
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 01 December 2020
Sec. Aquatic Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fphys.2020.530435
 Yukun Zhang1,2,3
Yukun Zhang1,2,3 Zhaoqun Liu1,2,3,4*
Zhaoqun Liu1,2,3,4* Yanan Zong1,2,3
Yanan Zong1,2,3 Yan Zheng1,2,3
Yan Zheng1,2,3 Yinan Li1,2,3
Yinan Li1,2,3 Zirong Han1,2,3
Zirong Han1,2,3 Lingling Wang1,2,3,4*
Lingling Wang1,2,3,4* Linsheng Song1,2,3,4
Linsheng Song1,2,3,4Engrailed is a transcription factor required in numerous species for important developmental steps such as neurogenesis, segment formation, preblastoderm organization, and compartment formation. Recent study has proved that engrailed is also a key gene related to shell formation in marine bivalves. In the present study, the expression pattern of an engrailed gene (Cgengrailed-1) in Pacific oyster Crassostrea gigas under CO2-driven acidification was investigated to understand its possible role in the regulation of shell formation and adaptation to ocean acidification (OA). The open reading frame (ORF) of Cgengrailed-1 was obtained, which was of 690 bp encoding a polypeptide of 229 amino acids with a HOX domain. Phylogenetic analysis indicated that the deduced amino acid sequence of Cgengrailed-1 shared high homology with other engraileds from Drosophila melanogaster, Mizuhopecten yessoensi, and Crassostrea virginica. The mRNA transcripts of Cgengrailed-1 were constitutively expressed in various tissues with the highest expression levels detected in labial palp and mantle, which were 86.83-fold (p < 0.05) and 75.87-fold (p < 0.05) higher than that in hepatopancreas. The mRNA expression of Cgengrailed-1 in mantle decreased dramatically after moderate (pH 7.8) and severe (pH 7.4) acidification treatment (0.75- and 0.15-fold of that in control group, p < 0.05). The results of immunofluorescence assay demonstrated that the expression level of Cgengrailed-1 in the middle fold of mantle increased significantly upon moderate and severe acidification treatment. Moreover, after the oyster larvae received acidification treatment at trochophore stage, the mRNA expression levels of Cgengrailed-1 increased significantly in D-shape larvae stages, which was 3.11- (pH 7.8) and 4.39-fold (pH 7.4) of that in control group (p < 0.05). The whole-mount immunofluorescence assay showed that Cgengrailed-1 was mainly expressed on the margin of shell gland, and the periostracum in trochophore, early D-shape larvae and D-shape larvae in both control and acidification treatment groups, and the intensity of positive signals in early D-shape larvae and D-shape larvae increased dramatically under acidification treatment. These results collectively suggested that the expression of Cgengrailed-1 could be triggered by CO2-driven acidification treatment, which might contribute to induce the initial shell formation in oyster larvae and the formation of periostracum in adult oyster to adapt to the acidifying marine environment.
Engrailed is a homeodomain transcription factor which is involved in plentiful species for prime developmental period, and its evolution is known to be closely related to the evolution of metazoan body plan (Loomis et al., 1996; Iwaki and Lengyel, 2002; Byrne et al., 2005). The first engrailed was characterized from Drosophila melanogaster (Eker, 1929). Since then, lots of homologues of engrailed have been identified from various animal groups including mammals, annelids, arthropods, insects, echinoderms, and chordates (Fjose et al., 1985; Joyner et al., 1985; Holland et al., 1997; Lowe and Wray, 1997; Ikuta, 2017). There are two members included in the engrailed family of vertebrates, engrailed-1 and engrailed-2 (Joyner and Martin, 1985; Matsuura et al., 2010). The sequence of engrailed contains a conserved HOX domain, which is regarded as an important DNA-binding site for transcriptional regulation in crucial developmental processes (Desplan et al., 1985; Patel et al., 1989). The HOX domain of engrailed can bind to DNA mainly relying on the helix-turn-helix structure (two α-helixes connected by a β-corner), in which one of the α-helix combines some hydrogen bonds and hydrophobic interactions to special chains and groups in the major groove of DNA (Piper et al., 1999). Engrailed is also able to bind directly to the eukaryotic translation initiation factor 4E, via a sequence located in the N-terminus (Nedelec et al., 2004).
Engrailed in invertebrates share similar sequence features with their homologues in mammals, which also contain a conserved HOX domain. Engraileds identified from invertebrates are mainly responsible for body segmentation (Tucker-Kellogg et al., 1997). For example, engrailed from arthropods controlled the liner boundary between the anterior and posterior compartments in the wings (Morata and Lawrence, 1975; Poole et al., 1985). Recently, several engraileds have also been characterized from molluscs, which are found to be involved in shell formation (Miyazaki et al., 2010). It was reported that engrailed participated in shell formation in larval and adult molluscs through separate ways. As for the larval of molluscs, engrailed might take part in shell biosynthesis process by establishing compartment boundaries between embryonic domains (Nederbragt et al., 2002). Molluscan engraileds were mainly expressed in shell formation cells around the shell anlagen (Moshel et al., 1998; Kin et al., 2009), and at the edge of the embryonic shell, all of which were implied as the boundary of initial shell in larvae (Lawrence and Struhl, 1996). Besides, engrailed and another regulatory gene dpp were proposed to control the development of molluscan shell by confining the boundary of the larval shell, whose expression level remained high until umbo stage (Kin et al., 2009; Huan et al., 2013). Unlike larval stage, engrailed took part in shell formation in adult molluscs by regulating matrix protein expressions in mantles (Takeuchi and Endo, 2006; Zhou et al., 2010). It was reported that engraileds in adult nautilus were expressed in the marginal cells of the shell field in mantle (Baratte et al., 2007). In snail Ilyanassa obsoleta, the engrailed was detected in the shell gland progenitor and progeny, and the expression of engrailed mRNA was also observed in the shell plate (Moshel et al., 1998). These findings evidence that engrailed is crucial for shell formation in both larval and adult molluscs, and the underlying molecular mechanism deserves a better exploration.
Oysters are benthic species native to the coastal area worldwide. In recent years, the shell formation process in oyster is severely inhibited owing to the OA caused by excessive emission of greenhouse gas such as CO2 and CH4 (Waldbusser and Salisbury, 2014; Wang et al., 2017b). In the present study, the cDNA sequence of an previously reported engrailed, named as Cgengrailed-1, was cloned from the Pacific oyster Crassostrea gigas to (1) understand the molecular feature of Cgengrailed-1, (2) investigate the expression level of Cgengrailed-1 during larval developmental stages and in various tissues of adult oysters, and (3) explore the alteration of Cgengrailed-1 expression in the oysters under CO2-driven acidification and its possible role in the regulation of shell formation and adaptation to ocean acidification (OA).
The Pacific oysters C. gigas (about 2-year old, averaging 150 mm in shell length) were collected from a local farm in Dalian, Liaoning Province, China, and maintained in the aerated seawater at 20°C for 7 days before processing. The eggs and sperms were scraped from different adult oysters and mixed together for fertilization. After fertilization, the developing embryos were continuously observed by microscope until the second polar body was formed about 90 min post fertilization. Then, the embryos were transferred into an aquaculture tank containing 100 L aerated seawater of 20°C (Zhang et al., 2019).
Before acidification treatment on oyster larvae, trochophore were collected at 12 h post fertilization (hpf) according to previous description (Kin et al., 2009), and randomly divided into three groups. The remaining larvae were then transferred into three tanks containing seawater at a pH level of 8.10 ± 0.05 (control group), 7.80 ± 0.05 (moderate acidification treatment group) and 7.40 ± 0.05 (severe acidification treatment group), respectively. After acidification treatment, the larvae were collected at 15 hpf (early D-shape larvae) and 19 hpf (D-shape larvae), respectively. Samples for RNA extraction were added with 1 mL of Trizol reagent (Invitrogen, United States) in each tube. Larvae for whole-mount immunofluorescence assay were first relaxed by gradual addition of 7.5% MgCl2, followed by fixation in fresh 4% paraformaldehyde (PFA) in 0.01 M phosphate buffer saline (PBS) (0.2 g KCl, 8.0 g NaCl, 2.9 g Na2HPO4⋅12H2O, 0.2 g KH2PO4, pH 7.40) at 4°C for 3 h and washed three times (15 min for each time) with pre-cold PBS. After fixation, the samples were dehydrated and stored in pure methanol at −20°C for the subsequent whole-mount immunofluorescence assay.
As for the acidification treatment on adult oysters, 27 oysters were equally divided into three groups. Oyster without any treatment were designated as the control group (pH 8.10 ± 0.05), while the other bathed in acidified seawater at a pH level of 7.80 ± 0.05 and 7.40 ± 0.05 were employed as the moderate acidification treatment group and severe acidification treatment group (Wang et al., 2016). The mantles of three groups were collected at 7th day post treatment and every mantle sample collected from three oysters as one replicate. In addition, there are three replicates were conducted for each assay group. Samples for RNA extraction were added with 1 mL of Trizol reagent in each tube (Invitrogen, United States) and frozen immediately in liquid nitrogen. Tissues for sectioning and immunofluorescence assay were firstly fixed with Bouin’s solution (formalin 10%, picric acid saturated solution in distilled water, and glacial acetic acid, 15:5:1) for 24 h and then dehydrated with gradient ethanol.
The pH value of the acidification treatment group was controlled using an acidometer (AiKB, Qingdao, China). Total alkalinity was determined by end-point titration of 25 mmol L–1 HCl on 50 mL samples. The pH value, total alkalinity and partial press of CO2 (pCO2) were measured in the experiment. The total alkalinity in the pH 8.1, pH 7.8 and pH 7.4 groups was 2849.1 ± 14.5, 2416.3 ± 125.1, and 2110.6 ± 75.3 μeq kg–1, respectively. And the partial pressure of CO2 was about 658.1 ± 11.0, 1217.3 ± 11.6, and 2268.4 ± 50.1 ppm in pH 8.1, pH 7.8 and pH 7.4 treatment groups, respectively (Zhang et al., 2019). All the animal-involving experiments of this study were approved by the Ethics Committee of Dalian Ocean University.
Total RNA was isolated from oyster tissues using Trizol reagent according to the standard protocol (Invitrogen). RNA concentration was measured on a NanoDrop reader (Saveen & Werner ApS, Denmark), and the integrity and purity of RNA was examined by running in 1.0% agarose gel. The total RNA was then treated with DNaseI (promega) to remove trace DNA contamination. The synthesis of the first-strand cDNA was carried out with Promega M-MLV RT with oligo (dT)-adaptor priming according to the manufactory’s protocol. The synthesis reaction was performed at 42°C for 1 h, terminated by heating at 95°C for 5 min. The cDNA mix was diluted to 1:40 and stored at −80°C for subsequent SYBR Green Fluorescent Quantificational real-time PCR.
An engrailed (CGI_10012208) was identified from oyster which was designated as Cgengrailed-1 (Huang et al., 2015). The open reading frame (ORF) of Cgengrailed-1 was cloned from cDNA library using specific primers (P1 and P2, Table 1). The PCR procedure of Cgengrailed-1 was as follows: 5 min at 95°C; 30 cycles at 94°C for 30 s, 56°C for 1 min, 72°C for 40 s, and 72°C for 10 min. The PCR product was gel-purified, cloned into PMD 19-T simple vector (Takara), and confirmed by DNA sequencing.
The homology searches of the cDNA sequence and protein sequence of Cgengrailed-1 was conducted with BLAST algorithm at the National Center for Biotechnology Information1. The deduced amino acid sequence was analyzed with the Expert Protein Analysis System2. The protein domain was predicted with the simple modular architecture research tool3 (SMART) version 5.1. Multiple sequence alignment of Cgengrailed-1 with other engraileds was created by the ClustalW multiple alignment program4 and multiple sequence alignment show program5.
The expression levels of Cgengrailed-1 in different development stages and different tissues were measured by an ABI PRISM 7500 Sequence Detection System with a total volume of 25.0 μL, containing 12.5 μL of SYBR Green Mix (Takara), 0.5 μL of each primer (10 μmol L–1) (P5 and P6, Table 1), 2.0 μL of cDNA, and 9.5 μL of DEPC-water. The oyster elongation factor (EF) was used as internal control (P7 and P8, Table 1). Dissociation curve analysis of amplification products was performed to confirm that only one PCR product was amplified and detected. The comparative average cycle threshold method was used to analyze the mRNA expression level of the immune-related genes according to previous research (Zhang et al., 2008). All data were given in terms of relative mRNA expression using the 2–Δ Δ Ct method (Yu et al., 2007).
The Cgengrailed-1 was induced and expressed through the prokaryotic expression system. The ORF fragment of Cgengrailed-1 was gained from the PCR reaction with pairs of primers (P3 and P4, Table 1). The PCR products and the pET-30a expression vector were linearized with double enzyme digestion, and then the PCR products and linearized expression vector were recombined to a complete vector through T4 ligase system. The DNA sequence combined to pET-30a expression vector was confirmed by DNA sequencing. The valid recombinant plasmid was extracted and transformed into Escherichia coli transetta DE3. Positive transformants were cultured in LB medium at 37°C, 120 rpm incubator. When the OD600 value of bacteria solution reached 0.4–0.6, the bacteria were incubated for additional 8 h with the induction of Isopropyl β-D-thiogalactoside (IPTG) at the final concentration of 1 mM. The recombinant protein of Cgengrailed-1 labeled with 6× His-tag at the C-terminal was purified by Ni+ affinity chromatography, and desalted by extensive dialysis. The purified objective protein (rCgengrailed-1) was concentrated and stored at −80°C (Zong et al., 2019).
Six-week-old mice were immunized with rCgengrailed-1 to acquire polyclonal antibodies following the description of previous study (Cheng et al., 2006). Briefly, 100 μL rCgengrailed-1 (0.3 mg mL–1) was emulsified with 100 μL complete Freund’s adjuvant (Sigma, United States) to immunize each mouse by subcutaneous implantation. The second and third injections were performed on the 14th and 21th day with incomplete Freund’s adjuvant (Sigma, United States). The fourth injection was performed with 100 μL rCgengrailed-1 on the 28th day. The anti-rCgengrailed-1 serum was obtained on the 36th day and stored at −80°C before use.
The specificity of polyclonal antibody against rCgengrailed-1 and natural Cgengrailed-1 (from mantles) was verified by western blot assay. Briefly, rCgengrailed-1 and natural Cgengrailed-1 was separated by 12% SDS-PAGE, and electrophoretically transferred onto a nitrocellulose membrane. The membrane was washed three times with TBS containing 0.1% Tween 20 (TBST), blocked by 5% skimmed milk (100 mL TBST, 5 g skimmed milk) at 4°C overnight, and then incubated with 1:500 diluted polyclonal antibodies against rCgengrailed-1 at 37°C for 3 h. After three times of washing with TBST, the membrane was incubated with 1:3,000 diluted secondary antibody Goat-anti-mouse IgG conjugated with HRP (ABclonal) at 37°C for 2 h. After the final three times of washing with TBST, the protein bands were developed by using Super ECL Detection Reagent (Sigma-Aldrich) for one minute, capture image and take picture by Amersham Imager 600 system (GE Healthcare, United States).
The whole-mount immunofluorescence assay was conducted as previously described (Thisse and Thisse, 2008; Liu et al., 2015) with minor modification. In brief, the larvae stored in pure methanol were rehydrated by PBS and washed three times with PBST (PBS with 0.1% Tween20). The shells of D-shape larvae were decalcified in 25 mmol–1 EDTA in PBS for 30 min, rinsed in PBST for 3 × 15 min, and blocked overnight in the blocking buffer (10% normal goat serum, 0.25% fetal bovine serum albumin, 1% TritonX-100 and 0.03% sodium azide in PBS). Then, the larvae were incubated with the primary antibodies (1:500 dilutions in blocking buffer) at 4°C for 3–4 days. These specimens were then washed for 3 × 15 min in PBST and incubated with goat anti-rat IgG conjugated to Alexa Fluor 488 (1:2,000 dilutions in blocking buffer; Invitrogen, Carlsbad, CA, United States) for 1 day at room temperature. After incubation in secondary antibodies, all the specimens were washed in PBST for 3 × 20 min and then mounted in 50% glycerol in PBS. The specimens incubated with pre-immune serum added with secondary antibodies were selected as negative control (Liu et al., 2015). All specimens were examined as whole-mount using the Zeiss Laser-Scanning Confocal Microscopy System LSM 710 (Zeiss, Jena, Germany).
The oyster mantles from control group and acidification treatment group were fixed in Bouin’s solution for 24 h and decolorized with 75% (v/v) ethanol. Then, the fixed samples were embedded in paraffin, and sectioned at 6 μm thickness. The slides were treated to remove the paraffin and rehydrated. Soak the slides in 0.01 M citrate buffer solution, conduct antigen renaturation in autoclave at 120° C for 6 min, and then cool it to room temperature naturally. After cooling down at room temperature, the slides were prepared for immunohistochemistry staining. The slides were first blocked in BSA, and Cgengrailed-1 polyclonal antibody was used as primary antibody (diluted at 1:500 in blocking buffer). Alexa Fluor 488 labeled goat-anti-mouse (diluted at 1:1,000, Abclonal, College Park, MD, United States) was used as secondary antibody. Finally, the slides were incubated with 4′, 6-diamidine-2-phenylindole dihydrochloride (DAPI, 10 mg mL–1, diluted in PBS by 1:2,000) (Roche, Nutley, NJ, United States) for 5 min. After three times of washing, 50% glycerol was added on the slides before observation under Axio Imager A2 microscope (Carl Zeiss, Germany).
All the data were shown as mean ± S.D. The two-samples Student’s tested were performed for the comparisons between groups. Multiple group comparisons were executed by one-way ANOVA and followed by a Turkey multiple group comparison tests using Statistical Package for Social Sciences (SPSS) 16.0 Software. The difference was considered as significant at p < 0.05.
The ORF of a previously reported engrailed gene (Cgengrailed-1, CGI_10012208) was identified from the Pacific oyster C. gigas, which was of 690 bp encoding a polypeptide of 229 amino acids (Figure 1A). The putative molecular weight of Cgengrailed-1 was 25.70 kDa and its theoretical isoelectric point (pI) was 9.62. SMART analysis showed that there was a HOX domain (from Glu141 to Asn203) and a low expression region in Cgengrailed-1 (Figure 1B).
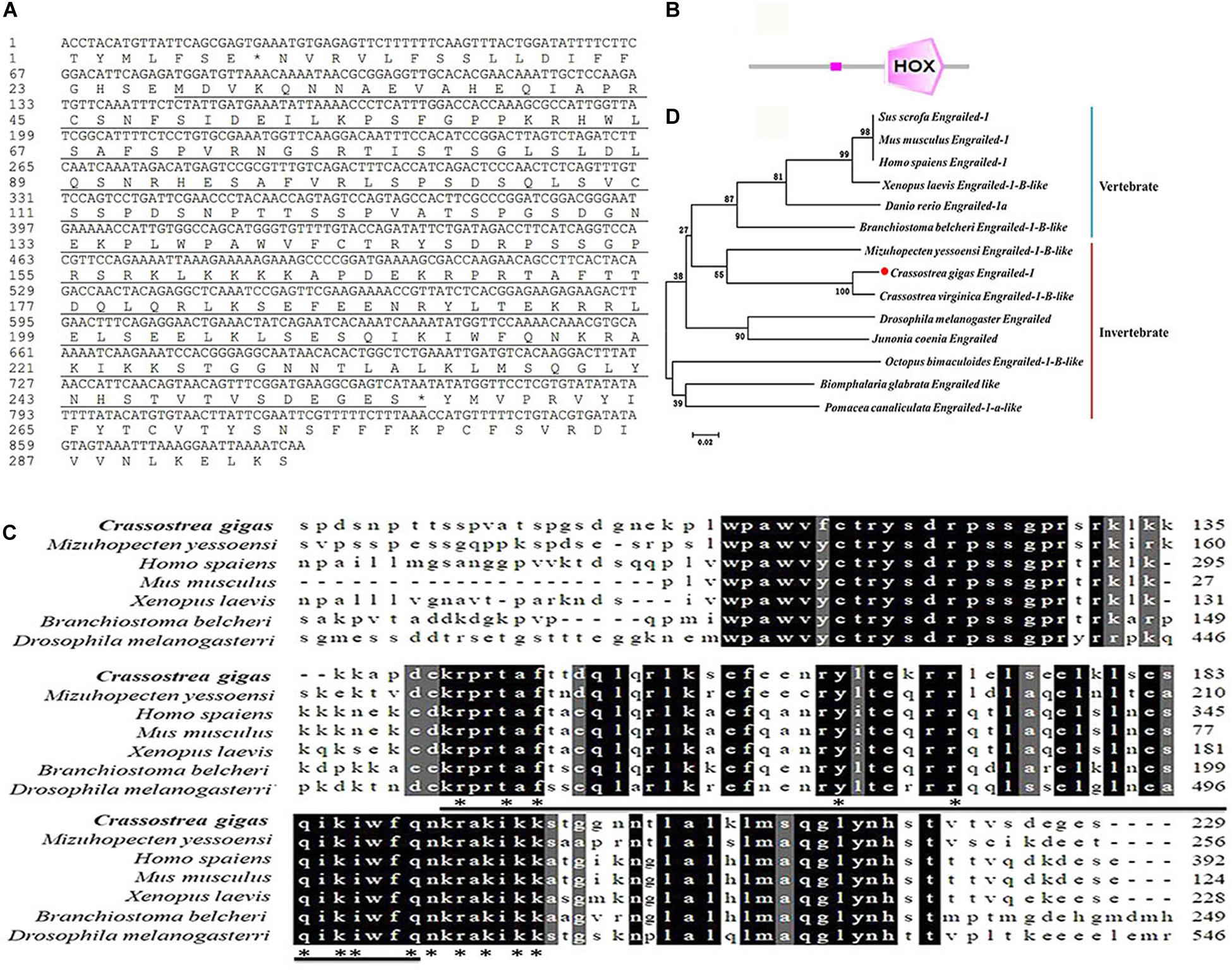
Figure 1. Bioinformatics analysis of Cgengrailed-1. (A) The cDNA sequence and the putative protein of Cgengrailed-1 which is marked by black underline. (B) The typical HOX domain predicted by SMAT analysis. (C) Multiple sequence alignment of the Cgengrailed-1. Multiple sequence alignment of Cgengrailed-1 with known engraileds from various species including C. gigas (XP_011414452.1), M. yessoensi (XP_021372813.1), Homo sapiens (NP_001417.3), Mus musculus (NP_034263.2), Xenopus laevis (XP_018094123.1), Branchiostoma belcheri (XP_019636788.1), and Drosophila melanogaster (NP_523700.2). (D) Phylogenetic analysis of Cgengrailed-1. Fourteen engrailed sequences from different species were obtained from GenBank on NCBI, including Sus scrofa (XP_003133330.2), Mus musculus (NP_034263.2), Homo sapiens (NP_001417.3), Xenopus laevis (XP_018094123.1), Danio rerio (NP_571120.2), Branchiostoma belcheri (XP_019636788.1), M. yessoensi (XP_021372813.1), C. gigas (XP_011414452.1), C. virginica (XP_022294249.1), Drosophila melanogaster (NP_523700.2), Junonia coenia (AAD08923.1), Octopus bimaculoides (XP_014774611.1), Biomphalaria glabrata (XP_013080601.1), and Pomacea canaliculata (XP_025083924.1). ClustalX software was used to perform the multiple sequence alignment of the fourteen engrailed sequences. The neighbor-joining (NJ) tree of Cgengrailed-1 was constructed with Mega 6.0 software.
Results of multiple sequences alignment revealed that the deduced amino acid sequence of Cgengrailed-1 shared 48.50% identity with that from Mizuhopecten yessoensi, 41.30% identity with that from Xenopus laevis, 40.50% identity with that from Branchiostoma belcheri, 37.90% identity with that from Mus musculus, 27.50% identity with that from Homo sapiens, and 19.60% identity with that from Drosophila melanogaster (Figure 1C). A total of 14 engraileds from various vertebrate and invertebrate species were employed in phylogenetic analysis. There were two major distinct branches in the phylogenetic tree, the vertebrate and invertebrate branches. Cgengrailed-1 was firstly gathered with that from C. virginica and then clustered with those from M. yessoensis and other molluscs, and finally clustered with those from vertebrates (Figure 1D).
The ORF fragment of Cgengrailed-1 was inserted into the pET-30a vector to build the recombinant plasmid and transferred into the expression bacterial strain (E. coli Transetta, DE3). After IPTG induction, the purified recombinant protein of rCgengrailed-1 was ascertained by the 12% SDS-PAGE through electrophoresis (Figure 2A). There is one single band about 35 kDa detected by SDS-PAGE, which was in consistent with the predicted molecular weight of rCgengrailed-1 (Figure 2A, lane 3). There was no corresponding band in the cell lysate without induction (Figure 2A).
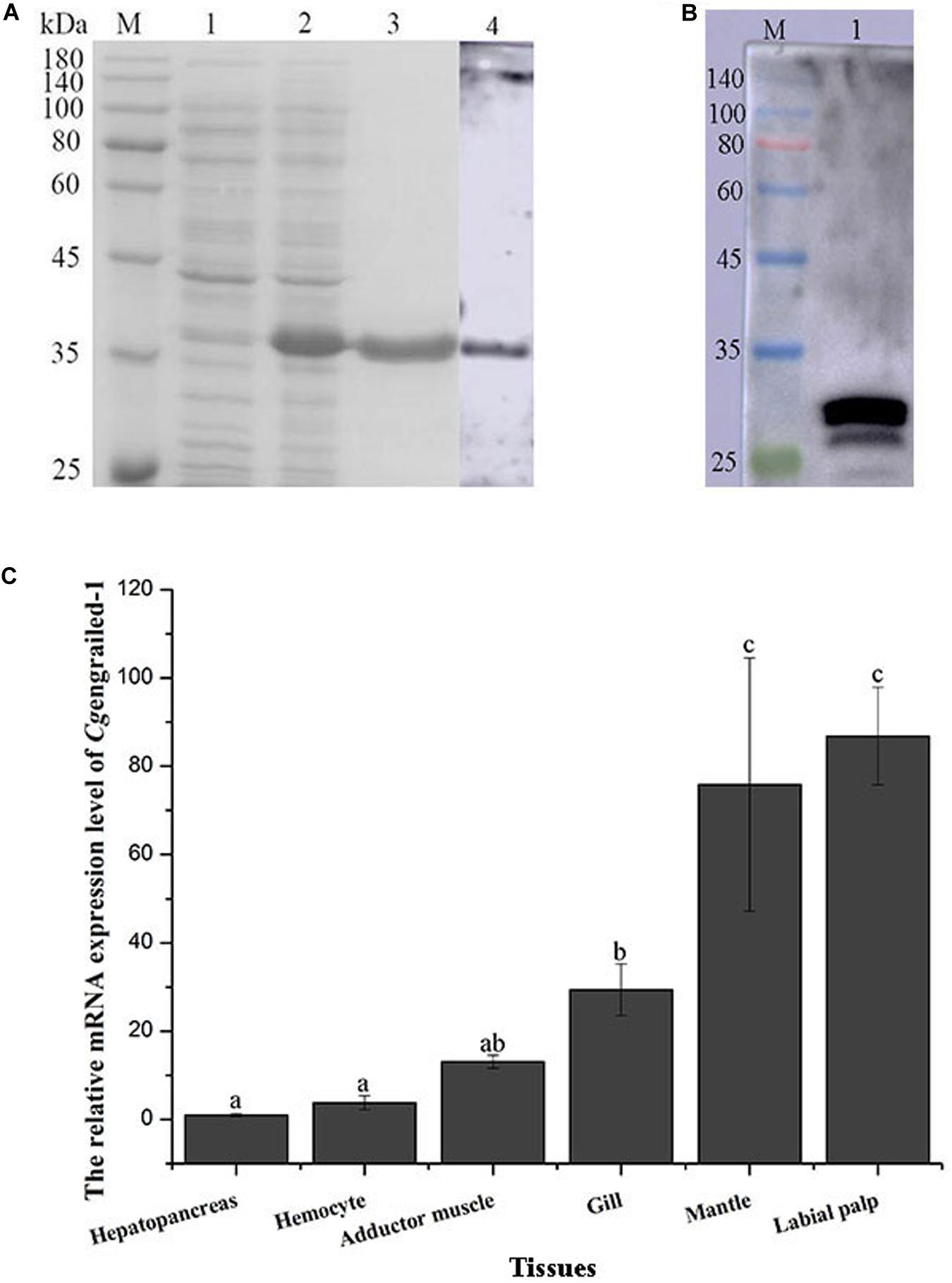
Figure 2. (A) SDS-PAGE and western blot analysis of rCgengrailed-1. Lane M: protein molecular standard; Lane 1: Proteins form E. coli without IPTG induction; Lane 2: positive transformant induced by IPTG; Lane 3: purified rCgengrailed-1; Lane 4: western blot analysis of the polyclonal antibody against rCgengrailed-1. (B) Western blot analysis of Cgengrailed-1 from mantle. Lane M: protein molecular standard; Lane 1: natural protein isolated from mantle. (C) Cgengrailed-1 mRNA expression level in different tissues. Each value was shown as mean ± SD (N = 3), and statistical significance (p < 0.05) was marked by different letters (a–c).
The purified rCgengrailed-1 was used to immune the female mice to prepare the polyclonal antibody. The polyclonal antibody specificity of the rCgengrailed-1 was verified by western blot. There was only a distinct band detected, whose molecular weight was in accordance with that of rCgengrailed-1 (Figure 2B). Meanwhile, no distinct band was observed in the negative control.
The expression level of Cgengrailed-1 mRNA in oyster tissues was detected by quantitative real-time PCR. The mRNA transcripts of Cgengrailed-1 were widely expressed in all the tested tissues including hepatopancreas, hemocytes, adductor muscle, gill, mantle and labial palp (Figure 2C). The highest expression level of Cgengrailed-1 transcripts was observed in labial palp, which was approximately 86.83-fold of that in hepatopancreas (p < 0.05). In addition, the expression level of Cgengrailed-1 transcripts in mantle and gill was 75.87-fold (p < 0.05) and 28.52-fold (p < 0.05) higher than that in hepatopancreas, respectively.
In the control (pH 8.1) group, the mRNA expression levels of Cgengrailed-1 decreased dramatically at early D-shape larvae and D-shape larvae stages, which were 0.64- and 0.15-fold compared to trochophore (p < 0.05). After the larvae received acidification treatment at trochophore stage, the mRNA expression levels of Cgengrailed-1 increased significantly in D-shape larvae stage, which was 3.11- (pH 7.8, moderate acidification treatment) and 4.39-fold (pH 7.4, severe acidification treatment) of that in control group (p < 0.05). No significant change of Cgengrailed-1 mRNA was observed in early D-shape larvae after acidification treatment, which was 1.12- (pH 7.8) and 1.23-fold (pH 7.4) of that in control group (p < 0.05, Figure 3).
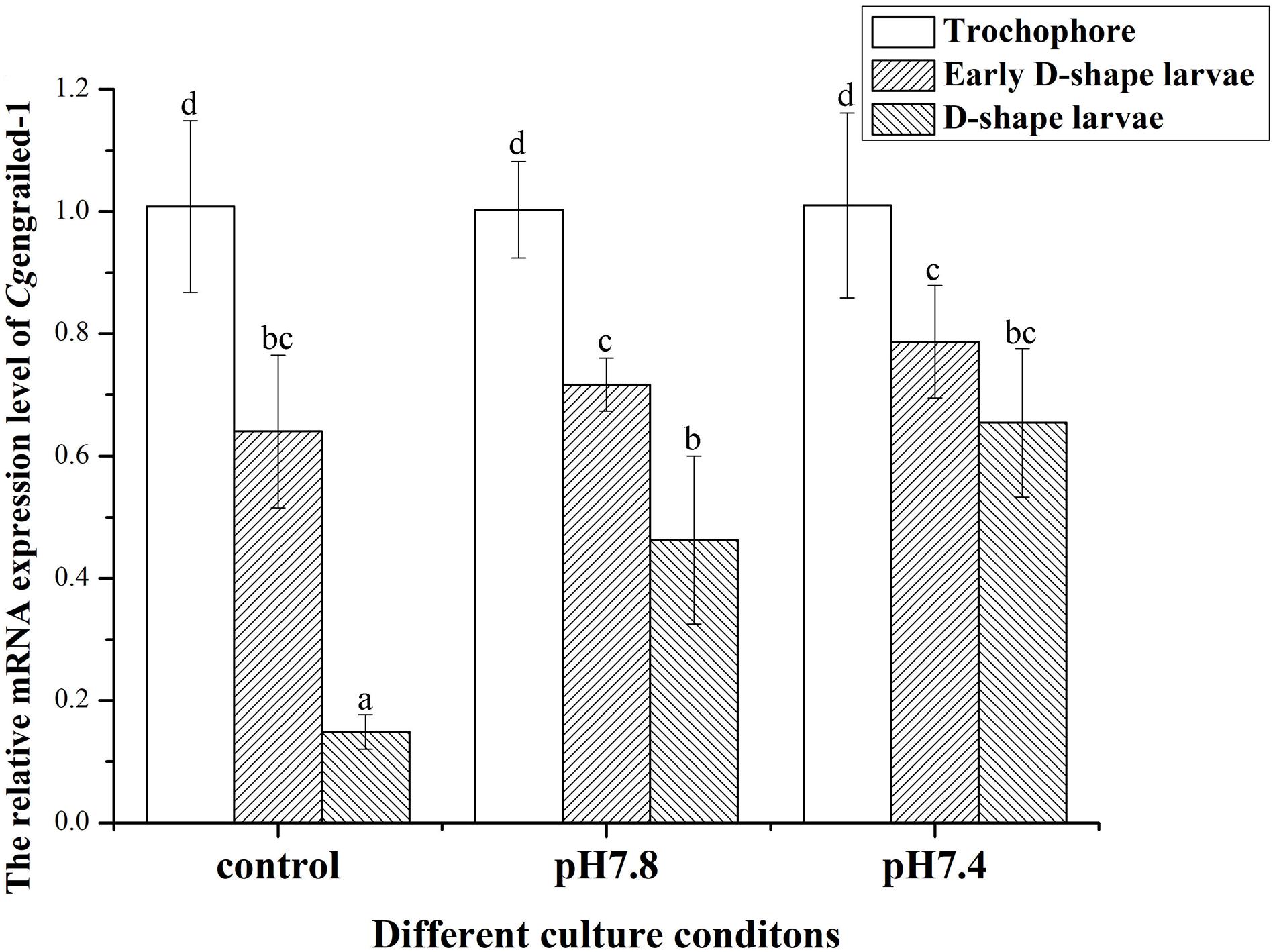
Figure 3. Changes of Cgengrailed-1 mRNA expression in trochophore, early D-shape larvae and D-shape larvae under acidification treatment. Each value was shown as mean ± SD (N = 3), and statistical significance (p < 0.05) was marked by different letters (a–c).
The expression of Cgengrailed-1 in oyster larvae in response to the acidification treatment was also measured by whole-mount immunofluorescence assay. In the control group (pH 8.1), the positive signals (the green point) were observed on the margin of shell gland or periostracum in trochophore, early D-shape larvae and D-shape larvae, and positive signals gradually decreased following the developmental progresses. After moderate or severe acidification treatment, the positive signals were observed on the same location of oyster larvae. No signal was detected in the negative group, indicating the specificity of polyclonal antibody and the reliability of the present results. In trochophore, lots of small and dense positive signals were observed on the dorsal edge of the periostracum. Once the oyster larvae received acidification treatment from trochophore stage, the positive signals in both early D-shape larvae and D-shape larvae were stronger than that in the control group (Figure 4).
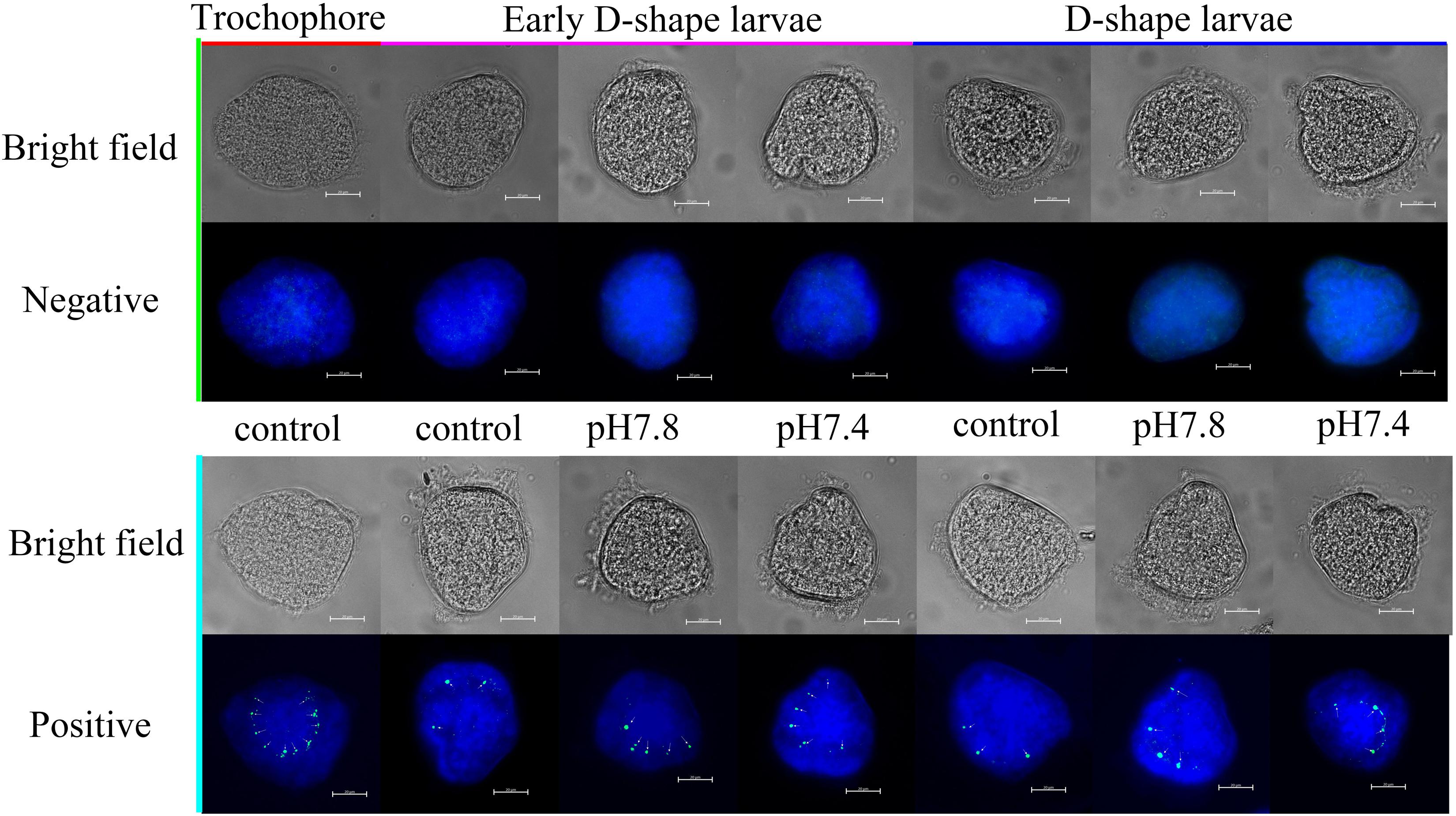
Figure 4. Whole-mount immunofluorescence assay of Cgengrailed-1 expression in oyster larvae. The expression of Cgengrailed-1 in oyster larvae at trochophore, early D-shape larvae and D-shape larvae stages upon moderate (pH 7.8) and severe (pH 7.4) acidification treatments were determined by whole-mount immunofluorescence assay. Mouse negative serum was employed in the negative control group. Nucleus was stained with DAPI and shown in blue; Green point indicated the positive signal which was depicted by little white arrows.
The expression level of Cgengrailed-1 mRNA in the mantle of adult oysters after different acidification treatment was determined by quantitative real-time PCR. After the adult oysters received moderate (pH 7.8) and severe acidification (pH 7.4) treatment for one week, the mRNA expression level of Cgengrailed-1 in mantle decreased significantly, which were 0.75- and 0.15-fold (p < 0.05) of that in the control (pH 8.1) group. The decreasing trend in pH 7.4 group was more obvious than that in pH 7.8 group (Figure 5).
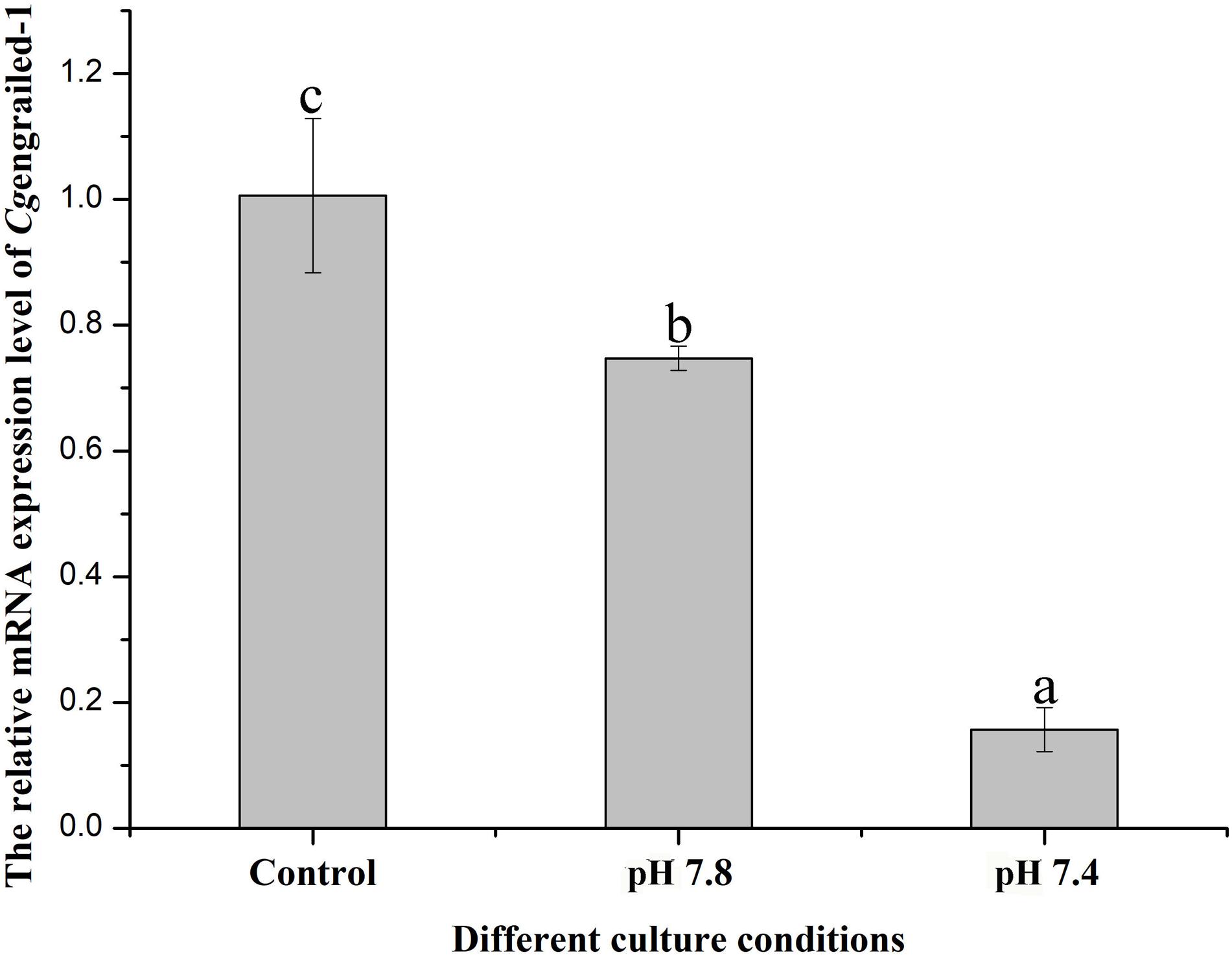
Figure 5. Changes of Cgengrailed-1 expression in adult oyster mantle upon acidification treatment. The mRNA expression of Cgengrailed-1 in mantle tissue of adult oyster upon moderate (pH 7.8) and severe (pH 7.4) acidification treatments were determined by quantitative real-time PCR. Each value was shown as mean ± SD (N = 3), and statistical significance (p < 0.05) was marked by different letters (a–c).
The expression of Cgengrailed-1 in mantle responsive to experimental acidification treatment was further examined by immunofluorescence assay. No positive signal was observed in the negative and control (pH 8.1) groups. When adult oysters received acidification treatment, positive signals were detected in the middle fold of mantle, and the signal intensity in moderate acidification treatment group was significantly stronger than that in severe acidification treatment group (Figure 6).
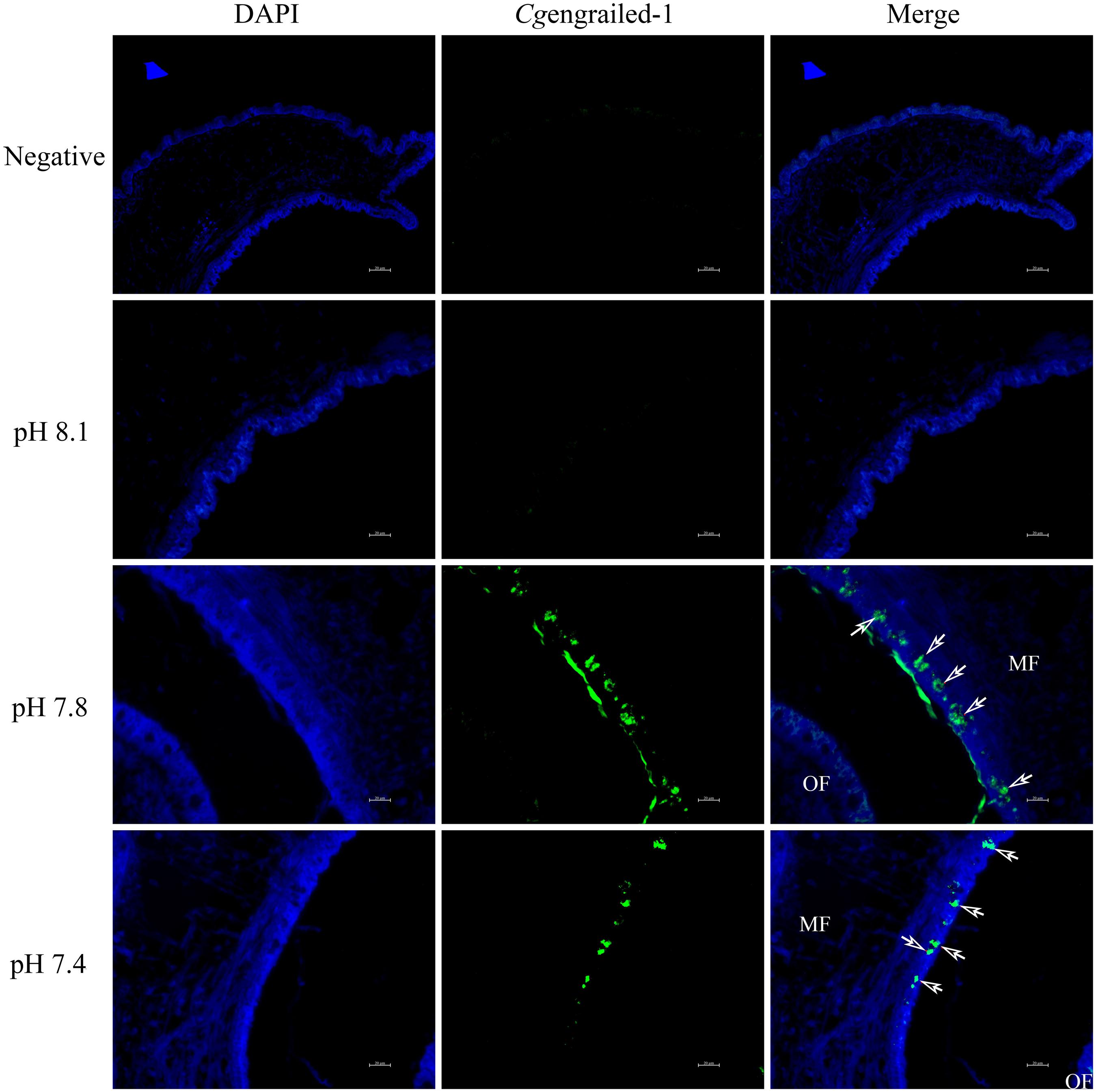
Figure 6. Detection of Cgengrailed-1 in oyster mantle of upon acidification treatment by immunofluorescence assay. Nucleus was stained with DAPI and shown in blue; anti-Cgengrailed-1 coupled to Alexa fluor-488 was shown in green. The white arrow indicated the positive signal. MF, middle fold of mantle edge; OF, outer fold of mantle edge.
Marine bivalves such as oysters and mussels secret matrix proteins and deposit calcium carbonate to form the calcified shells as a supporting skeleton to protect their soft bodies (Zhang et al., 2012; Hendriks et al., 2015), which is thought to be one of the key factors that trigger the expansion of bivalves at the dawn of the Cambrian times (Marie et al., 2012). However, they are now in great danger due to the OA caused by excessive burning of fossil fuels (Orr et al., 2005; Waldbusser et al., 2015; Wang et al., 2017a), and there is urgent need for a better understanding of the shell formation mechanism in bivalves and their stress responses and adaptation modes to CO2-driven acidification. Previous study proved that engrailed, a transcription factor related to the evolution of the metazoan body plan, was also responsible for shell formation in bivalves. However, it has never been explored whether OA affects such process by interrupting the expression of engrailed. In the present study, the expression patterns of a previously identified engrailed upon experimental acidification treatment in both larval and adult oysters were explored, hoping to illustrate the involvement of engrailed in oyster shell formation and acidification adaptation.
In this study, the ORF of Cgengrailed-1 was cloned from C. gigas, which was of 690 bp encoding a polypeptide of 229 amino acids. Cgengrailed-1 contained a typical HOX domain (from Glu141 to Asn203) and a low expression region without signal peptide and transmembrane domain. In vertebrates, engrailed is a member of the homeodomain transcription factors which possess a HOX domain and take part in many developmental processes through combining DNA (Brunet et al., 2005). There were several predicted DNA binding residues identified in Cgengrailed-1, including Arg (143), Thr (146), Phe (148), Tyr (165), Arg (171), Gln (184), Lys (186), Ile (187), Gln (190), Asn (191), Arg (193), Lys (195), Lys (197), and Lys (198). These residues regulate transcription via combining some special groups on the DNA sequence to affect segmentation, neurogenesis and metamorphosis (Morgan, 2006). According to previous study, engrailed may play vital roles in embryonic development and limb development via regulating the active state of key factor Gli in the hedgehog signal pathway (Riddle et al., 1993; Simonnet et al., 2004). The inferred amino acid sequences of Cgengrailed-1 shared higher sequence identity with its homologues from other invertebrates than in vertebrates. Phylogenetic analysis showed that Cgengrailed-1 shared high homology with engraileds from M. yessoensi and C. virginica. These results indicated that Cgengrailed-1 belonged to the engrailed family, which exhibited the function of a transcription factor.
Previous studies have reported that engraileds are responsible for the set-up of compartment boundaries between embryonic domains in molluscan species such as P. vulgate and C. gigas (Nederbragt et al., 2002; Huang et al., 2015). It was found that molluscan engraileds were mainly expressed in the shell forming cells (Moshel et al., 1998) around the shell anlagen (Kin et al., 2009), and at the edge of the embryonic shell, all of which were implied as the boundary of initial shell in larvae (Lawrence and Struhl, 1996). In the present study, results of the whole-mount immunofluorescence assay showed that Cgengrailed-1 was mainly expressed at the margin of shell gland or periostracum of oyster larvae, which was the exact region that the shell boundary formed (Huang et al., 2015). These results demonstrated that Cgengrailed-1 might be involved in the early shell formation progress in oyster larvae. In addition, after the trochophore of oyster received moderate and severe acidification treatment, the mRNA and protein expressions of Cgengrailed-1 altered significantly. Both moderate and severe acidification treatment could prompt the expression of Cgengrailed-1 mRNA, and the increase was greater in the severe acidification treatment group (pH 7.4). Results of whole-mount immunofluorescence assay revealed that the positive signals (indicating Cgengrailed-1 expression) were observed at the margin of periostracum in trochophore, early D-shape larvae and D-shape larvae under acidification treatment. These results suggested that Cgengrailed-1 was involved in larval shell formation in oyster, and both moderate and severe acidification treatment could significantly prompt the expression of Cgengrailed-1. In order to relieve the negative effects imposed by acidification treatment, oyster larvae tended to induce the expression of Cgengrailed-1 to sustain the formation of shell boundary.
In addition, it was found that engrailed participated in shell formation in adult molluscs through different ways from that in larval molluscs. The mRNA transcripts of Cgengrailed-1 were highly expressed in mantle comparing with other tissues. Severe acidification treatment could significantly trigger the expression of Cgengrailed-1 in mantle. Moreover, some dispersive signals (indicating Cgengrailed-1 expression) were detected at the epithelial cell of the mantle middle fold in moderate acidification treatment group, and the signals in pH 7.8 group were more intensive than those in pH 7.4 and control groups. The edge of oyster mantle consists of three folds, including inner fold, middle fold and outer fold (Supplementary Figure S1), and the middle fold was responsible for the formation of periostracum which was the major component of the molluscan shell (Saleuddin, 1979; Song et al., 2019). Previous study had constructed the biological model of periostracum secretion in bivalves, which indicated that the periostracum layer was initiated as a pellicle by basal cells located deep in the periostracal groove (Checa, 2000), and the outer epithelial cells of the middle fold in the mantle were responsible to lubricate and reinforce the pellicle in place (Waite, 1983). Some later research discovered that the outer epithelial cells of the middle fold also contained some membrane-bound vesicles (Richardson et al., 1981). In the present study, Cgengrailed-1 expression in the region close to periostracal groove was significantly increased upon acidification treatment, which suggested that Cgengrailed-1 was involved in the regulation of shell formation in adult oyster in response to acidification threat by modulating the synthesis of periostracum. These results indicated that Cgengrailed-1 might involve in shell formation in adult oyster by regulating the biosynthesis of periostracum in the epithelial cell in the middle fold of mantle. Acidification treatment could prompt the expression of Cgengrailed-1 in the middle fold of mantle to sustain the biosynthesis of periostracum in response to acidification stress.
In summary, the ORF of Cgengrailed-1 was cloned from oyster C. gigas, which contained a HOX domain and belonged to the homeodomain transcription factor family. Cgengrailed-1 was involved in the shell formation progress and the response of larval and adult oyster to the acidification stress through separate ways. The Cgengrailed-1 tended to increase expression to sustain the formation of shell boundary in oyster larvae, while it was prompted for the significant expression in the epithelial cell in the middle fold of mantle to trigger the biosynthesis of periostracum in adult oysters. Both larval and adult oysters were able to relieve the negative effects imposed by acidification treatment through increasing the expression of Cgengrailed-1.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by the Ethics Committee of Dalian Ocean University.
YKZ, ZL, and LW conceived and designed the experiments. YKZ, YNZ, YL, ZH, and YZ performed the experiments. YKZ and ZL analyzed the data. LW and LS contributed reagents, materials, and analysis tools. YKZ, ZL, LW, and LS wrote the manuscript. All the authors read and approved the final manuscript.
This research was supported by grants (41961124009, Nos. U1706204 and 31802339) from National Science Foundation of China, National Key R&D Program (2018YFD0900606), earmarked fund (CARS-49) from Modern Agro-industry Technology Research System, the Fund for Outstanding Talents and Innovative Team of Agricultural Scientific Research, Key R&D Program of Liaoning Province (2017203004 and 2017203001), AoShan Talents Cultivation Program Supported by Qingdao National Laboratory for Marine Science and Technology (No. 2017ASTCP-OS13), the Distinguished Professor of Liaoning (XLYC1902012), fund from Liaoning Department of Education (QL201901), Liaoning BaiQianWan Talents Program, and Research Foundation in Dalian Ocean University (HDYJ201806).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to all the laboratory members for their technical advice and helpful discussions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.530435/full#supplementary-material
Supplementary Figure 1 | The structure of the edge of oyster mantle. The outer fold was close to the edge of the shell and the inner fold was close to the gill tissue. The black arrow indicated the expression of Cgengrailed-1 protein. MF, middle fold of mantle edge; OF, outer fold of mantle edge; PG, periostracal groove.
Baratte, S., Andouche, A., and Bonnaud, L. (2007). Engrailed in cephalopods: a key gene related to the emergence of morphological novelties. Dev. Genes Evol. 217, 353–362. doi: 10.1007/s00427-007-0147-2
Brunet, I., Weinl, C., Piper, M., Trembleau, A., Volovitch, M., Harris, W., et al. (2005). The transcription factor Engrailed-2 guides retinal axons. Nature 438:94. doi: 10.1038/nature04110
Byrne, M., Cisternas, P., Elia, L., and Relf, B. (2005). Engrailed is expressed in larval development and in the radial nervous system of Patiriella sea stars. Dev. Genes Evol. 215, 608–617. doi: 10.1007/s00427-005-0018-7
Checa, A. (2000). A new model for periostracum and shell formation in Unionidae (Bivalvia, Mollusca). Tissue Cell 32, 405–416. doi: 10.1054/tice.2000.0129
Cheng, S., Zhan, W., Xing, J., and Sheng, X. (2006). Development and characterization of monoclonal antibody to the lymphocystis disease virus of Japanese flounder Paralichthys olivaceus isolated from China. J. Virol. Methods 135, 173–180. doi: 10.1016/j.jviromet.2006.03.016
Desplan, C., Theis, J., and O’farrell, P. H. (1985). The Drosophila developmental gene, engrailed, encodes a sequence-specific DNA binding activity. Nature 318, 630–635. doi: 10.1038/318630a0
Eker, R. (1929). The recessive mutant engrailed in Drosophila melanogaster. Hereditas 12, 217–222. doi: 10.1111/j.1601-5223.1929.tb02503.x
Fjose, A., Mcginnis, W. J., and Gehring, W. J. (1985). Isolation of a homoeo box-containing gene from engrailed region of Drosophila and the spatial distribution of its transcripts. Nature 313, 284–289. doi: 10.1038/313284a0
Hendriks, I. E., Duarte, C. M., Olsen, Y. S., Steckbauer, A., Ramajo, L., Moore, T. S., et al. (2015). Biological mechanisms supporting adaptation to ocean acidification in coastal ecosystems. Estuar. Coast Shelf Sci. 152, A1–A8.
Holland, L. Z., Kene, M., Williams, N. A., and Holland, N. D. (1997). Sequence and embryonic expression of the amphioxus engrailed (AmphiEn): the metameric pattern of transcription resembles that of its segment-polarity homolog in Drosophila. Development 124, 1723–1732.
Huan, P., Liu, G., Wang, H., and Liu, B. (2013). Identification of a tyrosinase gene potentially involved in early larval shell biogenesis of the Pacific oyster Crassostrea gigas. Dev. Genes Evol. 223, 389–394. doi: 10.1007/s00427-013-0450-z
Huang, X. H., Pin, H., and Liu, B. Z. (2015). Identification of two engrailed-homologs in Crassostrea gigas and their potential involvement in larval shell formation. Mar. Sci. 39, 14–20.
Ikuta, K. (2017). Expression of two engraileds in the embryo of Vargula hilgendorfii (Müller, 1890)(Ostracoda: Myodocopida). J. Crustacean Biol. 38, 23–26. doi: 10.1093/jcbiol/rux099
Iwaki, D. D., and Lengyel, J. A. (2002). A Delta–Notch signaling border regulated by Engrailed/Invected repression specifies boundary cells in the Drosophila hindgut. Mech. Dev. 114, 71–84. doi: 10.1016/s0925-4773(02)00061-8
Joyner, A. L., Kornberg, T., Coleman, K. G., Cox, D. R., and Martin, G. R. (1985). Expression during embryogenesis of a mouse gene with sequence homology to the Drosophila engrailed. Cell 43, 29–37. doi: 10.1016/0092-8674(85)90009-1
Joyner, A. L., and Martin, G. R. (1985). En-1 and En-2, two mouse genes with sequence homology to the Drosophila engrailed: expression during embryogenesis. Cell 43, 29–37. doi: 10.1101/gad.1.1.29
Kin, K., Kakoi, S., and Wada, H. (2009). A novel role for dpp in the shaping of bivalve shells revealed in a conserved molluscan developmental program. Dev. Biol. 329, 152–166. doi: 10.1016/j.ydbio.2009.01.021
Lawrence, P. A., and Struhl, G. (1996). Morphogens, compartments, and pattern: lessons from Drosophila? Cell 85, 951–961. doi: 10.1016/s0092-8674(00)81297-0
Liu, Z., Zhou, Z., Wang, L., Song, X., Chen, H., Wang, W., et al. (2015). The enkephalinergic nervous system and its immunomodulation on the developing immune system during the ontogenesis of oyster Crassostrea gigas. Fish Shellfish Immunol. 45, 250–259. doi: 10.1016/j.fsi.2015.03.041
Loomis, C. A., Harris, E., Michaud, J., Wurst, W., Hanks, M., and Joyner, A. L. (1996). The mouse Engrailed-1 gene and ventral limb patterning. Nature 382:360. doi: 10.1038/382360a0
Lowe, C. J., and Wray, G. A. (1997). Radical alterations in the roles of homeobox genes during echinoderm evolution. Nature 389, 718–721. doi: 10.1038/39580
Marie, B., Joubert, C., Tayale, A., Zanella-Cleon, I., Belliard, C., Piquemal, D., et al. (2012). Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc. Natl. Acad. Sci. U.S.A. 109, 20986–20991. doi: 10.1073/pnas.1210552109
Matsuura, M., Nishihara, H., Onimaru, K., Kokubo, N., Kuraku, S., Kusakabe, R., et al. (2010). Identification of four Engraileds in the Japanese lamprey, Lethenteron japonicum. Dev. Dyn. 237, 1581–1589. doi: 10.1002/dvdy.21552
Miyazaki, Y., Nishida, T., Aoki, H., and Samata, T. (2010). Expression of genes responsible for biomineralization of Pinctada fucata during development. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 155, 241–248. doi: 10.1016/j.cbpb.2009.11.009
Morata, G., and Lawrence, P. A. (1975). Control of compartment development by engrailed in Drosophila. Nature 255, 614–617. doi: 10.1038/255614a0
Morgan, R. (2006). Engrailed: complexity and economy of a multi-functional transcription factor. FEBS Lett. 580, 2531–2533. doi: 10.1016/j.febslet.2006.04.053
Moshel, S. M., Levine, M., and Collier, J. (1998). Shell differentiation and engrailed expression in the Ilyanassa embryo. Dev. Genes Evol 208, 135–141. doi: 10.1007/s004270050164
Nedelec, S., Foucher, I., Brunet, I., Bouillot, C., Prochiantz, A., and Trembleau, A. (2004). Emx2 homeodomain transcription factor interacts with eukaryotic translation initiation factor 4E (eIF4E) in the axons of olfactory sensory neurons. Proc. Natl. Acad. Sci. U.S.A. 101, 10815–10820. doi: 10.1073/pnas.0403824101
Nederbragt, A. J., Van Loon, A. E., and Dictus, W. J. (2002). Expression of Patella vulgata orthologs of engrailed and dpp-BMP2/4 in adjacent domains during molluscan shell development suggests a conserved compartment boundary mechanism. Dev. Biol. 246, 341–355. doi: 10.1006/dbio.2002.0653
Orr, J. C., Fabry, V. J., Aumont, O., Bopp, L., Doney, S. C., Feely, R. A., et al. (2005). Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 437, 681–686. doi: 10.1038/nature04095
Patel, N. H., Martin-Blanco, E., Coleman, K. G., Poole, S. J., Ellis, M. C., Kornberg, T. B., et al. (1989). Expression of engraileds in arthropods, annelids, and chordates. Cell 58, 955–968. doi: 10.1016/0092-8674(89)90947-1
Piper, D. E., Batchelor, A. H., Chang, C. P., Cleary, M. L., and Wolberger, C. (1999). Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96:587. doi: 10.1016/s0092-8674(00)80662-5
Poole, S. J., Kauvar, L. M., Drees, B., and Kornberg, T. (1985). Engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell 40, 37–43. doi: 10.1016/0092-8674(85)90306-x
Richardson, C. A., Runham, N. W., and Crisp, D. J. (1981). A histological and ultrastructural study of the cells of the mantle edge of a marine bivalve, Cerastoderma edule. Tissue Cell 13, 715–730. doi: 10.1016/s0040-8166(81)80008-0
Riddle, R. D., Johnson, R. L., Laufer, E., and Tabin, C. (1993). Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401–1416. doi: 10.1016/0092-8674(93)90626-2
Saleuddin, A. S. M. (1979). “Shell formation in molluscs with special reference to periostracum formation and regeneration,” in Pathways in Malacology, eds S. Van der Spoel, A. C. Van Bruggen, and J. Lever (Bohn: Springer), 47–81.
Simonnet, F., Deutsch, J., and Quéinnec, E. (2004). Hedgehog is a segment polarity gene in a crustacean and a chelicerate. Dev. Genes Evol. 214, 537–545. doi: 10.1007/s00427-004-0435-z
Song, X., Liu, Z., Wang, L., and Song, L. (2019). Recent advances of shell matrix proteins and cellular orchestration in marine molluscan shell biomineralization. Front. Mar. Sci. 6:41. doi: 10.3389/fmars.2019.00041
Takeuchi, T., and Endo, K. (2006). Biphasic and dually coordinated expression of the genes encoding major shell matrix proteins in the pearl oyster Pinctada fucata. Mar. Biotechnol. 8, 52–61. doi: 10.1007/s10126-005-5037-x
Thisse, C., and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3:59. doi: 10.1038/nprot.2007.514
Tucker-Kellogg, L., Rould, M. A., Chambers, K. A., Ades, S. E., Sauer, R. T., and Pabo, C. O. (1997). Engrailed (Gln50→ Lys) homeodomain–DNA complex at 1.9 Å resolution: structural basis for enhanced affinity and altered specificity. Structure 5, 1047–1054. doi: 10.1016/s0969-2126(97)00256-6
Waite, J. H. (1983). “Quinone-tanned scleroproteins,” in The Mollusca, Physiology, Vol. 4, eds A. S. M. Saleuddin and K. M. Wilbur (New York, NY: Academic Press), 467–504. doi: 10.1016/b978-0-12-751401-7.50018-1
Waldbusser, G. G., Hales, B., Langdon, C. J., Haley, B. A., Schrader, P., Brunner, E. L., et al. (2015). Saturation-state sensitivity of marine bivalve larvae to ocean acidification. Nat. Clim. Change 5:273. doi: 10.1038/nclimate2479
Waldbusser, G. G., and Salisbury, J. E. (2014). Ocean acidification in the coastal zone from an organism’s perspective: multiple system parameters, frequency domains, and habitats. Ann. Rev. Mar. Sci. 6, 221–247. doi: 10.1146/annurev-marine-121211-172238
Wang, X., Wang, M., Jia, Z., Qiu, L., Wang, L., Zhang, A., et al. (2017a). A carbonic anhydrase serves as an important acid-base regulator in pacific oyster Crassostrea gigas exposed to elevated CO2: implication for physiological responses of mollusk to ocean acidification. Mar. Biotechnol. 19, 22–35. doi: 10.1007/s10126-017-9734-z
Wang, X., Wang, M., Jia, Z., Song, X., Wang, L., and Song, L. (2017b). A shell-formation related carbonic anhydrase in Crassostrea gigas modulates intracellular calcium against CO2 exposure: implication for impacts of ocean acidification on mollusk calcification. Aquat. Toxicol. 189, 216–228. doi: 10.1016/j.aquatox.2017.06.009
Wang, X., Wang, M., Jia, Z., Wang, H., Jiang, S., Chen, H., et al. (2016). Ocean acidification stimulates alkali signal pathway: A bicarbonate sensing soluble adenylyl cyclase from oyster Crassostrea gigas mediates physiological changes induced by CO2 exposure. Aquat. Toxicol. 181, 124–135. doi: 10.1016/j.aquatox.2016.11.002
Yu, S., Liu, H., and Luo, L. (2007). Analysis of Relative Gene Expression Using Different Real-Time Quantitative PCR. Rome: AGRIS.
Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., Xu, F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54.
Zhang, H., Song, L., Li, C., Zhao, J., Wang, H., Qiu, L., et al. (2008). A novel C1q-domain-containing protein from Zhikong scallop Chlamys farreri with lipopolysaccharide binding activity. Fish Shellfish Immunol. 25, 281–289. doi: 10.1016/j.fsi.2008.06.003
Zhang, Y., Liu, Z., Song, X., Huang, S., Wang, L., and Song, L. (2019). The inhibition of ocean acidification on the formation of oyster calcified shell by regulating the expression of Cgchs1 and Cgchit4. Front. Physiol. 10:1034. doi: 10.3389/fphys.2019.01034
Zhou, Y. J., He, Z. X., Li, C. Z., Xiang, L., Zhu, F. J., Zhang, G. Y., et al. (2010). Correlations Among mRNA Expression Levels of Engrailed, BMP2 and Smad3 in Mantle Cells of Pearl Oyster Pinctada fucata. Prog. Biochem. Biophys. 37, 737–746. doi: 10.3724/sp.j.1206.2010.00040
Keywords: engrailed, Crassostrea gigas, shell formation, ocean acidification, stress response
Citation: Zhang Y, Liu Z, Zong Y, Zheng Y, Li Y, Han Z, Wang L and Song L (2020) The Increased Expression of an Engrailed to Sustain Shell Formation in Response to Ocean Acidification. Front. Physiol. 11:530435. doi: 10.3389/fphys.2020.530435
Received: 29 January 2020; Accepted: 10 November 2020;
Published: 01 December 2020.
Edited by:
Menghong Hu, Shanghai Ocean University, ChinaReviewed by:
Dahui Yu, Beibu Gulf University, ChinaCopyright © 2020 Zhang, Liu, Zong, Zheng, Li, Han, Wang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoqun Liu, bGl1emhhb3F1bkBkbG91LmVkdS5jbg==; Lingling Wang, d2FuZ2xpbmdsaW5nQGRsb3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.