
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol. , 22 August 2019
Sec. Invertebrate Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fphys.2019.01093
This article is part of the Research Topic Drosophila: A Versatile Model for Molecular, Physiological and Behavioral Studies View all 16 articles
 Yuri Prozzillo1,2
Yuri Prozzillo1,2 Francesca Delle Monache2
Francesca Delle Monache2 Diego Ferreri1,2
Diego Ferreri1,2 Stefano Cuticone1,2
Stefano Cuticone1,2 Patrizio Dimitri1,2*
Patrizio Dimitri1,2* Giovanni Messina1,2*
Giovanni Messina1,2*The Drosophila Yeti gene (CG40218) was originally identified by recessive lethal mutation and subsequently mapped to the deep pericentromeric heterochromatin of chromosome 2. Functional studies have shown that Yeti encodes a 241 amino acid protein called YETI belonging to the evolutionarily conserved family of Bucentaur (BCNT) proteins and exhibiting a widespread distribution in animals and plants. Later studies have demonstrated that YETI protein: (i) is able to bind both subunits of the microtubule-based motor kinesin-I; (ii) is required for proper chromosome organization in both mitosis and meiosis divisions; and more recently (iii) is a new subunit of dTip60 chromatin remodeling complex. To date, other functions of YETI counterparts in chicken (CENtromere Protein 29, CENP-29), mouse (Cranio Protein 27, CP27), zebrafish and human (CranioFacial Development Protein 1, CFDP1) have been reported in literature, but the fully understanding of the multifaceted molecular function of this protein family remains still unclear. In this review we comprehensively highlight recent work and provide a more extensive hypothesis suggesting a broader range of YETI protein functions in different cellular processes.
The eukaryotic genome is packaged into a highly condensed structure known as chromatin allowing cells to organize, compact and stabilize the genome into the nucleus. The first step of this process is achieved through nucleosome assembly which DNA into an 11 nm fiber that represents an approximate 6-fold level of compaction (Luger et al., 2012). Nucleosomes are octamers consisting of H2A/H2B and H3/H4 dimers which wrap around 147 bp of DNA. To gain access to DNA for replication, transcription and repair, nucleosomes must be shifted in different positions, removed or newly loaded to the DNA.
ATP-dependent protein complexes are the “molecular machines” having the task to specifically arrange the nucleosomal state. Thus far, all ATP-dependent chromatin-remodeling complexes identified contain a catalytic subunit which is part of the SWI2/SNF2 superfamily of ATPases and utilize a Swi2/Snf2-type ATPase domain consisting of an ATP-binding domain (DExx-domain), together with a helicase domain (HELICc-domain) (Clapier et al., 2017).
Four different classes (or families) of chromatin-remodeling complexes can be identified within chromatin remodeler ATPases superfamily: SWI/SNF, ISWI, CHD, and INO80 (Clapier et al., 2017). This classification is based on the presence of additional motifs outside the ATPase region. SWI/SNF members contain a bromodomain, ISWI members contain a SANT domain, while a
chromo- and a DNA-binding domain is found in CHD members. The INO80 class members do not contain any of these domains; instead, the ATPase domain is splitted into two segments (Lessard and Crabtree, 2010).
The process of chromatin remodeling generally refers to different changes in DNA–histone interaction within nucleosomes using the energy from ATP hydrolysis (Becker and Horz, 2002). They include repositioning of histone octamers in cis (along the same DNA template molecule) and in trans (from one DNA template molecule to another one), the loss of superhelical torsion of nucleosomal DNA, and the increase in accessibility to nucleosomal DNA for downstream processes as transcription. Interestingly, recent studies have shown that ATP-dependent chromatin remodeling may allow to change the histone composition of a nucleosome (Clapier and Cairns, 2009). Indeed, the yeast SWR1 complex, a member of the INO80 family, associates with Htz1, the homolog of mammalian histone variant H2A.Z. This complex can drive the ATP-dependent replacement of H2A–H2B with Htz1–H2B dimers (Krogan et al., 2003; Kobor et al., 2004; Mizuguchi et al., 2004; van Attikum and Gasser, 2005) by Swc1 ATPase subunit. In vivo, SWR1 catalyzes the incorporation of Htz1 into chromatin, which prevents the spreading of heterochromatin regions into regions of euchromatin (van Attikum and Gasser, 2005).
The human (SRCAP and P400/Tip60) and Drosophila (dTip60) orthologous chromatin remodeling complexes include proteins with high similarity to the subunits of two distinct chromatin-modifying complexes, the yeast NuA4 complex (Wang et al., 2018) harboring HAT (Histone Acetyl-transferase) activity, and the SWR1 ATP-dependent chromatin remodeling complex which catalyzes histones exchange (Table 1).
The dTip60 complex is made up by 14 core subunits (BAP55, dGAS41, dPontin, dReptin, Nipped-A, e(Pc), dYL1, dDMAP, Act87B, dMrg15, dMrgBP, dTRA1, dIng3, and dEaf6) and was found to be required for the replacement of acetylated phospho-H2A.V by unmodified H2A.V via Domino (Dom) ATPase (Kusch et al., 2004; March-Diaz and Reyes, 2009). In Drosophila, H2A.V is the only H2A variant and corresponds to mammalian H2A.X and H2A.Z (Baldi and Becker, 2013). Similarly to H2A.X, H2A.V is phosphorylated upon DNA double strand breaks to mark the DNA lesions, stimulate DNA repair machinery, and promote an easily accessible DNA conformation (Kusch et al., 2004).
The human P400/Tip60 complex highly resembles the dTip60 complex and contains the p400 protein, which also shows high sequence similarity to SWR1, DOMINO and SRCAP (Eissenberg et al., 2005; Table 1). Recent studies have demonstrated that several components of the P400/Tip60 complex are amplified or overexpressed in human neuroblastoma, glioblastoma, and colorectal cancer, while loss of function is mostly lethal, leading to cell growth arrest or cell death as well as genome instability (Yamada, 2012).
The YETI protein was found to be a new subunit of Drosophila Tip60 remodeling complex (Messina et al., 2014), arousing more and more curiosity because of a growing number of biological functions that it is supposed to perform.
Here, we comprehensively review emerging findings on YETI and its orthologous proteins in other species highlighting the broad-range involvement of these proteins in many biological processes.
Yeti, previously called CG40218, is an essential 1717-bp-long gene located in the heterochromatin of chromosome 2 of Drosophila melanogaster required for proper chromosome organization in both mitosis and meiosis (Hilliker, 1976; Corradini et al., 2003; Dimitri et al., 2003; Messina et al., 2014; Marsano et al., 2019). The evolutionary origin of Yeti gene is rather peculiar as such it has evolved from a euchromatic ancestor in drosophilids. The Yeti locus maps on euchromatin in two distantly related species, D. pseudoobscura and D. virilis indicating that over time (about 40 million of years, which is the estimated divergence time between D. melanogaster and D. virilis) it progressively moved closer to the heterochromatic pericentromeric regions (Moschetti et al., 2014; Caizzi et al., 2016). Interestingly, Yeti gene retained its original and robust organization in all analyzed species, with a short genomic region carrying a single short intron.
It is known that genes expressed at low levels harbor substantially shorter introns than those are expressed at high levels (Castillo-Davis et al., 2002). Therefore, despite of the genomic re-location, Yeti may have been under selective pressure maintaining a short size and ensuring high and efficient gene expression during early development (Moschetti et al., 2014).
YETI is a 241-aa-long protein belonging to BCNT protein superfamily which is characterized by a yeast-to-human highly conserved 82-amino acid domain located at the C-terminus (BCNT-C) (Takahashi et al., 1998; Figure 1).
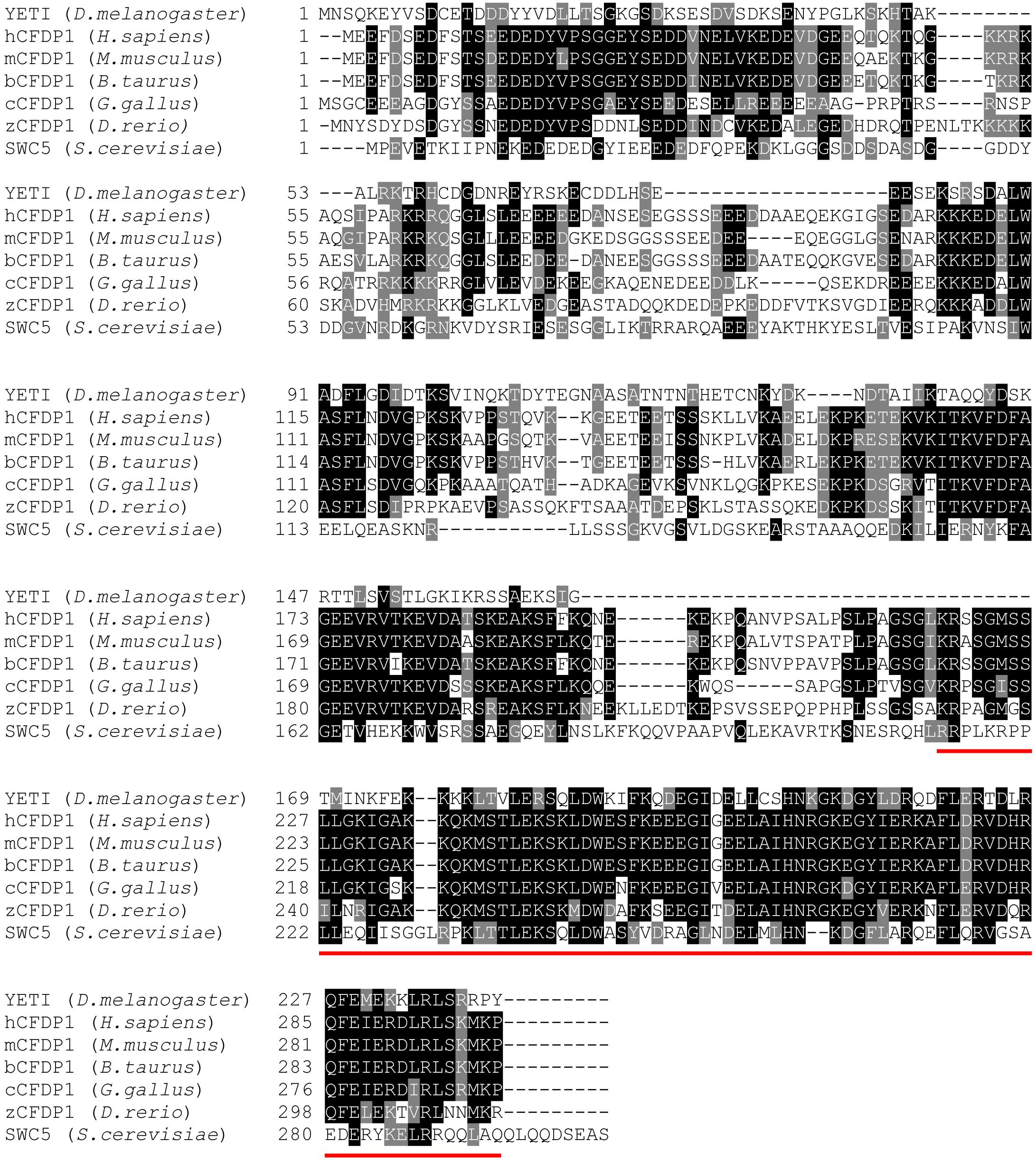
Figure 1. The YETI orthologous proteins. Blast alignments of YETI and its orthologous protein sequences from human (H. sapiens), mouse (M. musculus), bovine (B. taurus), chicken (G. gallus), Zebrafish (D. rerio) and yeast (S. cerevisiae). Note the high level of conservation (about 45% similarity) in the last 80 residues of the BCNT C-terminal domain (red line).
The YETI protein was originally identified as a both subunits kinesin-binding protein. In addition, Immunofluorescence experiments showed both a nuclear and cytoplasmic localization of V5 -tagged YETI in Drosophila S2 cells (Wisniewski et al., 2003). However, these experiments failed to clarify whether YETI is located within the nuclei or associated with the nuclear membrane.
In the last decade, YETI protein functions have been investigated more in depth (Messina et al., 2014). In particular, YETI was found to be a chromatin-associated protein member of the dTip60 complex (Figure 2), whose depletion leads to larval stage lethality accompanied by profound perturbations of higher-order chromatin organization due to the failure in loading of histone H2A.V, nucleosomal histones, and chromatin marks. Moreover, the BCNT domain of YETI was found to be involved in chromatin binding through the interaction with the H2A histone variant (H2A.V), Domino-A (DOM-A) ATPase, and HP1a protein (Ryu et al., 2014; Figure 3).
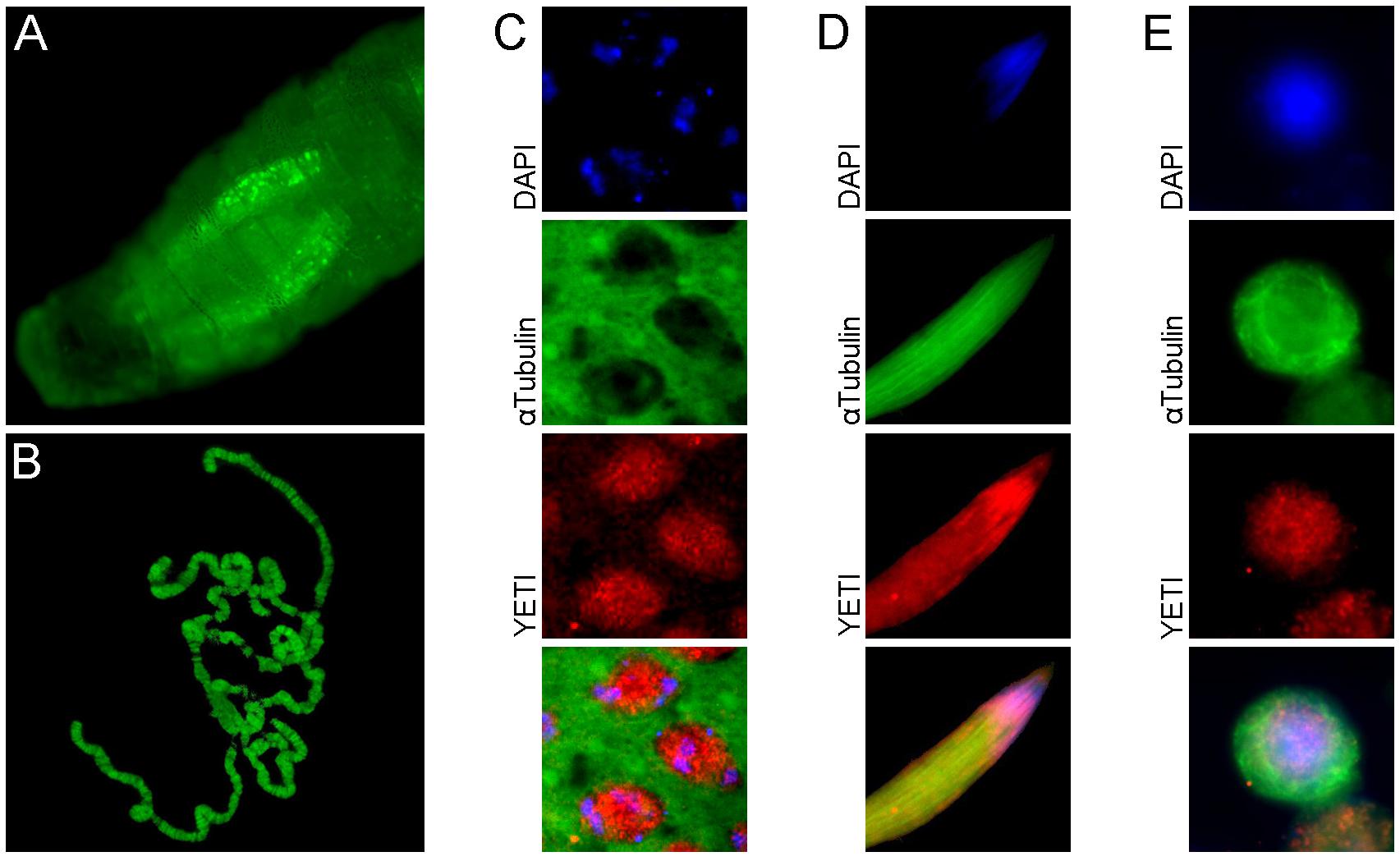
Figure 2. Subcellular YETI localization. (A) YETI::GFP fusion protein localizes in nuclei of salivary glands and (B) binds to polytene chromosomes. A nuclear immunolocalization of YETI protein is accordingly found in (C) spermatocytes, (D) elongated spermatids and (E) Drosophila S2 cells.
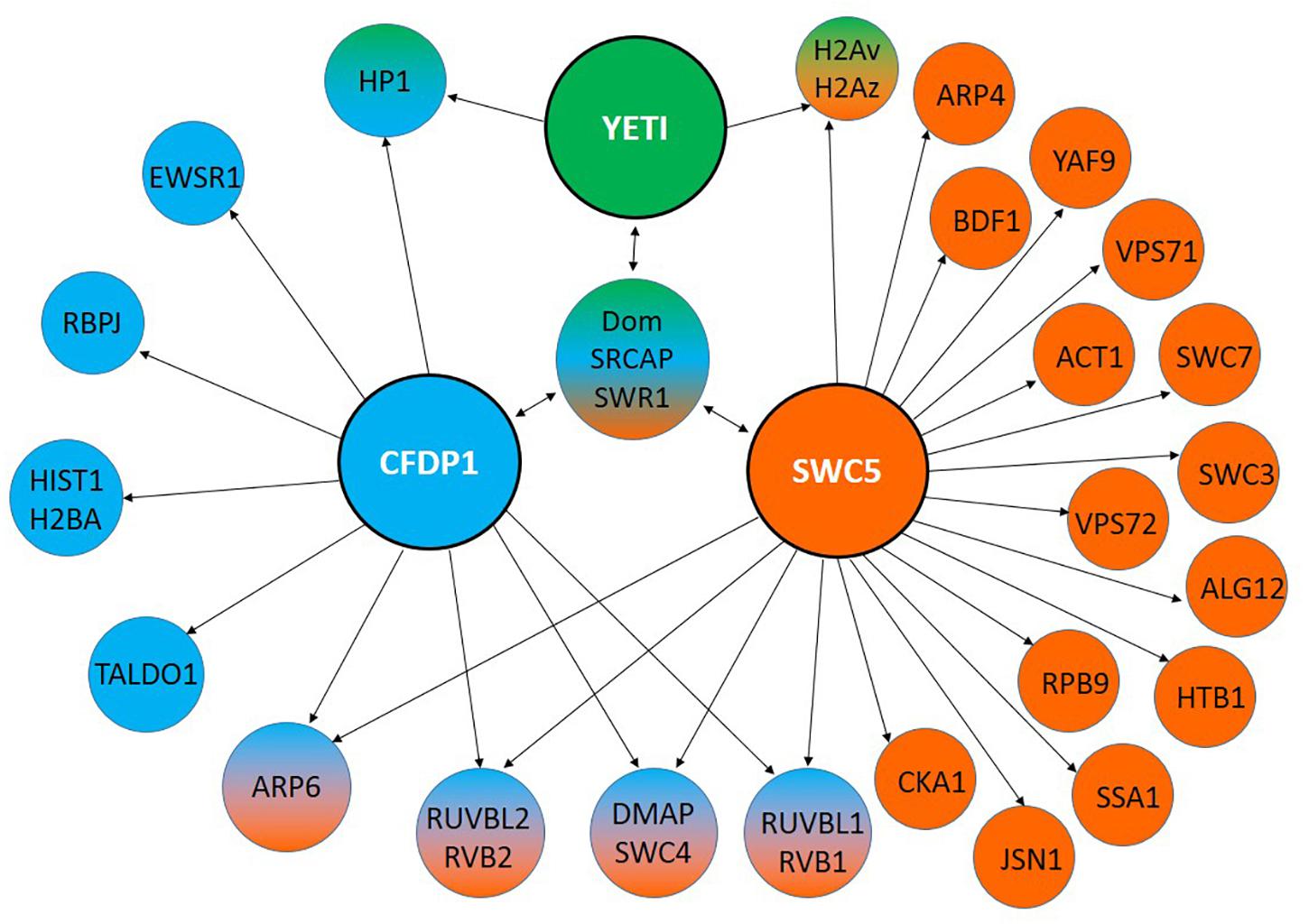
Figure 3. Interaction partners of YETI and its orthologs. YETI (D. melanogaster), hCFDP1 (H. sapiens) and SWR1 (S. cerevisiae) interacting proteins identified by using MIST (Molecular Interaction Search Tool). They refer only to physical interactions experimentally determined.
Taken together, these results strongly suggest that YETI may be directly implicated in the exchange of the variant H2A.V onto nucleosomes impacting on the dynamic changes of chromatin, and fulfilling its role as subunit of Tip60 chromatin remodeling complex, similarly to the yeast homolog SWC5 (Morillo-Huesca et al., 2010).
Furthermore, in a genetic interaction map of cell cycle regulators, YETI (and also the DOMINO ATPase) has been defined as an additional interesting connection in the proximity of Anaphase-promoting complex/Cyclosome (APC/C) (Billmann et al., 2016), suggesting a specific role in the regulation of chromatin and transcription during mitosis (Morrison and Shen, 2009; Tanenbaum et al., 2015). Moreover, there are emerging evidence indicating that YETI is involved in synapse assembly and function (Pazos Obregon et al., 2015).
In conclusion, YETI appears to be a multifaceted chromatin protein playing a key role in different cellular pathways.
Molecular insights of YETI orthologs come from other model organisms. For example, Swc5, the yeast ortholog protein, is a component of the SWR1 complex which catalyze the exchange of histone H2A with the H2A variant, Htz1 (Kobor et al., 2004; Mizuguchi et al., 2004). However, although Swc5 protein is not essential, Swc5 null-mutants are defective in chromosome maintenance, and show decreased resistance to hydroxyurea (Saccharomyces Genome Database) and increased heat sensitivity (Dastidar et al., 2012).
In vitro studies report that the binding of SWR1 complex to Htz1 is not dependent on Swc5 subunit, but Swc5 deleted mutants lack histone exchange function, indicating a key role of Swc5 during the histone replacement reaction (Wu et al., 2005). In vivo evidence supports this conclusion showing that the absence of Swc5 doesn’t affect complex assembly while it nearly dropped down the amount of Htz1 bound to chromatin, suggesting that Swc5 is required for the binding and destabilization of the H2A/H2B dimer (Morillo-Huesca et al., 2010). More in depth, SWR1 catalyzes H2A.Z in vitro replacement as one H2A.Z-H2B dimer at a time, generating heterotypic nucleosomes as intermediates and homotypic H2A.Z nucleosomes as final products. The SWR1 ATPase activity is specifically stimulated by H2A-containing nucleosomes, then free H2A.Z-H2B dimer leads to hyperstimulation of ATPase activity, eviction of nucleosomal H2A-H2B, and deposition of H2A.Z-H2B onto chromatin. In this way, SWR1 complex catalyzes nucleosomal conversion from H2A/H2A to Htz1/Htz1, with the heterotypic nucleosomes H2A/Htz1 as intermediates (Luk et al., 2010). Furthermore, a truncated Swc5 lacking the BCNT domain is not able to restore the reaction, indicating that BCNT domain is required for Htz1 deposition (Sun and Luk, 2017).
A 97 kDa BCNT protein (p97Bcnt) was first identified in bovine brain and found to be widely distributed in animals and plants (Nobukuni et al., 1997; Takahashi et al., 1998; Iwashita and Naoki, 2011; Messina et al., 2015).
Later on, the murine ortholog was isolated and cloned from a mouse E11 (Embrional day 11) λgt11 library using an antibody against Cem1, a tooth cementum-related protein marker of a distinct expression pattern of mouse craniofacial development (Diekwisch et al., 1999). It maps on chromosome 8 in the E1 region and is 85 kb long, with a 7 exons (888 nucleotides) open reading frame transcribing a single mRNA and encoding a 295 a.a. protein of 27 kDa weight (mCFDP1). Due to its expression pattern and protein MW, the gene was originally named craniofacial protein 27 (cp27) (Diekwisch et al., 1999).
Studies on the cp27 protein (later called mCFDP1) have shown specific interaction with NF-Y nuclear factor which bind CCAAT box (Luan et al., 2010; Ito et al., 2011), and with the TFII-I transcription factors involved in craniofacial development and osteo-differentiation (Makeyev and Bayarsaihan, 2011). Moreover, while RT-qPCR experiments revealed the presence of mCFDP1 in a pool of different genes related with transcription in osteo-differentiation (Bustos-Valenzuela et al., 2011), Northern blot analysis, combined with an in situ hybridization, have confirmed high level of its expression in developmental stage of mouse embryos, and in particular in neuroepithelium, cerebellum, heart, lung, liver, teeth, salivary glands, and periosteum of developing bones. These data have been also confirmed by immunohistochemical staining of developing tissues (Luan and Diekwisch, 2002).
Furthermore, cleft palate induction by persistent expression of PAX3 gene in neural crest correlates with cp27 downregulation (Wu et al., 2008).
The human BCNT ortholog maps to chromosome 16 in 16q22.2- q22.3, in proximity to several loci associated with inherited craniofacial disease genes (Diekwisch et al., 1999; Iwashita and Naoki, 2011). For this reason and for the expression pattern of the mouse ortholog it was called craniofacial development protein 1 gene (Cfdp1, OMIM number 608108). It is transcribed into two mRNAs differing by the alternative splicing-mediated inclusion or exclusion of exon encoding BCNT domain (Messina et al., 2017).
Independent experimental evidence suggests that the human CFDP1 (hCDFP1) protein is a subunit of the SNF2-related CBP activator protein (SRCAP) chromatin remodeling complex (Havugimana et al., 2012; Messina et al., 2017), which catalyzes the ATP-dependent replacement of canonical histone H2A with the H2A.Z variant (Monroy et al., 2001).
The first pioneer work aimed to address the mechanistic functions of hCFDP1 protein have been performed (Messina et al., 2016) by expressing human Cfdp1 transgene in D. melanogaster. The results of this experiment have shown that hCfdp1 expression lead to lethality and developmental and morphological defects, depending on the tissue-specific driver used. In particular, hCFDP1 binds to chromatin leading to abnormal polytene chromosome organization, decreased levels of H2A.V and H2A, brains and imaginal discs decreased size, developing of melanotic masses, prolonged larval stage and lethality. All these defects strongly resemble those produced by the lack of YETI (Messina et al., 2014). Thus, it is conceivable that hCFDP1 over-expression in D. melanogaster disrupts the physiological function of YETI resulting in a dominant-negative effect. Reciprocally, the Drosophila 3xFlag:YETI expressed in HeLa cells is able to go into the cell nucleus, bind to chromatin and produce a substantial decrease in the mitotic index with an important defects of cell cycle progression (Messina et al., 2016).
GST-pull down experiments have shown that both YETI and CFDP1 can undergo self-dimerization or hetero-dimerization. Thus, aberrant phenotypes could be due to the in vivo formation of YETI-CFDP1 non-functional hetero-dimers, together with a concomitant reduction of the amount of endogenous protein, YETI (drosophila cells) or CFDP1 (HeLa cells) (Messina et al., 2016; Messina et al., 2017).
Moreover, RNAi-mediated depletion of hCFDP1 in HeLa cells led to aberrant morphology and condensation defects of mitotic chromosomes impacting on their normal segregation during mitosis (Messina et al., 2017). These defects evoke those exhibited by Yeti null alleles in Drosophila melanogaster, suggesting a high-order chromatin organization maintenance function (Cenci et al., 2003; Messina et al., 2014).
Interestingly, the chromosome condensation defects observed in CFDP1 depleted cells resemble those found in cells lacking the MCPH1 gene encoding microcephalin, which is one of the causative genes of primary microcephaly (Yamashita et al., 2011; Messina et al., 2017).
Finally, in a human proteomic study, CFDP1 was found to interact with Ewing sarcoma related protein (EWSR1) whose mutations leads to Ewing’s sarcoma, a type of cancer that forms in bone or soft tissue (Bode-Lesniewska et al., 2019; Figure 3).
Among other vertebrates, data on YETI orthologs are only available from birds and fishes. In a recent study CENP-29, the chicken BCNT member, has been reported to be associated with kinetochores and therefore was suggested to play a role in chromosome segregation (Ohta et al., 2010).
In Danio rerio, the BCNT family gene, previously called rltpr (RGD motif, leucine-rich repeats, tropomodulin domain and proline-rich containing) and recently renamed zCfdp1, was found to be developmentally expressed in the eye, Central Nervous System and in branchial arches which lead to the craniofacial structures (Thisse and Thisse, 2004).
Recently, Celauro et al. (2017) have shown that zCFDP1 is rapidly produced after Maternal-To-Zygote transition and it is highly enriched in the head structures. Moreover, depletion of zCFDP1, induced by an ATG-blocking morpholino, produces drastic defects in craniofacial structures and bone mineralization.
Together, these results show that, not only that zCFDP1 is an essential protein required for proper embryonal development, but more importantly, they provide the first experimental evidence that it directly implicated in the morphogenesis of craniofacial territories in vertebrates. Moreover, based on the high level of identity (about 70%) (Figure 1), humans and zebrafish CFDP1 proteins, may be functionally conserved. It is then plausible that CFDP1, as a subunit of the SRCAP chromatin remodeling complex, participates to the epigenetic control of chromatin regions containing developmentally regulated genes, whose activation/silencing is crucial for proper differentiation of craniofacial structures and osteogenesis in zebrafish and likely in humans (Celauro et al., 2017).
As described above, the evolutionarily conserved Tip60 chromatin remodeling complex reveal multifaceted biological roles including transcriptional regulation, DNA repair, cell cycle progression, chromosome stability, stem cell maintenance and differentiation (Sapountzi et al., 2006; Tea and Luo, 2011).
As anticipated, depletion of YETI and CFDP1 in D. melanogaster and HeLa cells respectively affects mitotic index and led to chromosome segregation defects (Cenci et al., 2003; Messina et al., 2014, 2016). Moreover, the chicken YETI ortholog CENP-29 has been reported to be associated with kinetochores in chicken (Ohta et al., 2010). Finally, YETI protein has also been implicated in cell cycle control in both mitosis and meiosis (Billmann et al., 2016). According with these results, it has been reported that Myc, a well-known oncogenic player (Bellosta et al., 2005; Jonchere et al., 2017; Taylor et al., 2017), and the DOMINO ATPase interact each other and are required for normal asymmetric neuroblast (NB) division. Live-imaging analysis of dom-or myc-depleted NBs showed that spindle morphology is affected resulting in aberrant divisions (Rust et al., 2018). Similarly, Tip60 acetyl-transferase and Myc have been shown to co-regulate genes functionally involved in cell cycle and DNA replication (Zhao et al., 2018). Noteworthy, D. melanogaster mutants leading to symmetric division induce tumorigenesis (Albertson and Doe, 2003; Lee et al., 2006). Similarly, mutants lacking centrosomes and have severely impaired spindle asymmetry, over-proliferate and form tumors (Caous et al., 2015). Furthermore, mutations in genes encoding proteins directly or potentially implicated in chromatin biology are key players in human cancer and developmental disorders (Bickmore and van der Maarel, 2003; Fog et al., 2012; Harmacek et al., 2014).
Previous findings highlighted a role of YETI in spermatogenesis (Cenci et al., 2003). Accordingly, hCFDP1 has been found to physically interact with TALDO1 (Perl et al., 2006; Huttlin et al., 2015, 2017), an hallmark of human and murine spermatogenesis (Berkovits and Wolgemuth, 2013; Green et al., 2018), and HIST1H2BA (Figure 3), an intronless gene encoding a testis/sperm-specific member of the histone H2B family (Zalensky et al., 2002).
Taken together, the above-mentioned studies suggest that YETI and its orthologs, in addition to their canonical functions in chromatin remodeling, may play yet unexplored roles during both mitotic and meiotic cell division.
Most of the recent progresses in the study of the BCNT protein family originated from the functional analysis of a relatively small gene (Yeti) located in D. melanogaster constitutive heterochromatin, traditionally regarded as a transcriptionally inert or useless portion of the genome, albeit this prejudice has been debunked (Marsano et al., 2019). The fallout produced by those studies is relevant to basic research on epigenetics and to biomedical research too, as suggested by the experimental evidence showing that CFDP1 actively participates to craniofacial development in zebrafish. Craniofacial malformations are indeed developmental disorders of biomedical and clinical interest, because they represent the main cause of infant mortality and disability in humans. Therefore, it is reasonable to include the Cfdp1 gene in the screening test for human syndromes showing alteration of craniofacial development.
PD and GM conceived the concept of the study and wrote the manuscript. YP and GM designed and formatted the figures and table. All authors drafted the literature search, edited and reviewed the manuscript.
This work was supported by Ricerche Universitarie 2017 (MIUR) (PD) and “Teresa Ariaudo” Research Program 2018–2020 from Pasteur Institute, Cenci Bolognetti Foundation (GM).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge BOXSHADE (https://embnet.vital-it.ch/software/BOX_form.html) for the layout of the amino acid alignment (Figure 1).
Albertson, R., and Doe, C. Q. (2003). Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat. Cell. Biol. 5, 166–170. doi: 10.1038/ncb922
Baldi, S., and Becker, P. B. (2013). The variant histone H2A.V of Drosophila–three roles, two guises. Chromosoma 122, 245–258. doi: 10.1007/s00412-013-0409-x
Becker, P. B., and Horz, W. (2002). ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71, 247–273. doi: 10.1146/annurev.biochem.71.110601.135400
Bellosta, P., Hulf, T., Balla Diop, S., Usseglio, F., Pradel, J., Aragnol, D., et al. (2005). Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc. Natl. Acad. Sci. U.S.A. 102, 11799–11804. doi: 10.1073/pnas.0408945102
Berkovits, B. D., and Wolgemuth, D. J. (2013). The role of the double bromodomain-containing BET genes during mammalian spermatogenesis. Curr. Top. Dev. Biol. 102, 293–326. doi: 10.1016/B978-0-12-416024-8.00011-8
Bickmore, W. A., and van der Maarel, S. M. (2003). Perturbations of chromatin structure in human genetic disease: recent advances. Hum. Mol. Genet. 12(Suppl. 2), R207–R213.
Billmann, M., Horn, T., Fischer, B., Sandmann, T., Huber, W., and Boutros, M. (2016). A genetic interaction map of cell cycle regulators. Mol. Biol. Cell. 27, 1397–1407. doi: 10.1091/mbc.E15-07-0467
Bode-Lesniewska, B., Fritz, C., Exner, G. U., Wagner, U., and Fuchs, B. (2019). EWSR1-NFATC2 and FUS-NFATC2 gene fusion-associated mesenchymal tumors: clinicopathologic correlation and literature review. Sarcoma 2019:9386390. doi: 10.1155/2019/9386390
Bustos-Valenzuela, J. C., Fujita, A., Halcsik, E., Granjeiro, J. M., and Sogayar, M. C. (2011). Unveiling novel genes upregulated by both rhBMP2 and rhBMP7 during early osteoblastic transdifferentiation of C2C12 cells. BMC Res. Notes 4:370. doi: 10.1186/1756-0500-4-370
Caizzi, R., Moschetti, R., Piacentini, L., Fanti, L., Marsano, R. M., and Dimitri, P. (2016). Comparative genomic analyses provide new insights into the evolutionary dynamics of heterochromatin in Drosophila. PLoS Genet. 12:e1006212. doi: 10.1371/journal.pgen.1006212
Caous, R., Pascal, A., Rome, P., Richard-Parpaillon, L., Karess, R., and Giet, R. (2015). Spindle assembly checkpoint inactivation fails to suppress neuroblast tumour formation in aurA mutant Drosophila. Nat. Commun. 6:8879. doi: 10.1038/ncomms9879
Castillo-Davis, C. I., Mekhedov, S. L., Hartl, D. L., Koonin, E. V., and Kondrashov, F. A. (2002). Selection for short introns in highly expressed genes. Nat. Genet. 31, 415–418. doi: 10.1038/ng940
Celauro, E., Carra, S., Rodriguez, A., Cotelli, F., and Dimitri, P. (2017). Functional analysis of the cfdp1 gene in zebrafish provides evidence for its crucial role in craniofacial development and osteogenesis. Exp. Cell Res. 361, 236–245. doi: 10.1016/j.yexcr.2017.10.022
Cenci, G., Belloni, G., and Dimitri, P. (2003). 1(2)41Aa, a heterochromatic gene of Drosophila melanogaster, is required for mitotic and meiotic chromosome condensation. Genet. Res. 81, 15–24. doi: 10.1017/s0016672302006018
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
Clapier, C. R., Iwasa, J., Cairns, B. R., and Peterson, C. L. (2017). Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422. doi: 10.1038/nrm.2017.26
Corradini, N., Rossi, F., Verni, F., and Dimitri, P. (2003). FISH analysis of Drosophila melanogaster heterochromatin using BACs and P elements. Chromosoma 112, 26–37. doi: 10.1007/s00412-003-0241-9
Dastidar, R. G., Hooda, J., Shah, A., Cao, T. M., Henke, R. M., and Zhang, L. (2012). The nuclear localization of SWI/SNF proteins is subjected to oxygen regulation. Cell Biosci. 2:30. doi: 10.1186/2045-3701-2-30
Diekwisch, T. G., Marches, F., Williams, A., and Luan, X. (1999). Cloning, gene expression, and characterization of CP27, a novel gene in mouse embryogenesis. Gene 235, 19–30. doi: 10.1016/s0378-1119(99)00220-6
Dimitri, P., Corradini, N., Rossi, F., Verni, F., Cenci, G., Belloni, G., et al. (2003). Vital genes in the heterochromatin of chromosomes 2 and 3 of Drosophila melanogaster. Genetica 117, 209–215.
Eissenberg, J. C., Wong, M., and Chrivia, J. C. (2005). Human SRCAP and Drosophila melanogaster DOM are homologs that function in the notch signaling pathway. Mol. Cell. Biol. 25, 6559–6569. doi: 10.1128/mcb.25.15.6559-6569.2005
Fog, C. K., Galli, G. G., and Lund, A. H. (2012). PRDM proteins: important players in differentiation and disease. Bioessays 34, 50–60. doi: 10.1002/bies.201100107
Green, C. D., Ma, Q., Manske, G. L., Shami, A. N., Zheng, X., Marini, S., et al. (2018). A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell 46:e610. doi: 10.1016/j.devcel.2018.07.025
Hargreaves, D. C., and Crabtree, G. R. (2011). ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420. doi: 10.1038/cr.2011.32
Harmacek, L., Watkins-Chow, D. E., Chen, J., Jones, K. L., Pavan, W. J., Salbaum, J. M., et al. (2014). A unique missense allele of BAF155, a core BAF chromatin remodeling complex protein, causes neural tube closure defects in mice. Dev. Neurobiol. 74, 483–497. doi: 10.1002/dneu.22142
Havugimana, P. C., Hart, G. T., Nepusz, T., Yang, H., Turinsky, A. L., Li, Z., et al. (2012). A census of human soluble protein complexes. Cell 150, 1068–1081. doi: 10.1016/j.cell.2012.08.011
Hilliker, A. J. (1976). Genetic analysis of the centromeric heterochromatin of chromosome 2 of Drosophila melanogaster: deficiency mapping of EMS-induced lethal complementation groups. Genetics 83, 765–782.
Huttlin, E. L., Bruckner, R. J., Paulo, J. A., Cannon, J. R., Ting, L., Baltier, K., et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509. doi: 10.1038/nature22366
Huttlin, E. L., Ting, L., Bruckner, R. J., Gebreab, F., Gygi, M. P., Szpyt, J., et al. (2015). The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440. doi: 10.1016/j.cell.2015.06.043
Ito, Y., Zhang, Y., Dangaria, S., Luan, X., and Diekwisch, T. G. (2011). NF-Y and USF1 transcription factor binding to CCAAT-box and E-box elements activates the CP27 promoter. Gene 473, 92–99. doi: 10.1016/j.gene.2010.11.001
Iwashita, S., and Naoki, O. (2011). Bucentaur (Bcnt) Gene Family: Gene Duplication and Retrotransposons Insertion. Rijeka: InTech, 383–400.
Jonchere, V., Alqadri, N., Herbert, J., Dodgson, L., Mason, D., Messina, G., et al. (2017). Transcriptional responses to hyperplastic MRL signalling in Drosophila. Open Biol. 7:160306. doi: 10.1098/rsob.160306
Kobor, M. S., Venkatasubrahmanyam, S., Meneghini, M. D., Gin, J. W., Jennings, J. L., Link, A. J., et al. (2004). A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131. doi: 10.1371/journal.pbio.0020131
Krogan, N. J., Keogh, M. C., Datta, N., Sawa, C., Ryan, O. W., Ding, H., et al. (2003). A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565–1576. doi: 10.1016/s1097-2765(03)00497-0
Kusch, T., Florens, L., Macdonald, W. H., Swanson, S. K., Glaser, R. L., Yates, J. R., et al. (2004). Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306, 2084–2087. doi: 10.1126/science.1103455
Lee, C. Y., Andersen, R. O., Cabernard, C., Manning, L., Tran, K. D., Lanskey, M. J., et al. (2006). Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 20, 3464–3474. doi: 10.1101/gad.1489406
Lessard, J. A., and Crabtree, G. R. (2010). Chromatin regulatory mechanisms in pluripotency. Annu. Rev. Cell Dev. Biol. 26, 503–532. doi: 10.1146/annurev-cellbio-051809-102012
Luan, X., and Diekwisch, T. G. (2002). CP27 affects viability, proliferation, attachment and gene expression in embryonic fibroblasts. Cell Prolif. 35, 207–219. doi: 10.1046/j.1365-2184.2002.00238.x
Luan, X., Ito, Y., Zhang, Y., and Diekwisch, T. G. (2010). Characterization of the mouse CP27 promoter and NF-Y mediated gene regulation. Gene 460, 8–19. doi: 10.1016/j.gene.2010.03.014
Luger, K., Dechassa, M. L., and Tremethick, D. J. (2012). New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 13, 436–447. doi: 10.1038/nrm3382
Luk, E., Ranjan, A., Fitzgerald, P. C., Mizuguchi, G., Huang, Y., Wei, D., et al. (2010). Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143, 725–736. doi: 10.1016/j.cell.2010.10.019
Makeyev, A. V., and Bayarsaihan, D. (2011). Molecular basis of Williams-Beuren syndrome: TFII-I regulated targets involved in craniofacial development. Cleft Palate Craniofac. J. 48, 109–116. doi: 10.1597/09-093
March-Diaz, R., and Reyes, J. C. (2009). The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant 2, 565–577. doi: 10.1093/mp/ssp019
Marsano, R. M., Giordano, E., Messina, G., and Dimitri, P. (2019). A new portrait of constitutive heterochromatin: lessons from Drosophila melanogaster. Trends Genet. 35, 615–631. doi: 10.1016/j.tig.2019.06.002
Messina, G., Atterrato, M. T., Fanti, L., Giordano, E., and Dimitri, P. (2016). Expression of human Cfdp1 gene in Drosophila reveals new insights into the function of the evolutionarily conserved BCNT protein family. Sci. Rep. 6:25511. doi: 10.1038/srep25511
Messina, G., Atterrato, M. T., Prozzillo, Y., Piacentini, L., Losada, A., and Dimitri, P. (2017). The human cranio facial development protein 1 (Cfdp1) gene encodes a protein required for the maintenance of higher-order chromatin organization. Sci. Rep. 7:45022. doi: 10.1038/srep45022
Messina, G., Celauro, E., Atterrato, M. T., Giordano, E., Iwashita, S., and Dimitri, P. (2015). The bucentaur (BCNT) protein family: a long-neglected class of essential proteins required for chromatin/chromosome organization and function. Chromosoma 124, 153–162. doi: 10.1007/s00412-014-0503-8
Messina, G., Damia, E., Fanti, L., Atterrato, M. T., Celauro, E., Mariotti, F. R., et al. (2014). Yeti, an essential Drosophila melanogaster gene, encodes a protein required for chromatin organization. J. Cell Sci. 127, 2577–2588. doi: 10.1242/jcs.150243
Mizuguchi, G., Shen, X., Landry, J., Wu, W. H., Sen, S., and Wu, C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348. doi: 10.1126/science.1090701
Monroy, M. A., Ruhl, D. D., Xu, X., Granner, D. K., Yaciuk, P., and Chrivia, J. C. (2001). Regulation of cAMP-responsive element-binding protein-mediated transcription by the SNF2/SWI-related protein, SRCAP. J. Biol. Chem. 276, 40721–40726. doi: 10.1074/jbc.m103615200
Morillo-Huesca, M., Clemente-Ruiz, M., Andujar, E., and Prado, F. (2010). The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PLoS One 5:e12143. doi: 10.1371/journal.pone.0012143
Morrison, A. J., and Shen, X. (2009). Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 10, 373–384. doi: 10.1038/nrm2693
Moschetti, R., Celauro, E., Cruciani, F., Caizzi, R., and Dimitri, P. (2014). On the evolution of Yeti, a Drosophila melanogaster heterochromatin gene. PLoS One 9:e113010. doi: 10.1371/journal.pone.0113010
Nobukuni, T., Kobayashi, M., Omori, A., Ichinose, S., Iwanaga, T., Takahashi, I., et al. (1997). An Alu-linked repetitive sequence corresponding to 280 amino acids is expressed in a novel bovine protein, but not in its human homologue. J. Biol. Chem. 272, 2801–2807. doi: 10.1074/jbc.272.5.2801
Ohta, S., Bukowski-Wills, J. C., Sanchez-Pulido, L., Alves Fde, L., Wood, L., Chen, Z. A., et al. (2010). The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142, 810–821. doi: 10.1016/j.cell.2010.07.047
Pazos Obregon, F., Papalardo, C., Castro, S., Guerberoff, G., and Cantera, R. (2015). Putative synaptic genes defined from a Drosophila whole body developmental transcriptome by a machine learning approach. BMC Genomics 16:694. doi: 10.1186/s12864-015-1888-3
Perl, A., Qian, Y., Chohan, K. R., Shirley, C. R., Amidon, W., Banerjee, S., et al. (2006). Transaldolase is essential for maintenance of the mitochondrial transmembrane potential and fertility of spermatozoa. Proc. Natl. Acad. Sci. U.S.A. 103, 14813–14818. doi: 10.1073/pnas.0602678103
Rust, K., Tiwari, M. D., Mishra, V. K., Grawe, F., and Wodarz, A. (2018). Myc and the Tip60 chromatin remodeling complex control neuroblast maintenance and polarity in Drosophila. EMBO J. 37:e98659. doi: 10.15252/embj.201798659
Ryu, H. W., Lee, D. H., Florens, L., Swanson, S. K., Washburn, M. P., and Kwon, S. H. (2014). Analysis of the heterochromatin protein 1 (HP1) interactome in Drosophila. J. Proteom. 102, 137–147. doi: 10.1016/j.jprot.2014.03.016
Sapountzi, V., Logan, I. R., and Robson, C. N. (2006). Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 38, 1496–1509. doi: 10.1016/j.biocel.2006.03.003
Sun, L., and Luk, E. (2017). Dual function of Swc5 in SWR remodeling ATPase activation and histone H2A eviction. Nucleic Acids Res. 45, 9931–9946. doi: 10.1093/nar/gkx589
Takahashi, I., Nobukuni, T., Ohmori, H., Kobayashi, M., Tanaka, S., Ohshima, K., et al. (1998). Existence of a bovine line repetitive insert that appears in the cDNA of bovine protein BCNT in ruminant, but not in human, genomes. Gene 211, 387–394. doi: 10.1016/s0378-1119(98)00136-x
Tanenbaum, M. E., Stern-Ginossar, N., Weissman, J. S., and Vale, R. D. (2015). Regulation of mRNA translation during mitosis. Elife 4:e07957. doi: 10.7554/eLife.07957
Taylor, E., Alqadri, N., Dodgson, L., Mason, D., Lyulcheva, E., Messina, G., et al. (2017). MRL proteins cooperate with activated Ras in glia to drive distinct oncogenic outcomes. Oncogene 36, 4311–4322. doi: 10.1038/onc.2017.68
Tea, J. S., and Luo, L. (2011). The chromatin remodeling factor Bap55 functions through the TIP60 complex to regulate olfactory projection neuron dendrite targeting. Neural Dev. 6:5. doi: 10.1186/1749-8104-6-5
Thisse, B., and Thisse, C. (2004). Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. Available at: http://zfin.org
van Attikum, H., and Gasser, S. M. (2005). The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 6, 757–765. doi: 10.1038/nrm1737
Wang, X., Ahmad, S., Zhang, Z., Cote, J., and Cai, G. (2018). Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex. Nat. Commun. 9:1147. doi: 10.1038/s41467-018-03504-5
Wisniewski, T. P., Tanzi, C. L., and Gindhart, J. G. (2003). The Drosophila kinesin-I associated protein YETI binds both kinesin subunits. Biol. Cell 95, 595–602.
Wu, M., Li, J., Engleka, K. A., Zhou, B., Lu, M. M., Plotkin, J. B., et al. (2008). Persistent expression of Pax3 in the neural crest causes cleft palate and defective osteogenesis in mice. J. Clin. Invest. 118, 2076–2087. doi: 10.1172/JCI33715
Wu, W. H., Alami, S., Luk, E., Wu, C. H., Sen, S., Mizuguchi, G., et al. (2005). Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 12, 1064–1071. doi: 10.1038/nsmb1023
Yamada, H. Y. (2012). Human Tip60 (NuA4) Complex and Cancer. Oklahoma, OK: University of Oklahoma Health Sciences Center (OUHSC).
Yamashita, D., Shintomi, K., Ono, T., Gavvovidis, I., Schindler, D., Neitzel, H., et al. (2011). MCPH1 regulates chromosome condensation and shaping as a composite modulator of condensin II. J. Cell Biol. 194, 841–854. doi: 10.1083/jcb.201106141
Zalensky, A. O., Siino, J. S., Gineitis, A. A., Zalenskaya, I. A., Tomilin, N. V., Yau, P., et al. (2002). Human testis/sperm-specific histone H2B (hTSH2B). Molecular cloning and characterization. J. Biol. Chem. 277, 43474–43480. doi: 10.1074/jbc.m206065200
Keywords: Drosophila melanogaster, BCNT proteins, chromatin remodeling complex, YETI, epigenetic regulation
Citation: Prozzillo Y, Delle Monache F, Ferreri D, Cuticone S, Dimitri P and Messina G (2019) The True Story of Yeti, the “Abominable” Heterochromatic Gene of Drosophila melanogaster. Front. Physiol. 10:1093. doi: 10.3389/fphys.2019.01093
Received: 29 May 2019; Accepted: 08 August 2019;
Published: 22 August 2019.
Edited by:
Cristiano De Pitta, University of Padua, ItalyReviewed by:
René Massimiliano Marsano, University of Bari Aldo Moro, ItalyCopyright © 2019 Prozzillo, Delle Monache, Ferreri, Cuticone, Dimitri and Messina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrizio Dimitri, cGF0cml6aW8uZGltaXRyaUB1bmlyb21hMS5pdA==; Giovanni Messina, Z2lvdmFubmkubWVzc2luYUB1bmlyb21hMS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.