
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 18 September 2018
Sec. Avian Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.01298
 Wen Luo1,2
Wen Luo1,2 Xueyi Ren1,2
Xueyi Ren1,2 Jiahui Chen1,2
Jiahui Chen1,2 Limin Li1,2
Limin Li1,2 Shiyi Lu1,2
Shiyi Lu1,2 Tian Chen1,2
Tian Chen1,2 Qinghua Nie1,2
Qinghua Nie1,2 Xiquan Zhang1,2*
Xiquan Zhang1,2*Tumor protein 63 (TP63) comprises multiple isoforms and plays an important role during embryonic development. It has been shown that TP63 knockdown inhibits myogenic differentiation, but which isoform is involved in the underlying myogenic regulation remains uncertain. Here, we found that two transcripts of TP63, namely, TAp63α and ΔNp63α, are expressed in chicken skeletal muscle. These two transcripts have distinct expression patterns and opposite functions in skeletal muscle development. TAp63 has higher expression in skeletal muscle than in other tissues, and its expression is gradually upregulated during chicken primary myoblast differentiation. ΔNp63 can be expressed in multiple tissues and exhibits stable expression during myoblast differentiation. TAp63α overexpression inhibits myoblast proliferation, induces cell cycle arrest, and enhances myoblast differentiation. However, although ΔNp63α has no significant effect on cell proliferation, the overexpression of ΔNp63α inhibits myoblast differentiation. Using isoform-specific overexpression assays following RNA-sequencing, we identified potential downstream genes of TAp63α and ΔNp63α in myoblast. Bioinformatics analyses and experimental verification results showed that the differentially expressed genes (DEGs) between the TAp63α and control groups were enriched in the cell cycle pathway, whereas the DEGs between the ΔNp63α and control groups were enriched in muscle system process, muscle contraction, and myopathy. These findings provide new insights into the function and expression of TP63 during skeletal muscle development, and indicate that one gene may play two opposite roles during a single cellular process.
Tumor protein 63 is a p53 family member required for limb, craniofacial and epithelial development (Yang et al., 1999). Unlike for p53, several messenger RNAs are transcribed from the TP63 gene due to the use of two promoters and to alternative splicing (Lin et al., 2015). These mRNAs encode at least six TP63 isoforms (Guo and Mills, 2007). Isoforms with the N-terminal transactivation (TA) domain are referred to as the TA isoforms, and the N-terminal truncated (ΔN) isoform lacks the TA domain. It is well known that TP63 is involved in the formation of the epidermis. However, different TP63 isoforms perform different functions during epithelial development. ΔNp63 isoforms are important for maintaining the proliferative potential of the basal layer, whereas TAp63 isoforms contribute to late stage differentiation in mature keratinocytes (Candi et al., 2007). Different TP63 isoforms probably regulate gene sets that have completely distinct biological functions (Wu et al., 2003), and different isoforms may perform cell-type specific functions (Guo and Mills, 2007). For example, TAp63α promotes proliferation in the mouse epidermis (Koster et al., 2006), while it induces apoptosis in Hep3B cells (Gressner et al., 2005). ΔNp63 overexpression promotes HNSCC cell survival (Rocco et al., 2006), while it induces apoptosis in the non-small cell lung carcinoma cell line H1299 (Lo et al., 2006). Therefore, it is important to distinguish the different functions of TP63 isoforms during different cellular processes.
Skeletal muscle development is a complex process that is regulated at multiple levels. Many transcription factors and miRNAs are involved in the regulation of myogenesis (Braun and Gautel, 2011; Luo et al., 2013). It has been shown that p53 family members play a role in controlling myogenic differentiation (Cam et al., 2006). The p53 protein transactivates the RB gene, which plays a critical role in cell cycle exit in differentiated myocytes (Novitch et al., 1996). p63 and p73 induce the transcription of p57, maintain RB protein activity, and facilitate myogenic differentiation (Cam et al., 2006). However, the isoforms of TP63 have never been addressed in these studies. Which isoform is expressed during myogenic differentiation, and which isoform plays a major role in myogenic differentiation remain unclear. Recently, it was found that one of the TP63 isoforms, TAp63gamma, is involved in myogenic differentiation, and that the knockdown of TAp63 inhibited myotube formation (Cefalu et al., 2015). However, there are no results describing the expression and function of any other TP63 isoforms.
miR-203 is widely known as a skin-specific miRNA that plays an important role in epidermal development (Yi et al., 2008). miR-203 can regulate epidermal stratification and differentiation by directly repressing the expression of TP63 (Lena et al., 2008). However, in our previous work, we found that the “skin-specific miRNA” miR-203 could also be expressed in and function in the development of skeletal muscle (Luo et al., 2014). During muscle differentiation, miR-203 inhibits myoblast proliferation and differentiation by repressing c-Jun and MEF2C, respectively. In addition to c-Jun and MEF2C, TP63 was also found to be a direct target gene of miR-203 in skeletal muscle. Considering that TP63 has diverse transcripts and plays roles in muscle development in mammals, here, we explored its transcription, expression, and functional significance in chicken myoblast proliferation and differentiation. These results were important for understanding the function and regulation of TP63 isoforms in myogenesis.
This study was carried out in accordance with the principles of the Basel Declaration and recommendations of the Statute on the Administration of Laboratory Animal, the South China Agriculture University Institutional Animal Care and Use Committee. The protocol was approved by the South China Agriculture University Institutional Animal Care and Use Committee (approval ID: 2017046).
The embryonic and 7-week-old Xinghua female chickens were used in this study. For qPCR of TP63 in different tissues, the tissues were isolated from four 7-week-old Xinghua female chickens. For primary myoblast isolation, at least six embryos at embryo day 11 (E11) were used in each experiment. The sex of each embryos was determined by PCR with the sex-specific primers (Li et al., 2017).
Chicken embryo fibroblast cell line was cultured in high-glucose Dulbecco’s modified Eagle’s medium (Gibco) with 10% fetal bovine serum and 0.2% penicillin/streptomycin. The isolation and culture of chicken primary myoblasts were carried out as previously described (Li et al., 2017).
Total RNA was extracted from tissues or cells using RNAiso reagent (Takara, Otsu, Japan). The reverse transcription reaction for mRNA was performed with PrimeScript RT reagent Kit with gDNA Eraser (Takara) according to manufacturer’s manual. qPCR program was carried out in Bio-Rad CFX96 Real-Time Detection System (Bio-Rad, Hercules, CA, United States) with iTaqTM Universal SYBR® Green Supermix (Bio-Rad). All reactions were run in triplicate. The 2-ΔΔCt method was used to measure gene expression with β-actin as the reference gene (Kenneth and Thomas, 2001).
For 5′ RACE and 3′ RACE, total RNA isolated from chicken skeletal muscle and pooled total RNAs from different tissues were used. The detailed procedure was carried out according to previously described (Luo et al., 2015). All of the primers used in RACE were summarized in Supplementary Table S1.
The chicken primary myoblasts transfected with TAp63α, ΔNp63α, or GFP control overexpression vectors were harvested and total RNA was extracted using RNAiso reagent (Takara). Then the RNA samples were sent to Beijing Genomics Institute for RNA sequencing by using BGISEQ-500 (BGI, Wuhan, China). All the sequence data have been deposited in NCBI’s Gene Expression Omnibus (GEO1) and are accessible through GEO series accession number GSE114452.
Based on the TP63 mRNA sequence we obtained, primers for amplifying the TP63 3′ UTR region with predicted gga-miR-203 binding site were designed (Supplementary Table S1). The plasmid pmirGLO-TP63-3′UTR (wild-type) and pmirGLO-TP63-3′UTR-mutant (mutant with gga-miR-203 potential binding site deleted) were prepared for verification of target relationship between gga-miR-203 and TP63 mRNA. gga-miR-203 mimic (50 nM, RiboBio, Guangzhou, China) and pmirGLO-TP63-3′UTR (200 ng) were co-transfected into DF-1 cells (3 × 104 cells) by using Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s instructions. After 48 h, Luc-pair Duo-Luciferase Assay Kit 2.0 (GeneCopoeia, Rockville, MD, United States) was used to analyze the activities of luciferases. The luminescent signal was quantified using Synergy 2 Multi-mode Microplate Reader (Biotek, Winooski, VT, United States) and analyzed with Gene5 software (Biotek).
The TP63 overexpression vectors were constructed according to the user manual of Easy Ligation Kit (Sidansai, Shanghai, China). TAp63α and ΔNp63α coding sequences were amplified from chicken leg muscle cDNA by PCR. The PCR products were cloned into the pSDS-204 vector (Sidansai). The successful TAp63α and ΔNp63α overexpression vectors were confirmed by agarose gel electrophoresis and DNA sequencing.
Immunoblotting and immunofluorescence were performed as previously described (Luo et al., 2016). The following antibodies were used for immunoblotting: anti-MYOG (Biorbyt, Cambridge, United Kingdom), anti-MYOD (BD Biosciences, San Jose, CA, United States), anti-MyHC (Developmental Studies Hybridoma Bank, Iowa City, IA, United States) and anti-Tubulin (Bioworld, Minneapolis, MN, United States). The protein expression were presented as the ratio between indicated protein gray value and Tubulin gray value. We set the mean expression value of pSDS204-GFP group or si-NC group to 1, and the other group was a fold change comparing to the control group. Results are mean ± SEM from three independent experiments. The following antibody and reagent were used for immunofluorescence: anti-MyHC (DSHB), FITC-conjugated anti-mouse IgG (EarthOx, Millbrae, CA, United States), 4′6-diamidino-2-phenylindole (DAPI, Beyotime, Jiangsu, China).
After 48 h transfection of gene overexpression vectors, chicken primary myoblasts were collected and fixed in 75% ethanol overnight at -20°C. After ethanol fixation, the cells were stained with 50 μg/mL propidium iodide (Sigma) containing 10 μg/mL RNase A (Takara) and 0.2% (v/v) Triton X-100 (Sigma) for 30 min at 4°C. BD Accuri C6 flow cytometer (BD Biosciences) was subsequently used to analyze the cell cycle with Cell Cycle Analysis Kit (Thermo Fisher Scientific, Waltham, MA, United States), and the data analysis was performed using FlowJo 7.6 software (Verity Software House).
Primary myoblast were cultured in 96-well plates. A total of 10 μL of Cell counting kit-8 reagent (Dojindo, Kumamoto, Japan) was added into each well and incubated for 1 h. The assay was repeated at different time points of 12, 24, 36, 48 h after transfection. The absorbance was measured at 450 nm by a Model 680 Microplate Reader (Bio-Rad). All the data were acquired by averaging the results from six independent experiments.
Isoform-specific siRNAs against chicken TAp63α and ΔNp63α were all purchased from RiboBio (RiboBio, China). Target sequence of si-TAp63α is 5′-GGGACTTCCTGGAACAGCCAATATG-3′. Target sequence of si-ΔNp63α is 5′-CCGAGTCCTGTTATCTTCCAAGTAG-3′.
All data shown are mean ± SEM with at least three samples or cultures per group and three wells per culture. Well was considered the experimental unit for cell culture applications. We performed statistical analysis by using independent sample t-test through SPSS. We considered p < 0.05 to be statistically significant. ∗p < 0.05; ∗∗p < 0.01.
In our previous study (Luo et al., 2014), we found that the expression of TP63 was significantly downregulated when we transfected a gga-miR-203 mimic into chicken primary myoblasts. TargetScan (release 5.2) online software predicted that the TP63 mRNA is a direct target of gga-miR-203 (Figure 1A), and the predicted target site of gga-miR-203 in the 3′UTR of TP63 mRNA is highly conserved among vertebrates (Figure 1B). The dual-luciferase reporter gene assay confirmed that gga-miR-203 can directly bind to the predicted target site of gga-miR-203 in the 3′UTR of TP63 mRNA (Figure 1C). Considering that gga-miR-203 is a negative regulator of myogenic differentiation and that TP63 has been reported to play roles in myogenic differentiation, we next studied the roles of TP63 in chicken skeletal muscle differentiation. We synthesized a TP63 specific siRNA and found that this siRNA can efficiently inhibit TP63 expression (Figure 1D). Notably, TP63 knockdown significantly reduced the expression of MyHC (Figure 1E), which is a terminal myogenic differentiation marker gene. Therefore, these results suggested that TP63 is a gga-miR-203 target gene and is involved in myogenic differentiation.
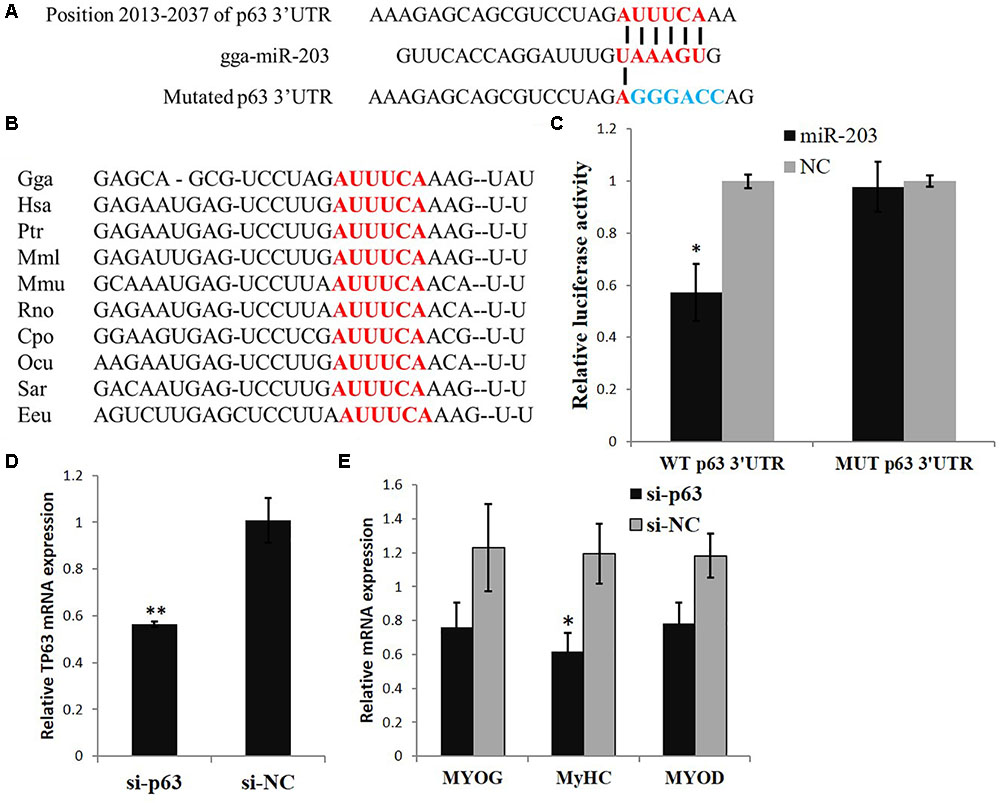
FIGURE 1. TP63 is a gga-miR-203 target gene and is involved in myogenic differentiation. (A) Predicted binding site and mutated site (blue) of gga-miR-203 in the 3′ UTR of TP63. (B) The predicted binding site of gga-miR-203 in the 3′UTR of TP63 is highly conserved among vertebrates. (C) Luciferase reporters were transfected into DF-1 cells with the gga-miR-203 mimic or negative control (NC) duplexes, and luciferase activity was determined 36 h after transfection. (D) TP63 specific siRNA transfection significantly inhibited TP63 mRNA expression in myoblast. (E) The expression of muscle differentiation marker genes in myoblast after the transfection of TP63-specific siRNA. The values are presented as the mean ± SEM (n = 3). An independent samples t-test was used to analyze the statistical differences between groups. ∗p < 0.05; ∗∗p < 0.01.
It is well-known that that TP63 gene has multiple transcripts in mammals. However, only one transcript has been identified in chickens; this transcript is short, without a 5′UTR or 3′UTR (NM_204351.1 and AB045224.1). To study the TP63 transcripts in chickens, we collected total mRNA from chicken embryonic skeletal muscle and pooled tissues, respectively. By using 5′ rapid-amplification of cDNA ends (RACE) and 3′ RACE, we found two TP63 transcripts existing in chicken tissues (accession number MH238465 and MH238464 in the NCBI database). One of the transcripts which we obtained from skeletal muscle mRNA, has a gene structure similar to that of the TP63 transcript in NCBI (NM_204351.1), but our transcript contained a 178 bp 5′UTR and a 2,721 bp 3′UTR sequence (Figure 2A). The other transcript was obtained from pooled tissues mRNA (Figure 2A). This transcript also had a 2,721 bp 3′UTR but the transcription start site was different from that of the first transcript (Figure 2A). By using ORFfinder, we obtained several potential ORFs in these two transcript (Supplementary Figure S1). The two longest ORFs among the predicted ORFs were then marked and subjected to BLAST analysis to find conserved proteins in the reference proteins database. The BLAST results showed that the ORF predicted from one of the chicken TP63 transcripts is conserved with TP63 isoform a (also known as TAp63α) in mice, whereas the other chicken TP63 transcript is conserved with TP63 isoform d (also known as ΔNp63α) in mice (Supplementary Figure S2). The BLAST search also showed that the chicken TAp63α and ΔNp63α have high percent identities to the homologous proteins in quail, ducks, humans, mice, rats, pigs, and cattle (Figures 2B,C). Amino acid alignment of the TAp63α and ΔNp63α proteins showed that these two proteins were strongly conserved among mammals and birds (Figures 2D,E). Therefore, these results suggested that TAp63α and ΔNp63α are two conserved transcripts expressed in chicken tissues.
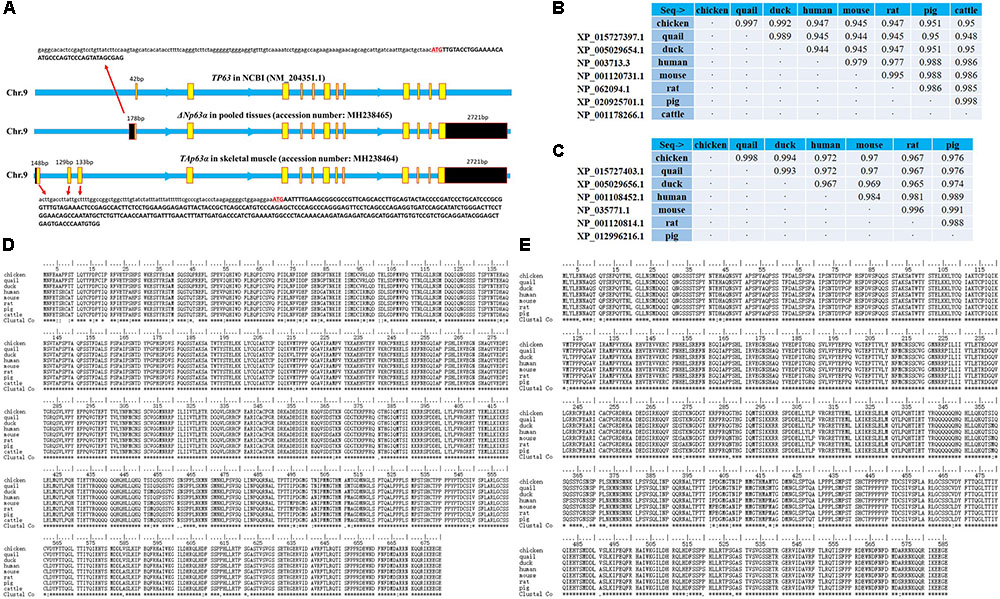
FIGURE 2. TAp63α and ΔNp63α are two conserved transcripts expressed in chicken tissues. (A) Genomic structure of the chicken TP63 gene. The yellow boxes indicate coding sequence regions, and the black boxes indicate the UTRs. The capital letters indicate coding sequences, and the lowercase letters indicate UTR sequences. (B) Percent identities of the chicken, quail, duck, human, mouse, rat, pig, and cow TAp63α amino acid sequences. (C) Percent identities of the chicken, quail, duck, human, mouse, rat, and pig ΔNp63α amino acid sequences. (D) Amino acid alignment of TAp63α proteins from chickens, quails, ducks, humans, mice, rats, pigs, and cattle. Conserved sequences are marked with asterisk within the Clustal Co sequences. (E) Amino acid alignment of ΔNp63α proteins from chickens, quails, ducks, humans, mice, rats, and pigs. Conserved sequences are marked with asterisks within the Clustal Co sequences. The data are the mean ± SEM of three cultures per group, and three wells per culture were assayed (n = 9/treatment group). An independent samples t-test was used to analyze the statistical differences between groups. ∗p < 0.05; ∗∗p < 0.01.
Next, we studied the expression of TAp63α and ΔNp63α in chicken tissues. Using TA- and ΔN-specific primers and a real-time polymerase chain reaction (qPCR) assay, we found that TAp63 has higher expression in skeletal muscle than in other tissues (Figure 3A), whereas ΔNp63 has high expression in bursal and thymus tissue and in skeletal muscle (Figure 3B). During myogenic differentiation, the expression of TAp63 was gradually upregulated, whereas the expression of ΔNp63 was relatively stable (Figure 3C). As Figures 1D,E show, our siRNA designed for TP63 was not isoform-specific (Supplementary Figure S3). To further study the functions of TAp63α and ΔNp63α in chickens, we constructed TAp63α and ΔNp63α overexpression vectors. Transfecting one of the TP63 overexpression vectors would upregulate the expression of that transcript without affecting the expression of the other transcript (Figure 3D). We then transfected these two vectors into chicken primary myoblasts, and induced the cells to differentiate. After 48 h, we found that TAp63α overexpression upregulated the mRNA and protein expression of MyHC (Figures 3E–G), which is a terminal marker of myogenic differentiation. However, ΔNp63α overexpression repressed MyHC expression (Figures 3E–G). Additionally, MyHC immunofluorescence showed that TAp63α and ΔNp63α have opposite effects on myotube formation (Figure 3H), as indicated by the quantification of myotube areas (Figure 3I). On the other hand, we used isoform-specific siRNAs to knockdown the expression of TAp63α and ΔNp63α (Figure 3J). TAp63α knockdown downregulated MyHC mRNA and protein expression, whereas ΔNp63α knockdown upregulated MyHC mRNA and protein expression (Figures 3K–M). Altogether, these results indicated that TAp63α and ΔNp63α play opposite roles in chicken myogenic differentiation.
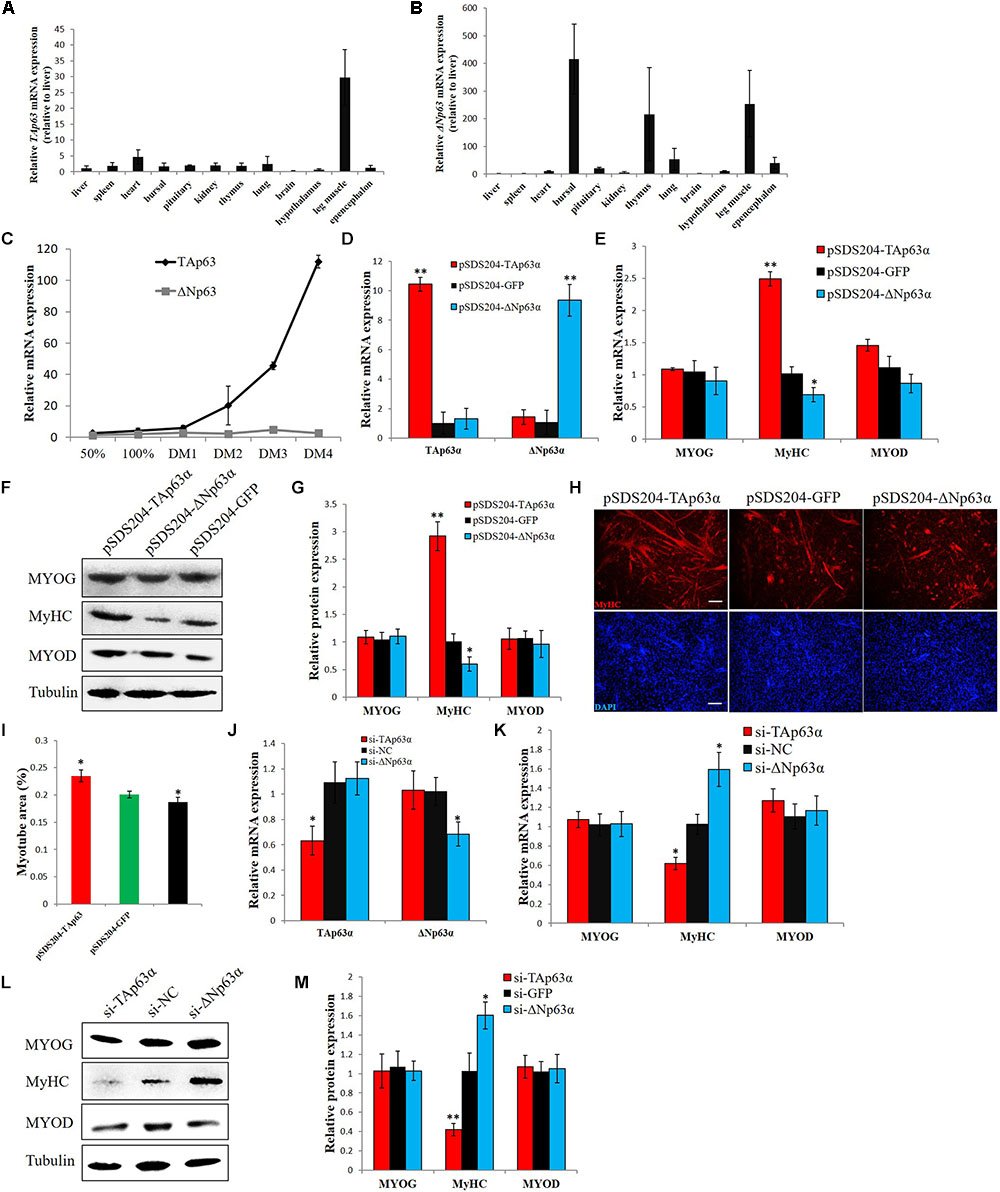
FIGURE 3. TAp63α and ΔNp63α play opposite roles in chicken myogenic differentiation. (A) qPCR for detecting the relative mRNA expression of TAp63 in 12 tissues from 7-week-old chickens. (B) qPCR for detecting the relative mRNA expression of ΔNp63 in 12 tissues from 7-week-old chickens. (C) Relative mRNA expression of TAp63 and ΔNp63 during chicken primary myoblast differentiation. (D) Relative mRNA expression of TAp63 and ΔNp63 after the transfection of pSDS204-TAp63α, pSDS204-ΔNp63α, or pSDS204-GFP. (E) Relative mRNA expression of muscle differentiation marker genes after the transfection of pSDS204-TAp63α, pSDS204-ΔNp63α, or pSDS204-GFP. (F) Protein expression of muscle differentiation marker genes after the transfection of pSDS204-TAp63α, pSDS204-ΔNp63α, or pSDS204-GFP. (G) Relative protein expression after transfection of pSDS204-TAp63α, pSDS204-ΔNp63α, or pSDS204-GFP. (H) MyHC immunostaining in primary myoblasts transfected with pSDS204-TAp63α, pSDS204-ΔNp63α, or pSDS204-GFP and differentiated for 48 h. The nuclei were visualized with DAPI. Bar, 100 μm. (I) Myotube area (%) at 48 h after the transfection of pSDS204-TAp63α, pSDS204-ΔNp63α, or pSDS204-GFP. (J) Relative mRNA expression of TAp63 and ΔNp63 after transfection of si-TAp63α, si-ΔNp63α, or si-NC. (K) Relative mRNA expression of muscle differentiation marker genes after transfection of si-TAp63α, si-ΔNp63α, or si-NC. (L) Western blotting results for muscle differentiation marker genes after the transfection of si-TAp63α, si-ΔNp63α, or si-NC. (M) Relative protein expression after the transfection of si-TAp63α, si-ΔNp63α, or si-NC. The data are the mean ± SEM of three cultures per group, and three wells per culture were assayed (n = 9/treatment group). An independent samples t-test was used to analyze the statistical differences between groups. ∗p < 0.05; ∗∗p < 0.01.
To study the downstream genes of TAp63α and ΔNp63α in chicken myoblast, we overexpressed these two transcripts in chicken primary myoblasts and collected the mRNA for RNA sequencing (RNA-seq). The RNA-seq results showed the successful overexpression of TAp63α and ΔNp63α in myoblasts (Figure 4A), and numerous differentially expressed genes (DEGs) between the groups (Figure 4B and Supplementary Table S2). From the gene expression heatmap we can see that the DEGs induced by TAp63α and ΔNp63α are very different (Figure 4B). TAp63α overexpression resulted in 1616 significantly DEGs, whereas ΔNp63α overexpression resulted in only 340 significantly DEGs (Figure 4C); furthermore, there were only 143 overlapping DEGs between these two groups (Figure 4C). GO analysis revealed that the TAp63α-induced DEGs are enriched in the cell cycle, DNA replication, and nucleotide binding terms (Figure 4D), whereas ΔNp63α-induced DEGs are enriched in the developmental process, regulation of biological process, and protein binding terms (Figure 4D). In addition, KEGG pathway analysis revealed that TAp63α-induced DEGs are enriched in the cell cycle pathway, whereas ΔNp63α-induced DEGs are enriched in the muscle development- or myopathy-related pathways (Figure 4E). Therefore, these results indicated that TAp63α and ΔNp63α regulate different set of genes in myoblasts.
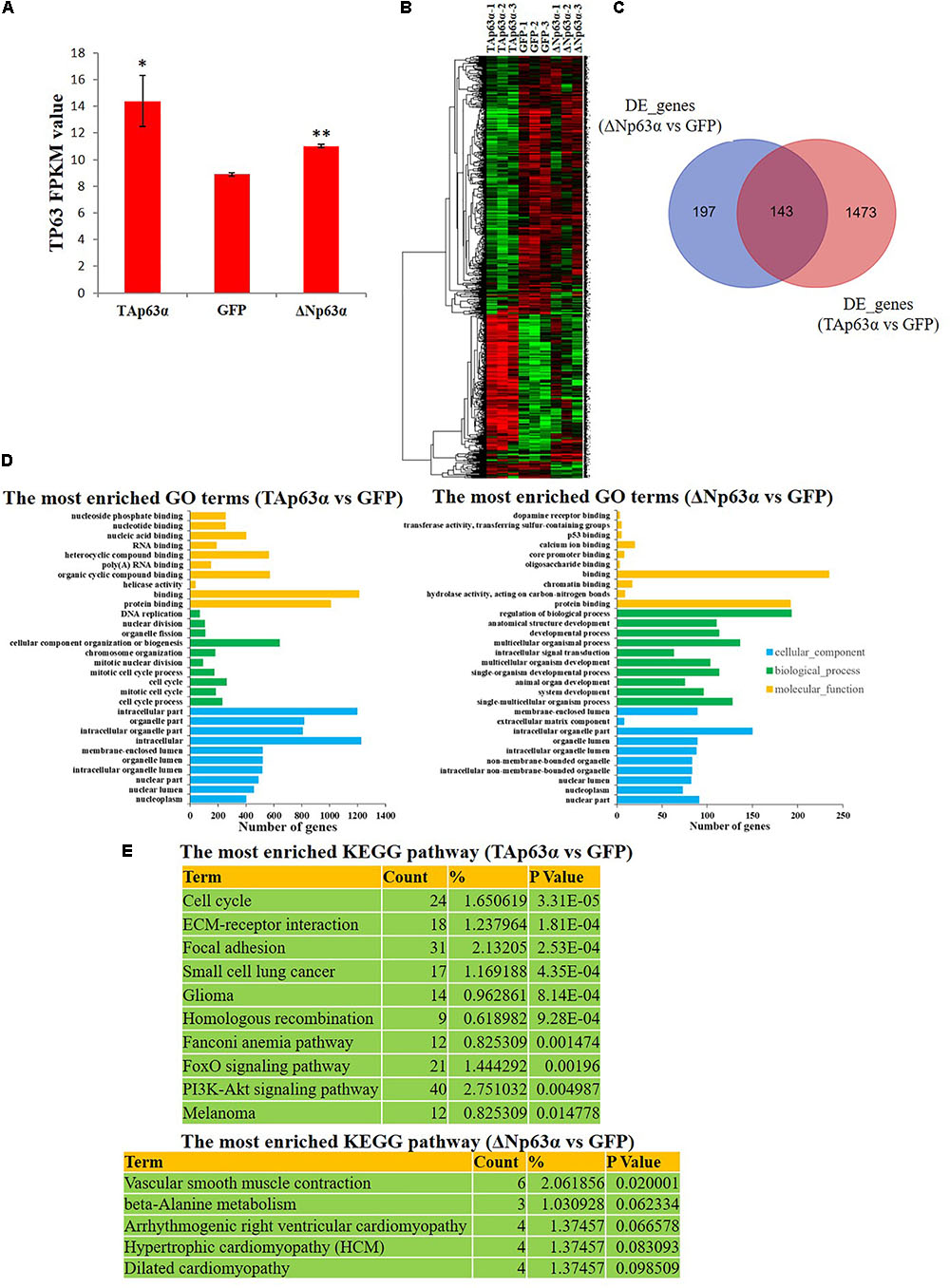
FIGURE 4. TAp63α and ΔNp63α regulate different sets of genes in myoblasts. (A) RNA-seq result showed the success of TAp63α and ΔNp63α overexpression in chicken primary myoblast. (B) Hierarchical clustering of differential expressed genes (DEGs) between TAp63α (TP), ΔNp63α (NP) and GFP (NC) overexpression groups. (C) The Venn diagram of DEGs between “ΔNp63α vs. NC” and “TAp63α vs. NC.” (D) GO enrichment for the DEGs between “TAp63α vs. NC” and “ΔNp63α vs. NC.” The y-axis represents GO terms and the x-axis represents number of genes (with p-value <0.05). (E) The most enriched KEGG pathway for the DEGs between “TAp63α vs. NC” and “ΔNp63α vs. NC.”
From the GO and KEGG pathway analysis results, we can see that the cell cycle is a potential target pathway of TAp63α. Many genes involved in the cell cycle pathway were differentially expressed in TAp63α-overexpressing myoblasts compared to control myoblasts (Figure 5A). Notably, TAp63α overexpression increased the number of cells in the G0/G1 stage and decreased the number of cells in the S stage (Figure 5B), whereas ΔNp63α overexpression decreased the number of cells in the G0/G1 stage (Figure 5B). The CCK-8 assay showed that TAp63α overexpression inhibited cell proliferation, whereas ΔNp63α overexpression had no significant effect on this process (Figure 5C). The qPCR results verified that the expression of many DEGs in the cell cycle pathway was significantly inhibited in TAp63α-overexpressing myoblasts but not in ΔNp63α-overexpressing cells (Figure 5D).
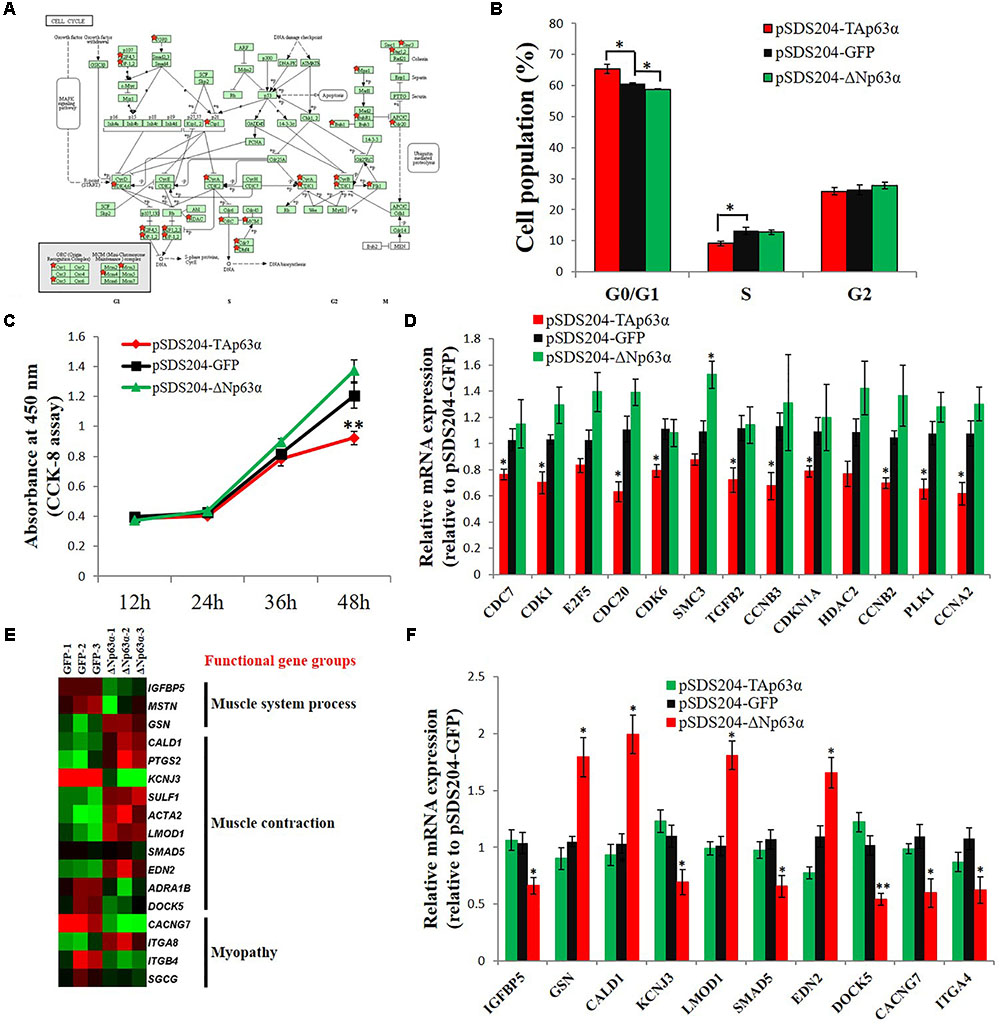
FIGURE 5. Identification of major downstream regulatory pathways and functional gene groups of TAp63α and ΔNp63α in myoblasts. (A) DEGs between TAp63α overexpression myoblast and NC myoblast were enriched in the cell cycle pathway. Red stars indicate the genes that are differentially expressed between TAp63α and NC group. (B) Myoblasts were transfected with pSDS204-TAp63α, pSDS204-ΔNp63α, or pSDS204-GFP, followed by 48 h culture, and the cell cycle phase was then analyzed. (C) CCK-8 assay was performed to access the effects of TAp63α and ΔNp63α overexpression on myoblast proliferation. (D) qPCR validation of the DEGs enriched in cell cycle pathway. (E) The heat-map of genes related to muscle system process, muscle contraction, and myopathy. (F) qPCR validation of the DEGs related to muscle system process, muscle contraction, and myopathy. The data are the mean ± SEM of three cultures per group, and three wells per culture were assayed (n = 9/treatment group). An independent samples t-test was used to analyze the statistical differences between groups. ∗p < 0.05; ∗∗p < 0.01.
From the GO and KEGG pathway analysis results, we found that many of the ΔNp63α downstream genes were involved in the muscle system process, muscle contraction, and myopathy (Figure 5E). Our qPCR results validated that ΔNp63α can regulate the expression of these genes (Figure 5F). Therefore, these results suggested that the cell cycle is a potential regulatory pathway targeted by TAp63α in myoblasts and that genes involved in muscle system process, muscle contraction, and myopathy were potential downstream targets of ΔNp63α in myoblasts.
In this study, we cloned the full-length cDNA of the chicken ΔNp63α, and found the full-length TAp63α transcript, which has never been reported in chickens. TAp63α and ΔNp63α have different expression patterns and perform different functions during myoblast differentiation. TAp63α inhibits myoblast proliferation and promotes myoblast differentiation by regulating cell cycle-related genes, whereas ΔNp63α inhibits myoblast differentiation by regulating genes related to muscle contraction, muscle system process, and myopathy (Figure 6).
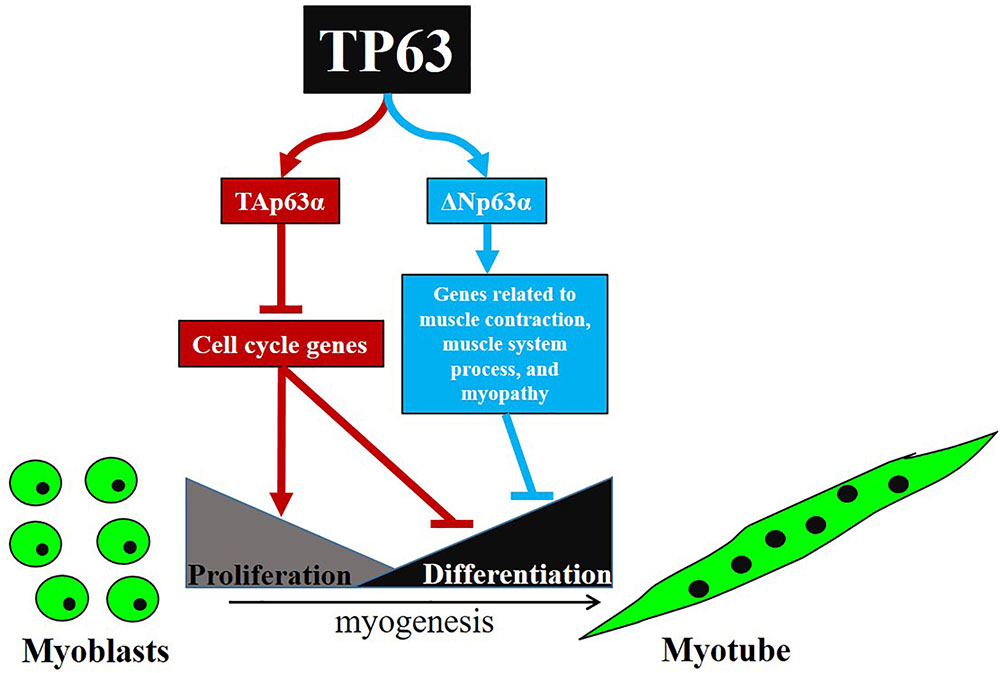
FIGURE 6. Schematic illustration for signaling pathways of myoblast proliferation and differentiation regulated by TP63 gene.
The TP63 gene has at least ten transcripts in humans, such as TAp63α, TAp63β, TAp63γ, TAp63δ, TAp63ε, ΔNp63α, ΔNp63β, ΔNp63γ, ΔNp63δ, and ΔNp63ε (Mangiulli et al., 2009). TA and ΔN represent 5′ variants, and α, β, γ, δ, and ε represent 3′ variants. However, we found only TAp63α and ΔNp63α in chicken tissues. The 5′RACE result identified the TA and ΔN transcripts, whereas 3′RACE identified only the α transcript. The other TP63 transcripts may also exist in chickens, but these transcripts may be expressed in different tissues with different time-course expression profiles. Because it is hard to design primers that can identify every single transcript, we used TA- and ΔN-specific primers to detect TAp63 and ΔNp63 in chicken tissues. Notably, TAp63 is mainly expressed in skeletal muscle, whereas ΔNp63 can be expressed in multiple tissues. The upregulation of TAp63 and the stable expression of ΔNp63 during chicken myoblast differentiation were consistent with the results in C2C7 (Cefalu et al., 2015). However, the isoform-specific expression of the TP63 transcripts during myoblast differentiation needs further investigation.
The TP63 transcripts encode the corresponding isoforms and play different roles in cellular processes. We found that TAp63α and ΔNp63α are not only differentially expressed but also play opposite roles in myogenic differentiation. Similarly, protein kinase C isoforms can play opposite roles in the proliferation, differentiation, and apoptosis of human HaCaT keratinocytes (Papp et al., 2004), and p38 isoforms exert opposite effects on MKK6-mediated VDR transactivation (Pramanik et al., 2003). These phenomena indicate that one gene can perform at least two different functions by expressing different isoforms during a single cellular process. However, the expression of these isoforms would be strictly controlled by gene expression regulation programs, such as alternative promoters, alternative splicing, and post-translational processing, so that the appropriate functional isoform is expressed at the appropriate time. In addition to playing opposite roles during a single cellular process, one TP63 isoform may influence the function of another during myogenic differentiation. For example, the upregulation of ΔNp63α inhibited myoblast differentiation, which was induced by TAp63α. A previous study showed that ΔNp63 can directly compete for TAp63 target promoters or sequester TAp63 to form inactive tetramers (Candi et al., 2007). Therefore, it is possible that the two isoforms compete for a sub-set of target genes during myogenic differentiation. In this case, identifying the target genes of these two isoforms is important in order to reveal the mechanism of action of TAp63α and ΔNp63α, as well as to confirm the interaction between these two isoforms.
TP63 is a well-known tumor suppressor gene that can regulate cell cycle progress and inhibit cancer cell proliferation (Benard et al., 2003). Here, we found that the TAp63α isoform is capable of inducing cell cycle arrest in myoblasts and is able to inhibit myoblast proliferation. Cell cycle arrest is important for myogenic differentiation. Myoblasts permanently exit from the cell cycle during terminal differentiation (Derer et al., 2002). The upregulation of TAp63α during myoblast differentiation may promote cell cycle arrest, therefore, facilitating the terminal differentiation of myoblasts. In addition, TP63 has been reported to play roles in the late stage of myogenic differentiation (Cefalu et al., 2015). The knockdown of TAp63gamma would affect the expression of genes related to myogenesis and skeletal muscle contractility (Cefalu et al., 2015). Our results also showed that TP63 isoforms could regulate myogenesis and muscle contraction and that the expression of many myogenic differentiation genes, such as MYH9, MYH10, RUNX1, ROCK1, ROCK5, MSTN, SMAD5, and CDKN1A, were significantly changed (Supplementary Table S2). Therefore, the TP63 gene is involved in skeletal muscle cell proliferation and differentiation.
TP63 is a conserved transcription factor with multiple binding sites in the genome (McDade et al., 2014). TP63 regulates the expression of downstream genes by through directly affecting the transcription of genes to whose promoter it binds (McDade et al., 2012). A better way to identify downstream genes of TP63 is via chromatin immunoprecipitation (ChIP)-related assays, such as ChIP-chip or ChIP-sequencing. However, there is no isoform-specific ChIP-grade antibody for TP63 in chickens. Furthermore, studies investigating the genome-wide binding of TP63 did not use isoform-specific antibodies (McDade et al., 2012, 2014). Not only will structural variations of protein isoforms affect protein function in cellular processes, but the binding sites will also be different (Kiselev et al., 2012). Therefore, it is important to develop isoform-specific antibodies for TP63 to better understand its genome-wide regulation in specific cell types. In this study, we used isoform-specific overexpression assays and identified a list of TAp63α- and ΔNp63α-specific potential downstream genes in myoblasts. Previous studies on TP63 in myogenesis used an siRNA strategy for functional investigation (Cam et al., 2006; Cefalu et al., 2015). The siRNAs designed to knockdown TP63 expression were not isoform-specific (Cam et al., 2006; Cefalu et al., 2015); therefore, it is hard to demonstrate the specific function of each TP63 isoforms in myogenesis. Here, we used an isoform-specific overexpression assay to investigate the function of TAp63α and ΔNp63α in myoblast proliferation and differentiation. Although this strategy is not optimal for screening the downstream target genes of TAp63α and ΔNp63α, our results identified the specific functions of these two isoforms in myoblast differentiation. In conclusion, TP63 is important for skeletal muscle development, and the isoforms of TP63, namely, TAp63α and ΔNp63α, play opposite roles in myoblast differentiation.
WL, QN, and XZ designed the experiments. WL and XZ wrote the manuscript. WL, XR, JC, LL, SL, and TC did the experiments.
This work was supported by Natural Scientific Foundation of China (31702105), Natural Scientific Foundation of China (31472090), the China Agriculture Research System (CARS-41-G03), and the Science and Technology Program of Guangzhou, China (201804020088).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer DLC and handling Editor declared their shared affiliation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01298/full#supplementary-material
DF-1, chicken embryo fibroblast cell line; DM, differentiation medium; GM, growth medium; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA, microRNA; NC, negative control; qPCR, quantitative polymerase chain reaction; TP63, tumor protein 63; UTR, untranslated region; w, week.
Benard, J., Douc-Rasy, S., and Ahomadegbe, J. C. (2003). TP53 family members and human cancers. Hum. Mutat. 21, 182–191. doi: 10.1002/humu.10172
Braun, T., and Gautel, M. (2011). Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 12, 349–361. doi: 10.1038/nrm3118
Cam, H., Griesmann, H., Beitzinger, M., Hofmann, L., Beinoraviciute-Kellner, R., Sauer, M., et al. (2006). p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell 10, 281–293. doi: 10.1016/j.ccr.2006.08.024
Candi, E., Dinsdale, D., Rufini, A., Salomoni, P., Knight, R. A., Mueller, M., et al. (2007). TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle 6, 274–285. doi: 10.4161/cc.6.3.3797
Cefalu, S., Lena, A. M., Vojtesek, B., Musaro, A., Rossi, A., Melino, G., et al. (2015). TAp63gamma is required for the late stages of myogenesis. Cell Cycle 14, 894–901. doi: 10.4161/15384101.2014.988021
Derer, W., Easwaran, H. P., Leonhardt, H., and Cardoso, M. C. (2002). A novel approach to induce cell cycle reentry in terminally differentiated muscle cells. FASEB J. 16, 132–133. doi: 10.1096/fj.01-0500fje
Gressner, O., Schilling, T., Lorenz, K., Schulze, S. E., Koch, A., Schulze-Bergkamen, H., et al. (2005). TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 24, 2458–2471. doi: 10.1038/sj.emboj.7600708
Guo, X., and Mills, A. A. (2007). p63, cellular senescence and tumor development. Cell Cycle 6, 305–311. doi: 10.4161/cc.6.3.3794
Kenneth, J. L., and Thomas, D. S. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Kiselev, Y., Eriksen, T. E., Forsdahl, S., Nguyen, L. H., and Mikkola, I. (2012). 3T3 cell lines stably expressing Pax6 or Pax6(5a)–a new tool used for identification of common and isoform specific target genes. PLoS One 7:e31915. doi: 10.1371/journal.pone.0031915
Koster, M. I., Lu, S. L., White, L. D., Wang, X. J., and Roop, D. R. (2006). Reactivation of developmentally expressed p63 isoforms predisposes to tumor development and progression. Cancer Res. 66, 3981–3986. doi: 10.1158/0008-5472.CAN-06-0027
Lena, A., Shalom-Feuerstein, R., Rivetti di Val Cervo, P., Aberdam, D., Knight, R., and Melino, G. (2008). miR-203 represses ‘stemness’ by repressing DNp63. Cell Death Differ. 15, 1187–1195. doi: 10.1038/cdd.2008.69
Li, G., Luo, W., Abdalla, B. A., Ouyang, H., Yu, J., Hu, F., et al. (2017). miRNA-223 upregulated by MYOD inhibits myoblast proliferation by repressing IGF2 and facilitates myoblast differentiation by inhibiting ZEB1. Cell Death Dis. 8:e3094. doi: 10.1038/cddis.2017.479
Lin, C., Li, X., Zhang, Y., Guo, Y., Zhou, J., Gao, K., et al. (2015). The microRNA feedback regulation of p63 in cancer progression. Oncotarget 6, 8434–8453. doi: 10.18632/oncotarget.3020
Lo, I. M., Di Costanzo, A., Calogero, R. A., Mansueto, G., Saviozzi, S., Crispi, S., et al. (2006). The Hay Wells syndrome-derived TAp63alphaQ540L mutant has impaired transcriptional and cell growth regulatory activity. Cell Cycle 5, 78–87. doi: 10.4161/cc.5.1.2268
Luo, W., Li, E., Nie, Q., and Zhang, X. (2015). Myomaker, regulated by MYOD, MYOG and miR-140-3p, promotes chicken myoblast fusion. Int. J. Mol. Sci. 16, 26186–26201. doi: 10.3390/ijms161125946
Luo, W., Li, G., Yi, Z., Nie, Q., and Zhang, X. (2016). E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop involved in myoblast proliferation and differentiation. Sci. Rep. 6:27904. doi: 10.1038/srep27904
Luo, W., Nie, Q., and Zhang, X. (2013). MicroRNAs involved in skeletal muscle differentiation. J. Genet. Genomics 40, 107–116. doi: 10.1016/j.jgg.2013.02.002
Luo, W., Wu, H., Ye, Y., Li, Z., Hao, S., Kong, L., et al. (2014). The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 5:e1347. doi: 10.1038/cddis.2014.289
Mangiulli, M., Valletti, A., Caratozzolo, M. F., Tullo, A., Sbisa, E., Pesole, G., et al. (2009). Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res. 37, 6092–6104. doi: 10.1093/nar/gkp674
McDade, S. S., Henry, A. E., Pivato, G. P., Kozarewa, I., Mitsopoulos, C., Fenwick, K., et al. (2012). Genome-wide analysis of p63 binding sites identifies AP-2 factors as co-regulators of epidermal differentiation. Nucleic Acids Res. 40, 7190–7206. doi: 10.1093/nar/gks389
McDade, S. S., Patel, D., Moran, M., Campbell, J., Fenwick, K., Kozarewa, I., et al. (2014). Genome-wide characterization reveals complex interplay between TP53 and TP63 in response to genotoxic stress. Nucleic Acids Res. 42, 6270–6285. doi: 10.1093/nar/gku299
Novitch, B. G., Mulligan, G. J., Jacks, T., and Lassar, A. B. (1996). Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J. Cell Biol. 135, 441–456. doi: 10.1083/jcb.135.2.441
Papp, H., Czifra, G., Bodo, E., Lazar, J., Kovacs, I., Aleksza, M., et al. (2004). Opposite roles of protein kinase C isoforms in proliferation, differentiation, apoptosis, and tumorigenicity of human HaCaT keratinocytes. Cell. Mol. Life Sci. 61, 1095–1105. doi: 10.1007/s00018-004-4014-2
Pramanik, R., Qi, X., Borowicz, S., Choubey, D., Schultz, R. M., Han, J., et al. (2003). p38 isoforms have opposite effects on AP-1-dependent transcription through regulation of c-Jun. The determinant roles of the isoforms in the p38 MAPK signal specificity. J. Biol. Chem. 278, 4831–4839. doi: 10.1074/jbc.M207732200
Rocco, J. W., Leong, C. O., Kuperwasser, N., DeYoung, M. P., and Ellisen, L. W. (2006). p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 9, 45–56. doi: 10.1016/j.ccr.2005.12.013
Wu, G., Nomoto, S., Hoque, M. O., Dracheva, T., Osada, M., Lee, C. C., et al. (2003). DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 63, 2351–2357.
Yang, A., Schweitzer, R., Sun, D., Kaghad, M., Walker, N., Bronson, R. T., et al. (1999). p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718. doi: 10.1038/19539
Keywords: TAp63α, ΔNp63α, chicken, myoblast differentiation, cell cycle
Citation: Luo W, Ren X, Chen J, Li L, Lu S, Chen T, Nie Q and Zhang X (2018) TP63 Transcripts Play Opposite Roles in Chicken Skeletal Muscle Differentiation. Front. Physiol. 9:1298. doi: 10.3389/fphys.2018.01298
Received: 15 May 2018; Accepted: 29 August 2018;
Published: 18 September 2018.
Edited by:
Sandra G. Velleman, The Ohio State University, United StatesReviewed by:
Takeshi Ohkubo, Ibaraki University, JapanCopyright © 2018 Luo, Ren, Chen, Li, Lu, Chen, Nie and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiquan Zhang, eHF6aGFuZ0BzY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.