
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 05 July 2018
Sec. Aquatic Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.00837
This article is part of the Research Topic The Physiological and Molecular Stress Responses in Aquatic Species: The Impact on Aquatic Animal Health View all 41 articles
The ubiquitin proteasome system (UPS) is a highly regulated mechanism of intracellular protein degradation and turnover. To evaluate the effect of crowding stress on the UPS in fish, grass carp (Ctenopharyngodon idellus) were randomly reared at low stocking density (LSD, 0.9 kg m−2) or high stocking density (HSD, 5.9 kg m−2) for 70 days. The expression of the genes regulating UPS, stress-related parameters, and profiles of amino acid in white muscle as well as growth rate of fish reared at two stocking densities were investigated. Fish exhibited significantly higher growth rate in the LSD group compared to the HSD group. Serum concentrations of cortisol, total protein, and glucose did not vary significantly in fish between two groups. There was no significant difference in the mRNA levels of nrf2, keap1, and hsp90 in white muscle of fish stocked at two densities at the endpoint of the experiment. In the UPS pathway, the expressions of ub, chip, psmc1 in the LSD group were significantly higher than those in the HSD group (P < 0.05). Ubiquitinated protein level and the content of 3-Methylhistidine elevated significantly in the LSD group (P < 0.05). The mRNA levels of mafbx, murf1, and s6k1 in the LSD group were significantly higher than those in HSD group (P < 0.05). These results illustrate that the fish cultured in lower stocking density would exhibit a greater growth rate and a fast protein turnover in muscle.
The ubiquitin proteasome system (UPS) is important for regulating protein degradation and function (Pickart and Eddins, 2004). The function of UPS is involved in various cellular processes, such as signal transduction, metabolic regulation, cell cycle control, development, apoptosis, protein quality control, and antigen presentation (Glickman and Ciechanover, 2002; Flick and Kaiser, 2012; Sommer and Wolf, 2014). The conjugation process of ubiquitinylation is a complex reaction that requires ubiquitin (ub), ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2), and ubiquitin protein ligase (E3) (Romn and Reznick, 2016). E3 ligases have a central role in coupling ubiquitin to the targeted protein and generate the specificity of the mechanism (Jackson and Eldridge, 2002). The UPS is an ATP-dependent proteolytic system and involves two main steps. First, targeted protein is polyubiquitinated with the specificity determined by the E3 ubiquitin ligase enzyme. Second, the ubiquitinated protein is recognized by 26S proteasome which degrades the substrate into peptides (Ciechanover, 1994).
Muscle RING finger 1 (MuRF1) and muscle atrophy Fbox (MAFbx/atrogin-1) have been regarded as central markers involved in protein degradation (Bodine et al., 2001). These two proteins, belong to ubiquitin E3 ligases, consistently highly express in atrophying muscle caused by cancer, diabetes, fasting, and limb immobilization (Sacheck et al., 2007). The growth of skeletal muscle depends on the balance between protein synthesis and degradation in teleost fish as well as other vertebrates. UPS is a major system for protein degradation in eukaryotic cells (Pickart and Eddins, 2004), and regulate the turnover of protein in rainbow trout (Oncorhynchus mykiss) (Cleveland and Evenhuis, 2010). A study revealed that specific growth rate was negatively correlated with 20S proteasome activity in the liver of rainbow trout (Dobly et al., 2004). Consequently, fish growth rate probably be related to the activity of UPS. As far as anabolism is concerned, mTOR (mammalian/mechanistic target of rapamycin complex) and S6K1 (ribosomal protein S6 kinase 1) are of importance in regulating protein synthesis and controlling cell growth (Huynh et al., 2017). The expression levels of MAFbx, MuRF1, and mTOR elevated significantly as IGF-1 increased in animals (Liu H.H. et al., 2014). It is implied that the growth enhancement is related to gene expression of mTOR, MAFbx, and MuRF1.
There are many external factors affecting growth, such as stocking density, food nutrition, temperature, and water quality (Siikavuopio and Sæther, 2006; Fraser and Rogers, 2007; Valenzuela et al., 2017). For example, previous study suggested that Nile tilapia (Oreochromis niloticus) obtained successful growth, maximum fertilization, and survival rate of fry up to 15% salinity (Malik et al., 2018). Also, Rahim et al. (2017) reported that good performances of black fin sea bream (Acanthopagrus berda) were closely associated with factors including food nutrition, temperature, and salinity. With the development of intensive aquaculture, fish reared at high stocking density exhibited reduced growth rate with lower resistance to diseases (Benjamín et al., 2008; VargasChacoff et al., 2011). In addition, high stocking density exerted negative effect on food conversion efficiency in fish (Beveridge et al., 1997). Excessive high stocking density often causes crowding stress and impairs fish welfare. Under the condition of environmental stress, the UPS can be activated to maintain cellular protein homeostasis, regulating a dynamic balance in protein synthesis and degradation processes. Especially, the ubiquitin ligases play important role in stress sensing, signaling, and activation of the hypoxia response pathway (Flick and Kaiser, 2012). Thermal stress resulted in a significant elevation of 20S proteasome activity in the liver and gill of notothenioid fishes (Todgham et al., 2017). And the downregulation of protein ubiquitination pathway was demonstrated in the liver of rainbow trout exposed to crowding stress (Rebl et al., 2017). Nrf2 is a master transcription factor of the cellular antioxidant response through keap1-nrf2 signaling pathway (Dodson et al., 2015). The misfolded or unfolded proteins during stress are refolded by chaperone systems first, when refolding is unsuccessful the UPS degrades proteins (Flick and Kaiser, 2012). HSP70 and HSP90 play an important roles in both refolding and selection of proteins for UPS through CHIP ubiquitin ligase (Benarroch, 2011).
To date, little is known about the response of UPS to crowding stress in fish. Grass carp (Ctenopharyngodon idellus) (Cypriniformes, Cyprinidae, Leuciscinae) (Figure 1) is a typical herbivorous fish, and the most popular cultured fish species in China (Jin et al., 2013). The aquaculture production of grass carp ranks top among the world’s freshwater aquaculture industries now (Xie et al., 2018). Its production reached more than 5 million tons and supplied nearly 20% of the total freshwater fish production in China (FAO Yearbook, 2016). The purpose of this study was to investigate the activity of UPS in fish reared at different stocking densities and to elucidate the relationship between crowding stress and muscular protein metabolism mediated by UPS.
All fish were collected from a fish farm in Hubei province, China. Before the start of the experiment, fish were reared in the freshwater tanks. We randomly distributed 140 individuals (initial body weight: 98.48 ± 6.00 g) into eight circular fiberglass tanks at two stocking densities (four tanks in each stocking density group): low stocking density (LSD, 0.9 kg m−2, 5 fishes per tank), and high stocking density (HSD, 5.9 kg m−2, 30 fishes per tank) for a period of 70 days. Water temperature (20 ± 0.7°C), pH (7.5 ± 0.3), dissolved oxygen (above 7.0 mg L−1), and ammonia concentration (less than 0.02 mg L−1) were maintained at the same water quality level in each group.
To gain an insight into the relation between stocking density and growth, we examined the body weight, weight gain rate (WGR), specific growth rate (SGR), feed conversion efficiency (FCR), and feed ration (FR) in each group during the experimental period. The growth performances were measured with eight fish (two fish per tank).
Wo, initial weight (g); Wt, final weight (g); T, feeding days; FI, feed intake (dry weight).
At the end of the experiment, fishes were randomly selected from each group (5 individuals per tank) and anesthetized with tricaine methanesulfonate (MS-222, 200 mg/L). Dorsal white muscle and blood of fish were collected for analysis. Muscle samples were immediately frozen in liquid nitrogen, and then stored at −80°C until RNA extraction. Each mix received from five fish acted as one sample used for subsequent experiment.
This study was approved by the Institutional Animal Care and Use Committees (IACUC) of Huazhong Agricultural University (HZAU) (Wuhan, China). The experiment was made according to the reporting of In Vivo Experiments (ARRIVE) guidelines and “Guidelines for Experimental Animals” of the Ministry of Science and Technology (Beijing, China). All efforts were made to minimize suffering of sampled fishes.
Serum biochemistry was analyzed to determine the level of fish stress. Serum cortisol level was analyzed using a commercial RIA kit (Coated Tube Cortisol 125I RIA Kit, BNIBT, and Beijing, China) according to the manufacturer’s instruction. Serum glucose (GLU) and total protein (TP) were measured by a commercial kit (Sysmex Wuxi Co., Let., China).
The RNA was extracted from 80 mg of muscle, homogenized by RNA isolater (Vazyme). RNA integrity was detected in 1.1% agarose gel electrophoresis, and the concentration was quantified spectrophotometrically. For each sample, 1 μg of total RNA was reverse transcribed in the presence of oligo-dT primers using a PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Dalian, China) according to the manufacturer’s instructions.
The 20S proteasome subunit alpha type 2 (psma2) was chosen as candidate gene representing an α-subunit, respectively. And 26S proteasome subunit ATPase 1 (psmc1) was chosen as a representative ATPase subunit in the 19S regulatory particle (Lupatsch et al., 2010). We searched for and downloaded ubiquitin (ub), murf1, mafbx, chip, nrf2, keap1, psma2, psmc1, s6k1, mtor, hsp70, and hsp90 genes’ sequences from National Center for Biotechnology Information (NCBI) databases. Primers were designed by Primer Premier 5. Sequences were initially analyzed using Blast search to confirm the correct genes were cloned.
Quantitative real-time (qRT)-PCR was performed on a Quant Studio 6 Flex Real-Time PCR Detection System (Applied Biosystems, United States). The 20 μL reaction mixture contained 10 μl HieffTM qPCR SYBR® Green Master Mix (YEASEN, Sanghai, China), 0.4 μL of each of the specific primers, 7.2 μL RNase-free water, and 2 μL cDNA template. The gene-specific primers (Table 1) were designed with Primer Premier 5.0. The reaction was performed in a Hard-Shell® High-Profile 96-well Semi-Skirted PCR plate (Bio-Rad, United States). The amplification protocol was as follows: 95°C for 5 min, then 40 cycles of 95°C for 10 s, 57°C for 20 s, and 72°C for 20 s, with a final cycle of 95°C for 15 s, 60°C for 60 s, 95°C for 15 s, as for melt curve analysis. The β-actin gene was chosen as a control for normalization of the qRT-PCR (Salem et al., 2006). The relative expression ratio (R) was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Total protein was isolated from muscle by RIPA Lysis Buffer (GBCBIO Technologies, China), and the concentration was determined with BCA Protein Assay Kit (GBCBIO Technologies, China). The concentrations of samples should be held same by PBS (0.67 μg μL−1). Then, the proteins were loaded on the Nitrocellulose Membrane (PALL Bio TraceTM NT) through the template of Bio-Dot® (Bio-Rad, United States). The membrane was kept in Blocking Buffer for 1 h to block nonspecific binding, after that, it was incubated overnight at 4°C with the primary antibody (diluted 1:1000). MAb to Polyubiquitinylated Conjugates (FK1) (Biomol, BML PW8805-0500) had been shown to recognize polyubiquitinylated proteins. After incubation with the secondary antibody which is labeled by FITC (anti-mouse, BOSTER, BA1101), the membrane was observed by Odyssey CLx (LI-COR, CLx-0813). We used ImageJ to compute the levels of ubiquitinated proteins.
The 3-MH provides a reliable index of the rate of myofibrillar protein breakdown in the musculature (Yong and Munro, 1978). Tyrosine is present in all myoblast, indirectly reflecting the degradation of total protein in the muscle. The contents of the free (3-MH) and Tyrosine (Tyr) of muscle were determined by High Performance Liquid Chromatography (HPLC) according to the National Standard of the People’s Republic of China (GB/T5009.124-2003).
All data were determined with one-way analysis of variance (ANOVA) following by Tukey’s HSD test with SPSS software. The differences were deemed statistically significant at P < 0.05.
Final weight (FW), final length (FL), weight gain rate (WGR), specific growth rate (SGR), feed conversion efficiency (FCR), and feed ration (FR) in each group are shown in Table 2. Compared to the HSD group, fish exhibit significantly higher growth rate in the LSD group (P < 0.05).
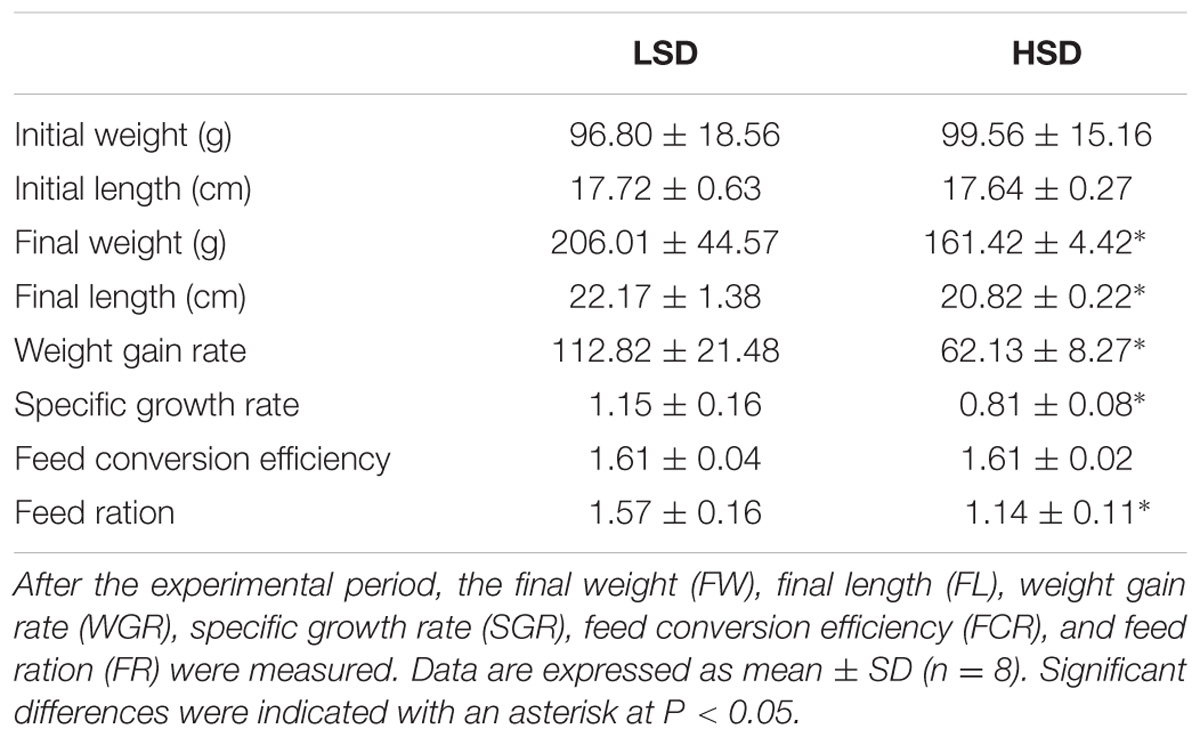
TABLE 2. Growth parameters of the grass carp kept at low (LSD, 0.9 kg m−2) or high (HSD, 5.9 kg m−2) stocking density for a period of 70 days.
There were no significant differences in serum levels of cortisol, TP and GLU between two groups (Table 3). Nrf2 and keap1 mRNA levels were not obvious differences between HSD and LSD. HSD did not cause a significantly increased in the expression of hsp90. The expression levels of hsp70 and chip in HSD group were significantly lower than those in LSD group (P < 0.05, Figure 2).
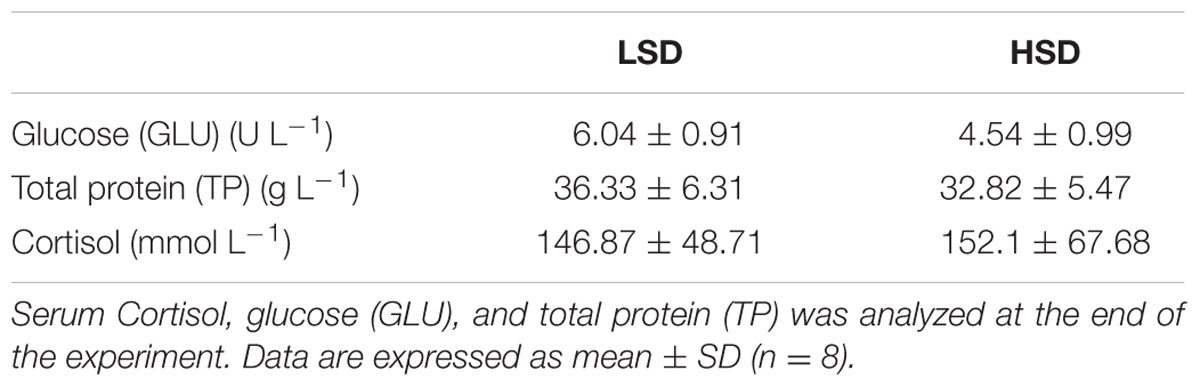
TABLE 3. Serum biochemical parameters of the grass carp kept at low (LSD, 0.9 kg m−2) or high (HSD, 5.9 kg m−2) stocking density for a period of 70 days.
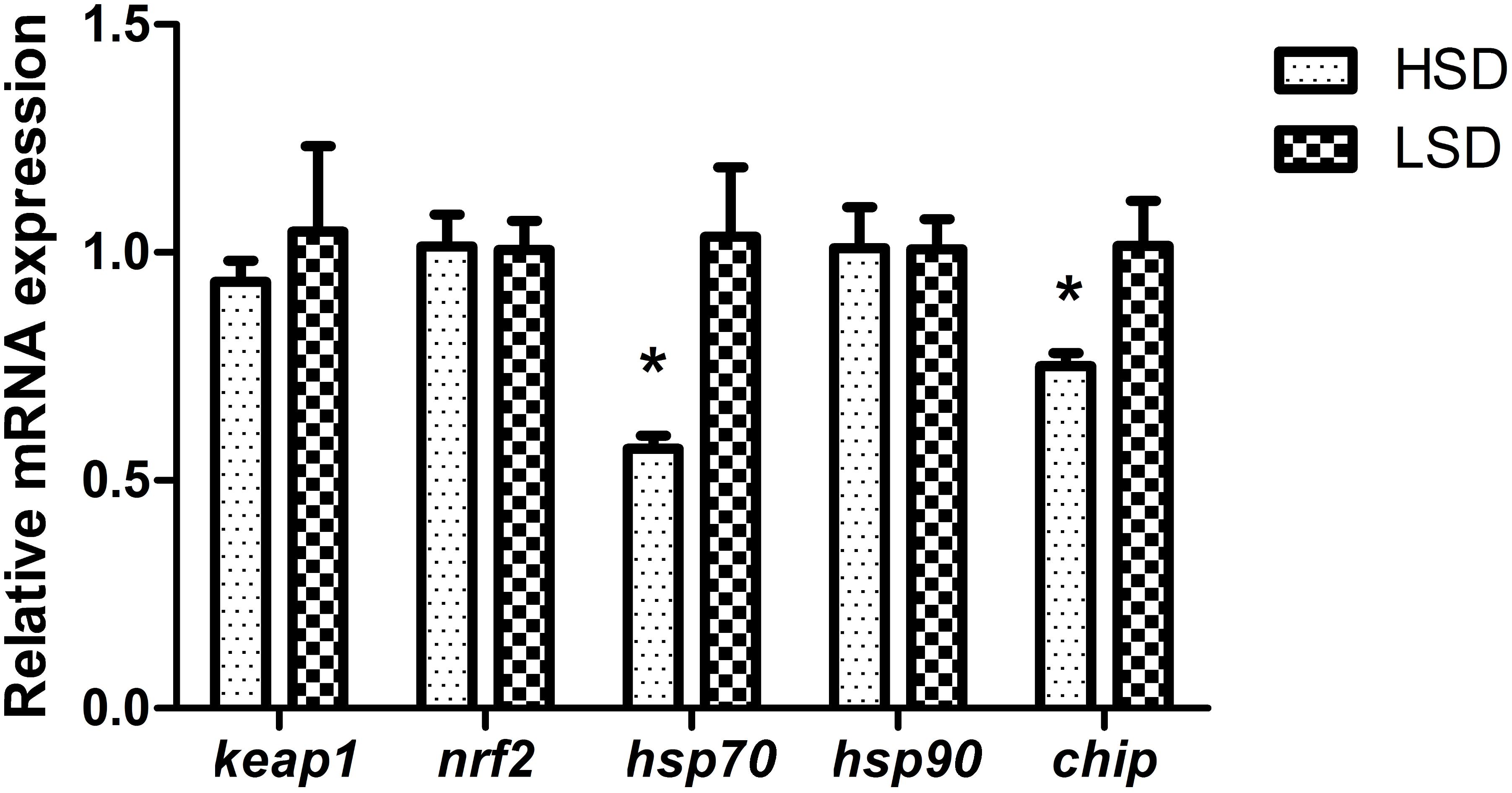
FIGURE 2. Expressions of keap1, nrf2, hsp70, hsp90, and chip in the white muscle of grass carp. Fish was randomly reared at low stocking density (LSD, 0.9 kg m−2) or high stocking density (HSD, 5.9 kg m−2) for 70 days, dorsal white muscle (5 individuals per tank) were collected for analysis. The expression in two densities with the expression level in LSD as 1. Significant differences were indicated with an asterisk at P < 0.05. Data are shown as means ± SEM (n = 4).
The expression levels of ubiquitin (ub) and psmc1 were significantly declined in the HSD group compared to the LSD group (P < 0.05, Figure 3A). The expression of psma2 was not significant difference in two groups. A significant high content of ubiquitinated proteins occurred in white muscle of fish in LSD group (P < 0.05, Figure 3B).
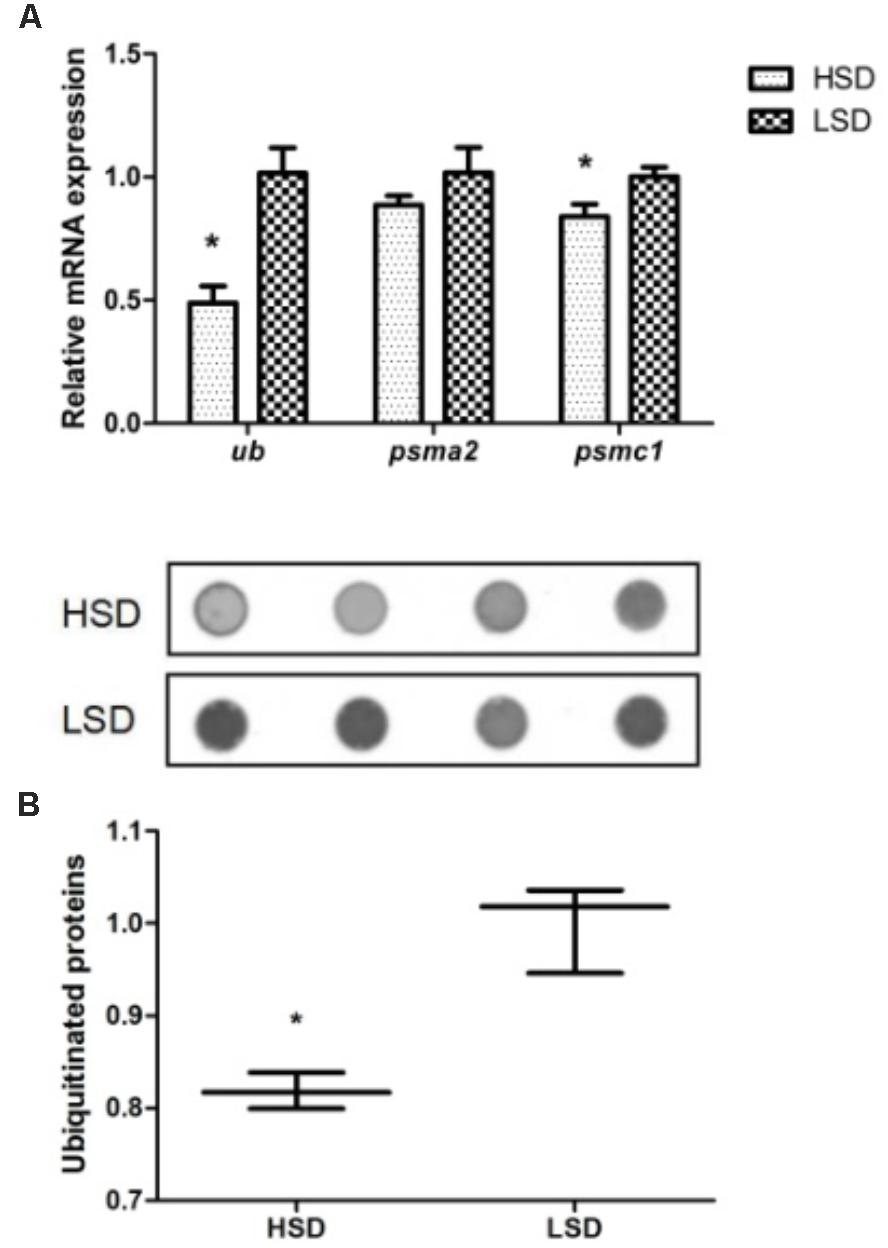
FIGURE 3. Expressions of ub, psma2, psmc1, and the ubiquitinated proteins in the white muscle of grass carp. (A) The mRNA expressions of ub, psma2, and psmc1. (B) Dot blot analysis about the ubiquitinated proteins. Dorsal white muscle (5 individuals per tank) were collected after the experimental period of 70 days. The expression in two densities with the expression level in LSD as 1. Significant differences were indicated with an asterisk at P < 0.05. Data are shown as means ± SEM (n = 4).
The mRNA levels of murf1, mafbx, and s6k1 decreased significantly in the HSD group (P < 0.05, Figure 4A). On the other hand, the stocking density did not modify the expression of mtor. The contents of 3-MH and Tyr were significantly higher in the LSD group than those in the HSD group (P < 0.05, Figure 4B).
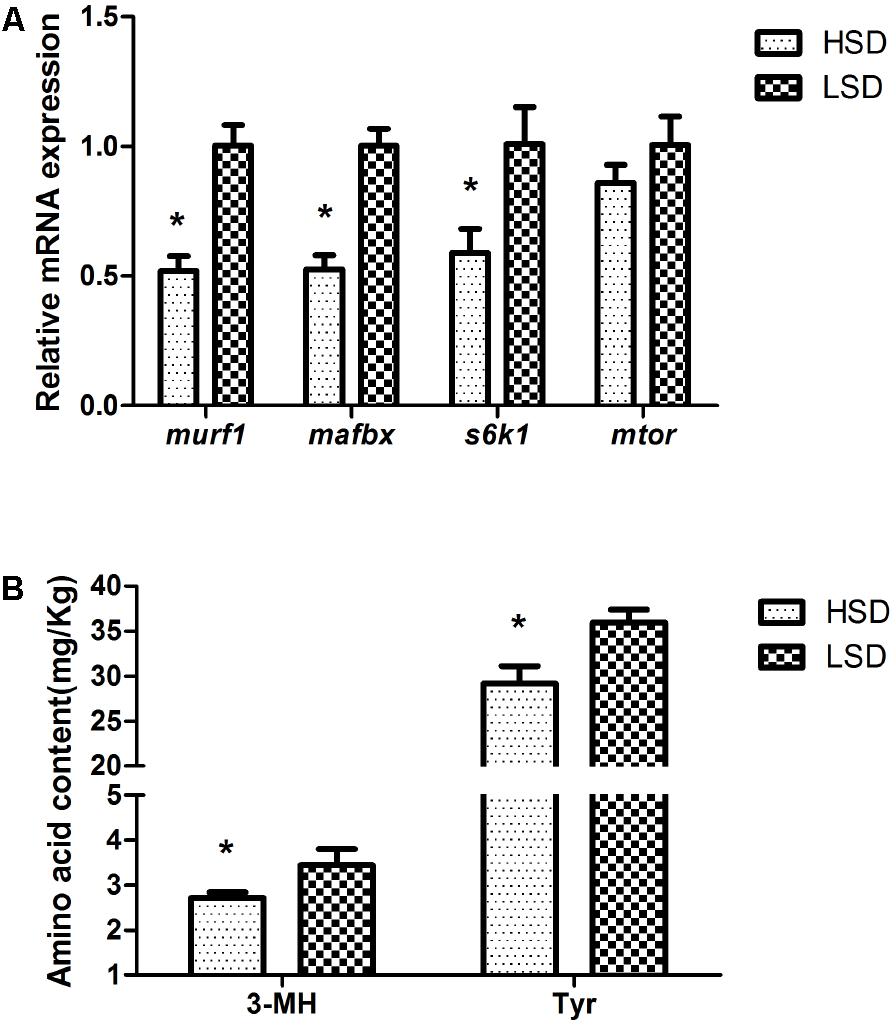
FIGURE 4. Expressions of murf1, mafbx, s6k1, mtor, and contents of 3-MH and Tyr in the white muscle of grass carp. (A) The mRNA expressions of murf1, mafbx, s6k1, and mtor. (B) Amino acid contents of 3-MH and Tyr. Dorsal white muscle (5 individuals per tank) were collected after the experimental period of 70 days. The expression in two densities with the expression level in LSD as 1. Significant differences were indicated with an asterisk at P < 0.05. Data are shown as means ± SEM (n = 4).
Growth is a good indicator of fish health. Rapid growth usually reflects abundant food supply and appropriate environmental conditions. The present study found that high stocking density had a negative effect on growth performance of grass carp. The similar findings have been reported in many fish species, such as Salminus brasiliensis, Megalobrama amblycephala, and Acipenser schrenckii (Braun et al., 2010; Li et al., 2012; Qi et al., 2016). Generally, the levels of serum cortisol and glucose as well as the expressions of hsp70, hsp90, and keap1-nrf2 signaling pathway can indicate the magnitude of fish stress (Lushchak, 2011). In this study, however, significant difference in the levels of either cortisol or glucose between groups was not found. The similar results can be observed in other fish species (Tort et al., 1996; Rotllant et al., 1997). In fish, nrf2 is a critical factor that regulates the expression of a group of genes encoding antioxidant enzymes (Lushchak, 2011). Under the salinity exposure, the gene expression of nrf2 in mosquito fish (Gambusia affinis) showed an obvious dose-effect (Bao et al., 2017). Nevertheless, not only the serum biochemical parameters levels but also the mRNA abundances of keap1, nrf2, and hsp90 did not change significantly between two groups at the end of the experiment in this study. Heat shock proteins are generally considered as characteristic biomarkers of cellular stress responses and contribute to numerous cellular signaling pathways (Benarroch, 2011; Kirschke et al., 2014). It indicates that denatured proteins accumulate upon exposure to stressors (Benarroch, 2011). In rainbow trout the mRNA levels of hsp70 remained at similar levels (Liu S. et al., 2014), and in senegalese sole (Solea senegalensis) it decreased after exposure to high stocking density (Salas-Leiton et al., 2010). In this study, the expression of hsp70 increased in the LSD group. A reason for the variable expression of hsp70 may be attributed to their pleiotropic functional spectra (Kirschke et al., 2014). The plasma cortisol concentration often surged in the early stage of stress, and then returned to the control after adaptation with a long-term (Rebl et al., 2017). No significant difference in stress-related parameters between two stocking density groups probably be associated with the duration of crowding stress. Grass carp in the HSD group might adapt to the crowding environment after 70-day culture period.
Proteolysis by the UPS is ATP-dependent, and Ub, E1, E2, E3, and 26S proteasome are required in this process. 26S proteasome is formed by 19S regulatory ATPase complex and 20S proteasome complex (Todgham et al., 2017). The stocking density did not affect the psma2 expression, while the higher mRNA levels of ub and psmc1 were observed in the LSD group, coupled with the elevation of the content of ubiquitinated proteins. This suggests strong activity of UPS in fish stocked at lower density. It has been demonstrated that the activity of the UPS was enhanced during the process of expenditure and atrophy of skeletal muscle (Dobly et al., 2004). And high expression of psmc1, mafbx, and murf1 appeared in the muscle of juvenile fine flounder (Paralichthys adspersus) at 7 weeks of crowding (Valenzuela et al., 2017).
E3 ligases, MuRF1 and MAFbx are regarded as the primary markers in the process of protein degradation (Bodine et al., 2001), and the enhanced expression of mafbx and murf1 activated the protein breakdown in mice (Choi et al., 2017). Actin and myosin are the contractile proteins of skeletal muscle, and 3-Methylhistidine (3-MH) can be detected in hydrolysates of these proteins (Yong and Munro, 1978). Consequently, the higher contents of 3-MH and Tyr coupled with higher mRNA abundances of mafbx and murf1 indicated more degraded proteins in fish muscle of the LSD group. Although more protein degradation occurred in white muscle, fish grew better in the LSD group. This means that synthetic metabolism is greater than catabolism. The tissues, such as heart, liver, and red muscle which contain relatively high abundance of proteins, showed high levels of mafbx. Furthermore, the relatively high level of metabolic activity of the tissues suggested that mafbx may influence the recycling of proteins involved in energy production or metabolic maintenance (Cleveland and Evenhuis, 2010).
The activation of mTOR positively mediates protein synthesis through the phosphorylation of S6K1, which can promote ribosomal protein expression and cell growth (Howell et al., 2013; Huynh et al., 2017). The high expressions of s6k1 and mtor in the LSD group suggest the activation of the protein synthesis in fish muscle. And hsp70 and chip also showed apparently high expression in the LSD group. In addition to a presumptive role in re-folding thermally damaged proteins, hsp70 seems to be part of a molecular chaperone complex. Hsp70 binds to polypeptides translating on ribosomes and regulates the protein biogenesis under normal cellular conditions (Frydman et al., 1994). Therefore, fish in the LSD group showed a high rate of protein synthesis. Previous research demonstrated that the expression of MAFbx, MuRF1, and mTOR were significantly increased in the IGF-1 treatment duck (Anas platyrhynchos) (Liu H.H. et al., 2014). It is inferred that IGF-1 can promote animal growth. The growing rats feed with a zein protein diet showed the increased expressions of mTOR, MAFbx, and MuRF1, and high protein metabolism (Luo et al., 2010). Therefore, not only protein synthesis, but also protein degradation manifested high activity in LSD.
The results demonstrated stocking density exerted potent effects on growth performance and UPS activity in muscle of grass carp. A greater growth rate occurred in the fish cultured at lower stocking density. The fish with a better growth performance exhibited a higher UPS activity in parallel with the activation of protein synthesis. These results illustrate that the fish cultured at low stocking density would exhibit a greater growth rate and a fast protein turnover in muscle.
YS and XL performed the experiment, analyzed the data, and wrote the manuscript. DL designed the experiment and polished the manuscript. JC cultured the fish and analyzed the growth data. RT and LL revised the manuscript. All authors reviewed the manuscript.
This study was supported by the Earmarked Fund for China Agriculture Research System (Project No. CARS-45), the National Natural Foundation of China (Project No. 31502140), and the Fundamental Research Funds for the Central Universities (Project No. 2662015PY119).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Mei Xiong, Weitong Xu, Haishan Wang, Xi Zhang, LL, RT, Xugang He, and Wei Chi for their valuable assistance in sample collection and biochemical measurements.
Bao, S., Nie, X. P., Ou, R. K., Chao, W., Ku, P., and Li, K. (2017). Effects of diclofenac on the expression of Nrf2 and its downstream target genes in mosquito fish (Gambusia affinis). Aquat. Toxicol. 188, 43–53. doi: 10.1016/j.aquatox.2017.04.008
Benarroch, E. E. (2011). Heat shock proteins: multiple neuroprotective functions and implications for neurologic disease. Neurology 76, 660–667. doi: 10.1212/WNL.0b013e31820c3119
Benjamín, C., Cláudia, A., Juanmiguel, M., Dinis, M. T., and Conceicao, L. E. C. (2008). High stocking density induces crowding stress and affects amino acid metabolism in Senegalese sole Solea senegalensis (Kaup, 1858) juveniles. Aquacult. Res. 39, 1–9. doi: 10.1111/j.1365-2109.2007.01845.x
Beveridge, M. C. M., Phillips, M. J., and Macintosh, D. J. (1997). Aquaculture and the environment: the supply of and demand for environmental goods and services by Asian aquaculture and the implications for sustainability. Aquacult. Res. 28, 797–807. doi: 10.1046/j.1365-2109.1997.00944.x
Bodine, S. C., Latres, E., Baumhueter, S., Lai, V. K. M., Nunez, L., Clarke, B. A., et al. (2001). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708. doi: 10.1126/science.1065874
Braun, N., de Lima, R. L., Baldisserotto, B., Dafre, A. L., and de Oliveira Nuñer, A. P. (2010). Growth, biochemical and physiological responses of Salminus brasiliensis with different stocking densities and handling. Aquaculture 301, 22–30. doi: 10.1016/j.aquaculture.2010.01.022
Choi, R. H., McConahay, A., Jeong, H. W., McClellan, J. L., Hardee, J. P., and Carson, J. A. (2017). Tribbles 3 regulates protein turnover in mouse skeletal muscle. Biochem. Biophys. Res. Commun. 493, 1236–1242. doi: 10.1016/j.bbrc.2017.09.134
Ciechanover, A. (1994). The ubiquitin–proteasome proteolytic pathway. Cell 79, 13–21. doi: 10.1016/0092-8674(94)90396-4
Cleveland, B. M., and Evenhuis, J. P. (2010). Molecular characterization of atrogin-1/F-box protein-32 (FBXO32) and F-box protein-25 (FBXO25) in rainbow trout (Oncorhynchus mykiss): expression across tissues in response to feed deprivation. Comp. Biochem. Physiol. B 157, 248–257. doi: 10.1016/j.cbpb.2010.06.010
Dobly, A., Martin, S. A. M., Blaney, S. C., and Houlihan, D. F. (2004). Protein growth rate in rainbow trout (Oncorhynchus mykiss) is negatively correlated to liver 20S proteasome activity. Comp. Biochem. Physiol. A 137, 75–85. doi: 10.1016/j.cbpb.2003.09.002
Dodson, M., Redmann, M., Rajasekaran, N. S., Darley-Usmar, V., and Zhang, J. H. (2015). Keap1-Nrf2 signalling and autophagy in protection against oxidative and reductive proteotoxicity. Biochem. J. 469, 347–355. doi: 10.1042/BJ20150568
Flick, K., and Kaiser, P. (2012). Protein degradation and the stress response. Semin. Cell Dev. Biol. 23, 515–522. doi: 10.1016/j.semcdb.2012.01.019
Fraser, K. P. P., and Rogers, A. D. (2007). Protein metabolism in marine animals: the underlying mechanism of growth. Adv. Mar. Biol. 52, 267–362. doi: 10.1016/S0065-2881(06)52003-6
Frydman, J., Nimmesgern, E., Ohtsuka, K., and Hartl, H. U. (1994). Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature 370, 111–117. doi: 10.1038/370111a0
Glickman, M. H., and Ciechanover, A. (2002). The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428. doi: 10.1152/physrev.00027.2001
Howell, J. J., Ricoult, S. J. H., Ben-Sahra, I., and Manning, B. D. (2013). A growing role for mTOR in promoting anabolic metabolism. Biochem. Soc. Trans. 41, 906–912. doi: 10.1042/BST20130041
Huynh, F. C., Nguyen, D., and Jones, F. E. (2017). Trastuzumab stimulation of ribosomal protein S6 kinase 1 (S6K1) predicts de novo trastuzumab resistance. Biochem. Biophys. Res. Commun. 483, 739–744. doi: 10.1016/j.bbrc.2016.12.072
Jackson, P. K., and Eldridge, A. G. (2002). The SCF ubiquitin ligase: an extended look. Mol. Cell. 9, 923–925. doi: 10.1016/S1097-2765(02)00538-5
Jin, Y., Tian, L. X., Zeng, S. L., Xie, S. W., Yang, H. J., Liang, G. Y., et al. (2013). Dietary lipid requirement on non-specific immune responses in juvenile grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 34, 1202–1208. doi: 10.1016/j.fsi.2013.01.008
Kirschke, E., Goswami, D., Southworth, D., Griffin, P. R., and Agard, D. A. (2014). Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157, 1685–1697. doi: 10.1016/j.cell.2014.04.038
Li, D., Liu, Z., and Xie, C. (2012). Effect of stocking density on growth and serum concentrations of thyroid hormones and cortisol in Amur sturgeon, Acipenser schrenckii. Fish Physiol. Biochem. 385, 511–520. doi: 10.1007/s10695-011-9531-y
Liu, H. H., Li, X. X., Sun, L. L., Wang, H. H., Zhang, R. P., Yang, C., et al. (2014). Effects of the regulation of follistatin mRNA expression by IGF-1 in duck (Anas platyrhynchos) skeletal muscle. Growth Horm. IGF Res. 24, 35–41. doi: 10.1016/j.ghir.2013.12.003
Liu, S., Gao, G., Palti, Y., Cleveland, B. M., Weber, G. M., and Rexroad, C. E. (2014). RNA-seq analysis of early hepatic response to handling and confinement stress in rainbow trout. PLoS One 9:e88492. doi: 10.1371/journal.pone.0088492
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, J., Chen, D., and Yu, B. (2010). Effects of different dietary protein sources on expression of genes related to protein metabolism in growing rats. Br. J. Nutr. 104, 1421–1428. doi: 10.1017/S000711451000231X
Lupatsch, I., Santos, G. A., Schrama, J. W., and Verreth, J. A. J. (2010). Effect of stocking density and feeding level on energy expenditure and stress responsiveness in European sea bass Dicentrarchus labrax. Aquaculture 298, 245–250. doi: 10.1016/j.aquaculture.2009.11.007
Lushchak, V. I. (2011). Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 101, 13–30. doi: 10.1016/j.aquatox.2010.10.006
Malik, A., Abbas, G., Ghaffar, A., Ferrando, S., Gallus, L., and Shah, S. S. A. (2018). Effect of Different Salinity Level on breeding, fertilization, hatching and survival of Nile Tilapia, Oreochromis niloticus (Linnaeus, 1758) in Captivity. Pakistan J. Zool. 50, 539–547. doi: 10.17582/journal.pjz/2018.50.2.539.547
Pickart, C. M., and Eddins, M. J. (2004). Ubiquitin structure, functions, mechanisms. Biochim. Biophys. Acta Mol. Cell Res. 1695, 55–72. doi: 10.1016/j.bbamcr.2004.09.019
Qi, C., Xie, C., Tang, R., Qin, X., Wang, D., and Li, D. (2016). Effect of stocking density on growth, physiological responses, and body composition of juvenile blunt snout bream, Megalobrama amblycephala. J. World Aquacult. Soc. 47, 358–368. doi: 10.1111/jwas.12278
Rahim, A., Abbas, G., Ferrando, S., Gallus, L., and Ghaffar, A. (2017). Assessment of growth performance and meat quality of black fin sea bream, Acanthopagrus berda (Forsskal, 1775) Reared in Brackish Water Ponds: a preliminary investigation. Pakistan J. Zool. 49, 869–876. doi: 10.17582/journal.pjz/2017.49.3.869.876
Rebl, A., Zebunke, M., Borchel, A., Bochert, R., Verleih, M., and Goldammer, T. (2017). Microarray-predicted marker genes and molecular pathways indicating crowding stress in rainbow trout (Oncorhynchus mykiss). Aquaculture 473, 355–365. doi: 10.1016/j.aquaculture.2017.03.003
Romn, O., and Reznick, A. Z. (2016). The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass. Free Radic. Biol. Med. 98, 218–230. doi: 10.1016/j.freeradbiomed.2015.12.031
Rotllant, J., Pavlidis, M., Kentouri, M., Abad, M. E., and Tort, L. (1997). Non-specific immune responses in the red porgy Pagrus pagrus after crowding stress. Aquaculture 156, 279–290. doi: 10.1016/S0044-8486(97)00075-6
Sacheck, J. M., Hyatt, J. P. K., Raffaello, A., Jagoe, R. T., Roy, R. R., Edgerton, V. R., et al. (2007). Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 21, 140–155. doi: 10.1096/fj.06-6604com
Salas-Leiton, E., Anguis, V., Martín-Antonio, B., Crespo, D., Planas, J. V., and Infante, C. (2010). Effects of stocking density and feed ration on growth and gene expression in the Senegalese sole (Solea senegalensis): potential effects on the immune response. Fish Shellfish Immunol. 28, 296–302. doi: 10.1016/j.fsi.2009.11.006
Salem, M., Kenney, P. B., Rexroad, C. E., and Yao, J. B. (2006). Microarray gene expression analysis in atrophying rainbow trout muscle: a unique nonmammalian muscle degradation model. Physiol. Genomics 28, 33–45. doi: 10.1152/physiolgenomics.00114.2006
Siikavuopio, S. I., and Sæther, B. S. (2006). Effects of chronic nitrite exposure on growth in juvenile Atlantic cod. Gadus morhua. Aquaculture 255, 351–356. doi: 10.1016/j.aquaculture.2005.11.058
Sommer, T., and Wolf, D. H. (2014). The ubiquitin-proteasome-system. Biochim. Biophys. Acta 1843:1. doi: 10.1016/j.bbamcr.2013.09.009
Todgham, A. E., Crombie, T. A., and Hofmann, G. E. (2017). The effect of temperature adaptation on the ubiquitin-proteasome pathway in notothenioid fishes. J. Exp. Biol. 220, 369–378. doi: 10.1242/jeb.145946
Tort, L., Sunyer, J. O., Gomez, E., and Molinero, A. (1996). Crowding stress induces changes in serum haemolytic and agglutinating activity in the gilthead sea bream Sparus aurata. Vet. Immunol. Immunopathol. 51, 179–188. doi: 10.1016/0165-2427(95)05502-9
Valenzuela, C. A., Zuloaga, R., Mercado, L., Einarsdottir, I. E., Bjornsson, B. T., Valdes, J. A., et al. (2017). Chronic stress inhibits growth and induces proteolytic mechanisms through two different non-overlapping pathways in the skeletal muscle of a teleost fish. Am. J. Physiol. Regul. 314, 102–113. doi: 10.1152/ajpregu.00009.2017
VargasChacoff, L., Calvo, A., Ruizjarabo, I., Villarroel, F., Munoz, J. L., Tinoco, A. B., et al. (2011). ). Growth performance, osmoregulatory and metabolic modifications in red porgy fry, Pagrus pagrus, under different environmental salinities and stocking densities. Aquacult. Res. 42, 1269–1278. doi: 10.1111/j.1365-2109.2010.02715.x
Xie, C., Li, J., Li, D., Shen, Y., Gao, Y., and Zhang, Z. (2018). “Grass carp: the fish that feeds half of china: success stories and modern trends,” in Aquaculture in China: Success Stories and Modern Trends, ed. J. Gui (Hoboken, NJ: Wiley-Blackwell), 95–120.
Yearbook, F. A. O. (2016). Fishery and Aquaculture Statistics. Beijing: Chinese Fisheries Year Book. Beijing: China Agriculture Press.
Keywords: ubiquitin proteasome system, protein metabolism, growth, fish, crowding stress
Citation: Sun Y, Liang X, Chen J, Tang R, Li L and Li D (2018) Change in Ubiquitin Proteasome System of Grass Carp Ctenopharyngodon idellus Reared in the Different Stocking Densities. Front. Physiol. 9:837. doi: 10.3389/fphys.2018.00837
Received: 30 March 2018; Accepted: 14 June 2018;
Published: 05 July 2018.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Alexssandro Geferson Becker, Universidade Federal do Paraná, BrazilCopyright © 2018 Sun, Liang, Chen, Tang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dapeng Li, ldp@mail.hzau.edu.cn
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.