
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 13 March 2020
Sec. Renal Physiology and Pathophysiology
Volume 11 - 2020 | https://doi.org/10.3389/fphys.2020.00209
This article is part of the Research TopicRenal Function in Acute and Chronic Kidney DiseasesView all 31 articles
 Lauren G. Douma1,2
Lauren G. Douma1,2 Kristen Solocinski1
Kristen Solocinski1 Sarah H. Masten1
Sarah H. Masten1 Dominique H. Barral1
Dominique H. Barral1 Sarah J. Barilovits1
Sarah J. Barilovits1 Lauren A. Jeffers3
Lauren A. Jeffers3 Kareme D. Alder4
Kareme D. Alder4 Ravi Patel1
Ravi Patel1 Charles S. Wingo1
Charles S. Wingo1 Kevin D. Brown2
Kevin D. Brown2 Brian D. Cain2
Brian D. Cain2 Michelle L. Gumz1,2*
Michelle L. Gumz1,2*Endothelin-1 (ET-1) is a peptide hormone that functions as a vasoconstrictor in the vasculature, whereas in the collecting duct of the kidney it exerts blood pressure-lowering effects via natriuretic actions. Aberrant ET-1 signaling is associated with several pathological states including hypertension and chronic kidney disease. ET-1 expression is regulated largely through transcriptional control of the gene that encodes ET-1, EDN1. Here we report a long, non-coding RNA (lncRNA) that appears to be antisense to the EDN1 gene, called EDN1-AS. Because EDN1-AS represents a potential novel mechanism to regulate ET-1 expression, we examined the regulation of EDN1-AS expression and action. A putative glucocorticoid receptor response (GR) element upstream of the predicted EDN1-AS transcription start site was identified using the ENCODE database and the UCSC genome browser. Two homozygous deletion clones of the element were generated using CRISPR/Cas9. This deletion resulted in a significant increase in the expression of EDN1-AS, which was associated with increased secretion of ET-1 peptide from HK-2 cells (two-fold increase in KO cells vs. CNTL, n = 7, P < 0.05). Phenotypic characterization of these CRISPR clones revealed a difference in cell growth rates. Using a standard growth assay, we determined that the KO1 clone exhibited a three-fold increase in growth over 8 days compared to control cells (n = 4, P < 0.01) and the KO2 clone exhibited a two-fold increase (n = 4, P < 0.01). These results support a role for EDN1-AS as a novel regulatory mechanism of ET-1 expression and cellular proliferation.
ET-1 has many functions including vascular constriction, nervous system activation, and renal sodium secretion. The kidney is both a source and a target of ET-1, and the kidney has the highest concentrations of ET-1 found in the body (Kohan et al., 2011). The mechanisms of ET-1 action in the pathology of chronic kidney disease (CKD) involve increased cell proliferation, inflammation, and elaboration of the extracellular matrix (Speed and Pollock, 2013; Kohan and Barton, 2014; Reichetzeder et al., 2014). Mice overexpressing ET-1 developed chronic renal failure (Theuring et al., 1998). In humans, excess ET-1 levels are seen in all stages of CKD (Dhaun et al., 2009), and there are marked increases in urinary ET-1 secretion in CKD (Grenda et al., 2007). ET-1 signaling works through two receptors, ETAR and ETBR, via paracrine and autocrine action (Dhaun and Webb, 2019). ETAR and ETBR exert largely opposite effects, and ET-1 stimulation of ETAR is primarily responsible for renal injury. Although regulation of ET-1 levels is widely attributed to control of transcription of the ET-1 gene (EDN1) (Stow et al., 2011), EDN1 mRNA is also regulated at the post-transcriptional level by miRNAs (Jacobs et al., 2013, 2014).
The ET-1 pathway is a therapeutic target for many diseases. The ET receptor blocker Macitentan improved morbidity and mortality in pulmonary arterial hypertension patients (Pulido et al., 2013) whereas studies of ET-1 blockers in the kidney have been less successful. The endothelin axis is an important target in CKD, but pharmacological manipulation of endothelin receptors is associated with adverse side effects that have led to termination of clinical trials (Kohan and Pollock, 2013; Yuan et al., 2015). The ASCEND trial using ET-1 receptor blockers for CKD therapy was abandoned due to increased incidence of congestive heart failure (Reichetzeder et al., 2014). More recently, promising results emerged from SONAR, a trial for the ETA antagonist Atrasentan, which utilized an enrichment protocol to mitigate fluid retention side effects (Heerspink et al., 2019). Atrasentan reduced the risk for renal events in patients with type 2 diabetes mellitus, although the trial was ended early due to a less than expected number of end points. Given the critical role of ET-1 in renal function and CKD, alternative approaches are needed to translate ET-1 pathway inhibition to the bedside. With this goal in mind, we sought to better understand EDN1 gene regulation in light of new findings regarding transcriptional control that continue to emerge from the Encyclopedia of DNA Elements (ENCODE).
Using the University of California-Santa Cruz (UCSC) Genome Browser to interrogate regulatory elements at the EDN1 locus, we identified a putative promoter downstream of the promoter coding sequence. We hypothesized that this promoter may drive expression of a long non-coding (lnc) RNA. Here we describe a novel lncRNA that is antisense with respect to the ET-1 transcript, EDN1-AS. We also identified EDN1-AS expression in multiple human cell types including kidney. Using a human kidney proximal tubule cell line (HK-2), we show that CRISPR-mediated deletion of a regulatory element within the EDN1-AS promoter resulted in increased levels of EDN1-AS. This effect was associated with increased secretion of ET-1 peptide and increased cell proliferation.
The human EDN1 chromatin state was analyzed using the UCSC Genome Browser1 (Karolchik et al., 2014). The EDN1-AS predicted promoter was identified using the Genome Segments and Broad Chromatin HMM tracks with HUVEC cell information selected. The Transcription Factor ChIP track and DNase Clusters track was also used to analyze transcriptional regulation of the predicted promoter site.
HMEC cells were cultured in MEGMTM Mammary Epithelial Cell Growth Medium with BulletKitTM (Lonza) and 10% charcoal stripped FBS. S9 cells were cultured in F12 Ham Kaighn’s modification (F12K) supplemented with 25 mM NaHCO3, 4 mM glutamine, 1% Penicillin/Streptomycin and 10% FBS. HK-2 cells were cultured in DMEM/Hamm’s F12 media supplemented with 10% FBS and 1% Penicillin/Streptomycin. HEK293 cells were cultured in DMEM containing 4.5 g/L glucose supplemented with 1% Penicillin/Streptomycin and 10% FBS. All cells were grown in a 37°C incubator, humidified at 5% CO2.
RNA was isolated from cells using TRIzol (Ambion) per manufacturer instructions. In general, 1 ml TRIzol was used per well in a 6-well plate. Total RNA was treated with DNase (Ambion) per manufacturer instructions to remove genomic DNA. RNA from an adult human female kidney was purchased from Life Technologies.
Human strand-specific EDN1-AS primers (SS1-6; Table 1) were designed to lay down at locations progressively closer to the 5′ end of the EDN1 gene for use in reverse transcriptase reactions. Reverse Transcriptase (RT) from Thermo Fisher was used as per manufacturer instructions. Oligo-dT primers were used as a positive control for any poly-adenylated tailed mRNA. All samples were used in –RT and + RT reactions. Primers have a complementary sequence to the EDN1 sense strand so they will only anneal to antisense RNA. PCR primers (PCR1 and PCR2) were designed to amplify the same region of cDNA regardless of the strand-specific primer used.
HK-2 cells were grown to ∼80% confluence in 6-well cell culture plates. To synchronize the circadian clock in all cells, 100 nM Dexamethasone was added for 30 min and then media was changed. After 24 h, cells were trypsinized and RNA was isolated every 2 h for 24 h.
Guide RNA (gRNA) design, cloning and cell engineering was done as previously outlined (Dükel et al., 2016). Briefly, gRNAs were designed using the online tool CC Top (2 Stemmer et al., 2015) using a ∼150 bp sequence of from the predicted human EDN1-AS promoter as input. The region was selected based on identified transcription factor binding sites hypothesized to be important to transcription regulation.
gRNA sequences were synthesized as complementary oligonucleotides and included overhangs for insertion into BsmB1-digested LentiCRISPRv2 plasmid (Addgene catalog # 52961). Oligonucleotides were 5′ phosphorylated and annealed as outlined by the Zhang lab3 and ligated to linearized LentiCRISPR v2 with Quick Ligase (NEB). Ligation reactions were transformed into Stbl3 bacteria following manufacturer instructions. Using this protocol, four (4) recombinant LentiCRISPR v2 plasmids (designated gRNA 1–4) were cloned and verified by automated Sanger sequencing.
HEK293 cells were co-transfected with the four LentiCRISPR + gRNA constructs (sgRNA 1, 2, 3, and 4) along with psPAX2 (Addgene #11260) and pMD2.G (Addgene #11259) for generation of pseudotyped lentiviral particles. Media containing virus particles was collected 72 h post-transfection and cleared viral supernatant was then added to HK-2 cells singly and pairwise (e.g., gRNA 1 + 2, 1 + 3, 1 + 4 etc.). 2 μg/ml puromycin was used for 2 weeks to select for cells transduced with virus.
After puromycin selection, cells were plated into the first of 4 columns of a 96 well plate in 200 μl of media. Cells were then serial diluted into the following columns. After growth time, wells were inspected for the presence of one to three colonies of cells. These wells were then trypsinized with 1 drop of 0.25% Trypsin per well and plated into 10 cm dishes. Cells were allowed to grow undisturbed for 1 week until colony formation. Cloning rings were dipped in sterile vacuum grease to create a seal and pressed down over the colony. Media was aspirated and 1 drop of Trypsin was added. After ∼5 min, trypsinized cells were removed and plated in 6-well plate and allowed to grow. Genomic DNA was isolated from these cells and used for genotyping PCR.
Gel-purified PCR products from PCR reactions using genotyping primers GT1 and GT2 were cloned into the TA TOPO vector according to the manufacturer’s instructions (Thermo Fisher). Cells with insert were identified by blue/white screening. Plasmids containing inserts were sent to GENEWIZ for sequencing using M13F and M13R primers. The resulting sequences were analyzed with Serial Cloner and sequences from NCBI and UCSC Genome Browser.
HK-2 cells were plated into 6-well Transwell plates (Corning) and grown to confluence. Media from HK-2 cell lines was spun down to remove cell debris and then frozen at −80°C until all samples were ready to use. ELISA for ET-1 was performed using the Human Endothelin-1 QuantiGlo ELISA Kit (R&D Systems) according to manufacturer instructions.
HK-2 cells were counted and 20,000 cells were plated into 6-well Transwell dishes. Cells were grown under normal conditions for 2, 4, 6, and 8 days. After the specified amount of growth time, cells were treated with Trypsin, diluted, and counted using the BioRad TC20 Automated Cell Counter.
The chromatin state of the human endothlien-1 gene (EDN1) gene was analyzed using the UCSC Genome Browser. The Genome Segments track and ChromHMM revealed a predicted promoter region with high confidence immediately downstream of the exon encoding the 3′ untranslated region (UTR) of EDN1 (about 800 bp long) in human umbilical vein endothelial cells (HUVEC) (Figure 1). This region also contained large clusters of DNase sensitivity sites, suggesting that the chromatin is in an open state and transcription factors could bind to this region (Figure 2). Using the UCSC Genome Browser transcription factor binding site database created using data from chromatin immunoprecipitation (ChIP) assays (ENCODE) (Karolchik et al., 2014), multiple transcription factors including C-JUN, CCAAT/enhancer-binding protein β (CEBPβ), and Forkhead Box A1 (FOXA1) were shown to bind at the location of the predicted promoter region. Additionally, Transcription Factor II D (TFIID), Upstream Transcription Factor 1 (USF1), and Glucocorticoid Receptor (GR) are predicted to bind to this region, and these transcription factors are associated with sites of active transcription. RNA polymerase II was also shown to bind to the predicted promoter region. Based on this information, we hypothesized that the predicted downstream promoter contained a transcription start site and potentially could be a promoter for an antisense RNA.
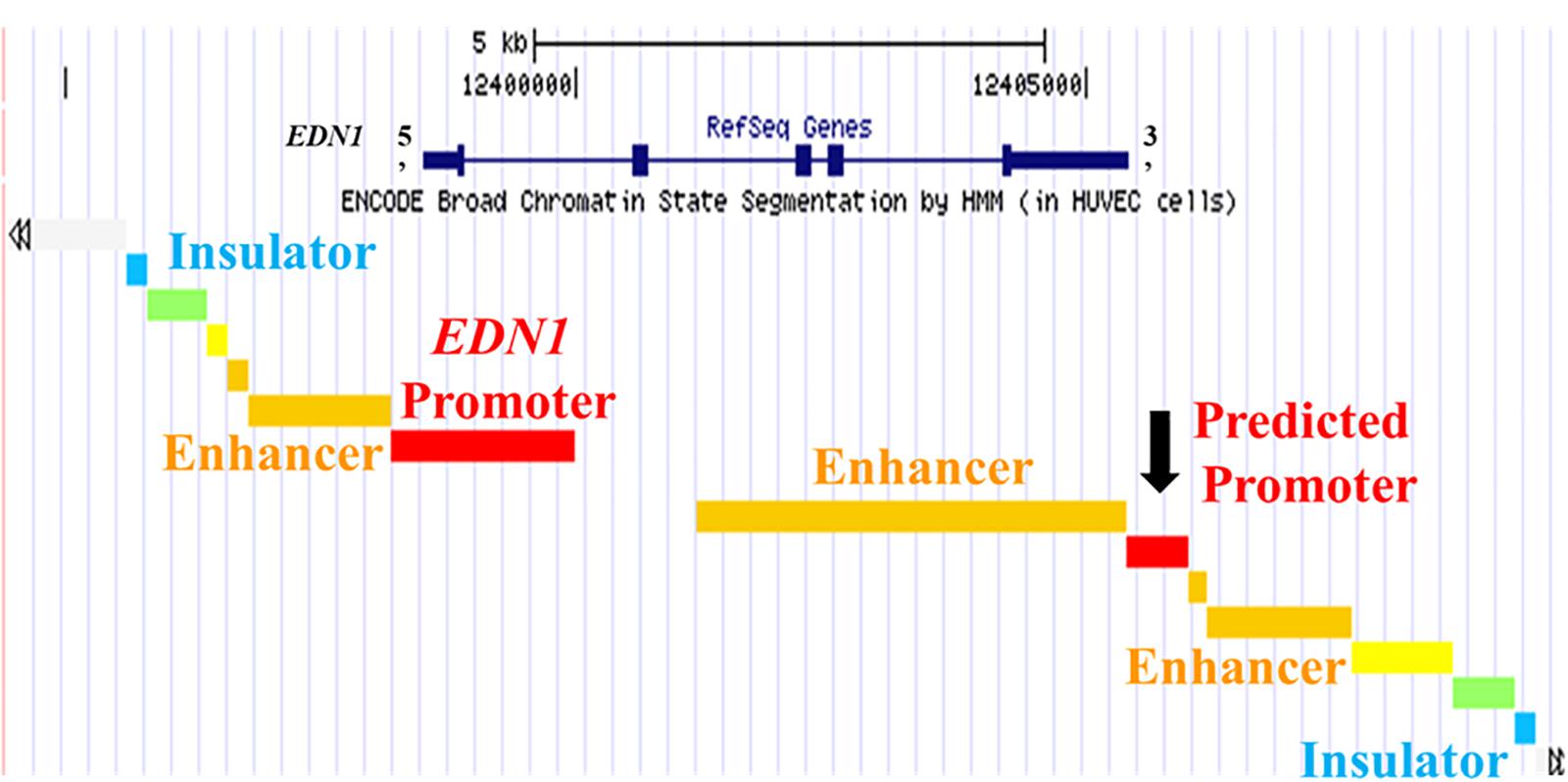
Figure 1. UCSC Genome Browser Predicts Promoter in 3′ UTR of EDN1 Gene. The University of California Santa Cruz (UCSC) Genome Browser with ENCODE broad chromatin state segmentation by HMM in human umbilical vein endothelial cells (HUVEC) cells shows a predicted promoter (black arrow) in the 3′ untranslated region (UTR) of the EDN1 gene. The EDN1 gene is shown at the top (navy blue) with boxes representing exons. The EDN1 gene position is aligned with the genome segments in the ENCODE broad chromatin state track.
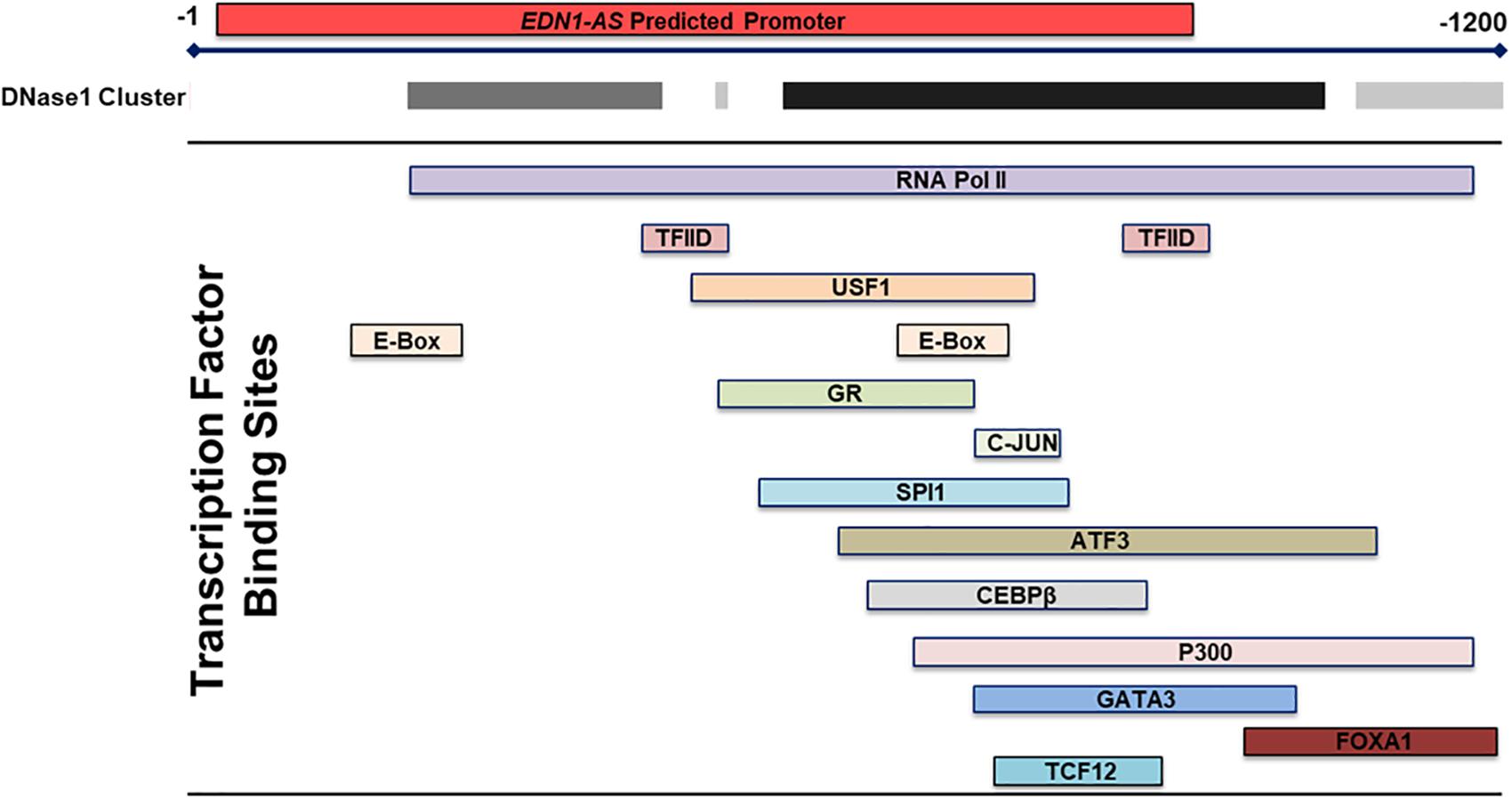
Figure 2. Transcription Factor Binding Sites within the Predicted Promoter in the 3′ UTR of EDN1. Using the transcription factor binding sites track within the UCSC Genome Browser, multiple transcription factor binding sites were identified by ChIP within the 3′ UTR EDN1 predicted promoter. Additionally, the DNase I cluster track showed multiple areas of high DNase sensitivity. The gray box indicates the extent of the hypersensitive region with darker boxes representing a stronger hypersensitivity signal. These data taken together suggests that this predicted promoter region is transcriptionally regulated and that the chromatin is in an open state.
To determine if an antisense RNA is being transcribed from the EDN1 downstream promoter, a strand-specific reverse transcriptase PCR (ssRT-PCR) strategy was designed (Figure 3). Primers for the reverse transcriptase reactions (RT) were designed to have a complementary sequence to the EDN1 sense strand, so the primers will only anneal to RNA transcribed from the antisense strand (Table 1). Multiple strand-specific primers were designed at varying distances from the predicted promoter toward the 5′ end of the EDN1 gene (SS1-6). Oligo dT primers were used as a positive control for the RT reactions. For each RT reaction, a –RT reaction was performed without the reverse transcriptase enzyme as a negative control to ensure the absence of genomic DNA contamination in our samples.
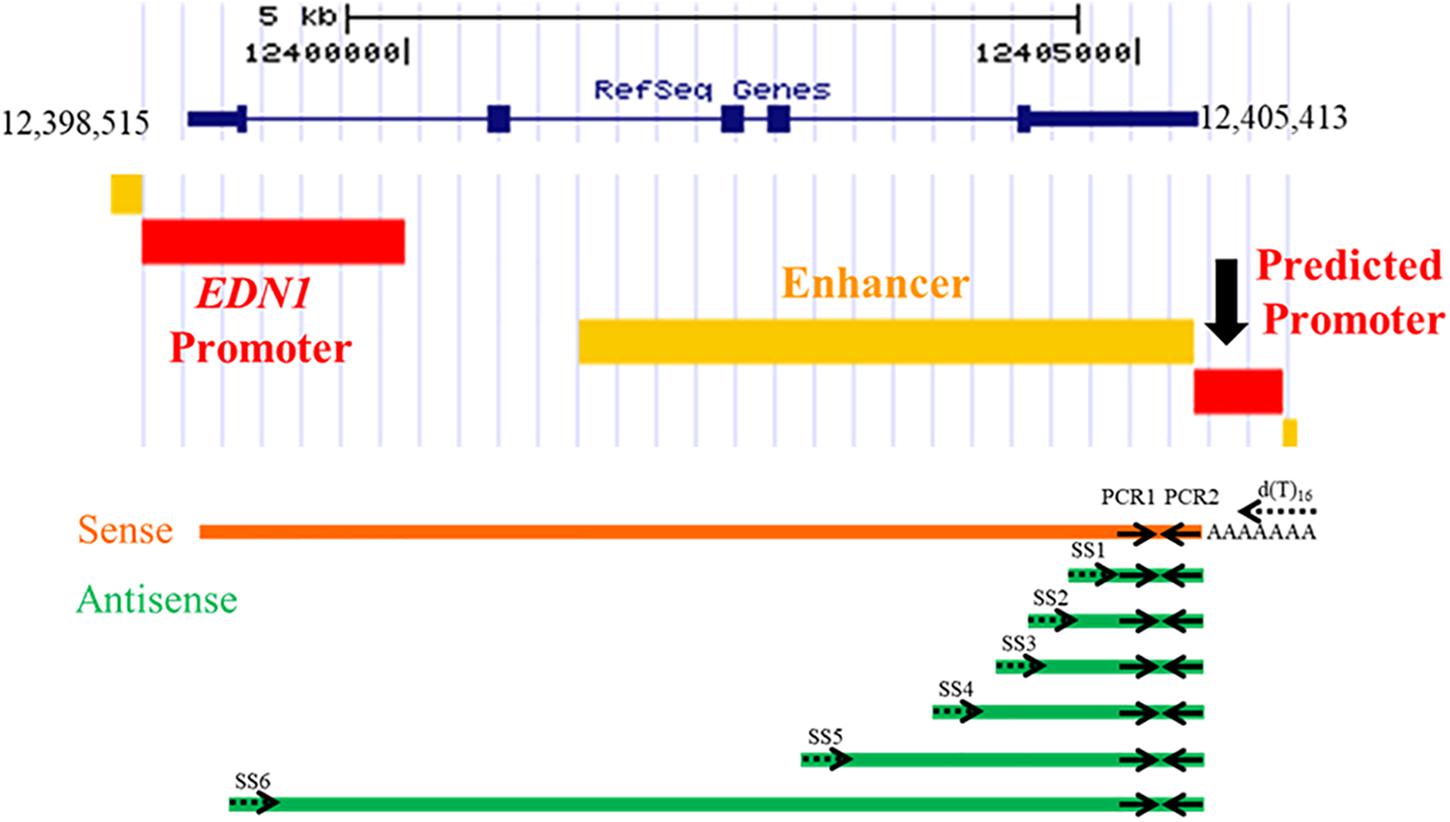
Figure 3. Strand-Specific Reverse Transcriptase PCR Reactions. Strand-specific reverse transcriptase (RT) PCR reactions were designed using primers that would only anneal to an antisense transcript to EDN1. A total of 6 different strand-specific primers were used in the RT reactions (SS1–SS6, dotted arrows), which were progressively further away from the predicted 3′ UTR promoter region. For a positive RT control, oligo dT primers were used as it is known EDN1 mRNA is poly-adenylated. After conversion of RNA into cDNA, PCR1 and PCR2 primers (solid arrows) were used in a PCR reaction to amplify any products generated. PCR1 and PCR2 amplify the same piece of cDNA regardless of the strand-specific primer used, so all bands visualized by gel electrophoresis are the same size (165 bp).
Total RNA was isolated from primary human mammary endothelial cells (HMEC), human kidney proximal tubule epithelial cells (HK-2), human bronchoendothelial cells (S9), and human embryonic kidney cells (HEK293). Additionally, human RNA from kidney tissue was obtained from Life Technologies. Each of the RNA samples was treated with DNase and then converted to cDNA using a strand-specific primer or an oligo dT primer. For every sample, both + RT and –RT reactions were performed. The resulting cDNA products were amplified in a PCR using the primers PCR1 and PCR2, which amplified the same region of cDNA regardless of the SS or oligo dT primer used. An antisense RNA transcript was detected in HMEC, HUVEC, HK-2, and S9 cell cultures (Figures 4A,B). The antisense transcript was also detected in human kidney tissue RNA samples (Figure 4C). Interestingly, this antisense RNA was not detected in HEK293 cells, but EDN1 mRNA was also not detected in these cells (Figure 4D). These results demonstrate that there is an RNA transcribed in an anti-sense direction within the EDN1 gene locus, and we have designated this transcript EDN1-AS. SS primers 4–6 (Figure 3) were designed to detect if the length of EDN1-AS spanned the entire EDN1 coding region (Figures 4B,C). The results indicate that transcription of EDN1-AS proceeds at least as far as the first exon of EDN1.
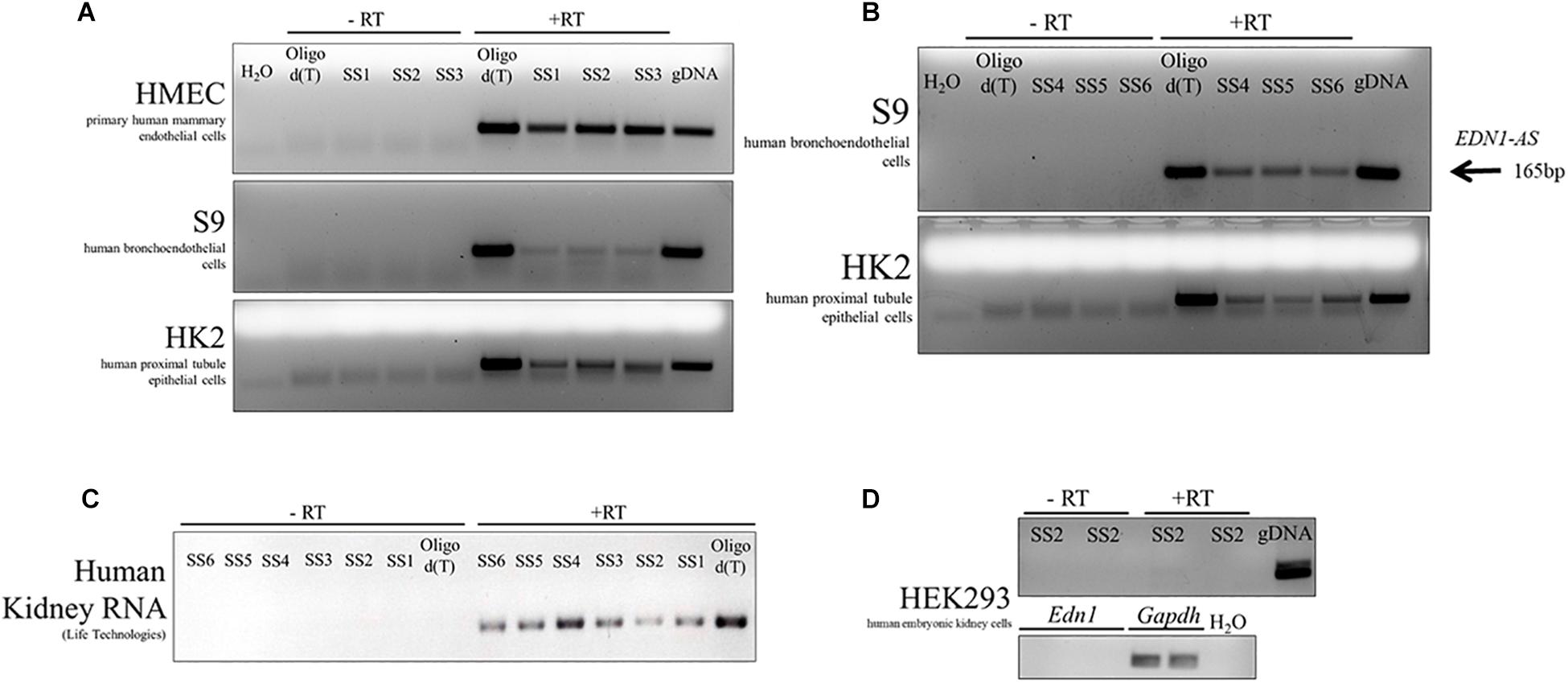
Figure 4. Identification of EDN1-AS in Human Cell Culture Models and Human Kidney RNA. Strand-specific reactions were used to test various human cell culture models for the presence of an antisense transcript originating from the predicted promoter region. This transcript has been named EDN1-AS. Strand-specific RT primers SS1-3 were used to test for an antisense transcript in primary human mammary endothelial cells (HMEC), human bronchoendothelial cells (S9), and in human proximal tubule epithelial cells (HK2) (A). Oligo dT primers were used as a positive control for the RT reactions. For each PCR reaction, water was used as a negative control and gDNA as a PCR positive control. For each of the strand-specific reactions, a no RT (-RT) control reaction was ran with the same cDNA sample. Strand-specific RT reactions using primers further away from the predicted promoter site (SS4–SS6) were designed to determine if the transcript spanned the entire length of the EDN1 gene using available samples from S9 and HK2 cells (B). All strand-specific RT primers (SS1–SS6) were used to determine if EDN1-AS could be detected in human kidney tissue RNA samples from Life Technologies (C). EDN1-AS and EDN1 transcripts could not be identified in human embryonic kidney cells (HEK293) (D). All gels are representative of at least n = 2 per cell line.
We and others have shown that the EDN1 gene is under the regulation of the circadian clock proteins (Hanai et al., 2005; Stow et al., 2012; Kozakai et al., 2014; Richards et al., 2014). Additionally, GR was shown to bind to the EDN1-AS promoter region and glucocorticoids have been shown to modulate the circadian clock (Balsalobre et al., 2000; Kino and Chrousos, 2010; Surjit et al., 2011). E-box response elements, to which circadian clock transcription factors bind, were also present in the predicted EDN1-AS promoter region. These data suggest that, similarly to the EDN1 gene, EDN1-AS may be under control of the circadian clock and may display a circadian rhythm of expression. To determine if EDN1-AS exhibits a circadian rhythm of expression, a 24-h expression profile of the antisense transcript was generated (Figure 5). The HK-2 cells were treated with dexamethasone for 30 min to synchronize their circadian clocks (Feillet et al., 2014). Twenty-four hours after the dexamethasone treatment, RNA was isolated from the HK-2 cells every 2 h for 24 h and the ssRT-PCR protocol was performed as described above. RT reactions with random hexamers were also performed on the same samples in order to detect the levels of GAPDH expression as a control. As seen in Figure 5, EDN1-AS expression varies over a 24- h period.
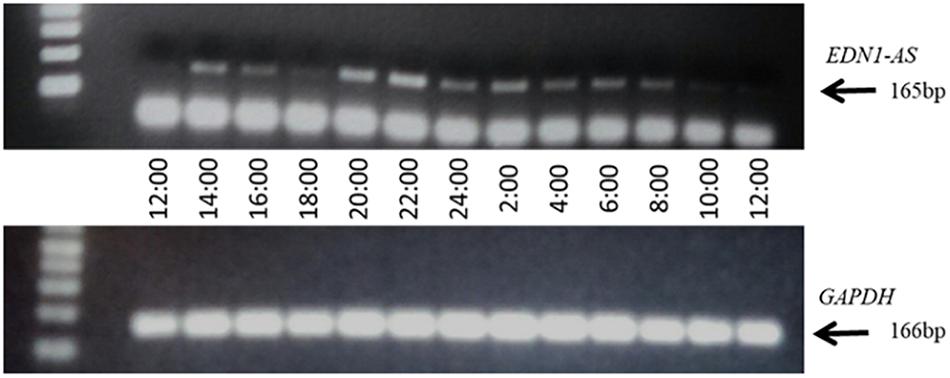
Figure 5. EDN1-AS Exhibits a Circadian Rhythm of Expression. HK2 cells were treated with 100 nM dexamethasone for 30 min, after which media was replaced and cells incubated for 24 h. After 24 h (noon), RNA was isolated every 2 h for 24 h total. This RNA was used in strand-specific RT PCR reactions with either SS2 primer (to measure EDN1-AS) or random hexamer reactions (to measure GAPDH). For the PCR reactions, PCR1 and PCR2 primers were used in EDN1-AS reactions and GAPDH Fwd and GAPDH Rev primers were used in the GAPDH reactions. Gel representative of n = 3 independent experiments.
Since the expression of EDN1-AS seems to be under circadian regulation, we wanted to investigate the effect of removing the E-box response element and GR binding site in the predicted promoter region of EDN1-AS. In order to create a KO of this regulatory region in HK-2 cells, we utilized CRISPR/Cas9 technology. Guide RNAs (gRNAs) were designed using the online tool CC Top (Stemmer et al., 2015; Table 2). gRNAs were selected based on location and predicted off-target sites. If possible, only gRNAs with no off-target sites or those with sites only in regions between genes were chosen. No gRNAs were used that had predicted off-target sites in exonic or intronic regions of any gene to minimize possible off-target effects. Various combinations of all the gRNAs were used in order to create a biallelic KO of the EDN1-AS regulatory region. The gRNA combination of gRNA 2 and 3 resulted in two HK-2 cell lines that had a homozygous deletion of an approximately 350 bp region within the predicted EDN1-AS promoter by genotyping PCR (Figure 6). Sanger sequencing using genomic DNA from these cell lines was used to confirm the KO region. In addition to the two homozygous KO cell lines, control (CNTL) cell lines that had undergone the CRISPR process, but did not have KO of the EDN1-AS regulatory region, were identified to use as control cells for future experiments.
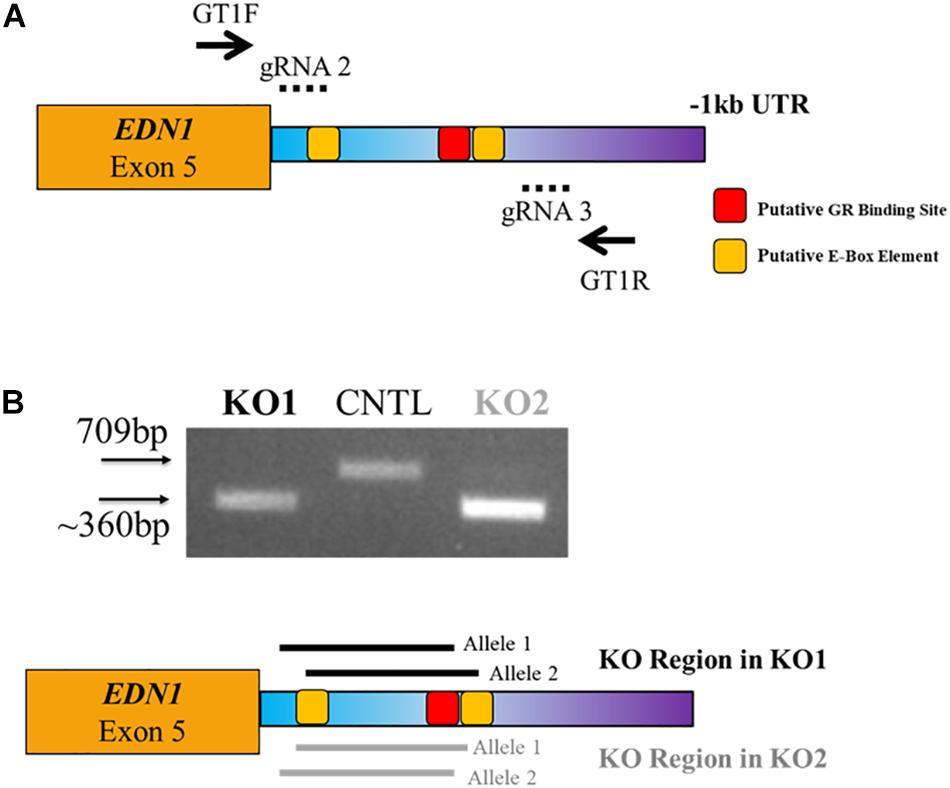
Figure 6. CRISPR/Cas9 Targeted Deletion of GR and E-Box Regulatory Elements within the EDN1-AS Predicted Promoter Region. CRISPR/Cas9 technology was utilized to create biallelic KO of a portion of the predicted Edn1AS promoter region, which contained GR and E-Box regulatory sites. Four guide RNAs (gRNAs) were designed to create deletion of this region (A). LentiCRISPR was used to deliver the gRNAs to HK2 cells. Puromycin was used to screen cells. Using genotyping primers, GT1F and GT1R (solid arrows), two clones were identified that had a KO of approximately 350 bp generated with gRNAs 2 and 3 (dotted lines) (B). Sequencing results reviled KO1 has a biallelic KO of 297 bp with 64 bp additional KO 5′ of this region on one allele and 78 bp KO 3′ of this region on the other allele (black solid lines). KO2 has a biallelic KO of 310 bp with 46 bp additional KO 3′ of this region on one allele and 71 bp KO 5′ of this region on the other allele (gray solid lines). Both KOs have a deletion of the 1st E-Box response element and the GR element. In addition, we identified control (CNTL) cells that had undergone the CRISPR process, but retained WT copes of the EDN1-AS regulatory region.
To determine how the KO of the EDN1-AS regulatory region containing the predicted GR and circadian clock binding sites affected the expression of EDN1-AS, we performed our ssRT-PCR protocol on the KO cell lines in addition to the CNTL cells. The CRISPR KO cells had higher expression of EDN1-AS compared to CNTL cells (Figure 7A). Secreted endothelin-1 peptide (ET-1) was measured to determine if the increase in expression of the EDN1-AS gene led to an increase in ET-1 peptide level. Briefly, KO and CNTL cells were plated into Transwell plates and media was collected from confluent cells. The media was then analyzed via an ET-1 ELISA to measure total ET-1 peptide levels. ET-1 peptide levels were normalized to total protein concentration measured by BCA assay. The CRISPR KO cells had a significant increase in total secreted ET-1 peptide levels compared to the CNTL cells (Figure 7B).
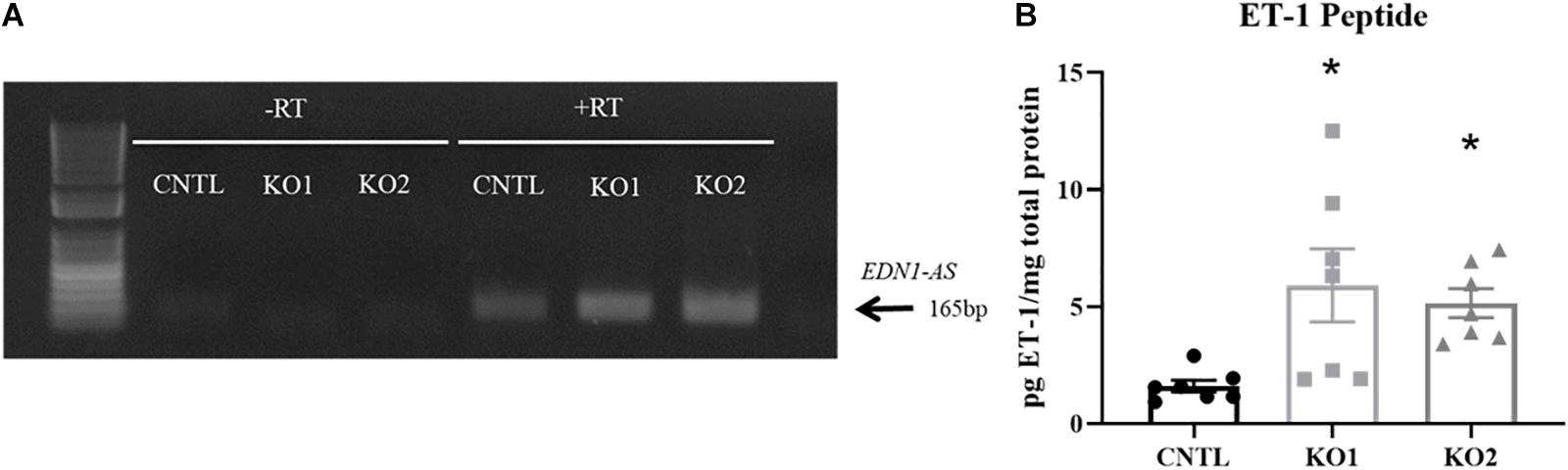
Figure 7. CRISPR/Cas9 KO of EDN1-AS Regulatory Elements Results in Increased EDN1-AS and EDN1 Expression. EDN1-AS expression levels were examined in CNTL and CRISPR/Cas9 KO cells by strand-specific RT PCR. For each reaction, 20 ng of cDNA was used in PCR reactions to amplify EDN1-AS. CRISPR/Cas9 KO cells had more EDN1-AS expression visualized by gel electrophoresis (A) (gel representative of n = 3 for CNTL and n = 2 for each KO cell line). To determine if these differences in EDN1-AS expression had an effect on EDN1 expression, the product of the EDN1 gene, endothelin-1 (ET-1) peptide was measured. Cells were placed in Transwells dishes and grown to confluence. Media from these cells was used in an endothelin-1 ELISA (R&D QuantiGlo ET-1 ELISA) to determine the amount of secreted ET-1 relativized to total protein concentration (B). One-way ANOVA was used to determine significant differences between each KO and the CNTL cell line, n = 7 for each cell line. *P < 0.05.
High expression of ET-1 in human proximal tubule cells, like HK-2 cells, is associated with increased cell proliferation (Zoja et al., 1995; Seccia et al., 2016). To determine if the increased ET-1 levels caused by KO of the EDN1-AS regulatory region affected cell proliferation, growth rates of the KO and CNTL cells were measured. Briefly, 20,000 cells were plated into 6-well dishes and counted after 2, 4, 6, and 8 days of growth. Both CRISPR KO cell lines grew significantly faster than the CNTL cells (Figure 8). Significant differences between individual KO cells and the CNTL cells total cell count were detected on day 6 and day 8.
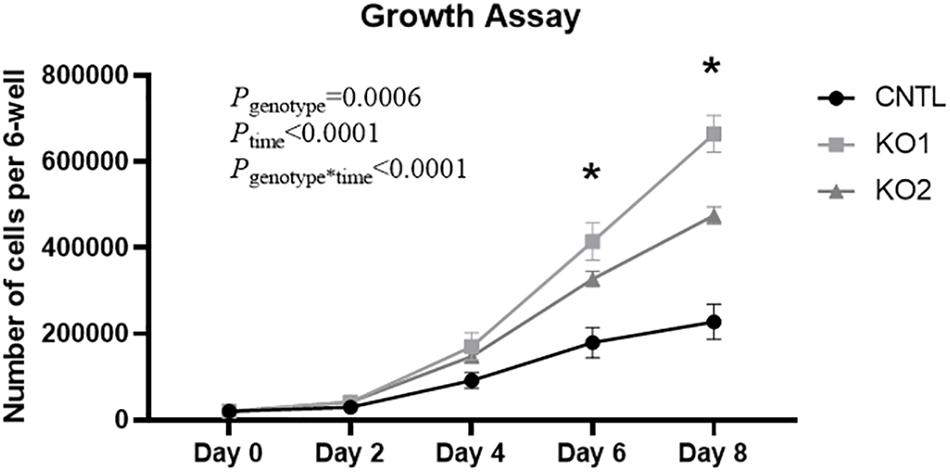
Figure 8. KO of EDN1-AS Regulatory Elements Results in Increased Growth Rate of HK2 Proximal Tubule Cells. Because high expression of ET-1 is known to lead to cell proliferation in kidney proximal tubule cells, growth rate of CNTL and CRISPR/Cas9 KO cells was measured. 20,000 cells were plated into a 6-well dish and allowed to grow under normal conditions for 2, 4, 6, or 8 days. An automated cell counter was used to determine the total amount of cells in each well. Two-way ANOVA with repeated measures was used to determine genotype differences. ∗P < 0.05 by T-test within 1 day denotes that both KO cell lines are significantly different from the CNTL cell line, n = 4.
Here we report the identification of a novel lncRNA, EDN1-AS, that is antisense to the ET-1 coding sequence. EDN1-AS is expressed in human mammary epithelial cells, human bronchoendothelial cells, human kidney proximal tubule cells, and in human kidney. Using a strand-specific RT-PCR approach, we demonstrated that EDN1-AS comprises the entire length of the EDN1 sense coding sequence. EDN1-AS appears to exhibit rhythmic expression in HK-2 cells. CRISPR-mediated deletion of regulatory sequence upstream of EDN1-AS containing a GR element and an E-box resulted in increased expression of EDN1-AS in HK-2 cells. This effect was associated with increased ET-1 peptide secretion from the cells and increased growth rates.
CRISPR-mediated knockout of the region containing the predicted GR element and E-box resulted in increased expression of EDN1-AS, suggesting that the deleted regulatory region contains a repressive element. Given that GR elements can be bound by both GR and MR and these transcription factors can mediate positive and negative effects on gene regulation (Koning et al., 2019), this result is not surprising. Deletion of a GR element in the promoter of αENaC revealed a role for repressive regulation (Lin et al., 1999). Likewise, circadian clock proteins such as PER1 interact with E-box elements to mediate positive and negative effects on gene expression (Cox and Takahashi, 2019). Promoter analysis of the gonadotropin-releasing hormone demonstrated a key role for the E-box elements and a repressive role for PER1 (Resuehr et al., 2007). The concept that PER1 may mediate negative regulation of EDN1-AS is consistent with our previous observations that PER1 in the kidney mediates repression of the sense EDN1 mRNA (Stow et al., 2012; Richards et al., 2014).
The non-coding RNA field has exploded in recent years due to the wealth of information provided by ENCODE. It is now estimated that more than 50% of the human genome is transcribed but only 1.2% of the genome encodes proteins (Ransohoff et al., 2018). Non-coding RNAs thus comprise a significant portion of the transcriptome, but our understanding of these RNA species is in its infancy. Some lncRNAs are natural antisense transcripts (NAT) because they arise by transcription of the strand opposite an mRNA (Khorkova et al., 2014). Our data suggest that EDN1-AS is a NAT with functional effects on ET-1 action. We estimate that the length of unprocessed EDN1-AS is at least 5 kb, based on the signal detected in HK2 and S9 cells and human kidney using SS6, which is located in the first exon of the sense EDN1 sequence (Figure 3). The ssRT-PCR results in Figure 4 demonstrate that EDN1-AS transcripts spanning the length of the EDN1 gene are present in human kidney and bronchoendothelial cells. Whether or not EDN1-AS is subject to splicing or other post-transcriptional regulation remains to be determined.
Our report is the first description of an antisense lncRNA associated with the EDN1 locus. According to LNCipedia, four lncRNAs have been identified near the EDN1 gene (Volders et al., 2019). However, all four of these are associated with the sense strand. Functional relevance of these lncRNAs has not been described. Interestingly, a SNP in PHACTR, a gene ∼400,000 bp downstream of EDN1, has been associated with regulation of ET-1 levels (Gupta et al., 2017). This region appears to interact with an area of open chromatin located in a “gene desert” between the EDN1 and PHACTR loci. These results, together with our current report, strongly suggest a new level of complexity in the regulation of EDN1.
A limitation of the current study is that the mechanism by which EDN1-AS affects ET-1 expression remains unclear. Many antisense RNAs act in an inhibitory manner, however, as the knowledge base regarding non-coding RNA increases, it is becoming increasingly clear that NATs act through myriad mechanisms (Wanowska et al., 2018). One example of a NAT that exerts positive effects on target mRNA expression is PTENP1, an antisense transcribed from the locus of the tumor suppressor gene PTEN. PTENP1 acts as a miRNA sponge for miR21, thus protecting the PTEN transcript from miRNA-mediated degradation (Wang et al., 2016). Another example of positive regulation by a NAT is BACE1-AS, which is transcribed from the β-amyloid cleaving enzyme 1 (BACE1) locus. BACE1-AS interacts with the BACE1 sense mRNA, masking a miRNA binding site and thus protecting BACE1 from miRNA-mediated degradation (Faghihi et al., 2010). BACE1-AS shows promise as a potential therapeutic in Alzheimer’s disease (Ge et al., 2020). Given that ET-1 is subject to regulation by miRNA (Jacobs et al., 2013), it is tempting to speculate that EDN1-AS may function as a positive regulator of EDN1 expression through a miRNA-protection mechanism. Future experiments are needed to test this hypothesis.
The lncRNAs are a newly recognized mechanism for gene regulation and are being explored as potential therapeutic targets in a variety of diseases (Li et al., 2018; Pecero et al., 2019; Wang et al., 2020), including diabetic nephropathy (Ge et al., 2019; Yang et al., 2019). A nucleic acid-based approach could allow greater specificity than pharmacological compounds, reducing side effects and allowing tissue-specific delivery (Sharma and Watts, 2015; Ku et al., 2016). Future studies are needed to determine whether or not manipulation of EDN1-AS could have therapeutic benefits. Although expression of EDN1-AS was confirmed in a variety of cell types and even in human kidney in the present study, the functional studies related to EDN1-AS are limited to HK-2 cells. In HK-2 cells, increased expression of EDN1-AS was associated with increased cell proliferation, which may have implications for manipulating EDN1-AS levels in a CKD setting.
The datasets generated for this study are available on request to the corresponding author.
LD, KS, LJ, CW, BC, KB, and MG conceived and designed research. LD, KS, SM, DB, SB, LJ, KA, RP, and KB performed experiments. LD, KS, SM, DB, and MG analyzed data. LD and MG interpreted results of experiments, prepared figures, and drafted manuscript. LD, SM, DB, SB, LJ, KB, BC, and MG edited and revised manuscript. LD, KS, SM, DB, SB, LJ, KA, RP, CW, KB, BC, and MG approved final version of manuscript.
This work was supported by the National Institutes of Health (NIH) R21AG052861 and 1R01DK109570-01A1 (MG), American Heart Association Grant-in-Aid (16GRNT31220009) (MG), University of Florida Department of Medicine Gatorade Trust (MG), American Heart Association Postdoctoral Fellowship Grants 18POST34030210 (LD), and NIH Grant T32-DK-104721 awarded to the University of Florida (UF) Division of Nephrology (LD).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Balsalobre, A., Brown, S. A., Marcacci, L., Tronche, F., Kellendonk, C., Reichardt, H. M., et al. (2000). Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289, 2344–2347. doi: 10.1126/science.289.5488.2344
Cox, K. H., and Takahashi, J. S. (2019). Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 63, R93–R102. doi: 10.1530/JME-19-0153
Dhaun, N., Lilitkarntakul, P., MacIntyre, I. M., Muilwijk, E., Johnston, N. R., Kluth, D. C., et al. (2009). Urinary endothelin-1 in chronic kidney disease and as a marker of disease activity in lupus nephritis. Am. J. Physiol. Ren. Physiol. 296, F1477–F14783. doi: 10.1152/ajprenal.90713.2008
Dhaun, N., and Webb, D. J. (2019). Endothelins in cardiovascular biology and therapeutics. Nat. Rev. Cardiol. 16, 491–502. doi: 10.1038/s41569-019-0176-173
Dükel, M., Streitfeld, W. S., Tang, T. C. C., Backman, L. R. F., Ai, L., May, W. S., et al. (2016). The breast cancer tumor suppressor TRIM29 is expressed via ATM-dependent signaling in response to hypoxia. J. Biol. Chem. 291, 21541–21552. doi: 10.1074/jbc.M116.730960
Faghihi, M. A., Zhang, M., Huang, J., Modarresi, F., Van der Brug, M. P., Nalls, M. A., et al. (2010). Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11:R56. doi: 10.1186/gb-2010-11-5-r56
Feillet, C., Krusche, P., Tamanini, F., Janssens, R. C., Downey, M. J., Martin, P., et al. (2014). Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc. Natl. Acad. Sci. U.S.A. 111, 9828–9833. doi: 10.1073/pnas.1320474111
Ge, X., Xu, B., Xu, W., Xia, L., Xu, Z., Shen, L., et al. (2019). Long noncoding RNA GAS5 inhibits cell proliferation and fibrosis in diabetic nephropathy by sponging miR-221 and modulating SIRT1 expression. Aging 11, 8745–8759. doi: 10.18632/aging.102249
Ge, Y., Song, X., Liu, J., Liu, C., and Xu, C. (2020). The combined therapy of berberine treatment with lncRNA BACE1-as depletion attenuates Aβ25-35 induced neuronal injury through regulating the expression of miR-132-3p in neuronal cells. Neurochem. Res. doi: 10.1007/s11064-019-02947-6 [Epub ahead of print].
Grenda, R., Wuhl, E., Litwin, M., Janas, R., Sladowska, J., Arbeiter, K., et al. (2007). Urinary excretion of endothelin-1 (ET-1), transforming growth factor- 1 (TGF- 1) and vascular endothelial growth factor (VEGF165) in paediatric chronic kidney diseases: results of the ESCAPE trial. Nephrol. Dial. Transplant. 22, 3487–3494. doi: 10.1093/ndt/gfm300
Gupta, R. M., Hadaya, J., Trehan, A., Zekavat, S. M., Roselli, C., Klarin, D., et al. (2017). A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell 170, 522–533.e15. doi: 10.1016/j.cell.2017.06.049
Hanai, S., Masuo, Y., Shirai, H., Oishi, K., Saida, K., and Ishida, N. (2005). Differential circadian expression of endothelin-1 mRNA in the rat suprachiasmatic nucleus and peripheral tissues. Neurosci. Lett. 377, 65–68. doi: 10.1016/j.neulet.2004.11.082
Heerspink, H. J. L., Parving, H. H., Andress, D. L., Bakris, G., Correa-Rotter, R., Hou, F. F., et al. (2019). Atrasentan and renal events in patients with type 2 diabetes and chronic kidney disease (SONAR): a double-blind, randomised, placebo-controlled trial. Lancet 393, 1937–1947. doi: 10.1016/S0140-6736(19)30772-X
Jacobs, M. E., Jeffers, L. A., Welch, A. K., Wingo, C. S., and Cain, B. D. (2014). MicroRNA regulation of endothelin-1 mRNA in renal collecting duct cells. Life Sci. 118, 195–199. doi: 10.1016/j.lfs.2014.03.003
Jacobs, M. E., Wingo, C. S., and Cain, B. D. (2013). An emerging role for microRNA in the regulation of endothelin-1. Front. Physiol. 4:22. doi: 10.3389/fphys.2013.00022
Karolchik, D., Barber, G. P., Casper, J., Clawson, H., Cline, M. S., Diekhans, M., et al. (2014). The UCSC genome browser database: 2014 update. Nucleic Acids Res. 42, D764–D770. doi: 10.1093/nar/gkt1168
Khorkova, O., Myers, A. J., Hsiao, J., and Wahlestedt, C. (2014). Natural antisense transcripts. Hum. Mol. Genet. 23, R54–R63. doi: 10.1093/hmg/ddu207
Kino, T., and Chrousos, G. P. (2010). Circadian CLOCK-mediated regulation of target-tissue sensitivity to glucocorticoids: implications for cardiometabolic diseases. Endocr. Dev. 20, 116–126. doi: 10.1159/000321232
Kohan, D. E., and Barton, M. (2014). Endothelin and endothelin antagonists in chronic kidney disease. Kidney Int. 86, 896–904. doi: 10.1038/ki.2014.143
Kohan, D. E., and Pollock, D. M. (2013). Endothelin antagonists for diabetic and non-diabetic chronic kidney disease. Br. J. Clin. Pharmacol. 76, 573–579. doi: 10.1111/bcp.12064
Kohan, D. E., Rossi, N. F., Inscho, E. W., and Pollock, D. M. (2011). Regulation of blood pressure and salt homeostasis by endothelin. Physiol. Rev. 91, 1–77. doi: 10.1152/physrev.00060.2009
Koning, A.-S. C. A. M., Buurstede, J. C., van Weert, L. T. C. M., and Meijer, O. C. (2019). Glucocorticoid and mineralocorticoid receptors in the brain: a transcriptional perspective. J. Endocr. Soc. 3, 1917–1930. doi: 10.1210/js.2019-2158
Kozakai, T., Sakate, M., Takizawa, S., Uchide, T., Kobayashi, H., Oishi, K., et al. (2014). Effect of feeding behavior on circadian regulation of endothelin expression in mouse colon. Life Sci. 118, 232–237. doi: 10.1016/j.lfs.2014.06.022
Ku, S. H., Jo, S. D., Lee, Y. K., Kim, K., and Kim, S. H. (2016). Chemical and structural modifications of RNAi therapeutics. Adv. Drug Deliv. Rev. 104, 16–28. doi: 10.1016/j.addr.2015.10.015
Li, Y., Liang, Y., Zhu, Y., Zhang, Y., and Bei, Y. (2018). Noncoding RNAs in cardiac hypertrophy. J. Cardiovasc. Transl. Res. 11, 439–449. doi: 10.1007/s12265-018-9797-x
Lin, H. H., Zentner, M. D., Ho, H. L., Kim, K. J., and Ann, D. K. (1999). The gene expression of the amiloride-sensitive epithelial sodium channel alpha-subunit is regulated by antagonistic effects between glucocorticoid hormone and ras pathways in salivary epithelial cells. J. Biol. Chem. 274, 21544–21554. doi: 10.1074/jbc.274.31.21544
Pecero, M. L., Salvador-Bofill, J., and Molina-Pinelo, S. (2019). Long non-coding RNAs as monitoring tools and therapeutic targets in breast cancer. Cell. Oncol. 42, 1–12. doi: 10.1007/s13402-018-0412-416
Pulido, T., Adzerikho, I., Channick, R. N., Delcroix, M., Galiè, N., Ghofrani, H.-A., et al. (2013). Macitentan and morbidity and mortality in pulmonary arterial hypertension. N. Engl. J. Med. 369, 809–818. doi: 10.1056/NEJMoa1213917
Ransohoff, J. D., Wei, Y., and Khavari, P. A. (2018). The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143–157. doi: 10.1038/nrm.2017.104
Reichetzeder, C., Tsuprykov, O., and Hocher, B. (2014). Endothelin receptor antagonists in clinical research — Lessons learned from preclinical and clinical kidney studies. Life Sci. 118, 141–148. doi: 10.1016/j.lfs.2014.02.025
Resuehr, D., Wildemann, U., Sikes, H., and Olcese, J. (2007). E-box regulation of gonadotropin-releasing hormone (GnRH) receptor expression in immortalized gonadotrope cells. Mol. Cell. Endocrinol. 278, 36–43. doi: 10.1016/j.mce.2007.08.008
Richards, J., Welch, A. K., Barilovits, S. J., All, S., Cheng, K. Y., Wingo, C. S., et al. (2014). Tissue-specific and time-dependent regulation of the endothelin axis by the circadian clock protein Per1. Life Sci. 118, 255–262. doi: 10.1016/j.lfs.2014.03.028
Seccia, T. M., Caroccia, B., Gioco, F., Piazza, M., Buccella, V., Guidolin, D., et al. (2016). Endothelin-1 drives epithelial-mesenchymal transition in hypertensive nephroangiosclerosis. J. Am. Heart Assoc. 5:721. doi: 10.1161/JAHA.116.003888
Sharma, V. K., and Watts, J. K. (2015). Oligonucleotide therapeutics: chemistry, delivery and clinical progress. Future Med. Chem. 7, 2221–2242. doi: 10.4155/fmc.15.144
Speed, J. S., and Pollock, D. M. (2013). Endothelin, kidney disease, and hypertension. Hypertens 61, 1142–1145. doi: 10.1161/HYPERTENSIONAHA.113.00595
Stemmer, M., Thumberger, T., Del Sol Keyer, M., Wittbrodt, J., and Mateo, J. L. (2015). CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10:e0124633. doi: 10.1371/journal.pone.0124633
Stow, L. R., Jacobs, M. E., Wingo, C. S., and Cain, B. D. (2011). Endothelin-1 gene regulation. FASEB J. 25, 16–28. doi: 10.1096/fj.10-161612
Stow, L. R., Richards, J., Cheng, K. Y., Lynch, I. J., Jeffers, L. A., Greenlee, M. M., et al. (2012). The circadian protein period 1 contributes to blood pressure control and coordinately regulates renal sodium transport genes. Hypertension 59, 1151–1156. doi: 10.1161/HYPERTENSIONAHA.112.190892
Surjit, M., Ganti, K. P., Mukherji, A., Ye, T., Hua, G., Metzger, D., et al. (2011). Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145, 224–241. doi: 10.1016/j.cell.2011.03.027
Theuring, F., Thöne-Reinecke, C., Vogler, H., Schmager, F., Rohmeiss, P., Slowinski, T., et al. (1998). Pathophysiology in endothelin-1 transgenic mice. J. Cardiovasc. Pharmacol. 31(Suppl. 1), S489–S491. doi: 10.1097/00005344-199800001-199800140
Volders, P.-J., Anckaert, J., Verheggen, K., Nuytens, J., Martens, L., Mestdagh, P., et al. (2019). LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 47, D135–D139. doi: 10.1093/nar/gky1031
Wang, L., Zhang, N., Wang, Z., Ai, D.-M., Cao, Z.-Y., and Pan, H.-P. (2016). Pseudogene PTENP1 functions as a competing endogenous RNA (ceRNA) to regulate PTEN expression by sponging miR-499-5p. Biochemistry 81, 739–747. doi: 10.1134/S0006297916070105
Wang, Y., Wu, S., Zhu, X., Zhang, L., Deng, J., Li, F., et al. (2020). LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J. Exp. Med. 217, pii: e20190950. doi: 10.1084/jem.20190950
Wanowska, E., Kubiak, M. R., Rosikiewicz, W., Makałowska, I., and Szcześniak, M. W. (2018). Natural antisense transcripts in diseases: from modes of action to targeted therapies. Wiley Interdiscip. Rev. RNA 9:1461. doi: 10.1002/wrna.1461
Yang, J., Shen, Y., Yang, X., Long, Y., Chen, S., Lin, X., et al. (2019). Silencing of long noncoding RNA XIST protects against renal interstitial fibrosis in diabetic nephropathy via microRNA-93-5p-mediated inhibition of CDKN1A. Am. J. Physiol. Renal Physiol. 317, F1350–F1358. doi: 10.1152/ajprenal.00254.2019
Yuan, W., Li, Y., Wang, J., Li, J., Gou, S., and Fu, P. (2015). Endothelin-receptor antagonists for diabetic nephropathy: a meta-analysis. Nephrology 20, 459–466. doi: 10.1111/nep.12442
Keywords: kidney, long non-coding RNA, proximal tubule cells, CRISPR, circadian rhythm
Citation: Douma LG, Solocinski K, Masten SH, Barral DH, Barilovits SJ, Jeffers LA, Alder KD, Patel R, Wingo CS, Brown KD, Cain BD and Gumz ML (2020) EDN1-AS, A Novel Long Non-coding RNA Regulating Endothelin-1 in Human Proximal Tubule Cells. Front. Physiol. 11:209. doi: 10.3389/fphys.2020.00209
Received: 20 December 2019; Accepted: 24 February 2020;
Published: 13 March 2020.
Edited by:
John D. Imig, Medical College of Wisconsin, United StatesReviewed by:
Noriaki Emoto, Kobe Pharmaceutical University, JapanCopyright © 2020 Douma, Solocinski, Masten, Barral, Barilovits, Jeffers, Alder, Patel, Wingo, Brown, Cain and Gumz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michelle L. Gumz, TWljaGVsbGUuR3VtekBtZWRpY2luZS51ZmwuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.