- 1Hunan Academy of Forestry, Changsha, China
- 2College of Life Science, Central South University of Forestry and Technology, Changsha, China
The entomopathogenic nematode Steinernema carpocapsae has been used for control of soil insects. However, S. carpocapse is sensitive to environmental factors, particularly temperature. We studied an S. carpocapse group that was shocked with high temperature. We also studied the transcriptome-level responses associated with temperature stress using a BGIseq sequencing platform. We de novo assembled the reads from the treatment and control groups into one transcriptome consisting of 43.9 and 42.9 million clean reads, respectively. Based on the genome database, we aligned the clean reads to the Nr, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases and analyzed the differentially expressed genes (DEGs). Compared with the control, the heat-shocked group had significant differential expression of the heat shock protein (HSP) family, antioxidase [glutathione S-transferases (GSTs) and superoxide dismutase (SOD)], monooxygenase (P450), and transcription factor genes (DAF-16 and DAF-2). These DEGs were demonstrated to be part of the Longevity pathway and insulin/insulin-like signaling pathway. The results revealed the potential mechanisms, at the transcriptional level, of S. carpocapsae under thermal stress.
Introduction
Traditional chemical pesticides are used to reduce agricultural losses caused by insect pests. However, chemical pesticide residues pose hazards to both humans and the environment (Thomas, 2018). Biological control can be an effective method to reduce pesticide residues, and entomopathogenic nematodes (EPNs) can be useful biocontrol agents. EPNs can help manage damaging pests in many farming systems (Cruz-Martinez et al., 2017; Dlamini et al., 2019), and they have been commercialized in many countries. The EPN Steinernema carpocapsae interacts with symbiotic bacteria (Xenorhabdus nematophila) and parasitizes soil insect pests (Richards and Goodrich-Blair, 2009; Labaude and Griffin, 2018). Nematodes in the infective juvenile stage (dauers) search for insect hosts in the soil and enter through wounds or natural orifices, such as the mouth, spiracle, and anus (Dlamini et al., 2019). The symbiotic Xenorhabdus bacteria are released into the insect hemocoel and propagate in vivo (Khandelwal and Banerjee-Bhatnagar, 2003; Khandelwal et al., 2004). Also, a toxic venom protein is directly synthesized in S. carpocapse (Balasubramanian et al., 2010) and released in vitro (Lu et al., 2017). This is a new component of the lethal mechanism of S. carpocapsae. S. carpocapsae plays a key role in delivering the toxins to the host during pathogenesis (Chang et al., 2019). S. carpocapsae are sensitive to many environmental factors, such as temperature, UV, and humidity (Kepenekci et al., 2015; Salame and Glazer, 2015; El-Borai et al., 2016). It is probable that temperature is the most important factor impacting the efficacy of EPNs (Odendaal et al., 2016; Wilson et al., 2016). Therefore, understanding how S. carpocapsae resists thermal stress may help improve the ability of EPNs to serve as biological control agents.
The transcriptome can be used to study the responses of an organism, at the transcriptional level, following experimental treatments. In nematodes, toxic chemicals activate the immune response and can change their fecundity and life span. RNA-Seq analyses demonstrated that some differentially expressed genes (DEGs) were related to the immune response (Martis et al., 2017; Di et al., 2018). Changes of gene expression, in different life cycles, were detected by transcriptome sequencing (Seiml-Buchinger and Matveeva, 2019). The transcriptomes and genomes of Steinernema spp. have been characterized (Dillman et al., 2015; Rougon-Cardoso et al., 2016; Yaari et al., 2016; Serra et al., 2019). However, no studies have focused on the specific genes and pathways involved in the heat shock reactions of S. carpocapsae.
We studied changes in the transcriptome related to heat shock treatment and analyzed the pathways and genes related to different temperature treatments. This study demonstrates the heat shock response mechanisms of S. carpocapsae at the molecular level. The results may promote the practical use of S. carpocapsae in biocontrol.
Materials and Methods
Nematode Cultures
The S. carpocapsae (strain All) was supplied by the Guangdong Institute of Applied Biological Resources, Guangdong Province, China (Han et al., 1997). We used Galleria mellonella larvae as the hosts of S. carpocapsae (Kaya and Stock, 1997). The infected hosts were cultured in a 25°C incubator. These nematodes were selected as the controls and original nematodes for the heat shock tests. After the culture period, the infective juveniles (IJs) were introduced into sterile distilled water in a culture dish in preparation for the next step.
Heat Treatment and Sample Collection
We measured the nematode heat resistance based on procedures used in a previous study (Yaari et al., 2016). We exposed the nematodes to 32°C for 3 h. They were then transferred to 23°C for 1 h to recover and finally exposed to 35°C for 5 h. We then selected 10 drops with 10-μl suspensions in each drop and calculated the average nematode mortality. Each drop contained 80–100 nematodes. The surviving nematodes were cultured in the host in normal condition, and the offspring were tested in the same manner. As the survival rate exceeded 50%, the high temperature used in the experiment was increased from 35 to 38°C with successive generations of nematodes. The nematodes whose survival rate exceeded 50% at 38°C were collected as the treatment group. Samples for RNA extraction were taken from all instars. Both the heat shock treatment and the control group were prepared in three replicates. Approximately 2,000–2,200 nematodes, representing all growth stages, were collected for each replicate.
RNA Isolation and Transcriptome Sequencing
Total RNA for sequencing was isolated using the RNeasy® plus Micro Kit according to manufacturer’s instructions (Qiagen, Hilden, Germany). A NanoVue spectrophotometer (GE Healthcare Bio-Science, Uppsala, Sweden) was used to assess the quality of total RNA. The degradation and quality of the RNA were tested by electrophoresis on a 1.0% agarose gel. To isolate poly(A) mRNA, magnetic beads with oligo (dT) were used. After the fragmentation of the mRNA under the effect of the fragmentation buffer, the cDNA was synthesized using the template mRNA fragments. These short cDNA fragments were resolved and purified by ethidium bromide buffer for end reparation and connected with adapters. After the cDNA samples were selected for PCR amplification and quantified and qualified using an Agilent 2100 Bioanaylzer and ABI StepOnePlus Real-Time PCR System (Agilent Technologies, Santa Clara, CA, United States). The library was sequenced using an BGIseq PE150 system (Shenzhen, China). The cDNA library construction and sequencing were performed by Beijing Genomics Institution (Shenzhen, China).
De novo Assembly and Bioinformatics Analysis
The adaptors and low-quality sequences from the raw reads were removed using FASTX1. SOAP2 (version 2.2.1)2 was used to filter out potential rRNA reads, and the data were filtered to be clean reads. The filtered clean reads were mapped to the reference genome, S. carpocapsae, using HISAT2 (Hierarchical Indexing for Spliced Alignment of Transcripts)3 (Rougon-Cardoso et al., 2016). The assembled de novo transcriptome was implemented using Trinity (Grabherr et al., 2011). The reads with overlap length were combined to structure the contigs and then mapped back to contigs. Trinity connected the contigs from the same transcript that could not be extended on either end and formed the defined unigenes. The unigenes obtained were distributed into two groups: singletons and clusters. With an E-value cutoff of 10–5, the sequences were compared based on the databases of Nr and Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The software Blast2GO was used to analyze the gene annotation with Gene Ontology (GO) terms4 with default parameters. The KEGG database was used to perform pathway annotation analysis5. Selected from the Nr annotation, the interesting genes were analyzed using NCBI BLASTx6.
Analysis of the Differential Gene Expressions
After the clean reads were aligned to the genome sequence, the expression level of each sample was calculated using RSEM (Li and Dewey, 2011) based on the RPKM (Reads Per Kilobase per Million mapped reads) value. Next, significance levels (P-values) of DEGs between treatment and control groups were calculated using edgeR (Robinson et al., 2010), an R package7 recommended for use in cases without biological or technical replicates. According to the recommendation of the edgeR manual8, all of the parameters were set. By controlling the false discovery rate (FDR), two methods were used to correct significant levels (P-values): the Benjamini and Hochberg and Storey and Tibshirani methods (Benjamini and Hochberg, 1995; Storey and Tibshirani, 2003). For the accuracy of the improved DEGs, we defined the fold change of gene expression ≥ 2 and adjusted the P ≤ 0.001 as the criteria for significant differences in gene expression levels. To deeply extract the DEGs and pathway data that were related to the treatment and control, GO and KEGG pathway cluster analyses were conducted using Blast2GO and KOBAS, respectively. After revision by FDR, the notable levels were defined as those with a P ≤ 0.001.
Substantiation of the RNA-Seq Data and Gene Expression by Quantitative Real-Time PCR
To convey the results of the transcriptome, quantitative real-time PCR (qPCR) was conducted for the key genes in the heat reaction, including SOD and the heat shock proteins (HSPs). Following alignment using NCBI BLASTx, the isolated sequences were preliminarily confirmed, and primers of these genes are designed using Primer 3.09 and delineated in Supplementary Table S2. The expression abundances of mRNA were detected by qPCR using ABI StepOne PlusTM Real-Time PCR System (Life Technologies, Woodlands, Singapore) with GoTaq® qPCR master mix (Promega, Madison, WI, United States). The templates for qPCR were the same as those for the RNA-Seq. qPCR detection was based on a 20-μl reaction, composed of 10 μl master mix, 7 μl ddH2O, 1 μl template cDNA, and 1 μl of each primer. The cycling program is as follows: 95°C for 2 min, 40 cycles of 95°C for 15 s, and 60°C for 30 s, 60°C for 15 s, and 95°C for 30 s. The relative expression was calculated using the 2–Δ Δ Ct method (Pfaffl, 2001) with Actin set as the reference gene (Yan et al., 2011). Fold changes were clarified after the relative expression values were standardized by the lowest value.
Results
Isolation of the Differentially Expressed Genes Between the Heat-Shock Treatment and Control
The raw data were filtered after sequencing, and the low-quality reads were discarded. We obtained 43.9 and 42.9 million clean reads with an average of approximately 96.6 and 96.1% Q20 bases for the treatment and control groups, respectively. The filtered clean data were mapped to the S. carpocapsae genome. The percentage of mapping was 82.2% in the control and 88.9% in the treatment group. A total of 72.3% (15,850) and 65.5% (14,354) of all the genes (21,917) in the treatment and control groups were obtained as expressed genes (FPKM > 1). For functional annotation, the sequences were aligned to the database, including Nr, AnimalTFDB 2.0, GO, and KEGG. The number of annotated genes in the TFDB database was 2,206, as well as 13,319, 12,525, and 6,902 genes in the KEGG, Nr, and GO databases, respectively.
A total of 7,891 determined genes have been significantly expressed between the treatment and control groups, including 4,902 upregulated and 2,989 downregulated DEGs (Supplementary Figure S1). The top 10 most upregulated and downregulated genes are shown in Supplementary Table S1. Among them, the top three upregulated DEGs were ladderlectin-like protein, proline-rich extension-like protein, and synapse-associated protein. The three most downregulated DEGs were WD repeat domain 39, followed by decaprenyl-diphosphate synthase and lipase-like protein. In addition, some longevity regulating pathway and phosphoinositide 3-kinase (PI3K)-Akt signaling pathway genes were conspicuously expressed, such as insulin-like receptor and nematode cuticle collagen domain protein. Finally, several DEGs were detected as hypothetical or predicted proteins, indicating that their functions are not known and merit investigation.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Analysis of the Differentially Expressed Genes
We classified significant enriched GO terms involved in the heat shock reaction by their determination as DEGs of the treatment and control groups. All of the DEGs were categorized into three GO types: biological process, cellular component, and molecular function, and these contained 19, 15, and 10 terms, respectively (Figure 1). In the biological process category, the top three terms with the largest number of DEGs included cellular process (874), metabolic process (723), and biological regulation (488). Among the cellular component category, the membrane (1,214), membrane part (1,143), and cell (988) terms contained the greatest number of DEGs. Binding, catalytic activity, and transporter activity were the top three terms in the molecular function category. Among all of these terms, the three least common terms were pigmentation (biological process), virion (cellular component), and virion part (cellular component), which all included four DEGs.
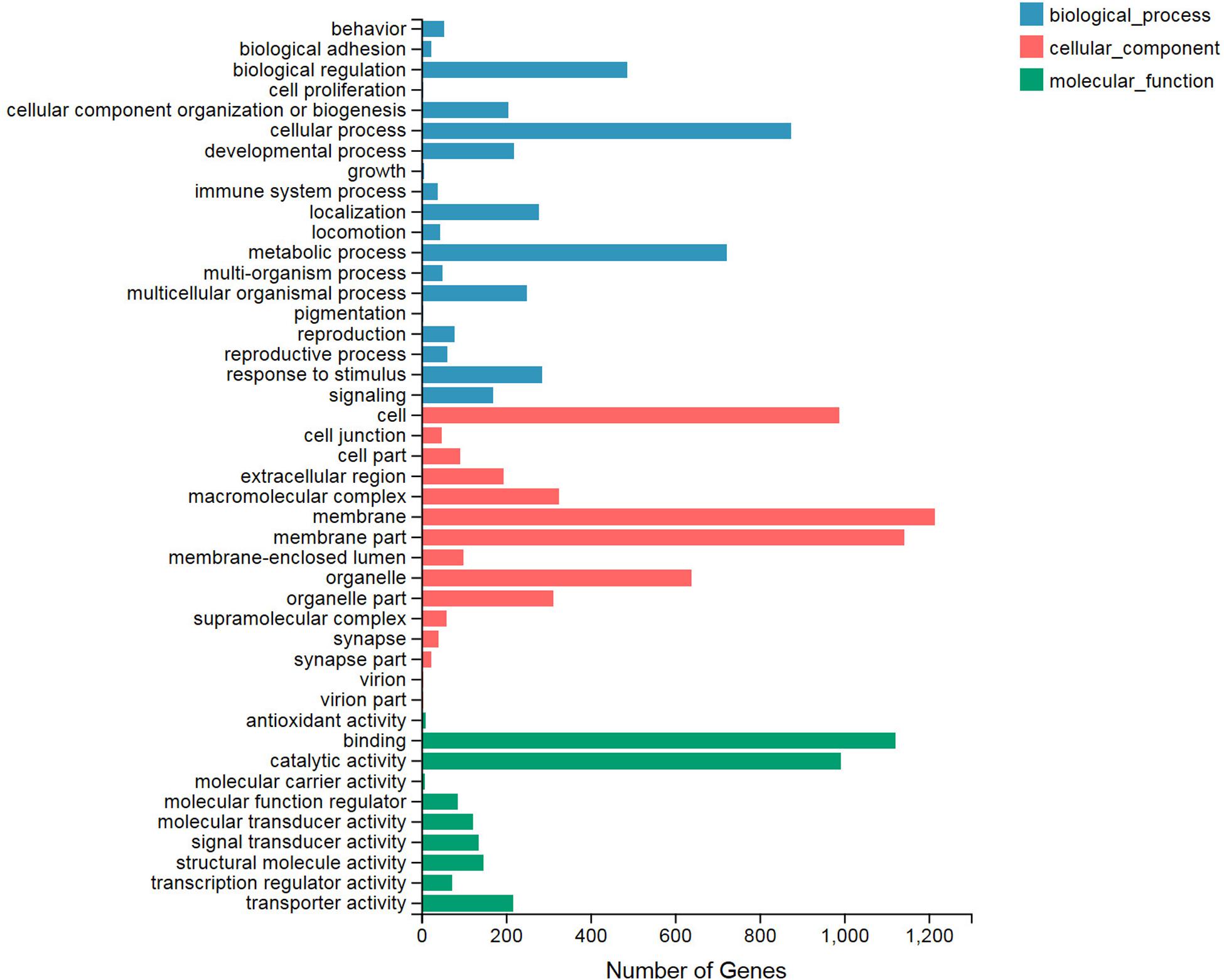
Figure 1. Gene Ontology (GO) functional enrichment analysis for the differentially expressed genes (DEGs). All of the GO terms are categorized into three clusters: biological process (blue), cellular component (red), and molecular function (green). The X axis indicates the number of DEGs. The Y axis represents the GO terms.
In the KEGG pathway enrichment analysis, 44 terms were classified to six categories: cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, and organismal systems (Figure 2). Among them, signal transduction (791), global and overview maps (637), and cancers: specific types (620) were top three DEG terms. These terms were categorized to environmental information processing, metabolism, and human diseases, respectively. The term “biosynthesis of other secondary metabolites” included four DEGs, while the lipid metabolism term included 229 DEGs.
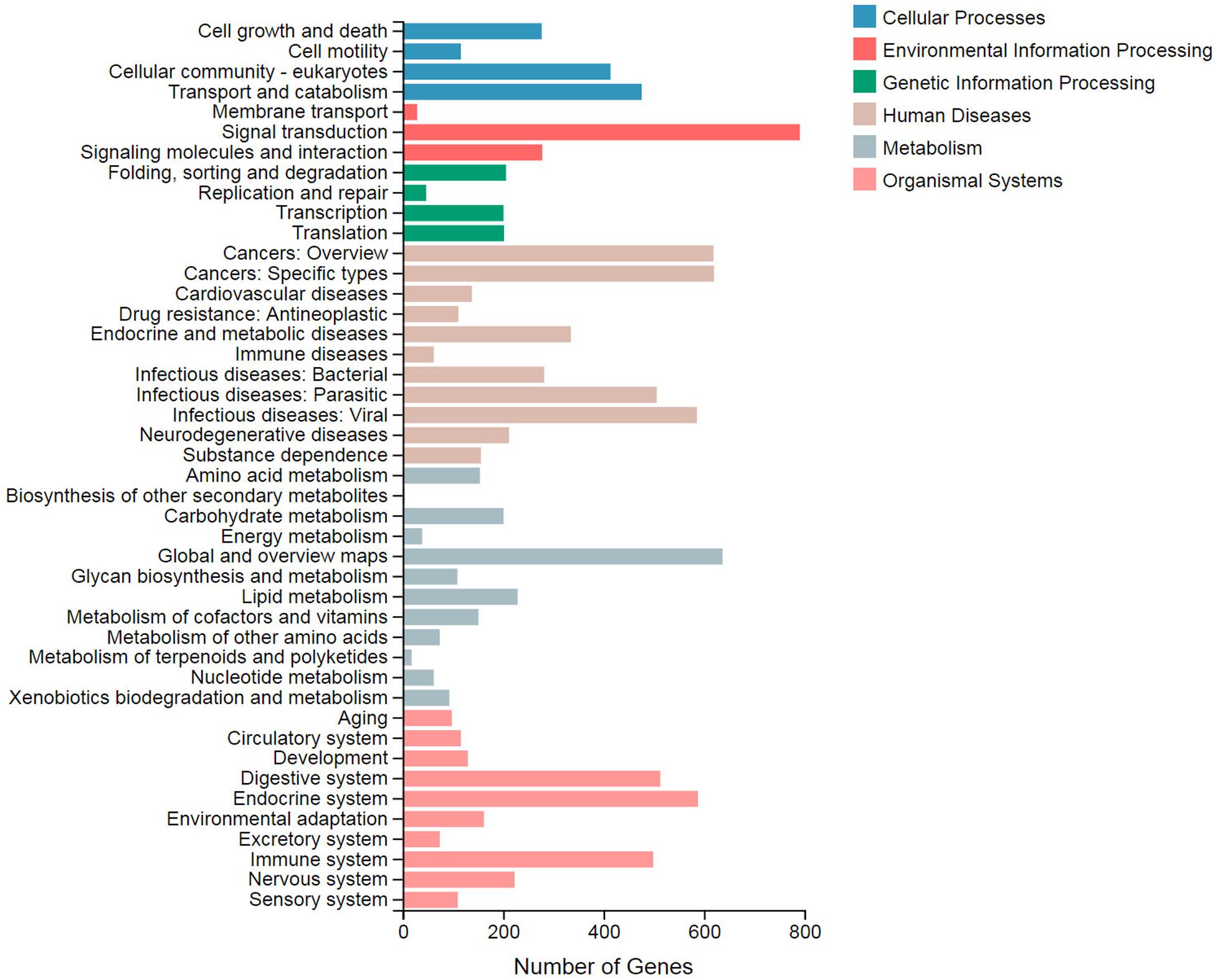
Figure 2. Statistics of the Kyoto Encyclopedia of Genes and Genomes (KEGG) classification of the differentially expressed genes (DEGs). The X axis displays the number of DEGs. The Y axis displays the second KEGG pathway terms. All of the second pathway terms are categorized to six pathway classifications indicated in different colors.
Analysis of Heat Shock-Related Differentially Expressed Genes
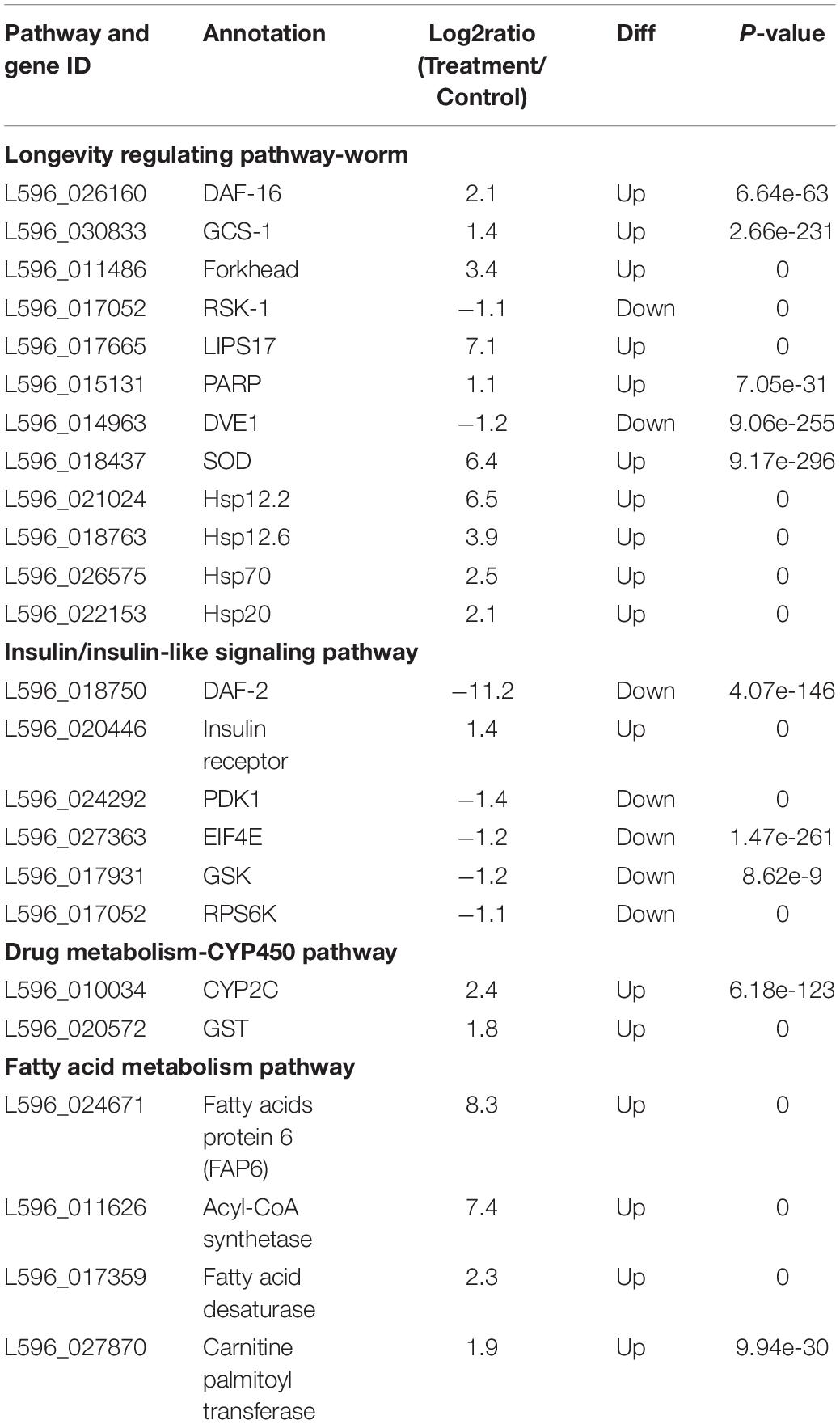
Annotation of the DEGs showed that some genes were clearly associated with the heat shock response. After isolation, some DEGs were identified as heat shock-related DEGs (HRDs) to comprehensively understand the stress response of the HRDs to heat shock. Based on the preliminary categories of heat shock genes in nematodes in previous studies (Yaari et al., 2016), we isolated and identified four pathways: Longevity regulating pathway, IIS (insulin/insulin-like signaling) pathway, Drug metabolism-CYP450 pathway, and Fatty acid metabolism pathway (Table 1). Among the four pathways, 25 HRDs were selected, including 17 upregulated and eight downregulated. The top three most upregulated HRDs were Fatty acids protein 6 (8.3), Acyl-CoA synthetase (7.4), and LIPS17 (7.1). DAF-2 (-11.2) was the most downregulated HRD.
Validation of the RNA-Seq Results by a Quantitative Real-Time PCR Analysis of 10 Heat Shock-Related Differentially Expressed Genes
Ten HRDs, which were clustered to different groups and key pathways, were isolated to validate the RNA-Seq results using qPCR. The expression trends of all 10 selected HRDs determined by qPCR were consistent with those detected by RNA-Seq, indicating that the transcriptome analysis was reliable (Figure 3).
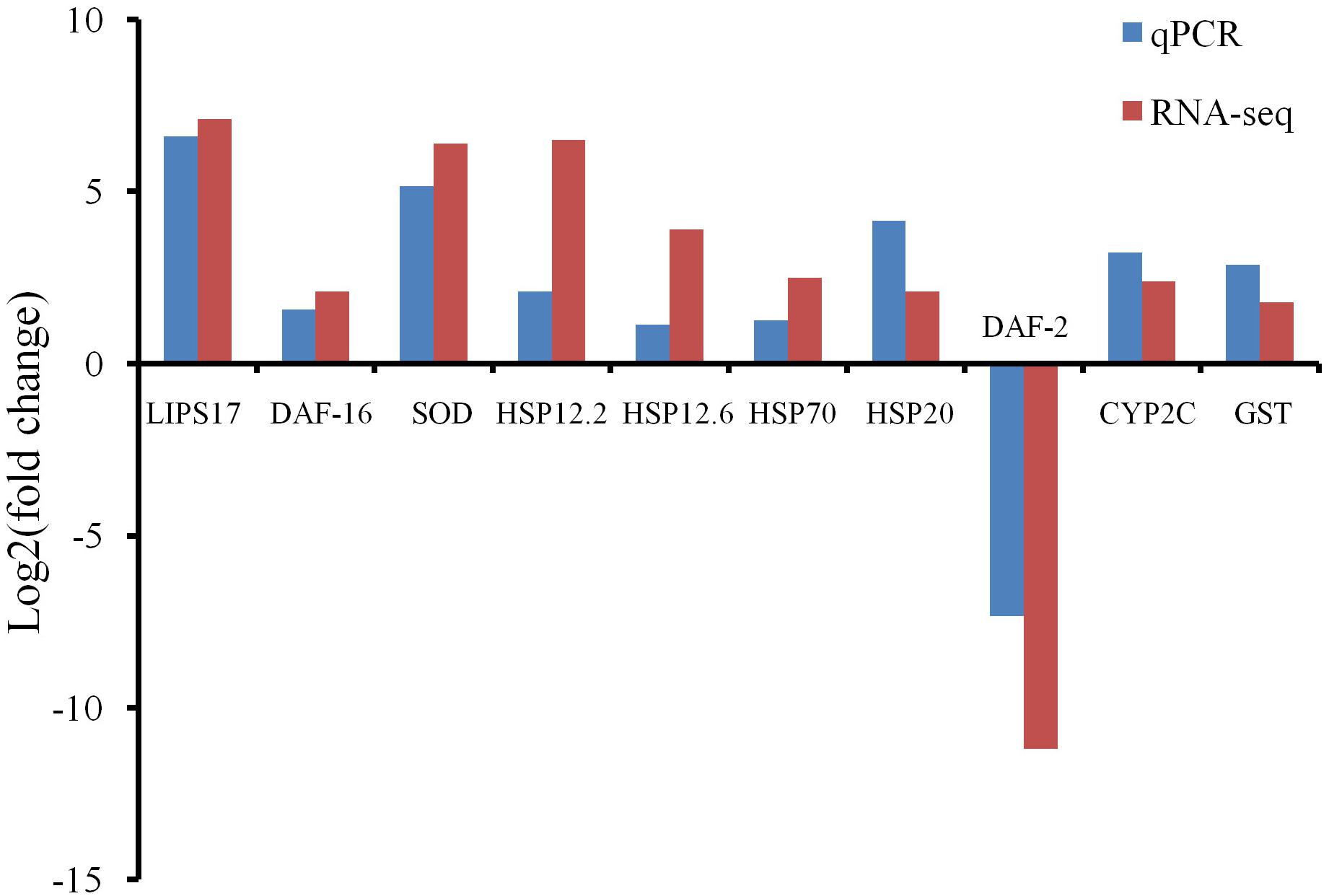
Figure 3. Quantitative real-time PCR (qPCR) detection for RNA-Seq results confirmation. Comparison of the RNA-Seq and qPCR results in expression level fold change of 10 selected heat shock-related differentially expressed genes.
Discussion
Transcriptome technology has been used to obtain detailed information on the expression of genes in other species of nematodes. In Bursaphelenchus xylophilus and Pelodera strongyloides, the average clean reads of each treatment were 50.8 and 47.5 million, respectively (Wang et al., 2019; Zhang et al., 2019). In previous studies on the transcriptome of S. carpocapsae, nearly 15 million clean reads were obtained, and 95% of the reads were mapped to the genome using the Illumina platform. In addition, the 454 platform characterized 9,274 sequences in S. riobrave (Yaari et al., 2016). In our study, the mapping rates were 82.2% in the control and 88.9% in the treatment groups, which indicated the reliability of RNA-Seq. In B. xylophilus and P. strongyloides, the clean reads exceeded 42 million, which is far more than the number of reads documented in previous studies.
In Caenorhabditis elegans, alteration of lipid metabolism was involved in the stress reaction mechanisms (Zhang et al., 2010; Reis et al., 2011; Barros et al., 2012). When C. elegans became stressed, the distribution of nutrition can be affected in the nematode (Kelli et al., 2017). Some chemicals produced by plants can activate both fat consumption and heat stress tolerance, delineating the possible relationship between these two mechanisms (Hirano and Sakamoto, 2019). Although the organic compounds in lipid metabolism were not measured in this study, the DEGs in lipid metabolism and transportation were detected, suggesting a possible modification of nutrient allocation. It is not known if these changes affect energy redistribution.
Heat shock stress has often been studied. In C. elegans, the AFD neuron is the sensor that perceives increasing temperatures (Prahlad et al., 2008; Tatum et al., 2015). The thermal stress excites the FLP-6 neuropeptidergic signaling in the AFD neuron (Lee and Kenyon, 2009). The secretion of FLP-6 represses insulin-like peptide INS-7 and thus affects the downstream genes via the IIS pathway (Chen et al., 2016). INS-7 collaborates with DAF-2 in the downstream part of this pathway (Kawano et al., 2006; Sato et al., 2014). Silencing of DAF-2 stimulated DAF-16 expression and thus increased longevity in C. elegans (Murphy and Hu, 2013; Sonani et al., 2014). The KEGG analysis, based on our transcriptome data, indicated that the DAF-2 and DAF-16 were categorized in the IIS and Longevity regulation pathway. The expressions of daf-2 and daf-16 were found to be downregulated and upregulated, suggesting a negative regulation (repressor) relationship between these two genes. These results suggested that DAF-2 and DAF-16 could play a role in the upstream heat shock reaction in S. carpocapsae. These two factors will be studied using RNA interference in future studies.
GSTs enzymes are involved in the detoxification of metabolites. They are found in most species, including microorganisms, plants, and animals. In nematodes, GSTs enable detoxification of the endogenous chemicals secreted by the hosts (Brophy and Pritchard, 1994; Campbell et al., 2001; Espada et al., 2016; Matouskova et al., 2016). DAF-16 can upregulate the expression of GSTs and help extend the life span of C. elegans under thermal stress (Shanmugam et al., 2017). This demonstrated that GSTs are critical enzymes that protect the worms from a stressful environment. Similarly, thermal stress increased the expression of GSTs in S. carpocapsae, as well as the expression of DAF-16. Thus, GSTs, under the regulation of DAF-16, are likely involved in the heat shock reaction process.
Heat shock proteins are important enzymes that protect nematodes from protein denaturation at high temperatures (Kim et al., 2017). In the heat shock response, hsps are primarily controlled by two regulators, heat shock factor-1 (hsf-1) and daf-16. In the early stages of nematode development, HSF-1 is negatively regulated by daf-16-dependent longevity genes (DDL-1&2), which belong to the IIS pathway (Chiang et al., 2012). In contrast, DAF-16 is modulated by DAF-2 in the adult stage (Volovik et al., 2012; Murphy and Hu, 2013). HSP12.6 and HSP16.2 are key enzymes in resistance to high temperatures (Kim et al., 2017; Hirano and Sakamoto, 2019). Moreover, HSP70 interacts with other proteins to recover protein stability via disaggregation (Nillegoda et al., 2015). The hsps, including hsp12.2, hsp12.6, hsp70, and hsp20, were upregulated in S. carpocapsae after the heat shock (Table 1), suggesting that these genes function in anti-thermal stress. A previous study revealed that the anti-thermal stress process in nematodes was activated by the neuron sensor AFD and finally operated by GSTs and HSPs.
In summary, we performed a comparative transcriptomic analysis of an S. carpocapsae line exposed to high-temperature treatment. Many DEGs were identified and transcriptionally validated to be key factors involved in the mechanism of anti-heat shock in S. carpocapsae. These included GSTs, P450s, HSPs, SODs, and transcription factor genes. Their roles and cooperative relationships, as well as the association between nutrient distribution and anti-thermal mechanisms, require further investigation. The gene expression profiling and differentially expressed identification responses related to heat shock tolerance in S. carpocapsae provide information for further functional analysis of the molecular mechanisms involved.
Data Availability Statement
The datasets generated for this study can be found in the NCBI SRA accession PRJNA635371.
Author Contributions
Y-FX, D-HZ, and ZH contributed to the conception and design of the study. Y-FX organized the database and wrote sections of the manuscript. X-DW performed the statistical analysis. W-HZ wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Hunan Forestry Science and Technology Innovation Projects (XLK201974 and XLK201972).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00721/full#supplementary-material
FIGURE S1 | A volcano plot of differentially expressed genes. The red and blue points indicate notable up- and down-regulation, respectively. The gray points mean that no genes varied significantly.
TABLE S1 | Top 10 most up- and down-regulated differentially expressed genes.
TABLE S2 | Primers used in qPCR detection for RNA-seq results confirmation.
Footnotes
- ^ http://hannonlab.cshl.edu/fastx_toolkit/
- ^ http://soap.genomics.org.cn/
- ^ http://www.ccb.jhu.edu/software/hisat
- ^ http://www.geneontology.org
- ^ http://www.genome.jp/kegg
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://www.bioconductor.org/
- ^ http://www.bioconductor.org/packages/release/bioc/manuals/edgeR/man/edgeR.pdf
- ^ http://frodo.wi.mit.edu/
References
Balasubramanian, N., Toubarro, D., and Simoes, N. (2010). Biochemical study and in vitro insect immune suppression by a trypsin-like secreted protease from the nematode Steinernema carpocapsae. Parasite Immunol. 32, 165–175. doi: 10.1111/j.1365-3024.2009.01172.x
Barros, A. G. D., Liu, J., Lemieux, G. A., Mullaney, B. C., and Ashrafi, K. (2012). “Analyses of C. elegans fat metabolic pathways,” in Caenorhabditis Elegans: Cell Biology and Physiology, 2nd Edn, eds J. H. Rothman and A. Singson (San Diego, CA: Elsevier Acdemic Press), 383–407. doi: 10.1016/b978-0-12-394620-1.00013-8
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Brophy, P. M., and Pritchard, D. I. (1994). Parasitic helminth glutathione S-transferases: an update on their potential as targets for immuno- and chemotherapy. Exp. Parasitol. 79, 89–96. doi: 10.1006/expr.1994.1067
Campbell, A. M., Teesdale-Spittle, P. H., Barrett, J., Liebau, E., Jefferies, J. R., and Brophy, P. M. (2001). A common class of nematode glutathione S-transferase (GST) revealed by the theoretical proteome of the model organism Caenorhabditis elegans. Comp. Biochem. Phys. B. 128, 701–708. doi: 10.1016/s1096-4959(00)00360-2
Chang, D. Z., Serra, L., Lu, D. H., Mortazavi, A., and Dillman, A. R. (2019). A core set of venom proteins is released by entomopathogenic nematodes in the genus Steinernema. PLoS Pathog. 15:e7626. doi: 10.1371/journal.ppat.1007626
Chen, Y. C., Chen, H. J., Tseng, W. C., Hsu, J. M., Huang, T. T., Chen, C. H., et al. (2016). A C. elegans thermosensory circuit regulates longevity through crh-1/CREB-dependent flp-6 neuropeptide signaling. Dev. Cell 39, 209–223. doi: 10.1016/j.devcel.2016.08.021
Chiang, W. C., Ching, T. T., Lee, H. C., Mousigian, C., and Hsu, A. L. (2012). HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell 148, 322–334. doi: 10.1016/j.cell.2011.12.019
Cruz-Martinez, H., Ruiz-Vega, J., Matadamas-Ortiz, P. T., Cortes-Martinez, C. I., and Rosas-Diaz, J. (2017). Formulation of entomopathogenic nematodes for crop pest control - a review. Plant Prot. Sci. 53, 15–24. doi: 10.17221/35/2016-pps
Di, R., Zhang, H. Z., and Lawton, M. A. (2018). Transcriptome analysis of C. elegans reveals novel targets for DON cytotoxicity. Toxins 10:262. doi: 10.3390/toxins10070262
Dillman, A. R., Macchietto, M., Porter, C. F., Rogers, A., Williams, B., Antoshechkin, I., et al. (2015). Comparative genomics of Steinernema reveals deeply conserved gene regulatory networks. Genome Biol. 16:200. doi: 10.1186/s13059-015-0746-6
Dlamini, B. E., Addison, P., and Malan, A. P. (2019). A review of the biology and control of Phlyctinus callosus (Schonherr) (Coleoptera: Curculionidae), with special reference to biological control using entomopathogenic nematodes and fungi. Afr. Entomol. 27, 279–288. doi: 10.4001/003.027.0279
El-Borai, F., Killiny, N., and Duncan, L. W. (2016). Concilience in entomopathogenic nematode responses to water potential and their geospatial patterns in Florida. Front. Microbiol. 7:356. doi: 10.3389/fmicb.2016.00356
Espada, M., Jones, J. T., and Mota, M. (2016). Characterization of glutathione S-transferases from the pine wood nematode, Bursaphelenchus xylophilus. Nematology 18:697. doi: 10.1163/15685411-00002985
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–U130. doi: 10.1038/nbt.1883
Han, R. C., Li, L. Y., and Pang, X. F. (1997). Modelling of the culture parameters for production of Steinernema carpocapsae and Heterorhabditis bacteriophora in solid cultures. Nat. Enemies Insects 19, 75–83.
Hirano, N., and Sakamoto, K. (2019). Linalool odor stimulation improves heat stress tolerance and decreases fat accumulation in nematodes. Biosci. Biotechnol. Biochem. 83, 148–154. doi: 10.1080/09168451.2018.1518699
Kawano, T., Nagatomo, R., Kimura, Y., Gengyo-Ando, K., and Mitani, S. (2006). Disruption of ins-11, a Caenorhabditis elegans insulin-like gene, and phenotypic analyses of the gene-disrupted animal. Biosci. Biotechnol. Biochem. 70, 3084–3087. doi: 10.1271/bbb.60472
Kaya, H. K., and Stock, S. P. (1997). “Techniques in insect nematology,” in Manual of Techniques in Insect Pathology, ed. L. A. Lacey (San Diego, CA: Academic Press), 281–324. doi: 10.1016/b978-012432555-5/50016-6
Kelli, H. M., Corrigan, F. E., Heinl, R. E., Dhindsa, D. S., Hammadah, M., Samman-Tahhan, A., et al. (2017). Relation of changes in body fat distribution to oxidative stress. Am. J. Cardiol. 120, 2289–2293. doi: 10.1016/j.amjcard.2017.08.053
Kepenekci, I., Hazir, S., and Ozdem, A. (2015). Evaluation of native entomopathogenic nematodes for the control of the European cherry fruit fly Rhagoletis cerasi L. (Diptera: Tephritidae) larvae in soil. Turk. J. Agric. For. 39, 74–79. doi: 10.3906/tar-1403-96
Khandelwal, P., and Banerjee-Bhatnagar, N. (2003). Insecticidal activity associated with the outer membrane vesicles of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 69, 2032–2037. doi: 10.1128/aem.69.4.2032-2037.2003
Khandelwal, P., Bhatnagar, R., Choudhury, D., and Banerjee, N. (2004). Characterization of a cytotoxic pilin subunit of Xenorhabdus nematophila. Biochem. Biophys. Res. Commun. 314, 943–949. doi: 10.1016/j.bbrc.2003.12.187
Kim, S. J., Beak, S. M., and Park, S. K. (2017). Supplementation with triptolide increases resistance to environmental stressors and lifespan in C. elegans. J. Food Sci. 82, 1484–1490. doi: 10.1111/1750-3841.13720
Labaude, S., and Griffin, C. T. (2018). Transmission success of entomopathogenic nematodes used in pest control. Insects 9:72. doi: 10.3390/insects9020072
Lee, S. J., and Kenyon, C. (2009). Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans. Curr. Biol. 19, 715–722. doi: 10.1016/j.cub.2009.03.041
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Lu, D. H., Macchietto, M., Chang, D., Barros, M. M., Baldwin, J., Mortazavi, A., et al. (2017). Activated entomopathogenic nematode infective juveniles release lethal venom proteins. PLoS Pathog. 13:e6302. doi: 10.1371/journal.ppat.1006302
Martis, M. M., Tarbiat, B., Tyden, E., Jansson, D. S., and Hoglund, J. (2017). RNA-Seq de novo assembly and differential transcriptome analysis of the nematode Ascaridia galli in relation to in vivo exposure to flubendazole. PLoS One 12:e185182. doi: 10.1371/journal.pone.0185182
Matouskova, P., Vokral, I., Lamka, J., and Skalova, L. (2016). The role of xenobiotic-metabolizing enzymes in anthelmintic deactivation and resistance in helminths. Trends Parasitol. 32, 481–491. doi: 10.1016/j.pt.2016.02.004
Murphy, C. T., and Hu, P. J. (2013). Insulin/insulin-like growth factor signaling in C. elegans. WormBook 26, 1–43. doi: 10.1895/wormbook.1.164.1
Nillegoda, N. B., Kirstein, J., Szlachcic, A., Berynskyy, M., Stank, A., Stengel, F., et al. (2015). Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524, 247–251. doi: 10.1038/nature14884
Odendaal, D., Addison, M. F., and Malan, A. P. (2016). Entomopathogenic nematodes for the control of the codling moth (Cydia pomonella L.) in field and laboratory trials. J. Helminthol. 90, 615–623. doi: 10.1017/s0022149x15000887
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Prahlad, V., Cornelius, T., and Morimoto, R. I. (2008). Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320, 811–814. doi: 10.1126/science.1156093
Reis, R. J. S., Xu, L. L., Lee, H., Chae, M., Thaden, J. J., Bharill, P., et al. (2011). Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging 3, 125–147. doi: 10.18632/aging.100275
Richards, G. R., and Goodrich-Blair, H. (2009). Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell. Microbiol. 11, 1025–1033. doi: 10.1111/j.1462-5822.2009.01322.x
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rougon-Cardoso, A., Flores-Ponce, M. I, Erendira Ramos-Aboites, H., Eduardo Martinez-Guerrero, C., Hao, Y. J., Cunha, L., et al. (2016). The genome, transcriptome, and proteome of the nematode Steinernema carpocapsae: evolutionary signatures of a pathogenic lifestyle. Sci. Rep. 6:37536. doi: 10.1038/srep37536
Salame, L., and Glazer, I. (2015). Stress avoidance: vertical movement of entomopathogenic nematodes in response to soil moisture gradient. Phytoparasitica 43, 647–655. doi: 10.1007/s12600-015-0488-8
Sato, K., Yoshiga, T., and Hasegawa, K. (2014). Activated and inactivated immune responses in Caenorhabditis elegans against Photorhabdus luminescens TT01. Springerplus 3:274. doi: 10.1186/2193-1801-3-274
Seiml-Buchinger, V. V., and Matveeva, E. M. (2019). Expression and functional characteristics of genes in the Globodera rostochiensis Woll. potato cyst nematode at different stages of its life cycle. Biol. Bull. 46, 225–233. doi: 10.1134/s1062359019030142
Serra, L., Macchietto, M., Macias-Munoz, A., McGill, C. J., Rodriguez, I. M., Rodriguez, B., et al. (2019). Hybrid assembly of the genome of the entomopathogenic nematode Steinernema carpocapsae identifies the X-chromosome. G3 9, 2687–2697. doi: 10.1534/g3.119.400180
Shanmugam, G., Mohankumar, A., Kalaiselvi, D., Nivitha, S., Murugesh, E., Shanmughavel, P., et al. (2017). Diosgenin a phytosterol substitute for cholesterol, prolongs the lifespan and mitigates glucose toxicity via DAF-16/FOXO and GST-4 in Caenorhabditis elegans. Biomed. Pharmacother. 95, 1693–1703. doi: 10.1016/j.biopha.2017.09.096
Sonani, R. R., Singh, N. K., Awasthi, A., Prasad, B., Kumar, J., and Madamwar, D. (2014). Phycoerythrin extends life span and health span of Caenorhabditis elegans. Age 36:9717. doi: 10.1007/s11357-014-9717-1
Storey, J. D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445. doi: 10.1073/pnas.1530509100
Tatum, M. C., Ooi, F. K., Chikka, M. R., Chauve, L., Martinez-Velazquez, L. A., Steinbusch, H. W. M., et al. (2015). Neuronal serotonin rlease triggers the heat shock response in C. elegans in the absence of temperature increase. Curr. Biol. 25, 163–174. doi: 10.1016/j.cub.2014.11.040
Thomas, M. B. (2018). Biological control of human disease vectors: a perspective on challenges and opportunities. Biocontrol 63, 61–69. doi: 10.1007/s10526-017-9815-y
Volovik, Y., Maman, M., Dubnikov, T., Bejerano-Sagie, M., Joyce, D., Kapernick, E. A., et al. (2012). Temporal requirements of heat shock factor-1 for longevity assurance. Aging Cell 11, 491–499. doi: 10.1111/j.1474-9726.2012.00811.x
Wang, B. W., Hao, X., Xu, J. Y., Ma, Y., and Ma, L. (2019). Transcriptome-based analysis reveals a crucial role of BxGPCR17454 in low temperature response of pine wood nematode (Bursaphelenchus xylophilus). Int. J. Mol. Sci. 20:2898. doi: 10.3390/ijms20122898
Wilson, M. J., Wilson, D. J., Rodgers, A., and Gerard, P. J. (2016). Developing a strategy for using entomopathogenic nematodes to control the African black beetle (Heteronychus arator) in New Zealand pastures and investigating temperature constraints. Biol. Control 93, 1–7. doi: 10.1016/j.biocontrol.2015.11.002
Yaari, M., Doron-Faigenboim, A., Koltai, H., Salame, L., and Glazer, I. (2016). Transcriptome analysis of stress tolerance in entomopathogenic nematodes of the genus Steinernema. Int. J. Parasitol. 46, 83–95. doi: 10.1016/j.ijpara.2015.08.011
Yan, X., De Clercq, P., Han, R. C., Jones, J., Chen, S. L., and Moens, M. (2011). Osmotic responses of different strains of Steinernema carpocapsae. Nematology 13, 845–851. doi: 10.1163/138855411x554181
Zhang, M. L., Heikkinen, L., Knott, K. E., and Wong, G. (2019). De novo transcriptome assembly of a facultative parasitic nematode Pelodera (syn. Rhabditis) strongyloides. Gene 710, 30–38. doi: 10.1016/j.gene.2019.05.041
Keywords: Steinernema carpocapsae, transcriptome, heat shock response, differentially expressed gene, entomopathogenic nematode
Citation: Xie Y-F, Wang X-D, Zhong W-H, Zhu D-H and He Z (2020) Transcriptome Profile Changes Associated With Heat Shock Reaction in the Entomopathogenic Nematode, Steinernema carpocapsae. Front. Physiol. 11:721. doi: 10.3389/fphys.2020.00721
Received: 03 February 2020; Accepted: 01 June 2020;
Published: 10 July 2020.
Edited by:
Hadley Wilson Horch, Bowdoin College, United StatesReviewed by:
Richou Han, Guangdong Institute of Applied Biological Resources, ChinaAdler Ray Dillman, University of California, Riverside, United States
Copyright © 2020 Xie, Wang, Zhong, Zhu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dao-Hong Zhu, ZGFvaG9uZ3podUB5ZWFoLm5ldA==; Zhen He, aGV6aGVuNTMxQHNpbmEuY29t
 Yi-Fei Xie
Yi-Fei Xie Xiu-Dan Wang2
Xiu-Dan Wang2 Dao-Hong Zhu
Dao-Hong Zhu