., Zhu, M., Xie, X., Ding, Y., Shao, X., et al. (2020). miR-485 suppresses inflammation and proliferation of mesangial cells in an in vitro model of diabetic nephropathy by targeting NOX5. Biochem. Biophys. Res. Commun. 521, 984–990. doi: 10.1016/j.bbrc.2019.11.020
Xie, C., Wu, W., Tang, A., Luo, N., and Tan, Y. (2019). lncRNA GAS5/miR-452-5p reduces oxidative stress and pyroptosis of high-glucose-stimulated renal tubular cells. Diabetes Metab. Syndrome Obes. Targets Ther. 12:2609. doi: 10.2147/dmso.s228654
Xu, B. H., Sheng, J., You, Y. K., Huang, X. R., Ma, R. C. W., Wang, Q., et al. (2020a). Deletion of Smad3 prevents renal fibrosis and inflammation in type 2 diabetic nephropathy. Metabolism 103:154013. doi: 10.1016/j.metabol.2019.154013
Xu, J., Xiang, P., Liu, L., Sun, J., and Ye, S. (2020b). Metformin inhibits extracellular matrix accumulation, inflammation and proliferation of mesangial cells in diabetic nephropathy by regulating H19/miR-143-3p/TGF−β1 axis. J. Pharmacy Pharmacol. 72, 1101–1109. doi: 10.1111/jphp.13280
Yagi, S., Hirosawa, M., and Shiota, K. (2012). DNA methylation profile: a composer-, conductor-, and player-orchestrated mammalian genome consisting of genes and transposable genetic elements. J. Reproduct. Deve. 58, 265–273. doi: 10.1262/jrd.2011-030
Yang, C., Chen, X. C., Li, Z. H., Wu, H. L., Jing, K. P., Huang, X. R., et al. (2020). SMAD3 promotes autophagy dysregulation by triggering lysosome depletion in tubular epithelial cells in diabetic nephropathy. Autophagy Epub ahead of print,
Yang, L. (2015). Splicing noncoding RNAs from the inside out. Wiley Interdisciplinary Rev. RNA 6, 651–660. doi: 10.1002/wrna.1307
Yao, T., Zha, D., Gao, P., Shui, H., and Wu, X. (2019). MiR-874 alleviates renal injury and inflammatory response in diabetic nephropathy through targeting toll-like receptor-4. J. Cell. Physiol. 234, 871–879. doi: 10.1002/jcp.26908
Yao, T., Zha, D., Hu, C., and Wu, X. (2020). Circ_0000285 promotes podocyte injury through sponging miR-654-3p and activating MAPK6 in diabetic nephropathy. Gene 747:144661. doi: 10.1016/j.gene.2020.144661
Yi, H., Peng, R., Zhang, L. Y., Sun, Y., Peng, H. M., Liu, H. D., et al. (2017). LincRNA-Gm4419 knockdown ameliorates NF-kappa B/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis. 8:e2583. doi: 10.1038/cddis.2016.451
You, H., Gao, T., Cooper, T. K., Brian Reeves, W., and Awad, A. S. (2013). Macrophages directly mediate diabetic renal injury. Am. J. Physiol. R. Physiol. 305, F1719–F1727.
Yu, R., Zhang, Y., Lu, Z., Li, J., Shi, P., and Li, J. (2019). Long-chain non-coding RNA UCA1 inhibits renal tubular epithelial cell apoptosis by targeting microRNA-206 in diabetic nephropathy. Arch. Physiol. Biochem. Epub ahead of print,
Yu, Y., Jia, Y. Y., Wang, M., Mu, L., and Li, H. J. (2021). PTGER3 and MMP-2 play potential roles in diabetic nephropathy via competing endogenous RNA mechanisms. BMC Nephrol. 22:27.
Yuan, E. F., Yang, Y., Cheng, L., Deng, X., Chen, S. M., Zhou, X., et al. (2019). Hyperglycemia affects global 5-methylcytosine and 5-hydroxymethylcytosine in blood genomic DNA through upregulation of SIRT6 and TETs. Clin. Epigenet. 11, 1–9.
Zha, F., Qu, X., Tang, B., Li, J., Wang, Y., Zheng, P., et al. (2019). Long non-coding RNA MEG3 promotes fibrosis and inflammatory response in diabetic nephropathy via miR-181a/Egr-1/TLR4 axis. Aging (Albany NY) 11:3716. doi: 10.18632/aging.102011
Zhan, J. F., Huang, H. W., Huang, C., Hu, L. L., and Xu, W. W. (2020). Long non-coding RNA NEAT1 regulates pyroptosis in diabetic nephropathy via mediating the miR-34c/NLRP3 Axis. Kidney Blood Pres. Res. 45, 589–602. doi: 10.1159/000508372
Zhang, C., Xiao, C., Wang, P., Xu, W., Zhang, A., Li, Q., et al. (2014). The alteration of Th1/Th2/Th17/Treg paradigm in patients with type 2 diabetes mellitus: Relationship with diabetic nephropathy. Hum. Immunol. 75, 289–296. doi: 10.1016/j.humimm.2014.02.007
Zhang, H., Nair, V., Saha, J., Atkins, K. B., Hodgin, J. B., Saunders, T. L., et al. (2017). Podocyte-specific JAK2 overexpression worsens diabetic kidney disease in mice. Kidney Int. 92, 909–921. doi: 10.1016/j.kint.2017.03.027
Zhang, M., Zhao, S., Xu, C., Shen, Y., Huang, J., Shen, S., et al. (2020a). Ablation of lncRNA MIAT mitigates high glucose-stimulated inflammation and apoptosis of podocyte via miR-130a-3p/TLR4 signaling axis. Biochem. Biophys. Res. Commun. 533, 429–436. doi: 10.1016/j.bbrc.2020.09.034
Zhang, P., Sun, Y., Peng, R., Chen, W., Fu, X., Zhang, L., et al. (2019a). Long non-coding RNA Rpph1 promotes inflammation and proliferation of mesangial cells in diabetic nephropathy via an interaction with Gal-3. Cell Death Dis. 10, 1–16.
Zhang, R., Qin, L., and Shi, J. (2020b). MicroRNA-199a-3p suppresses high glucose-induced apoptosis and inflammation by regulating the IKKβ/NF−κB signaling pathway in renal tubular epithelial cells. Int. J. Mol. Med. 46, 2161–2171. doi: 10.3892/ijmm.2020.4751
Zhang, Y. L., Wang, J. M., Yin, H., Wang, S. B., He, C. L., and Liu, J. (2020c). DACH1, a novel target of miR-218, participates in the regulation of cell viability, apoptosis, inflammatory response, and epithelial-mesenchymal transition process in renal tubule cells treated by high-glucose. REN Fail 42, 463–473. doi: 10.1080/0886022x.2020.1762647
Zhang, Y. Y., Tang, P. M., Niu, Y., Córdoba, G. A., Alexandra, C., Huang, X. R., et al. (2020d). Long non-coding RNA LRNA9884 promotes acute kidney injury via regulating NF-kB-mediated transcriptional activation of MIF. Front. Physiol. 11:1399. doi: 10.3389/fphys.2020.590027
Zhang, Y. Y., Tang, P. M. K., Tang, P. C. T., Xiao, J., Huang, X. R., Yu, C., et al. (2019b). LRNA9884, a novel smad3-dependent long noncoding RNA, promotes diabetic kidney injury in db/db mice via enhancing MCP-1–dependent renal inflammation. Diabetes 68, 1485–1498. doi: 10.2337/db18-1075
Zhong, X., Chung, A. C. K., Chen, H., Dong, Y., Meng, X., Li, R., et al. (2013). miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia 56, 663–674. doi: 10.1007/s00125-012-2804-x
Zhong, X., Zhang, L., Li, Y., Li, P., Li, J., and Cheng, G. (2018). Kaempferol alleviates ox-LDL-induced apoptosis by up-regulation of miR-26a-5p via inhibiting TLR4/NF-κB pathway in human endothelial cells. Biomedicine Pharmacother. 108, 1783–1789. doi: 10.1016/j.biopha.2018.09.175
Zhou, H., Ni, W.-J., Meng, X.-M., and Tang, L.-Q. (2021). MicroRNAs as regulators of immune and inflammatory responses: potential therapeutic targets in diabetic nephropathy. Front. Cell Dev. Biol. 8:1837. doi: 10.3389/fcell.2020.618536
Zhou, L., Xu, D. Y., Sha, W. G., Shen, L., Lu, G. Y., Yin, X., et al. (2015). High glucose induces renal tubular epithelial injury via Sirt1/NF-kappaB/microR-29/Keap1 signal pathway. J. Trans. Med. 13, 1–12.
Keywords: diabetic nephropathy, epigenetics, DNA methylation, histone modifications, non-coding RNAs, inflammation
Citation: Shao B-Y, Zhang S-F, Li H-D, Meng X-M and Chen H-Y (2021) Epigenetics and Inflammation in Diabetic Nephropathy. Front. Physiol. 12:649587. doi: 10.3389/fphys.2021.649587
Received: 05 January 2021; Accepted: 12 April 2021;
Published: 05 May 2021.
Edited by:
Matthew A. Bailey, University of Edinburgh, United KingdomReviewed by:
Laura Denby, University of Edinburgh, United KingdomPatrick Ming-Kuen Tang, The Chinese University of Hong Kong, China
Marpadga Reddy, Beckman Research Institute, City of Hope, United States
Copyright © 2021 Shao, Zhang, Li, Meng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Ming Meng, bWVuZ3hpYW9taW5nQGFobXUuZWR1LmNu; Hai-Yong Chen, aGFpeW9uZ0Boa3UuaGs=
†These authors have contributed equally to this work
 Bao-Yi Shao
Bao-Yi Shao Shao-Fei Zhang2†
Shao-Fei Zhang2† Xiao-Ming Meng
Xiao-Ming Meng Hai-Yong Chen
Hai-Yong Chen