
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 20 February 2019
Sec. Integrative Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fphys.2019.00133
This article is part of the Research Topic Circadian Plasticity – A Collaboration Between Neuronal and Glial Oscillators View all 11 articles
 Paola Cusumano1†
Paola Cusumano1† Milena Damulewicz2†
Milena Damulewicz2† Elena Carbognin1†
Elena Carbognin1† Laura Caccin1
Laura Caccin1 Antonietta Puricella3
Antonietta Puricella3 Valeria Specchia3
Valeria Specchia3 Maria Pia Bozzetti3
Maria Pia Bozzetti3 Rodolfo Costa1
Rodolfo Costa1 Gabriella M. Mazzotta1*
Gabriella M. Mazzotta1*Circadian clocks control and synchronize biological rhythms of several behavioral and physiological phenomena in most, if not all, organisms. Rhythm generation relies on molecular auto-regulatory oscillations of interlocked transcriptional-translational feedback loops. Rhythmic clock-gene expression is at the base of rhythmic protein accumulation, though post-transcriptional and post-translational mechanisms have evolved to adjust and consolidate the proper pace of the clock. In Drosophila, BELLE, a conserved DEAD-box RNA helicase playing important roles in reproductive capacity, is involved in the small RNA-mediated regulation associated to the piRNA pathway. Here, we report that BELLE is implicated in the circadian rhythmicity and in the regulation of endogenous transposable elements (TEs) in both nervous system and gonads. We suggest that BELLE acts as important element in the piRNA-mediated regulation of the TEs and raise the hypothesis that this specific regulation could represent another level of post-transcriptional control adopted by the clock to ensure the proper rhythmicity.
Circadian clocks have evolved in most, if not all, organisms to time their physiology and behavior to the most appropriate phases of the day–night cycles (Young and Kay, 2001; Dubruille and Emery, 2008; Peschel and Helfrich-Forster, 2011). They control and synchronize the biological rhythms of a vast array of behavioral and physiological phenomena, including visual sensitivity, hormone secretion, locomotion, hatching, eclosion, ecdysteroid synthesis, sperm release, gene expression, and sleep wake cycle (Sandrelli et al., 2008; Allada and Chung, 2010; Schiesari et al., 2011). Biological clocks are self-sustained in the absence of environmental cues. Rhythm generation occurs in single cells and is based on molecular auto-regulatory oscillations of interlocked transcriptional-translational feedback loops. In Drosophila, the transcription factors CLOCK (CLK) and CYCLE (CYC) activate, as heterodimer, the expression of the clock genes period (per) and timeless (tim). The correspondent proteins, PER and TIM, are post-translational regulated and eventually accumulate in the cytoplasm and enter the nucleus, where they ultimately inhibit their own transcription by repressing CLK-CYC activity. Two other interlocked feedback loops regulating the expression of the Clock gene and CLK/CYC mediated transcription, respectively, contribute to the robustness of the circadian oscillations (reviewed in Ozkaya and Rosato, 2012).
Rhythmic clock-gene expression is important to generate rhythmic protein accumulation; however, cycling protein levels do not depend only on cycling mRNAs but post-transcriptional and post-translational mechanisms have also evolved to adjust and consolidate the proper pace of the clock (Kojima et al., 2011). Among these, alternative splicing, mRNA nuclear export, polyadenilation, regulation of translation and degradation, play fundamental roles in the post-transcriptional regulation of circadian gene expression (Majercak et al., 1999, 2004; Collins et al., 2004; Boothroyd et al., 2007; Kojima et al., 2012; Huang et al., 2013; MacGregor et al., 2013; Robles et al., 2014; Montelli et al., 2015).
In recent years, a role for small, non-coding RNAs (miRNA) as post-transcriptional regulators of circadian rhythmicity in Drosophila has been also established. They regulate the proper development and maintenance of circadian rhythms acting either directly on the expression of clock genes or indirectly on signaling outputs (reviewed in Xue and Zhang, 2018).
Another class of small RNA, the piRNAs, has recently been associated to the age-dependent rhythmicity (Kuintzle et al., 2017). In particular, putative primary piRNA transcripts corresponding to transposons exhibit a de novo oscillation in old flies and circadian clock components appear to regulate these late rhythmicity (Kuintzle et al., 2017). piRNAs are small molecules protecting animal cells from the de-regulation of transposons and other repetitive genetic elements, preserving genome stability (Li et al., 2009; Thomson and Lin, 2009; Le Thomas et al., 2014). The piRNA biogenesis starts with the transcription of genomic clusters dispersed in the genome, producing the primary piRNA precursor transcripts (Brennecke et al., 2007). The piRNA pathway was first discovered in the gonads (Vagin et al., 2006; Aravin et al., 2007; Brennecke et al., 2007; Specchia et al., 2008, 2010, 2017; Specchia and Bozzetti, 2009; Bozzetti et al., 2015; Iwasaki et al., 2015; Sahin et al., 2016), but later it was observed in the nervous system of both Drosophila and mice (Lee et al., 2011; Perrat et al., 2013).
belle emerged in a screen for genes involved in the RNAi pathway: it binds to Ago1 and Ago2, involved in the small RNA-mediated regulation (Kim et al., 2005; Ulvila et al., 2006; Zhou et al., 2008). Very recently, a role of belle in the regulation on specific P-derived constructs in the ovary has been described (Lo et al., 2016), and therefore it is now considered a “bona fide” piRNA gene. belle encodes a DEAD-box RNA helicase: it is the closest paralog of vasa and plays roles in the reproductive system allowing fertility in Drosophila melanogaster. Like other members of the DEAD box family, Vasa and BELLE are supposed to have ATPase, RNA binding, and RNA unwinding activities (Liang et al., 1994; Sengoku et al., 2006). BELLE is essential for both male and female reproductive capacity and its function in gonads is conserved during evolution. It localizes at the perinuclear region, called “nuage,” of the germ cells in male and female gonads (Johnstone et al., 2005; Kibanov et al., 2011), where the majority of key components regulating transposable elements (TEs) are located (Lim and Kai, 2007; Klattenhoff and Theurkauf, 2008; Kibanov et al., 2011; Nagao et al., 2011).
Here, we have identified for BELLE important roles as putative circadian clock component and regulator of TEs in brain and in gonads. These apparently unrelated functions suggest the hypothesis that BELLE might be a key element in small RNA (piRNA)-mediated regulation of the TEs and that this regulatory pathway is involved in circadian rhythmicity.
The following strains of Drosophila melanogaster were used: w1118, CantonS, yw;tim-Gal4 (Emery et al., 1998), UAS-belleRNAi (VDRC #6299), GMR-Gal4 (BDSC #8440), ninaE-Gal4 (BDSC #1104), repo-Gal4 (BDSC #7415), pdf-Gal4 (BDSC #6900), Eaat1-Gal4 (BDSC #8849), Lama-Gal4 (BDSC #35543), per0 (Smith and Konopka, 1981), yw;P{EPgy2}belEY 08943 (BDSC #19945); P{PZ}belcap-1ry506/TM3, ryRK Sb1 Ser1 (BDSC #11778), bel::GFP (Morin et al., 2001) (Kyoto Stock Centre #ZCL2696), UAS-CD8-GFP (BDSC #5137). Co-IP was performed with yw; tim-GAL4/+; UAS-HAcry/+ (Dissel et al., 2004). Flies were maintained on a standard cornmeal medium under LD 12:12 regime and at constant 23°C.
Head extracts from overexpressing HACRY flies raised in 12:12 light:dark cycles and collected at Zeitgeber Time 24 (ZT24), before lights on, and after a 15-min light pulse were subjected to coimmunoprecipitation and Mass Spectrometry analysis as previously described (Mazzotta et al., 2013). Immunocomplexes were analyzed by Western blot using the following antibodies: rabbit polyclonal anti-BELLE (1:2000) (Johnstone et al., 2005) and mouse anti-HA (1:5000; Sigma Aldrich, St. Louis, MO, United States). Anti-rabbit IgG HRP (1:3000; BioRad Laboratories, Hercules, CA, United States) and anti-mouse IgG HRP (1:5000; Sigma Aldrich, St. Louis, MO, United States) were used as secondary antibodies.
RNA was extracted using the reagent Trizol (Invitrogen) as indicated in manufacturer’s instructions. For the analysis of belle expression, around 50 heads from flies reared in LD cycles for at least 3 days and collected at the indicated time points were used. For the analysis of transposons 30 mg of male or female gonadal tissues or heads were used. 1 μg of total RNA was used as a template for oligonucleotide dT-primed reverse transcription using SuperScriptIII RNaseH-reverse transcriptase (Invitrogen), according to manufacturer’s instructions. qRT-PCR was performed in the 7500 Real-TimePCR System (Applied Biosystem) using SYBR green (Celbio) according to the manufacturer’s instruction. For quantification of the transcripts, we used the 2ΔΔCt method as previously reported (Specchia et al., 2010). The primers used are listed in Supplementary Table S1.
For the analysis of BELLE localization, heads from 7 days old males were fixed in 4% paraformaldehyde in phosphate buffer saline (PBS; pH 7.4) for 4 h, then they were washed in PBS twice and cryoprotected by incubation in 12.5% sucrose for 10 min and in 25% sucrose at 4°C overnight. Material was embedded in Tissue-Tek, frozen in liquid nitrogen, and cryostat 20 μm sections were cut. The sections were washed in PBS for 30 min and five times in phosphate buffer (PB) with an addition of 0.2% Triton X-100 (PBT). After that, sections were incubated in 5% normal goat serum (NGS) with an addition of 0.5% bovine serum albumin (BSA) for 30 min first at room temperature, and then incubated with primary antibodies anti-REPO (mouse, 1:500, Hybridoma Bank), anti-Ebony (rabbit, 1:750, a gift from Dr. B. T. Hovemann, Ruhr Universität Bochum) and anti-GFP (rabbit, 1:1000, Novus Biologicals) for 24 h. Afterward, sections were washed six times in PBT/BSA, and blocked in 5% NGS for 45 min. After that, goat anti-mouse (conjugated with Cy3) and goat anti-rabbit (conjugated with Alexa 488) secondary antibodies (both 1:500, Jackson Immuno Research) were applied overnight in 4°C. Finally, sections were washed twice in BSA, six times in PBT, and twice in PBS. Then, cryosections were mounted in Vectashield medium (Vector) and examined with a Zeiss Meta 510 Laser Scanning Microscope. About 30 brains or cryosections were checked.
For the analysis of PER in clock-neurons, flies were synchronized by LD cycles at least for 3 days and then collected under LD or DD conditions at the indicated time points. Whole flies were quickly fixed for 2 h in PFA 4%. About 10–12 brains were dissected in PBS and then treated as previously described (Vanin et al., 2012). Average fluorescence values are reported.
One young male was mated to three control virgin females, and three virgin females were mated with three control males. Ten individual males and 30 females were tested for each genotype. After 4 days, the crosses were transferred to a fresh vial. The parental flies were removed from the last vial after an additional 4 days. The number of the adult progeny from each vial was counted.
Locomotor activity of individual flies was recorded for 3 days in LD and 7 days in DD using the Drosophila Activity Monitoring System (TriKinetics, Inc., Waltham, MA, United States). Data were collected every 5 min and then analyzed in 30 min bins using spectral analysis and autocorrelation (Zordan et al., 2007). Morning anticipations was detected fly-by-fly examining the activity mean of 3 days under LD conditions (Vanin et al., 2012) and the Morning Index was calculated as in Seluzicki et al. (2014).
For comparisons between two measurements a two-tailed Student’s t-test was used to show the significance level of the replicated experiments. Expression profile’s significance was evaluated by non-parametric ANOVA, Kruskal–Wallis test (p-value < 0.05) whereas putative period and phase of oscillation were identified by RAIN non-parametric test for independent samples (adjusted p-value < 0.05) (Thaben and Westermark, 2014).
In an experiment aimed at the identification of new genes involved in the circadian machinery in Drosophila, a coimmunoprecipitation assay followed by mass spectrometry analysis was performed on transgenic flies overexpressing a hemagglutinin (HA)-tagged form of dCRY (HACRY) raised in 12:12 light:dark cycles and collected at Zeitgeber Time 24 (ZT24), before lights on, and after a 15-min light pulse (Mazzotta et al., 2013).
A ∼85-kDa species was observed in the sample in the dark that was not present in the negative control (Figure 1A). This protein band was digested in-gel, and the peptide mixtures analyzed by liquid chromatography–mass spectrometry (LC-MS)/MS using an ESI-QTOF mass spectrometer (Wilm et al., 1996). Analysis of the MS/MS data using the MASCOT software yielded the identification of BELLE (Supplementary Table S2), a DEAD box RNA helicase involved in RNA metabolism at several levels (Linder and Jankowsky, 2011).
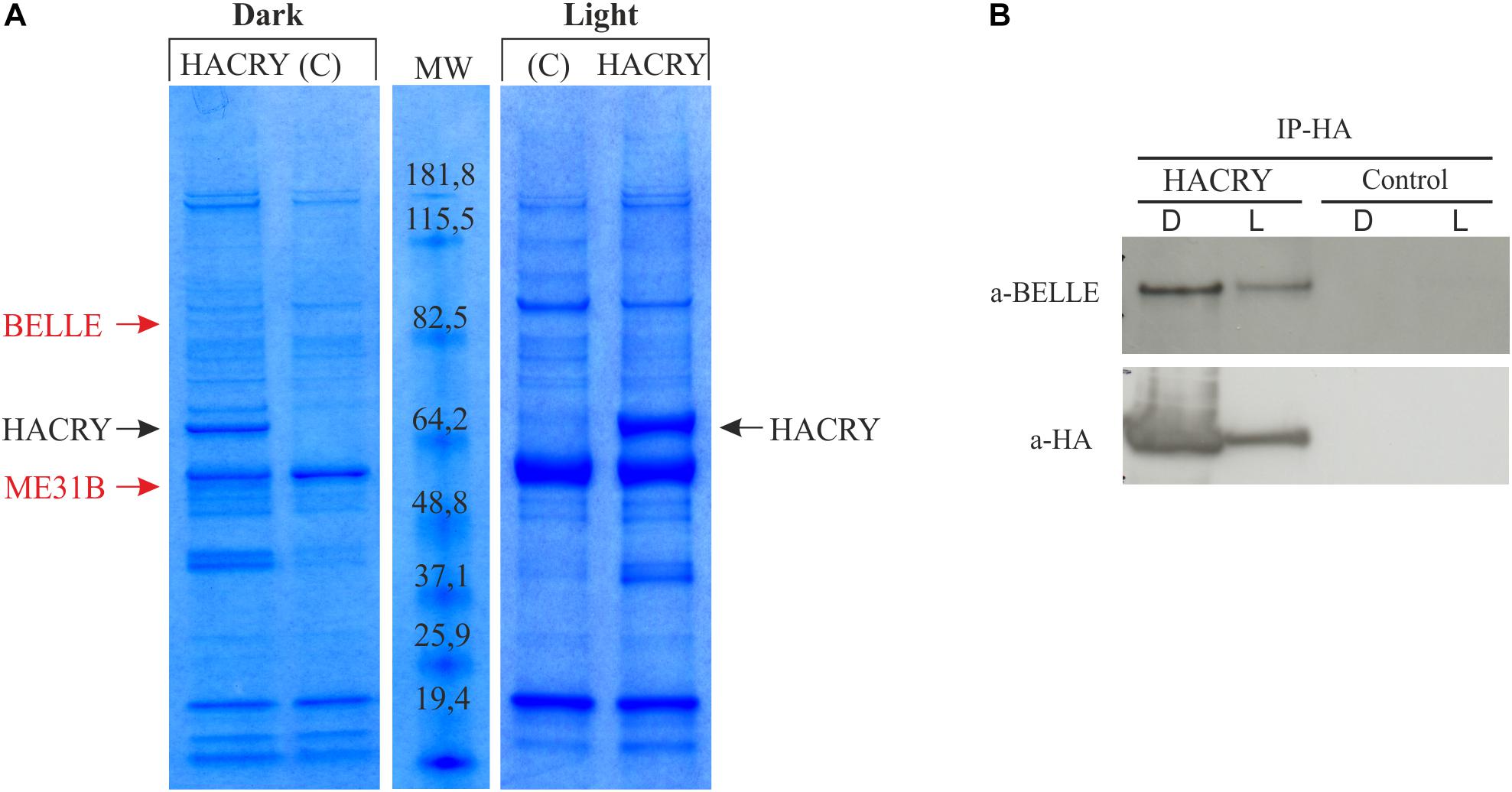
Figure 1. dCRY interacts with BELLE. (A) Coomassie blue-stained gel of heads protein extracts coimmunoprecipitated with an anti-HA antibody. HACRY-overexpressing flies (HACRY, yw;tim-GAL4/+; UASHAcry/+) and relative controls (C, yw;tim-GAL4) were reared in 12:12 light: dark and collected in the dark (ZT24) and in the light (ZT24 + 15-min light pulse). Molecular masses of markers are indicated (BenchMark Pre-Stained Protein Ladder; Invitrogen). MW, molecular weight. Bands corresponding to HACRY are indicated in black, while stained proteins excised and characterized by mass spectrometry are indicated in red. (B) Co-immunoprecipitation and Western blot confirming the interaction between HACRY and BELLE in HACRY-overexpressing flies (HACRY, yw;tim-GAL4/+; UASHAcry/+). tim-GAL4 flies were used as control. Heads were collected as in (A). Membranes were probed with anti-BELLE and anti-HA antibodies.
The presence of BELLE in the complex with HACRY was also confirmed by Western blot with an anti-BELLE antibody (Johnstone et al., 2005). By this procedure, BELLE was also detected after a 15 min of light pulse (Figure 1B). The same blot was hybridized also with an anti-HA antibody, in order to assess the specificity of the interaction (Figure 1B). The signal corresponding to BELLE was more intense in head collected in the dark; in fact after 15 min of light pulse the amount of BELLE was lower, in line with that of HACRY that decreased due to the well-known light-dependent degradation of CRY (Peschel et al., 2009).
It is known that BELLE is expressed in germ cells throughout all developmental stages and in larval neurons (Johnstone et al., 2005; Rolls et al., 2007), but little information was available regarding its expression in the adult fly head.
In order to characterize BELLE expression in the fly brain, we performed an immunocytochemistry experiment on flies carrying a GFP exon trap in belle (bel::GFP) (Morin et al., 2001). Flies were maintained in standard light–dark conditions, brains were dissected and an anti-GFP antibody was used to detect BEL::GFP expression. We observed BELLE expression in clock neurons and in glial cells in the optic lobe, with a predominantly cytoplasmic localization (Figures 2, 3). In clock neurons, BELLE was expressed in either the small or the large ventral lateral neurons (s-LNvs and l-LNvs respectively) (Figure 2), PDF-expressing cells that act in promoting and governing the circadian locomotor behavior. In glia we observed BELLE expression in the epithelial glia and a weak labeling also in the lamina (Figures 3D–F): position and shape of these GFP- labeled cells suggest they may be R1-R6 photoreceptor terminals (Figures 3D–F); however, anti-REPO staining showed glia cells nuclei at the top of each structure (Figure 3G). Observed columnar shape is also similar to anti-EBONY staining (Figure 3H), which is a marker for epithelial glia (Richard et al., 2002), suggesting that BELLE may be located in epithelial glia in the lamina. Moreover, we observed immunostaining for BEL::GFP also in giant inner chiasm glia, which connects the medulla with the lobula complex in the optic lobe (Kremer et al., 2017). In these cells, both BELLE and PER are expressed (Supplementary Figure S1).
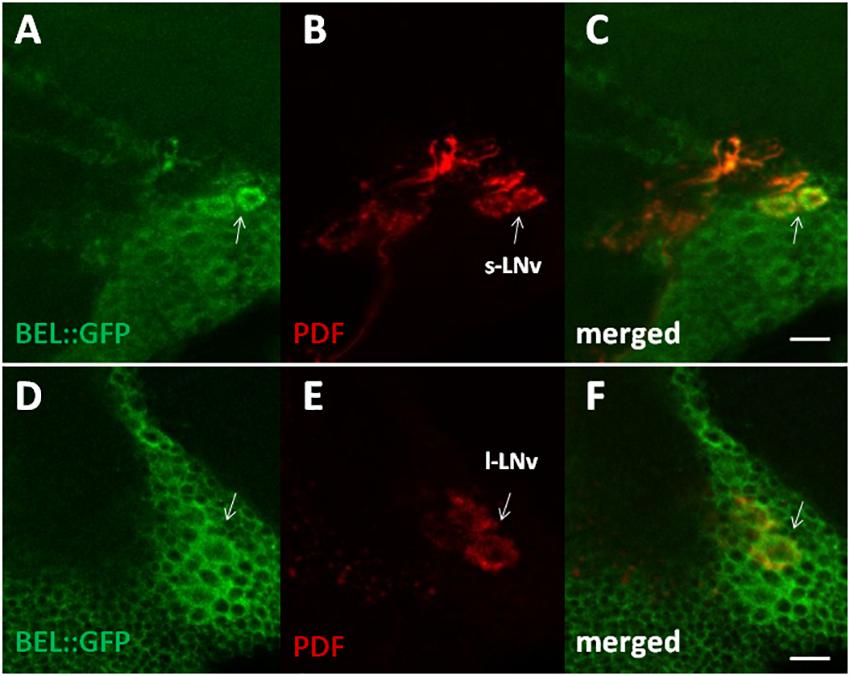
Figure 2. BELLE localizes in the main pacemaker cells in the brain of Drosophila melanogaster. Confocal images of whole mount of adult brain expressing BELLE fused with green fluorescent protein (bel::GFP). Scale bar: 10 μm. (A,D) Anti-GFP immunostaining (BEL::GFP). (B,E) Anti-PDF was used to label PDF-expressing ventral lateral neurons. Depends on optical section small LNv (s-LNv) or large LNv (l-LNv) are visualized. (C,F) BEL::GFP and PDF immune staining co-localize in the area of LNv cell bodies.
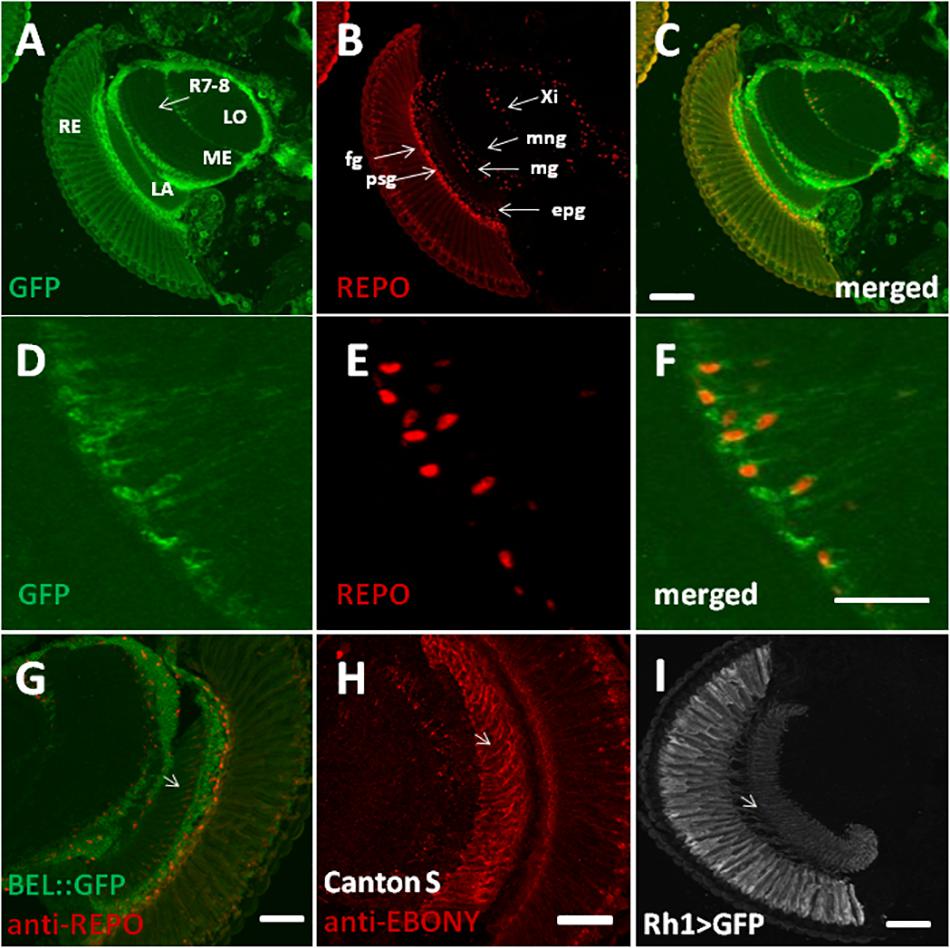
Figure 3. BELLE localizes in the visual system of Drosophila melanogaster. Confocal images of horizontal cryosection of adult brain expressing BELLE fused with green fluorescent protein (bel::GFP). Anti-REPO was used to label all glia nuclei. (A) BEL::GFP signal is localized in the optic neuropils: lamina (LA), medulla (ME), and lobula (LO). Weak signal is visible in R7 and R8 photoreceptor terminals (R7-8) in the medulla. Signal in the retina (RE) is probably due to autofluorescence. (B) Anti-REPO staining allowed recognized different glia types: fenestrated glia (fg), pseudocartridge glia (psg), epithelial glia (epg), medulla glia (mg), marginal glia (mng), and inner chiasm glia (Xi). (C) Merged signal showed localization of BELLE in glial cells. Bar 50 μm. (D–F) Higher magnification of inner chiasm glia showed that BELLE is localized mostly in the cytoplasm. Bar 20 μm. (G–I) Localization of BELLE in the first optic neuropile, called lamina, analyzed on confocal images of horizontal cryosection of adult brain. Bar: 50 μm. (G) Anti-GFP and Anti-REPO immunostaining showing that BEL::GFP signal is higher in the distal lamina (close to retina), where interneurons cell bodies are located. Weak labeling is also visible across the lamina, forming regular columns. Glia nuclei staining overlaps with observed BELLE location, suggesting that this signal originates from epithelial glia. (H) Section of wild-type CantonS brain stained with anti-EBONY to label epithelial glial cells in the lamina and their specific columnar shape (see Richard et al., 2002). (I) Anti-GFP staining on section of Rh1 > GFP brains showing R1–R6 photoreceptors terminals in the lamina.
In order to characterize the temporal expression of belle, mRNA and protein levels were analyzed during the 24 h. Wild-type flies white1118 were synchronized by 12:12 LD cycles and collected every 3 h, either in LD regime or at the third day after being transferred to constant darkness (DD).
belle expression appeared to oscillate both in LD and in DD conditions. In particular, in LD cycles the level of differential expression during the 24 h was just below the limit of significance (Kruskal–Wallis p-value 0.06); nevertheless, the algorithm RAIN suggested the presence of a rhythmic expression pattern (adjusted p-value 2,61E-02) with a peak at ZT12 (Figure 4A). In constant darkness belle mRNA showed differential expression around the 24 h (Kruskal–Wallis p-value 0.03), and a peak at CT63 was identified (RAIN p-value 1,92E-05) (Figure 4B). These results are in line with a previous microarray study showing a peak of expression for belle at ZT17 (Claridge-Chang et al., 2001). In per0 mutant, belle oscillation is maintained in LD cycles, although with a peak at ZT3 (Kruskal–Wallis p-value 0.02, RAIN p-value 4,91E-07) (Figure 4A), while it was lost in constant conditions (Kruskal–Wallis p-value 0.609) (Figure 4B).
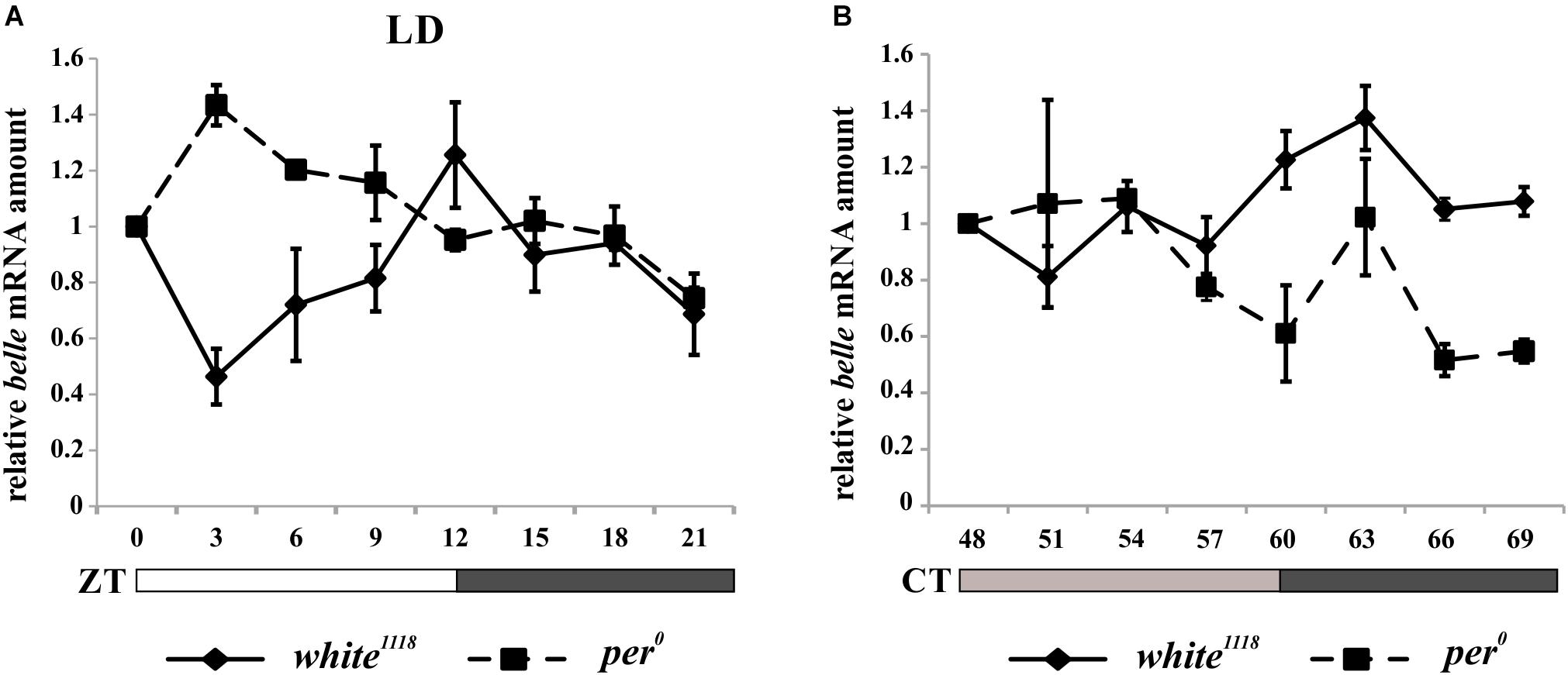
Figure 4. belle mRNA expression shows circadian features. qRT-PCR on heads from white1118 and per0 flies reared in 12.12 LD (A) or over the third day in DD (B). Relative abundance of belle mRNA was defined as a ratio with rp49. Values represent mean levels ± SEM of three biological replicates. The value corresponding to CT 54 in per0 flies represents mean level of two biological replicates.
The analysis of BELLE in fly heads by western blot showed no significant protein oscillation, neither in LD nor in DD (Kruskal–Wallis p-values 0.9 and 0.8, respectively) (Supplementary Information and Supplementary Figure S2). This result is in line with that obtained by co-immunoprecipitation (Figure 1B): the lower amount of BELLE pulled down in light compared to dark uniquely depends on the light-mediated degradation of CRY.
In order to unravel a possible involvement of belle in the generation of circadian rhythmicity in Drosophila, we depleted the expression of the gene by RNAi in both glia and clock neurons.
When we knocked down belle with the pan-glial driver repo-Gal4 (Awasaki and Ito, 2004) and lama-Gal4, a driver for glial precursor cells, lamina precursor cells and lamina neurons (Apitz and Salecker, 2016), we observed developmental lethality. The same result was obtained by KD in all canonical clock neurons and clock relevant glial cells expressing timeless (with a tim-Gal4) (Emery et al., 1998). This result is in agreement to what reported for the null allele of belle (Johnstone et al., 2005). When we depleted belle in PDF expressing neurons and a restricted subset of glial cells (with pdf-Gal4 and Eaat1-Gal4, respectively) (Renn et al., 1999; Rival et al., 2004), flies were viable and endogenous rhythmicity was not altered (Table 1); only a slight impairment in the morning anticipation of locomotor activity in LD was observed (Table 2). In contrast, RNAi against belle in the photoreceptor cells (GMR-Gal4 and ninaE-Gal4) did not affect either vitality or behavior (Supplementary Table S3).
We then decided to investigate a possible role for belle in the circadian machinery using two viable mutant lines generated by p-element insertion in the regulatory region (belle cap-1 and belle EY 08943) (Bellen et al., 2004). Flies were entrained for 3 days in light dark cycles at constant temperature (23°) followed by 7 days of constant darkness. Either mutant exhibited an impairment of the locomotor behavior (Figure 5): in LD conditions, both lines presented the canonical bimodal profile, but a severe loss of the morning anticipation of the locomotor activity was displayed (Figure 5A and Table 3). A high percentage of flies showed also an arrhythmic behavior in DD, although no defects were monitored concerning the period, which was comparable to wild-type (Figure 5B and Table 4).
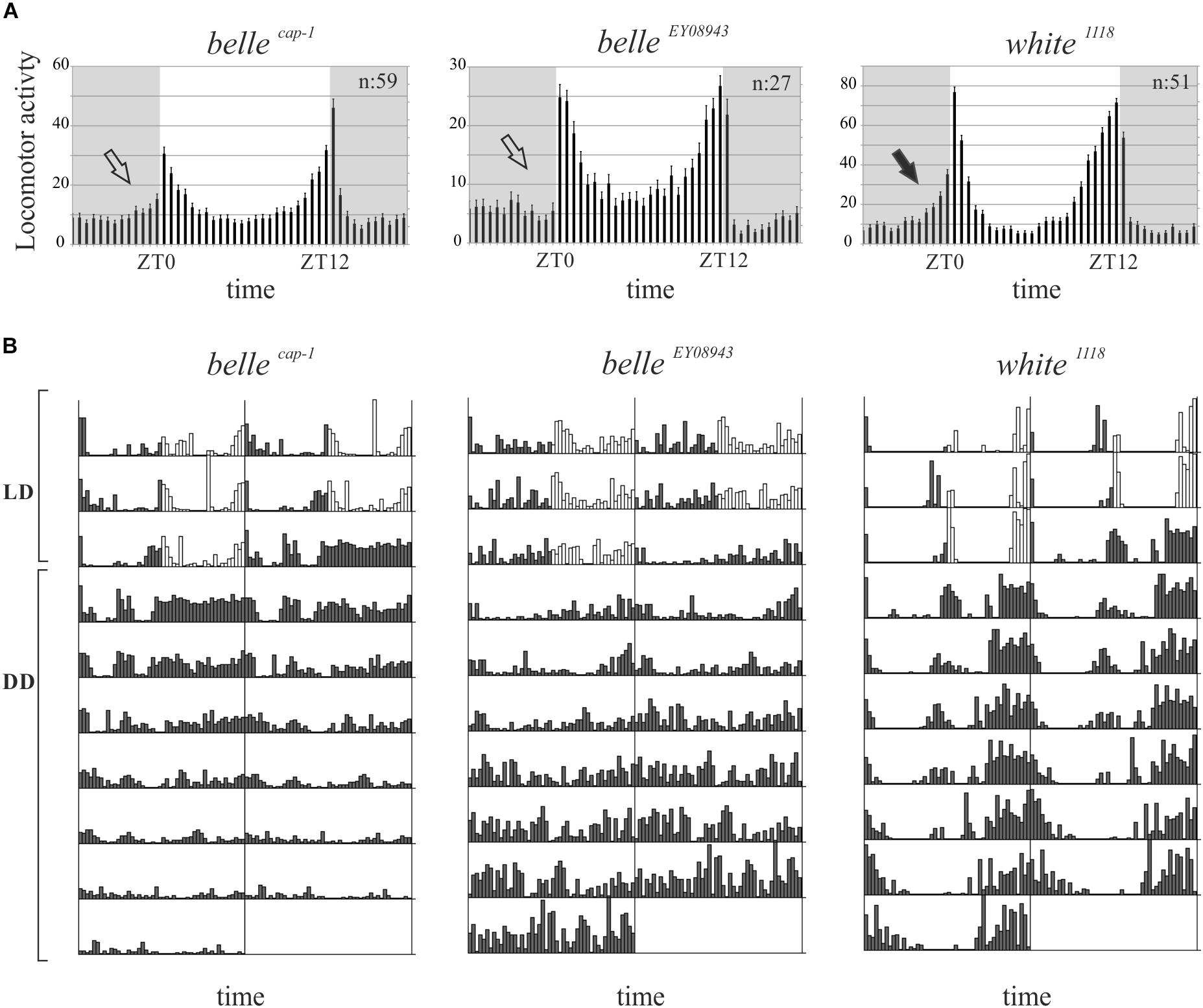
Figure 5. BELLE affects circadian locomotor activity. (A) Activity means of flies over 3 days of LD cycles ± SEM. N: number of flies; gray boxes: dark phase. Belle mutants lost the morning anticipation (empty arrows) compared to control (black arrow). ZT0: lights-on, ZT12: lights-off. (B) Representative double-plotted locomotor activity of a single fly recorded for 3 days under LD and 7 days under DD conditions (gray bars: dark phase; white bars: light phase). LD: light–dark cycles; DD: constant darkness.
We performed PER staining on brains dissected every 4 h from flies collected under LD cycles and during the 5th day of DD conditions, in order to assess whether the impairment of the circadian locomotor activity of the belle mutants could be due to an altered PER cycling in the pacemaker neurons. PDF expressing cells were identified by co-staining for this peptide.
Mutant lines showed defects in some clusters of neurons. Under LD conditions, belle cap-1 flies exhibited a significant reduction of oscillation amplitude in both s-LNvs and l-LNvs, particularly marked at ZT0 (Figure 6). Similarly, in constant darkness, a general decrease of PER staining was observed compared to wild-type, which was particularly enhanced during the subjective night/beginning of the subjective day (Figure 7). Quantification of PER levels indicated that the greatest effect occurs in the small PDF cells (s-LNvs) (Figure 7), those cells that normally show high PER expression and also govern the behavioral phenotype in constant conditions (Grima et al., 2004). Besides a decrease of its levels, in these cells the accumulation of PER is slow down, with a peak delayed of about 4 h with respect to control (Figure 7). In large PDF neurons (l-LNvs) PER cycling is dampened, similarly to control and in accordance to previous observations (Grima et al., 2004).
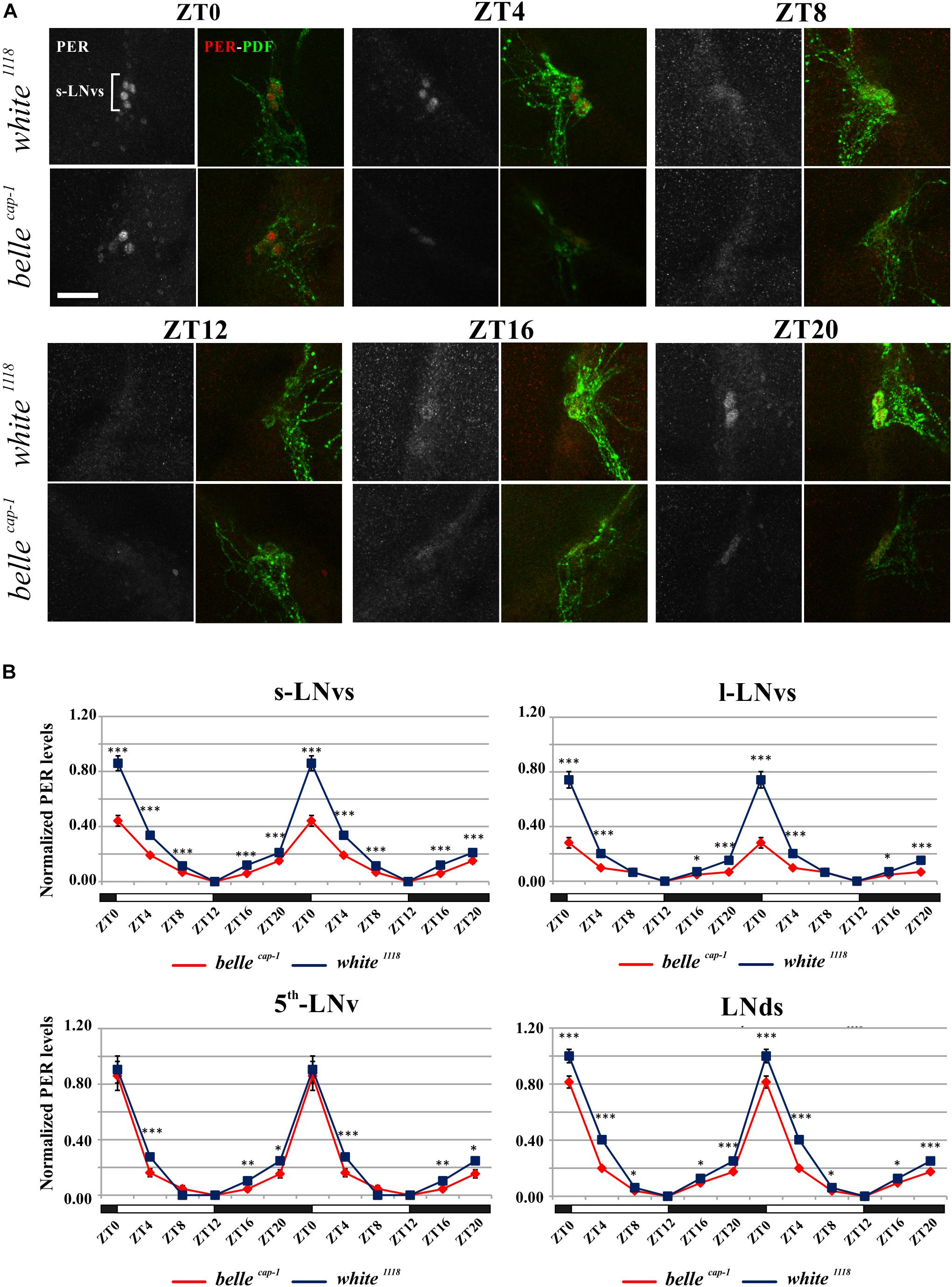
Figure 6. Altered PER expression in belle cap-1 mutant in entrainment conditions. PER oscillations in the clock Lateral Neurons of belle cap-1 mutant flies in LD cycles. Flies were collected at the indicated time points after 3 days in LD and stained for PER and PDF. (A) Representative confocal stack images of PER staining in s-LNvs. belle cap-1 mutants displayed lower PER expression in circadian pacemaker neurons, compared to control (white 1118). Scale bar: 25 μm. (B) PER quantification in clock Lateral Neurons. Peak value was set to 1 and the rest of the values were normalized accordingly. Data are shown as double-plotted. t-Test: ∗∗∗p < 0.005, ∗∗p < 0.01, ∗p < 0.05.
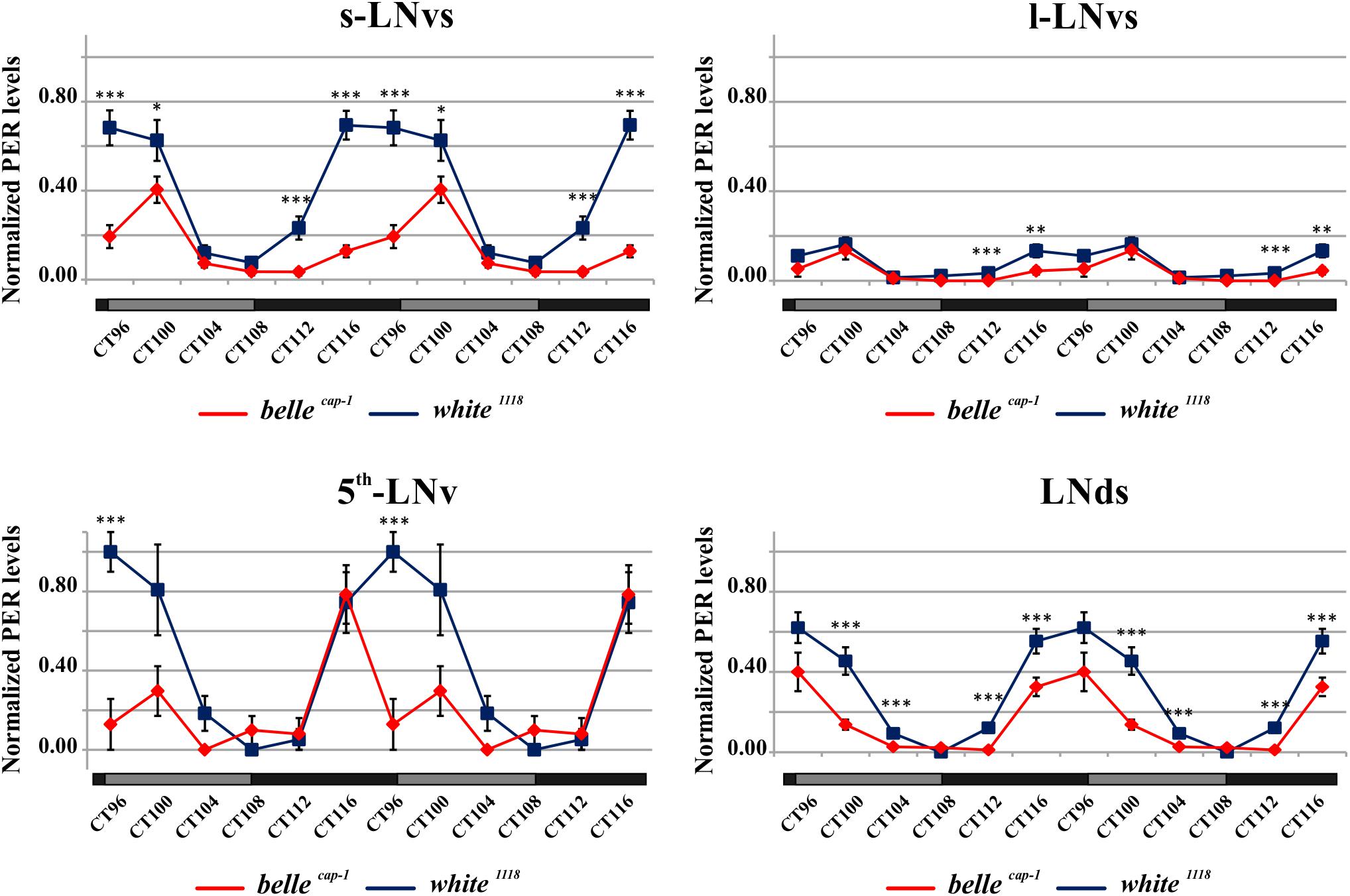
Figure 7. Altered PER expression in belle cap-1 mutant in constant conditions. PER quantifications in the clock Lateral Neurons of belle cap-1 mutant flies in constant darkness. Flies were first entrained for 2 days in LD and then analyzed after 4 days of DD. Only arrhythmic flies were collected every 4 h and stained for PER expression during the 5th day of DD. Peak value was set to 1 and the rest of the values were normalized accordingly. Data are shown as double-plotted. t-Test: ∗∗∗p < 0.005, ∗∗p < 0.01, ∗p < 0.05.
The belEY 08943 mutant exhibited a less striking phenotype: a significant difference was observed in the PDF positive s-LNvs only in constant darkness, where the oscillation of PER was 4 h delayed compared to control (Supplementary Figure S3), similarly to what described in belcap-1 flies (see above).
Interestingly, both belle mutants were characterized by a reduction in the number of l-LNvs: a high percentage of brains, in fact, presented three neurons instead of the canonical four (83.93, 76.28, and 6.78% in bel cap-1, bel EY 08943, and white 1118, respectively).
As BELLE co-localizes with its paralog Vasa, a key component of the piRNA pathway-mediated regulation of the TEs, in ovaries and testes (Johnstone et al., 2005), we decided to further investigate about a possible involvement of belle in the piRNA-mediated regulation of TEs elements.
We analyzed the RNA expression of specific transposons in heads of adult belle EY 08943 and belle cap1 flies. In belle EY 08943 two of them, roo and blood, showed a significant reduction in their expression compared to control (Figure 8, left panel). In addition, R1 also exhibited a slight reduction in the RNA amount, though not significant. In belle cap-1 the TE transcripts were greatly reduced, also compared to belle EY 08943, and even R1 exhibited a significant reduction compared to control (Figure 8, right panel). These results are in disagreement with other piRNA mutants, that normally show an increased amount of the TE transcripts (Vagin et al., 2006; Brennecke et al., 2007; Ghildiyal and Zamore, 2009; Khurana and Theurkauf, 2010; Specchia et al., 2010; Piacentini et al., 2014; Bozzetti et al., 2015). Nevertheless, based on our observations, a role for belle in the regulation of the TEs expression can still be hypothesized.
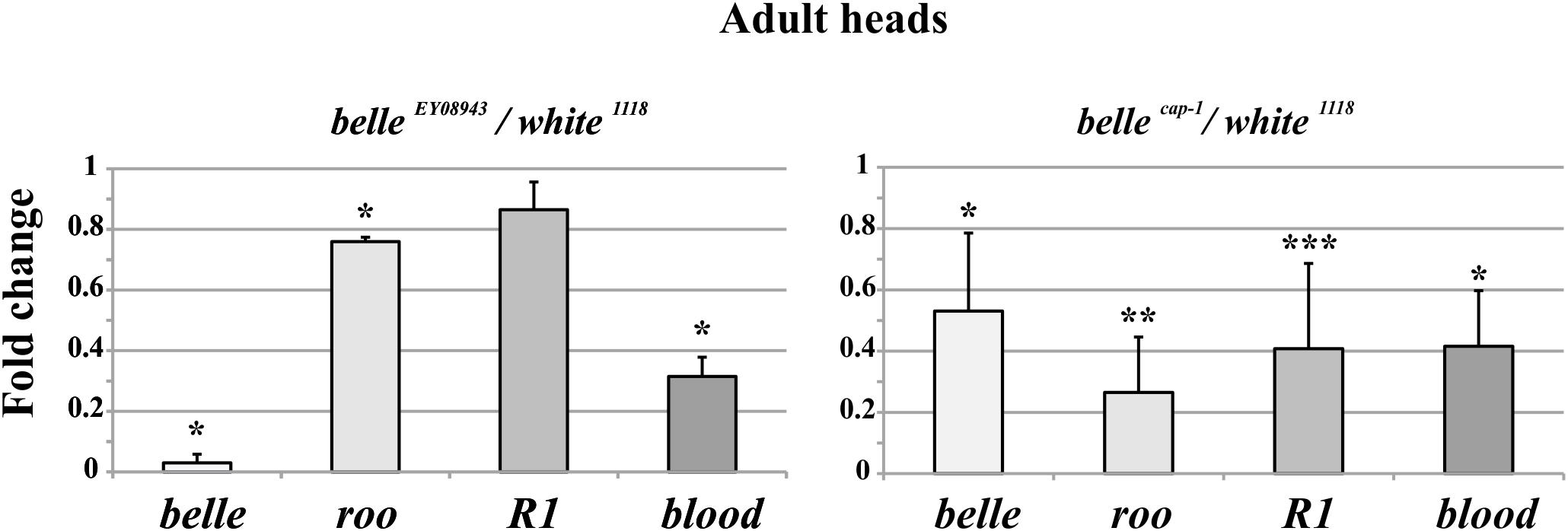
Figure 8. Altered TEs expression in belle mutant heads. qRT-PCR on heads from belle EY 08943, belle cap-1, and white 1118 flies reared in 12:12 LD cycles and collected at ZT3. Represented are relative amount of belle and three transposable elements (TEs) (roo, R1, and blood) in belle EY 08943 (A) and in belle cap-1 (B) compared to white 1118 control, to which an arbitrary value of 1 was assigned. Represented are mean ± SD of three biological replicates. ∗∗∗p < 0.005, ∗∗p < 0.01, ∗p < 0.05.
In order to better elucidate this involvement, we decided to investigate belle function in the gonads, where the role of the different components of the piRNA pathway is almost entirely known (Vagin et al., 2006; Brennecke et al., 2007; Klattenhoff and Theurkauf, 2008; Li et al., 2009; Malone et al., 2009; Khurana and Theurkauf, 2010; Senti and Brennecke, 2010; Czech et al., 2013; Handler et al., 2013; Muerdter et al., 2013; Iwasaki et al., 2015). Our group has identified some key components of the piRNA pathway, like aubergine, hsp83, dFmr1 (Specchia et al., 2008, 2010, 2017; Specchia and Bozzetti, 2009; Bozzetti et al., 2015) and demonstrated that TEs and repetitive sequences like Stellate are transcriptionally activated as a consequence of mutations in these piRNA genes (Aravin et al., 2001, 2003; Nishida et al., 2007; Specchia and Bozzetti, 2009; Bozzetti et al., 2012, 2015; Malone et al., 2015; Sahin et al., 2016).
We first tested the expression of some transposons in gonads: a dramatic reduction of transposons transcript was observed in ovaries from belle mutants in comparison to wild-type (Figure 9B and Supplementary Figure S4), similarly to what observed in heads. A similar reduction was also observed in belle EY 08943 testes, with the exception of roo, whose transcript increases in belle EY 08943 compared to control (Figure 9A).
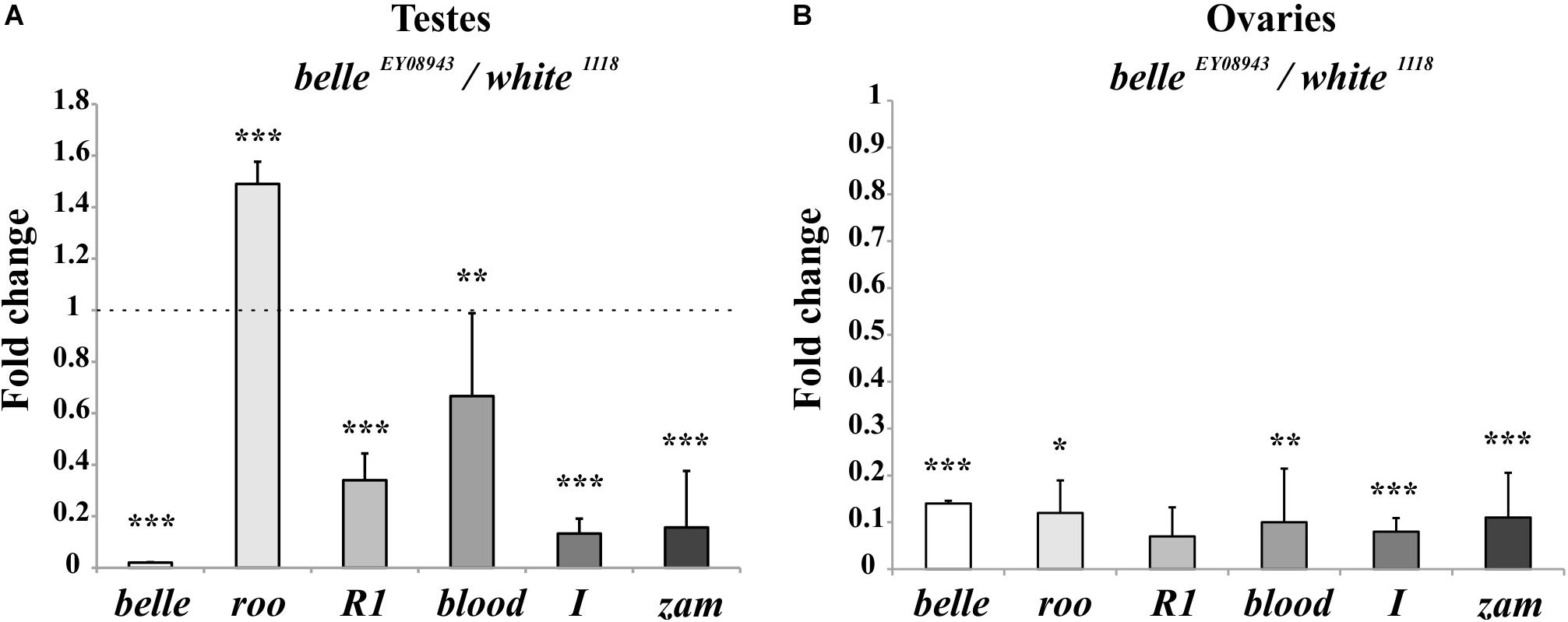
Figure 9. Altered TEs expression in belle mutant gonads. qRT-PCR on ovaries (A) and testes (B) from belle EY 08943 and white 1118 flies reared in 12:12 LD cycles and collected at ZT3. Represented relative mRNA levels of belle and five TEs (roo, R1, blood, I-element, and ZAM) in belle EY 08943 compared to white 1118, control, to which an arbitrary value of 1 was assigned. Represented are mean ± SD of three biological replicates. ∗∗∗p < 0.005, ∗∗p < 0.01, ∗p < 0.05.
We have also performed a fertility test, showing that mutant males are completely sterile with respect to controls (average progeny of 128), while females exhibit a severe reduction of fertility (average progeny of 6,6 in mutants with respect to 43,4 in control) (Supplementary Figure S5). This observation is in agreement to what reported for other alleles of belle (Johnstone et al., 2005).
Aiming at finding new molecular component of the clock machinery in Drosophila, we have identified the DEAD-box RNA helicase BELLE as interactor of CRY. DEAD (Asp-Glu-Ala-Asp)-box RNA helicases are highly conserved proteins known to play central roles in essentially every stages of RNA metabolism, in both nucleus and cytoplasm (Linder and Fuller-Pace, 2013). BELLE has been implicated in splicing and translational regulation (Worringer et al., 2009; Ihry et al., 2012) and, more recently, in miRNA and siRNA pathways (Pek and Kai, 2011). It is essential for viability, organismal growth, and fertility (Johnstone et al., 2005).
The involvement of a RNA helicase in the circadian clock is not uncommon. In Neurospora crassa, in fact, two members of DEAD-box RNA helicase family play important roles in the circadian machinery: FRH (Frequency-interacting RNA Helicase), that regulates the stability of FREQUENCY (FRQ), thus acting on both negative and positive circadian feedback (Cheng et al., 2005; Guo et al., 2010; Shi et al., 2010; Lauinger et al., 2014), and PRD-1 (PERIOD-1), ortholog of human DDX5 and DDX17, specifically regulating circadian rhythmicity also in FRQ-less oscillators (Emerson et al., 2015; Adhvaryu et al., 2016).
BELLE has been extensively studied for its crucial role in the Drosophila gonads, where it is essential for viability and fertility (Johnstone et al., 2005; Kibanov et al., 2011). However the presence of BELLE has been reported also in Drosophila neurons, specifically in the neuronal cell body (Beckham et al., 2008), and proteomic data describe its expression in adult fly heads (Aradska et al., 2015).
We have depicted BELLE expression in both PDF expressing clock neurons and in the glial cells in the optic lobe. It is reported that adult glial cells contain a molecular clock and evidences about their role in the modulation of circadian rhythmicity are continuously accumulating (Siwicki et al., 1988; Zerr et al., 1990; Ewer et al., 1992; Pyza and Gorska-Andrzejak, 2004; Suh and Jackson, 2007; Damulewicz et al., 2013; Ng and Jackson, 2015; Ng et al., 2016). Nevertheless, the signaling between glia and the clock neurons that control the locomotor behavior is not yet fully understood. Many PER-expressing glial cells, such as epithelial glial, are located in proximity of clock neurons or their processes (Suh and Jackson, 2007), suggesting a direct communication. Epithelial glia affects rhythmic behavior also by regulating neurotransmitter metabolism through Ebony (Suh and Jackson, 2007). BELLE is mostly localized in the cytoplasm: this observation is in accordance with previous reports of a predominantly cytoplasmic distribution for BELLE in fly gonads (Johnstone et al., 2005) and its localization in cytoplasmic processing bodies (P-bodies), where it is supposed to act as translational repressor (Beckham et al., 2008). At the transcriptional level, we observed a clear daily rhythm for belle mRNA either in LD and DD; the phase of the oscillation is altered in per0 flies in LD, while it is abolished in constant conditions, suggesting that the circadian clock controls the expression of this helicase.
These results are perfectly in line with previously reported observations: in fact, a microarray study showed a peak of expression for belle at ZT17 (Claridge-Chang et al., 2001) and, in a chromatin immunoprecipitation (ChIP) analysis, belle was found to be a direct target of CLK in fly heads (Abruzzi et al., 2011). The peak-time of belle mRNA in the late day/early night is in agreement with a direct control of CLOCK in the expression of this gene, since it is reported that CLK-CYC bind to E-box of their target genes in the late day (reviewed in Peschel and Helfrich-Forster, 2011).
The effect of down-regulation of belle in glia and in TIM-expressing cells, lethality and arrested growth at larval stage, respectively, is in agreement with previously reported observation (Johnstone et al., 2005). In fact, strong alleles of belle are zygotic lethal and larvae arrest their development at stage L1, which is prolonged up to 5 days (Johnstone et al., 2005). This larval growth arrest phenotype is common to other genes encoding for translation factors, such as elongation factors elF4A and elF4E (Galloni and Edgar, 1999; Lachance et al., 2002), for which a lethality was observed also when knocked down in TIM cells (Bradley et al., 2012). The down-regulation of belle in a restricted subset of clock neurons or glial cells resulted in the loss of the morning anticipation in about 60% of flies reared in entrainment conditions, suggesting a possible role for BELLE in the regulation of morning activity. This phenotype was much stronger in two viable allele of belle: in LD cycles, in fact, a high proportion of flies were devoid of morning anticipatory, further indicating that the small ventral lateral neurons, that constitute the morning oscillator (Grima et al., 2004), are affected in these flies. In constant conditions the percentage of rhythmic flies in the two belle mutants was significantly lower compared to wild-type, reinforcing the fact that the activity of lateral ventral neurons, that drive also the endogenous behavioral rhythmicity (Helfrich-Forster, 1998; Renn et al., 1999; Lin et al., 2004), are impaired by belle mutation.
We sought to analyze PER cycling in clock neurons of belle mutants, in order to determine whether an altered PER expression could account for the behavioral defects of these flies. In fact PER oscillation in circadian pacemaker neurons is a hallmark of a functioning molecular oscillator, and an impairment of LNvs activity results in a de-synchronization of PER cycling within different groups of circadian neurons (Renn et al., 1999; Peng et al., 2003; Lin et al., 2004).
Although with a different magnitude between the two mutants, a general decrease of PER staining was observed compared to control. In entrainment conditions, in belle cap-1 flies the amplitude of PER oscillation was significantly reduced in both s-LNvs and l-LNvs, and the difference was particularly marked at ZT0. In constant conditions, s-LNvs displayed the stronger effect, with a PER reduction more pronounced at the end of subjected night/beginning of subjective day, when the protein is expressed at maximum levels. Moreover, in these cells the accumulation of PER was also slow down, reaching maximum levels about 4 h later than the control. The belle EY 08943 mutant exhibited a less striking phenotype: only in constant darkness PDF positive s-LNvs displayed a 4 h delay compared to control. Altered PER oscillation in PDF-positive LNvs may account for the defects in the morning anticipation of the locomotor activity of belle mutants. These flies did not display particular defects in PDF projections/arborizations, but a reduction in the number of large ventral lateral neurons (l-LNvs) was observed. The altered PER cycling in clock neurons is reminiscent of the phenotype observed in mutants for RNA binding proteins involved at different levels in the post-transcriptional regulation of circadian clock, such as the translational factors NAT1 (Bradley et al., 2012) and ATAXIN2 (ATX2) (Lim and Allada, 2013; Zhang et al., 2013). ATX-2 is a RNA-binding protein that very recently has been shown to sustain circadian behavior in Drosophila functioning as both activator and repressor of translation, and this switch is mediated by complex formation with two specific factors, LSM12 and ME31B/DDX6, respectively (Lim and Allada, 2013). Lsm12 mutant flies exhibited a dampened PER cycling in circadian pacemaker neurons, that resulted in a lengthening of the locomotor activity period (Lee et al., 2017). ME31B is the homolog of human DDX6, a RNA helicase belonging to the same family of DEAD-box ATP-dependent helicase of BELLE, and flies in which the expression of this gene was depleted exhibit very poor rhythmicity (Lee et al., 2017). All these similarities led us to hypothesize that also BELLE could play a role in the post-transcriptional control of the circadian clock in Drosophila, probably in mechanisms in which the aforementioned factors are also involved. This idea is supported by the fact that we have found ME31B in our Co-IP and MS analysis in the same complex as BELLE in fly heads (Figure 1 and Supplementary Table S2), and that the two proteins belongs to the same translational repression complex that in Drosophila embryo is responsible for both repression and degradation of non-localized nanos mRNA, in a mechanism that involves also the Piwi-interacting RNA (piRNA) machinery (Gotze et al., 2017).
BELLE has a well-studied role in the Drosophila gonads (Johnstone et al., 2005; Kibanov et al., 2011; Kotov et al., 2016). In this study we have observed that loss of belle function impinges on the expression of TEs either in the brain or in the gonads, with an impact on the fertility of both males and females. We have observed a reduction of the transposons’ transcription in belle mutant, in comparison to wild-type; notably, other piRNA mutants normally exhibit an increased TEs’ expression (Aravin et al., 2007; Specchia et al., 2008, 2017; Specchia and Bozzetti, 2009; Senti and Brennecke, 2010). This observation well-agrees with the recently reported data on the effect of belle mutants in the silencing of P-transposon-derived constructs in ovaries, depending on de novo piRNAs generated from the transgenes in the region of their insertion (Lo et al., 2016). Our data led us to infer that belle regulates the transposons probably via the piRNA-mediated pathway. This hypothesis is somehow supported by the fact that Vasa and Spindle-E, two members of the same DEAD box protein family, have a well-recognized role in the piRNA pathway in ovaries and testes, though their loss of function results in an up-regulation of TEs (Lasko, 2013; Iwasaki et al., 2015; Specchia et al., 2017), as opposite to that of belle. They also co-localize at the “nuage” of the germline where, Argonaute, Tudor and many other factors are also located for an efficient TE silencing (Lasko and Ashburner, 1990; Lim and Kai, 2007; Lasko, 2013; Bozzetti et al., 2015). However, in contrast to the majority of piRNA-related genes (Nishida et al., 2007; Specchia et al., 2008, 2010, 2017; Specchia and Bozzetti, 2009; Bozzetti et al., 2015; Sahin et al., 2016), belle seems not to be involved in the crystal-Stellate regulatory pathway, since mutant flies did not exhibited Stellate-made crystalline aggregates in the spermatocytes (Supplementary Information and Supplementary Figure S6). This observation well-agrees with the demonstration that belle EY 08943 mutant does not show an increase of RNA for TEs and repetitive sequences like Stellate.
A possible role of BELLE in the piRNA pathway is only starting to be elucidated: our experiments suggest that it might act in maintaining precise levels of TE RNAs, probably regulating the activity of other piRNA components.
The involvement of belle in both circadian rhythmicity and piRNA mediated transposon regulation suggests association between these two biological processes. This hypothesis is supported by indirect, though reasonable, observations.
First of all, circadian components have been linked to piRNA factors in the gonads. In fact, the mammalian CLOCK and BMAL1 transcription factors have been associated to the chromatoid body (CB) (Peruquetti et al., 2012), a cytoplasmic electron dense structure in the male germline with similarities to Drosophila polar granules and “nuage” of the germ cells (Grivna et al., 2006), where many components of different RNA metabolism pathways, like piRNAs, are located (Tanaka et al., 2000; Kotaja et al., 2006; Nagamori et al., 2011).
Secondly, a piRNA gene plays essential roles in neuronal development and circadian regulation. Our group has recently identified dFmr1, a gene with well-characterized functions in development of nervous system (Schenck et al., 2002; Napoli et al., 2008; Aitken and Lorsch, 2012; Doll and Broadie, 2016) and in regulation of circadian rhythmicity (Dockendorff et al., 2002; McBride et al., 2005; Santos et al., 2014), as a piRNA gene playing a crucial role in the gonads, where it ensures fertility and piRNA-regulated genome stability (Bozzetti et al., 2015; Specchia et al., 2017). Nevertheless, a role of this gene in the piRNA-mediated regulation in the nervous system has not been reported yet. dFmr1 mutants display an altered rhythmicity in both eclosion and locomotor activity, due to an effect on the output factor CREB (cAMP response element binding protein) rather than the molecular oscillator (Dockendorff et al., 2002).
Third, circadian transposon regulation ensures genome integrity during aging. It is well-known that TEs activation induces an age-dependent loss of neuronal functions in Drosophila brain (Li et al., 2013). Very recently, a set of putative primary piRNA transcripts fully overlapping transposons in antisense were shown to display a clock-controlled de novo oscillation in old flies, raising the hypothesis that circadian piRNA expression could represent a new strategy adopted by the clock to preserve genome integrity during aging (Kuintzle et al., 2017).
A clear involvement of the piRNA pathway in the circadian rhythmicity has not been demonstrated so far. However, we have described an emerging role of belle in both circadian rhythmicity and transposon regulation, that leads to the attractive hypothesis that piRNA-mediated regulation could be another level of post-transcriptional control adopted by the clock to ensure the proper rhythmicity.
RC, GMM, and MPB conceived and supervised the study. PC, MD, EC, LC, VS, and AP performed experiments and analyzed the data. PC, MD, GMM, RC, VS, and MPB discussed results and wrote the manuscript.
This work was funded by grants from: University of Padova to GMM (Grant No. CPDA099390/09) and PC (“Progetto Giovani 2010”_Grant GRIC101061), European Community (6th Framework Project EUCLOCK No. 018741), National Research Council of Italy and MIUR (EPIGEN Flagship Project – Subproject 4), INsecTIME Marie Curie Initial Training Network (Grant No. PITN-GA-2012-316790) to RC, TELETHON (Grant No. GG14181) to MPB, Polish National Science Centre (Narodowe Centrum Nauki, NCN _ Grant No. UMO-2014/15/D/NZ3/05207) to MD.
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537), Vienna Drosophila Resource Center (VDRC) and Kyoto Stock Centre were used in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Moyra Mason (University of Padova) for her work on CoIP and MS; Federica Sandrelli (University of Padova) for her initial help with locomotor activity analysis; Alberto Biscontin (University of Padova) for help with statistical analyses; Prof. Paul Lasko (Department of Biology, McGill University, Montreal, QC, Canada) for anti-BELLE antibody.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00133/full#supplementary-material
Abruzzi, K. C., Rodriguez, J., Menet, J. S., Desrochers, J., Zadina, A., Luo, W., et al. (2011). Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 25, 2374–2386. doi: 10.1101/gad.174110.111
Adhvaryu, K., Firoozi, G., Motavaze, K., and Lakin-Thomas, P. (2016). PRD-1, a component of the circadian system of Neurospora crassa, is a member of the DEAD-box RNA Helicase family. J. Biol. Rhythms 31, 258–271. doi: 10.1177/0748730416639717
Aitken, C. E., and Lorsch, J. R. (2012). A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19, 568–576. doi: 10.1038/nsmb.2303
Allada, R., and Chung, B. Y. (2010). Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol. 72, 605–624. doi: 10.1146/annurev-physiol-021909-135815
Apitz, H., and Salecker, I. (2016). Retinal determination genes coordinate neuroepithelial specification and neurogenesis modes in the Drosophila optic lobe. Development 143, 2431–2442. doi: 10.1242/dev.135004
Aradska, J., Bulat, T., Sialana, F. J., Birner-Gruenberger, R., Erich, B., and Lubec, G. (2015). Gel-free mass spectrometry analysis of Drosophila melanogaster heads. Proteomics 15, 3356–3360. doi: 10.1002/pmic.201500092
Aravin, A. A., Hannon, G. J., and Brennecke, J. (2007). The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761–764. doi: 10.1126/science/.1146484
Aravin, A. A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., et al. (2003). The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337–350. doi: 10.1016/S1534-5807(03)00228-4
Aravin, A. A., Naumova, N. M., Tulin, A. V., Vagin, V. V., Rozovsky, Y. M., and Gvozdev, V. A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027. doi: 10.1016/S0960-9822(01)00299-8
Awasaki, T., and Ito, K. (2004). Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr. Biol. 14, 668–677. doi: 10.1016/j.cub.2004.04.001
Beckham, C., Hilliker, A., Cziko, A. M., Noueiry, A., Ramaswami, M., and Parker, R. (2008). The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol. Biol. Cell 19, 984–993. doi: 10.1091/mbc.E07-09-0954
Bellen, H. J., Levis, R. W., Liao, G., He, Y., Carlson, J. W., Tsang, G., et al. (2004). The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761–781. doi: 10.1534/genetics.104.026427
Boothroyd, C. E., Wijnen, H., Naef, F., Saez, L., and Young, M. W. (2007). Integration of light and temperature in the regulation of circadian gene expression in Drosophila. PLoS Genet. 3:e54. doi: 10.1371/journal.pgen.0030054
Bozzetti, M. P., Fanti, L., Di Tommaso, S., Piacentini, L., Berloco, M., Tritto, P., et al. (2012). The “special” crystal-stellate system in Drosophila melanogaster reveals mechanisms underlying piRNA pathway-mediated canalization. Gen. Res. Int. 2012:324293. doi: 10.1155/2012/324293
Bozzetti, M. P., Specchia, V., Cattenoz, P. B., Laneve, P., Geusa, A., Sahin, H. B., et al. (2015). The Drosophila fragile X mental retardation protein participates in the piRNA pathway. J. Cell Sci. 128, 2070–2084. doi: 10.1242/jcs.161810
Bradley, S., Narayanan, S., and Rosbash, M. (2012). NAT1/DAP5/p97 and atypical translational control in the Drosophila circadian oscillator. Genetics 192, 943–957. doi: 10.1534/genetics.112.143248
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. doi: 10.1016/j.cell.2007.01.043
Cheng, P., He, Q., Wang, L., and Liu, Y. (2005). Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 19, 234–241. doi: 10.1101/jad.1266805
Claridge-Chang, A., Wijnen, H., Naef, F., Boothroyd, C., Rajewsky, N., and Young, M. W. (2001). Circadian regulation of gene expression systems in the Drosophila head. Neuron 32, 657–671. doi: 10.1016/S0896-6273(01)00515-3
Collins, B. H., Rosato, E., and Kyriacou, C. P. (2004). Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc. Natl. Acad. Sci. U.S.A. 101, 1945–1950. doi: 10.1073/pnas.0308240100
Czech, B., Preall, J. B., McGinn, J., and Hannon, G. J. (2013). A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol. Cell. 50, 749–761. doi: 10.1016/j.molcel.2013.04.007
Damulewicz, M., Rosato, E., and Pyza, E. (2013). Circadian regulation of the Na+/K+-ATPase alpha subunit in the visual system is mediated by the pacemaker and by retina photoreceptors in Drosophila melanogaster. PLoS One 8:e73690. doi: 10.1371/journal.pone.0073690
Dissel, S., Codd, V., Fedic, R., Garner, K. J., Costa, R., Kyriacou, C. P., et al. (2004). A constitutively active cryptochrome in Drosophila melanogaster. Nat. Neurosci. 7, 834–840. doi: 10.1038/nn1285
Dockendorff, T. C., Su, H. S., McBride, S. M., Yang, Z., Choi, C. H., Siwicki, K. K., et al. (2002). Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron 34, 973–984. doi: 10.1016/S0896-6273(02)00724-9
Doll, C. A., and Broadie, K. (2016). Neuron class-specific requirements for fragile X mental retardation protein in critical period development of calcium signaling in learning and memory circuitry. Neurobiol. Dis. 89, 76–87. doi: 10.1016/j.nbd.2016.02.006
Dubruille, R., and Emery, P. (2008). A plastic clock: how circadian rhythms respond to environmental cues in Drosophila. Mol. Neurobiol. 38, 129–145. doi: 10.1007/s12035-008-8035-y
Emerson, J. M., Bartholomai, B. M., Ringelberg, C. S., Baker, S. E., Loros, J. J., and Dunlap, J. C. (2015). period-1 encodes an ATP-dependent RNA helicase that influences nutritional compensation of the Neurospora circadian clock. Proc. Natl. Acad. Sci. U.S.A. 112, 15707–15712. doi: 10.1073/pnas.1521918112
Emery, P., So, W. V., Kaneko, M., Hall, J. C., and Rosbash, M. (1998). CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679. doi: 10.1016/S0092-8674(00)81637-2
Ewer, J., Frisch, B., Hamblen-Coyle, M. J., Rosbash, M., and Hall, J. C. (1992). Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J. Neurosci. 12, 3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992
Galloni, M., and Edgar, B. A. (1999). Cell-autonomous and non-autonomous growth-defective mutants of Drosophila melanogaster. Development 126, 2365–2375.
Ghildiyal, M., and Zamore, P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Gen. 10, 94–108. doi: 10.1038/nrg2504
Gotze, M., Dufourt, J., Ihling, C., Rammelt, C., Pierson, S., Sambrani, N., et al. (2017). Translational repression of the Drosophila nanos mRNA involves the RNA helicase Belle and RNA coating by Me31B and Trailer hitch. RNA 23, 1552–1568. doi: 10.1261/rna.062208.117
Grima, B., Chelot, E., Xia, R., and Rouyer, F. (2004). Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873. doi: 10.1038/nature02935
Grivna, S. T., Beyret, E., Wang, Z., and Lin, H. (2006). A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714. doi: 10.1101/gad.1434406
Guo, J., Cheng, P., and Liu, Y. (2010). Functional significance of FRH in regulating the phosphorylation and stability of Neurospora circadian clock protein FRQ. J. Biol. Chem. 285, 11508–11515. doi: 10.1074/jbc.M109.071688
Handler, D., Meixner, K., Pizka, M., Lauss, K., Schmied, C., Gruber, F. S., et al. (2013). The genetic makeup of the Drosophila piRNA pathway. Mol. Cell 50, 762–777. doi: 10.1016/j.molcel.2013.04.031
Helfrich-Forster, C. (1998). Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J. Comp. Physiol. A 182, 435–453. doi: 10.1007/s003590050192
Huang, Y., Ainsley, J. A., Reijmers, L. G., and Jackson, F. R. (2013). Translational profiling of clock cells reveals circadianly synchronized protein synthesis. PLoS Biol. 11:e1001703. doi: 10.1371/journal.pbio.1001703
Ihry, R. J., Sapiro, A. L., and Bashirullah, A. (2012). Translational control by the DEAD Box RNA helicase belle regulates ecdysone-triggered transcriptional cascades. PLoS Genet. 8:e1003085. doi: 10.1371/journal.pgen.1003085
Iwasaki, Y. W., Siomi, M. C., and Siomi, H. (2015). PIWI-interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433. doi: 10.1146/annurev-biochem-060614-034258
Johnstone, O., Deuring, R., Bock, R., Linder, P., Fuller, M. T., and Lasko, P. (2005). Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev. Biol. 277, 92–101. doi: 10.1016/j.ydbio.2004.09.009
Khurana, J. S., and Theurkauf, W. (2010). piRNAs, transposon silencing, and Drosophila germline development. J. Cell Biol. 191, 905–913. doi: 10.1083/jcb.201006034
Kibanov, M. V., Egorova, K. S., Ryazansky, S. S., Sokolova, O. A., Kotov, A. A., Olenkina, O. M., et al. (2011). A novel organelle, the piNG-body, in the nuage of Drosophila male germ cells is associated with piRNA-mediated gene silencing. Mol. Biol. Cell 22, 3410–3419. doi: 10.1091/mbc.E11-02-0168
Kim, D. S., Gusti, V., Pillai, S. G., and Gaur, R. K. (2005). An artificial riboswitch for controlling pre-mRNA splicing. RNA 11, 1667–1677. doi: 10.1261/rna.2162205
Klattenhoff, C., and Theurkauf, W. (2008). Biogenesis and germline functions of piRNAs. Development 135, 3–9. doi: 10.1242/dev.006486
Kojima, S., Sher-Chen, E. L., and Green, C. B. (2012). Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 26, 2724–2736. doi: 10.1101/gad.208306.112
Kojima, S., Shingle, D. L., and Green, C. B. (2011). Post-transcriptional control of circadian rhythms. J. Cell Sci. 124, 311–320. doi: 10.1242/jcs.065771
Kotaja, N., Bhattacharyya, S. N., Jaskiewicz, L., Kimmins, S., Parvinen, M., Filipowicz, W., et al. (2006). The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl. Acad. Sci. U.S.A. 103, 2647–2652. doi: 10.1073/pnas.0509333103
Kotov, A. A., Olenkina, O. M., Kibanov, M. V., and Olenina, L. V. (2016). RNA helicase Belle (DDX3) is essential for male germline stem cell maintenance and division in Drosophila. Biochim. Biophys. Acta 1863, 1093–1105. doi: 10.1016/j.bbamcr.2016.02.006
Kremer, M. C., Jung, C., Batelli, S., Rubin, G. M., and Gaul, U. (2017). The glia of the adult Drosophila nervous system. Glia 65, 606–638. doi: 10.1002/glia.23115
Kuintzle, R. C., Chow, E. S., Westby, T. N., Gvakharia, B. O., Giebultowicz, J. M., and Hendrix, D. A. (2017). Circadian deep sequencing reveals stress-response genes that adopt robust rhythmic expression during aging. Nat. Commun. 8:14529. doi: 10.1038/ncomms14529
Lachance, P. E., Miron, M., Raught, B., Sonenberg, N., and Lasko, P. (2002). Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol. Cell Biol. 22, 1656–1663. doi: 10.1128/MCB.22.6.1656-1663.2002
Lasko, P. (2013). The DEAD-box helicase Vasa: evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim. Biophys. Acta 1829, 810–816. doi: 10.1016/j.bbagrm.2013.04.005
Lasko, P. F., and Ashburner, M. (1990). Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 4, 905–921. doi: 10.1101/gad.4.6.905
Lauinger, L., Diernfellner, A., Falk, S., and Brunner, M. (2014). The RNA helicase FRH is an ATP-dependent regulator of CK1a in the circadian clock of Neurospora crassa. Nat. Commun. 5:3598. doi: 10.1038/ncomms4598
Le Thomas, A., Toth, K. F., and Aravin, A. A. (2014). To be or not to be a piRNA: genomic origin and processing of piRNAs. Genome Biol. 15:204. doi: 10.1186/gb4154
Lee, E. J., Banerjee, S., Zhou, H., Jammalamadaka, A., Arcila, M., Manjunath, B. S., et al. (2011). Identification of piRNAs in the central nervous system. RNA 17, 1090–1099. doi: 10.1261/rna.2565011
Lee, J., Yoo, E., Lee, H., Park, K., Hur, J. H., and Lim, C. (2017). LSM12 and ME31B/DDX6 define distinct modes of posttranscriptional regulation by ATAXIN-2 protein complex in Drosophila circadian pacemaker neurons. Mol. Cell 66, 129–140.e7. doi: 10.1016/j.molcel.2017.03.004
Li, C., Vagin, V. V., Lee, S., Xu, J., Ma, S., Xi, H., et al. (2009). Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137, 509–521. doi: 10.1016/j.cell.2009.04.027
Li, W., Cressy, M., Qin, H., Fulga, T., Van Vactor, D., and Dubnau, J. (2013). MicroRNA-276a functions in ellipsoid body and mushroom body neurons for naive and conditioned olfactory avoidance in Drosophila. J. Neurosci. 33, 5821–5833. doi: 10.1523/JNEUROSCI.4004-12.2013
Liang, L., Diehl-Jones, W., and Lasko, P. (1994). Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120, 1201–1211.
Lim, A. K., and Kai, T. (2007). Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 104, 6714–6719. doi: 10.1073/pnas.0701920104
Lim, C., and Allada, R. (2013). ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science 340, 875–879. doi: 10.1126/science.1234785
Lin, Y., Stormo, G. D., and Taghert, P. H. (2004). The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J. Neurosci. 24, 7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004
Linder, P., and Fuller-Pace, F. V. (2013). Looking back on the birth of DEAD-box RNA helicases. Biochim. Biophys. Acta 1829, 750–755. doi: 10.1016/j.bbagrm.2013.03.007
Linder, P., and Jankowsky, E. (2011). From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516. doi: 10.1038/nrm3154
Lo, P. K., Huang, Y. C., Poulton, J. S., Leake, N., Palmer, W. H., Vera, D., et al. (2016). RNA helicase Belle/DDX3 regulates transgene expression in Drosophila. Dev. Biol. 412, 57–70. doi: 10.1016/j.ydbio.2016.02.014
MacGregor, D. R., Gould, P., Foreman, J., Griffiths, J., Bird, S., Page, R., et al. (2013). HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 is required for circadian periodicity through the promotion of nucleo-cytoplasmic mRNA export in Arabidopsis. Plant Cell 25, 4391–4404. doi: 10.1105/tpc.113.114959
Majercak, J., Chen, W. F., and Edery, I. (2004). Splicing of the period gene 3’-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol. Cell. Biol. 24, 3359–3372. doi: 10.1128/MCB.24.8.3359-3372.2004
Majercak, J., Sidote, D., Hardin, P. E., and Edery, I. (1999). How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230. doi: 10.1016/S0896-6273(00)80834-X
Malone, C. D., Brennecke, J., Dus, M., Stark, A., McCombie, W. R., Sachidanandam, R., et al. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535. doi: 10.1016/j.cell.2009.03.040
Malone, C. D., Lehmann, R., and Teixeira, F. K. (2015). The cellular basis of hybrid dysgenesis and stellate regulation in Drosophila. Curr. Opin. Genet. Dev. 34, 88–94. doi: 10.1016/j.gde.2015.09.003
Mazzotta, G., Rossi, A., Leonardi, E., Mason, M., Bertolucci, C., Caccin, L., et al. (2013). Fly cryptochrome and the visual system. Proc. Natl. Acad. Sci. U.S.A. 110, 6163–6168. doi: 10.1073/pnas.1212317110
McBride, S. M., Choi, C. H., Wang, Y., Liebelt, D., Braunstein, E., Ferreiro, D., et al. (2005). Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron 45, 753–764. doi: 10.1016/j.neuron.2005.01.038
Montelli, S., Mazzotta, G., Vanin, S., Caccin, L., Corra, S., De Pitta, C., et al. (2015). period and timeless mRNA Splicing Profiles under Natural Conditions in Drosophila melanogaster. J. Biol. Rhythms 30, 217–227. doi: 10.1177/0748730415583575
Morin, X., Daneman, R., Zavortink, M., and Chia, W. (2001). A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 98, 15050–15055. doi: 10.1073/pnas.261408198
Muerdter, F., Guzzardo, P. M., Gillis, J., Luo, Y., Yu, Y., Chen, C., et al. (2013). A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol. Cell 50, 736–748. doi: 10.1016/j.molcel.2013.04.006
Nagamori, I., Cruickshank, V. A., and Sassone-Corsi, P. (2011). Regulation of an RNA granule during spermatogenesis: acetylation of MVH in the chromatoid body of germ cells. J. Cell Sci. 124, 4346–4355. doi: 10.1242/jcs.096461
Nagao, A., Sato, K., Nishida, K. M., Siomi, H., and Siomi, M. C. (2011). Gender-specific hierarchy in nuage localization of PIWI-interacting RNA factors in Drosophila. Front. Genet. 2:55. doi: 10.3389/fgene.2011.00055
Napoli, I., Mercaldo, V., Boyl, P. P., Eleuteri, B., Zalfa, F., De Rubeis, S., et al. (2008). The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134, 1042–1054. doi: 10.1016/j.cell.2008.07.031
Ng, F. S., and Jackson, F. R. (2015). The ROP vesicle release factor is required in adult Drosophila glia for normal circadian behavior. Front Cell Neurosci. 9:256. doi: 10.3389/fncel.2015.00256
Ng, F. S., Sengupta, S., Huang, Y., Yu, A. M., You, S., Roberts, M. A., et al. (2016). TRAP-seq Profiling and RNAi-based genetic screens identify conserved glial genes required for adult Drosophila behavior. Front. Mol. Neurosci. 9:146. doi: 10.3389/fnmol.2016.00146
Nishida, K. M., Saito, K., Mori, T., Kawamura, Y., Nagami-Okada, T., Inagaki, S., et al. (2007). Gene silencing mechanisms mediated by aubergine piRNA complexes in Drosophila male gonad. RNA 13, 1911–1922. doi: 10.1261/rna.744307
Ozkaya, O., and Rosato, E. (2012). The circadian clock of the fly: a neurogenetics journey through time. Adv. Genet. 77, 79–123. doi: 10.1016/B978-0-12-387687-4.00004-0
Pek, J. W., and Kai, T. (2011). DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc. Natl. Acad. Sci. U.S.A. 108, 12007–12012. doi: 10.1073/pnas.1106245108
Peng, Y., Stoleru, D., Levine, J. D., Hall, J. C., and Rosbash, M. (2003). Drosophila free-running rhythms require intercellular communication. PLoS Biol. 1:E13. doi: 10.1371/journal.pbio.0000013
Perrat, P. N., DasGupta, S., Wang, J., Theurkauf, W., Weng, Z., Rosbash, M., et al. (2013). Transposition-driven genomic heterogeneity in the Drosophila brain. Science 340, 91–95. doi: 10.1126/science.1231965
Peruquetti, R. L., de Mateo, S., and Sassone-Corsi, P. (2012). Circadian proteins CLOCK and BMAL1 in the chromatoid body, a RNA processing granule of male germ cells. PLoS One 7:e42695. doi: 10.1371/journal.pone.0042695
Peschel, N., Chen, K. F., Szabo, G., and Stanewsky, R. (2009). Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr. Biol. 19, 241–247. doi: 10.1016/j.cub.2008.12.042
Peschel, N., and Helfrich-Forster, C. (2011). Setting the clock–by nature: circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett. 585, 1435–1442. doi: 10.1016/j.febslet.2011.02.028
Piacentini, L., Fanti, L., Specchia, V., Bozzetti, M. P., Berloco, M., Palumbo, G., et al. (2014). Transposons, environmental changes, and heritable induced phenotypic variability. Chromosoma 123, 345–354. doi: 10.1007/s00412-014-0464-y
Pyza, E., and Gorska-Andrzejak, J. (2004). Involvement of glial cells in rhythmic size changes in neurons of the housefly’s visual system. J. Neurobiol. 59, 205–215. doi: 10.1002/neu.10307
Renn, S. C., Park, J. H., Rosbash, M., Hall, J. C., and Taghert, P. H. (1999). A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802. doi: 10.1016/S0092-8674(00)81676-1
Richard, A., Rybak, J., Stortkuhl, K., Meinertzhagen, I., and Hovemann, B. (2002). Ebony protein in the Drosophila nervous system: optic neuropile expression in glial cells. J. Comp. Neurol. 452, 93–102. doi: 10.1002/cne.10360
Rival, T., Soustelle, L., Strambi, C., Besson, M. T., Iché, M., and Birman, S. (2004). Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr. Biol. 14, 599–605. doi: 10.1016/j.cub.2004.03.039
Robles, M. S., Cox, J., and Mann, M. (2014). In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 10:e1004047. doi: 10.1371/journal.pgen.1004047
Rolls, M. M., Satoh, D., Clyne, P. J., Henner, A. L., Uemura, T., and Doe, C. Q. (2007). Polarity and intracellular compartmentalization of Drosophila neurons. Neural Dev. 2:7. doi: 10.1186/1749-8104-2-7
Sahin, H. B., Karatas, O. F., Specchia, V., Tommaso, S. D., Diebold, C., Bozzetti, M. P., et al. (2016). Novel mutants of the aubergine gene. Fly 10, 81–90. doi: 10.1080/19336934.2016.1174355
Sandrelli, F., Costa, R., Kyriacou, C. P., and Rosato, E. (2008). Comparative analysis of circadian clock genes in insects. Insect Mol. Biol. 17, 447–463. doi: 10.1111/j.1365-2583.2008.00832.x
Santos, A. R., Kanellopoulos, A. K., and Bagni, C. (2014). Learning and behavioral deficits associated with the absence of the fragile X mental retardation protein: what a fly and mouse model can teach us. Learn. Mem. 21, 543–555. doi: 10.1101/lm.035956.114
Schenck, A., Van de Bor, V., Bardoni, B., and Giangrande, A. (2002). Novel features of dFMR1, the Drosophila orthologue of the fragile X mental retardation protein. Neurobiol. Dis. 11, 53–63. doi: 10.1006/nbdi.2002.0510
Schiesari, L., Kyriacou, C. P., and Costa, R. (2011). The hormonal and circadian basis for insect photoperiodic timing. FEBS Lett. 585, 1450–1460. doi: 10.1016/j.febslet.2011.02.026
Seluzicki, A., Flourakis, M., Kula-Eversole, E., Zhang, L., Kilman, V., and Allada, R. (2014). Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol. 12:e1001810. doi: 10.1371/journal.pbio.1001810
Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S., and Yokoyama, S. (2006). Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125, 287–300. doi: 10.1016/j.cell.2006.01.054
Senti, K. A., and Brennecke, J. (2010). The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet. 26, 499–509. doi: 10.1016/j.tig.2010.08.007
Shi, M., Collett, M., Loros, J. J., and Dunlap, J. C. (2010). FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock. Genetics 184, 351–361. doi: 10.1534/genetics.109.111393
Siwicki, K. K., Eastman, C., Petersen, G., Rosbash, M., and Hall, J. C. (1988). Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1, 141–150. doi: 10.1016/0896-6273(88)90198-5
Smith, R. F., and Konopka, R. J. (1981). Circadian clock phenotypes of chromosome aberrations with a breakpoint at the per locus. Mol. Gen. Genet. 183, 243–251. doi: 10.1007/BF00270625
Specchia, V., Benna, C., Mazzotta, G. M., Piccin, A., Zordan, M. A., Costa, R., et al. (2008). aubergine gene overexpression in somatic tissues of aubergine(sting) mutants interferes with the RNAi pathway of a yellow hairpin dsRNA in Drosophila melanogaster. Genetics 178, 1271–1282. doi: 10.1534/genetics.107.078626
Specchia, V., and Bozzetti, M. P. (2009). Different aubergine alleles confirm the specificity of different RNAi pathways in Drosophila melanogaster. Fly 3, 170–172. doi: 10.4161/fly.8054
Specchia, V., D’Attis, S., Puricella, A., and Bozzetti, M. P. (2017). dFmr1 plays roles in small RNA pathways of Drosophila melanogaster. Int. J. Mol. Sci. 18:E1066. doi: 10.3390/ijms18051066
Specchia, V., Piacentini, L., Tritto, P., Fanti, L., D’Alessandro, R., Palumbo, G., et al. (2010). Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 463, 662–665. doi: 10.1038/nature08739
Suh, J., and Jackson, F. R. (2007). Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron 55, 435–447. doi: 10.1016/j.neuron.2007.06.038
Tanaka, S. S., Toyooka, Y., Akasu, R., Katoh-Fukui, Y., Nakahara, Y., Suzuki, R., et al. (2000). The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 14, 841–853. doi: 10.1101/gad.14.7.841
Thaben, P. F., and Westermark, P. O. (2014). Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29, 391–400. doi: 10.1177/07487304145530
Thomson, T., and Lin, H. (2009). The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 25, 355–376. doi: 10.1146/annurev.cellbio.24.110707.175327
Ulvila, J., Parikka, M., Kleino, A., Sormunen, R., Ezekowitz, R. A., Kocks, C., et al. (2006). Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J. Biol. Chem. 281, 14370–14375. doi: 10.1074/jbc.M513868200
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., and Zamore, P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324. doi: 10.1126/science.1129333
Vanin, S., Bhutani, S., Montelli, S., Menegazzi, P., Green, E. W., Pegoraro, M., et al. (2012). Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature 484, 371–375. doi: 10.1038/nature10991
Wilm, M., Shevchenko, A., Houthaeve, T., Breit, S., Schweigerer, L., Fotsis, T., et al. (1996). Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379, 466–469. doi: 10.1038/379466a0
Worringer, K. A., Chu, F., and Panning, B. (2009). The zinc finger protein Zn72D and DEAD box helicase Belle interact and control maleless mRNA and protein levels. BMC Mol. Biol. 10:33. doi: 10.1186/1471-2199-10-33
Xue, Y., and Zhang, Y. (2018). Emerging roles for microRNA in the regulation of Drosophila circadian clock. BMC Neurosci. 19:1. doi: 10.1186/s12868-018-0401-8
Young, M. W., and Kay, S. A. (2001). Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2, 702–715. doi: 10.1038/35088576
Zerr, D. M., Hall, J. C., Rosbash, M., and Siwicki, K. K. (1990). Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J. Neurosci. 10, 2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990
Zhang, Y., Ling, J., Yuan, C., Dubruille, R., and Emery, P. (2013). A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science 340, 879–882. doi: 10.1126/science.1234746
Zhou, R., Hotta, I., Denli, A. M., Hong, P., Perrimon, N., and Hannon, G. J. (2008). Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol. Cell. 32, 592–599. doi: 10.1016/j.molcel.2008.10.018
Keywords: RNA helicase BELLE, Drosophila, post-transcriptional control, circadian clock, piRNA-mediated transposons regulation
Citation: Cusumano P, Damulewicz M, Carbognin E, Caccin L, Puricella A, Specchia V, Bozzetti MP, Costa R and Mazzotta GM (2019) The RNA Helicase BELLE Is Involved in Circadian Rhythmicity and in Transposons Regulation in Drosophila melanogaster. Front. Physiol. 10:133. doi: 10.3389/fphys.2019.00133
Received: 31 October 2017; Accepted: 04 February 2019;
Published: 20 February 2019.
Edited by:
Geoffrey A. Head, Baker Heart and Diabetes Institute, AustraliaReviewed by:
Francesco Cappello, Università degli Studi di Palermo, ItalyCopyright © 2019 Cusumano, Damulewicz, Carbognin, Caccin, Puricella, Specchia, Bozzetti, Costa and Mazzotta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriella M. Mazzotta, Z2FicmllbGxhLm1henpvdHRhQHVuaXBkLml0
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.