
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 09 January 2019
Sec. Aquatic Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.01895
This article is part of the Research Topic Molecular Physiology in Molluscs View all 20 articles
 Nan Chen1,2
Nan Chen1,2 Zekun Huang1,3,4
Zekun Huang1,3,4 Chengkuan Lu3,4
Chengkuan Lu3,4 Yawei Shen1,3,4
Yawei Shen1,3,4 Xuan Luo1,3,4
Xuan Luo1,3,4 Caihuan Ke1,3,4
Caihuan Ke1,3,4 Weiwei You1,3,4*
Weiwei You1,3,4*The Pacific abalone Haliotis discus hannai is one of the most economically important mollusks in China. Even though it has been farmed in southern China for almost 20 years, summer mortality remains the most challengeable problem for Pacific abalone aquaculture recently. Here, we determined the different heat tolerance ability for five selective lines of H. discus hannai by measuring the cardiac performance and Arrhenius breakpoint temperature (ABT). The Red line (RL) and Yangxia line (YL) were determined as the most heat-sensitive and most heat-tolerant line, respectively. Heart rates for RL were significantly lower than those of the YL at the same temperature (p < 0.05). The differentially expressed genes (DEGs), which were enriched in several pathways including cardiac muscle contraction, glutathione metabolism and oxidative phosphorylation, were identified between RL and YL at control temperature (20°C) and heat stress temperature (28.5°C, the ABT of the RL) by RNA-seq method. In the RL, 3370 DEGs were identified between the control and the heat-stress temperature, while only 1351 DEGs were identified in YL between these two temperature tests. Most of these DEGs were enriched in the pathways such as protein processing in endoplasmic reticulum, nucleotide binding and oligomerization domain (NOD) like receptor signaling, and ubiquitin mediated proteolysis. Notably, the most heat-tolerant line YL used an effective heat-protection strategy based on moderate transcriptional changes and regulation on the expression of key genes.
The Pacific abalone (Haliotis discus hannai) is one of the most economically important mollusk species in China. The Pacific abalone is endemic to the coastal areas of Eastern Asia, including Northern China, Korea, and Japan (Li et al., 2007; Li J. et al., 2012; Chen N. et al., 2017). The large-scale Pacific abalone farming began in the late 1980s to cope with the decreasing of wild resources and increasing market demand (Guo et al., 1999; Li et al., 2007). Originally, most abalone farms were situated in coastal areas within or close to the range of wild abalone. However, new varieties of the Pacific abalone had been successfully introduced to locations outside of the endemic range since 2000, including subtropical areas such as Fujian province (Deng et al., 2008). In 2016, China produced nearly 140,000 tons of abalone, and Fujian accounted for 80% of that total production (Guo and Zhao, 2017). However, the water temperature in Fujian is much higher than that in the natural habitat of the Pacific abalone, which put tremendous pressure on abalone. The warmer temperature not only reduce oxygen solubility but increase the mollusk’s metabolic rate (Hamburger et al., 1994; Hoegh-Guldberg and Bruno, 2010; Vaquer-Sunyer and Duarte, 2011), which may trigger a rising number of oxygen deficient. The immune response of abalone could also be affected by the elevated temperature associated with increased in its susceptibility to disease outbreak (Cheng et al., 2004; Dang et al., 2012). Especially in summer, these influences would be more serious and the mass mortality occurred frequently (Liang et al., 2014). Therefore, it is important to explore how the abalone respond to the heat stress and whether different strategies are taken to adapt to the high temperature.
How do organisms response to the environmental change is one of the basic question for environmental physiology. Recently, advances in biotechnology have allowed experiments aimed at assessing this question to become increasingly reductionist, by relating changes in environment to genome-scale phenomena (Evans, 2015). Transcriptomics has been one of the popular methods because it can be used to compare the gene expression of different tissues and development stages with various treatment, even without the need for a reference genome (Martin and Wang, 2011; Shiel et al., 2015). The different trait related transcriptome studies had been widely performed on abalone (Franchini et al., 2011; van der Merwe et al., 2011; Valenzuela-Munoz et al., 2013; Choi et al., 2015; Picone et al., 2015; Valenzuela-Miranda et al., 2015; Harney et al., 2016). However, previous work on heat tolerance in the abalone was limited. Transcriptome-wide scans of the red abalone (H. rufescens) from three environmentally distinct regions were analyzed, and some loci associated with the heat response were identified (Wit and Palumbi, 2013). In the green lip abalone (H. laevigata), transcriptome analyses indicated that the heat shock protein 70 (HSP 70) gene family was strongly associated with heat response (Shiel et al., 2015). The related genes and transposable elements that respond quickly and effectively to summer heat stress (20°C) have been identified in heat-resistant green lip abalone line (Shiel et al., 2017). In southern China, ambient water temperatures can be over 28°C in summer, which was much higher than those published abalone heat test on red and green lip abalones. It would be very interesting to conduct the transcirptome analyses for the Pacific abalone under such high temperature and analyze whether there are similar or different heat response mechanism on different abalone species.
Different environments may exert strong selection pressures on different populations, leading to local adaptations to particular conditions (Hereford, 2009; North et al., 2011; Wit and Palumbi, 2013). Typically, such natural selection should produce phenotypes sensitive or resistant to a particular environmental stressor. In ectothermic organisms, physiological performance is sensitive to ambient temperature variation and is closely linked with the organismal heat tolerance (Portner et al., 2006; Clark et al., 2008). The Arrhenius breakpoint temperature (ABT) of cardiac performance has been used as an indicator for heat tolerance in porcelain crabs (Petrolisthes) (Stillman and Somero, 1996), marine snail (Tegula) (Stenseng et al., 2005), limpets (Cellana) (Dong and Williams, 2011; Han et al., 2013), and scallops (Xing et al., 2016). The ABT also has been shown to be a credible, non-invasive indicator of heat tolerance in abalone (Chen et al., 2016; Alter et al., 2017).
The study’s objective was to identify differences in patterns of gene expression between heat-sensitive and heat-tolerant abalones when subjected to heat stress on subtropical area. We first aimed to assess heat tolerance with ABT measurements in various selective lines of H. discus hannai. Based on this assessment, we were able to identify heat-sensitive and heat-tolerant abalone lines, and to conduct transcriptome analyses. We also aimed to identify genes or pathways associated with heat response in abalones by comparing gene expression patterns between sensitive and tolerant lines. The results might provide clues as to how abalone cope with life at high temperature, increase our understanding of the thermal response patterns of the Pacific abalone and highlighted potentially important genes or pathways for future study.
Five Pacific abalone selective lines were used in this research for the heat tolerance assessment: Yangxia line (YL), Dongshan line (DL), Red line (RL), Changdao line (CL) and Japanese line (JL). 50 individuals with the same size (4.5–6 cm) from each line were selected, labeled and cultured in a recirculating system for 7 days acclimation. Temperature, dissolved oxygen and salinity were kept at 20°C, 6 mg/L and 32, respectively. All abalones were fed once each day with fresh seaweed and all the residual food particles and fecal debris were removed 12 h before the start of the experiment.
Sixteen individuals of each line were randomly selected, and crystal dishes (diameter = 20.0 cm, height = 9.5 cm) were used for abalone to settle. Fresh seawater was added into the dishes, and the dissolved oxygen concentration was kept by delivering air from a compressor to each dish. To measure real-time body temperature, a fine thermometer was inserted between the foot and the crystal dish. The crystal dish was immersed in a water bath, and the temperature of the seawater in the dish was increased using the water bath on the rate of approximately 0.1°C min-1.
The non-invasive method was used for heart rate measurement (Depledge and Andersen, 1990; as modified by Chelazzi et al., 1999; Dong and Williams, 2011). To detect heart beats, an infrared sensor was glued to each shell above the heart (Krazy Glue, Westerville, OH, United States). The fluctuations of heart beat were amplified, filtered and recorded by an infrared signal amplifier (AMP03, Newshift, Leiria, Portugal) and Powerlab (8/35, ADInstruments, March-Hugstetten, Australia). All heart rate data were monitored and analyzed with LabChart v8.0. The ABT was defined as the temperature at which the heart rate decreased dramatically. To determine the ABT, we used regression analyses to generate the best fit lines on both side of a putative break point (Stillman and Somero, 1996). To construct Arrhenius plots, heart rates were transformed to the natural logarithm of beats min-1. Temperatures are shown as 1000/K (Kelvin temperatures). To test the significance of differences in ABT and maximum heart rate among lines, we used one-way analyses of variance (ANOVAs) in SPSS v17.0. We considered p < 0.05 statistically significant.
Based on mean ABT for each line, we could distinguish the heat-sensitive lines from the heat-tolerant lines. The ABT of the most sensitive line was selected as the thermal stressor. We randomly selected an additional 24 specimens, 12 from the most heat-tolerant line and 12 from the most heat-sensitive line. Half of all specimens (6 per line) were randomly assigned to the control group. All others were assigned to experimental group. For heat exposure, the abalones were transferred to the experimental tank from the acclimation condition (20°C) directly, in which the temperature was set as ABT of most sensitive line prior to the experiment. The heat stress lasted for 2 h and the control group was kept at 20°C at the same time. All the abalones were dissected at the end of experiments and gills were immediately frozen in liquid nitrogen and stored at -80°C until use.
Total RNA was extracted from the gills of the 24 samples using Trizol reagent (Gibco BRL, United States). To check the purity and integrity of RNA, the Nanophotometer® spectrophotometer (IMPLEN, Westlake Village, CA, United States) and RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States) were used. Based on these results, we selected three individuals with the best RNA quality of six samples from each of the four groups (sensitive line/tolerant line × heat stress/control) for transcriptomic analyses. Library preparation and sequencing were performed by Novogene (Beijing, China). We obtained an average of ∼52.8 million raw reads per specimen (maximum: 63.0 million raw reads; minimum: 44.8 million raw reads).
Raw reads were cleaned by removing ambiguous reads (those with > 10% ambiguous nucleotides) and low-quality reads (those consist of more than half bases whose quality scores < 10) to achieve clean reads. All clean data were assembled with Trinity r20140413p1 (Grabherr et al., 2011), with min_kmer_cov set to 2 and other parameters set to defaults. The longest transcript for each gene was considered to represent this gene and named unigene. Unigenes were used for all subsequent annotations.
To comprehensively annotate the assemblies, transcripts were compared to several public databases: Nt (NCBI non-redundant nucleotide sequences), Nr (NCBI non-redundant protein sequences), and Swiss-Prot using BLAST+ v2.2.28+ (e-value = 1e-5; Altschul et al., 1990); GO (Gene Ontology) with BLAST2GO (e-value = 1e-10; Götz et al., 2008); and KO (KEGG Orthologs) with KAAS r140224 (e-value = 1e-10; Mao et al., 2005; Moriya et al., 2007).
The de novo assembled transcriptome was used as a reference for read mapping and gene expression profiling. To identify differential gene expression, the sequence reads for each specimen were mapped back to the assembled transcriptome using RSEM v1.2.1.5 (bowtie2 set mismatch = 0; Li and Dewey, 2011). The gene expression levels were based on FPKM values (expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced). We compared differential gene expression between pairs of groups with DESeq v1.10.1 in R (Anders and Huber, 2012). The strict filter (p-value < 0.05 and |log2 (fold change)| > 1) was set as the threshold for significant differential expression. We performed four analyses to identify differential gene expression between the two lines or stress conditions: (I) the heat-sensitive line vs. the heat-tolerant line at the control temperature; (II) the heat-sensitive line vs. the heat-tolerant line at the heat stress temperature; (III) the heat-sensitive line at the control temperature vs. the heat stress temperature; and (IV) the heat-tolerant line at the control temperature vs. the heat stress temperature.
Gene Ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GOseq v1.10.0 in R packages (Young et al., 2010). GOseq is based on Wallenius non-central hyper-geometric distribution, which adjusts for DEG gene length bias. We used KOBAS v2.0.12 (Mao et al., 2005) to test the statistically significant enrichment of DEGs in KEGG pathways.
The further validation for the RNA-seq results was conducted by real-time quantitative PCR (RT-PCR) with all the 24 individuals which were prepared in Section “Heat Stress Experiments.” The expression levels of 10 genes which selected from intriguing pathways were quantified. Based on the reference unigene sequences, we designed gene-specific primers with Primer Premier v5.0 (Supplementary Table S1). First-strand cDNA was synthesized with a PrimeScript RT reagent Kit (TaKaRa, Dalian, China), and qRT-PCR was performed with a ThermoDyNAmo Flash SYBR Green qPCR Kit (Thermo Scientific, United States) on an ABI 7500 Fast Real-Time PCR system (Applied Biosystems, United States), following the manufacturer’s instructions. Gene expression was normalized based on the expression of 18S and β-actin. Relative gene expression was then calculated using the 2-ΔΔCT method.
The cardiac performance at different temperatures were shown by the Arrhenius plots (Figure 1A). The cardiac performance could be divided into two phases. Abalone heart rates initially increased slowly with rising water temperature. As water temperature continued to rise, incidents of arrhythmia were observed. Heart rates decreased abruptly as soon as the water temperature exceeded a critical value (the ABT). The ABTs for the YL, DL, RL, JL, and CL were 31.9 ± 0.7, 29.5 ± 1.7, 28.5 ± 1.8, 29.5 ± 1.2, and 30.4 ± 0.6°C, respectively (Figure 1B) using regression analysis. The ABT of the YL was significantly higher than that of the DL, RL, and JL. The ABT of the CL was also significantly higher than that of the RL. The YL was the most tolerant to heat stress, while the RL was most sensitive. Therefore, the individuals from YL and RL were selected for transcriptome analysis, and 28.5°C (RL’s ABT) was set as heat stress temperature. Heart capacity also differed between the RL and YL. Heart rates of RL specimens were lower than those of YL at the same temperature. The maximum of heart rates was also significantly different (Duncan’s post hoc analysis, p = 0.001; RL: 54.75 ± 6.90 beats min-1 and YL: 75.14 ± 7.29 beats min-1).
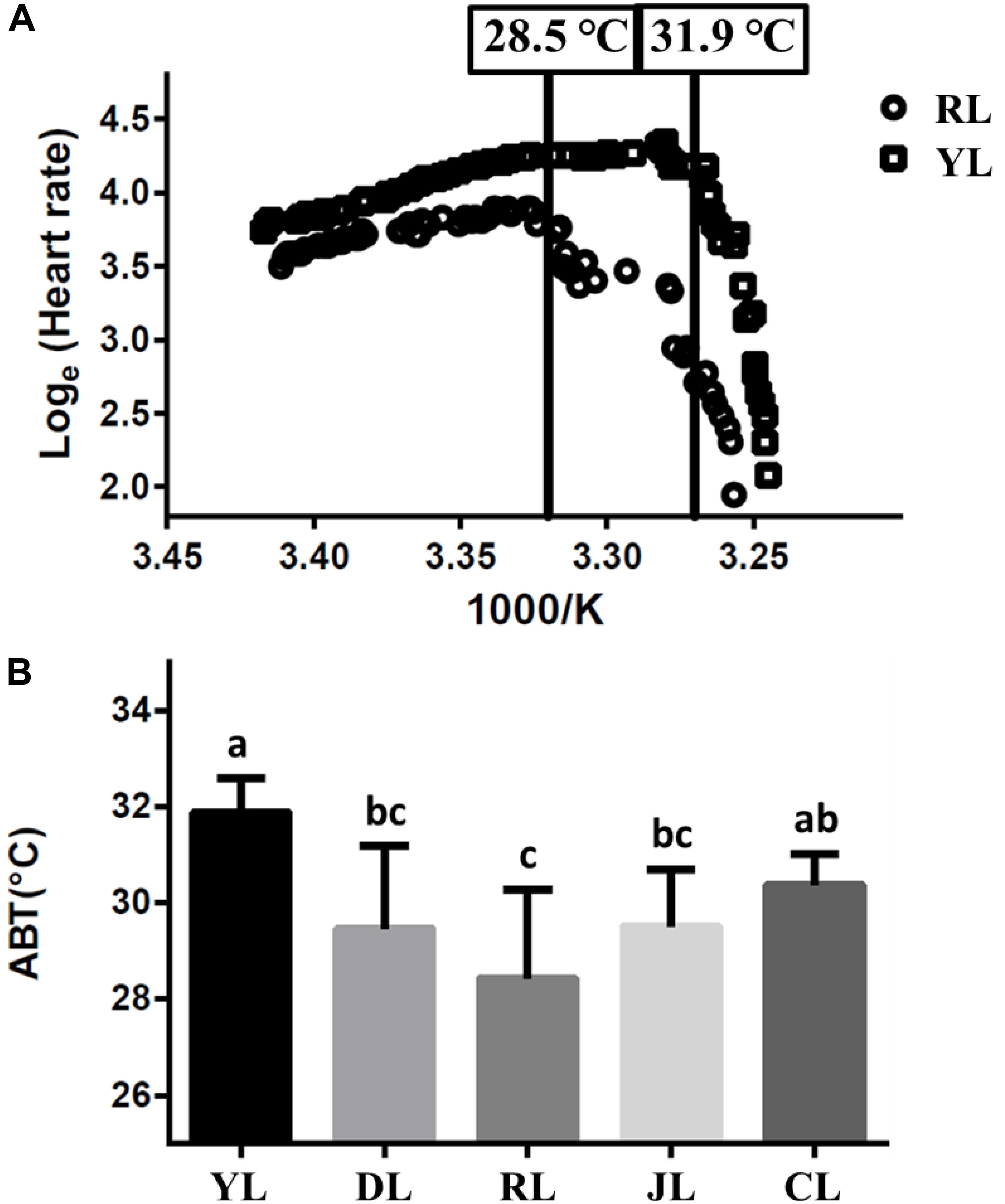
Figure 1. The Arrhenius breakpoint temperature (ABT) for different Pacific abalone lines. (A) The Arrhenius plots for YL and RL. (B) The ABTs for the five selective lines. Bars labeled with different letters are significantly different (p < 0.05).
After filtering, a total of 43.2 to 60.3 million clean reads for each sample were obtained. The Trinity assembly generated 227,066 transcirpts and 161,135 unigenes, with the minimum length of 201 bp and the maximum length of 17 kb. Only 27.26% of all unigenes could be annotated in one or more databases. Approximately 70% of all reads were successfully mapped back to the reference transcriptome (68.12–72.33%) (Supplementary Table S2).
We identified 535 DEGs between the heat-tolerant and heat-sensitive lines at the control temperature (20°C), and 492 DEGs at the heat stress condition. Of these DEGs, 150 were differentially expressed at both temperatures (Figure 2A). However, more genes were differentially expressed between the two temperatures as compared to those between the two lines. In the YL, 1351 DEGs were identified between the control and heat stress temperature: 1121 were up-regulated at the heat stress temperature as compared to the control, and 230 were down-regulated. Many more DEGs were identified in the RL. In the RL, 2217 DEGs were up-regulated at the heat stress temperature as compared to the control, and 1153 genes were down-regulated (Figure 2B). Of these DEGs, 1046 genes were differentially expressed in both lines.
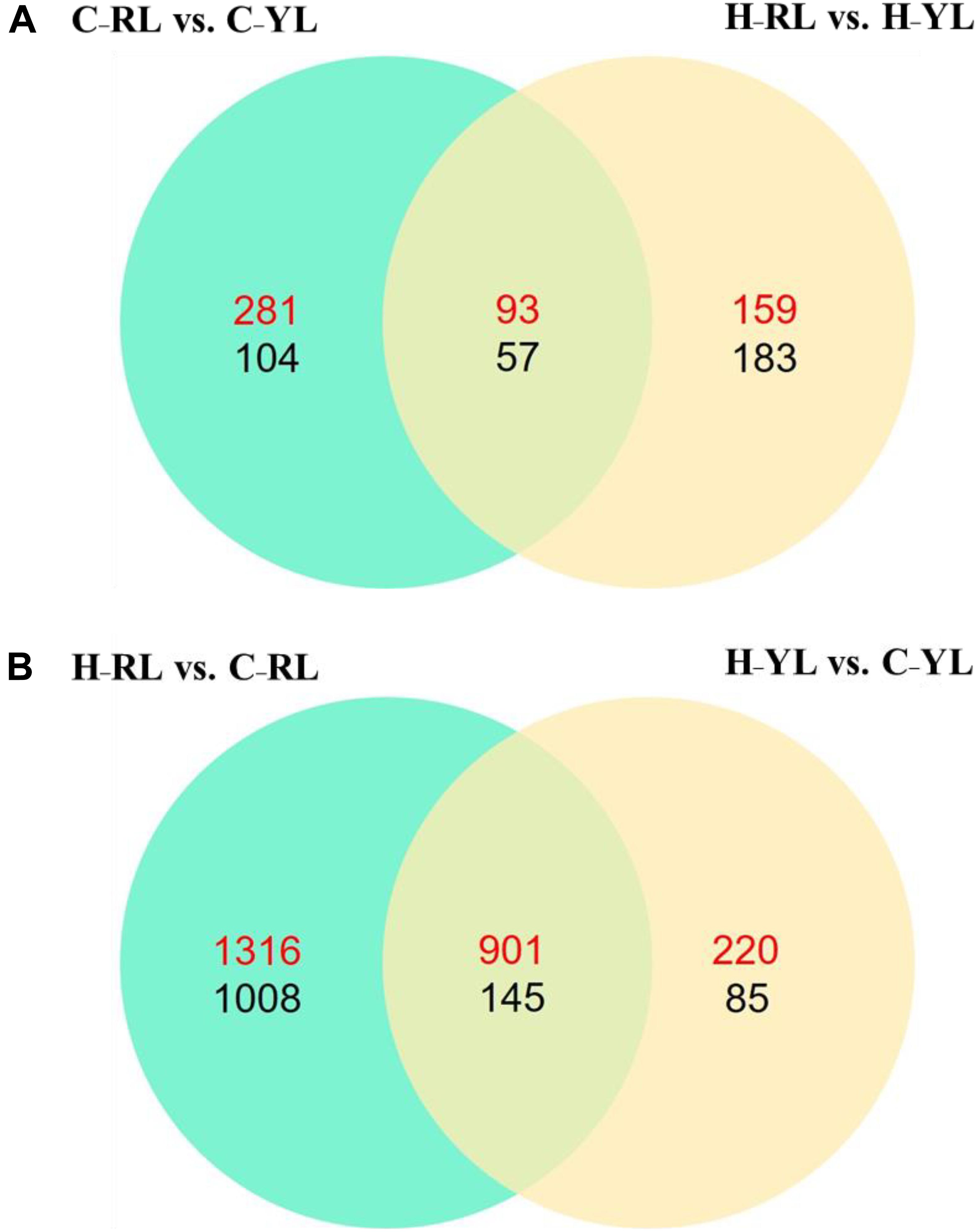
Figure 2. The Venn diagram of significant differently expressed genes. (A) Comparison between different lines. (B) Comparison between control temperature and heat stress temperature. C_RL: RL at control temperature; C_YL: YL at control temperature; H_RL: RL at heat stress temperature; H_YL: YL at heat stress temperature. Up-regulated genes were numbered in red and down-regulated genes were numbered in black.
The genes differentially expressed between the RL and YL were enriched in several interesting GO terms, including organic substance metabolic process, macromolecule metabolic process, hydrolase activity, and binding (Figure 3). Between control and heat stress treatment, the DGEs were significantly enriched in protein binding, ion binding, biological regulation, and intracellular membrane-bounded organelle for RL (Figure 4A) and transcription factor complex, intracellular membrane-bounded organelle, protein complex for YL (Figure 4B). Of these, nearly twice as many genes were enriched for each term in the RL as compared to YL (Supplementary Table S3).
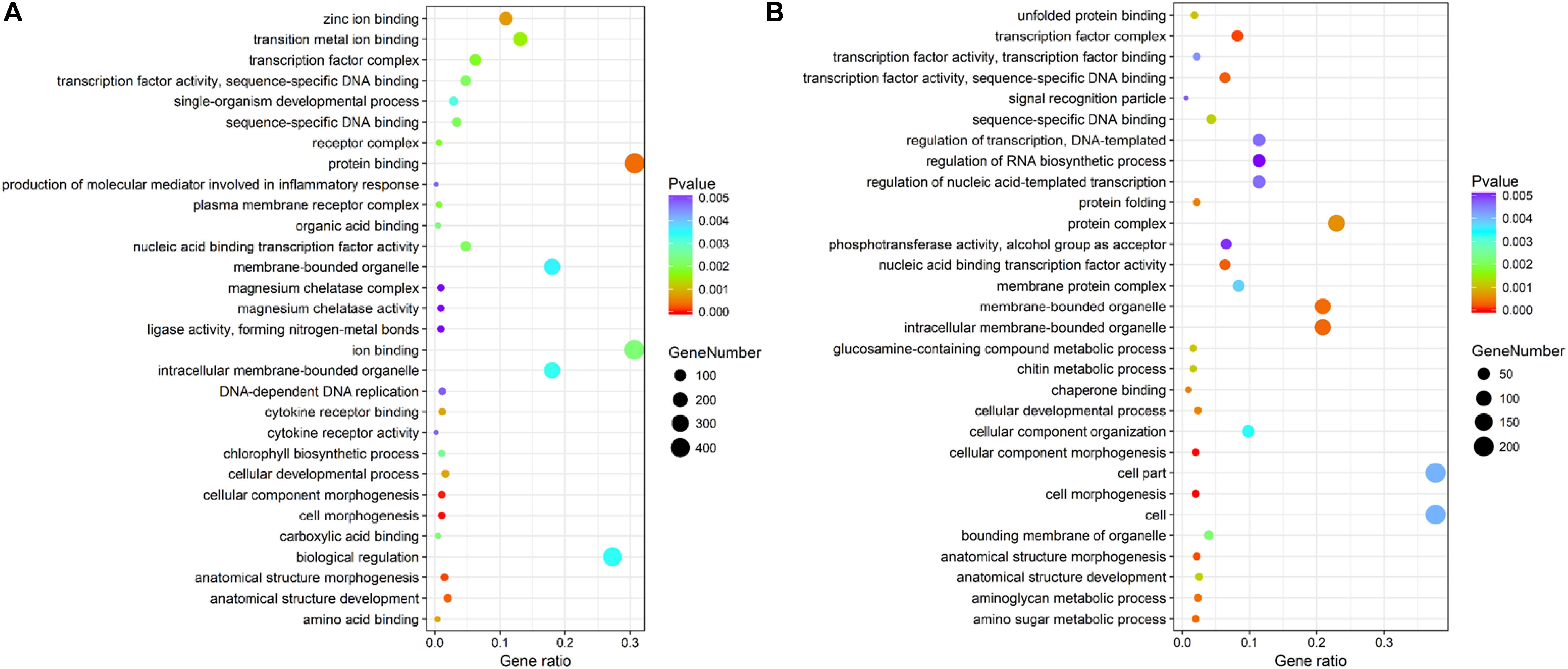
Figure 4. Top 30 GO terms enrichment statistics between control and heat stress temperature. (A) Terms enrichment in RL. (B) Terms enrichment in YL.
Within KEGG pathway analysis, the results showed that the DEGs between the RL and YL were most enriched in the following five pathways: pancreatic secretion, cardiac muscle contraction, protein digestion and absorption, oxidative phosphorylation, and Parkinson’s disease (Figure 5). Between the control and heat stress temperature, DEGs were most significantly enriched in the protein processing in endoplasmic reticulum pathway: 40 RL and 29 YL DEGs were enriched in this pathway. Several other pathways were also significant up-regulated, including ubiquitin mediated proteolysis, apoptosis, nucleotide binding, and oligomerization domain (NOD-like) receptor signaling pathway and TNF signaling pathway (Figures 6A,B).
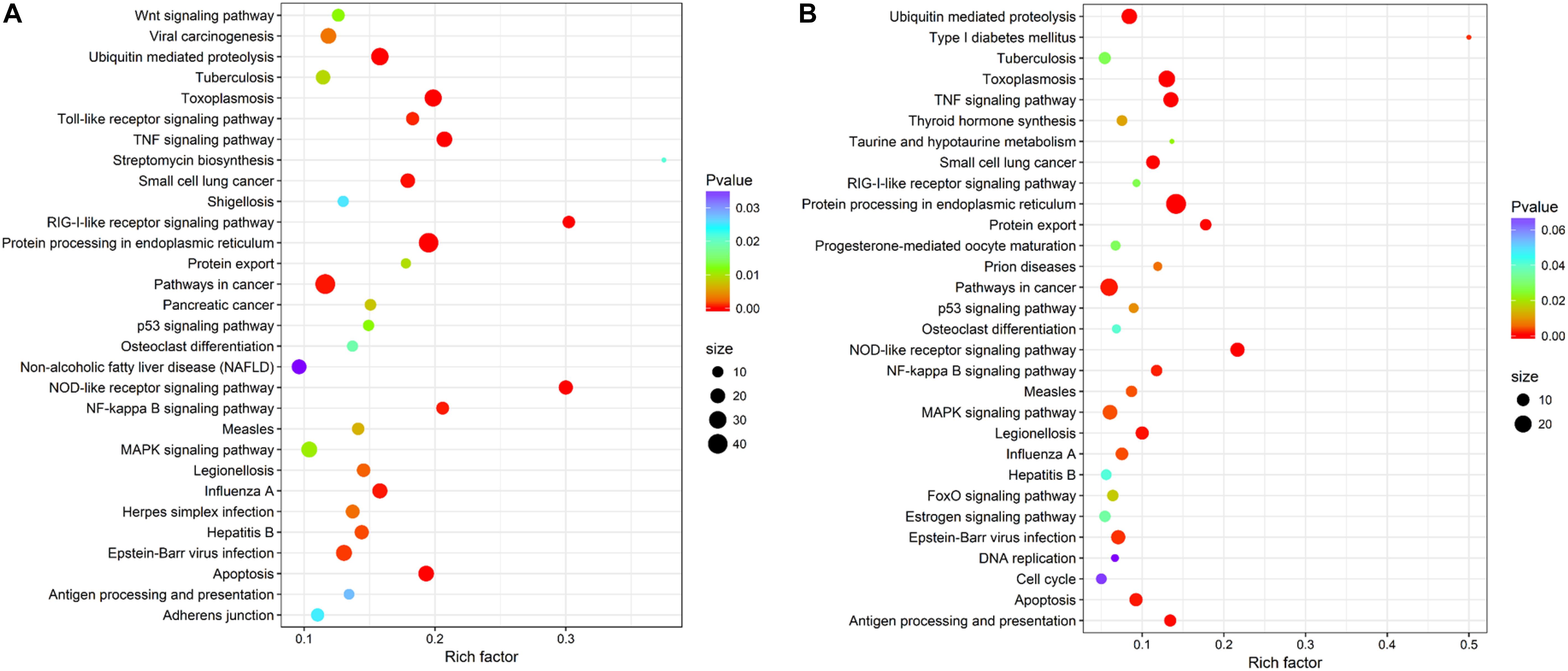
Figure 6. Top 30 pathways enrichment statistics between control and heat stress temperature. (A) Pathway enrichment in the RL. (B) Pathway enrichment in the YL.
Several genes in the HSP gene family were enriched in multiple pathways. Indeed, the expression levels of 35 HSP genes were significantly different between control and heat stressed specimens (Figure 7 and Table 1). Of these, 20 HSP genes were differentially expressed in both abalone lines, 11 were differentially expressed in the RL only, and 4 were differentially expressed in the YL only. The degree of up-regulation also differed among the differentially expressed HSP genes: from 1.6-fold (log2 Fold Change) (DnaJ homolog subfamily C member 7) to 11.9-fold (small heat shock protein 26) in RL and 2.9-fold (stress-70 protein, mitochondrial) to 11.4-fold (Heat shock 70 kDa protein) in YL.
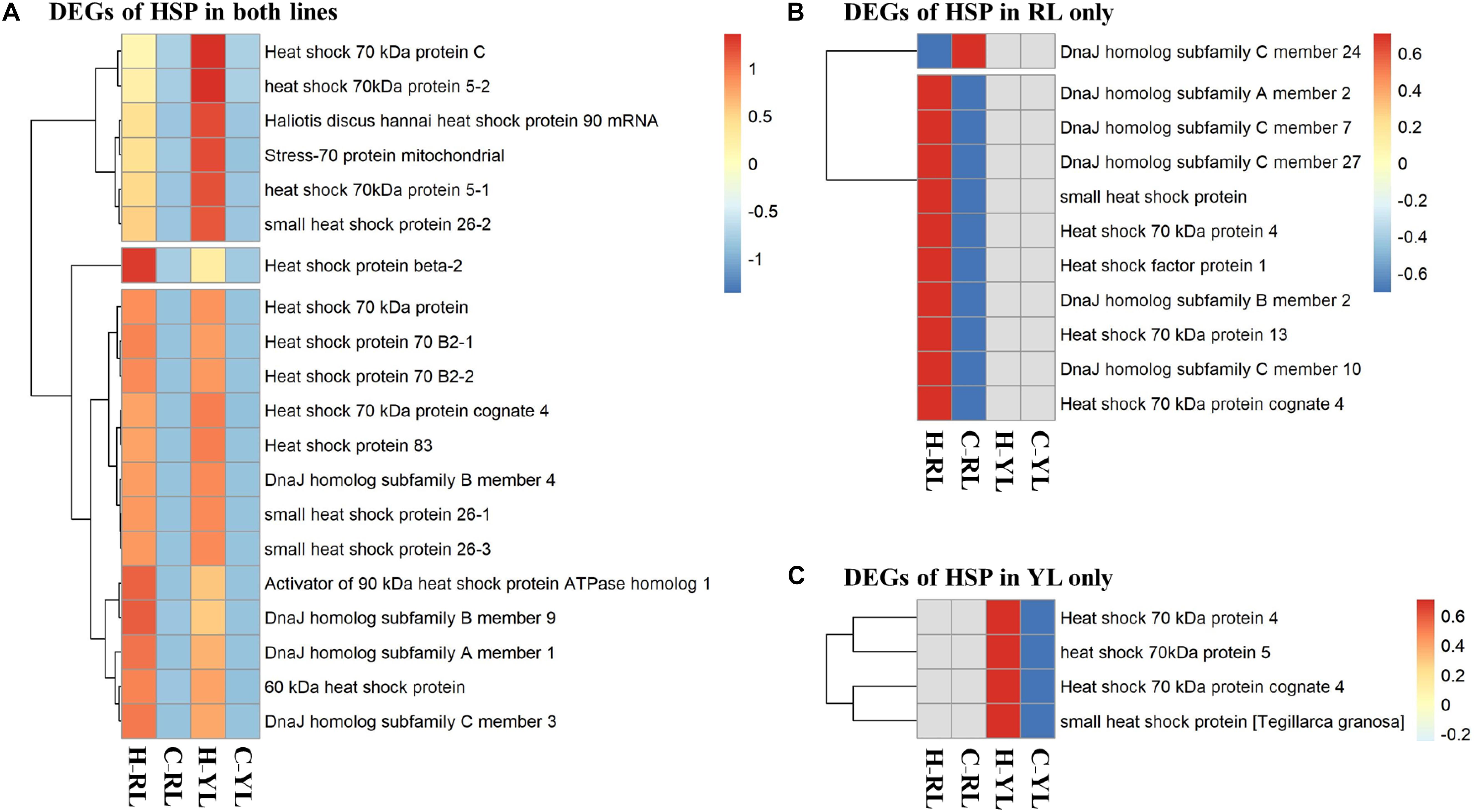
Figure 7. Heatmaps of differentially expressed heat shock protein (HSP) genes. (A) Genes differentially expressed in both lines. (B) Genes differentially expressed in the RL only. (C) Genes differentially expressed in the YL only.
Ubiquitination serves as a versatile post-translational modification in all eukaryotic species. As shown in Figure 8, a total of 32 genes that involved in this pathway expressed differently. Of these, 16 genes and 30 genes were up regulated significantly in YL and RL, respectively.
Heat stress can result in oxidative stress as the formation of reactive oxygen species (ROS). There were several genes (including catalase, superoxide dismutase, peroxiredoxin 5) related to antioxidant system that annotated in this study (Figure 9) and only one catalase gene (c92672_g3) was up-regulated at heat stress temperature of both lines.
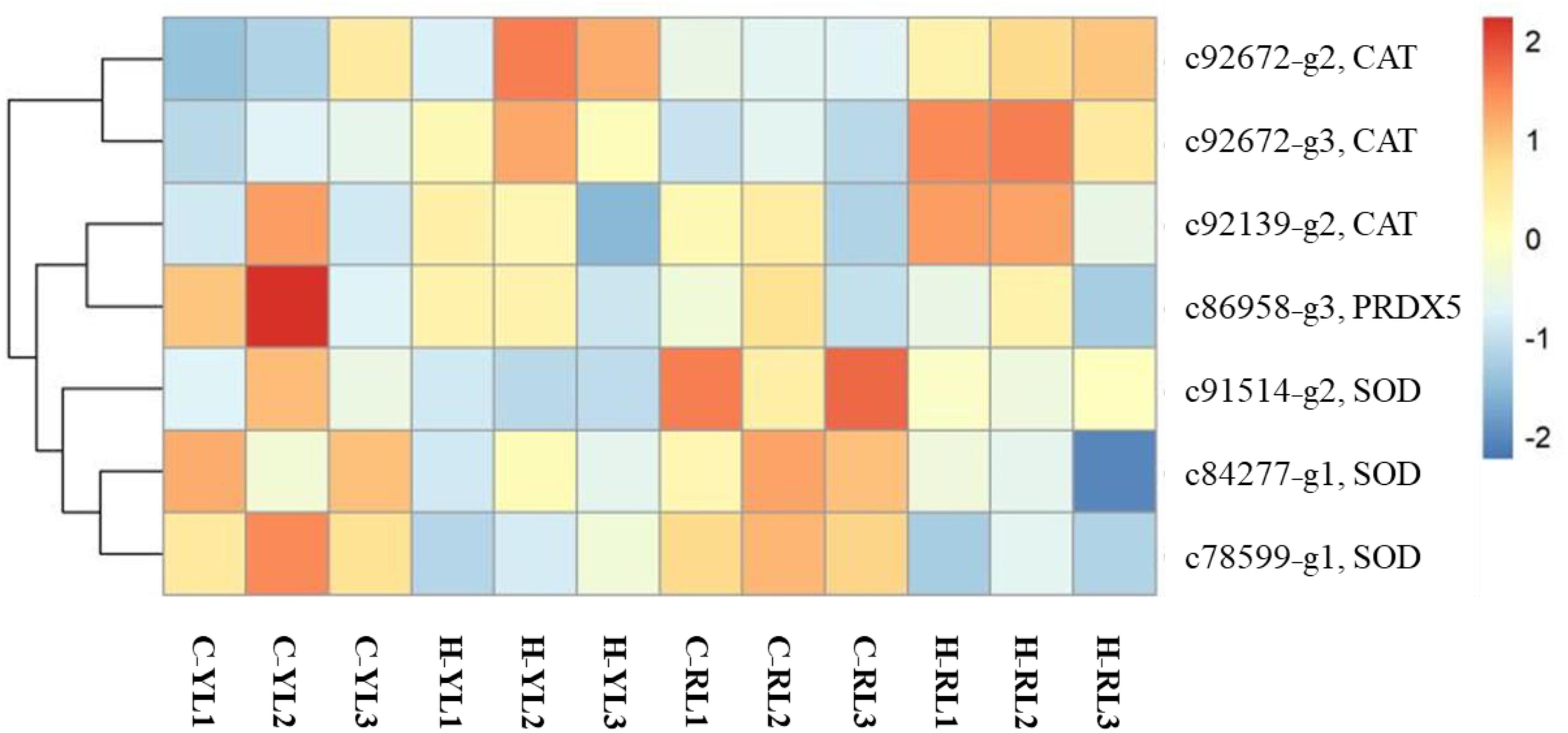
Figure 9. Heatmap of differentially expressed antioxidant-related genes. CAT, catalase; SOD, superoxide dismutase; PRDX5, peroxiredoxin 5.
We used 10 genes that were differentially expressed between the control and heat stress temperatures for qRT-PCR validation. These DEGs included HSP90 (c_95969_g1), HSP70 (c_92061_g1) and cyclin B (c_86983_g1). The patterns of gene expression indicated by the qRT-PCR and RNA-seq analyses were similar (Figure 10).
In ectothermic animals, temperature is the main variable which controls the rate of most metabolic processes (Morash and Alter, 2015). Increased temperatures can diminish oxygen solubility and change the metabolic rates (Hoegh-Guldberg and Bruno, 2010; Vaquer-Sunyer and Duarte, 2011). It thus becomes difficult for the organism to obtain sufficient oxygen to sustain aerobic scope, both with respect to providing metabolic fuel and to removing metabolic waste products at high temperature (Somero, 2012). Aerobic scope drops when the maximum metabolic rate fails to keep pace with the routine metabolic rate as temperature increases (Portner and Knust, 2007; Casselman et al., 2012). For many marine ectotherms, heat-stress tolerance is based on the ability to maintain normal aerobic scope in elevated water temperatures (Portner et al., 2006; Portner, 2010; Han et al., 2013). In gastropods, this capacity is largely dependent on heart function; the ABT of cardiac performance has thus been used as an effective indicator of heat tolerance in gastropods (Han et al., 2013; Chen et al., 2016).
We found that the five tested different Pacific abalone lines had different levels of heat tolerance. There were nearly 3.5°C difference between the most temperature-tolerant line (YL) and the most temperature-sensitive line (RL). At high water temperature, even a 1°C difference would affect abalone survival (Chen et al., 2016). The ABT for RL was 28.5 ± 1.8°C, which means RL abalone’s cardiac function is approaching the limit at this temperature. Thus, at 28.5°C, cardiac function in an RL individual is approaching the maximum, while the individuals of YL could maintain normal oxygen supply and cardiac function at the same temperature. The cardiac muscle contraction pathway was significantly down-regulated when compared control of RL to control of YL by RNA-seq results. Only six genes, including myosin light chain 4 (MYL 4) were involved in this pathway, MYL 4 is particularly notable because this gene regulates several critical biological processes, including heart contraction force, muscle organ development, and cardiac muscle contraction. The MYL 4 protein is down-regulated in failing human hearts, as compared to normal human hearts (Li W. et al., 2012). To verify the difference in cardiac function, further study of the expression level of myosin family gene in the abalone heart is necessary.
Between the RL and the YL, 535 DEGs were identified at the control temperature, and 492 DEGs were identified at the heat-stress temperature. However, more genes were differentially expressed between the control and heat-stress temperatures: 1351 DEGs in YL and 3370 DEGs in RL. This increase transcriptome fluctuation in the more temperature-sensitive line has been shown in other organisms, including copepod (Tigriopus californicus; Schoville et al., 2012), coral (Acropora hyacinthus; Barshis et al., 2013), snail (Chlorostoma funebralis; Gleason and Burton, 2015), and redband trout (Oncorhynchus mykiss gairdneri; Chen Z. et al., 2017). Indeed, the increased heat-tolerance seems to be based on the moderate transcriptional changes under heat stress (Bita et al., 2011). Here, twice as many RL DEGs were enriched per GO term as compared to YL DEGs. However, it is possible that this discrepancy is due to the higher level of physiological stress experienced by the less thermally tolerant individuals. Thus, we speculated that the more effective protection strategy was not only based on fine-tuned but the more effective expression response in the heat-tolerant abalone line. In this research, we observed a higher degree change of gene expression in the YL, as compared to the RL in some key genes including HSP family.
The different physiological stress might also be indicated by the down-regulated pathways: cell cycle and DNA replication. In face of stress condition, cells often delayed the DNA replication and cell division in favor of cytoprotective functions (Jonas et al., 2013). More genes (Table 2) were down-regulated in sensitive line vs. tolerant line (9 vs. 5), which mean the cells of RL might be subjected to more severe suppression.
Genes in the “Protein processing in endoplasmic reticulum” pathway, including many HSP genes, were significantly up-regulated. HSPs are highly conserved proteins that act as molecular chaperones to protect normal proteins from degeneration, catalyze the folding of normal proteins and the refolding of abnormal proteins, remove irreversibly damaged proteins, and help maintain cellular homeostasis (Snyder et al., 2001; Farcy et al., 2007; Liang et al., 2014). The molecular chaperones (HSP70, HSP90, and other HSPs) are one of the most varied gene families affected by heat stress in mollusks (Lang et al., 2009; Lockwood et al., 2010; Chapman et al., 2011). The rapid response of HSPs to heat stress is an important part of the molluscan protection mechanism (Zhao and Jones, 2012). There were fewer HSP DEGs in the YL (24) as compared to the RL (31), but the average fold change (log2 fold change) was higher in the YL (7.0) than in the RL (5.4). This indicated that the heat response strategy used by the YL might be more effective.
The HSP40 gene family is another important group, which primary function is to act as co-chaperones for HSP70 (Gleason and Burton, 2015). HSP40s increase the HSP70 ATP-hydrolysis rate and strengthen the HSP70 activity fields (Misselwitz et al., 1998; Genevaux et al., 2007; Mayer, 2013). In thermotolerant cells, this interaction might increase protein thermostability and accelerate heat-damage recovery (Gebauer et al., 1997; Terada et al., 1997; Gleason and Burton, 2015). The gastropod C. funebralis exhibited the opposite reaction to heat stress (Gleason and Burton, 2015): 66.7% of all annotated HSP70s showed a significantly higher fold change in heat-sensitive lines, as compared to heat-tolerant lines, while the expression levels of 60% of all HSP40s were higher in the heat-tolerant lines. Here, more than 70% of common HSP70 and HSP40 genes that differentially expressed in both lines showed a higher fold change in the YL, as compared to the RL. This might be because C. funebralis inhabits intertidal areas, where water temperature fluctuation is more drastic (Gleason and Burton, 2015) than in the subtidal habitat of the abalone. Some gene families, such as HSP60, are pre-adapted in heat-tolerant lines (Gleason and Burton, 2015), which might serve as a preparative defense against frequent heat stress events. In addition, significant up-regulation of HSP40 combined with reasonable up-regulation of HSP70 expression might reduce the need for the heat-tolerant lines to up-regulate HSP70 as drastically as the heat-sensitive populations do. For abalones, ambient water temperature does not fluctuate so frequently. Thus, the higher fold change of HSP gene family would be help to reduce the heat-damage more quickly in tolerant lines, which offered a more effective protection strategy.
Genes involved in the ubiquitin-mediated proteolysis pathway were considered as key transcription factors that respond to abiotic stresses, acting to clear irreparably damaged proteins (Glickman and Ciechanover, 2002; Kültz, 2005; Lyzenga and Stone, 2012). The up-regulation of genes involved in proteolysis has been reported in Mytilus trossulus (Lockwood et al., 2010), Saccharina japonica (Liu et al., 2015), and Pyropia haitanensis (Wang et al., 2018b). The RL and YL showed similar induction of many genes involved in this pathway. However, the less DEGs occurred in YL may also indicate these two lines endured different physiological stress. More irreparably damaged protein may be accumulated in RL. Therefore, for RL, more genes may be need to participate in this response that was helpful to transfer more ubiquitin to damaged protein and target them for degradation in time.
Heat stress can induce the overproduction of ROS (Abele et al., 2002; Heise et al., 2003; Yang et al., 2010; Cui et al., 2011). ROS are toxic to the cell as they cause damage to macromolecules (Kültz, 2005). Then, mitigating oxidative stress by increasing synthesis of antioxidants can increase thermal tolerance (Dilly et al., 2012). It has been demonstrated that the genes of antioxidant defense system were up regulated in the study of blue mussel (M. galloprovincialis and M. trossulus) (Lockwood et al., 2010), coral (Acropora hyacinthus) (Barshis et al., 2013), snail (C. funebralis) (Gleason and Burton, 2015) and P. haitanensis (Wang et al., 2018a). However, few genes related to antioxidant system could be annotated in this study (Figure 9). This might be explained by the stress exposure time. Wang et al. (2018b) found that the expression of antioxidant related genes showed gradual up-regulation with sustained high temperature in P. haitanensis. The significant fold changes were observed at 2- or 6-day time points. The heat stress last a total of 5.5 h in study of snail (C. funebralis) (Gleason and Burton, 2015) and it was more than 4 h in blue mussel (M. galloprovincialis and M. trossulus) (Lockwood et al., 2010). While, it was just 2 h in this study. Therefore, more stress time points were necessary for future studying the response of antioxidant system.
Besides acute responsiveness, the thermally tolerant snail (C. funebralis) individuals appeared to also utilize the “preadaptation” to cope with heat stress (Gleason and Burton, 2015). Some genes, including HSP60, partly of superoxide dismutase (SOD), glutathione peroxidase (GPx), and ubiquitin showed higher constitutive expression in tolerant populations and less up-regulated following heat stress compared with sensitive ones. While, as to these gene or related families, the higher constitutive expression was not found in the tolerant population (YL). Therefore, the preadaptation could not be the abalone’s main strategy for coping with the heat stress. We speculated the better tolerance of YL may mostly rely on its precise coping strategy, which indicated by fine-tuning of transcirptome and stronger ability to regulate key genes.
Our results suggest that the cardiac performance of thermally tolerant lines was less disrupted by high water temperatures stress. In addition, the heat-tolerant line employed a more effective strategy to cope with heat stress than the heat-sensitive line. This strategy included moderate changes in gene transcription and more effective regulation (such as more powerful to up-regulate the HSP family genes). Overall, our results provide insight into the different heat-response strategies employed by heat-tolerant and heat-sensitive abalone lines and these findings might provide some suggestions for further studies of heat-response mechanisms in mollusks. The key genes or pathways would be great indicator for our follow-up work. Combined these with the results with other studies, such as genome-wide association study (GWAS), would provide fundamental information for breeding of better heat-tolerant line.
The methods were carried out in accordance with the approved guidelines by Laboratory Animal Management and Ethics Committee of Xiamen University. All experimental procedures involving abalones were performed according to the Regulations for the Administration of Affairs Concerning Experimental Animals (Xiamen University, China; revised in November 2014).
CK and WY conceived and designed the experiments. NC, CL, and YS performed the experiments. NC and ZH analyzed the data. XL offered reagents and experiment animals. NC, CK, and WY wrote the manuscript. All authors has reviewed the manuscript.
This work was supported by grants from National Natural Science Foundation of China (Grant No. U1605213), Earmarked Fund for Modern Agro-Industry Technology Research System (Grant No. CARS-49), Key S & T Program of Fujian Province (Grant Nos. 2016NZ01010006 and 2016NZ0001-4), Shandong Province (Grant Nos. LJNY201709 and 2016GGH4513), Xiamen Southern Oceanographic Center (Grant No. 18GZY012HJ02), and China Postdoctoral Science Foundation (Grant No. 2017M622082).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01895/full#supplementary-material
Abele, D., Heise, K., Portner, H. O., and Puntarulo, S. (2002). Temperature-dependence of mitochondrial function and production of reactive oxygen species in the intertidal mud clam Mya arenaria. J. Exp. Biol. 205, 1831–1841.
Alter, K., Andrewartha, S. J., Morash, A. J., Clark, T. D., Hellicar, A. D., Leon, R. I., et al. (2017). Hybrid abalone are more robust to multi-stressor environments than pure parental species. Aquaculture 478, 25–34. doi: 10.1016/j.aquaculture.2017.04.035
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anders, S., and Huber, W. (2012). Differential Expression of RNA-Seq Data at the Gene Level–the DESeq Package. Heidelberg: European Molecular Biology Laboratory (EMBL).
Barshis, D. J., Ladner, J. T., Oliver, T. A., Seneca, F. O., Traylor-Knowles, N., and Palumbi, S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U.S.A. 110, 1387–1392. doi: 10.1073/pnas.1210224110
Bita, C. E., Zenoni, S., Vriezen, W. H., Mariani, C., Pezzotti, M., and Gerats, T. (2011). Temperature stress differentially modulates transcription in meiotic anthers of heat-tolerant and heat-sensitive tomato plants. BMC Genomics 12:384. doi: 10.1186/1471-2164-12-384
Casselman, M. T., Anttila, K., and Farrell, A. P. (2012). Using maximum heart rate as a rapid screening tool to determine optimum temperature for aerobic scope in Pacific salmon Oncorhynchus spp. J. Fish Biol. 80, 358–377. doi: 10.1111/j.1095-8649.2011.03182.x
Chapman, R. W., Mancia, A., Beal, M., Veloso, A., Rathburn, C., and Blair, A. (2011). The transcriptome responses of the eastern oyster, Crassostrea virginica, to environmental conditions. Mol. Ecol. 20, 1431–1449. doi: 10.1111/j.1365-294X.2011.05018.x
Chelazzi, G., Williams, G. A., and Gray, D. R. (1999). Field and laboratory measurement of heart rate in a tropical limpet, Cellana grata. J. Mar. Biol. Assoc. U.K. 79, 749–751. doi: 10.1017/S0025315498000915
Chen, N., Luo, X., Gu, Y., Han, G., Dong, Y., You, W., et al. (2016). Assessment of the thermal tolerance of abalone based on cardiac performance in Haliotis discus hannai, H. gigantea and their interspecific hybrid. Aquaculture 465, 258–264. doi: 10.1016/j.aquaculture.2016.09.004
Chen, N., Luo, X., Lu, C., Ke, C., and You, W. (2017). Effects of artificial selection practices on loss of genetic diversity in the Pacific abalone, Haliotis discus hannai. Aquac. Res. 48, 4923–4933. doi: 10.1111/are.13311
Chen, Z., Farrell, A. P., Matala, A., and Narum, S. R. (2017). Mechanisms of thermal adaptation and evolutionary potential of conspecific populations to changing environments. Mol. Ecol. 27, 659–674. doi: 10.1111/mec.14475
Cheng, W., Hsiao, I. S., Hsu, C. H., and Chen, J. C. (2004). Change in water temperature on the immune response of Taiwan abalone Haliotis diversicolor supertexta and its susceptibility to Vibrio parahaemolyticus. Fish Shellfish Immunol. 17, 235–243. doi: 10.1016/j.fsi.2004.03.007
Choi, M., Kim, G., Kim, J., and Lim, H. K. (2015). Differentially-expressed genes associated with faster growth of the Pacific abalone, Haliotis discus hannai. Int. J. Mol. Sci. 16, 27520–27534. doi: 10.3390/ijms161126042
Clark, M. S., Fraser, K. P. P., and Peck, L. S. (2008). Lack of an HSP70 heat shock response in two Antarctic marine invertebrates. Polar Biol. 31, 1059–1065. doi: 10.1007/s12192-008-0014-8
Cui, Y., Du, Y., Lu, M., and Qiang, C. (2011). Antioxidant responses of Chilo suppressalis (Lepidoptera: Pyralidae) larvae exposed to thermal stress. J. Therm. Biol. 36, 292–297. doi: 10.1016/j.jtherbio.2011.04.003
Dang, V. T., Speck, P., and Benkendorff, K. (2012). Influence of elevated temperature on the immune response of abalone, Haliotis rubra. Fish Shellfish Immunol. 32, 732–740. doi: 10.1016/j.fsi.2012.01.022
Deng, Y. W., Liu, X., Wu, F. C., and Zhang, G. F. (2008). Experimental evaluation of heterobeltiosis and heterosis between two populations of Pacific abalone. Acta Oceanol. Sin. 27, 112–119.
Depledge, M. H., and Andersen, B. B. (1990). A computer-aided physiological monitoring-system for continuous, long-term recording of cardiac activity in selected invertebrates. Comp. Biochem. Physiol. Part A Physiol. 96, 473–477. doi: 10.1016/0300-9629(90)90664-E
Dilly, G. F., Young, C. R., Lane, W. S., Pangilinan, J., and Girguis, P. R. (2012). Exploring the limit of metazoan thermal tolerance via comparative proteomics: thermally induced changes in protein abundance by two hydrothermal vent polychaetes. Proc. R. Soc. B 279, 3347–3356. doi: 10.1098/rspb.2012.0098
Dong, Y. W., and Williams, G. A. (2011). Variations in cardiac performance and heat shock protein expression to thermal stress in two differently zoned limpets on a tropical rocky shore. Mar. Biol. 158, 1223–1231. doi: 10.1007/s00227-011-1642-6
Evans, T. G. (2015). Considerations for the use of transcriptomics in identifying the ‘genes that matter’ for environmental adaptation. J. Exp. Biol. 218, 1925–1935. doi: 10.1242/jeb.114306
Farcy, E., Serpentini, A., Fievet, B., and Lebel, J. M. (2007). Identification of cDNAs encoding HSP70 and HSP90 in the abalone Haliotis tuberculata: transcriptional induction in response to thermal stress in hemocyte primary culture. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 146, 540–550. doi: 10.1016/j.cbpb.2006.12.006
Franchini, P., van der Merve, M., and Roodt-Wilding, R. (2011). Transcriptome characterization of the South African abalone Haliotis midae using sequencing-by-synthesis. BMC Res. Notes 4:59. doi: 10.1186/1756-0500-4-59
Gebauer, M., Zeiner, M., and Gehring, U. (1997). Proteins interacting with the molecular chaperone hsp70/hsc70: physical associations and effects on refolding activity. FEBS Lett. 417, 109–113. doi: 10.1016/S0014-5793(97)01267-2
Genevaux, P., Georgopoulos, C., and Kelley, W. L. (2007). The hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions. Mol. Microbiol. 66, 840–857. doi: 10.1111/j.1365-2958.2007.05961.x
Gleason, L. U., and Burton, R. S. (2015). RNA-seq reveals regional differences in transcriptome response to heat stress in the marine snail Chlorostoma funebralis. Mol. Ecol. 24, 610–627. doi: 10.1111/mec.13047
Glickman, M. H., and Ciechanover, A. (2002). The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428. doi: 10.1152/physrev.00027.2001
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, X. M., Ford, S. E., and Zhang, F. S. (1999). Molluscan aquaculture in China. J. Shellfish Res. 18, 19–31.
Guo, Y. F., and Zhao, W. W. (2017). China Fishery Statistical Yearbook, 2017. Beijing: China agriculture press.
Hamburger, K., Dall, P. C., and Lindegaard, C. (1994). Energy metabolism of Chironomus anthracinus (Diptera: Chironomidae) from the profundal zone of Lake Esrom, Denmark, as a function of body size, temperature and oxygen concentration. Hydrobiologia 294, 43–50. doi: 10.1007/BF00017624
Han, G. D., Zhang, S., Marshall, D. J., Ke, C. H., and Dong, Y. W. (2013). Metabolic energy sensors (AMPK and SIRT1), protein carbonylation and cardiac failure as biomarkers of thermal stress in an intertidal limpet: linking energetic allocation with environmental temperature during aerial emersion. J. Exp. Biol. 216, 3273–3282. doi: 10.1242/jeb.084269
Harney, E., Dubief, B., Boudry, P., Basuyaux, O., Schilhabel, M. B., Huchette, S., et al. (2016). De novo assembly and annotation of the European abalone Haliotis tuberculata transcriptome. Mar. Genomics 28, 11–16. doi: 10.1016/j.margen.2016.03.002
Heise, K., Puntarulo, S., Portner, H. O., and Abele, D. (2003). Production of reactive oxygen species by isolated mitochondria of the Antarctic bivalve Laternula elliptica (King and Broderip) under heat stress. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 134, 79–90. doi: 10.1016/S1532-0456(02)00212-0
Hereford, J. (2009). A quantitative survey of local adaptation and fitness trade-offs. Am. Nat. 173, 579–588. doi: 10.1086/597611
Hoegh-Guldberg, O., and Bruno, J. F. (2010). The impact of climate change on the world’s marine ecosystems. Science 328, 1523–1528. doi: 10.1126/science.1189930
Jonas, K., Liu, J., Chien, P., and Laub, M. T. (2013). Proteotoxic stress induces a cell-cycle arrest by stimulating lon to degrade the replication initiator DnaA. Cell 154, 623–636. doi: 10.1016/j.cell.2013.06.034
Kültz, D. (2005). Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 67, 225–257. doi: 10.1146/annurev.physiol.67.040403.103635
Lang, R. P., Bayne, D. J., Camara, M. D., Cunningham, C., Jenny, M. J., and Langdon, C. J. (2009). Transcriptome profiling of selectively bred Pacific oyster Crassostrea gigas families that differ in tolerance of heat shock. Mar. Biotechnol. 11, 650–668. doi: 10.1007/s10126-009-9181-6
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, J., He, Q., Sun, H., and Liu, X. (2012). Acclimation-dependent expression of heat shock protein 70 in Pacific abalone (Haliotis discus hannai Ino) and its acute response to thermal exposure. Chin. J. Oceanol. Limnol. 30, 146–151. doi: 10.1007/s00343-012-1026-x
Li, W., Rong, R., Zhao, S., Zhu, X., Zhang, K., Xiong, X., et al. (2012). Proteomic analysis of metabolic, cytoskeletal and stress response proteins in human heart failure. J. Cell. Mol. Med. 16, 59–71. doi: 10.1111/j.1582-4934.2011.01336.x
Li, Q., Shu, J., Yu, R., and Tian, C. (2007). Genetic variability of cultured populations of the Pacific abalone (Haliotis discus hannai Ino) in China based on microsatellites. Aquac. Res. 38, 981–990. doi: 10.1111/j.1365-2109.2007.01764.x
Liang, S., Luo, X., You, W., Luo, L., and Ke, C. (2014). The role of hybridization in improving the immune response and thermal tolerance of abalone. Fish Shellfish Immunol. 39, 69–77. doi: 10.1016/j.fsi.2014.04.014
Liu, F., Wang, W., Sun, X., Liang, Z., and Wang, F. (2015). Conserved and novel heat stress-responsive microRNAs were identified by deep sequencing in Saccharina japonica (Laminariales, Phaeophyta). Plant Cell Environ. 38, 1357–1367. doi: 10.1111/pce.12484
Lockwood, B. L., Sanders, J. G., and Somero, G. N. (2010). Transcriptomic response to heat stress in invasive and native blue mussels (genus Mytilus): molecular correlates of invasive success. J. Exp. Biol. 213, 3548–3558. doi: 10.1242/jeb.046094
Lyzenga, W. J., and Stone, S. L. (2012). Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 63, 599–616. doi: 10.1093/jxb/err310
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Martin, J. A., and Wang, Z. (2011). Next-generation transcriptome assembly. Nat. Rev. Genet. 12, 671–682. doi: 10.1038/nrg3068
Mayer, M. P. (2013). Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 38, 507–514. doi: 10.1016/j.tibs.2013.08.001
Misselwitz, B., Staeck, O., and Rapoport, T. A. (1998). J proteins catalytically activate hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell 2, 593–603. doi: 10.1016/S1097-2765(00)80158-6
Morash, A., and Alter, K. (2015). Effects of environmental and farm stress on abalone physiology: perspectives for abalone aquaculture in the face of global climate change. Rev. Aquac. 7, 1–27.
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
North, A., Pennanen, J., Ovaskainen, O., and Laine, A. L. (2011). Local adaptation in a changing world: the roles of gene-flow, mutation, and sexual reproduction. Evolution 65, 79–89. doi: 10.1111/j.1558-5646.2010.01107.x
Picone, B., Rhode, C., and Roodt-Wilding, R. (2015). Transcriptome profiles of wild and cultured South African abalone, Haliotis midae. Mar. Genomics 20, 3–6. doi: 10.1016/j.margen.2015.01.002
Portner, H. O. (2010). Oxygen- and capacity-limitation of thermal tolerance: a matrix for integrating climate-related stressor effects in marine ecosystems. J. Exp. Biol. 213, 881–893. doi: 10.1242/jeb.037523
Portner, H. O., Bennett, A. F., Bozinovic, F., Clark, A., Lardies, M. A., Lucassen, M., et al. (2006). Trade-offs in thermal adaptation: the need for a molecular to ecological integration. Physiol. Biochem. Zool. 79, 295–313. doi: 10.1086/499986
Portner, H. O., and Knust, R. (2007). Climate change affects marine fishes through the oxygen limitation of thermal tolerance. Science 315, 95–97. doi: 10.1126/science.1135471
Schoville, S. D., Barreto, F. S., Moy, G. W., Wolff, A., and Burton, R. S. (2012). Investigating the molecular basis of local adaptation to thermal stress: population differences in gene expression across the transcriptome of the copepod Tigriopus californicus. BMC Evol. Biol. 12:170. doi: 10.1186/1471-2148-12-170
Shiel, B. P., Hall, N. E., Cooke, I. R., Robinson, N. A., and Strugnell, J. M. (2015). De novo characterization of the greenlip abalone transcriptome (Haliotis laevigata) with a focus on the heat shock protein 70 (HSP70) family. Mar. Biotechnol. 17, 23–32. doi: 10.1007/s10126-014-9591-y
Shiel, B. P., Hall, N. E., Cooke, I. R., Robinson, N. A., and Strugnell, J. M. (2017). Epipodial tentacle gene expression and predetermined resilience to summer mortality in the commercially important greenlip abalone, Haliotis laevigata. Mar. Biotechnol. 19, 191–205. doi: 10.1007/s10126-017-9742-z
Snyder, M., Girvetz, E., and Mulder, E. (2001). Induction of marine mollusk stress proteins by chemical or physical stress. Arch. Environ. Contam. Toxicol. 41, 22–29. doi: 10.1007/s002440010217
Somero, G. N. (2012). The physiology of global change: linking patterns to mechanisms. Annu. Rev. Mar. Sci. 4, 39–61. doi: 10.1146/annurev-marine-120710-100935
Stenseng, E., Braby, C. E., and Somero, G. N. (2005). Evolutionary and acclimation-induced variation in the thermal limits of heart function in congeneric marine snails (Genus Tegula): implications for vertical zonation. Biol. Bull. 208, 138–144. doi: 10.2307/3593122
Stillman, J., and Somero, G. (1996). Adaptation to temperature stress and aerial exposure in congeneric species of intertidal porcelain crabs (genus Petrolisthes): correlation of physiology, biochemistry and morphology with vertical distribution. J. Exp. Biol. 199, 1845–1855.
Terada, K., Kanazawa, M., Bukau, B., and Mori, M. (1997). The human DnaJ homologue dj2 facilitates mitochondrial protein import and luciferase refolding. J. Cell Biol. 139, 1089–1095. doi: 10.1083/jcb.139.5.1089
Valenzuela-Miranda, D., Del Rio-Portilla, M., and Gallardo-Escarate, C. (2015). Characterization of the growth-related transcriptome in California red abalone (Haliotis rufescens) through RNA-Seq analysis. Mar. Genomics 24, 199–202. doi: 10.1016/j.margen.2015.05.009
Valenzuela-Munoz, V., Araya-Garay, J. M., and Gallardo-Escarate, C. (2013). SNP discovery and high resolution melting analysis from massive transcriptome sequencing in the California red abalone Haliotis rufescens. Mar. Genomics 10, 11–16. doi: 10.1016/j.margen.2012.12.003
van der Merwe, M., Franchini, P., and Roodt-Wilding, R. (2011). Differential growth-related gene expression in abalone (Haliotis midae). Mar. Biotechnol. 13, 1125–1139. doi: 10.1007/s10126-011-9376-5
Vaquer-Sunyer, R., and Duarte, C. M. (2011). Temperature effects on oxygen thresholds for hypoxia in marine benthic organisms. Glob. Change Biol. 17, 1788–1797. doi: 10.1111/j.1365-2486.2010.02343.x
Wang, W., Lin, Y., Teng, F., Ji, D., Xu, Y., Chen, C., et al. (2018a). Comparative transcriptome analysis between heat-tolerant and sensitive Pyropia haitanensis strains in response to high temperature stress. Algal Res. 29, 104–112. doi: 10.1016/j.algal.2017.11.026
Wang, W., Teng, F., Lin, Y., Ji, D., Xu, Y., Chen, C., et al. (2018b). Transcriptomic study to understand thermal adaptation in a high temperature-tolerant strain of Pyropia haitanensis. PLoS One 13:e0195842. doi: 10.1371/journal.pone.0195842
Wit, P., and Palumbi, S. R. (2013). Transcriptome-wide polymorphisms of red abalone (Haliotis rufescens) reveal patterns of gene flow and local adaptation. Mol. Ecol. 22, 2884–2897. doi: 10.1111/mec.12081
Xing, Q., Li, Y., Guo, H., Yu, Q., Huang, X., Wang, S., et al. (2016). Cardiac performance: a thermal tolerance indicator in scallops. Mar. Biol. 163:244. doi: 10.1007/s00227-016-3021-9
Yang, L., Huang, H., and Wang, J. (2010). Antioxidant responses of citrus red mite, Panonychus citri (McGregor) (Acari: Tetranychidae), exposed to thermal stress. J. Insect Physiol. 56, 1871–1876. doi: 10.1016/j.jinsphys.2010.08.006
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Keywords: Pacific abalone, heat stress, cardiac performance, ABT, transcriptome
Citation: Chen N, Huang Z, Lu C, Shen Y, Luo X, Ke C and You W (2019) Different Transcriptomic Responses to Thermal Stress in Heat-Tolerant and Heat-Sensitive Pacific Abalones Indicated by Cardiac Performance. Front. Physiol. 9:1895. doi: 10.3389/fphys.2018.01895
Received: 10 July 2018; Accepted: 17 December 2018;
Published: 09 January 2019.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Li Li, Institute of Oceanology (CAS), ChinaCopyright © 2019 Chen, Huang, Lu, Shen, Luo, Ke and You. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiwei You, d3d5b3VAeG11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.