
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Physiol. , 29 July 2020
Sec. Red Blood Cell Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fphys.2020.00588
This article is part of the Research Topic The Future of Physiology: 2020 and Beyond, Volume I View all 8 articles
 Anna Bogdanova1*
Anna Bogdanova1* Lars Kaestner2,3*
Lars Kaestner2,3*  the European Red Cell Society (ERCS)
the European Red Cell Society (ERCS)Why should we take interest in studying red blood cells? This mini review attempts to answer this question and highlights the problems that authors find most appealing in this dynamic research area. It addresses the early career scientists who are just starting their independent journey and facing tough times. Despite unlimited access to information, the exponential development of computational and intellectual powers, and the seemingly endless possibilities open to talented and ambitious early career researchers, they soon realize that the pressure of imminent competition for financial support is hard. They have to hit deadlines, produce data, publish, report, teach, manage, lead groups, and remain loving family members at the same time. Are these countless hardships worth it? We think they are. Despite centuries of research, red blood cells remain a mysterious and fascinating study objects. These cells bring together experts within the family of the European Red Cell Society and beyond. We all share our joy for the unknown and excitement in understanding how red cells function and what they tell us about the microenvironments and macroenvironments they live in. This review is an invitation to our colleagues to join us on our quest.
Red blood cells have power over us, no doubt. Making up over 50% of our cells (2 × 1013 cells), these cells provide us with energy to live, think, and create (Bianconi et al., 2013; Lew and Tiffert, 2017). Each day, we lose 1.7 × 1011 cells and make the same number anew (Lew and Tiffert, 2017). However, the deeper we delve into the red blood cell universe, the humbler we feel, as we still have no ultimate answer to “the Question of Life, the Universe, and Everything.”
Are all the 1.7 × 1011 cells we produced today the same? Do they differ from the cells we made yesterday when we went hiking in the mountains or were swimming in the lake? How do the heat waves associated with climate change affect these cells? How does microgravity affect them when, e.g., hitchhiking through the galaxy? How do the cells change as we get older and older with the increasing life expectancy?
Do the red cells produced today pass away on the same day and from the same cause? For humans, the causes of death and lifespan of individual cells seem to be somewhat random (Kaestner and Minetti, 2017), and the investigations to be done are complex, as we cannot trace the life cycle of each individual cell back as detectives do. We do not know exactly what forces red blood cells to die and how they exactly cease to exist. Some of us acknowledge oxidation-induced clustering of band 3 proteins in the membrane as a signal that tags cells with antibodies against these clusters as “labeled for removal” (Lutz, 2012; Lutz and Bogdanova, 2013). Others describe red cell death as an “apoptosis-like” process and call it eryptosis (Lang et al., 2006, 2008). The “Nomenclature Committee on Cell Death,” specializing in terminology, warns us for translation of the process as well as the word “eryptosis” to the way the enucleated cells pass away (Galluzzi et al., 2018). Is there one or more than one way to die? This question remains unclear and must still be resolved.
What do we need to learn about red blood cells? As in Adams’ novels, experts and early career scientists witness dynamic developments in the field that leave us both excited and thrilled. We seem to know much about the major function of red blood cells, which is gas transport, but there is much more to what these cells are doing. This makes it difficult to limit the number of ultimate questions to just one (Figure 1).
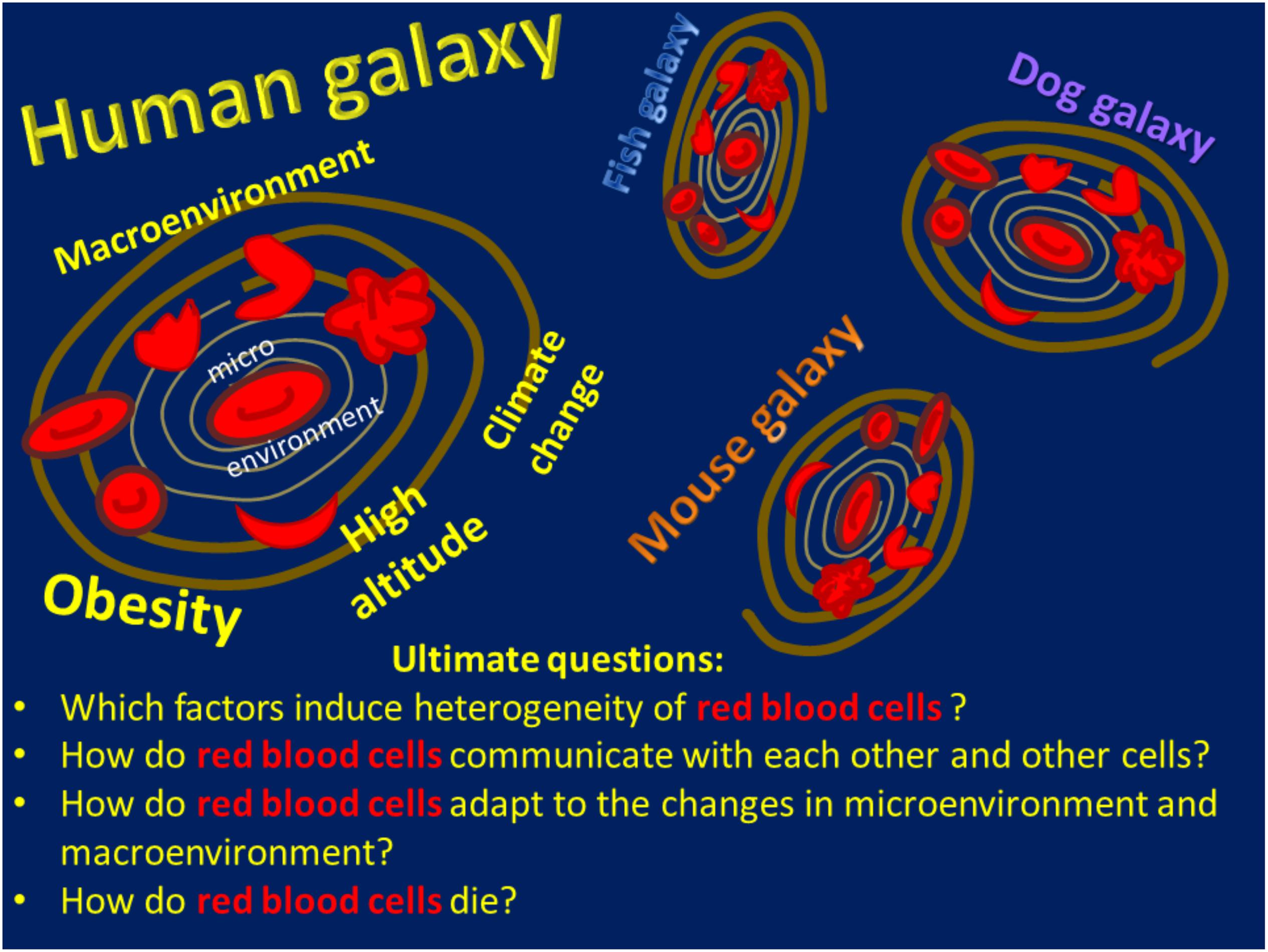
Figure 1. A schematic of the Red Blood Cell Universe as the authors see it, including a list of ultimate questions.
There are more than 100 years of evidence for the active participation of red blood cells in blood coagulation (Duke, 1983; Andrews and Low, 1999; Steffen et al., 2011; Byrnes and Wolberg, 2017). This concept, however, did not mature enough to enter the textbooks. How many million years will it take?
There are some indications that red blood cells may sense the changes in plasma levels of hormones, such as insulin (Pedersen et al., 1982; Zhang et al., 2015), catecholamines (Hasan et al., 2017) and cortisol (Farese and Plager, 1962), sex hormones (Koefoed and Brahm, 1994), and erythropoietin (Trial et al., 2001; Mihov et al., 2009). If so, what happens to the cells as hormones interact with the receptors on the red blood cell membrane or cytosolic components?
Red blood cells are famous because they are widely used as a perfect cell model for studying cell membranes (Agre and Parker, 1989; Bernhardt and Ellory, 2003; Yawata, 2003). Earlier, all cells looked the same to the observers, and their properties were studied “en masse” e.g. using radioactive isotopes, rubbing cells between two plates to examine their viscosity and deformability, by inflating or deflating them, and by applying all possible approaches to whole blood samples. Mean volume and hemoglobin content values were assigned to them. Only desperate experts, such as Marcel Bessis, were photographing red blood cells for their beauty and turning their appearance into art by means of scanning electron microscopy (Bessis, 1974). If we want to study cells of similar densities and, eventually, ages, we may apply centrifugal force to produce fractions of such cells (Figure 2). Everyone that has once done such an experiment appreciates that each red cell has a certain density and joins one, but not the other group of cells of certain density, producing a striped pattern in centrifuged samples. Why certain densities are favored and others avoided is a question that needs answering. Our current understanding of red blood cell shapes includes their individual appearance and properties, their dynamic shape transitions, and their “shape memory” (Fischer, 2004; Tomaiuolo, 2014; Lanotte et al., 2016; Cordasco and Bagchi, 2017; Kihm et al., 2018). These studies make use of cellular and molecular biophysics, sophisticated in vivo imaging techniques for microfluidic channels and even blood vessels, and a great deal of artificial intelligence – even coming close to “Deep Thought,” the supercomputer in Adams’ story – but there is one crucial difference: patients do not have a million years to wait for the answer.

Figure 2. Red blood cells may have different densities, as shown on this continuous Percoll density gradient, but some densities are favored, and others are mysteriously abandoned.
Most red blood cell researchers are deeply devoted to developing novel tools that may reveal so far unnoticed sides of cells we all like to study. Benefits are include protein profiling of red blood cells using omics approaches (Nemkov et al., 2018). Technologies use stem cells for production and manipulation of red blood cells (Hansen et al., 2019). We may follow red blood cells running through the blood vessels of living hosts (Hertz et al., 2019; Slovinski et al., 2019) and model and evaluate responses of red blood cells to mechanical shear forces – the ones they are exposed to in our microvasculature (Lizarralde Iragorri et al., 2018; Moura et al., 2019). We can detect electric currents that ions mediate passing through red blood cell membranes in hundreds of individual cells at the same time (Rotordam et al., 2019). As progress in research is fast, sometimes careful examination of the possible pitfalls and sources of artifacts is required (Minetti et al., 2013).
There is even more to explore in the universe of pathophysiology. For some patients, we do not have an answer as to what causes a red blood cell defect and only know the symptoms. Even when the molecular cause of the disease is clear, the links between the defective protein (a mismatch in amino acid composition or a dysregulated production program) and disease severity are often unknown. This is the case for sickle cell disease, hereditary spherocytosis, and Gardos channelopathy, just to name a few examples. Some patients get scientists involved into a thrilling quest for the actual cause of disease, those that were identified by hematologists as carriers of “idiopathic hemolytic anemia.” New tools are currently in development that will enable the diagnosis of “newly identified” diseases (Kaestner and Bianchi, 2020).
Even more mysterious cases are described when defects in red blood cells come along with neurodegenerative symptoms. One such disease was named “acanthocytosis” (from the Greek word “acantha,” meaning “thorn”) because of the spiked thorny appearance of red blood cells (De Franceschi et al., 2014; Adjobo-Hermans et al., 2015). In fact, neurons and erythroid progenitor cells in the bone marrow were recently shown to share common gene regulatory pathways defining their fate and properties (Kinney et al., 2019).
The heart and blood also have much in common (Kaestner, 2013). The mortality of patients with myocardial infarction (acute coronary syndrome) and those undergoing valve replacement surgery may be predicted based on the degree of variance in red blood cell shapes and sizes (Ghaffari et al., 2016; Duchnowski et al., 2017; Abrahan et al., 2018). Furthermore, red blood cells were recently shown to function as actors, not passive witnesses, in cardiovascular diseases, contributing to the regulation of redox state and vascular tone and activating protective or disruptive signaling cascades in the myocardium and blood vessels (Pernow et al., 2019). Can red blood cells be regarded as deputies for other organs of our body, such as the brain and the heart?
One more exciting and rapidly developing area aims at revolutionizing blood donations and transfusions. Instead of relying on people readily offering their blood for the others to use, researchers are producing, so far in very small amounts, red blood cells of the type needed for each individual patient in a test tube (Shah et al., 2014; Hawksworth et al., 2018). However, it will take some time until cell culture can upscale to provide enough red blood cells for transfusion. Therefore, before this happens, we still have to rely on blood donations and do our best to improve the red blood cell storage conditions (D’Alessandro and Seghatchian, 2017) and to manage and reduce damage of cells during lesions (Yoshida et al., 2019). Furthermore, each patient may decide in the future to use his/her own red blood cells as transport containers to deliver toxic drugs to the location in the body where they are supposed to act without poisoning the host (Villa et al., 2016; Sun et al., 2017). Nature itself has chosen to modify components of red blood cells to protect hosts from Plasmodium infection causing a deadly disease that claimed over 400 000 lives in 2018 alone, malaria (Weatherall, 2008; Timmann et al., 2012; Malaria Genomic Epidemiology et al., 2015). This evolutionary selection has taken ages to occur and may now be of use to the development of protective strategies for the human population, as the spread of Plasmodium further to the north will follow the increase in atmospheric temperatures.
The universe of red blood cells spreads far beyond the cells that function in Homo sapiens. In agreement with the Hitchhiker’s Guide, we have learned much about RBCs in the true rulers of the Earth, mice. These furry fellows give us a chance to study the mechanisms of diseases and to design new therapies for mice and humans. Our knowledge of the red blood cells in other species, including our pets and other tamed and wild, warm- and cold-blooded creatures that attend veterinary clinics from time to time, is rather fragmentary and requires more attention (Figure 1).
The ultimate “Answer to the Ultimate Question of Life, the Universe, and Everything” may only be given as we keep working and using our brains along with artificial intelligence. The next edition of “The Guide to Red Blood Cells” is on the way, and the motto for the early career scientists in the area stands as stated by Adams: “Don’t Panic.”
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abrahan, L. L. T., Ramos, J. D. A., Cunanan, E. L., Tiongson, M. D. A., and Punzalan, F. E. R. (2018). Red cell distribution width and mortality in patients with acute coronary syndrome: a meta-analysis on prognosis. Cardiol. Res. 9, 144–152. doi: 10.14740/cr732w
Adjobo-Hermans, M. J., Cluitmans, J. C., and Bosman, G. J. (2015). Neuroacanthocytosis: observations, theories and perspectives on the origin and significance of acanthocytes. Tremor Other Hyperkinet. Mov. 5:328.
Agre, P., and Parker, J. C. (eds) (1989). Red Blood Cell Membranes: Structure, Function, Clinical Implication. New York, NY: Marcel Dekker, Inc.
Andrews, D. A., and Low, P. S. (1999). Role of red blood cells in thrombosis. Curr. Opin. Hematol. 6, 76–82.
Bernhardt, I., and Ellory, J. C. (eds) (2003). Red Cell Membrane Transport in Health and Disease. Berlin: Springer Verlag.
Bianconi, E., Piovesan, A., Facchin, F., Beraudi, A., Casadei, R., Frabetti, F., et al. (2013). An estimation of the number of cells in the human body. Ann. Hum. Biol. 40, 463–471. doi: 10.3109/03014460.2013.807878
Byrnes, J. R., and Wolberg, A. S. (2017). Red blood cells in thrombosis. Blood 130, 1795–1799. doi: 10.1182/blood-2017-03-745349
Cordasco, D., and Bagchi, P. (2017). On the shape memory of red blood cells. Phys. Fluids 29:041901. doi: 10.1063/1.4979271
D’Alessandro, A., and Seghatchian, J. (2017). Hitchhiker’s guide to the red cell storage galaxy: omics technologies and the quality issue. Transfus. Apher. Sci. 56, 248–253. doi: 10.1016/j.transci.2017.03.006
De Franceschi, L., Bosman, G. J., and Mohandas, N. (2014). Abnormal red cell features associated with hereditary neurodegenerative disorders: the neuroacanthocytosis syndromes. Curr. Opin. Hematol. 21, 201–209. doi: 10.1097/moh.0000000000000035
Duchnowski, P., Hryniewiecki, T., Stoklosa, P., Kusmierczyk, M., and Szymanski, P. (2017). Red cell distribution width as a prognostic marker in patients undergoing valve surgery. J. Heart Valve Dis. 26, 714–720.
Duke, W. W. (1983). The relation of blood platelets to hemorrhagic disease. By W.W. Duke. JAMA 250, 1201–1209. doi: 10.1001/jama.250.9.1201
Farese, R. V., and Plager, J. E. (1962). The in vitro red blood cell uptake of C-14-cortisol; studies of plasma protein binding of cortisol in normal and abnormal states. J. Clin. Invest. 41, 53–60. doi: 10.1172/jci104466
Fischer, T. M. (2004). Shape memory of human red blood cells. Biophys. J. 86, 3304–3313. doi: 10.1016/s0006-3495(04)74378-7
Galluzzi, L., Vitale, I., Aaronson, S. A., Abrams, J. M., Adam, D., Agostinis, P., et al. (2018). Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death. Differ. 25, 486–541.
Ghaffari, S., Pourafkari, L., Sepehrvand, N., Aslanabadi, N., Faridi, L., Tajlil, A., et al. (2016). Red cell distribution width is a predictor of ST resolution and clinical outcome following thrombolysis in acute ST elevation myocardial infarction. Thromb. Res. 140, 1–6. doi: 10.1016/j.thromres.2016.02.008
Hansen, M., Von Lindern, M., Van Den Akker, E., and Varga, E. (2019). Human-induced pluripotent stem cell-derived blood products: state of the art and future directions. FEBS Lett. 593, 3288–3303. doi: 10.1002/1873-3468.13599
Hasan, S., Mosier, M. J., Szilagyi, A., Gamelli, R. L., and Muthumalaiappan, K. (2017). Discrete beta-adrenergic mechanisms regulate early and late erythropoiesis in erythropoietin-resistant anemia. Surgery 162, 901–916. doi: 10.1016/j.surg.2017.06.001
Hawksworth, J., Satchwell, T. J., Meinders, M., Daniels, D. E., Regan, F., Thornton, N. M., et al. (2018). Enhancement of red blood cell transfusion compatibility using CRISPR-mediated erythroblast gene editing. EMBO Mol Med 10: e8454.
Hertz, L., Ruppenthal, S., Simionato, G., Quint, S., Kihm, A., Abay, A., et al. (2019). The evolution of erythrocytes becoming red in respect to fluorescence. Front. Physiol. 10:753. doi: 10.3389/fphys.2019.00753
Kaestner, L. (2013). Calcium Signalling: Approaches and Findings in the Heart and Blood. Berlin: Springer.
Kaestner, L., and Bianchi, P. (2020). Trends in the development of diagnostic tools for red blood cell related diseases and anaemias. Front. Physiol. 11:387. doi: 10.3389/fphys.2020.00387
Kaestner, L., and Minetti, G. (2017). The potential of erythrocytes as cellular aging models. Cell Death. Differ. 24, 1475–1477. doi: 10.1038/cdd.2017.100
Kihm, A., Kaestner, L., Wagner, C., and Quint, S. (2018). Classification of red blood cell shapes in flow using outlier tolerant machine learning. PLoS Comput. Biol. 14:e1006278. doi: 10.1371/journal.pcbi.1006278
Kinney, M. A., Vo, L. T., Frame, J. M., Barragan, J., Conway, A. J., Li, S., et al. (2019). A systems biology pipeline identifies regulatory networks for stem cell engineering. Nat. Biotechnol. 37, 810–818. doi: 10.1038/s41587-019-0159-2
Koefoed, P., and Brahm, J. (1994). The permeability of the human red cell membrane to steroid sex hormones. Biochim. Biophys. Acta 1195, 55–62. doi: 10.1016/0005-2736(94)90009-4
Lang, F., Gulbins, E., Lerche, H., Huber, S. M., Kempe, D. S., and Foller, M. (2008). Eryptosis, a window to systemic disease. Cell Physiol. Biochem. 22, 373–380. doi: 10.1159/000185448
Lang, F., Lang, K. S., Lang, P. A., Huber, S. M., and Wieder, T. (2006). Mechanisms and significance of eryptosis. Antioxid. Redox. Signal. 8, 1183–1192. doi: 10.1089/ars.2006.8.1183
Lanotte, L., Mauer, J., Mendez, S., Fedosov, D. A., Fromental, J. M., Claveria, V., et al. (2016). Red cells’ dynamic morphologies govern blood shear thinning under microcirculatory flow conditions. Proc. Natl. Acad. Sci. U.S.A. 113, 13289–13294. doi: 10.1073/pnas.1608074113
Lew, V. L., and Tiffert, T. (2017). On the mechanism of human red blood cell longevity: roles of Calcium, the Sodium Pump, PIEZO1, and Gardos Channels. Front. Physiol. 8:977. doi: 10.3389/fphys.2017.00977
Lizarralde Iragorri, M. A., El Hoss, S., Brousse, V., Lefevre, S. D., Dussiot, M., Xu, T., et al. (2018). A microfluidic approach to study the effect of mechanical stress on erythrocytes in sickle cell disease. Lab Chip 18, 2975–2984. doi: 10.1039/c8lc00637g
Lutz, H. U. (2012). Naturally occurring autoantibodies in mediating clearance of senescent red blood cells. Adv. Exp. Med. Biol. 750, 76–90. doi: 10.1007/978-1-4614-3461-0_6
Lutz, H. U., and Bogdanova, A. (2013). Mechanisms tagging senescent red blood cells for clearance in healthy humans. Front. Physiol. 4:387. doi: 10.3389/fphys.2013.00387
Malaria Genomic Epidemiology, N., Band, G., Rockett, K. A., Spencer, C. C., and Kwiatkowski, D. P. (2015). A novel locus of resistance to severe malaria in a region of ancient balancing selection. Nature 526, 253–257. doi: 10.1038/nature15390
Mihov, D., Vogel, J., Gassmann, M., and Bogdanova, A. (2009). Erythropoietin activates nitric oxide synthase in murine erythrocytes. Am. J. Physiol. Cell Physiol. 297, C378–C388.
Minetti, G., Egee, S., Morsdorf, D., Steffen, P., Makhro, A., Achilli, C., et al. (2013). Red cell investigations: art and artefacts. Blood Rev. 27, 91–101. doi: 10.1016/j.blre.2013.02.002
Moura, P. L., Lizarralde Iragorri, M. A., Francais, O., Le Pioufle, B., Dobbe, J. G. G., Streekstra, G. J., et al. (2019). Reticulocyte and red blood cell deformation triggers specific phosphorylation events. Blood Adv. 3, 2653–2663. doi: 10.1182/bloodadvances.2019000545
Nemkov, T., Reisz, J. A., Xia, Y., Zimring, J. C., and D’alessandro, A. (2018). Red blood cells as an organ? How deep omics characterization of the most abundant cell in the human body highlights other systemic metabolic functions beyond oxygen transport. Expert Rev. Proteomics 15, 855–864. doi: 10.1080/14789450.2018.1531710
Pedersen, O., Beck-Nielsen, H., Sorensen, N. S., and Svejgaard, A. (1982). Heterogeneity of insulin receptors in patients with untreated insulin-dependent diabetes mellitus. J. Clin. Endocrinol. Metab. 55, 30–39. doi: 10.1210/jcem-55-1-30
Pernow, J., Mahdi, A., Yang, J., and Zhou, Z. (2019). Red blood cell dysfunction: a new player in cardiovascular disease. Cardiovasc. Res. 115, 1596–1605. doi: 10.1093/cvr/cvz156
Rotordam, M. G., Fermo, E., Becker, N., Barcellini, W., Bruggemann, A., Fertig, N., et al. (2019). A novel gain-of-function mutation of Piezo1 is functionally affirmed in red blood cells by high-throughput patch clamp. Haematologica 104, e179–e183. doi: 10.3324/haematol.2018.201160
Shah, S., Huang, X., and Cheng, L. (2014). Concise review: stem cell-based approaches to red blood cell production for transfusion. Stem Cells Transl. Med. 3, 346–355. doi: 10.5966/sctm.2013-0054
Slovinski, A. P., Hajjar, L. A., and Ince, C. (2019). Microcirculation in Cardiovascular diseases. J. Cardiothorac. Vasc. Anesth. 33, 3458–3468.
Steffen, P., Jung, A., Nguyen, D. B., Muller, T., Bernhardt, I., Kaestner, L., et al. (2011). Stimulation of human red blood cells leads to Ca2+-mediated intercellular adhesion. Cell Calcium 50, 54–61. doi: 10.1016/j.ceca.2011.05.002
Sun, Y., Su, J., Liu, G., Chen, J., Zhang, X., Zhang, R., et al. (2017). Advances of blood cell-based drug delivery systems. Eur. J. Pharm. Sci. 96, 115–128.
Timmann, C., Thye, T., Vens, M., Evans, J., May, J., Ehmen, C., et al. (2012). Genome-wide association study indicates two novel resistance loci for severe malaria. Nature 489, 443–446. doi: 10.1038/nature11334
Tomaiuolo, G. (2014). Biomechanical properties of red blood cells in health and disease towards microfluidics. Biomicrofluidics 8:051501. doi: 10.1063/1.4895755
Trial, J., Rice, L., and Alfrey, C. P. (2001). Erythropoietin withdrawal alters interactions between young red blood cells, splenic endothelial cells, and macrophages: an in vitro model of neocytolysis. J. Investig. Med. 49, 335–345. doi: 10.2310/6650.2001.33899
Villa, C. H., Anselmo, A. C., Mitragotri, S., and Muzykantov, V. (2016). Red blood cells: Supercarriers for drugs, biologicals, and nanoparticles and inspiration for advanced delivery systems. Adv. Drug Deliv. Rev. 106, 88–103. doi: 10.1016/j.addr.2016.02.007
Weatherall, D. J. (2008). Genetic variation and susceptibility to infection: the red cell and malaria. Br. J. Haematol. 141, 276–286. doi: 10.1111/j.1365-2141.2008.07085.x
Yoshida, T., Prudent, M., and D’alessandro, A. (2019). Red blood cell storage lesion: causes and potential clinical consequences. Blood Transfus 17, 27–52.
Keywords: red blood cell, senescence, Clearance, adaptation, Comparative, function, morphology
Citation: Bogdanova A and Kaestner L (2020) Early Career Scientists’ Guide to the Red Blood Cell – Don’t Panic!. Front. Physiol. 11:588. doi: 10.3389/fphys.2020.00588
Received: 02 January 2020; Accepted: 11 May 2020;
Published: 29 July 2020.
Edited by:
Geoffrey A. Head, Baker Heart and Diabetes Institute, AustraliaReviewed by:
Jeanne Elise Hendrickson, Yale University School of Medicine, United StatesCopyright © 2020 Bogdanova and Kaestner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Bogdanova, YW5uYWJAYWNjZXNzLnV6aC5jaA==; Lars Kaestner, bGFyc19rYWVzdG5lckBtZS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.