- 1CAS Key Laboratory of Tropical Marine Bio-Resources and Ecology, Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Southern Marine Science and Engineering Guangdong Laboratory, Guangzhou, China
- 4Sansha Marine and Fisheries Bureau, Sansha, China
The giant triton snail (Charonia tritonis), an endangered gastropod species of ecological and economic importance, is widely distributed in coral reef ecosystems of the Indo-West Pacific region and the tropical waters of the South China Sea. Limited research on molecular mechanisms can be conducted because the complete genomic information on this species is unavailable. Hence, we performed transcriptome sequencing of the C. tritonis foot muscle and mantle using the Illumina HiSeq sequencing platform. In 109,722 unigenes, we detected 7,994 (3,196 up-regulated and 4,798 down-regulated) differentially expressed genes (DEGs) from the C. tritonis foot muscle and mantle transcriptomes. These DEGs will provide valuable resources to improve the understanding of molecular mechanisms involved in biomineralization of C. tritonis. In the Gene Ontology (GO) database, DEGs were clustered into three main categories (biological processes, molecular functions, and cellular components) and were involved in 50 functional subcategories. The top 20 GO terms in the molecular function category included sulfotransferase activity, transferring sulfur-containing groups, and calcium ion binding, which are terms considered to be related to biomineralization. In KEGG classifications, transcriptomic DEGs were mainly enriched in glycosaminoglycan biosynthesis-chondroitin sulfate/dermatan sulfate, and sulfur metabolism pathway, which may be related to biomineralization. The results of qPCR showed that three of the eight genes examined were significantly up-regulated in the mantle. The phylogenetic tree of BMP1 suggested a significant divergence between homologous genes in C. tritonis. Our results improve the understanding of biomineralization in C. tritonis and provide fundamental transcriptome information to study other molecular mechanisms such as reproduction.
Introduction
The giant triton snail (Charonia tritonis) is an ecologically and commercially important species that belongs to Mollusca, Gastropoda, Ranellidae, and mainly inhabits in the sandy bottom of coral reefs in the Indo-West Pacific region, as well as the tropical waters of the South China Sea (Beu, 1970; Aungtonya, 1994). C. tritonis preys on echinoderms especially the crown-of-thorns starfish (COTS, Acanthaster planci), which is considered to have importantly potential outcome on the biodiversity of coral reefs and their biological communities (Morton, 2012; Doxa et al., 2013a,b; Kang et al., 2016). As this mollusk is highly valued for its decorative shell, overfishing has driven this organism near to extinction in most of its area of habitats (Zhang et al., 2013). Despite the important ecological value of C. tritonis, basic information on this organism is very limited (Hall et al., 2017).
Some preliminary studies of C. tritonis have been proceeded in connection with the growth, reproduction processes and egg-laying patterns, as well as analysis in development and morphological of early life history stages, feeding behavior in captivity, and the important to limit population outbreaks of COTS in environments (Zhang et al., 2013). The next-generation transcriptome sequencing and in silico protein analysis of salivary glands were firstly reported by Bose et al. (2017b), and identified numerous secreted proteins including putative toxin- and feeding- related protein families (Bose et al., 2017a). And a cerebral ganglia transcriptome of C. tritonis were sequenced using Illumina HiSeq 2500 sequencing platform, with an emphasis on the bioinformatic analysis of neuropeptide genes with mollusk reproduction (Bose et al., 2017b). However, to date, no studies on biomineralization genes and pathways have been reported in C. tritonis. Mollusk shells are an exoskeleton mainly constituted of a large set of calcium carbonate and organic macromolecules, including proteins, glycoproteins, polysaccharides and sometimes lipids, which are secreted by mantle tissue during the biomineralization process. Numerous biomineralizing genes were found to be responsible for shell formation in various molluscan species, such as genome and multi-omic analysis of biomineralization in Pinctada fucata martensii, genomic databases and proteomic in-depth analysis of shell matrix construction-related genes in the limpet Lottia gigantea, transcriptome analysis for potential biomineralization-related genes in Tectus pyramis. The next-generation sequencing technology has been significantly applied to non-model organisms because it is efficient and cost-effective, which provided an efficient way to identify putatively functional genes and explain molecular mechanisms involved in the processes of biomineralization in mollusks (Moreira et al., 2015; Shi et al., 2018, 2019).
In this study, we first performed de novo transcriptome sequencing of the foot muscle and mantle tissues of C. tritonis using the Illumina sequence platform, which will provide transcriptome resources for this mollusk. Additionally, the comparison with foot muscle and mantle tissue transcription in C. tritonis allows to investigate and identify the biomineralization-related genes and pathway at molecular level, so as to provide further insight into molecular mechanisms involved in biomineralization for mollusk.
Materials and Methods
Sample Collection
The adult C. tritonis were obtained from Nansha archipelagic waters of the South China Sea. Tissue samples (foot muscle and mantle) were separated and quickly frozen in liquid nitrogen for 24 h, and then stored at −80°C until total RNA extraction.
Total RNA Extraction, Illumina Sequencing, and Library Construction
Total RNA was isolated from each sample of C. tritonis using a Mollusk RNA kit (Omega, Atlanta, GA, United States) according to the manufacturer’s instructions. RNA integrity was detected by 1% agarose gel and the purity of the RNA was identified by using NanoDrop (Thermo Fisher Scientific, Waltham, MA, United States). Then, the total RNA concentration, the RNA integrity number (RIN), 28S/18S, and the fragment length distribution were measured using Agilent 2100 Bioanalyzer and Agilent RNA 6000 Nano Kit, (Agilent Technologies, CA, United States). The sample mRNA was enriched from total RNA using the oligo (dT) method and fragmented using a suitable reagent under high temperature. First-strand cDNA was synthesized using fragmented mRNA as a template. The reaction system was then configured to synthesize two-strand cDNA. The cDNA fragments were purified using purification kits and resolved with elution buffer for end reparation and single nucleotide A (adenine) addition. The cDNA fragments were connected to adapters and those of a suitable size were selected as templates for PCR amplification. The quality of the library constructed with PCR products was assessed by an Agilent 2100 Technologies Bioanalyzer (Santa Clara, CA, United States) and the ABI StepOnePlus Real-Time PCR System (Foster City, CA, United States). High-throughput sequencing was performed using Illumina HiSeq 2000 Technology at BGI Shenzhen (Shenzhen, China).
De novo Assembly and Functional Annotation
Clean reads were obtained from raw reads by removing adaptor-polluted reads, reads with unknown bases (N) at more than 5%, and low-quality reads. High-quality clean reads were de novo assembled to transcripts using Trinity software (v2.0.6). Transcripts were clustered to unigenes with Tgicl software (v2.0.6). Unigenes obtained from each sample were clustered again using Tgicl, and the final unigenes for downstream analysis were named “All-Unigenes.” Unigenes were aligned to the NCBI non-redundant protein (NR), NCBI nucleotide (NT), Clusters euKaryotic Ortholog Groups (KOG), Kyoto Encyclopedia of Genes and Genomes (KEGG), and SwissProt databases to conduct functional annotation using BLAST1 (E-value ≤ 1e–5). InterProScan5 (v5.11-51.0) was used to obtain InterPro annotations. Unigenes with NR annotations were further analyzed with Blast2GO (v2.5.0) to obtain their Gene Ontology (GO) annotations.
Analysis of Functional Enrichment of Differentially Expressed Genes
The clean reads of each sample were mapped to unigenes with Bowtie2 software (v2.2.5) and the gene expression levels were calculated using RSEM (v1.2.12). Fragments per kilobase of transcript per million (FPKM) mapped reads were used to evaluate the expression level of unigenes based on read counts and transcript lengths. Differentially expressed genes (DEGs) were detected by the PossionDis algorithm based on comparing the FPKM mapped reads between samples with thresholds of | log2FC| ≥ 2 and false discovery rate (FDR) ≤ 0.001. All identified DEGs were mapped to GO and KEGG annotations to classify. GO and KEGG enrichment analyses were performed with the phyper package in R. The p-value was calculated in a hypergeometric test and then the p-value results were adjusted based on the FDR. p < 0.05 was considered significant (∗), and p < 0.01 was considered highly significant (∗∗).
Screening of Transcripts Involved in Biomineralization
Based on the functional annotation of DEGs in the mantle transcriptome of C. tritonis and the representative biomineralization genes in the mantle of mollusks published in the database, a total of 24 putative biomineralization-related transcripts were screened by searching BLASTx.
Validation of DEGs by Quantitative Real-Time PCR
Quantitative real-time RT-PCR (qRT-PCR) was performed using 24 DEGs annotated as biomineralization-related genes in the mantle and foot muscle tissues from three C. tritonis. Tissues were ground in a homogenizer (IKA, Staufen, Germany). Total RNA was extracted from the foot muscles and mantles of mature C. tritonis (three individuals from each tissue) using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States), after which the quality was checked as described above and then a ReverTra Ace qPCR RT master mix was used with a gDNA Remover kit (Toyobo, Japan) to synthesize cDNA template. The primers were designed by the software Primer Premier version 6.02. Primers for amplification of target genes and the 18SrRNA internal reference gene are listed in Supplementary Table 1. The qRT-PCR analysis was conducted by using a Roche LightCycler 480 Real-time PCR System (Roche, Switzerland) with SYBR Green Real-time PCR Mix (Toyobo, Japan) according to the manufacturer’s protocols and using the following reaction conditions: 95°C for 10 s, followed by 50 cycles of 95°C for 10 s, 57°C for 15 s, and 72°C for 15 s. At the end of the reaction, a melting curve was generated to confirm an accurate amplification of the target and the reference gene 18S amplicon. Crossing point (Cp) was recorded to calculate the relative expression of target genes using the 2–ΔΔCt transformation. All results are expressed as the mean ± SD. The SPSS 23.0 (SPSS, Chicago, IL, United States) was used for statistical analysis. A p-value of <0.05 indicated a significant difference and p < 0.01 indicated a highly significant difference.
Results
Illumina Sequencing and de novo Assembly
A total of 13.23 Gb raw sequencing data were generated through the Illumina HiSeq 2000 platform. After filtering, 44.07 M clean reads with a total of 6.61 Gb clean bases were acquired with 92.83% Q30 bases from C. tritonis foot muscle, and the 44.12 M clean reads with a total of 6.62 Gb clean bases were acquired with 92.79% Q30 bases for C. tritonis mantle (Table 1).
Based on high-quality clean reads, 183,823 transcripts were assembled with an average length of 419 bp from the C. tritonis foot muscle data, and 168,326 transcripts were assembled with an average length of 430 bp from the C. tritonis mantle data. After clustering, 87,443 unigenes with a mean length of 419 bp and an N50 of 476 bp were generated from the foot muscle. A total of 81,100 unigenes were assembled from the mantle, with a mean length of 597 bp and N50 of 843 bp. The GC percentage of the unigenes was 43.26% in the foot muscle and 43.78% in the mantle. The unigenes of these two tissues were clustered using Tgicl and the final unigenes for downstream analyses were named “All-Unigenes.” The length distribution of All-Unigenes is shown in (Supplementary Figure 1). The above results show that the quality of transcriptome sequencing and assembly was credible to subsequent analysis.
Functional Annotation
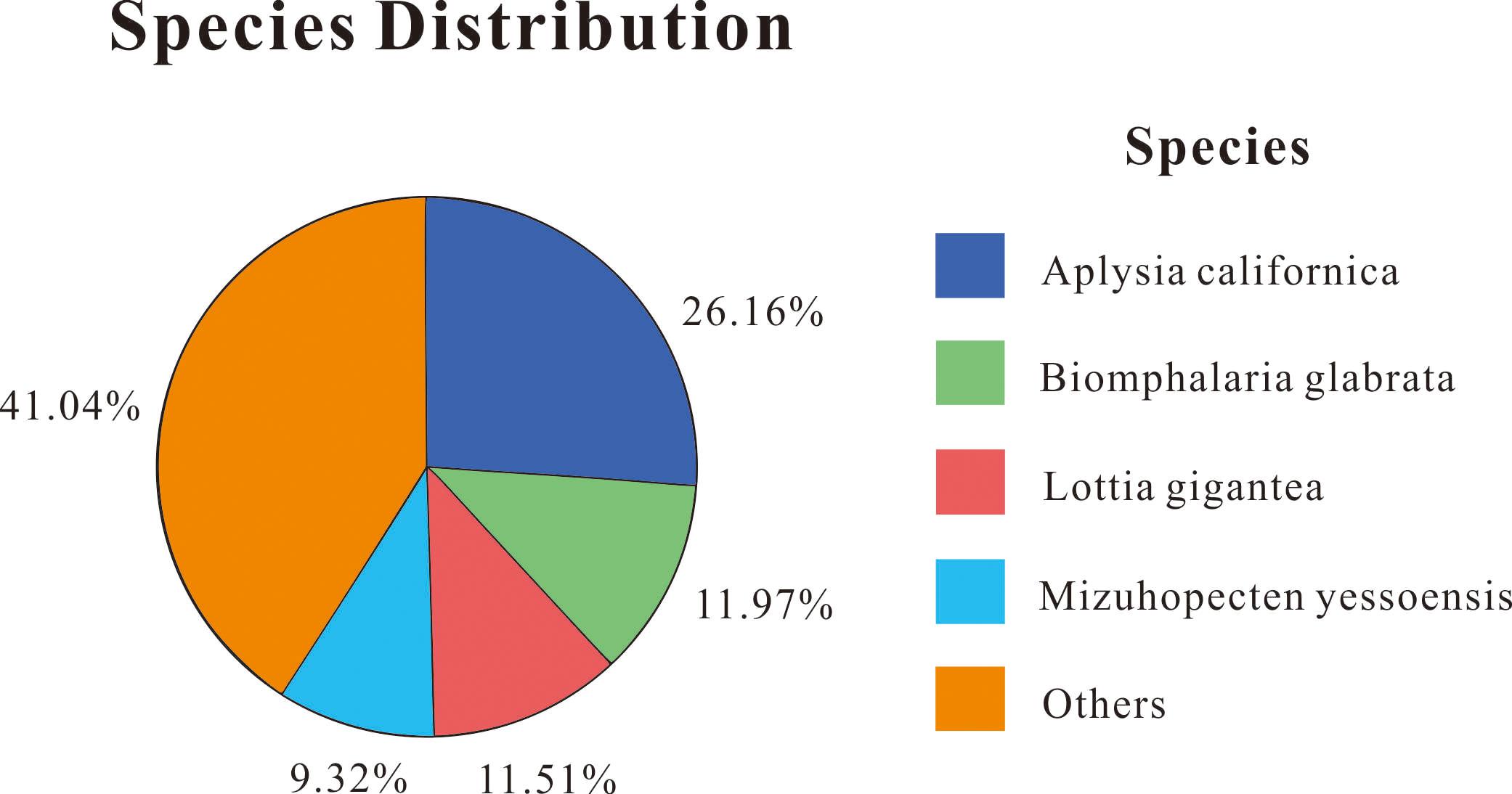
Out of 109,722 unigenes, 46,239 (42.14%) obtained annotations in at least one public database, which included NR, NT, SwissProt, KEGG, KOG, InterPro, and GO, using BLAST (E-value ≤ 1 × 10–5). The most unigenes were annotated in the NT database (27,059, 24.66%), followed by NR (26,050, 23.74%), KEGG (18,705, 17.05%), InterPro (17,617, 16.06%), Swiss-Prot (16,944, 15.44%), KOG (15,319, 13.96%), and GO (9,788, 8.92%). Based on the NR database function annotation results, the best match of the species distribution for each sequence is shown in Figure 1. Among these annotated species, the largest number of unigenes matched with Aplysia californica (26.16%), followed by Biomphalaria glabrata (11.97%), Lottia gigantean (11.51%), and Mizuhopecten yessoensis (9.32%). In addition, 41.04% unigene sequences could not be matched to known genes (Figure 1).
According to GO annotation, 9,788 unigenes were categorized into 58 GO terms in the biological process, cellular component, and molecular function ontologies. In the biological process category, most of the unigenes were clustered into 26 classifications. The categories that contained the most annotated genes were “cellular process” (3,682, 27.88%), “metabolic process” (2,613, 19.79%), and “biological regulation” (1,358, 10.28%). The most represented categories among the cellular component category were “cell” (3,371, 18.03%), “cell part” (3,314, 17.72%), and “membrane” (3,313, 17.72%). In the molecular function category, the matched sequences were clustered into 12 classifications, and a majority of unigenes were divided into three subcategories “binding” (4,669, 45%), “catalytic activity” (3,830, 36.89%), and “transporter activity” (565, 5.44%). A total of 15,319 unigenes were divided into 25 functional classifications in the KOG database. The categories with the highest proportion of unigenes were “general function prediction only” (3,528, 23.03%) and “signal transduction mechanisms” (3,273, 21.37%). There were 18,705 unigenes significantly matched in the KEGG database and were assigned to 336 KEGG pathways involving six categories (cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, and organismal systems). Among these pathways, the most enriched unigenes were “metabolic pathways” (2,516, 13.45%; ko01100), “pathways in cancer” (993, 5.31%; ko05200), and the “calcium signaling pathway” (782, 4.18%; ko04020). The KEGG classifications of unigenes are listed in Supplementary Figure 2.
Functional Annotation of DEGs
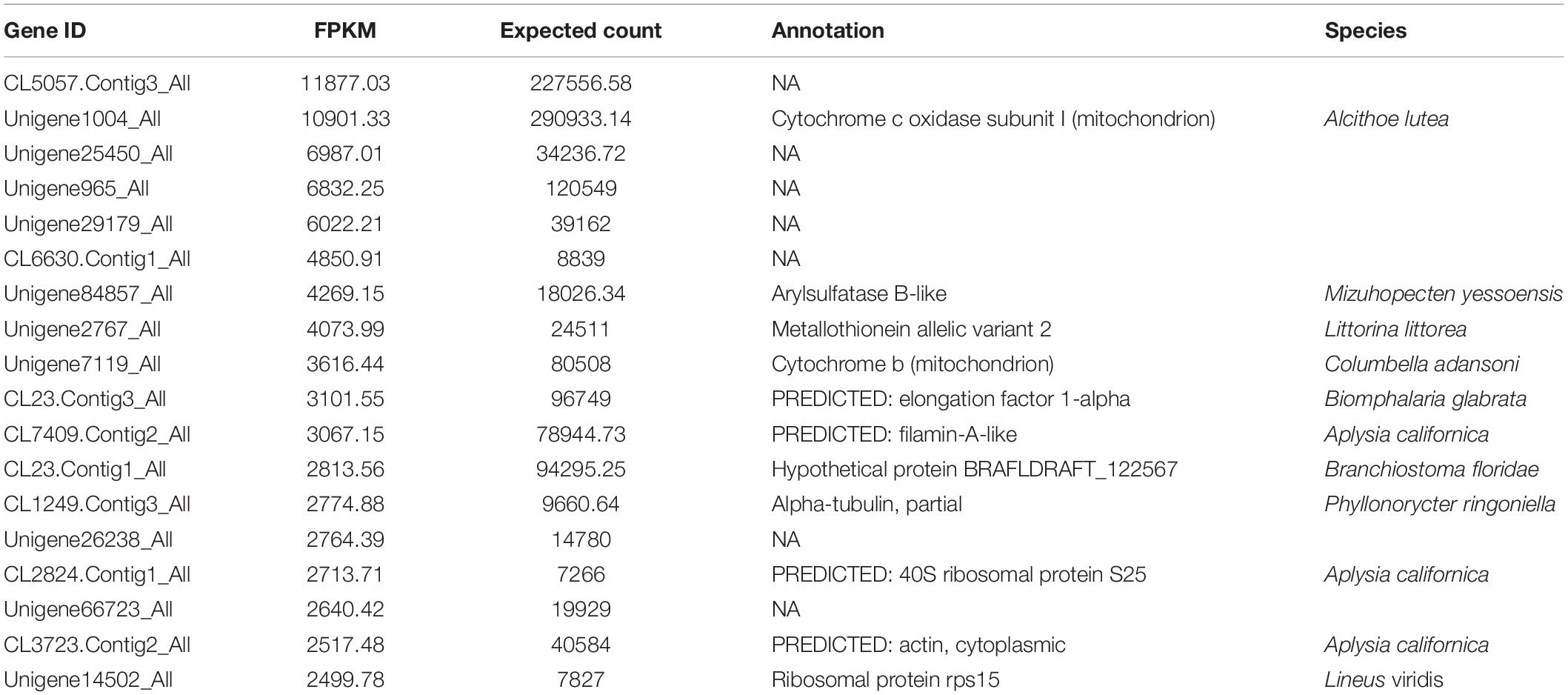
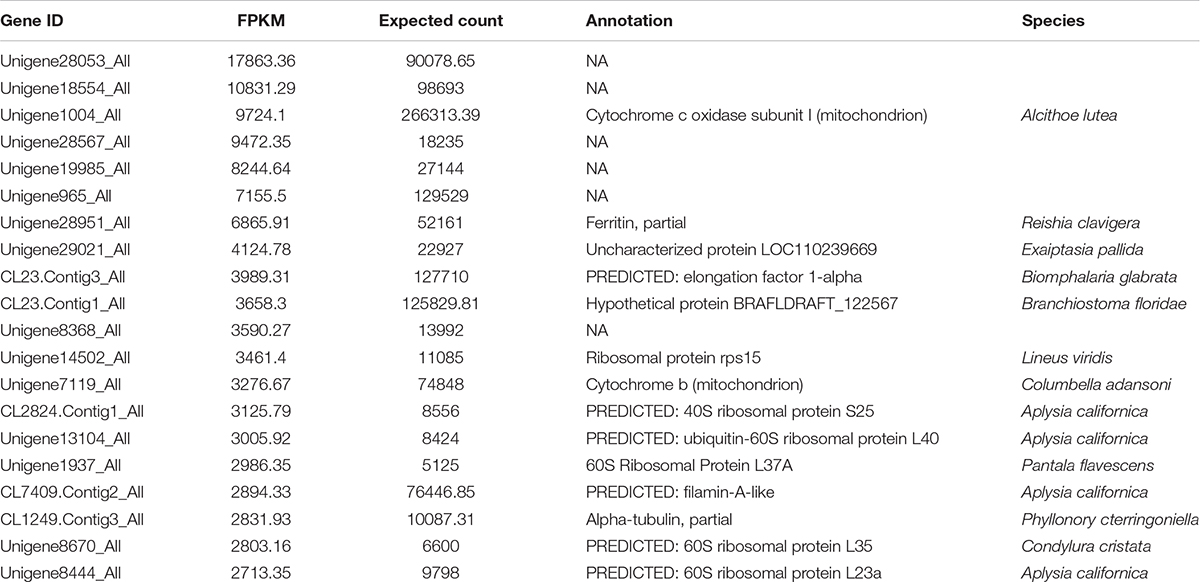
Based on sequence similarities, a total of 44,353 unigenes were found to be conserved between the foot muscle and mantle transcriptomes (Figure 2A); 19,502 unigenes for foot muscle and 16,239 unigenes for mantle, specifically. Some of the differentially expressed unigenes may be responsible for the unique features of each of these species. A total of 7,994 DEGs (3,196 up-regulated and 4,798 down-regulated) were detected in C. tritonis foot muscle compared with mantle tissue (Figure 2B). The top 20 most highly expressed genes of the foot muscle and mantle are similar, as shown in Tables 2A,B, respectively. It is noteworthy that highly expressed unique sequences in the mantle were annotated as ferritin, which was probably involved in biomineralization.
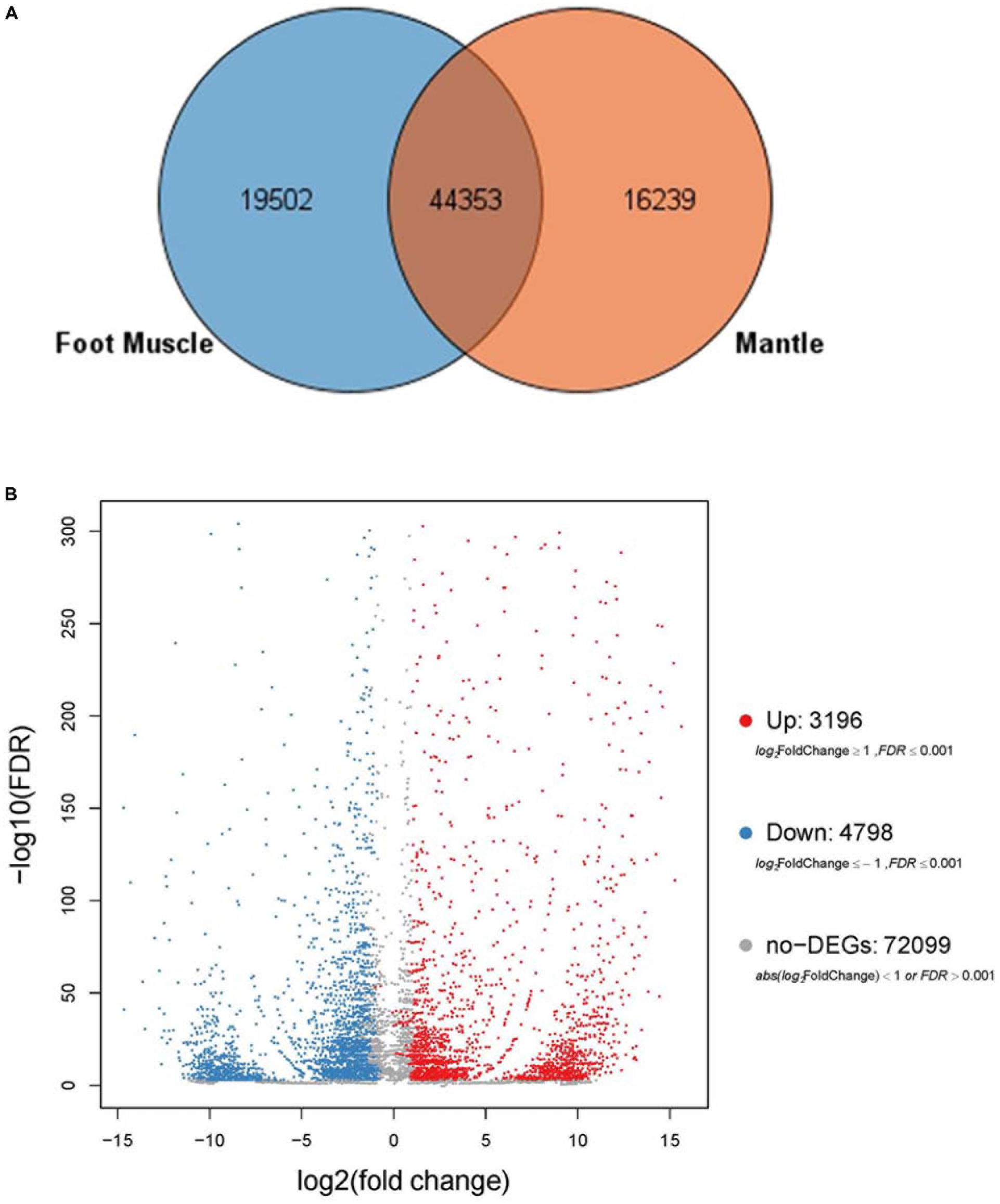
Figure 2. (A) Venn diagrams for number of comparisons of transcripts among foot muscle and mantle tissue. (B) Volcano plot of differentially expressed genes (DEGs) in foot muscle and mantle from C. tritonis.
Based on the gene expression level, GO enrichment analysis was performed to understand the functions of the exclusive DEGs in tissues and to search significantly enriched GO terms. All the DEGs were divided into three clusters categories (i.e., molecular biological function, cellular component, and biological process.). The vast majority of genes were involved in binding (623, 43.44%) and catalytic activity functions (522, 36.40%) in the molecular function category. In the cellular component class, most of the genes in the two tissues were related to the membrane (544, 22.25%), membrane part (501, 20.50%), and cell (378, 15.46%). In the biological process class, most genes were categorized as being related to cellular processes (423, 27.67%), metabolic processes (321, 21%), and biological regulation (155, 10.14%) (Supplementary Table 2). To investigate this further, GO-term enrichment was performed to gain insight into the potential functions of orthogroups shared exclusively between the two tissues using DEGs (Up-regulated and down-regulated genes) separately. The main functional classes of the listed up-regulated genes were consistent with the functional classification from down-regulated genes in GO categories (Figures 3A,B). Although similar compositions were observed for both tissues in the three major categories using GO assignments, there were some categories of biological processes that were differentially expressed between foot muscle and mantle, including GO terms linked to cell killing, detoxification, and molecular carrier activity. This finding indicates that some of the genes may be related to tissue-specific roles.
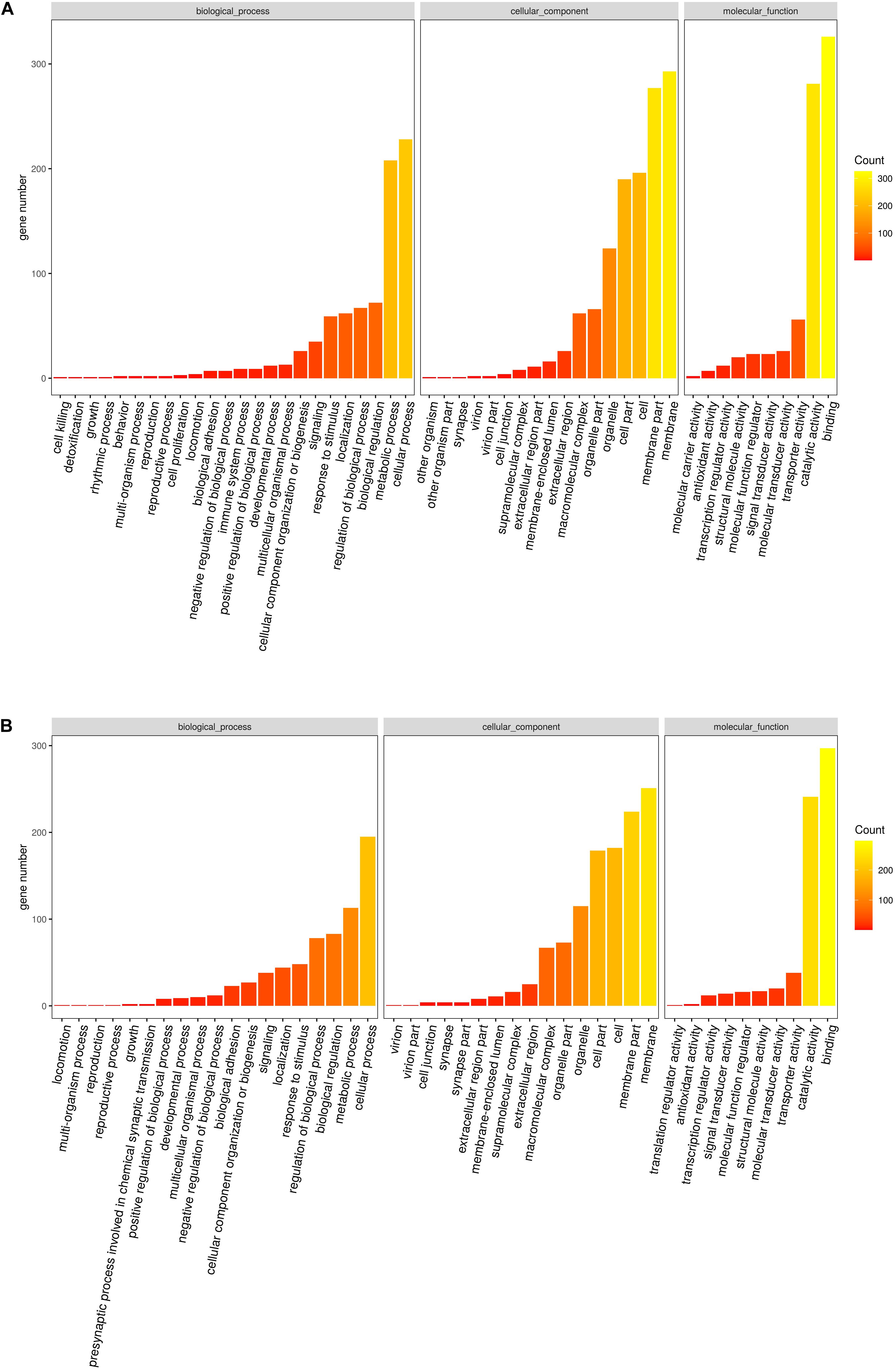
Figure 3. (A) Statistically common enriched Gene Ontology (GO) terms between foot muscle and mantle tissues for the up-regulated unigenes. (B) Statistically common enriched Gene Ontology (GO) terms between foot muscle and mantle tissues for the down-regulated unigenes.
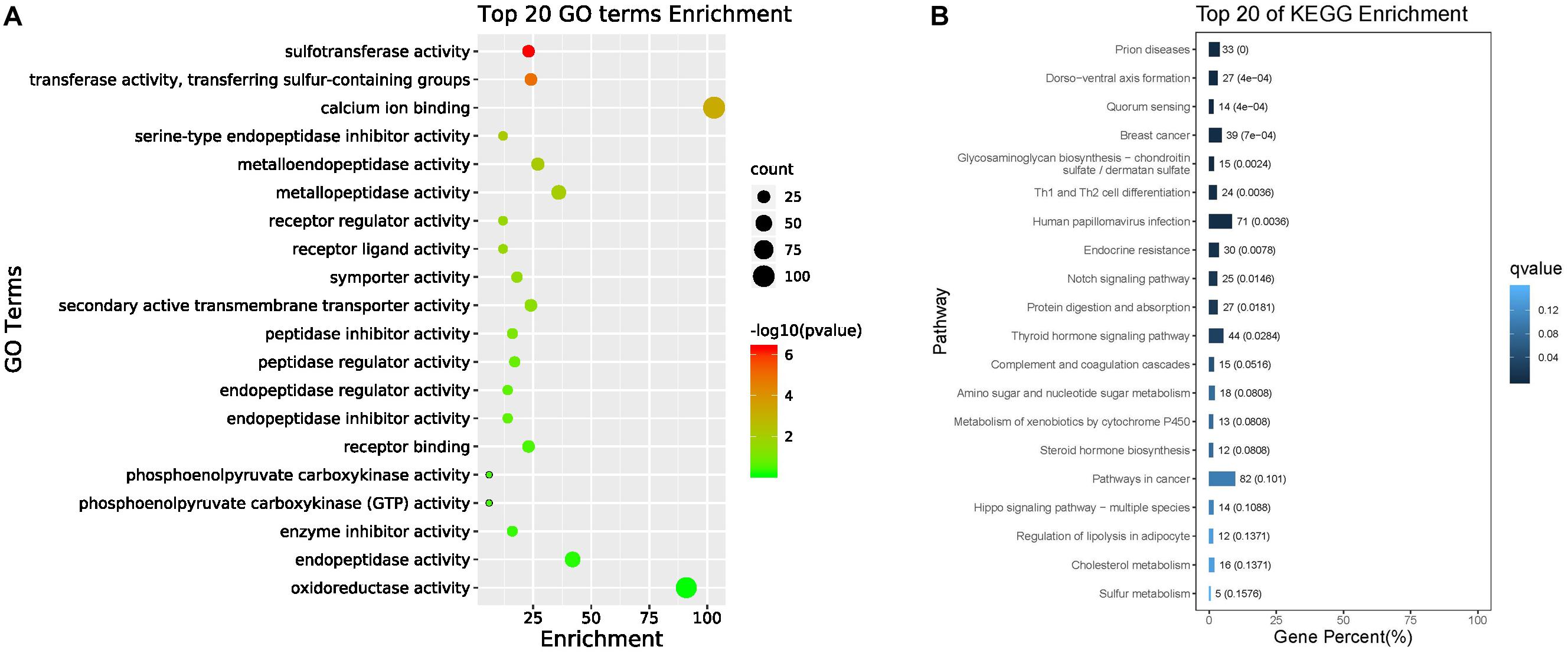
The top 20 enriched GO terms related to DEGs in the molecular function category are listed in Figure 4A. The results revealed that some DEGs that were significantly enriched (FDR ≤ 0.01) in GO terms, including sulfotransferase activity, transferase activity, transferring sulfur-containing groups, and calcium ion binding, may be correlated with biomineralization. The up-regulated unigenes in the mantle were again mapped to reference pathways in the KEGG database based on function. KEGG pathway analysis indicated that DEGs significantly enriched in glycosaminoglycan biosynthesis-chondroitin sulfate/dermatan sulfate (ko00532, p = 0.000034), and sulfur metabolism (ko00920, p = 0.009006) may be related to biomineralization (Figure 4B).
qPCR Validation of Genes Related to Biomineralization
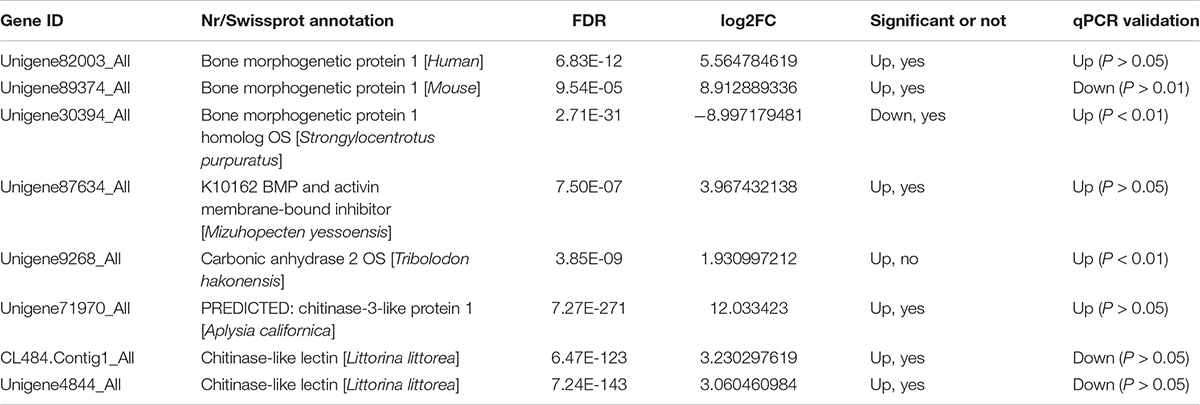
Twenty-four candidate sequences encoding members of the bone morphogenetic family, carbonic anhydrase, and chitinase were found in the transcriptome of C. tritonis as potentially involved in biomineralization. The biomineralization-related genes were examined by qPCR in the foot muscle and mantle (Table 3). Overall, bone morphogenetic protein 1 (Unigene82003_All), bone morphogenetic protein 1 homolog OS (Unigene30394_All), and carbonic anhydrase 2 OS (Unigene9268_All) were highly expressed in mantle tissue, whereas bone morphogenetic protein 1 (Unigene89374_All) was extremely significantly down-regulated in mantle according to qRT-PCR results, but a higher expression was found in transcriptome sequencing. Chitinase-3-like protein 1 (Unigene71970_All) was up-regulated in the mantle, while the other Chitinase-like lectin (CL484.Contig1_All, Unigene4844_All) showed lower expression in the mantle according to transcriptome sequencing qRT-PCR (Figure 5).
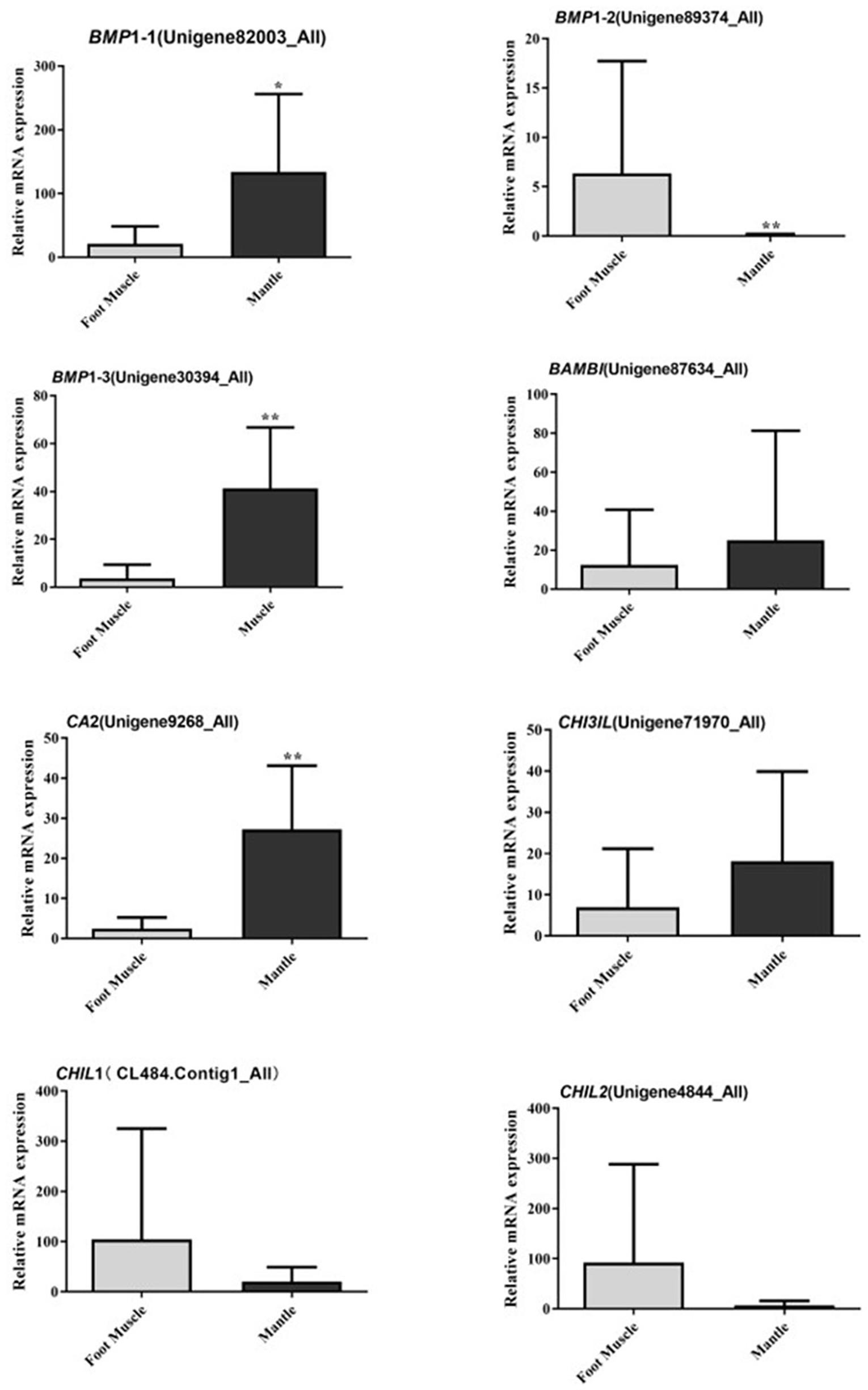
Figure 5. Relative mRNA expression profiles of eight selected biomineralization-related genes from mantle and foot muscle tissues of C. tritonis. [A p-value < 0.05 was considered significant (*), and p < 0.01 was considered highly significant (**)].
Phylogenetic Tree of Bone Morphogenetic Protein Gene Families
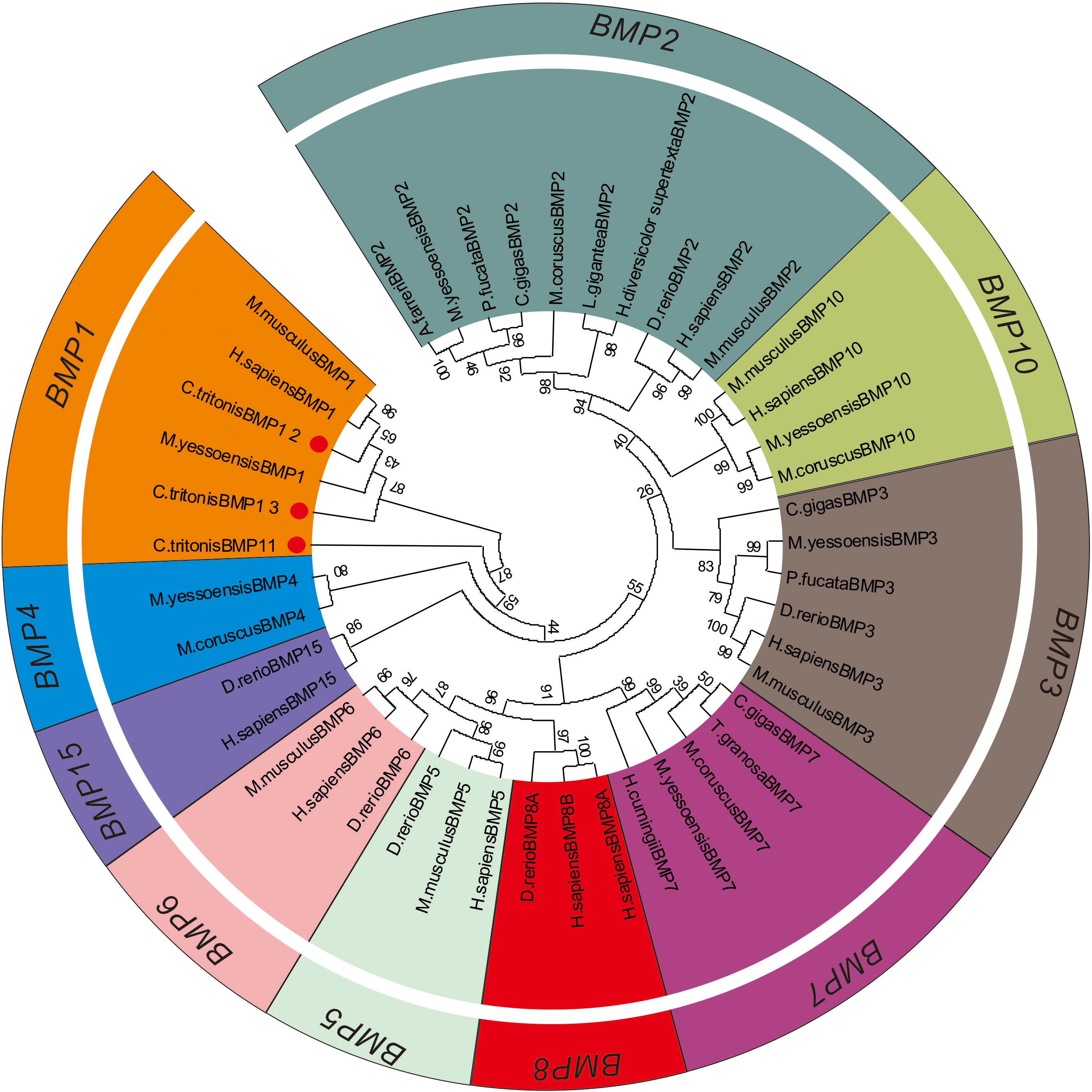
A phylogenetic tree was constructed to identify the evolutionary history of C. tritonis bone morphogenetic proteins (BMP) based on deduced amino acid sequences of BMP from C. tritonis and other species (Supplementary Table 3). The results showed that three sequences of C. tritonis BMP (BMP1-1, BMP1-2, and BMP1-3) and BMP1 from other species were clustered into one branch, implying that C. tritonis BMP may be a category of the BMP1 subfamily. At the same time, BMP1-1 (Unigene82003_All) had a close genetic relationship with BMP1-3 (Unigene30394_All), but not with BMP1-2 (Unigene89374_All) in C. tritonis (Figure 6).
Discussion
In this study, the comparative transcriptome of C. tritonis foot muscle and mantle were generated and assembled using the Illumina HiSeq sequencing platform. A total of 13.23 Gb bases were assembled into 109,722 unigenes (mean length = 640 bp), of which 46,239 were annotated, including 3,196 annotated significantly up-regulated genes and 4,798 significantly down-regulated genes in mantle [| log2(fold change)| ≥ 2 and FDR < 0.001]. Two reasons may explain the low-annotation rate: (1) At present, there is no research report on the genome sequencing of C. tritonis, and there is a lack of available C. tritonis transcriptome information in public databases. (2) The accuracy of next-generation sequencing technology greatly depends on the reference genome (Joubert et al., 2010; Kim et al., 2017; Lavelle et al., 2018).
According to the results of the GO terms enrichment analysis, in the molecular function ontology, most DEGs were enriched in sulfotransferase activity, transferase activity, transferring sulfur-containing groups, and calcium ion binding pathways. It is suggested that sulfotransferase catalyzes the transfer of sulfonic acid groups, which can affect the synthesis of chondroitin sulfate and then regulate biomineralization (Furuhashi et al., 2009; Sun et al., 2015; Zhang et al., 2020). KEGG enrichment analysis of the DEGs revealed major involvement in glycosaminoglycan biosynthesis-chondroitin sulfate/dermatan sulfate (ko00532), which performs biomineralization (Shi et al., 2013; Moreira et al., 2015; Li et al., 2017; Mao et al., 2018). Despite reports of many biomineralization-related genes in gastropods, very little is known about the basis of biomineralization in C. tritonis (Bootsma et al., 1988; Liu et al., 2011). Hence, we analyzed all of the unigenes known to be annotated with biomineralization.
Bone morphogenetic proteins are multi-functional growth factors that belong to the members of the transforming growth factor-beta (TGFβ) superfamily (Steiglitz et al., 2004; Cao and Chen, 2005; Boraldi et al., 2019). As is well known, BMPs are ubiquitous in a broad range of species and fulfill a variety of metabolic roles including osteogenesis. In the BMP family, BMP1 is a metalloprotease that plays an important role in the synthesis of collagen, the core substance of bones, by cleaving and activating lysyl oxidase (LOX) in the extracellular matrix, which promotes collagen cross-linking (Ge et al., 2006; Kokabu et al., 2012). In previous experiments, the knockout of BMP1 resulted in osteogenesis imperfecta in mice (Steiglitz et al., 2004; Maruhashi et al., 2010; Muir et al., 2014; Zhang et al., 2017). A phylogenetic analysis of BMP sequences between C. tritonis and other species was performed, and C. tritonis BMP1 were grouped in BMP1 clusters with other species, which suggested that C. tritonis BMP1 may belong to the BMP1 subfamily. At the same time, our data indicated that C. tritonis BMP1 genes displayed diverse expression patterns in different tissues, BMP1-1 (Unigene82003_All) and BMP1-3 (Unigene30394_All) genes were significantly up-regulated in the mantle compared with the foot muscle, indicating that BMPs play important roles in the shell formation. While BMP1-2 (Unigene89374_All) had a high expression in foot muscle tissue, suggesting a significant divergence between homologous genes in C. tritonis. The BMPs amino acid sequences clustering analysis of C. tritonis versus other species. A phylogenetic analysis revealed that C. tritonis BMP, in this study, created two separate clades: BMP1-1 had a close genetic relationship with BMP1-3, but not with BMP1-2, suggesting that the functions of these genes differentiated during evolution.
Carbonic anhydrase (CA) is believed to play crucial physiological roles in the biomineralization of mollusks due to their catalytic activity (Song et al., 2014; Le Roy et al., 2016; Cardoso et al., 2019; Zebral et al., 2019). In our analysis, CA was significantly up-regulated in the mantle, indicated that CA might participate in the mineralization of C. tritonis. This is the first report of BMPs and CA in C. tritonis, thus further study on BMPs and CA function needs to be undertaken in C. tritonis. Chitinase plays an important role in the metabolism of chitin, which is the major organic matrix protein of the shell, suggesting that chitinase may be involved in the mineralization of mollusks (Proespraiwong et al., 2010; Okada et al., 2013). Unlike in other mollusks, chitinase-related genes were not expressed at a higher level in the mantle. This distinct expression pattern suggested that chitin may play diverse roles in the foot muscle of C. tritonis. In previous studies, it has been revealed that ferritin, a transport protein ubiquitous in many organisms, was expressed exclusively in some tissues, especially the radula of marine mollusks, such as Acanthopleura hirtosa, Patella laticostata, and Mytilus edulis, and was considered to be potentially involved in biomineralization (Saunders et al., 2009; Narayanan et al., 2020). Ferritin was also found exclusively expressed in the mantle of C. tritonis, suggesting that it plays a similar role in biomineralization in C. tritonis. To our knowledge, this is the first time that biomineralization-related genes have been reported in C. tritonis. Overall, our analyses provide further insight into the biomineralization of C. tritonis.
This study is the first to represent transcriptomes of mantle and foot muscle tissues from C. tritonis, which provides transcriptomic resources for future genetic or genomic studies on this mollusk. Transcriptome comparison assists in better understanding of the molecular mechanisms of biomineralization in C. tritonis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA695322).
Author Contributions
GZ: experiment, writing, and editing. WL: reviewing. HZ: supervision. MH: project administration and approval. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA13020206), the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0402), Sansha Marine and Fisheries Bureau and the Science and Technology Planning Project of Guangdong Province, China, and Guangdong Provincial Key Laboratory of Applied Marine Biology (2020B1212060058).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.632518/full#supplementary-material
Footnotes
References
Aungtonya, A. N. C. (1994). The indo-pacific trumpet triton snail, Charonia tritonis L.: morphometrics of a species on the verge of local extinction. Phuket Mar. Biol. Cent. Spec. Publ. 13, 137–140.
Beu, A. G. (1970). The Mollusca of the Genus Charonia (Family Cymatiidae). Trans. R. Soc. N.Z. 11, 205–223.
Bootsma, N., Macey, D. J., Webb, J., and Talbot, V. (1988). Isolation and characterization of ferritin from the hepatopancreas of the mussel Mytilus edulis. Biol. Metals 1, 106–111. doi: 10.1007/bf01138068
Boraldi, F., Lofaro, F. D., Accorsi, A., Ross, E., and Malagoli, D. (2019). Toward the molecular deciphering of Pomacea canaliculata immunity: first proteomic analysis of circulating hemocytes. Proteomics 19:e1800314.
Bose, U., Suwansa-Ard, S., Maikaeo, L., Motti, C. A., Hall, M. R., and Cummins, S. F. (2017a). Neuropeptides encoded within a neural transcriptome of the giant triton snail Charonia tritonis, a crown-of-thorns starfish predator. Peptides 98, 3–14. doi: 10.1016/j.peptides.2017.01.004
Bose, U., Wang, T., Zhao, M., Motti, C. A., Hall, M. R., and Cummins, S. F. (2017b). Multiomics analysis of the giant triton snail salivary gland, a crown-of-thorns starfish predator. Sci. Rep. 7:6000.
Cao, X., and Chen, D. (2005). The BMP signaling and in vivo bone formation. Gene 357, 1–8. doi: 10.1016/j.gene.2005.06.017
Cardoso, J. C. R., Ferreira, V., Zhang, X., Anjos, L., Felix, R. C., Batista, F. M., et al. (2019). Evolution and diversity of alpha-carbonic anhydrases in the mantle of the Mediterranean mussel (Mytilus galloprovincialis). Sci. Rep. 9:10400.
Doxa, C. K., Divanach, P., and Kentouri, M. (2013a). Consumption rates and digestibility of four food items by the marine gastropod Charonia seguenzae (Aradas & Benoit, 1870). J. Exp. Mar. Biol. Ecol. 446, 10–16. doi: 10.1016/j.jembe.2013.04.019
Doxa, C. K., Kentouri, M., and Divanach, P. (2013b). Feeding of Charonia seguenzae (Aradas & Benoit, 1870) on natural prey and alternative foods. J. Molluscan Stud. 79, 76–78. doi: 10.1093/mollus/eys029
Furuhashi, T., Schwarzinger, C., Miksik, I., Smrz, M., and Beran, A. (2009). Molluscan shell evolution with review of shell calcification hypothesis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 154, 351–371. doi: 10.1016/j.cbpb.2009.07.011
Ge, G., Zhang, Y., Steiglitz, B. M., and Greenspan, D. S. (2006). Mammalian tolloid-like 1 binds procollagen C-proteinase enhancer protein 1 and differs from bone morphogenetic protein 1 in the functional roles of homologous protein domains. J. Biol. Chem. 281, 10786–10798. doi: 10.1074/jbc.m511111200
Hall, M. R., Motti, C., and Kroon, F. (2017). The Potential Role of the Giant Triton Snail, Charonia tritonis (Gastropoda: Ranellidae) in Mitigating Populations of the Crown-of-Thorns Starfish. Integrated Pest Management of Crown-of-Thorns Starfish. Report to the National Environmental Science Programme. Cairns: Reef and Rainforest Research Centre Limited.
Joubert, C., Piquemal, D., Marie, B., Manchon, L., Pierrat, F., Zanella-Cleon, I., et al. (2010). Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: focus on biomineralization. BMC Genomics 11:613. doi: 10.1186/1471-2164-11-613
Kang, K., Kim, M., Hong, H., Cha, G., and Sui, Z. (2016). Feeding broodstocks different starfish diets affect growth and survival of larvae of trumpet shell (Charonia lampas sauliae Reeve 1844). J. Ocean Univ. China 15, 861–865. doi: 10.1007/s11802-016-3040-7
Kim, M. A., Rhee, J. S., Kim, T. H., Lee, J. S., Choi, A. Y., Choi, B. S., et al. (2017). Alternative splicing profile and sex-preferential gene expression in the female and male pacific abalone haliotis discus hannai. Genes (Basel) 8:99. doi: 10.3390/genes8030099
Kokabu, S., Gamer, L., Cox, K., Lowery, J., Tsuji, K., Raz, R., et al. (2012). BMP3 suppresses osteoblast differentiation of bone marrow stromal cells via interaction with Acvr2b. Mol. Endocrinol. 26, 87–94. doi: 10.1210/me.2011-1168
Lavelle, C., Smith, L. C., Bisesi, J. H. Jr., Yu, F., Silva-Sanchez, C., Moraga-Amador, D., et al. (2018). Tissue-based mapping of the fathead minnow (Pimephales promelas) transcriptome and proteome. Front Endocrinol. (Lausanne) 9:611. doi: 10.3389/fendo.2018.00611
Le Roy, N., Jackson, D. J., Marie, B., Ramos-Silva, P., and Marin, F. (2016). Carbonic anhydrase and metazoan biocalcification: a focus on molluscs. Key Eng. Mater. 672, 151–157. doi: 10.4028/www.scientific.net/kem.672.151
Li, H., Zhang, B., Fan, S., Liu, B., Su, J., and Yu, D. (2017). Identification and differential expression of biomineralization genes in the mantle of pearl Oyster Pinctada fucata. Mar. Biotechnol. (NY) 19, 266–276.
Liu, C. L., Liang, L. K., Chen, P., Liu, C. S., and Chen, X. G. (2011). Ferritin location and iron biomineralization paths on the cusp of the major lateral tooth of chiton acanthochiton rubrolineatus. Acta Hydrobiologica Sinica 1788, 911–917.
Mao, J., Zhang, W., Zhang, X., Tian, Y., Wang, X., Hao, Z., et al. (2018). Transcriptional changes in the Japanese scallop (Mizuhopecten yessoensis) shellinfested by Polydora provide insights into the molecular mechanism of shell formation and immunomodulation. Sci. Rep. 8:17664.
Maruhashi, T., Kii, I., Saito, M., and Kudo, A. (2010). Interaction between periostin and BMP-1 promotes proteolytic activation of lysyl oxidase. J. Biol. Chem. 285, 13294–13303. doi: 10.1074/jbc.m109.088864
Moreira, R., Pereiro, P., Canchaya, C., Posada, D., Figueras, A., and Novoa, B. (2015). RNA-Seq in Mytilus galloprovincialis: comparative transcriptomics and expression profiles among different tissues. BMC Genomics 16:728. doi: 10.1186/s12864-015-1817-5
Morton, B. (2012). Foregut anatomy and predation by Charonia lampas (Gastropoda: Prosobranchia: Neotaenioglossa) attacking Ophidiaster ophidianus (Asteroidea: Ophidiasteridae) in the açores, with a review of triton feeding behaviour. J. Nat. Hist. 46, 2621–2637. doi: 10.1080/00222933.2012.724721
Muir, A. M., Ren, Y., Butz, D. H., Davis, N. A., Blank, R. D., Birk, D. E., et al. (2014). Induced ablation of Bmp1 and Tll1 produces osteogenesis imperfecta in mice. Hum. Mol. Genet. 23, 3085–3101. doi: 10.1093/hmg/ddu013
Narayanan, S., Shahbazian-Yassar, R., and Shokuhfar, T. (2020). In situ visualization of ferritin biomineralization via graphene liquid cell-transmission electron microscopy. ACS Biomater. Sci. Eng. 6, 3208–3216. doi: 10.1021/acsbiomaterials.9b01889
Okada, Y., Yamaura, K., Suzuki, T., Itoh, N., Osada, M., and Takahashi, K. G. (2013). Molecular characterization and expression analysis of chitinase from the Pacific oyster Crassostrea gigas. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 165, 83–89. doi: 10.1016/j.cbpb.2013.03.008
Proespraiwong, P., Tassanakajon, A., and Rimphanitchayakit, V. (2010). Chitinases from the black tiger shrimp Penaeus monodon: phylogenetics, expression and activities. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 156, 86–96. doi: 10.1016/j.cbpb.2010.02.007
Saunders, M., Kong, C., Shaw, J. A., Macey, D. J., and Clode, P. L. (2009). Characterization of biominerals in the radula teeth of the chiton, Acanthopleura hirtosa. J. Struct. Biol. 167, 55–61. doi: 10.1016/j.jsb.2009.03.003
Shi, Y., Liu, W., and He, M. (2018). Proteome and transcriptome analysis of ovary, intersex gonads, and testis reveals potential key sex reversal/differentiation genes and mechanism in Scallop Chlamys nobilis. Mar. Biotechnol. (NY) 20, 220–245. doi: 10.1007/s10126-018-9800-1
Shi, Y., Xu, M., Huang, J., Zhang, H., Liu, W., Ou, Z., et al. (2019). Transcriptome analysis of mantle tissues reveals potential biomineralization-related genes in Tectus pyramis Born. Comp. Biochem. Physiol. Part D Genomics Proteomics 29, 131–144. doi: 10.1016/j.cbd.2018.11.010
Shi, Y., Yu, C., Gu, Z., Zhan, X., and Wang, Y. (2013). Characterization of the pearl oyster (Pinctada martensii) Mantle transcriptome unravels biomineralization genes. Mar. Biotechnol. 15, 175–187. doi: 10.1007/s10126-012-9476-x
Song, X., Wang, X., Li, L., and Zhang, G. (2014). Identification two novel nacrein-like proteins involved in the shell formation of the Pacific oyster Crassostrea gigas. Mol. Biol. Rep. 41, 4273–4278. doi: 10.1007/s11033-014-3298-z
Steiglitz, B. M., Ayala, M., Narayanan, K., George, A., and Greenspan, D. S. (2004). Bone morphogenetic protein-1/Tolloid-like proteinases process dentin matrix protein-1. J. Biol. Chem. 279, 980–986. doi: 10.1074/jbc.m310179200
Sun, X., Yang, A., Wu, B., Zhou, L., and Liu, Z. (2015). Characterization of the mantle transcriptome of yesso scallop (Patinopecten yessoensis): identification of genes potentially involved in biomineralization and pigmentation. PLoS One 10:e0122967. doi: 10.1371/journal.pone.0122967
Zebral, Y. D., Fonseca, J., Marques, J. A., and Bianchini, A. (2019). Carbonic Anhydrase as a biomarker of global and local impacts: insights from calcifying animals. Int. J. Mol. Sci. 20:3092. doi: 10.3390/ijms20123092
Zhang, H., Jani, P., Liang, T., Lu, Y., and Qin, C. (2017). Inactivation of bone morphogenetic protein 1 (Bmp1) and tolloid-like 1 (Tll1) in cells expressing type I collagen leads to dental and periodontal defects in mice. J. Mol. Histol. 48, 83–98. doi: 10.1007/s10735-016-9708-x
Zhang, H., Yi, X., Guan, Y., Shi, Y., Ou, Z., and He, M. (2020). The role of the chondroitin sulfate synthase-1 gene in the immune response of the pearl oyster Pinctada fucata. Fish. Sci. 86, 487–494. doi: 10.1007/s12562-020-01420-6
Keywords: biomineralization, transcriptome, Charonia tritonis, foot muscle, mantle tissue
Citation: Zhang G, Xu M, Zhang C, Jia H, Zhang H, He M and Liu W (2021) Comparative Transcriptomic and Expression Profiles Between the Foot Muscle and Mantle Tissues in the Giant Triton Snail Charonia tritonis. Front. Physiol. 12:632518. doi: 10.3389/fphys.2021.632518
Received: 26 November 2020; Accepted: 03 February 2021;
Published: 24 February 2021.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Hong Yu, Ocean University of China, ChinaLinbao Zhang, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (CAFS), China
Qiang Chen, University of Macau, China
Copyright © 2021 Zhang, Xu, Zhang, Jia, Zhang, He and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenguang Liu, bHdnQHNjc2lvLmFjLmNu
 Gege Zhang
Gege Zhang Meng Xu1,2,3
Meng Xu1,2,3 Wenguang Liu
Wenguang Liu