
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 23 May 2017
Sec. Craniofacial Biology and Dental Research
Volume 8 - 2017 | https://doi.org/10.3389/fphys.2017.00320
 Ryouichi Satou1
Ryouichi Satou1 Masaki Sato2
Masaki Sato2 Maki Kimura2
Maki Kimura2 Yoichi Ishizuka1
Yoichi Ishizuka1 Masakazu Tazaki2
Masakazu Tazaki2 Naoki Sugihara1
Naoki Sugihara1 Yoshiyuki Shibukawa2*
Yoshiyuki Shibukawa2*Circadian rhythms are essential for health and regulate various physiological functions. These rhythms are regulated by a negative-feedback loop involving clock genes in the suprachiasmatic nucleus (SCN) and peripheral tissues. The rate of secretion of salivary substances, ions, and water follows a circadian rhythm, however, the relationship between the molecular mechanism of salivary secretion and peripheral circadian rhythm is not yet clear. Anoctamin 1 (ANO1, also known as TMEM16A) and Aquaporin 5 (AQP5) play an important role in the transport of ions and water in the submandibular glands (SGs). We examined the interaction between the rhythmic expression pattern of the clock genes, Ano1 and Aqp5, in rat whole SGs as well as isolated acinar and ductal cells. Circadian rhythmic expression for Bmal1, Per1, Per2, Clock, Cry1, Cry2, Rorα, and Rev-erbα mRNAs, also called the clock genes, was observed in rat SGs by semi-quantitative RT-PCR analysis. We also observed rhythmic patterns in Ano1 and Aqp5 mRNA expression. The expression of ANO1 protein also showed circadian rhythm, as confirmed by western blot analysis. We could not observe any time delay between the peak expression of ANO1 protein and its mRNA. Expression levels of the clock gene mRNAs in the ductal cells was higher than that in acinar cells, however, rhythmic oscillations were observed in both. Our results suggest that SGs have peripheral clocks, and rhythmic expressions of Ano1 and Aqp5 along with the clock genes, may play an important role in the circadian regulation of salivary secretion.
Circadian rhythm is essential for driving various physiological and metabolic functions in most living organisms, and coordination of their behavioral and biological processes such as sleep/wake cycles, body temperature, hormone secretion, blood pressure, and salivary secretion (Reppert and Weaver, 2002). Disturbance of the circadian rhythms can cause major systemic conditions such as cancers, central nervous system diseases, cardiovascular disorders, metabolic syndromes, and mental illness (Hastings et al., 2007). The rhythm is regulated by a master clock and other peripheral biological clocks. The master clock generates 24-h circadian rhythms in all mammals. It is located in the suprachiasmatic nucleus (SCN) of the hypothalamus and orchestrates the regulation of peripheral clocks. Several other peripheral clocks are regulated independently by endogenous self-oscillating transcription factors called the clock genes. Molecular regulation by the clock genes in the circadian oscillator is based on interconnected transcriptional/translational feedback loops (Reppert and Weaver, 2002; Suárez-Trujillo and Casey, 2016). Among them, aryl hydrocarbon receptor nuclear translocator-like protein 1 (Arntl/Bmal1), period (Per), circadian locomotor output cycles kaput (Clock), and cryptochrome circadian clock (Cry) are termed as the core clock genes. Retinoic acid receptor-related orphan receptor alpha (Rorα) and nuclear receptor subfamily 1 group D member 1 (Rev-erbα) regulate the transcription of Bmal1. The key transcription factors, CLOCK and BMAL1, form heterodimers that bind to the enhancer box (E-box) sequences and activate transcription of the Per and Cry genes. The PER and CRY proteins subsequently repress the transcription at their own promoters through a negative feedback loop, effected by acting on the CLOCK–BMAL1 complex (Shearman et al., 2000; Sato et al., 2014). This feedback loop controls the master and the peripheral clocks in most tissues.
Salivary flow rate or the secretion rate of salivary substances such as Na+, , Cl−, K+, and α-amylase also follows a circadian rhythm (Dawes, 1972). Especially, it is known that unstimulated salivary flow rate is extremely low during sleep (Dawes, 1972). Recent studies have shown the circadian expression rhythm of clock gene (Per1, Per2, Bmal1, Cry1, deleted in esophageal cancer 1[Dec1], Dec2, D-site of albumin promoter binding protein [Dbp] and Rev-erbα) and amylase 1 in SGs (Furukawa et al., 2005). The localization of core clock proteins (Bmal1, Per2, and Clock) and Bmal1 and Per2 mRNAs in the mucous acini and striated ducts of salivary glands was also determined by in situ hybridization (Zheng et al., 2012). Moreover, light and food entrainment control the phase of submaxillary Per1 expression (Vujović et al., 2008). These studies suggest that not only acinar but also ductal cells play an important role in circadian oscillation of salivary secretion. Although the findings imply that clock genes influence the physiological functions in the salivary glands, detailed rhythmic and temporal expression patterns of the clock genes in salivary gland cells, that is, acinar and ductal cells, remain to be investigated. The major salivary glands, SGs as well as the parotid and sublingual glands normally contribute over 90% to the total volume of unstimulated saliva. Percentage contributions of salivary glands during unstimulated saliva are as follows: 65% from SGs, 20% from parotid, and <10% from sublingual and minor glands (Humphrey and Williamson, 2001). SGs are mainly composed of two epithelial cell types: the acinar cells, which secrete water, ions, and the salivary proteins; and the ductal cells, which modulate the ionic composition of the saliva (Humphrey and Williamson, 2001).
There are transcellular and paracellular transport pathways for the secretion of water and ions in the SGs, which are driven by changes in transmembrane osmosis and water channel gating (Turner and Sugiya, 2002). Recent studies have shown that Anoctamin 1 (ANO1) and Aquaporin 5 (AQP5) play an important role in water and ion transport in SGs (Ma et al., 1999; Yang et al., 2008). ANO1 and AQP5 are localized on the apical membrane of the SGs (Yang et al., 2008). ANO1, is a transmembrane protein that functions as a Ca2+-activated chloride channel (CaCC). CaCCs control the apical Cl− efflux, which is essential for the vectorial transport of electrolytes and water in the bronchiolar epithelial cells, pancreatic acinar cells, proximal kidney tubule epithelium, retina, dorsal root ganglion sensory neurons, airways, and salivary glands (Caputo et al., 2008; Yang et al., 2008; Ferrera et al., 2010). AQPs are channel proteins that regulate the transmembrane movement of water in response to osmotic gradients for driving the salivary secretions. In SGs, AQP5 is one of the major aquaporins expressed on the apical membrane of the acinar and intercalated ductal cells (Delporte and Steinfeld, 2006; Matsuzaki et al., 2012). Although Ano1 and Aqp5 are key genes required for water and ion secretion in the SGs, their exact contribution to the regulation of circadian rhythm in salivary secretion remains to be validated. In the present study, we examined the interaction of temporal rhythmic expression patterns among the clock genes Ano1 and Aqp5, in rat SG acinar and ductal cells.
All animals were treated in accordance with the Guiding Principles for the Care and Use of Animals in the Field of Physiological Sciences, approved by the Council of the Physiological Society of Japan and the American Physiological Society. All animal experiments were carried out in accordance with the Guidelines for the Treatment of Experimental Animals at Tokyo Dental College. All the experimental protocols were approved by the Ethics Committee of Tokyo Dental College (No. 280901).
Six to eight weeks old, male Wistar rats (Charles River Laboratories Japan, Inc., Tsukuba, Japan) were used. Only male rats were chosen, to avoid the effect of sex-related hormonal differences on the circadian rhythm. Animals were housed with 12:12-h light/dark cycle (lights on from 08:00 to −20:00) with food and water available ad libitum. Before all experiments, we confirmed that rats are acclimated to these light/dark conditions by monitoring the daily pattern of wheel-running activity for 14 days (data not shown). All experiments were performed in accordance with Zeitgeber time (ZT, referring to an objective time scale wherein ZT0 is set as the time of lights on and, ZT12 is set as the time of lights off.), with ZT0 set as 08:00.
Submandibular acinar and ductal cells were isolated as previously described (Nezu et al., 2000; Sakai et al., 2002) with modification. Detailed methods are described in the Appendix. SGs were dissociated from anesthetized rats at each time point (ZT0 to ZT48). All cell isolation protocols were carried out within 2 h, and cells obtained (acinar and ductal populations) were immediately subjected to mRNA expression analysis or measurement of kallikrein activity.
Kallikrein activity was measured (Supplementary Figure 1 and Supplementary Table 3) according to the method by Geiger et al. (1980), using Nα-benzoyl-dl-arginine p-nitroanilide. Detailed methods are described in the Appendix.
Both sides of SGs were dissociated from anesthetized rats at ZT0, ZT6, ZT12, ZT18, ZT24, ZT30, ZT36, ZT42, and ZT48, and total RNA was immediately extracted by a modified acid guanidium phenol-chloroform method following SG dissociation. For cell isolation, both sides of SGs were dissociated at ZT0 and at 6-h intervals from ZT0 to ZT24. Isolated acinar and duct-like cells were then subjected to total RNA extraction within 2 h after whole SG dissection. For these samples, sqPCR analysis (Thermal Cycler Dice, TaKaRa Bio) was performed. For all series of sqPCR analyses, the same quantity of total RNA (50 ng) was used. Expression level of the internal reference gene, β-actin, was measured using One Step SYBR® PrimeScript® RT-PCR Kit II (Perfect Real Time, TaKaRa Bio), using probes labeled with 6-carboxyfluorescein (6-FAM). The primers used are described in Supplementary Tables 1, 2. sqPCR analysis was performed using the comparative Ct method (2−ΔΔCt, where Ct implies cycle threshold). This method was used to assess relative expression level of mRNA normalized to β-actin, using the Thermal Cycler Dice real time system software version 5.11.
Both sides of SGs were dissected at ZT0, ZT6, ZT12, ZT18, ZT24, ZT30, ZT36, ZT42, and ZT48, and immediately stored in liquid nitrogen. The tissue was homogenized in ice cold radioimmunoprecipitation assay (RIPA) lysis buffer containing 20 mM Tris-HCl (pH 8.0), 137 mM NaCl, 1% Nonidet-P40, 2 mM EDTA, 10% glycerol (pH 8.0), 10 μL/mL protease inhibitor cocktail, and 10 μL/mL phenylmethylsulfonyl fluoride, incubated for 5 min and centrifuged at 5,000 g for 10 min at 4°C. The supernatant was separated and its protein concentration was calculated using the DC protein assay kit (Bio-Rad, Richmond, CA) based on the Lowry method, with BSA (2 mg/mL) as the protein standard. For each sample, 30 μg protein was electrophoresed on 10% SDS-PAGE gel, transferred to a polyvinylidene-difluoride (PVDF) membrane and analyzed using the Trans-Blot SD semi-dry electrophoretic transfer cell (Bio-Rad). Membranes were blocked with 5% skimmed milk powder in PBS-Tween (0.1% PBS-T) for 1 h. The membrane was cut into two pieces, and then probed overnight at 4°C separately with anti-ANO1 (1:500, ab53212; Abcam, Cambridge, UK) or anti-β-actin (1:10,000, GTX110564; GeneTex, Iryine, CA, USA) antibodies. Excess primary antibody was removed by washing with PBS-T, and membrane was incubated with horse-radish peroxidase (HRP)-conjugated polyclonal goat anti-rabbit immunoglobulins (1:1,000, P0448; Dako, California, US) for 1 h at room temperature. All antibodies were diluted in the skimmed milk buffer. Protein bands were visualized with the ECL chemiluminescence WB Detection Reagents (GE Health Care, Little Chalfont, UK), and documented using the Image Quant LAS-4000 (GE Health Care). Quantification of bands was performed by using Image Quant TL 7.0 software (GE Health Care).
Student's t-test was performed to examine the differences between two groups. All results were represented as mean ± SD, and differences were considered to be significant at p < 0.05. Circadian rhythms during 24- and 48-h periods were statistically analyzed by one-way analysis of variance (ANOVA), and differences were considered significant at p < 0.05. In addition, rhythmicity was determined by CircWave version 1.4 (Oster et al., 2006) (p < 0.05), and the significance (p < 0.05) of rhythmicity was evaluated at a 95% confidence level (α = 0.05).
Temporal relative expression profiles of the mRNAs of the clock genes, Ano1 and Aqp5 in the SGs were examined every 6 h from ZT0 to ZT48 (three experiments, with 27 rats in total; Figure 1). Relative expression level of Bmal1 mRNA was significantly higher at ZT0, ZT24, and ZT48, whereas it was lower at ZT12 and ZT36 (Figure 1A). Per1 mRNA was significantly upregulated at ZT12 and ZT36 (Figure 1B). Per2 mRNA showed significantly higher expression at ZT12 and ZT36, and lower expression at ZT6 and ZT30 (Figure 1C). Bmal1 expression was in antiphase with the temporal expression pattern of Per2 with 12 h phase difference. Clock mRNA also showed significant rhythmic expression (one-way ANOVA, p < 0.01), however a clear phase variation in its expression peaks could not be observed (Figure 1D). The phase of expression of Cry1 mRNAs also deviated by 6–12 h from the phase of Per2 expression peaks (Figure 1E). Cry2 mRNA showed significant upregulation at ZT12 and ZT36 and lower expression at ZT0, ZT24, and ZT48 (Figure 1F). Relative expression level of Rorα mRNA was significantly higher at ZT12 and ZT36 but lower, at ZT0, ZT24, and ZT48 (Figure 1G). Rev-erbα mRNA was significantly upregulated at ZT12 and ZT30, but showed lower expression at ZT0, ZT24, and ZT48 (Figure 1H). In addition, temporal mRNA expression profiles of water and ion secretion related plasma membrane proteins in the saliva, Ano1, and Aqp5, showed rhythmicity, with significantly higher expression observed at ZT12 and ZT36, and lower expression at ZT0, ZT24, and ZT48 (Figures 1I,J; p < 0.05). The expression levels of Bmal1, Per1, Per2, Clock, Cry1, Cry2, Rorα, Rev-erbα, Ano1, and Aqp5 were considered rhythmic by CircWave (Figure 1).
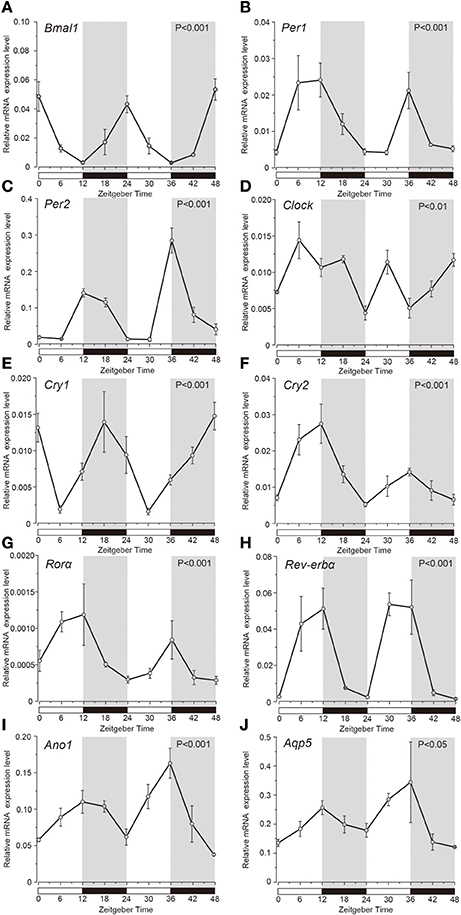
Figure 1. Temporal relative mRNA expression profiles of clock-gene in whole SGs. At the indicated Zeitgeber time (ZT: lights on at ZT0, ZT24; lights off at ZT12, ZT36), SGs were extracted from Wistar rats housed under a 12:12-h light/dark cycle for 14 days). Figure showing gene expression of (A) Bmal1, (B) Per1, (C) Per2, (D) Clock, (E) Cry1, (F) Cry2, (G) Rorα, (H) Rev-erbα, (I) Ano1, and (J) Aqp5, at 6 h intervals in ZT. The horizontal white and black bars indicate light and dark phases (shown by gray), respectively. The mRNA levels were normalized to the expression of β-actin mRNA and represented as the means ± SDs of three replicates per time-point (n = 3). P-values were calculated by one-way ANOVA and significance at p < 0.05. Rhythmicity was determined using CircWave (p < 0.05) at a 95% confidence level (α = 0.05). Clock genes (A–H) and Ano1 (I) and Aqp5 (J) showed rhythmic mRNA expression patterns.
Western blot analysis revealed the circadian oscillation of ANO1 expression in the rat SGs. Temporal ANO1 protein expression was examined every 6 h from ZT0 to ZT48 (three experiments with 27 rats). A single band (~110–120 kDa) was detected for ANO1 on the SDS-PAGE gel (Figure 2A). ANO1 expression was normalized with ß-actin, a constitutively expressed internal control, every 6 h for a 48 h period. The circadian expression of ANO1 showed significant oscillation patterns peaking at ZT12 and ZT36 (ANOVA, p < 0.01) (Figure 2B). ANO1 expression was considered rhythmic by CircWave (p < 0.05, α = 0.05).
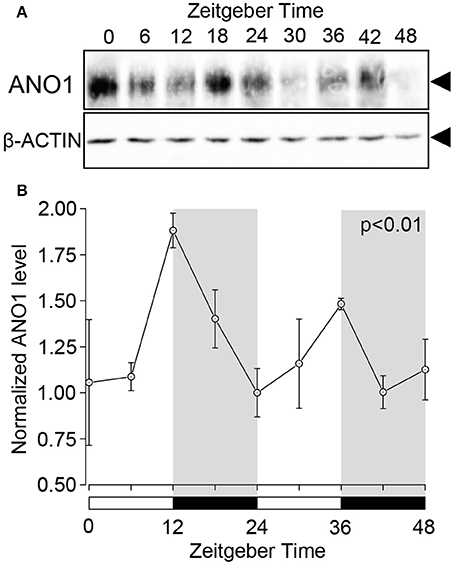
Figure 2. Circadian expression of ANO1 protein in rat SGs. (A) SGs were extracts at ZT0, ZT6, ZT12, ZT18, ZT24, ZT30, ZT36, ZT42, and ZT48 from three rats per time-point and subjected to western blot analysis. Total protein was separated on SDS-PAGE and probed with rabbit polyclonal ANO1 antibody (1:500). The relative density of ANO1 from the western blot was normalized against β-actin and plotted (1, density at ZT24). (B) Graph showing the temporal expression profile of ANO1 protein. Each point represents the mean ± SD from three independent experiments (n = 3). The horizontal white and black bars represent the light and dark phase (shown in gray) respectively. P-values were calculated by one-way ANOVA and significant differences observed at p < 0.01. Rhythmicity was determined using CircWave (p < 0.05) at a 95% confidence level (α = 0.05). The circadian expression of ANO1 showed significant oscillation patterns peaking at ZT12 and ZT36 (B).
Expression profile of the clock genes in the isolated acinar and ductal cells was examined by sqPCR from three experiments with a total of 15 rats. We observed rhythmic mRNA expression patterns for Bmal1 (Figures 3A, 4A), Per1 (Figures 3B, 4B), Per2 (Figures 3C, 4C), Cry1 (Figures 3E, 4E), Cry2 (Figures 3F, 4F), and Rev-erbα (Figures 3H, 4H) in both the acinar and ductal cells. Rhythmic expression was not observed in Clock mRNA pattern in the acinar cells (Figure 3D). However, the same was observed in the ductal cells, with ZT0 and ZT24 showing the highest, and ZT12 showing the lowest expression (Figure 4D). Rorα mRNA expression in ductal cells (Figure 4G) also showed no rhythmic expression, and similar results were observed in acinar cells (Figure 3G). The phases showing peak expression of Bmal1 (Figures 3A, 4A), Per1 (Figures 3B, 4B), Cry1 (Figures 3E, 4E), Cry2 (Figures 3F, 4F), and Rev-erbα (Figures 3H, 4H) mRNAs were almost identical in the acinar and ductal cells, however, Per2, Clock, and Rev-erbα showed different expression patterns in these cells (Figures 3C,D,G, 4C,D,G). Per2 mRNA displayed highest expression in acinar and ductal cells at ZT18 and ZT12, respectively (Figures 3C, 4C). Rorα mRNA expression showed significantly high expression at ZT6 in acinar cells, whereas rhythmic expression was not observed in ductal cells (Figures 3G, 4G).
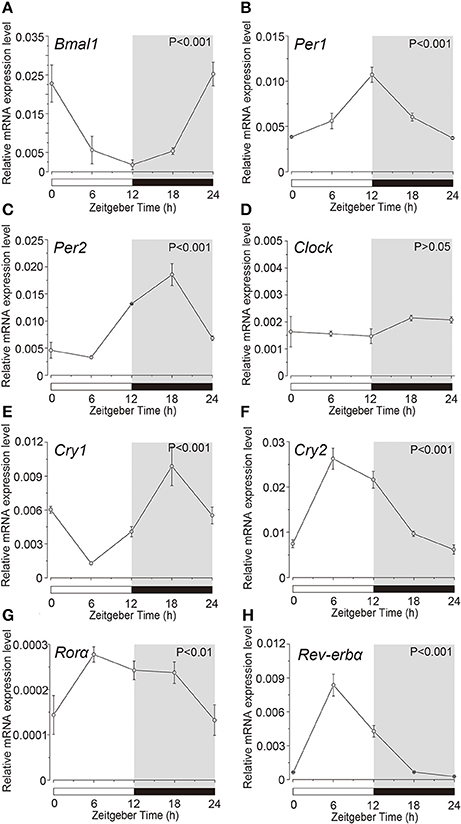
Figure 3. Circadian mRNA expression profiles of clock genes in acinar cells. Figure showing circadian rhythm in clock gene expression pattern in acinar cells by sqPCR. Relative mRNA expressions of (A) Bmal1, (B) Per1, (C) Per2, (D) Clock, (E) Cry1, (F) Cry2, (G) Rorα, and (H) Rev-erbα during 24 h period, at 6 h intervals in ZT, in the acinar cells. The horizontal white and black bars indicate light and dark phase (shown by gray). The mRNA levels were normalized to the expression of β-actin mRNA and expressed as means ± SD of three independent experiments per time-point (n = 3). P-values were calculated by one-way ANOVA and statistical differences at p < 0.05. Rhythmicity was also determined using CircWave (p < 0.05) at a 95% confidence level (α = 0.05). We observed rhythmic mRNA expression patterns for clock genes with the exception of Clock mRNA in acinar cells. Note that we used the time separating SG as a time (ZT) to evaluate temporal expression of clock gene mRNA.
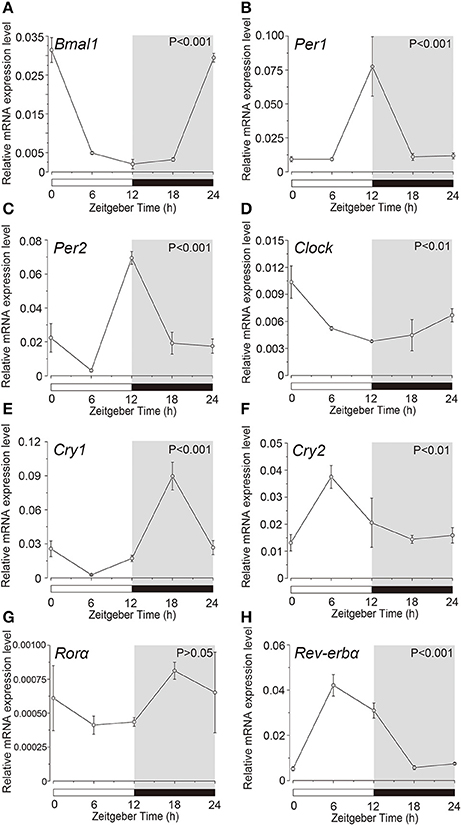
Figure 4. Circadian mRNA expression profiles of clock genes in ductal cells. Figure shows the circadian rhythm of clock gene expression pattern in acinar cells by sqPCR. Relative mRNA expression of (A) Bmal1, (B) Per1, (C) Per2, (D) Clock, (E) Cry1, (F) Cry2, (G) Rorα, and (H) Rev-erbα during a 24-h time period at 6-h intervals according to ZT in ductal cells. The horizontal white and black bars indicate light and dark phases (shown as gray), respectively. The mRNA levels were normalized to the expression of β-actin mRNA and expressed as means ± SDs of three independent experiments per time-point (n = 3). P-values were calculated by one-way ANOVA and results with at p < 0.05 were considered significant. Rhythmicity was also determined using CircWave (p < 0.05) at a 95% confidence level (α = 0.05). Temporal expression profiles of clock genes in the ductal cells showed circadian rhythms, except for Rorα. Note that we used the time separating SG as a time (ZT) to evaluate temporal expression of clock gene mRNA.
The peak expression of Bmal1 in ductal cells was higher than that in the acinar cells; however, the difference was not significant (Figure 5A). Additionally, the mRNA expression of Per1, Per2, Clock, Cly1, Cry2, and Rorα in ductal cells was significantly higher than that in the acinar cells (Figures 5B–G). The peak expression of Rev-erbα in acinar cells was slightly higher than that in ductal cells; however, this difference was not significant (Figure 5H).
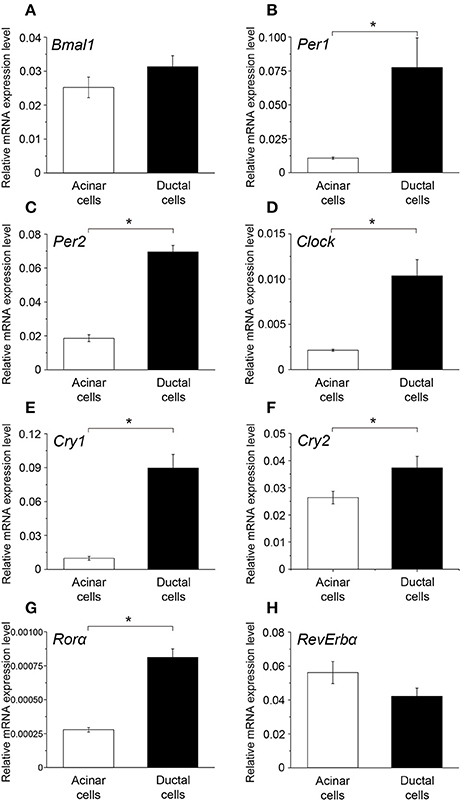
Figure 5. Comparison of the peak mRNA expression of clock genes between acinar and ductal cells. Bar graph showing the peak mRNA expression level of (A) Bmal1, (B) Per1, (C) Per2, (D) Clock, (E) Cry1, (F) Cry2, (G) Rorα, and (H) Rev-erbα in the acinar (white column) and ductal cells (black column). The mRNA levels were expressed as means ± SDs of three independent experiments (n = 3). Statistical analyses was performed by Student's t-test and statistical differences at p < 0.05 are indicated as “*” (B–G).
We proved that Bmal1, Per1, Per2, Clock, Cry1, Cry2, Rorα, and Rev-erbα show rhythmic circadian expressions in SGs (Figure 1). As previously shown, the phases and peaks of Bmal1, Per1, and Per2 expression exhibited opposite rhythms, which were shifted 12 h and repeated every 24 h. The Cry1 expression peaks were shifted compared to those of Per1, Per2, and Bmal1. Rorα and Rev-erbα showed rhythmic circadian expression similar to that of Per2. The phases and peaks in their expression profiles were similar to those in the SCN and other organs (Reppert and Weaver, 2002; Ueda et al., 2005). Our results were consistent with previous studies in SGs (Furukawa et al., 2005; Zheng et al., 2012). These results suggest that rat SGs have peripheral clocks and molecular clock mechanism with negative feedback loops. Although, Clock expression in whole SGs showed circadian oscillations, the peaks could not be distinguished clearly. The indistinct expression pattern may be caused by the phase differences in the Clock expression in each acinar and ductal cell individually (see below). It has been reported that temporal expression profile of Clock gene in the SCN did not show any circadian rhythm, however the peripheral clocks in organs such as liver and kidney, showed circadian rhythm in Clock expression (Ueda et al., 2005). This is in consensus with our results.
The expression of Ano1 and Aqp5 mRNA in whole SGs showed circadian rhythm, which was synchronous with Per2 expression (Figure 1). In recent studies, ANO1, CaCC, was found to express highly in the luminal membrane of the acinar cells in the SGs (Caputo et al., 2008; Yang et al., 2008). CaCC controls the apical efflux of Cl− ions, which drives the transport of electrolytes and water secretion in the SGs. Transfection with mouse Ano1 siRNA significantly reduced the peak salivary flow rate induced by muscarinic-cholinergic stimulation (Yang et al., 2008). The Activation of CaCC caused an increase in intracellular Ca2+ concentration, which also showed circadian rhythm (Ikeda et al., 2003). In addition, Aqp5 knock-out mice showed decrease in expression of the tight junction proteins and water permeability, and a significant increase in the volume of the acinar cells compared to the wild type (Krane et al., 2001; Kawedia et al., 2007). In the Aqp5 knock-out mice, saliva production by muscarinic-cholinergic stimulation was reduced by more than 60% (Yang et al., 2008). In the present study, Aqp5 mRNA expression showed similar temporal patterns and peak times as that of Ano1. These results suggest, that the circadian rhythm in water secretion in SGs may be associated with water permeability, which is controlled by the circadian oscillation of Ano1 and Aqp5 expression. Rats displayed an increased intake of water and food during the nocturnal period (ZT12–ZT24). The expression profile of Ano1 and Aqp5 also correlated with their feeding and drinking behavior (Boulos and Terman, 1980; Mistlberger, 2009).
We showed that ANO1 protein expression displayed rhythmic circadian oscillations. There was no time-lag between the peak time of protein and mRNA expression (Figure 2). BMAL1 protein was transcribed from Bmal1 mRNA without any significant time-lag (Tamaru et al., 2000). PER2 protein expression rhythm showed 4 to 6-h phase delay compared to the mRNA expression rhythm (Field et al., 2000). The result indicating that Ano1 mRNA translated without any delay owing to factors such as phosphorylation-dependent proteolysis or shuttling by nuclear export signals.
In this study, we successfully isolated almost pure populations of acinar and ductal cells from SGs to analyze clock genes mRNA expression in each individual cell. Kallikrein, a serine protease, has been found to be localized in striated duct cells, but not in acinar cells in rat SGs (Simson et al., 1979). Immunoreactive Egf, Egfr, Hgf, Fgf2, Tgf-α, Tgf-β1, and Igf1 are localized in ductal cells (Simson et al., 1979; Motoko et al., 1992; Amano and Iseki, 1993, 2001). In contrast, Ano1 is present in acinar cells, but not in ductal cells in SGs (Yang et al., 2008). In the present study, the isolated duct-like cells showed higher kallikrein activity (Supplementary Figure 1) and Egf, Egfr, Hgf, Fgf2, Tgf-α Tgf-β1, and Igf1 expression compared with Ano1-positive acinar-like cells (Supplementary Figure 2), indicating that we could isolate almost pure populations of acinar and ductal cells. In addition, we could not find any significant differences in Aqp5 expression between acinar and ductal-like cells (data not shown). These results were consistent with the finding that Aqp5 was expressed in both the apical plasma membrane in acinar cells and cells in the striated duct (Matsuzaki et al., 2012).
Acinar cells showed rhythmic expression of Bmal1, Per1, Per2, Cry1, Cry2, Rorα, and Rev-erbα, but not Clock (Figure 3). In ductal cells, mRNA expression of Bmal1, Per1, Per2, Clock, Cry1, Cry2, and Rev-erbα displayed a circadian rhythm, whereas Rorα did not (Figure 4). The temporal expression pattern of Bmal1, Per1, Cry1, Cry2, and Rev-erbα in both the acinar and ductal cells, isolated from the SGs, had very similar circadian rhythms. The rhythmic expression pattern of Clock was indistinct in whole SGs and acinar cells. However, ductal cells showed rhythmic Clock mRNA expression, similar in pattern to Bmal1. The obscurity in Clock rhythmic expression in whole SGs might be due to overlapping cycles of mRNA expression in individual acinar and ductal cells. Phase difference in mRNA expression for Per2 and Rorα was also observed between acinar and ductal cells. The peak mRNA expression time for Per2 in whole SGs, corresponded with that in the isolated ductal cells. The peak time of expression for Rorα in whole SGs did not coincide with that in acinar and ductal cells. Variable oscillatory expressions of Per1, a paralog of Per2, has been observed in individual cell in the SCN (Yamaguchi, 2003). These results suggest that temporal expression patterns of clock gene mRNAs in whole SGs are established by the combination of their expression profiles in both the acinar and ductal cells. Per1, Per2, Clock, Cry1, Cry2, and Rorα showed high expression in ductal cells (Figure 5). The mRNA expression levels of Bmal1 and Rev-erbα were not significantly different. These results are also consistent with previous studies demonstrating the localization of Per2 mRNA expression in mice SGs by in situ hybridization (Zheng et al., 2012). Our results also confirmed the localization of mRNAs encoding clock genes in acinar and ductal cells. In addition, we revealed that there were broad differences in clock gene expression levels between acinar and duct cells.
To confirm the relationship between clock genes and Ano1 and Aqp5 expression, we studied their DNA sequences. The promoters of rat Ano1 and Aqp5 have an enhancer box (E-box) binding sequence. The E-box is a DNA response element that acts as a protein-binding site and is necessary for the binding of BMAL1-CLOCK heterodimer to the promoter region. E-box plays a major role in regulating the transcription of Per2 and Cry1 (Bozek et al., 2009). A recent study has identified the E-box protein-binding sequence on the Aqp5 promoter in humans and mice (Zheng et al., 2012). In rats, the E-box element has been identified from −160 to +69 on the Aqp5 promoter (Nomura et al., 2007). The existence of E-box in the promoter region is a characteristic of clock-controlled genes (CCGs), which show rhythmic expression and contain clock gene binding motifs, in the promoter (Bozek et al., 2009). Thus, Ano1 and Aqp5 may be putative CCGs and key targets of the BMAL1-CLOCK heterodimer in SGs.
In conclusion, we showed circadian rhythmic expression of Bmal1, Per1, Per2, Clock, Cry1, Cry2, Rorα, and Rev-erbα mRNAs in not only whole SGs, but also isolated acinar and ductal cells. From our personal communications, Bmal1 mRNA also showed rhythmic expression in a salivary ductal cell line (He et al., 1998). Although we could not exclude the possibility of contributions of a master clock in the suprachiasmatic nucleus to circadian rhythmic salivation, cells in the salivary gland may have independent peripheral clocks. Further studies are needed to clarify the involvement of the suprachiasmatic nucleus in the salivation rhythm by using in vivo experiments, such as monitoring of salivary secretion following section of nervous input to the SGs. Clock genes may also regulate the circadian rhythmic oscillatory expression of Ano1, and Aqp5 mRNA and ANO1 protein, and may play an important role in the physiological nocturnal salivary secretion including water and electrolyte secretion. In addition, predominant rhythmic expression of clock genes in ductal cells may play an important role in modifying the ionic composition of the saliva, by reabsorbing and/or secreting ions during salivation.
RS, YS, and NS conceived and designed the experiments. RS performed all the experiments. RS and YS conceived the ideas, and wrote the paper. RS, YS, MS, MK, MT, YI, and NS contributed to the analysis of results and preparation of the figures and tables. All authors read and approved the results and the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We sincerely thank the Oral Health Science Center, Tokyo Dental College, for allowing us to use its facilities. RS acknowledges The Iwadare Scholarship Foundation for the award. We would like to thank Editage (www.editage.jp) for English language editing.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2017.00320/full#supplementary-material
Amano, O., and Iseki, S. (1993). Expression, localization and developmental regulation of insulin-like growth factor I mRNA in rat submandibular gland. Arch. Oral Biol. 38, 671–677. doi: 10.1016/0003-9969(93)90006-8
Amano, O., and Iseki, S. (2001). Expression and localization of cell growth factors in the salivary gland: a review. Kaibogaku Zasshi 76, 201–212.
Boulos, Z., and Terman, M. (1980). Food availability and daily biological rhythms. Neurosci. Biobehav. Rev. 4, 119–131. doi: 10.1016/0149-7634(80)90010-X
Bozek, K., Relógio, A., Kielbasa, S. M., Heine, M., Dame, C., Kramer, A., et al. (2009). Regulation of clock-controlled genes in mammals. PLoS ONE 4:e4882. doi: 10.1371/journal.pone.0004882
Caputo, A., Caci, E., Ferrera, L., Pedemonte, N., Barsanti, C., Sondo, E., et al. (2008). TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 322, 590–594. doi: 10.1126/science.1163518
Dawes, C. (1972). Circadian rhythms in human salivary flow rate and composition. J. Physiol. 220, 529–545. doi: 10.1113/jphysiol.1972.sp009721
Delporte, C., and Steinfeld, S. (2006). Distribution and roles of aquaporins in salivary glands. Biochim. Biophys. Acta 1758, 1061–1070. doi: 10.1016/j.bbamem.2006.01.022
Ferrera, L., Caputo, A., and Galietta, L. J. V. (2010). TMEM16A protein: a new identity for Ca2+-dependent Cl− channels. Physiology 25, 357–363. doi: 10.1152/physiol.00030.2010
Field, M. D., Maywood, E. S., O'Brien, J. A., Weaver, D. R., Reppert, S. M., and Hastings, M. H. (2000). Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron 25, 437–447. doi: 10.1016/S0896-6273(00)80906-X
Furukawa, M., Kawamoto, T., Noshiro, M., Honda, K. K., Sakai, M., Fujimoto, K., et al. (2005). Clock gene expression in the submandibular glands. J. Dent. Res. 84, 1193–1197. doi: 10.1177/154405910508401219
Geiger, R., Fink, E., Lottspeich, F., Henschen, A., and Fritz, H. (1980). Human urinary kallikrein—Biochemistry and physiological studies. Fresenius Z. Für Anal. Chem. 301, 172–173. doi: 10.1007/BF00467808
Hastings, M., O'Neill, J. S., and Maywood, E. S. (2007). Circadian clocks: regulators of endocrine and metabolic rhythms. J. Endocrinol. 195, 187–198. doi: 10.1677/JOE-07-0378
He, X., Kuijpers, G. A., Goping, G., Kulakusky, J. A., Zheng, C., Delporte, C., et al. (1998). A polarized salivary cell monolayer useful for studying transepithelial fluid movement in vitro. Pflüg. Arch. Eur. J. Physiol. 435, 375–381. doi: 10.1007/s004240050526
Humphrey, S. P., and Williamson, R. T. (2001). A review of saliva: normal composition, flow, and function. J. Prosthet. Dent. 85, 162–169. doi: 10.1067/mpr.2001.113778
Ikeda, M., Sugiyama, T., Wallace, C. S., Gompf, H. S., Yoshioka, T., Miyawaki, A., et al. (2003). Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron 38, 253–263. doi: 10.1016/S0896-6273(03)00164-8
Kawedia, J. D., Nieman, M. L., Boivin, G. P., Melvin, J. E., Kikuchi, K.-I., Hand, A. R., et al. (2007). Interaction between transcellular and paracellular water transport pathways through Aquaporin 5 and the tight junction complex. Proc. Natl. Acad. Sci. U.S.A. 104, 3621–3626. doi: 10.1073/pnas.0608384104
Krane, C. M., Melvin, J. E., Nguyen, H.-V., Richardson, L., Towne, J. E., Doetschman, T., et al. (2001). Salivary acinar cells from aquaporin 5-deficient mice have decreased membrane water permeability and altered cell volume regulation. J. Biol. Chem. 276, 23413–23420. doi: 10.1074/jbc.M008760200
Ma, T., Song, Y., Gillespie, A., Carlson, E. J., Epstein, C. J., and Verkman, A. S. (1999). Defective secretion of saliva in transgenic mice lacking aquaporin-5 water channels. J. Biol. Chem. 274, 20071–20074. doi: 10.1074/jbc.274.29.20071
Matsuzaki, T., Susa, T., Shimizu, K., Sawai, N., Suzuki, T., Aoki, T., et al. (2012). Function of the membrane water channel aquaporin-5 in the salivary gland. Acta Histochem. Cytochem. 45, 251–259. doi: 10.1267/ahc.12018
Mistlberger, R. E. (2009). Food-anticipatory circadian rhythms: concepts and methods. Eur. J. Neurosci. 30, 1718–1729. doi: 10.1111/j.1460-9568.2009.06965.x
Motoko, I., Koichi, U., Chiyonori, I., Toshio, Y., Tadami, K., and Tohru, T. (1992). Epidermal growth factor in salivary glands -epidermal growth factor and its receptor in human salivary glands. Soc. Pract. Otolaryngol. 85, 805–814.
Nezu, A., Tanimura, A., and Tojyo, Y. (2000). Isolation of rat submandibular ductal cells by Percoll centrifugation. Higash Nippon Dent. J. 19, 15–22. Available online at: http://ci.nii.ac.jp/els/contentscinii_20170509121051.pdf?id=ART0000535761
Nomura, J., Hisatsune, A., Miyata, T., and Isohama, Y. (2007). The role of CpG methylation in cell type-specific expression of the aquaporin-5 gene. Biochem. Biophys. Res. Commun. 353, 1017–1022. doi: 10.1016/j.bbrc.2006.12.126
Oster, H., Damerow, S., Hut, R. A., and Eichele, G. (2006). Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J. Biol. Rhythms 21, 350–361. doi: 10.1177/0748730406293053
Reppert, S. M., and Weaver, D. R. (2002). Coordination of circadian timing in mammals. Nature 418, 935–941. doi: 10.1038/nature00965
Sakai, T., Michikawa, H., Furuyama, S., and Sugiya, H. (2002). Methacholine-induced cGMP production is regulated by nitric oxide generation in rabbit submandibular gland cells. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 132, 801–809. doi: 10.1016/S1096-4959(02)00102-1
Sato, M., Matsuo, T., Atmore, H., and Akashi, M. (2014). Possible contribution of chronobiology to cardiovascular health. Front. Physiol. 4:409. doi: 10.3389/fphys.2013.00409
Shearman, L. P., Sriram, S., Weaver, D. R., Maywood, E. S., Chaves, I., Zheng, B., et al. (2000). Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019. doi: 10.1126/science.288.5468.1013
Simson, J. A., Spicer, S. S., Chao, J., Grimm, L., and Margolius, H. S. (1979). Kallikrein localization in rodent salivary glands and kidney with the immunoglobulin-enzyme bridge technique. J. Histochem. Cytochem. 27, 1567–1576. doi: 10.1177/27.12.391993
Suárez-Trujillo, A., and Casey, T. M. (2016). Serotoninergic and circadian systems: driving mammary gland development and function. Front. Physiol. 7:301. doi: 10.3389/fphys.2016.00301
Tamaru, T., Isojima, Y., Yamada, T., Okada, M., Nagai, K., and Takamatsu, K. (2000). Light and glutamate-induced degradation of the circadian oscillating protein BMAL1 during the mammalian clock resetting. J. Neurosci. 20, 7525–7530. Available online at: http://www.jneurosci.org/content/20/20/7525.long
Turner, R. J., and Sugiya, H. (2002). Understanding salivary fluid and protein secretion. Oral Dis. 8, 3–11. doi: 10.1034/j.1601-0825.2002.10815.x
Ueda, H. R., Hayashi, S., Chen, W., Sano, M., Machida, M., Shigeyoshi, Y., et al. (2005). System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192. doi: 10.1038/ng1504
Vujović, N., Davidson, A. J., and Menaker, M. (2008). Sympathetic input modulates, but does not determine, phase of peripheral circadian oscillators. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R355–R360. doi: 10.1152/ajpregu.00498.2007
Yamaguchi, S. (2003). Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412. doi: 10.1126/science.1089287
Yang, Y. D., Cho, H., Koo, J. Y., Tak, M. H., Cho, Y., Shim, W.-S., et al. (2008). TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 455, 1210–1215. doi: 10.1038/nature07313
Keywords: aquaporin, circadian rhythm, clock gene, secretion, chloride channel, transport
Citation: Satou R, Sato M, Kimura M, Ishizuka Y, Tazaki M, Sugihara N and Shibukawa Y (2017) Temporal Expression Patterns of Clock Genes and Aquaporin 5/Anoctamin 1 in Rat Submandibular Gland Cells. Front. Physiol. 8:320. doi: 10.3389/fphys.2017.00320
Received: 02 February 2017; Accepted: 03 May 2017;
Published: 23 May 2017.
Edited by:
Petros Papagerakis, University of Michigan, United StatesReviewed by:
Li Zheng, Ohio State University at Columbus, United StatesCopyright © 2017 Satou, Sato, Kimura, Ishizuka, Tazaki, Sugihara and Shibukawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoshiyuki Shibukawa, eXNoaWJ1a2FAdGRjLmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.