- State Key Laboratory of Rice Biology, Ministry of Agriculture Key Lab of Molecular Biology of Crop Pathogens and Insects, Institute of Insect Science, Zhejiang University, Hangzhou, China
Vitellogenin (Vg) is precursor of vitellin. Here, we identified a Vg (NlVg) and two Vg-likes (NlVg-like1 and NlVg-like2) in the brown planthopper, Nilaparvata lugens. Phylogenetic analyses showed that NlVg-like1 and NlVg-like2 are not clustered with the conventional insect Vgs associated with vitellogenesis. Temporo-spatial expression analyses showed that the NlVg and NlVg-like2 transcript levels increased significantly 24 h after emergence and were primarily expressed in female adults. However, NlVg-like1 was expressed during all stages, and in both genders. Tissue-specific analyses showed that all three genes were most highly expressed in the fat body. The injection of double-stranded RNA targeting NlVg showed that NlVg is essential not only for oocyte development but also for nymph development. The knockdown of NlVg-like1 in female adults resulted in failure to hatch or death before eggshell emergence in 18% of offspring embryos, suggesting that NlVg-like1 plays an important role during late embryogenesis. Approximately 65% of eggs laid by females that were treated with double-stranded RNA targeting NlVg-like2 failed to hatch, indicating that NlVg-like2 plays a role in nutrition absorption during oocyte, or embryonic development. Our results illustrate the structural and functional differences among the Vg and Vg-like genes and provide potential targets for RNA-interference-based insect pest management strategies.
Introduction
The brown planthopper (BPH), Nilaparvata lugens, a phloem-feeding hemipteran insect, has emerged as a major pest for rice plants in Asian countries. With 24–33 ovarioles per ovary, the BPH has a robust capacity for producing offspring, and each female adult can produce more than 400 offspring throughout the lifetime of them under appropriate conditions (Chen et al., 1979). The strong fecundity of the BPH represents a primary cause for the serious economic losses associated with the BPH and is regulated by ovary development and environmental conditions. A key process associated with the reproductive success of oviparous species is the biosynthesis of vitellogenin (VTG, Vg) during oogenesis and its accumulation in oocytes (Raikhel, 1992).
Vg belongs to the large lipid transfer protein (LLTP) superfamily, which includes microsomal triglyceride transfer protein (MTP), apolipoprotein (APO), and Vg. This superfamily originates from a series of duplications of Vg, and the LTTP superfamily has been hypothesized to be the oldest gene family (Avarre et al., 2007). Several studies have verified the structural and functional relationships among LLTP superfamily members, which have diverse functions. In general, Vgs generally contain a vitellogenin_N domain at the N-terminus, which is also known as the lipoprotein N-terminal domain (LPD_N); a 1943 domain (DUF1943) of unknown function; and a C-terminal von Willebrand factor type D domain (VWD). These domains can be found across an extensive range of proteins (Banaszak et al., 1991; Li et al., 2003; Roth et al., 2013). APOs contain the LPD_N, DUF1081, and VWD domains, whereas MTPs only contain the LPD_N domain. The primary yolk protein precursor family of Vgs (average of 1,300–2,000 amino acids aa long) act as the major energy sources during the embryonic period. This family contains both conventional Vgs and Vg-like protein families, including the immune-related protein family in insects (IP), a protein family of unknown function in fishes (UP), and novel hymenopteran Vg classes (Vg-like A, Vg-like B, and Vg-like C) (Morandin et al., 2014). The APO family (average of 3,200–4,400 aa long, which represent the longest sequence lengths among all LLTPs) includes proteins that are indispensable during the formation and secretion of very low–density lipoproteins, and play important roles during cholesterol and plasma triacylglycerol circulation and metabolism. The MTP family (average, 800–1,000 aa long, which represents the shortest sequence lengths among all LLTPs) transports neutral lipids between membranes in vitro and participates in the formation of nascent lipoprotein particles (Wu et al., 2013).
Vgs are large, oligomeric glycolipophosphoproteins, composed of at least 2 subunits (apoproteins), which are synthesized in the fat body (Hagedorn and Kunkel, 1979) as either single, or multiple precursors. Vgs are cleaved proteolytically into the major Vg polypeptide subunits, which are secreted into the hemolymph, where they accumulate via pinocytosis to form vitellin in the developing oocyte (Raikhel and Dhadialla, 1992). Vgs are stored as Vn in crystalline form after incorporation into oocytes and provide amino acids, lipids, fats, carbohydrates, vitamins, microelements, and other nutrients to the maturing oocyte (Tufail and Takeda, 2008). Although the fat body is the exclusive site of Vg synthesis in most insects (Valle, 1993), Vgs are also produced by the ovarian follicular epithelium in some Coleoptera, and Diptera species (Belles, 1998). More surprisingly, in some species, Vg is present not only in females but also in males (Trenczek and Engels, 1986; Piulachs et al., 2003).
The conserved structural domains of Vgs give rise to multiple functions. In addition to the primary function of egg yolk formation, Vg is associated with other functions, including the sexual division of foraging, the specialization of labor, and pleiotropic effects in the female castes of social Hymenoptera. Vg has also been associated with the regulation of hormonal dynamics, longevity, and gustatory responsiveness (Engels, 1974; Amdam and Omholt, 2003; Guidugli et al., 2005; Nelson et al., 2007; Munch and Amdam, 2010). Moreover, Vg can protect some species from oxidative stress, such as the honey bee, nematodes (Caenorhabditis elegans), and fish (Anguilla japonica) (Ando and Yanagida, 1999; Nakamura et al., 1999; Seehuus et al., 2006). In addition, Vg plays important roles during immunity in amphioxus (Zhang et al., 2005), fish (Li et al., 2008, 2009; Liu et al., 2009; Garcia et al., 2010), bay scallop (Wu et al., 2015), mud crab (Yang et al., 2016), and noble scallop (Zhang et al., 2016).
To date, Vg has been studied extensively in numerous species, including both vertebrates and invertebrates, and has been cloned from seven insect orders: Blattodea, Orthoptera, Coleoptera, Diptera, Lepidoptera, Hemiptera, and Hymenoptera. Vg copy numbers are highly variable among different species, ranging from one to several (Morandin et al., 2014). Specifically, the mosquito (Aedes aegypti), and the ant (Linepithema humile) (Corona et al., 2013) both have five copies (Romans et al., 1995), which are the largest number of Vg genes among insect species, whereas the honeybee (Apis mellifera) (Piulachs et al., 2003; Amdam et al., 2012), the ant (Camponotus floridanus) (Corona et al., 2013), and the silkworm (Bombyx mori) (Yano et al., 1994) have only a single copy, which is the lowest copy number. These copy number differences have been attributed to gene duplication events, which are thought to be a crucial source of evolutionary novelties that result in new gene expression patterns and functions (Lynch and Conery, 2000), indicating that copy number variations for Vg genes may be associated with insect adaptation and evolution (Lynch and Force, 2000; Garcia et al., 2010). More importantly, ancient duplications may result in different physiological effects among different species (Innan and Kondrashov, 2010). For example, the white perch (Morone americana) has three types of Vg proteins: VtgAa, VtgAb, and VtgC, which have different gene expression patterns in the ovary, plasma, and liver during pre-, early-, mid-, and post-vitellogenic oocyte growth, and appear to be associated with different functions. The relative levels between VtgAa and VtgAb are associated with egg buoyancy (determining whether eggs are neutrally buoyant or demersal adhesive), and the VtgC protein content may affect nutrition supplementation during early embryonic development (Schilling et al., 2015).
Due to the various gene duplication events, cleavage sites, and temporo-spatial expression patterns of Vg proteins, a new and diverse class of Vg genes has emerged in several insects over time, termed the Vg-like genes. For example, three Vg homologs, Vg-likeA, Vg-likeB, and Vg-likeC, which are orthologous to the conventional Vgs, have emerged in Hymenoptera (Morandin et al., 2014). The conserved protein domains in these genes vary and are unlikely to be associated with vitellogenesis. In particular, Vg-likeA plays an important role in determining longevity of winter bees. Vg-likeB is linked to the oxidative stress response. The function of Vg-likeC remains generally unknown, although it does not appear to be associated with anti-aging functions (Salmela et al., 2016). In the potato psyllid, Bactericera cockerelli, BcVg1-like encodes a conventional Vg, whereas BcVg6-like appears to be associated with lipid movement (a lipid transporter during metamorphosis), protection from reactive oxygen species, and immune defense (Ibanez et al., 2017, 2018). There was only one Vg-like gene in the bed bug, Cimex lectularius, and it may have acquired a novel function that differs from that of ClVg; however, the function of the ClVg-like gene remains unclear and requires further study (Moriyama et al., 2016).
Vg is not only expressed in female adults but can also be detected in other stages in many insect species, and many insects possess several Vg-like genes. However, the biological functions of Vg in nymphs and the functions of Vg-like genes in all stages remain largely unclear. In this study, we identified three Vg or Vg-like genes in N. lugens. To better understand the evolution and functions of the Vg and Vg-like genes in this phloem-feeding, destructive rice pest, the complete open reading frame (ORF) sequences of NlVg, NlVg-like1, and NlVg-like2 from N. lugens were cloned, analyzed, and compared with known insect Vgs, Vg-like genes, and other LLTP family sequences to investigate their phylogenetic relationships. Then, the three genes were characterized functionally to enable a better understanding of the roles played by these novel Vgs in the phloem-feeding species and to determine whether these genes represent potential targets for BPH management based on RNA interference (RNAi) technologies.
Materials and Methods
Insects
The N. lugens colony used for this study was obtained from local rice fields on the Huajiachi Campus of Zhejiang University, Hangzhou, China. The colony was maintained on fresh rice seedlings at 26 ± 0.5°C, with 50 ± 5% relative humidity, under a 16 h light:8 h dark photoperiod. Because ovary development is delayed in the long-winged morphs of many insects, including N. lugens, and short-winged morphs often have higher fecundity than long-winged morphs (Zera and Denno, 1997), short-winged BPHs were used in this study, unless otherwise specified.
Identification and Analyses of Vg and Vg-like Genes
The cDNA sequences of Vg and the Vg-like genes were obtained from transcriptome databases and used as query sequences against N. lugens genome databases (GenBank accession number AOSB00000000) using the National Center for Biotechnology Information (NCBI)-basic local alignment search tool (BLAST)-2.5.01 (using BLASTn, default parameters) to obtain the genome sequences.
The exon-intron organization of Vg and Vg-like genes were predicted using Splign (https://www.ncbi.nlm.nih.gov/sutils/splign/splign.cgi). The domain structures of Vg and Vg-like proteins were predicted using the Simple Modular Architectural Research Tool (SMART; http://smart.embl-heidelberg.de/) and a specialized BLAST tool for identifying conserved domains within sequences that can be found on the NCBI website (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
To investigate the evolutionary relationships among the Vg and Vg-like genes identified in the present study, the gene sequences were used to search for homologous genes in the NCBI non-redundant database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic analyses were performed using the deduced amino acid sequences of the identified Vg and Vg-like proteins and Crustacea Vgs and Crustacea APOs that were selected from different representative species among 15 orders. Multiple sequence alignments were performed using CLUSTALX (Larkin et al., 2007). Phylogenetic trees were constructed using MEGA7 (Kumar et al., 2016), using the maximum likelihood method and bootstraps with 1,000 replications.
Isolation of Total RNA and Reverse Transcription-PCR
Total RNA was isolated from whole insects at various developmental stages or from tissue samples, using a TRIzol Total RNA Isolation Kit (TaKaRa, Dalian, China) according to the manufacturer's protocol. Developmental samples were collected from eggs (every 24 h after the eggs were laid, n = 100), first-instar nymphs (every 24 h after molting, n = 80), second-instar nymphs (every 12 h after molting, n = 60), third-instar nymphs (every 12 h after molting, n = 40), fourth-instar nymphs (every 12 h after molting, n = 20), fifth-instar nymphs (every 12 h after molting, n = 15), female adults (every 12 h after molting, n = 15), and male adults (every 12 h after molting, n = 15). Similarly, tissue samples, including the abdominal integument (n = 50), gut (n = 50), fat body (n = 30), and ovaries (n = 30) were dissected from females 48–72 h after emergence; the testes (n = 40) were dissected from males 48–72 h after emergence.
The concentration of total RNA was measured using a NanoDrop 2,000/2,000 c spectrophotometer (Thermo Fisher Scientific, Bremen, Germany), and 1,000 ng RNA was used for reverse transcription (RT) in a 10-μL reaction with a ReverTra Ace qPCR RT Master Mix and genomic DNA (gDNA) Remover Kit (ToYoBo, Osaka, Japan), according to the manufacturer's recommendations. The synthesized cDNAs were used as templates to amplify the NlVg, NlVg-like1, and NlVg-like2 coding sequences, using Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Jiangsu, China) according to the manufacturer's protocol. The PCR procedure was as follows: pre-denaturation at 95°C for 3 min, 35 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 15 s, and an additional extension at 72°C for 5 min. The PCR products were cloned into a pEASY–Blunt Cloning Vector (TransGen, Beijing, China) and sequenced. Table S1 lists the primers used during the PCR amplifications.
Real-Time qPCR
To investigate the developmental and tissue-specific expression patterns of NlVg, NlVg-like1, and NlVg-like2, real-time qPCR was conducted using gene-specific primer pairs designed by Primer Premier 5 (Table S1) and the cDNA was prepared as described above. Each 20-μL reaction contained 2 μL 10-fold diluted cDNA, 0.6 μM each primer, and 10 μL SYBR Premix Ex Taq (Bio-Rad, Hercules, CA, USA), and was conducted on a CFX96 Real-Time PCR Detection System (Bio-Rad) under the following conditions: initial denaturation at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s and 60°C for 30 s. The relative RT-qPCR data were analyzed with CFX Manager 3.1 (Bio-Rad). The N. lugens 18S rRNA housekeeping gene (Nl18S) (GenBank accession number JN662398.1) was used as the reference gene. All RT-qPCR data are presented as relative mRNA expression levels; RT-qPCR was performed for each gene based on independent RNA sample preparation, and consisted of three technical replicates and three biological replicates. The relative quantitative method [2−ΔΔCt: 2−(Cttarget−CtNl18S)timex−(Cttarget−CtNl18S)time0], where timex is any time point and time 0 represents the 1 × expression of the target gene normalized to Nl18S; the Ct (cycle threshold) (Schmittgen and Livak, 2008) was used to evaluate the quantitative variation.
dsRNA Preparation
To minimize non-target silencing, a unique region of each NlVg or NlVg-like gene was chosen for the dsRNA design. The coding sequences of NlVg, NlVg-like1, NlVg-like2, and GFP (control) were cloned into the pMD-19T vector (TaKaRa). PCR-generated DNA templates were then used to synthesize the dsRNA, which contained T7 promoter sequences at each end. Table S1 shows the primers used for dsRNA synthesis. We used a MEGAscript T7 Transcription Kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions to produce the specific dsRNA of each gene. The PCR was performed using Green Taq Mix (Vazyme) in 50-μL reactions, and was as follows: pre-denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, and additional extension at 72°C for 7 min. Subsequently, dsRNA was purified using LiCl precipitation, resuspended using nuclease-free water, and quantified spectrophotometrically using a NanoDrop 2,000/2,000 c spectrophotometer (Thermo Fisher Scientific, Bremen, Germany). The dsRNA products' quality and size were verified by 2% agarose gel electrophoresis.
Effects of RNAi on N. lugens
RNAi was performed as described previously (Liu et al., 2010; Xu et al., 2015). Briefly, newly emerged adult female BPHs (within 2 h) were used for microinjections. First, the female BPHs were anesthetized with carbon dioxide for 15–20 s. Then, ~50 ng dsRNA was injected into each BPH using a FemtoJet (Eppendorf-Netheler-Hinz, Hamburg, Germany). Post-injection, the female BPHs were placed on 4–5-leaf–stage rice seedlings for 5 h, and then an equal or greater number of male BPHs were placed in the same jar. Fifty injected adult female BPHs were used for each treatment, with three replicates performed.
The efficiency of RNA silencing was evaluated by real-time quantitative PCR (RT-qPCR), 72 h after injection. The female BPHs from day 4 to day 9 after emergence were dissected. Approximately 20 females per treatment were maintained and counted daily until day 8 after injection. For dissection, the insects were anesthetized on ice and placed in a Petri dish, and then the cuticle was carefully removed, and the tissues were removed using forceps and placed in a drop of phosphate-buffered saline (PBS: 0.9% NaCl, 0.02% KCl, 10 mM Na2HPO4, 2 mM KH2PO4, [pH 7.4]). Images of the insects and of their internal reproductive systems were captured with a DFC320 digital camera (Leica, Germany) attached to a Leica S8 APO stereomicroscope, using the LAS V3.8 digital imaging system (Leica, Germany).
Western Blotting
BPH nymphs of all stages (0–12 h first, second, third, four, fifth instar) and 3-day-old females, as well as second- and fifth-instar nymphs and adult females that had been treated with dsNlVg and dsGFP in their early stages (0–12 h) were homogenized in radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific), quantified using the bicinchoninic acid (BCA) method (catalog no. 23227, Thermo Fisher Scientific), and electrophoresed via sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The optimized western blotting protocol consisted of the following steps: First, the polyvinylidene fluoride (PVDF) membrane (Merck KGaA, Darmstadt, Germany) was activated with methanol for 30 s. Beginning from the bottom up with a foam pad, the transfer box was assembled using filter paper, gel, membrane, another layer of filter paper, and foam pad. Then, protein samples were transferred to the PVDF membrane in buffer (2.8 g Tris base, 2.9 g glycine, 200 mL methanol in 800 mL ddH2O [double-distilled water]) (Thermo Fisher Scientific) at 60 V for 2 h 20 min. Next, the PVDF membrane was blocked with blocking buffer (5% skim milk powder in 1 × TTBS; 1 × TTBS; 10 × TBS [Tris-buffered saline] with 1% Tween 20) at room temperature for 1 h 30 min. The primary antibodies against NlVg and β-actin (1:10,000)prepared in our laboratory (Xu et al., 2015; Zhuo et al., 2018) were then added to the blocking buffer at room temperature for 2 h 30 min or at 4°C overnight. The PVDF membrane was washed with 1 × TTBS three times for 15 min each. Then, the secondary antibody (goat anti-mouse; HuaBio, Hangzhou, China) was added with fresh blocking buffer (0.5% skim milk powder in 1 × TTBS) at room temperature for 2 h 30 min. The PVDF membrane was washed with 1 × TBS three times for 10 min each. The chemiluminescence apparatus (Bio-Rad) was prepared 20 min in advance, and the PVDF membrane was visualized with color-developing agent (Bio-Rad); a standard protein ruler was used (catalog no. 26616, Thermo Fisher Scientific). Samples were collected from 0 to 12 h first-instar nymphs (n = 80), second-instar nymphs (n = 60), third-instar nymphs (n = 40), fourth-instar nymphs (n = 20), fifth-instar nymphs (n = 15), and 3 day-old females (n = 15). Similarly, samples treated with dsNlVg and dsGFP in their early stages (0–12 h): second-instar nymphs (n = 60), and fifth-instar nymphs (n = 15), and adult females (n = 15).
Triglyceride Assay
The triglyceride content of the BPHs was measured at 550 nm using a triglyceride assay kit (Applygen Technologies, Beijing, China). Total protein concentration was estimated using the BCA method (catalog no. 23227, Thermo Fisher Scientific) according to the manufacturer's instructions and is expressed as μmol triglyceride per μg total protein.
Statistical Analysis
Data are presented as mean ± SEM from three independent biological replicates, uNless otherwise noted. Statistical analysis was performed using GRAPHPAD PRISM 7.0 (GraphPad Software, La Jolla, CA, USA). Differences between two groups were compared using a two-tailed unpaired t-test (Tukey's multiple comparisons test for multigroups) at the significance levels of *P < 0.05, **P < 0.01, ***P < 0.001. One-way analysis of variance (ANOVA) was applied for comparing the differences among more than two samples.
Results
Sequencing and Phylogenetic Analysis
Three Vg homologs were identified from the BPH transcriptome (Xue et al., 2010) and genome (Xue et al., 2014), using the Vg amino acid sequences from A. mellifera, Ae. aegypti, and Acyrthosiphon pisum as queries, and were tentatively named NlVg (2,072 aa), NlVg-like1 (1,414 aa), and NlVg-like2 (1,987 aa) (Figure 1, Figures S1–S3). The coding sequences of the three genes were validated through cloning and sequencing (GenBank accession numbers: AB353856, MK779307, and MK779308). Comparisons among the transcriptome and genome sequences of the three genes revealed that NlVg, NlVg-like1, and NlVg-like2 contain 25, 22, and 18 exons, respectively (Figure S4). All three putative proteins contain the LPD_N and VWD domains, two large domains that are usually found in conventional Vgs and other LLTPs (Wu et al., 2013). In addition, NlVg and NlVg-like1 contain the unknown function motif 1943 (DUF1943) (Figure 1), which is also found in conventional Vgs from other insect species.
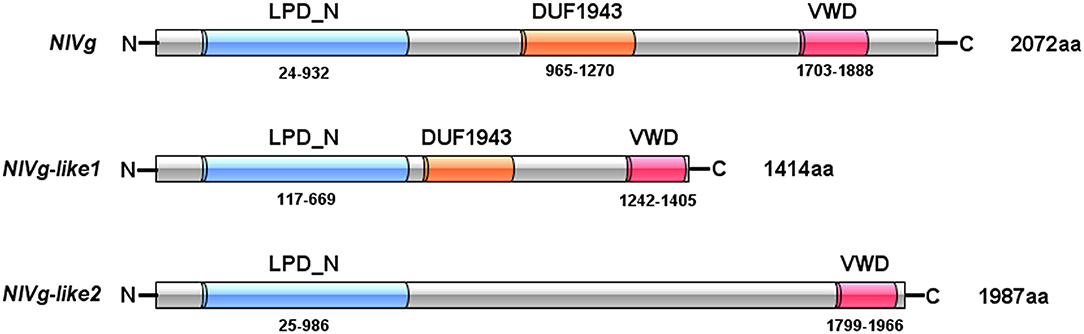
Figure 1. Domain architecture of NlVg, and NlVg-like1, and NlVg-like2. All of these proteins contain the LPD_N and VWD domains, whereas only NlVg-like2 has no predicted DUF1943.
Using the maximum likelihood method, NlVg, NlVg-like1, and NlVg-like2 were compared phylogenetically with conventional Vgs, the APO family, the MTP family, three new hymenopteran Vg classes (Vg-likeA, Vg-likeB, Vg-likeC), IP, and UP (Figure 2). NlVg appeared to belong to the conventional insect Vgs, grouping with hemipterans, whereas NlVg-like1 and NlVg-like2 were divided into different groups. Specifically, NlVg-like1 was closely related to B. cockerelli Vg-likeB and clustered with a described group termed Hemiptera Vg-likeB (Morandin et al., 2014; Ibanez et al., 2018), which is supposed to be a sister clade of hymenopteran Vg-likeB. Interestingly, NlVg-like2 was located with other Vg-like genes belonging to hemimetaboly and was placed within the clade composed of Vg-like proteins from western flower thrip (Frankliniella occidentalis), German cockroach (Blattella germanica), and two termite species (Zootermopsis nevadensis and Cleaveius secundus). Hence, these three genes diverged from the common ancestor, implying that they may have different functions.
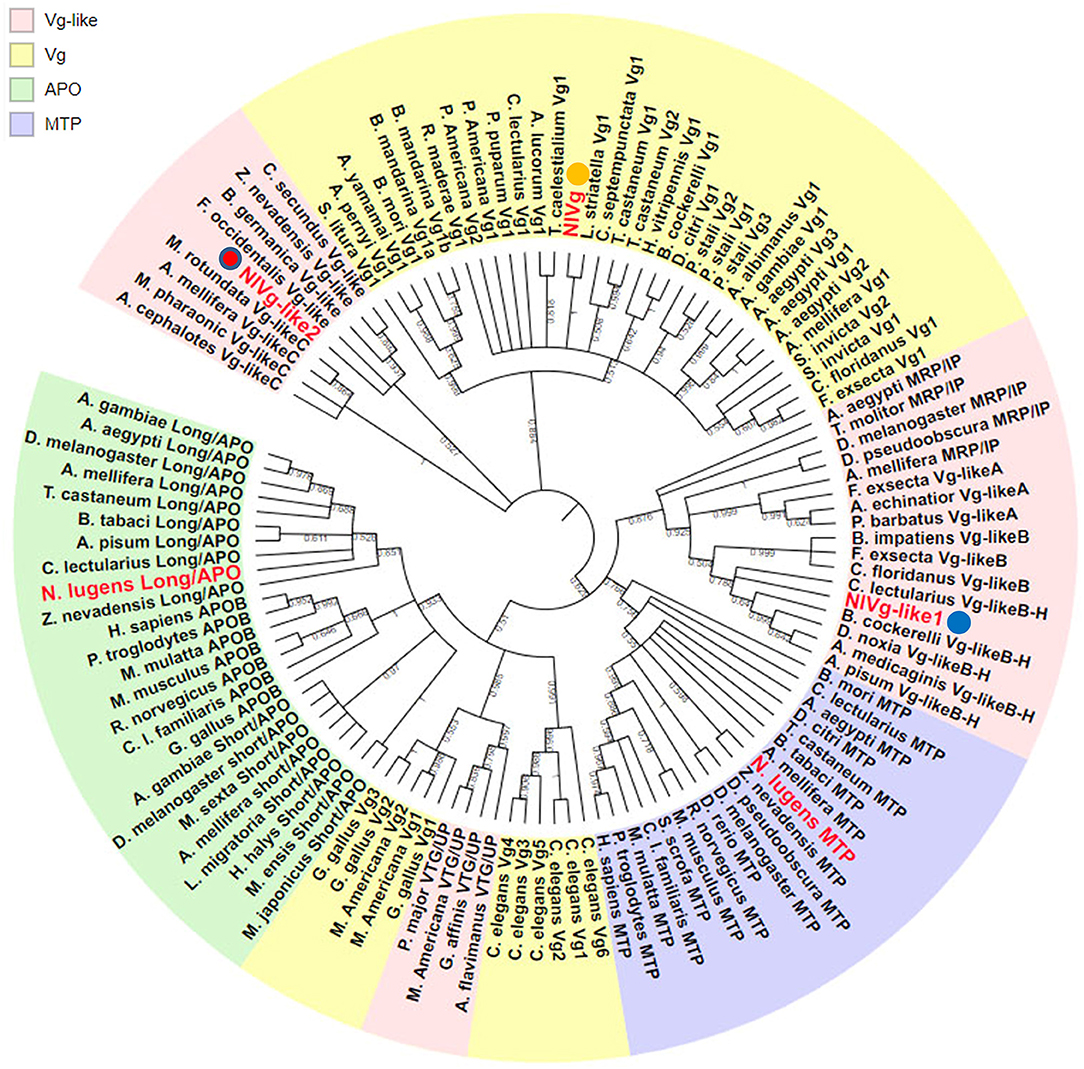
Figure 2. Phylogenetic relationship of NlVg, NlVg-like1, and NlVg-like2 with members of the LLTP superfamily. Phylogenetic analysis of NlVg and NlVg-like proteins involved other members of the LLTP superfamily, including conventional Vgs, APO family, MTP family, Vg-like A, Vg-like B, Vg-like C, IP, and UP. Cladograms show bootstrap values; only branches with ≥50% support are shown. The best-fit amino acid sequences of 119 proteins from 14 different orders in five classes were chosen for alignment and analysis. Numbers at the nodes denote posterior probabilities (support values > 50% are shown). Table S2 lists the detailed information of each protein sequence used.
Temporospatial Expression Patterns
RT-qPCR results showed that the transcription levels of both NlVg and NlVg-like2 increased significantly 24 h after emergence and remained at high levels throughout the adult stage. The two genes were expressed almost exclusively in females and were expressed at low or nearly undetectable levels in eggs and nymphs. As ovarian maturation generally occurs 3 days after eclosion, the expression patterns of NlVg and NlVg-like2 were consistent with oocyte development (Figure 3A). Western blotting also detected high levels of NlVg expression in females. Surprisingly, NlVg could also be detected in nymphs (Figure 3B). Contrary to NlVg-like2 and NlVg, which were only detected in females, NlVg-like1 transcripts were detected in both males and females, with high transcript levels that peaked at ~0 h in female adults. However, female adults had higher levels of NlVg-like1 mRNA than males (Figure 3A).
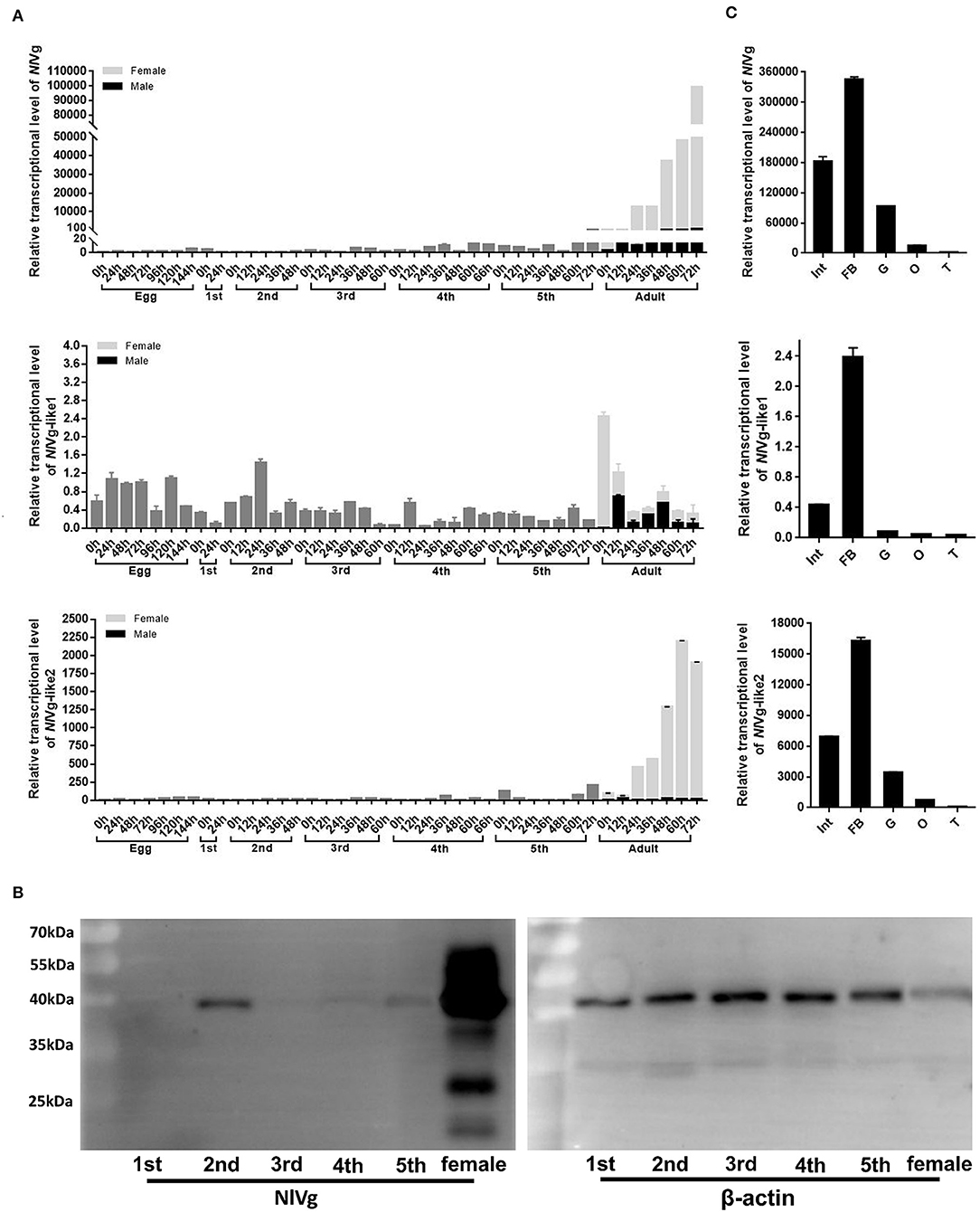
Figure 3. Temporospatial expression patterns of NlVg, NlVg-like1, and NlVg-like2. (A) Temporal expression pattern of the three genes. Samples were collected from different developmental stages of eggs, nymphs (first, second, third, fourth, fifth instar), and adults (female and male). Nl18S was used as the internal control gene. The x-axis represents the time points at which insects were collected; the y-axis represents the relative transcript level of the target gene. Results are the mean ± SEM of three biological replicates with their respective three technical replications. (B) Western blotting of NlVg in nymphs (first, second, third, fourth, fifth instar), and females. Approximately 8 μg protein from each nymph sample and 4 μg protein from each female adult sample was used. β-Actin was used as the positive control. (C) Spatial expression patterns of the three genes in BPH tissues. Total RNA was extracted from the following tissues: abdominal integument (Int, n = 50), fat body (FB, n = 30), gut (G, n = 50), and ovary (O, n = 30), which were dissected from the females 48–120 h after emergence; testes (T, n = 40) were dissected from the males. Nl18S was used as the internal control gene. Data are the relative mRNA levels of NlVg, NlVg-like1, and NlVg-like2. Results are the mean ± SEM of three biological replicates with their respective three technical replications. Different letters indicate statistical differences by the HSD-test at the 5% level of significance in each histogram.
The tissue-specific expression profiles of these three genes were analyzed by RT-qPCR in five tissues (integument, gut, fat body, ovaries and testes) (Figure 3C). Different levels of transcripts for the three genes were detected in almost all the tissues; transcription levels for all three genes were highest in the fat body, followed by those in the integument, gut, and the two reproductive organs.
Effects of NlVg, NlVg-like1, and NlVg-like2 on Reproductivity and Embryogenesis
Based on the developmental expression patterns of NlVg, NlVg-like1, and NlVg-like2, RNAi experiments were conducted on newly emerged (0–2 h) females before ovary maturation to observe the phenotypes regarding oviposition and the embryonic development of the next generation. NlVg-like1, NlVg-like2, and NlVg mRNA levels were measured at 72 h post-injection, and RNAi efficiently downregulated their expression levels by 82.8, 98.8, and 95.8%, respectively (Figure S5A). The injection of NlVg double-stranded RNA (dsRNA) led to remarkable phenotypes. The body sizes of female adults injected with NlVg dsRNAs (dsNlVg) became substantially larger at day 7 than those of females treated with dsRNAs for green fluorescent protein (dsGFP, control), with the obvious stretching of the lateral and intersegmental membranes of the abdomen. However, no obvious external morphological changes were detected in female adults that were treated with dsRNA against NlVg-like1 or NlVg-like 2 (dsNlVg-like1 or dsNlVg-like2, respectively; Figure 4A). Subsequently, we found that the dsNlVg-treated females were full of hypertrophied fat bodies that stuck to the in walls of the swollen abdomens, in contrast to the thinner abdomens containing smaller fat bodies observed for females in the other two experimental groups and the control group.
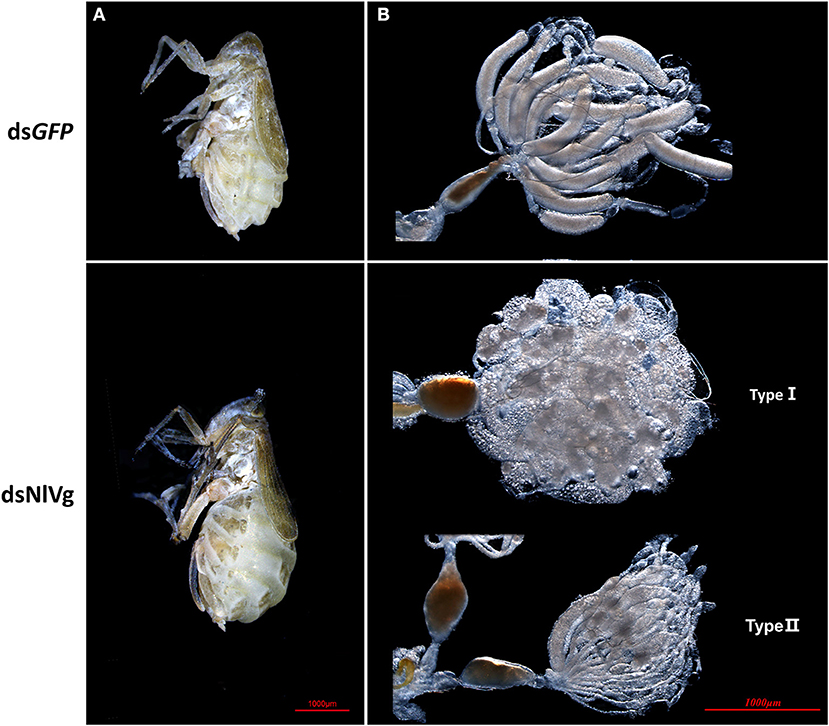
Figure 4. Knockdown of NlVg leads to oocyte malformation. (A) Female BPHs at day 7 after emergence (n = 120). dsNlVg-treated BPHs (n = 120) had larger abdomens compared to the control, with greatly stretched intersegmental membranes in the abdomen. (B) Oocytes derived from dsGFP and dsNlVg-treated female adults. At 9 days after emergence, oocytes in the dsNlVg-treated group were ball-shaped (type I) or filiform (type II) compared with the replete banana-shaped oocytes in the dsGFP-treated group.
To observe ovary development, we dissected the ovaries of virgin female adults at day 9 after dsRNA injection. No obvious oocyte malformations were observed in females that had been treated with dsNlVg-like1 and dsNlVg-like2; every ovariole contained one or two matured and replete banana-shaped oocytes. However, NlVg knockdown severely inhibited oocyte growth in the ovarioles, and the oocytes were misshapen, with irregular edges (ball-shaped or filiform), instead of banana-shaped, with smooth chorion (Figure 4B). This malformation indicated that NlVg is required for oogenesis and oocyte maturation, and nearly all dsNlVg-treated females failed to produce offspring (Figure 5A). Females that were treated with dsNlVg-like1 could oviposit normally after mating with wild-type males; however, only 82% of the laid eggs successfully hatched, compared with the 98% hatchability observed for the dsGFP-treated group (Figure 6A, Video S1). Females that were treated with dsNlVg-like2 also laid eggs normally, but the eggs had significantly decreased hatchability compared with the control group. In addition to only 31.11% of eggs hatching successfully (Figure 5B), the malformed phenotypes of the abnormal eggs varied greatly. Some embryos showed no eyespots; in other eggs, the eyespots appeared near the posterior poles of the eggs, suggesting that blastokinesis did not successfully complete (Figure 6B).
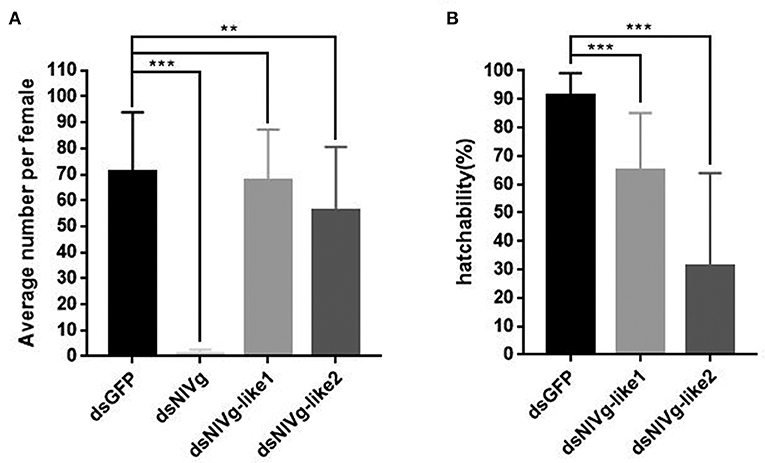
Figure 5. The fecundity (amount of eggs) per female after injection of dsNlVg, dsNlVg-like1, dsNlVg-like2, or dsGFP (n = 34). Three days after injection, females were paired with male BPHs of the same age and allowed to lay eggs for 3 days. (A) Average number of eggs per female. Eggs with eyespots after 4 days were identified as developed eggs. (B) Hatchability (%) shows the percentage of hatchlings in fertilized eggs. Results are the mean ± SEM of 34 independent experiments. **P < 0.01, ***P < 0.001 (Tukey's multiple comparisons test).
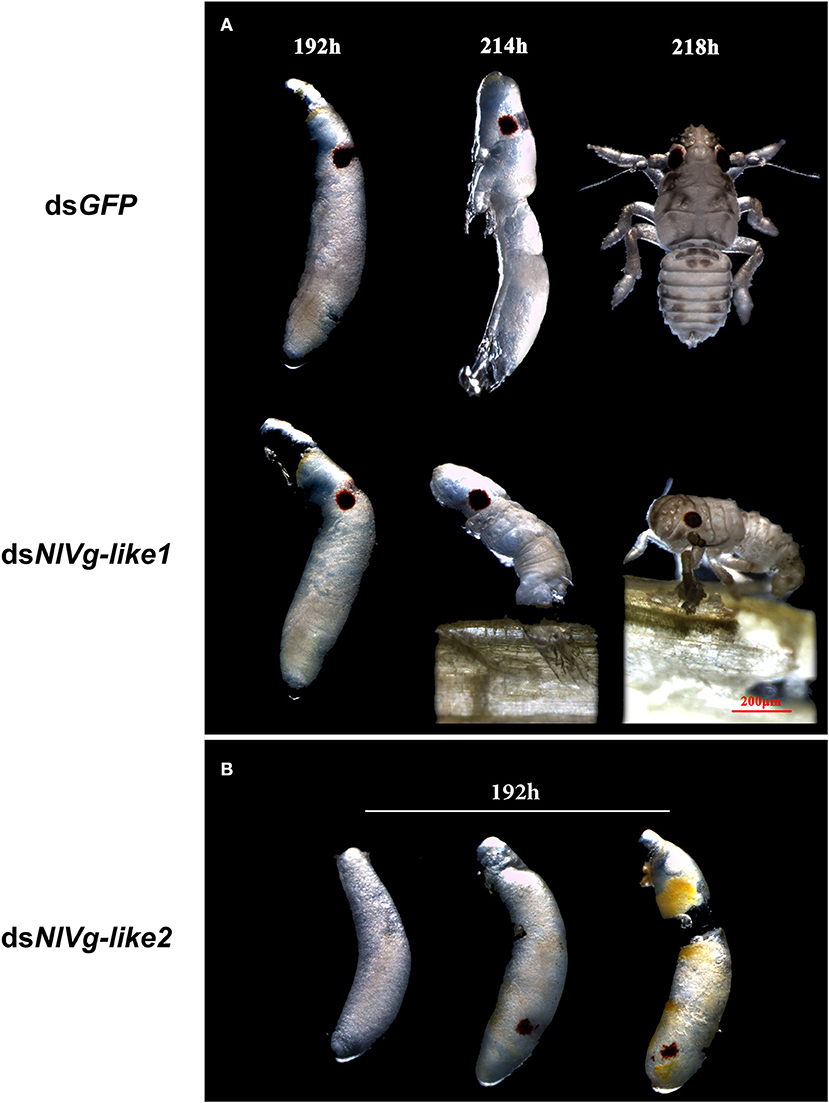
Figure 6. Embryonic development after injection of dsNlVg-like1, dsNlVg-like2, or dsGFP (observed every 24 h after the eggs were laid, n = 100). Most first-instar nymphs hatched from the shells at 214–218 h after egg formation. (A) Phenotypes of dsNlVg-like1 group (n = 2,000): 18% of hatchlings failed to exit the egg after 218 h and died before leaving the eggshell, compared to only 2% of individuals in the control that failed the hatching process. (B) Phenotypes of abnormal eggs from the dsNlVg-like2 group (n = 300). Malformed eggs arrested at 0–96 h before eyespot formation or embryo eversion, with abnormal development of most organs. Results are the mean ± SEM of 34 independent experiments.
Because NlVg-like1 was highly expressed in males, fifth-instar male BPHs (within 2 h after molting) were injected with dsNlVg-like1 to determine its functions in males. No obvious malformations were observed. When wild-type females mated with males that were treated with dsNlVg-like1, no obvious differences were observed for either fecundity or hatchability.
Effects of NlVg, NlVg-like1, and NlVg-like2 on Nymphal Development
Because reduced expression levels of NlVg, dsNlVg-like1, and dsNlVg-like2 were also detected in the nymph stages, 0–12-h second-instar nymphs (representing early-instars) and 0–12-h fifth-instar nymphs (representing late-instars) were treated with dsNlVg, dsNlVg-like1, dsNlVg-like2, and dsGFP to determine the possible functions of NlVg, NlVg-like1, and NlVg-like2 in BPH nymphs. RT-qPCR confirmed that the dsRNAs suppressed the target genes efficiently (Figure S5A). Western blotting showed that NlVg could be weakly detected in the second-instar nymphs and was completely absent from the fifth-instar nymphs and female adults following dsNlVg treatment, whereas signals could be detected in all three dsGFP-treated groups (Figure S5B). dsNlVg treatment in the nymphs caused a lethal phenotype, with thinner body shapes compared with those of the control group (Figure 7A). Moreover, the dsNlVg-treated BPHs had significantly decreased triglyceride contents compared with the control BPHs (Figure 7B), suggesting that NlVg plays a vital role during the normal growth and development of the BPH. The survival rate of treated early-instar nymphs was <30% at 10 days after injection, compared with ~95% survival rate of dsGFP-treated BPHs (Figure 8). Intriguingly, dsNlVg-like1 and dsNlVg-like2 did not lead to high mortality rates or distinct physiological malfunctions in the nymph stages (Figure 8).
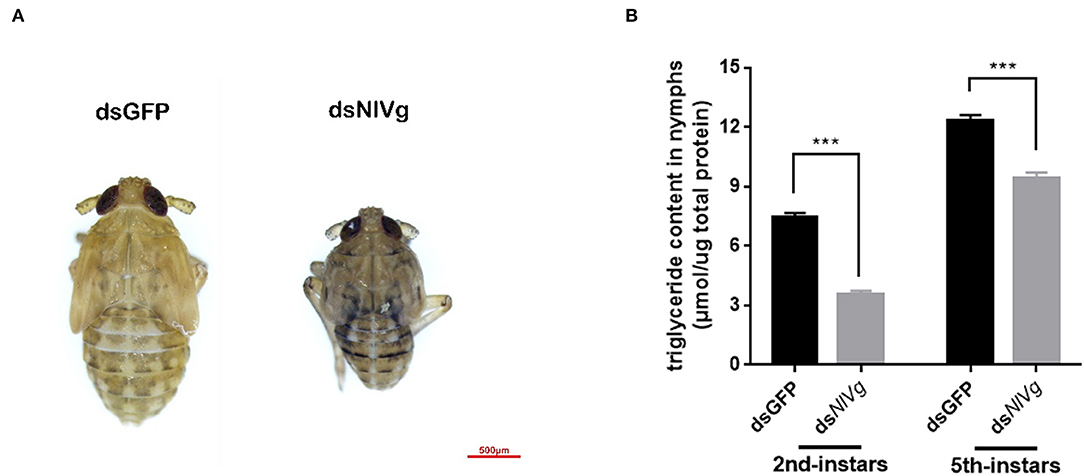
Figure 7. Effects of NlVg on nymphal development. (A) Injection of dsNlVg in nymphs (n = 100) caused a lethal phenotype, with thinner body shape compared to the control. Results are the mean ± SEM of three biological replicates with their respective three technical replications. (B) Triglyceride content (μmol triglyceride/μg total protein) in dsNlVg-treated BPHs. BPH instars (72 h after injection, n = 10) were used. dsGFP was used as the negative control. The values shown are the means ± SEM. The triglyceride content was calculated based on six biological replicates with their respective three technical replications. ***P < 0.001 (two-tailed unpaired t-test) as compared to dsGFP.
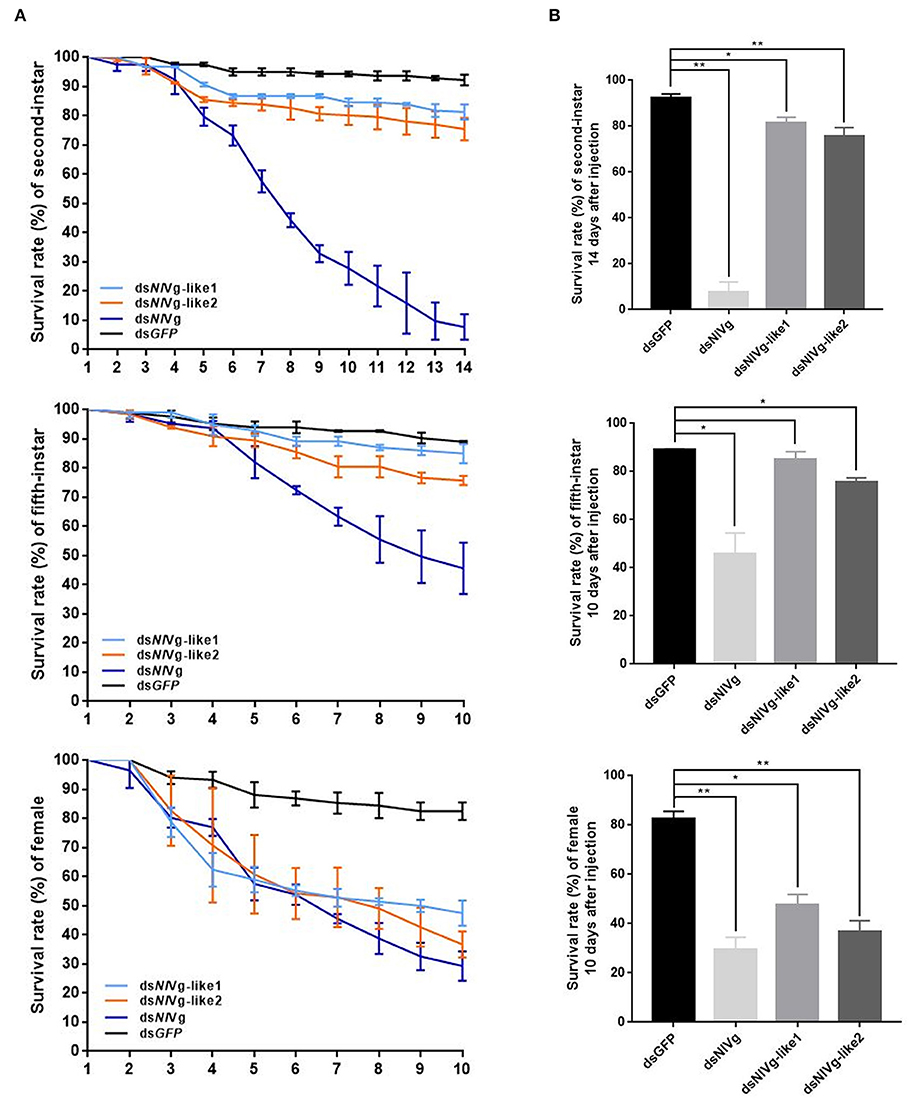
Figure 8. (A) Survival rate of BPHs 10 days after knockdown of three genes in late-instar nymphs and females (14 days after injection in early-instar nymphs). dsNlVg-like1, dsNlVg-like2, dsNlVg, or dsGFP were injected into early-stage second- and fifth-instar BPH nymphs and early-stage newly emerged female adults (0–12 h). dsGFP was used as the negative control for the non-specific effects of dsRNA; n = 100. The x-axis represents the days after injection; the y-axis represents the survival rate of the target gene. The values were calculated from three biological replicates with their respective three technical replications. (mean ± SEM). Light blue line, dsNlVg-like1; orange line, dsNlVg-like2; dark blue line, dsNlVg; black line, dsGFP. Results are the mean ± SEM of three independent experiments. (B) Survival rate 10 days after dsRNA injection in late-instar nymphs and females (14 days in early-instar). Results are the mean ± SEM of three biological replicates with their respective three technical replications. *P < 0.05, **P < 0.01 (Tukey's multiple comparisons test).
Discussion
For several decades, Vg has been extensively studied due to its variable copy numbers and multiple functions in several oviparous species, including insects. To date, more than one Vg and Vg-like transcript has been identified in hemipteran insects, such as B. cockerelli, Bemisia tabaci, Plautia stali, and Riptortus clavatus (Shinoda et al., 1996; Ibanez et al., 2018), whereas only one Vg gene has been reported in Cimex lectularius, BPH, and cicada (Tufail et al., 2010; Nagaba et al., 2011). The present study reveals that the BPH harbors more novel Vg genes than previously described, which we have named NlVg-like1 and NlVg-like2. Some insects have multiple Vg genes with highly similar sequences and conserved domains, and they all participate in vitellogenesis. However, we found that the exon–intron organizations, conserved domains, and expression profiles of these three Vg genes varied greatly, which hinted that NlVg orthologs may have evolved different functions after duplicating. For example, the DUF1943 domain, which has been identified in Vgs from many insect species, including NlVg and NlVg-like1, was not found in NlVg-like2. DUF1943 not only interacts with lipoteichoic acid (LTA) and lipopolysaccharide (LPS) found in the outer membranes of bacteria (Sun et al., 2013), but it can also enhance macrophage phagocytosis of bacteria, playing a role in the antibacterial defense of zebrafish (Du et al., 2017). Similar cases have been described for insect Vgs, such as in Apis mellifera (decreased oxidative stress), Anopheles gambiae (anti-Plasmodium response), and B. cockerelli (immunity and lipid transportation) (Seehuus et al., 2006; Rono et al., 2010; Ibanez et al., 2018). Moreover, in animals, such as amphioxus, Vgs are considered to be immunocompetent molecules that are associated with host defense (Zhang et al., 2005; Shi et al., 2006; Li et al., 2009; Liu et al., 2009; Garcia et al., 2010); therefore, we wondered whether the different domains in NlVg play different roles in their parental molecule, allowing the protein to play dual roles in immune defense, and nutritional supply.
Vg uptake is essential for ovarian development. Previous work examining Vg or VG-like genes in hemipterous insects has primarily focused on sequence and protein structural analyses of Vg, the Vg expression pattern, phylogenetic analyses of Vg, the regulation of Vg by juvenile hormone (JH) III, 20E, or ecdysteroids, and the Vg receptor (Khalil et al., 2011; Lu et al., 2015; Ibanez et al., 2017). Functional analyses using RNAi have seldom been utilized in hemipterans. Here, we observed three different phenotypes after conducting RNAi experiments in newly emerged female BPHs. In particular, NlVg downregulation suppressed ovary development and stretched the fat body-filled abdomen. The phenotypes after NlVg RNAi may result not only from reduced Vg synthesis but also from nutritional stagnation in the fat body. A similar phenotype was found in the bedbug, C. lectularius, a hemipterous insect (Moriyama et al., 2016), in which hypertrophied fat bodies, atrophied ovaries and drastically reduced egg production were observed after the downregulation of ClVg in adult females.
Although the dsNlVg-like1 group had similar reproductive abilities as the control group, egg hatchability (only 65.13%) was considerably lower in the dsGFP group (91.41%), with 17.84% of embryos failing to break away from the eggshell in the dsNlVg-like1 group, compared with 1.54% in the control group, suggesting that NlVg-like1 may be related to the later period of embryonic development. NlVg-like2 knockdown decreased the number of offspring and led to lower egg hatchability compared those for other groups. The phenotypes of the abnormal eggs showed that the downregulation of these genes may result in the insufficient accumulation of Vg during oogenesis (Zhang et al., 2005; Shi et al., 2006) or may prevent lipid uptake by the ovary, affecting the nutrition and energy accumulated in the fat body, which modulates the rate of egg development (Arrese and Soulages, 2010).
Many studies have illustrated that RNAi-mediated crop protection can be considered to be a potential strategy for insect pest control. Nowadays, with the development of genome-scale high-throughput screening and the identification of homologous genes, the use of sprayed dsRNA reagents and the integration of dsRNAs that target insect genes into transgenic plants have been verified to induce RNAi effects in targeted insects. Since 1990, lepidopteran pests have been controlled by Bacillus thuringiensis (Bt) toxin transgenic plants (Shelton et al., 2002; Naranjo, 2011). However, no Bt toxin currently exists with adequate insecticidal effects against phloem sap-sucking insects, such as planthoppers, aphids and plant bugs. Hence, novel approaches to combat these sucking pests must be considered. The most destructive insect pest for rice crops, BPHs damage rice directly by sucking plant sap, which can transmit viruses, and they have the robust capacity to produce offspring. When BPH nymphs were fed with rice plants that express dsRNAs, RNAi of targeted genes could be triggered and the gene transcription levels were suppressed, although no remarkable lethal phenotypes were observed (Zha et al., 2011), suggesting that suitable target genes should be strictly selected when designing dsRNA-transgenic plants. NlVg and NlVg-like genes may impact the strategies used by the agricultural industry to combat crop pests, such as BPH, because these genes are critical for reproduction, oocyte maturation and nymph development. The identification of these genes could be of tremendous value for future RNAi applications via the development of transgenic plants that target phloem feeders. Moreover, these genes may provide potential targets for BPH management. RNAi-mediated crop protection could not only represent a potential novel technology for pest control but could also be used as a complementary approach to other control methods. However, further intensive studies are required to improve the efficiencies of dsRNA applications mediated by oral delivery and dsRNA spraying methods to increase the feasibility of RNAi-mediated crop protection techniques.
In conclusion, NlVg is a conventional insect Vg that is involved not only in egg yolk formation and supporting embryogenesis, but also in the normal development of BPH nymphs; the NlVg-like1 gene plays an important role in late embryogenesis, ensuring that individuals hatch properly, and the NlVg-like2 gene may play roles in oocyte nutrition absorption and in embryonic development. Nevertheless, the specific mechanism behind the NlVg-like1 and NlVg-like2 genes in development requires further research. These results suggest the potential of NlVg and NlVg-like genes as the promising target for RNAi-based population management of the BPH.
Data Availability
The datasets for this manuscript are not publicly available because the genetic sequences studied in this manuscript have been submitted to GenBank and will be released to the public database as soon as they are processed. Requests to access the datasets should be directed to C-XZ, Y2h4emhhbmdAemp1LmVkdS5jbg==.
Author Contributions
C-XZ and YS designed the experiments. YS carried out the main experiments. Y-ZC helped to perform the experiments. All authors contributed to critical analysis. YS and Y-HL analyzed experimental results. YS and C-XZ wrote the manuscript and all authors approved the final manuscript for publication.
Funding
This work was supported by the National Natural Science Foundation of China (31630057 and 31471765).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.01181/full#supplementary-material
Figure S1. Nucleotide and deduced amino acid sequence of the NlVg coding region. Blue, the location of primers designed for RT-qPCR; red, the location of primers designed for dsRNA synthesis; yellow frame, the position of lipoprotein N-terminal domain (LPD_N) domain; blue frame, unknown function motif 1943 (DUF1943) domain; greed frame, Von Willebrand factor type D domain (VWD); red frame, the site of the promoter or terminator.
Figure S2. Nucleotide and deduced amino acid sequence of the NlVg-like1 coding region. Blue, the location of primers designed for RT-qPCR; red, the location of primers designed for dsRNA synthesis; yellow frame, the position of lipoprotein N-terminal domain (LPD_N) domain; blue frame, unknown function motif 1943 (DUF1943) domain; greed frame, Von Willebrand factor type D domain (VWD); red frame, the site of the promoter or terminator.
Figure S3. Nucleotide and deduced amino acid sequence of the NlVg coding region. Blue, the location of primers designed for RT-qPCR; red, the location of primers designed for dsRNA synthesis; yellow frame, the position of lipoprotein N-terminal domain (LPD_N) domain; blue frame, unknown function motif 1943 (DUF1943) domain; greed frame, Von Willebrand factor type D domain (VWD); red frame, the site of the promoter or terminator.
Figure S4. Exon–intron organization in 3 Nilaparvata lugens vitellogenin (NlVg) and vitellogenin-like genes (NlVg-like1 and NlVg-like2).
Figure S5. Effection of RNA interference. (A) The values effect of RNA interference using the dsRNA molecules targeting the specific regions of NlVg, NlVg-like1, and NlVg-like2. BPHs were treated dsRNAs at early-instar (second-instar), late-instar (fifth-instar) nymphs and female; n = 100. Real-time qPCR analysis of dsRNA-treated insects showed that the target gene transcripts were inhibited effectively 3 days after injection (n = 10). Double-stranded RNA for green fluorescent protein were injected in the control group. N. lugens 18S rRNA was used as the internal control genes. Mean ± SEM from three biological replicates with their respective three technical replications. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed unpaired t-test). (B) Western blotting analysis of NlVg after dsNlVg or dsGFP RNAi treatment at the early-instar (2nd-instar), late-instar (5th-instar) nymphs and females. ~8 μg of protein from each nymph sample, and 4 μg of protein from female adult sample was used. β-actin was used as the positive control.
Table S1. Primers used in this work.
Table S2. Sequences used in the phylogenetic analysis.
Video S1. The hatching process of offspring in dsNlVg-like1-treated group.
References
Amdam, G. V., Fennern, E., and Havukainen, H. (2012). “Vitellogenin in honey bee behavior and lifespan,” in Honeybee Neurobiology and Behavior, eds D. E. C. Giovanni Galizia, and M. Giurfa (Dordrecht: Springer), 17–29. doi: 10.1007/978-94-007-2099-2_2
Amdam, G. V., and Omholt, S. W. (2003). The hive bee to forager transition in honeybee colonies: the double repressor hypothesis. J. Theor. Biol. 223, 451–464. doi: 10.1016/S0022-5193(03)00121-8
Ando, S., and Yanagida, K. (1999). Susceptibility to oxidation of copper-induced plasma lipoproteins from Japanese eel: protective effect of vitellogenin on the oxidation of very low density lipoprotein. Compar. Biochem. Physiol. C Toxicol. Pharmacol. 123, 1–7. doi: 10.1016/S0742-8413(98)10137-8
Arrese, E. L., and Soulages, J. L. (2010). Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 55, 207–225. doi: 10.1146/annurev-ento-112408-085356
Avarre, J. C., Lubzens, E., and Babin, P. J. (2007). Apolipocrustacein, formerly vitellogenin, is the major egg yolk precursor protein in decapod crustaceans and is homologous to insect apolipophorin II/I and vertebrate apolipoprotein B. BMC Evol. Biol. 7:3. doi: 10.1186/1471-2148-7-3
Banaszak, L., Sharrock, W., and Timmins, P. (1991). Structure and function of a lipoprotein - lipovitellin. Annu. Rev. Biophys. Biophys. Chem. 20, 221–246. doi: 10.1146/annurev.bb.20.060191.001253
Belles, X. (1998). “Endocrine effectors in insect vitellogenesis,” in Recent Advances in Arthropod Endocrinology, ed. G. M. Coast, Webster. (Cambridge: Cambridge University, 71–90.
Chen, J. C., Cheng, S. N., Yan, L. M., and Yin, H. T. (1979). Ovarial development of the brown planthopper (Nilaparvata-Lugens Stal) and its relation to migration. Acta Entomol. Sinica 22, 280–288.
Corona, M., Libbrecht, R., Wurm, Y., Riba-Grognuz, O., Studer, R. A., and Keller, L. (2013). Vitellogenin underwent subfunctionalization to acquire caste and behavioral specific expression in the harvester ant pogonomyrmex barbatus. PLoS Genet. 9:e1003730. doi: 10.1371/journal.pgen.1003730
Du, X. Y., Wang, X., Wang, S., Zhou, Y., Zhang, Y., and Zhang, S. C. (2017). Functional characterization of Vitellogenin_N domain, domain of unknown function 1943, and von Willebrand factor type D domain in vitellogenin of the non-bilaterian coral Euphyllia ancora: Implications for emergence of immune activity of vitellogenin in basal metazoan. Dev. Comp. Immunol. 67, 485–494. doi: 10.1016/j.dci.2016.10.006
Engels, W. (1974). Occurrence and significance of vitellogenins in female castes of social hymenoptera. Am. Zool. 14, 1229–1237. doi: 10.1093/icb/14.4.1229
Garcia, J., Munro, E. S., Monte, M. M., Fourrier, M. C. S., Whitelaw, J., Smail, D. A., et al. (2010). Atlantic salmon (Salmo salar L.) serum vitellogenin neutralises infectivity of infectious pancreatic necrosis virus (IPNV). Fish Shellfish Immunol. 29, 293–297. doi: 10.1016/j.fsi.2010.04.010
Guidugli, K. R., Nascimento, A. M., Amdam, G. V., Barchuk, A. R., Omholt, S., Simoes, Z. L. P., et al. (2005). Vitellogenin regulates hormonal dynamics in the worker caste of a eusocial insect. FEBS Lett. 579, 4961–4965. doi: 10.1016/j.febslet.2005.07.085
Hagedorn, H. H., and Kunkel, J. G. (1979). Vitellogenin and vitellin in insects. Annu. Rev. Entomol. 24, 475–505. doi: 10.1146/annurev.en.24.010179.002355
Ibanez, F., Levy, J., and Tamborindeguy, C. (2017). Identification and expression analyses of vitellogenin in Bactericera cockerelli (Sulc). J. Insect Physiol. 98, 205–213. doi: 10.1016/j.jinsphys.2016.11.009
Ibanez, F., Tang, X., and Tamborindeguy, C. (2018). Bactericera cockerelli vitellogenin-6 like, a vitellogenin without a direct reproductive function? Insect Mol. Biol. 27, 166–176. doi: 10.1111/imb.12361
Innan, H., and Kondrashov, F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genetics 11, 97–108. doi: 10.1038/nrg2689
Khalil, S. M., Donohue, K. V., Thompson, D. M., Jeffers, L. A., Ananthapadmanaban, U., Sonenshine, D. E., et al. (2011). Full-length sequence, regulation and developmental studies of a second vitellogenin gene from the American dog tick, dermacentor variabilis. J. Insect Physiol. 57, 400–408. doi: 10.1016/j.jinsphys.2010.12.008
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, A. K., Sadasivam, M., and Ding, J. L. (2003). Receptor-ligand interaction between vitellogenin receptor (VtgR) and vitellogenin (Vtg), implications on low density lipoprotein receptor and apolipoprotein B/E - The first three ligand-binding repeats of VtgR interact with the amino-terminal region on Vtg. J. Biol. Chem. 278, 2799–2806. doi: 10.1074/jbc.M205067200
Li, Z. J., Zhang, S. C., and Liu, Q. H. (2008). Vitellogenin functions as a multivalent pattern recognition receptor with an opsonic activity. PLoS ONE 3:e1940. doi: 10.1371/journal.pone.0001940
Li, Z. J., Zhang, S. C., Zhang, J., Liu, M., and Liu, Z. H. (2009). Vitellogenin is a cidal factor capable of killing bacteria via interaction with lipopolysaccharide and lipoteichoic acid. Mol. Immunol. 46, 3232–3239. doi: 10.1016/j.molimm.2009.08.006
Liu, Q. H., Zhang, S. C., Li, Z. J., and Gao, C. R. (2009). Characterization of a pattern recognition molecule vitellogenin from carp (Cyprinus carpio). Immunobiology 214, 257–267. doi: 10.1016/j.imbio.2008.10.003
Liu, S., Ding, Z., Zhang, C., Yang, B., and Liu, Z. (2010). Gene knockdown by intro-thoracic injection of double-stranded RNA in the brown planthopper, Nilaparvata lugens. Insect Biochem. Mol. Biol. 40, 666–671. doi: 10.1016/j.ibmb.2010.06.007
Lu, K., Shu, Y., Zhou, J., Zhang, X., Zhang, X., Chen, M., et al. (2015). Molecular characterization and RNA interference analysis of vitellogenin receptor from Nilaparvata lugens (Stal). J. Insect Physiol. 73, 20–29. doi: 10.1016/j.jinsphys.2015.01.007
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Lynch, M., and Force, A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473.
Morandin, C., Havukainen, H., Kulmuni, J., Dhaygude, K., Trontti, K., and Helantera, H. (2014). Not only for egg yolk-functional and evolutionary insights from expression, selection, and structural analyses of formica ant vitellogenins. Mol. Biol. Evol. 31, 2181–2193. doi: 10.1093/molbev/msu171
Moriyama, M., Hosokawa, T., Tanahashi, M., Nikoh, N., and Fukatsu, T. (2016). Suppression of bedbug's reproduction by RNA interference of vitellogenin. PLoS ONE 11:0153984. doi: 10.1371/journal.pone.0153984
Munch, D., and Amdam, G. V. (2010). The curious case of aging plasticity in honey bees. FEBS Lett. 584, 2496–2503. doi: 10.1016/j.febslet.2010.04.007
Nagaba, Y., Tufail, M., Inui, H., and Takeda, M. (2011). Hormonal regulation and effects of four environmental pollutants on vitellogenin gene transcription in the giant water bFfigug, Lethocerus deyrollei (Hemiptera: Belostomatidae). J. Insect Conserv. 15, 421–431. doi: 10.1007/s10841-010-9315-1
Nakamura, A., Yasuda, K., Adachi, H., Sakurai, Y., Ishii, N., and Goto, S. (1999). Vitellogenin-6 is a major carbonylated protein in aged nematode, Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 264, 580–583. doi: 10.1006/bbrc.1999.1549
Naranjo, S. E. (2011). Impacts of Bt transgenic cotton on integrated pest management. J. Agric. Food Chem. 59, 5842–5851. doi: 10.1021/jf102939c
Nelson, C. M., Ihle, K. E., Fondrk, M. K., Page, R. E., and Amdam, G. V. (2007). The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 5, 673–677. doi: 10.1371/journal.pbio.0050062
Piulachs, M. D., Guidugli, K. R., Barchuk, A. R., Cruz, J., Simoes, Z. L. P., and Belles, X. (2003). The vitellogenin of the honey bee, Apis mellifera: structural analysis of the cDNA and expression studies. Insect Biochem. Mol. Biol. 33, 459–465. doi: 10.1016/S0965-1748(03)00021-3
Raikhel, A. S. (1992). “Vitellogenesis in Mosquitoes,” in Advances in Disease Vector Research, ed F. H. Kerry (New York, NY: Springer), 1–39. doi: 10.1007/978-1-4612-2910-0_1
Raikhel, A. S., and Dhadialla, T. S. (1992). Accumulation of yolk proteins in insect oocytes. Annu. Rev. Entomol. 37, 217–251. doi: 10.1146/annurev.en.37.010192.001245
Romans, P., Tu, Z. J., Ke, Z. X., and Hagedorn, H. H. (1995). Analysis of a vitellogenin gene of the mosquito, aedes-aegypti and comparisons to vitellogenins from other organisms. Insect Biochem. Mol. Biol. 25, 939–958. doi: 10.1016/0965-1748(95)00037-V
Rono, M. K., Whitten, M. M. A., Oulad-Abdelghani, M., Levashina, E. A., and Marois, E. (2010). The major yolk protein vitellogenin interferes with the anti-plasmodium response in the malaria mosquito anopheles gambiae. PLoS Biol. 8:e1000434. doi: 10.1371/journal.pbio.1000434
Roth, Z., Weil, S., Aflalo, E. D., Manor, R., Sagi, A., and Khalaila, I. (2013). Identification of receptor-interacting regions of vitellogenin within evolutionarily conserved beta-sheet structures by using a peptide array. Chembiochem 14, 1116–1122. doi: 10.1002/cbic.201300152
Salmela, H., Stark, T., Stucki, D., Fuchs, S., Freitak, D., Dey, A., et al. (2016). Ancient duplications have led to functional divergence of vitellogenin-like genes potentially involved in inflammation and oxidative stress in honey bees. Genome Biol. Evol. 8, 495–506. doi: 10.1093/gbe/evw014
Schilling, J., Loziuk, P. L., Muddiman, D. C., Daniels, H. V., and Reading, B. J. (2015). Mechanisms of egg yolk formation and implications on early life history of white perch (Morone americana). PLoS ONE 10:e0143225. doi: 10.1371/journal.pone.0143225
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Seehuus, S. C., Norberg, K., Gimsa, U., Krekling, T., and Amdam, G. V. (2006). Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 103, 962–967. doi: 10.1073/pnas.0502681103
Shelton, A. M., Zhao, J. Z., and Roush, R. T. (2002). Economic, ecological, food safety, and social consequences of the deployment of bt transgenic plants. Annu. Rev. Entomol. 47, 845–881. doi: 10.1146/annurev.ento.47.091201.145309
Shi, X. D., Zhang, S. C., and Pang, Q. X. (2006). Vitellogenin is a novel player in defense reactions. Fish Shellfish Immunol. 20, 769–772. doi: 10.1016/j.fsi.2005.09.005
Shinoda, T., Miura, K., Taylor, D., and Chinzei, Y. (1996). Vitellogenins and vitellins in the bean bug, Riptortus clavatus (Hemiptera: Alydidae): purification, immunological identification, and induction by juvenile hormone. Arch. Insect Biochem. Physiol. 31, 395–412. doi: 10.1002/(SICI)1520-6327(1996)31:4<395::AID-ARCH3>3.0.CO;2-V
Sun, C., Hu, L. L., Liu, S. S., Gao, Z., and Zhang, S. C. (2013). Functional analysis of domain of unknown function (DUF) 1943, DUF1944 and von Willebrand factor type D domain (VWD) in vitellogenin2 in zebrafish. Dev. Comp. Immunol. 41, 469–476. doi: 10.1016/j.dci.2013.07.005
Trenczek, T., and Engels, W. (1986). Occurrence of vitellogenin in drone honeybees (Apis-Mellifica). Int. J. Invert. Reprod. Develop. 10, 307–311. doi: 10.1080/01688170.1986.10510254
Tufail, M., Naeemullah, M., Elmogy, M., Sharma, P. N., Takeda, M., and Nakamura, C. (2010). Molecular cloning, transcriptional regulation, and differential expression profiling of vitellogenin in two wing-morphs of the brown planthopper, Nilaparvata lugens Stal (Hemiptera: Delphacidae). Insect Mol. Biol. 19, 787–798. doi: 10.1111/j.1365-2583.2010.01035.x
Tufail, M., and Takeda, M. (2008). Molecular characteristics of insect vitellogenins. J. Insect Physiol. 54, 1447–1458. doi: 10.1016/j.jinsphys.2008.08.007
Valle, D. (1993). Vitellogenesis in insects and other groups - a review. Mem. Inst. Oswaldo Cruz 88, 1–26. doi: 10.1590/S0074-02761993000100005
Wu, B., Liu, Z. H., Zhou, L. Q., Ji, G. D., and Yang, A. G. (2015). Molecular cloning, expression, purification and characterization of vitellogenin in scallop Patinopecten yessoensis with special emphasis on its antibacterial activity. Dev. Comp. Immunol. 49, 249–258. doi: 10.1016/j.dci.2014.12.004
Wu, L. T., Hui, J. H. L., and Chu, K. H. (2013). Origin and evolution of yolk proteins: expansion and functional diversification of large lipid transfer protein superfamily. Biol. Reprod. 88:102. doi: 10.1095/biolreprod.112.104752
Xu, H. J., Xue, J., Lu, B., Zhang, X. C., Zhuo, J. C., He, S. F., et al. (2015). Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519, 464–467. doi: 10.1038/nature14286
Xue, J., Zhou, X., Zhang, C. X., Yu, L. L., Fan, H. W., Wang, Z., et al. (2014). Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 15:521. doi: 10.1186/s13059-014-0521-0
Xue, J. A., Bao, Y. Y., Li, B. L., Cheng, Y. B., Peng, Z. Y., Liu, H., et al. (2010). Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS ONE 5:0014233. doi: 10.1371/journal.pone.0014233
Yang, Y. N., Zheng, B. Q., Bao, C. C., Huang, H. Y., and Ye, H. H. (2016). Vitellogenin2: spermatozoon specificity and immunoprotection in mud crabs. Reproduction 152, 235–243. doi: 10.1530/REP-16-0188
Yano, K., Sakurai, M. T., Watabe, S., Izumi, S., and Tomino, S. (1994). Structure and expression of messenger-rna for vitellogenin in bombyx-mori. Biochim. Biophys. Acta Gene Struct. Expres. 1218, 1–10. doi: 10.1016/0167-4781(94)90094-9
Zera, A. J., and Denno, R. F. (1997). Physiology and ecology of dispersal polymorphism in insects. Annu. Rev. Entomol. 42, 207–230. doi: 10.1146/annurev.ento.42.1.207
Zha, W., Peng, X., Chen, R., Du, B., Zhu, L., and He, G. (2011). Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS ONE 6:e20504. doi: 10.1371/journal.pone.0020504
Zhang, Q., Lu, Y. Q., Zheng, H. P., Liu, H. L., and Li, S. K. (2016). Differential immune response of vitellogenin gene to Vibrio anguillarum in noble scallop Chlamys nobilis and its correlation with total carotenoid content. Fish Shellfish Immunol. 50, 11–15. doi: 10.1016/j.fsi.2016.01.001
Zhang, S. C., Sun, Y. N., Pang, Q. X., and Shi, X. D. (2005). Hemagglutinating and antibacterial activities of vitellogenin. Fish Shellfish Immunol. 19, 93–95. doi: 10.1016/j.fsi.2004.10.008
Keywords: Nilaparvata lugens, vitellogenin, vitellogenin-like, nymph, embryogenesis
Citation: Shen Y, Chen Y-Z, Lou Y-H and Zhang C-X (2019) Vitellogenin and Vitellogenin-Like Genes in the Brown Planthopper. Front. Physiol. 10:1181. doi: 10.3389/fphys.2019.01181
Received: 14 April 2019; Accepted: 02 September 2019;
Published: 18 September 2019.
Edited by:
Guy Smagghe, Ghent University, BelgiumReviewed by:
Kai Lu, Fujian Agriculture and Forestry University, ChinaLiang Sun, Tea Research Institute, Chinese Academy of Agricultural Sciences, China
Copyright © 2019 Shen, Chen, Lou and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan-Xi Zhang, Y2h4emhhbmdAemp1LmVkdS5jbg==
 Yan Shen
Yan Shen Yuan-Zhi Chen
Yuan-Zhi Chen Yi-Han Lou
Yi-Han Lou Chuan-Xi Zhang
Chuan-Xi Zhang