
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 14 February 2020
Sec. Aquatic Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fphys.2020.00102
This article is part of the Research TopicPhysiological Responses in Aquatic Organisms Adapted to Extreme or Changing EnvironmentsView all 21 articles
Parts of this article's content have been modified or rectified in:
Erratum: LPS Modulates the Expression of Iron-Related Immune Genes in Two Antarctic Notothenoids
 Danixa Pamela Martínez1,2†
Danixa Pamela Martínez1,2† Carmen Sousa3†
Carmen Sousa3† Ricardo Oyarzún1,2,4
Ricardo Oyarzún1,2,4 Juan Pablo Pontigo1
Juan Pablo Pontigo1 Adelino V. M. Canario3
Adelino V. M. Canario3 Deborah Mary Power3
Deborah Mary Power3 Luis Vargas-Chacoff1,2*
Luis Vargas-Chacoff1,2* Pedro Miguel Guerreiro3*
Pedro Miguel Guerreiro3*The non-specific immunity can induce iron deprivation as a defense mechanism against potential bacterial pathogens, but little information is available as to its role in Antarctic fish. In this study the response of iron metabolism related genes was evaluated in liver and head kidney of the Antarctic notothenoids Notothenia coriiceps and Notothenia rossii 7 days after lipopolysaccharide (LPS) injection. Average plasma Fe2+ concentration was unaffected by treatment in any of the species. The gene expression response to LPS varied between tissues and species, being stronger in N. coriiceps and more prominent in the head kidney than liver. The reaction to LPS was marked by increased individual variability in most genes analyzed, even when the change in expression was not statistically significant, suggesting different individual sensitivity and coping responses in these wild fish. We found that iron related genes had an attenuated and homogenous response to LPS but there was no detectable relationship between plasma Fe2+ and gene expression. However, overall in both tissues and species LPS exposure set a multilevel response that concur to promote intracellular accumulation of iron, an indication that Antarctic Notothenoids use innate nutritional immunity as a resistance mechanism against pathogens.
The vertebrates, including fishes, have developed innate and an adaptive immune systems composed both cellular and humoral components to respond to pathogens (Barandica and Tort, 2008). Both systems can be communicated through soluble mediators known as cytokines that regulate the magnitude of the immune response, controlling pathogen growth, promoting inflammation and triggering the adaptive immune response (Sompayrac, 2012). The main organs with immunological function in fish are the thymus, spleen, liver, head kidney and mucosa-associated lymphoid tissue (MALT). They can be grouped into primary (thymus and head kidney) and secondary (spleen, liver, and MALT) immune organs according to their participation in the production, maturation, activation and proliferation of immune components (Soulliere and Dixon, 2017).
During inflammation, the innate immune system can induce several antimicrobial mechanisms, including the depletion of iron available to the pathogen at the systemic and cellular levels (Johnson and Wessling-Resnick, 2012). This defense mechanism, known as nutritional immunity, consists of the removal of iron from the circulation and its subsequent sequestration inside the cell (Collins, 2008). Proinflammatory cytokines such as IL-6 stimulate the transcription of hepcidin, triggering and potentiating the hypoferric response of inflammation (Nemeth et al., 2004a). Several bacterial pathogens obtain the iron they need for their survival and replication from an external source (Ratledge and Dover, 2000; Pulgar et al., 2015; Bethke et al., 2016, 2018; Calquín et al., 2018). For this reason, eukaryotic organisms must effectively control iron homeostasis, through the regulation of the proteins involved in its metabolism (Skaar, 2010).
In fish, iron is mainly taken up by the intestine (Bury and Martin, 2003) where it can enter as heme iron, Fe2+ and Fe3+ (Bury and Martin, 2003). Duodenal cytochrome B is necessary to convert Fe3+ to Fe2+ (Bury and Martin, 2003). The pool of Fe2+ interacts with the divalent metal transporter present on the apical side of the enterocytes (Gunshin et al., 1997; Andrews, 1999). Once inside the cell, the iron can be stored in the cytoplasmic ferritin (the portion of iron that is not for immediate use) (Torti and Torti, 2002), directed toward the mitochondria (for biosynthesis of the Fe-S cluster and heme group) or used in other routes of iron metabolism (Hentze et al., 2004).
The exit of iron from the enterocytes is regulated by ferroportin (Donovan et al., 2000). However, hepcidin can induce the degradation of ferroportin, controlling the release of iron from the basolateral membrane into the bloodstream (Nemeth et al., 2004b). The export of iron also depends on the presence of a copper-associated oxidase, hephaestin in intestinal cells or of ceruloplasmin in non-intestinal cells (Harris et al., 1999; Vulpe et al., 1999). The ferroxidase allows the iron to be loaded on to the cytoplasmic transferrin to be directed to other tissues that need it (Hentze et al., 2004). Iron uptake can be then mediated by transferrin receptor 1 (TfRC1) that is expressed in all tissues (Cheng et al., 2004) or by transferrin receptor 2 (TfRC2), yet not described in fish, that in mammals is found in hepatocytes, duodenal crypt cells and erythroid cells, suggesting a more specialized role of this receptor in iron metabolism (Kawabata et al., 1999). In fish, such as Salmo salar and Eleginops maclovinus, the expression of the main proteins involved in iron metabolism appears to be modulated by the presence of live bacterial pathogens (Pulgar et al., 2015; Martínez et al., 2017a).
Research on Antarctic fish immune responses has mainly addressed the effect of thermal stress on the metabolism of carbohydrates and antioxidant defense system (Machado et al., 2014; Klein et al., 2017; Souza et al., 2018), or analyzed the level of transcriptional response of immune genes after stimulation with Poly I:C and with heat-killed cells, E. coli (Ahn et al., 2016) and Psychrobacter sp. (Buonocore et al., 2016). Furthermore, cDNAs of iron metabolism proteins have been cloned, e.g. ceruloplasmin (Cp), transferrin (Tf), ferritin (Fth1) and divalent metal transporter 1 (Dmt1) (Scudiero et al., 2007, 2009), but no specific information is available on how they are regulated before an infectious process. Furthermore, there is no information on nutritional immunity associated with iron in Antarctic fish or if this type of immune response could be activated by the detection of pathogen-associated molecular patterns (PAMPs), which activate the immune system, but do not require iron. Therefore, the objective of the present study was to evaluate the transcriptional response of immune genes associated with iron in Antarctic Notothenoid fish (Notothenia coriiceps and Notothenia rossii) stimulated with a bacterial PAMPs (specifically LPS, a lipopolysaccharide that integrates the cell wall of gram-negative bacteria).
Notothenia coriiceps (30 ± 2.4 cm of length and 384 ± 93 g of weight) and N. rossii (30 ± 4.2 cm of length and 312 ± 124.7 g of weight) were captured by hook-and-line from a boat, from 5–20 m deep in the waters near the Chinese Great Wall Station, at King George Island (GPS coordinates: 62°13′S, 58°58′W), in the Antarctic Peninsula, during late January 2017. Upon fishing fish were initially randomly placed in six 200 L flow-through plastic tanks for up to 3 days and then measured, weighed, tagged with opercular metal tags and allocated in groups with similar biomass and size distribution in the same flow-through system (two tanks per species; n = 7–8 per tank). Fish were anesthetized in phenoxyethanol (0.02 ml/L) before all experimental manipulations and left undisturbed for 1 week before the experiments to acclimate. At the beginning of the assay (day 0), fish were anesthetized as above and one group per species was injected intraperitoneally (IP) with saline (1.1% NaCl, 0.2 vol.% body weight) and acted as control, while the other group was IP-injected with LPS (1.5 mg/ml LPS in 1.1% NaCl, 0.2% vol body weight, to an effective dosage of 3 mg/kg). This procedure was repeated in day 2, when fish received a second injection of either saline (control) or LPS. At day 7 all fish were lethally anesthetized, blood was collected in heparinized syringes (ammonium salt heparin, 1000 U/ml, Sigma-Aldrich), and fish were sacrificed by cervical section to collect tissues. The head kidney and liver were dissected aseptically, immediately placed in RNAlater (Sigma-Aldrich), placed at 4°C for 24 h and then stored at −20°C. The blood was centrifuged at 10,000 g for 5 min at 4°C to obtain the plasma, which was subsequently stored at −80°C. Water parameters in tanks were monitored twice daily and were approximately constant throughout the experiment – temperature (2.1 ± 0.5°C), salinity (29 ± 0.5 ppt) and oxygen (10.5 ± 1.0 ppm). Two N. coriiceps died in the LPS group and no mortality was observed for N. rossii.
For determination of iron, 1 ml of 5% nitric acid (Sigma-Aldrich) was added to the blood plasma (110 μl), followed by sonication for 10 min and centrifugation at 16,000 g for 5 min at room temperature. The supernatant was removed to a new 15 ml falcon tube and the Fe2+ fraction was measured at 259.940 nm in an Agilent Microwave Plasma Atomic Emission Spectroscope (4200 MP-AES, Agilent Technologies). Only Fe2+ was measured due to limitations of plasma volume and of the spectrometer which could not measure the Fe3+ emission wavelength. The Agilent MP-AES Expert software was used to calculate concentration based on a 14 points standard curve (range 4.47E-05 mM–4.47E-08 mM) with automatic background subtraction provided by a blank (5% nitric acid), in accordance with the manufacturer’s instructions. The limits of detection (LOD) and limit of quantification (LOQ) were calculated from three and ten times the standard deviation of 15 consecutive blank measurements, respectively (Li et al., 2013). The values obtained were 3.77E-05 mM to LOD and 1.26E-04 mM to LOQ. The presence of interference was analyzed through spiking samples with 8.95E-04 mM (50 ppb) and 1.79E-03 mM (100 ppb) standards to the plasma samples and was found not to be significant. Recoveries were calculated from the same spiked plasma samples and estimated at 105%.
Total RNA was extracted from 50 mg of tissue using Trizol (Ambion), following the manufacturer’s instructions. The RNA pellets were dissolved in diethylpyrocarbonate water (DEPC, Sigma-Aldrich) and stored at −80°C. RNA was quantified spectrophotometrically at 260 nm (NanoDrop Technologies) and quality was checked using electrophoresis in 1% agarose gels. Total RNA (2 μg) was used as a template for reverse transcription reactions to synthesize cDNA using MMLV reverse transcriptase (Promega) and the oligo-dT primer (Invitrogen).
The cDNAs were diluted to 100 ng (2 μL) and used as a template for quantitative polymerase chain reaction (qPCR) of ferritin heavy chain (Fth1), ferritin middle chain (Fm), ferroportin (Fp), transferrin (Tf), general identification as transferrin receptor (TfRC) on NCBI database, however, with higher similarity with transferrin receptor type 1 (TfRC1) based in ENSEMBL and CCMAR Sea Genomics databases, general nomenclature as hepcidin-like (Hamp) identified on public NCBI database, however, has more similarity to hepcidin type 2 variant 4 (Hamp2) based on CCMAR Sea Genomics database, ceruloplasmin (Cp), interleukin-6 receptor alpha chain (IL-6Rα), IL-6 receptor beta chain (IL-6Rβ), and 18s ribosomal RNA (18S). The primers for interleukin-6 were not considered because they did not work very well. These primers were designed based on N. coriiceps species because is the only genome partially published and available on public NCBI database1 and further confirmed by blast against of several known genome of fish species that are phylogenetically similar such as Danio rerio, Oryzias melastigma, Oreochromis niloticus, Gasterosteus aculeatus, Tetraodon nigroviridis, Xiphophorus maculatus by public ENSEMBL database2 and Dicentrarchus labrax and Sparus aurata fish species by public CCMAR Sea Genomics database3. The qPCR reaction was performed according to a published protocol (Martínez et al., 2017b), specifically: 95°C for 10 min, followed by 40 cycles at 90°C for 10 s, 60°C for 15 s, and 72°C for 15 s. Melting curve analysis of the amplified products was performed at the end of each PCR to confirm that only one PCR product was amplified and detected. Expression levels were analyzed using the comparative Ct method (2–ΔΔCT) (Livak and Schmittgen, 2001). Data were expressed as the fold change in gene expression normalized to an endogenous reference gene (housekeeping 18s) and relative to the untreated control. The primers used are listed in Table 1. PCR efficiencies were calculated according to the equation: E = 10 [–1/slope] (Rasmussen, 2001).
Data is shown as mean ± standard error (SE) of the mean. Normality and homogeneity of the variances were verified using the Shapiro-Wilk test. The logarithmic transformation was used if data did not follow a normal distribution. The Student’s t-test was used to compare the effect of LPS versus control for iron and gene expression data. The level of statistical significance was 5%. Because several comparisons did not achieve statistical significance and an increase in variability was observed in the LPS treated samples, a Principal Component Analysis (PCA) with rotation based on the Varimax equation with Kaiser Normalization was carried out order to determine the global sample distribution between control and LPS mean centered expression data in liver and head kidney of the two species. The software SPSS (IBM SPSS version 25) was used for the statistical analysis, applying univariate analysis of variance (ANOVA), means, box’s M and Fisher’s transformation to produce a correlation and covariance matrix for discriminant analysis. A Principal Component Regression based on a linear regression on principal components score was applied considering the component score coefficient matrix and component score covariance matrix to discriminate between control and LPS treatment groups. The RStudio software (version 1.0.143) was used to confirm PCA data and produce the confidence ellipses (95%), starting from centroid position for each tissue and species based on component regression of two dimensions (Principal components 1 and 2).
The concentration of plasma iron (Fe2+) varied between 1.53 and 10.8 μM in N. coriiceps and 1.55–23.8 μM in N. rossii, with no statistical difference between control and LPS treatment (Figures 1A,B).
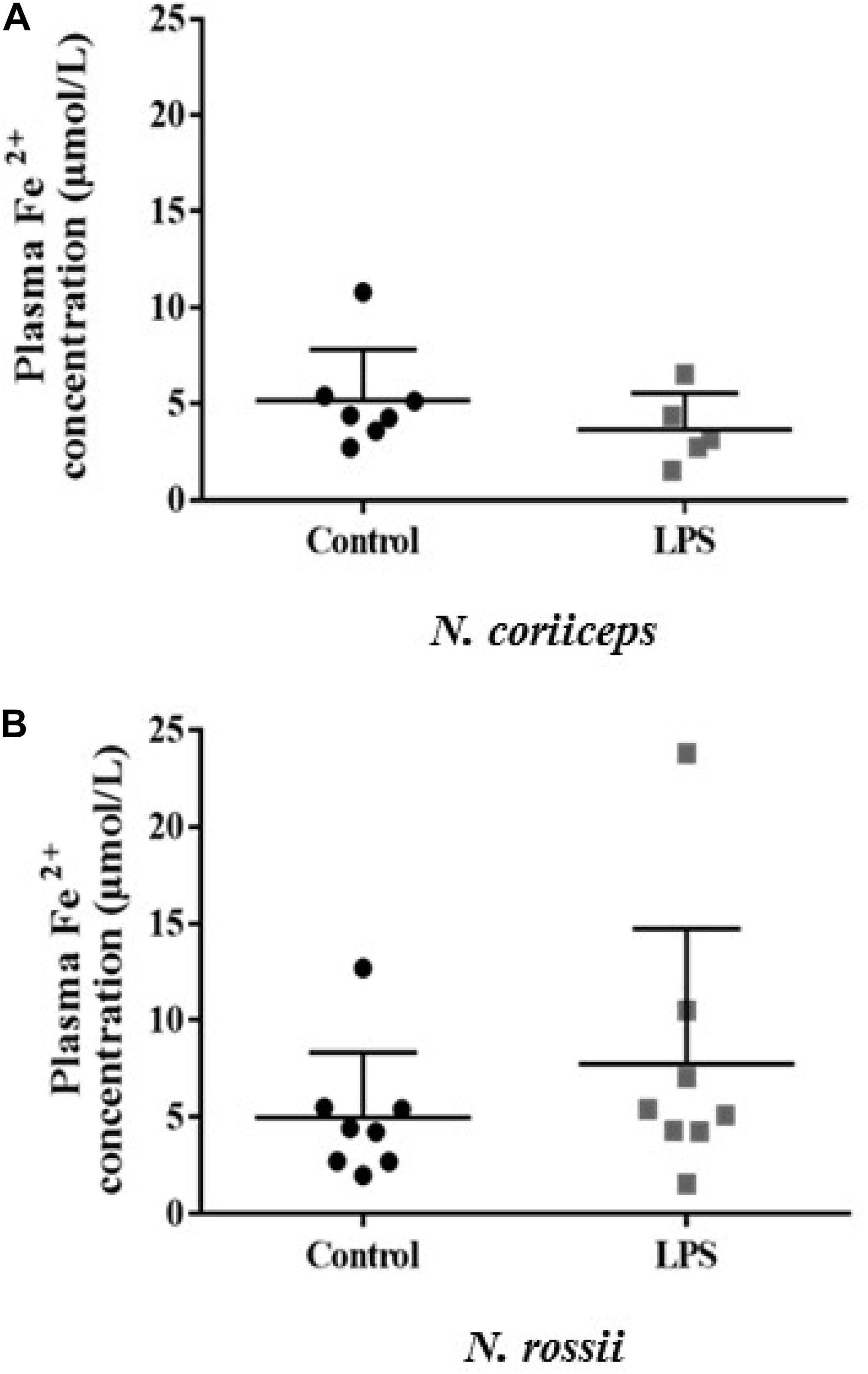
Figure 1. Iron concentration (Fe2+) in plasma at day 7 (5 days after last injection) in fish receiving two injections (on day 0 and day 2) of saline solution (Control) or LPS. (A) N. coriiceps (n = 7 and 5, respectively) (B) N. rossii (n = 8). The final concentration was calculated considering the dilution factor (9×) and molar mass (55.847 g mol–1). No statistical differences were observed among the different treatments (Student’s t-test, P > 0.05).
The iron-related immune gene expression response to LPS varied with tissues and species. N. coriiceps was more responsive to LPS than N. rossii. In N. coriiceps of the nine genes analyzed, 4 were significantly upregulated (p < 0.05) in the liver and 7 in the head kidney, 3 of which were common to both tissues – Fth1, Tf and Hamp2 (Figures 2A–I). In contrast, in N. rossii only Hamp2 was significantly upregulated in the liver and 3 genes were significantly upregulated in head kidney – Fth1, Tf and Cp (Figures 2A–I). Fp was significantly downregulated in N. coriiceps liver while Cp and TfRC1 were significantly downregulated in N. rossii liver (Figures 2C,E,G).
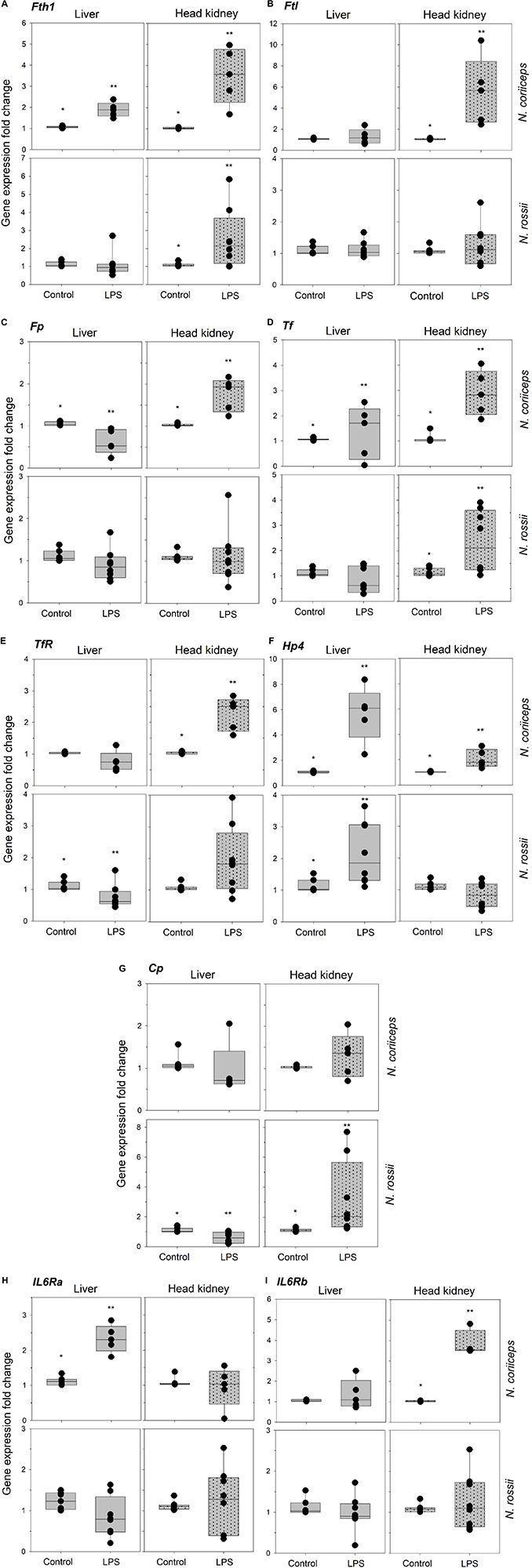
Figure 2. Gene expression of ferritin heavy chain (Fth1) (A), ferritin middle chain (Fm) (B), ferroportin (Fp) (C), transferrin (Tf) (D), transferrin receptor 1(TfRC1) (E), hepcidin 2 (Hamp2) (F), ceruloplasmin (Cp) (G), interleukin 6-receptor alpha chain (IL6Rα) (H), interleukin 6-receptor beta chain (IL6Rβ), (I) in liver and head-kidney of N. coriiceps (NC) and N. rossii (NR) injected with serum saline (Control) or LPS, on day 7 (5 days after last injection). Relative expression was calculated by the 2–ΔΔCT method using the 18s ribosomal protein as the internal reference gene. Each gray bar value is the mean ± SEM and the black dot values correspond to each individual fish (n = 5–7 for N. coriiceps and n = 8 for N. rossii). Asterisks indicate statistical differences between control and LPS treated fish, for each tissue and species (Student’s t-test, P < 0.05).
Since the response to LPS was relatively weak but an increase in gene expression variability was apparent, even when not statistically significant, we sought to determine if there was an association of the gene expression responses with the experimental groups using a PCA for each tissue and species. In N. coriiceps liver, 78% of the gene expression variance was explained by the two first principal components (PC), 42% for PC1 and 36% for PC2. In head kidney, the two first PC explained 85% of the gene expression variance, 60.5% for PC1 and 25.2% for PC2. In N. rossii liver, 3 PC were required to explain almost 75% of the gene expression variance, 34.1% for PC1, 22.9% for PC2, and 18.3% for PC3. In head kidney, 3 PC explained 80% of the gene expression variance, 40.8% for PC1, 23% for PC2, and 15.7% for PC3.
From the component coefficient matrix and the rotated component matrix (Table 2), the differentiation between genes and treatment groups occurred mainly along PC1 for both tissues and species. However, the load factors of the different genes, regardless of treatment, vary widely for each PC, depending on tissue and species. The PCA analysis of gene expression in N. coriiceps liver shows that PC1 was strongly and positively correlated (>0.75) with Fth1, Hamp2, IL6Rα and moderately (>0.5) with Tf, and inversely correlated with Fp, while PC2 directly correlates with Cp, ILR6β, TfRC1, Fm and less with Fp. In N. coriiceps head kidney, PC1 was strongly or moderately correlated (>0.75 or >0.5) to most genes tested with exception IL6Rα, and PC2 correlated positively with IL6Rα and less with Cp, and inversely with Fm. As indicated, above, in N. rossii dispersion was higher and three components were required to explain at least 75% of the distribution, but the same three genes appear to be responsible for determining the PC1 in both liver and head kidney. Thus, in liver, Fm, Fp and ILR6β were the most loading factors on PC1, Fth1 and TfRC1 on PC2, while PC3 was mostly positively correlated with IL6Rα (>0.5) and inversely with Hamp2. In head kidney PC1 correlates strongly (>0.75) with Fm, Fp and ILR6β and moderately (>0.5) with Fth1 and Cp. PC2 was mostly determined by Tf and IL6Rα and PC3 directly by Fth1 and TfRC1 (>0.5) and inversely by Hamp2 (<0.5).
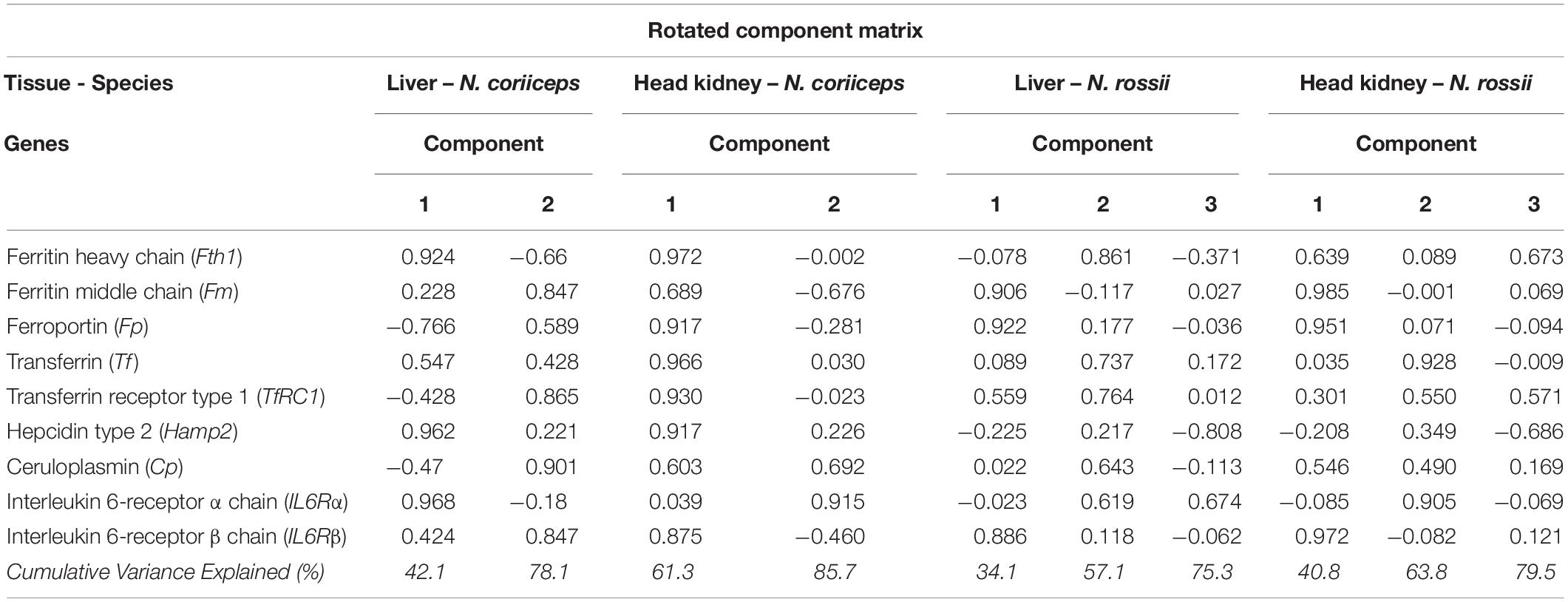
Table 2. Principal Component Analysis by rotation method (Varimax with Kaiser Normalization) applied in different iron-related genes in liver and head kidney of N. coriiceps and N. rossii.
A Principal Component Regression used the first 2 PC variables as regressors to determine the variance between the group clusters. For both immune tissues of N. coriiceps the clusters of control and LPS groups aggregated wide apart and the variance of control individuals was low contrasting with the LPS treated fish which showed a much higher variance, spreading mostly along the PC2 axis (Figures 3A,B). In N. rossii the clusters of the control and LPS groups were closer together, including a slight overlapping, in both liver and head kidney expression distribution, with control centroids on the edge of the LPS 95% confidence ellipses. Nonetheless, and as for N. coriiceps, individuals of the control group clustered close together, as in N. coriiceps, while individuals of LPS group show high dispersion, along both PC1 and PC2 axis (Figures 3C,D). Overall, the analysis explains the allocation of all control individuals into the same treatment group for both species, whereas for LPS 82% of N. coriiceps and 85% of N. rossii were correctly classified as belonging to the same treatment group.
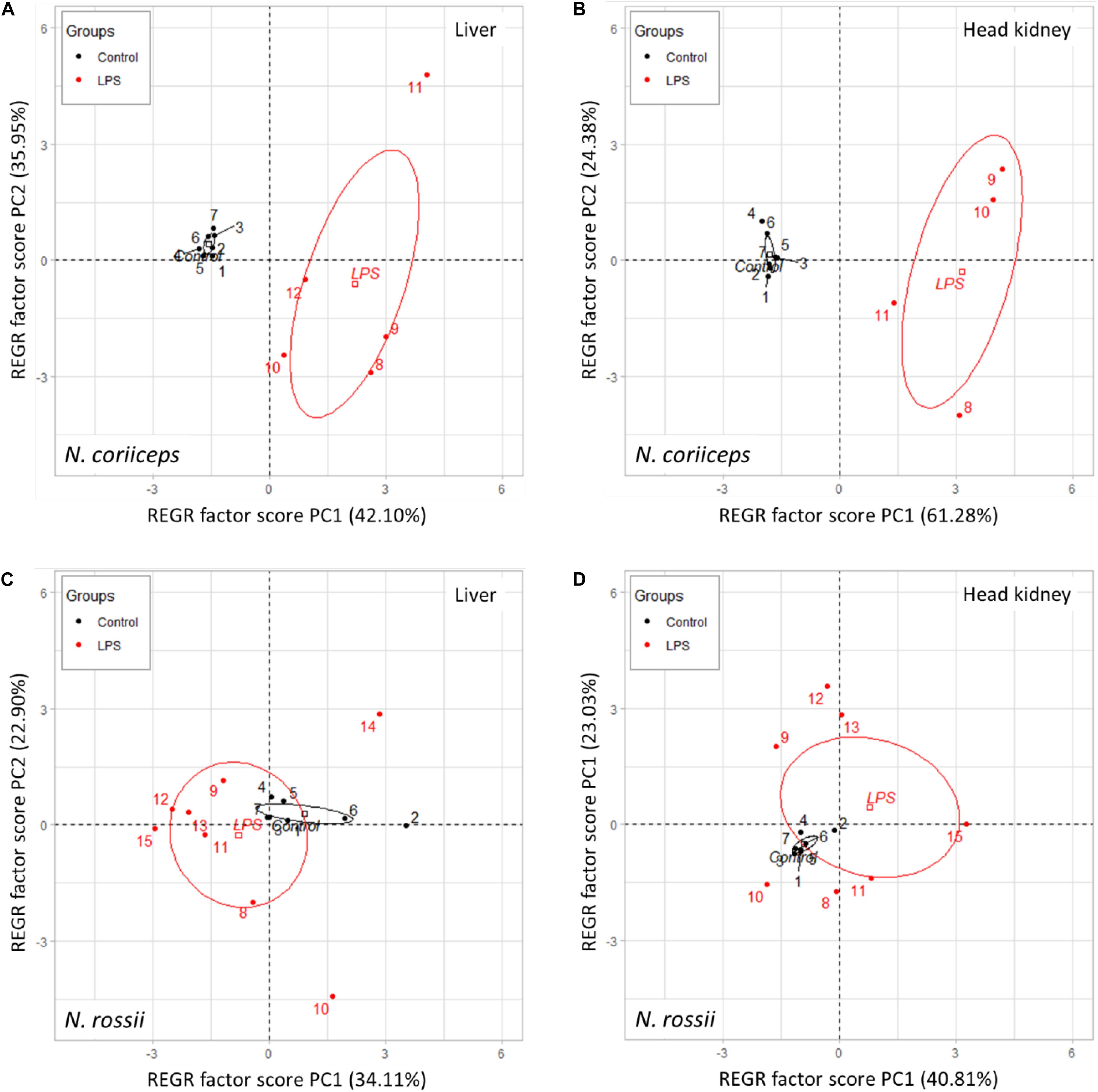
Figure 3. Scatterplot of principal component regression on two dimensions (PC1 and PC2) showing the confidence ellipses (95%) and respective centroids between control and LPS groups, in liver (A) and head kidney (B) of N. coriiceps and in liver (C) and head kidney (D) of N. rossii.
This study found that stimulation with lipopolysaccharide, a gram-negative cell wall component, had no effect on circulating Fe2+ levels, of two closely Notothenioid species, N. coriiceps and N. rossii, but caused modifications in the expression of iron metabolism related immune genes, both in liver and in head kidney. This gene response showed important differences in magnitude between species, and the effects of LPS were more attenuated and variable in N. rossii when compared to N. coriiceps.
In the present study no significant changes were observed in plasma Fe2+ between the two Notothenia species and in response to LPS. Since Fe2+ concentrations were within the range of measurements made in other teleosts (Congleton and Wagner, 1991; Welker et al., 2007; Kopp et al., 2011, 2014; Eslamloo et al., 2012; Hedayati et al., 2016), it suggests potentially similar requirements and homeostatic mechanisms between Antarctic and temperate fish. Although it has been shown in many vertebrate species that bacterial infection leads to a depletion of circulating iron (reviewed by Skaar, 2010; Johnson and Wessling-Resnick, 2012), most studies in fish showed either acute or week-lasting effects of LPS administration over the expression of iron-chelating or iron-transporting mechanisms but did not measure the ion itself (Prieto-Álamo et al., 2009; Liu et al., 2012; Martínez et al., 2017a) and thus the effectiveness and dynamics of LPS alone in lowering plasma iron is far from resolved. In rainbow trout, Oncorhynchus mykiss, LPS injection (1 mg/kg) caused a significant decrease in plasma iron within 48–72 h post-injection but circulating levels returned to normal by 96–120 h (Congleton and Wagner, 1991) and similar kinetics were observed in Atlantic salmon, Salmo salar, injected an identical dosage (Langston et al., 2001). It is therefore also possible that an effect in Fe2+ plasma concentrations may have been missed in our time-frame and dosage, which followed previous studies on temperate fish (Nayak et al., 2011; Guzman-Villanueva et al., 2014; Seppola et al., 2015; Chen et al., 2016) but may less adequate in these Antarctic species. In the present study fish were injected twice, at 0 and 48 h, and sampled at 120 h after the last injection, a design chosen taken in consideration that low temperature habitats may contribute to a slower innate immune system response, as it has been described in cold-water fish when compared to temperate fish (Tort et al., 2003; MacKenzie et al., 2008; Magnoni et al., 2015; Bonneaud et al., 2016; Chen et al., 2016; Abram et al., 2017; Martínez et al., 2017a).
The overall gene expression response, albeit not as vigorous as in other vertebrates (Collins, 2008; Johnson and Wessling-Resnick, 2012; Bethke et al., 2016), also suggests that an iron related immune response is active, much alike what it was seen in the sub-Antarctic Notothenioid Eleginops maclovinus (Martínez et al., 2017a), the Senegalese sole, Solea senegalensis (Prieto-Álamo et al., 2009) or the roughskin sculpin, Trachidermus fasciatus (Liu et al., 2012), among other fish species.
Several genes with important roles in iron metabolism responded in the liver and head kidney in one or both species. Interestingly, there were differences in responsiveness between the two species, with more genes responding and at a higher level in N. coriiceps. The reason for this is not immediately apparent but the two species, although captured in the same general area, tend to segregate with N. rossii in open and deeper channels and N. coriiceps usually under rocks in shallower waters (Jones et al., 2009; Kandalski et al., 2018; Barrera-Oro et al., 2019), which could lead to different exposure to potential pathogens and different immune responsiveness. Some N. coriiceps specimens can be found near the surface in low tide, hiding in rock crevices surrounded by algal debris, thus possibly more exposed to microorganisms. Additionally, while N. coriiceps lives in these coastal shallow waters throughout its life, in the case of N. rossii only the juveniles appear to use these areas, with adults moving into deeper and open waters (Jones et al., 2009), and a life stage difference can thus account for some of the variance in the responses.
The liver is a major organ for iron storage and the site of synthesis for many proteins involved in iron metabolism. Although we have not determined iron levels in liver, the overall changes in gene expression in both species seem to favor iron accumulation in liver cells although this is more evident in N. coriiceps. In N. coriiceps liver upregulation of Fth1 and Tf may promote binding of intracellular iron while upregulation of Hamp2 and downregulation of Fp should inhibit its export (Zahringer et al., 1976; Torti and Torti, 2002; Neves et al., 2009). In N. rossii liver the export machinery seemed to be downregulated (increased Hamp2 and Cp and reduced TfRC1 transcription) but the net effect is expected to be in the same direction and would reduce iron availability for extracellular bacteria.
The fish head kidney is involved in the immune response, but it is also the major haematopoietic organ, thus a likely responder to LPS and an important site for iron trafficking. In fact, expression of head kidney hepcidin genes has been described as a crucial regulator of erythropoiesis during anemia in fish (Joao et al., 2016). In our experiment, the tendency for intracellular accumulation of iron in the head kidney is apparent in N. rossii (by upregulation of Tf, Fth1 and Cp) and also in N. coriiceps (upregulation of Tf, Fth1, Fm, and Hamp2) although in the later species iron turnover rate may be increased (by upregulation of TfRC1 and Cp, possibly involved in extracellular transport). Overall this is consistent with previous reports which suggested an iron deficiency condition after LPS stimulation of the European sea bass, Dicentrarchus labrax (Neves et al., 2009) and the sub-Antarctic Eleginops maclovinus (Martínez et al., 2017a) and up-regulation of the same iron-related genes during experimentally induced anemia in fish (Joao et al., 2016).
Interleukins are modulators of haematopoiesis, inflammation and immune responses. It has been further shown in fish that IL-6 is induced in macrophages during sepsis, and suggested that this may concur to reduce iron availability by induction of hepcidin, as a means to limit the spread of infection (Costa et al., 2011). Interleukin-6 could not be analyzed despite several attempts to isolate the IL-6 sequence in both species, based on the N. coriiceps genome (available in NCBI database, under ID: GCF_000735185.1) (Shin et al., 2014) and in our transcriptomic data on N. rossii (unpublished). However, the two interleukin-6 receptor genes were analyzed, IL-6Rα was highly expressed in liver and IL-6Rβ was highly expressed in head kidney of N. coriiceps. Interestingly, no significant change in gene expression response occurred in N. rossii. The differential response of IL-6R genes between the two species is consistent with iron metabolism-related genes response which was higher in N. coriiceps. This may suggest that also in these species IL-6 cytokines are involved in the induction of the synthesis of the iron regulatory hepcidin during hypoferremia inflammation (Berczi and Szentivanyi, 2003; Nemeth et al., 2004a; Shi and Camus, 2006; Roy et al., 2016). As indicated above, either the timing or a lower general immune response may explain the observations of low or poor immune responses. Although unlikely because of their close phylogeny, it is also possible that the two species may employ alternative pathways in iron metabolism defense. Indeed, species differences in physiology seems to exist. N. coriiceps appear to have lower thermal tolerance than N. rossii based on endocrine, metabolic and antioxidant parameters (Kandalski et al., 2018). Also, the more active N. rossii seems to have higher antioxidant defense system (ROS scavenge) enzyme activity than N. coriiceps, which could be related to their habits and different metabolic rates (Klein et al., 2017).
The PCA analysis also reflects the differences on the response among species, with a clearer separation between control and LPS-treated fish in N. coriiceps than in N. rossii. However, the large variability in responses between individuals, and specific genes in each tissue, especially in the LPS groups makes it difficult to identify which are the main genes reacting to challenge, and thus determining the different PC. These differences, however, show that overall, in both species, the weight of genes determining PC1 appears to be stronger for head kidney than for in liver, where PC2 has a relatively larger weight, which may show that more factors can be involved in the response in the latter tissue. Still, in both tissues of N. coriiceps, the PC1 is mostly determined by Fth1 and Hamp2, while Fm and Cp appear relevant for PC2. In N. rossii PC1 in liver and head kidney is mainly dependent on the weight of Fm, Fp and also IL6Rβ, while the determination of PC2 is shared by more genes but with lower relative importance. Thus, it seems that iron-binding proteins and transmembrane iron-transporters are responding but that magnitude, time-frame and the repertoire of mechanisms vary widely between species and individuals upon challenge.
Indeed, regardless of the changes in the mean expression level of the genes tested, a striking effect of the LPS injection was the increase in the variability of the individual response, when compared to the fish injected with saline. This is observable when looking at the individual gene expression variation and becomes obvious when a PCA was performed. From the analysis it is obvious that some individuals clearly responded to LPS by mounting an immune/iron-related reaction, but not others, although control and LPS-treated fish generally group together in the respective 95% confidence cluster. This lack of synchronicity in response to a seemingly stressful process has been observed in many species, especially when using wild-caught specimens, with likely variations in their life-history experiences (Robertson et al., 2016), nutrition state (Ahola et al., 2017), physiological conditions (Killen et al., 2017) or stress response norms (Balasch and Tort, 2019). In our case, fish were acclimated before the experiments and treated equally thereafter, but changes in size, age and possibly on social condition (Filby et al., 2010) still existed. Additionally, this variability was also observed on the nature of the genes that responded and not only on the magnitude of the response. This could reflect differences in sensitivity or differences in baseline iron, resulting in an asynchronous response. As indicated above for plasma Fe2+ our initial assumption was that a slower metabolism at low temperature could delay the response to LPS compared to temperate teleosts, but this assumption may have been proven wrong. It is possible that an acute response to LPS may have been missed early in the relatively long timeframe of this experiment. Further studies, using several shorter time points, and including iron transport should help to understand innate nutritional immunity in Antarctic fish.
In summary, despite a less than robust immune response of Antarctic Notothenoids marked by considerable individual variability we have shown that overall LPS promotes the mobilization of genes important for iron retention in liver and head-kidney which should contribute to lower iron levels in extracellular fluids and fight bacterial infection through iron starvation.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
The animal collection and experimentation were approved by the Portuguese Environment Agency, under the regulations set by the Treaty of Madrid for scientific investigation on Antarctica. The experiments performed were complied with the EU and Portuguese regulations for animal experimentation and the Guidelines of the Comisión Nacional de Investigación Cientifica y Tecnológica de Chile (CONICYT) and the Universidad Austral de Chile for the use of animals.
DM, CS, PG, AC, DP, JP, RO, and LV-C contributed to analyzing the data, working on the original manuscript and several revisions, and final version. PG, LV-C, and AC designed the experimental assay. CS and PG carried out the experiments and collected the samples. DM, CS, JP, and RO performed the lab analysis.
This study was carried out with logistic and financial support of the Portuguese Polar Program (FACC PROPOLAR 2016-2017) and funded by Portuguese Foundation for Science and Technology (FCT) through projects FCT-NSFC/0002/2016, PTDC/BIAANM/3484/2014, and CCMAR/Multi/04326/2019, and a Ph.D. fellowship SFRH/BD/120040/2016 to CS. This study was also supported by Chilean projects FONDAP-IDEAL grant no. 15150003, FONDECYT REGULAR grant no. 1160877, VIDCA UACH and INACH.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge Pedro Palma and Vera Gomes for help with iron measurements, Stefan Fernandes for help with RStudio software, and the 2017 crew of the Great Wall Chinese Antarctic Station for their hospitability and logistic support.
Abram, Q. H., Dixon, B., and Katzenback, B. A. (2017). Impacts of low temperature on the teleost immune system. Biology (Basel) 6:39. doi: 10.3390/biology6040039
Ahn, D. H., Kang, S., and Park, H. (2016). Transcriptome analysis of immune response genes induced by pathogen agonists in the Antarctic bullhead notothen Notothenia coriiceps. Fish Shellfish Immunol. 55, 315–322. doi: 10.1016/j.fsi.2016.06.004
Ahola, A. J., Lassenius, M. I., Forsblom, C., Harjutsalo, V., Lehto, M., and Groop, P. H. (2017). Dietary patterns reflecting healthy food choices are associated with lower serum LPS activity. Sci. Rep. 7, 1–9. doi: 10.1038/s41598-017-06885-7
Andrews, N. C. (1999). The iron transporter DMT1. Int. J. Biochem. Cell Biol. 31, 991–994. doi: 10.1016/s1357-2725(99)00065-5
Balasch, J. C., and Tort, L. (2019). Netting the stress responses in fish. Front. Endocrinol. (Lausanne) 10:62. doi: 10.3389/fendo.2019.00062
Barandica, L., and Tort, L. (2008). Neuroendocrinología e inmunologiía de la respuesta al estreés en peces. Rev. Acad. Colomb. Cienc. 32, 267–284.
Barrera-Oro, E., Moreira, E., Seefeldt, M. A., Valli Francione, M., and Quartino, M. L. (2019). The importance of macroalgae and associated amphipods in the selective benthic feeding of sister rockcod species Notothenia rossii and N. coriiceps (Nototheniidae) in West Antarctica. Polar Biol. 42, 317–334. doi: 10.1007/s00300-018-2424-0
Bethke, J., Poblete-Morales, M., Irgang, R., Yáñez, A., and Avendaño-Herrera, R. (2016). Iron acquisition and siderophore production in the fish pathogen Renibacterium salmoninarum. J. Fish Dis. 39, 1275–1283. doi: 10.1111/jfd.12456
Bethke, J., Yáñez, A. J., and Avendaño-Herrera, R. (2018). Comparative genomic analysis of two Chilean Renibacterium salmoninarum isolates and the type strain ATCC 33209T. Genome Biol. Evol. 10, 1816–1822. doi: 10.1093/gbe/evy138
Bonneaud, C., Wilson, R. S., and Seebacher, F. (2016). Immune-challenged fish up-regulate their metabolic scope to support locomotion. PLoS One 11:e0166028. doi: 10.1371/journal.pone.0166028
Buonocore, F., Bernini, C., Coscia, M. R., Giacomelli, S., de Pascale, D., Randelli, E., et al. (2016). Immune response of the Antarctic teleost Trematomus bernacchii to immunization with Psychrobacter sp. (TAD1). Fish Shellfish Immunol. 56, 192–198. doi: 10.1016/j.fsi.2016.07.009
Bury, N., and Martin, G. (2003). Iron acquisition by teleost fish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 135, 97–105. doi: 10.1016/s1532-0456(03)00021-8
Calquín, P., Ruiz, P., Oliver, C., Sánchez, P., Haro, R., Oliva, H., et al. (2018). Physiological evidence that Piscirickettsia salmonis produces siderophores and uses iron from different sources. J. Fish Dis. 41, 553–558. doi: 10.1111/jfd.12745
Chen, Y., Zhou, S., Jiang, Z., Wang, X., and Liu, Y. (2016). Chemokine receptor CXCR3 in turbot (Scophthalmus maximus): cloning, characterization and its responses to lipopolysaccharide. Fish Physiol. Biochem. 42, 659–671. doi: 10.1007/s10695-015-0167-1
Cheng, Y., Zak, O., Aisen, P., Harrison, S. C., Walz, T., and York, N. (2004). Structure of the human transferrin receptor-transferrin complex. Cell 116, 565–576. doi: 10.1016/s0092-8674(04)00130-8
Collins, H. L. (2008). Withholding iron as a cellular defence mechanism – friend or foe? Eur. J. Immunol. 38, 1803–1806. doi: 10.1002/eji.200838505
Congleton, J. L., and Wagner, E. J. (1991). Acute-phase hypoferremic response to in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. A Physiol. 98, 195–200. doi: 10.1016/0300-9629(91)90519-i
Costa, M. M., Maehr, T., Diaz-Rosales, P., Secombes, C. J., and Wang, T. (2011). Bioactivity studies of rainbow trout (Oncorhynchus mykiss) interleukin-6: effects on macrophage growth and antimicrobial peptide gene expression. Mol. Immunol. 48, 1903–1916. doi: 10.1016/j.molimm.2011.05.027
Donovan, A., Brownlie, A., Zhou, Y., Shepard, J., Pratt, S. J., Moynihan, J., et al. (2000). Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403, 776–781. doi: 10.1038/35001596
Eslamloo, K., Falahatkar, B., and Yokoyama, S. (2012). Effects of dietary bovine lactoferrin on growth, physiological performance, iron metabolism and non-specific immune responses of Siberian sturgeon Acipenser baeri. Fish Shellfish Immunol. 32, 976–985. doi: 10.1016/j.fsi.2012.02.007
Filby, A. L., Paull, G. C., Bartlett, E. J., Van Look, K. J. W., and Tyler, C. R. (2010). Physiological and health consequences of social status in zebrafish (Danio rerio). Physiol. Behav. 101, 576–587. doi: 10.1016/j.physbeh.2010.09.004
Gunshin, H., Mackenzie, B., Berger, U. V., Gunshin, Y., Romero, M. F., Boron, W. F., et al. (1997). Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 388, 482–488. doi: 10.1038/41343
Guzman-Villanueva, L. T., Ascencio-Valle, F., Macias-Rodriguez, M. E., and Tovar-Ramirez, D. (2014). Effects of dietary β-1,3/1,6-glucan on the antioxidant and digestive enzyme activities of Pacific red snapper (Lutjanus peru) after exposure to lipopolysaccharides. Fish Physiol. Biochem. 40, 827–837. doi: 10.1007/s10695-013-9889-0
Harris, Z. L., Durley, A. P., Man, T. K., and Gitlin, J. D. (1999). Targeted gene disruption reveals an essential role for ceruloplasmin in cellular iron efflux. Proc. Natl. Acad. Sci. U.S.A. 96, 10812–10817. doi: 10.1073/pnas.96.19.10812
Hedayati, A., Hoseini, S. M., and Hoseinifar, S. H. (2016). Response of plasma copper, ceruloplasmin, iron and ions in carp, Cyprinus carpio to waterborne copper ion and nanoparticle exposure. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 179, 87–93. doi: 10.1016/j.cbpc.2015.09.007
Hentze, M. W., Muckenthaler, M. U., and Andrews, N. C. (2004). Balancing acts: molecular control of mammalian iron metabolism. Cell 117, 285–297.
Joao, N., Caldas, C., Ramos, M. F., and Rodrigues, P. N. S. (2016). Hepcidin-dependent regulation of erythropoiesis during anemia in a teleost fish, Dicentrarchus labrax. PLoS One 11:e0153940. doi: 10.1371/journal.pone.0153940
Johnson, E. E. E., and Wessling-Resnick, M. (2012). Iron metabolism and the innate immune response to infection. Microbes Infect. 14, 207–216. doi: 10.1016/j.micinf.2011.10.001
Jones, C., Damerau, M., Deitrich, K., Driscoll, R., Kock, K. H., Kuhn, K., et al. (2009). Demersal Finfish Survey of the South Orkney Islands. AMLR 2008/2009 Field Season Report: Objectives, Accomplishments and Tentative. La Jolla, CA: Antarctiv Marine Living Resources, 49–66.
Kandalski, P. K., de Souza, M. R. D. P., Herrerias, T., Machado, C., Zaleski, T., Forgati, M., et al. (2018). Effects of short-term thermal stress on the plasma biochemical profiles of two Antarctic nototheniid species. Rev. Fish Biol. Fish. 28, 925–940. doi: 10.1007/s11160-018-9535-0
Kawabata, H., Yang, R., Hirama, T., Vuong, P. T., Kawano, S., Gombart, A. F., et al. (1999). Molecular cloning of transferrin receptor 2:a new member of the transferrin receptor-like family. J. Biol. Chem. 274, 20826–20832. doi: 10.1074/jbc.274.30.20826
Killen, S. S., Marras, S., Nadler, L., and Domenici, P. (2017). The role of physiological traits in assortment among and within fish shoals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160233. doi: 10.1098/rstb.2016.0233
Klein, R. D., Rosa, C. E., Colares, E. P., Robaldo, R. B., Martinez, P. E., and Bianchini, A. (2017). Antioxidant defense system and oxidative status in Antarctic fishes: the sluggish rockcod Notothenia coriiceps versus the active marbled notothen Notothenia rossii. J. Therm. Biol. 68, 119–127. doi: 10.1016/j.jtherbio.2017.02.013
Kopp, R., Lang, Š, Brabec, T., and Mareš, J. (2014). The influence of physicochemical properties of water on plasma indices in brook trout (Salvelinus fontinalis, Mitchill) reared under conditions of intensive aquaculture. Acta Vet. Brno 82, 427–433. doi: 10.2754/avb201382040427
Kopp, R., Mareš, J., Lang, Š, Brabec, T., and Ziková, A. (2011). Assessment of ranges plasma indices in rainbow trout (Oncorhynchus mykiss) reared under conditions of intensive aquaculture. Acta Univ. Agric. Silvic. Mendelianae Brun. 59, 181–188. doi: 10.11118/actaun201159060181
Langston, A. L., Johnstone, R., and Ellis, A. E. (2001). The kinetics of the hypoferraemic response and changes in levels of alternative complement activity in diploid and triploid Atlantic salmon, following injection of lipopolysaccharide. Fish Shellfish Immunol. 11, 333–345. doi: 10.1006/fsim.2000.0319
Li, W., Simmons, P., Shrader, D., Herrman, T. J., and Dai, S. Y. (2013). Talanta microwave plasma-atomic emission spectroscopy as a tool for the determination of copper, iron, manganese and zinc in animal feed and fertilizer. Talanta 112, 43–48. doi: 10.1016/j.talanta.2013.03.029
Liu, Y., Yu, S., Chai, Y., and Zhu, Q. (2012). Transferrin gene expression in response to LPS challenge and heavy metal exposure in roughskin sculpin (Trachidermus fasciatus). Fish Shellfish Immunol. 32, 223–229. doi: 10.1016/j.fsi.2011.10.023
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔct Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Machado, C., Zaleski, T., Rodrigues, E., Carvalho, C., dos, S., Cadena, S. M. S. C., et al. (2014). Effect of temperature acclimation on the liver antioxidant defence system of the Antarctic nototheniids Notothenia coriiceps and Notothenia rossii. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 172–173, 21–28. doi: 10.1016/j.cbpb.2014.02.003
MacKenzie, S., Balasch, J. C., Novoa, B., Ribas, L., Roher, N., Krasnov, A., et al. (2008). Comparative analysis of the acute response of the trout, O. mykiss, head kidney to in vivo challenge with virulent and attenuated infectious hematopoietic necrosis virus and LPS-induced inflammation. BMC Genomics 9:141. doi: 10.1186/1471-2164-9-141
Magnoni, L. J., Roher, N., Crespo, D., Krasnov, A., and Planas, J. V. (2015). In vivo molecular responses of fast and slow muscle fibers to Lipopolysaccharide in a teleost fish, the rainbow trout (Oncorhynchus mykiss). Biology (Basel) 4, 67–87. doi: 10.3390/biology4010067
Martínez, D., Oyarzún, R., Pontigo, J., Romero, A., Yáñez, A., and Vargas-Chacoff, L. (2017a). Nutritional immunity triggers the modulation of iron metabolism genes in the sub-Antarctic Notothenioid Eleginops maclovinus in response to Piscirickettsia salmonis. Front. Immunol. 8:1153. doi: 10.3389/fimmu.2017.01153
Martínez, D., Oyarzún, R., Vargas-Lagos, C., Pontigo, J. P., Soto-Dávila, M., Saravia, J., et al. (2017b). Identification, characterization and modulation of ferritin-H in the sub-Antarctic Notothenioid Eleginops maclovinus challenged with Piscirickettsia salmonis. Dev. Comp. Immunol. 73, 88–96. doi: 10.1016/j.dci.2017.03.015
Nayak, S. K., Swain, P., Nanda, P. K., Mohapatra, D., and Behera, T. (2011). Immunomodulating potency of lipopolysaccharides (LPS) derived from smooth type of bacterial pathogens in Indian major carp. Vet. Microbiol. 151, 413–417. doi: 10.1016/j.vetmic.2011.03.021
Nemeth, E., Rivera, S., Gabayan, V., Keller, C., Taudorf, S., Pedersen, B. K., et al. (2004a). IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J. Clin. Invest. 113, 1271–1276. doi: 10.1172/jci200420945
Nemeth, E., Tuttle, M., Powelson, J., Vaughn, M., Donovan, A., McVey-Ward, D., et al. (2004b). Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306, 2090–2093. doi: 10.1126/science.1104742
Neves, V., Wilson, J. M., and Rodrigues, P. N. S. (2009). Transferrin and ferritin response to bacterial infection: the role of the liver and brain in fish. Dev. Comp. Immunol. 33, 848–857. doi: 10.1016/j.dci.2009.02.001
Prieto-Álamo, M. J., Abril, N., Osuna-Jiménez, I., and Pueyo, C. (2009). Solea senegalensis genes responding to lipopolysaccharide and copper sulphate challenges: large-scale identification by suppression subtractive hybridization and absolute quantification of transcriptional profiles by real-time RT-PCR. Aquat. Toxicol. 91, 312–319. doi: 10.1016/j.aquatox.2008.11.001
Pulgar, R., Hödar, C., Travisany, D., Zuñiga, A., Domínguez, C., Maass, A., et al. (2015). Transcriptional response of Atlantic salmon families to Piscirickettsia salmonis infection highlights the relevance of the iron-deprivation defence system. BMC Genomics 16:495. doi: 10.1186/s12864-015-1716-9
Ratledge, C., and Dover, L. G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Micribiol. 54, 881–941. doi: 10.1146/annurev.micro.54.1.881
Robertson, S., Bradley, J. E., and Maccoll, A. D. C. (2016). Measuring the immune system of the three-spined stickleback – investigating natural variation by quantifying immune expression in the laboratory and the wild. Mol. Ecol. Resour. 16, 701–713. doi: 10.1111/1755-0998.12497
Roy, S., Kumar, V., Kumar, V., and Behera, B. K. (2016). Acute phase proteins and their potential role as an indicator for fish health and in diagnosis of fish diseases. Protein Pept. Lett. 24, 78–89. doi: 10.2174/0929866524666161121142221
Scudiero, R., Trinchella, F., and Parisi, E. (2009). Iron metabolism genes in Antarctic notothenioids: a review. Mar. Genomics 1, 79–85. doi: 10.1016/j.margen.2008.12.001
Scudiero, R., Trinchella, F., Riggio, M., and Parisi, E. (2007). Structure and expression of genes involved in transport and storage of iron in red-blooded and hemoglobin-less antarctic notothenioids. Gene 397, 1–11. doi: 10.1016/j.gene.2007.03.003
Seppola, M., Mikkelsen, H., Johansen, A., Steiro, K., Myrnes, B., and Nilsen, I. W. (2015). Ultrapure LPS induces inflammatory and antibacterial responses attenuated invitro by exogenous sera in Atlantic cod and Atlantic salmon. Fish Shellfish Immunol. 44, 66–78. doi: 10.1016/j.fsi.2015.01.018
Shi, J., and Camus, A. C. (2006). Hepcidins in amphibians and fishes: antimicrobial peptides or iron-regulatory hormones? Dev. Comp. Immunol. 30, 746–755. doi: 10.1016/j.dci.2005.10.009
Shin, S. C., Ahn, D. H., Kim, S. J. C., Pyo, C. W., Lee, H., Kim, M. K., et al. (2014). The genome sequence of the Antarctic bullhead notothen reveals evolutionary adaptations to a cold environment. Genome Biol. 15:468. doi: 10.1186/s13059-014-0468-1
Skaar, E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6:e1000949. doi: 10.1371/journal.ppat.1000949
Souza, M. R. D. P., de, Herrerias, T., Zaleski, T., Forgati, M., Kandalski, P. K., Machado, C., et al. (2018). Heat stress in the heart and muscle of the Antarctic fishes Notothenia rossii and Notothenia coriiceps: carbohydrate metabolism and antioxidant defence. Biochimie 146, 43–55. doi: 10.1016/j.biochi.2017.11.010
Tort, L., Balasch, J. C., and Mackenzie, S. (2003). Fish immune system. A crossroads between innate and adaptive responses. Inmunologia 22, 277–286.
Torti, F. M., and Torti, S. V. (2002). Regulation of ferritin genes and protein. Blood 99, 3505–3516. doi: 10.1182/blood.v99.10.3505
Vulpe, C. D., Kuo, Y. M., Murphy, T. L., Cowley, L., Askwith, C., Libina, N., et al. (1999). Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat. Genet. 21, 195–199. doi: 10.1038/5979
Welker, T. L., Lim, C., Yildirim-Aksoy, M., and Klesius, P. H. (2007). Growth, immune function, and disease and stress resistance of juvenile Nile tilapia (Oreochromis niloticus) fed graded levels of bovine lactoferrin. Aquaculture 262, 156–162. doi: 10.1016/j.aquaculture.2006.09.036
Keywords: Notothenia coriiceps, Notothenia rossii, iron metabolism, Antarctic fish, nutritional immunity
Citation: Martínez DP, Sousa C, Oyarzún R, Pontigo JP, Canario AVM, Power DM, Vargas-Chacoff L and Guerreiro PM (2020) LPS Modulates the Expression of Iron-Related Immune Genes in Two Antarctic Notothenoids. Front. Physiol. 11:102. doi: 10.3389/fphys.2020.00102
Received: 26 September 2019; Accepted: 28 January 2020;
Published: 14 February 2020.
Edited by:
Ignacio Ruiz-Jarabo, University of Cádiz, SpainReviewed by:
María Jesús Delgado, Complutense University of Madrid, SpainCopyright © 2020 Martínez, Sousa, Oyarzún, Pontigo, Canario, Power, Vargas-Chacoff and Guerreiro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luis Vargas-Chacoff, bHVpcy52YXJnYXNAdWFjaC5jbA==; Pedro Miguel Guerreiro, cG1nZ0B1YWxnLnB0
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.