
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 23 June 2020
Sec. Reproduction
Volume 11 - 2020 | https://doi.org/10.3389/fphys.2020.00561
 Shimeng Huang1,2†
Shimeng Huang1,2† Zhenhua Wu2†
Zhenhua Wu2† Xiongkun Yuan2
Xiongkun Yuan2 Na Li2
Na Li2 Tiantian Li2
Tiantian Li2 Junjun Wang2
Junjun Wang2 Crystal L. Levesque3
Crystal L. Levesque3 Cuiping Feng1*
Cuiping Feng1*Impaired intestinal function is frequently detected in newborns with intrauterine growth restriction (IUGR), whereas the mechanism between transcriptome profiles and small intestinal dysfunction is still unclear. Therefore, this study was conducted by using IUGR neonatal piglets to uncover the mechanism underlying intestinal dysfunction. Neonatal piglets with IUGR and normal birth weight (NBW) were sacrificed at birth. Transcriptomic sequencing was performed on jejunum samples and generated 18,997 and 17,531 genes in NBW and IUGR groups, respectively. A total of 10 differentially expressed genes (DEGs) were identified; of note, only seven were mapped to the genome reference database, with two up-regulated (HSF4 and NR1H4; heat shock transcription factor 4 and nuclear receptor subfamily 1 group H member 4, respectively) and five down-regulated (SLC35C1, BTNL3, BPI, NLRP6, and SLC5A8; Solute carrier family 35 member C1, butyrophilin like 3, bactericidal permeability increasing protein, NLR family pyrin domain containing 6, and solute carrier family 5 member 8, respectively). Combining an enrichment analysis and reverse transcriptase–quantitative polymerase chain reaction validation of DEGs, our results proved the lipid metabolism disorder, intestinal dysfunction, and inflammatory response in IUGR piglets. Here, IUGR piglets presented lower concentration of glucose and triglyceride and higher concentration of total cholesterol and low-density lipoprotein cholesterol in plasma, compared with NBW piglets. Histological analysis revealed decreased mucins and increased apoptosis in both jejunum and ileum for IUGR piglets. Collectively, we found that IUGR induced intestinal dysfunction by altering lipid metabolism, intestinal barrier, and inflammatory response in neonatal piglets at birth, which provides new insights into the prevention and treatment of IUGR that protects against metabolic disorders and inflammatory-related diseases.
Intrauterine growth restriction (IUGR) refers to impaired growth and development of the mammalian embryo/fetus or its organs during pregnancy, which is defined by fetal or birth weight more than 2 standard deviations (SDs) below the mean body weight for gestational age (Wu et al., 2008; Wang et al., 2010). As a crucial health problem, IUGR not only causes neonatal mortality and morbidity in 5–10% human neonates worldwide, but also leads to adverse effects on postnatal growth and threatens long-term health (Widdowson, 1971; Sangild et al., 2006; Tomasz et al., 2007). Besides, IUGR is a multifactor disease associated with inadequate nutrient intake, environmental stress, and malfunction of the placenta or uterus (McMillen and Robinson, 2005).
Because of the severe prevalence of IUGR across other mammalian species (Wu et al., 2006) and the similarity of gastrointestinal tract with respect to structure, development, and metabolism (Wang et al., 2008; Ferenc et al., 2014), piglets are often applied to the study of the IUGR syndrome of human neonates. Previous studies have reported that IUGR hinders the development of piglet small intestine (SI) by changing the expression of proteins and genes related to cellular signaling, redox balance, protein synthesis, and lipid metabolism (Wu et al., 2006; Wang et al., 2008, 2010, 2018; Li et al., 2017; Huang et al., 2020). Small intestine (SI) is a crucial organ for food digestion and nutrient absorption, as well as a natural defensive barrier against endogenous pathogens and dietary contaminations (Hsueh et al., 2003; Sangild et al., 2006). The abnormal alterations during SI development may trigger feeding intolerance, fat absorption disorder, and digestive disease in newborns (Bernstein et al., 2000; Lee et al., 2001). Furthermore, certain studies detected morphological changes of SI in postnatal piglets, including intestinal length, villous height, and crypt depth, as well as decreased body weight, which may be associated with the symptoms of IUGR (Xu et al., 1994; Baserga et al., 2004). There are several studies focused on liver and islet transcriptome diversity between growing pigs and rat, respectively, but no study has compared differences in the SI transcriptome between the normal birth weight (NBW) piglets and IUGR piglets at birth.
Analysis on differences in the transcriptome between NBW and IUGR neonatal piglets at birth can help us better understand the underlying mechanism of IUGR-induced intestinal dysfunction, metabolic disorders, and inflammatory development and seek appropriate strategies for the prevention and treatment of the disease. Therefore, this study is conducted to (1) perform RNA-seq to identify the genes responsible for the intestinal alteration; (2) conduct reverse transcriptase–quantitative polymerase chain reaction (RT-qPCR) validation and histological analysis on jejunum to detect differentially expressed genes (DEGs) expression levels and mucosal barrier alterations between groups, respectively, and verify transcriptomic findings.
All experiments were carried out with the approval of China Agricultural University Animal Care and Use Committee (CAU20170114-1, Beijing, China).
In the current study, a total of nine multiparous sows (Yorkshire, 2–4 parities) were selected and raised in a commercial pig breeding farm in Sichuan province, China. During the experimental period, the selected sows were fed the same commercial feed and had free access to clean water. Porcine neonates (Landrace × Yorkshire) were spontaneously delivered from sows, and their body weights were recorded at birth. IUGR was identified when the piglet’s body weight was 2 SDs below the mean body weight of the total population. Then, one IUGR with the mean body weight of 0.92 ± 0.04 kg and one NBW with the mean body weight of 1.44 ± 0.05 kg piglets were selected in each of nine litters.
For sampling, a total of 18 male piglets (nine IUGR and nine NBW) were weighed and then killed by a jugular puncture after anesthesia without suckling, as we described previously (Wang et al., 2008, 2018). Blood samples were drawn into heparin-treated tubes via anterior vena cava puncture, and piglets were killed by jugular exsanguination. Plasma was obtained by centrifugation (3,000 revolutions/min for 15 min at 4°C) and was then stored at −80°C. Tissues from all selected IUGR and NBW neonatal piglets were collected. The SI in the neonatal piglet was defined as the portion of the digestive tract from the pylorus of the stomach to the ileocecal valve, with the first 10-cm segment being duodenum. The jejunum and ileum are the distal two parts of the SI. The jejunum begins at the duodenojejunal flexure and the ileum ends at the ileocecal junction. The subsequent 40 and 60% of the SI length below the duodenum are the jejunum and the ileum, respectively (Wang et al., 2008, 2018). The intestine was dissected free of the mesentery and placed on a chilled stainless steel plate. Segments (5 and 10 cm in length) were obtained, respectively, from the midjejunum and midileum (Wang et al., 2008; Yi et al., 2018). Meanwhile, the segments of 10 cm were opened longitudinally and carefully flushed with ice-cold phosphate-buffered saline. Then, the mucosa was collected by scraping using a microscope slide at 4°C and rapidly stored in liquid nitrogen of −80°C until further analysis.
All experiments were carried out with three biological replicates, and each biological replicate contained three piglets from the same group. The jejunum mucosa was collected randomly from three independent piglets at the same group and then pooled together for the following measurement of RNA extraction until further RNA-seq. For RNA extraction, samples were frozen directly in liquid nitrogen after harvest and then ground into powder.
Total RNA was extracted from intestinal sample using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions, and RNA-seq was performed by Majorbio BioTech Co., Shanghai, China. RNA purity and concentration were measured using a Nano Photometer® spectrophotometer (IMPLEN, Los Angeles, CA, United States) and a Qubit® RNA Assay Kit in a Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States). RNA integrity was assessed using an RNA Nano 6000 Assay Kit in a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States). Only samples with RNA integrity number >8 were used for sequencing. Illumina HiSeq 2500 platform was applied to construct RNA libraries and generate reads of 125-bp long paired-end (Illumina, San Diego, CA).
Clean data were obtained by removing reads containing adapters and greater than 10% of poly(N) and low-quality reads (>50% of the bases had Phred quality scores <10) from the raw data. All the downstream analyses were based on the high-quality clean data. Sus scrofa reference genome and gene model annotation files were downloaded from the NCBI database. Index of the reference genome was built using Bowtie v2.0.6 (Broad Institute, Cambridge, MA, United States), and paired-end clean reads were aligned to the reference genome using TopHat v2.0.14. Cufflinks v2.2.1 was applied to assemble mapped reads from each library and identify mRNA transcripts from the TopHat2 alignment results using the reference annotation based on transcript assembly method (Trapnell et al., 2012).
Cuffdiff v2.1.1 was used to calculate FPKM (fragment per kilo base of exon model per million mapped reads) scores for transcripts in each library (Trapnell et al., 2012). Differentially expressed genes were identified through pairwise comparisons between every two stages by DESeq2. Also, we analyzed the expression clustering by systematic analysis for all DEGs using the R package DESeq (Benjamini and Hochberg, 1995). A false discovery rate (FDR) was preset to a number no larger than 0.01, following the procedure of Benjamini-Hochberg (Calafat et al., 1998), which was used to determine the threshold of P-value in multiple tests of differential expression genes. The statistical significance of gene expression differences was evaluated using P < 0.05 and a fold change ≥ 1.5 as a threshold. In addition, we analyzed the expression clustering by systematic analysis for all DEGs using the Heatmaps software package in R (ggplot2 package). Ten DEGs with the most significant alteration were presented as a volcano plot. The log2-transformed fold changes are plotted for abscissa and the -log10-transformed Padjust for ordinate.
Gene Ontology (GO) analysis of DEGs was carried out using Cytoscape software (version 3.4.0; Cytoscape Consortium, San Diego, CA, United States) with the BiNGO application. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of DEGs was performed with KOBAS software using a hypergeometric test. GO terms and KEGG pathways with a Q <0.05 were considered significantly enriched.
To validate the repeatability and reproducibility of DEGs obtained from RNA-seq, five DEGs were quantified by RT-qPCR. Primers were designed based on primer Premier 7.0 and synthesized by Shanghai Generay Biotech Co., Ltd. (Shanghai, China) which are shown in Supplementary Table 1. RNA extraction, cDNA synthesis, and RT-qPCR were conducted according to the method as described previously (Huang et al., 2020). Total RNAs from midjejunum and midileum segment were extracted using Trizol Reagent (Invitrogen) following the manufacturer’s protocol. The RT-qPCR was performed according to the SYBR Premix Ex TaqTM II instructions (Takara, Tokyo, Japan). The reactions were performed on a LightCycler® System (Roche, Mannheim, Germany) as follows: 95°C for 1 min, followed by 40 cycles of 95°C for 10 s, and 10 s at the annealing temperature (Tm). Melting curve analysis was performed from 65 to 95°C with increments of 0.5°C. The relative mRNA levels of target samples to that of control samples were calculated according to 2–ΔΔCt method (Bustin et al., 2009), in which the difference in Ct values (ΔCt) between the target gene and the reference gene/internal control (β-actin) was calculated for normalization, and the ΔCt of the different samples was compared directly (ΔΔCt). Each RNA sample was analyzed in nine biological replicate. Data were expressed as least-square means ± standard error of the mean (SEM).
The concentrations of glucose, triglyceride (TG), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C) in plasma were assayed by Hitachi automatic biochemistry analyzer 7160 (Hitachi High-Tech Corporation, Tokyo, Japan) with Maccura chemical reagents (Maccura Biotechnology Co., Ltd., Chengdu, China).
The midjejunum and midileum were collected, fixed in 4% paraformaldehyde (4°C), dehydrated in graded alcohol, and embedded in paraffin wax. Then, the embedded tissues were cut into 4-μm-thick sections and stained with Alcian blue (AB) and periodic acid–Schiff for morphological analysis. After deparaffinization and rehydration were performed, the slices were treated with AB G8X (Servicebio, G1049) for 5 min and washed with running tap water for 2 min. Afterward, slices were treated with periodic acid (Servicebio, G1049) for 5 min and subjected to Schiff’s staining (Servicebio, G1049) for 30 min in the dark. Afterward, the slices were stained with hematoxylin, dehydrated, mounted, and developed. The images were evaluated using the Image J software (US National Institutes of Health, Bethesda, MD, United States).
For gut apoptosis detection, it was detected by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining of sections of jejunal and ileal tissues using an In Situ Cell-Death Detection Kit (Roche, Basel, Switzerland) following the manufacturer’s instructions. Images were captured using an Olympus BS43 microscope (Olympus, Tokyo, Japan) and processed using DP73 version software (Olympus). The software was used to select the green fluorescent cell nuclei with the same label as the unified standard for judging all photopositive cells. The DAPI blue nuclei with the same label were selected as the total cells, and the TUNEL-positive cell number per field of intestinal epithelial cells was analyzed. Cell apoptosis was observed by green fluorescence microscopy (200 × magnification).
The minimum sample size of three samples per group (n = 3 in the study) was calculated using RNA-seq in the liver of adult pigs (Shen et al., 2018). Litter variation (P > 0.05) was tested using linear regression model (mixed effects). A logarithmic transformation was applied to the fold change of gene abundance in order to generate Figure 1B. Gene expression abundance of NBW sample at birth was used as the denominator, by which the gene expression abundance of all samples was compared. To compare the differences among the groups, one-way analysis of variance and Duncan post-hoc test for multiple comparisons were used for normally distributed data, whereas the Kruskal–Wallis test was used for non–normally distributed data. All analyses were performed using SAS 9.0 statistical software (SAS, Cary, NC, United States). Data are expressed as means with their SEM, and values of P < 0.05 were considered significant.
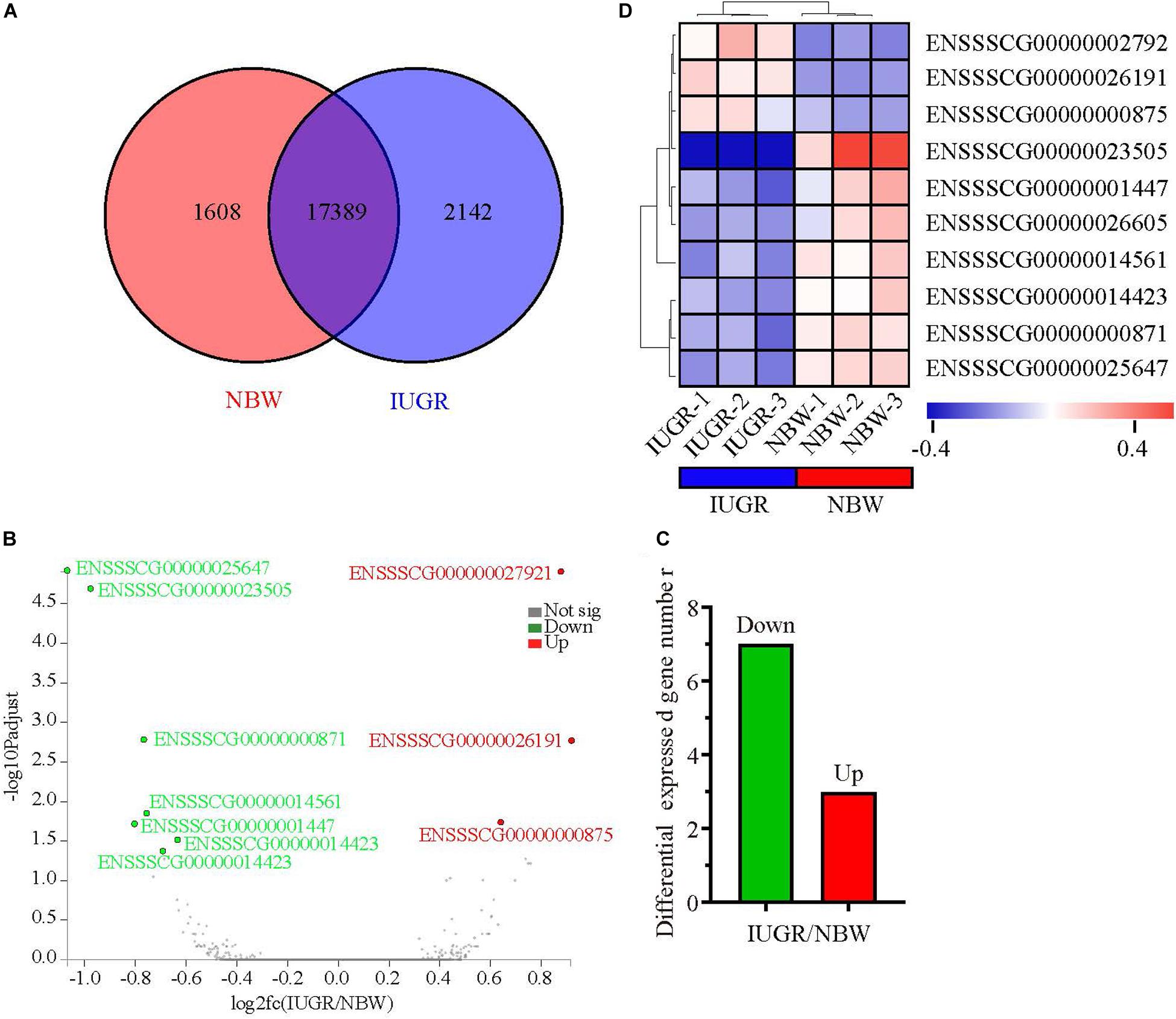
Figure 1. Genome-wide distribution of differentially expressed genes between NBW and IUGR piglets at birth. (A) Venn diagram of expressed genes between NBW and IUGR piglets. (B) Volcano plots of the most significantly altered genes. (C) Heatmap diagram of differentially expressed genes between NBW and IUGR piglets. (D) Distribution of up-regulated genes in NBW and IUGR piglets.
A total of 247,167,220 raw reads were generated from RNA-seq. Subsequently, 23 million clean reads were obtained after the low-quality and adaptor sequences were filtered out. Approximately 80.58–81.99% of all reads were mapped to the reference genome, and 74.74–75.96% of all reads were unique and aligned to the University of California, Santa Cruz pig reference genome (S. scrofa) using the TopHat2 package (Table 1). It was considered to be an expressed transcript and included for subsequent analysis. Therefore, 18,997 and 19,531 known transcripts were identified as being expressed in NBW piglets and IUGR groups, respectively. To explore the global transcriptional changes, we identified a total of 17,389 transcripts coexpressed in both groups, whereas 1,608 and 2,142 genes were differentially expressed in the NBW and IUGR groups, respectively (Figure 1A). Based on the threshold of P < 0.05 and fold change ≥ 1.5, 10 DEGs with the most significant alteration were identified and presented as a volcano plot, with seven down-regulated and three up-regulated in the IUGR group (Figures 1B,C and Table 2). Interestingly, only seven genes were mapped to the pig genome database (PiGenome), with two up-regulated (HSF4 and NR1H4) and five down-regulated (SLC35C1, BTNL3, BPI, NLRP6, and SLC5A8). The heatmap of the hierarchical clustering analysis revealed the significantly differential expressions between the two groups and indicated the high reproducibility of these DEGs (Figure 1D).
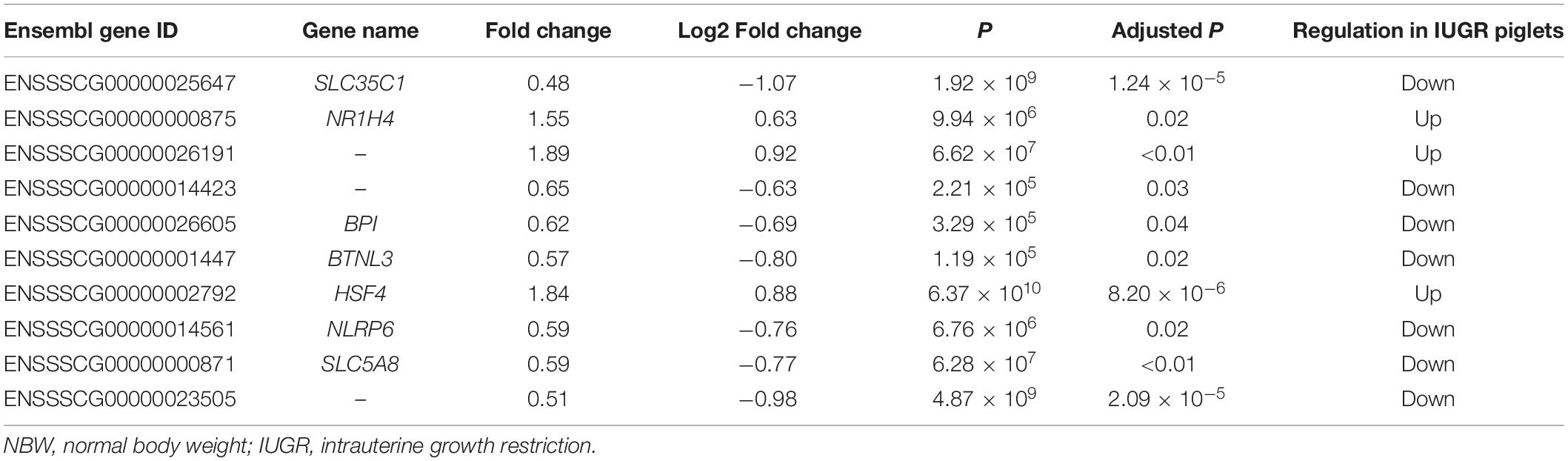
Table 2. Differentially expressed genes of IUGR piglets in relative to NBW piglets with a fold change ≥ 1.5 and P ≤ 0.05.
A total of 10 genes including NR1H4, BPI, HSF4, NLRP6, SLC5A8, SLC35C1, BTNL3, ENSSSCG00000026191, ENSSSCG00000014423, and ENSSSCG00000023505 were selected for RT-qPCR analysis. As shown in Figure 2, no significant change was observed for SLC5A8 mRNA levels between NBW and IUGR groups. Conversely, the mRNA levels of NR1H4, HSF4, and ENSSSCG00000026191 were significant up-regulated (P < 0.05) in the IUGR group. Meanwhile, the gene expressions of BPI, NLRP6, SLC35C1, BTNL3, ENSSSCG00000014423, and ENSSSCG00000023505 were lower (P < 0.05) in the IUGR piglets compared to NBW piglets.
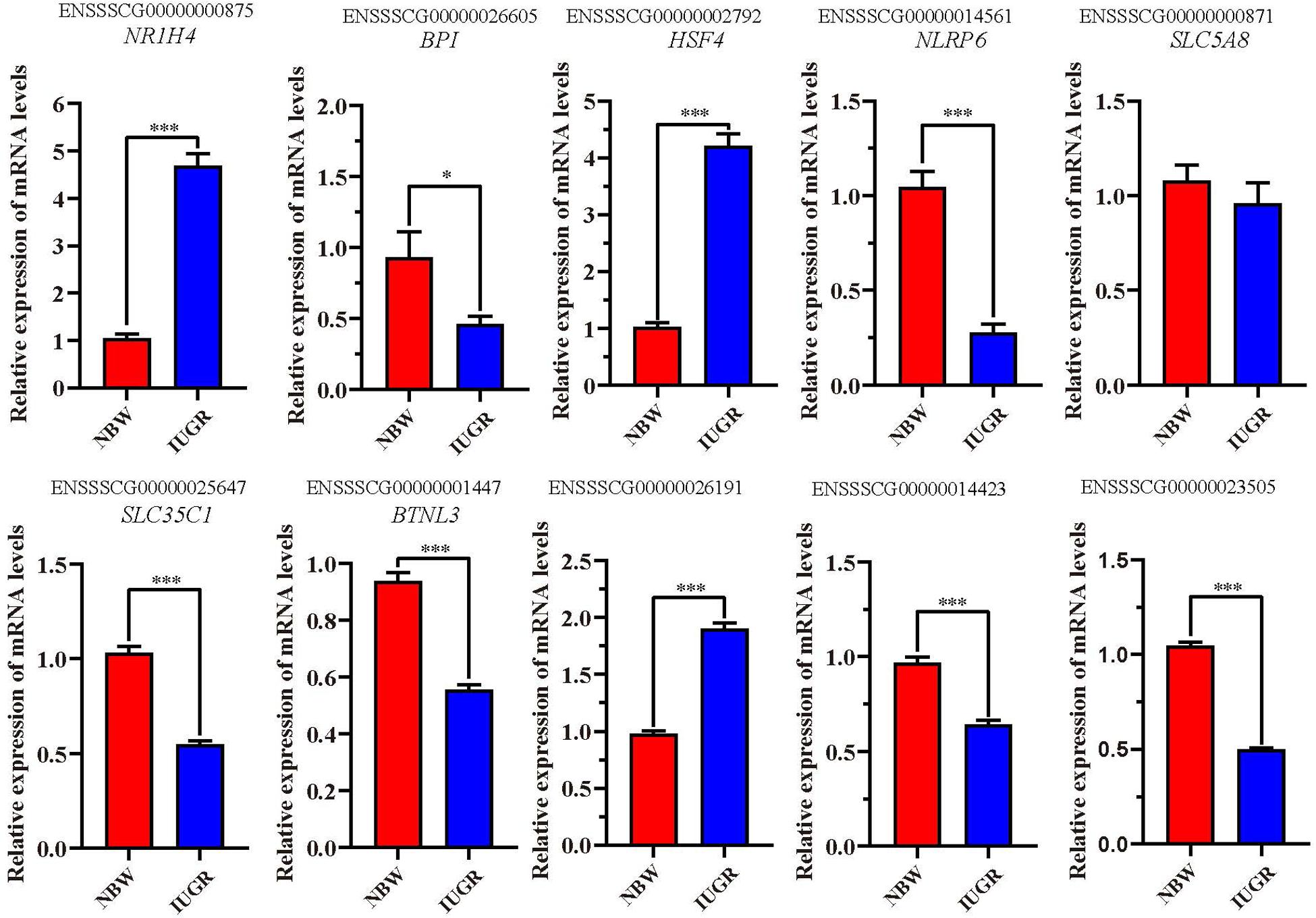
Figure 2. Reverse transcriptase–quantitative polymerase chain reaction validation of the selected genes. Data are shown as mean ± SEM. ∗P < 0.05, ∗∗∗P < 0.001. n = 9 per group. NBW, normal body weight; IUGR, intrauterine growth restriction.
In order to determine the potential functions of these 10 DEGs, the GO enrichment analysis was performed. Therefore, our results showed that more genes were involved in biological processes, followed by cellular components and molecular functions (Figure 3A). As presented in Figure 3B, most GO terms were related to two main classes of biological process: inflammatory and immune activity (such as negative regulation of interleukin 6 production, negative regulation of tumor necrosis factor production, negative regulation of IκB jinase/nuclear factor κB signaling, and negative regulation of inflammatory response) and metabolism (such as regulation of ammonia assimilation cycle, intracellular bile acid receptor signaling pathway, regulation of nitrogen cycle metabolic process, and positive regulation of glutamate metabolic process). In addition, the enrichment ratio of GO terms related to metabolism was higher than those related to inflammatory and immune activity. Interestingly, our results showed that the significantly overrepresented GO term was the response to the bacterium (P < 0.05).
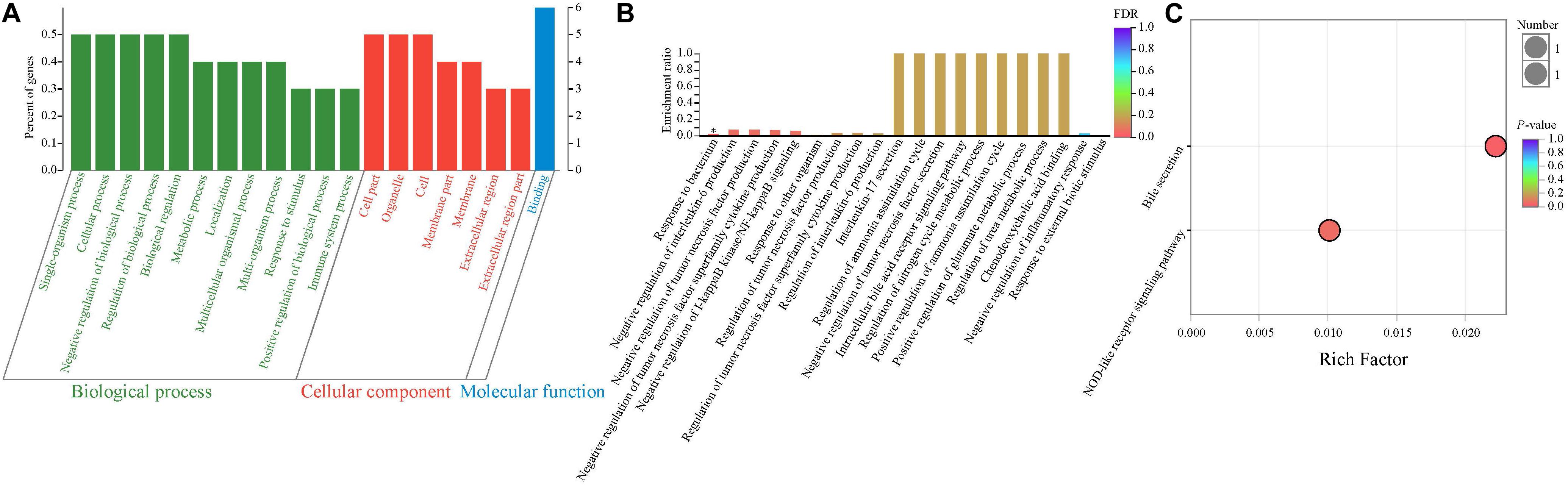
Figure 3. Functional enrichment analysis of differentially expressed genes between NBW and IUGR piglets. (A) Gene Ontology categories enriched for differentially expressed genes. The abscissa represents the secondary classification terms of GO, the left ordinate represents the percent of genes enriched in the secondary classification, and the right ordinate represents the number of genes. These three colors represent three categories: green represents the biological process, red represents the cellular component, and blue represents the molecular function. (B) Gene Ontology enrichment analysis enriched for differentially expressed genes. Significantly enriched GO terms were selected based on a FDR < 0.05. (C) KEGG pathway enrichment analysis of differentially expressed genes.
Next, the KEGG pathway enrichment analysis was performed. As shown in Figure 3C, the DEGs were significantly enriched in the bile secretion pathway and NOD-like receptor signaling pathway (P < 0.05), which are related to metabolism and immune and inflammatory activity, respectively.
In the analysis of lipid metabolites in plasma, IUGR group exhibited lower concentrations of glucose (P < 0.05) and TG (P < 0.01) and higher concentrations of TC (P < 0.05), HDL-C, and LDL-C, P < 0.05) compared with the normal group. Details are presented in Table 3.
Periodic acid–Schiff and AB staining analysis revealed that the IUGR group presented fewer positive cells in both jejunum (P < 0.005) and ileum (P < 0.05) compared with the NBW group (Figures 4, 5). Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling histological analysis revealed that IUGR group presented more apoptotic cells in both jejunum (P < 0.001) and ileum (P < 0.05) compared with the NBW group (Figure 6).
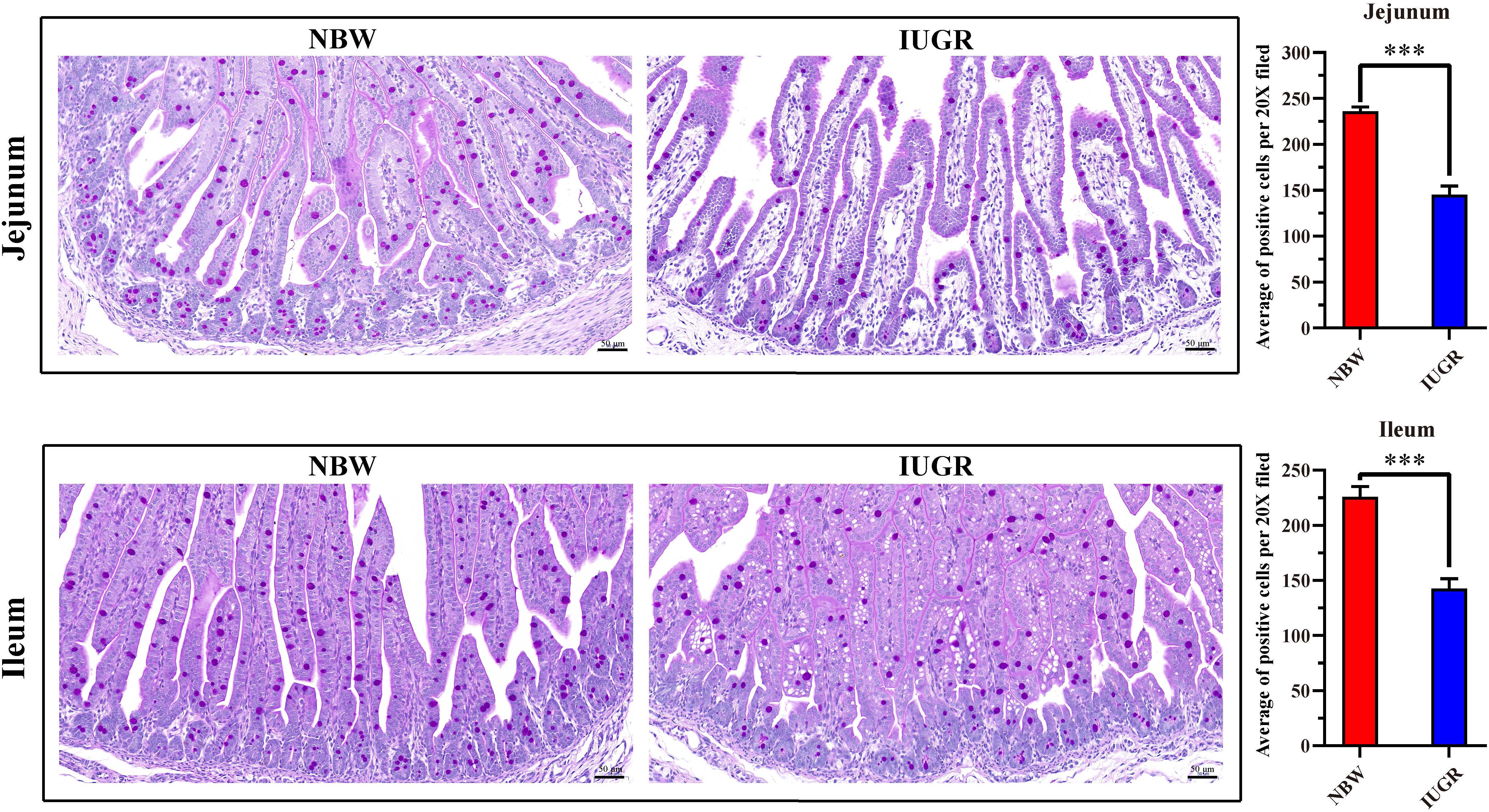
Figure 4. Periodic acid–Schiff staining on jejunum and ileum in NBW and IUGR piglets. Data are shown as mean ± SEM. ∗∗∗P < 0.001. n = 3 per group. NBW, normal body weight; IUGR, intrauterine growth restriction. Bar = 50 μm.
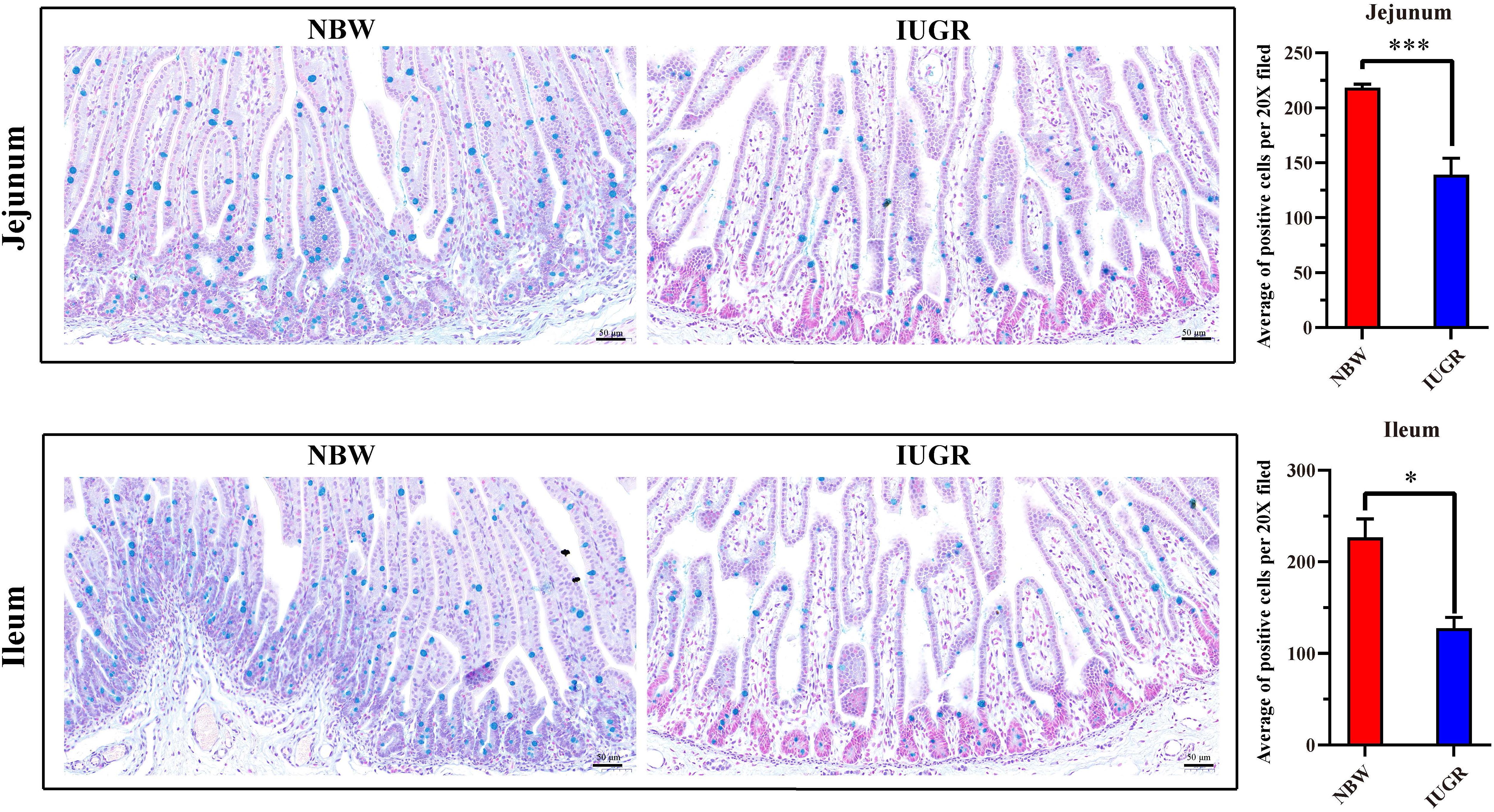
Figure 5. Alcian blue staining on jejunum and ileum in NBW and IUGR piglets. Data are shown as mean ± SEM. ∗P < 0.05, ∗∗∗P < 0.001. n = 3 per group. NBW, normal body weight; IUGR, intrauterine growth restriction. Bar = 50 μm.
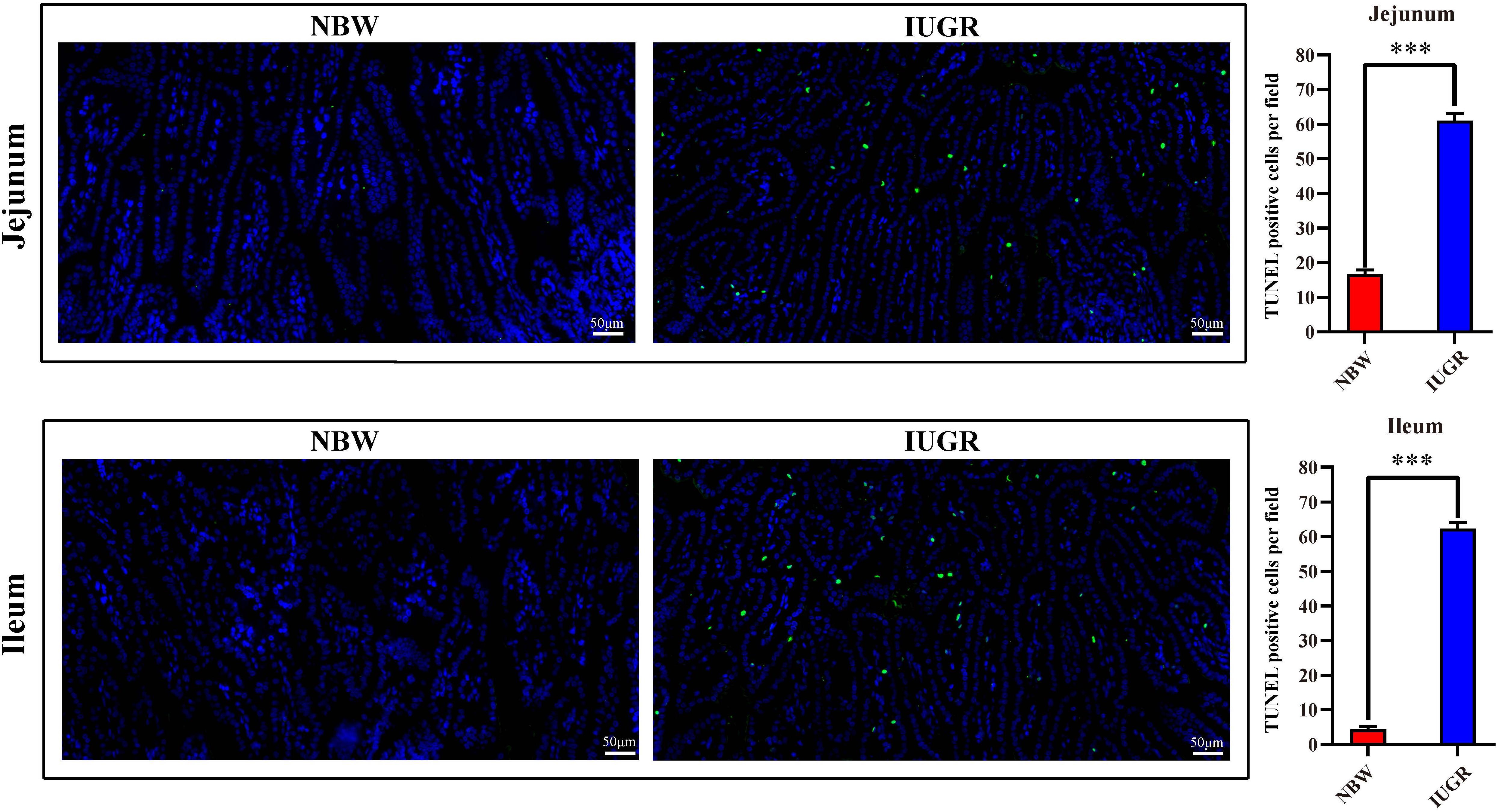
Figure 6. Apoptosis analysis on jejunum and ileum by TUNEL in NBW and IUGR piglets. Data are shown as mean ± SEM. ∗∗∗P < 0.001. n = 3 per group. TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; NBW, normal body weight; IUGR, intrauterine growth restriction. Bar = 50 μm.
To validate intestinal dysfunction results, gene mRNA expressions of interest were detected to perform RT-qPCR in both jejunum and ileum. As shown in Figure 7, the mRNA levels of occludin, zonula occludens-1 (ZO-1), Mucin1, and Mucin4 were significantly down-regulated (P < 0.01) in jejunum and ileum tissues from IUGR piglets. Besides, Mucin2 was remarkably down-regulated (P < 0.001) in the jejunum from IUGR group. Conversely, we observed that the expression of proinflammatory genes interleukin 6 (IL-6) and interleukin-10 (IL-10) was significantly increased (P < 0.001) in jejunum and ileum from IUGR piglets.
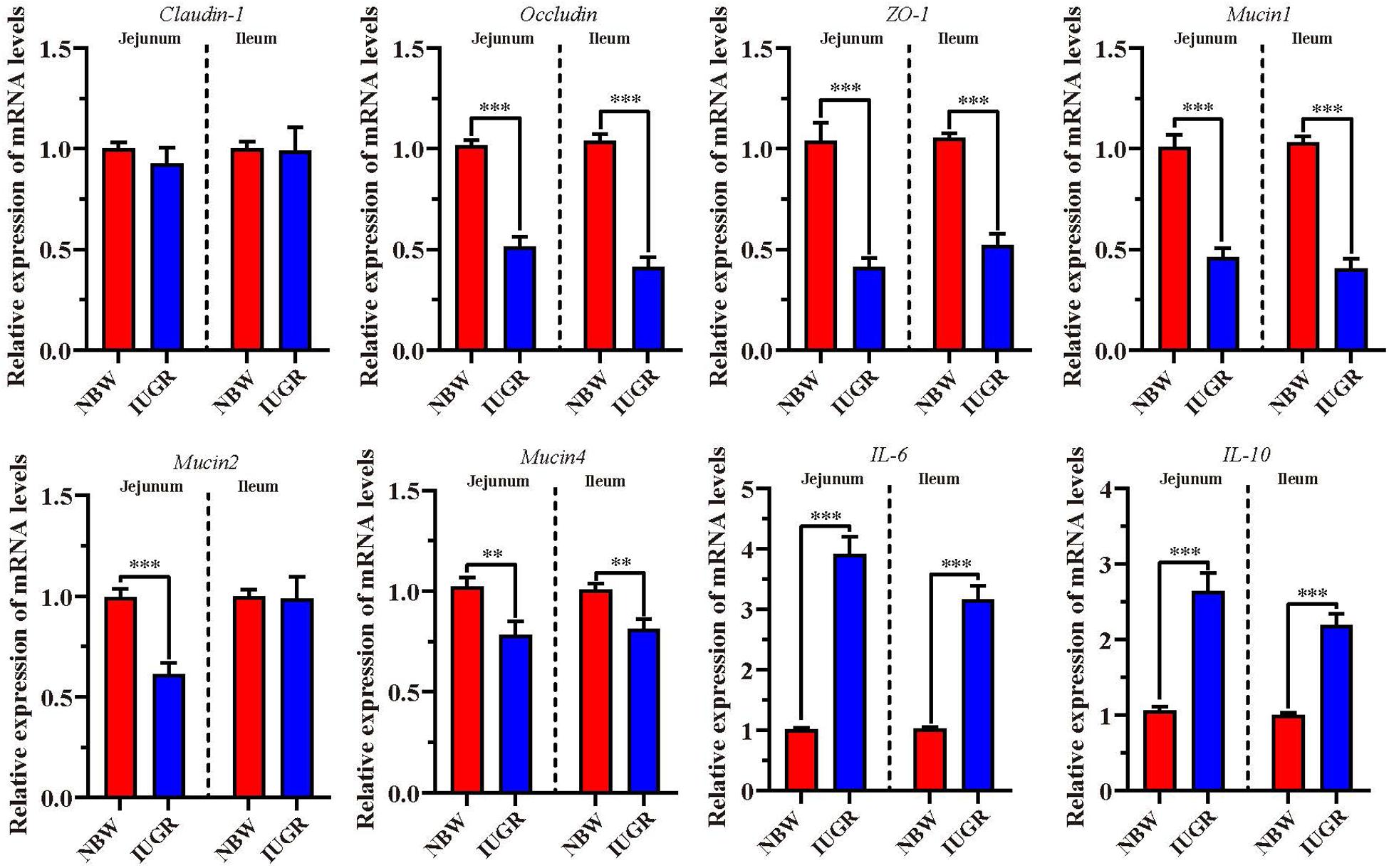
Figure 7. Analysis of genes involved in intestinal barrier and proinflammatory cytokines. Data are shown as mean ± SEM. ∗∗P < 0.01, ∗∗∗P < 0.001. n = 9 per group. NBW, normal body weight; IUGR, intrauterine growth restriction.
Intrauterine growth restriction is a global disease associated with high mortality and morbidity in both human and animals, especially in pig neonates, which triggers great financial loss in the swine industry. Previous studies have detected that IUGR affects the development of infant intestine, leading to intestinal dysfunction and metabolic disorders (Wang et al., 2008, 2018; Li et al., 2017; Huang et al., 2020). Therefore, the current study was performed with the purpose to understand the mechanism of IUGR on neonatal intestine at birth.
By utilizing RNA-seq, we identified 10 significantly altered genes between the groups, which are involved in the GO terms and pathways related to metabolism and inflammatory and immune activity. Among the 10 DEGs, only seven were mapped to the genome reference database, with two up-regulated (HSF4 and NR1H4) and five down-regulated (SLC35C1, BTNL3, BPI, NLRP6, and SLC5A8).
As a member of the NR-family, NR1H4 (or FXR) is expressed predominantly in the intestine and plays a major role in maintaining bile acid (BA) homeostasis (Cariou and Staels, 2007; Fiorucci et al., 2007; Zhang and Edwards, 2008), which is important for the regulation of lipid, glucose, and energy metabolism (Lee et al., 2006; Cariou and Staels, 2007; Staels and Kuipers, 2007; Zhang and Edwards, 2008). In addition, the effect of BA to modulate TG metabolism is primarily dependent on the activation of FXR, which changes the transcription of the genes related to fatty acid and TG synthesis and lipoprotein metabolism (Sirvent et al., 2004; Watanabe et al., 2004). According to our previous studies, low birth weight or IUGR may damage lipid metabolism and intestinal barrier and aggravate malnutrition and metabolic dysfunction in IUGR piglets during early life (Wang et al., 2008; Huang et al., 2019, 2020). In the present study, our result has shown that the concentrations of TC and LDL-C were increased, and the levels of glucose and TG were decreased in IUGR piglets, compared with the NBW piglets. NR1H4 was demonstrated to reduce plasma glucose and TC in FXR-null mice (Lambert et al., 2003; Ma et al., 2006; Zhang et al., 2006). Hence, in the current study, we observed that IUGR group presented significantly higher expression of NR1H4 and lower concentration of glucose and TG compared with the NBW group, suggesting that IUGR affects intestinal homeostasis by regulating lipid metabolism and explaining the IUGR symptoms such as metabolic disorder and low birth weight in neonates.
In addition, we have observed the significant alteration of immunity and inflammation-related genes between the NBW and IUGR piglets. The Nod-like receptor, NLRP6, is recognized as inflammasome that is highly expressed in small and large intestine (Deo and Bandiera, 2008; Hofmann and Hagey, 2008; Russell, 2009). It has been demonstrated to promote intestinal homeostasis and prevent inflammatory disease such as colitis and colitis associated to tumorigenesis (D’Inca et al., 2011). Consistent with our results, expression of NLRP6, occludin, ZO-1, Mucin1, and Mucin4 was shown to decrease upon IUGR in the SI, suggestive of intestinal dysfunction and inflammatory disease in IUGR piglets at birth. Chen et al. (2011) created a chemically induced colitis model in mice and found that the deficiency of NLRP6 increased the susceptibility of injury and inflammation within the colon. Additionally, Al-Azemi et al. (2017) observed a decrease of anti-inflammatory cytokines in IUGR group compared with the normal controls, and D’Inca et al. (2011) detected that the expression of proinflammatory cytokine IL-6 was modified in the intestine of IUGR piglets at birth, indicating the involvement of inflammatory activity in the mechanism of IUGR. Thus, NLRP6, IL-6, and IL-10 exhibited significantly decreased expression level in IUGR group compared with the NBW group in our study. Because the intestinal function is closely linked to proinflammatory cytokines and inflammasome activation, the beneficial effects of NLRP6 on intestinal homeostasis and inflammatory disease may add value to the use of NLRP6 as a therapeutic target for dysregulation of inflammasome activation-associated intestine diseases.
BPI, an important antimicrobial polypeptide (Canny and Levy, 2008), plays a cardinal role in hindering the invasion of Gram-negative bacteria and inflammatory activity (Weiss et al., 1978). According to previous studies, BPI protein helps the host body in fighting with gram-negative bacteria by targeting its opsonization, by neutralizing LPS-mediated inflammatory responses, and through microbicidal activity (Canny and Levy, 2008; Palmer et al., 2011). We have shown that expression of BPI gene was significantly lower in IUGR piglets compared to NBW piglets, which we confirmed with RNA-seq and RT-qPCR analysis; similar results were reported in LBW newborns compared to NBW newborns (Singh et al., 2013). BPI dysregulation in intestinal epithelium is conceded to be associated with various inflammatory diseases, such as Crohn disease, ulcerative colitis, and infectious enteritis (Balakrishnan and Chakravortty, 2017). In agreement with our findings, the down-regulation of genes (BPI, occludin, ZO-1, and Mucin1) and up-regulation of genes (IL-6 and IL-10) detected in IUGR piglets are suggestive of various inflammatory diseases and intestinal epithelium dysregulation in IUGR piglets.
Besides, the expression levels of BTNL3, SLC5A8, and SLC31C1 were also altered in IUGR piglets. First, the butyrophilin-like (BTNL) genes are part of the immunoglobulin superfamily (Blazquez et al., 2018). Consistent with the recent studies, the role of BTNL3 in the homing and maintenance of a semi-activated state on Vγ4 + γδ T cells in the human gut might be relevant for the onset of gut autoimmune diseases such as ulcerative colitis and inflammatory bowel disease (Lebrero-Fernandez et al., 2016; Blazquez et al., 2018). In this study, a down-regulation of BTNL3 mRNA expression levels was observed in IUGR piglets at birth. Recent studies have shown the expression of BTNL3 in tumors and its correlation with patient prognosis, immune response, and inflammatory bowel disease (Blazquez et al., 2018). Compared to NBW piglets at birth, similar results showed that the modulation of BTNL3 seems of interest in the impaired intestinal barrier and inflammatory response in IUGR piglets. Secondly, SLC5A8 is regarded as a Na+-coupled high-affinity transporter for short-chain fatty acid, which is a vital factor for mucosal immune system (Gurav et al., 2015). Consistent with previous studies (Dong et al., 2014; Ferenc et al., 2014; Shalom-Paz et al., 2017), all these findings demonstrate that IUGR disturbs intestinal homeostasis by regulating the genes responsible for immune and inflammatory system, which predisposes the neonates to pathogens and increases the risk of mortality and morbidity. Finally, SLC35C1, also known as GDP-fucose transporter 1, is a member of the solute carrier (SLC) group protein (Fredriksson et al., 2008; Deng et al., 2020). In the previous study, SLC35C1 is a negative regulator of the Wnt signaling pathway in gastrointestinal cancer and colon cancer (Petretti et al., 2000; Carvalho et al., 2010; Deng et al., 2020). Down-regulation of SLC35C1 in colon cancer induces Wnt pathway activity, whose overactivation has been proved to be a hallmark of colon cancer (Deng et al., 2020). Interestingly, this group found that SLC35C1 mRNA expression was decreased in IUGR piglets. This study also offers the potential to explore drugs that can restore SLC35C1 to prevent intestinal diseases.
Additionally, heat shock transcription factors (HSFs) are highly conserved and are required for normal cell growth and development in addition to their central importance in stress adaptation, survival, and disease (Morimoto, 1993, 1998). HSF4 is a member of the heat shock transcription factor family that contains conserved DNA binding and trimerization domains (Morimoto, 1993; Nakai et al., 1997). Recent studies showed that HSF4 is expressed in many tissues and is required for mammalian cellular proliferation and differentiation (Min et al., 2004; Mitsuaki et al., 2004; Gao et al., 2017). Notably, Jin et al. (2012) observed HSF4 is positively involved in tumorigenesis that is induced following disruption of the p53 gene. Here, our analysis of RNA-seq and RT-qPCR showed that HSF4 mRNA and cell apoptosis are up-regulated in IUGR group, revealing the existence of an inverse relationship between HSF4 and IUGR.
As noted previously, our data further suggest that the alteration of SI transcriptional profiles is critical for this intestinal dysfunction and metabolic disorders in IUGR piglets at birth. Moreover, histological analysis revealed that mucosal epithelial cells were damaged. Here, our results showed that IUGR piglets presented fewer positive cells in both jejunum and ileum, compared with the NBW group. Mucus covers the intestinal surface and serves as the first line of innate defense, with mucins forming the basic skeleton. Alteration of mucins expression is considered to be related to inflammatory bowel diseases and colorectal cancer (Andrianifahanana et al., 2006). A lack of mucins has been observed in a model of mucosal injury (Lin et al., 2005), as well as in neonates with necrotizing enterocolitis (Mattar et al., 2003; Vieten et al., 2005), which is a disease of increased risk for IUGR neonates. Interestingly, consistent with previous studies (Canny and Levy, 2008; Wlodarska et al., 2014; Lebrero-Fernandez et al., 2016; Seregin et al., 2017) and our findings, expression of NLRP6, BPI, BTNL3, occludin, ZO-1, Mucin1, and Mucin4 affects mucus secretion and intestinal barrier and contributes to intestinal homeostasis. Therefore, in the histological analysis on jejunum and ileum, we observed significantly increased apoptosis in IUGR group compared with the NBW group, indicating impairment of the intestinal mucosa in IUGR, which may explain the intestinal dysfunction and related syndromes in IUGR neonates.
The present study uncovers that IUGR modulates lipid metabolism, intestinal barrier, and inflammatory activity, as well as impairs the intestinal mucosa in neonatal piglets. These findings may elucidate the primary mechanisms responsible for impaired growth in IUGR neonates and aid in the development of new nutritional intervention strategies.
The datasets generated for this study can be found in the NCBI BioProject accession PRJNA598528 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA598528).
The animal study was reviewed and approved by the China Agricultural University Animal Care and Use Committee. Written informed consent was obtained from the owners for the participation of their animals in this study.
CF, JW, and SH designed the experiments. SH, NL, TL, and ZW conducted the experiment, performed the analysis of samples, and analyzed the data. SH, NL, XY, and ZW carried out the experiments and collected the samples. SH, CL, CF, and JW wrote the manuscript. All authors read and approved the final manuscript.
This study was funded by the Beijing Municipal Natural Science Foundation (S170001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Mianyang New-hope Livestock Farming Co., Ltd., in Sichuan Province, China, for the assistance in this study. We thank Dr. Shuai Zhang for his assistance in manuscript revision and statistical analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00561/full#supplementary-material
BPI, Bactericidal Permeability Increasing Protein; HSFs, Heat-Shock Transcription Factors; IUGR, Intrauterine Growth Restriction; NBW, Normal Birth Weight; NLRP6, NLR Family Pyrin Domain Containing 6; NR1H4, Nuclear Receptor Subfamily 1 Group H Member 4; DEGs, Differentially Expressed Genes; SI, Small Intestine; SD, Standard Deviation; SLC35C1, Solute Carrier Family 35 Member C1; SLC5A8, Solute Carrier Family 5 Member 8; SEM, Standard Error of the Mean; RABT, Reference Annotation Based On Transcript; FPKM, Fragment Per Kilo Base Of Exon Model Per Million Mapped Reads; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; AB, Alcian Blue; TG, Triglyceride; TC, Total Cholesterol; Tm, Annealing Temperature; TUNEL, Terminal Deoxynucleotidyl Transferase-Mediated Dutp Nick-End-Labeling; HDL-C, High Density Lipoprotein Cholesterol; LDL-C, Low Density Lipoprotein Cholesterol; IL-6, Interleukin-6; IL-10, Interleukin-10; FDR, False Discovery Rate; NCBI, National Center for Biotechnology Information; ZO-1, Zonula occludens-1.
Al-Azemi, M., Raghupathy, R., and Azizieh, F. (2017). Pro-inflammatory and anti-inflammatory cytokine profiles in fetal growth restriction. Clin. Exp. Obstet. Gynecol. 44, 98–103.
Andrianifahanana, M., Moniaux, N., and Batra, S. K. (2006). Regulation of mucin expression: mechanistic aspects and implications for cancer and inflammatory diseases. Biochim. Biophys. Acta 1765, 189–222. doi: 10.1016/j.bbcan.2006.01.002
Balakrishnan, A., and Chakravortty, D. (2017). Epithelial cell damage activates bactericidal/permeability increasing-protein (BPI) expression in intestinal epithelium. Front. Microbiol. 8:1567. doi: 10.3389/fmicb.2017.01567
Baserga, M., Bertolotto, C., Maclennan, N. K., Hsu, J. L., Pham, T., Laksana, G. S., et al. (2004). Uteroplacental insufficiency decreases small intestine growth and alters apoptotic homeostasis in term intrauterine growth retarded rats. Early Hum. Dev. 79, 93–105. doi: 10.1016/j.earlhumdev.2004.04.015
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bernstein, I. M., Horbar, J. D., Badger, G. J., Ohlsson, A., and Golan, A. (2000). Morbidity and mortality among very-low-birth-weight neonates with intrauterine growth restriction. Am. J. Obst. Gynecol. 182, 198–206. doi: 10.1016/s0002-9378(00)70513-8
Blazquez, J. L., Benyamine, A., Pasero, C., and Olive, D. (2018). New insights into the regulation of gammadelta T Cells by BTN3A and Other BTN/BTNL in tumor immunity. Front. Immunol. 9:1601. doi: 10.3389/fimmu.2018.01601
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Calafat, J., Janssen, H., Tool, A., Dentener, M. A., Knol, E. F., Rosenberg, H. F., et al. (1998). The bactericidal/permeability-increasing protein (BPI) is present in specific granules of human eosinophils. Blood 91, 4770–4775. doi: 10.1214/aos/1013699998
Canny, G., and Levy, O. (2008). Bactericidal/permeability-increasing protein (BPI) and BPI homologs at mucosal sites. Trends Immunol. 29, 541–547. doi: 10.1016/j.it.2008.07.012
Cariou, B., and Staels, B. (2007). FXR: a promising target for the metabolic syndrome? Trends Pharmacol. Sci. 28, 236–243. doi: 10.1016/j.tips.2007.03.002
Carvalho, A. S., Harduin-Lepers, A., Magalhães, A., Machado, E., Mendes, N., Costa, L. T., et al. (2010). Differential expression of alpha-2,3-sialyltransferases and alpha-1,3/4-fucosyltransferases regulates the levels of sialyl Lewis a and sialyl Lewis x in gastrointestinal carcinoma cells. Int. J. Biochem. Cell Biol. 42, 80–89. doi: 10.1016/j.biocel.2009.09.010
Chen, G. Y., Liu, M., Wang, F., Bertin, J., and Nunez, G. (2011). A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 186, 7187–7194. doi: 10.4049/jimmunol.1100412
Deng, M., Chen, Z., Tan, J., and Liu, H. (2020). Down-regulation of SLC35C1 induces colon cancer through over-activating Wnt pathway. J. Cell Mol. Med. 24, 3079–3090. doi: 10.1111/jcmm.14969
Deo, A. K., and Bandiera, S. M. (2008). Biotransformation of lithocholic acid by rat hepatic microsomes: metabolite analysis by liquid chromatography/mass spectrometry. Drug Metab. Dispos. 36, 442–451. doi: 10.1124/dmd.107.017533
D’Inca, R., Gras-Le Guen, C., Che, L., Sangild, P. T., and Le Huerou-Luron, I. (2011). Intrauterine growth restriction delays feeding-induced gut adaptation in term newborn pigs. Neonatology 99, 208–216. doi: 10.1159/000314919
Dong, L., Zhong, X., Ahmad, H., Li, W., Wang, Y., Zhang, L., et al. (2014). Intrauterine growth restriction impairs small intestinal mucosal immunity in neonatal piglets. J. Histochem. Cytochem. 62, 510–518. doi: 10.1369/0022155414532655
Ferenc, K., Pietrzak, P., Godlewski, M. M., Piwowarski, J., Kilianczyk, R., Guilloteau, P., et al. (2014). Intrauterine growth retarded piglet as a model for humans–studies on the perinatal development of the gut structure and function. Reprod. Biol. 14, 51–60. doi: 10.1016/j.repbio.2014.01.005
Fiorucci, S., Rizzo, G., Donini, A., Distrutti, E., and Santucci, L. (2007). Targeting farnesoid X receptor for liver and metabolic disorders. Trends Mol. Med. 13, 298–309. doi: 10.1016/j.molmed.2007.06.001
Fredriksson, R., Nordstrom, K. J., Stephansson, O., Hagglund, M. G., and Schioth, H. B. (2008). The solute carrier (SLC) complement of the human genome: phylogenetic classification reveals four major families. FEBS Lett. 582, 3811–3816. doi: 10.1016/j.febslet.2008.10.016
Gao, M., Huang, Y., Wang, L., Huang, M., Liu, F., Liao, S., et al. (2017). HSF4 regulates lens fiber cell differentiation by activating p53 and its downstream regulators. Cell Death Dis. 8:e3082. doi: 10.1038/cddis.2017.478
Gurav, A., Sivaprakasam, S., Bhutia, Y. D., Boettger, T., Singh, N., and Ganapathy, V. (2015). Slc5a8, a Na+-coupled high-affinity transporter for short-chain fatty acids, is a conditional tumour suppressor in colon that protects against colitis and colon cancer under low-fibre dietary conditions. Biochem. J. 469, 267–278. doi: 10.1042/bj20150242
Hofmann, A. F., and Hagey, L. R. (2008). Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell Mol. Life Sci. 65, 2461–2483. doi: 10.1007/s00018-008-7568-7566
Hsueh, W., Caplan, M. S., Qu, X. W., Tan, X. D., De Plaen, I. G., and Gonzalez-Crussi, F. (2003). Neonatal necrotizing enterocolitis: clinical considerations and pathogenetic concepts. Pediatr. Dev. Pathol. 6, 6–23. doi: 10.1007/s10024-002-0602-z
Huang, S., Liu, C., Li, N., Wu, Z., Li, T., Han, D., et al. (2019). Membrane proteomic analysis reveals the intestinal development is deteriorated by intrauterine growth restriction in piglets. Funct. Integr. Genom. 20, 277–291. doi: 10.1007/s10142-019-00714-y
Huang, S.-M., Wu, Z.-H., Li, T.-T., Liu, C., Han, D.-D., Tao, S.-Y., et al. (2020). Perturbation of the lipid metabolism and intestinal inflammation in growing pigs with low birth weight is associated with the alterations of gut microbiota. Sci. Tot. Environ. 719:137382. doi: 10.1016/j.scitotenv.2020.137382
Jin, X., Eroglu, B., Cho, W., Yamaguchi, Y., Moskophidis, D., and Mivechi, N. F. (2012). Inactivation of heat shock factor Hsf4 induces cellular senescence and suppresses tumorigenesis in vivo. Mol. Cancer Res. 10, 523–534. doi: 10.1158/1541-7786.MCR-11-0530
Lambert, G., Amar, M. J., Guo, G., Brewer, H. B. Jr., and Gonzalez, F. J. (2003). The farnesoid X-receptor is an essential regulator of cholesterol homeostasis. J. Biol. Chem. 278, 2563–2570. doi: 10.1074/jbc.M209525200
Lebrero-Fernandez, C., Wenzel, U. A., Akeus, P., Wang, Y., Strid, H., Simren, M., et al. (2016). Altered expression of Butyrophilin (BTN) and BTN-like (BTNL) genes in intestinal inflammation and colon cancer. Immun. Inflamm. Dis. 4, 191–200. doi: 10.1002/iid3.105
Lee, F. Y., Lee, H., Hubbert, M. L., Edwards, P. A., and Zhang, Y. (2006). FXR, a multipurpose nuclear receptor. Trends Biochem. Sci. 31, 572–580. doi: 10.1016/j.tibs.2006.08.002
Lee, M. J., Conner, E. L., Charafeddine, L., Woods, J. R. Jr., and Del Priore, G. (2001). A critical birth weight and other determinants of survival for infants with severe intrauterine growth restriction. Ann. N. Y. Acad. Sci. 943, 326–339. doi: 10.1111/j.1749-6632.2001.tb03813.x
Li, N., Wang, W., Wu, G., and Wang, J. (2017). Nutritional support for low birth weight infants: insights from animal studies. Br. J. Nutr. 117, 1390–1402. doi: 10.1017/S000711451700126X
Lin, J., Peng, L., Itzkowitz, S., Holzman, I. R., and Babyatsky, M. W. (2005). Short-chain fatty acid induces intestinal mucosal injury in newborn rats and down-regulates intestinal trefoil factor gene expression in vivo and in vitro. J. Pediatr. Gastroenterol. Nutr. 41, 607–611. doi: 10.1097/01.mpg.0000179659.09210.ff
Ma, K., Saha, P. K., Chan, L., and Moore, D. D. (2006). Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Invest. 116, 1102–1109. doi: 10.1172/jci25604
Mattar, A. F., Coran, A. G., and Teitelbaum, D. H. (2003). MUC-2 mucin production in Hirschsprung’s disease: possible association with enterocolitis development. J. Pediatr. Surg. 38, 417–421. doi: 10.1053/jpsu.2003.50071
McMillen, I. C., and Robinson, J. S. (2005). Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol. Rev. 85, 571–633. doi: 10.1152/physrev.00053.2003
Min, J. N., Zhang, Y., Moskophidis, D., and Mivechi, N. F. (2004). Unique contribution of heat shock transcription factor 4 in ocular lens development and fiber cell differentiation. Genesis 40, 205–217. doi: 10.1002/gene.20087
Mitsuaki, F., Izu, H., Seki, K., Fukuda, K., Nishida, T., Yamada, S., et al. (2004). HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 23, 4297–4306. doi: 10.1038/sj.emboj.7600435
Morimoto, R. I. (1993). Cells in stress: transcriptional activation of heat shock genes. Science 5, 1409–1410. doi: 10.1126/science.8451637
Morimoto, R. I. (1998). Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 15, 3788–3796. doi: 10.1101/gad.12.24.3788
Nakai, A. T. M., Kawazoe, Y., Inazawa, J., Morimoto, R. I., and Nagata, K. (1997). HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol. Cell Biol. 17, 469–481. doi: 10.1128/mcb.17.1.469
Palmer, C. D., Guinan, E. C., and Levy, O. (2011). Deficient expression of bactericidal/permeability-increasing protein in immunocompromised hosts: translational potential of replacement therapy. Biochem. Soc. Trans. 39, 994–999. doi: 10.1042/BST0390994
Petretti, T., Kemmner, W., Schulze, B., and Schlag, P. M. (2000). Altered mRNA expression of glycosyltransferases in human colorectal carcinomas and liver metastases. Gut 46, 359–366. doi: 10.1136/gut.46.3.359
Russell, D. W. (2009). Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 50(Suppl.), S120–S125. doi: 10.1194/jlr.R800026-JLR200
Sangild, P. T., Siggers, R. H., Schmidt, M., Elnif, J., Bjornvad, C. R., Thymann, T., et al. (2006). Diet- and colonization-dependent intestinal dysfunction predisposes to necrotizing enterocolitis in preterm pigs. Gastroenterology 130, 1776–1792. doi: 10.1053/j.gastro.2006.02.026
Seregin, S. S., Golovchenko, N., Schaf, B., Chen, J., Eaton, K. A., and Chen, G. Y. (2017). NLRP6 function in inflammatory monocytes reduces susceptibility to chemically induced intestinal injury. Mucosal Immunol. 10, 434–445. doi: 10.1038/mi.2016.55
Shalom-Paz, E., Weill, S., Ginzberg, Y., Khatib, N., Anabusi, S., Klorin, G., et al. (2017). IUGR induced by maternal chronic inflammation: long-term effect on offspring’s ovaries in rat model-a preliminary report. J. Endocrinol. Invest. 40, 1125–1131. doi: 10.1007/s40618-017-0681-683
Shen, L., Gan, M., Zhang, S., Ma, J., Tang, G., Jiang, Y., et al. (2018). Transcriptome analyses reveal adult metabolic syndrome with intrauterine growth restriction in pig models. Front. Genet. 9:291. doi: 10.3389/fgene.2018.00291
Singh, V. V., Chauhan, S. K., Rai, R., Kumar, A., Singh, S. M., and Rai, G. (2013). Decreased pattern recognition receptor signaling, interferon-signature, and bactericidal/permeability-increasing protein gene expression in cord blood of term low birth weight human newborns. PLoS One 8:e62845. doi: 10.1371/journal.pone.0062845
Sirvent, A., Claudel, T., Martin, G., Brozek, J., Kosykh, V., Darteil, R., et al. (2004). The farnesoid X receptor induces very low density lipoprotein receptor gene expression. FEBS Lett. 566, 173–177. doi: 10.1016/j.febslet.2004.04.026
Staels, B., and Kuipers, F. (2007). Bile acid sequestrants and the treatment of type 2 diabetes mellitus. Drugs 67, 1383–1392. doi: 10.2165/00003495-200767100-200767101
Tomasz, D., Daniel, G., Rafal, R., Agnieszka, S., Jaroslaw, S., Romuald, Z., et al. (2007). Urinary excretion rates of 8-oxoGua and 8-oxodG and antioxidant vitamins level as a measure of oxidative status in healthy, full-term newborns. Free Radic. Res. 41, 997–1004. doi: 10.1080/10715760701468757
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Prot. 7, 562–578. doi: 10.1038/nprot.2012.016
Vieten, D., Corfield, A., Carroll, D., Ramani, P., and Spicer, R. (2005). Impaired mucosal regeneration in neonatal necrotising enterocolitis. Pediatr. Surg. Int. 21, 153–160. doi: 10.1007/s00383-004-1312-1316
Wang, J., Chen, L., Li, D., Yin, Y., Wang, X., Li, P., et al. (2008). Intrauterine growth restriction affects the proteomes of the small intestine, liver, and skeletal muscle in newborn pigs. J. Nutr. 138, 60–66. doi: 10.1093/jn/138.1.60
Wang, X., Wu, W., Lin, G., Li, D., Wu, G., and Wang, J. (2010). Temporal proteomic analysis reveals continuous impairment of intestinal development in neonatal piglets with intrauterine growth restriction. J. Proteome Res. 9, 924–935. doi: 10.2527/jas.2015-8928
Wang, X., Zhu, Y., Feng, C., Lin, G., Wu, G., Li, D., et al. (2018). Innate differences and colostrum-induced alterations of jejunal mucosal proteins in piglets with intra-uterine growth restriction. Br. J. Nutr. 119, 734–747. doi: 10.1017/S0007114518000375
Watanabe, M., Houten, S. M., Wang, L., Moschetta, A., Mangelsdorf, D. J., Heyman, R. A., et al. (2004). Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Invest. 113, 1408–1418. doi: 10.1172/JCI21025
Weiss, J., Elsbach, P., Olsson, I., and Odeberg, H. (1978). Purification and characterization of a potent bactericidal and membrane active protein from the granules of human polymorphonuclear leukocytes. J. Biol. Chem. 253, 2664–2672.
Widdowson, E. M. (1971). Intra-uterine growth retardation in the pig. I. Organ size and cellular development at birth and after growth to maturity. Biol. Neonate 19, 329–340. doi: 10.1159/000240427
Wlodarska, M., Thaiss, C. A., Nowarski, R., Henao-Mejia, J., Zhang, J. P., Brown, E. M., et al. (2014). NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 156, 1045–1059. doi: 10.1016/j.cell.2014.01.026
Wu, G., Bazer, F. W., Datta, S., Johnson, G. A., Li, P., Satterfield, M. C., et al. (2008). Proline metabolism in the conceptus: implications for fetal growth and development. Amino Acids 35, 691–702. doi: 10.1007/s00726-008-0052-57
Wu, G., Bazer, F. W., Wallace, J. M., and Spencer, T. E. (2006). Board-invited review: intrauterine growth retardation: implications for the animal sciences. J. Anim. Sci. 84, 2316–2337. doi: 10.2527/jas.2006-2156
Xu, R. J., Mellor, D. J., Birtles, M. J., Reynolds, G. W., and Simpson, H. V. (1994). Impact of intrauterine growth retardation on the gastrointestinal tract and the pancreas in newborn pigs. J. Pediatr. Gastroenterol. Nutr. 18, 231–240. doi: 10.1097/00005176-199402000-00018
Yi, D., Li, B., Hou, Y., Wang, L., Zhao, D., Chen, H., et al. (2018). Dietary supplementation with an amino acid blend enhances intestinal function in piglets. Amino Acids 50, 1089–1100. doi: 10.1007/s00726-018-2586-2587
Zhang, Y., and Edwards, P. A. (2008). FXR signaling in metabolic disease. FEBS Lett. 582, 10–18. doi: 10.1016/j.febslet.2007.11.015
Keywords: intrauterine growth restriction, intestinal dysfunction, inflammatory, lipid metabolism, neonatal piglets, transcriptomics
Citation: Huang S, Wu Z, Yuan X, Li N, Li T, Wang J, Levesque CL and Feng C (2020) Transcriptome Differences Suggest Novel Mechanisms for Intrauterine Growth Restriction Mediated Dysfunction in Small Intestine of Neonatal Piglets. Front. Physiol. 11:561. doi: 10.3389/fphys.2020.00561
Received: 11 December 2019; Accepted: 04 May 2020;
Published: 23 June 2020.
Edited by:
Marc Yeste, University of Girona, SpainReviewed by:
Ryan Dilger, University of Illinois at Urbana-Champaign, United StatesCopyright © 2020 Huang, Wu, Yuan, Li, Li, Wang, Levesque and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cuiping Feng, YnlkZmNwQGhvdG1haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.