- State Key Laboratory of Crop Stress Biology for Arid Areas, Key Laboratory of Crop Pest Integrated Pest Management on the Loess Plateau of Ministry of Agriculture, Northwest A&F University, Yangling, China
Carboxylesterase (CarE) is an important class of detoxification enzymes involved in insecticide resistance. However, the molecular mechanism of CarE-mediated insecticide resistance in Rhopalosiphum padi, a problematic agricultural pest, remains largely unknown. In the present study, an isoprocarb-resistant (IS-R) strain and a cyhalothrin-resistant (CY-R) strain were successively selected from a susceptible (SS) strain of R. padi. The enzyme activity indicated that enhanced carboxylesterase activity contributes to isoprocarb and cyhalothrin resistance. The expression levels of putative CarE genes were examined and compared among IS-R, CY-R, and SS strains, and only the R. padi carboxylesterase gene (RpCarE) was significantly over expressed in both the IS-R and CY-R strains compared to the SS strain. The coding region of the RpCarE gene was cloned and expressed in Escherichia coli. The purified RpCarE protein was able to catalyze the model substrate, α-naphtyl acetate (Kcat = 5.50 s-1; Km = 42.98 μM). HPLC assay showed that the recombinant protein had hydrolase activity against isoprocarb and cyhalothrin. The modeling and docking analyses consistently indicated these two insecticide molecules fit snugly into the catalytic pocket of RpCarE. Taken together, these findings suggest that RpCarE plays an important role in metabolic resistance to carbamates and pyrethroids in R. padi.
Introduction
The bird cherry-oat aphid, Rhopalosiphum padi, is a serious worldwide wheat pest. R. padi causes serious damage to wheat by directing extracting nourishment and transmitting barley yellow dwarf virus (Du et al., 2007; Wang et al., 2016). In China, aphid control is dependent primarily on the application of chemical insecticides, including carbamates, and pyrethroids. Unfortunately, the extensive application of insecticides has resulted in the development of resistance in field populations of R. padi (Chen et al., 2007; Zuo et al., 2016). Thus, uncovering the resistance mechanism and its molecular basis is essential to better control this important pest.
The two main mechanisms predominantly responsible for insecticide resistance are target site insensitivity and metabolic resistance due to elevated levels of insecticide detoxifying enzymes (Li et al., 2007; Liu, 2015; Barres et al., 2016). Biochemical assays have demonstrated the resistance to carbamates and organophosphates caused by insensitive acetylcholinesterase (ACE) in a lot of insect species (Hemingway et al., 2004; Bass et al., 2014). A S431F substitution of ACE-1 in Myzus persicae resistant clones was correlated with the resistance of the species to pirimicarb (Nabeshima et al., 2003). The A302S in ACE-1 was found to be associated with organophosphates resistance in Aphis gossypii (Li and Han, 2004). The substitutions in ACE disturbed both the space and hydrophobicity, thus, preventing pirimicarb from interacting with the active site in the resistant M. persicae and A. gossypii (Nabeshima et al., 2003; Andrews et al., 2004). Mutations in the voltage-gated sodium channel, a trans-membrane ion channel that plays an essential role in the initiation and propagation of action potentials in neurons, are involved in resistance to pyrethroids (Zlotkin, 1999). L1014F and M918T initially identified in the housefly and, respectively, referred as kdr and super-kdr mutations are two most common mutations in the voltage-gated sodium channel (Williamson et al., 1996; Franck et al., 2012). The kdr mutation can cause moderate resistance to DTT and pyrethroids, whereas the super-kdr mutation is usually linked to the kdr and shown to significantly enhance the resistance phenotypic expression due to kdr (Vais et al., 2000; Eleftherianos et al., 2008; Franck et al., 2012). The kdr or super-kdr mutations have been indentified in the pyrethroid resistant strains of several aphid species (Eleftherianos et al., 2008; Marshall et al., 2012; Foster et al., 2014). The metabolic resistance has evolved by the amplification, overexpression and coding sequence variation of three major detoxification enzymes, i.e., cytochrome P450 monooxygenases (P450s), carboxylesterases (CarE), and glutathione-S-transferases (GSTs) (Hemingway and Ranson, 2000; Hemingway et al., 2004; Li et al., 2007; Ramsey et al., 2010).
In recent years, R. padi had developed resistance to some insecticides (Zuo et al., 2016; Zhang et al., 2017). Regional susceptibility analyses of 12 R. padi field populations has shown that the populations varied in their resistance levels to the tested insecticides, with the highest resistant ratio of 13.6, 18.2, 13.1, and 12.1 to imidacloprid, bifenthrin, decamethrin, and abamectin, respectively (Zuo et al., 2016). Both insecticide target site mutations and metabolic resistance were found in this aphid. Acetylcholinesterase gene mutations might contribute to the resistance of this pest to organophosphate and carbamate insecticides (Chen et al., 2007; Bettaibi et al., 2016). Significant higher GST activity was found in two field populations collected in China (Zhang et al., 2017). In the imidacloprid resistant strain of R. padi, the CYP6CY3-1 and CYP6CY3-2 were significantly overexpressed (Wang et al., 2018). So far, information about CarE from R. padi and its role in insecticide resistance is still unavailable.
Carboxylesterases (CarEs, EC 3.1.1.1) are a superfamily of metabolic enzymes that hydrolyse carboxylic ester bonds with the addition of water and are thought to play important physiological roles in xenobiotic metabolism (Campbell et al., 2003; Wheelock et al., 2005; Birner-Gruenberger et al., 2012). The overexpression of CarEs has been associated with carbamate and pyrethroid resistance in several insects (Wheelock et al., 2005). Carboxylesterase E4, which is produced by resistant M. persicae, both hydrolyses and sequesters the insecticide, leading to carbamate and pyrethriod resistance in these aphids (Devonshire and Moores, 1982; Lan et al., 2005). Elevated esterase hydrolysis activity is related to cyhalothrin resistance in Aphis glycines (Xi et al., 2015).
In the present work, isoprocarb-resistant (IS-R) and cyhalothrin-resistant (CY-R) strains and a relatively susceptible (SS) strain were obtained from the same field population by successive selection with or without insecticide. CarE activity was evaluated among the IS-R, CY-R, and SS strains, and the expression patterns of seven putative CarE or CarE-likes genes were studied by RT-PCR, with the target gene (RpCarE) being confirmed and cloned. We heterologously expressed RpCarE in Escherichia coli cells and purified the fusion protein. Moreover, we measured the activity of the fusion protein against the standard substrate (α-naphthyl acetate) and examined the hydrolase activity against isoprocarb and cyhalothrin. Homology modeling and insecticide docking studies were also conducted to interpret the substrate metabolic detoxification. The current results will contribute to understanding of the mechanism of insecticide resistance mediated by RpCarE.
Materials and Methods
Insects
The susceptible strain (SS) originated from a field R. padi population collected in 2013 from Gansu Province, China. The isoprocarb resistant strain (IS-R) and cyhalothrin resistant strain (CY-R) were successively selected by exposing isoprocarb or cyhalothrin for more than 60 generations. The IS-R strain showed an LC50 of 33.436 mg L-1 for isoprocarb, with an ∼32.4-fold increased resistance compared with the SS (LC50 of 1.032 mg L-1 for isoprocarb). The CY-R strain (LC50 of 8.858 mg L-1 for cyhalothrin) displayed ∼27.8-fold increased resistance compared to the SS (LC50 of 0.319 mg L-1 for cyhalothrin) (Supplementary Table S1). During the insecticide selection, the toxicity of insecticides was evaluated every four generations, and the evaluated LC50 values were used as the selection concentration for the following four generations. Resistance ratio = LC50 of resistant strain/LC50 of susceptible strain. All insects were reared on seedlings of wheat (cultivar “Xiaoyan 22”) in mesh cages (41 cm × 41 cm × 41 cm) in the laboratory at 23 ± 1°C, 70% relative humidity and a photoperiod of L16:D8.
Chemicals
The insecticides used for bioassays included isoprocarb (95% purity, Anhui Huaxing Chemical industry Co. Ltd., China) and cyhalothrin (96% purity, Yancheng Nongbo Bio-technology Co. Ltd., China).
α-naphthol (α-N) and fast blue RR salt were products of Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China). α-naphthyl acetate (α-NA) was purchased from Solarbio (Beijing, China). All other chemicals were of analytical grade and purchased from commercial suppliers.
Carboxylesterase Activity Assays
Carboxylesterase (CarE) activity was determined by the method of Han et al. (2012) and Li et al. (2016) with modification. All aphids tested were fed on seedlings (three-leaf stage) of wheat cultivar “Xiaoyan 22” under the aforementioned condition, and did not contact with any insecticides from the first instar to the adult before enzyme activity assays. Ten 9-days old apterous adult specimens were taken from each of the three strains (IS-R, CY-R, and SS), homogenized on ice in 1 mL of pre-chilled PBS (0.1 mol L-1, pH 7.5, containing 1.0 × 10-3 mol L-1 EDTA) and centrifuged at 4°C and 12,000 × g for 10 min. The enzyme source was supernatant of the homegenized specimens. Supernatants were used for testing. Protein concentrations were determined using the Bradford method with bovine serum albumin as the standard (Bradford, 1976). The assay mixture contained 100 μL of substrate solution (10 mM α-NA and 3 mM Fast Blue RR salt, pH 6.0) and 100 μL of enzyme solution. After incubation at 30°C for 10 min, assays were conducted at 30°C in 96-well microplates, and absorbance was measured at 450 nm in a microplate reader (M200 PRO, Tecan, Männedorf, Switzerland). The experiment was repeated three times.
Screening of R. padi CarE Genes Associated With Insecticide Resistance
Putative CarE genes were systematically searched based on R. padi transcriptome data (Duan et al., 2017) and re-sequenced via PCR amplification. Sequences of the seven CarE genes obtained were deposited in GenBank database. The GenBank accession numbers for each sequence are: CL2012, MH561903; CL3077, MH561904; CL869, MH561905; U11937, MH561906; U14486, MH561907; U4474, MH561908; and U6896, MH561909. Ten apterous adult aphids from each of the three strains (IS-R, CY-R, and SS) were subjected to total RNA extraction using Trizol (Invitrogen, Carlsbad, CA, United States) and Direct-zol RNA miniprep kit (Zymo, Irvine, CA, United States) according to the manufacturer’s protocol, including a DNAse treatment. First-strand cDNA was synthesized using the M-MLV reverse transcriptase cDNA Synthesis Kit (Promega, Madison, WI, United States). RT-qPCR was performed in a final volume of 20 μL, including 10 μL of FastStart Essential DNA Green Master (Roche, NJ, United States), 0.8 μL of each specific primer (Table 1), 2 μL of cDNA template, and 6.4 μL of RNase-free water. The reaction was performed with the thermal cycler program: 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 58°C for 20 s, and 72°C for 20 s. A melting curve was determined (ramping from 55°C to 95°C by 0.5°C every 5 s) to confirm the amplification of specific PCR products. The R. padi β-actin and EF-1α (elongation factor 1α) genes were used as the internal control (Table 1). RT-qPCR was performed on a LightCycler Nano System (Roche, Mannheim, Germany), and the relative expression level was calculated using the 2-ΔΔCt method (Pfaffl, 2001). qPCR experiments were repeated using three biological replicates using aphids from the same generation, and each replicate was performed at least three times.
Cloning of Carboxylesterase From R. padi
Based on the above-mentioned screening of CarE genes from R. padi transcriptome data (Duan et al., 2017), a carboxylesterase gene (CL2012, hereinafter referred as RpCarE)) which was over-expreesed in the resistant strains was confirmed by qPCR, and the coding region was cloned by RT-PCR. Pre-analysis showed the 5′ sequences of RpCarE from R. padi transcriptome were sufficient for protein expression. Gene-specific primes were used for 3′-RACE (RpCarE-3R1 and RpCarE-3R2) to clone the 3′ sequences of the gene. To confirm the accuracy of the RpCarE linked from the 3′-RACE results, a specific primer pair (RpCarE-CF and RpCarE-CR) was designed to amplify the full coding region of the gene. The primers used for RACE are shown in Table 1. The amplification reaction mix contained 2 units of Taq DNA polymerase (5 U/μL, Sangon Biotech Co. Ltd., Shanghai, China), 100 μM dNTPs, 4 mM MgCl2, 0.4 μM of forward and reverse primers, and 1 μL of template DNA. All purified PCR products were cloned into pGEM-T easy vectors (Promega, Madison, WI, United States) and transformed into Escherichia coli DH5α competent cells (Takara, Kyoto, Japan). To ensure that the correct cDNA sequences were obtained, three positive clones from each sample were randomly chosen for bidirectional sequencing on an Applied Biosystems 3730 automated sequencer (Applied Biosystems, Foster City, CA, United States).
Sequence Analysis
Sequence identification and similarities were analyzed using BLAST1. Amino acid sequences of RpCarE and homologs from other insect species were aligned using ClustalW2 software (Larkin et al., 2007). The molecular weight (MW) and theoretical isoelectric point (pI) of RpCarE were calculated using the ExPASy (Gasteiger et al., 2003). Signal peptides were predicted using the Signal P4.1 software (Petersen et al., 2011).
Protein Expression/Purification and Western Blot Analysis
The encoding region of RpCarE was amplified from the pGEM-T/RpCarE plasmid with gene specific primers (RpCarE -BamHI: CGCGGATCCATGGAAGTGGTCATCGAACAAGGT; RpCarE-HindIII: CCCAAGCTTTTAAACAATGGATTCTTTTATTAA) introducing BamHI and HindIII restriction sites (underlined). Amplicon was purified and digested with corresponding restriction enzymes and subcloned into pET-28a (Novagen, Merck, Germany). The plasmid was verified by restriction digestion and nucleotide sequencing. The expression of RpCarE in the E. coli BL-21 (DE-3) strain (Takara, Kyoto, Japan) was induced using Isopropyl β-D-thiogalactoside (IPTG, with final concentration of 0.4 mM). Cultured cells were harvested by centrifugation (6,000 × g, 20 min, 4°C). The resulting pellet was re-suspended in 50 mM Tris-HCl (pH 8.0) and lysed by sonication (Sonics Vibra-Cell, 130 w, 30% power). Cell lysate was centrifuged at 12,000 × g for 30 min, and the supernatant was incubated with 2 mL of cOmplete His-Tag Purification Resin (Roche) for 1 h with shaking, and protein was eluted using lysis buffer supplemented with 50 mM imidazole. Protein concentrations were determined using the Bradford method with bovine serum albumin as the standard (Bradford, 1976).
Recombinant RpCarE was analyzed using 12% SDS-PAGE and stained with Coomassie Brilliant Blue R-250. For western blot, separated proteins were electrotransferred to a PVDF membrane (Millipore), and the membrane was subsequently blocked with 5% skim milk powder in TBST for 1 h. Target proteins were verified with mouse anti-His mouse monoclonal antibodies (CWBIO) followed by staining with goat-anti-mouse IgG (CWBIO). Blots were visualized using WesternBright ECL kit (Advasta), and signal was detected using a chemiluminescence imaging system (Clinx Science Instruments, Shanghai, China).
Determination of Enzymatic Activity
The kinetics of purified RpCarE protein against α-NA was determined using the method of Li et al. (2016) with modification. Reactions were carried out in a 96-well microplate, with each well containing 100 μL of appropriately diluted purified protein in Tris-HCl buffer and 100 μL of α-NA in buffer (3 mM Fast Blue RR salt and increasing concentrations of α-NA). The formation of α-N was recorded at 450 nm for 5 min in a microplate reader (M200 PRO, Tecan, Männedorf, Switzerland) and quantified using α-N standard curves. The values for kcat and Km were estimated using the “Hyper32” hyperbolic regression software (Xie et al., 2014).
HPLC Analysis of Insecticide Metabolism
Assays for the hydrolysis of isoprocarb and cyhalothrin were carried out by monitoring substrate loss with a Hitachi D-2000 Elite HPLC system (Hitachi High-Technologies Corporation, Tokyo, Japan) as described by Li et al. (2016), with some modifications. First, 100 μL of isoprocarb and cyhalothrin (each in 100 μM) was incubated separately with 100 μL of recombinant RpCarE protein (0.2 mg mL-1). Insecticides with heat-inactivated enzyme served as controls. Incubation reactions were carried out at 30°C for 1 h and stopped by addition of 100 μL acetonitrile. Samples were centrifuged at 12,000 × g for 10 min before transferring the supernatant to HPLC vials. Ten microliter of the supernatant were injected onto a reverse-phase Symmetry C18 column (250 mm × 4.6 mm, 5 μm, Waters Crop., Milford, MA, United States) with a flow rate of 1 mL min-1 at 30°C for 30 min. Reactions were run with a mobile phase (80% acetonitrile: 20% water) and monitored at 215 and 230 nm for isoprocarb and cyhalothrin, respectively.
RpCarE Modeling and Substrate Docking
The molecular model of RpCarE was created as described by Zhang (2008) and Elzaki et al. (2017) using the I-TASSER on-line server2. Isoprocarb and cyhalothrin molecules were docked into the active site of RpCarE using the Surflex-Dock (SFXC) function in the SYBYLx2.0 software (Tripos, St. Louis, MO, United States). Final figures were prepared using the PyMOL program (DeLano, 2002).
Statistical Analysis
The significance of the differences in mRNA levels and enzyme activities was determined by non-parametric Mann–Whitney U-test with the level of significance at P < 0.05. Data analyses were performed using SPSS Version 21.0 software (SPSS Inc., Chicago, IL, United States).
Results
Determination of CarE Activity
Carboxylesterase activities of different strains were determined using α-NA as a substrate. IS-R and CY-R strains exhibited significantly higher carboxylesterase activity (2.20- and 2.05-fold) compared with the susceptible strain (P < 0.05) (Figure 1 and Supplementary Table S2).
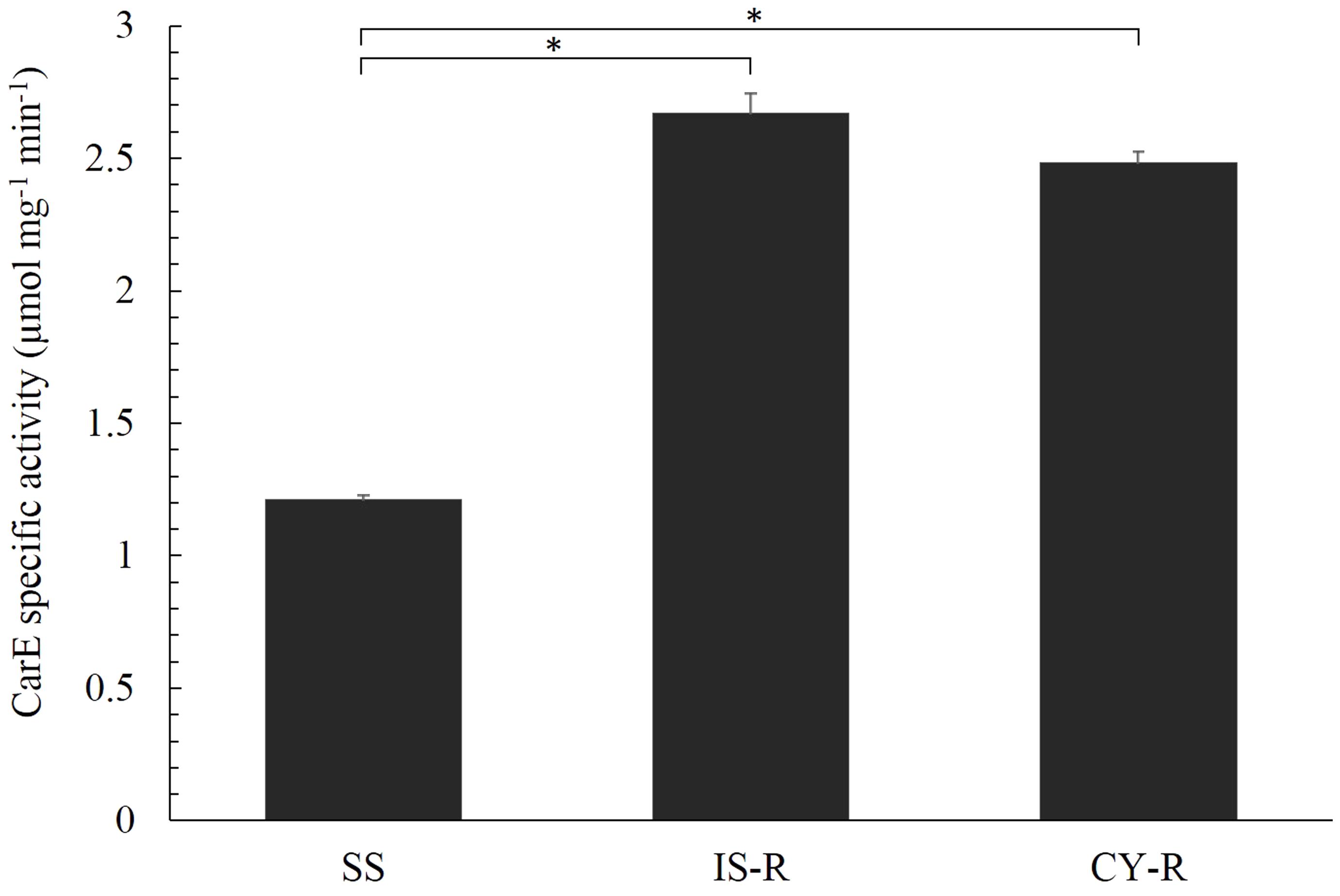
FIGURE 1. Specific activity of R. padi carboxylesterase for α-NA in IS-R, CY-R, and SS strains. The results of three replicates were averaged from three different protein preparations. Vertical bars represent the standard error of the mean from three independent replicates. Asterisk indicates significant differences by Mann–Whitney U-test (P < 0.05).
CarE Gene mRNA Level Determination
Seven putative carboxylesterase genes were obtained from the transcriptome database of R. padi (Table 1), and compared among susceptible and resistant strains. Relative expression levels of CL2102 were 4.99- and 2.73-fold higher in IS-R and CY-R than in the susceptible strain, respectively (Figure 2). Relative expression analysis of the seven carboxylesterase genes revealed that CL2102 was moderately abundant in the isoprocarb resistant and cyhalothrin resistant strains of R. padi, whereas the other six carboxylesterase genes was lowly abundant in the two resistant strains.
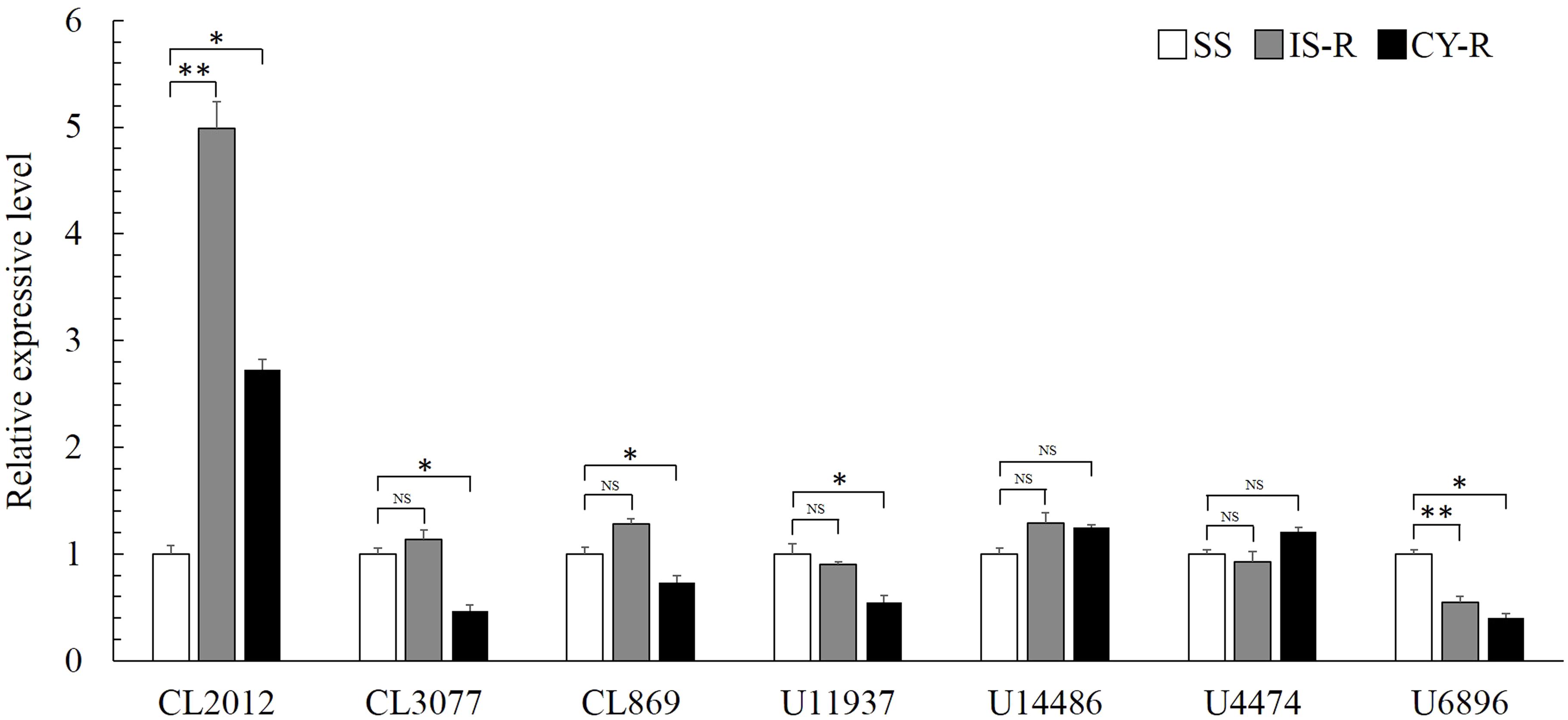
FIGURE 2. Comparison of carboxylesterase gene mRNA levels among the isoprocarb resistant (IS-R), cyhalothrin resistant (CY-R), and susceptible (SS) strains. Values are plotted as the mean ± SE of three repeats. Vertical bars represent the standard error of the mean of three independent replicates. Asterisk indicates significant differences by Mann–Whitney U-test (∗P < 0.05, ∗∗P < 0.01).
cDNA Amplification
To obtain the complete sequence, gene-specific primers for CL2102 were designed for amplification of the 3′ cDNA ends. The resulting carboxylesterase cDNA sequence contains an open reading frame (ORF) of 1,581 bp that encodes a putative protein containing 526 amino acid residues. Its calculated molecular weight is 59.22 kDa, and the predicted isoelectric point is 6.61. No signal peptide was detected upon amino acid sequencing. Homology analysis against the already published genes in GenBank indicated that carboxylesterase shares high identity with M. persicae FE4-like esterase (XP_022165140; 80% identity), Acyrthosiphon pisum FE4 esterase (XP_001951456; 80% identity), A. glycines carboxylesterase (AEI70326; 78% identity), and A. gossypii carboxylesterase (BAE66715; 78% identity). The alignment shows that this R. padi carboxylesterase has highly conserved residues that form an atalytic triad (S185-H432-E312) (Figure 3).
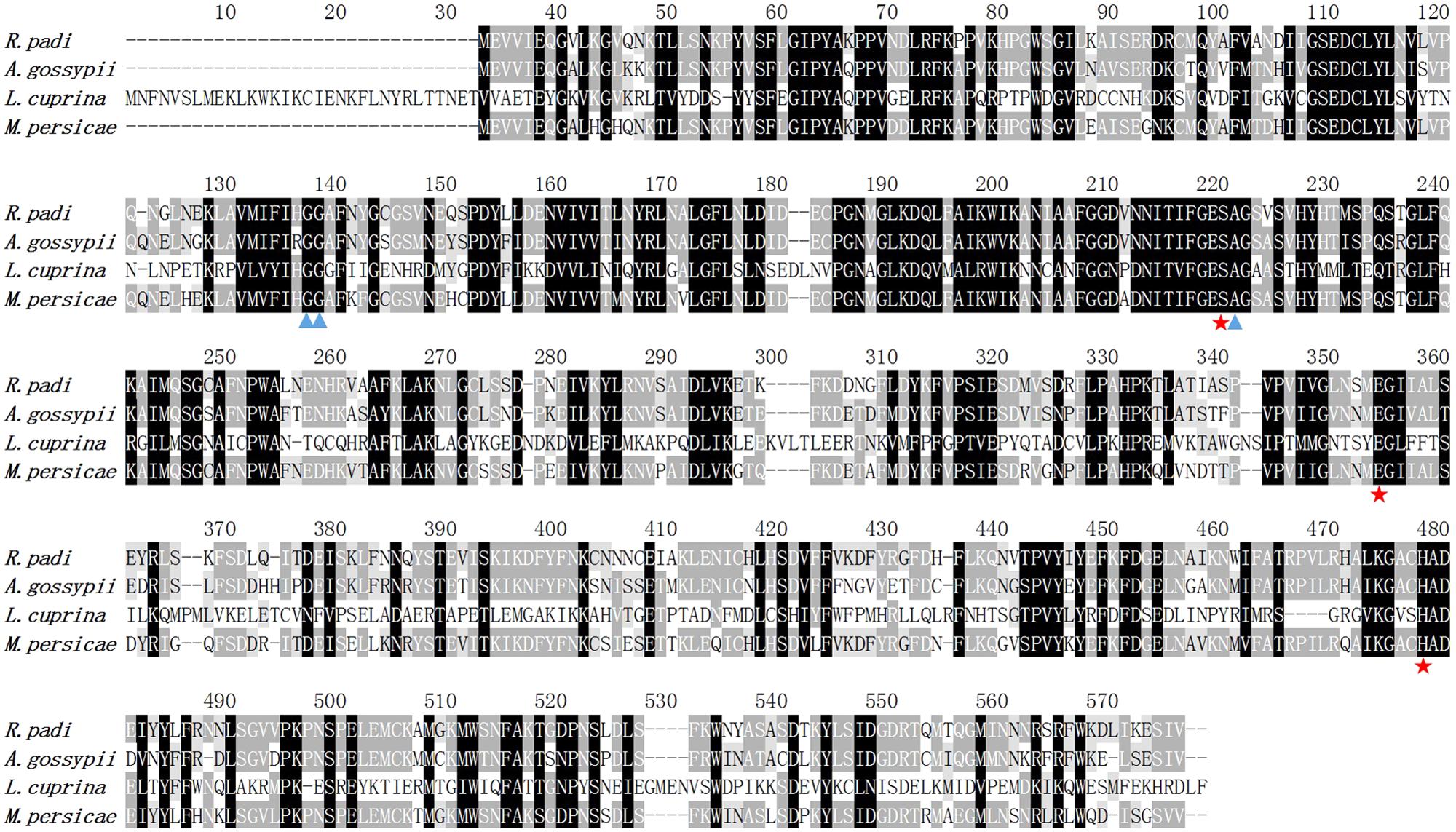
FIGURE 3. Alignment of the deduced amino acid sequences of R. padi carboxylesterase with the previously reported carboxylesterases from A. gossypii (GenBank accession number XP_022165140), E3 from Lucilia cuprina (GenBank accession number XP_022165140), and FE4-like from M. persicae (GenBank accession number XP_022165140).
Expression of RpCarE in E. coli and Verification via Western Blot
To functionally express RpCarE, its coding sequence was inserted into the pET-28a expression vector and expressed in the BL-21 (DE-3) E. coli strain. Cells harvested 12 h after IPTG induction were considered to be the most suitable for protein expression and isolation based on the Coomassie blue-stained sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) images (data not shown). Immunoreactivity of the target proteins for His-mouse antibodies in the western blot are shown in Figure 4. The fusion protein with a His6 tag migrated as a single band with a molecular mass of ∼60 kDa, which matched the calculated molecular mass of 59.22 kDa. These results indicate that RpCarE was successfully expressed in E. coli.
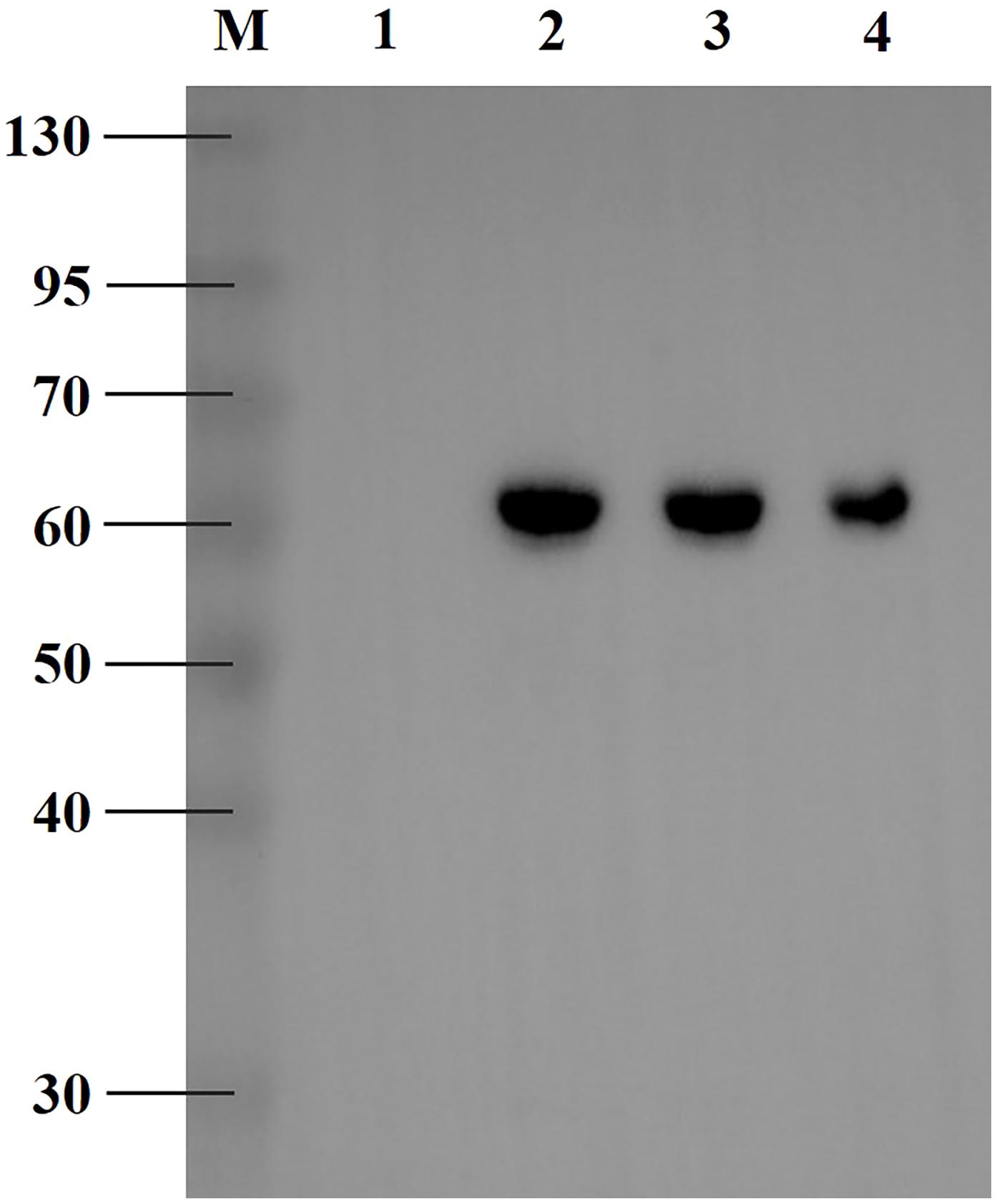
FIGURE 4. Western blot analysis of exogenously expressed RpCarE. The RpCarE protein was verified via His-mouse antibody. M, protein molecular markers; line 1, total proteins from uninduced E. coli cells; line 2, total proteins from induced E. coli cells; line 3, soluble proteins from induced E. coli cells; line 4, purified fusion protein.
RpCarE Activity for α-Naphthyl Acetate
The activity of the purified recombinant enzyme toward α-NA is shown in Table 2. RpCarE exhibited high catalytic efficiency with a Km of 42.98 μM, a Kcat of 5.50 s-1 and a specific activity of 4.43 μM s-1(μM-1⋅protein). These results suggest that the purified recombinant protein (RpCarE) has a high affinity and turnover of substrate.

TABLE 2. Kinetic parameters for the purified R. padi carboxylesterase toward the α-naphthyl acetate.
RpCarE Metabolism of Isoprocarb and Cyhalothrin
Specific activities of the purified recombinant enzyme against isoprocarb and cyhalothrin were assessed by measuring substrate depletion. As shown in Figure 5, the fusion protein exhibited significant activity toward both isoprocarb and cyhalothrin.
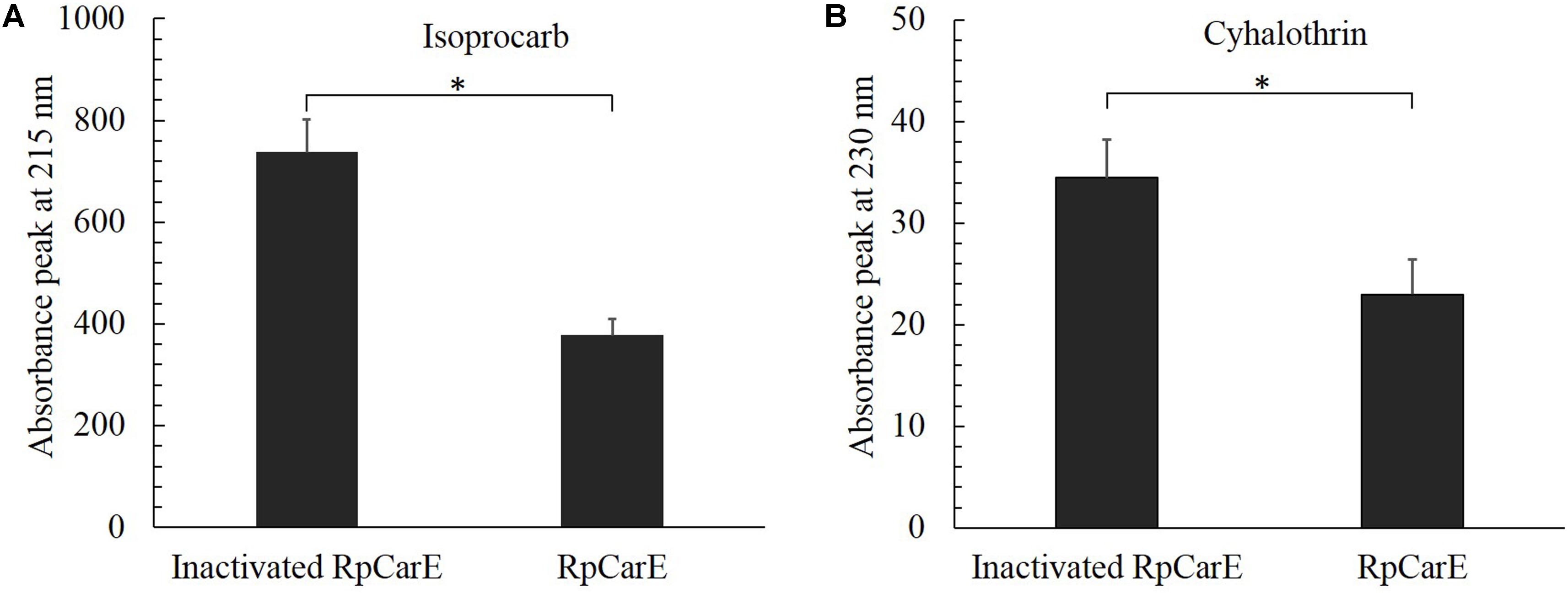
FIGURE 5. Hydrolytic activity of purified fusion RpCarE protein against isoprocarb (A) and cyhalothrin (B). Insecticide metabolism was analyzed using a Hitachi D-2000 Elite HPLC system. Isoprocarb and cyhalothrin were monitored via their absorption at 215 and 230 nm, respectively, and quantified via their peak integration. A control was prepared using heat-inactivated RpCarE. Values are shown as the mean ± SE of three repeats. Vertical bars represent the standard error of the mean of three independent replicates. Asterisk indicates significant differences by Mann–Whitney U-test (P < 0.05).
The Binding Mode of Insecticides to RpCarE
To better understand the underlying mechanism causing RpCarE to metabolize insecticides, a molecular docking simulation was conducted using the homology mode of RpCarE with insecticides, including isoprocarb and cyhalothrin. The docking experiments were conducted using the Surflex-Dock (SFXC) program from the SYBYL X2.0 software. The RpCarE protein model was generated based on the crystal structure of Lucilia cuprina α-E7. RpCarE contains several conserved CarE characteristics, such as the canonical catalytic triad (serine, glutamate, and histidine) and the oxyanion hole (alanine and glycine) (Figure 3). Modeling and docking analyses showed that isoprocarb and cyhalothrin fit snugly into the catalytic pocket of RpCarE (Figure 6). The Asn-108 residue was predicted to anchor isoprocarb by hydrogen donors (Figure 6A). Tyr-109 and Glu-116 were the major determinants in cyhalothrin binding, positioning the molecule close to catalytic triads (Ser185-His432-Glu312) (Figure 6B). These results indicate that the active site pocket of RpCarE is ideally shaped for isoprocarb and cyhalothrin and is able to effectively metabolize these insecticides.
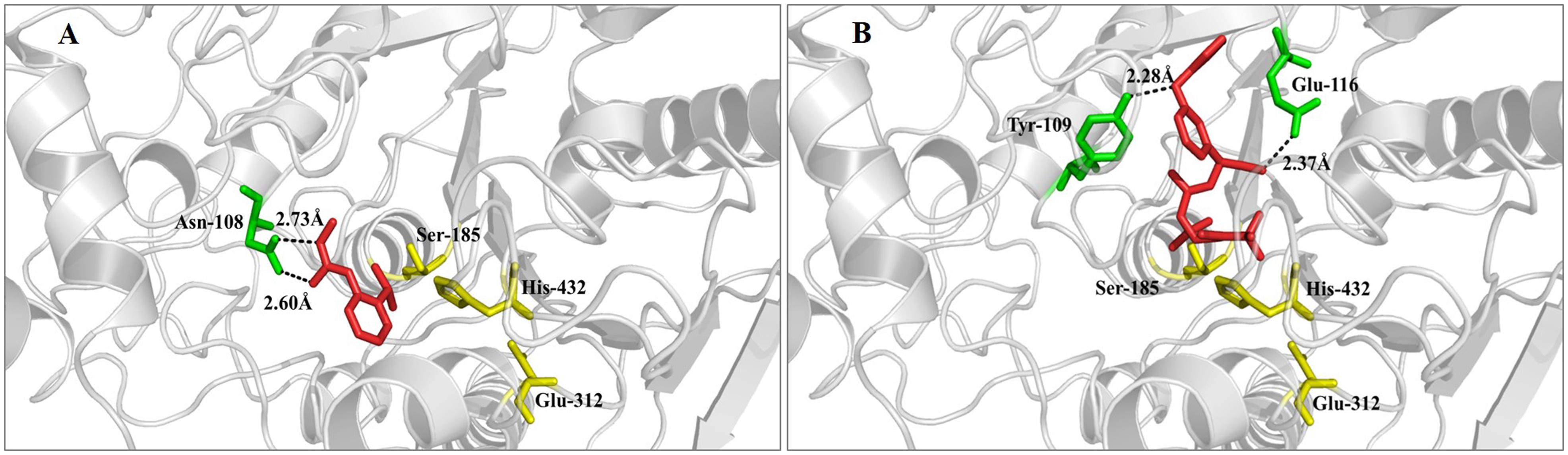
FIGURE 6. Binding models of the complex formed by RpCarE and insecticides. (A) Isoprocarb-RpCarE, (B) cyhalothrin-RpCarE. Insecticides are displayed as red sticks. Residues of the catalytic triad (serine, glutamate, and histidine) are represented as yellow sticks. Other residues that interact with the substrate are shaded green.
Discussion
Carboxylesterases, an important detoxification enzyme family, are involved in mediating the metabolic resistance to carbamate, organophosphate and pyrethroid insecticides in many insect species (Wheelock et al., 2005). To reveal the CarE-mediated metabolism of isoprocarb and cyhalothrin in R. padi, one isoprocarb resistance strain (IS-R), and one cyhalothrin resistance strain (CY-R) were obtained by successive selection with insecticides. CarE activities were measured in IS-R, CY-R and susceptible strains of R. padi, yielding significantly higher activities in the IS-R and CY-R strains than in the SS strain (Figure 1), suggesting a correlation between CarE and isoprocarb and cyhalothrin resistance. Furthermore, the hydrolytic activity of a R. padi CarE (RpCarE) to isoprocarb and cyhalothrin suggest that RpCarE is involved in resistance to the chemicals. In the early 1970s, resistant strains of M. persicae were shown to possess improved hydrolytic activity against the esterase standard substrate α-NA (Needham and Sawicki, 1971). Further study revealed that EF4 accounts for the broad spectrum of resistance to carbamate and pyrethroid insecticides (Devonshire and Moores, 1982). CarE activity in A. gossypii was determined using several standard substrates and was shown to be significantly higher in omethoate, deltamethrin, and malathion resistant strains than in susceptible strains (Cao et al., 2008a,b; Pan et al., 2009). There was a significant difference in carboxylesterase activity between a beta-cyperthrin-resistant strain (4,419-fold) and a susceptible strain of Musca domestica (Zhang et al., 2007).
Based on sequence alignment and conserved motifs, seven carboxylesterase and carboxylesterase-like genes were identified. The relative expression patterns of candidate genes were determined in the insecticide resistant and susceptible strains. Among the seven CarE genes, only CL2012 (RpCarE) was found to have significantly increased expression levels in both the IS-R and CY-R compared to the SS. A homology search of CL2012 showed maximum similarity (86%) to A. gossypii carboxylesterase. Therefore, we proposed that the CL2012 gene, as the carboxylesterase gene of R. padi, might participate in isoprocarb and cyhalothrin resistance. Twenty-eight CarE and CarE-like genes were identified in the transcriptome of Laodelphax striatellus, and LsCarE1 was significantly overexpressed in the chlorpyifos-resistant and chlorpyifos-relaxed selection strains by 32.06- and 8.6-fold, respectively. Additionally, overexpressed LsCarE1 mediated chlorpyifos resistance in L. striatellus (Zhang et al., 2012). In the deltamethrin-resistant strain of A. gossypii, the transcript levels of carboxylesterase were significantly enhanced, and this up-regulation was responsible for the development of resistance to deltamethrin (Cao et al., 2008a). The enhanced activity of carboxylesterases of M. persicae confers broad spectrum resistance to organophosphate, carbamate and pyrethroid (Bass et al., 2014). The M. persicae field population resistant to methomyl and omethoate exhibited a higher carboxylesterases activity compared to the laboratory susceptible strain (Tang et al., 2017). Three populations of the cowpea aphid (Aphis craccivora) collected in Egypt showed higher carboxylesterases which may cause the resistance of the species to organophosphates, carbamates, and neonicotinoids (Fouad et al., 2016). CarE enzyme activity were positively correlated with resistance level to chlorpyrifos, deltamethrin, and methomyl in the field populations of Sitobion avenae collect in wheat fields (Zhang et al., 2017).
There are a conserved catalytic triad (Ser, His, and Glu) and an oxyanion hole for carboxylesterases (Stok et al., 2004; Wheelock et al., 2005). During hydrolysis of carboxylesterases, a nucleophilic attack occurs by Ser on the carbon of the carbonyl group, which is next transferred to the His from the catalytic Ser. The protonated His is, in turn, stabilized via a hydrogen bond to the Glu (Stok et al., 2004). The Ser nucleophile attacks the substrate and forms the first tetrahedral intermediate, which is stabilized by two Gly residues in the oxyanion hole (Wheelock et al., 2005). The intermediate rapidly collapses to produce the acyl-enzyme complex, which then undergoes attacks by an His-activated water molecule and forms the second tetrahedral intermediate. The acid component of the substrate is released after the rapid rearrangement (Stok et al., 2004; Wheelock et al., 2005). To further investigate the role of RpCarE in resistance to isoprocarb and cyhalothrin in R. padi, the full coding region was cloned and characterized. The deduced amino acid sequence of RpCarE was found to exhibit striking homology to CarEs in other insects, including the highly conserved catalytic triad (Ser, His, and Glu) and an oxyanion hole consisting of backbone amide group of Ala, Gly, and Gly which indicate that RpCarE can function as an active esterase (Tsubota and Shiotsuki, 2010; Jackson et al., 2013).
RpCarE was cloned into pET-28a and expressed the in E. coli. The molecular mass was similar to the carboxylesterase E4 in M. persicae (60 kDa) (Lan et al., 2005) and differed from that of A. gossypii (65 kDa) (Gong et al., 2017). The purified fusion protein displayed significant hydrolase activity against the model substrate α-naphthyl acetate with a Kcat of 5.50 s-1, suggesting RpCarE was active and successfully expressed in the E. coli strain. Fusion proteins of carboxylesterase 001D from Helicoverpa armigera expressed in E. coli showed enzyme activities against α-NA with a Kcat between 0.35 and 2.29 s-1 (Li et al., 2016). HPLC showed that purified RpCarE can metabolize the two insecticide substrates, isoprocarb and cyhalothrin, in vitro, indicating that the carboxylesterase is involved in the detoxification of isoprocarb and cyhalothrin in R. padi. Many studies have demonstrated that carboxylesterases can metabolize organophosphorus, carbamate and pyrethroid insecticides in pest insects (Bass and Field, 2011; Coppin et al., 2012; Li et al., 2013). The E4 carboxylesterase degraded 64% of carbaryl and 80% of malathion within 2.5 and 1.25 h, respectively (Lan et al., 2005). All recombinant expressed wild type and mutant A. gossypii carboxylesterases can hydrolyse paraoxon and parathion to varying degrees (Gong et al., 2017). Li et al. (2016) showed that the H. armigera carboxylesterase 001D exhibited low but measurable hydrolase activity toward β-cypermethrin and fenvalerate. Fifty percent of malathion and 89% of malathion were hydrolysed by recombinant D1CarE5 within 25 and 100 min, respectively, (Xie et al., 2013).
Our modeling and docking data were in accordance with our metabolism analyses. Some residues created a hydrophobic interface at the binding cavity, and critical residues anchored the insecticide molecule. In our study, isoprocarb and cyhalothrin were docked into the predicted active site pocket and were further stabilized by hydrogen bonds with Asn-108, Tyr-109, and Glu-116. These analyses predicted that RpCarE is capable of hydrolysing isoprocarb and cyhalothrin. L. cuprina α-E (LcαE7), the template for RpCarE model building, showed a strong hydrophobic binding ability. Molecular modeling of LcαE7 indicated that some mutations in the enzyme were responsible for organophosphates resistance of L. cuprina (Yan et al., 2009). LcαE7 could sequester the insecticide molecule and slowly detoxify it; this high affinity gave LcαE7 an important function in insecticide resistance (Jackson et al., 2013). The A. gossypii CarE protein models based on the crystal structure of LcαE7 showed that some of non-synonymous mutations (H104R, A128V, T333P, and K484R) affected the active site pocket and binding energy, resulted in different binding affinity between A. gossypii CarE with insecticide compounds (Gong et al., 2017).
In summary, we cloned a carboxylesterase gene from R. padi and overexpressed it in both isoprocarb-resistant and cyhalothrin-resistant strains. RpCarE was successfully expressed in E. coli and exhibited hydrolytic activity toward the model substrate α-NA and to isoprocarb and cyhalothrin. These results suggest that RpCarE is involved in resistance to isoprocarb and cyhalothrin in R. padi. Further studies will be required to determine the detailed role of RpCarE as well as to clarify whether other metabolic enzymes (e.g., P450s and GST) are involved in metabolic resistance or whether target site mutations can play a role in the insecticide resistance of R. padi resistant strains.
Author Contributions
KW and MC designed the research and wrote the paper. KW, YH, and XL performed research. KW analyzed data.
Funding
This work was funded by the National Natural Science Foundation of China (Grants # 31772160 and 31471766).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00992/full#supplementary-material
Footnotes
References
Andrews, M. C., Callaghan, A., Field, L. M., Williamson, M. S., and Moores, G. D. (2004). Identification of mutations conferring insecticide-insensitive AChE in the cotton-melon aphid, Aphis gossypii Glover. Insect Mol. Biol. 13, 555–561. doi: 10.1111/j.0962-1075.2004.00517.x
Barres, B., Micoud, A., Corio-Costet, M. F., Debieu, D., Fillinger-David, S., Walker, A. S., et al. (2016). Trends and challenges in pesticide resistance detection. Trends Plant Sci. 21, 834–853. doi: 10.1016/j.tplants.2016.06.006
Bass, C., and Field, L. M. (2011). Gene amplification and insecticide resistance. Pest Manag. Sci. 67, 886–890. doi: 10.1002/ps.2189
Bass, C., Puinean, A. M., Zimmer, C. T., Denholm, I., Field, L. M., Foster, S. P., et al. (2014). The evolution of insecticide resistance in the peach potato aphid, Myzus persicae. Insect Biochem. Mol. Biol. 51, 41–51. doi: 10.1016/j.ibmb.2014.05.003
Bettaibi, K., Mezghani-Khemakhem, M., Bouktila, D., Charaabi, K., Raboudi, F., Makni, H., et al. (2016). A novel method for molecular targeting of insecticide resistance in Rhopalosiphum padi L. (Homoptera: Aphididae). Int. J. Pest Manag. 62, 284–287. doi: 10.1080/09670874.2016.1185190
Birner-Gruenberger, R., Bickmeyer, I., Lange, J., Hehlert, P., Hermetter, A., Kollroser, M., et al. (2012). Functional fat body proteomics and gene targeting reveal in vivo functions of Drosophila melanogaster α-Esterase-7. Insect Biochem. Mol. Biol. 42, 220–229. doi: 10.1016/j.ibmb.2011.12.004
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Campbell, P. M., Robin, D. Q., Court, L. N., Dorrian, S. J., Russell, R. J., and Oakeshott, J. G. (2003). Developmental expression and gene/enzyme identifications in the alpha esterase gene cluster of Drosophila melanogaster. Insect Mol. Biol. 12, 459–471. doi: 10.1046/j.1365-2583.2003.00430.x
Cao, C. W., Zhang, J., Gao, X. W., Liang, P., and Guo, H. L. (2008a). Differential mRNA expression levels and gene sequences of carboxylesterase in both deltamethrin resistant and susceptible strains of the cotton aphid, Aphis gossypii. Insect Sci. 15, 209–216.
Cao, C. W., Zhang, J., Gao, X. W., Liang, P., and Guo, H. L. (2008b). Overexpression of carboxylesterase gene associated with organophosphorous insecticide resistance in cotton aphids, Aphis gossypii (Glover). Pestic. Biochem. Physiol. 90, 175–180. doi: 10.1016/j.pestbp.2007.11.004
Chen, M. H., Han, Z. J., Qiao, X. F., and Qu, M. J. (2007). Mutations in acetylcholinesterase genes of Rhopalosiphum padi resistant to organophosphate and carbamate insecticides. Genome 50, 172–179. doi: 10.1139/G07-021
Coppin, C. W., Jackson, C. J., Sutherland, T., Hart, P. J., Devonshire, A. L., Russell, R. J., et al. (2012). Testing the evolvability of an insect carboxylesterase for the detoxification of synthetic pyrethroid insecticides. Insect Biochem. Mol. Biol. 42, 343–352. doi: 10.1016/j.ibmb.2012.01.004
DeLano, W. L. (2002). The PyMOL Molecular Graphics System. Available at: http://pymol.org
Devonshire, A. L., and Moores, G. D. (1982). A carboxylesterase with broad substrate specificity causes organophosphorus, carbamate and pyrethroid resistance in peach-potato aphids (Myzus persicae). Pestic. Biochem. Physiol. 18, 235–246. doi: 10.1016/0048-3575(82)90110-9
Du, Z. Q., Li, L., Liu, L., Wang, X. F., and Zhou, G. (2007). Evaluation of aphid transmission abilities and vector transmission phenotypes of barley yellow dwarf viruses in China. J. Plant Pathol. 89, 251–259.
Duan, X. L., Wang, K., Su, S., Tian, R. Z., Li, Y. T., and Chen, M. H. (2017). De novo transcriptome analysis and microsatellite marker development for population genetic study of a serious insect pest, Rhopalosiphum padi (L.) (Hemiptera: Aphididae). PLoS One 12:e0172513. doi: 10.1371/journal.pone.0172513
Eleftherianos, I., Foster, S. P., Williamson, M. S., and Denholm, I. (2008). Characterization of the M918T sodium channel gene mutation associated with strong resistance to pyrethroid insecticides in the peach-potato aphid, Myzus persicae (Sulzer). Bull. Entomol. Res. 98, 183–191. doi: 10.1017/S0007485307005524
Elzaki, M. E. A., Miah, M. A., Wu, M., Zhang, H., Pu, J., Jiang, L., et al. (2017). Imidacloprid is degraded by CYP353D1v2, a cytochrome P450 overexpressed in a resistant strain of Laodelphax striatellus. Pest Manag. Sci. 73, 1358–1363. doi: 10.1002/ps.4570
Foster, S. P., Paul, V. L., Slater, R., Warren, A., Denholm, I., Field, L. M., et al. (2014). A mutation (L1014F) in the voltage-gated sodium channel of the grain aphid, Sitobion avenae, is associated with resistance to pyrethroid insecticides. Pest Manag. Sci. 70, 1249–1253. doi: 10.1002/ps.3683
Fouad, E. A., Abou-Yousef, H. M., Abdallah, I. S., and Kandil, M. A. (2016). Resistance monitoring and enzyme activity in three field populations of cowpea aphid (Aphis craccivora) from Egypt. Crop Prot. 81, 163–167. doi: 10.1016/j.cropro.2015.12.015
Franck, P., Siegwart, M., Olivares, J., Toubon, J. F., and Lavigne, C. (2012). Multiple origins of the sodium channel kdr mutations in codling moth populations. PLoS One 7:e43543. doi: 10.1371/journal.pone.0043543
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gong, Y. H., Ai, G. M., Li, M., Shi, X. Y., Diao, Q. Y., and Gao, X. W. (2017). Functional characterization of carboxylesterase gene mutations involved in Aphis gossypii resistance to organophosphate insecticides. Insect Mol. Biol. 26, 702–714. doi: 10.1111/imb.12331
Han, Y., Wu, S., Li, Y., Liu, J. W., Campbell, P. M., Farnsworth, C., et al. (2012). Proteomic and molecular analyses of esterases associated with monocrotophos resistance in Helicoverpa armigera. Pestic. Biochem. Physiol. 104, 243–251. doi: 10.1016/j.pestbp.2012.09.005
Hemingway, J., Hawkes, N. J., McCarroll, L., and Ranson, H. (2004). The molecular basis of insecticide resistance in mosquitoes. Insect Biochem. Mol. Biol. 34, 653–665. doi: 10.1016/j.ibmb.2004.03.018
Hemingway, J., and Ranson, H. (2000). Insecticide resistance in insect vectors of human disease. Annu. Rev. Entomol. 45, 371–391. doi: 10.1146/annurev.ento.45.1.371
Jackson, C. J., Liu, J. W., Carr, P. D., Younus, F., Coppin, C., Meirelles, T., et al. (2013). Structure and function of an insect α-carboxylesterase (αEsterase7) associated with insecticide resistance. Proc. Natl. Acad. Sci. U.S.A. 11, 10177–10182. doi: 10.1073/pnas.1304097110
Lan, W. S., Cong, J., Jiang, H., Jiang, S. R., and Qiao, C. L. (2005). Expression and characterization of carboxylesterase E4 gene from peach–potato aphid (Myzus persicae) for degradation of carbaryl and malathion. Biotechnol. Lett. 27, 1141–1146. doi: 10.1007/s10529-005-8464-x
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, F., and Han, Z. (2004). Mutations in acetylcholinesterase associated with insecticide resistance in the cotton aphid, Aphis gossypii Glover. Insect Biochem. Mol. Biol. 34, 397–405. doi: 10.1016/j.ibmb.2004.02.001
Li, X., Schuler, M. A., and Berenbaum, M. R. (2007). Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52, 231–253. doi: 10.1146/annurev.ento.51.110104.151104
Li, Y., Farnsworth, C. A., Coppin, C. W., Teese, M. G., Liu, J. W., Scott, C., et al. (2013). Organophosphate and pyrethroid hydrolase activities of mutant esterases from the cotton bollworm Helicoverpa armigera. PLoS One 8:e77685. doi: 10.1371/journal.pone.0077685
Li, Y., Liu, J., Lu, M., Ma, Z., Cai, C., Wang, Y., et al. (2016). Bacterial Expression and Kinetic Analysis of Carboxylesterase 001D from Helicoverpa armigera. Int. J. Mol. Sci. 17:493. doi: 10.3390/ijms17040493
Liu, N. (2015). Insecticide resistance in mosquitoes: impact, mechanisms, and research directions. Annu. Rev. Entomol. 60, 537–559. doi: 10.1146/annurev-ento-010814-020828
Marshall, K. L., Moran, C., Chen, Y., and Herron, G. A. (2012). Detection of kdr pyrethroid resistance in the cotton aphid, Aphis gossypii (Hemiptera: Aphididae), using a PCR-RFLP assay. J. Pestic. Sci. 37, 169–172. doi: 10.1584/jpestics.D11-017
Nabeshima, T., Kozaki, T., Tomita, T., and Kono, Y. (2003). An amino acid substitution on the second acetylcholinesterase in the pirimicarb-resistant strains of the peach potato aphid, Myzus persicae. Biochem. Biophys. Res. Commun. 307, 15–22. doi: 10.1016/S0006-291X(03)01101-X
Needham, P. H., and Sawicki, R. M. (1971). Diagnosis of resistance to organophosphorus insecticides in Myzus persicae (Sulz.). Nature 230:125. doi: 10.1038/230125a0
Pan, Y., Guo, H., and Gao, X. (2009). Carboxylesterase activity, cDNA sequence, and gene expression in malathion susceptible and resistant strains of the cotton aphid, Aphis gossypii. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 152, 266–270. doi: 10.1016/j.cbpb.2008.12.002
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Ramsey, J. S., Rider, D. S., Walsh, T. K., De Vos, M., Gordon, K. H. J., Ponnala, L., et al. (2010). Comparative analysis of detoxification enzymes in Acyrthosiphon pisum and Myzus persicae. Insect Mol. Biol. 19, 155–164. doi: 10.1111/j.1365-2583.2009.00973.x
Stok, J. E., Goloshchapov, A., Song, C., Wheelock, C. E., Derbel, M. B. H., Morisseau, C., et al. (2004). Investigation of the role of a second conserved serine in carboxylesterases via site-directed mutagenesis. Arch. Biochem. Biophys. 430, 247–255. doi: 10.1016/j.abb.2004.06.020
Tang, Q. L., Ma, K. S., Hou, Y. M., and Gao, X. W. (2017). Monitoring insecticide resistance and diagnostics of resistance mechanisms in the green peach aphid, Myzus persicae (Sulzer) (Hemiptera: Aphididae) in China. Pestic. Biochem. Physiol. 143, 39–47. doi: 10.1016/j.pestbp.2017.09.013
Tsubota, T., and Shiotsuki, T. (2010). Genomic and phylogenetic analysis of insect carboxyl/cholinesterase genes. J. Pestic. Sci. 35, 310–314. doi: 10.1584/jpestics.R10-04
Vais, H., Williamson, M. S., Goodson, S. J., Devonshire, A. L., Warmke, J. W., Usherwood, P. N., et al. (2000). Activation of Drosophila sodium channels promotes modification by deltamethrin: reductions in affinity caused by knock-down resistance mutations. J. Gen. Physiol. 115, 305–318. doi: 10.1085/jgp.115.3.305
Wang, K., Peng, X., Zuo, Y., Li, Y., and Chen, M. (2016). Molecular cloning, expression pattern and polymorphisms of NADPH-cytochrome P450 reductase in the bird cherry-oat aphid Rhopalosiphum padi (L.). PLoS One 11:e0154633. doi: 10.1371/journal.pone.0154633
Wang, K., Zhang, M., Huang, Y., Yang, Z., Su, S., and Chen, M. (2018). Characterisation of imidacloprid resistance in the bird cherry-oat aphid, Rhopalosiphum padi, a serious pest on wheat crops. Pest Manag. Sci. 74, 1457–1465. doi: 10.1002/ps.4834
Wheelock, C. E., Shan, G., and Ottea, J. (2005). Overview of carboxylesterases and their role in the metabolism of insecticides. J. Pestic. Sci. 30, 75–83. doi: 10.1584/jpestics.30.75
Williamson, M. S., Martinez-Torres, D., Hick, C. A., Castells, N., and Devonshire, A. L. (1996). “Analysis of sodium channel gene sequences in pyrethroid-resistant houseflies,” in Molecular Genetics and Evolution of Pesticide Resistance, ed. T. M. Brown (Washington, DC: American Chemical Society), 53–61.
Xi, J., Pan, Y., Bi, R., Gao, X., Chen, X., Peng, T., et al. (2015). Elevated expression of esterase and cytochrome P450 are related with lambda–cyhalothrin resistance and lead to cross resistance in Aphis glycines matsumura. Pestic. Biochem. Physiol. 118, 77–81. doi: 10.1016/j.pestbp.2014.12.002
Xie, J., Zhao, Y., Zhang, H., Liu, Z., and Lu, Z. (2014). Improving methyl parathion hydrolase to enhance its chlorpyrifos-hydrolysing efficiency. Lett. Appl. Microbiol. 58, 53–59. doi: 10.1111/lam.12155
Xie, Z., Bo, X., Ding, J., Liu, L., Zhang, X., Li, J., et al. (2013). Heterologous expression and characterization of a malathion-hydrolyzing carboxylesterase from a thermophilic bacterium, Alicyclobacillus tengchongensis. Biotechnol. Lett. 35, 1283–1289. doi: 10.1007/s10529-013-1195-5
Yan, S. G., Cui, F., and Qiao, C. L. (2009). Structure, function and applications of carboxylesterases from insects for insecticide resistance. Protein Pept. Lett. 16, 1181–1188. doi: 10.2174/092986609789071243
Zhang, L., Gao, X., and Liang, P. (2007). Beta-cypermethrin resistance associated with high carboxylesterase activities in a strain of house fly, Musca domestica (Diptera: Muscidae). Pestic. Biochem. Physiol. 89, 65–72. doi: 10.1016/j.pestbp.2007.03.001
Zhang, L. P., Lu, H., Guo, K., Yao, S. M., and Cui, F. (2017). Insecticide resistance status and detoxification enzymes of wheat aphids Sitobion avenae and Rhopalosiphum padi. Sci. China Life Sci. 60, 927–930. doi: 10.1007/s11427-017-9105-x
Zhang, Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40
Zhang, Y., Wang, L., Guo, H., Li, G., Zhang, Z., Xie, L., et al. (2012). A transcriptome-based screen of carboxylesterase-like genes that are involved in chlorpyrifos resistance in Laodelphax striatellus (Fallén). Pestic. Biochem. Physiol. 104, 224–228. doi: 10.1016/j.pestbp.2012.08.006
Zlotkin, E. (1999). The insect voltage-gated sodium channel as target of insecticides. Annu. Rev. Entomol. 44, 429–455. doi: 10.1146/annurev.ento.44.1.429
Keywords: carboxylesterase, Rhopalosiphum padi, isoprocarb, pyrethroid, metabolism resistance
Citation: Wang K, Huang Y, Li X and Chen M (2018) Functional Analysis of a Carboxylesterase Gene Associated With Isoprocarb and Cyhalothrin Resistance in Rhopalosiphum padi (L.). Front. Physiol. 9:992. doi: 10.3389/fphys.2018.00992
Received: 03 April 2018; Accepted: 06 July 2018;
Published: 25 July 2018.
Edited by:
Su Wang, Beijing Academy of Agriculture and Forestry Sciences, ChinaReviewed by:
Bruno Gomes, Liverpool School of Tropical Medicine, United KingdomPin-Jun Wan, China National Rice Research Institute (CAAS), China
Copyright © 2018 Wang, Huang, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maohua Chen, bWFvaHVhLmNoZW5AbndzdWFmLmVkdS5jbg==
 Kang Wang
Kang Wang Yanna Huang
Yanna Huang Xinyu Li
Xinyu Li Maohua Chen
Maohua Chen