
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Physiol., 22 June 2017
Sec. Mitochondrial Research
Volume 8 - 2017 | https://doi.org/10.3389/fphys.2017.00436
This article is part of the Research TopicCoenzyme Q Redox State and Cellular HomeostasisView all 7 articles
Coenzyme Q is a lipid that participates to important physiological functions. Coenzyme Q is synthesized in multiple steps from the precursor 4-hydroxybenzoic acid. Mutations in enzymes that participate to coenzyme Q biosynthesis result in primary coenzyme Q deficiency, a type of mitochondrial disease. Coenzyme Q10 supplementation of patients is the classical treatment but it shows limited efficacy in some cases. The molecular understanding of the coenzyme Q biosynthetic pathway allowed the design of experiments to bypass deficient biosynthetic steps with analogs of 4-hydroxybenzoic acid. These molecules provide the defective chemical group and can reactivate endogenous coenzyme Q biosynthesis as demonstrated recently in yeast, mammalian cell cultures, and mouse models of primary coenzyme Q deficiency. This mini review presents how the chemical properties of various analogs of 4-hydroxybenzoic acid dictate the effect of the molecules on CoQ biosynthesis and how the reactivation of endogenous coenzyme Q biosynthesis may achieve better results than exogenous CoQ10 supplementation.
Coenzyme Q (CoQ, compound 1 on Figure 1), also known as ubiquinone, is a lipid conserved from proteobacteria to humans. CoQ is composed of a benzoquinone ring that is attached to a polyisoprenyl tail of various length (six isoprenyl units in Saccharomyces cerevisiae hence CoQ6, ten units in humans, hence CoQ10). The benzoquinone ring is redox active and exchanges two electrons and two protons between the oxidized and reduced forms of CoQ, which play numerous roles in cellular physiology (Bentinger et al., 2010; Wang and Hekimi, 2016).
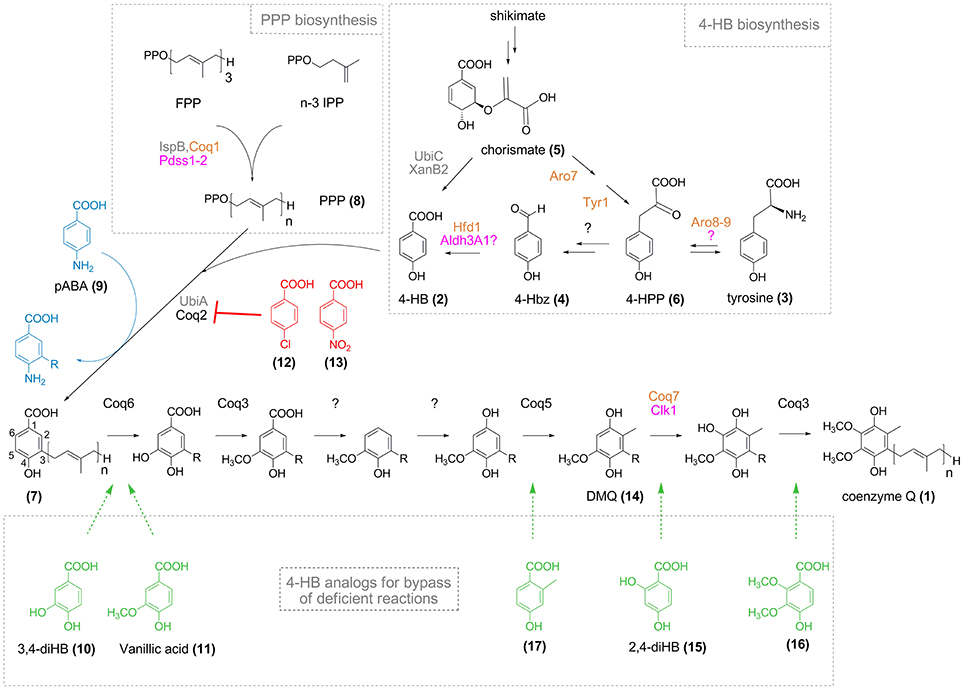
Figure 1. CoQ biosynthesis and effect of 4-HB analogs. Names for enzymes of E. coli, S. cerevisiae, and humans are in gray, orange, and pink, respectively (in black when common to S. cerevisiae and humans). Polyprenyl pyrophosphate (PPP, 8, n = 6–10) synthesized from farnesyl pyrophosphate (FPP) and n-3 isopentenyl pyrophosphate (IPP) is conjugated to 4-HB (2) by Coq2/UbiA to yield polyprenyl-4-HB (7). The numbering of the aromatic carbon atoms is shown on polyprenyl-4-HB and is uniformly applied to the different intermediates cited in the text, although IUPAC nomenclature might be different depending on the substitution pattern. Since the order of some reactions is different in bacteria (Aussel et al., 2014), only the eukaryotic pathway from polyprenyl-4-HB to CoQ is shown. R corresponds to the polyprenyl moiety (n = 6–10). 4-Chlorobenzoic acid (12) and 4-nitrobenzoic acid (13) (red) are inhibitors of Coq2/UbiA, whereas pABA (9, blue) is prenylated and progress to different stages of the CoQ biosynthetic pathway depending on the organisms (see text for details). 4-HB analogs (in green) have potential (16, 17) or proven (10, 11, 15) capacities to bypass the biosynthetic steps indicated by the dashed arrows.
In eukaryotic cells, the biosynthesis of CoQ takes place at the mitochondrial inner membrane (Wang and Hekimi, 2013a) and also possibly in the Golgi apparatus (Mugoni et al., 2013). 4-hydroxybenzoic acid (4-HB, 2) is the precursor of the benzoquinone ring of CoQ. 4-HB is first prenylated by Coq2 (UbiA in bacteria) and then, a total of seven reactions—one decarboxylation, three hydroxylation, and three methylation—produce the fully substituted benzoquinone ring of CoQ (Figure 1). Even though the structure of CoQ was established almost 60 years ago (Lester et al., 1958; Morton, 1958), the identity of the enzymes that catalyze the decarboxylation reaction and one of the three hydroxylation reaction is still elusive in eukaryotes (Kawamukai, 2016). In addition, the pathway that converts tyrosine (3) into 4-HB is poorly characterized and the last reaction, the oxidation of 4-hydroxybenzaldehyde (4-Hbz, 4) to 4-HB was only recently elucidated in S. cerevisiae (Payet et al., 2016; Stefely et al., 2016).
Primary CoQ10 deficiency, caused by mutations in genes involved in CoQ biosynthesis, is a rare condition with a heterogeneous clinical spectrum. Mutations in PDSS1, PDSS2, COQ2, COQ4, COQ6, COQ7, COQ9, ADCK3, and ADCK4 have been identified to date (Acosta et al., 2016). CoQ deficiency represents one of the few mitochondrial disorder that is treatable (Hirano et al., 2012), although not all patients respond to oral CoQ10 supplementation (Hirano et al., 2012). The success of the treatment is influenced by the advance of the disease at the time when CoQ10 supplementation is initiated (Acosta et al., 2016). The poor bioavailability of CoQ10 also contributes to the variable results of CoQ10 supplementation. Indeed, the lipophilic nature of CoQ10 is thought to limit its distribution in the human body and its transport to the mitochondrial inner-membrane (Bentinger et al., 2003; Lopez et al., 2010; Hirano et al., 2012).
This mini-review will discuss how various 4-HB analogs impact CoQ biosynthesis and how some of them bypass altered biosynthetic steps, as first demonstrated in S. cerevisiae by my group and that of Catherine Clarke (Ozeir et al., 2011; Xie et al., 2012). I will also present the potential benefits and limitations of using analogs of 4-HB over CoQ10 supplementation to treat CoQ10 deficiency linked to mutations in specific genes.
4-HB together with 4-Hbz have been suspected early on as potential precursors of the benzoquinone ring of CoQ in animals, yeast, and bacteria (Parson and Rudney, 1964). In Escherichia coli, 4-HB is produced by a chorismate pyruvate-lyase reaction catalyzed by UbiC (Nichols and Green, 1992; Siebert et al., 1992), which substrate is chorismic acid (5), an intermediate of the shikimate pathway that feeds the biosynthesis of aromatic amino acids (Lawrence et al., 1974). However, many proteobacteria that synthesize CoQ lack an ubiC homolog and the widespread xanB2 gene was recently shown to encode a bifunctional enzyme that converts chorismate into either 4-HB or 3-HB (Zhou et al., 2013). Animals do not possess the shikimate pathway and derive 4-HB from tyrosine and phenylalanine (Olson et al., 1965; Olson, 1966), via a pathway that remains putative (Clarke, 2000), but potentially implicates para-coumarate (Xie et al., 2015). In the plant Arabidopsis thaliana, phenylalanine and tyrosine independently contribute to the synthesis of 4-HB (Block et al., 2014). Only the pathway originating from phenylalanine has been partially characterized and involves β-oxidation in peroxisomes (Block et al., 2014).
Unlike bacteria, S. cerevisiae does not produce 4-HB in a single step from chorismate. Instead, pathways from shikimate or exogenous tyrosine converge at 4-hydroxyphenyl pyruvate (4-HPP, 6), which is further converted to 4-Hbz via uncharacterized steps (Payet et al., 2016) (Figure 1). As a final reaction, 4-Hbz is oxidized to 4-HB by the aldehyde dehydrogenase Hfd1 (Payet et al., 2016; Stefely et al., 2016). Since the human homolog ALDH3A1 complements the defects of yeast Δhfd1 cells for CoQ biosynthesis and respiratory growth (Payet et al., 2016; Stefely et al., 2016), the oxidation of 4-Hbz to 4-HB may also take place in humans, as suspected from early results in animals (Parson and Rudney, 1964). The elucidation of the human pathway from tyrosine to 4-HB is important as mutations in participating genes may result in CoQ deficiency, which may be compensated by supplying 4-HB. Indeed, exogenous 4-HB rescues the levels of CoQ in mutants that disrupt 4-HB biosynthesis in bacteria, yeast, and plants (Zhou et al., 2013; Block et al., 2014; Payet et al., 2016; Stefely et al., 2016).
4-HB enters the CoQ biosynthetic pathway via the prenylation of the position 3 catalyzed by Coq2 in eukaryotes and UbiA in bacteria that yield 3-polyprenyl-4-hydroxybenzoic acid (7). The polyprenyl pyrophosphate (PPP, 8) is formed by Coq1/Pdss1-Pdss2 (Kawamukai, 2016) (Figure 1). Aromatic compounds are substrates of UbiA if their carbon C3 is activated by a carboxylic acid moiety on C1 and a group on C4 that is electron- and hydrogen bond-donor, like hydroxyl or amine groups (Wessjohann and Sontag, 1996). Electron-withdrawing groups on C4, such as nitro and chloro, led to loss of activity of UbiA (Wessjohann and Sontag, 1996; Brandt et al., 2009). Interestingly, substituents in position 5 and 6 of 4-HB are tolerated.
These structural requirements are also applicable with the rat Coq2 enzyme since para-aminobenzoic acid (pABA, 9), 3,4-dihydroxybenzoic acid (3,4-diHB, 10), and 4-hydroxy-3-methoxybenzoic acid (vanillic acid, 11) were prenylated in cell free extracts, whereas chlorobenzoic acid (12) inhibited the prenyl transferase reaction (Alam et al., 1975; Nambudiri et al., 1977). Furthermore, 4-nitrobenzoic acid (4-NB, 13) was shown to be a competitive inhibitor of Coq2 and to decrease CoQ biosynthesis in mammalian cell cultures in a dose- and time-dependent manner (Forsman et al., 2010). 4-NB was subsequently used to evaluate how different residual levels of CoQ impact various cellular parameters (Quinzii et al., 2012), contributing to a better understanding of the pathomechanisms underlying primary CoQ deficiency. Altogether, these studies demonstrate that the active site of Coq2-UbiA can accommodate several 4-HB analogs which will act as substrates or inhibitors depending on their chemical properties.
The crystal structures of two UbiA homologs from archaeal thermophiles have recently been reported (Cheng and Li, 2014; Huang et al., 2014). Both membrane-embedded proteins present nine transmembrane helices. In one structure, a lateral portal delineated by two transmembrane domains was proposed to open to the membrane and allow access to the PPP molecule (Cheng and Li, 2014). Two conserved aspartate-rich motifs are located in a central cavity and coordinate two Mg2+ ions involved in binding the PPP analogs used in co-crystallization experiments. Unfortunately, no details are available regarding the coordination of 4-HB in the active site. Indeed, in one enzyme, 4-HB binding could not be detected by isothermal titration calorimetry and tentative 4-HB modeling clashed with the position occupied by the PPP analog (Huang et al., 2014) whereas in the other enzyme, 4-HB was modeled so that its carboxyl group contacts Arg43 (Cheng and Li, 2014). Yet, the mutation of the corresponding arginine residue in E. coli UbiA did not completely abolish activity (Cheng and Li, 2014), raising doubts about the involvement of this residue in the coordination of 4-HB.
pABA fulfills the requirements for prenylation by the Coq2-UbiA prenyltransferases and labeling experiments demonstrated that pABA is converted to CoQ in S. cerevisiae (Marbois et al., 2010; Pierrel et al., 2010). Consequently, the C4 amino group of pABA must be replaced by a C4 hydroxyl group and we recently reported that the monooxygenase Coq6 catalyzes this reaction in addition to the previously reported C5-hydroxylation (Ozeir et al., 2015). The L382E mutation in Coq6 severely impairs the C4-deamination reaction but globally maintains the C5-hydroxylation (Ozeir et al., 2015). Δcoq9 cells accumulate C4-aminated intermediates of the CoQ pathway (Xie et al., 2012; He et al., 2015) but Coq9 plays an indirect role in the C4-deamination as its absence impacts the C-terminal region of Coq6, which is important for the C4-deamination but quite dispensable for the C5-hydroxylation (Ozeir et al., 2015).
So far, pABA was reported to be a precursor of CoQ only in S. cerevisiae. E. coli, the plant Arabidopsis, or mammalian cells do not incorporate pABA into CoQ (Block et al., 2014; Xie et al., 2015). In E. coli, prenylated pABA is decarboxylated and hydroxylated to yield 2-amino-3-octaprenylphenol, which was suggested to be a “dead-end” product (Xie et al., 2015). In mammalian cell cultures, pABA decreases CoQ levels (Gonzalez-Aragon et al., 2005; Xie et al., 2015) and unidentified prenylated compounds were previously detected (Alam et al., 1975; Forsman et al., 2010). Indeed, results from my group confirmed a strong decrease of cellular CoQ9 in Chinese Hamster Ovary (CHO) cells and NIH/3T3 fibroblasts (derived from Swiss mouse embryo tissue) treated with pABA (Figure 2). In addition, both cell lines accumulate a major redox compound, which was identified as 4-imino-6-demethoxycoenzyme Q9 (IDMQ9) by high resolution mass spectrometry. IDMQ9 can be reduced to 4-amino-6-demethoxycoenzyme Q9 (ADMQ9) (Figure 2), similarly to IDMQ6 that was previously detected in yeast Δcoq9 cells (Xie et al., 2012). These results suggest that the C4-deamination is not taking place in mammalian cells and that Clk1-Coq7 cannot hydroxylate IDMQ or ADMQ (I-ADMQ). The properties of I-ADMQ are not known but they may have antioxidant capacities and may also participate to mitochondrial respiration as demonstrated for the closely related molecule demethoxy-coenzyme Q (DMQ, 14; Wang and Hekimi, 2013b). Therefore, pABA may not be the most appropriate agent to induce CoQ10 deficiency in cell lines and study the consequential physiological impacts (Gonzalez-Aragon et al., 2005; Duberley et al., 2013, 2014). Instead, 4-NB must be considered for such purposes since it does not form any prenylated products (Forsman et al., 2010; Quinzii et al., 2012).
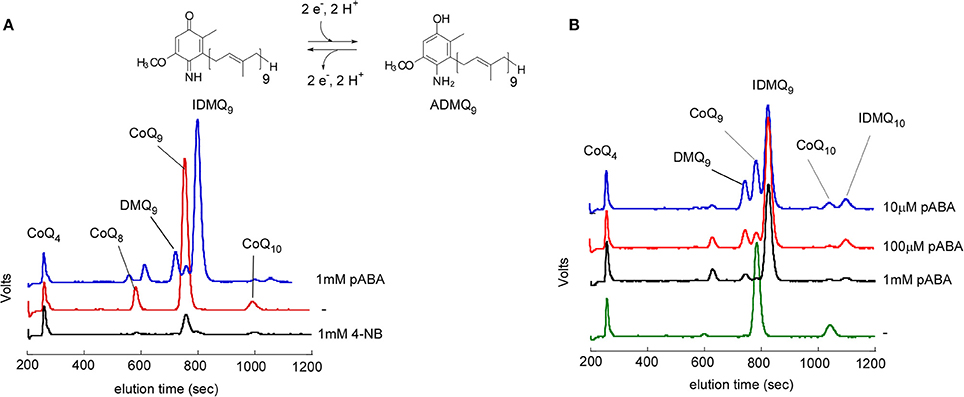
Figure 2. Effect of pABA on mammalian cell lines. Analysis of cellular lipid extracts by high performance liquid chromatography coupled to electrochemical detection with a precolumn electrode set in oxidizing mode. NIH/3T3 fibroblasts (A) and CHO cells (B) were grown for one week in the absence (−) or with the indicated concentrations of pABA or 4-NB. CoQ4 was used as an internal standard. The peaks corresponding to CoQ9 and CoQ10, 4-imino-6-demethoxyubiquinone 9 (IDMQ9) and IDMQ10, 6-demethoxyubiquinone 9 (DMQ9) are indicated. The chemical structure of IDMQ9 and ADMQ9 are shown.
Mammalian cells do not synthesize pABA contrary to microorganisms, which derive pABA from the shikimate pathway (Botet et al., 2007). pABA is frequently found in nutritional supplements but is rapidly and efficiently eliminated from the human body (Sharma et al., 2014), limiting the risk of interference with CoQ biosynthesis. Accordingly, pABA did not perturb CoQ biosynthesis in mice. Indeed the levels of CoQ9 were maintained in various tissues—heart, brain, lung, spleen, kidney, liver, and skeletal muscle—of C57BL/6 mice injected intraperitoneally with pABA (50 mg/kg/day) for 4 weeks and I-ADMQ9 were not detected (unpublished results). Thus, pABA seems to interfere with mammalian CoQ biosynthesis only in cell cultures.
The possibility to bypass a deficient step in CoQ biosynthesis by providing the defective chemical group within a synthetic analog of 4-HB was first demonstrated with 3,4-diHB and vanillic acid (Ozeir et al., 2011). Both compounds restored CoQ biosynthesis in S. cerevisiae cells impaired in C5-hydroxylation because of mutations in coq6 (Ozeir et al., 2011; Doimo et al., 2014). When working with knock-out S. cerevisiae mutants, the effect of 4-HB analogs is dependent upon overexpression of Coq8 to stabilize several Coq proteins that are instable in Δcoq strains (Padilla et al., 2009; Xie et al., 2012; He et al., 2014). 2,4-Dihydroxybenzoic acid (2,4-diHB, 15) bypassed a C6-hydroxylation defect and allowed CoQ6 biosynthesis in Δcoq7 yeast cells overexpressing Coq8 (Xie et al., 2012). 2,4-diHB increased CoQ10 levels in fibroblasts with an homozygous V141E mutation in COQ7 (Freyer et al., 2015) but was inefficient with the L111P mutation (Wang et al., 2017).
Besides cell cultures, 2,4-diHB was also efficient in mice with an inducible deletion of Coq7 (also called Mclk1). Addition of 2,4-diHB to the drinking water shortly after induction of the Mclk1 deletion increased CoQ9 levels in heart, kidney, and skeletal muscle and markedly rescued the mutant phenotypes, including mitochondrial respiration, blood lactate levels, and lifespan (Wang et al., 2015). In this model, dietary CoQ10 supplementation was almost without an effect, likely because of poor tissue uptake except by the liver (Wang et al., 2015). Interestingly, 2,4-diHB was also efficient when provided as a late treatment, i.e., when the symptoms had already developed. Oral 2,4-diHB also proved efficient in the Coq9R239X mouse, as it increased the kidney CoQ9 content (Luna-Sanchez et al., 2015). Coq9R239X mice have decreased CoQ9 and accumulate DMQ9 (Garcia-Corzo et al., 2013) as the Coq7 reaction is impeded since Coq9 interacts with Coq7 and is thought to facilitate its function (Lohman et al., 2014). Together, these results suggest that 2,4-diHB may be beneficial to patients with primary CoQ deficiency due to COQ7 or COQ9 mutations.
Such bypass strategies may be applicable to other cases of primary coenzyme Q deficiency. COQ6 patients (Heeringa et al., 2011) may respond to vanillic acid or 3,4-diHB (Doimo et al., 2014). Although disease-causing mutations in COQ3 and COQ5 genes have not been reported yet, 2,3-dimethoxy-4-hydroxybenzoic acid (16) and 2-methyl-4-hydroxybenzoic acid (17) may, in theory, bypass the requirement for Coq3 and Coq5, respectively. The possibility for the former compound to be prenylated by Coq2 is supported by the in vitro prenylation of the related 2,3,4-trihydroxybenzoic acid (2,3,4-tri-HB) by UbiA (Wessjohann and Sontag, 1996). Altogether, several reactions of the CoQ biosynthetic pathway are potentially amenable to bypass. However, it is impossible to circumvent the C1 decarboxylation or C1 hydroxylation reactions with analogs bearing a hydroxyl group on C1, since the presence of a carboxyl group at C1 of the benzene ring is indispensable for prenylation (Alam et al., 1975; Wessjohann and Sontag, 1996).
To replenish CoQ levels in CoQ deficient cells and organisms, the use of bypass 4-HB analogs may be advantageous over CoQ10 supplementation for the following reasons. (i) 4-HB analogs will allow to preserve the endogenous ratio between the major and minor isoforms of CoQ. Indeed, many species have a prominent CoQ isoform (CoQ9 in rodents, CoQ10 in humans) but also synthesize minor isoforms (CoQ10 in rodents, CoQ9 in humans). The ratio of both isoforms varies significantly depending on the organs (Turunen et al., 2004), yet, it remains unknown whether these varying ratios have any physiological consequences. (ii) Thanks to their hydrophilic nature, 4-HB analogs may have a superior bioavailability than exogenously supplied CoQ10, which accumulates efficiently in the liver but not in other organs (Miles, 2007). Indeed, CoQ10 supplementation of CoQ deficient mouse models did not increase CoQ10 content in kidney (Saiki et al., 2008) or heart or muscle (Wang et al., 2015). However, a new formulation of CoQ10 demonstrated improved bioavailability as it increased CoQ10 levels in all tested organs of Coq9R239X mice, although to a limited extent for most of them (Garcia-Corzo et al., 2014). (iii) 4-HB analogs may achieve higher CoQ levels in organs than CoQ10 supplementation. 2,4-diHB doubled the kidney CoQ9 content of Coq9R239X mice (Luna-Sanchez et al., 2015) and tripled that of Mclk1-deficient mice (Wang et al., 2015), although WT levels were not reached in either model. For comparison, CoQ10 supplementation yielded a ~50% increase of total kidney CoQ (CoQ9+CoQ10) in the former model (Garcia-Corzo et al., 2014) and none in the latter (Wang et al., 2015). (iv) CoQ produced from 4-HB analogs should distribute normally between subcellular compartments whereas exogenously supplied CoQ10 has difficulties to reach mitochondria and their inner membrane (Bentinger et al., 2003). (v) Short chains analogs of CoQ like idebenone and decylubiquinone have been reported to increase superoxide production (Genova et al., 2003), but 4-HB analogs are not expected to have such effect since they don't have a redox-active benzoquinone moiety.
The successful restoration of endogenous CoQ biosynthesis by 4-HB analogs depends on several factors. (i) The 4-HB analogs must outcompete endogenous 4-HB in the prenylation reaction catalyzed by Coq2. Thus, the Km of Coq2 for the analog should be in the same range as that for 4-HB or the analog should be substantially more abundant than 4-HB. (ii) Except for the defective CoQ biosynthetic step, all other enzymatic reactions must be maintained. However, many CoQ biosynthetic proteins form a complex in human cells (Floyd et al., 2016) and the abundance of several Coq proteins decreased in multiple tissues of the Coq9R239X mice (Lohman et al., 2014; Luna-Sanchez et al., 2015), reflecting the instability of an incompletely assembled complex, as already observed in yeast (Hsieh et al., 2007; Xie et al., 2012). (iii) The Coq enzymes must be able to modify unnatural substrates with extra chemical groups. For example, Coq6, which usually hydroxylates 3-polyprenyl-4-hydroxybenzoic acid (7), has to hydroxylate 3-polyprenyl-2,4-dihydroxybenzoic acid in 2,4-diHB treated cells. (iv) The 4-HB analogs should be retained in the body unlike pABA and must be non-toxic, like vanillic acid which is licensed as a food additive (Gitzinger et al., 2012). However, other analogs may not be as innocuous, since control mice treated with 2,4-diHB gained less body weight than untreated mice (Wang et al., 2015) and high doses of 2,3,4-tri-HB were toxic in cell lines (Herebian et al., 2017).
As demonstrated in yeast, mice, and human cell cultures, 4-HB analogs can bypass deficiencies in some steps of CoQ biosynthesis. 4-HB itself could be used to compensate for defects in the tyrosine to 4-HB pathway. These strategies are only possible thanks to a detailed molecular and genetic understanding of the CoQ biosynthetic pathway and efforts must continue to elucidate the steps that remain uncharacterized to date. In specific cases of primary CoQ deficiency, providing 4-HB analogs to reactivate the endogenous production of CoQ may represent a therapeutic alternative to CoQ10 supplementation. Further, investigations with animal models will establish whether this approach is realistic.
The author confirms being the sole contributor of this work and approved it for publication.
This work was supported by Fondation pour la Recherche Médicale (grant number “DPM20121125553”) and by Agence Nationale de la Recherche (grant pABACoQ “ANR-11-JSV8-002”).
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
I would like to thank the numerous colleagues with whom I had fruitful interactions over the years. I apologize to the authors whose work could not be discussed due to space limitation.
Acosta, M. J., Fonseca, L. V., Desbats, M. A., Cerqua, C., Zordan, R., Trevisson, E., et al. (2016). Coenzyme Q biosynthesis in health and disease. Biochim. Biophys. Acta 1857, 1079–1085. doi: 10.1016/j.bbabio.2016.03.036
Alam, S. S., Nambudiri, A. M., and Rudney, H. (1975). 4-Hydroxybenzoate: polyprenyl transferase and the prenylation of 4-aminobenzoate in mammalian tissues. Arch. Biochem. Biophys. 171, 183–190. doi: 10.1016/0003-9861(75)90022-3
Aussel, L., Pierrel, F., Loiseau, L., Lombard, M., Fontecave, M., and Barras, F. (2014). Biosynthesis and physiology of coenzyme Q in bacteria. Biochim. Biophys. Acta 1837, 1004–1011. doi: 10.1016/j.bbabio.2014.01.015
Bentinger, M., Dallner, G., Chojnacki, T., and Swiezewska, E. (2003). Distribution and breakdown of labeled coenzyme Q(10) in rat. Free Radic. Biol. Med. 34, 563–575. doi: 10.1016/S0891-5849(02)01357-6
Bentinger, M., Tekle, M., and Dallner, G. (2010). Coenzyme Q-biosynthesis and functions. Biochem. Biophys. Res. Commun. 396, 74–79. doi: 10.1016/j.bbrc.2010.02.147
Block, A., Widhalm, J. R., Fatihi, A., Cahoon, R. E., Wamboldt, Y., Elowsky, C., et al. (2014). The origin and biosynthesis of the benzenoid moiety of ubiquinone (Coenzyme Q) in Arabidopsis. Plant Cell 26, 1938–1948. doi: 10.1105/tpc.114.125807
Botet, J., Mateos, L., Revuelta, J. L., and Santos, M. A. (2007). A chemogenomic screening of sulfanilamide-hypersensitive Saccharomyces cerevisiae mutants uncovers ABZ2, the gene encoding a fungal aminodeoxychorismate lyase. Eukaryot. Cell 6, 2102–2111. doi: 10.1128/EC.00266-07
Brandt, W., Brauer, L., Gunnewich, N., Kufka, J., Rausch, F., Schulze, D., et al. (2009). Molecular and structural basis of metabolic diversity mediated by prenyldiphosphate converting enzymes. Phytochemistry 70, 1758–1775. doi: 10.1016/j.phytochem.2009.09.001
Cheng, W., and Li, W. (2014). Structural insights into ubiquinone biosynthesis in membranes. Science 343, 878–881. doi: 10.1126/science.1246774
Clarke, C. F. (2000). New advances in coenzyme Q biosynthesis. Protoplasma 213, 134–147. doi: 10.1007/BF01282151
Doimo, M., Trevisson, E., Airik, R., Bergdoll, M., Santos-Ocana, C., Hildebrandt, F., et al. (2014). Effect of vanillic acid on COQ6 mutants identified in patients with coenzyme Q deficiency. Biochim. Biophys. Acta 1842, 1–6. doi: 10.1016/j.bbadis.2013.10.007
Duberley, K. E., Abramov, A. Y., Chalasani, A., Heales, S. J., Rahman, S., and Hargreaves, I. P. (2013). Human neuronal coenzyme Q10 deficiency results in global loss of mitochondrial respiratory chain activity, increased mitochondrial oxidative stress and reversal of ATP synthase activity: implications for pathogenesis and treatment. J. Inherit. Metab. Dis. 36, 63–73. doi: 10.1007/s10545-012-9511-0
Duberley, K. E., Heales, S. J., Abramov, A. Y., Chalasani, A., Land, J. M., Rahman, S., et al. (2014). Effect of Coenzyme Q supplementation on mitochondrial electron transport chain activity and mitochondrial oxidative stress in Coenzyme Q deficient human neuronal cells. Int. J. Biochem. Cell Biol. 50, 60–63. doi: 10.1016/j.biocel.2014.02.003
Floyd, B. J., Wilkerson, E. M., Veling, M. T., Minogue, C. E., Xia, C., Beebe, E. T., et al. (2016). Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol. Cell. 63, 621–32. doi: 10.1016/j.molcel.2016.06.033
Forsman, U., Sjoberg, M., Turunen, M., and Sindelar, P. J. (2010). 4-Nitrobenzoate inhibits coenzyme Q biosynthesis in mammalian cell cultures. Nat. Chem. Biol. 6, 515–517. doi: 10.1038/nchembio.372
Freyer, C., Stranneheim, H., Naess, K., Mourier, A., Felser, A., Maffezzini, C., et al. (2015). Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2,4-dihydroxybensoic acid. J. Med. Genet. 52, 779–783. doi: 10.1136/jmedgenet-2015-102986
Garcia-Corzo, L., Luna-Sanchez, M., Doerrier, C., Garcia, J. A., Guaras, A., Acin-Perez, R., et al. (2013). Dysfunctional Coq9 protein causes predominant encephalomyopathy associated with CoQ deficiency. Hum. Mol. Genet. 22, 1233–1248. doi: 10.1093/hmg/dds530
Garcia-Corzo, L., Luna-Sanchez, M., Doerrier, C., Ortiz, F., Escames, G., Acuna-Castroviejo, D., et al. (2014). Ubiquinol-10 ameliorates mitochondrial encephalopathy associated with CoQ deficiency. Biochim. Biophys. Acta 1842, 893–901. doi: 10.1016/j.bbadis.2014.02.008
Genova, M. W., Pich, M. M., Biondi, A., Bernacchia, A., Falasca, A., Bovina, C., et al. (2003). Mitochondrial production of oxygen radical species and the role of coenzyme Q as an antioxidant. Exp. Biol. Med. 228, 506–513. doi: 10.1177/15353702-0322805-14
Gitzinger, M., Kemmer, C., Fluri, D. A., El-Baba, M. D., Weber, W., and Fussenegger, M. (2012). The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Res. 40:e37. doi: 10.1093/nar/gkr1251
Gonzalez-Aragon, D., Buron, M. I., Lopez-Lluch, G., Herman, M. D., Gomez-Diaz, C., Navas, P., et al. (2005). Coenzyme, Q., and the regulation of intracellular steady-state levels of superoxide in HL-60 cells. Biofactors 25, 31–41. doi: 10.1002/biof.5520250105
He, C. H., Black, D. S., Nguyen, T. P., Wang, C., Srinivasan, C., and Clarke, C. F. (2015). Yeast Coq9 controls deamination of coenzyme Q intermediates that derive from para-aminobenzoic acid. Biochim. Biophys. Acta 1851, 1227–1239. doi: 10.1016/j.bbalip.2015.05.003
He, C. H., Xie, L. X., Allan, C. M., Tran, U. C., and Clarke, C. F. (2014). Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim. Biophys. Acta 1841, 630–644. doi: 10.1016/j.bbalip.2013.12.017
Heeringa, S. F., Chernin, G., Chaki, M., Zhou, W., Sloan, A. J., Ji, Z., et al. (2011). COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Invest. 121, 2013–2024. doi: 10.1172/JCI45693
Herebian, D., Seibt, A., Smits, S. H. J., Bunning, G., Freyer, C., Prokisch, H., et al. (2017). Detection of 6-demethoxyubiquinone in CoQ10 deficiency disorders: insights into enzyme interactions and identification of potential therapeutics. Mol. Genet. Metab. S1096-7192(17)30100-2. doi: 10.1016/j.ymgme.2017.05.012
Hirano, M., Garone, C., and Quinzii, C. M. (2012). CoQ(10) deficiencies and MNGIE: two treatable mitochondrial disorders. Biochim. Biophys. Acta 1820, 625–631. doi: 10.1016/j.bbagen.2012.01.006
Hsieh, E. J., Gin, P., Gulmezian, M., Tran, U. C., Saiki, R., Marbois, B. N., et al. (2007). Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch. Biochem. Biophys. 463, 19–26. doi: 10.1016/j.abb.2007.02.016
Huang, H., Levin, E. J., Liu, S., Bai, Y., Lockless, S. W., and Zhou, M. (2014). Structure of a membrane-embedded prenyltransferase homologous to UBIAD1. PLoS Biol. 12:e1001911. doi: 10.1371/journal.pbio.1001911
Kawamukai, M. (2016). Biosynthesis of coenzyme Q in eukaryotes. Biosci. Biotechnol. Biochem. 80, 23–33. doi: 10.1080/09168451.2015.1065172
Lawrence, J., Cox, G. B., and Gibson, F. (1974). Biosynthesis of ubiquinone in Escherichia-coli-k-12 - biochemical and genetic characterization of a mutant unable to convert chorismate into 4-hydroxybenzoate. J. Bacteriol. 118, 41–45.
Lester, R. L., Crane, F. L., and Hatefi, Y. (1958). Coenzyme Q - a new group of quinones. J. Am. Chem. Soc. 80, 4751–4752. doi: 10.1021/ja01550a095
Lohman, D. C., Forouhar, F., Beebe, E. T., Stefely, M. S., Minogue, C. E., Ulbrich, A., et al. (2014). Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, E4697–E4705. doi: 10.1073/pnas.1413128111
Lopez, L. C., Quinzii, C. M., Area, E., Naini, A., Rahman, S., Schuelke, M., et al. (2010). Treatment of CoQ(10) deficient fibroblasts with ubiquinone, CoQ analogs, and vitamin C: time- and compound-dependent effects. PLoS ONE 5:e11897. doi: 10.1371/journal.pone.0011897
Luna-Sanchez, M., Diaz-Casado, E., Barca, E., Tejada, M. A., Montilla-Garcia, A., Cobos, E. J., et al. (2015). The clinical heterogeneity of coenzyme Q(10) deficiency results from genotypic differences in the Coq9 gene. EMBO Mol. Med. 7, 670–687. doi: 10.15252/emmm.201404632
Marbois, B., Xie, L. X., Choi, S., Hirano, K., Hyman, K., and Clarke, C. F. (2010). para-aminobenzoic acid is a precursor in coenzyme Q(6) biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 285, 27827–27838. doi: 10.1074/jbc.M110.151894
Miles, M. V. (2007). The uptake and distribution of coenzyme Q(10). Mitochondrion 7, S72–S77. doi: 10.1016/j.mito.2007.02.012
Mugoni, V., Postel, R., Catanzaro, V., De Luca, E., Turco, E., Digilio, G., et al. (2013). Ubiad1 is an antioxidant enzyme that regulates eNOS activity by CoQ10 synthesis. Cell 152, 504–518. doi: 10.1016/j.cell.2013.01.013
Nambudiri, A. M. D., Brockman, D., Alam, S. S., and Rudney, H. (1977). Alternate routes for ubiquinone biosynthesis in rats. Biochem. Biophys. Res. Commun. 76, 282–288. doi: 10.1016/0006-291X(77)90723-9
Nichols, B. P., and Green, J. M. (1992). Cloning and sequencing of Escherichia coli ubiC and purification of chorismate lyase. J. Bacteriol. 174, 5309–5316. doi: 10.1128/jb.174.16.5309-5316.1992
Olson, R. E., Dialamieh, G. H., Bentley, R., Springer, C. M., and Ramsey, V. G. (1965). Studies on Coenzyme Q. Pattern of labeling in Coenzyme Q9 after administration of isotopic acetate and aromatic amino acids to rats. J. Biol. Chem. 240, 514–523.
Ozeir, M., Muhlenhoff, U., Webert, H., Lill, R., Fontecave, M., and Pierrel, F. (2011). Coenzyme Q biosynthesis: Coq6 Is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem. Biol. 18, 1134–1142. doi: 10.1016/j.chembiol.2011.07.008
Ozeir, M., Pelosi, L., Ismail, A., Mellot-Draznieks, C., Fontecave, M., and Pierrel, F. (2015). Coq6 Is responsible for the C4-deamination reaction in coenzyme Q biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 290, 24140–24151. doi: 10.1074/jbc.M115.675744
Padilla, S., Tran, U. C., Jimenez-Hidalgo, M., Lopez-Martin, J. M., Martin-Montalvo, A., Clarke, C. F., et al. (2009). Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis. Cell. Mol. Life Sci. 66, 173–186. doi: 10.1007/s00018-008-8547-7
Parson, W. W., and Rudney, H. (1964). Biosynthesis of benzoquinone ring of ubiquinone from p-hydroxybenzaldehyde + p-hydroxybenzoic acid in rat kidney azotobacter vinelandii + bakers yeast. Proc. Natl. Acad. Sci. U.S.A. 51, 444–450. doi: 10.1073/pnas.51.3.444
Payet, L. A., Leroux, M., Willison, J. C., Kihara, A., Pelosi, L., and Pierrel, F. (2016). Mechanistic details of early steps in coenzyme Q biosynthesis pathway in yeast. Cell Chem. Biol. 23, 1241–1250. doi: 10.1016/j.chembiol.2016.08.008
Pierrel, F., Hamelin, O., Douki, T., Kieffer-Jaquinod, S., Muhlenhoff, U., Ozeir, M., et al. (2010). Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 17, 449–459. doi: 10.1016/j.chembiol.2010.03.014
Quinzii, C. M., Tadesse, S., Naini, A., and Hirano, M. (2012). Effects of inhibiting CoQ(10) Biosynthesis with 4-nitrobenzoate in human fibroblasts. PLoS ONE 7:e30606. doi: 10.1371/journal.pone.0030606
Saiki, R., Lunceford, A. L., Shi, Y., Marbois, B., King, R., Pachuski, J., et al. (2008). Coenzyme Q10 supplementation rescues renal disease in Pdss2kd/kd mice with mutations in prenyl diphosphate synthase subunit 2. Am. J. Physiol. Renal Physiol. 295, F1535–F1544. doi: 10.1152/ajprenal.90445.2008
Sharma, R. S., Joy, R. C., Boushey, C. J., Ferruzzi, M. G., Leonov, A. P., and McCrory, M. A. (2014). Effects of Para-Aminobenzoic Acid (PABA) form and administration mode on PABA recovery in 24-hour urine collections. J. Acad. Nutr. Diet. 114, 457–463. doi: 10.1016/j.jand.2013.07.045
Siebert, M., Bechthold, A., Melzer, M., May, U., Berger, U., Schroder, G., et al. (1992). Ubiquinone biosynthesis. Cloning of the genes coding for chorismate pyruvate-lyase and 4-hydroxybenzoate octaprenyl transferase from Escherichia coli. FEBS Lett 307, 347–350. doi: 10.1016/0014-5793(92)80710-X
Stefely, J. A., Kwiecien, N. W., Freiberger, E. C., Richards, A. L., Jochem, A., Rush, M. J., et al. (2016). Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat. Biotechnol. 34, 1191–1197. doi: 10.1038/nbt.3683
Turunen, M., Olsson, J., and Dallner, G. (2004). Metabolism and function of coenzyme Q. Biochim. Biophys. Acta 1660, 171–199. doi: 10.1016/j.bbamem.2003.11.012
Wang, Y., and Hekimi, S. (2013a). Molecular genetics of ubiquinone biosynthesis in animals. Crit. Rev. Biochem. Mol. Biol. 48, 69–88. doi: 10.3109/10409238.2012.741564
Wang, Y., and Hekimi, S. (2013b). Mitochondrial respiration without ubiquinone biosynthesis. Hum. Mol. Genet. 22, 4768–4783. doi: 10.1093/hmg/ddt330
Wang, Y., and Hekimi, S. (2016). Understanding ubiquinone. Trends Cell Biol. 26, 367–378. doi: 10.1016/j.tcb.2015.12.007
Wang, Y., Oxer, D., and Hekimi, S. (2015). Mitochondrial function and lifespan of mice with controlled ubiquinone biosynthesis. Nat. Commun. 6, 6393. doi: 10.1038/ncomms7393
Wang, Y., Smith, C., Parboosingh, J. S., Khan, A., Innes, M., and Hekimi, S. (2017). Pathogenicity of two COQ7 mutations and responses to 2,4-dihydroxybenzoate bypass treatment. J. Cell. Mol. Med. doi: 10.1111/jcmm.13154. [Epub ahead of print].
Wessjohann, L., and Sontag, B. (1996). Prenylation of benzoic acid derivatives catalyzed by a transferase from Escherichia coli overproduction: method development and substrate specificity. Angew. Chem. 35, 1697–1699. doi: 10.1002/anie.199616971
Xie, L. X., Ozeir, M., Tang, J. Y., Chen, J. Y., Kieffer-Jaquinod, S., Fontecave, M., et al. (2012). Over-expression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J. Biol. Chem. 287, 23571–23581. doi: 10.1074/jbc.M112.360354
Xie, L. X., Williams, K. J., He, C. H., Weng, E., Khong, S., Rose, T. E., et al. (2015). Resveratrol and para-coumarate serve as ring precursors for coenzyme Q biosynthesis. J. Lipid Res. 56, 909–919. doi: 10.1194/jlr.M057919
Keywords: coenzyme Q, CoQ deficiency, mitochondrial disease, 4-hydroxybenzoic acid, para-aminobenzoic acid, biosynthesis, chemical analogs, bioavailability
Citation: Pierrel F (2017) Impact of Chemical Analogs of 4-Hydroxybenzoic Acid on Coenzyme Q Biosynthesis: From Inhibition to Bypass of Coenzyme Q Deficiency. Front. Physiol. 8:436. doi: 10.3389/fphys.2017.00436
Received: 28 March 2017; Accepted: 08 June 2017;
Published: 22 June 2017.
Edited by:
Alberto Sanz, Newcastle University, United KingdomReviewed by:
Iain P. Hargreaves, Liverpool John Moores University, United KingdomCopyright © 2017 Pierrel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fabien Pierrel, ZmFiaWVuLnBpZXJyZWxAdW5pdi1ncmVub2JsZS1hbHBlcy5mcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.