- State Key Laboratory of Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
Background: Molting is a crucial physiological behavior during arthropod growth. In the past few years, molting as well as chitin biosynthesis triggered by molting, is subject to regulation by miRNAs. However, how many miRNAs are involved in insect molting at the genome-wide level remains unknown.
Results: We deeply sequenced four samples obtained from nymphs at the 2nd−3rd and 4th−5th instars, and then identified 61 miRNAs conserved in the Arthropoda and 326 putative novel miRNAs in the brown planthopper Nilaparvata lugens, a fearful pest of rice. A total of 36 mature miRNAs with significant different expression levels at the genome scale during molting, including 19 conserved and 17 putative novel miRNAs were identified. After comparing the expression profiles, we found that most of the targets of 36 miRNAs showing significantly differential expression were involved in energy and hormone pathways. One of the 17 putative novel miRNAs, nlu-miR-173 was chosen for functional study. nlu-miR-173 acts in 20-hydroxyecdysone signaling through its direct target, N. lugens Ftz-F1(NlFtz-F1), a transcription factor. Furthermore, we found that the transcription of nlu-miR-173 was promoted by Broad-Complex (BR-C), suggesting that its involvement in the 20-hydroxyecdysone pathway contributes to proper molting function.
Conclusion: We provided a comprehensive resource of miRNAs associated with insect molting and identified a novel miRNA as a potential target for pest control.
Introduction
Insects which are the hugest group of animals sometimes are human disease vectors and even more agricultural pests in the nature (Zhang et al., 2009; Liu et al., 2018). Over thousands of years, humans have developed necessary measures to prevent disease and pests from ourselves and plants. Although pesticides have played a pivotal role in both agriculture and public health, their widespread use has also been linked to the development of insecticide resistance and environmental issues (Soko et al., 2015). Thus, pest control urgently requires seeking novel types of pesticides such as insect growth regulators or novel targets in insects for pesticide development. Molting is an important developmental behavior during arthropod growth, and the genes involved in insect molting are usually used as effective targets for pest control (Retnakaran et al., 2003; Guerrero and Rosell, 2005; Soko et al., 2015; Niwa and Niwa, 2016). Chitin, the second most rich polysaccharide in biomass after cellulose (Tharanathan and Kittur, 2003; Kurita, 2006; Zakrzewski et al., 2014), is one of the most important substances during molting (Zhu et al., 2016). In recent years, microRNAs have been discovered to be involved in various developmental processes of animals. However, little miRNAs have been reported to act in insect molting or chitin biosynthesis (Belles, 2017). In Drosophila melanogaster, two miRNA family, Let-7-Complex and miR-34 function as a key factor to control insect metamorphosis (Sempere et al., 2003). In Bombyx mori, miRNAs with intact expression profiles during the whole life cycle have been established (Liu et al., 2009; Pan et al., 2017). In Helicoverpa armigera, miR-24 shows a negative correlation with chitinase transcript expression lavel in fifth-instar larvae (Agrawal et al., 2013). In Nilaparvata lugens, The conserved miRNAs miR-8-5p and miR-2a-3p targets two genes in the chitin biosynthesis pathway responsed by 20-hydroxyecdysone signaling (Chen et al., 2013). Chitin synthase A repressed by microRNA and dsRNA injection in vivo shows to be good for pest control (Li et al., 2017). These results indicate that the miRNAs involved in insect molting are largely unknown.
The brown planthopper N. lugens is one fearful pest of rice. Since its genome was published in 2014 (Xue et al., 2014), N. lugens has become a model hemipteran insect for scientific study such as migration, genetics, insecticide resistance, and adaptive evolution in insects. In this study, we provide the first resource for the dynamic profiles of miRNAs during insect molting, and we identify a novel miRNA, nlu-miR-173, that regulates insect molting by targeting the transcription factor NlFtz-F1.
Materials and Methods
Insects and Cells
The brown planthopper N. lugens used in our work has been reared steadily on susceptible rice varieties in our laboratory (Chen et al., 2013).
Drosophila S2 cells were cultured in Schneider's medium (Gibco) supplemented with 10% fetal bovine serum at 27 °C.
miRNA Sequencing and Prediction
Solexa sequencing was performed as described previously by BGI (Shenzhen, China) (Chen et al., 2013). Eighteen or more nucleotides sequences were blasted into the genome of N. lugens (version 6.1) with their entire length by SOAP (Li et al., 2008). First, miRBase miRNA Database (v20.0) was used for known miRNAs identification by BLAST search. Then, other sequences which have hairpin structures can be used to predict novel miRNAs using MIREAP by genome mapping.
Target Genes Prediction
We predicted targets for 36 miRNAs using both miRanda and RNAhybrid programs by a same criterion that six or more bases perfectly matched in the seed region (2–7 nt). In addition, higher filters were required by RNAhybrid, for example, free energy should be < -25 and the 3′ ends of sequences should have complementary bases.
Semi-quantitative and Quantitative PCR of miRNAs
The expression levels of 40 predicted miRNAs during the molting of 2nd−3rd and 4th−5th instars were detected by semi-quantitative PCR. The forward primers was miRNAs themselves and the reverse primer was a linker primer according to the specification of mir-X™ miRNA first-strand synthesis kit (TaKaRa, Japan).
The expression levels of Ftz-F1, nlu-mir-173, BR-C, and E74 (Ecdysone-induced protein 74EF). Figures 4, 7 were detected by qPCR using LightCycler 480 system. All the primers details were shown in Table S5. U6 was used as an internal control for miRNA assay and Actin was for gene assay. SYBR Premix Ex Taq was used (TaKaRa, Japan).
Co-expression of miRNAs and Their Targets in S2 Cells
We use pGL3-Basic and pRL-null vectors modified by the Pac promoter to detect luciferase signals in S2 cells. Details were related similarly to the previous work (Chen et al., 2013).
Promoter Assay
The promoter assay was related similarly to the previous work (Chen et al., 2013).
20E Assay
An Ecdysone ELISA Kit (Biosense, China) was used to quantify the content of ecdysteroids. Ten individuals per sample were selected and considered one set for the ecdysteroid detection. Insects were cleaned, weighed and homogenized in 70% methanol. Supernatants were collected by 12,000 g for 10 min at 4°C, and the pellets were repeated twice aboved. Supernatants overall were dried and resoluted between 70% methanol and hexane to remove non-polar lipids. The hexane phase was throwed and the lower methanolic phase was dried, then re-dissolved in 100 μl methanol.
Feeding and Injection on Nymphs
The feeding and injection experiments were improved by our laboratory. The suitable glass capillary (1B100F-4, WPI, USA) was used to injected into the abdomen of early fifth-instar adults which was on narcosis by CO2. Fifty nanoliters of liquid was injected in our experiments. In the feeding experiment, we placed the same age of third-instar individuals into a chamber for miRNA mimics or inhibitor feeding. After 7 days with continuous feeding, the survival rates were recorded. Other details were related similarly to the previous work (Chen et al., 2013).
Chitin Content Analysis
The method of assay was amended slightly on the basis of that mentioned by Arakane et al. (2004).
Statistical Analysis
The asterisk indicated the significant differences between control and treatment groups by t-test. Significant differences were marked by letters from different groups of survival rates (p < 0.05, LSR, SPSS). In qPCR assay, each bar indicates the mean ± SEM with three independent experiments, and asterisks indicate significant differences at the p < 0.05 level.
Results
Deep-sequencing of microRNAs in N. lugens
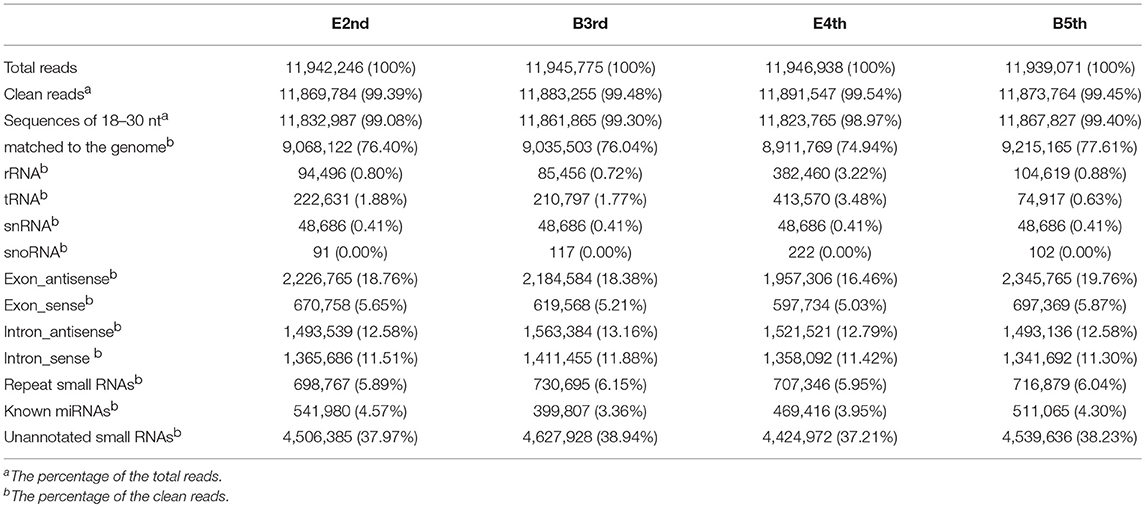
Instar transition is the best period to detect the dynamic changes in the miRNA levels during molting. Four time periods, the end of the 2nd instar (E2nd), the beginning of the 3rd instar (B3rd), the end of the 4th instar (E4th) and the beginning of the 5th instar (B5th), were used to represent two periods of molting, namely, the 2nd−3rd and 4th−5th molting periods. N. lugens individuals at the above four periods were chosen and sequenced for their small RNAs using Solexa high-throughput. After removing unrelated data, we obtained 11,869,784, 11,883,255, 11,891,547, and 11,873,764 clean reads from the E2nd, B3rd, E4th, and B5th larvae, respectively (Table 1). Using a filtration series, we finally obtained 541,980, 399,807, 469,416, and 511,065 small RNA sequences, respectively. These were used as candidates for identifying known miRNAs by mapping with miRNA database in miRBase (Table 1). At last, we identified 619, 581, 616, and 557 conserved miRNA sequences in the four samples, respectively. The others were further mapped to the N. lugens genome to predict novel miRNAs. We obtained 183, 158, 151, and 168 putative novel miRNAs (Table S1). In total, we found 61 miRNA families conserved in the Arthropoda (Table S1), and 326 putative novel miRNAs at the genome-wide scale in N. lugens.
Differentially Expressed miRNAs During Molting
We compared the expression levels of putative novel miRNA profiles during the 2nd−3rd and 4th−5th instar molting periods (Figure 1) and found 20 putative novel miRNAs with significantly different expression in larval molting of N. lugens by the criterion of log2 defined as having a ratio of absolute values ≥1.0 (Figure 2). In addition, 20 miRNAs that are conserved in arthropods were also identified (Figure 2). As shown in the profiles, only 7 miRNAs (miR-87a-3p, miR-133, miR-2796-5p, miR-71-3p, miR-124, nlu-miR-186, and nlu-miR-213) had different expression levels during both molting periods. Furthermore, more miRNAs showed differential expression during the 4th−5th molting period, which may be due to increased complexity of the physiological activities during this period. To confirm the expression changes of 40 miRNAs determined using Solexa sequencing, an independent experiment using semi-quantitative PCR was performed. The results showed that only four miRNAs were unsuccessfully cloned (miR-317, nlu-miR-150, nlu-miR-26, and nlu-miR-55) (Figure 2), indicating a strong consistency between the fold-change values obtained via sequencing and the semi-quantitative PCR.
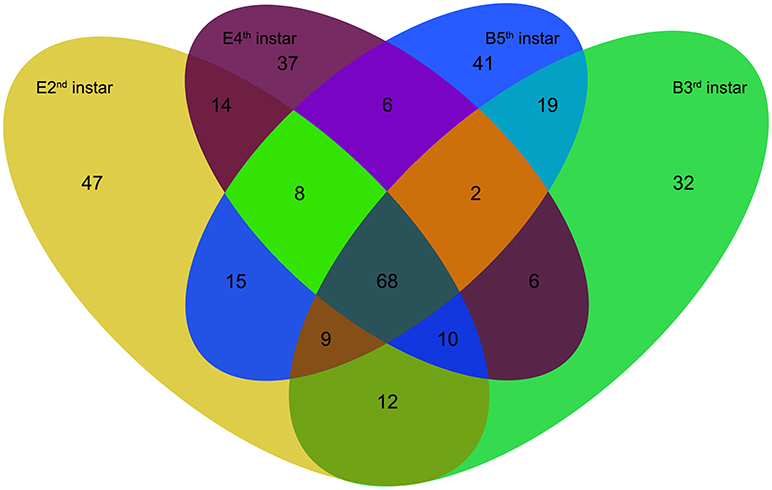
Figure 1. The numbers of putative novel miRNAs in four samples of N. lugens. E2nd instar, the miRNA numbers acquired from the sample at the end of the 2nd instar; B3rd instar, the miRNA numbers acquired from the sample at the beginning of the 3rd instar; E4th instar, the miRNA numbers acquired from the sample at the end of the 4th instar; B5th instar, the miRNA numbers acquired from the sample at the beginning of the 5th instar. The overlapping numbers indicate the common miRNAs in different samples.
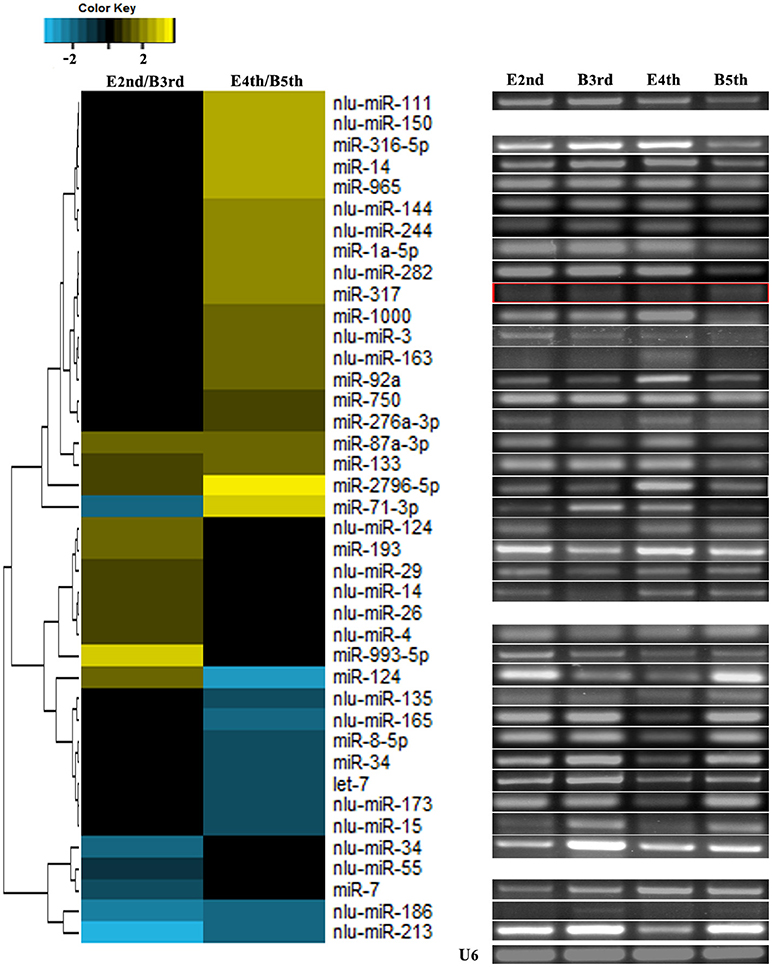
Figure 2. Validation of expression changes of miRNAs during molting. The left heatmap indicates the sequencing data of miRNA expression ratios. The bar represents the scale of the ratios of expression levels (log 2). The right electrophoretograms indicate the semi-quantitative PCR of miRNAs, using U6 as an internal control. E2nd/B3rd, the expression level in the sample at the end of the 2nd instar/the expression level in the sample at the beginning of the 3rd instar; E4th/B5th, the expression level in the sample at the end of the 4th instar/the expression level in the sample at the beginning of the 5th instar. The empty and red boxes indicate false positive results.
Potential Targets of miRNAs Involved in Molting
We predicted target genes for 36 miRNAs using both miRanda and RNAhybrid except for four false positive miRNAs, and 173 targets for 19 conserved miRNAs and 108 targets for 17 putative novel miRNAs with high filters both in two programs were found (Table S2). In total, 122 genes possibly targeted by the identified 36 miRNAs were annotated in help of the genome, with an average of 3.4 targets per miRNA. A high amount of the potential targets were involved in signal transduction, cellular structure, transcription, and translation. Moreover, targets involved in cuticular and hormone regulation, energy and other metabolic pathways were overrepresented (Table S3).
Because target prediction can produce false positives, RNA sequencing was also used to determine the gene expression patterns at the four time periods (E2nd, B3rd, E4th, and B5th). Using the transcriptome data of N. lugens (data not shown), 539 unigenes demonstrated lower expression levels at the beginning of the 3rd instar (B3rd) than at the end of the 2nd instar (E2nd), whereas 211 unigenes were up-regulated at B3rd compared to B2nd. Similarly, 841 unigenes demonstrated lower expression levels at the beginning of the 5th instar (B5th) than at the end of the 4th instar (E4th), whereas 830 unigenes were up-regulated at B5th compared to E4th. To exclude the possibility that different genes participate in molting at different instars, we chose genes that were differentially expressed during both time periods. A total of 82 unigenes up-regulated and 414 unigenes down-regulated during molting were identified at the whole-genome level. Furthermore, Gene Ontology (GO) abundance analysis of these genes revealed terms related to catalytic activity (Figure S1A), oxidation-reduction processes (Figure S1B), and structural molecule activity, including the structural constituents of the chitin-based cuticle, that were all statistically significant (Figure S1C).
We chose negative correlations between miRNAs and targets for further study through small RNA sequencing and RNA sequencing (Figure 3, Table S4). Most of the targets showing significant differential expression (defined as having a ratio of absolute values ≥1.5) were involved in energy and hormone functions. For example, trehalase was a target of miR-8-5p and is a link between the hormone and chitin biosynthesis pathways according to our previous work (Chen et al., 2013). In total, we identified a total of 12 miRNAs (let-7, miR-34, miR-1000, miR-133, miR-14, miR-71-3p, miR-965, miR-87, miR-8, nlu-miR-213, nlu-miR-186, and nlu-miR-173) and target molecules showing significantly different expression levels as potential factors during molting. Among the above potential miRNAs involved in insect molting, we chose nlu-miR-173 for functional study because of its target (NlFtz-F1) and species specificity.

Figure 3. Expression profiles of miRNAs and their targets during molting. The bar represents the scale of ratios of expression levels (log 2). E2nd/B3rd, the expression level in the sample at the end of the 2nd instar/the expression level in the sample at the beginning of the 3rd instar; E4th/B5th, the expression level in the sample at the end of the 4th instar/the expression level in the sample at the beginning of the 5th instar. The genes behind the miRNA indicate that these are the potential targets of this miRNA. A single asterisk indicates a significant difference at the p < 0.05 level. A double asterisk indicates a significant difference at the p < 0.01 level.
NlFtz-F1 Is a Target of nlu-miR-173 in Response to 20-hydroxyecdysone Signaling
To validate NlFtz-F1 as a target of nlu-miR-173, the expression of luciferase from the constructs bearing the 3′ UTR sequences of NlFtz-F1 was detected after co-transfection of nlu-miR-173 and NlFtz-F1 in cells. Because of unavailable cell lines in N. lugens, we use S2 cells for identification between miRNA and its target referring to the previous work in Chen et al. (2013). The construct with co-transfection of nlu-miR-173 decreased by 44.60%, while the construct without co-transfection of nlu-miR-173 also showed a little reduced expression of luciferase relative to the construct without the 3′ UTR sequences co-expressed with nlu-miR-173(Figure 4A). This indicated that the 3′ UTR sequences of NlFtz-F1 may include other seed regions for other miRNAs. In vivo, the transcription level (Figure 4B) and protein level (Figure 4C) of NlFtz-F1 were decreased after mimics of nlu-miR-173 injection, confirmed that NlFtz-F1 was the target of nlu-miR-173.
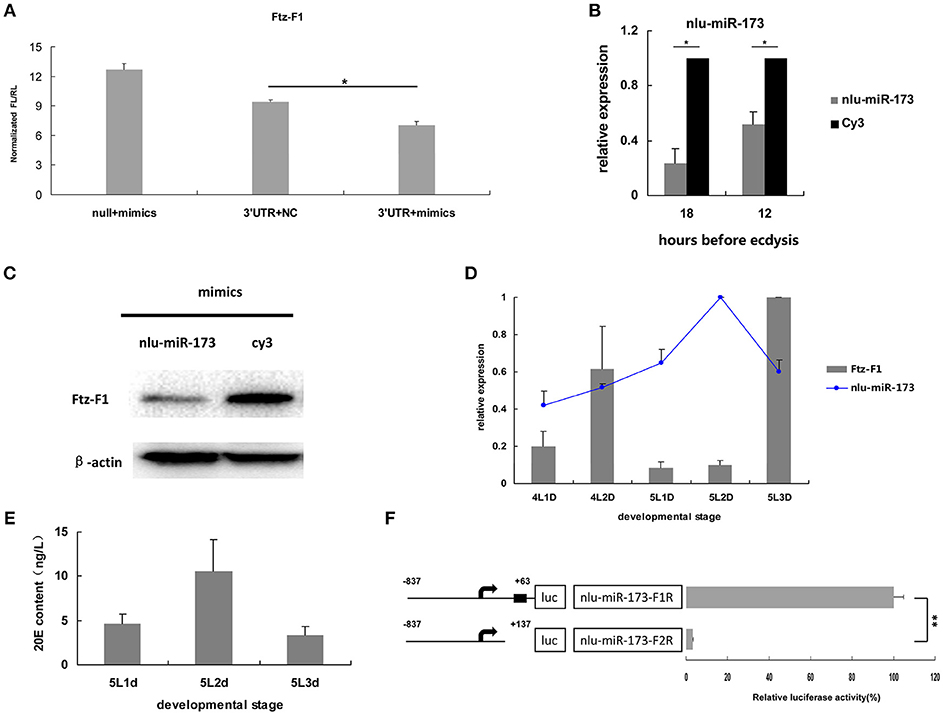
Figure 4. NlFtz-F1 is a target of nlu-miR-173 in response to 20-hydroxyecdysone signaling. (A) Co-transfection of nlu-miR-173 and its target NlFtz-F1. NC represents an unrelated small RNA sequence, and null represents a reporter without the 3′ UTR sequence. A single asterisk indicates a significant differences between the control group and the treated group at the p < 0.05 level. Each point represents the mean ± SEM from three independent experiments. (B) Transcription analysis. All mRNA levels are shown relative to the level of β-actin. A single asterisk indicates a significant difference between the two groups at the p < 0.05 level. Each point represents the mean ± SEM (n = 3). (C) Western blot. Protein extracts (5 μg) from the injected nymphs were loaded onto 12% SDS-PAGE gels. The gels were immunostained with anti-Ftz-F1 serum. β-Actin was used as a control. (D) Expression levels of nlu-miR-173 and NlFtz-F1. 4L1D: Nymphs on the 1st day of the 4th instar. All miRNA levels are shown relative to the U6 level, all mRNA levels are expressed relative to the level of β-actin, and the expression level of nlu-miR-173 is normalized to 5L2D, and that of NlFtz-F1 is normalized to 5L3D. (E) 20E content. (F) Luciferase assays used to test the BR-C binding sites in the promoters of nlu-miR-173. Black boxes means binding sites for BR-C. Each point represents the mean ± SEM from three independent experiments. A double asterisk indicates a significant difference between the 2 days at the p < 0.01 level. The highest luciferase level of a segment is designated as 100%.
Besides, the expression levels of nlu-miR-173 and NlFtz-F1 were detected in vivo using qPCR. The results showed that the expression level of nlu-miR-173 decreased largely on the last day and increased on the previous day during the 5th instar, whereas the target gene NlFtz-F1 showed the opposite pattern (Figure 4D). Combined with the similar expression pattern of 20E (Figure 4E), these findings suggested that NlFtz-F1 and nlu-miR-173 may be in response to 20-hydroxyecdysone signaling. The transcription factor BR-C is a key gene in 20-hydroxyecdysone signaling. We use several programs to analyze whether the promoter of nlu-miR-173 was regulated by BR-C. The luciferase activity assay showed that the shorter segment lacking the BR-C binding site was significantly lower than the longer one (Figure 4F). This indicated that NlFtz-F1 is the target of nlu-miR-173 in response to 20-hydroxyecdysone signaling.
nlu-miR-173 Is Required for Molting and Chitin Biosynthesis Through NlFtz-F1
To determine the developmental functions of nlu-miR-173 in N. lugens, in vivo nlu-miR-173 mimics or inhibitors were fed on the 3rd nymphs. The death rates of nymphs increased to 84.92 and 47.22% after 7 days of continuous feeding of nlu-miR-173 mimics and inhibitors, respectively, especially a huge mortality of the group of nlu-miR-173 mimics was shown. However, there is no obvious difference between the control and unrelated miRNA (Cy3)-feeding groups (Figure 5A). Individuals fed with nlu-miR-173 mimics or inhibitors exhibited similar phenotypes of molting failure (Figure 6B), abdominal defects (Figure 6C), and wing defects (Figure 6D) at rates of 28.15 and 27.50%, 23.70 and 10.37%, 14.07 and 10.12%, respectively(Figure 6). In addition, the chitin contents changed significantly by nlu-miR-173 mimics or inhibitors injection (Figure 5B). Together, these results indicated that nlu-miR-173 through its target NlFtz-F1 is required for molting and chitin biosynthesis.
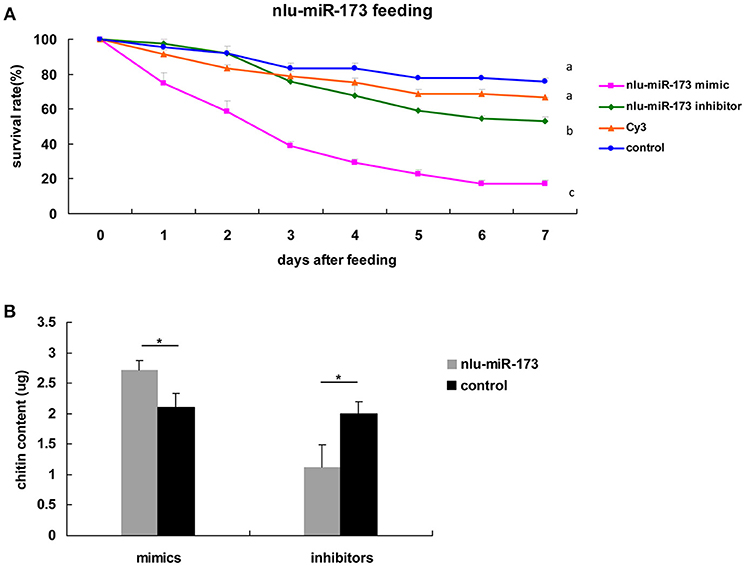
Figure 5. Nlu-miR-173 is required for molting and chitin biosynthesis through NlFtz-F1. (A) Survival rates with feeding of the miRNA mimics or the inhibitor. Each point represents the mean ± SEM from three independent experiments. Different letters indicate significant differences (p < 0.05, LSR, SPSS). (B) Mimics or an inhibitor of nlu-miR-173 changed the chitin content of N. lugens. A single asterisk indicates a significant difference between the two groups at the p < 0.05 level. Each point represents the mean ± SEM (n = 3).
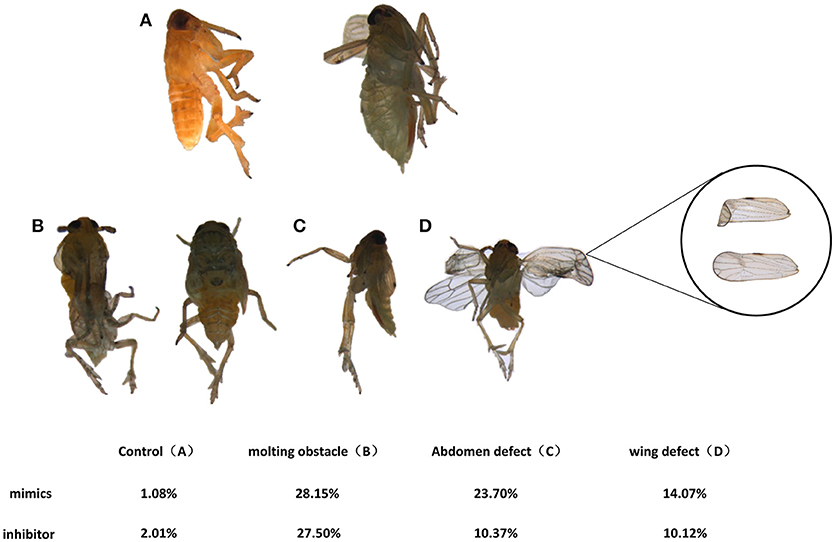
Figure 6. Phenotypes effected by feeding on nymphs. (A) Normal adults. Individuals in the mimic-feeding group died with phenotypes of molting failure (B), abdominal defects (C) and wing defects (D) at rates of 23.70, 28.15, and 14.07%, respectively, whereas individuals in the inhibitor-feeding group died with those phenotypes at rates of 10.37, 27.50, and 10.12%, respectively.
To reveal the mechanism that 20E signaling regulated molting and chitin biosynthesis partly through nlu-miR-173 modified by the activity of BR-C and E74 (Ecdysone-induced protein 74EF), we first detected the expression levels of nlu-miR-173 and NlFtz-F1 in vivo after 4th-instar nymphs injection by 20E, we found that the expression level of nlu-miR-173 was greatly increased but NlFtz-F1 was decreased (Figures 7A,B). However, when BR-C or E74 of dsRNAs was injected 24 h before 20E injection, the expression levels of BR-C, E74, or nlu-miR-173 was not increased as above (Figures 7C–F), indicating that activity of BR-C and E74 are essential for molting regulated by nlu-miR-173. Further, injection of dsRNAs targeting BR-C and E74 24 h prior to 20E injection also decreased the expression level of NlFtz-F1 (Figures 7G,H), showing that NlFtz-F1 could also be regulated directly by 20E signaling as a transcription factor.
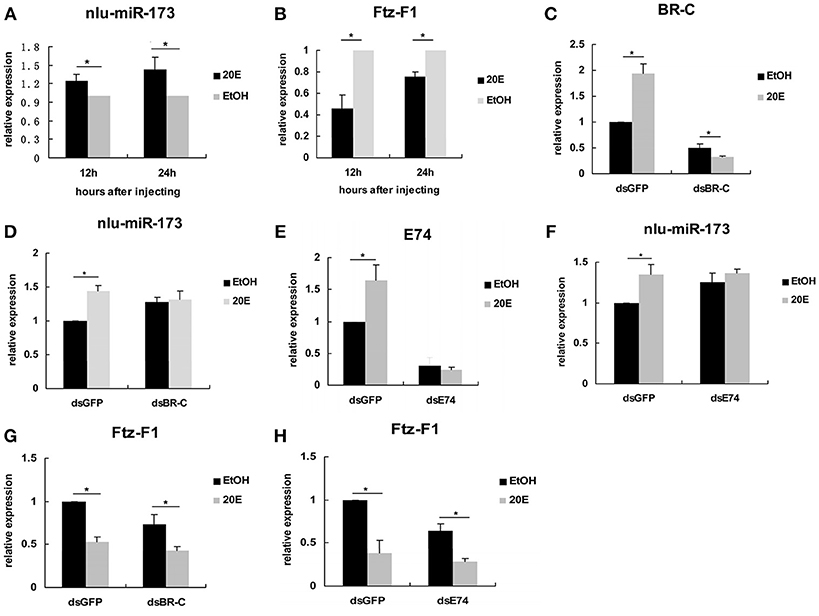
Figure 7. 20E signaling regulates nlu-miR-173 through BR-C and E74. (A,B) The expression levels of nlu-miR-173 and Ftz-F1 after 20E injection. (C–H) The expression levels of nlu-miR-173, Ftz-F1, BR-C and E74 after 20E and dsRNA injection. All expression levels are shown relative to the levels of U6 (nlu-miR-173) or β-actin (BR-C, E74, and Ftz-F1), and the expression levels are normalized to those of the dsGFP, dsBR-C, or dsE74 group injected with ethanol. A single asterisk indicates a significant difference between the two groups at the p < 0.05 level. Each point represents the mean ± SEM (n = 3).
Discussion
Using the genomic data for N. lugens as well as high-throughput sequencing, we have identified candidate miRNAs involved in insect molting. Moreover, we identified the novel miRNA nlu-miR-173 and showed its involvement in the regulation of molting and chitin biosynthesis in N. lugens through its target NlFtz-F1. This is the first to demonstrate the potential function of miRNAs to act in arthropods at the genome-wide level as regulators of molting and chitin biosynthesis.
Computational Prediction of miRNAs and their Potential Targets on a Genome-wide Scale
Benefiting from the high-throughput sequencing technology coupled with bioinformatic methods, increasing numbers of miRNAs have been identified many species of insects (Legeai et al., 2010; Liu et al., 2010; Wei et al., 2011; Chilana et al., 2013; Tan et al., 2014). Generally, there are hundreds of known miRNAs in any given insect species, such as Aedes aegypti, B. mori, D. melanogaster, and Tribolium castaneum with 124, 563, 466, and 430 known mature miRNAs, respectively, according to miRBase 21.0. Recently, a thousand of miRNAs were discovered in Locusta migratoria based on both genome searching and small RNA sequencing during different stages and tissues (Wang et al., 2015). In our work, we succeed to found more miRNAs involved in the coordination of molting and chitin biosynthesis by examining stage-specific expression of miRNAs. Based on our previous data and the results in this study, we obtained a total of 326 putative novel miRNAs in N. lugens, which significantly extends the list of miRNAs in insect species (Table S6). To predict the putative novel miRNAs, miRNA precursors with hairpin structure and bioinformatic searches based on alignments with the available genome sequences or transcriptomes were performed. Among all previous identified miRNAs in a species, the predicated novel miRNAs usually occupy a small proportion (ranging from 5 to 30%) in most samples (Jagadeeswaran et al., 2010; Wang et al., 2013; Hong et al., 2014; Huang et al., 2014; Zhang et al., 2014b; Hu et al., 2015; Shan et al., 2015; Xu et al., 2015). In previous work, we have acquired 688 conserved and 71 putative novel miRNAs by sequencing from adults and nymphs of N. lugens (Chen et al., 2012). However, there was no genome data to obtain the accuracy of miRNA identification in N. lugens at that time. Only a little of the 326 putative novel miRNAs identified here overlapped with the previous paper, demonstrating that the quantity of identified miRNAs and the proportion of putative novel miRNAs increased greatly in our work. Moreover, we successfully cloned 36 miRNAs out of 40 selected miRNAs (Figure 2), including 19 conserved and 17 putative novel miRNAs. Considering the limitations of PCR-based methods and the low transcription levels of miRNAs, the predicted rate of false positives would actually be lower than 10%. It showed that our method aboved to predict species-specific miRNAs is referential.
In past few years, it has become routine to predict the targets of all putative miRNAs in silico after insect miRNA profiles are reported (Li et al., 2014; Zhang et al., 2014a; Zhou et al., 2014; Jain et al., 2015; Shan et al., 2015; Tariq et al., 2015; Xu et al., 2015; Ling et al., 2017; Wu et al., 2017). However, a large of miRNA targets cannot be verificated by experimental assay. Some studies have reported that only a few miRNAs predicted in silico are eventually validated by in vivo experiments (Sultan et al., 2014; Suh et al., 2015). False positive and false negative results are unavoidable when conducting genome-wide target prediction due to the precision necessary. We explored the hotmap of miRNAs and their targets in Figure 3, using both miRNA and transcriptome RNA sequencing during molting. It is believed that most miRNAs and their targets appeared a negatively correlation (Lomate et al., 2014). To verify the accuracy of target prediction, a conserved miRNA (miR-34) and a putative novel miRNA (nlu-miR-173) were selected for target validation in vitro. We identified JHBP and FTZ-F1 as the respective targets of miR-34 (Figure S2) and nlu-miR-173 (Figure 4) in cells, revealing the high precision of our prediction. Moreover, we also detected a subset of miRNAs that showed both negative and positive correlations with their predicted targets. Recently, some studies has revealed that miRNAs can trigger gene expression via different mechanisms post-transcriptionally, indicating that miRNA and targets also can showed a positive correlation (Vasudevan et al., 2007; Mortensen et al., 2011). This suggests that miRNAs may have different actions toward their targets. However, validation of the relationship between miRNAs and genes showing differential expression between the two molting periods requires more experimental evidence.
Function of miRNAs in Insect Molting
Recent studies have shown key roles for miRNAs during insect developmental transitions. In D. melanogaster, let-7 and miR-34 mutants led to serious metamorphic obstacles (Sempere et al., 2003), and miR-8 may have a key factor in eclosion (Hua et al., 2012). In B. mori, let-7 was found its importance in molting and metamorphosis by transgenic miRNA sponge (miR-SP) technology and GAL4/UAS system in silkworm (Lin et al., 2014). Then, CRISPR/Cas9 system mediated miR-14 disruption led to an obvious phenotype in ecdysteriod titers (Liu et al., 2018). In Bactrocera dorsalis, ecdysone signaling pathway was triggered by Let-7 through target gene BdE75 (Peng et al., 2017). In Blattela germanica, miR-252-3p and miR-2 may play important functions during metamorphosis (Rubio et al., 2012; Jesus et al., 2015). In N. lugens, the survival rate and chitin content was affected by miR-8-5p and miR-2a-3p through their targets, membrane-bound trehalase (Tre-2) and phosphoacetylglucosamine mutase (PAGM) (Chen et al., 2013). However, most researchers have focused on this line of research at the level of individual conserved miRNAs. To our knowledge, molting-related miRNAs in insects investigated at the omics level have only been reported in cockroaches, without addressing putative novel miRNAs due to the lack of a complete genome sequence (Rubio et al., 2012). We are the first to detect the molting-related miRNAs on a genome-wide scale, and we identified 19 conserved and 17 putative novel miRNAs with differential expression during molting periods, significantly extending the research on insect molting-related miRNAs.
As the interaction between ecdysone and juvenile hormone (JH) is important for molting and metamorphosis (Zhou et al., 1998; Wang et al., 2012), there is accumulating evidence of a link between hormone and miRNAs in insect development and metamorphosis. In D. melanogaster, Let-7-Complex can be stimulated by the 20E through BR-C whereas miR-34 is repressed by 20E and can be activated by JH (Sempere et al., 2003). Moreover, miR-8-5p and miR-2a-3p are also repressed by BR-C and E74 in N. lugens (Chen et al., 2013). Thus, miRNAs can regulate the expression and function of genes in the hormone cascade, for instance, miR-14 via its target EcR controls a steroid hormone signaling in Drosophila (Varghese and Cohen, 2007), and miR-34 prevents neurodegeneration in the adult brain by repressing E74A (Liu et al., 2012). These studies have revealed a regulatory network of miRNAs in the 20E and JH pathways. In B. mori, BmEcR-B could be affected by bmo-miR-281 (Jiang et al., 2013). Moreover, FTZ-F1 and E74 targeted by Let-7, play crucial factors in the ecdysone pathway (Lin et al., 2014). As shown in our in vitro study, miR-34 was activated by BR-C directly because of its bind regions of BR-C in the promoter (Figure S2D), and the target gene JHBP was down-regulated by miR-34 mimics transfection in S2 cells (Figure S2B), suggested that 20E signaling coordinates with the early response gene BR-C to suppress JHBP through miR-34. Besides, JHBP also could be activated by BR-C directly in Figure S2C. Thus, miR-34 and nlu-miR-173 act as a molecular link that coordinates the two physiological hormone pathways. Further in vivo research revealed that the targeting of NlFTZ-F1 by nlu-miR-173 is a key factor in the action of 20E and acts as a positive regulator of chitin synthesis in N. lugens. As a nuclear receptor-type transcription factor, NlFTZ-F1 could be regulated directly by 20E signaling and suppressed via nlu-miR-173 activated by the 20E. Our present work also reveals that miRNAs and their targets can affect chitin biosynthesis in response to hormone signaling, which reveals hormone-miRNA-gene crosstalk in chitin biosynthesis. In short, there is hormone-miRNA-gene crosstalk that affects chitin biosynthesis and molting (Figure 8).
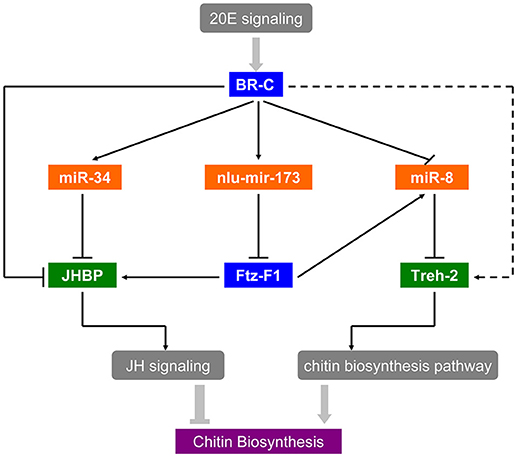
Figure 8. Hormone-miRNA-gene crosstalk in insect chitin biosynthesis. Solid arrows in the picture represent the experimental interactions and broken arrows represent the supposed interactions.
Potential Regulatory Mechanism of Ftz-F1 and nlu-miR-173
Mounting evidence suggests that Ftz-F1 acts importantly responsed by the 20E and JH signaling pathways during molting (Broadus et al., 1999; Bernardo and Dubrovsky, 2012; Cho et al., 2013; Borras-Castells et al., 2017). Ftz-F1 is induced after the decrease in the 20-hydroxyecdysone level (Woodard et al., 1994; Yamada et al., 2000; Hiruma and Riddiford, 2001), and the function of Ftz-F1 during metamorphosis in several insects was reported. In D. melanogaster, Ftz-F1 is required during each stage from embryogenesis, larval ecdysis to pupation (Yamada et al., 2000; Sultan et al., 2014). In several other insect species, silencing Ftz-F1 caused severe failure of ecdysis or pupation and larval lethality such as T. castaneum (Tan and Palli, 2008), L. Decemlineata (Liu et al., 2014), and B. Germanica (Cruz et al., 2008). Our work is keeping with the finding during the larval period, Ftz-F1 is induced just before larval ecdysis. The normal expression patterns of nlu-miR-173 and its target NlFtz-F1 ensure that the molting system operates regularly. Induction of NlFtz-F1 is prevented by the overexpression of nlu-miR-173, which interrupts the 20E signaling cascade, thereby disrupting the developmental transition, which leads to ecdysis defects and chitin accumulation in N. lugens. Our analyses show that nlu-miR-173 is a novel miRNA in N. lugens. In B. mori, Ftz-F1 is a target gene of let-7 (Lin et al., 2014). We need infer that NlFtz-F1 regulated by nlu-miR-173 may be specific to N. lugens or hemimetabolous insects that will be studied in the further. This suggests that let-7 and nlu-miR-173 both target NlFtz-F1, indicating that both highly conserved miRNAs and newly evolved miRNAs use this pathway to control development and metamorphosis to achieve common and lineage-specific functions. These results indicate that the comparison of putative novel miRNAs and their targets with conserved miRNAs may help explain the long-term evolution of species, including mechanisms regulating developmental transition in holometabolous and hemimetabolous insects.
Author Contributions
JC and WZ wrote the paper. JH reviewed the paper. JC and TL did most of the experiments. RP did the bioinformatics works and XY did some of works.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to Prof. Yueqing Chen and Prof. Songshan Jiang (Sun Yat-sen University, Guangzhou, China) for their suggestions and reading of the manuscript. This work was supported by the National Natural Science Foundation of China (31171900, 31301716), and the Science and Technology Planning Project of Guangdong Province, China (2016A050502021).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2018.01854/full#supplementary-material
Table S1. All known and novel miRNA sequences identified during molting.
Table S2. All genes differentially expressed in four samples during molting.
Table S3. Targets of all known and putative novel miRNAs in four samples during molting.
Table S4. The expression levels of miRNAs and targets in Figure 3.
Table S5. Primers used in this study.
Table S6. A list of the novel miRNAs in N. lugens.
Figure S1. The GO assay of genes differentially expressed during molting. (A) cellular component, (B) biological process, and (C) molecular function. The color scale shows the p-value cutoff levels, the more statistically significant, the darker, and redder the color.
Figure S2. Hormone-miRNA-gene crosstalk genes identification. (A,C,D) Luciferase assays of miR-8,miR-34, and JHBP. Black and white boxes indicate binding sites for BR-C and Ftz-F1. Double asterisks show a significant difference at the p < 0.01 level. The highest luciferase level of a segment is designated as 100%. (B) Target identification of miR-34 and JHBP in S2 cells.
References
Agrawal, N., Sachdev, B., Rodrigues, J., Sree, K. S., and Bhatnagar, R. K. (2013). Development associated profiling of chitinase and microRNA of Helicoverpa armigera identified chitinase repressive microRNA. Sci. Rep. 3:2292. doi: 10.1038/srep02292
Arakane, Y., Hogenkamp, D. G., Zhu, Y. C., Kramer, K. J., Specht, C. A., Beeman, R. W., et al. (2004). Characterization of two chitin synthase genes of the red flour beetle, Tribolium castaneum, and alternate exon usage in one of the genes during development. Insect Biochem. Mol. Biol. 34, 291–304. doi: 10.1016/j.ibmb.2003.11.004
Belles, X. (2017). MicroRNAs and the evolution of insect metamorphosis. Annu. Rev. Entomol. 62, 111–125. doi: 10.1146/annurev-ento-031616-034925
Bernardo, T. J., and Dubrovsky, E. B. (2012). The Drosophila juvenile hormone receptor candidates methoprene-tolerant (MET) and germ cell-expressed (GCE) utilize a conserved LIXXL motif to bind the FTZ-F1 nuclear receptor. J. Biol. Chem. 287, 7821–7833. doi: 10.1074/jbc.M111.327254
Borras-Castells, F., Nieva, C., Maestro, J. L., Maestro, O., Belles, X., and Martín, D. (2017). Juvenile hormone biosynthesis in adult Blattella germanica requires nuclear receptors Seven-up and FTZ-F1. Sci. Rep. 7:40234. doi: 10.1038/srep40234
Broadus, J., Mccabe, J. R., Endrizzi, B., Thummel, C. S., and Woodard, C. T. (1999). The Drosophila βFTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol. Cell 3, 143–149. doi: 10.1016/S1097-2765(00)80305-6
Chen, J., Liang, Z., Liang, Y., Pang, R., and Zhang, W. (2013). Conserved microRNAs miR-8-5p and miR-2a-3p modulate chitin biosynthesis in response to 20-hydroxyecdysone signaling in the brown planthopper, Nilaparvata lugens. Insect Biochem. Mol. Biol. 43, 839–848. doi: 10.1016/j.ibmb.2013.06.002
Chen, Q., Lu, L., Hua, H., Zhou, F., Lu, L., and Lin, Y. (2012). Characterization and comparative analysis of small RNAs in three small RNA libraries of the brown planthopper (Nilaparvata lugens). PLoS ONE7:e32860. doi: 10.1371/journal.pone.0032860
Chilana, P., Sharma, A., Arora, V., Bhati, J., and Rai, A. (2013). Computational identification and characterization of putative miRNAs in Heliothis virescens. Bioinformation 9, 79–83. doi: 10.6026/97320630009079
Cho, K. H., Daubnerová, I., Park, Y., Zitnan, D., and Adams, M. E. (2013). Secretory competence in a gateway endocrine cell conferred by the nuclear receptor βFTZ-F1 enables stage-specific ecdysone responses throughout development in Drosophila. Dev. Biol. 385, 253–262. doi: 10.1016/j.ydbio.2013.11.003
Cruz, J., Nieva, C., Mané-Padrós, D., Martín, D., and Bellés, X. (2008). Nuclear receptor BgFTZ-F1 regulates molting and the timing of ecdysteroid production during nymphal development in the hemimetabolous insect Blattella germanica. Dev. Dyn. 237, 3179–3191. doi: 10.1002/dvdy.21728
Guerrero, A., and Rosell, G. (2005). Biorational approaches for insect control by enzymatic inhibition. Curr. Med. Chem. 12, 461–469. doi: 10.2174/0929867053363126
Hiruma, K., and Riddiford, L. M. (2001). Regulation of transcription factors MHR4 and βFTZ-F1 by 20-hydroxyecdysone during a larval molt in the tobacco hornworm, Manduca sexta. Dev. Biol. 232, 265–274. doi: 10.1006/dbio.2001.0165
Hong, S., Qin, G., Wang, W., Hu, S., Fang, F., LV, Y., et al. (2014). Identification of differentially expressed microRNAs in Culex pipiens and their potential roles in pyrethroid resistance. Insect Biochem. Mol. Biol. 55, 39–50. doi: 10.1016/j.ibmb.2014.10.007
Hu, W., Criscione, F., Liang, S., and Tu, Z. (2015). MicroRNAs of two medically important mosquito species: Aedes aegypti and Anopheles stephensi. Insect Mol. Biol. 24, 240–252. doi: 10.1111/imb.12152
Hua, J. V., Narry, K., and Seogang, H. (2012). Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila. Genes Dev. 26, 1427–1432. doi: 10.1101/gad.192872.112
Huang, Y., Dou, W., Liu, B., Wei, D., Liao, C. Y., Smagghe, G., et al. (2014). Deep sequencing of small RNA libraries reveals dynamic expression patterns of microRNAs in multiple developmental stages of Bactrocera dorsalis. Insect Mol. Biol. 23, 656–667. doi: 10.1111/imb.12111
Jagadeeswaran, G., Zheng, Y., Sumathipala, N., Jiang, H., Arrese, E. L., Soulages, J. L., et al. (2010). Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics 11, 564–572. doi: 10.1186/1471-2164-11-52
Jain, S., Rana, V., Tridibes, A., Sunil, S., and Bhatnagar, R. K. (2015). Dynamic expression of miRNAs across immature and adult stages of the malaria mosquito Anopheles stephensi. Parasites Vec. 8, 1–20. doi: 10.1186/s13071-015-0772-y
Jesus, L., Raúl, M. E., and Xavier, B. (2015). MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 112, 3740–3745. doi: 10.1073/pnas.1418522112
Jiang, J., Ge, X., Li, Z., Wang, Y., Song, Q., Stanley, D. W., et al. (2013). MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform B in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 43, 692–700. doi: 10.1016/j.ibmb.2013.05.002
Kurita, K. (2006). Chitin and chitosan: functional biopolymers from marine crustaceans. Mar. Biotechnol. 8, 203–226. doi: 10.1007/s10126-005-0097-5
Legeai, F., Rizk, G., Walsh, T., Edwards, O., Gordon, K., Lavenier, D., et al. (2010). Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acyrthosiphon pisum. BMC Genomics 11:281. doi: 10.1186/1471-2164-11-281
Li, J., Cai, Y., Ye, L., Wang, S., Che, J., You, Z., et al. (2014). MicroRNA expression profiling of the fifth-instar posterior silk gland of Bombyx mori. BMC Genomics 15:410. doi: 10.1186/1471-2164-15-410
Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. doi: 10.1093/bioinformatics/btn025
Li, T., Chen, J., Fan, X., Chen, W., and Zhang, W. (2017). MicroRNA and dsRNA targeting chitin synthase A reveal a great potential for pest management of the hemipteran insect Nilaparvata lugens. Pest Manag. Sci. 73, 1529–1537. doi: 10.1002/ps.4492
Lin, L., Xie, G., Li, Z., Zeng, B., Xu, J., Aslam, A. F. M., et al. (2014). MicroRNA Let-7 regulates molting and metamorphosis in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 53, 13–21. doi: 10.1016/j.ibmb.2014.06.011
Ling, L., Kokoza, V. A., Zhang, C., Aksoy, E., and Raikhel, A. S. (2017). MicroRNA-277 targets insulin-like peptides 7 and 8 to control lipid metabolism and reproduction in Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 114, E8017–E8024. doi: 10.1073/pnas.1710970114
Liu, N., Landreh, M., Cao, K., Abe, M., Hendriks, G. J., Kennerdell, J. R., et al. (2012). The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 482, 519–523. doi: 10.1038/nature10810
Liu, S., Li, D., Li, Q., Zhao, P., Xiang, Z., and Xia, Q. (2010). MicroRNAs of Bombyx mori identified by Solexa sequencing. BMC Genomics 11:148. doi: 10.1186/1471-2164-11-148
Liu, S., Zhang, L., Li, Q., Zhao, P., Duan, J., Cheng, D., et al. (2009). MicroRNA expression profiling during the life cycle of the silkworm (Bombyx mori). BMC Genomics 10:455. doi: 10.1186/1471-2164-10-455
Liu, X.-P., Fu, K.-Y., Lü, F.-G., Meng, Q.-W., Guo, W.-C., and Li, G.-Q. (2014). Involvement of FTZ-F1 in the regulation of pupation in Leptinotarsa decemlineata. Insect Biochem. Mol. Biol. 55, 51–60. doi: 10.1016/j.ibmb.2014.10.008
Liu, Z., Ling, L., Xu, J., Zeng, B., Huang, Y., Shang, P., et al. (2018). MicroRNA-14 regulates larval development time in Bombyx mori. Insect Biochem. Mol. Biol. 93, 57–65. doi: 10.1016/j.ibmb.2017.12.009
Lomate, P. R., Mahajan, N. S., Kale, S. M., Gupta, V. S., and Giri, A. P. (2014). Identification and expression profiling of Helicoverpa armigera microRNAs and their possible role in the regulation of digestive protease genes. Insect Biochem. Mol. Biol. 54, 129–137. doi: 10.1016/j.ibmb.2014.09.008
Mortensen, R. D., Serra, M., Steitz, J. A., and Vasudevan, S. (2011). Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs). Proc. Natl. Acad. Sci. U.S.A. 108, 589–607. doi: 10.1073/pnas.1105401108
Niwa, Y. S., and Niwa, R. (2016). Transcriptional regulation of insect steroid hormone biosynthesis and its role in controlling timing of molting and metamorphosis. Dev. Growth Differ. 58, 94–105. doi: 10.1111/dgd.12248
Pan, Z. H., Wu, P., Gao, K., Hou, C. X., Qin, G. X., Geng, T., et al. (2017). Identification and characterization of two putative microRNAs encoded by Bombyx mori cypovirus. Virus Res. 233, 86–94. doi: 10.1016/j.virusres.2017.03.009
Peng, W., Zheng, W. W., Tariq, K., Yu, S. N., and Zhang, H. Y. (2017). MicroRNA Let-7 targets the ecdysone signaling pathway E75 gene to control larval-pupal development in Bactrocera dorsalis. Insect Sci. doi: 10.1111/1744-7917.12542. [Epub ahead of print].
Retnakaran, A., Krell, P., Feng, Q., and Arif, B. (2003). Ecdysone agonists: mechanism and importance in controlling insect pests of agriculture and forestry. Arch. Insect Biochem. Physiol. 54, 187–199. doi: 10.1002/arch.10116
Rubio, M., Horna, A., and Belles, X. (2012). MicroRNAs in metamorphic and non-metamorphic transitions in hemimetabolan insect metamorphosis. BMC Genomics 13:386. doi: 10.1186/1471-2164-13-386
Sempere, L. F., Sokol, N. S., Dubrovsky, E. B., Berger, E. M., and Ambros, V. (2003). Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev. Biol. 259, 9–18. doi: 10.1016/S0012-1606(03)00208-2
Shan, Q., Breuker, C. J., and Holland, P. W. (2015). A diversity of conserved and novel ovarian microRNAs in the speckled wood (Pararge aegeria). PLoS ONE 10:e142243. doi: 10.1371/journal.pone.0142243
Soko, W., Chimbari, M. J., and Mukaratirwa, S. (2015). Insecticide resistance in malaria-transmitting mosquitoes in Zimbabwe: a review. Infect. Dis. Poverty 4:46. doi: 10.1186/s40249-015-0076-7
Suh, Y. S., Bhat, S., Hong, S. H., Shin, M., Bahk, S., Cho, K. S., et al. (2015). Genome-wide microRNA screening reveals that the evolutionary conserved miR-9a regulates body growth by targeting sNPFR1/NPYR. Nat. Commun. 6:7693. doi: 10.1038/ncomms8693
Sultan, A. R., Oish, Y., and Ueda, H. (2014). Function of the nuclear receptor FTZ-F1 during the pupal stage in Drosophila melanogaster. Dev. Growth Differ. 56, 245–253. doi: 10.1111/dgd.12125
Tan, A., and Palli, S. R. (2008). Identification and characterization of nuclear receptors from the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 38, 430–439. doi: 10.1016/j.ibmb.2007.09.012
Tan, L., Yu, J. T., Tan, M. S., Liu, Q. Y., Wang, H. F., Zhang, W., et al. (2014). Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer's disease. J. Alzheimers Dis. 40, 1017–1027. doi: 10.3233/JAD-132144
Tariq, K., Peng, W., Saccone, G., and Zhang, H. (2015). Identification, characterization and target gene analysis of testicular microRNAs in the oriental fruit fly Bactrocera dorsalis. Insect Mol. Biol. 25, 32–43. doi: 10.1111/imb.12196
Tharanathan, R. N., and Kittur, F. S. (2003). Chitin–the undisputed biomolecule of great potential. Crit. Rev. Food Sci. Nutr. 43, 61–87. doi: 10.1080/10408690390826455
Varghese, J., and Cohen, S. M. (2007). microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 21, 2277–2282. doi: 10.1101/gad.439807
Vasudevan, S., Tong, Y., and Steitz, J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934. doi: 10.1126/science.1149460
Wang, H.-B., Ali, S. M., Moriyama, M., Iwanaga, M., and Kawasaki, H. (2012). 20-hydroxyecdysone and juvenile hormone analog prevent precocious metamorphosis in recessive trimolter mutants of Bombyx mori. Insect Biochem. Mol. Biol. 42, 102–108. doi: 10.1016/j.ibmb.2011.11.002
Wang, Y., Jiang, F., Wang, H., Song, T., Wei, Y., Yang, M., et al. (2015). Evidence for the expression of abundant microRNAs in the locust genome. Sci. Rep. 5:13608. doi: 10.1038/srep13608
Wang, Y. L., Yang, M. L., Jiang, F., Zhang, J. Z., and Kang, L. (2013). MicroRNA-dependent development revealed by RNA interference-mediated gene silencing of LmDicer1 in the migratory locust. Insect Sci. 20, 53–60. doi: 10.1111/j.1744-7917.2012.01542.x
Wei, L. Q., Yan, L. F., and Wang, T. (2011). Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol. 12:R53. doi: 10.1186/gb-2011-12-6-r53
Woodard, C. T., Baehrecke, E. H., and Thummel, C. S. (1994). A molecular mechanism for the stage specificity of the Drosophila prepupal genetic response to ecdysone. Cell 79, 607–615. doi: 10.1016/0092-8674(94)90546-0
Wu, W., Xiong, W., Li, C., Zhai, M., Li, Y., Ma, F., et al. (2017). MicroRNA-dependent regulation of metamorphosis and identification of microRNAs in the red flour beetle, Tribolium castaneum. Genomics 109, 362–373. doi: 10.1016/j.ygeno.2017.06.001
Xu, L. N., Ling, Y. H., Wang, Y. Q., Wang, Z. Y., Hu, B. J., Zhou, Z. Y., et al. (2015). Identification of differentially expressed microRNAs between Bacillus thuringiensis Cry1Ab-resistant and-susceptible strains of Ostrinia furnacalis. Sci. Rep. 5:15461. doi: 10.1038/srep15461
Xue, J., Zhou, X., Zhang, C. X., Yu, L. L., Fan, H. W., Wang, Z., et al. (2014). Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 15:521. doi: 10.1186/s13059-014-0521-0
Yamada, M.-A., Murata, T., Hirose, S., Lavorgna, G., Suzuki, E., and Ueda, H. (2000). Temporally restricted expression of transcription factor betaFTZ-F1: significance for embryogenesis, molting and metamorphosis in Drosophila melanogaster. Dev. 127, 5083–5092.
Zakrzewski, A. C., Weigert, A., Helm, C., Adamski, M., Adamska, M., Bleidorn, C., et al. (2014). Early divergence, broad distribution, and high diversity of animal chitin synthases. Genome Biol. Evol. 6, 316–325. doi: 10.1093/gbe/evu011
Zhang, X., Yun, Z., Jagadeeswaran, G., Ren, R., Sunkar, R., and Jiang, H. (2014b). Identification of conserved and novel microRNAs in Manduca sexta and their possible roles in the expression regulation of immunity-related genes. Insect Biochem. Mol. Biol. 47, 12–22. doi: 10.1016/j.ibmb.2014.01.008
Zhang, X., Zheng, Y., Cao, X., Ren, R., Yu, X. Q., and Jiang, H. (2014a). Identification and profiling of Manduca sexta microRNAs and their possible roles in regulating specific transcripts in fat body, hemocytes, and midgut. Insect Biochem. Mol. Biol. 62, 11–22. doi: 10.1016/j.ibmb.2014.08.006
Zhang, Y., Zhou, X., Ge, X., Jiang, J., Li, M., Jia, S., et al. (2009). Insect-specific microRNA involved in the development of the silkworm Bombyx mori. PLoS ONE 4:e4677. doi: 10.1371/journal.pone.0004677
Zhou, B., Hiruma, K., Jindra, M., Shinoda, T., Segraves, W. A., Malone, F., et al. (1998). Regulation of the transcription factor E75 by 20-hydroxyecdysone and juvenile hormone in the epidermis of the tobacco hornworm, Manduca sexta, during larval molting and metamorphosis. Dev. Biol. 193, 127–138. doi: 10.1006/dbio.1997.8798
Zhou, G., Wang, T., Lou, Y., Cheng, J., Zhang, H., and Xu, J. H. (2014). Identification and characterization of microRNAs in small brown planthopper (Laodephax striatellus) by next-generation sequencing. PLoS ONE 9:e103041. doi: 10.1371/journal.pone.0103041
Keywords: nlu-miR-173, NlFtz-F1, molting, Nilaparvata lugens, pest control
Citation: Chen J, Li TC, Pang R, Yue XZ, Hu J and Zhang WQ (2018) Genome-Wide Screening and Functional Analysis Reveal That the Specific microRNA nlu-miR-173 Regulates Molting by Targeting Ftz-F1 in Nilaparvata lugens. Front. Physiol. 9:1854. doi: 10.3389/fphys.2018.01854
Received: 19 September 2018; Accepted: 07 December 2018;
Published: 20 December 2018.
Edited by:
Jin-Jun Wang, Southwest University, ChinaReviewed by:
Yifan Zhai, Shandong Academy of Agricultural Sciences, ChinaXiao-Yue Hong, Nanjing Agricultural University, China
Copyright © 2018 Chen, Li, Pang, Yue, Hu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Qing Zhang, bHNzendxQG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work
 Jie Chen†
Jie Chen† Rui Pang
Rui Pang Wen Qing Zhang
Wen Qing Zhang