- 1Department of Microbiology, Punjab Agricultural University, Ludhiana, India
- 2Regional Research Station, Punjab Agricultural University, Faridkot, India
- 3Department of Entomology, Punjab Agricultural University, Ludhiana, India
- 4Department of Vegetable Sciences, Punjab Agricultural University, Ludhiana, India
Whitefly Bemisia tabaci is a global invasive pest that causes substantial losses to agricultural crops worldwide either by direct feeding or vectoring numerous plant viruses. Management with insecticides remains a big challenge due to its rapid resistance development potential as well as the impact of these chemicals on non-target organisms. Thus, in search of alternate and novel pest management strategies RNA interference (RNAi) has come up as potential future tool in this direction. The present study targets nine potential genes (Aquaporin (AQP), Calcitonin (CAL), CyclophilinB (CYCP), Knottin-1 (k-1), Heat shock proteins (Hsp20, Hsp40 and Hsp70), SWItch/Sucrose Non-fermentable (SNF7) and inhibitor of apoptosis (IAP) of whitefly that have been implicated to play a role in various vital physiological functions and virus transmission. The RNAi mediated knockdown efficiency of these genes has been improved through the conjugation of respective target gene dsRNA with CQD (carbon quantum dots) nanoparticles or simultaneous knockdown of dsRNA specific gut nucleases. The studies revealed that feeding of dsRNA (40 μg/ml sucrose diet) of the target gene(s) either conjugated with CQD or along with dsRNA against dsRNase (dsdsRNase) (40 μg/ml sucrose diet) enhanced the RNAi efficiency by 24–89% compared to whiteflies fed with naked dsRNA of the same target gene. The studies provide insights about the functional role of various genes in whitefly, which can possibly be exploited for the management of this pest in the future.
Introduction
Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) is a cosmopolitan pest capable of feeding on hundreds of plant species and transmits several major plant viruses (Brown, 2000; Navas-Castillo et al., 2011; Rosen et al., 2015). B. tabaci has gained the status of key pest of economic importance due to wider host range, dispersal behavior, fecundity, competency in virus transmission, and insecticide resistance (Ahmad et al., 2002; Boykin et al., 2007). During 2015–2016, the massive infestation of whitefly in north India resulted in huge loss to cotton crop. There was complete failure of all the management strategies including the mainstay chemical insecticides to control this pest due to resistance development or substandard pesticides. Biotechnological tools in recent times have been successful in providing pest management solutions such transgenic Bt-cotton expressing Cry proteins against bollworms. Such biotechnological tool RNA interference (RNAi) has emerged as a revolutionary technology for exploring novel targets in insects from that can be potentially exploited for their management. The major advantage of this technology over present transgenic is that the chances of insects developing resistance to dsRNA are very rare. RNAi is an antiviral mechanism that leads to activation of defense response, which recognize the corresponding homologous, double-stranded RNA (dsRNA) and targets it for enzymatic degradation (Mello and Conte, 2004). The mechanism involves the introduction of exogenous or experimental dsRNA in the host cell (Winston et al., 2002) followed by the cleavage of long dsRNA into 21–25 bp siRNA (Bernstein et al., 2001) by Dicer enzyme which is RNaseIII type nuclease. The duplex complex of siRNA is recruited by a multi-protein complex called RNA-induced silencing complex (RISC) where passenger strand tagged for degradation and guide strand of siRNA along with argonaute proteins bind to complementary mRNA in a sequence specific manner and leads to its degradation (Scott et al., 2013). This results in the modulation of target protein by depletion of target mRNA either by transcriptional or at a post-transcriptional level. Thus, RNAi has the potential to identify novel genes whose knockdown is detrimental to the target insect. RNAi has been well established in many insect species such as pea aphid (Acyrthosiphon pisum) (Jaubert-Possamai et al., 2007), leafhopper (Nilaparvata lugens) (Zha et al., 2011), flour beetle (Tribolium castaneum) (Whyard et al., 2009), kissing bug (Rhodnius prolixus) (Taracena et al., 2015), African malaria mosquito (Anopheles gambiae) (X Zhang et al., 2010), Cotton leafhopper (Amrasca biguttula biguttula) (Singh et al., 2018), cotton mealybug (Phenacoccus solenopsis) (Singh et al., 2019), Fruitfly (Drosophila melanogaster) (Schwedes and Carney, 2012). Recently the corn plants expressing dsRNA against reproductive genes of Diabrotica virgifera virgifera has demonstrated the potential of this technology through RNAi based transgenic plants (Niu et al., 2017). Alternatively, dsRNA-based sprays under curtailed conditions have also been proven to be effective against different insects (Palli, 2014; Cagliari et al., 2019).
RNAi has been well established in whitefly either through injection (Ghanim et al., 2007; Luan et al., 2011), oral route (Vyas et al., 2017) or by expressing their homologous double stranded RNAs in plants (Malik et al., 2016). Delivery of siRNA/dsRNA against genes like, α-tubulin, V-ATPase, Cyp315a1, EcR, E75, Cyp18a, jhe, P450 CYP6CM1,Hsp70, RPL9, Glutathione S-transferase GST gene and ADP/ATP translocase has been studied in case of B. tabaci (Luan et al., 2013; Upadhyay et al., 2013; Li J. et al., 2015). The dsRNA expressed in entomopathogenic fungi, Isaria fumosorosea has also been reported to induce silencing of target genes in the insect host (Chen et al., 2015). In all these studies the extent of knockdown achieved across targeted genes ranged between 60 and 90% compared to controls. This is because the success and efficacy of RNAi depends upon several factors which include presence of core RNAi machinery, dose of dsRNA, the mode of RNAi delivery (Roignant et al., 2003), and the genes being targeted (Kitzmann et al., 2013). RNAi response is highly variable among insect species with lepidopteran being toward the lowest side and coleopterans showing highest efficiency (Shukla et al., 2016; Singh et al., 2017). The variability may be due to several factors, but majorly an impaired or slow cellular uptake of dsRNA in the gut and degradation of dsRNA in the insect body lead to deficient RNAi response. In hemipteran insects the dsRNA specific gut nucleases play a key role in the degradation of ingested dsRNA before it reaches the target cell (Arimatsu et al., 2007; Luo et al., 2017).
In order to develop an efficient RNAi-based method to control whiteflies it is important to protect the dsRNA from proteolytic degradation in the insect gut. Conjugation of target gene dsRNA with nanoparticles can protect the dsRNA against nucleolytic degradation and prolong the stability of dsRNA in the gut long enough to allow enough cellular uptake by the midgut cells (Xiang et al., 2012; Li M. et al., 2015; Dhandapani et al., 2019). Various studies have evidenced the potential usage of nanoparticles to improve RNAi in insects through feeding which are otherwise refractory to RNAi. Targeting dsRNA incorporated with guanylated nanoparticles against chitinase synthase B resulted in improved efficiency of RNAi in Spodoptera exigua (Christiaens et al., 2018) and S. frugiperda (Parsons et al., 2018) chitosan coated dsRNA resulted in 40% downregulation of axon guidance gene semaphorin-1a in Aedes aegypti (Zhang et al., 2015). The other alternative to nanoparticles is the simultaneous or consecutive silencing of dsRNases specific genes along with the target gene to restrict synthesis of dsRNses in the gut, which will in turn help to improve the RNAi efficiency (Spit et al., 2017; Guan et al., 2018). Thus, the current studies explore both the strategies i.e., conjugation of dsRNA with CQD nanoparticles and knockdown of dsRNA specific gut nucleases (dsRNases) for enhancement of RNAi of genes associated with various physiological functions and validated the same through various assays.
Materials and Methods
Whitefly Culture Maintenance and Collection
Whitefly haplotype Asia-II-1 was reared on cotton (Gossypium hirsutum) plants and maintained in an insect-proof glass house at 26°C with a relative humidity of 60 ± 70% and 14:10 h light and dark period.
Extraction of Total RNA and cDNA Synthesis
Total RNA from whitefly was extracted using Trizol reagent (Sigma Aldrich) as per manufacturer’s protocol. For this 15–20 whiteflies were homogenized in 1 ml of trizol reagent. The DNA traces were removed from the total RNA by treating it with 1U of DNase I (Fermentas, ThermoFisher Scientific) for 30 min at 37°C followed by enzyme inactivation using EDTA and incubation at 70°C for 10 min. A total of 500 ng RNA was used for cDNA synthesis with RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific) as per manufacturer’s instructions. The cDNA was stored at –20°C until further use.
In vitro dsRNA Synthesis
The target gene sequences were pulled out from whitefly genome database1. Selection of genes was done on the basis of their functional role i.e., osmoregulation (Aquaporin (AQP) (Benoit et al., 2014), Calcitonin (CAL) (Furuya et al., 2000; Cohen, 2012; Zandawala, 2012), virus transmission (Cyclophilin B (CYCP) (Kanakala and Ghanim, 2016), Knottin (k-1) (Hariton Shalev et al., 2016), Heat shock proteins i.e., Hsp20, Hsp40 and Hsp70 (Gotz et al., 2012; Gorovits and Czosnek, 2017) and other genes associated with vital processes i.e., SNF7 (SWItch/Sucrose Non-Fermentable) and IAP (inhibitor of apoptosis) (Blitvich et al., 2002). The expression of these genes was validated using gene specific primers (designed using Primer3 software) (Table 1) in qRT-PCR performed using LightCycler® 96 System (Roche life sciences, Mannheim, German) with SYBR-Green detection (SYBR® Premix Ex TaqTMII, Takara). The reaction was as follows: 30 s at 94°C followed by 40 cycles consisting of 10 s at 94°C, 30 s at 55°C, and 20 s at 72°C. The template for dsRNA (300–500 bp) against respective gene was amplified from cDNA using gene specific primers having T7 promoter sequence (5′-TAATACGACTCACTATAGGG-3′) at the 5′ and 3′ends of both reverse and forward primer. Amplicons were gel purified using Macherey-NagelTM NucleoSpinTM Gel and PCR Clean-up Kit as per manufacturer’s protocol and used as template for in vitro transcription using TranscriptAid T7 High Yield Transcription Kit (Fermentas) as per kit manual. The dsRNA quality and quantity was determined by agarose gel electrophoresis and Nanodrop quantification.
dsRNA Degradation Studies
To study the impact of whitefly gut nucleases specifically dsRNases on orally delivered dsRNA, about 14 mg of whiteflies (∼N = 300) were taken in 200 μl of PBS (Phosphate buffer saline) and macerated softly to extract the hemolymph and gut juices in the buffer. Total protein was extracted in 100 μl of PBS with the help of 900 μl of 10% TCA (Trichloroacetic acid), followed by centrifugation at 8000 rpm for 10 min. The pellet was washed with three parts of ethyl ether and one part of ethanol to remove unwanted fat molecules and finally dissolved in 200 μl of 0.1 N NaOH. The total proteins were quantified with Folin Phenol reagent method (Lowry et al., 1951). The total protein concentration was estimated from crude hemolymph using spectrophotometer (Eppendorf) which were further serially diluted to half till four concentrations using nuclease free water followed by incubation of the dsRNA (1 μg) at 37°C for 1, 3, and 5 h. The degradation was estimated on the basis of integrity of residual bands on gel electrophoresis.
Oral Delivery of in vitro Synthesized dsRNA
Adult whiteflies were collected by directly tapping leaf over 50 ml falcon tube which was cut down at the bottom and covered with muslin cloth (Upadhyay et al., 2011). Adult whiteflies released in tube were subjected to 2–3 h starvation prior to feeding on artificial diet. The dsRNA (400 ng) against different target genes (AQP, CAL, SNF7, IAP, Hsp20, Hsp40, Hsp70, k-1, and CYCP) was incorporated in 20% sucrose diet and sandwiched between two sterile layers of Parafilm M on the top side of 50 ml falcon tube. Whiteflies (30–35) in three different biological replicates were given the feeding access to mixture of dsRNA and sucrose diet for 48 h. Similar quantity of dsRNA against green fluorescent protein gene (dsGFP) was used as control.
Post 48 h of feeding accesses to dsRNA-sucrose diet mixture, about 25 whiteflies were collected in Trizol (Sigma-Aldrich) from each biological replicate followed by RNA isolation and cDNA synthesis as described in earlier section. The cDNA was diluted 10 times before setting the reaction and each qPCR reaction consisted of 1 μl cDNA, 0.1 μL of each primer (10 μM), and 5 μl of SYBR® Premix Ex TaqTM II in a total volume of 10 μl. The PCR parameters consisted of one cycle at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. The reactions were carried out in a LightCycler® 96 Real-Time PCR System (Roche Applied Science). The relative quantification of genes was done using △△Ct method followed by Students’ t-test for testing the level of significance (p = 0.05). The expression data was normalized with β-tubulin as housekeeping gene and compared with dsGFP fed whiteflies as control.
Enhancing RNAi
Knockdown of Gut Nucleases and Conjugation of dsRNA With CQD Nanoparticles
Gut dsRNases are major bottleneck for efficient RNAi in hemipteran insects as these are considered to be responsible for degradation of orally ingested dsRNA (Arimatsu et al., 2007; Allen and Walker, 2012; Liu et al., 2012). Protection against these nucleases in insect gut can be achieved either by known down of the gene(s) encoding for dsRNases or conjugation dsRNA with some nanoparticles. In the first approach dsRNA against dsRNases (dsdsRNases) was synthesized as per earlier described methodologies and fed (400 ng/μl sucrose diet) to the whitefly adults along with the target gene in three biological replicates. The second approach involved the preparation dsRNA-CQD (carbon quantum dots) nano-conjugate with target gene. For preparation of CQD, 9 ml of polyethylene glycol (M. W. 200; PEG-200) was mixed with 3 ml of water followed by addition of 100 mg of polyethylenimine (PEI) in 2 ml ddH2O. The mixture was heated in a microwave for nearly 3 min at 800 W till it turns light golden yellow. The cooled down PEG-PEI solution (100 μl) was mixed with 40 μg of target gene dsRNA in 100 μl of sodium sulfate solution followed by 4°C overnight incubation. Next day, the tubes were centrifuged at 12,000 rpm for 10 min and pellets were dissolved in 40 μl dd water. To confirm the conjugation of dsRNA with CQD, serial dilutions (1: 100; 1:200; 1:500 and 1:800) of this final CQD preparation were conjugated with the fixed amount of dsRNA (40 μg) and subjected to comparative gel retardation assay. It was anticipated that at a certain concentration CQD won’t be sufficient to retain/conjugate the dsRNA. Thus this dsRNA will appear down the agarose gel as visible in 1:500 and 1:800 dilution of CQD compared to the higher concentrations (1:100 and 1:200), which retained the dsRNA in the well. Thus it was confirmed that the CQD formed a conjugate with the dsRNA.
Conjugation of dsdsRNases and Target Genes With CQD Nanoparticles
The role of dsRNases and efficacy of the CQD particles was further confirmed by conjugating the dsRNA against dsRNase gene and feeding it along with target genes (Hsp 40 and IAP) CQD-dsRNA conjugate. The CQD nanoparticles were prepared as described earlier and conjugated with 40 μg of dsdsRNase each separately with equal amount of dsHsp40 and dsIAP. The similar concentration of dsRNA against GFP was used to prepare dsGFP-CQD conjugate, which served as control. The conjugation of dsRNA and CQD prepared in each experiment was confirmed through Gel Retardation Assay (Das et al., 2015). Adult whiteflies (25–30) in four biological replicates were allowed to feed on different dsRNA-CQD conjugates incorporated in 20% sucrose diet. The total RNA from whiteflies in each treatment was isolated 48 h post feeding and reverse transcribed as per earlier described methodology. The relative expression of all the targeted gene (Hsp40, IAP, dsRNase) in each treatment was quantified by qRT-PCR with respective gene specific primers using 2–ΔΔCT method after normalization with ß-tubulin as housekeeping gene (Kaur et al., 2019).
Validating the Impact of dsRNA Mediated Knockdown of dsRNase Gene on dsRNA Degradation in Hemolymph and Gut Juices
The gut nucleases such as dsRNases are known to hamper the RNAi efficiency by degrading the dsRNA in the insect gut. This was reconfirmed by the knockdown of dsRNase gene in whiteflies followed by extraction of haemolymph and gut juices and its consequent impact on dsRNA incubated with this extract. The whiteflies were allowed to feed on dsRNA (400 ng/μl of sucrose diet) against dsRNase gene synthesized as per earlier described protocols. Post 48 h of feeding accesses, the whiteflies were slightly macerated and processed for haemolymph and gut juice extraction as described in dsRNA degradation studies. The integrity of the dsRNA was assessed as per earlier described methodology.
Estimation of Water Loss
The water loss after feeding of dsAQP and dsCAL was qualitatively estimated on water sensitive paper (Teejet Technologies, United States) disk (5 cm diameter), which turns blue on contact with any type of aqueous solution. The fluid excreted by the whitefly when in contact with this paper is estimated by the number of blue dots appearing on it. Whiteflies were released on the water sensitive paper 48 h post feeding of dsRNA for evaluating the silencing effect of these genes on fluid excretion. The qualitative assessment of the fluid excretion was done by the comparative evaluation of number of blue dots appearing on water sensitive paper in treatment and control. The quantitative estimation of fluid excretion was done using water loss monitoring system consisting of RM-8 Flow Multiplexer, RH-300 Relative Humidity analyzer, SS-4 gas analyzer sub-sampler pump and mass flow meter, RC-chambers and UI-3 A/D interface (Sable Systems International, United States). The control and target gene dsRNA (dsAQP and dsCAL (400 ng/μl sucrose diet) were fed to whiteflies in three biological replicates for 48 h followed by releasing counted number of insects in pre-conditioned (with N2 gas) RC chamber for 1 h. The amount of fluid excretion was sensed by the equipment and quantified use ExpeData software (Sable Systems International, United States).
dsRNA Dose Mortality Relationship Calculated Using Probit Analysis
The dose-mortality relationship was calculated using serial concentrations of dsRNA against SNF7 (dsSNF7) and IAP (dsIAP) genes. Different dsRNA doses against these genes, i.e., 100, 200, 400, and 800 ng/μl of sucrose diet along with dsGFP control were fed to adult whiteflies (∼100 in number) in three different replicates. The number of dead and live whiteflies were recorded 48 h post feeding and the data were subjected to Probit analysis (Higuchi et al., 1993) in POLO Plus software to calculate the LD50 values.
Impact of Knockdown of Hsp20, Hsp40, Hsp70, k-1, and CYCP on Titer and Transmission of Cotton Leaf Curl Virus (CLCuV)
The impact of knockdown of Hsp20, Hsp40, Hsp70, k-1, and CYCP genes was evaluated through virus titer in whitefly and its transmission efficiency in cotton plants. The Cotton leaf curl Rajasthan virus (CLCuRv) (AB-2; KM098115.1) one of the most prevalent virus species (Geminiviridae: Begomovirus) in the region was maintained on susceptible cotton cv. RST9 at Regional Research Station, Faridkot and used as a source of inoculum in the study. The experiment was conducted in two separate batches. The first batch of viruliferous whiteflies (20–30) were allowed to feed for 48 h separately on dsRNA (400 ng/μl of diet) against different genes incorporated in sucrose diet as per earlier described methodology. The viral load in B. tabaci was quantified using qRTPCR with virus specific primers qCLCUV_F1 (5′CGTCGACCTGTTGATAAACCTC3′) and qCLCUV_R1 (5′GCATATTGACCACCGGTAACAG3′) with two technical replicates for each of the four biological replicates. After 48 h post feeding total RNA from each treatment was isolated and reverse transcribed into cDNA, which was used to quantify the transcripts using ΔCT method as per as per earlier described methodology. The whitefly β-tubulin gene (Gene ID: Bta0035 from whitefly genome with primers β-tublin-F: 5′- CACTGGTACGTAGGAGAAGGTA -3′ and β-tubulin-R: 5′-ACTGAGTCCATGCCAACTTC -3) was used as a reference gene for normalization of expression data (Kaur et al., 2019). To determine the implication of each gene in the transmission of CLCuV, the second batch of viruliferous B. tabaci (10 whiteflies/plant) were transferred on virus free cotton plants (three- four leaf stage) maintained in insect proof cages after feeding access of 48 h to each target gene dsRNA (400 ng/μl of diet) and sucrose diet mixture. Whiteflies allowed to feed only on 20% sucrose diet served as a control for this experiment. The experiment was conducted with four individual plants representing each replicate. Plants were continuously monitored for appearance of disease symptoms after 5 days of inoculation access period. After the appearance of early symptoms (formation of green islands) the top most young leaf was harvested in liquid nitrogen wrapped in aluminum foil and grounded to fine powder using a pestle and mortar and processed for total RNA isolation using NucleoSpin® RNA Plant kit (Ref#740949.50) as per user guidelines. After DNase treatment for half an hour at 37°C and inactivation of enzyme at 70°C degrees. RNA purity and yield was analyzed using a spectrophotometer and 500 ng of total RNA was used for cDNA synthesis using PrimeScriptTM 1st strand cDNA Synthesis Kit (Cat no. 6110A). For quantification of the expression level of CLCuV, amplification was performed using ten-fold diluted cDNA as mentioned above using coat protein specific primers. The expression was normalized using RNA helicase gene (Table 1).
Statistical Analysis
The relative expression was calculated using 2–ΔΔCT method (Livak and Schmittgen, 2001) using suitable housekeeping gene for normalization. The comparative expression of target genes and GFP control was compared using Student t-test at p < 0.05. The expression data was presented means ±SEM.
Results and Discussion
dsRNA Degradation Studies and Enhancing RNAi Efficiency
The whitefly crude hemolymph comprised of 0.3 μg/ml of total protein which was further diluted to make 0.15 μg/ml, 0.07 μg/ml and 0.035 μg/ml concentration. Incubation of dsRNA with hemolymph and gut juices resulted in its degradation as indicated by the gel retardation assay (Figures 1A–C, The level of degradation was directly dependent on the concentration of hemolymph and gut juices as well as the period of incubation. The maximum degradation was observed in crude mixture (0.3 μg/ml of total protein), however, retention of integrity of dsRNA in higher dilutions signified the role of dsRNA specific nucleases in RNAi. The time of exposure was also a critical factor and directly proportional to dsRNA degradation. Prolonged period (5 h) of incubation resulted in more exposure of dsRNA to dsRNases consequently resulting in its degradation. When hemolymph was isolated from dsRNases knockdown whitefly, the incubated dsRNA didn’t showed notable degradation of dsRNA till 5 h (Figures 1D–F), which signifies the role of nucleases (dsRNases) in the hindrance of RNAi in whitefly. Various earlier reports also suggests the degradation of dsRNA in the insect belonging to lepidoptera, hemiptera, coleoptera and orthoptera orders due the presence of dsRNA specific nucleases in the gut or hemolymph (Singh et al., 2017; Peng et al., 2018; Prentice et al., 2019; Singh et al., 2019; Song et al., 2019). The conjugation of dsRNA with CQD was confirmed through gel retardation assay, which clearly indicated that an optimum concentration of CQD was required to conjugate the dsRNA (Figure 2A). The higher dilutions (1:500 and 1:800) of original CQD concentration were not sufficient to retain the dsRNA in the well of agarose gel and thus it was visualized down the gel equivalent to the naked dsRNA. Thus confirming that optimum concentration of CQD successfully conjugated with dsRNA and retained it in the agarose well. It is presumed that conjugation of dsRNA with CQD and its incubation in hemolymph and gut juices successfully protected the dsRNA from nucleolytic degradation. The gel retardation assay clearly indicated the retention of dsRNA-CQD conjugate in the agarose gel well (Figure 2A). The gut nucleases in hemipteran insects are reported as major bottleneck in efficient RNAi, as these enzymes hamper the concentration of dsRNA reaching the cell (Huvenne and Smagghe, 2010; Joga et al., 2016). Besides hemipteran, the degradation of dsRNA by dsRNases have also been reported from other insect orders (Almeida Garcia et al., 2017; Singh et al., 2017; Prentice et al., 2019; Song et al., 2019). Our studies successfully demonstrated the degradation of dsRNA in hemolymph and gut juices of whitefly. Further the conjugation of dsRNA with CQD was helpful in protecting dsRNA against the degradation by these nucleases. Protecting the dsRNA against dsRNases gene with CQDs helped in increasing the knockdown efficiency of this gene from 45.0 to 79.08% (Figures 2C,D). Furthermore, conjugation of dsRNA against dsRNase gene with CQD and administering it to whiteflies along with dsRNA against Hsp40 and IAP resulted in higher knockdown efficiency i.e., 99.4% and 99.6% of both the genes, respectively, compared to control (Figures 2G,K). Although RNAi is a well implemented strategy in whiteflies for gene knockdown (Vyas et al., 2017) but due to some major factors such as degradation of dsRNA due to some nucleases poses constraint to sensitivity of the process in vivo (Luo et al., 2017). The uptake of dsRNA in hemiperetan insect’s cells needs validation about the systemic spread of RNAi, which may occur either through SID-1 receptors or clathrin mediated endocytosis. Clathrin-endocytosis mediated uptake of dsRNA toward cellular membrane is a very slow cellular process which can enhanced by nanoparticle mediated delivery of dsRNA (Joga et al., 2016). Nanoparticles act as molecular carriers for dsRNA delivery thereby increasing the persistence of dsRNA and cellular uptake (Das et al., 2015). Several nanoparticles such as liposomes, guanylated polymers, quantum dots, branched amphiphilic peptide capsules (BAPCs), core shell nanoparticles have been used in various insects to enhance the RNAi effect (Avila et al., 2018; Zhang et al., 2015; Christiaens et al., 2018). The association of between nanoparticles and target dsRNA is mediated by electrostatic interaction which involves binding of amino group of nanoparticle with the phosphate group of dsRNA (Kunte et al., 2020). The conjugation between dsRNA and CQD particles may be due to polyethyleneimine (PEI) which is a cationic polymer and possess binding affinity to the nucleic acid to form a particulate complex which results in a decrease in the size of target dsRNA resulting in faster transfection efficiency and improved stability (Tros de Ilarduya et al., 2010). Another plausible explanation may be the ability of CQD particles to induce buffering causing osmotic rupture of endosomes and leading to internalization of dsRNA into the cytoplasm of the gut cells (Ma, 2014). CQD nanoparticles have been used for efficient delivery of dsRNA targeted against glyceraldehyde-3-phosphate dehydrogenase gene (G3PDH) to induce gene knockdown and systemic delivery of dsRNA against rice striped stem borer (Chilo suppressalis) (Wang et al., 2019). These nanoparticles have also been demonstrated as efficient carriers for dsRNA with high retention capability and delivery leading to gene silencing and mortality in A. aegypti (Das et al., 2015). Our studies have systematically demonstrated the degradation of dsRNA in whitefly body fluids due to the presence of dsRNA specific nucleases, which have been earlier characterized from B. tabaci (Luo et al., 2017). Both conjugation of dsRNA with CQDs and knockdown of dsRNases proved efficient ways for improving RNAi efficiency of any target gene. The conjugation of dsRNA with nanoparticles like CQD will boost up gene function studies by enhancing the RNAi efficiency, which in turn will be helpful to visualize the exact impact of particular gene knockdown on whitefly. On the other hand, the enhancement of RNAi through knockdown of gut nucleases can be used in future pest management strategies based on dsRNA sprays and transgenic plants. The dsRNA against dsRNases gene can be sprayed or expressed in transgenic plant along with target gene dsRNA to have an efficient whitefly management strategy.
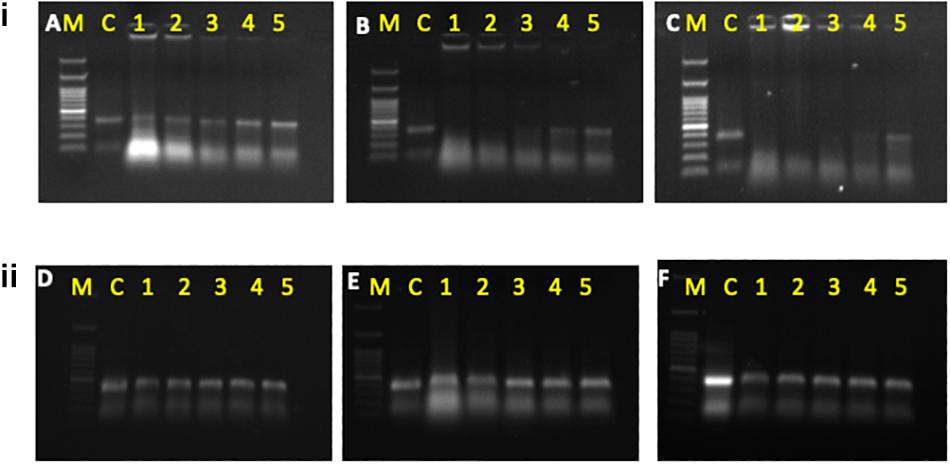
Figure 1. (i) Degradation of dsRNAat different concentrations of hemolymph at different time intervals i.e., 1 h (A), 3 h (B), and 5 h (C). (ii) Degradation of dsRNA at different concentrations of hemolymph isolated from dsRNases knockdown whitefly insects at different time intervals i.e., l h (D), 3 h (E), and 5 h (F) M: 100 bp ladder, C, control dsRNA; 1, crude hemolymph (CH); 2, 1:1 dilution of CH; 3, 1:25 of CH; 4, 1:50 of CH; 5, l:100 of CH.
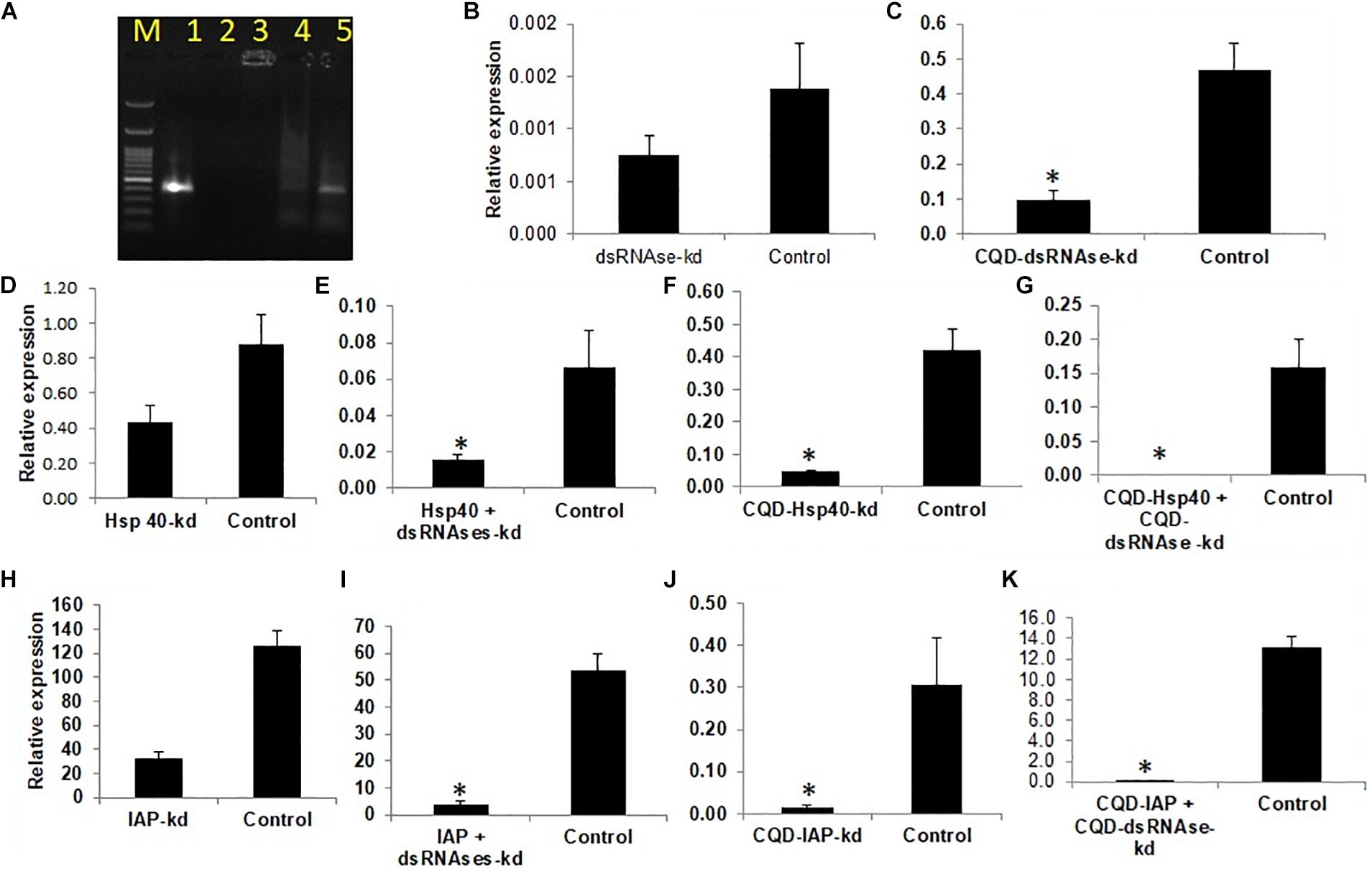
Figure 2. (A) Affinity of CQD nanoparticles at different dilutions with fived concentration of dsRNA.M -100 bp ladder 1- Control, 2-1:100,3-1:200,4- 1:500,5- 1:800, (B) dsRNAses knockdown- 45% (C) CQD-dsRNAses-79.08% (D) Hsp40 knockdown 61.7% (E) Hsp40 + dsRNAses knockdown -77.1% (F) CQD- Hsp40 knockdown 88.9% (G) CQD- Hsp40 + CQD-dsRNAse knockdown 99.4% (H) IAP knockdown 74.3% (I) IAP + dsRNAses knockdown- 92.54% (J) CQD-IAP knockdown 95.2% (K) CQD-IAP + CQD-dsRNAse knockdown 99.6%. The error bar represents the standard deviation (n = 3) and * represents significant differences in mRNA transcripts compared to the control (P ≤ 0.05, Student’s t-test).
Knockdown of Osmoregulatory Genes
Two genes Aquaporion (AQP) and Calcitonin (CAL) were targeted to functionally validate their role in osmoregulation in whitefly through their dsRNA mediated knockdown. Feeding of dsAQP and dsCAL led to 1.9 and 14.7 fold reduction mRNA level of AQP and CAL in whitefly, respectively (Figures 3A,B). Further enhancing RNAi by feeding dsdsRNase along with dsAQP and dsCAL in each treatment resulted in 2.9 and 22.5 fold reduction in mRNA levels of AQP and CAL, respectively (Figures 3D,E). Aquaporins are trans-membrane proteins which have a role in excretion of fluids and heat tolerance (Cohen, 2012; Benoit et al., 2014). On the other hand, Calcitonin is a neuropeptide which is responsible for the transportation of salt and water with the help of Malpighian tubules in the insects (Zandawala, 2012). The qualitative assessment of water loss in AQP and CAL silenced whiteflies was quantified on water sensitive paper, which revealed least fluid excretion (0.039 spots/mm2) in AQP + dsRNAses knockdown followed by sole AQP knockdown (0.08 spots/mm2) compared to GFP control (0.25 spots/mm2) [Figures 4(1–3)]. Similarly, the blue spots calculated in CAL + dsRNAses knockdown were 0.03 spots/mm2, while 0.08 spots/mm2 were observed in CAL alone knockdown whiteflies were as compared to 0.16 spots/mm2 in GFP control [Figures 4(4–6)]. The quantitative fluid loss estimated through humidity analyzer revealed that the knockdown of dsAQP and dsCAL led to excretion of 123 and 86 nl/min per whitefly compared to 203.15 nl/min per whitefly in dsGFP control whiteflies [Figures 4(7–8)]. Earlier studies with knockdown of AQP in Cimex lectularius also observed in 50% reduction in water excretion (Tsujimoto et al., 2017). The knockdown of AQP and CAL in Phenacoccus solenopsis also witnessed reduction in fluid loss compared to GFP control (Singh et al., 2019). Since Calcitonins are responsible for diuresis in insects (Furuya et al., 2000), the RNAi mediated knockdown of calcitonin receptor in Aedes aegypti led to significant reduction in fluid excretion as compared to dsGFP control insects (Kwon et al., 2012) and enhanced mosquito desiccation resistance (Drake et al., 2015).
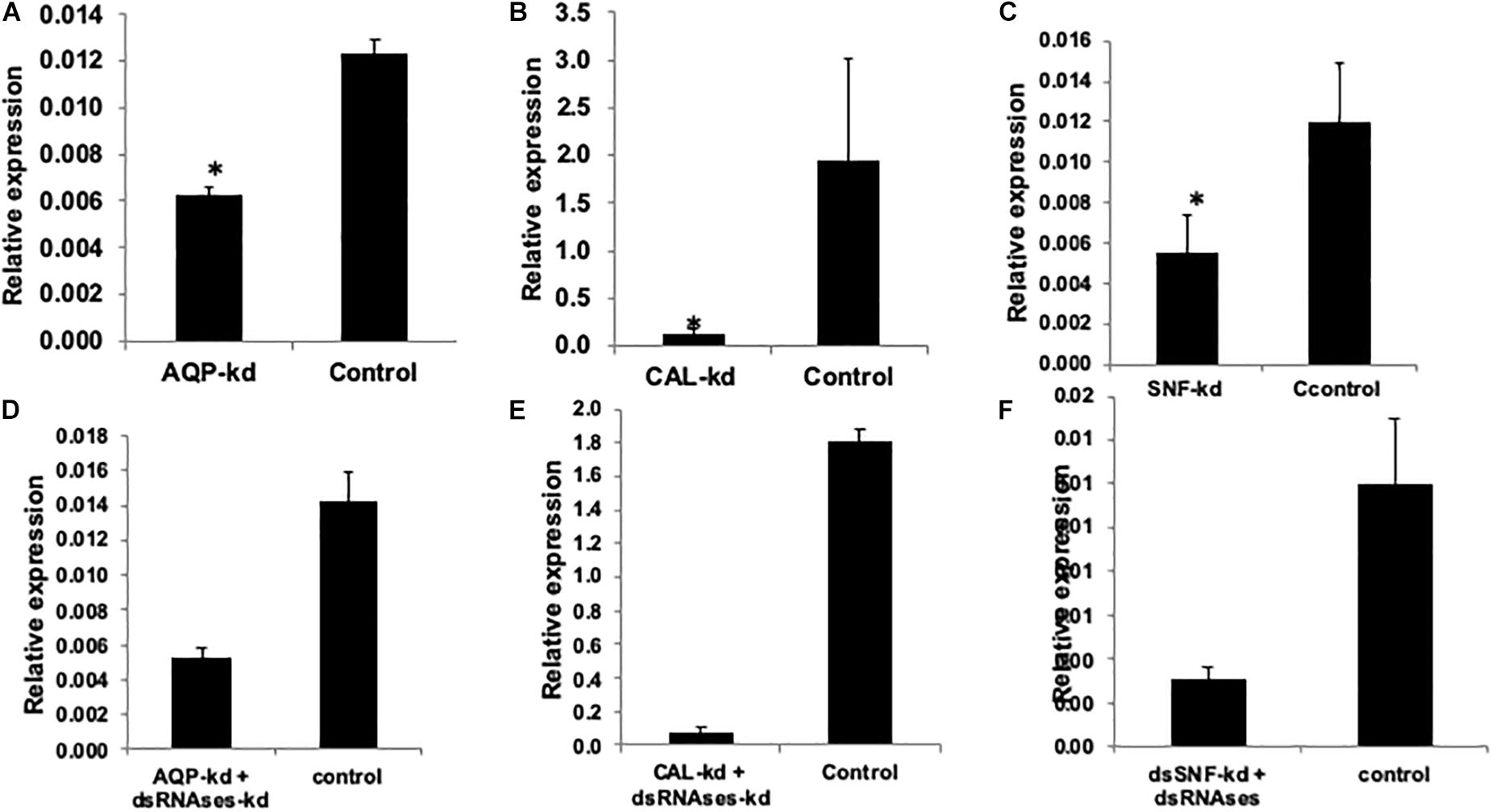
Figure 3. Relative expression of AQP, CAL, SNF, and IAP compared to GFP control when normalized with tubutin. (A) AQP knockdown (36.9%), (B) CAL knockdown (93.27%), (C) SNF 53.2%, (D) AQP + dsRNAses knockdown (62.7%), (E) CAL + dsRNAses knockdown (95.57%) (F) SNF + dsRNAses 74.16% The error bar represents the standard deviation (n = 3) and *represents significant differences in mRNA transcripts compared to the control (P ≤ 0.05, Student’s t-test).
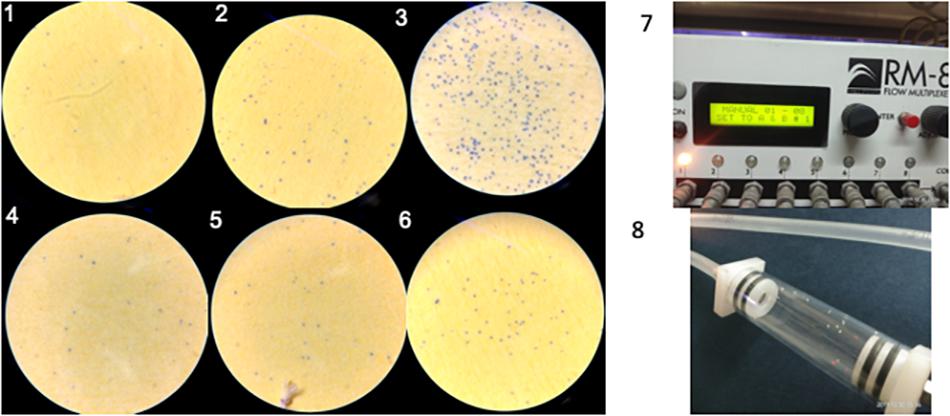
Figure 4. (1–3) Water spots calculated in AQP + dsRNAses knockdown ts 0.039 spots/mm2, AQP knockdown is 0.08 spots/mm2 as compared to GFP control is 0.25 spots/mm2, respectively. (4–6) water spots calculated in CAL + dsRNAses knockdown is 0.03 spots/mm2, CAL knockdown is 0.08 spots/mm2 as compared to GFP control is 0.16 spots/mm2, respectively. (7) Water vapor analyzer (RM-S flow multiplexer) to study water loss from the whitefly. (8) whitefly released in the channel of RM-8 flow multiplexer to estimate water loss.
Dose-Mortality Relationship Vis-a-Vis Knockdown of SNF7 and IAP
Feeding of 400 ng/μl of dsSNF7 and dsSNF7 + dsdsRNase caused 53.2 and 74.1% decrease in expression of SNF7 gene compared to GFP control (Figures 3C,F). The knockdown of IAP further demonstrated significant suppression in mRNA levels of this gene with dsIAP + dsdsRNaseII (92.54%) and dsIAP-CQD nanoconjugate (95.2%) when compared to naked dsIAP (74.3%) and GFP control (Figures 2H–J). The lethal effects of dsIAP and dsSNF estimated through dose-mortality relationship suggested that 100 ng/μl dose of any of dsRNA against these gene could not cause any mortality of whitefly. The LD50 of dsIAP and dsSNF was 1023.9 and 1062.1 ng, respectively, while 4713.6 ng of IAP and 10834 ng of dsSNF was required for causing mortality in 90% of the treated whitefly population (LD90). SNF7 is responsible for relieving major obstacle during transcription occurring due to nucleosome genome packaging. The SWI-SNF complex triggers chromatin remodeling via conformational or positional changes of nucleosomes, thus providing the access of transcriptional machinery to target genes (van Attikum et al., 2004; Nguyen et al., 2017). Additionally, SNF7 complexes have potential role in DNA double-strand breaks repair and nucleotide excisions (van Attikum et al., 2004). Earlier reports on bed bugs (Basnet and Kamble, 2018) and mealybug bug (Singh et al., 2019) suggested induction of mortality when injected with SNF7 specific dsRNAs. However, the amount of dsRNA against a particular gene that may cause a certain level of mortality may strictly depend its vitality or role that gene plays in an organism. There was not much variation between LD50 dose of dsIAP and dsSNF7 but the difference between LD90 doses of two genes was almost half for dsIAP compared to dsSNF. IAP is known to block apoptosis in mammalian cells and its functional analysis of this gene has been studied in many insect species (Q. Li et al., 2005; Singh et al., 2018, 2019). IAP proteins provide a fundamental restriction to apoptosis (Vandergaast et al., 2015), thus their knockdown in insects or insect cell lines has resulted in their mortality at variable doses (Rodrigues et al., 2017; Cao et al., 2018).
Impact of Knockdown of Hsp’s, k-1, and CYCP Genes in Whitefly on Virus Transmission Efficiency in Cotton Plants
Comparative knockdown effect of different genes associated with virus transmission was evaluated through feeding of respective naked dsRNA as well as its conjugation with Carbon Quantum Dots (CQDs) and simultaneous knockdown dsRNA specific (dsRNase) gut nuclease gene. As described earlier CQD-dsRNA conjugation and dsRNase gene knockdown was attempted with the aim to provide protection to dsRNA against nucleolytic degradation in the whitefly gut and its enhanced stability to allow sufficient cellular uptake. The qRT-PCR results clearly indicated that the feeding with target gene dsRNA (40 μg/ml sucrose diet) either conjugated with CQD or along with dsRNA against dsRNase (dsdsRNase) (40 μg/ml sucrose diet) enhanced the RNAi efficiency compared to whiteflies fed with only dsRNA (naked dsRNA against the same target gene) as well as control (dsRNA against green fluorescent protein gene- dsGFP). Feeding whitefly with dsHsp40, dsHsp40 + dsdsRNase and dsHsp40-CQD resulted in 61.7, 77.1, and 70.0% knockdown of mRNA levels compared to control whiteflies, respectively (Figures 2D–F). The results further suggested that among CQD conjugated dsRNA and parallel knockdown of dsRNAse, the RNAi efficiency of latter had a numerical edge over CQD-dsRNA conjugate in case Hsp40 genes. Feeding whiteflies with dsHsp20 and dsHsp20 + dsdsRNase resulted in 61 and 92.2% reduction in mRNA levels when compared to GFP control (Figure 5A). The knockdown of Hsp70 with naked dsRNA and dsHsp70 + dsRNAse caused 65.5 and 95.9% reduction in mRNA levels, which was significantly lower as compared to GFP control (Figure 5B). The coadministration of dsHsp70 along with dsdsRNaes resulted in significant reduction (88.3%) in the expression of Hsp70 as compared to naked dsHsp70. For the past few years RNA interference (RNAi) has emerged as a promising alternative technology to suppress crop pests and its application toward pest management is already close to a reality (Palli, 2014; Zotti and Smagghe, 2015). However, the success of RNAi depends upon the persistence of double-stranded RNA in insect hemolymph. Nonetheless several insect species mainly lepidopteran insects have been observed totally refractory to RNAi due to degradation of exogenous dsRNA (Garbutt and Reynolds, 2012; Shukla et al., 2016). Many studies suggest the role of gut nucleases specifically dsRNases in poor RNAi response in insects (Singh et al., 2019). The success rate of RNAi in earlier studies on B. tabaci leads to partial knockdown of the target genes thereby posing major constrained for RNAi in order to be used as a potent strategy for the management of whiteflies (Ghanim et al., 2007; Upadhyay et al., 2011; Raza et al., 2016). Several studies from the past evidenced that gut nucleases are the major factor limiting the efficacy of orally delivered dsRNA in insects (Arimatsu et al., 2007; Liu et al., 2012).
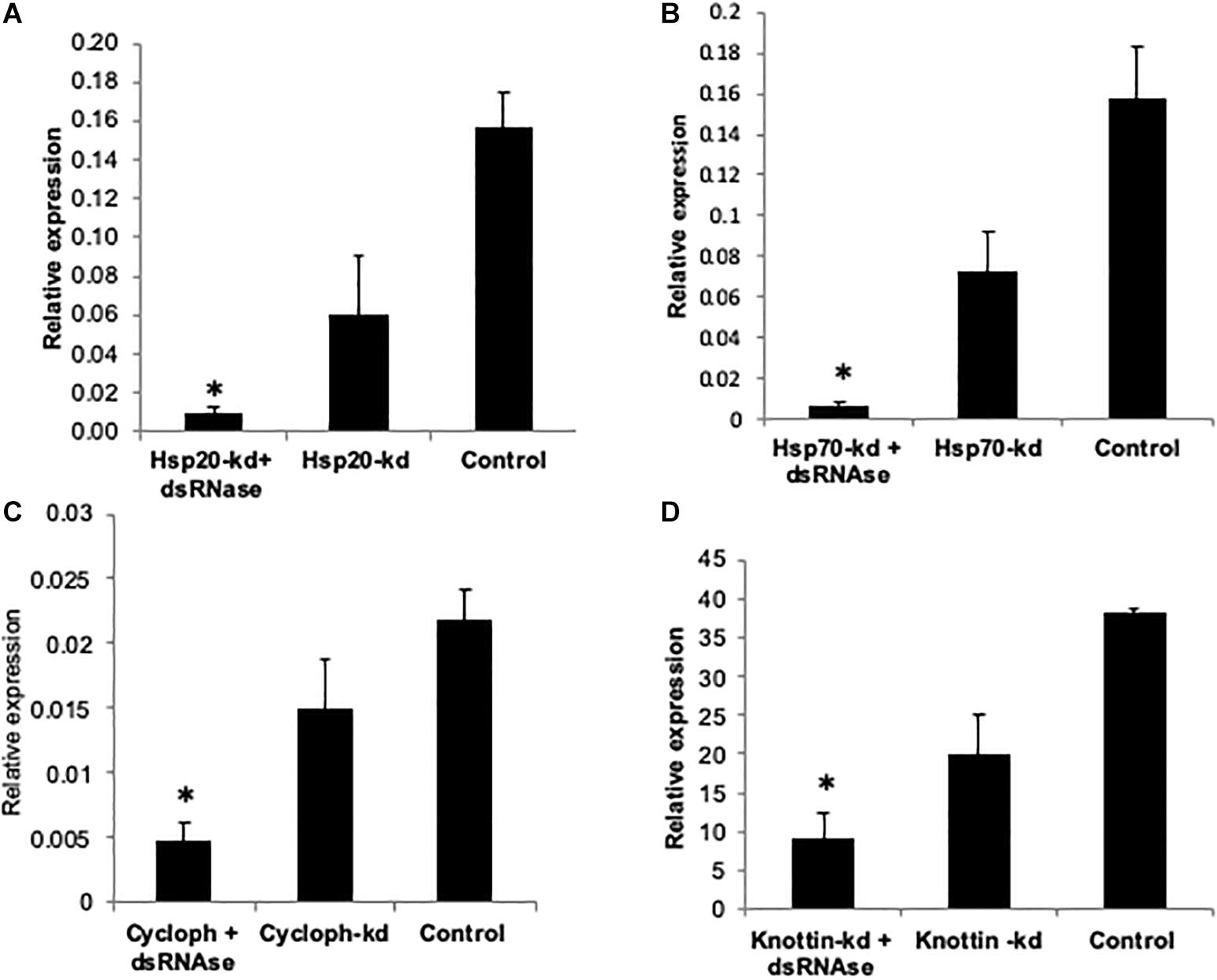
Figure 5. Relative expression of (A) Hsp20 (B) Hsp70 (C) Cyclophitin (D) Knottin-1, compared to GFP control when normalized with tubulin. The error bar represent the standard deviation (n = 3) and ∗represents significant differences in mRNA transcripts compared to the control (P ≤ 0.05, Student’s t-test).
The results showed that knockdown of dsRNAse caused significant reduction in transcripts of target genes compared to dsGFP control and naked dsRNA against the target genes. Earlier reports also suggest that in phloem feeding insects such as B. tabaci effectiveness and success of RNAi against specific genes can be improved by combination with RNAi against the dsRNase genes (Luo et al., 2017). Our results also indicate that dsRNase may be considered as a major barriers in limiting the efficacy of RNAi. Parallel to results, the depletion of nuclease leads to enhanced RNAi response in Colorado potato beetle Leptinotarsa decemlineata thereby increasing stability of oral delievered dsRNA (Spit et al., 2017). Similarly in migratory locust Locusta migratoria, RNAi mediated reduction of gene transcripts encoding dsRNAase significantly enhanced the RNAi process by oral delivery (Song et al., 2017, 2019). Altogether, RNAi-mediated knockdown of dsRNase genes emerged as a promising strategy for improving durability and efficacy of RNAi in various insect species (Scott et al., 2013; Wang et al., 2016). It has been also demonstrated that conjugation with different types of nanoparticles can help to protect the dsRNA in the insect gut thereby enhancing the RNAi efficiency of the target gene (Mysore et al., 2013; Zhang et al., 2015; Christiaens et al., 2018). However, the use of nanoparticles practically under field conditions have many regulatory issues and high input cost issues. Thus alternate strategy of dsRNases knockdown to enhance the RNAi efficiency of the target gene hold more potential and is feasible both in cases of dsRNA based sprays or plants expressing dsRNA. Keeping this in view the knockdown studies with other genes were done with parallel knockdown of dsRNases gene along with virus transmission associated genes. Hsp’s belong to multicomponent and multifunctional highly conserved molecular chaperone family involved in protein quality control. These prevent aggregation of damaged proteins, transportation, folding and unfolding, assembly and disassembly of multi-structured unit and assist degradation of misfolded proteins under stressed conditions (Lindquist, 1986; Parsell and Lindquist, 1993; Gregersen et al., 2001). This family of proteins is universally present in most of the organisms and has been classified and nomenclatured based on their molecular weight into Hsp100, Hsp90, Hsp70, Hsp60, Hsp40, and small Hsps (size >30 kDA), which is represented by the numeric prefix in the name of respective Hsp (Pelham, 1986). Hsp70 comprises of ATP dependent most conserved family of proteins which are also known to play a major role in the biology of host and mediate circulative transmission of Begomoviruses (Mayer, 2005). Microarray based transcriptome analyses revealed that Hsp70 was found to be overexpressed in response to virus acquisition and retention in whitefly (Gotz et al., 2012). The knockdown effect of these genes was also related to the virus titer in the knockdown host vector and its transmission efficiency to the cotton plants. Downregulation of Hsp70, Hsp40 and Hsp20 resulted in significant increase i.e., 3.1, 1.5, and 1.2 fold in virus titer within the host vector compared to the control viruliferous whitefly (Figures 6A–C). Transmission efficiency of whitefly vector fed with dsRNA against Hsp genes assessed by the relative amount of virus titer in cotton leaves at 14 dpi showed significantly variation with the silencing of target genes. The amount of viral transcripts associated with test plants were measured 14 days post-inoculation. Whiteflies silenced with Hsp70 showed significantly increased (2.2 fold) transmission efficiency when compared to control plants (Figure 7). Numerically higher transmission efficiency i.e., 1.2 and 1.7 fold was observed with the knockdown of Hsp40 and Hsp20 in whitefly, which otherwise was statistically at par with control (Figures 7A,B).
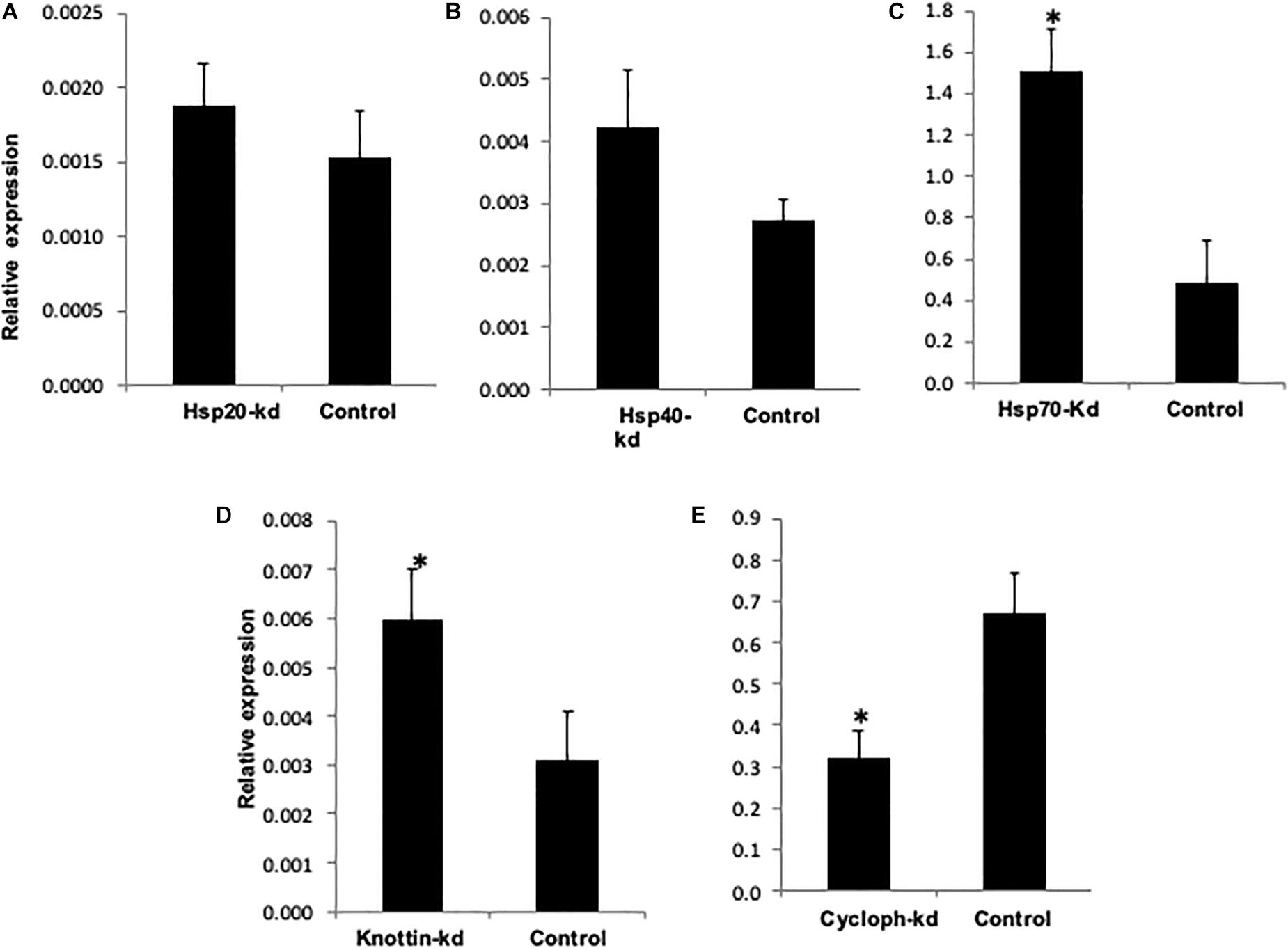
Figure 6. Relative amount of CLCuV (normalized to the host β- tubutin gene) after 48 h of feeding in (A) Hsp20 knocked out whiteflies. (B) Hsp40 knocked out whiteflies. (C) Hsp70 knockout whiteflies. (D) Knottin-1 knocked out whiteflies. (E) Cychphilin knocked out whiteflies. *represents significant differences in mRNA transcripts compared to the control (P ≤ 0.05, Student’s t-test).
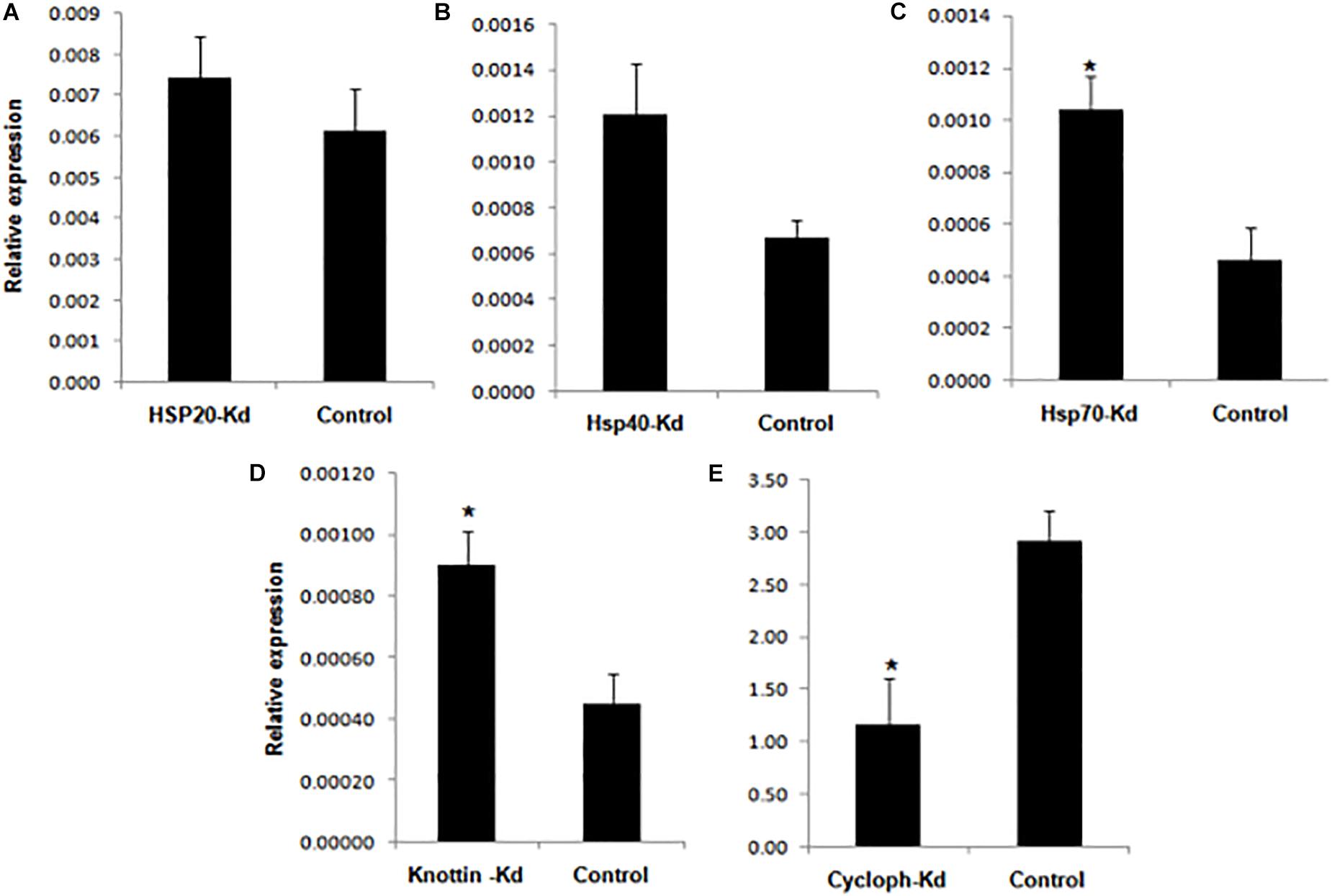
Figure 7. Relative amount of CLCuV (normalized to the cotton RNA helicase gene after 14 dpi with whiteflies fed with dsRNA + dsRNAase corresponding to (A) Hsp20 (B) Hsp4G (C) Hsp70 (D) Knottin-1 (E) Cyclophilin. *represents significant differences in mRNA transcripts compared to the control (P ≤ 0.05, Student’s t-test).
Knottins comprises structural family of proteins containing small disulfide rich proteins with a knotted appearance (Gelly et al., 2004). Whiteflies fed with dsk-1 and combination of dsk-1 + dsRNAse against k-1 showed 47.6 and 76% reduction in expression of k-1, respectively, 48 h post-treatment compared to the GFP control (Figure 5D). The knockdown of k-1 resulted in 1.9 fold increase in virus titer in the whitefly compared to the control whiteflies (Figure 6D). Further, the transmission efficiency of Cotton leaf curl disease (CLCuD) in cotton plants with these whiteflies was 2.0 fold higher as compared to dsGFP fed control whiteflies (Figure 7D). It may be possible that Hsp’s and k-1 are involved in maintaining the amount of virus associated with whitefly vector to a level that may not be deleterious to host. Present study supports the fact that knockdown of Hsp and knottin-1 might have led to the disruption of circulatory route of virion particle, which increased number of copies throughout the insect body and ultimately higher virus transmission efficiency in the inoculating test plant. In this study, inhibition of Hsp70 expression resulted in a 55% increase in CLCuV transmission by B. tabaci thereby confirming previous results reporting inhibitory role of this gene against begomoviruses and act by minimizing the potential long-term harmful effects of the virus on the whitefly (Gotz et al., 2012). The role of Hsp70 isoforms has also been reported in animal viruses like Dengue virus entry, replication and biogenesis in mosquito (Welsch et al., 2009). The knockdowns of Hsp40 and Hsp20 have also shown similar implication on virus titer in vector as well as its transmission in cotton host. Activity of both Hsp70 and Hsp40 is interdependent as Hsp40 function as cochaperon acting as DnaJ dimer which stimulates the ATPase activity of Hsp70 (Wickner et al., 1991; Jordan and McMacken, 1995). Hsp20 belongs to the family of a 16-kDa protein also predicted to be involved in virus interaction (Ohnesorge and Bejarano, 2009). The downregulation of all these three genes result in increased virus titer in whitefly and increased virus transmission efficiency in the host plant. Knottins are found to be in abundance with diverse biological function. Silencing of k-1 also led to significant increase in virus titer in vector B. tabaci which ultimately increased the percentage transmission by 50% in cotton plants at 14 dpi compared to control. These results are in accordance with previous report which used leaf mediated dip assay to target k-1 gene in whiteflies and found that its downregulation led to increased amount of virus with in the whiteflies and tomato plants (Hariton Shalev et al., 2016). Hence, this study supports the earlier reports, which suggested that Hsp and k-1 modulate virus titer in such a way which limits the virus titer to such a level so that it does not lead to any harmful effect inside whitefly body.
Cyclophilins belongs to a peptidyl–prolyl cis–trans isomerase (PPIases or Cyps) family of proteins which plays important role in protein refolding, maturation, cell signaling and gene transcription (Wang and Heitman, 2005; Hanes, 2015). The whiteflies fed with dsCYCP and ds + dsRNAse + CYCP, showed ∼31.1% and 74.5% reduced mRNA levels of CYCP compared to the GFP control (Figure 5C). In comparison to Hsp70, Hsp40, Hsp20, and k-1 lower number of viral transcripts were observed in CYCP knocked out whiteflies (Figure 6E). However, contrary to cotton plants infected with silenced Hsp20, Hsp40, Hsp70, and k -1 whitefly which showed increased viral load, the plants infected with CYCP silenced whiteflies contained lower number of viral transcripts then the untreated plants (Figure 7E). The obtained results are consistent with previous reports in which knockdown of cyclophilin (CYCP) in B. tabaci showed lower efficiency to transmit tomato leaf curl virus (Kanakala and Ghanim, 2016; Kanakala et al., 2019).
Data Availability Statement
The datasets generated for this study will not be made publicly available. There is no data set associated with this manuscript.
Author Contributions
SS conceptualized and designed the experimental setup. RK and MG conducted the experiments, wrote the manuscript, and analyzed the data. NJ and AS contributed to data analysis and manuscript preparation.
Funding
This research was performed at the Regional Research Station PAU Faridkot. The authors acknowledge the support of Ministry of Science and Technology, New Delhi and Punjab Agricultural University, Ludhiana for the conduct of this research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Ahmad, M., Arif, M. I., Ahmad, Z., and Denholm, I. (2002). Cotton whitefly (Bemisia tabaci) resistance to organophosphate and pyrethroid insecticides in Pakistan. Pest Manag. Sci. 58, 203–208. doi: 10.1002/ps.440
Allen, M. L., and Walker, W. B. (2012). Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J. Insect Physiol. 58, 391–396. doi: 10.1016/j.jinsphys.2011.12.014
Almeida Garcia, R., Lima Pepino Macedo, L., Cabral do Nascimento, D., Gillet, F. X., Moreira-Pinto, C. E., Faheem, M., et al. (2017). Nucleases as a barrier to gene silencing in the cotton boll weevil, Anthonomus grandis. PLoS One 12:e0189600. doi: 10.1371/journal.pone.0189600
Arimatsu, Y., Kotani, E., Sugimura, Y., and Furusawa, T. (2007). Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 37, 176–183. doi: 10.1016/j.ibmb.2006.11.004
van Attikum, H., Fritsch, O., Hohn, B., and Gasser, S. M. (2004). Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788. doi: 10.1016/j.cell.2004.11.033
Avila, L. A., Chandrasekar, R., Wilkinson, K. E., Balthazor, J., Heerman, M., Bechard, J., et al. (2018). Delivery of lethal dsRNAs in insect diets by branched amphiphilic peptide capsules. J. Control. Release 273, 139–146. doi: 10.1016/j.jconrel.2018.01.010
Basnet, S., and Kamble, S. T. (2018). Silencing of four genes involved in chromatin remodeling by RNA interference adversely affects fecundity of bed bugs (hemiptera: cimicidae). J. Med. Entomol. 55, 1440–1445. doi: 10.1093/jme/tjy112
Benoit, J. B., Hansen, I. A., Attardo, G. M., Michalková, V., Mireji, P. O., Bargul, J. L., et al. (2014). Aquaporins are critical for provision of water during lactation and intrauterine progeny hydration to maintain tsetse fly reproductive success. PLoS Negl. Trop. Dis. 8:e2517. doi: 10.1371/journal.pntd.0002517
Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. doi: 10.1038/35053110
Blitvich, B. J., Blair, C. D., Kempf, B. J., Hughes, M. T., Black, W. C., Mackie, R. S., et al. (2002). Developmental- and tissue-specific expression of an inhibitor of apoptosis protein 1 homologue from Aedes triseriatus mosquitoes. Insect Mol. Biol. 11, 431–442. doi: 10.1046/j.1365-2583.2002.00352.x
Boykin, L. M., Shatters, R. G., Rosell, R. C., McKenzie, C. L., Bagnall, R. A., De Barro, P., et al. (2007). Global relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) revealed using Bayesian analysis of mitochondrial COI DNA sequences. Mol. Phylogenet. Evol. 44, 1306–1319. doi: 10.1016/j.ympev.2007.04.020
Brown, J. K. (2000). Molecular markers for the identification and global tracking of whitefly vector-Begomovirus complexes. Virus Res. 71, 233–260. doi: 10.1016/s0168-1702(00)00221-5
Cagliari, D., Dias, N. P., Galdeano, D. M., Dos Santos, E. Á., Smagghe, G., and Zotti, M. J. (2019). Management of pest insects and plant diseases by non-transformative RNAi. Front. Plant Sci. 10:1319. doi: 10.3389/fpls.2019.01319
Cao, M., Gatehouse, J. A., and Fitches, E. C. (2018). A Systematic study of RNAi effects and dsRNA stability in Tribolium castaneum and Acyrthosiphon pisum, following injection and ingestion of analogous dsRNAs. Int. J. Mol. Sci. 19:E1079. doi: 10.3390/ijms19041079
Chen, X., Li, L., Hu, Q., Zhang, B., Wu, W., Jin, F., et al. (2015). Expression of dsRNA in recombinant Isaria fumosorosea strain targets the TLR7 gene in Bemisia tabaci. BMC Biotechnol. 15:64. doi: 10.1186/s12896-015-0170-8
Christiaens, O., Tardajos, M. G., Martinez Reyna, Z. L., Dash, M., Dubruel, P., and Smagghe, G. (2018). Increased RNAi efficacy in Spodoptera exigua via the formulation of dsRNA with guanylated polymers. Front. Physiol. 9:316. doi: 10.3389/fphys.2018.00316
Cohen, E. (2012). Roles of aquaporins in osmoregulation, desiccation and cold hardiness in insects. Entomol. Ornithol. Herpetol. S1:001. doi: 10.4172/2161-0983.S1-001
Das, S., Debnath, N., Cui, Y., Unrine, J., and Palli, S. R. (2015). Chitosan, carbon quantum dot, and silica nanoparticle mediated dsRNA delivery for gene silencing in Aedes aegypti: a comparative analysis. ACS Appl. Mater. Interfaces 7, 19530–19535. doi: 10.1021/acsami.5b05232
Dhandapani, R. K., Gurusamy, D., Howell, J. L., and Palli, S. R. (2019). Development of CS-TPP-dsRNA nanoparticles to enhance RNAi efficiency in the yellow fever mosquito, Aedes aegypti. Sci. Rep. 9:8775. doi: 10.1038/s41598-019-45019-z
Drake, L. L., Rodriguez, S. D., and Hansen, I. A. (2015). Functional characterization of aquaporins and aquaglyceroporins of the yellow fever mosquito, Aedes aegypti. Sci. Rep. 5:7795. doi: 10.1038/srep07795
Furuya, K., Milchak, R. J., Schegg, K. M., Zhang, J., Tobe, S. S., Coast, G. M., et al. (2000). Cockroach diuretic hormones: characterization of a calcitonin-like peptide in insects. Proc. Natl. Acad. Sci. U.S.A. 97, 6469–6474. doi: 10.1073/pnas.97.12.6469
Garbutt, J. S., and Reynolds, S. E. (2012). Induction of RNA interference genes by double-stranded RNA; implications for susceptibility to RNA interference. Insect Biochem. Mol. Biol. 42, 621–628. doi: 10.1016/j.ibmb.2012.05.001
Gelly, J.-C., Gracy, J., Kaas, Q., Le-Nguyen, D., Heitz, A., and Chiche, L. (2004). The KNOTTIN website and database: a new information system dedicated to the knottin scaffold. Nucleic Acids Res. 32, D156–D159. doi: 10.1093/nar/gkh015
Ghanim, M., Kontsedalov, S., and Czosnek, H. (2007). Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius). Insect Biochem. Mol. Biol. 37, 732–738. doi: 10.1016/j.ibmb.2007.04.006
Gorovits, R., and Czosnek, H. (2017). The involvement of heat shock proteins in the establishment of tomato yellow leaf curl virus infection. Front. Plant Sci. 8:355. doi: 10.3389/fpls.2017.00355
Gotz, M., Popovski, S., Kollenberg, M., Gorovits, R., Brown, J. K., Cicero, J. M., et al. (2012). Implication of Bemisia tabaci heat shock protein 70 in Begomovirus-whitefly interactions. J. Virol. 86, 13241–13252. doi: 10.1128/JVI.00880-12
Gregersen, N., Bross, P., Andrese, B. S., Pedersen, C. B., Corydon, T. J., and Bolund, L. (2001). The role of chaperone-assisted folding and quality control in inborn errors of metabolism: protein folding disorders. J. Inherit. Metab. Dis. 24, 189–212.
Guan, R.-B., Li, H.-C., Fan, Y.-J., Hu, S.-R., Christiaens, O., Smagghe, G., et al. (2018). A nuclease specific to lepidopteran insects suppresses RNAi. J. Biol. Chem. 293, 6011–6021. doi: 10.1074/jbc.RA117.001553
Hanes, S. D. (2015). Prolyl isomerases in gene transcription. Biochim. Biophys. Acta 1850, 2017–2034. doi: 10.1016/j.bbagen.2014.10.028
Hariton Shalev, A., Sobol, I., Ghanim, M., Liu, S.-S., and Czosnek, H. (2016). The whitefly Bemisia tabaci Knottin-1 gene is implicated in regulating the quantity of tomato yellow leaf curl virus ingested and transmitted by the insect. Viruses 8:E205. doi: 10.3390/v8070205
Higuchi, R., Fockler, C., Dollinger, G., and Watson, R. (1993). Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology 11, 1026–1030. doi: 10.1038/nbt0993-1026
Huvenne, H., and Smagghe, G. (2010). Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol. 56, 227–235. doi: 10.1016/j.jinsphys.2009.10.004
Jaubert-Possamai, S., Le Trionnaire, G., Bonhomme, J., Christophides, G. K., Rispe, C., and Tagu, D. (2007). Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol. 7:63. doi: 10.1186/1472-6750-7-63
Joga, M. R., Zotti, M. J., Smagghe, G., and Christiaens, O. (2016). RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front. Physiol. 7:553. doi: 10.3389/fphys.2016.00553
Jordan, R., and McMacken, R. (1995). Modulation of the ATPase activity of the molecular chaperone DnaK by peptides and the DnaJ and GrpE heat shock proteins. J. Biol. Chem. 270, 4563–4569. doi: 10.1074/jbc.270.9.4563
Kanakala, S., and Ghanim, M. (2016). Implication of the whitefly Bemisia tabaci cyclophilin B protein in the transmission of tomato yellow leaf curl virus. Front. Plant Sci. 7:1702. doi: 10.3389/fpls.2016.01702
Kanakala, S., Kontsedalov, S., Lebedev, G., and Ghanim, M. (2019). Plant-mediated silencing of the whitefly Bemisia tabaci cyclophilin B and heat shock protein 70 impairs insect development and virus transmission. Front. Physiol. 10:557. doi: 10.3389/fphys.2019.00557
Kaur, R., Gupta, M., Singh, S., and Pandher, S. (2019). Evaluation and validation of experimental condition-specific reference genes for normalization of gene expression in Asia II-I Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Gene Expr. Patterns 34:119058. doi: 10.1016/j.gep.2019.119058
Kitzmann, P., Schwirz, J., Schmitt-Engel, C., and Bucher, G. (2013). RNAi phenotypes are influenced by the genetic background of the injected strain. BMC Genomics 14:5. doi: 10.1186/1471-2164-14-5
Kunte, N., McGraw, E., Bell, S., Held, D., and Avila, L.-A. (2020). Prospects, challenges and current status of RNAi through insect feeding. Pest Manag. Sci. 76, 26–41. doi: 10.1002/ps.5588
Kwon, H., Lu, H.-L., Longnecker, M. T., and Pietrantonio, P. V. (2012). Role in diuresis of a calcitonin receptor (GPRCAL1) expressed in a distal-proximal gradient in renal organs of the mosquito Aedes aegypti (L.). PLoS One 7:e50374. doi: 10.1371/journal.pone.0050374
Li, J., Li, X., Bai, R., Shi, Y., Tang, Q., An, S., et al. (2015). RNA interference of the P450 CYP6CM1 gene has different efficacy in B and Q biotypes of Bemisia tabaci. Pest Manag. Sci. 71, 1175–1181. doi: 10.1002/ps.3903
Li, M., Schlesiger, S., Knauer, S. K., and Schmuck, C. (2015). A tailor-made specific anion-binding motif in the side chain transforms a tetrapeptide into an efficient vector for gene delivery. Angew. Chem. Int. Ed. Engl. 54, 2941–2944. doi: 10.1002/anie.201410429
Li, Q., Liston, P., and Moyer, R. W. (2005). Functional analysis of the inhibitor of apoptosis (iap) gene carried by the entomopoxvirus of Amsacta moorei. J. Virol. 79, 2335–2345. doi: 10.1128/JVI.79.4.2335-2345.2005
Lindquist, S. (1986). The heat-shock response. Annu. Rev. Biochem. 55, 1151–1191. doi: 10.1146/annurev.bi.55.070186.005443
Liu, J., Swevers, L., Iatrou, K., Huvenne, H., and Smagghe, G. (2012). Bombyx mori DNA/RNA non-specific nuclease: expression of isoforms in insect culture cells, subcellular localization and functional assays. J. Insect Physiol. 58, 1166–1176. doi: 10.1016/j.jinsphys.2012.05.016
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951). Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275.
Luan, J.-B., Ghanim, M., Liu, S.-S., and Czosnek, H. (2013). Silencing the ecdysone synthesis and signaling pathway genes disrupts nymphal development in the whitefly. Insect Biochem. Mol. Biol. 43, 740–746. doi: 10.1016/j.ibmb.2013.05.012
Luan, J.-B., Li, J.-M., Varela, N., Wang, Y.-L., Li, F.-F., Bao, Y.-Y., et al. (2011). Global analysis of the transcriptional response of whitefly to tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J. Virol. 85, 3330–3340. doi: 10.1128/JVI.02507-10
Luo, Y., Chen, Q., Luan, J., Chung, S. H., Van Eck, J., Turgeon, R., et al. (2017). Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem. Mol. Biol. 88, 21–29. doi: 10.1016/j.ibmb.2017.07.005
Ma, D. (2014). Enhancing endosomal escape for nanoparticle mediated siRNA delivery. Nanoscale 6, 6415–6425. doi: 10.1039/c4nr00018h
Malik, H. J., Raza, A., Amin, I., Scheffler, J. A., Scheffler, B. E., Brown, J. K., et al. (2016). RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci. Rep. 6:38469. doi: 10.1038/srep38469
Mayer, M. P. (2005). Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 153, 1–46. doi: 10.1007/s10254-004-0025-5
Mello, C. C., and Conte, D. (2004). Revealing the world of RNA interference. Nature 431, 338–342. doi: 10.1038/nature02872
Mysore, K., Flannery, E. M., Tomchaney, M., Severson, D. W., and Duman-Scheel, M. (2013). Disruption of Aedes aegypti olfactory system development through chitosan/siRNA nanoparticle targeting of semaphorin-1a. PLoS Negl. Trop. Dis. 7:e2215. doi: 10.1371/journal.pntd.0002215
Navas-Castillo, J., Fiallo-Olivé, E., and Sánchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Nguyen, T. T., Savory, J. G. A., Brooke-Bisschop, T., Ringuette, R., Foley, T., Hess, B. L., et al. (2017). Cdx2 regulates gene expression through recruitment of Brg1-associated switch-sucrose non-fermentable (SWI-SNF) chromatin remodeling activity. J. Biol. Chem. 292, 3389–3399. doi: 10.1074/jbc.M116.752774
Niu, X., Kassa, A., Hu, X., Robeson, J., McMahon, M., Richtman, N. M., et al. (2017). Control of western corn rootworm (Diabrotica virgifera virgifera) reproduction through plant-mediated RNA interference. Sci. Rep. 7:12591. doi: 10.1038/s41598-017-12638-3
Ohnesorge, S., and Bejarano, E. R. (2009). Begomovirus coat protein interacts with a small heat-shock protein of its transmission vector (Bemisia tabaci). Insect Mol. Biol. 18, 693–703. doi: 10.1111/j.1365-2583.2009.00906.x
Palli, S. R. (2014). RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Curr. Opin. Insect Sci. 6, 1–8. doi: 10.1016/j.cois.2014.09.011
Parsell, D. A., and Lindquist, S. (1993). The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27, 437–496. doi: 10.1146/annurev.ge.27.120193.002253
Parsons, K. H., Mondal, M. H., McCormick, C. L., and Flynt, A. S. (2018). Guanidinium-functionalized interpolyelectrolyte complexes enabling RNAi in resistant insect pests. Biomacromolecules 19, 1111–1117. doi: 10.1021/acs.biomac.7b01717
Pelham, H. R. (1986). Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell 46, 959–961. doi: 10.1016/0092-8674(86)906938
Peng, Y., Wang, K., Fu, W., Sheng, C., and Han, Z. (2018). Biochemical comparison of dsRNA degrading nucleases in four different insects. Front. Physiol. 9:624. doi: 10.3389/fphys.2018.00624
Prentice, K., Smagghe, G., Gheysen, G., and Christiaens, O. (2019). Nuclease activity decreases the RNAi response in the sweetpotato weevil Cylas puncticollis. Insect Biochem. Mol. Biol. 110, 80–89. doi: 10.1016/j.ibmb.2019.04.001
Raza, A., Malik, H. J., Shafiq, M., Amin, I., Scheffler, J. A., Scheffler, B. E., et al. (2016). RNA interference based approach to down regulate osmoregulators of whitefly (Bemisia tabaci): potential technology for the control of whitefly. PLoS One 11:e0153883. doi: 10.1371/journal.pone.0153883
Rodrigues, T. B., Dhandapani, R. K., Duan, J. J., and Palli, S. R. (2017). RNA interference in the Asian Longhorned Beetle: identification of Key RNAi Genes and Reference Genes for RT-qPCR. Sci. Rep. 7:8913. doi: 10.1038/s41598-017-08813-1
Roignant, J.-Y., Carré, C., Mugat, B., Szymczak, D., Lepesant, J.-A., and Antoniewski, C. (2003). Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9, 299–308. doi: 10.1261/rna.2154103
Rosen, R., Kanakala, S., Kliot, A., Cathrin Pakkianathan, B., Farich, B. A., Santana-Magal, N., et al. (2015). Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol. 15, 1–8. doi: 10.1016/j.coviro.2015.06.008
Schwedes, C. C., and Carney, G. E. (2012). Ecdysone signaling in adult Drosophila melanogaster. J. Insect Physiol. 58, 293–302. doi: 10.1016/j.jinsphys.2012.01.013
Scott, J. G., Michel, K., Bartholomay, L. C., Siegfried, B. D., Hunter, W. B., Smagghe, G., et al. (2013). Towards the elements of successful insect RNAi. J. Insect Physiol. 59, 1212–1221. doi: 10.1016/j.jinsphys.2013.08.014
Shukla, J. N., Kalsi, M., Sethi, A., Narva, K. E., Fishilevich, E., Singh, S., et al. (2016). Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 13, 656–669. doi: 10.1080/15476286.2016.1191728
Singh, I. K., Singh, S., Mogilicherla, K., Shukla, J. N., and Palli, S. R. (2017). Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 7:17059. doi: 10.1038/s41598-017-17134-2
Singh, S., Gupta, M., Pandher, S., Kaur, G., Goel, N., and Rathore, P. (2019). Using de novo transcriptome assembly and analysis to study RNAi in Phenacoccus solenopsis Tinsley (Hemiptera: Pseudococcidae). Sci. Rep. 9:13710. doi: 10.1038/s41598-019-49997-y
Singh, S., Gupta, M., Pandher, S., Kaur, G., Rathore, P., and Palli, S. R. (2018). Selection of housekeeping genes and demonstration of RNAi in cotton leafhopper, Amrasca biguttula biguttula (Ishida). PLoS One 13:e0191116. doi: 10.1371/journal.pone.0191116
Song, H., Fan, Y., Zhang, J., Cooper, A. M., Silver, K., Li, D., et al. (2019). Contributions of dsRNAses to differential RNAi efficiencies between the injection and oral delivery of dsRNA in Locusta migratoria. Pest Manag. Sci. 75, 1707–1717. doi: 10.1002/ps.5291
Song, H., Zhang, J., Li, D., Cooper, A. M. W., Silver, K., Li, T., et al. (2017). A double-stranded RNA degrading enzyme reduces the efficiency of oral RNA interference in migratory locust. Insect Biochem. Mol. Biol. 86, 68–80. doi: 10.1016/j.ibmb.2017.05.008
Spit, J., Philips, A., Wynant, N., Santos, D., Plaetinck, G., and Vanden Broeck, J. (2017). Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 81, 103–116. doi: 10.1016/j.ibmb.2017.01.004
Taracena, M. L., Oliveira, P. L., Almendares, O., Umaña, C., Lowenberger, C., Dotson, E. M., et al. (2015). Genetically modifying the insect gut microbiota to control Chagas disease vectors through systemic RNAi. PLoS Negl. Trop. Dis. 9:e0003358. doi: 10.1371/journal.pntd.0003358
Tros de Ilarduya, C., Sun, Y., and Düzgüneş, N. (2010). Gene delivery by lipoplexes and polyplexes. Eur. J. Pharm. Sci. 40, 159–170. doi: 10.1016/j.ejps.2010.03.019
Tsujimoto, H., Sakamoto, J. M., and Rasgon, J. L. (2017). Functional characterization of Aquaporin-like genes in the human bed bug Cimex lectularius. Sci. Rep. 7:3214. doi: 10.1038/s41598-017-03157-2
Upadhyay, S. K., Chandrashekar, K., Thakur, N., Verma, P. C., Borgio, J. F., Singh, P. K., et al. (2011). RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 36, 153–161. doi: 10.1007/s12038-011-9009-1
Upadhyay, S. K., Dixit, S., Sharma, S., Singh, H., Kumar, J., Verma, P. C., et al. (2013). siRNA machinery in whitefly (Bemisia tabaci). PLoS One 8:e83692. doi: 10.1371/journal.pone.0083692
Vandergaast, R., Mitchell, J. K., Byers, N. M., and Friesen, P. D. (2015). Insect inhibitor-of-apoptosis (IAP) proteins are negatively regulated by signal-induced N-terminal degrons absent within viral IAP proteins. J. Virol. 89, 4481–4493. doi: 10.1128/JVI.03659-14
Vyas, M., Raza, A., Ali, M. Y., Ashraf, M. A., Mansoor, S., Shahid, A. A., et al. (2017). Knock down of whitefly gut gene expression and mortality by orally delivered gut gene-specific dsRNAs. PLoS One 12:e0168921. doi: 10.1371/journal.pone.0168921
Wang, K., Peng, Y., Pu, J., Fu, W., Wang, J., and Han, Z. (2016). Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 77, 1–9. doi: 10.1016/j.ibmb.2016.07.007
Wang, K., Penga, Y., Chen, J. J., Peng, Y., Wang, X., Shen, Z., et al., (2019). Comparison of efficacy of RNAi mediated by various nanoparticles in the rice striped stem borer (Chilo suppressalis). Pestic. Biochem. Physiol. (in press). doi: 10.1016/j.pestbp.2019.10.005
Wang, P., and Heitman, J. (2005). The cyclophilins. Genome Biol. 6:226. doi: 10.1186/gb-2005-6-7-226
Welsch, S., Miller, S., Romero-Brey, I., Merz, A., Bleck, C. K. E., Walther, P., et al. (2009). Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5, 365–375. doi: 10.1016/j.chom.2009.03.007
Whyard, S., Singh, A. D., and Wong, S. (2009). Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 39, 824–832. doi: 10.1016/j.ibmb.2009.09.007
Wickner, S., Hoskins, J., and McKenney, K. (1991). Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature 350, 165–167. doi: 10.1038/350165a0
Winston, W. M., Molodowitch, C., and Hunter, C. P. (2002). Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459. doi: 10.1126/science.1068836
Xiang, S., Tong, H., Shi, Q., Fernandes, J. C., Jin, T., Dai, K., et al. (2012). Uptake mechanisms of non-viral gene delivery. J. Control. Release 158, 371–378. doi: 10.1016/j.jconrel.2011.09.093
Zandawala, M. (2012). Calcitonin-like diuretic hormones in insects. Insect Biochem. Mol. Biol. 42, 816–825. doi: 10.1016/j.ibmb.2012.06.006
Zha, W., Peng, X., Chen, R., Du, B., Zhu, L., and He, G. (2011). Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS One 6:e20504. doi: 10.1371/journal.pone.0020504
Zhang, X., Mysore, K., Flannery, E., Michel, K., Severson, D. W., Zhu, K. Y., et al. (2015). Chitosan/interfering RNA nanoparticle mediated gene silencing in disease vector mosquito larvae. J. Vis. Exp. 97:52523. doi: 10.3791/52523
Zhang, X., Zhang, J., and Zhu, K. Y. (2010). Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 19, 683–693. doi: 10.1111/j.1365-2583.2010.01029.x
Keywords: nanoparticles, gut nucleases, knockdown, Bemisia tabaci, virus transmission, mortality, osmoregulation
Citation: Kaur R, Gupta M, Singh S, Joshi N and Sharma A (2020) Enhancing RNAi Efficiency to Decipher the Functional Response of Potential Genes in Bemisia tabaci AsiaII-1 (Gennadius) Through dsRNA Feeding Assays. Front. Physiol. 11:123. doi: 10.3389/fphys.2020.00123
Received: 25 November 2019; Accepted: 03 February 2020;
Published: 02 March 2020.
Edited by:
Gianluca Tettamanti, University of Insubria, ItalyReviewed by:
S. Kanakala, North Carolina State University, United StatesLing Tian, South China Agricultural University, China
Copyright © 2020 Kaur, Gupta, Singh, Joshi and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satnam Singh, c2F0bmFtQHBhdS5lZHU=
†These authors have contributed equally to this work
 Ramandeep Kaur1†
Ramandeep Kaur1† Satnam Singh
Satnam Singh Abhishek Sharma
Abhishek Sharma