
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 19 June 2018
Sec. Invertebrate Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fphys.2018.00745
This article is part of the Research Topic Physiological Adaptations of Insects Exposed to Different Stress Conditions, Volume I View all 38 articles
A pyramid strategy combining the crystal (Cry) 1A and 2A toxins in Bacillus thuringiensis (Bt) crops are active against many species of insects and nematode larvae. It has been widely used to delay pest adaption to genetically modified plants and broaden the insecticidal spectrum in many countries. Unfortunately, Cry2A can also bind with the specific receptor proteins of Cry1A. ATP-binding cassette (ABC) transporters can interact with Cry1A toxins as receptors in the insect midgut, and ABC transporter mutations result in resistance to Bt proteins. However, there is limited knowledge of the ABC transporters that specifically bind to Cry2Ab. Here, we cloned the ABCC1 gene in Helicoverpa armigera, which expressed at all larval stages and in nine different tissues. Expression levels were particularly high in fifth-instar larvae and Malpighian tubules. The two heterologously expressed HaABCC1 transmembrane domain peptides could specifically bind to Cry2Ab with high affinity levels. Moreover, transfecting HaABCC1 into the Spodoptera frugiperda nine insect cell significantly increased its mortality when exposed to Cry2Ab in vitro, and silencing HaABCC1 in H. armigera by RNA interference significantly reduced the mortality of larvae exposed to Cry2Ab in vivo. Altogether current results suggest that HaABCC1 serves as a functional receptor for Cry2Ab.
The crystal (Cry) proteins produced by Bacillus thuringiensis (Bt) are specifically toxic to some insect pests, such as Lepidoptera, Diptera, and Coleoptera, while they are almost harmless to non-target organisms (Sanahuja et al., 2011; Pardo-López et al., 2013; Comas et al., 2014; Nicolia et al., 2014). To reduce the use of chemical insecticides, Bt proteins, such as Cry1Ac and Cry2Ab, have been used worldwide as bio-pesticide sprays or expressed in genetically modified (GM) plants to control certain insect pests (Sanahuja et al., 2011; James, 2016). The hectares of Bt crops worldwide increased from 1.1 million in 1996 to 98.5 million in 2016, Bt corn, cotton, and soybean accounted for >99% of this total amount, with a cumulative total of more than 830 million (James, 2016).
The high selection pressure of Bt could lead to the rapid evolution of insect resistance. Cases of pest resistance to Bt proteins produced by GM crops increased from 3 in 2005 to 16 in 2016 (Pardo-López et al., 2013; Jakka et al., 2016; Tabashnik and Carrière, 2017). Bt cotton (expressing the Cry1Ac protein) has been planted in China since 1997, and recent bioassay data showed that the percentage of resistant Helicoverpa armigera collected from fields in north China increased from 0.93% (2010) to 5.5% (2013) (Jin et al., 2015). To delay pest adaption to GM crops, some different resistance management strategies have been used, including trait pyramiding (Xue et al., 2008; Brévault et al., 2013; Jin et al., 2015). The “pyramid” strategy has been widely adopted to replace the first generation Bt crops. For example, the transgenic cotton which can produce Cry1Ac and Cry2Ab is the only type of Bt cotton grown in Australia. And it’s also the predominant type of Bt cotton grown in India and the United States (Tabashnik et al., 2013a; Fabrick et al., 2014).
However, the occurrence of cross-resistance sometimes weakens these advantageous characteristics of Cry1A + Cry2A “pyramid” strategy (Tabashnik et al., 2013b; Wei et al., 2015; Welch et al., 2015). Although Cry1A and Cry2A were predicted to have different structures and different mode of actions in the target lepidopteran pests because of their low amino acid homology (Hernández-Rodríguez et al., 2008; Caccia et al., 2010), but studies have indicated that they share some of the same receptors. Several important functional Cry1A receptors, such as cadherin (CAD), amionpeptidase-N (APN), or alkaline phosphatase (ALP), they have been identified as binding protein or reported to play vital functional role in the toxicity of Cry2A (Onofre et al., 2017; Yuan et al., 2017; Zhao et al., 2017). Further research showed although CAD was a functional receptor for both the Cry2Aa and Cry1Ac toxins in Spodoptera exigua, but Cry1Ac and Cry2Aa toxins did not compete for the same binding sites and may bind to diverse CAD protein epitopes (Qiu et al., 2015). Thus, the same receptors but different binding sites may associate with the asymmetrical cross-resistance between Cry1A and Cry2A (Tabashnik et al., 2009; Wei et al., 2015). So, it is urgent to find the receptor of Cry2Ab.
Recently, some ATP-binding cassette (ABC) transporters are involved in the resistance of insect to Bt toxins. For example, ABCC2 mutations cause the resistance of H. armigera and Heliothis virescen to Cry1Ac (Gahan et al., 2010; Xiao et al., 2014). The Cry1Ac-resistance of Plutella xylostella is closely related to the reduced expression of PxABCC2, PxABCC3, and PxABCG1 in the midgut (Guo et al., 2015a,b). Meanwhile, high levels of resistance to Bt Cry2Ab toxin have been verified to be genetically linked with loss of function mutations of an ABC transporter gene (ABCA2) in two Lepidopteran insects, H. armigera and H. punctigera (Tay et al., 2015). Moreover, two HaABCA2 knockout strains created from the susceptible SCD strain by using the CRISPR/Cas9 genome editing system display high levels of resistance to Cry2Ab (>100-fold) compared with the original SCD strain (Wang et al., 2017). In addition, the binding experiments showed there is more than one receptor of Cry2Ab in insect midgut (Wei et al., 2016). Functional ABC transporter family proteins contain four core domains: two membrane-spanning domains (transmembrane domains, TMs), each built from six membrane-spanning α-helices, alternating with two nucleotide-binding domains (NBDs) located on the cytosolic side (Dassa and Bouige, 2001; Linton, 2007). All of these indicated that other ABC transporters may also involve in the mode of action of Cry2Ab.
ABCC1 has four core domains like other ABC transporter proteins. Additionally, ABCC1 involved in the toxicity of Cry2Ab to H. armigera by itraq data (unpublished data). However, whether ABCC1 serves as a functional receptor for Cry2Ab in H. armigera remained unclear. Here, we discovered Cry2Ab could bind to the heterologously expressed peptides with high affinity levels, transfecting HaABCC1 into the Sf9 cell line significantly increased its mortality rate when exposed to Cry2Ab in vitro, and silencing HaABCC1 in H. armigera by RNA interference (RNAi) significantly reduced the mortality rates of larvae exposed to Cry2Ab in vivo. Based on our results, we propose that ABCC1 in H. armigera is involved in the action mode of Cry2Ab.
Susceptible H. armigera (96S strain) was collected from the cotton fields in Xinxiang County, Henan Province, China in 1996. The larvae of this colony were reared in the laboratory on an artificial diet without exposure to any Bt toxins or insecticides (Liang et al., 1999).
We collected samples from nine developmental stages of H. armigera: egg, first- to sixth-instar larva, pupa, adult (male and female). For each biological replicate, we collected samples from 200 eggs and 5–20 individuals from other developmental stages. The following different tissues: head, foregut, midgut, hindgut, Malpighian tubule, peritrophic membrane, hemolymph, fat body, and cuticle from fifth-instar larvae were collected, and tissues dissected from 25 larvae served as one biological replication. All collected samples were quickly frozen in liquid nitrogen and stored at -80°C for subsequent RNA extraction. Four biological replicates were prepared for each treatment.
The activated Bt toxin Cry2Ab was purchased from Envirologix Inc. (Portland, ME, United States). Activated Cry2Ab protein was biotinylated using the EZ-Link Sulfo N-hydroxysuccinimide Liquid Chromatography (LC) Biotinylation Kit (Pierce, FL, United States) with a 1:20 molar ratio (Cry protein:biotin) following the manufacturer’s instructions. Biotinylated Cry2Ab proteins were separated on a 4–20% SDS–polyacrylamide gel, transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Corp., Billerica, MA, United States) (150 mA, 1 h) and were treated in 5% (w/v) bovine serum albumin (BSA) diluted in phosphate buffered saline (PBS; 135 mM NaCl, 2 mM KCl, 10 mM Na2HPO4, and 1.7 mM KH2PO4, pH 7.4) containing 0.5% (v/v) Tween-20 (PBST) at 4°C overnight. The membranes were washed three times with PBST, and streptavidin horseradish peroxidase was used to detect biotinylated toxins. The blots were developed with the Easysee Western Blot Kit (Transgen, Beijing, China) and observed in a multifunction laser imager (TYPhoon 9410, GE Healthcare, United States).
TRIzol reagent (Invitrogen, CA, United States) was used to extract total RNA from the midguts of 25 fifth-instar larvae of H. armigera according to the manufacturer’s instructions. Then, contaminating genomic DNA was removed by treating with DNase I (TakaRa, Japan). The 260/280 and 260/230 ratios measured using a NanoDrop 3300 (Thermo Fisher, MA, United States) were used to evaluate the purity of the total RNA, and a 1% agarose gel was used to determine the integrity. Then, the first-strand cDNA was synthesized immediately from 1 μg of total RNA using a SuperScriptTM III First-Strand Synthesis Kit (Invitrogen) following the manufacturer’s instructions and stored at -20°C for further use.
Parts of the ABCC1 gene were obtained from the previous transcriptome sequencing data in our laboratory. Rapid amplification of cDNA ends (RACE) was used to obtain the full-length cDNA. RACE-ready cDNAs were amplified using a SMARTScribeTM RACE cDNA Amplification Kit (Clontech, Mountain View, CA, United States) according to the manufacturer’s instructions. Gene-specific primers were designed based on the fragments of the ABCC1 gene. The primers were designed by Primer Premier 5.0, and UPM was used as the universal primer (Supplementary Table S1). RACE PCR was used under the following conditions: Initial denaturation at 94°C for 4 min, followed by five cycles at 94°C for 30 s and 72°C for 2.5 min, followed by another five cycles at 94°C, for 30 s, 70°C for 30 s, and 72°C for 2.5 min. Subsequently, 25 cycles were performed at 94°C for 30 s, 68°C for 30 s, and 72°C for 2.5 min, followed by 72°C for 10 min. To ensure the entire open reading frame (ORF) was amplified, specific primers that included initiation and stop codons were designed and used to amplify the entire ORF sequence. PCR products were separated by 1% agarose gel electrophoresis, and the expected bands were gel-purified and cloned into the pEasy-T3 vector (TransGen). The cloned fragments were then sequenced. The full sequence was obtained and submitted to GenBank (accession no. KY796050).
The NCBI BLAST database was used to analyze the homology of the ABCC1 gene with other ABC sequences (Supplementary Table S2). The molecular weights and isometric points of the proteins were predicted using the ExPaSy proteomics server website1. The TM helices were analyzed using TMHMM Server v.2.02. The protein domains were predicted using the ExPaSy-PROSITE3. The N-terminal signal peptide positions were determined using SignalP 4.1 Server4. NetNGlyc was analyzed using NetNGlyc 1.0 Server5. The NetOGlyc were analyzed using NetOGlyc 4.0 Server6. The percentage of amino acid sequence identity was calculated using ClustalW, and the phylogenetic tree was constructed in MEGA 7.0, using the neighbor-joining method (Tamura et al., 2013).
Based on the nucleotide sequence of the gene encoding HaABCC1, we designed two pairs of primers with EcoRV restriction sites contained in forward primers and HindIII restriction sites contained in reverse primers, to clone two fragments of HaABCC1 that incorporated the potential toxin-binding regions, which we termed TMD1 and TMD2, respectively. The primers used for the PCR-amplification of TMD1 and TMD2 fragment were TM1-EcoRV-F and TM1-HindIII-R, and TM2-EcoRV-F and TM2-HindIII–R, respectively (Supplementary Table S4).
Then, the synthesized cDNA was used as the template to amplify the two partial fragments by PCR. The PCR program included denaturation at 95°C for 3 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 90 s, and final extension at 72°C for 10 min. After purified by AxyPrep DNA Gel Extraction Kit (Axygen Scientific, CA, United States), the PCR products were cloned into the pEASY-T3 vector (TransGen Biotech), and then transformed into competent cell, Escherichia coli Trans1-T1 cells (TransGen Biotech). The recombinant plasmids were double-digested with EcoRV and HindIII (TaKaRa, Dalian, China) for 3 h at 37°C, and the products were subcloned into the expression vector pET32a (Novagen, United States), which was digested with the same restriction enzymes to generate the two pETHaABCC1 recombinant plasmids, which were transfected into E. coli BL21 (DE3) cells (Tiangen, Beijing, China) for protein expression.
The two HaABCC1 proteins fragments were tested by gradient SDS–polyacrylamide gel electrophoresis (PAGE) 4–20% (Genscript Biology Co., China). Then, the two HaABCC1 proteins fragments were identified by LC-MS/MS (Q-TOF) at the Beijing Protein Institute (Beijing, China).
The expressed HaABCC1 protein fragments were purified using a micro-protein PAGE recovery kit (Sangon Biotech., Shanghai, China). The two HaABCC1 fragments were tested for purity using a 4–20% gradient SDS–PAGE (Genscript Biology Co., NJ, United States).
For the ligand blot analysis, 10 μg purified HaABCC1 fragments were separated using 4–20% gradient SDS-PAGE gels (Genscript Biology Co., NJ, United States) and then electro-transferred onto PVDF filters (Millipore Corp.). After blocking, the membrane was incubated with biotinylated Cry2Ab toxin (5 μg/ml) in blocking buffer for 1 h at room temperature and then washed three times (15 min each) with PBST. Then, the PVDF membrane was incubated with horseradish peroxidase-conjugated anti-rabbit IgG (ZSGB-BIO, Beijing, China) as a secondary antibody in blocking buffer (1:10,000) for 1 h at room temperature. After additional washing, the membrane was developed using an EasySee Western Blot Kit (TransGen) and exposed on an ImageQuant LAS4000mini system (GE Healthcare, Japan).
Total RNA extraction and cDNA synthesis of samples from different tissues and developmental stages were performed as described above. The relative mRNA expression levels of the ABCC1 gene were analyzed by qRT-PCR. The primers can be found in Supplementary Table S3. Each TaqMan qRT-PCR (Tiangen) reaction was performed in a total volume of 20 μl containing the following components: 1 μl template cDNA, 10 μl 2 × SuperReal PreMix (probe), 0.6 μl each 10 μM primer, 0.4 μl 10 μM probe, 0.2 μl 50 × ROX Reference Dye and 7.2 μl RNase-Free ddH2O. All qRT-PCR reactions were performed in 96-well optical plates in an ABI 7500 Real-time PCR System (Applied Biosystems). qRT-PCR was performed for 40 cycles of 95°C for 3 s and 60°C for 32 s. Both β-actin (GenBank EU527017) and GAPDH (GenBank JF417983) genes were used internal references, and the expression level of the target gene (ABCC1) for each treatment was normalized with the geometric mean of the expression of the two reference genes (β-actin and GAPDH) (Liu et al., 2015; Zhang et al., 2016). To check reproducibility, six biological replicates were analyzed with each biological replicate consisting of three technical repeats.
The Spodoptera frugiperda 9 (Sf9) cell line was cultured in Sf-900 II SFM medium (GIBCO/BRL/Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone-QB perbio, Logan, UT, United States), 50 U/ml penicillin, 50 mg/ml streptomycin, and 12 mg/ml gentamycin (Invitrogen) in an incubator at 28°C (Cordova et al., 2006).
The whole ORF of ABCC1 was cloned into pAc5.1b vector (Huayueyang Biotechnology Co., Ltd, Beijing, China). First, we designed gene-specific primers that contained two restriction enzyme sites. The primers for the PCR-amplification of the ABCC1 ORF were ORF-PmeI-F and ORF-StuI-R (Supplementary Table S4). The PCR reaction and conditions were described in Section 2.4.2. PCR products were cloned into pEasy-T3 vector (TransGen), followed by restriction enzyme digestion with PmeI and StuI (TaKaRa). The pAc5.1b vector was also digested with PmeI and StuI for 3 h at 37°C. The double-enzyme digestion products were electrophoresed on a 1% low-melting point agarose gel (Invitrogen) and the PCR products were purified by AxyPrep DNA gel extraction kit. Then, the ABCC1 gene was ligated into the pAc5.1b vector for 3 min at 22°C using T4 DNA ligase (Thermo Fisher). The ligation product was transfected into E. coli Trans1-T1 cells (TransGen), and individual clones were selected from an LB plate.
First, the activated Cry2Ab was precipitated by adding 4 M acetic acid dropwise to the activation reaction tube to adjust the pH to 8.0. Then, the tube was incubated at 4°C for 30 min and centrifuged at 12,000 × g for 20 min. The activated Cry2Ab pellets were washed three times using 40 ml ice cold ddH2O, dissolved in 15 ml Sf-900 II SFM media (GIBCO/BRL/Life Technologies) and centrifuged at 12,000 × g for 5 min. The upper supernatant was transferred into a clean tube and used as the stock solution for cytotoxicity assays. Finally, the concentrations of activated Cry2Ab toxin in the stock solution were estimated by electrophoresis of 15 μl 200 μg/ml BSA solution as well as 15 μl of Cry2Ab stock solution using SDS–PAGE, and the intensity of the corresponding bands was quantified with Image J software (NIH, v1.46).
Sf9 cells were seeded onto a 12-well plate (∼9 × 105 cells/well), allowing cells to attach overnight (12 h). Then, cells were transiently (5.5 h) transfected with (∼2 mg/well) pAc-ABCC1 plasmid using Cellfectin (Invitrogen; 8 μl per well). The empty pAc5.1b vector was used as the control. The transfection mixture was removed and replaced with 1.5 ml of supplemented medium (Sf-900 II SFM media containing antibiotics and serum). The cells were incubated at 28°C for 60 h. Then, the cells were collected and the cell concentration was measured with a hemocytometer using Trypan blue. In a 96-well micro-plate, 100 μL of cells (∼10,000 cells) were reseeded allowing cells to attach for at least 2.5 h. Using activated Cry2Ab toxin (0.0091 mg/ml) to treat the cells, the mortality was calculated after 5 h (Wei et al., 2016).
RNA interference with small interfering RNA (siRNA) was conducted by microinjection to study the role of HaABCC1 in the susceptibility of H. armigera larvae to the Bt toxin Cry2Ab. The HaABCC1 siRNA and enhanced green fluorescent protein (EGFP) siRNA (negative control) used were sequence specific and custom synthesized (Invitrogen). The sequences 5’-GGAUGUACCUGGUGGGCAUTT-3’ and 5’-GCGUUGGGAAGUCAAGUUUTT-3’designed based on the gene-specific TMD region to avoid potential off-target effects were used as siHaABCC1 and siEGFP, respectively.
Two microgram siRNA (siHaABCC1 and siEGFP) by 5 μl-microsyringe (Hamilton, Bonaduz, Switzerland) or 2 μg DEPC water (blank control) was microinjected in the abdomen of the newly emerged third-instar larvae. The injection point was sealed immediately with geoline. In addition, a third parallel non-treated control was performed. Each treatment included 24 individuals and replicated three times. To calculate the RNAi efficiency by qRT-PCR as the above described, 10 larvae were randomly selected for testing at 48 h after the injection.
To evaluate the susceptibility of H. armigera to Cry2Ab after RNAi, diet overlay bioassays were used (Hernández-Rodríguez et al., 2008). The toxin was dissolved and diluted in PBS. At the beginning, we distributed 1 ml liquid artificial diet into each well of 24-well plates (TianJin Xiangyushun Co., TianJin, China). After the diet was solidified, 75 μl solution of Bt toxin Cry2Ab (105 μg/cm2) and PBS (control) were overlaid on the surface of the artificial diet on two 24-well plates, respectively. Finally, within 48 h of microinjection, 24 third-instar larvae were placed on the surface of the dried diet in the 24-well plates. Each treatment included 24 larvae, and there were three replicates. After 5 days, the numbers of dead larvae were recorded (Zhou et al., 2010).
Cell mortality was calculated as the description in Wei et al. (2016). Significant differences among the different treatments were analyzed by one-way analysis of variance (ANOVA), followed by Tukey’s honestly significance difference (HSD) test for mean comparison. All statistical analysis was performed with SPSS v.18.0 (SPSS Inc., Chicago, IL, United States) at P < 0.05 level of significance.
The ORF of HaABCC1 (GenBank KY796050) was 4,545 bp, and it encoded predicted proteins of 1,515 amino acid, with a predicted molecular masses of 169.75 kDa. It predicted isoelectric point was 6.68. The protein sequence analysis revealed that HaABCC1 had no signal peptide. Additionally, it had 14 TM helices, 14 NetNGlyc sites, 16 NetOGlyc sites, and four domains for HaABCC1 (Figure 1). In total, 33 ABCC1–3 sequences from various species, including those from the current study and those available in GenBank, were aligned and used to construct a phylogenetic tree (Figure 2). The phylogenetic tree showed that the ABCC1, ABCC2, and ABCC3 proteins were closely related in different lepidopteran insects. HaABCC1 was most closely to SlitABCC1, AtraABCC1, PpolABCC1, and PxutABCC1.
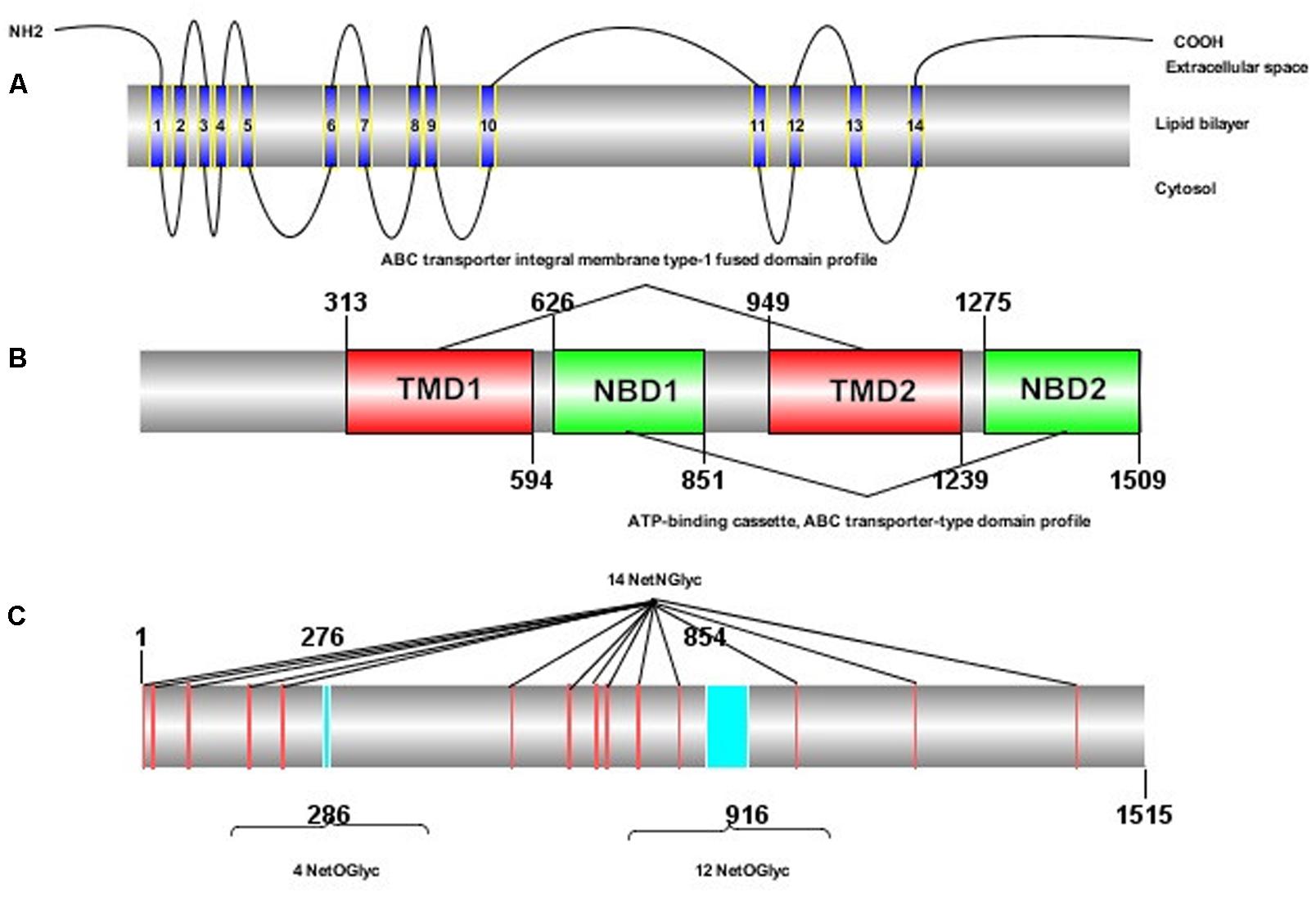
FIGURE 1. Protein structure of HaABCC1 in H. armigera. (A) Transmembrane structure of the HaABCC1 protein. The protein contains 14 transmembrane segments. (B) Domains of the HaABCC1 protein. The protein contains two ATP-binding cassette (ABC) transporter integral membrane type-1 fused domain profiles (TMD) and two ABC transporter-type domain profiles (NBD). (C) NetNGlyc and NetOGlyc sites of the HaABCC1 protein. The protein contains 14 NetNGlyc sites and 16 NetOGlyc sites.
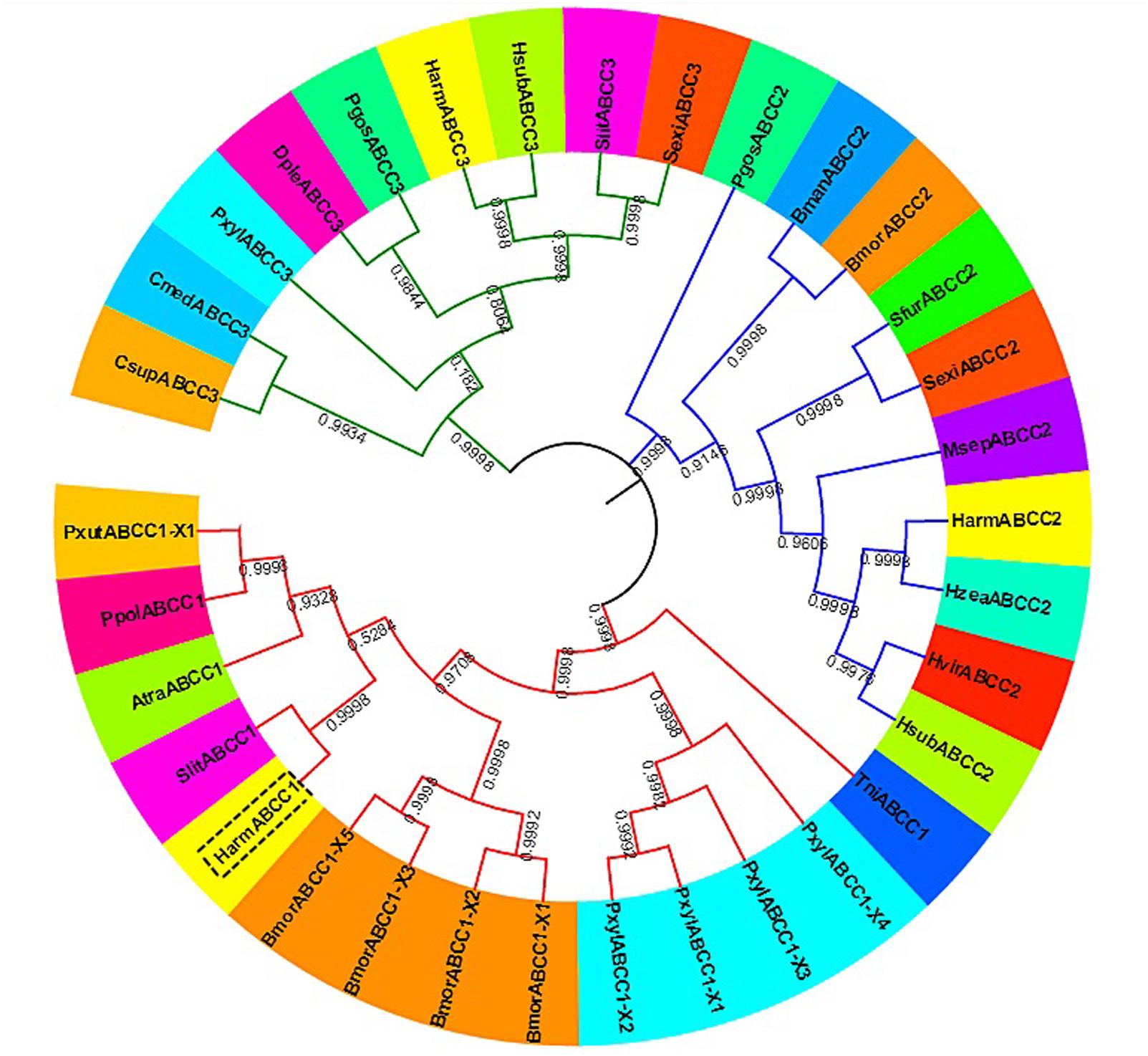
FIGURE 2. An unrooted phylogenetic tree showing the phylogenetic relationships of ABCC1–3 genes from different insect species. The tree was generated by a ClustalW alignment of the full-length amino acid sequences of ABCC1–3 using the neighbor-joining method in MEGA 7.0. Bootstrap values are expressed as percentages of 1,000 replications. GenBank accession numbers or Gene IDs are displayed in parentheses in Supplementary Table S2.
HaABCC1 was expressed in all developmental stages but peaked in the fourth- and fifth-instar larvae (p < 0.001) (Figure 3A). Meanwhile, Expression levels of HaABCC1 were markedly different among the tissues (p < 0.001) (Figure 3B). The results showed it expressed in midgut, but the highest expression was detected in the Malpighian tubules, for other tissues, the expression levels of HaABCC1 were relatively low and not significantly different.
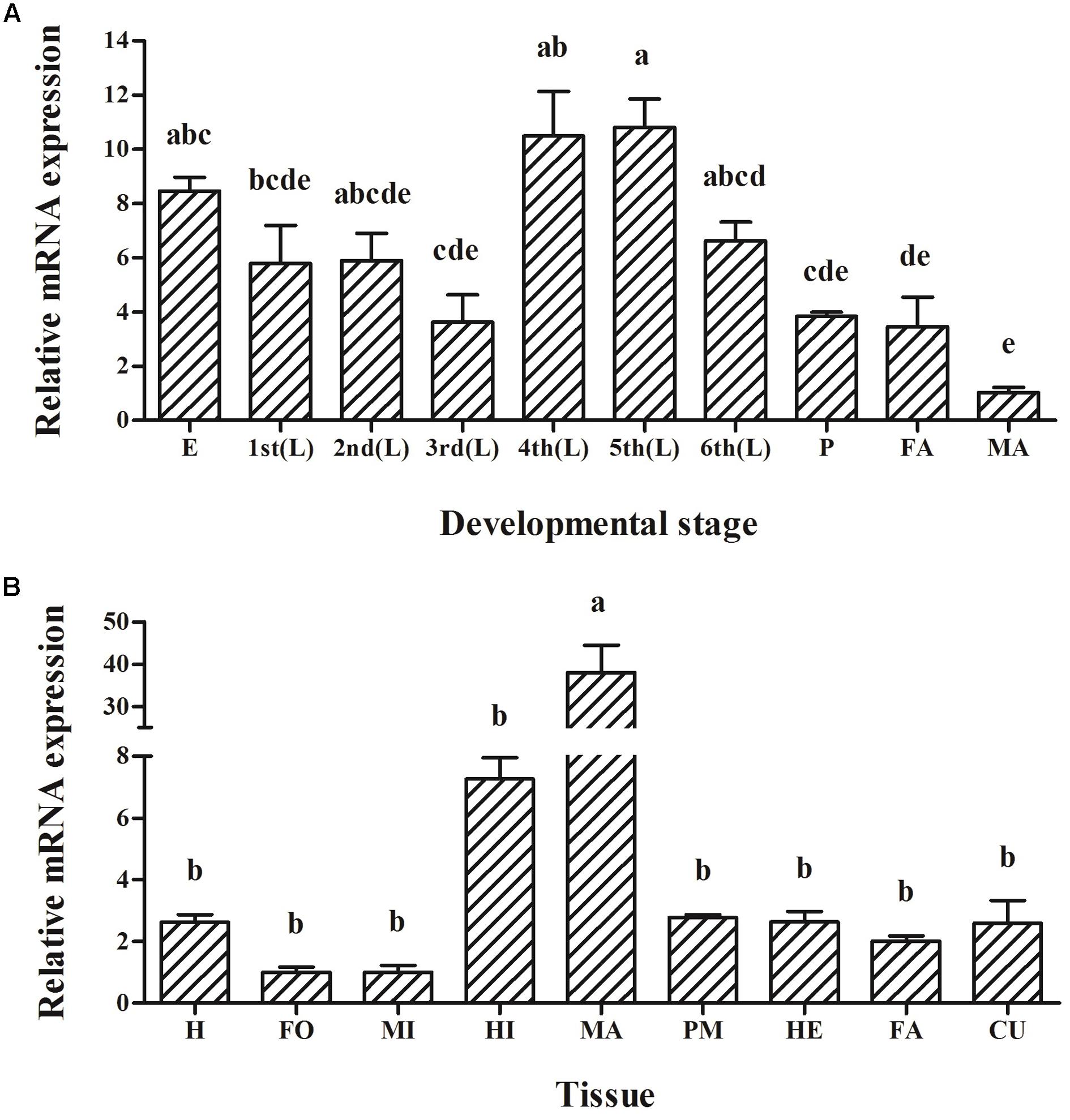
FIGURE 3. Expression of the HaABCC1 gene in different developmental stages and tissues of H. armigera larvae as determined by a RT-qPCR analysis. (A) Relative expression levels of HaABCC1 in eggs (E), first-instar larvae (1st), second-instar larvae (2nd), third-instar larvae (3rd), fourth-instar larvae (4th), fifth-instar larvae (5th), sixth-instar larvae (6th), pupae (P), female adults (FA), and male adults (MA). (B) Relative expression levels of HaABCC1 in head (H), foregut (FO), midgut (MI), hindgut (HI), Malpighian tubule (MA), peritrophic membrane (PM), hemolymph (HE), fat body (FA), and cuticle (CU) from fifth-instar larvae. Values shown are means and standard errors. Different letters indicate significant expression differences among different tissues or developmental stages based on three biological replications and four technical repeats (p < 0.05).
Two cDNA fragments, TMD1 and TMD2, of the H. armigera gene HaABCC1 (924-bp and 882-bp) were cloned and expressed in E. coli BL21 (DE3) cells. In prediction, these cDNA fragments were translated to peptides of 308 and 294 amino acid residues, respectively, of which the molecular weights were 34 and 33 kDa, respectively. All proteins extracted from E. coli BL21 (DE3) cells were confirmed by SDS–PAGE (Figure 4A), and the recombinant proteins (HaABCC1-pET32a construct) were present in the pelleted inclusion bodies. The molecular masses of these proteins were consistent with the predicted molecular weights. The expressed HaABCC1 protein fragments were purified, and the LC-MS/MS (Q-TOF) indicated that they were parts of the HaABCC1 protein. To test for specific interactions between the Cry2Ab toxins and HaABCC1, a ligand blot analysis was conducted, and the two expressed HaABCC1 fragments were bound to the activated Cry2Ab. TMD2 had a higher binding affinity than TMD1 (Figure 4B).
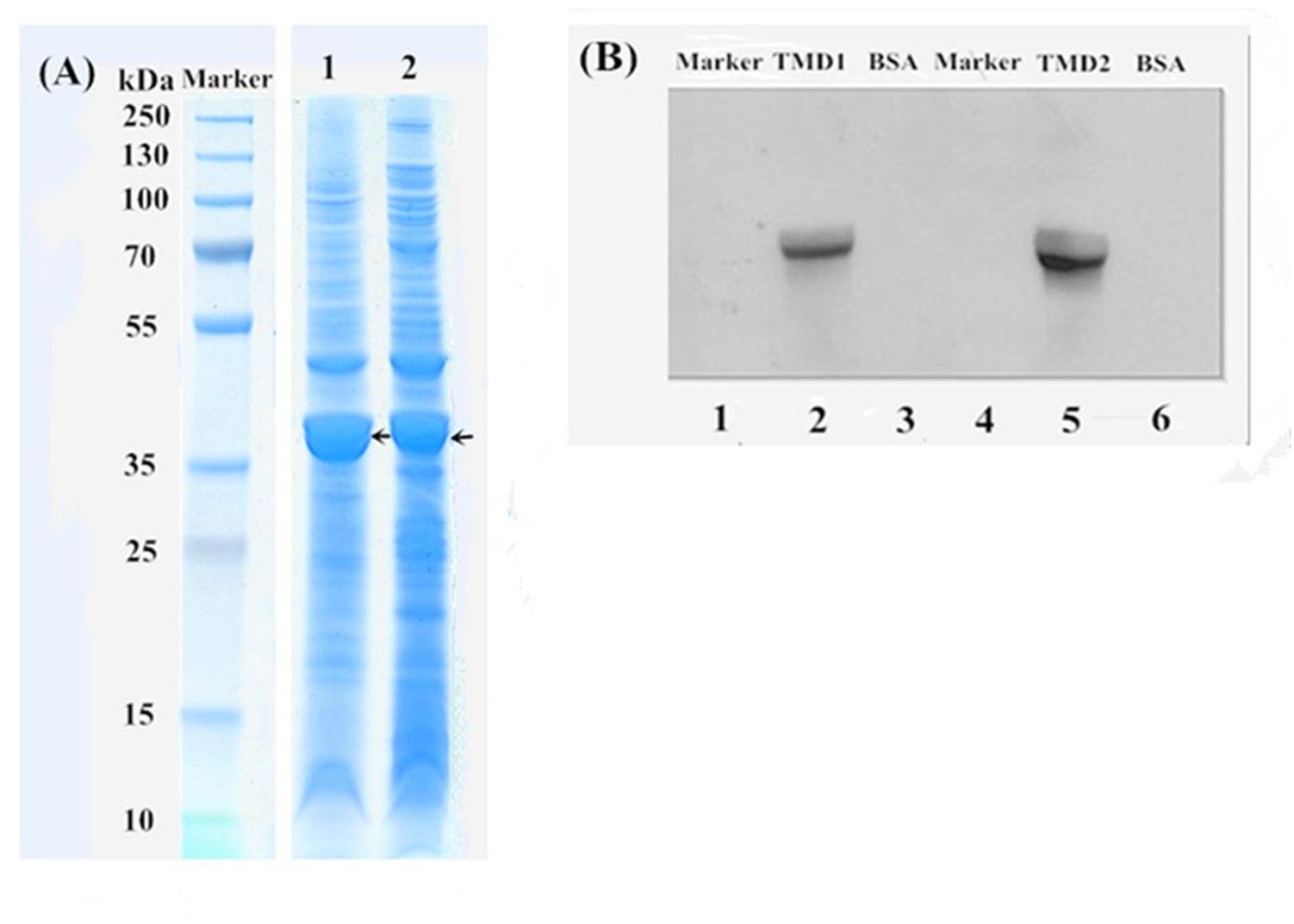
FIGURE 4. SDS–PAGE and a ligand blot analysis of the HaABCC1 fragments. (A) Marker, protein marker; Lane 1, TMD1 fragment after ultrasound treatment; Lane 2, TMD2 fragment after ultrasound treatment. (B) Lanes 1 and 4, protein marker; Lanes 3 and 6, bovine serum albumin (BSA), as a control; Lane 2, TMD1 fragment bound with Cry2Ab; Lane 5 TMD2 fragment bound with Cry2Ab.
HaABCC1 was expressed after be transfected into Sf9 cells (Figure 5A). The additional expression of HaABCC1 led to the Cry2Ab susceptibility significantly increasing, with 41.25% cells dying after being treated by 0.0091 mg/ml Cry2Ab. The mortality rate was significantly greater than that of the control (2.35%) (Figure 5B).
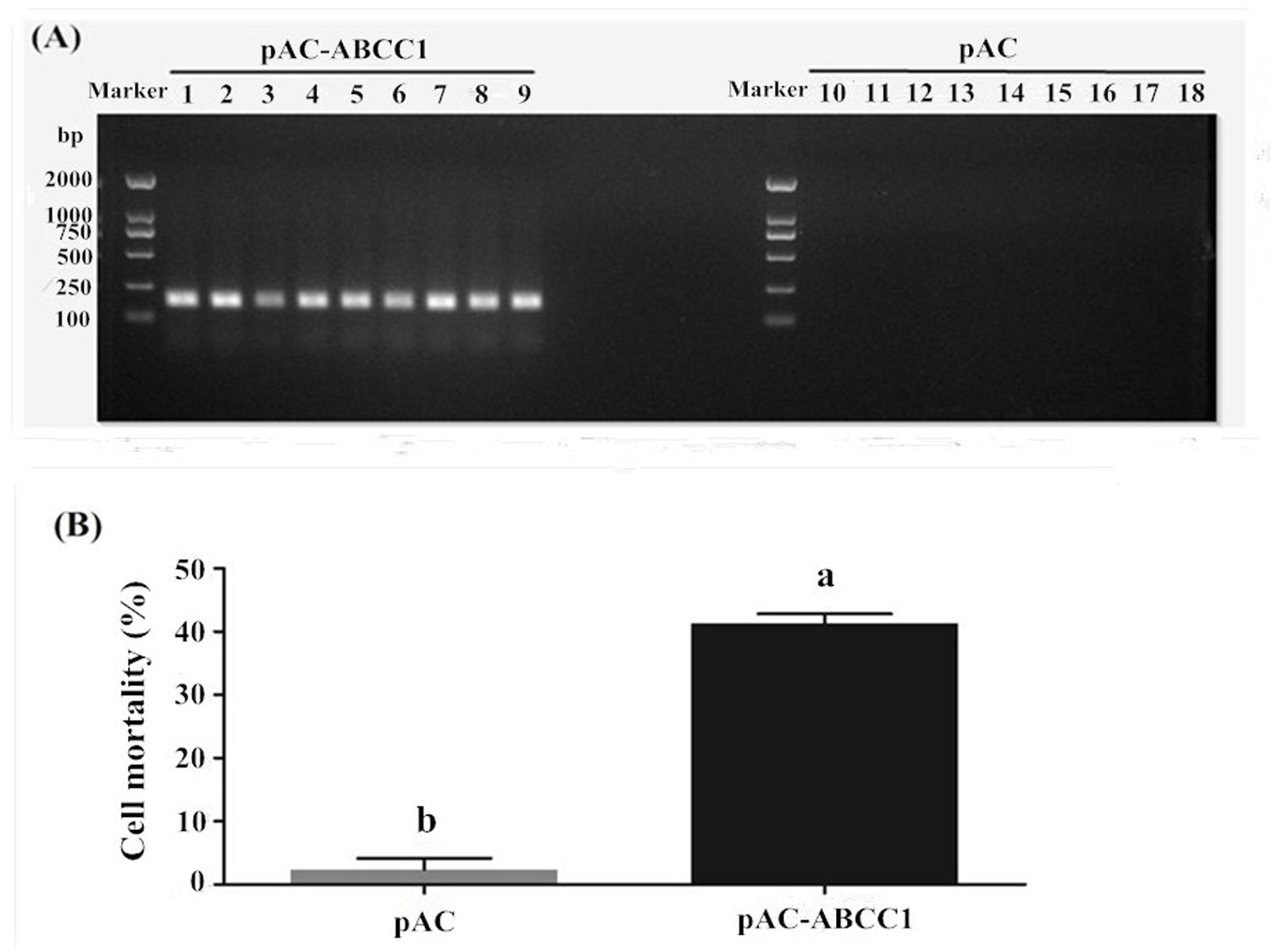
FIGURE 5. Effects of pAc-ABCC1 plasmid transfection in Sf9 cells on their mortality after being treated with activated Cry2Ab. (A) Confirmation of ABCC1 expression in cells transfected with pAc-ABCC1 by PCR. Lines 1–9: pAc-ABCC1 plasmid transfected into the Sf9 cell line (3 biological replicates × 3 technical replicates). Lines 10–18: pAC empty plasmid transfected into the Sf9 cell line. (B) The mortality rate of Sf9 cells exposed to Cry2Ab was after being transfected with the pAc-ABCC1 or pAC empty plasmid. Different letters indicate significant differences between treatments (p < 0.05).
To further explore the potential function of the HaABCC1 gene in the action mode of Cry2Ab against H. armigera, we used in vivo RNAi to knockdown HaABCC1 expression by injecting siABCC1 into early third-instar larvae. The siABCC1 sequence was complementary to the internal gene-specific TMD region of the HaABCC1 mRNA. The qPCR analysis showed that there were no significant differences among controls (injections of DEPC water and siEGFP, and the non-treated control). Additionally, the injection of siABCC1 into larvae significantly reduced HaABCC1 transcript levels by 54.0, 49.4, and 52.2% relative to the non-treated, DEPC water- and siEGFP-injected larvae, respectively (p < 0.001) (Figure 6A). When larvae pretreated with siRNA of HaABCC1 for 2 days were transferred to a diet containing Cry2Ab, the larval mortality rate of the Cry2Ab-treated group decreased by 70.0, 70.7, and 65.7% relative to non-treated, DEPC water- and siEGFP-injected larvae, respectively. Moreover, there were no significant differences among the three controls (p > 0.05) (Figure 6B).
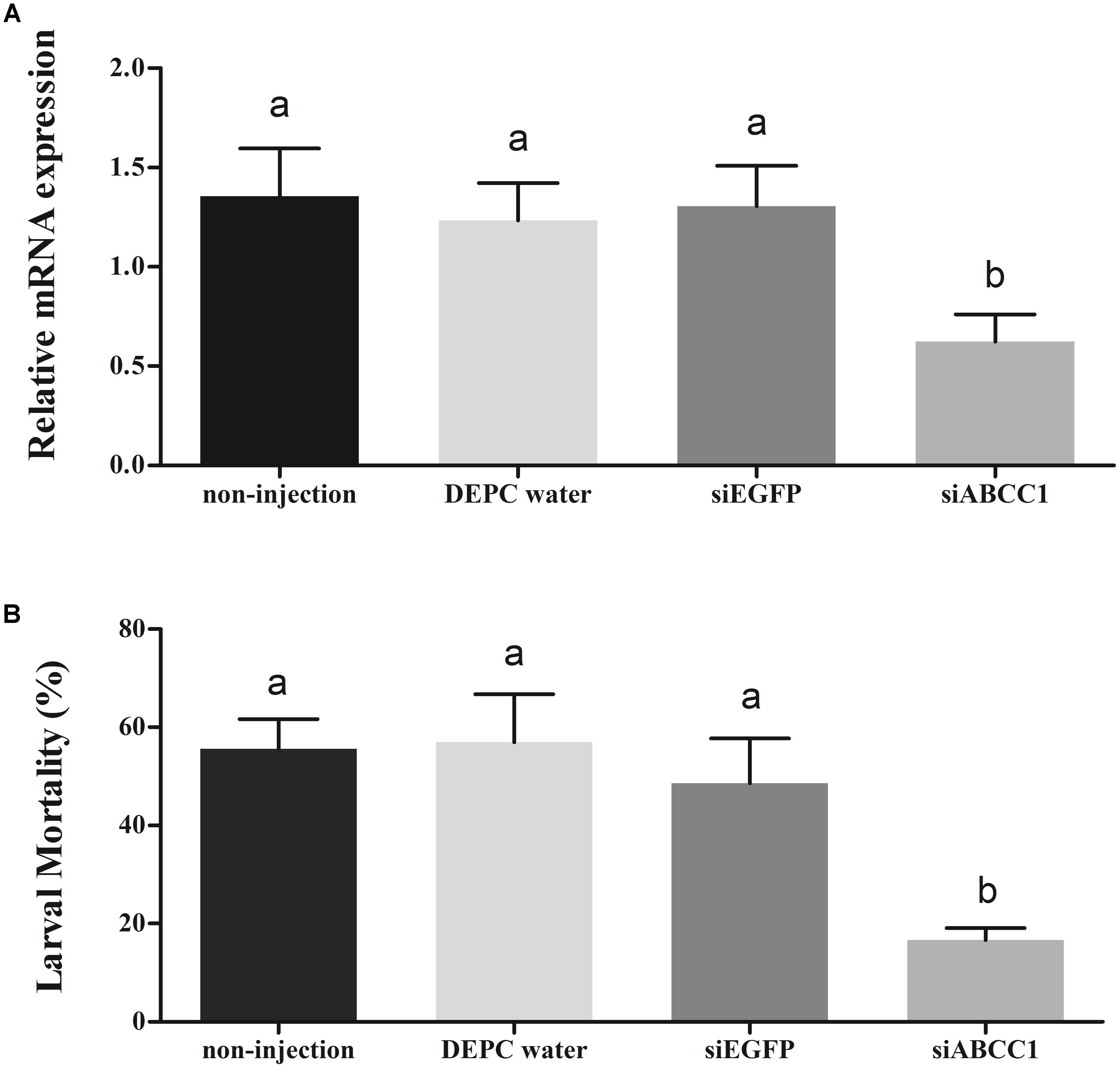
FIGURE 6. Silencing of HaABCC1 expression and its effects on H. armigera susceptibility to Cry2Ab. (A) Effects of the injection of H. armigera larvae with DEPC water, siRNA-targeting (si) EGFP, or siABCC1 on the relative expression of HaABCC1 at 48 h post-RNAi treatment. A non-injected control was also used. Different letters indicate significant differences among treatments (p < 0.05). (B) Susceptibility of H. armigera third-instar larvae to active Cry2Ab toxin as affected by prior injection with DEPC water, siEGFP, or siABCC1. A non-injected control was also used. Different letters indicate significant differences among treatments (p < 0.05).
Recently, ABC transporters, especially members of the ABCA, ABCB, ABCC, ABCG, and ABCH subfamilies, have become a focus of research in arthropods because of their important roles in xenobiotic transport and insecticide resistance (Dermauw and Van Leeuwen, 2014; Merzendorfer, 2014; Xiao et al., 2014; Guo et al., 2015a,b; Tay et al., 2015). The HaABCC1 in our study had a structure similar to those of other ABC transporters, containing two extracellular domains that were present as long loops between helices TM I and TM II, 14 NetNGlyc sites and 16 NetOGlyc sites (Figure 1). Moreover, its sequence was similar to SlitABCC1, AtraABCC1, PpolABCC1, and PxutABCC1 (Figure 2). HaABCC1 expression was widespread in the H. armigera larvae (Figures 3, 4), with the highest expression level detected in fourth- and fifth-instar larvae, and in Malpighian tubules, which are part of the excretory and osmoregulatory systems in insect larvae. It was confirmed that ABCC1 was expressed highest in Malpighian tubules in H. armigera larvae (Bretschneider et al., 2016). Thus, HaABCC1 may have functions similar to those of ABC transporters involved in Bt intoxication.
For Cry1A toxins, a sequential mode of action had been proposed, and some specific and saturable binding membrane targets on the midgut are important for the toxicity (Bravo et al., 2011; Heckel, 2012). The toxin first binds to membrane-bound glycosylated proteins, such as aminopeptidases, ALP, and other glycoproteins, and then binds to the 12-CAD domain protein, resulting in processing and accelerated oligomerization (Bravo et al., 2011; Vachon et al., 2012). Cry2A proteins have three domain structures comparable to those of Cry1A toxins (Pigott and Ellar, 2007; Hernández-Rodríguez et al., 2008), making them likely to act in similar ways as pore-forming toxins. Upon activation, Cry2A toxins can bind to the glycosylated loops of TMD1 and/or TMD2 in ABC transporters of Helicoverpa species, and this binding can form the basis of oligomerization and bring the pre-pore structures close to the TMDs for pore insertion (Hernández-Rodríguez et al., 2008; Gahan et al., 2010). Additionally, other proteins may be involved in Cry2Ab binding and pore formation, particularly because mammalian ABCs may occur in multi-protein complexes in the membrane (Kaminski et al., 2006). In our study, we also found the two expressed HaABCC1 fragments (TMD1 and TMD2) could bind with the activated Cry2Ab as assessed by ligand blotting experiments (Figure 4B). We hypothesized that ABCC1 may also provide binding and pore insertion functions in the action mode of Cry2Ab, like other ABC transporters.
In fact, some ABC transporters, like ABCC2, ABCC3, and ABCG1 etc. have been proved as functional receptors for Cry1A (Xiao et al., 2014; Guo et al., 2015a,b; Tanaka et al., 2016). For example, the over-expression of SlABCC3 strongly increases the susceptibility of Trichoplusia ni Hi5 cells to Cry1A, which suggested that ABCC3 was also a functional receptor of Cry1A toxins (Chen et al., 2015). Additionally, the ABC transporter mutations have been identified as being associated with Bt resistance. ABCC2 in H. virescens were first reported as being involved in Cry1Ac resistance based on quantitative trait locus (QTL) mapping results that investigated the insertion of a premature stop codon in ABCC2 (Gahan et al., 2010). Then, different deletions, point mutations, truncations and spliceosome variants in ABC transporter orthologs were subsequently reported as being associated with the resistance of P. xylostella, T. ni, and H. armigera to Cry1Ac or Cry2Ab (Baxter et al., 2011; Xiao et al., 2014; Tay et al., 2015), Bombyx mori to Cry1Ab (Atsumi et al., 2012), and Ostrinia nubilalis survival on transgenic Cry1Fa maize (Coates and Siegfried, 2015). Furthermore, the changes in ABCC2, ABCC3, or ABCG1 transcript levels or their down-regulation were also linked to Cry1A or Cry1Ca toxin-resistance in S. exigua, P. xylostella, and Ostrinia furnacalis (Park et al., 2014; Guo et al., 2015a,b; Zhang et al., 2017). Here, we provided evidences that HaABCC1 encoded a functional receptor for Cry2Ab in H. armigera. We tested the susceptibility changes in in vitro experiments after transfecting HaABCC1 into Sf9 cells, and the transfected cells were more susceptible to Cry2Ab (Figure 5B). To confirm this view, we knocked down HaABCC1 expression using RNAi technology, and the larval mortality significantly decreased (Figure 6B). The specific and saturable binding to membranes in Helicoverpa species has been shown for Cry2Ab (Hernández-Rodríguez et al., 2008, 2013), and the resistance to Cry2Ab is also associated with a loss of binding between receptors and Cry2Ab (Caccia et al., 2010). However, the function of ABCC1 in the resistance evolution of H. armigera to Bt requires further research. Meanwhile, ABCC1 and ABCC2 are belonging to ABC transporter C family members. ABCC2 is a functional receptor of Cry1Ac and the mutations of it caused the resistance to Cry1Ac. But here we first report ABCC1 is a functional receptor of Cry2Ab. So, whether ABCC1 (or ABCC2) have function in mode of action of Cry1Ac (or Cry2Ab), and caused the cross-resistance between Cry1Ac and Cry2Ab, then reduced the benefit of Cry1A + Cry2A “pyramid” strategy, which are also requires further study.
This is the first report to demonstrate that ABCC1 is associated with Cry2Ab toxicity in H. armigera. The results presented here indicated that HaABCC1 could bind with the Cry2Ab toxin and had important roles in the action modes of Cry2Ab. We suggested that ABCC1 serves as a functional receptor in H. armigera for Cry2Ab. Our findings provide mechanistic insights into the interactions between ABCC1 and Cry toxins.
GL and LC designed the study. LC prepared experimental materials, performed the experiments, and analyzed the data. GL, LC, and JW wrote the manuscript. CL, WZ, BW, and LN contributed to completing the experimental contents. All authors have read and approved the manuscript for publication.
This research was supported by the Key Project for Breeding Genetically Modified Organisms (Grant No. 2016ZX08011–002), the National Natural Science Funds of China (Grant No. 31621064), and the State Key Laboratory for Biology of Plant Diseases and Insect Pests (Grant No. SKLOF201708).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00745/full#supplementary-material
Atsumi, S., Miyamoto, K., Yamamoto, K., Narukawa, J., Kawai, S., Sezutsu, H., et al. (2012). Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 109, E1591–E1598. doi: 10.1073/pnas.1120698109
Baxter, S. W., Badenespérez, F. R., Morrison, A., Vogel, H., Crickmore, N., Kain, W., et al. (2011). Parallel evolution of Bacillus thuringiensis toxin resistance in lepidoptera. Genetics 189, 675–679. doi: 10.1534/genetics.111.130971
Bravo, A., Likitvivatanavong, S., Gill, S. S., and Soberón, M. (2011). Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 41, 423–431. doi: 10.1016/j.ibmb.2011.02.006
Bretschneider, A., Heckel, D. G., and Vogel, H. (2016). Know your ABCs: characterization and gene expression dynamics of ABC transporters in the polyphagous herbivore Helicoverpa armigera. Insect Biochem. Mol. Biol. 72, 1–9. doi: 10.1016/j.ibmb.2016.03.001
Brévault, T., Heuberger, S., Zhang, M., Ellers-Kirk, C., Ni, X., Masson, L., et al. (2013). Potential shortfall of pyramided transgenic cotton for insect resistance management. Proc. Natl. Acad. Sci. U.S.A. 110, 5806–5811. doi: 10.1073/pnas.1216719110
Caccia, S., Hernández-Rodríguez, C. S., Mahon, R. J., Downes, S., James, W., Bautsoens, N., et al. (2010). Binding site alteration is responsible for field-isolated resistance to Bacillus thuringiensis Cry2A insecticidal proteins in two Helicoverpa species. PLoS One 5:e9975. doi: 10.1371/journal.pone.0009975
Chen, Z., Fei, H., Xiao, Y., Liu, C., Li, J., Yang, Y., et al. (2015). Endogenous expression of a Bt toxin receptor in the Cry1Ac-susceptible insect cell line and its synergistic effect with cadherin on cytotoxicity of activated Cry1Ac. Insect Biochem. Mol. Biol. 59, 1–17. doi: 10.1016/j.ibmb.2015.01.014
Coates, B. S., and Siegfried, B. D. (2015). Linkage of an ABCC transporter to a single QTL that controls Ostrinia nubilalis larval resistance to the Bacillus thuringiensis Cry1Fa toxin. Insect Biochem. Mol. Biol. 63, 86–96. doi: 10.1016/j.ibmb.2015.06.003
Comas, C., Lumbierres, B., Pons, X., and Albajes, R. (2014). No effects of Bacillus thuringiensis maize on nontarget organisms in the field in southern Europe: a meta-analysis of 26 arthropod taxa. Transgenic Res. 23, 135–143. doi: 10.1007/s11248-013-9737-0
Cordova, D., Benner, E. A., Sacher, M. D., Rauh, J. J., Sopa, J. S., Lahm, G. P., et al. (2006). Anthranilic diamides: a new class of insecticides with a novel mode of action, ryanodine receptor activation. Pestic. Biochem. Physiol. 84, 196–214. doi: 10.1016/j.pestbp.2005.07.005
Dassa, E., and Bouige, P. (2001). The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 152, 211–229. doi: 10.1016/S0923-2508(01)01194-9
Dermauw, W., and Van Leeuwen, T. (2014). The ABC gene family in arthropods: comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 45, 89–110. doi: 10.1016/j.ibmb.2013.11.001
Fabrick, J. A., Ponnuraj, J., Singh, A., Tanwar, R. K., Unnithan, G. C., Yelich, A. J., et al. (2014). Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to Bt cotton in India. PLoS One 9:e97900. doi: 10.1371/journal.pone.0097900
Gahan, L. J., Pauchet, Y., Vogel, H., and Heckel, D. G. (2010). An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 6:e1001248. doi: 10.1371/journal.pgen.1001248
Guo, Z., Kang, S., Zhu, X., Xia, J., Wu, Q., Wang, S., et al. (2015a). Down-regulation of a novel ABC transporter gene (Pxwhite) is associated with Cry1Ac resistance in the diamondback moth, Plutella xylostella (L.). Insect Biochem. Mol. Biol. 59, 30–40. doi: 10.1016/j.ibmb.2015.01.009
Guo, Z., Kang, S., Zhu, X., Xia, J., Wu, Q., Wang, S., et al. (2015b). The novel ABC transporter ABCH1 is a potential target for RNAi-based insect pest control and resistance management. Sci. Rep. 5:13728. doi: 10.1038/srep13728
Heckel, D. G. (2012). Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pestic. Biochem. Physiol. 104, 103–110. doi: 10.1016/j.pestbp.2012.05.007
Hernández-Rodríguez, C. S., Hernández-Martínez, P., Rie, J. V., Escriche, B., and Ferré, J. (2013). Shared midgut binding sites for Cry1A.105, Cry1Aa, Cry1Ab, Cry1Ac and Cry1Fa proteins from Bacillus thuringiensis in two important corn pests, Ostrinia nubilalis and Spodoptera frugiperda. PLoS One 8:e68164. doi: 10.1371/journal.pone.0068164
Hernández-Rodríguez, C. S., Vliet, A. V., Bautsoens, N., Rie, J. V., and Ferré, J. (2008). Specific binding of Bacillus thuringiensis Cry2A insecticidal proteins to a common site in the midgut of Helicoverpa species. Appl. Environ. Microbiol. 74, 7654–7659. doi: 10.1128/AEM.01373-08
Jakka, S. R. K., Gong, L., Hasler, J., Banerjee, R., Sheets, J. J., Narva, K., et al. (2016). Field-evolved mode 1 resistance of the fall armyworm to transgenic Cry1Fa-expressing corn associated with reduced Cry1Fa toxin binding and midgut alkaline phosphatase expression. Appl. Environ. Microbiol. 82, 1023–1034. doi: 10.1128/AEM.02871-15
James, C. (2016). Global status of commercialized biotech/GM Crops: 2016 International Service for the Acquisition of Agri-Biotech Applications Brief No 52. Uthaca, NY: ISAAA.
Jin, L., Zhang, H., Lu, Y., Yang, Y., Wu, K., Tabashnik, B. E., et al. (2015). Large-scale test of the natural refuge strategy for delaying insect resistance to transgenic Bt crops. Nat. Biotechnol. 33, 169–174. doi: 10.1038/nbt.3100
Kaminski, W. E., Piehler, A., and Wenzel, J. J. (2006). ABCA-subfamily transporters: structure, function and disease. Biochim. Biophys. Acta 1762, 510–524. doi: 10.1016/j.bbadis.2006.01.011
Liang, G., Tan, J., and Guo, Y. (1999). An improvement in the technique of artificial rearing cotton bollworm. Plant Prot. 15–17.
Linton, K. J. (2007). Structure and function of ABC transporters. Physiology 22, 122–130. doi: 10.1152/physiol.00046.2006
Liu, S., Wang, M., and Li, X. (2015). Overexpression of tyrosine hydroxylase and dopa decarboxylase associated with pupal melanization in Spodoptera exigua. Sci Rep. 5:11273. doi: 10.1038/srep11273
Merzendorfer, H. (2014). Chapter one - ABC transporters and their role in protecting insects from pesticides and their metabolites. Adv. Insect Physiol. 46, 1–72. doi: 10.1016/B978-0-12-417010-0.00001-X
Nicolia, A., Manzo, A., Veronesi, F., and Rosellini, D. (2014). An overview of the last 10 years of genetically engineered crop safety research. Crit. Rev. Biotechnol. 34, 77–88. doi: 10.3109/07388551.2013.823595
Onofre, J., Gaytán, M. O., Peña-Cardeña, A., García-Gómez, B. I., Pacheco, S., Gómez, I., et al. (2017). Identification of Aminopeptidase-N2 as a Cry2Ab binding protein in Manduca sexta. Peptides 98, 93–98. doi: 10.1016/j.peptides.2017.01.006
Pardo-López, L., Soberon, M., and Bravo, A. (2013). Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 37, 3–22. doi: 10.1111/j.1574-6976.2012.00341.x
Park, Y., González-Martínez, R. M., Navarro-Cerrillo, G., Chakroun, M., Kim, Y., Ziarsolo, P., et al. (2014). ABCC transporters mediate insect resistance to multiple Bt toxins revealed by bulk segregant analysis. BMC Biol. 12:46. doi: 10.1186/1741-7007-12-46
Pigott, C. R., and Ellar, D. J. (2007). Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 71, 255–281. doi: 10.1128/MMBR.00034-06
Qiu, L., Hou, L., Zhang, B., Liu, L., Li, B., Deng, P., et al. (2015). Cadherin is involved in the action of Bacillus thuringiensis toxins Cry1Ac and Cry2Aa in the beet armyworm, Spodoptera exigua. J. Invertebr. Pathol. 127, 47–53. doi: 10.1016/j.jip.2015.02.009
Sanahuja, G., Banakar, R., Twyman, R. M., Capell, T., and Christou, P. (2011). Bacillus thuringiensis: a century of research, development and commercial applications. Plant Biotechnol. J. 9, 283–300. doi: 10.1111/j.1467-7652.2011.00595.x
Tabashnik, B. E., Brévault, T., and Carrière, Y. (2013a). Insect resistance to Bt crops: lessons from the first billion acres. Nat. Biotechnol. 31, 510–521. doi: 10.1038/nbt.2597
Tabashnik, B. E., Fabrick, J. A., Unnithan, G. C., Yelich, A. J., Masson, L., Zhang, H., et al. (2013b). Efficacy of genetically modified Bt toxins alone and in combinations against pink bollworm resistant to Cry1Ac and Cry2Ab. PLoS One 8:e80496. doi: 10.1371/journal.pone.0080496
Tabashnik, B. E., and Carrière, Y. (2017). Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 35, 926–935. doi: 10.1038/nbt.3974
Tabashnik, B. E., Unnithan, G. C., Masson, L., Crowder, D. W., Li, X., and Carriére, Y. (2009). Asymmetrical cross-resistance between Bacillus thuringiensis toxins Cry1Ac and Cry2Ab in pink bollworm. Proc. Natl. Acad. Sci. U.S.A. 106, 11889–11894. doi: 10.1073/pnas.0901351106
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanaka, S., Miyamoto, K., Noda, H., Endo, H., Kikuta, S., and Sato, R. (2016). Single amino acid insertions in extracellular loop 2 of Bombyx mori ABCC2 disrupt its receptor function for Bacillus thuringiensis Cry1Ab and Cry1Ac but not Cry1Aa toxins. Peptides 78, 99–108. doi: 10.1016/j
Tay, W. T., Mahon, R. J., Heckel, D. G., Walsh, T. K., Downes, S., James, W. J., et al. (2015). Insect resistance to Bacillus thuringiensis toxin Cry2Ab is conferred by mutations in an ABC transporter subfamily a protein. PLoS Genet. 11:e1005534. doi: 10.1371/journal.pgen.1005534
Vachon, V., Laprade, R., and Schwartz, J. L. (2012). Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins: a critical review. J. Invertebr. Pathol. 111, 1–12. doi: 10.1016/j.jip.2012.05.001
Wang, J., Wang, H., Liu, S., Liu, L., Tay, W. T., Walsh, T. K., et al. (2017). CRISPR/Cas9 mediated genome editing of Helicoverpa armigera with mutations of an ABC transporter gene HaABCA2 confers resistance to Bacillus thuringiensis Cry2A toxins. Insect Biochem. Mol. Biol. 87, 147–153. doi: 10.1016/j.ibmb.2017.07.002
Wei, J., Guo, Y., Liang, G., Wu, K., Zhang, J., Tabashnik, B. E., et al. (2015). Cross-resistance and interactions between Bt toxins Cry1Ac and Cry2Ab against the cotton bollworm. Sci. Rep. 5:7714. doi: 10.1038/srep07714
Wei, J., Zhang, M., Liang, G., Wu, K., Guo, Y., Ni, X., et al. (2016). APN1 is a functional receptor of Cry1Ac but not Cry2Ab in Helicoverpa zea. Sci. Rep. 6:19179. doi: 10.1038/srep19179
Welch, K. L., Unnithan, G. C., Degain, B. A., Wei, J., Zhang, J., Li, X., et al. (2015). Cross-resistance to toxins used in pyramided Bt crops and resistance to Bt sprays in Helicoverpa zea. J. Invertebr. Pathol. 132, 149–156. doi: 10.1016/j.jip.2015.10.003
Xiao, Y., Zhang, T., Liu, C., Heckel, D. G., Li, X., Tabashnik, B. E., et al. (2014). Mis-splicing of the ABCC2 gene linked with Bt toxin resistance in Helicoverpa armigera. Sci. Rep. 4:6184. doi: 10.1038/srep06184
Xue, J., Liang, G., Crickmore, N., Li, H., He, K., Song, F., et al. (2008). Cloning and characterization of a novel Cry1A toxin from Bacillus thuringiensis with high toxicity to the Asian corn borer and other lepidopteran insects. FEMS Microbiol. Lett. 280, 95–101. doi: 10.1111/j.1574-6968.2007.01053.x
Yuan, X., Zhao, M., Wei, J., Zhang, W., Wang, B., Myint, K. M., et al. (2017). New insights on the role of alkaline phosphatase 2 from Spodoptera exigua (Hübner) in the action mechanism of bt toxin Cry2Aa. J. Insect Physiol. 98, 101–107. doi: 10.1016/j.jinsphys.2016.12.004
Zhang, D., Xiao, Y., Dhiloo, K. H., Soberon, M., Bravo, A., and Wu, K. (2016). Molecular cloning, expression, and identification of bre genes involved in glycosphingolipids synthesis in Helicoverpa armigera (lepidoptera: noctuidae). J. Econ. Entomol. 109:tow040. doi: 10.1093/jee/tow040
Zhang, T., Coates, B. S., Wang, Y., Wang, Y., Bai, S., Wang, Z., et al. (2017). Down-regulation of aminopeptidase N and ABC transporter subfamily G transcripts in Cry1Ab and Cry1Ac resistant Asian corn borer, Ostrinia furnacalis (lepidoptera: crambidae). Int. J. Biol. Sci. 13, 835–851. doi: 10.7150/ijbs.18868
Zhao, M., Yuan, X., Wei, J., Zhang, W., Wang, B., Khaing, M., et al. (2017). Functional roles of cadherin, aminopeptidase-N and alkaline phosphatase from Helicoverpa armigera (Hübner) in the action mechanism of Bacillus thuringiensis Cry2Aa. Sci. Rep. 7:46555. doi: 10.1038/srep46555
Keywords: Helicoverpa armigera, HaABCC1, functional receptors, binding, RNAi
Citation: Chen L, Wei J, Liu C, Zhang W, Wang B, Niu L and Liang G (2018) Specific Binding Protein ABCC1 Is Associated With Cry2Ab Toxicity in Helicoverpa armigera. Front. Physiol. 9:745. doi: 10.3389/fphys.2018.00745
Received: 19 March 2018; Accepted: 28 May 2018;
Published: 19 June 2018.
Edited by:
Bin Tang, Hangzhou Normal University, ChinaReviewed by:
Yuan-Xi Li, Nanjing Agricultural University, ChinaCopyright © 2018 Chen, Wei, Liu, Zhang, Wang, Niu and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gemei Liang, Z21saWFuZ0BpcHBjYWFzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.