
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 25 January 2021
Sec. Invertebrate Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fphys.2020.618983
This article is part of the Research TopicInsect Olfactory Proteins (From Gene Identification to Functional Characterization), Volume IIView all 20 articles
A correction has been applied to this article in:
Corrigendum: Identification and functional characterization of two putative pheromone receptors in the potato tuber moth, Phthorimaea operculella
 Xiaoli He1
Xiaoli He1 Yajie Cai1
Yajie Cai1 Jinglei Zhu1,2
Jinglei Zhu1,2 Mengdi Zhang2
Mengdi Zhang2 Yadong Zhang1
Yadong Zhang1 Yang Ge1
Yang Ge1 Zengrong Zhu1
Zengrong Zhu1 Wenwu Zhou1*
Wenwu Zhou1* Guirong Wang2*
Guirong Wang2* Yulin Gao2*
Yulin Gao2*Pheromones are a kind of signal produced by an animal that evoke innate responses in conspecifics. In moth, pheromone components can be detected by specialized olfactory receptor neurons (OSNs) housed in long sensilla trichoids on the male antennae. The pheromone receptors (PRs) located in the dendrite membrane of OSNs are responsible for pheromone sensing in most Lepidopteran insects. The potato tuber moth Phthorimaea operculella is a destructive pest of Solanaceae crops. Although sex attractant is widely used in fields to monitor the population of P. operculella, no study has been reported on the mechanism the male moth of P. operculella uses to recognize sex pheromone components. In the present study, we cloned two pheromone receptor genes PopeOR1 and PopeOR3 in P. operculella. The transcripts of them were highly accumulated in the antennae of male adults. Functional analysis using the heterologous expression system of Xenopus oocyte demonstrated that these two PR proteins both responded to (E, Z)-4,7–13: OAc and (E, Z, Z)-4,7,10–13: OAc, the key sex pheromone components of P. operculella, whilst they responded differentially to these two ligands. Our findings for the first time characterized the function of pheromone receptors in gelechiid moth and could promote the olfactory based pest management of P. operculella in the field.
The potato tuber moth, Phthorimaea operculella (Lepidoptera: Gelechiidae), is one of the main pests affecting potatoes around the world (Rondon, 2020). It reduces potato production either via mining and damaging leaves in fields or via burrowing and destroying tubers in storage. The sex pheromone glands of P. operculella females produce a blend of odors that can influence the behavior of males. Two major pheromone components trans-4, cis-7-tridecadienyl acetate ((E, Z)-4,7–13: OAc) and tran-4, cis-7, cis-10-tridecatrienyl acetate ((E, Z, Z)-4,7,10–13: OAc) have been identified from this blend (Roelofs et al., 1975; Persoons et al., 1976). The structures of these components are quite similar, with the only difference in their double bond numbers. In field trapping studies, P. operculella males flew to lure sources containing either single or mixed compounds, while (E, Z, Z)-4,7,10–13: OAc attracted more male moths than (E, Z)-4,7–13: OAc, indicating the differentiated recognition of these components in male antennae. Moreover, when both compounds were present the number of moths attracted also increased significantly. While pheromone-based technologies have been widely used as part of sustainable control strategies, it remains unclear how these two pheromones are detected by P. operculella.
Insects use pheromones for mate recognition, and these chemical cues are mainly perceived by olfactory sensory neurons (OSNs), which are housed in olfactory sensilla on the antenna (Laurent, 1999; Steinbrecht, 1999). Pheromone detection is mediated by the heteromeric ligand-gated ion channels in the cell membrane of OSN dendrites. These channels are formed by the combination of specific pheromone receptors (PRs) and the olfactory co-receptor (Orco; Sato et al., 2008). Previous studies indicated that PRs belong to the olfactory receptor (OR) family, and they are thought to constitute a monophyletic clade in the phylogeny of insect OR for a long time (Liu et al., 2018b). However, recent studies found that except classical clade, there are multiple PR clades that have evolved independently (Yuvaraj et al., 2018). For example, Bastin-Heline et al. (2019) found SlitOR5, a PR from the cotton leafworm Spodoptera littoralis, grouped with a novel PR clade. With a typical structure of seven transmembrane domains, PRs are more abundantly expressed in male antennae (He et al., 2014; Zhang and Löfstedt, 2015; Ke et al., 2017), whilst some PRs are also found in female antennae and other tissues such as the wings and ovipositor (Chang et al., 2015; Li et al., 2020).
In Lepidopteran insects, PR was first functionally characterized in Bombyx mori using the heterologous expression system of Xenopus oocyte (Sakura et al., 2004), after that more than 60 PRs have been studied in over 30 moth species across 10 families by using different systems, such as in Xenopus oocytes (Wang et al., 2011), HEK293 cells (Forstner et al., 2009), transgenic Drosophila (Montagné et al., 2012), and more recently, in specific insects via CRISPR/Cas techniques (An et al., 2020). These PRs are predominately identified from moths causing crop damages, such as Manduca sexta (Große, 2010), Spodoptera exigua (Liu et al., 2013a), Spodoptera litura (Zhang et al., 2015a), Plutella xylostella (Sun et al., 2013; Liu et al., 2018b), Ostinia furnacalis (Liu et al., 2018a), Athetis lepigone (Zhang et al., 2018), Sesamia inferens (Zhang et al., 2015b), and Athetis dissimilis (Liu et al., 2019; Guo et al., 2020). Based on these studies, PRs were further grouped into three types: (1) narrowly tuned to a single component of the sex pheromones; (2) tuned to one component with high specificity, but still sensitive to other components at higher doses; and (3) broadly tuned to a wide range of pheromone components (Liu et al., 2018b).
Sex pheromone components have been characterized in P. operculella and other gelechiid insects and widely used in the management of pests, while the molecular mechanism of sex pheromone component perception is still largely unknown (Persoons et al., 1976). In the present study, we cloned two pheromone receptor genes PopeOR1 and PoprOR3 in P. operculella. Their transcripts were highly accumulated in the antennae of male adults. Their expression patterns were measured in different tissues of male and female adults by using the quantitative real-time PCR (qPCR). The function of these two candidate PRs in the detection of the key pheromones (E, Z)-4,7–13: OAc and (E, Z, Z)-4,7,10–13: OAc were further characterized using the heterologous expression system of Xenopus oocyte. Our results provide insights into the mechanism of pheromone perception in a gelechiid insect.
Phthorimaea operculella used for this study were collected from a suburban area in Yunnan Province in 2017 and the larvae were reared on potatoes at 26 ± 1°C on a 16:8 h (light/dark) photoperiod cycle and 70 ± 5% relative humidity at the Institute of Plant Protection, Chinese Academy of Agricultural Science (Beijing, China). The adults were fed with a 10% honey solution for three generations.
Total RNA was isolated from tissues using Trizol Reagent (Invitrogen) according to the manufacturer’s protocol. Total RNA was dissolved in RNase-free water and gel electrophoresis was used to verify its quality. The concentration of RNA was determined by NanoDrop-2000 (Thermo Scientific, Waltham, MA, USA). cDNA was synthesized from 1 μg of total RNA using a RevertAid Frist Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania). The cDNA products were stored at −20°C until use.
Two candidate PR genes, PopeOR1 and PopeOR3, and the Orco gene (PopeOrco) were identified from the transcriptome of P. operculella (unpublished data). Their full length cDNAs were cloned with specific primers (Supplementary Table 1) designed by primer5.0 (PREMIER Biosoft International, CA, United States). The open-reading frames (ORFs) of these three genes were predicted using the ORF Finder.1 The PCR reaction was performed in a 50 μl system containing 25 μl of TransStartPfu PCR SuperMix (TransGen Biotech, Beijing, China), 22 μl of ddH2O, 1 μl of cDNA template, and 1 μl of forward and reverse primers (10 μM). The PCR conditions were: 95°C for 2 min; 35 cycles of 98°C for 10 s, 55°C for 30 s, 72°C for 1.4 min; 72°C for 10 min. The PCR products were verified on a 1.2% agarose gel, and the band was recovered and purified by an AxyPrep™ DNA gel extraction kit (YMbio, Beijing, China). Purified fragments were cloned in the pEASY®-Blunt3 cloning vector (TransGen Biotech, Beijing, China) and then transformed into Trans5α chemically competent cells (TransGen Biotech, Beijing, China). The transformants were incubated on LB-Agar plates containing 100 μg ml−1 of ampicillin. The target DNA products were sequenced by Novogene (Beijing, China).
To construct the phylogenic tree, OR genes from H. virescens, S. littoralis (Walker et al., 2019), M. sexta, and B. mori were used, and the MEGA7 program was used for phylogenic analysis (Tamura et al., 2011). The phylogenic tree was constructed by the neighbor-joining method with a bootstrap test using 500 replications. The Genbank accession numbers for all the OR genes used are shown in Supplementary Table 2, and all the P. operculella OR proteins used for the phylogenetic analysis are presented in Supplementary Material.
Transmembrane domains of the two candidate PRs were predicted by TMHMM Server Version 2.0,2 and the amino acid sequences of PopeOR1 and PopeOR3 were aligned by the DNAMAN 8.0 software (Lynnon Biosoft, San Ramon, CA, United States).
To determine the tissue expression profiles of the two candidate PR genes, female antennae (FA), male antennae (MA), heads without antennae (H), thoraxes (T), abdomens (AB), legs (L), wings (W), and genitalia (G) from 3-day-old unmated adult moths were collected between the 6th and 8th hours of the dark period, and were immediately frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted and cDNA was synthesized as mentioned above. The qPCR analysis was conducted using an ABI Prime 7,500 Detection System (Applied Biosystem, United States). The qPCR reaction was performed in a 20 μl system containing 10 μl of SYBR Green PCR Master Mix (Biomed, Beijing, China), 0.4 μl of each primer (10 μM), 0.4 μl of ROX Reference Dye II, 1 μl of cDNA template, and 7.8 μl of nuclease-free water. The thermal cycling parameters were: 95°C for 1 min, 40 cycles of 95°C for 10 s, 55°C for 5 s, and 72°C for 15 s. DNase were used to eliminate the DNA contamination of the RNAs samples. The actin gene was used to standardize the target gene expression (Sun et al., 2013). For each tissue, three biological replicates were measured with three technical replicates for each replicate, gene expression levels were analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001). The sequences of the primer pairs used in this analysis are listed in Supplementary Table 1. The SPSS 20.0 software (IBM, Endicatt, NY, United States) was used for data analysis, the statistical comparison of the expression of the PRs was assessed using one-way ANOVA followed by Tukey’s honest significant differences (HSD) test (p < 0.05), data were presented as mean ± SEM.
Primers containing the Kozak consensus sequence and restriction enzyme cutting site (Apa I and Not I) were designed to amplify the open-reading frame (ORFs) of PopeOR1, PopeOR3, and PopeOrco. The products were then cloned into pT7Ts vectors with the primers listed in Supplementary Table 1 (Wang et al., 2011). The extracted plasmids were linearized by digestion with SamI, and used as templates to synthesis cRNAs by using T7 polymerase of the mMESSAGE mMACHINE®T7 Kit (Thermo Fisher Scientific, Waltham, MA, United States). The purified cRNAs were diluted with nuclease-free water at a concentration of 2 μg/μl and stored at −80°C until use.
The pheromones (E, Z)-4,7–13: OAc and (E, Z, Z)-4,7,10–13: OAc were purchased from Nimrod Inc. (Changzhou, China) with 95% minimum purity. The stock solution was prepared in dimethyl sulfoxide (DMSO) at 1 M concentration and stored at −20°C. Prior to the two-electrode voltage clamp electrophysiological recording experiments, the stock solutions were diluted with Ringer’s buffer (96 mM of NaCl, 2 mM of KCl, 5 mM of MgCl2, 0.8 mM of CaCl2, and 5 mM of HEPES; pH=7.6) into the concentration of 10−4 M Ringer’s buffer containing 0.1% DMSO was used as the negative control. All chemicals were freshly prepared for the experiments.
Each of the two candidate PRs were co-expressed with the PopeOrco in Xenopus oocytes for 3–4 d, and the ligand sensitivity was detected using a two electrode voltage-clamp recording as previously reported (Lu et al., 2007; Wang et al., 2010). Healthy, matured Xenopus oocytes (stage V-VII) were treated with 2 mg/ml of collagenase in washing buffer (96 mM of NaCl, 2 mM of KCl, 5 mM of MgCl2, 5 mM of HEPES; pH 7.6) for 1–2 h at room temperature. Equal amounts of PR and Orco cRNA (27.6 ng) were microinjected into the oocytes (Wang et al., 2011). The oocytes were then incubated at 18°C for 4–7 d in 1× Ringer’s solution (96 mM of NaCl, 2 mM of KCl, 5 mM of MgCl2, 0.8 mM of CaCl2, and 5 mM of HEPES; pH=7.6) supplemented with dialyzed horse serum (5%), tetracycline (50 μg/ml), streptomycin (100 μg/ml), and sodium pyruvate (550 μg/ml). Whole-cell currents were recorded from the injected Xenopus oocytes in an OC-725 two-electrode voltage clamp amplifier (Warner Instruments, United States) at a holding potential of −80 mV. Oocytes were exposed to a concentration series of different pheromone components from low to high with an interval between exposures that allowed the current to return to baseline. To avoid the sequential effect of two sex pheromone components on the candidate PRs, the experiment was repeated by reversing the order of component stimulation. Oocytes containing PopeOR1/Orco and PopeOR3/Orco were injected with 1× Ringer’s buffer solution containing 0.1% DMSO to be used as negative control, respectively. All experiments were repeated 5 times on different oocytes. The Digidata 1440A and pCLAMP 10.2 software were used to collect and analyze the data (Axon Instruments Inc., United States). Dose-response curves were obtained and analyzed using GraphPad Prism 5 (GraphPad Software Inc., United States). A statistical comparison of the response of the oocytes to the candidate ligands was assessed using student’s t-test with the SPSS 10.0.1 software (IBM, Endicatt, NY, United States).
The full-length sequences of the three candidate PR genes were cloned, based on the nomenclature of ORs in other Lepidopteran insects, they were named PopeOR1, PopeOR2, and PopeOR3. In addition, PopeOR2 was likely a pheromone co-receptor gene of P. operculella, and was further named PopeOrco. The lengths of the complete ORF region of PopeOR1, PopeOR3, and PopeOrco were 1,302, 1,215, and 1,419 bp, which encode 434, 405, and 473 amino acid residues, respectively. Consistent with other insect ORs, the predicted PopeOR1 and PopeOR3 proteins contain seven putative transmembrane domains (Figure 1) with a predicted extracellular C-terminus and an intracellular N-terminus. Phylogenic analysis showed that these two PRs were grouped into the same branch with PRs from other Lepidopteran species, and this branch was independent from other ORs which may mainly respond to general environmental odors. In the phylogenic tree, PopeOR1 and PopeOR3 clustered with PRs of other Lepidopteran insects including BmorOR1, BmorOR3, BmorOR6, HvirOR11, HvirOR13, and SlitOR11, SlitOR13. The PopeOrco gene (PopeOR2) clustered into the Lepidopteran Orco group with HvirOrco, SlitOrco, and BmorOrco (Figure 2).
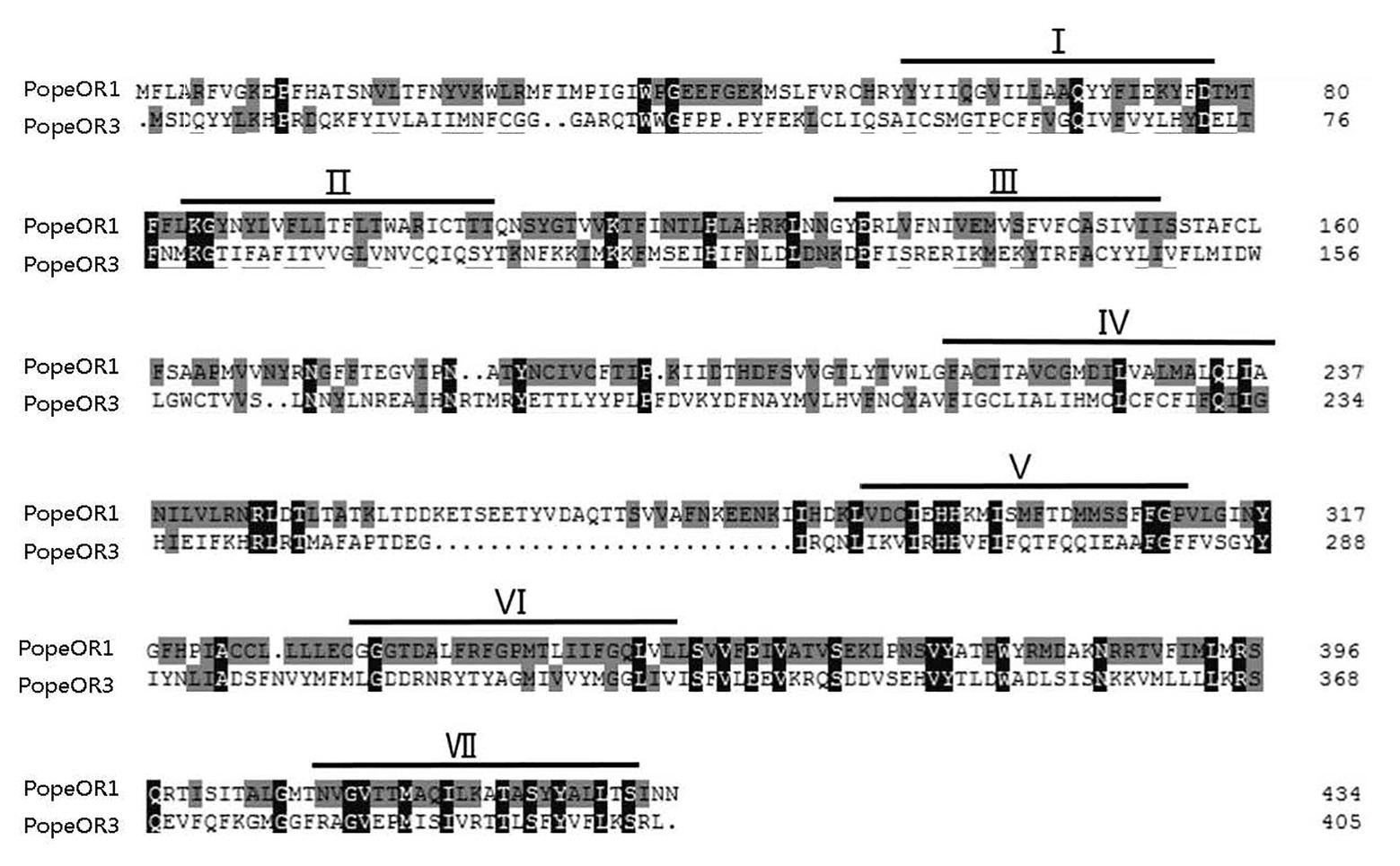
Figure 1. Alignment of the amino acid sequences of the two candidate PRs in Phthorimaea operculella. Identical amino acids are marked with gray and black shading. Predicted seven transmembrane domains (TMD1-TMD7) are indicated by bold lines.
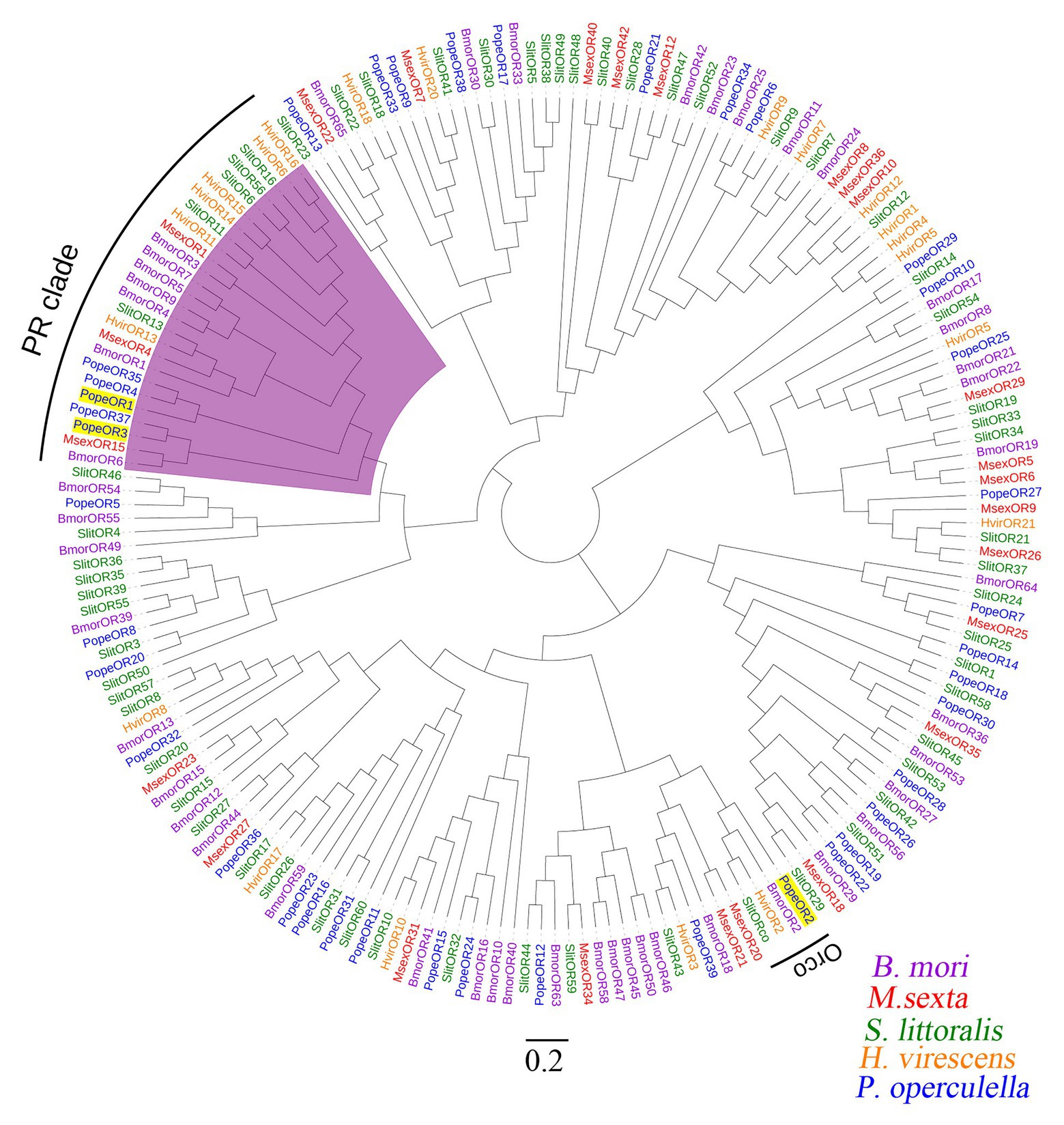
Figure 2. An unrooted neighbor-joining tree of the two candidate PRs of P. operculella together with the ORs from four other Lepidopteran species; B. mori: Bombyx mori (purple), M. sexta: Manduca sexta (red), S. littoralis: Spodoptera littoralis (green), and H. virescens: Heliothis virescens (orange) was constructed. The clade in purple indicates the pheromone receptor clade. The two candidate PRs and Orco of P. operculella are marked with yellow shading.
The qPCR was performed to evaluate the transcription levels of two candidate PRs in different tissues of adults (Figure 3). As expected, the transcripts of both PR genes were highly accumulated in the male antennae. The transcript of PopeOR1 was not expressed in other tissues including heads (without antennae), thoraxes, abdomens, legs, wings, and genitalia. For PopeOR3, its transcript was also hardly detectable in heads (without antennae), thoraxes, abdomens, legs, and genitalia, but not in the wings.
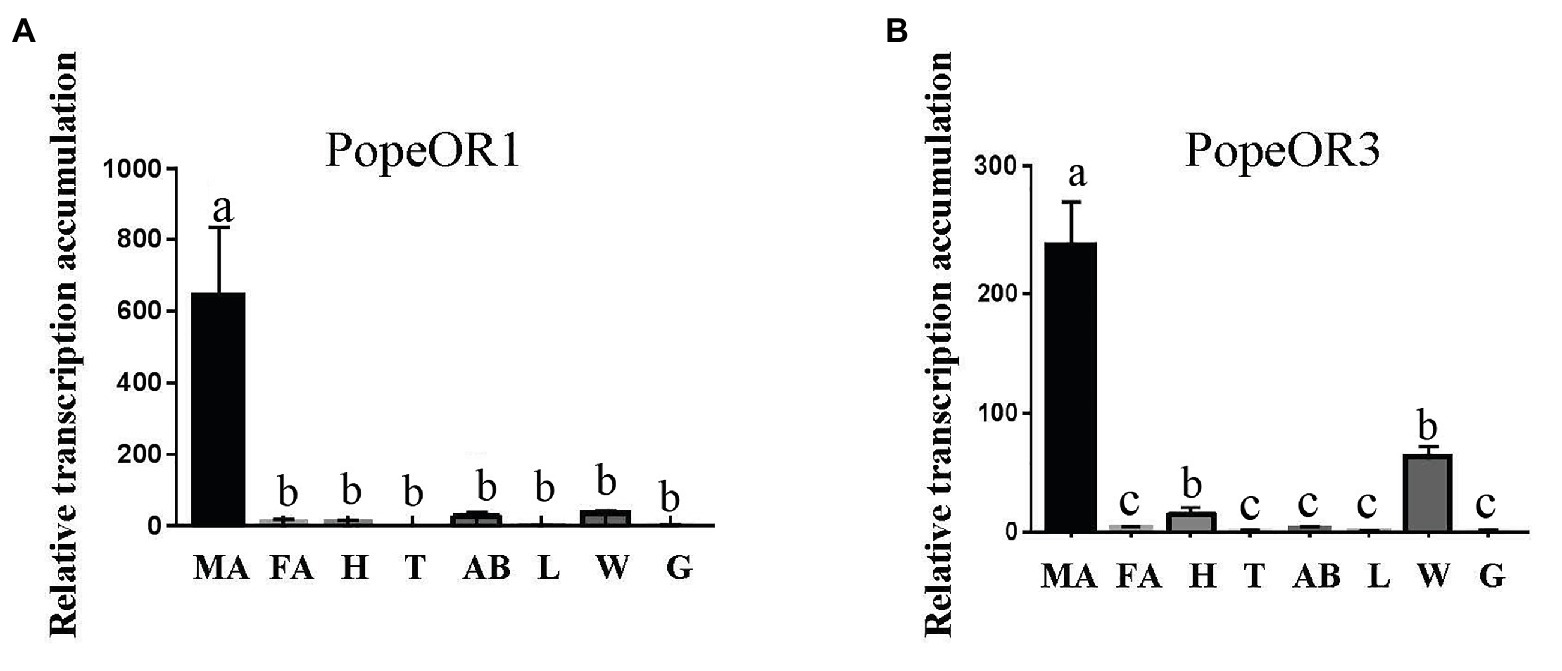
Figure 3. Expression profiles of PopeOR1 (A) and PopeOR3 (B) genes in different tissues. MA, male antennae; FA, female antennae; H, heads (without antennae); T, thoraxes; AB, abdomens; L, legs; W, wings, G, genitalia. Error bars indicate standard error of three biological replicates. Different letters within the same figure mean that the values are significantly different under one-way ANOVA followed by Tukey’s honest significant differences (HSD) test (p < 0.05).
The heterologous expression system of Xenopus oocyte and the voltage clamp recording technique were used to explore the function of the two candidate PRs. Both PopeOR1/PopeOrco and PopeOR3/PopeOrco were successfully activated by two pheromone components (E, Z)-4,7–13: OAc and (E, Z, Z)-4,7,10–13: OAc. PopeOR1/PopeOrco responded to (E, Z)-4,7–13: OAc and (E, Z, Z)-4,7,10–13: OAc with the responses of 313.7 ± 28.26 and 137.6 ± 19.22 nA, respectively (Figures 4A,B). For PopeOR3/PopeOrco, a strong response was observed when bound to (E, Z, Z)-4,7,10–13: OAc (155.7 ± 20.26 nA), while weak responses were found when bound to (E, Z)-4,7–13: OAc (29.41 ± 6.87 nA; Figures 4I,J). By reversing the order of the stimulation from the two components, the PopeOR1/Orco showed decreased responses on (E, Z)-4,7–13: OAc with a current value of 45.4 ± 3.207 nA, but enhanced responses on (E, Z, Z)-4,7,10–13: OAc with a current value of 215.5 ± 34.07 nA (Figures 4C,D), while changing the order of the supply of components had no impact on the sensitivity of PopeOR3/PopeOrco to them (Figures 4K,L). Oocytes injected with the buffer did not respond to any of the test compounds (Figures 4O,P).
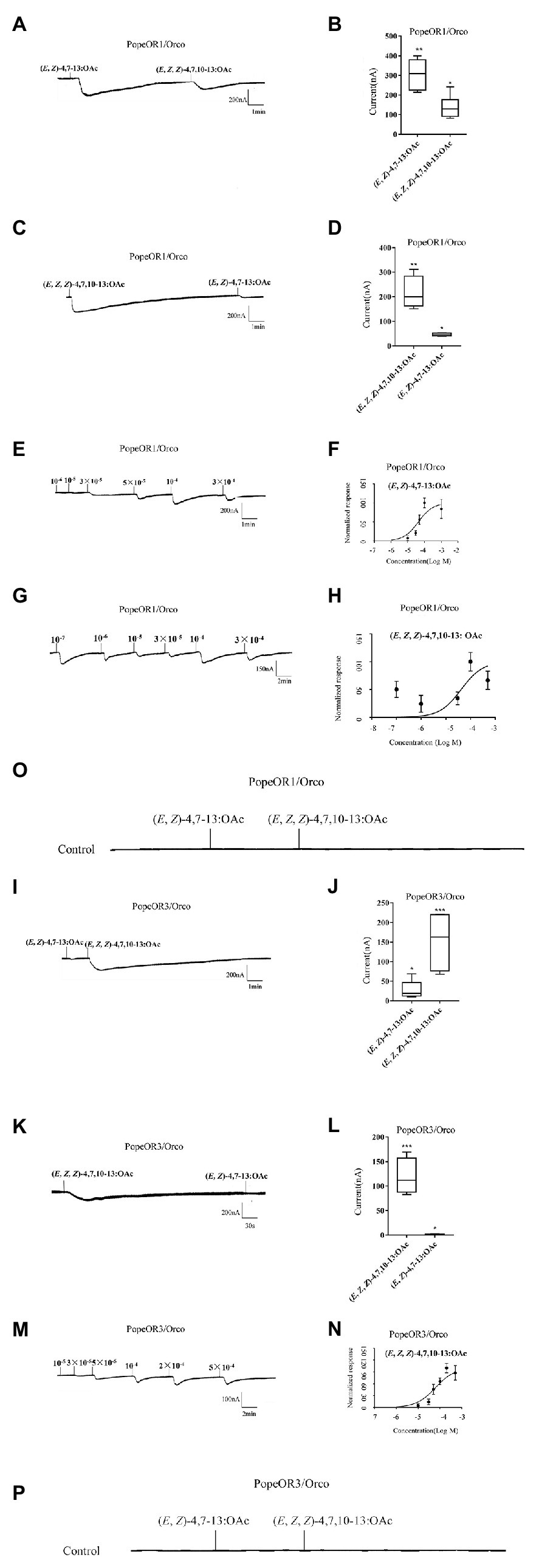
Figure 4. ContinuedFunctional characterization of PopeOR1 and PopeOR3 in Xenopus oocytes. (A,I) Inward current responses of PopeOR1/Orco and PopeOR3/Orco expressed Xenopus oocytes in response to a 10−4 mol/L solution of tested compounds. (B,J) Boxplots of tested compounds response profile of PopeOR1/Orco and PopeOR3/Orco expressed Xenopus oocytes. (C,K) Inward current responses of PopeOR1/Orco and PopeOR3/Orco expressed Xenopus oocytes in response to a 10−4 mol/L solution of tested compounds (reverse stimuli order). (D,L) Boxplots of tested compounds response profile of PopeOR1/Orco and PopeOR3/Orco expressed Xenopus oocytes (reverse stimuli order). (E) PopeOR1/Orco expressed Xenopus oocytes stimulated by a series of concentrations of (E, Z)-4,7–13: OAc. (F) Dose-response curves of PopeOR1/Orco expressed Xenopus oocytes to (E, Z)-4,7–13: OAc. Responses were normalized by setting the maximal response to 100. EC50 values were calculated to be 7.3 × 10−5 M. (G) PopeOR1/Orco expressed Xenopus oocytes stimulated by a serious of concentrations of (E, Z, Z)-4,7,10–13: OAc. (H) Dose-response curves of PopeOR1/Orco expressed Xenopus oocytes to (E, Z, Z)-4,7,10–13: OAc. Responses were normalized by setting the maximal response to 100. EC50 values were calculated to be 3.9 × 10−5 M. (M) PopeOR3/Orco expressed Xenopus oocytes stimulated by a range of concentrations of (E, Z, Z)-4,7,10–13: OAc. (N) Dose-response curves of PopeOR3/Orco expressed Xenopus oocytes to (E, Z, Z)-4,7,10–13: OAc. Responses were normalized by setting the maximal response to 100. EC50 values were calculated to be 8.9 × 10−5 M. Error bars indicate SEM (n = 6). (O,P) Negative control of PopeOR1/Orco and PopeOR3/Orco expressed Xenopus oocytes stimulated by tested compounds. Significance is indicated by asterisk.
In the dose-response comparison experiments, PopeOR1/PopeOrco responded to (E, Z)-4,7–13: OAc at the concentration of 10−6 M, and the response peak occurred at 10−4 M, with an EC50 value of 7.3 × 10−5 M (Figures 4E,F); PopeOR1/PopeOrco responded to (E, Z, Z)-4,7,10–13: OAc at the concentration of 10−7 M, and the response peak occurred at 10−4 M, with an EC50 value of 3.9 × 10−5 M (Figures 4G,H). PopeOR3/PopeOrco responded to (E, Z, Z)-4,7,10–13: OAc at the concentration of 10−6 M and the response peak occurred at 2 × 10−4 M, with the calculated EC50 of 8.9 × 10−5 M (Figures 4M,N).
Pheromone communication is widely used by Lepidopteran insects to find cognate individuals of the opposite sex (Vogt, 2005). Of all the protein families involved in pheromone perception, PRs play critical roles in determining the specificity and sensitivity of the recognition of the chemical mating signals (Liu et al., 2018b), which is considered an important basis for interspecies isolation and intraspecies choice (Groot et al., 2006). In the current study, we identified and characterized two candidate PRs from P. operculella, an important pest of the Solanaceae crop.
The expression analysis suggested that both the two PR genes in P. operculella were almost only expressed in male antennae. The males of Lepidopteran insects are the main receivers for pheromone cues. It is therefore reasonable to see the sex-specific expression of PR genes in P. operculella and many other Lepidopteran species (Krieger et al., 2004; Sakura et al., 2004; Wang et al., 2011; Liu et al., 2013b, 2018a). Moreover, it is interesting to see that PopeOR3 is also expressed in the wings of male adults. A recent study into Helicoverpa assulta found that an odorant receptor gene, HassOR31 which was highly expressed in the ovipositor rather than in antennae, was related to the determination of oviposition in host plants in females. This finding reveals that some ORs located outside the antenna might also have a functional role in vivo (Li et al., 2020). Therefore, further studies are needed to uncover the function of PopeOR3 in the wings of P. operculella.
The heterologous expression in Xenopus oocytes and electrophysiological recording were widely used in investigating the function of insect PRs. In the phylogenic tree, PopeOR1 and PopeOR3 clustered with the PRs from other insects, including BmorOR1, BmorOR3, HvirOR6, and SlitOR6, whose functions in pheromone detection have been confirmed (Sakura et al., 2005; Wang et al., 2011; Cheng et al., 2017), indicating their potential roles as PRs. When co-expressed with PopeOrco, both PopeOR1 and PopeOR3 showed strong responses to the (E, Z, Z)-4,7,10–13: OAc, whereas only PopeOR1 had strong responses to (E, Z)-4,7–13: OAc. Thus, it is likely that PopeOR1 is more sensitive to female-produced pheromones than PopeOR3. Moreover, we found that PopeOR1 had a higher affinity to (E, Z, Z)-4,7,10–13: OAc (with an EC50 value of 3.9 × 10−5 M) than to (E, Z)-4,7–13: OAc (with an EC50 value of 7.3 × 10−5 M). A previous field study found that sole sex pheromones can attract males, and (E, Z, Z)-4,7,10–13: OAc was more attractive to P. operculella males than (E, Z)-4,7–13: OAc (Persoons et al., 1976). Our findings may partially explain this phenomenon. Despite the fact that single sex pheromone compounds can attract males, a field study revealed that when (E, Z)-4,7–13: OAc and (E, Z, Z)-4,7,10–13: OAc are mixed with a ratio of 1:4, the combination showed the highest attraction (Persoons et al., 1976). This implied that in the field the two pheromone compounds should stimulate the OSN together simultaneously and that co-stimulation of the two sex pheromone components would enhance the overall response of the neurons. A previous study on the PRs of noctuid moth Spodoptera littoralis indicated that two different PRs can detect the same pheromone compound with a different sensitivity (de Fouchier et al., 2015). In this study, we noticed that PopeOR1 and PopeOR3 showed different affinities to the same sex pheromone compound (E, Z, Z)-4,7,10–13: OAc. One possible reason is that these two PRs might be located on different sites of the male antennae, which contributes to their distinct affinities to the single same sex pheromone compound. Another possibility is that these two receptors are expressed by two different types of OSNs, in which case they are able to show different affinities for the same sex pheromone compound.
Although we identified the roles of PopeOR1 and PopeOR3 in the detection of two key pheromones in vitro, since we only tested two pheromone compounds in the present study, we supposed that these PRs might also detect other pheromone components untested here. Further studies in testing a wider range of pheromone compounds on this insect and also the identification of other candidate PR genes in the genome of P. operculella could provide more information on the pheromone detection of this pest. Moreover, while the Xenopus oocytes system is widely used to analyze the function of PRs, recent findings suggested that PRs were more sensitive to pheromones when pheromone binding proteins were present (Chang et al., 2015). It might be interesting to see the in vivo biological functions of PopeOR1 and PopeOR3 by using techniques such as CRISPR/Cas in P. operculella.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Ethics Committee of Institute of Insect Sciences, Key Laboratory of Biology of Crop Pathogens and Insects of Zhejiang Province, Key Laboratory of Molecular Biology of Crop Pathogens and Insects, Ministry of Agriculture, State Key Laboratory of Rice Biology, Zhejiang University.
WZ, GW, and YuG conceived and designed the experiments. XH and JZ wrote the manuscript. XH, YC, JZ, MZ, YZ, and YaG performed the experiments. WZ, GW, and YuG provided valuable suggestions and helped to revise the manuscript. All authors discussed the results and approved the final manuscript.
This work was supported by the National Nature Science Foundation of China (Grant No. 31701798); the Key Research and Development Program of Zhejiang Province (Grant No. 2019C04007) and the National Key Research and Development Program (Grant No. 2018YFD0200802).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.618983/full#supplementary-material
An, X. K., Khashaveh, A., Liu, D. F., Xiao, Y., Wang, Q., Wang, S. N., et al. (2020). Functional characterization of one sex pheromone receptor (AlucOR4) in Apolygus lucorum (Meyer-Dür). J. Insect Physiol. 120:103986. doi: 10.1016/j.jinsphys.2019.103986
Bastin-Heline, L., de Fouchier, A., Cao, S., Koutroumpa, F., Caballero-Vidal, G., Robakiewicz, S., et al. (2019). A novel lineage of candidate pheromone receptors for sex communication in moth. eLife 8:e49826. doi: 10.7554/eLife.49826
Chang, H., Liu, Y., Yang, T., Pelosi, P., Dong, S., and Wang, G. (2015). Pheromone binding proteins enhance the sensitivity of olfactory receptors to sex pheromones in Chilo suppressalis. Sci. Rep. 5:13093. doi: 10.1038/srep13093
Cheng, T., Wu, J., Wu, Y., Chilukuri, R. V., Huang, L., Yamamoto, K., et al. (2017). Genomic adaptation to polyphagy and insecticides in a major east Asian noctuid pest. Nat. Ecol. Evol. 1, 1747–1756. doi: 10.1038/s41559-017-0314-4
de Fouchier, A., Sun, X., Monsempes, C., Mirabeau, O., Jacquin-Joly, E., and Montagné, N. (2015). Evolution of two receptors detecting the same pheromone compound in crop pest moth of the genus Spodoptera. Front. Ecol. Evol. 3:95. doi: 10.3389/fevo.2015.00095
Forstner, M., Breer, H., and Krieger, J. (2009). A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth: Antheraea polyphemus. Int. J. Biol. Sci. 5, 745–757. doi: 10.7150/ijbs.5.745
Groot, A., Horovitz, J. L., Hamiton, J., Santangelo, R. G., Schal, C., and Gould, F. (2006). Experimental evidence for interspecific directional selection on moth pheromone communication. Proc. Natl. Acad. Sci. U. S. A. 103, 5858–5863. doi: 10.1073/pnas.0508609103
Große, W. (2010). Sex-specific odorant receptors of the tobacco hornworm, Manduca sexta. Front. Cell. Neurosci. 4:22. doi: 10.3389/fncel.2010.00022
Guo, J. M., Liu, X. L., Liu, S. R., Wei, Z. Q., Han, W. K., Guo, Y., et al. (2020). Functional characterization of sex pheromone receptors in the fall armyworm (Spodoptera frugiperda). Insects 11:193. doi: 10.3390/insects11030193
He, P., Zhang, J., Li, Z. Q., Zhang, Y. N., Yang, K., Dong, S. L., et al. (2014). Functional characterization of an antennal esterase from the noctuid moth, Spodoptera exigua. Arch. Insect Biochem. Physiol. 86, 85–99. doi: 10.1002/arch.21164
Ke, Y., Huang, L. Q., Chao, N., and Wang, C. Z. (2017). Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. eLife 6:e29100. doi: 10.7554/eLife.29100
Krieger, J., Grosse, W. E., Gohl, T., Dewer, Y. M., Raming, K., and Breer, H. (2004). Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc. Natl. Acad. Sci. U. S. A. 101, 11845–11850. doi: 10.1073/pnas.0403052101
Laurent, G. (1999). A systems perspective on early olfactory coding. Science 286, 723–728. doi: 10.1126/science.286.5440.72
Li, R. T., Huang, L. Q., Dong, J. F., and Wang, C. Z. (2020). A moth odorant receptor highly expressed in the ovipositor is involved in detecting host-plant volatiles. eLife 9:e53706. doi: 10.7554/elife.53706.sa2
Liu, W., Jiang, X. C., Cao, S., Yang, B., and Wang, G. R. (2018a). Functional studies of sex pheromone receptors in asian corn borer, Ostrinia furnacalis. Front. Physiol. 9:591. doi: 10.3389/fphys.2018.00591
Liu, Y., Liu, Y., Jiang, X., and Wang, G. (2018b). Cloning and functional characterization of three new pheromone receptors from the diamondback moth, Plutella xylostella. J. Insect Physiol. 107, 14–22. doi: 10.1016/j.jinsphys.2018.02.005
Liu, Y., Liu, C., Lin, K., and Wang, G. (2013a). Functional specificity of sex pheromone receptors in the cotton bollworm, Helicoverpa armigera. PLoS One 8:e62094. doi: 10.1371/journal.pone.0062094
Liu, C., Liu, Y., Walker, W. B., Dong, S., and Wang, G. (2013b). Identification and functional characterization of sex pheromone receptors in beet armyworm, Spodoptera exigua (Hubner). Insect Biochem. Mol. Biol. 43, 747–754. doi: 10.1016/j.ibmb.2013.05.009
Liu, X. L., Sun, S. J., Khuhro, S. A., Elzaki, M. E. A., Yan, Q., and Dong, S. L. (2019). Functional characterization of pheromone receptors in the moth, Athetis dissimilis (Lepidoptera: Noctuidae). Pestic. Biochem. Physiol. 158, 69–76. doi: 10.1016/j.pestbp.2019.04.011
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, T., Qiu, Y. T., Wang, G., Kwon, J. Y., Rutzler, M., Kwon, H. W., et al. (2007). Odor coding in the maxillary palp of the malaria vector mosquito, Anopheles gambiae. Curr. Biol. 17, 1533–1544. doi: 10.1016/j.cub.2007.07.062
Montagné, N., Chertemps, T., Brigaud, I., Francois, A., Francois, M. C., de Fouchier, A., et al. (2012). Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur. J. Neurosci. 36, 2588–2596. doi: 10.1111/j.1460-9568.2012.08183
Persoons, C. J., Voerman, S., Verwiel, P. E. J., Ritter, F. J., Nooyen, W. J., and Minks, A. K. (1976). Sex pheromone of the potato tuberworm moth, Phthorimaea operculella: isolation, dentification and field evaluation. Entomol. Exp. Appl. 20, 289–300. doi: 10.1111/j.1570-7458.1976.tb02645.x
Roelofs, W. L., Kochansky, J. P., Carde, R. T., Kennedy, G. G., and Corbin, V. L. (1975). Sex pheromone of the potato tuberworm moth, Phthorimaea operculella. Ithaca, NY: Cornell Research Foundation, Inc. U.S. Patent No 4,010,255.
Rondon, S. I. (2020). Decoding phthorimaea operculella (Lepidoptera: Gelechiidae) in the new age of change. J. Integr. Agric. 19, 316–324. doi: 10.1016/S2095-3119(19)62740-1
Sakura, T., Nakagawa, T., Hidefumi, M., Mori, H., Endo, Y., Tanoue, S., et al. (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth, Bombyx mori. Proc. Natl. Acad. Sci. U. S. A. 101, 16653–16658. doi: 10.1073/pnas.0407596101
Sakura, T., Nakagawa, T., Mitsuno, H., Mori, H., Endo, Y., and Tanoue, S. (2005). Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307, 1638–1642. doi: 10.1126/science.1106267
Sato, K., Pellegrino, M., Nakagawa, T., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1009. doi: 10.1038/nature06850
Steinbrecht, S. A. (1999). Atlas of arthropod sensory receptor-dynamic morphology in relation to function. Tokyo: Springer, 155–176.
Sun, M., Liu, Y., Walker, W. B., Liu, C., Lin, K., Gu, S., et al. (2013). Identification and characterization of pheromone receptors and interplay between receptors and pheromone binding proteins in the diamondback moth, Plutella xyllostella. PLoS One 8:e62098. doi: 10.1371/journal.pone.0062098
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Vogt, R. G. (2005). Molecular basis of pheromone detection in insects. Comp. Mol. Insect. Sci. 3, 735–803. doi: 10.1016/B0-44-451924-6/00047-8
Walker, W. B., Roy, A., Anderson, P., Schlyter, F., and Larsson, M. C. (2019). Transcriptome analysis of gene families involved in chemosensory function in Spodoptera littoralis (Lepidoptera: noctuidae). BMC Genomics 20:428. doi: 10.1186/s12864-019-5815-x
Wang, G., Carey, A. F., Carlson, J. R., and Zwiebel, L. J. (2010). Molecular basis of odor coding in the malaria vector mosquito, Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 107, 4418–4423. doi: 10.1073/pnas.0913392107
Wang, G., Vásquez, G. M., Schal, C., Zwiebel, L. J., and Gould, F. (2011). Functional characterization of pheromone receptors in the tobacco budworm, Heliothis virescens. Insect Mol. Biol. 20, 125–133. doi: 10.1111/j.1365-2583.2010.01045.x
Yuvaraj, J. K., Andersson, M. N., Corcoran, J. A., Anderbrant, O., and Löfstedt, C. (2018). Functional characterization of odorant receptors from Lampronia capitella suggests a non-ditrysian origan of the lepidopteran pheromone receptor clade. Insect Biochem. Mol. Biol. 100, 39–47. doi: 10.1016/j.ibmb.2018.06.002
Zhang, Y., Du, L. X., Xu, J. W., Wang, B., Zhang, X. Q., Yan, Q., et al. (2018). Functional characterization of four sex pheromone receptors in the newly discovered maize pest Athetis lepigone. J. Insect Physiol. 113, 59–66. doi: 10.1016/j.jinsphys.2018.08.009
Zhang, D. D., and Löfstedt, C. (2015). Moth pheromone receptors: gene sequences, function, and evolution. Front. Ecol. Evol. 3:105. doi: 10.3389/fevo.2015.00105
Zhang, J., Yan, S., Liu, Y., Jacquin-Joly, E., Dong, S., and Wang, G. (2015a). Identification and functional characterization of sex pheromone receptors in the common cutworm (Spodoptera litura). Chem. Senses 40, 7–16. doi: 10.3389/fphys.2018.00591
Keywords: Phthorimaea operculella, pheromone receptor, Xenopus oocytes, pheromone communication, sexual pheromone
Citation: He X, Cai Y, Zhu J, Zhang M, Zhang Y, Ge Y, Zhu Z, Zhou W, Wang G and Gao Y (2021) Identification and Functional Characterization of Two Putative Pheromone Receptors in the Potato Tuber Moth, Phthorimaea operculella. Front. Physiol. 11:618983. doi: 10.3389/fphys.2020.618983
Received: 19 October 2020; Accepted: 18 December 2020;
Published: 25 January 2021.
Edited by:
Peng He, Guizhou University, ChinaReviewed by:
Ya-Nan Zhang, Huaibei Normal University, ChinaCopyright © 2021 He, Cai, Zhu, Zhang, Zhang, Ge, Zhu, Zhou, Wang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenwu Zhou, d2Vud3V6aHpvdUB6anUuZWR1LmNu; Guirong Wang, d2FuZ2d1aXJvbmdAY2Fhcy5jbg==; Yulin Gao, Z2FveXVsaW5AY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.