- Key Laboratory of Entomology and Pest Control Engineering, College of Plant Protection, Southwest University, Chongqing, China
Laodelphax striatellus is naturally infected with the Wolbachia strain wStri, which induces strong cytoplasmic incompatibility of its host. MicroRNAs (miRNAs) are a class of endogenous non-coding small RNAs that play a critical role in the regulation of gene expression at post-transcriptional level in various biological processes. Despite various studies reporting that Wolbachia affects the miRNA expression of their hosts, the molecular mechanism underlying interactions between Wolbachia and their host miRNAs has not been well understood. In order to better understand the impact of Wolbachia infection on its host, we investigated the differentially expressed miRNAs between Wolbachia-infected and Wolbachia-uninfected strains of L. striatellus. Compared with uninfected strains, Wolbachia infection resulted in up-regulation of 18 miRNAs and down-regulation of 6 miRNAs in male, while 25 miRNAs were up-regulated and 15 miRNAs were down-regulated in female. The target genes of these differentially expressed miRNAs involved in immune response regulation, reproduction, redox homeostasis and ecdysteroidogenesis were also annotated in both sexes. We further verified the expression of several significantly differentially expressed miRNAs and their predicted target genes by qRT-PCR method. The results suggested that Wolbachia appears to reduce the expression of genes related to fertility in males and increase the expression of genes related to fecundity in females. At the same time, Wolbachia may enhance the expression of immune-related genes in both sexes. All of the results in this study may be helpful in further exploration of the molecular mechanisms by which Wolbachia affects on its hosts.
Introduction
Wolbachia is a maternally inherited endosymbiotic bacteria that infects with 40% of terrestrial arthropod species (Zug and Hammerstein, 2012). It draws attention by manipulating the reproduction of host in arthropod species. For example, cytoplasmic incompatibility (CI) is the best-known reproductive phenotype and result in early embryonic lethality when males infected by Wolbachia mating with uninfected females or females carrying different Wolbachia strains (Laven, 1957; Yen and Barr, 1971). In addition to reproductive regulation, a growing body of researches have shown that Wolbachia also affect many aspects of arthropod host, including the expression of immune genes in many arthropod hosts (Kambris et al., 2009; Thomas et al., 2011; Pan et al., 2012; Rances et al., 2012; Ye et al., 2013; Joshi et al., 2017). In fact, Wolbachia can affects the immune response of insects to a variety of pathogens, including infections against bacteria, viruses, and parasites (Kambris et al., 2009, 2010; Moreira et al., 2009; Bian et al., 2010; Mousson et al., 2010; Hughes et al., 2011; Wong et al., 2011; Pan et al., 2012; Eleftherianos et al., 2013; Gupta et al., 2017; Ant et al., 2018; Monsanto-Hearne and Johnson, 2018; Thomas et al., 2018).
MicroRNAs (miRNAs) exist in invertebrates and vertebrates, which play an important role in the regulation of gene expression at post-transcriptional level by combining with target mRNA complementarily (Lee et al., 1993; Reinhart et al., 2000; Lagos-Quintana et al., 2001). It takes part in lots of important physiological processes, including immune response (Lee and Hyun, 2014), responding to bacterial infection (Lourenco et al., 2013), reproduction (Zhang et al., 2016), and germline stem cell differentiation (Eun et al., 2013). In fact, some studies also showed that insect miRNAs are widely involved in host-microorganism interactions (Asgari, 2011; Wu et al., 2013; Slonchak et al., 2014; Li et al., 2015, 2018; Liu et al., 2015; Xing et al., 2016).
In recent years, Wolbachia regulating host genes expression through disturbing the expression of host miRNAs were reported (Hussain et al., 2011; Osei-Amo et al., 2012; Zhang et al., 2013, 2014; Mayoral et al., 2014; Rong et al., 2014). In Aedes aegypti, Wolbachia induce the expression of aae-miR-2940, which up-regulates the expression of arginine methyltransferase 3 and metalloprotease gene, as well as down-regulates the expression of DNA methyl-transferase gene, which was critical for its colonization and efficient maintenance of its density in host (Hussain et al., 2011; Zhang et al., 2013, 2014). In Ae. aegypti cells, Argonaute 1 distribution to the nucleus was blocked by Wolbachia via upregulating the expression of miR-981 (Hussain et al., 2013). Researchers also found Wolbachia induced the expression of aae-miR-12 to downregulate the expression of monocarboxylate transporter MCT1 and DNA replication licensing factor MCM6 genes that were critical for its persistence in Ae. aegypti cell line (Osei-Amo et al., 2012). In Tetranychus urticae, Wolbachia infection significantly suppresses expression of miRNAs, and the target genes of Wolbachia-responsive miRNAs involve in lysosome function and apoptosis in both sexes, in the meantime, it may regulate reproduction in females (Rong et al., 2014). All of these studies suggest that miRNAs may play a key role in Wolbachia-host interaction.
The small brown planthopper (SBPH), Laodelphax striatellus, is one of the most serious agricultural pests that feeds on the phloem sap of several important crops, such as rice, wheat and corn. It is also an insect vector, and transmits plant viruses by feeding on healthy and diseased plants (Zhang et al., 2007; Li et al., 2011). SBPH is naturally infected with the Wolbachia strain wStri, which induces CI strongly (Noda et al., 2001). In this study, we inquired the effects of Wolbachia on the genes expression of L. striatellus by comparing miRNA expression levels in infected females (FI) and uninfected females (FUI), and infected males (MI) and uninfected males (MUI). In the meantime, based on differentially expressed miRNAs, we also predicted the target genes of these miRNAs. Revealing the Wolbachia-induced microRNA will help us to further understand the interactions between Wolbachia and its host.
Materials and Methods
Laodelphax striatellus
The L. striatellus that naturally infected Wolbachia were collected from Nanjing, Jiangsu province of China, in 2011. After rearing in laboratory for several generations, some individuals of this population were treated with the tetracycline hydrochloride solution (0.1%) that was added to the rice seedlings for 3–4 generations until no Wolbachia was detected by diagnostic PCR detection. Then, both Wolbachia-uninfected and Wolbachia-infected L. striatellus used in this study were long-term kept in our laboratory. These lines were reared in clear plastic cups (150 mm in height and 110 mm in diameter) which were covered with gauze and contained rice seedlings. Then these planthoppers were maintained in an artificial climate chamber (temperature: 27 ± 1°C, relative humidity: 60 ± 10%, and under 16 h light: 8 h dark photoperiod). Infection status of each strain was confirmed by using PCR method to detected the wsp gene of Wolbachia with 81F (TGG TCC AAT AAG TGA TGA AGA AAC) and 691R (AAA AAT TAA ACG CTA CTC CA) primers (Braig et al., 1998).
Small RNA Library Construction and Illumina Sequencing
In order to avoid the impact of mating, the 5-th-instar nymphs were single reared in glass test tubes (180 mm in height and 18 mm in diameter) which were covered with gauze and contained rice seedlings, and were observed every 8 h. Since the 3-day-old adults have shown sexual maturity and have the ability to mate, and the density of Wolbachia shows an increase in adult bodies from 0 to 4 days (Noda et al., 2001), so the 3-day-old adults were selected for RNA extraction. After they emerged, 3-day-old females (5 individuals) and males (10 individuals) adults were collected in 1.5 mL centrifuge tube, respectively. All samples were quickly frozen in liquid nitrogen, then stored at −80°C for RNA extractions. Total RNA was extracted from every sample using Trizol Reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions (recommended protocol). To ensure that the use of qualified samples for sequencing, RNA integrity was evaluated using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, United States).
A total amount of 2.5 μg RNA per sample was used for sequencing. The sequencing libraries were created using NEB Next Ultra small RNA Sample Library Prep Kit (NEB, United States) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. The PAGE gel was used for electrophoretic fragment screening purposes, and the small RNA library obtained as the pieces was recovered by gelatinization. The clustering of the index-coded samples was operated on a cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS (Illumia), then the library preparations were sequenced on an Illumina Hiseq 2500 platform. The dataset of this study has been deposited in the Sequence Read Archive (SRA) database of NCBI with accession number PRJNA530287.
Bioinformatics Analysis of Sequencing Data
To obtain clean reads from raw reads having low-quality, raw reads of fastq format were firstly processed through in-house perl scripts. In this step, clean reads were obtained by removing reads containing base N (N is an unrecognized base) content of 10% or more, with 5′ adapter contaminants, without 3′ adapter, and low-quality reads, and cut off the 3′ adapter sequence from raw data. And reads were trimmed and cleaned by removing the sequences smaller than 18 nt or longer than 30 nt. At the same time, Q20, Q30, GC-content and sequence duplication level of the clean data were calculated. Using Bowtie software (v1.0.0) (Langmead et al., 2009), sequence alignment of clean reads with Silva database,1 GtRNAdb database,2 Rfam database3 and Repbase database,4 filtering ribosomal RNA (rRNA), transport RNA (tRNA), nuclear small RNA (snRNA), nucleolar small RNA (snoRNA) and other ncRNAs and repeats, obtain unannotated readings containing miRNA. Sequence alignment of unannotated reads using Bowtie software to obtain positional information on the reference gene, which is a map read. In the known miRNA identification, the mapped reads were aligned with the sequence of the mature miRNA in miRBase5 database. In the novel miRNA identification, based on the biometric characteristics of miRNA, the miRDeep2 tool (v2.0.5) (Friedlander et al., 2012) was used to obtain possible precursor sequences. And the precursor structure energy information and miRNA secondary structure for the prediction of novel miRNAs. For the name of the novel miRNAs which first reported in L. striatellus, the code “lst-miRn” followed by a number assigned to the novel miRNA as number designator was used (Li et al., 2015).
Differential Expression Analysis of Between Wolbachia-Uninfected and Wolbachia-Infected L. striatellus
The expression levels of miRNAs between Wolbachia-uninfected and Wolbachia-infected L. striatellus [FI and FUI (control), MI and MUI (control)] were analyzed using the IDEG6 in this study (Romualdi et al., 2003). The expression of miRNA in the four libraries was normalized to transcripts per million (TPM) on the basis of the following formula: Normalized expression = actual miRNA count/total count of clean reads × 106. The p value was adjusted using q value (Storey, 2003). The q value < 0.005 and |log2 (fold change)| ≥ 1 was set as the threshold for significantly differential expression.
Verification the Expression of Wolbachia-Responsive miRNAs and Target Genes Prediction via qRT-PCR
In order to further verify the expression of each miRNA, reverse transcription was carried out using the miRNA cDNA Synthesis Kit (Kang Wei Century). The miRNA was quantified according to the instructions of the miRNA qPCR Assay Kit (Kang Wei Century) following the program: 95°C for 10 min, 45 cycles of 95°C for 15 s, 64°C for 1 min, and the U6 gene was used as an internal reference gene. Specific forward primers were designed based on mature sequence (Supplementary Table S1), and the reverse primer was provided by the kit. For the target gene, specific primers were designed based on CDs sequences of related L. striatellus genes on line6 (Supplementary Table S2), reverse transcription was performed using the Prime ScriptTM RT (TaKaRa) kit. The quantitative real time polymerase chain reaction (qRT-PCR) was using NovoStart® SYBR qPCR SuperMix Plus reaction kit (Novoprotein, China) following the program: 95°C for 1 min, 39 cycles of 95°C for 20 s, 60°C for 1 min. The relative expression levels were normalized by L. striatellus ARF (ADP-ribosylation factor-like protein 2) gene which was a stably expressed internal reference gene, and it was recommended to be used in qPCR in L. striatellus (He et al., 2014). All reaction quantitative reactions were carried out in a fluorescence quantitative gradient PCR instrument qTOWER3.0 Real-Time System (Analytik Jena, Germany). Three biologic replicates were performed for each experiment. The relative expression level of each gene was calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001). Relative transcriptional levels were analyzed with the SPSS v.22.0 software (SPSS, Chicago, IL, United States), independent t test was used to analysis the differences between groups. The p value of <0.05 was considered statistically significant.
MicroRNA Target Prediction and Function Analysis
Target genes of Wolbachia-responsive miRNAs were predicted using our transcriptome data of L. striatellus by miRanda (v3.3a) (Betel et al., 2008) and RNAhybrid (v2.1.1) (Rehmsmeier et al., 2004) based on the gene sequence information of known miRNAs and newly predicted miRNAs. Target gene were annotated by aligning their sequences with NR (Deng et al., 2006), Swiss-Prot (Apweiler et al., 2004), GO (Ashburner et al., 2000), KEGG (Kanehisa et al., 2004), and Pfam (Eddy, 1998) databases using BLAST software.
Results
Overview Over sRNAs Sequencing
To identify Wolbachia-responsive miRNAs in L. striatellus, four sRNA libraries (FUI, FI, MUI, and MI) were constructed using high-throughput Illumina sequencing platform. In total, 17,772,698; 28,351,576; 24,916,579 and 24,311,953 raw reads were obtained, respectively. For the raw data, the reads shorter than 18 nt or longer than 30 nt were discarded. After removing low-quality sequences, 16,616,711; 26,020,481; 23,702,808 and 22,471,936 clean reads were obtained in the FUI, FI, MUI, and MI libraries, respectively, for further analysis. In all four libraries, the ratio of rRNA reads was less than 10%. The lowest rRNA read ratio appeared in the infected male sequencing library (MI, 3.69%), and overall, the proportion of rRNA read were lower in the male sequencing libraries than in the female (Supplementary Table S3). Subsequently, the proportions of common and specific sRNAs between pairs of libraries were further analyzed (Supplementary Figure S1). For the total reads, the results indicated that the number of sRNAs shared by any two libraries accounted for more than 75% of the total sRNAs, while the library-specific sRNAs accounted for only 5.56–16.55%. After removing the redundant read, the Uniq-sRNA shared between the two samples accounted for 11.45–16.14%, while the library-specific sRNA type accounted for more than 25% in all comparative groups.
Identification and Analysis of miRNAs From Sequencing Libraries
To identify conserved and novel miRNAs in L. striatellus, the filtered small RNA sequences were analyzed. After compared with known miRNAs from miRBase database and predicted novel miRNA using miRDeep2 software, 152 miRNAs were annotated in the four sequencing libraries, which include 49 known and 103 novel miRNAs (Table 1). The length distribution of all miRNAs in the four libraries was 18–25 nt, and it was mainly distributed in 21–24 nt, with 22 nt as the largest number of miRNAs (Figure 1), which in accordance with the typical sizes of Dicer processing products (Ambros et al., 2003). The miRNA name, the mature sequences, the precursor sequence and the number of reads of each miRNA were shown in Supplementary Table S4. Familial analysis of known miRNAs and novel miRNAs of the four libraries based on sequence similarity, a total of 17 novel miRNAs and 49 known miRNAs were divided into 30 families.
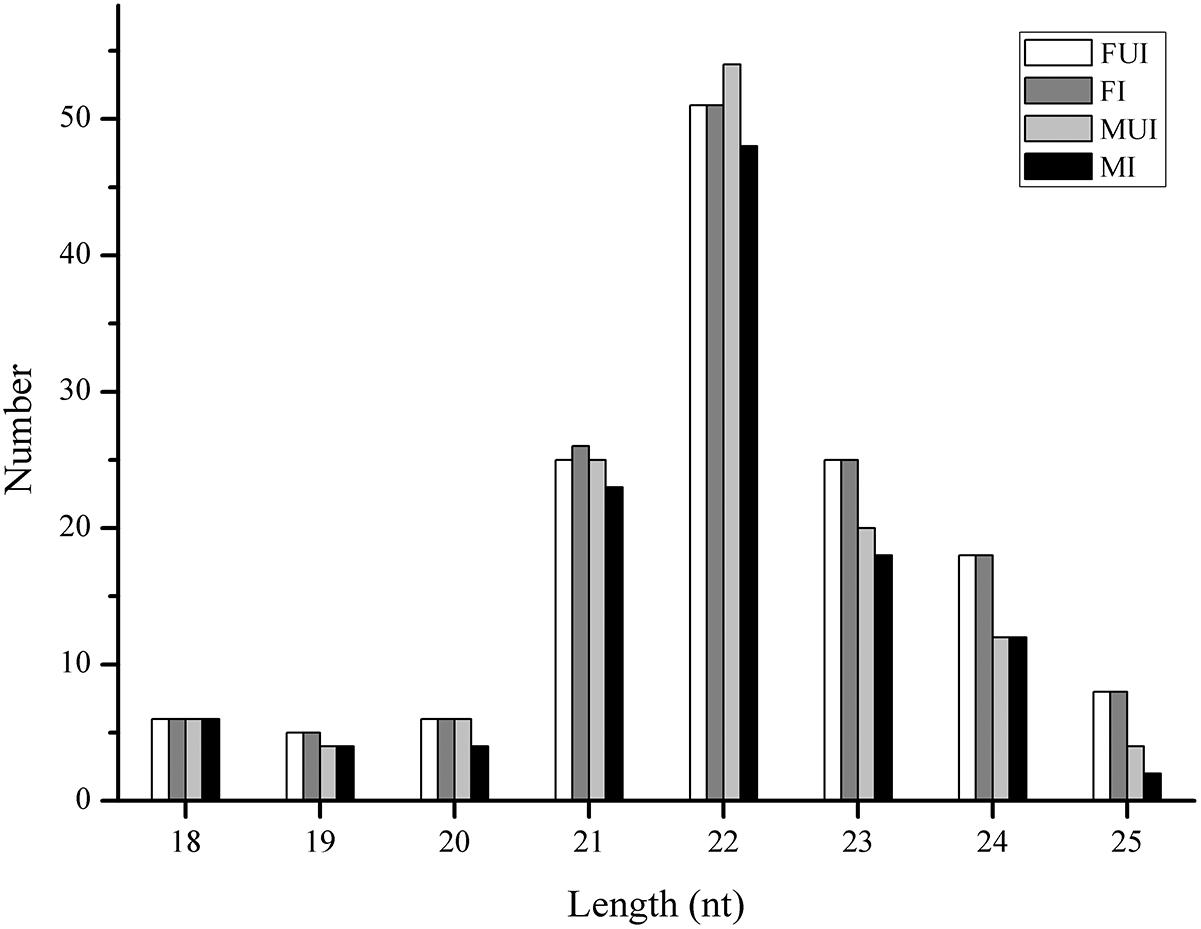
Figure 1. Length distribution of miRNAs in the four sequencing libraries of L. striatellus. FUI, female uninfected; FI, female infected; MUI, male uninfected; MI, male infected; nt, nucleotides.
Totally, 104 miRNAs were shared in the four libraries (Figure 2). For females, 48 known and 91 novel miRNAs were shared, and there were 6 and 5 specifically expressed miRNAs in Wolbachia infected and uninfected females, respectively. Meanwhile, for males, 49 known and 62 novel miRNAs were shared, and there were 6 and 16 specifically expressed miRNAs in Wolbachia infected and uninfected males, respectively.
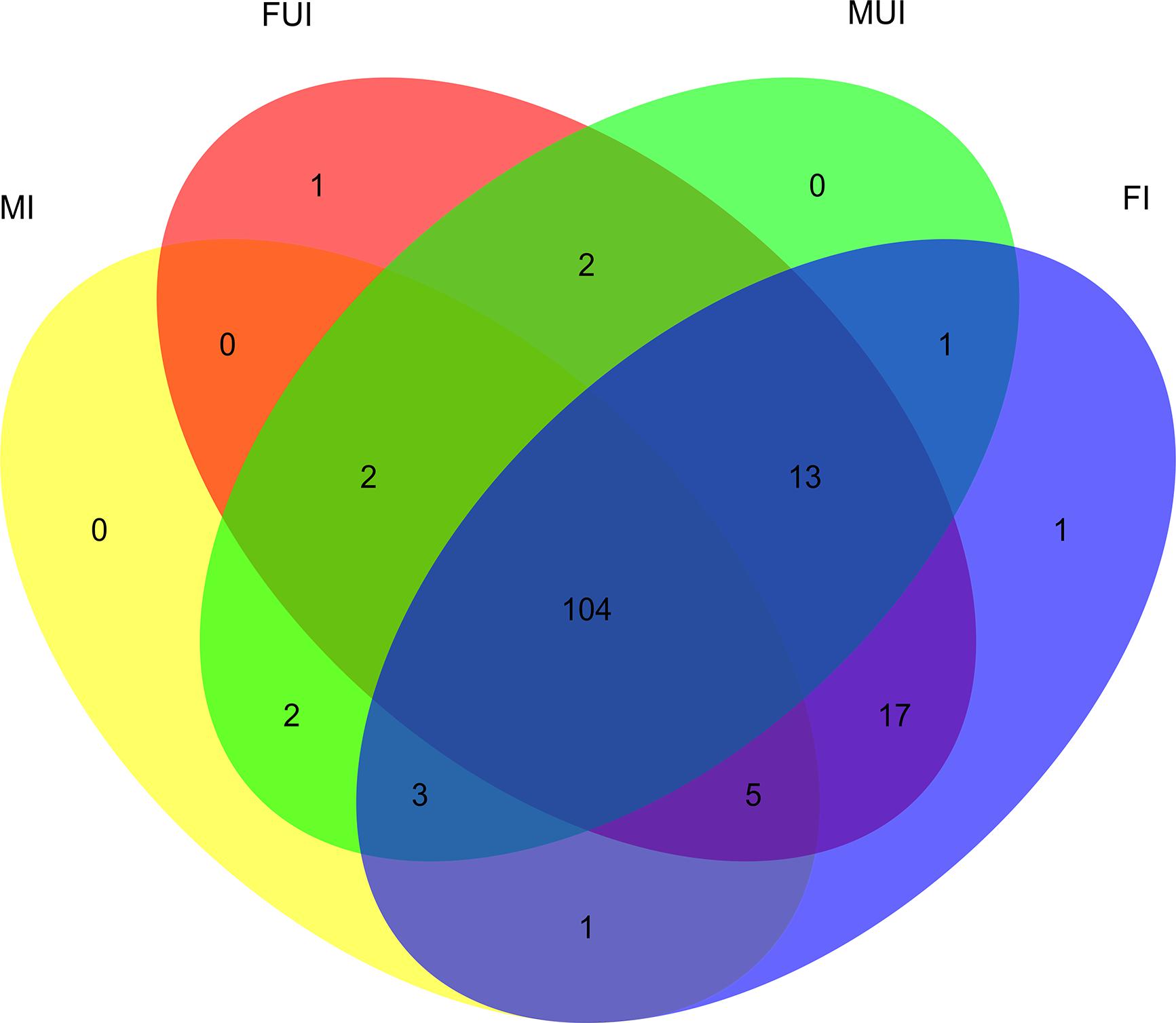
Figure 2. Venn diagram of unique and commonly expressed miRNAs among four sequencing libraries of L. striatellus (FI, FUI, MI, and MUI). Yellow oval represents MI; red oval represents FUI; green oval represents MUI; blue oval represents FI.
Abundance of miRNAs
In order to clarify the highly abundant miRNAs in every library, top twenty most abundant miRNAs expressed in each of libraries were shown in Table 2 (accounting for more than 90% of total miRNA reads), 16 of which expressed in the four libraries were shared, including two newly identified miRNAs (lst-miR-n74-5p and lst-miR-n97-5p). The two most abundantly expressed miRNAs of the four sequenced libraries were lst-miR-1-3p and lst-miR-184-3p.
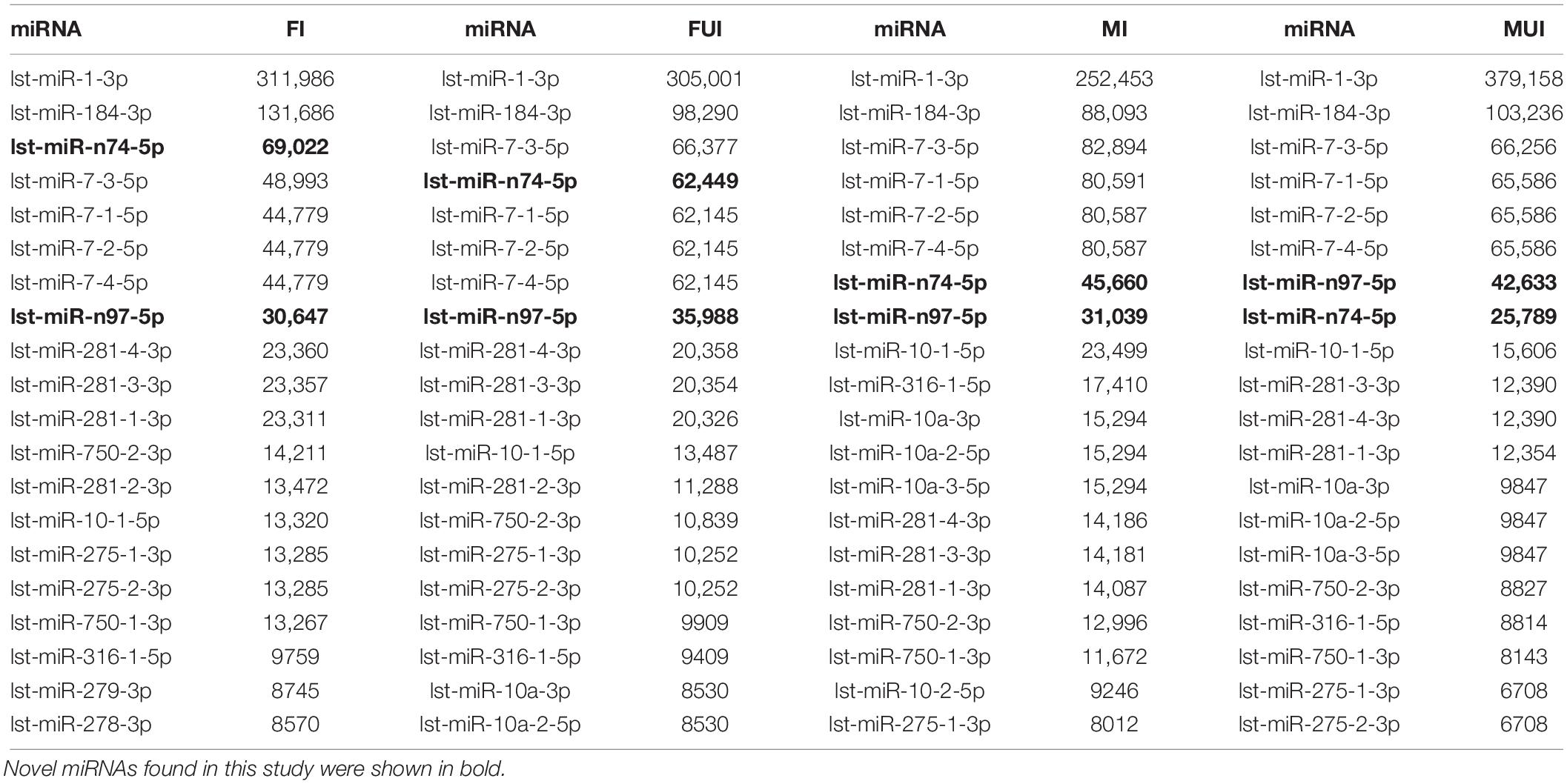
Table 2. Top 20 most abundant miRNAs expressed in the four sequencing libraries of L. striatellus (TPM data were shown).
Differentially Expressed miRNA of L. striatellus in Response to Wolbachia Infection
To reveal differentially expressed miRNAs in response to Wolbachia-infection, the expression levels of these miRNA were compared in infected and uninfected females and males of L. striatellus, respectively. A volcano plot of the different expressed miRNAs was shown in Figure 3. Overall, compared with uninfected individuals, there were 51 differently expressed miRNAs including 4 known and 47 novel miRNAs in Wolbachia-infected females and males (Figure 4). Compared with the uninfected females, 25 miRNAs were up-regulated and 15 miRNAs were down-regulated in Wolbachia infected females. By contrast, 18 miRNAs were up-regulated and 6 miRNAs were down-regulated in MI compared with the MUI (Figure 5A). Apparently, Wolbachia induced more differentially expressed miRNAs in females than males (about 1.67-fold), suggesting that Wolbachia infection may have a broader effect on females (Figure 5). In T. urticae, Wolbachia infection also induced more differentially expressed miRNAs in females (Rong et al., 2014).
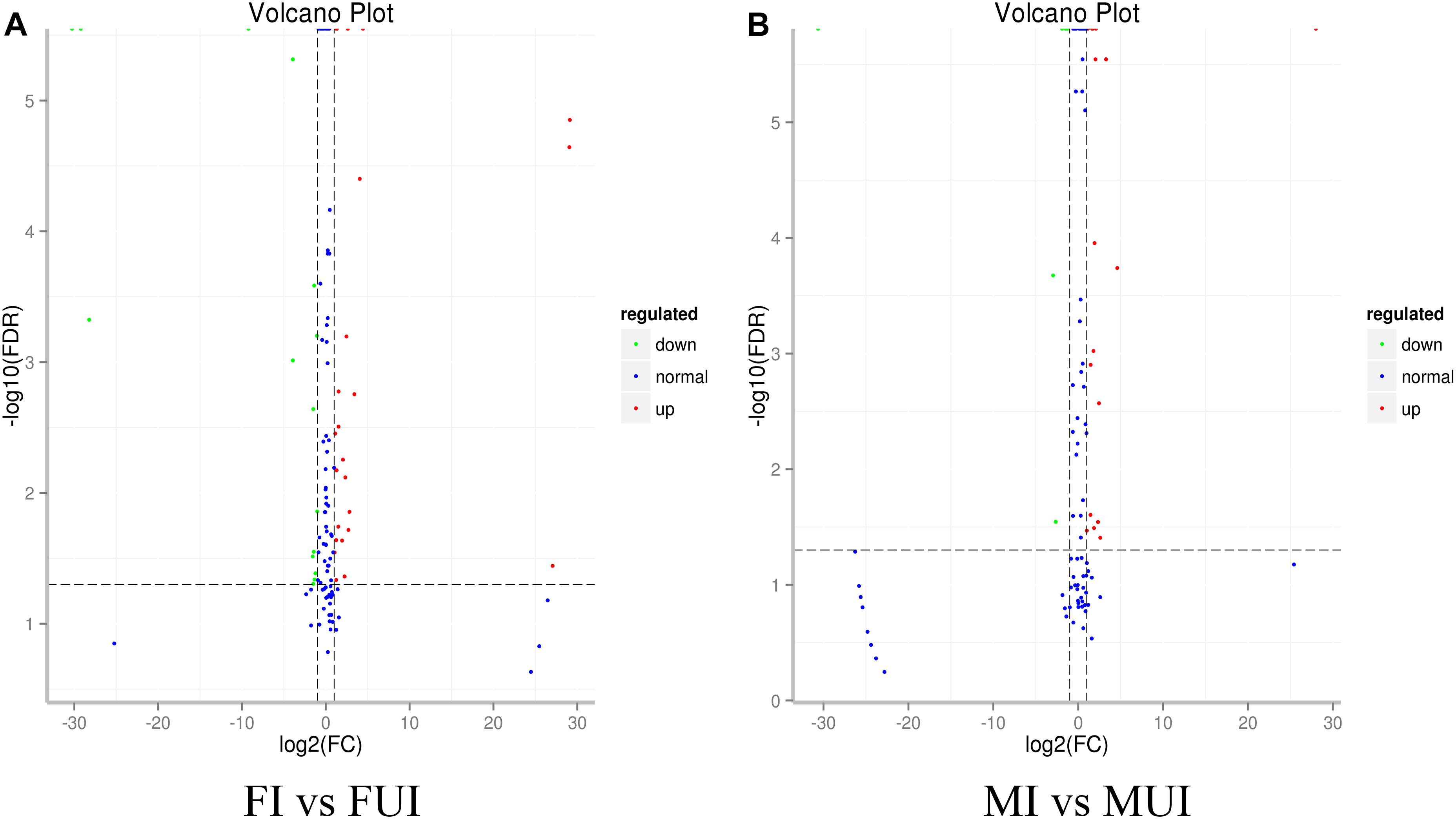
Figure 3. Volcano maps of differential miRNAs analysis between Wolbachia-infected and Wolbachia-uninfected L. striatellus (A: Comparison between females; B: Comparison between males). Each point represents a miRNA, red dots indicate significantly up-regulated miRNAs, and green dots indicate significantly down-regulated miRNAs, blue dots indicate no significant difference in miRNAs.
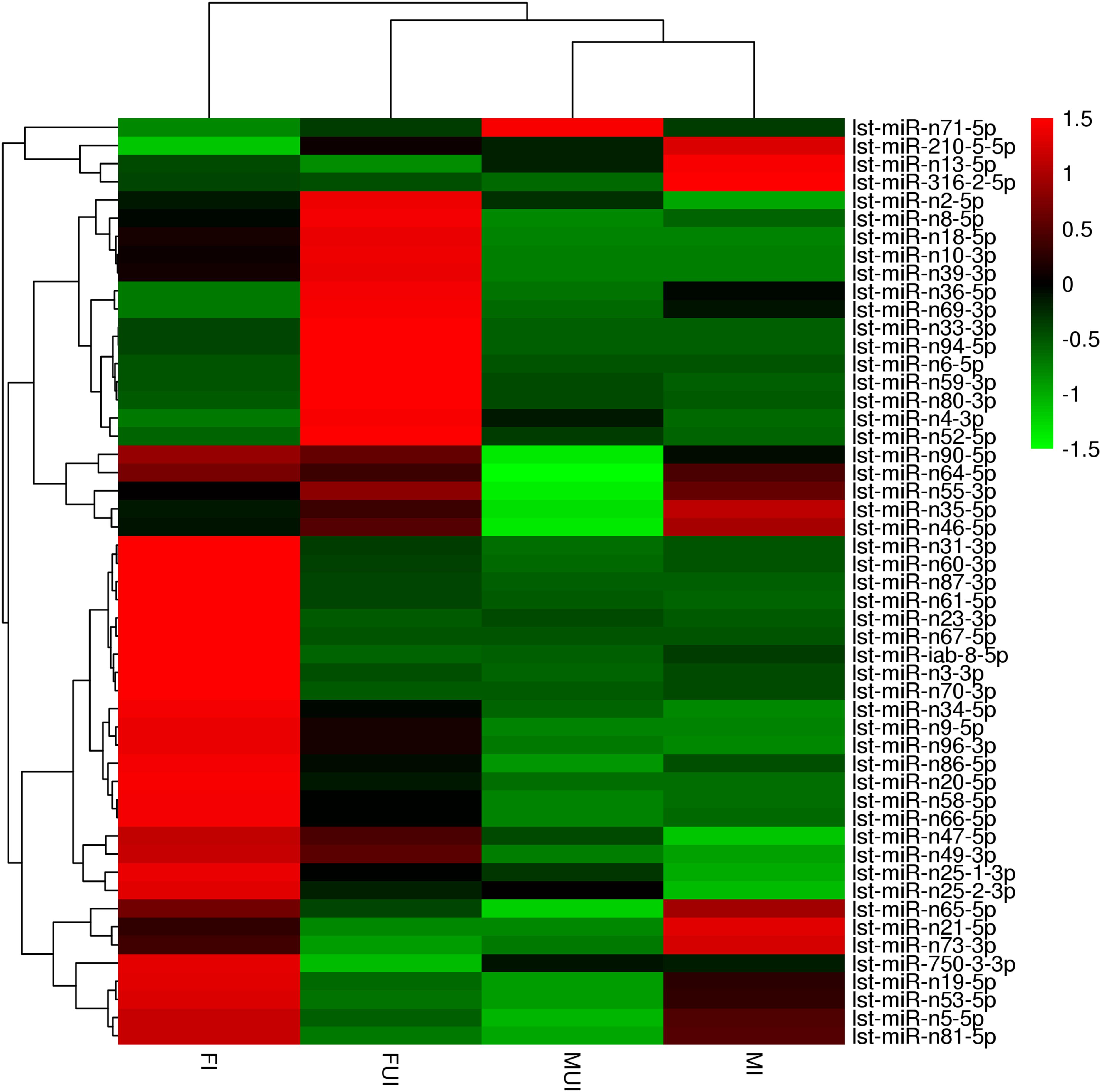
Figure 4. Heat map showing the differentially expressed miRNAs in four libraries (FUI, FI, MUI and MI) of L. striatellus. The fold-change ratios of the miRNAs indicated by the different colors, red indicates higher levels of miRNAs and green indicates lower levels of miRNAs.
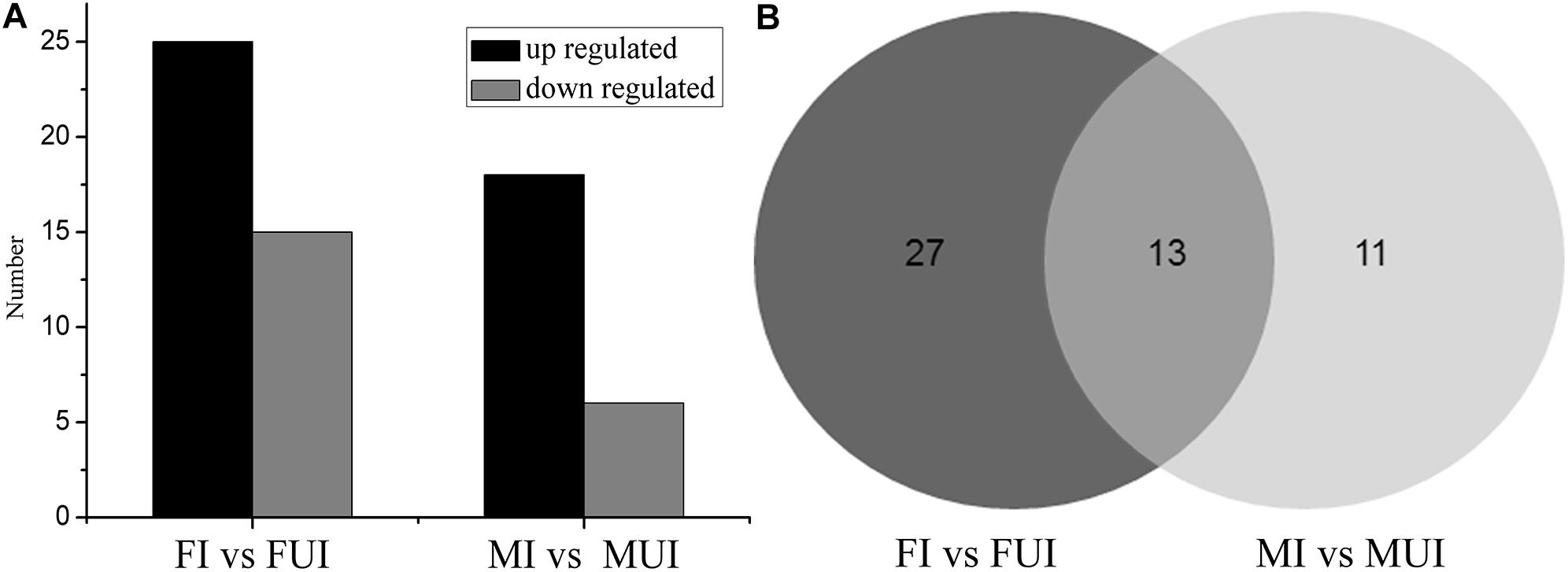
Figure 5. The statistics of differentially expressed miRNAs in two comparisons of L. striatellus (FI vs. FUI and MI vs. MUI). (A) Contrast between up-regulated and down-regulated differentially expressed miRNAs in two comparisons of L. striatellus. (B) Venn diagram of differentially expressed miRNAs that were shared and unique in two comparisons of L. striatellus.
Interestingly, 13 differentially expressed miRNAs were shared in the comparisons of female and male (Figure 5B and Table 3). Among them, three miRNAs were down-regulated and 8 miRNAs were up-regulated in both females and males after infection with Wolbachia. The miRNA lst-miR-n21-5p showed the highest degree of up-regulation in Wolbachia-infected females and males, and both lst-miR-n4-3p and lst-miR-n52-5p showed the highest degree of down-regulation in Wolbachia-infected females and males. Besides, lst-miR-n25-2-3p showed up-regulation in females, but was down-regulated in males. On the contrary, lst-miR-n8-5p showed down-regulation in females, but was up-regulated in males. These data show that the miRNAs in response to Wolbachia-infected in both sexes of L. striatellus which might have connections and differences.
Target Gene Prediction, and GO and KEGG Analyses of Differentially Expressed miRNAs
To better understand the function of Wolbachia-responsive miRNAs, 926 and 799 predicted target genes were predicted for the 44 differentially expressed miRNAs in female and male comparisons, respectively (Supplementary Tables S5, S6).
The GO enrichment analysis indicated that the predicted target genes related to metabolic process (198 genes in FI vs. FUI and 169 genes in MI vs. MUI), catalytic activity (186 genes in FI vs. FUI and 161 genes in MI vs. MUI), cellular process (160 genes in FI vs. FUI and 122 genes in MI vs. MUI), binding (148 genes in FI vs. FUI and 112 genes in MI vs. MUI), single-organism process (119 genes in FI vs. FUI and 91 genes in MI vs. MUI) were the most enriched categories in both comparisons (Figure 6A). Notably, most of categories in GO enrichment showed that the predicted target gene number in the female comparison were more than in the male comparison. As mentioned above, these results also suggested that Wolbachia infection had a broader impact on females than males. In view of previous research showed the reproduction and immunity of host might be affected by Wolbachia, we also found predicted genes related to immune system process, reproductive process, reproduction, developmental process, response to stimulus, antioxidant activity and growth were enriched in female and male comparisons (Figure 6A).
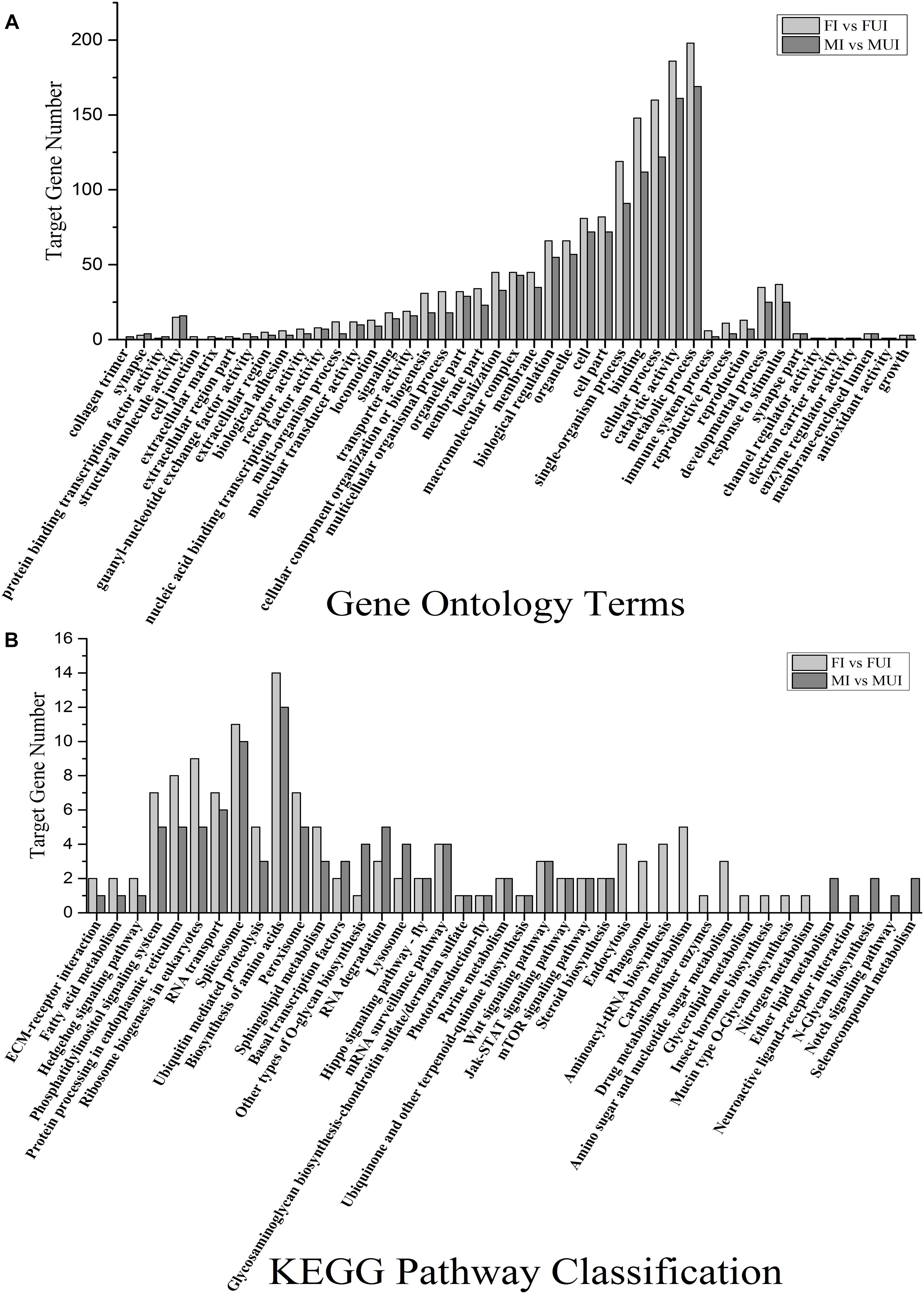
Figure 6. Gene ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) classification of the target genes of the two comparisons in L. striatellus. The y-axis showed the number of genes, and the x-axis showed the GO category (A) and the KEGG classification (B), respectively.
KEGG pathways analysis showed that the predicted target genes were annotated to 36 and 31 KEGG pathways in female and male comparisons, respectively. Among them, 26 KEGG pathways were shared in both female and male comparisons (Figure 6B), and the predicted target genes related to biosynthesis of amino acids and spliceosome were the most enriched pathways in both comparisons. In this study, we observed that target genes related to reproduction (mTOR signaling pathway), immune (lysosome, spliceosome, peroxisome, and Jak-STAT signaling pathway), and sphingolipid metabolism and steroid biosynthesis were enriched in both female and male comparisons. Intriguingly, some pathways such as endocytosis, phagosome, aminoacyl-tRNA biosynthesis, carbon metabolism, amino sugar and nucleotide sugar metabolism, glycerolipid metabolism and insect hormone biosynthesis were only enriched in the female comparison. Meanwhile, other pathways such as ether lipid metabolism, neuroactive ligand-receptor interaction, N-glycan biosynthesis, notch signaling pathway and selenocompound metabolism were only enriched in the male comparison.
qRT-PCR Validations of Differently Expressed miRNA and Their Predicted Target Genes
The quantitative real time polymerase chain reaction was conducted to further validate the expression pattern of differentially expressed miRNAs. The expression patterns of several miRNAs by Illumina sequencing and qRT-PCR were shown in Figures 7A–D. We found that the miRNA lst-miR-n52-5p was significantly down-regulated while another miRNA lst-miR-n21-5p was significantly up-regulated in both Wolbachia-infected females and males. In addition, the miRNA lst-miR-n6-5p was also significantly down-regulated in Wolbachia infected females. Although high-throughput sequencing of small RNAs could reveal a lot of useful information, it could also produce some inaccurate results. For instance, in this study, the Illumina sequencing results of these two miRNAs lst-miR-n49-3p and lst-miR-n5-5p in males were inconsistent with their qRT-PCR results. However, the expression patterns of their predicted target genes were opposite to the expression of their corresponding miRNAs (including lst-miR-n49-3p and lst-miR-n5-5p) except LsDscam (Figures 7E,F). We preliminarily speculate that LsDscam may not be the real target gene of the miRNA lst-miR-n52-5p. In summary, our RNA sequencing results could reflect the effects of Wolbachia infection on the expression of miRNAs and their corresponding target genes in the L. striatellus.
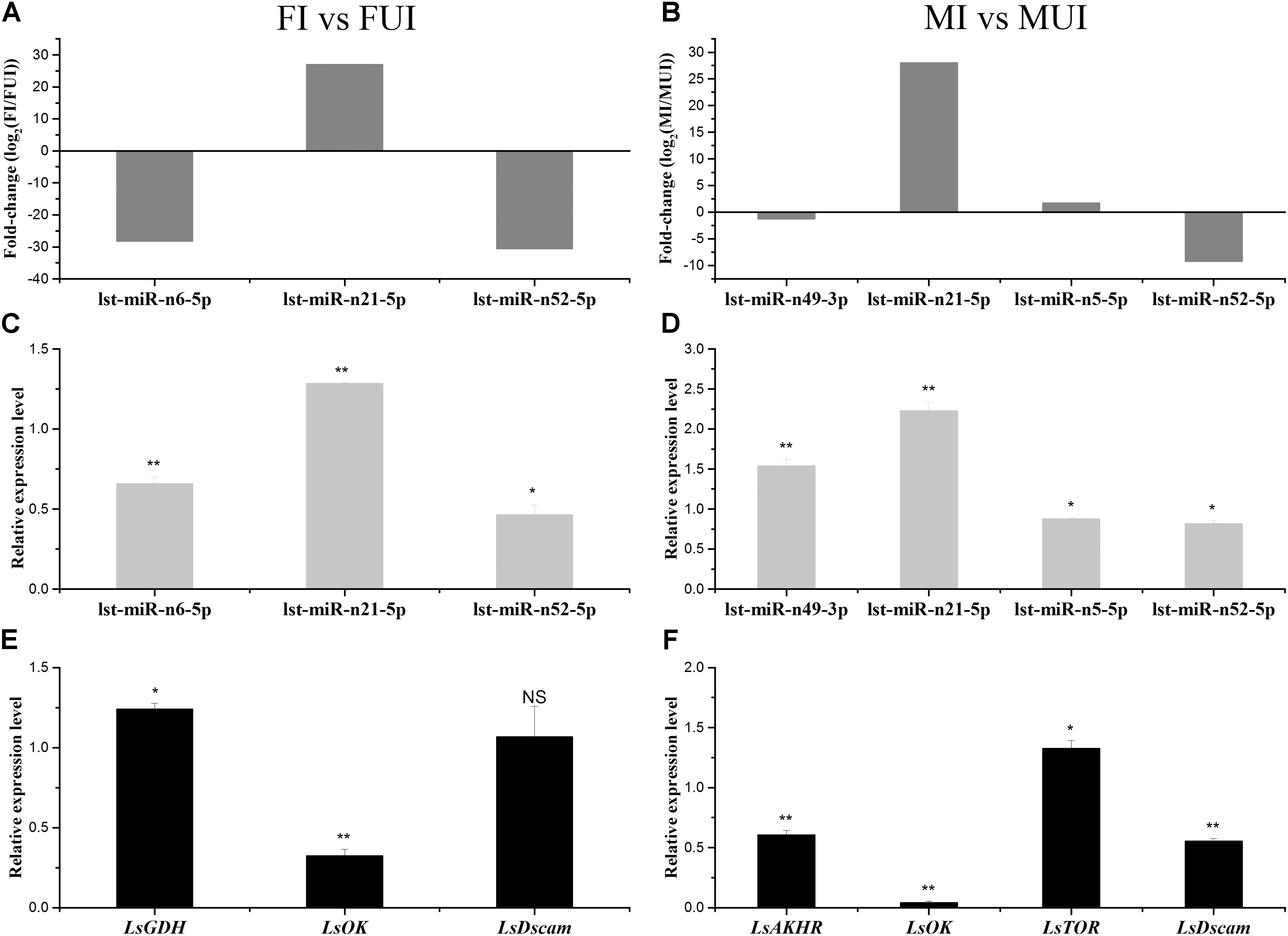
Figure 7. Expression patterns of five differentially expressed miRNAs and their predicted target genes in response to Wolbachia infection of L. striatellus. For the comparisons FI vs. FUI and MI vs. MUI, the relative expression levels of differentially expressed miRNAs were identified by Illumina sequencing (A,B) and qRT-PCR methods (C,D), respectively. (E,F) The expression profiles of corresponding target gene of differentially expressed miRNAs were verified using qRT-PCR methods. The corresponding mRNA under each miRNA is its predicted target gene. GDH, glutamate dehydrogenase; OK, orcokinin; Dscam, Down syndrome cell adhesion molecule-like protein; AKHR, adipokinetic hormone receptor; TOR, target of rapamycin. The error bars indicate standard errors of averages from three biological replicates (NS: not significant; *P < 0.05; ∗∗P < 0.01).
Discussion
At present very little is known about the effects of Wolbachia-infection on host miRNA expression. In the present study, we constructed and sequenced four sRNA libraries from both sexes of Wolbachia-infected and Wolbachia-uninfected L. striatellus. The results showed that Wolbachia infection caused a change in the expression of miRNA in the L. striatellus. These differentially expressed miRNAs may be involved in multiple aspects of the biological characteristics of the host. For instance, miR-210 play a role in mitotic progression and modulating circadian outputs (He et al., 2013; Cusumano et al., 2018). We also found the miRNA lst-miR-210-5p was up-regulated in Wolbachia-infected male L. striatellus, which targets a ubiquitin-protein ligase E3A gene (Figure 4 and Supplementary Table S5). The ubiquitin-protein ligase E3A plays an essential role in the regulation of the circadian system in mammals and flies (Gossan et al., 2014). Although the role of these differentially expressed miRNAs were based solely on their predicted target genes, and even many of them were newly discovered, we still discussed several important differentially expressed miRNAs that supposedly affected multiple biologic aspects of its host below.
miRNAs in Response to Wolbachia May Be Involved in Ecdysteroidogenesis of L. striatellus
Existing research result indicated that Wolbachia might be involved in ecsysteroidogenesis (Negri, 2011). For instance, in filarial worms, Wolbachia might play a critical role in host embryogenesis and molting (Casiraghi et al., 2002; Arumugam et al., 2008). In Eurema hecabe, removing Wolbachia resulting phenotypic defects were similar to knock-out of ecdysone receptor (EcR) gene in Blattella germanica and Drosophila melanogaster (Negri, 2011). Here we showed that the miRNA lst-miR-n21-5p was up-regulated in Wolbachia-infected females and males of L. striatellus which target a orcokinin (OK) gene (Figures 7C,D). The OK gene encoded a kind of neuropeptide that has been identified in a variety of arthropods, and this gene was down-regulated in both Wolbachia-infected females and males, with especially lower expression level in Wolbachia-infected males (Figures 7E,F). In Rhodnius prolixus, 20-hydroxyecdysone (20E) could regulate the expression level of OK gene (Wulff et al., 2018), and the OK gene was also involved in the neuronal regulation of ecdysteroidogenesis in Bombyx mori (Yamanaka et al., 2011). We hypothesized that Wolbachia interferes with the pathway involved in ecdysteroidogenesis by regulating expression of lst-miR-n21-5p in L. striatellus.
In addition, our results also showed that lst-miR-n3-3p whose predicted target gene, the Halloween gene Shade (Shd, cyp314a1) was notably up-regulated in Wolbachia-infected females of L. striatellus. The Shd gene was a cytochrome P450 monooxygenase (CYP) which catalyzes the conversion of ecdysone into active 20E. In L. striatellus, knock-down of the Shd gene could decrease expression level of EcR gene, and also significantly decrease the titer of 20E (Jia et al., 2013; Zhai et al., 2017). Similarly, nuclear hormone receptor E75 gene, a crucial 20E response gene that affects ecdysteroid titer was also downregulated in Wolbachia-infected females and males of T. urticae (Rong et al., 2014). Taken together, our data seem to support previous hypothesis that there might be a link between Wolbachia and ecdysteroid signaling (Negri, 2011).
Wolbachia-Responsive miRNA May Involve in Immune Response of L. striatellus
It was theorized that Wolbachia can activate the immune system of host insect. We found that miRNA lst-miR-n52-5p was down-regulated in both Wolbachia-infected female and male of L. striatellus. Although the results of qRT-PCR indicated that LsDscam might not be the real target gene of this miRNA, we also found another predicted target gene, hexamerin gene which was one gene of the haemocyanin protein family (Figure 4 and Supplementary Tables S5, S6). In adults of Riptortus pedestris, that was infected with gut symbiont Burkholderia, the hexamerin-a and hexamerin-b proteins were highly expressed compared to uninfected individuals (Lee et al., 2017). The hexamerin gene was up-regulated in Spiroplasma citri-infected Circulifer haematoceps, and RNAi knockdown of hexamerin gene resulted in significant reduction in phenoloxidase-like activity, as well as increased mortality of S. citri-infected leafhoppers (Eliautout et al., 2016). In Ae. aegypti, the transcripts of three prophenoloxidase genes were up-regulated by Wolbachia infection (Kambris et al., 2009), phenoloxidase activity was also found significantly elevated in Wolbachia-infected females of D. melanogaster (Thomas et al., 2011). Therefore, it could be suspected that Wolbachia might enhance the expression level of hexamerin gene, and consequently, activity the phenoloxidase of L. striatellus through down-regulating the expression of lst-miR-n52-5p.
Female Fecundity May Be Regulated by miRNAs That Response to Wolbachia
Previous studies have shown that Wolbachia could enhance the fecundity of female hosts (Mazzetto et al., 2015; Rahimi-Kaldeh et al., 2017; Guo et al., 2018). In this study, we found that several target genes of differently expressed miRNAs responsing to Wolbachia infection were associated with female fecundity. The miRNA lst-miR-n36-5p was notably down-regulated in Wolbachia-infected females which targets a gene coding for vitellogenin-6 (vg6) (Figure 4 and Supplementary Table S5). In insect, the vg gene was highly expressed in the female fat body, and it could significantly affect on oviposition and egg hatchability (Ali et al., 2017; Zhang et al., 2017). Interestingly, lst-miR-n10-3p which targets nuclear hormone receptor Fushi tarazu-factor 1 beta (βFTZ-F1) gene was also observed down-regulated in Wolbachia-infected females (Figure 4 and Supplementary Table S5). In Drosophila, knockdown of βFTZ-F1 gene could prevent juvenile hormone (JH) activation, whereas overexpression enhanced the activation of JH (Dubrovsky et al., 2011). We speculate that Wolbachia appears to enhance the fecundity of L. striatellus by down-regulating the expression of lst-miR-n36-5p and lst-miR-n10-3p in female hosts.
In L. striatellus, it has already confirmed that Wolbachia infection can increase the fecundity of females (Guo et al., 2018). The results reported in this study might help to further reveal the cause of this phenomenon.
Male Fertility May Be Regulated by miRNAs in Response to Wolbachia
Although cytoplasmic incompatibility is the most famous reproductive phenotype caused by Wolbachia in arthropod, the mechanism of this phenomenon is still unclear. However, more and more researches about Wolbachia infection affecting on fertility of male host have been reported (Liu et al., 2014; Ju et al., 2017). In this study, we also annotated several differently expressed miRNAs whose predicted target gene were concerned with male fertility. For instance, the miRNA lst-miR-n5-5p that targets a gene coding target of rapamycin (TOR) was observed down-regulated in Wolbachia-infected males (Figures 7D,F). A growing number of studies show that the mammalian target of rapamycin (mTOR) signaling pathway plays a crucial role in spermatogenesis (Schell et al., 2016; Serra et al., 2017). Another fertility-related miRNA gene, lst-miR-n13-5p was also observed up-regulated in Wolbachia-infected males, and it targets a gene coding juvenile hormone esterase (JHE) (Figure 4 and Supplementary Table S6). JHE play an important role in the regulation of JH titer (Mackert et al., 2008; Tsubota et al., 2010). In D. melanogaster, the researchers also found that over-expression of the Juvenile hormone-inducible protein 26 (JhI-26) gene in Wolbachia-uninfected males resulted in a significant reduction in egg hatching rates after mating with Wolbachia-uninfected females, and that Wolbachia-infected females could rescue egg hatching (Liu et al., 2014). This result show that the occurrence of fertility might related to the changes of juvenile hormone levels. It is possible that Wolbachia down-regulates the expression of jhe gene to interfere the JH pathway by up-regulate the lst-miR-n13-5p in males, thereby possibly induce paternal defects in fertility. In addition, the miRNA lst-miR-n81-5p was also up-regulated in Wolbachia-infected males, it targets a sperm-associated antigen 6 (Spag6) gene (Figure 4 and Supplementary Table S6). The Spag6 initially found in human testis and it was essential for sperm motility and male fertility (Sapiro et al., 2002). The down-regulation of lst-miR-n81-5p maybe in associated with decreased fertility.
Redox Homeostasis of L. striatellus May Be Regulated by miRNAs in Response to Wolbachia
It was considered that Wolbachia regulated redox homeostasis to maintain their relationship with host (Zug and Hammerstein, 2015). The miRNA lst-miR-n23-3p was likely to involve in oxidation-reduction reactions and showed up-regulated in Wolbachia-infected females which targets a gene coded mitochondrial manganese superoxide dismutase (mMnSOD) (Figure 4 and Supplementary Table S5). Similarly, Wolbachia infection significantly reduced SOD activity in the larvae of D. melanogaster (Wang et al., 2012). The miRNA lst-miR-n47-5p was found down-regulated in Wolbachia-infected males that targets a gene coded thioredoxin reductase (TrxR) (Figure 4 and Supplementary Table S6). The expression of TrxR gene was up-regulated in varying degrees after M. anisopliae and E. coli infection of Helicoverpa armigera (Zhang et al., 2015). Interestingly, silencing TrxR gene caused a significant decrease in the native bacterial load of the ticks in both the midgut and salivary glands (Budachetri and Karim, 2015). In L. striatellus, native Wolbachia may utilize mMnSOD and TrxR to better coexist with its host.
Conclusion
In summary, this study provides the first information on miRNAs of L. striatellus responding to Wolbachia. Our results suggest that Wolbachia may manipulate the physiological processes of L. striatellus by using miRNAs of host, and these results could contribute to further insight into the mechanisms of Wolbachia-host interaction. However, there were no confirmed targets for L. striatellus miRNAs in our research, future research is necessary to confirm interaction between these miRNAs and their predicted gene targets.
Data Availability
Publicly available datasets were analyzed in this study. This data can be found at https://www.ncbi.nlm.nih.gov/sra.
Ethics Statement
The research project was conducted on insect pest species that are not subjected to any specific ethical issue and legislation.
Author Contributions
K-JZ, LL, and HL conceived and designed the study. LL and K-JZ performed the experiments, analyzed the data, and drafted the manuscript. XR, Y-YL, and HL participated in manuscript drafted and modification. All authors read and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (Grant 31401801), the Fundamental and Advanced Research Program of Chongqing (Grant 2014jcyjA80009), and the China Postdoctoral Science Foundation (Grant 2014M562270).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank associate professor Yuan Guo-Rui at College of Plant Protection, Southwest University for the helpful discussion of these data and the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00928/full#supplementary-material
FIGURE S1 | Venn diagram illustrating the numbers and percentages of total and unique reads between various comparisons of L. striatellus. Total sRNA, total number of sRNA reads; Unique sRNA, numbers of sRNA types.
TABLE S1 | The primers used for reverse transcription quantitative PCR (RT-qPCR) of miRNA.
TABLE S2 | The primers used for reverse transcription quantitative PCR (RT-qPCR) of predicted target genes.
TABLE S3 | Statistics of sequencing data in the four libraries of L. striatellus.
TABLE S4 | Description of the 49 conserved and 103 newly miRNAs identified in L. striatellus.
TABLE S5 | Predicted target genes that may be reverse regulated by differentially expressed miRNAs in the female comparison.
TABLE S6 | Predicted target genes that may be reverse regulated by differentially expressed miRNAs in the male comparison.
Footnotes
- ^ http://www.arb-silva.de/
- ^ http://lowelab.ucsc.edu/GtRNAdb/
- ^ http://rfam.xfam.org/
- ^ http://www.girinst.org/repbase/
- ^ http://www.mirbase.org/
- ^ http://bioinfo.ut.ee/primer3/
References
Ali, M. W., Zhang, Z. Y., Xia, S., and Zhang, H. (2017). Biofunctional analysis of vitellogenin and vitellogenin receptor in citrus red mites, Panonychus citri by RNA interference. Sci. Rep. 7:16123. doi: 10.1038/s41598-017-16331-3
Ambros, V., Bartel, B., Bartel, D. P., Burge, C. B., Carrington, J. C., Chen, X. M., et al. (2003). A uniform system for microRNA annotation. RNA. 9, 277–279. doi: 10.1261/rna.2183803
Ant, T. H., Herd, C. S., Geoghegan, V., Hoffmann, A. A., and Sinkins, S. P. (2018). The Wolbachia strain wAu provides highly efficient virus transmission blocking in Aedes aegypti. PLoS Pathog 14:e1006815. doi: 10.1371/journal.ppat.1006815
Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., et al. (2004). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 32, D115–D119. doi: 10.1093/nar/gkh131
Arumugam, S., Pfarr, K. M., and Hoerauf, A. (2008). Infection of the intermediate mite host with Wolbachia-depleted Litomosoides sigmodontis microfilariae: impaired L1 to L3 development and subsequent sex-ratio distortion in adult worms. Int. J. Parasitol. 38, 981–987. doi: 10.1016/j.ijpara.2007.12.006
Asgari, S. (2011). Role of microRNAs in insect host-microorganism interactions. Front. Physiol. 2:48. doi: 10.3389/fphys.2011.00048
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Betel, D., Wilson, M., Gabow, A., Marks, D. S., and Sander, C. (2008). The microRNA.org resource: targets and expression. Nucleic Acids Res. 36, D149–D153. doi: 10.1093/nar/gkm995
Bian, G. W., Xu, Y., Lu, P., Xie, Y., and Xi, Z. Y. (2010). The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog 6:e1000833. doi: 10.1371/journal.ppat.1000833
Braig, H. R., Zhou, W. G., Dobson, S. L., and O’Neill, S. L. (1998). Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J. Bacteriol. 180, 2373–2378.
Budachetri, K., and Karim, S. (2015). An insight into the functional role of thioredoxin reductase, a selenoprotein, in maintaining normal native microbiota in the Gulf Coast tick (Amblyomma maculatum). Insect Mol. Biol. 24, 570–581. doi: 10.1111/imb.12184
Casiraghi, M., McCall, J. W., Simoncini, L., Kramer, L. H., Sacchi, L., Genchi, C., et al. (2002). Tetracycline treatment and sex-ratio distortion: a role for Wolbachia in the moulting of filarial nematodes? Int. J. Parasitol. 32, 1457–1468. doi: 10.1016/S0020-7519(02)00158-3
Cusumano, P., Biscontin, A., Sandrelli, F., Mazzotta, G. M., Tregnago, C., De Pitta, C., et al. (2018). Modulation of miR-210 alters phasing of circadian locomotor activity and impairs projections of PDF clock neurons in Drosophila melanogaster. Plos Genet. 14:e1007500. doi: 10.1371/journal.pgen.1007500
Deng, Y., Li, J., Wu, S., Zhu, Y., Chen, Y., and He, F. (2006). Integrated nr database in protein annotation system and its localization. Comput. Eng. 32, 71–74. doi: 10.1109/INFOCOM.2006.241
Dubrovsky, E. B., Dubrovskaya, V. A., Bernardo, T., Otte, V., DiFilippo, R., and Bryan, H. (2011). The Drosophila FTZ-F1 nuclear receptor mediates juvenile hormone activation of E75A gene expression through an intracellular pathway. J. Biol. Chem. 286, 33689–33700. doi: 10.1074/jbc.M111.273458
Eddy, S. R. (1998). Profile hidden Markov models. Bioinformatics. 14, 755–763. doi: 10.1093/bioinformatics/14.9.755
Eleftherianos, I., Atri, J., Accetta, J., and Castillo, J. C. (2013). Endosymbiotic bacteria in insects: guardians of the immune system? Front. Physiol. 4:46. doi: 10.3389/fphys.2013.00046
Eliautout, R., Dubrana, M. P., Vincent-Monegat, C., Vallier, A., Braquart-Varnier, C., Poirie, M., et al. (2016). Immune response and survival of Circulifer haematoceps to Spiroplasma citri infection requires expression of the gene hexamerin. Dev. Comp. Immunol. 54, 7–19. doi: 10.1016/j.dci.2015.08.007
Eun, S. H., Stoiber, P. M., Wright, H. J., McMurdie, K. E., Choi, C. H., Gan, Q., et al. (2013). MicroRNAs downregulate bag of marbles to ensure proper terminal differentiation in the Drosophila male germline. Development. 140, 23–30. doi: 10.1242/dev.086397
Friedlander, M. R., Mackowiak, S. D., Li, N., Chen, W., and Rajewsky, N. (2012). MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52. doi: 10.1093/nar/gkr688
Gossan, N. C., Zhang, F., Guo, B., Jin, D., Yoshitane, H., Yao, A., et al. (2014). The E3 ubiquitin ligase UBE3A is an integral component of the molecular circadian clock through regulating the BMAL1 transcription factor. Nucleic Acids Res. 42, 5765–5775. doi: 10.1093/nar/gku225
Guo, Y., Hoffmann, A. A., Xu, X. Q., Zhang, X., Huang, H. J., Ju, J. F., et al. (2018). Wolbachia-induced apoptosis associated with increased fecundity in Laodelphax striatellus (Hemiptera: Delphacidae). Insect Mol. Biol. 27, 796–807. doi: 10.1111/imb.12518
Gupta, V., Vasanthakrishnan, R. B., Siva-Jothy, J., Monteith, K. M., Brown, S. P., and Vale, P. F. (2017). The route of infection determines Wolbachia antibacterial protection in Drosophila. Proc. Biol. Sci. 284:20170809. doi: 10.1098/rspb.2017.0809
He, J., Wu, J., Xu, N., Xie, W., Li, M., Li, J., et al. (2013). MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucleic Acids Res. 41, 498–508. doi: 10.1093/nar/gks995
He, X. T., Liu, C. C., Li, Z. Q., Zhang, Z., Li, G. Q., Li, F., et al. (2014). Validation of reference genes for quantitative real-time PCR in Laodelphax striatellus. J. Integr. Agr. 13, 811–818. doi: 10.1016/S2095-3119(13)60515-8
Hughes, G. L., Koga, R., Xue, P., Fukatsu, T., and Rasgon, J. L. (2011). Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog 7:e1002043. doi: 10.1371/journal.ppat.1002043
Hussain, M., Frentiu, F. D., Moreira, L. A., O’Neill, S. L., and Asgari, S. (2011). Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 108, 9250–9255. doi: 10.1073/pnas.1105469108
Hussain, M., O’Neill, S. L., and Asgari, S. (2013). Wolbachia interferes with the intracellular distribution of Argonaute 1 in the dengue vector Aedes aegypti by manipulating the host microRNAs. RNA Biol. 10, 1868–1875. doi: 10.4161/rna.27392
Jia, S., Wan, P. J., Zhou, L. T., Mu, L. L., and Li, G. Q. (2013). Knockdown of a putative halloween gene shade reveals its role in ecdysteroidogenesis in the small brown planthopper Laodelphax striatellus. Gene. 531, 168–174. doi: 10.1016/j.gene.2013.09.034
Joshi, D., Pan, X., McFadden, M. J., Bevins, D., Liang, X., Lu, P., et al. (2017). The maternally inheritable Wolbachia wAlbB induces refractoriness to Plasmodium berghei in Anopheles stephensi. Front. Microbiol. 8:366. doi: 10.3389/fmicb.2017.00366
Ju, J. F., Hoffmann, A. A., Zhang, Y. K., Duan, X. Z., Guo, Y., Gong, J. T., et al. (2017). Wolbachia-induced loss of male fertility is likely related to branch chain amino acid biosynthesis and iLvE in Laodelphax striatellus. Insect Biochem. Mol. Biol. 85, 11–20. doi: 10.1016/j.ibmb.2017.04.002
Kambris, Z., Blagborough, A. M., Pinto, S. B., Blagrove, M. S., Godfray, H. C., Sinden, R. E., et al. (2010). Wolbachia stimulates immune gene expression and inhibits plasmodium development in Anopheles gambiae. PLoS Pathog 6:e1001143. doi: 10.1371/journal.ppat.1001143
Kambris, Z., Cook, P. E., Phuc, H. K., and Sinkins, S. P. (2009). Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326, 134–136. doi: 10.1126/science.1177531
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280. doi: 10.1093/nar/gkh063
Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. doi: 10.1126/science.1064921
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome. Biol. 10, R25. doi: 10.1186/gb-2009-10-3-r25
Laven, H. (1957). Vererbung Durch Kerngene und das Problem der ausserkaryotischen Vererbung bei Culex Pipiens. II. Ausserkaryotische Vererbung. Z Indukt. Abstamm Vererbungsl. 88, 478–516. doi: 10.1007/Bf00309428
Lee, G. J., and Hyun, S. (2014). Multiple targets of the microRNA miR-8 contribute to immune homeostasis in Drosophila. Dev. Comp. Immunol. 45, 245–251. doi: 10.1016/j.dci.2014.03.015
Lee, J. B., Park, K. E., Lee, S. A., Jang, S. H., Eo, H. J., Jang, H. A., et al. (2017). Gut symbiotic bacteria stimulate insect growth and egg production by modulating hexamerin and vitellogenin gene expression. Dev. Comp. Immunol. 69, 12–22. doi: 10.1016/j.dci.2016.11.019
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y
Li, H. R., Wei, X. Y., Ding, T. B., and Chu, D. (2018). Genome-wide profiling of Cardinium-Responsive microRNAs in the exotic whitefly, Bemisia tabaci (Gennadius) biotype Q. Front. Physiol. 9:1580. doi: 10.3389/Fphys.2018.01580
Li, J. M., Zhou, Y. R., Sun, Z. T., Wang, X., Xie, L., and Chen, J. P. (2015). Identification and profiling of conserved and novel microRNAs in Laodelphax striatellus in response to rice black-streaked dwarf virus (RBSDV) infection. Genom. Data 3, 63–69. doi: 10.1016/j.gdata.2014.08.010
Li, L., Li, H., Dong, H., Wang, X., and Zhou, G. (2011). Transmission by Laodelphax striatellus Fallen of rice black-streaked dwarf virus from frozen infected rice leaves to healthy plants of rice and maize. J. Phytopathol. 159, 1–5. doi: 10.1111/j.1439-0434.2010.01713.x
Liu, C., Wang, J. L., Zheng, Y., Xiong, E. J., Li, J. J., Yuan, L. L., et al. (2014). Wolbachia-induced paternal defect in Drosophila is likely by interaction with the juvenile hormone pathway. Insect Biochem. Mol. Biol. 49, 49–58. doi: 10.1016/j.ibmb.2014.03.014
Liu, Y., Zhou, Y., Wu, J., Zheng, P., Li, Y., Zheng, X., et al. (2015). The expression profile of Aedes albopictus miRNAs is altered by dengue virus serotype-2 infection. Cell Biosci. 5, 16. doi: 10.1186/s13578-015-0009-y
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-delta delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lourenco, A. P., Guidugli-Lazzarini, K. R., Freitas, F. C., Bitondi, M. M., and Simoes, Z. L. (2013). Bacterial infection activates the immune system response and dysregulates microRNA expression in honey bees. Insect Biochem. Mol. Biol. 43, 474–482. doi: 10.1016/j.ibmb.2013.03.001
Mackert, A., do Nascimento, A. M., Bitondi, M. M., Hartfelder, K., and Simoes, Z. L. (2008). Identification of a juvenile hormone esterase-like gene in the honey bee, Apis mellifera L.-expression analysis and functional assays. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 150, 33–44. doi: 10.1016/j.cbpb.2008.01.004
Mayoral, J. G., Etebari, K., Hussain, M., Khromykh, A. A., and Asgari, S. (2014). Wolbachia infection modifies the profile, shuttling and structure of microRNAs in a mosquito cell line. PLoS One 9:e96107. doi: 10.1371/journal.pone.0096107
Mazzetto, F., Gonella, E., and Alma, A. (2015). Wolbachia infection affects female fecundity in Drosophila suzukii. B Insectol 68, 153–157.
Monsanto-Hearne, V., and Johnson, K. N. (2018). Wolbachia-mediated protection of Drosophila melanogaster against systemic infection with its natural viral pathogen Drosophila C virus does not involve changes in levels of highly abundant miRNAs. J. Gen. Virol. 99, 827–831. doi: 10.1099/jgv.0.001064
Moreira, L. A., Iturbe-Ormaetxe, I., Jeffery, J. A., Lu, G., Pyke, A. T., Hedges, L. M., et al. (2009). A Wolbachia symbiont in Aedes aegypti limits infection with dengue. Cell 139, 1268–1278. doi: 10.1016/j.cell.2009.11.042
Mousson, L., Martin, E., Zouache, K., Madec, Y., Mavingui, P., and Failloux, A. B. (2010). Wolbachia modulates Chikungunya replication in Aedes albopictus. Mol Ecol 19, 1953–1964. doi: 10.1111/j.1365-294X.2010.04606.x
Negri, I. (2011). Wolbachia as an “infectious” extrinsic factor manipulating host signaling pathways. Front. Endocrinol. 2:115. doi: 10.3389/fendo.2011.00115
Noda, H., Koizumi, Y., Zhang, Q., and Deng, K. (2001). Infection density of Wolbachia and incompatibility level in two planthopper species. Insect Biochem. Mol. Biol. 31, 727–737. doi: 10.1016/S0965-1748(00)00180-6
Osei-Amo, S., Hussain, M., O’Neill, S. L., and Asgari, S. (2012). Wolbachia-induced aae-miR-12 miRNA negatively regulates the expression of MCT1 and MCM6 genes in Wolbachia-infected mosquito cell line. PLoS One 7:e50049. doi: 10.1371/journal.pone.0050049
Pan, X., Zhou, G., Wu, J., Bian, G., Lu, P., Raikhel, A. S., et al. (2012). Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 109, E23–E31. doi: 10.1073/pnas.1116932108
Rahimi-Kaldeh, S., Ashouri, A., Bandani, A., and Tomioka, K. (2017). The effect of Wolbachia on diapause, fecundity, and clock gene expression in Trichogramma brassicae (Hymenoptera: Trichogrammatidae). Dev. Genes. Evol. 227, 401–410. doi: 10.1007/s00427-017-0597-0
Rances, E., Ye, Y. H., Woolfit, M., McGraw, E. A., and O’Neill, S. L. (2012). The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathog 8:e1002548. doi: 10.1371/journal.ppat.1002548
Rehmsmeier, M., Steffen, P., Hochsmann, M., and Giegerich, R. (2004). Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517. doi: 10.1021/rna.5248604
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. doi: 10.1038/35002607
Romualdi, C., Bortoluzzi, S., D’Alessi, F., and Danieli, G. A. (2003). IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics. 12, 159–162. doi: 10.1152/physiolgenomics.00096.2002
Rong, X., Zhang, Y. K., Zhang, K. J., and Hong, X. Y. (2014). Identification of Wolbachia-responsive microRNAs in the two-spotted spider mite. BMC Genomics. 15:1122. doi: 10.1186/1471-2164-15-1122
Sapiro, R., Kostetskii, I., Olds-Clarke, P., Gerton, G. L., Radice, G. L., and Strauss III, J. F. (2002). Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol. Cell Biol. 22, 6298–6305. doi: 10.1128/mcb.22.17.6298-6305.2002
Schell, C., Kretz, O., Liang, W., Kiefer, B., Schneider, S., Sellung, D., et al. (2016). The rapamycin-sensitive complex of mammalian target of rapamycin is essential to maintain male fertility. Am. J. Pathol. 186, 324–336. doi: 10.1016/j.ajpath.2015.10.012
Serra, N. D., Velte, E. K., Niedenberger, B. A., Kirsanov, O., and Geyer, C. B. (2017). Cell-autonomous requirement for mammalian target of rapamycin (Mtor) in spermatogonial proliferation and differentiation in the mouse. Biol. Reprod. 96, 816–828. doi: 10.1093/biolre/iox022
Slonchak, A., Hussain, M., Torres, S., Asgari, S., and Khromykh, A. A. (2014). Expression of mosquito microRNA aae-miR-2940-5p is downregulated in response to west nile virus infection to restrict viral replication. J. Virol. 88, 8457–8467. doi: 10.1128/Jvi.00317-14
Storey, J. D. (2003). The positive false discovery rate: a bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035. doi: 10.1214/aos/1074290335
Thomas, P., Kenny, N., Eyles, D., Moreira, L. A., O’Neill, S. L., and Asgari, S. (2011). Infection with the wMel and wMelPop strains of Wolbachia leads to higher levels of melanization in the hemolymph of Drosophila melanogaster. Dev. Comp. Immunol. 35, 360–365. doi: 10.1016/j.dci.2010.11.007
Thomas, S., Verma, J., Woolfit, M., and O’Neilll, S. L. (2018). Wolbachia-mediated virus blocking in mosquito cells is dependent on XRN1-mediated viral RNA degradation and influenced by viral replication rate. PLoS Pathog 14:e1006879. doi: 10.1371/journal.ppat.1006879
Tsubota, T., Minakuchi, C., Nakakura, T., Shinoda, T., and Shiotsuki, T. (2010). Molecular characterization of a gene encoding juvenile hormone esterase in the red flour beetle. Insect Mol. Biol. 19, 527–535. doi: 10.1111/j.1365-2583.2010.01019.x
Wang, L., Zhou, C., He, Z., Wang, Z. G., Wang, J. L., and Wang, Y. F. (2012). Wolbachia infection decreased the resistance of Drosophila to lead. PLoS One 7:e32643. doi: 10.1371/journal.pone.0032643
Wong, Z. S., Hedges, L. M., Brownlie, J. C., and Johnson, K. N. (2011). Wolbachia-mediated antibacterial protection and immune gene regulation in Drosophila. PLoS One 6:e25430. doi: 10.1371/journal.pone.0025430
Wu, P., Han, S. H., Chen, T., Qin, G. X., Li, L., and Guo, X. J. (2013). Involvement of microRNAs in infection of Silkworm with Bombyx mori Cytoplasmic Polyhedrosis Virus (BmCPV). PLoS One 8:e68209. doi: 10.1371/journal.pone.0068209
Wulff, J. P., Capriotti, N., and Ons, S. (2018). Orcokinins regulate the expression of neuropeptide precursor genes related to ecdysis in the hemimetabolous insect Rhodnius prolixus. J. Insect Physiol. 108, 31–39. doi: 10.1016/j.jinsphys.2018.05.006
Xing, S., Du, J., Gao, S., Tian, Z., Zheng, Y., Liu, G., et al. (2016). Analysis of the miRNA expression profile in an Aedes albopictus cell line in response to bluetongue virus infection. Infect. Genet. Evol. 39, 74–84. doi: 10.1016/j.meegid.2016.01.012
Yamanaka, N., Roller, L., Zitnan, D., Satake, H., Mizoguchi, A., Kataoka, H., et al. (2011). Bombyx orcokinins are brain-gut peptides involved in the neuronal regulation of ecdysteroidogenesis. J. Comp. Neurol. 519, 238–246. doi: 10.1002/cne.22517
Ye, Y. H., Woolfit, M., Rances, E., O’Neill, S. L., and McGraw, E. A. (2013). Wolbachia-associated bacterial protection in the mosquito Aedes aegypti. PLoS Negl. Trop. Dis. 7:e2362. doi: 10.1371/journal.pntd.0002362
Yen, J. H., and Barr, A. R. (1971). New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature 232, 657–658. doi: 10.1038/232657a0
Zhai, Y. F., Zhang, Z. M., Gao, H. H., Chen, H., Sun, M., Zhang, W. Q., et al. (2017). Hormone signaling regulates nymphal diapause in Laodelphax striatellus (Hemiptera: Delphacidae). Sci Rep 7:13370. doi: 10.1038/S41598-017-13879-Y
Zhang, G., Hussain, M., and Asgari, S. (2014). Regulation of arginine methyltransferase 3 by a Wolbachia-induced microRNA in Aedes aegypti and its effect on Wolbachia and dengue virus replication. Insect Biochem. Mol. Biol. 53, 81–88. doi: 10.1016/j.ibmb.2014.08.003
Zhang, G., Hussain, M., O’Neill, S. L., and Asgari, S. (2013). Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 110, 10276–10281. doi: 10.1073/pnas.1303603110
Zhang, S., Li, L., Wang, X., and Zhou, G. (2007). Transmission of Rice stripe virus acquired from frozen infected leaves by the small brown planthopper (Laodelphax striatellus Fallen). J. Virol. Methods. 146, 359–362. doi: 10.1016/j.jviromet.2007.05.028
Zhang, S., Li, Z., Nian, X., Wu, F., Shen, Z., Zhang, B., et al. (2015). Sequence analysis, expression profiles and function of thioredoxin 2 and thioredoxin reductase 1 in resistance to nucleopolyhedrovirus in Helicoverpa armigera. Sci. Rep. 5:15531. doi: 10.1038/srep15531
Zhang, T., Zhang, G., Zeng, F., Mao, J., Liang, H., and Liu, F. (2017). Molecular cloning of the vitellogenin gene and the effects of vitellogenin protein expression on the physiology of Harmonia axyridis (Coleoptera: Coccinellidae). Sci. Rep. 7:13926. doi: 10.1038/s41598-017-14339-3
Zhang, Y., Zhao, B., Roy, S., Saha, T. T., Kokoza, V. A., Li, M., et al. (2016). microRNA-309 targets the Homeobox gene SIX4 and controls ovarian development in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 113, E4828–E4836. doi: 10.1073/pnas.1609792113
Zug, R., and Hammerstein, P. (2012). Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7:e38544. doi: 10.1371/journal.pone.0038544
Keywords: Laodelphax striatellus, Wolbachia, microRNA, endosymbiont, insect-symbiont interaction
Citation: Liu L, Zhang K-J, Rong X, Li Y-Y and Liu H (2019) Identification of Wolbachia-Responsive miRNAs in the Small Brown Planthopper, Laodelphax striatellus. Front. Physiol. 10:928. doi: 10.3389/fphys.2019.00928
Received: 10 April 2019; Accepted: 09 July 2019;
Published: 24 July 2019.
Edited by:
Peng He, Guizhou University, ChinaReviewed by:
Kai Lu, Fujian Agriculture and Forestry University, ChinaXiaoli Bing, Nanjing Agricultural University, China
Haijian Huang, Nanjing Agricultural University, China
Ary Hoffmann, The University of Melbourne, Australia
Copyright © 2019 Liu, Zhang, Rong, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai-Jun Zhang, a2p6aGFuZ0Bzd3UuZWR1LmNu; Huai Liu, bGl1aHVhaUBzd3UuZWR1LmNu
†These authors have contributed equally to this work
 Lei Liu
Lei Liu Kai-Jun Zhang
Kai-Jun Zhang Xia Rong
Xia Rong Huai Liu
Huai Liu
