- 1Guangdong Province Key Laboratory for Aquatic Economic Animals, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
- 2Guangdong Provincial Engineering Technology Research Center for Healthy Breeding of Important Economic Fish, Guangzhou, China
- 3Southern Marine Science and Engineering Guangdong Laboratory, Zhuhai, China
How organisms display many different biochemical, physiological processes through genes expression and regulatory mechanisms affecting muscle growth is a central issue in growth and development. In Siniperca chuatsi, the growth-related genes and underlying relevant mechanisms are poorly understood, especially for difference of body sizes and compensatory growth performance. Muscle from 3-month old individuals of different sizes was used for transcriptome analysis. Results showed that 8,942 different expression genes (DEGs) were identified after calculating the RPKM. The DEGs involved in GH-IGF pathways, protein synthesis, ribosome synthesis and energy metabolisms, which were expressed significantly higher in small individuals (S) than large fish (L). In repletion feeding and compensatory growth experiments, eight more significant DEGs were used for further research (GHR2, IGFR1, 4ebp, Mhc, Mlc, Myf6, MyoD, troponin). When food was plentiful, eight genes participated in and promoted growth and muscle synthesis, respectively. Starvation can be shown to inhibit the expression of Mhc, Mlc and troponin, and high expression of GHR2, IGFR1, and 4ebp inhibited growth. Fasting promoted the metabolic actions of GHR2, IGFR1, and 4ebp rather than the growth-promoting actions. MyoD can sense and regulate the hunger, which also worked with Mhc and Mlc to accelerate the compensatory growth of S. chuatsi. This study is helpful to understand the regulation mechanisms of muscle growth-related genes. The elected genes will contribute to the selective breeding in future as candidate genes.
Introduction
Muscle growth is regulated by the gene expression nets of muscle cell proliferation and protein metabolism (Riley et al., 2002). In vertebrates, several growth-related genes have been identified, including MRFs, IGF, IGFR, GH, GHR (De-Santis and Jerry, 2007; Devlin et al., 2009). Interestingly, for mammals, muscle growth occurs mainly by hypertrophy, with little proliferation. Unlike mammals, fish muscle growth is achieved by hyperplasia and hypertrophy (Rowlerson and Veggetti, 2001). Furthermore, in large and fast-growing fish, both hyperplasia and hypertrophy contribute to muscle growth and continue into a large body size, whereas small and slow-growing fish largely rely on hypertrophy and the rate of muscle fiber recruitment is rather low (Veggetti et al., 1993; Steinbacher et al., 2010; Weatherley et al., 2010). Thus, the same genes may have different expression and regulatory mechanisms in different fish. Studies on genes expression of different sizes and compensatory growth prove fruitful for comprehending growth regulatory mechanisms of fish (Chauvigné et al., 2003; Nebo et al., 2013; Picha et al., 2014; Tian et al., 2016).
Body size is an obvious and vital characteristic of fish, which is controlled by cell number and cell size (Brogiolo et al., 2001), and this process is tightly modulated by growth-related genes and nutrition. An adequate re-feeding following a period of starvation or unfavorable environmental conditions can result in accelerated growth, which is called compensatory growth and has been widely studied in vertebrates (Ali et al., 2003; Chauvigné et al., 2003; Montserrat et al., 2007; Nebo et al., 2013). Especially, it has been reported that fish has the capacity of compensatory growth (Ali et al., 2003). In the meantime, transcriptome analysis can identify growth-related genes and expand knowledge in body size and compensatory growth, promoting the enhanced rate of food utilization and cut costs in aquaculture industry.
Transcriptome and RNA-sequencing (RNA-seq) have been applied to a substantial amount of fish biology studies, including zebrafish (Collins et al., 2012), channel catfish (Liu et al., 2012), European sea bass (Sarropoulou et al., 2012), and rainbow trout (Palstra et al., 2013). Many biological processes, including development, immune, stress response, and adaptive evolution can be mapped, annotated and understood by RNA-seq.
Siniperca chuatsi is an important commercial fresh water fish in China. At present, the research on S. chuatsi mainly focuses on aquaculture, disease and immunity, nevertheless the growth-related genes and mechanisms are not well-understood (Wu et al., 2015; Huang et al., 2017; Dou et al., 2018). To this end, different size muscle of S. chuatsi was subjected to transcriptome analysis by RNA-seq and the mechanisms of growth-related genes by compensatory growth in this study. The results would be helpful to find significantly molecule markers for selective breeding.
Materials and Methods
Ethics Statement
Mandarin fish experiments were approved by the Institutional Animal Care and Ethics Committee of Sun Yat-sen University and performed in accordance with the guidelines for experimental animals established by this committee.
Experimental Animals
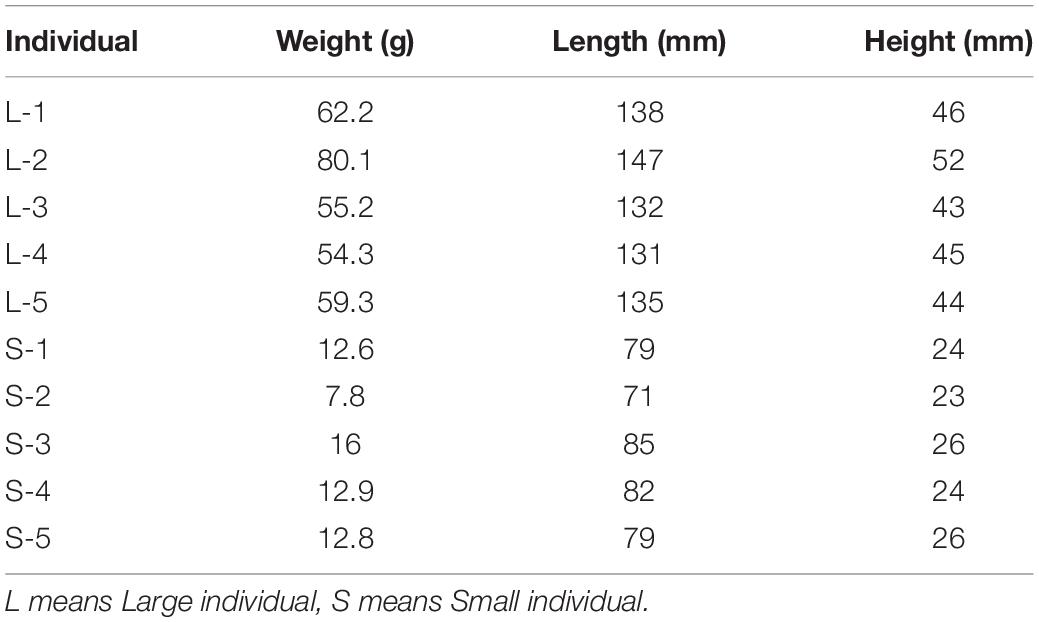
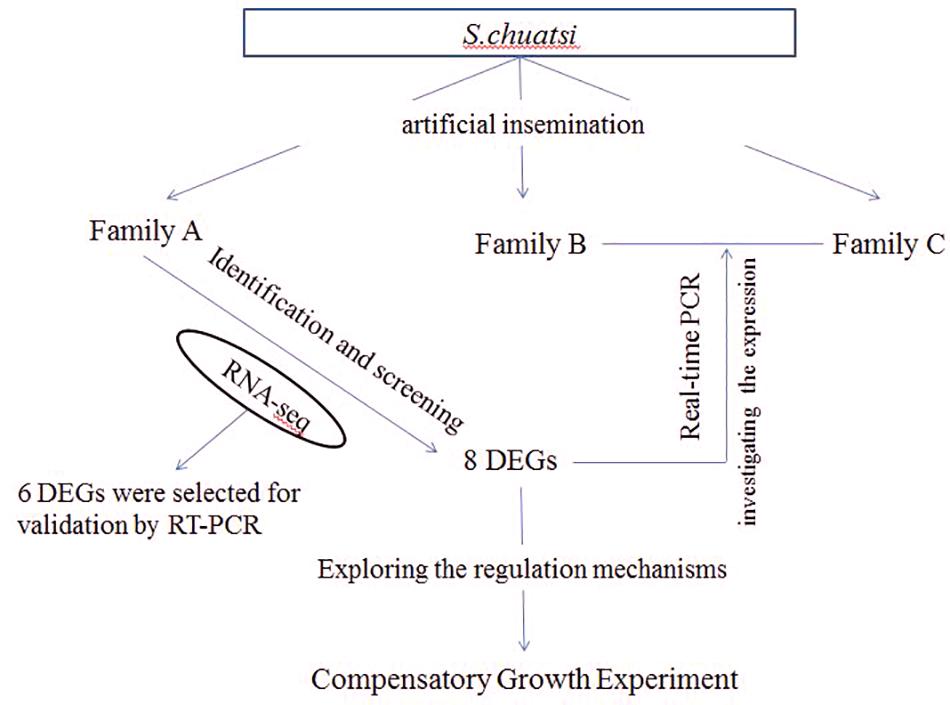
In this study, all of S. chuatsi came from Guangdong Foshan Bairong Aquatic Breeding, Co., Ltd. Three full-sib families A, B, and C were constructed by artificial insemination (Figure 8). Each family contained 80 S. chuatsi in a separate tank. The experimental fish were fed under uniform conditions; they were provided adequate live fish as bait twice daily at 0900 and 1800 h. The water temperature was maintained at 25–26°C. After feeding 3 months in family A, 5S and 5L was performed for RNA-seq. For B and C families, 6L and 6S fish were selected from each family at 3- and 5-month old for real-time PCR (One of the six fish was chosen as a standby) (Figure 8).
Tissue Sampling
All of the above experimental fish were anesthetized with tricaine methanesulfonate (MS-222, 100 mg/L) and sacrificed via decapitation for subsequent sampling, and white muscle was removed immediately. The tissues were frozen in liquid nitrogen quickly, and stored at −80°C until use. Furthermore, each experiment was performed with three independent biological replicates.
RNA Extraction and Library Construction
For RNA-seq, total RNA was extracted from white muscle in Family A with E.Z.N.A. total RNA kit II and detected with the concentration and quality. To acquire the entire transcriptome information, and to find out the growth-related DEGs, the muscle samples of family A were mixed with equal amount and then were divided into two RNA pools. PolyA mRNA was isolated by Beads with Oligo (dT) after total RNA was collected and interrupted to short fragments. Random hexamer-primer was used to synthesize the first-strand cDNA using the Qiaquick PCR Purification Kit (Qiagen). The second-strand cDNA was synthesized using buffer, dNTPs, RNaseH and DNA polymerase I, respectively (Invitrogen). Subsequently, short fragments were purified, enriched for end reparation and adding polyA, connected with sequencing adapters. After that, the suitable fragments were selected using agarose gel electrophoresis for the PCR amplification as templates. At last, the two cDNA library could be sequenced in BGI-Shenzhen using Illumina HiSeqTM 2000.
Illumina Reads Processing and Assembly
Clean reads were screened from raw reads gained from sequencing machines by removing adaptors, unknown nucleotides larger than 5% and low quality reads (which the percentage of low Q-value ≤ 10 base was more than 20%) which would negatively affect following bioinformatics analysis. Firstly, program-Trinity combines reads with certain length of overlap to form longer fragments without N, which are called contigs. Then, these contigs were taken into further process of sequence cluster with sequence clustering software to form longer sequences without N, which are defined as unigenes.
Functional Annotation and Classification
The unigenes sequences were firstly aligned with a series of public databases, such as the non-redundant protein database, the Cluster of Orthologous Groups (COG) of protein database, the Kyoto Encyclopedia of Genes and GenBank non-redundant (NR) database and Swiss-Prot database, using BLASTx (E-valueb10-5) and BLAST (E-valueb10-10), respectively. According to the Nr annotation information, the obtained unigene was enriched with Gene Ontology (GO) by Blast2GO program. And then unigenes were classified into different GO functional cluster using WEGO software. GO has three ontologies: molecular function, cellular component and biological process. The basic unit of GO is GO-term, and every GO-term belongs to a type of ontology.
Differential Genes Expression Analysis
The genes expression was calculated by the numbers of reads that mapped to the reference sequence and every gene. All genes expression levels were calculated by using the formula RPKM method and the RPKM of genes can be used for comparing between different samples. Different expression genes were carried out GO function analysis to understand its function. False discovery rate (FDR) was used to determine the threshold of P-value which corresponds to differential gene expression test in multiple tests. FDR ≤ 0.001 and the absolute value of log2Ratio ≥ 1 is used as the threshold to judge the significance of gene expression difference.
The Synthesis of cDNA and Real-Time PCR Detection
cDNA was synthesized from muscle in B and C families with PrimeScript®RT reagent Kit with gDNA Eraser (Perfect Real Time). The specific primers for the RT-PCR were designed based on selected growth-related genes using the Primer Premier5.0. Corresponding primers were designed to vertify the results (Table 5). 6 DEGs were selected as the verification genes to confirm the reliability of data obtained by RNA-seq. And 18SrRNA was used as a reference gene which was no evident difference in different samples of S. chuatsi. The primers were listed in Table 5. Quantitative Fluorescence PCRassay was performed with LightCycler® 480 II Real-Time PCR System (Roche), and the kits were SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (TaKaRa, Japan), with 384-well plates. In each reaction system, there were 10 ul including the first-strand cDNA, 250 nM primers, and 5 μl SYBR Green PCR Master Mix (TaKaRa, Japan). Each reaction was repeated three times. And the relative expression levels were calculated by the 2–ΔCt method.
Fasting and Re-feeding Experiments
Since transcriptome results represented gene expression at a certain time, change of gene expression over time cannot be known. So we researched the change of gene expression and its mechanism in compensatory growth experiments. A total of 80 experimental fish from one family (300 ± 30 g, body weight) were randomly divided into control and experimental groups that each included four repeated subgroups, and they were provided live fish as bait. Each subgroup contained 20 in a separate tank. The water temperature was maintained at 25–26°C, and the compensatory growth experiment lasted for 6 weeks. During an initial 3 weeks of fasting, food was withheld from the experimental group, while that of the control group was maintained at a normal level. After this, both the experimental group and the control group were fed continuously during the following 3 weeks. Seven sampling time points (0–6) were set throughout the experiment. At each sampling time point, six S. chuatsi in a pond were randomly selected from each group and sampled following anesthesia with MS-222 (100 mg/L). Sampling time points 0, 1, 2, and 3 represented S. chuatsi that had been fasted for 0, 1, 2, and 3 weeks, respectively. Sampling time points 4, 5, and 6 represented the fish that were re-fed continuously for 1, 2, and 3 weeks after starvation, respectively. The fish were fed twice a day and weighed every week, and weights were used to calculated The Specific growth rate (SGR). SGR means ((In W 2 -W 1)/(T 2 -T 1) × 100), where W 2 is the weight at the end of the growth interval and W 1 is the weight at the beginning of the growth interval, while T 2 -T 1 represents the duration (days) of the growing interval. The white muscle samples collected from S. chuatsi was rapidly frozen in liquid nitrogen and stored at −80°C and samples were used for real-time PCR analysis.
Statistical Analyses
SPSS version 18.0 was used for the analyses in the study, Tukey’s test was applied to compare the significant differences of the expression of the growth-related genes. The level of significance for all statistical tests was set at 5% (P < 0.05).
Results
Sequencing and Annotation
The different weight of S. chuatsi was shown in Table 1. The brief information of Illumina deep sequencing was shown in Table 2. A total of 91,264,026 high quality clean reads (48963598 belongs L, 46617484 belongs S) were obtained (Table 2). After screening, 39005 unigenes were annotated into four databases, including Nr (38833, 52.9%), Swissprot (337479, 46.0%), COG (10926, 14.8%), and KEGG (19791, 26.9%) (Figure 1). So, we annotated the results by Nr database which defined the maximum number of unigenes.

Table 2. The summary information of deep sequencing and assembly for small and large Siniperca chuatsi.
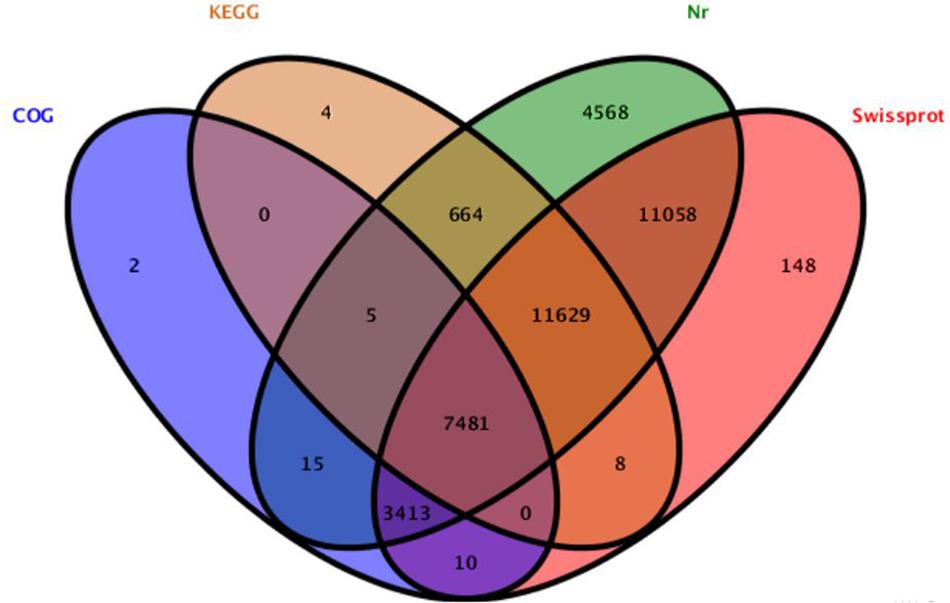
Figure 1. Detection of homologous genes in public databases. The numbers of annotated all-unigenes were indicated in the ellipses respectively.
Analysis of RNA-seq Data and Confirmation
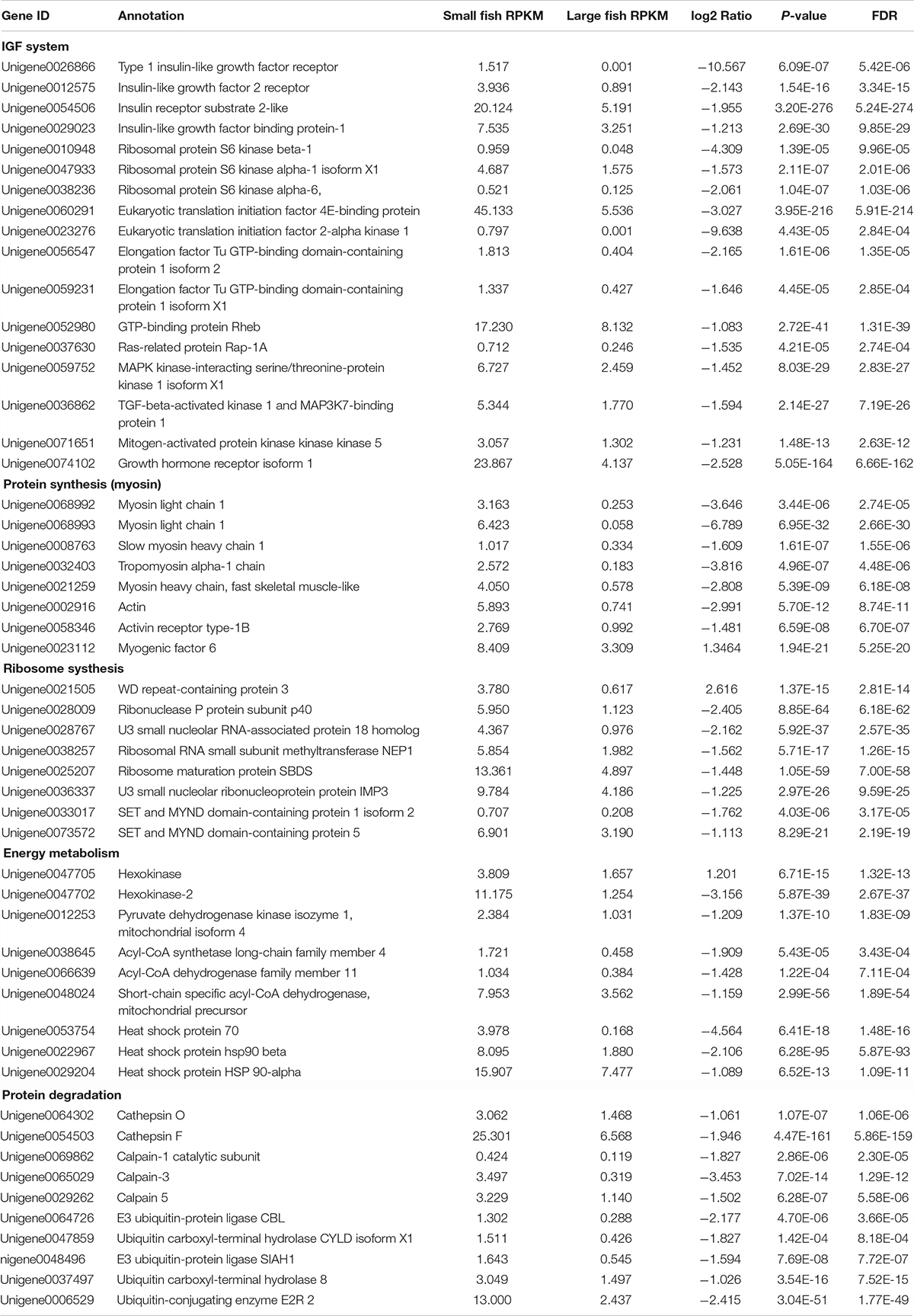
By GO terms enrichment analysis, the biological process, cellular component and molecular function have 1530, 1737, and 1883 genes respectively (Figure 2), which illustrated the dominant groups was cellular process in biological process, cell and cell part in cellular component and binding in molecular function (Figure 2). 8,942 DEGs were identified after calculating the RPKM. Among them, 1084 unigenes were up-regulated in L, while 7858 unigenes were up-regulated in S (Figure 3). The results showed that more genes were up-regulated in S compared to L (Figure 3), including GH-IGF pathways receptors and downstream genes (GHR2, IGFR1, 4ebp, P70S6K, transcriptional elongation factor, etc.), muscle synthesis and contraction genes (Mhc, Mlc, troponin, etc.) and energy metabolism genes (Glucose-6 phosphate dehydrogenase, etc.) (Table 3). Thus, eight genes were selected for further research by real-time PCR, involving GHR2, IGFR1, 4ebp, MHC, Mlc, MyoD, Myf6, and troponin.
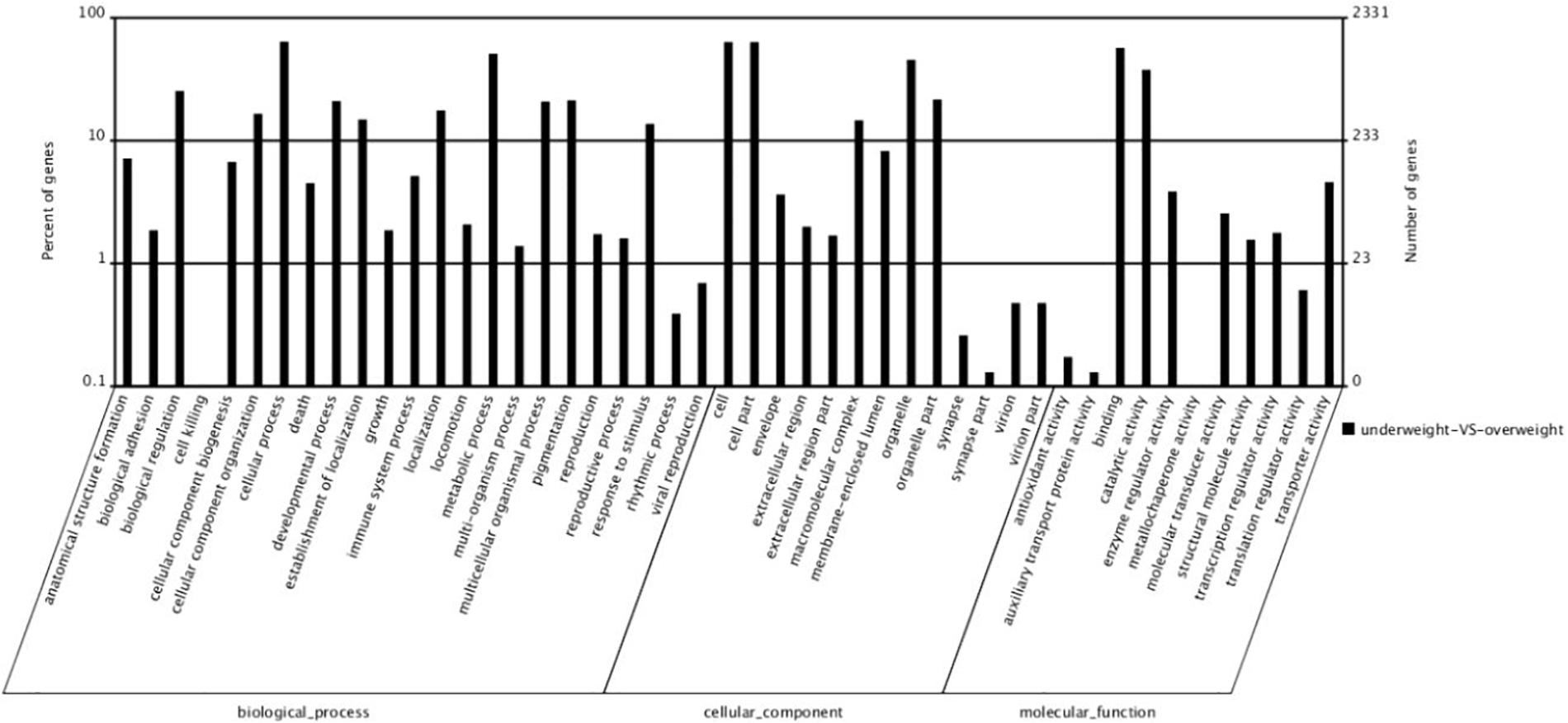
Figure 2. The number of different expression genes that the genes of large fish compared to the genes of small fish.
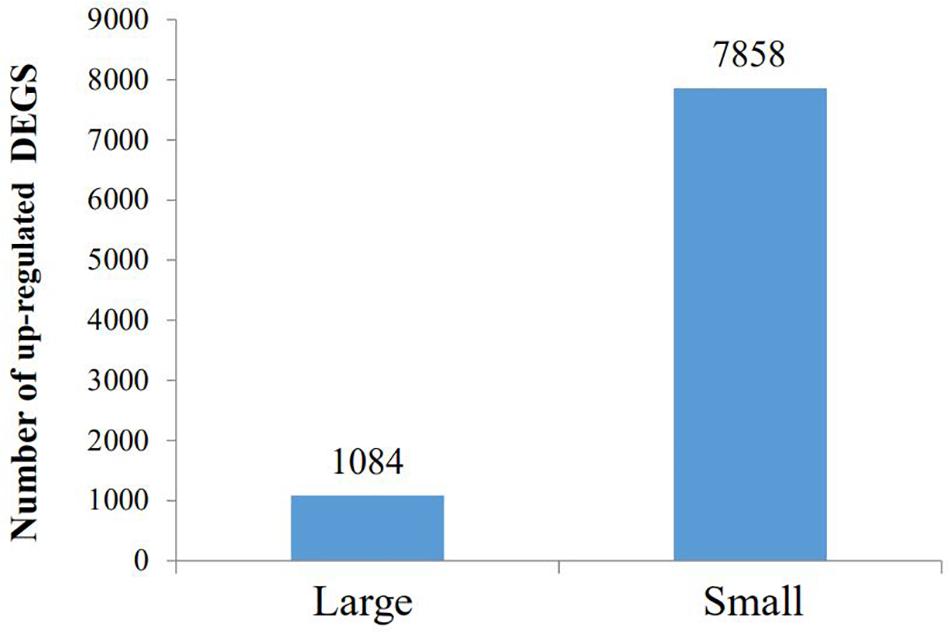
Figure 3. The GO analysis of different expression genes. The 5150 differentially expressed genes were divided into three groups, respectively, biological process (1530), cellular component (1737), and molecular function (1883).
KEGG analysis obtained 25 pathways, among which the most differentially expressed genes were protein processing in endoplasmic reticulum (119, 3.77%), ubiquitin mediated proteolysis (101, 3.2%) and RNA transport (91, 2.88%), respectively (Table 4). The results were confirmed by RT-PCR, proving that the genes expression difference and transcriptome results had the same trend (Figure 4).
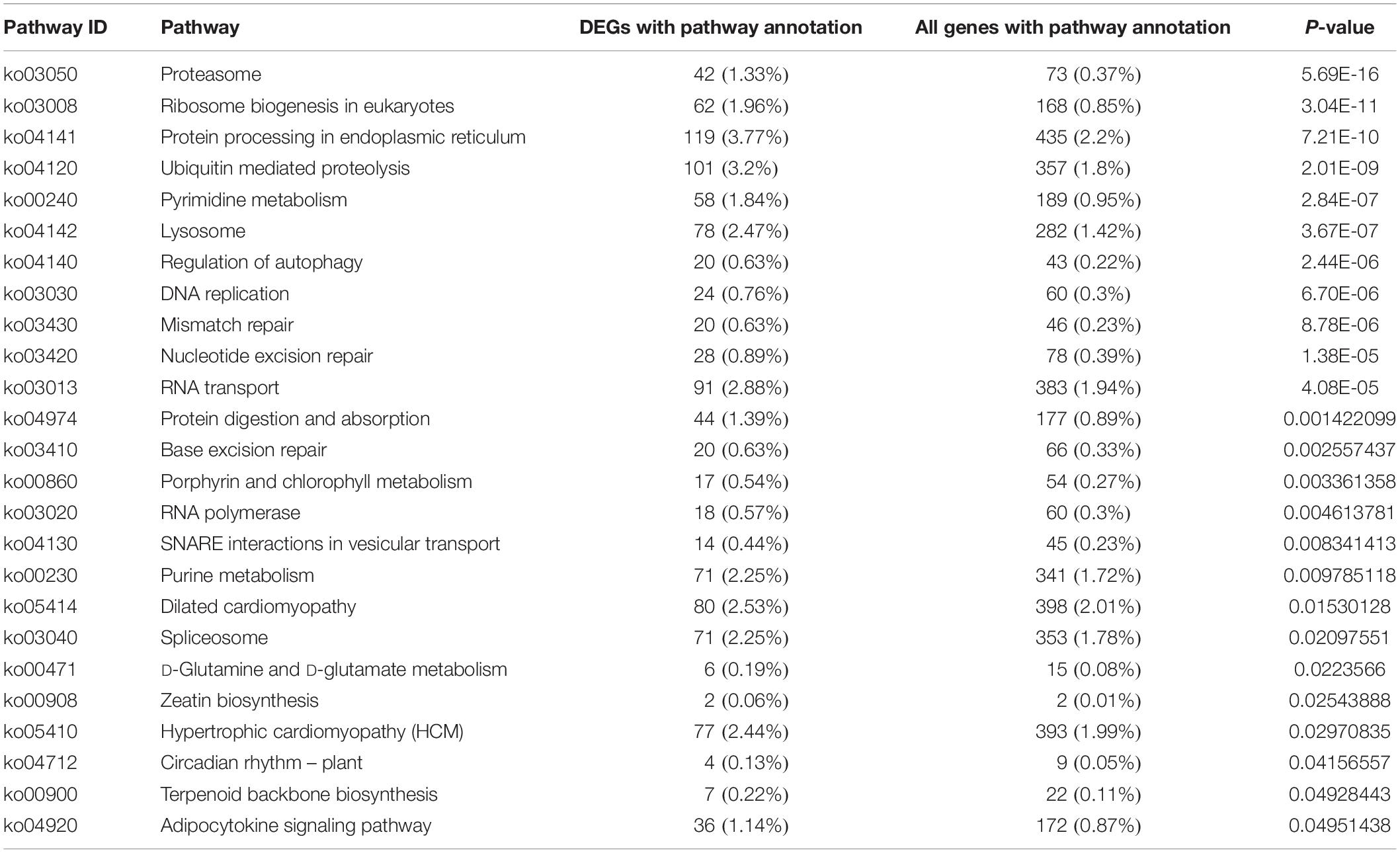
Table 4. The pathway analysis of different expression genes between small and large in Siniperca chuatsi.
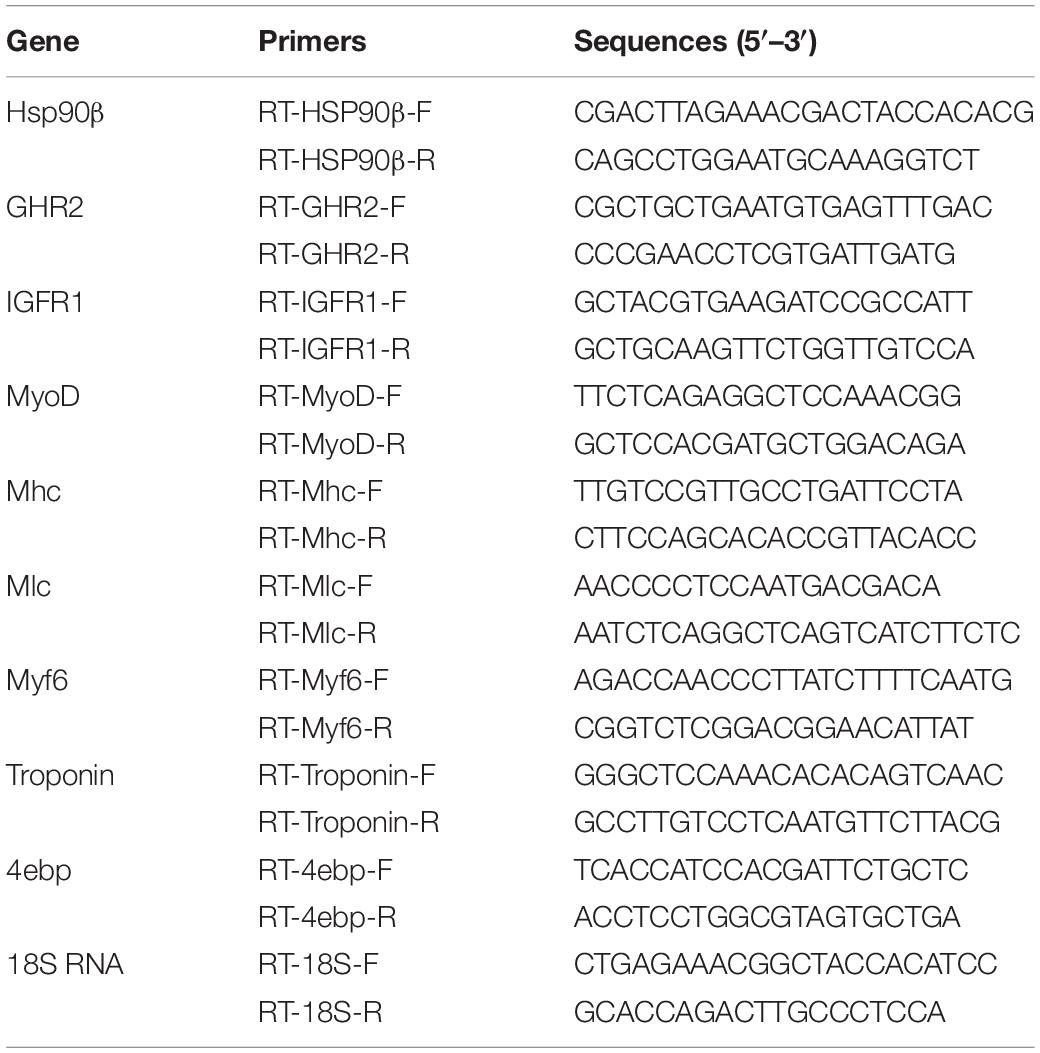
Table 5. Primer sequences for Hsp90β, GHR2, IGFR1, MyoD, Mhc, Mlc, Myf6, Troponin, 4ebp, and 18S RNA.
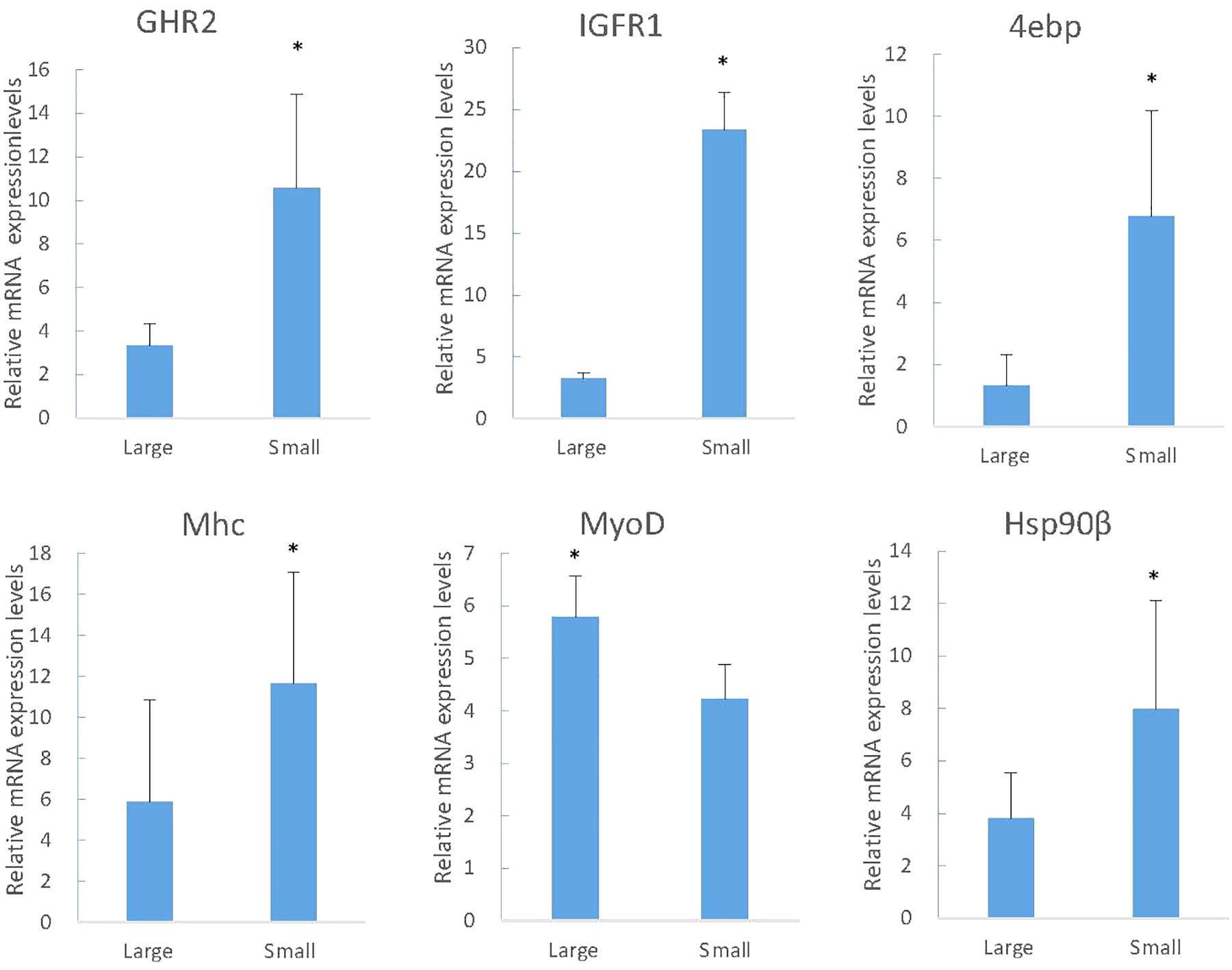
Figure 4. The RT-PCR analysis of gene expression in large and small fish. The y-axis indicates relative expression level between the samples of large and small fish using the Relative Quantitation. IGFR1, insulin-like growth factor receptor -1; GHR1, growth factor receptor-1; 4EBP, eukaryotic translation initiation factor 4E-binding protein; Mhc, fast skeletal muscle myosin heavy chain; Hsp90β, heat shock protein hsp90 beta (Hsp 90β); MyoD, myoblast determination protein. Significant differences at the P < 0.05 level are indicated by * above the columns.
The Expression Growth-Related Genes in Different Periods
The muscle growth-related genes screened in RNA-Seq were studied in different family and period to identify the expression and regulatory mechanisms. In two periods of two families, the expression of MyoD in L was just above S, but Myf6 was just the opposite. For the expression of IGFR1 and 4ebp in S fish was higher. Along with the increased growth time (5-month old), the expression of Mhc and Mlc in S was significantly higher than that in L (Figure 5, Mhc and Mlc).
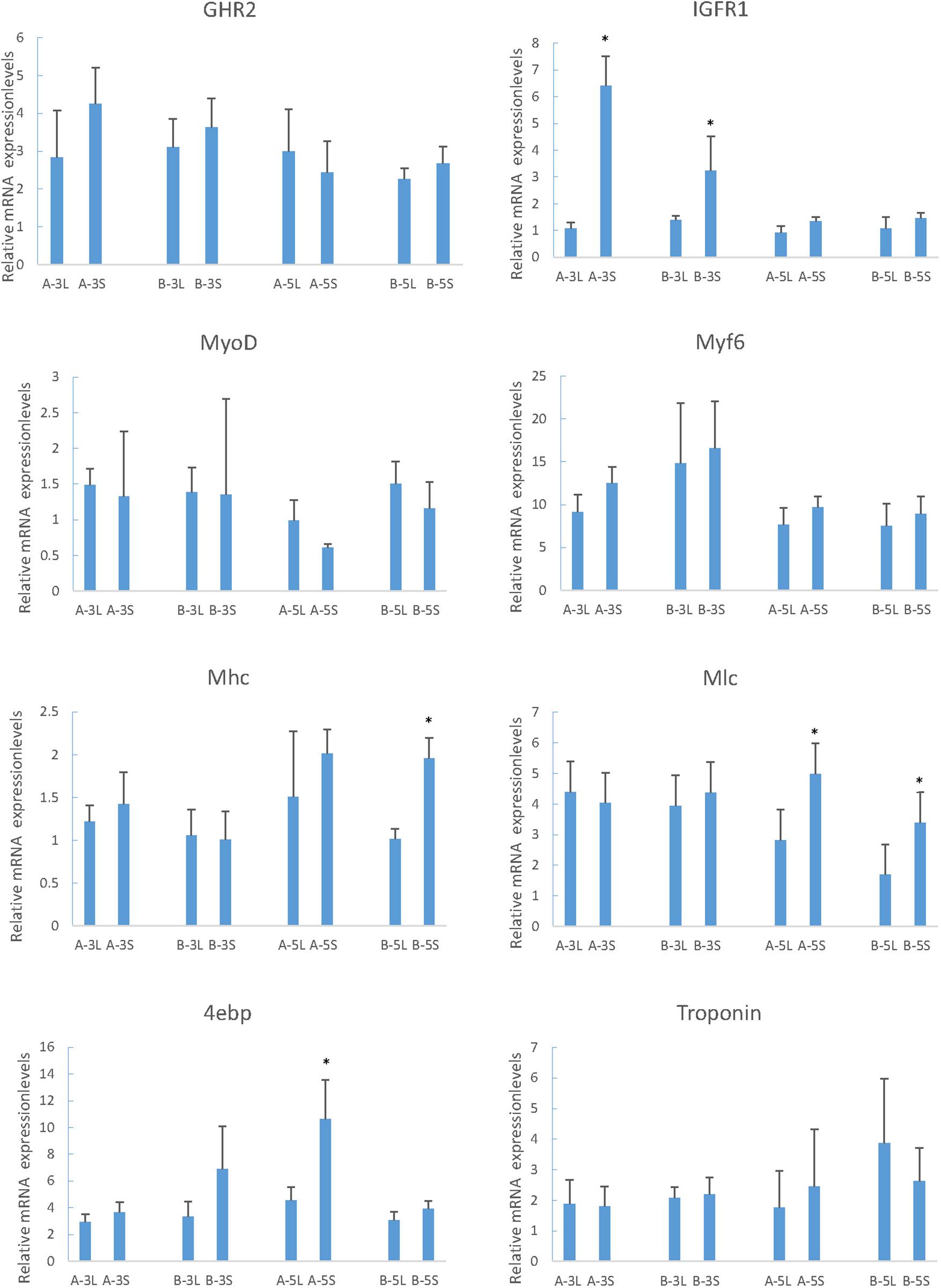
Figure 5. Relative expression of growth-related genes in mandarin fish with different growth traits. 18S was used as a housekeeping gene. All data are represented as the mean ± SEM (n = 6). * Indicates a significant (p < 0.05) difference between overweight and underweight. A-3L, large fish of 3-month old from A family; A-3S, small fish of 3 months from A family; B-3L, large fish of 3-month old from B family; B-3S, small fish of 3-month old from B family; A-5L, large fish of 5- month old from A family; A-5S, small fish of 5-month old from A family; B-5L, Large fish of 5-month old from B family; B-5S, small fish of 5-month old from B family.
Changes in Body Weight During Compensatory Growth
The SGR in different time intervals was calculated (Figure 6A) and the weight of fish in two groups was reflected at seven time points and curves were drawn (Figure 6B). In the first week, the weight of the experimental group decreased markedly following the induction of starvation, as indicated by a negative SGR (−0.98) which reached the lowest (−2.62) in the second week. In the third week, the body weight loss slowed down (SGR −0.64). During the following week of re-feeding, the weight of the experimental group increased rapidly and resulted in a positive of SGR (2.99), which was significantly higher than (P < 0.01) that in the control group. The elevated SGR that characterizes compensatory growth subsequently declined back to low level during the 2 weeks of realimentation, but the SGR of experimental groups was higher than (P > 0.05) that in the control group (Figure 6A). The bodyweight difference of experiment and controls ultimately was not significant difference, indicating that the experimental groups achieved complete compensatory growth after re-feeding.
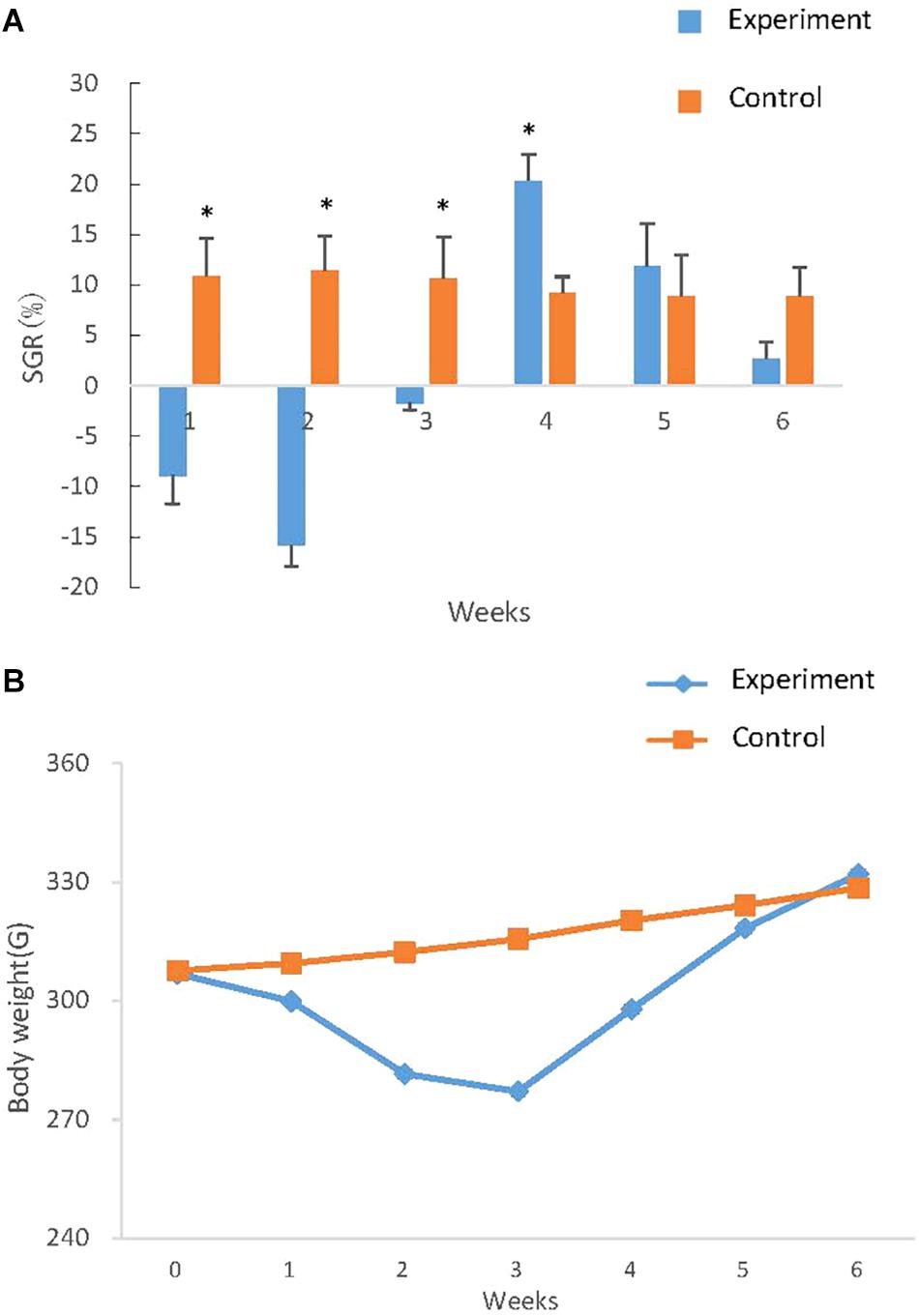
Figure 6. Growth curve and SGR of S. chuatsi during compensatory growth. (A) Growth curve of S. chuatsi during compensatory growth. Fish in experimental and control groups were weighted at six time points and the weights were subjected to curve drawn. In each time point, 6 S. chuatsi were random selected and weighted. Data are given as mean ± standard deviation (SD). (B) SGR of S. chuatsi during compensatory growth. SGRs were calculated for control and experimental group during the six time intervals: the first 3 weeks (1, 2, 3 weeks) means 1, 2, 3 weeks of fasting respectively, the last 3 weeks (4, 5, 6 weeks) means 1, 2, 3 weeks of re-feeding. Asterisks represent significant differences between groups at each time intervals (P < 0.05) that calculated by t- test.
Effect of Re-feeding to Expression of Related-Growth Genes
During starvation, the expression of GHR2, IGFR1 and 4ebp was up-regulated, and the difference of expression increased with the increase of starvation time, and the expression was close to that of the control group following re-feeding (Figure 7). The expression levels of Mhc, Mlc, troponin, and MyoD in starvation revealed the descent tendency, however, there was no significant difference in Myf6 expression (Figure 7).
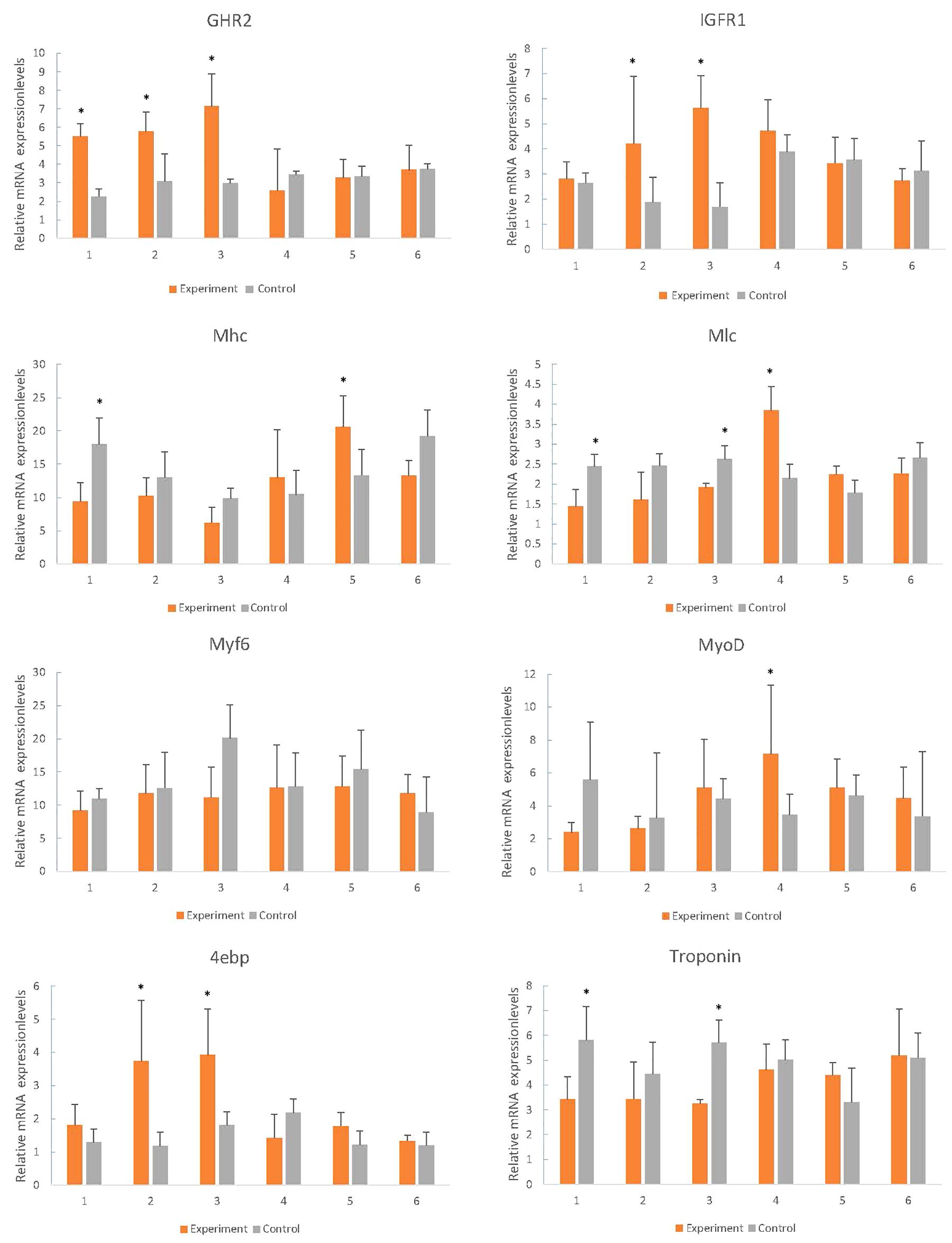
Figure 7. Effects of fasting and re-feeding on muscle expression of growth-related genes in mandarin fish. After acclimated for a month, fish were divided into two groups (n = 40/group). As for fed or re-fed group, fish were fed or fasted for 21 days, and at the end of experiment, fish were fed or re-fed for 6 h before sampling; and in the fasted group, fish had been fasted before sampling. Times of sampling: times 1, 2, 3 means 1, 2, 3 weeks of fasting respectively, times 4, 5, 6 means 1, 2, 3 weeks after re-feeding. Each bar represents the mean ± SEM (n = 6). Significant differences at the P < 0.05 level are indicated by * above the columns.
Discussion
The growth and development of vertebrates are mainly mediated through the GH-IGF system. Especially in teleost fish, the multiple forms of GHRs, and IGFRs have been described. However, they have distinct functions and expression patterns in various fish tissues (Saera-Vila et al., 2005). In tilapia, GHR2 participates in growth and metabolism and is highly expressed in liver and muscle, while GHR1 expression is higher in liver (Pierce et al., 2007). In seabream and rainbow trout, GHR2 expression is much higher than GHR1 in numerous tissues such as muscle, pituitary, kidney, and gonad (Very et al., 2005; Jiao et al., 2006). Interestingly, although GHR expression is affected by GH in many other fish, the opposite effects are found may due to the different expression levels. The growth responses are enhanced in loaches and GH-transgenic salmon and restrained in tilapia and carp (Zhang et al., 1990; Devlin et al., 1994; Hernández et al., 1997; Nam et al., 2001). The weight and thick muscle fibers of GH/GHR-transgenic zebrafish is lower than the other genotypes (Silva et al., 2015). In our experiments, however, both the enhancement and inhibition of growth occurred under different conditions, respectively. With the change of time, the expression of GHR2 among different sizes of individuals showed no significant difference (Figure 5, GHR2), indicating that the stable expression of GHR2 was crucial to promoting growth and development. During starvation, high expression of GHR on the surface of muscle cells can save protein consumption (Pierce et al., 2007), which also has been reported in gilthead sea bream and rainbow trout (Saera-Vila et al., 2005; Gabillard and Kamangar, 2006). Therefore, in S. chuatsi, GHR2 appears to inhibit growth through significant high expression during starvation (Figure 7, GHR2) and plays a physiologically significant role in muscle metabolism.
IGFRs can bind to IGF-1, IGF-2 and insulin, and the affinity for insulin is much lower than for IGFs in all species (Wood and Duan, 2005). Furthermore, in contrast to the situation in mammals, the IGFR1 of fish muscle is greater abundance than insulin receptors (IR) (Wood and Duan, 2005), indicating that IGFR1 is more relevant to the regulation of muscle function than IR in fish (Reinecke et al., 2005), involving promoting growth, enhancing protein, increasing cell proliferation and reducing protein degradation (Radaelli et al., 2003). IGFR1 was highly expressed in S at 3-month old (Figure 5, IGFR1) (P < 0.05), suggesting IGFR1 played a more essential role for S over L in muscle, such as participating in and promoting the growth of muscle. Interestingly, prolonged fasting results in significantly reduced IGF-1 mRNA in the liver and muscle of numerous fish species (Pedroso et al., 2006; Peterson and Waldbieser, 2009; Chen et al., 2019), whereas IGFR1 increased significantly (Yakar et al., 2005; Montserrat et al., 2007; Chen et al., 2019). In this study, the IGFR1 expression markedly rose during fasting (Figure 7, IGFR1), illustrating that IGFR1 expression will increase to restrict IGF-I during starvation, which is probably related to the anti-apoptotic effects reported in mammals (Yakar et al., 2005).
Phosphorylation of protein kinase B (PKB) is controlled by the metabolic pathway regulated by IGFR1, and PKB can mediate the AKT/mTOR/p70S6K pathway to facilitate protein synthesis and cell growth (Jiang et al., 1999; Bodine et al., 2001; Sasai and Miyazu, 2010). Meanwhile, mTOR can regulate translation by 4ebp (Nissim and Nahum, 2004), and the phosphorylation of 4ebp releases eIF4E to stimulate translation initiation (Stephan et al., 2006). eIF4E is able to enhance the translation of mRNAs, implicating in cell proliferation and growth (Mamane et al., 2004), but it is hypothesized that overexpression of eIF4E leads to the deregulation of translational and cellular homeostasis (Mamane et al., 2006). However, the family of 4ebps can inhibit the assembly of eIF4F complex (Mamane et al., 2006). Thus, 4ebp inhibits cell growth and reverts the transformed phenotype of eIF4E-overexpressing cell (Mamane et al., 2006), which may save energy and prevent the malignant transformation of cells during starvation by significant expression (Figure 7, 4ebp). In conclusion, we speculate that the expression of IGFR1 will promote the expression of 4ebp through the above-mentioned pathways to cope with the food restriction. In compensatory growth experiments, the expression of IGFR1 and 4ebp was consistent (Figure 7, IGFR1 and 4ebp), which proved the hypothesis. But further research is needed on specific regulatory mechanisms. It has been reported that fasting promotes the metabolic actions of GH rather than the growth-promoting actions (Norbeck et al., 2007), however, relevant aspects of researches in GHR2, IGFR1 and 4ebp are less. In the study, the functions of GHR2, IGFR1 and 4ebp also changed from growth promotion to growth inhibition in starvation, thus highlighting the metabolic functions.
Mhc expression is significantly related with growth rate (Dhillon et al., 2010; Churova et al., 2015; Overturf and Hardy, 2015), which also affects myofiber hyperplasia and indeterminate growth (Biga and Goetz, 2006). In this study, Mhc was highly expressed in S at 5-month old (Figure 5, Mhc) (P < 0.05), and transcriptome results also showed that the growth rate of small fish was high, suggesting that Mhc can promote the growth of S through its high expression. In larval stage of S. chuatsi, the high expression of Mlc regulates muscle formation and early development (Chu et al., 2011), which also influences the tail shaft swimming character at posterior area, needing more muscle fiber and protein (Patel et al., 1996). The significantly high expression of Mlc at 5-month old in S (Figure 5, Mlc) proved that muscle protein and muscle fiber synthesize quickly to meet physiological requirement. Taken together, as important components of myosin, Mhc and Mlc played a more positive role in S comparing to L at 5-month old. Interestingly, although they all enhanced the growth of S. chuatsi, the function time and action modes of myosin (Mlc, Mhc), GHR2, IGRF1 and 4ebp were different. GHR2 was expressed stably to promote growth (Figure 5, GHR2), and IGFR1 had a more obvious promotion effects in S at 3-month old (Figure 5, IGFR1). Furthermore, both myosin (Mlc, Mhc) and 4ebp facilitated more markedly in S at 5-month old (Figure 5, Mlc, Mhc, and 4ebp), showing all of them had their own temporal and spatial expression patterns in S. chuatsi.
Myosin, the primary protein of muscles, amounts to 50% of the muscle proteins (Watabe and Ikeda, 2006). Troponin as a complex protein is largely expressed in muscle and plays a vital role in muscle contraction (Fu et al., 2009; Huang and Zhang, 2009). During fasting conditions, the markedly down-regulated of Mhc and Mlc indicated that muscle protein synthesis decreased (Figure 7, Mhc and Mlc), and muscle proteins may be used as the major energy source to maintain basic metabolism, which limited growth and was consistent with the observed reduced weight (Figure 6). In the meantime, because muscle breakdown and muscle protein synthesis were inhibited, muscle contraction and motor function were restrained, which was manifested as a significant decrease in troponin expression (Figure 7, Troponin). After re-feeding, significant up-regulation of Mhc and Mlc was observed (Figure 7, Mhc and Mlc), showing that muscle protein was no longer used as the major energy source, and synthesis of muscle and rapid compensatory growth occurred, which resulted in increased weight (He et al., 2015). As the normal synthesis and growth of muscle, muscle contraction was no longer inhibited and expression levels returned to normal (Figure 7, Troponin).
MyoD is associated with myogenesis during developmental condition which plays a regulatory role in muscle hypertrophy and muscle mass (Ahammad et al., 2015; Huang et al., 2018). Myf6 participates in myofiber differentiation by recruiting structural proteins (Xie et al., 2010). From 3- to 5-month old, MyoD and Myf6 expression had a downward trend (Figure 5, MyoD and Myf6), which agreed with the experiment phenomenon of Zhu et al. (2016). It suggested that 3-month old may be a rapid growth stage for S. chuatsi, accompanying with an intense myoblast differentiation, which primarily contributed to the muscle growth (Zhu et al., 2016). What puzzled us is that both MyoD and Myf6 belong to the MRFs family, but the trends of expression results were opposite in L and S (Figure 5, MyoD and Myf6), and there was no significant difference, which may be influenced by the complex regulatory systems in vivo. It has been reported that the reduction in muscle weight may stimulate MRFs transcription (Johansen and Overturf, 2005). And in mammals, satellite cells can express MyoD and Myf5 in response to muscle damage (Smith et al., 1994; Hawke and Garry, 2001). Interestingly, during the starvation period, the expression of MyoD fell to the lowest level and then increased following the weight loss. After re-feeding, the expression gradually decreased from the highest level, and during the whole process, the expression had a certain delayed (Figure 7, MyoD). But, the expression of Myf6 was stable throughout the process. These phenomena indicated that MyoD had sensory and regulatory effects on the weight loss and was conducive to the muscle growth and recovery for S. chuatsi.
Conclusion
Numerous DEGs were identified, and several significant DEGs were chosen to explore the expression in compensatory growth. Fasting promoted the metabolic actions (growth inhibiting) of GHR2, IGFR1, and 4ebp rather than the growth-promoting actions. MyoD can sense and regulate the hunger, which also worked with Mhc and Mlc to accelerate the compensatory growth of S. chuatsi. This study expands our understanding of the mechanism of compensatory growth, and the elected genes will contribute to the selective breeding in future as candidate genes.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found in the DRYAD database: https://doi.org/10.5061/dryad.wh70rxwjx.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Ethics Committee of Sun Yat-sen University.
Author Contributions
GL conceived the study. GL and XL designed the experiments and drafted the work or revised it critically for important content. XL, SZ, HL, and SB collected the samples. XL did the experiments. XL, SZ, and DG prepared the figures and tables. YZ, GW, XC, XZ, SL, YS, and HY made other contributions. The manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This research was supported by the National Key R&D Program of China (2018YFD0901203), Guangdong Basic and Applied Basic Research Foundation (2019B1515120072), the Research in Spreading and Breeding Feedstuff of a New Species of Siniperca chuatsi × Siniperca scherzeri (Yuenong 2019A2), and the Results Transformation and Achievement Extension of Siniperca chuatsi × Siniperca scherzeri. None of the funding bodies participated in the design of the study, collection, analysis, and interpretation of data, or writing the manuscript.
Acknowledgments
We would like to thank the aquatic breeding farm in Foshan for providing mandarin fish and local fishermen for assistance in collection of samples, GL for his invaluable feedback on drafts of this manuscript, DG and HL for their assistance with mandarin fish husbandry. This manuscript has been released as a pre-print at Research Square (Liu et al., 2020).
Abbreviations
4ebp, eukaryotic translation initiation factor 4E-binding protein; DEGs, differentially expressed genes; GH, growth hormone; GHR, GH receptors; GO, gene ontology; Hsp90 β, heat shock protein hsp90 beta; IGF, insulin-like growth factor; IGFR, insulin-like growth factor receptors; KEGG, kyoto encyclopedia of genes and genomes; L, large individuals; Mhc, myosin heavy chain; Mlc, myosin light chain; MRFs, myogenic regulatory factors; MyoD, myoblast determination protein; Myf6, myogenic factor 6; RNA-seq, RNA-sequencing; S, small individuals; SGR, specific growth rate.
References
Ahammad, A. K. S., Asaduzzaman, M., Asakawa, S., Watabe, S., and Kinoshita, S. (2015). Regulation of gene expression mediating indeterminate muscle growth in teleosts. Mech. Dev. 137, 53–65. doi: 10.1016/j.mod.2015.02.006
Ali, M. A., Nicieza, A. G., and Wootton, R. J. (2003). Compensatory growth in fishes: a response to growth depression. Fish Fish. 4, 147–190. doi: 10.1046/j.1467-2979.2003.00120.x
Biga, P. R., and Goetz, F. W. (2006). Zebrafish and giant danio as models for muscle growth: determinate vs. indeterminate growth as determined by morphometric analysis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 291, R1327–R1337.
Bodine, S. C., Stitt, T. N., Gonzalez, M., Kline, W. O., Stover, G. L., Bauerlein, R., et al. (2001). Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3, 1014–1019. doi: 10.1038/ncb1101-1014
Brogiolo, W., Stocker, H., Ikeya, T., Rintelen, F., Fernandez, R., and Hafen, E. (2001). An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213–221. doi: 10.1016/s0960-9822(01)00068-9
Chauvigné, F., Gabillard, J. C., Weil, C., and Rescan, P. Y. (2003). Effect of refeeding on IGFI, IGFII, IGF receptors, FGF2, FGF6, and myostatin mRNA expression in rainbow trout myotomal muscle. Gen. Comp. Endocrinol. 132, 209–215. doi: 10.1016/s0016-6480(03)00081-9
Chen, X., Wang, G., Lu, X., Xu, P., Zeng, S., Chen, Z., et al. (2019). Molecular characterization and expression profiles of two insulin-like growth factor 1 receptors during fasting and re-feeding in Siniperca chuatsi. Fish. Sci. 85, 349–360. doi: 10.1007/s12562-019-01290-7
Chu, W. Y., Chen, J., Zhou, R. X., Zhao, F. L., and Meng, E. (2011). Characterization and ontogenetic expression analysis of the myosin light chains from the fast white muscle of mandarin fish Siniperca chuatsi. J. Fish Biol. 78, 1225–1238. doi: 10.1111/j.1095-8649.2011.02929.x
Churova, M. V., Meshcheryakova, O. V., Veselov, A. E., and Nemova, N. N. (2015). Activity of enzymes involved in the energy and carbohydrate metabolism and the level of some molecular-genetic characteristics in young salmons (Salmo salar L.) with different age and weight. Ontogenez 46:304.
Collins, J. E., White, S., Searle, S. M. J., and Stemple, D. L. (2012). Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome Res. 22, 2067–2078. doi: 10.1101/gr.137901.112
De-Santis, C., and Jerry, D. R. (2007). Candidate growth genes in finfish — Where should we be looking? Aquaculture 272, 22–38. doi: 10.1016/j.aquaculture.2007.08.036
Devlin, R. H., Dionne, S., Tymchuk, W. E., Rise, M. L., and Benjamin, G. (2009). Domestication and growth hormone transgenesis cause similar changes in gene expression in coho salmon (Oncorhynchus kisutch). Proc. Natl. Acad. Sci. U.S.A. 106, 3047–3052. doi: 10.1073/pnas.0809798106
Devlin, R. H., Timothy, Y., Yesaki, C. A., Biagi, E. M., Donaldson, P., and Swanson, W. K. C. (1994). Extraordinary salmon growth. Nature 371, 209–210. doi: 10.1038/371209a0
Dhillon, R. S., Esbaugh, A. J., Wang, Y. S., and Tufts, B. L. (2010). Characterization and expression of a myosin heavy-chain isoform in juvenile walleye Sander vitreus. J. Fish Biol. 75, 1048–1062. doi: 10.1111/j.1095-8649.2009.02376.x
Dou, Y., He, S., Liang, X. F., Cai, W., Wang, J., Shi, L., et al. (2018). Memory function in feeding habit transformation of mandarin fish (Siniperca chuatsi). Int. J. Mol. Sci. 19:1254. doi: 10.3390/ijms19041254
Fu, C. Y., Lee, H. C., and Tsai, H. J. (2009). The molecular structures and expression patterns of zebrafish troponin I genes. Gene Expr. Patterns 9, 348–356. doi: 10.1016/j.gep.2009.02.001
Gabillard, J., and Kamangar, N. B. (2006). Coordinated regulation of the GH/IGF system genes during refeeding in rainbow trout (Oncorhynchus mykiss). J. Endocrinol. 191, 15–24. doi: 10.1677/joe.1.06869
Hawke, T. J., and Garry, D. J. (2001). Myogenic satellite cells: physiology to molecular biology. J. Appl. Physiol. 91, 534–551. doi: 10.1152/jappl.2001.91.2.534
He, L., Pei, Y., Jiang, Y., Li, Y., Liao, L., Zhu, Z., et al. (2015). Global gene expression patterns of grass carp following compensatory growth. BMC Genomics 16:184. doi: 10.1186/s12864-015-1427-2
Hernández, O., Guillén, I., Estrada, M. P., Cabrera, E., Pimentel, R., Piña, J. C., et al. (1997). Characterization of transgenic tilapia lines with different ectopic expression of tilapia growth hormone. Mol. Mar. Biol. Biotechnol. 6:364.
Huang, R., Zhang, J., Zhu, G., He, J., and Xie, J. (2017). The core ubiquitin system of mandarin fish, Siniperca chuatsi, can be utilized by infectious spleen and kidney necrosis virus. Fish Shellfish Immunol. 70, 293–301. doi: 10.1016/j.fsi.2017.09.017
Huang, W., and Zhang, R. X. (2009). Myofibrillogenesis in the developing zebrafish heart: a functional study of tnnt2. Dev. Biol. 331, 237–249. doi: 10.1016/j.ydbio.2009.04.039
Huang, Y., Wen, H., Zhang, M., Hu, N., Si, Y., Li, S., et al. (2018). DNA methylation status of MyoD and IGF-I gene correlated with its muscle growth during different development stages of Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 219, 33–43. doi: 10.1016/j.cbpb.2018.02.005
Jiang, B. H., Aoki, M., Zheng, J. Z., Li, J., and Vogt, P. K. (1999). Myogenic signaling of phosphatidylinositol 3-kinase requires the serine-threonine kinase Akt/protein kinase B. Proc. Natl. Acad. Sci. U.S.A. 96, 2077–2081. doi: 10.1073/pnas.96.5.2077
Jiao, B., Huang, X., Chan, C. B., Zhang, L., Wang, D., and Cheng, C. H. (2006). The co-existence of two growth hormone receptors in teleost fish and their differential signal transduction, tissue distribution and hormonal regulation of expression in seabream. J. Mol. Endocrinol. 36, 23–40. doi: 10.1677/jme.1.01945
Johansen, K. A., and Overturf, K. (2005). Quantitative expression analysis of genes affectingmuscle growth during development of rainbow trout(Oncorhynchus mykiss). Mar. Biotechnol. 7, 576–587. doi: 10.1007/s10126-004-5133-3
Liu, S., Zhang, Y., Zhou, Z., Waldbieser, G., Sun, F., Lu, J., et al. (2012). Efficient assembly and annotation of the transcriptome of catfish by RNA-Seq analysis of a doubled haploid homozygote. BMC Genomics 13:595. doi: 10.1186/1471-2164-13-595
Liu, X. G., Liu, S., Zeng, S., Wang, G. P., Lai, H., Zhao, X. P., et al. (2020). Identifying muscle growth-related genes of mandarin fish (Siniperca chuatsi) by transcriptome, and exploring the expression in different sizes and compensatory growth. Res. Square
Mamane, Y., Emmanuel, P., Liwei, R., Kaori, Y., Wee, L. L., and Nahum, S. (2004). eIF4E–from translation to transformation. Oncogene 23, 3172–3179. doi: 10.1038/sj.onc.1207549
Mamane, Y., Petroulakis, E. L. O., and Sonenberg, N. (2006). mTOR, translation initiation and cancer. Oncogene 25, 6416–6422.
Montserrat, N., Gabillard, J. C., Capilla, E., Navarro, M. I, and Gutiérrez, J. (2007). Role of insulin, insulin-like growth factors, and muscle regulatory factors in the compensatory growth of the trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 150, 462–472. doi: 10.1016/j.ygcen.2006.11.009
Nam, Y. K., Noh, J. K., Cho, Y. S., Cho, H. J., Cho, K. N., Kim, C. G., et al. (2001). Dramatically accelerated growth and extraordinary gigantism of transgenic mud loach Misgurnus mizolepis. Transgenic Res. 10, 353–362. doi: 10.1111/j.1749-7345.2001.tb00461.x
Nebo, C., Portella, M. C., Carani, F. R., De Almeida, F. L. A., Padovani, C. R., Carvalho, R. F., et al. (2013). Short periods of fasting followed by refeeding change the expression of muscle growth-related genes in juvenile Nile tilapia (Oreochromis niloticus). Comp. Biochem. Physiol. B 164, 268–274. doi: 10.1016/j.cbpb.2013.02.003
Norbeck, L. A., Kittilson, J. D., and Sheridan, M. A. (2007). Resolving the growth-promoting and metabolic effects of growth hormone: differential regulation of GH–IGF-I system components. Gen. Comp. Endocrinol. 151, 332–341. doi: 10.1016/j.ygcen.2007.01.039
Overturf, K., and Hardy, R. W. (2015). Myosin expression levels in trout muscle: a new method for monitoring specific growth rates for rainbow trout Oncorhynchus mykiss (Walbaum) on varied planes of nutrition. Aquac. Res. 32, 315–322. doi: 10.1046/j.1365-2109.2001.00582.x
Palstra, A. P., Sergi, B., Erik, B., Brittijn, S. A., Magnoni, L. J., Henkel, C. V., et al. (2013). Deep RNA sequencing of the skeletal muscle transcriptome in swimming fish. PLoS One 8:e53171. doi: 10.1371/journal.pone.0053171
Patel, J. R., Diffee, G. M., and Moss, R. L. (1996). Myosin regulatory light chain modulates the Ca2+ dependence of the kinetics of tension development in skeletal muscle fibers. Biophys. J. 70, 2333–2340. doi: 10.1016/s0006-3495(96)79799-0
Pedroso, F. L., Jesus-Ayson, E. G. T., Cortado, H. H., Hyodo, S., and Ayson, F. G. (2006). Changes in mRNA expression of grouper (Epinephelus coioides) growth hormone and insulin-like growth factor I in response to nutritional status. Gen. Comp. Endocrinol. 145, 237–246. doi: 10.1016/j.ygcen.2005.09.001
Peterson, B. C., and Waldbieser, G. C. (2009). Effects of fasting on IGF-I, IGF-II, and IGF-binding protein mRNA concentrations in channel catfish (Ictalurus punctatus). Domest. Anim. Endocrinol. 37, 74–83. doi: 10.1016/j.domaniend.2009.03.004
Picha, M. E., Biga, P. R., Galt, N., Mcginty, A. S., Gross, K., Hedgpeth, V. S., et al. (2014). Overcompensation of circulating and local insulin-like growth factor-1 during catch-up growth in hybrid striped bass (Morone chrysops × Morone saxatilis) following temperature and feeding manipulations. Aquaculture 428-429, 174–183. doi: 10.1016/j.aquaculture.2014.02.028
Pierce, A. L., Fox, B. K., Davis, L. K., Visitacion, N., Kitahashi, T., Hirano, T., et al. (2007). Prolactin receptor, growth hormone receptor, and putative somatolactin receptor in Mozambique tilapia: tissue specific expression and differential regulation by salinity and fasting. Gen. Comp. Endocrinol. 154, 31–40. doi: 10.1016/j.ygcen.2007.06.023
Radaelli, G., Domeneghini, C., Arrighi, S., Bosi, G., Patruno, M., and Funkenstein, B. (2003). Localization of IGF-I, IGF-I receptor, and IGFBP-2 in developing Umbrina cirrosa (Pisces: Osteichthyes). Gen. Comp. Endocrinol. 130, 232–244. doi: 10.1016/s0016-6480(02)00609-3
Reinecke, M., Björnsson, B. T., Dickhoff, W. W., McCormick, S. D., Navarro, I., Power, D. M., et al. (2005). Growth hormone and insulin-like growth factors in fish: where we are and where to go. Gen. Comp. Endocrinol. 142, 20–24. doi: 10.1016/j.ygcen.2005.01.016
Riley, L. G., Richman, N. H., Tetsuya, H., and Grau Gordon, E. (2002). Activation of the growth hormone/insulin-like growth factor axis by treatment with 17 alpha-methyltestosterone and seawater rearing in the tilapia, Oreochromis mossambicus. Gen. Comp. Endocrinol. 127, 285–292. doi: 10.1016/s0016-6480(02)00051-5
Rowlerson, A., and Veggetti, A. (2001). 5 – cellular mechanisms of post-embryonic muscle growth in aquaculture species. Fish Physiol. 18, 103–140.
Saera-Vila, A., Calduch-Giner, J. A., and Pérez-Sánchez, J. (2005). Duplication of growth hormone receptor (GHR) in fish genome: gene organization and transcriptional regulation of GHR type I and II in gilthead sea bream (Sparus aurata). Gen. Comp. Endocrinol. 142, 193–203. doi: 10.1016/j.ygcen.2004.11.005
Sarropoulou, E., Galindo-Villegas, J., García-Alcázar, A., Kasapidis, P., and Mulero, V. (2012). Characterization of European sea bass transcripts by RNA SEQ after oral vaccine against V. anguillarum. Mar. Biotechnol. 14, 634–642. doi: 10.1007/s10126-012-9466-z
Sasai, N., and Miyazu, A. N. (2010). Involvement of PI3K/Akt/TOR pathway in stretch-induced hypertrophy of myotubes. Muscle Nerve 41, 100–106. doi: 10.1002/mus.21473
Silva, A. C., Almeida, D. V., Nornberg, B. F., Figueiredo, M. A., Romano, L. A., and Marins, L. F. (2015). Effects of double transgenesis of somatotrophic axis (GH/GHR) on skeletal muscle growth of zebrafish (Danio rerio). Zebrafish 12, 408–413. doi: 10.1089/zeb.2015.29001.sil
Smith, C. K., Janney, M. J., and Allen, R. E. (1994). Temporal expression of myogenic regulatory genes during activation, proliferation, and differentiation of rat skeletal muscle satellite cells. J. Cell. Physiol. 159:379. doi: 10.1002/jcp.1041590222
Steinbacher, P., Haslett, J. R., Six, M., Gollmann, H. P., Sänger, A. M., and Stoiber, W. (2010). Phases of myogenic cell activation and possible role of dermomyotome cells in teleost muscle formation. Dev. Dyn. 235, 3132–3143. doi: 10.1002/dvdy.20950
Stephan, W., Robbie, L., and Hall, M. N. (2006). TOR signaling in growth and metabolism. Cell 124, 471–484.
Tian, C., Li, L., Liang, X. F., He, S., Guo, W., Lv, L., et al. (2016). Identification of differentially expressed genes associated with differential body size in mandarin fish (Siniperca chuatsi). Genetica 144, 445–455. doi: 10.1007/s10709-016-9913-2
Veggetti, A., Mascarello, F., Scapolo, P. A., Rowlerson, A., and Carnevali, M. D. C. (1993). Muscle growth and myosin isoform transitions during development of a small teleost fish, Poecilia reticulata (Peters) (Atheriniformes, Poeciliidae): a histochemical, immunohistochemical, ultrastructural and morphometric study. Anat. Embryol. 187, 353–361.
Very, N. M., Kittilson, J. D., Norbeck, L. A., and Sheridan, M. A. (2005). Isolation, characterization, and distribution of two cDNAs encoding for growth hormone receptor in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 140, 615–628. doi: 10.1016/j.cbpc.2004.12.008
Watabe, S., and Ikeda, D. (2006). Diversity of the pufferfish Takifugu rubripes fast skeletal myosin heavy chain genes. Comp. Biochem. Physiol. D Genomics Proteomics 1, 28–34. doi: 10.1016/j.cbd.2005.12.001
Weatherley, A. H., Gill, H. S., and Lobo, A. F. (2010). Recruitment and maximal diameter of axial muscle fibres in teleosts and their relationship to somatic growth and ultimate size. J. Fish Biol. 33, 851–859. doi: 10.1111/j.1095-8649.1988.tb05532.x
Wood, A. W., and Duan, C. H. (2005). Insulin-like growth factor signaling in fish. Int. Rev. Cytol. 243, 215–285. doi: 10.1016/s0074-7696(05)43004-1
Wu, N., Zhang, X.-Y., Huang, B., Zhang, N., Zhang, X.-J., Guo, X., et al. (2015). Investigating the potential immune role of fish NCAMs: molecular cloning and expression analysis in mandarin fish. Fish Shellfish Immunol. 46, 765–777. doi: 10.1016/j.fsi.2015.08.006
Xie, S. Q., Mason, P. S., Stickland, N. C., Wilkes, D., Goldspink, G., and Fauconneau, B. (2010). Lower environmental temperature delays and prolongs myogenic regulatory factor expression and muscle differentiation in rainbow trout (Onchrhynchus mykiss) embryos. Differentiation 68, 106–114. doi: 10.1046/j.1432-0436.2001.680204.x
Yakar, S., Pennisi, P., Kim, C. H., Zhao, H., Toyoshima, Y., Gavrilova, O., et al. (2005). Studies involving the GH-IGF axis: lessons from IGF-I and IGF-I receptor gene targeting mouse models. J. Endocrinol. Invest. 28, 19–22.
Zhang, P., Hayat, M., Joyce, C., Gonzalezvillaseñor, L. I., Lin, C. M., Dunham, R. A., et al. (1990). Gene transfer, expression and inheritance of pRSV-rainbow trout-GH cDNA in the common carp, Cyprinus carpio (Linnaeus). Mol. Reprod. Dev. 25:3. doi: 10.1002/mrd.1080250103
Keywords: mandarin fish, RNA-seq, compensatory growth, muscle growth-related genes, gene expression
Citation: Liu X, Zeng S, Liu S, Wang G, Lai H, Zhao X, Bi S, Guo D, Chen X, Yi H, Su Y, Zhang Y and Li G (2020) Identifying the Related Genes of Muscle Growth and Exploring the Functions by Compensatory Growth in Mandarin Fish (Siniperca chuatsi). Front. Physiol. 11:553563. doi: 10.3389/fphys.2020.553563
Received: 19 April 2020; Accepted: 31 August 2020;
Published: 25 September 2020.
Edited by:
Samad Rahimnejad, University of South Bohemia in České Budějovice, CzechiaReviewed by:
Maeli Dal-Pai-Silva, São Paulo State University, BrazilPin Nie, Institute of Hydrobiology, Chinese Academy of Sciences, China
Tiziano Verri, University of Salento, Italy
Copyright © 2020 Liu, Zeng, Liu, Wang, Lai, Zhao, Bi, Guo, Chen, Yi, Su, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guifeng Li, bGlndWlmQG1haWwuc3lzdS5lZHUuY24=
 Xuange Liu
Xuange Liu Shuang Zeng1,2,3
Shuang Zeng1,2,3 Yong Zhang
Yong Zhang Guifeng Li
Guifeng Li