
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 22 January 2019
Sec. Avian Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.01892
Chicken mesenchymal stem cells (MSCs) can be used as an avian culture model to better understand osteogenic, adipogenic, and myogenic pathways and to identify unique bioactive nutrients and molecules which can promote or inhibit these pathways. MSCs could also be used as a model to study various developmental, physiological, and therapeutic processes in avian and other species. MSCs are multipotent stem cells that are capable of differentiation into bone, muscle, fat, and closely related lineages and express unique and specific cell surface markers. MSCs have been isolated from numerous sources including human, mouse, rabbit, and chicken with potential clinical and agricultural applications. MSCs from chicken compact bones have not been isolated and characterized yet. In this study, MSCs were isolated from compact bones of the femur and tibia of day-old male broiler chicks to investigate the biological characteristics of the isolated cells. Isolated cells took 8–10 days to expand, demonstrated a monolayer growth pattern and were plastic adherent. Putative MSCs were spindle-shaped with elongated ends and showed rapid proliferation. MSCs demonstrated osteoblastic, adipocytic, and myogenic differentiation when induced with specific differentiation media. Cell surface markers for MSCs such as CD90, CD105, CD73, CD44 were detected positive and CD31, CD34, and CD45 cells were detected negative by PCR assay. The results suggest that MSCs isolated from broiler compact bones (cBMSCs) possess similar biological characteristics as MSCs isolated from other chicken tissue sources.
Global production of chicken meat has dramatically increased, and the consumer demand for high quality poultry meat has continuously risen in recent years (Scanes, 2007; Mir et al., 2017). However, genetic selection for rapid growth and high feed efficiency causes skeletal disorders, excess fat accumulation, and muscle degeneration in broilers, which are important health and economic issues for the poultry industry (Velleman et al., 2003; Fleming, 2008; Kim et al., 2011; Fornari et al., 2014). Thus, identifying innovative methods to minimize these issues in broilers is important for the sustainability of poultry production. Studies on chicken mesenchymal stem cells (MSCs) can provide critical insight for skeletal development, muscle growth, and fat accumulation in poultry. MSCs are multipotent, plastic adherent cells that have been isolated from several species, including human, mouse, rat, dog, cat, sheep, and chickens (Crigler et al., 2007; Dumas et al., 2008; Kar et al., 2016; Kumar et al., 2016; Li et al., 2016). The ability of MSCs to differentiate into lineage specific tissue types (Pittenger et al., 1999; Freeman et al., 2015), their homing potential (Sohni and Verfaillie, 2013), and tissue repair potential (Melo et al., 2016) has generated increasing interest in utilizing MSCs to study mechanisms in basic biology, as well as clinical therapeutics. Chicken MSCs have significant agricultural applications as they can be utilized to better understand the mechanisms that drive osteogenic, myogenic, and adipogenic differentiation and potentially be used to identify novel compounds that drive these processes to the more desirable muscle and bone phenotypes. MSCs derived from bone marrow were first described as fibroblast-like colony forming cells, which maintained differentiation potential, and proliferation (Friedenstein et al., 1987). The Mesenchymal and Tissue Stem Cell Committee of the International Society for Cellular Therapy (ISCT) has defined these minimal criteria for MSCs: (a) the capacity to adhere to a plastic surface under standard culture conditions, (b) have a multilineage differentiation property, and (c) display the presents and absence of surface antigens (Dominici et al., 2006).
One of the major challenges of isolating chicken MSCs utilizing bone marrow from non-compact bone sources is the requirement of multiple purification steps to remove non-MSC cell types (e.g., hematopoietic stem cells, blood cells) (Khatri et al., 2009). These purification approaches often lead to cytotoxicity, changes in functionality of MSCs, and low MSC yields (Wee et al., 2013). However, there are no reports of MSCs isolated from compact bones of the chickens, which may overcome these limitations as there are fewer contaminating cell types (Pittenger et al., 1999). There are two main stem cell populations that are the main residents of bone marrow: hematopoietic stem cells and MSCs (Pittenger et al., 1999; Zhu et al., 2010). One significant drawback of isolating MSCs from bone marrow is the contamination of blood cells and hematopoietic stem cells. Various techniques have been used to purify or enrich the MSC population isolated from bone marrow including preferential attachment to culture plastic, density gradient centrifugation (Yamamoto et al., 2015), use of ficole to filter out blood cells, antibody-based cell sorting (Van Vlasselaer et al., 1994; El-Sayed et al., 2015), low and high density culture techniques (Eslaminejad and Nadri, 2009), and frequent media change (Soleimani and Nadri, 2009). However, these methods used to purify MSCs isolated from bone marrow have a number of negative effects. Use of preferential attachment to cell culture plates could yield phenotypically and functionally heterogenous cell population (Tremain et al., 2001). Use of immune depletion techniques downregulated many genes involved in cell proliferation and cell cycle progression (Baddoo et al., 2003). Exposure of MSCs isolated from immunodepleted cells to insulin-like growth factor or leukemia inhibitory factor reversibly inhibited the ability of cells to differentiate into adipocyte, chondrocyte and osteoblasts in vitro (Baddoo et al., 2003). Use of low density culture yielded only about 27 fibrobalstoid colonies of 5 or more cells from a total of 200 culture disc (Wang and Wolf, 1990). Cell sorting approaches to isolate multi-lineage MSCs from hematopoietic cells resulted in reduced clonogenicity and limited osteogenic potentials in isolated MSCs (Van Vlasselaer et al., 1994). Isolation of MSCs from compact bones could be an easy and economic isolation technique which can avoid the use of other purification techniques during isolation and reduce the chances of hematopoietic cells contamination in the isolated cultures (Guo et al., 2006; Zhu et al., 2010).
In this study, we present for the first time, an effective, simple, and economical method for isolation and characterization of MSCs from compact bones (cBMSCs) of day old chickens. cBMSC are a robust and highly proliferative cell population that meet the ISCT MSC criteria. These cells open the door for future in vitro studies of critically important osteogenic, adipogenic, and myogenic pathways in avian species and for the identification of novel bioactive nutrients and molecules which promote skeletal health, muscular growth, and efficient feed utilization in poultry.
All experiments were performed in accordance with the guidelines for the use of animal in research as stated by the Institutional Animal Care and Use Committee at the University of Georgia. The protocol was approved by the Institutional Animal Care and Use Committee at the University of Georgia.
cBMSCs were isolated by using a modified approach of the previously described methods in human trabecular and murine compact bones (Tuli et al., 2003; Zhu et al., 2010). Femurs and tibia bones from both legs were obtained from the day-old chicks after cervical dislocation. The birds were soaked in alcohol for 2 min after cervical dislocation. Legs were removed from hip joint and metacarpal (Figures 1A–C). Dissected legs were kept in Dulbecco's Modified Eagle's medium (DMEM) (Mediatech Inc.,VA, USA) containing 10% Fetal Bovine Serum (FBS) (Mediatech Inc.,VA, USA), 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.292 mg/mL L-glutamine (Thermo Fisher Scientific, MA, USA) until connective tissues and muscles were completely removed. Muscles and connective tissues around tibia and femurs were removed immediately using a scalpel and micro-dissecting scissors in a bio-safety cabinet (Figure 1C). The cleaned tibia and femurs were placed in washing buffer containing Phosphate-Buffer Saline (PBS) (Mediatech Inc., VA, USA) and 2% FBS. The epiphysis of the bones were removed to expose the bone marrow cavity. Bone marrow inside the bone was flushed four times with washing buffer in a syringe to remove the bone marrow and hematopoietic cells adhered to the compact bones (Figure 1D). The bones were cracked with a scalpel and washed three more times with washing buffer to make sure that all the bone marrow cells were washed. The bones appeared whitish in color after the wash (Figure 1D). The bones were transferred to new cell culture dishes with 5 ml of digestion media (DMEM containing 100 IU/ml penicillin and 100 ug/ml streptomycin, 0.25% collagenase (Sigma-Aldrich, MO, USA), and 20% FBS). The bones were chopped to smaller fragments of about 3 mm3 (Figures 1E,F). Bone fragments were suspended in a 50-ml tube that contained digestion media. The bone fragments were digested in a shaking water bath for 60 min at 37°C at 180 rpm. The digestion media containing bone fragments were filtered with 40 μm sterile filter. Bone fragments in the filter were rinsed with 5 ml of 10% DMEM. Filtered contents were centrifuged at 1,200 rpm for 10 min. The supernatant was discarded and the cell pellet was disrupted with 20 ml 10% DMEM, and cells were plated in two 100-mm cell culture dishes. Cultures were incubated at 37°C in a humidified incubator containing 5% CO2. Half of the media was replaced by a fresh media at 12 h, complete media was changed at 24 h to remove the non-adherent cells. After that, media was changed once every 2–3 days. These cells were labeled as P0. Once the cells reached 95% confluently, the cells were washed twice with 5 ml 1X PBS, dissociated with 0.1% Trypsin-EDTA (Mediatech Inc., USA) for 2 min and subcultured at a ratio of 25,000 cells/ cm2 in 100-mm cell culture dishes. This passage was marked as P1, subsequent cultures were named as P2, P3, P4… Pn consecutively. P4 cells were used for cBMSCs differentiation experiments.
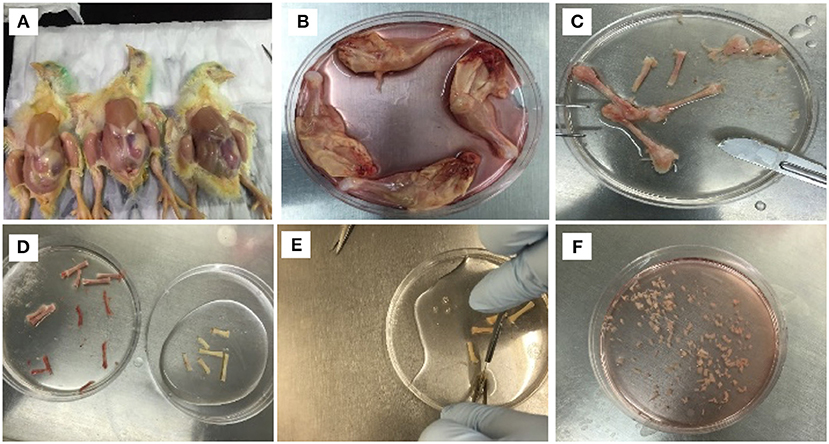
Figure 1. Isolation procedure of MSCs from compact bones of chick (cBMSCs). (A) Chicks were soaked in alcohol for leg dissections (B) and legs dissected and soaked in DMEM with 10% FBS. (C) Muscles were separated to obtain femur and tibia. (D) Epiphysis was dissected, and bone marrow was flushed with PBS containing 2% FBS. (E) After washing, the bones appeared white in color. (F) Bones were chopped in 1–3 mm3 and digested in digestion media.
cBMSCs were observed daily to understand their morphology and growth characteristics. Cells were observed under a microscope daily, and pictures of cells were taken at different time points. Cells were observed for morphological features and capacity to adhere to plastic plates.
Self-renewal capacity and colony forming potential in isolated MSCs were evaluated by plating cells at low densities in 100-mm cell culture dishes. Colonies formed from single cells were counted at P4 and P8. At both passage, 25 cells/cm2, 50 cells/cm2, and 75 cells/cm2 were plated in 100-mm cell culture dishes and incubated for 10 days. On day 10, cells were fixed and stained with 1% crystal violet (Sigma-Aldrich, MO, USA) in 100% methanol for 30 min. Colonies were defined as more than 50 fibroblastic cells in a cluster. The number of colonies were scored, and microscopic pictures were obtained.
Cells at P2, P4, and P8 were dissociated by 0.25% trypsin and plated in 24-well plates with a density of 1 × 104 cells /ml. The number of cells were counted by a viability detection method using a trypan blue exclusion test on 4 wells daily for 7 days. Each well was counted three times, and mean value was calculated. Growth curves were plotted for each passage. Population doubling time (PDT) was calculated by formula reported by Aliborzi et al. (2016). PDT was calculated using the formula PDT = T ln2/ln (Xei/Xbi), where T means incubation time in hours, Xbi is the number of cells at the beginning of the incubation and Xei corresponds to the number of cells at the end of incubation.
cBMSCs at P4 were seeded at a density of 20,000 cells /cm2 in 24-well plates for Alizarin Red (Gregory et al., 2004), Alkaline Phosphate (ALP) (Parhami et al., 1997), and Von Kossa stain (VK) (Parhami et al., 1997), and in 6-well plates for measurement of osteogenic gene regulation. Cells were cultured in basal media containing DMEM, 10% FBS, 100 IU/ml penicillin, and 100 μg/ml streptomycin until 90% confluent. On confluency, cells were treated with osteogenic media (OM) containing DMEM with 10−7M dexamethasone (DXA) (Sigma-Aldrich, MO, USA), 10 mM β-glycerophosphate (Sigma-Aldrich, MO, USA), 50 μg/ml ascorbate (Sigma-Aldrich, MO, USA), and 5% FBS for osteogenic induction. Cells cultured in DMEM basal media with 10% FBS were used as negative control. Fresh media was replaced with the old media in the culture plate in every 2–3 days. Cells were stained with Alizarin Red and VK for detection of mineralization and ALP for detection of osteogenic differentiation on 7 and 14 days of treatment. Cells plated in 6-well plates were harvested at 72 h for osteogenic gene expression analysis using Quantative Reverse Transcription Polymerase Chain Reaction (qRT-PCR). ALP, Runt-related transcription factor 2 (RUNX2), Bone Morphogenetic Protein (BMP2), Bone Sialoprotein (BSP), and Bone Gamma-Carboxyglutamate Protein (BGLAP) were analyzed to detect osteogenic differentiation of cBMSCs.
cBMSCs at P4 were placed at a density of 20,000 cells/cm2 in 24-well plates and 6-well plates. At confluency, cells were treated with adipogenic cocktail (DMI containing 500 nM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine and 20 mg/mL insulin) (Cedarlane, NC, USA) + 300 μM Oleic acid. The induced cells were observed under an inverted microscope for fat vacuole deposition. After 96 h of treatment with adipogenic media, cells cultured in 24-well plates were harvested and stained with Oil Red O using a procedure described by Kim et al. (2009). Cells treated in 6-well plates were harvested at 48 h for adipogenic gene expression using qRT-PCR. Key adipogenic gene, PPARγ, FABP2, c/EBPα, and c/EBPβ were measured for adipogenic differentiation of cBMSCs.
cBMSCs were cultured in 6-well plates at a density of 20,000 cells/cm2 and treated with myogenic medium (MM) containing DMEM, 5% horse serum (Thermo Fisher Scientific, MA, USA), 50 μM hydrocortisone (Sigma-Aldrich, MO, USA), and 0.1 μM dexamethasone when confluent. Cells cultured in 6-well plates were harvested after 72 h for key myogenic gene expressions such as MyoD, Pax7, Myf5, and Myogenin using qRT-PCR (Quantative Reverse Transcription Polymerase Chain Reaction to detect multilineage gene expression).
Total RNA was extracted from harvested cell with QIAzol Lysis reagent (Qiagen, MD, USA) following the manufacturer's protocol. Two micrograms of RNA was reverse-transcribed to make cDNA using High-Capacity cDNA Reverse Transcription Kits (Thermo Fisher Scientific, MA, USA). Template, multiscript reverse transcriptase enzyme, random primers, dNTPs, and buffers were subjected to thermocycling at 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min in a veriti 96-well thermal cycler (Thermo Fisher Scientific, MA USA). Specific primers were used to quantify the osteogenic, myogenic, and adipogenic mRNA expression in harvested cells by qRT-PCR. iTaqTM Universal SYBR Green Supermix (Bio-Rad, CA, USA), primers and cDNA templates were subjected to qRT-PCR at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, annealing temperature was variable according to primers for 20 s, and 72°C for 15 s, followed by 95°C for 15 s and a melt curve stage in a StepOneTM Real-Time PCR machine (Thermo Fisher Scientific, MA, USA). All samples were prepared in duplicated wells in 96-well plates. Relative abundance of mRNA between the samples were analyzed by the ΔΔCT method. GAPDH was used as a housekeeping gene. The values were reported as fold changes of the mRNA expression of the target genes in differentiation groups compared to the control group. Primers for each gene were designed and checked for target identity using the National Center for Biotechnology Information (NCBI). Primers used for qRT-PCR assays and their sequences are presented in Table 1.
Cells were plated in 6-well plates and harvested when confluent at P2, 4, 6, and 8 to analyze cell surface markers. RNA was isolated from the harvested cells, and cDNA was transcribed following the method described above. Synthesized cDNA was subjected to PCR amplification using DreamTaq Green PCR Master Mix (Thermo Fisher Scientific, MA, USA) following manufacturer's protocol. Samples were subjected to thermocycling at 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, the annealing temperature of primers for 30 s, and 72°C for 30 s, followed by 72°C for 7 min. PCR products obtained were separated by 1.5% agarose gel electrophoresis to visualize the bands detected using SYBR Safe DAN gel stain (Thermo fisher Scientific, MA, USA). mRNA expression of cell surface markers, CD44, CD29, CD45, CD90, CD105, and CD34, was used to characterize cBMSCs population at P2, P4, and P8. GAPDH was used as an internal control. Primer sequences for cells surface markers such as CD29, CD34, CD44, CD71 were used to characterize cBMSCs as reported (Khatri et al., 2009; Bai et al., 2013). Primers for each gene were designed and checked for target identity using the National Center for Biotechnology Information (NCBI). Primers used for PCR assays and their sequences are presented in Table 1.
Cells were plated onto glass four-chamber slides (BD Bioscience, San Jose, CA, http://www.bdbiosciences.com) and fixed with 4% paraformaldehyde for 15 min. Antibodies were directed against CD44 (1:100; Bio Rad) and CD45 (1:100; SouthernBioTech). Primary antibodies were diluted in blocking solution and incubated for 1 h at room temperature. Primary antibodies were detected using secondary antibodies conjugated to Alexa Fluor 488 (1:1,000; Life Technologies) and incubated for 1 h at room temperature. Cells were then washed before mounting with Prolong Gold with DAPI (Life Technologies). Cell observations were made using the Olympus Ix81 (Olympus, Tokyo, http://www.olympus-global.com) with Disc-Spinning Unit and Slide Book Software (Intelligent Imaging Innovations, Santa Monica, CA, http://www.intelligent-imaging.com). Quantification was performed by imaging 5 random fields for each antibody in triplicate and utilizing the cell counter plugin on ImageJ 1.0 software (Schneider et al., 2012).
Statistical analysis was performed using the general linear model procedure of the Statistical Analysis System (SAS) software (SAS institute, 2011). Mean separation test was conducted using Tukey test and P ≤ 0.05 was considered as statistically significant among the groups.
cBMSCs were isolated from 1-day-old broiler compact bones of the tibia and femur (Figure 1). cBMSCs at P0 were round for an initial 2 days and appeared to be spindle-shaped after 3 days of culture (Figure 2A). Cells formed distinct colonies and were passaged 10–12 days after isolation (Figure 2B). Cells were passaged at a ratio of 20,000 cells/cm2 for subsequent passage once reaching confluence (Figure 2C). Cells adhered to the plates, were spindle shaped, divided rapidly, and reached confluency within 2–3 days consistent with an MSC phenotype and growth pattern. This rapid proliferation and spindle shaped morphology were consistently observed in subsequent passage 4, 6, and 8 (Figures 2D–F). Cells were passaged up to P12 to observe morphological characteristics. Cells at P4 were used for further analyses of proliferation potential, immunocytochemistry, qRT-PCR, and multilineage differentiation capacity.
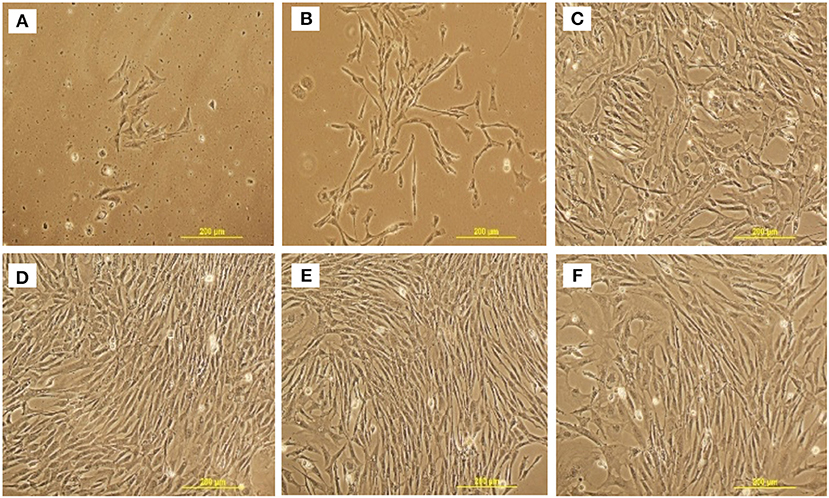
Figure 2. Representative cell morphology at different stages and passage of chicken MSCs isolated from compact bones. (A) passage 0; day 4, (B) passage 0; day 10, (C) passage 2, (D) passage 4, (E) passage 6, and (F) passage 8.
As cBMSCs have the ability to grow and self-renew, a CFU assay was used to assess the proliferation ability and colony forming potential of isolated cBMSCs. cBMSCs formed distinct colonies of cells when seeded at lower densities, indicating that they have the potential to self-renew (Figure 3). Cells plated at densities of 25 cells/cm2, 50 cells/cm2, and 75 cells/cm2 formed 90, 156, and 209 colonies at P4 (Figure 3A) and 75, 106, and 144 colonies at P8, respectively (Figure 3B).
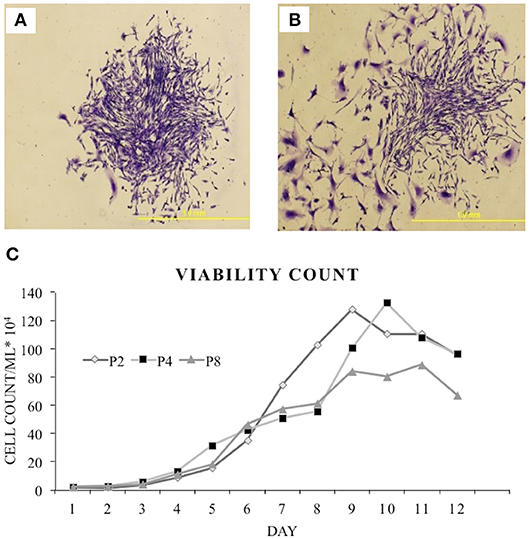
Figure 3. Colony forming assay. Formation of colony from chicken compact bone derived MSCs stained with 1% crystal violet in methanol: (A) Passage 4 and (B) passage 8. (C) Growth curve of the isolated chicken BMSCs. Growth curve of P2, P4, and P8 were all sigmodal in shape that showed latent phase, log phase, and plateau phase.
A growth kinetics assay with Trypan blue exclusion was performed on cBMSCs by quantifying the number of cells over 12 day period at P2, 4, and 8. Growth and proliferation potential of cBMSCs were similar at P2, 4, and 8. Cells initially had a latent phase of 1–3 days, a logarithmic growth phase for 4–7 days, and reached a plateau phase in about 9–11 days in P2 and 4 and in about 8–9 days in passage 8 (Figure 3C). Thus, this result showed that the cell viability of P8 was overall lower than one of P2 or P4, whereas P2 and P4 showed similar cell viability. PDT of the passage 2, 4, and 8 were 66, 76, and 74 h, respectively. Comparing the growth curve and PDT, cBMSCs showed robust and comparable proliferation rates across passages.
cBMSCs were evaluated at P2, P4, and P8 for MSC marker gene expression. cBMSCs at all 3 passages revealed positive results for mRNA transcripts of CD90, CD105, CD73, CD44, and CD29 (Figure 4). CD45 and CD34 mRNA were not expressed in cBMSCs at all 3 passages. In addition, immunofluorescence revealed that chicken cBMSCs expressed CD44 (98.5% ± 1.5) but not CD45 (0%) (Figure 5) further indicating that these cells are cBMSCs (Khatri et al., 2009).
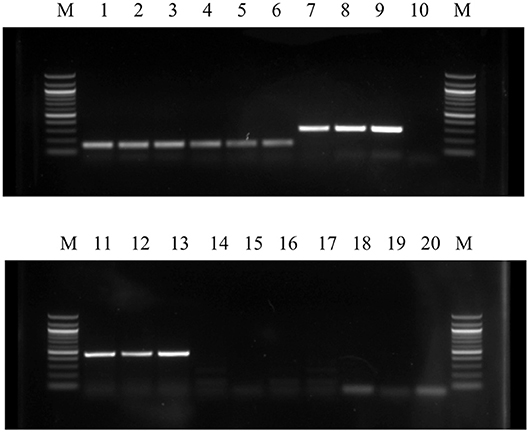
Figure 4. Qualitative expression of cell surface markers of cBMSCs were detected by RT-PCR in all three passages (P2, P4, and P8). High level of expression of cell surface markers CD90, CD105, CD44, CD29, and GAPDH as well as lack of expression of CD 45 and CD34 mRNA in MSCs indicates that the cells were homogenous population or mesenchymal stem cells. M- Marker; Lane 1- P2 GAPDH; Lane 2- P4 GAPDH; Lane 3- P8 GAPDH; Lane 4- P2 CD44; Lane 5- P4 CD44; Lane 6- P8 CD44; Lane 7- P2 CD29; Lane 8- P4 CD29; Lane 9- P8 CD29; Lane 10- Control; Lane 11- P2 CD90; Lane 12- P4 CD90; Lane 13- P8 CD90; Lane 14- P2 CD34; Lane 15- P4 CD34; Lane 16- P8 CD34; Lane 17- P2 CD45; Lane 18- P4 CD45; Lane 19- P8 CD45; and Lane 20- Control.
cBMSCs treated with specific differentiation media in 6-well plates were harvested for osteogenic, adipogenic, and myogenic gene expression using qRT-PCR. cBMSCs treated with OM expressed higher levels of Runx2, BMP, BSP, and BGLAP mRNA relative to non-treated control cells, indicating osteogenic differentiation of cBMSCs (Figure 6A). cBMSCs treated with adipogenic media expressed a higher level of FABP4 and PPARγ mRNA in comparison to non-treated control cell at 48 h post-treatment. However, c/EBPα, and c/EBPβ were not significantly different between treatments at 48 h post- treatment (Figure 6B). Cells subjected to MM expressed higher levels of Myogenin and MyoD mRNA expression which are early differentiation markers of myogenesis. However, Myf5, and Pax7 were not significantly different between the treated and control cells (Figure 6C). These results demonstrate that cBMSCs are capable of osteogenic, adipogenic, and myogenic differentiation.
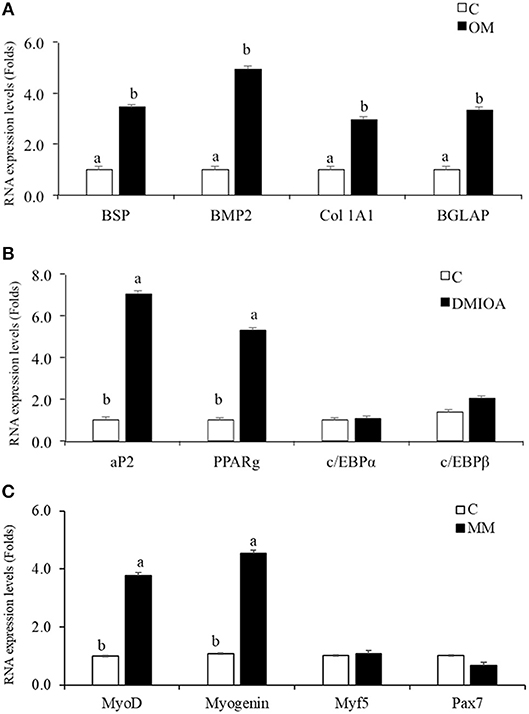
Figure 6. Comparative analysis of lineage differentiation specific gene expression in cBMSc. Relative gene expression of osteogenic, adipogenic, and myogenic markers were analyzed between induced cells and control cells (C) using qRT-PCR. (A) BSP, Collagen Type 1 Alpha 2 (Col 1A2), BMP2, and BGLAP mRNA expression were analyzed in cells treated with osteogenic media (OM) (containing DMEM with 10−7M dexamethasone (DXA), 10 mM β-glycerophosphate, 50 μg/ml ascorbate, and 5% FBS) and control media (C) for 72 h; (B) FABP2, PPARγ, c/EBPα, and c/EBPβ expression were analyzed in cells treated with adipogenic media (AM) (containing 500 nM dexamethasone, 0.5 μM 3-isobutyl-1-methylxanthine, and 20 mg/mL insulin and 300 μM OA) and control media (C) for 48 h (C) MyoD, Myogenin, Myf5, and Pax7 mRNA expression were analyzed in cells treated with myogenic media (MM) (containing DMEM, 5% horse serum, 50 μM hydrocortisone, and 0.1 μM dexamethasone) and control media (C) for 72 h. GAPDH was used as a housekeeping gene. Different letters (a,b) indicate significant difference at P < 0.05.
To further assess the multipotent differentiation capacity of cBMSCs, cells were differentiated under osteogenic, and adipogenic conditions. To first assess osteogenic differentiation potential, MSCs were treated with OM for 2 wk. At the end of wk1 and wk2, calcification deposits of cells treated with OM were detected with cells staining positive for Alizarin Red and Von Kossa, both of which were negative in control cells not treated with OM differentiation media. Differentiated cells stained with Alizarin red were bright orange-red in color (Figure 7B), whereas undifferentiated cells were not (Figure 7A). Differentiated cells stained with Von Kossa stain showed brown to black mineralized deposits, indicating increased mineralization of differentiated cells (Figures 7C,D). Cells treated with OM showed positive alkaline phosphatase activity compared to untreated cBMSCs. Undifferentiated cBMSCs were colorless or faint blue (Figure 7E), whereas differentiated cBMSCs stained dark blue-violet (Figure 7F). Cells treated with adipogenic media displayed an adipocyte phenotype with the appearance of cytoplasmic lipid vacuoles detected by oil red O staining (Figure 8B). The control cells did not show any adipocyte formation (Figure 8A).
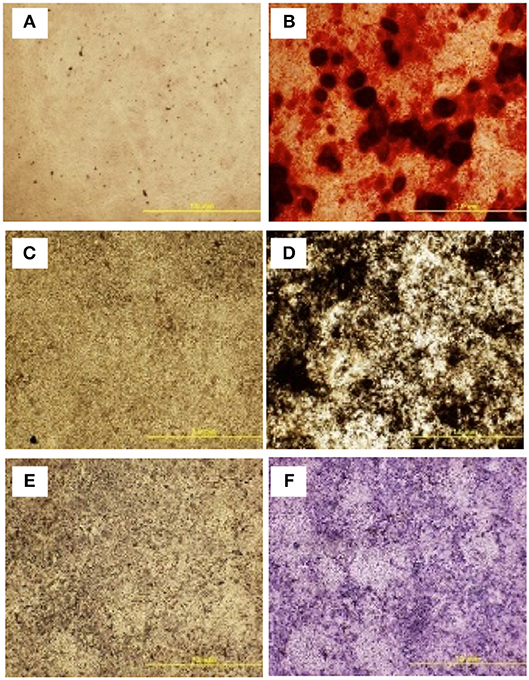
Figure 7. Multilineage differentiation potential of cBMSCs. On confluency, cBMSCs were treated with osteogenic media (OM) containing DMEM with 10−7M dexamethasone (DXA) (Sigma Aldrich, MO, USA), 10 mM β-glycerophosphate (Sigma Aldrich, MO, USA), 50 μg/ml ascorbate (Sigma Aldrich, MO, USA), and 5% FBS for osteogenic induction. Cells cultured in DMEM basal media with 10% FBS were used as negative control. Both cells were stained with Alizarin red stain and Von Kossa stain at day 14. Alizarin Red stain in (A) control cells (B) OM treated cells. Von Kossa stain in (C) control cells and (D) OM treated cells. Alkaline Phosphate assay in (E) control cells and (F) OM treated cells.
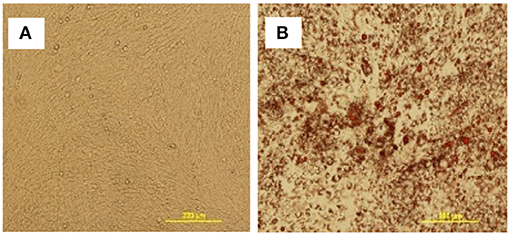
Figure 8. Adipogenic differentiation of cBMSCs. cBMSCs were treated with adipogenic cocktail (DMI containing 500 nM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine, and 20 mg/mL insulin) and 300 μM of OA (DMIOA) when cells were confluent. Cells cultured in 10% DMEM were used as negative control. Differentiation of adipocyte was detected by Oil Red O stain in (A) control cells vs. (B) induced cells at 96 h.
In this study, we have for the first time characterized MSCs from compact bones of broiler chickens. MSCs isolated from compact bones displayed spindle-shaped cells that proliferate rapidly and arrange in a whirlpool pattern similar to MSCs isolated from murine compact bones (Zhu et al., 2010; Short and Wagey, 2013). Cells were able to self-renewal forming colonies from single cells and display osteogenic, adipogenic, and myogenic differentiation (Dominici et al., 2006). This study also demonstrated that cBMSCs possessed the critical 3 MSC defining characteristics of rapid proliferation, adherence to plastic, and multilineage differentiation as described by ICST (Dominici et al., 2006).
The growth curve of the cBMSCs isolated from compact bones in this study is similar to the growth curve of the MSCs isolated from non-compact bone marrow of chicken (Khatri et al., 2009; Bai et al., 2013). In the present study, cells had a latency phase of 1–3 days with a logarithmic phase and a plateau phase in about 7–8 days. PDT of the cBMSCs were 66, 76, and 74 h for P2, P4, and P6, respectively, which was different from the PDT of MSCs isolated from mouse which was 20 h for primary culture and 80 h at passage 3 (Nadri and Soleimani, 2007). Also, in guinea pig, PDT of cells isolated from bone marrow were 62.9, 65.6, and 91.4 h at P 2, 5, and 8, respectively (Aliborzi et al., 2016). These reports indicate that the growth potential of the MSCs could vary depending on the passage rate as well as tissue source of MSCs isolation and species of origin.
cBMSC demonstrated multilineage differentiation potential when cultured in osteogenic, adipogenic, and myogenic differentiation conditions. cBMSCs treated with OM promoted osteogenic differentiation of MSCs by increasing osteogenic gene transcripts. Similarly, in previous reports, treatment of MSCs with OM resulted in osteogenic differentiation of MSCs isolated from bone marrow in poultry (Bhuvanalakshmi et al., 2014) and in humans (Noth et al., 2002; Klepsch et al., 2013). Osteogenic differentiation of cells was characterized by staining the treated cells with Alizarin Red, Von Kossa, and ALP. These cytochemistry tests are routinely used to characterize the osteogenic differentiation of MSCs (Donato et al., 2016; Shi et al., 2016). ALP, Alizarin Red and Von Kossa stain both indicate mineralization of MSCs through osteogenic differentiation (Parhami et al., 1997; Gregory et al., 2004). Differentiated osteoblasts expressed a high ALP activity compared to undifferentiated MSCs which expressed very weak activity. Undifferentiated MSCs do not have extracellular calcium deposits, whereas differentiated osteoblasts do. Calcium deposit is an excellent way to detect if there is osteogenic differentiation of MSCs when treated with OM (Bellows et al., 1986; Wang et al., 2006). Silver ions in Von Kossa stain reacts with anions (phosphates, sulfates, or carbonates) of calcium salts in the cells and the reduction of silver salts forms a brown to black stain. However, Von Kossa stain alone cannot unequivocally confirm osteogenic differentiation of cells and calcium deposits as AgNo3 could be displaced by SO4 ions in any metal (Bonewald et al., 2003). Another test done to confirm calcium deposits along with Von Kossa stain is the Alizarin Red test. Alizarin Red reacts with calcium cations to form an orange-red chelate thus confirming the deposits of Ca in cells (Wang et al., 2006). Further osteogenic differentiation was confirmed in cBMSCs by analyzing key osteogenic genes using qRT-PCR. BGLAP, BSP, BMP2, and ColA1 mRNA was highly expressed in cells treated with OM which indicates osteogenic differentiation capacity of MSCs isolated in this study. Treatment with OM induced similar osteogenic gene expression and expressed positive ALP, Alizarin Red, and Von Kossa stain in MSCs derived from bone marrow of chicken (Bai et al., 2013), mouse (Nadri and Soleimani, 2007), bovine (de Moraes et al., 2016), and human (Kulterer et al., 2007; Honda et al., 2013). In this study, differentiated cells transformed from elongated to shorter cuboidal cells and formed mineralized nodules, which was also reported in MSCs derived from bone marrow of chickens (Khatri et al., 2009).
Adipogenic differentiation of cBMSCs was induced by treating cells with adipogenic cocktail DMI and OA in this study. The characteristics and molecular mechanism of adipocyte differentiation in murine preadipocyte cell line 3T3-L1 has been extensively studied (Ntambi and Young-Cheul, 2000). It has been reported that preadipocytes after differentiation from MSCs stay in growth arrest stage, which reenters into the cell cycle and undergoes mitotic division in response to an adipogenic cocktail, which then terminally differentiates into adipocytes (Tang et al., 2003). However, in human adipose precursor cells derived from adipose tissue and bone marrow, MSCs do not undergo cell division during differentiation (Entenmann and Hauner, 1996; Lehmann et al., 1997). The exact mechanism of adipogenic differentiation by addition of DMI and AO in chicken MSCs is not known. In our study, addition of DMI and OA induced adipogenic differentiation of cBMSCs which was detected by accumulation of lipid vacuole within the cells and an increase in the master regulator of adipogenesis, PPARγ. This is in agreement with human (Tontonoz et al., 1994; Neubauer et al., 2004; Scott et al., 2011) and mouse (Scott et al., 2011) adipogenic differentiation studies. Similarly, DMI and OA increased PPARγ, and FABP4 mRNA expression during an early stage of adipogenesis in pre-adipocyte cells isolated from abdominal adipose tissue of 10-day old broiler chick (Matsubara et al., 2005). However, no increase in c/EBPα and c/EBPβ mRNA expression at 48 h in cMSCs has been observed in the current study. This result is not in agreement with studies with mouse 3T3-L1 cells and hen preadipocytes (Ntambi and Young-Cheul, 2000; Tang et al., 2003; Regassa and Kim, 2013). Since c/EBPβ is an early adipogenic transcription factor, this gene would have been expressed before 48 h. Typically, c/EBPα and PPARγ expression increases during adipogenesis because of the positive feedback loop between c/EBPα and PPARγ (Wu et al., 1999). There are some differences in adipogenic mechanisms among species and cell sources. Hen preadipocytes showed significant c/EBPα and c/EBPβ mRNA expression at 48 h during adipogenic differentiation (Regassa and Kim, 2013). Further studies are necessary to understand uniqueness of cMSCs for adipogenic mechanisms. Myogenic regulatory factors orchestrate the differentiation of MSCs into muscles cells. MyoD and Myogenin are believed to be early differentiation markers of myogenic differentiation, and Myf5 and MRF4 are believed to regulate terminal differentiation and cell fusion (Perez-Serrano et al., 2017). Similar to muscle development, MyoD and Myogenin are main muscle-specific transcription factors that are expressed during myogenic differentiation of MSCs (Gang et al., 2004, 2008). In this study, cBMSCs expressed higher MyoD and Myogenin mRNA levels when subjected to myogenic media. This indicates that cBMSCs are capable of differentiating into osteogenic, myogenic, and adipogenic lineages when subjected to appropriate differentiation conditions.
Because availability of stem cell specific markers is limited in poultry, researchers have to rely on reports of cell surface markers in mammalian species. Use of markers to verify MSC identity is an important quality control step to reduce experimental variability and obtain a homogenous population of MSCs. Our study detected the presence of mesenchymal cell surface markers CD90, CD105, CD73, CD44, and CD29 and lack of hematopoietic cell surface markers CD45 and CD34 mRNA expression at all passage studies. Furthermore, immunocytochemistry confirmed that chicken MSCs were positive for the MSC marker CD44 and negative for the hematopoietic stem cell marker CD45. This demonstrates that the characteristic immunophenotype of cBMSCs is consistent with previously reported chicken MSCs (Khatri et al., 2009; Bai et al., 2013; Intarapat and Stern, 2013).
Despite significant progress in our understanding of MSC biology based on human and murine cells, much of the information pertaining to avian MSCs remains poorly defined including identity and functionality. In the present study, we showed that MSCs can be isolated from the compact bone of day-old broilers, cultured, and characterized. The adherence, morphology, differentiation potential, and specific markers are comparable with the MSCs derived from other source and animals, making them a suitable model for various research applications. Establishment of primary MSC culture can have a huge potential impact on our understanding of chicken development and nutrition with increased ease of availability and a better defined and characterized isolation and culture conditions. Chicken compact bone derived MSCs could be used as an ideal stem cell source for various biological research because of its easy purification, amplification, multipotency, and maintenance. cBMSCs can be a cell culture model in avian species to understand osteogenic, adipogenic, and myogenic differentiation mechanism exerted by different bioactive/nutrient compounds, which can promote skeletal health, muscular growth, fat development, and efficient feed utilization in poultry.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. WK and FW conceived and designed this study. RA contributed to isolation of cBMSCs, cell differentiation, qRT-PCR, PCR for cell surface markers, and data analyses. CC and EW contribute to cell isolation, immunocytochemistry, and data analyses. The paper was written through contribution and critical review of the manuscript by all authors (RA, CC, EW, FW and WK).
USDA-NIFA (#2017-67015-26586).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ALP, Alkaline Phosphatase Assay; BMP, Bone Morphogenic Protein; BSP, Bone Sialoprotein; BGLAP, Bone Carboxyglutamate Protein; MSCs, Mesenchymal Stem Cells; cBMSCs, Chicken Compact Bone Derived Mesenchymal Stem Cells; OA, Oleic Acid; DMI-Dexamethasone, 3, isobutyl-1-methylxanthine, and Insulin; FBS, Fetal Bovine Serum; DMIOA, Dexamethasone, 3-isobutyl-1-methylxanthine, Insulin, and Oleic Acid; qRT-PCR, Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction; FABP4, Fatty Acid Binding Protein 4; PPARγ2, Peroxisome Proliferator-Activated Receptor gamma; DMEM, Dulbecco's Modified Eagle's Medium; SAS, Statistical Analysis System.
Aliborzi, G., Vahdati, A., Mehrabani, D., Hosseini, S. E., and Tamadon, A. (2016). Isolation, characterization and growth kinetic comparison of bone marrow and adipose tissue mesenchymal stem cells of guinea pig. Int. J. Stem Cells 9, 115–123. doi: 10.15283/ijsc.2016.9.1.115
Baddoo, M., Hill, K., Wilkinson, R., Gaupp, D., Hughes, C., Kopen, G. C., et al. (2003). Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J. Cell Biochem. 89, 1235–1249. doi: 10.1002/jcb.10594
Bai, C., Hou, L., Ma, Y., Chen, L., Zhang, M., and Guan, W. (2013). Isolation and characterization of mesenchymal stem cells from chicken bone marrow. Cell Tissue Bank 14, 437–451. doi: 10.1007/s10561-012-9347-8
Bellows, C. G., Aubin, J. E., Heersche, J. N., and Antosz, M. E. (1986). Mineralized bone nodules formed in vitro from enzymatically released rat calvaria cell populations. Calcif Tissue Int. 38, 143–154. doi: 10.1007/BF02556874
Bhuvanalakshmi, G., Arfuso, F., Dharmarajan, A., and Warrier, S. (2014). Multifunctional properties of chicken embryonic prenatal mesenchymal stem cells- pluripotency, plasticity, and tumor suppression. Stem Cell Rev. 10, 856–870. doi: 10.1007/s12015-014-9530-3
Bonewald, L. F., Harris, S. E., Rosser, J., Dallas, M. R., Dallas, S. L., Camacho, N. P., et al. (2003). von Kossa staining alone is not sufficient to confirm that mineralization in vitro represents bone formation. Calcif Tissue Int. 72, 537–547. doi: 10.1007/s00223-002-1057-y
Crigler, L., Kazhanie, A., Yoon, T. J., Zakhari, J., Anders, J., Taylor, B., et al. (2007). Isolation of a mesenchymal cell population from murine dermis that contains progenitors of multiple cell lineages. FASEB. J. 21, 2050–2063. doi: 10.1096/fj.06-5880com
de Moraes, C. N., Maia, L., Dias, M. C., Dell'Aqua, C. P., da Mota, L. S., Chapwanya, A., et al. (2016). Bovine endometrial cells: a source of mesenchymal stem/progenitor cells. Cell Biol. Int. 40, 1332–1339. doi: 10.1002/cbin.10688
Dominici, M., Le Blanc, K., Mueller, I., Slaper-Cortenbach, I., Marini, F., Krause, D., et al. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8, 315–317. doi: 10.1080/14653240600855905
Donato, T. A., Martinez, E. F., and Arana-Chavez, V. E. (2016). Effects of TGF-beta1 on mineralization mediated by rat calvaria-derived osteogenic cells. Microsc. Res. Tech. 79, 1139–1146. doi: 10.1002/jemt.22768
Dumas, A., Le Drevo, M. A., Moreau, M. F., Guillet, C., Basle, M. F., and Chappard, D. (2008). Isolation of osteoprogenitors from murine bone marrow by selection of CD11b negative cells. Cytotechnology 58, 163–171. doi: 10.1007/s10616-009-9184-1
El-Sayed, K. M., Paris, S., Graetz, C., Kassem, N., Mekhemar, M., Ungefroren, H., et al. (2015). Isolation and characterisation of human gingival margin-derived STRO-1/MACS(+) and MACS(-) cell populations. Int. J. Oral. Sci. 7, 80–88. doi: 10.1038/ijos.2014.41
Entenmann, G., and Hauner, H. (1996). Relationship between replication and differentiation in cultured human adipocyte precursor cells. Am. J. Physiol. 270, C1011–C1016. doi: 10.1152/ajpcell.1996.270.4.C1011
Eslaminejad, M. B., and Nadri, S. (2009). Murine mesenchymal stem cell isolated and expanded in low and high density culture system: surface antigen expression and osteogenic culture mineralization. In vitro Cell Dev. Biol. Anim. 45, 451–459. doi: 10.1007/s11626-009-9198-1
Fleming, R. H. (2008). Nutritional factors affecting poultry bone health. Proc. Nutr. Sc. 67, 177–183. doi: 10.1017/S0029665108007015
Fornari, M. B., and Zanella, R. (2014). Unraveling the associations of osteoprotegerin gene with production traits in a paternal broiler line. SpringPlus 3, 682–690. doi: 10.1186/2193-1801-3-682
Freeman, B. T., Jung, J. P., and Ogle, B. M. (2015). Single-cell RNA-seq of bone marrow-derived mesenchymal stem cells reveals unique profiles of lineage priming. PLoS ONE 10:e0136199. doi: 10.1371/journal.pone.0136199
Friedenstein, A. J., Chailakhyan, R. K., and Gerasimov, U. V. (1987). Bone marrow osteogenic stem cells: in vitro cultivation and transplantation in diffusion chambers. Cell Tissue Kinet. 20, 263–272. doi: 10.1111/j.1365-2184.1987.tb01309.x
Gabriel, J. E., Alvares, L. E., Gobet, M. C., de Paz, C. C. P., Packer, I. U., Macari, M., et al. (2003). Expression of MyoD, myogenin, myostatin and Hsp70 transcripts in chicken embryos submitted to mild cold or heat. J. Therm. Biol. 28, 261–269. doi: 10.1016/S0306-4565(02)00085-2
Gang, E. J., Bosnakovski, D., Simsek, T., To, K., and Perlingeiro, R. C. (2008). Pax3 activation promotes the differentiation of mesenchymal stem cells toward the myogenic lineage. Exp. Cell Res. 314, 1721–1733. doi: 10.1016/j.yexcr.2008.02.016
Gang, E. J., Jeong, J. A., Hong, S. H., Hwang, S. H., Kim, S. W., Yang, I. H., et al. (2004). Skeletal myogenic differentiation of mesenchymal stem cells isolated from human umbilical cord blood. Stem Cells 22, 617–624. doi: 10.1634/stemcells.22-4-617
Gregory, C. A., Gunn, W. G., Peister, A., and Prockop, D. J. (2004). An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal. Biochem. 329, 77–84. doi: 10.1016/j.ab.2004.02.002
Guo, Z., Li, H., Li, X., Yu, X., Wang, H., Tang, P., et al. (2006). In vitro characteristics and in vivo immunosuppressive activity of compact bone-derived murine mesenchymal progenitor cells. Stem Cells 24, 992–1000. doi: 10.1634/stemcells.2005-0224
Honda, Y., Ding, X., Mussano, F., Wiberg, A., Ho, C. M., and Nishimura, I. (2013). Guiding the osteogenic fate of mouse and human mesenchymal stem cells through feedback system control. Sci. Rep. 3:3420. doi: 10.1038/srep03420
Intarapat, S., and Stern, C. D. (2013). Chick stem cells: current progress and future prospects. Stem Cell Res. 11, 1378–1392. doi: 10.1016/j.scr.2013.09.005
Kar, S., Mitra, S., and Banerjee, E. R. (2016). Isolation and culture of embryonic stem cells, mesenchymal stem cells, and dendritic cells from humans and mice. Methods Mol. Biol. 1516, 145–152. doi: 10.1007/7651_2015_315
Khatri, M., O'Brien, T. D., and Sharma, J. M. (2009). Isolation and differentiation of chicken mesenchymal stem cells from bone marrow. Stem Cells Dev. 18, 1485–1492. doi: 10.1089/scd.2008.0223
Kim, W. K., Meliton, V., Park, K. W., Hong, C., Tontonoz, P., Niewiadomski, P., et al. (2009). Negative regulation of Hedgehog signaling by liver X receptors. Mol. Endocrinol. 23, 1532–1543. doi: 10.1210/me.2008-0453
Kim, W. K., Bloomfield, S. A., and Ricke, S. C. (2011). Effects of age, vitamin D3, and fructooligosaccharides on bone growth and skeletal integrity of broiler chicks. Poult. Sci. 90, 2425–2432. doi: 10.3382/ps.2011-01475
Klepsch, S., Jamnig, A., Trimmel, D., Schimke, M., Kapferer, W., Brunauer, R., et al. (2013). Isolation of mesenchymal stem cells from human bone and long-term cultivation under physiologic oxygen conditions. Methods Mol. Biol. 976, 99–109. doi: 10.1007/978-1-62703-317-6_8
Kulterer, B., Friedl, G., Jandrositz, A., Sanchez-Cabo, F., Prokesch, A., Paar, C., et al. (2007). Gene expression profiling of human mesenchymal stem cells derived from bone marrow during expansion and osteoblast differentiation. BMC Genom. 8, 1–15. doi: 10.1186/1471-2164-8-70
Kumar, K., Agarwal, P., Das, K., Mili, B., Madhusoodan, A. P., Kumar, A., et al. (2016). Isolation and characterization of mesenchymal stem cells from caprine umbilical cord tissue matrix. Tissue Cell 48, 653–658. doi: 10.1016/j.tice.2016.06.004
Lehmann, J. M., Lenhard, J. M., Oliver, B. B., Ringold, G. M., and Kliewer, S. A. (1997). Peroxisome proliferator-activated receptors alpha and gamma are activated by indomethacin and other non-steroidal anti-inflammatory drugs. J. Biol. Chem. 272, 3406–3410. doi: 10.1074/jbc.272.6.3406
Li, H., Ghazanfari, R., Zacharaki, D., Lim, H. C., and Scheding, S. (2016). Isolation and characterization of primary bone marrow mesenchymal stromal cells. Ann. N. Y. Acad. Sci. 1370, 109–118. doi: 10.1111/nyas.13102
Matsubara, Y., Sato, K., Ishii, H., and Akiba, Y. (2005). Changes in mRNA expression of regulatory factors involved in adipocyte differentiation during fatty acid induced adipogenesis in chicken. Comparative Compar. Biochem. Physiol. Physiol. Part A 141, 108–115. doi: 10.1016/j.cbpb.2005.04.013
Melo, F. R., Bressan, R. B., Forner, S., Martini, A. C., Rode, M., Delben, P. B., et al. (2016). Transplantation of human skin-derived mesenchymal stromal cells improves locomotor recovery after spinal cord injury in rats. Cell. Molecul. Molecul. Neurobiol. Neurobiol. 37, 941–947. doi: 10.1007/s10571-016-0414-8
Mir, N. A., Rafiq, A., Kumar, F., Singh, V., and Shukla, V. (2017). Determinants of broiler chicken meat quality and factors affecting them: a review. J. Food Sci. Technol. 54, 2997–3009. doi: 10.1007/s13197-017-2789-z
Nadri, S., and Soleimani, M. (2007). Isolation murine mesenchymal stem cells by positive selection. In vitro Cell Dev. Biol. Anim. 43, 276–282. doi: 10.1007/s11626-007-9041-5
Neubauer, M., Fischbach, C., Bauer-Kreisel, P., Lieb, E., Hacker, M., Tessmar, J., et al. (2004). Basic fibroblast growth factor enhances PPARgamma ligand-induced adipogenesis of mesenchymal stem cells. FEBS Lett. 577, 277–283. doi: 10.1016/j.febslet.2004.10.020
Noth, U., Osyczka, A. M., Tuli, R., Hickok, N. J., Danielson, K. G., and Tuan, R. S. (2002). Multilineage mesenchymal differentiation potential of human trabecular bone-derived cells. J. Orthop. Res. 20, 1060–1069. doi: 10.1016/S0736-0266(02)00018-9
Ntambi, J. M., and Young-Cheul, K. (2000). Adipocyte differentiation and gene expression. J. Nutr. 130, 3122s−3126s. doi: 10.1093/jn/130.12.3122S
Parhami, F., Morrow, A. D., Balucan, J., Leitinger, N., Watson, A. D., Tintut, Y., et al. (1997). Lipid oxidation products have opposite effects on calcifying vascular cell and bone cell differentiation. A possible explanation for the paradox of arterial calcification in osteoporotic patients. Arterioscler Thromb. Vasc. Biol. 17, 680–687. doi: 10.1161/01.ATV.17.4.680
Perez-Serrano, R. M., Gonzalez-Davalos, M. L., Lozano-Flores, C., Shimada, A., Antaramian, A., Varela-Echavarria, A., et al. (2017). PPAR agonists promote the differentiation of porcine bone marrow mesenchymal stem cells into the adipogenic and myogenic lineages. Cells Tissues Organs. 203, 153–172. doi: 10.1159/000447628
Pittenger, M. F., Mackay, A. M., Beck, S. C., Jaiswal, R. K., Douglas, R., Mosca, J. D., et al. (1999). Multilineage potential of adult human mesenchymal stem cells. Science 284, 143–147. doi: 10.1126/science.284.5411.143
Regassa, A., and Kim, W. K. (2013). Effects of oleic acid and chicken serum on the expression of adipogenic transcription factors and adipogenic differentiation in hen preadipocytes. Cell Biol. Int. 37, 961–971. doi: 10.1002/cbin.10122
Scanes, C. G. (2007). The global importance of poultry. Poult. Sci. 86, 1057–1058. doi: 10.1093/ps/86.6.1057
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012. 9, 671–675. doi: 10.1038/nmeth.2089
Scott, M. A., Nguyen, V. T., Levi, B., and James, A. W. (2011). Current methods of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev. 20, 1793–1804. doi: 10.1089/scd.2011.0040
Shi, Y., Wang, F., Tiwari, S., Yesilbas, M., Steubesand, N., Weitkamp, J. T., et al. (2016). Role of myeloid early endothelial progenitor cells in bone formation and osteoclast differentiation in tissue construct based on hydroxyapatite poly(ester-urethane) scaffolds. J. Orthop. Res. 34, 1922–1932. doi: 10.1002/jor.23222
Short, B., and Wagey, R. (2013). Isolation and culture of mesenchymal stem cells from mouse compact bone. Methods Mol. Biol. 946, 335–347. doi: 10.1007/978-1-62703-128-8_21
Sławinska, A., Brzezinska, J., Siwek, M., and Elminowska-Wenda, G. (2013). Expression of myogenic genes in chickens stimulated in ovo with light and temperature. Reprod. Biol. 13, 161–165. doi: 10.1016/j.repbio.2013.04.003
Sohni, A., and Verfaillie, C. M. (2013). Mesenchymal stem cells migration homing and tracking. Stem Cells Int. 2013:130763. doi: 10.1155/2013/130763
Soleimani, M., and Nadri, S. (2009). A protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. Nat. Protoc. 4, 102–106. doi: 10.1038/nprot.2008.221
Tang, Q. Q., Otto, T. C., and Lane, M. D. (2003). Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 44–49. doi: 10.1073/pnas.0137044100
Tontonoz, P., Hu, E., and Spiegelman, B. M. (1994). Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 79, 1147–1156. doi: 10.1016/0092-8674(94)90006-X
Tremain, N., Korkko, J., Ibberson, D., Kopen, G. C., DiGirolamo, C., and Phinney, D. G. (2001). MicroSAGE analysis of 2,353 expressed genes in a single cell-derived colony of undifferentiated human mesenchymal stem cells reveals mRNAs of multiple cell lineages. Stem Cells 19, 408–418. doi: 10.1634/stemcells.19-5-408
Tuli, R., Tuli, S., Nandi, S., Wang, M. L., Alexander, P. G., Haleem-Smith, H., et al. (2003). Characterization of multipotential mesenchymal progenitor cells derived from human trabecular bone. Stem Cells 21, 681–693. doi: 10.1634/stemcells.21-6-681
Usui, M., Xing, L., Drissi, H., Zuscik, M., O'Keefe, R., Chen, D., et al. (2008). Murine and chicken chondrocytes regulate osteoclastogenesis by producing RANKL in response to BMP2. J. Bone Min. Res. 23, 314–325. doi: 10.1359/jbmr.071025
Van Vlasselaer, P., Falla, N., Snoeck, H., and Mathieu, E. (1994). Characterization and purification of osteogenic cells from murine bone marrow by two-color cell sorting using anti-Sca-1 monoclonal antibody and wheat germ agglutinin. Blood 84, 753–763.
Velleman, S. G., Anderson, J. W., Coy, C. S., and Nestor, K. E. (2003). Effect of selection for growth rate on muscle damage during turkey breast muscle development. Poult. Sci. 82, 1069−1074. doi: 10.1093/ps/82.7.1069
Wang, Q. R., and Wolf, N. S. (1990). Dissecting the hematopoietic microenvironment. VIII. Clonal isolation and identification of cell types in murine CFU-F colonies by limiting dilution. Exp. Hematol. 18, 355–359.
Wang, Y. H., Liu, Y., Maye, P., and Rowe, D. W. (2006). Examination of mineralized nodule formation in living osteoblastic cultures using fluorescent dyes. Biotechnol. Prog. 22, 1697–1701. doi: 10.1002/bp060274b
Wee, A. S., Lim, C. K., Merican, A. M., Ahmad, T. S., and Kamarul, T. (2013). Total cell pooling in vitro: an effective isolation method for bone marrow-derived multipotent stromal cells. In vitro Cell Dev. Biol. Anim. 49, 424–432. doi: 10.1007/s11626-013-9626-0
Wu, Z., Rosen, E. D., Brun, R., Hauser, S., Adelmant, G., Troy, A. E., et al. (1999). Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipognesis and insulin sensitivity. Mol. Cell 3, 151–158. doi: 10.1016/S1097-2765(00)80306-8
Yamamoto, Y., Itoh, S., Yamauchi, Y., Matsushita, K., Ikeda, S., Naruse, H., et al. (2015). Density gradient centrifugation for the isolation of cells of multiple lineages. J. Cell Biochem. 116, 2709–2714. doi: 10.1002/jcb.25270
Keywords: mesenchymal stem cells, isolation, broiler, biological characteristics, pluripotency
Citation: Adhikari R, Chen C, Waters E, West FD and Kim WK (2019) Isolation and Differentiation of Mesenchymal Stem Cells From Broiler Chicken Compact Bones. Front. Physiol. 9:1892. doi: 10.3389/fphys.2018.01892
Received: 10 August 2018; Accepted: 14 December 2018;
Published: 22 January 2019.
Edited by:
Sami Dridi, University of Arkansas, United StatesReviewed by:
Takeshi Ohkubo, Ibaraki University, JapanCopyright © 2019 Adhikari, Chen, Waters, West and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woo Kyun Kim, d2traW1AdWdhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.