- Instituto de Investigaciones Marinas, Consejo Superior de Investigaciones Científicas, Pontevedra, Spain
One of the main factors limiting molluscs production is the presence of pathogens and diseases. Disease agent transfer via transfers of live molluscs has been a major cause of disease outbreaks and epizootics. Because of that, the European Union has adopted several decisions and directives, the last in 2006 (2006/88/EC) to control movements of marine organisms over the European countries. Once the disease is established in a determined area its eradication is a complicated task because life cycle of pathogens are not completely known and only a good and early diagnosis of the disease could be the most appropriate way to deal with it. Besides, molluscs do not have an adaptive immune response and vaccination strategies are not possible. Molluscs listed diseases under EU legislation are mainly protozoan parasites, that's why histological techniques are recognized for their diagnosis. However, molecular techniques are being increasingly used primarily as confirmatory techniques of the presence of the pathogens but also in disease monitoring programs. Research perspectives are mainly focussed in the optimization, of the already described techniques to gain in sensitivity and sensibility and in the development of new molecular biology techniques (quantitative real time PCRs), that are faster and easier to apply and that allow a positive diagnosis even in early stages of infection. However, molecular tools detect DNA sequences of the pathogen which does not imply that pathogen is viable in the cell host and the infection is established. Consequently, it needs to be validated against other techniques, such as histology or in situ hybridization, so that its reliability can be determined.
Introduction
Aquaculture is a dynamic activity worldwide that over the past few decades has doubled its production. According to FAO statistics (FAO, 2014), the world aquaculture production in 2012 was 66, 63 million tons (not including aquatic algae and non-food products).
Asian countries are the main producers of molluscs being China by far the leading producer. In the European Union, the marine bivalve industry has grown to be very important for many regions contributing substantially to social and economic activity in the coastal zones. European aquaculture is focused on a limited number of species (oysters and mussels mainly) being raised at an industrial level. France, Spain, and Italy are the main bivalve producing countries. France is leading the oyster, Crassotrea gigas, production in Europe (80.000 tons/year). The mussels Mytilus edulis and Mytilus galloprovincialis are the bivalve species with the highest production output in Europe (491.000 tons/year). Spain is the third world producer and the top European producer with an output of nearly 300.000 tons/year.
There is a global trend in the growth of human consumption of seafood. Seafood constitutes an important and essential part of the diet of many people in the world and the need for increasing production will persist as the human population expands. In some countries, seafood is recognized as an important and healthy part of the human diet. Most of the demand for seafood is and will continue to be for finfish, but the production and harvest of molluscs, particularly bivalves, will also be an important rising demand. While the harvest of natural bivalve stocks will continue being significant, many wild stocks are probably already being harvested at or near maximum sustainable limits, and in some places some stocks may even be overharvested. Because of it, aquaculture is a good alternative to the harvest of wild stocks.
Bivalves are ideal animals for aquaculture: they are herbivores that require no additional feeding apart from the natural algae content of seawater and generally minimum husbandry. Although they have been cultured for hundreds of years, advances in culture technology in recent years have led to significantly higher production. Continued improvements in culture methodology and technology will be still required to meet increasing demand and also to make bivalve culture economically more attractive to both investors and people who wish to become shellfish farmers. An examination of the FAO marine aquaculture production data (FAO 2001–2014), shows that the number of bivalve species being cultured is increasing annually, and that more and more developing countries are becoming involved. Toward the end of the 19th century reliable techniques for culturing bivalves were developed. This allowed a rapid growth in production that was featly augmented about a century later by the development of hatchery techniques for seed production.
In agreement with the World Organization of Animal Health (OIE)1, during last decades, the world production of molluscs has been adversely affected by numerous diseases and due to his severe impact in the economic and socioeconomic development in many countries, some of these diseases have turned into a primary restriction for the development and the sustainability of the culture of molluscs. The transport of alive molluscs transferring infectious agents has been the principal reason of outbreaks of diseases and epizooties. Bearing this in mind, one of the very few ways to reduce the impact of such pathogens on commercially exploited bivalves is to establish effective programs to prevent the transfer of infected stocks. The risk associated with transfer of molluscs is particularly serious when they occur over long distances or overseas. However, a serious limiting factor is the lack of scientific information on even basic biology of molluscs pathogens.
Historically, infectious diseases have seriously affected the marine bivalve industry in Europe in various occasions. For example, in the early 1970's the Portuguese oyster, Crassostrea angulata, was dramatically affected by an irido-like virus (Marteil, 1976) leading to the almost extermination of the oysters in the European Atlantic waters. It was speculated that uncontrolled transfers of C. gigas introduced this irido virus to C. angulata, which was highly susceptible. In the late 1970s and early 1980s, mass mortalities attributed to Bonamiosis and marteiliosis caused declines in oyster production in Europe which favored the development of different management strategies to counteract the threat of both pathogens (Herrbach, 1971; Grizel et al., 1974; Alderman, 1979; Pichot et al., 1980; Bannister and Key, 1982; Van Banning, 1982; Polanco et al., 1984; Grizel, 1985; Friedman et al., 1989; Barber and Davis, 1994; Friedman and Perkins, 1994). Since 2008 massive mortality outbreaks affecting Crassostrea gigas has also being reported in different Member States attributed to the presence of the newly described oyster herpes virus μvar (Segarra et al., 2010) and a new Marteilia species, Marteilia cochillia, which is also causing a collapse in the cockle, Cerastoderma edule, fisheries in Galicia and Catalonia (NW and NE of Spain respectively; Carrasco et al., 2013; Villalba et al., 2014).
The control of farmed shellfish health is one of the key elements to maintain the competitiveness and to increase the sustainability of the European industry. Because of that, the European Union has adopted the Council Directive 2006/88/EC “on animal health requirements for aquaculture animals and products thereof and on the prevention and control of certain diseases in aquatic animals” to control movements over the European countries stablishing two categories of notifiable diseases including the most relevant molluscs diseases classifying them as exotic or non exotic diseases and making emphasis in the prevention, the control and the eradication of the aquatic animal diseases. Exotic diseases are those that are not presented in the European countries and if detected they must be eradicated (Bonamia exitiosa, Microcytos mackini, and Prekinsus marinus). Non exotic-diseases are generally confined to certain areas within the EU (Bonamia ostreae and Marteilia refringens). This Directive also leaves an open frame to include newly described pathogens as “emerging diseases” if they are consider to be a threat to the European aquaculture as it has been the case of the oyster herpes virus μvar (Commission regulation 175/2010)2. In Table 1 it is summarized the main molluscs species produced in Europe and the EU listed pathogens detected on its specie.
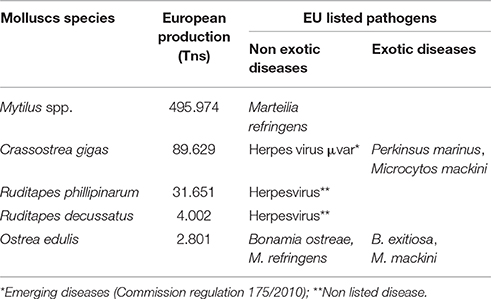
Table 1. Main molluscs species produced in Europe (FAO, 2014) and detected pathogens controlled under EU directive.
The effectiveness of any treatment against diseases in molluscs is limited by the physiology of the animals as well as by the characteristics of the culture system (Berthe, 2008). Molluscs are usually reared in the open sea which strongly limits the potential use of chemotherapy, because of the quantity of product to be used, its impact on the environment and the obvious risk of re-infection. Moreover, molluscs lack and adaptive immune response for what they cannot produce antibodies (Bayne, 2003; Ottaviani, 2011) and vaccination strategies are not possible. Therefore, the eradication of a molluscs disease is a very difficult task mainly because there is not enough knowledge on the life cycle of the parasites. There have been same attempts to eradicate molluscs pathogens with no success. In the 1980's bonamiosis was introduced to the Dutch flat oyster stocks with devasting consequences. Attempts to eliminate the disease by cleaning sites and restocking with disease free animals failed. Van Banning (1990, 1991) demonstrated that, when an oyster bed was cleaned and fallowed, the naïve oysters reintroduced into the areas, quickly developed the infection, suggesting that a reservoir of infection had persisted in the areas. The pathogen, B. ostreae, was thought to be eradicated from the Limfjord in Denmark due to the extreme climatic conditions like ice winters over several consecutive years, either by direct elimination of the parasite or by elimination of the host, previously imported French oysters, thus preventing the spread of Bonamia parasites to the naïve flat oyster population in the Limfjord (Madsen et al., 2013). Nevertheless, B. ostreae was detected for the first time in the naïve European flat oysters from Denmark in autumn 2014 although no increased mortalities were found in connection with this finding (Madsen, 2015).
Because of that, health management policies in areas where disease outbreaks occur are focused in the importance to reduce the incidence or the severity of a disease (Grizel et al., 1986) using different strategies; changes in culture management procedures (Korringa, 1950; Lauckner, 1983; Andrews and Ray, 1988; Montes et al., 2003), increasing the tolerance of the individuals to disease through stimulating the immune system (Macey and Coyne, 2005; Xue et al., 2008; Van Hai et al., 2009), producing tolerant strains by genetic handling (Ford and Haskin, 1987; Martin et al., 1993; Gaffney and Bushek, 1996; Hand et al., 1998; Nell et al., 2000; Culloty et al., 2004; Nell and Perkins, 2006; Samain et al., 2007; Villalba et al., 2007; Dove et al., 2013a,b; Lynch et al., 2014) or replacing susceptible species with resistant ones (Grizel and Héral, 1991; Man et al., 1991). However, the introduction of foreign species is a risky task that could introduce new pathogens, predators, or competitors that may perturb the equilibrium of the ecosystem and affect adjacent areas or countries (Carriker, 1992; Shatkin et al., 1997).
Taking into account all these considerations, the effectivity of health management measures to avoid economic losses and to avoid the spread of the disease to a disease free areas are directly related with the early diagnosis of the diseases using sensitive and rapid diagnostic techniques, the collection of diagnosis data, and the establishment of diagnostic laboratories using standardized procedures with samples collected according to defined rules.
Lately, there has also been tentative studies on predictive models using epidemiological principles to design surveillance plans to evaluate the most probable time of the year when a pathogen can be detected (Lupo et al., 2014; Pande et al., 2015). Besides, the development of molecular biology tools applied to diseases processes in marine bivalve molluscs is giving us large information mainly in the study of immunity and host–pathogen interactions in molluscan bivalves that it is being applied in the development of disease resistant strains and the development of new diagnostic tools (Gomez-Chiarri et al., 2015).
From Histopathology to Molecular Tools
The methods of diagnosis of the diseases of the aquatic animals has evolved rapidly. Historically, most of the descriptions of molluscs pathogens are based on structural and ultra-structural studies. Traditionally, the diagnosis of molluscs diseases has primarily been achieved using histological methods and transmission electron microscopy (Azevedo et al., 1990; Anderson et al., 1994; Hervio et al., 1996; Longshaw et al., 2001; Howard et al., 2004). Histology provides a large amount of information not only of the general health of a shellfish but also the detection of a wide range of pathogens specially protozoan parasites associated to mortalities or alterations in the quality of cultured populations, or lesions associated to the interaction of the pathogen with the molluscs immune system. However, histopathology is time consuming and requires professional training and therefore many pathogens are difficult to detect by this method if there are low numbers of parasites present within infected tissues. It can also be difficult to definitively diagnose infections based exclusively on parasite species morphology criteria when there are similar morphological characteristics between species (Hine et al., 2001; Abollo et al., 2008). This is especially relevant when dealing with microcell parasites. Abollo et al. (2008) described slight differences between B. ostreae and B. exitiosa detected in the European flat oyster, Ostrea edulis, when observed at the microscope. B. ostreae shows a peripheral nucleus and scant cytoplasm while B. exitiosa shows a central nucleus, sometimes subcentral but rarely peripheral, and the cytoplasm larger than the one observed in B. ostreae.
To overcome these problems, from the 90s to nowadays, there has been a rapid increase of the use of molecular techniques for diagnosing bivalve infectious agents. Since 1997–2011 there have been published more than 2000 research papers trying to detect, identify or quantify parasites or diseases organisms carried out by ecto-parasites using DNA based tools (Hunt, 2011). These techniques are in comparison with many traditional methods, more rapid, sensitive, and profitable, and they do not demand the presence of too specialized personnel. DNA is a useful molecule to target for diagnostic procedures because its sequence does not usually vary with the life stage or developmental phase of the pathogen or with the host or tissue location (Mialhe et al., 1995).
The amplification of DNA fragments of a pathogen and subsequent sequence analysis can help us to confirm infections in different hosts (Abollo et al., 2008; Grijalva-Chon et al., 2015). This is of particular interest considering that, apparently, pathogens such as M. refringens have been detected in different molluscs species not showing a tight host specificity (Berthe et al., 1999, 2004). The European Directive 2006/88/EC has stablished a list of susceptible species for each of the notifiable pathogens, and based on molecular data this list should be actualized. That is the case of the protozoan parasite B. exitiosa listed in the Directive as an exotic disease. The pathogen was known to infect Ostrea chilensis in New Zealand (Hine et al., 2001; Berthe and Hine, 2004) and Ostrea angasi in Australia (Corbeil et al., 2006a) causing mass mortalities. However, molecular analyses have led to detection of B. exitiosa naturally infecting the European flat oyster O. edulis in Galician (NW Spain) coastal waters (Abollo et al., 2008) for the first time. Since this detection, the parasite has also been reported from different European countries infecting O. edulis (Narcisi et al., 2010; Arzul et al., 2012; Carrasco et al., 2012a; Longshaw et al., 2013; Batista et al., 2016).
Nevertheless, legislation is not flexible and is slow reacting to new pathological situations and because of that, at present time, B. exitiosa is still considered as an exotic disease to the EU.
It is also important to emphasize the potential of these new detection tools for elucidating the life-cycle of the parasites, crucial for the control and prevention of the diseases. These techniques offer the advantages of high sensitivity and high specificity, and possible rapid screening of aquatic organisms for the presence of the pathogen even in water samples. In that way, PCR was first used to screen every species sampled in the claire ponds for the presence of the parasite M. refringens. The copepod Paracartia (Acartia) grani as potential hosts (Audemard et al., 2002). Carrasco et al. (2007) also detected the presence of the pathogen in six different zooplankton taxa including copepods and larval stages of decapod crustaceans, although their role as intermediate host have not already being investigated. More recently, Boyer et al. (2013) and Arzul et al. (2014) confirmed by in situ hybridization the presence of M. refringens in the copepod Paracartia grani and Paracartia latisetosa respectively. Although it is not a notifiable disease for the EU, based also on molecular tools, the parasite Marteilia sydneyi, the aetiological agent of QC disease in the Sydney rock oyster (Saccostrea glomerata), was also identified in the epithelium of the intestine of two specimens of the polychaete worm, Nephtys australiensis (Adlard and Nolan, 2015), its detection was also confirmed by in situ hybridization. These new findings have contributed to a better knowledge of the disease.
Molecular tools can also circumvent problems inherent in study of organisms for which no in vitro culture medium is available as the case of molluscs viruses. The lack of established cell lines from mollucs difficults their isolation and subsequent study.
The Other Side of Using Molecular Tools as Diagnostic Techniques
However, as review by Burreson (2008), the detection of the pathogen's DNA by the polymerase chain reaction (PCR) indicates the presence of the infectious agent but it does not confirm that the pathogen is in a viable form causing an actual infection and no information is given about the tissue localization of the pathogen. Because of that, it is important to confirm the results by histology or in situ- hybridization (ISH) to verify that the pathogen really affects the mollusc. ISH, first developed by Joseph G. Gall (Gall and Pardue, 1969), is the most adequate technique for this purpose because it offers the advantages of the histology and the specificity of the molecular tools, but it is time consuming and its use is not recommended as a technique in surveillance programs.
In addition, the routine use of DNA based diagnostic techniques is hampered by a number of problems, which may result in false positive or false negative results. The design of these diagnostic techniques for molluscs diseases requires the search of molecular markers of certain specie-specific regions of the genome of the targeted pathogen (López-Flores et al., 2007a). Not all regions of the DNA are equally useful as targets for probes and/or PCR primers. rRNA genes are useful targets for diagnostic tests because there are many copies in the genome, which can help to ensure good sensitivity, and they offer a mosaic of conserved and variable regions which allow analyses at various levels of resolution. As a general rule, regions of the small-subunit and large–subunit ribosomal RNA genes (18 and 28S) are used to design probes specific as a genus level while regions in the intergenic spacers and internal transcribed spacers (IGS and ITS1-2) are most commonly used to design probes at an specie-specific level. These intergenic regions do not have a codificating function and they show a greater divergence even among close related species (López-Flores et al., 2007b). Efforts are being made to obtain sequences of the same regions from related organisms to optimize the chance of developing probes and PCR primers with the desired specificity. The development of species specific molecular diagnostic tools will be facilitated as sequences for more genes and pathogens become known. However, in order to minimize the possibility of species-specific molecular diagnostics failing to detect a particular strain of a pathogen, as many strains as possible from a wide geographic range should be sequenced. This is an important issue specially when the target strain of pathogen is pathogenic, while close relatives are harmless.
On the other hand, false-negatives results can be obtained from sampling error due to small abundance of pathogen DNA especially when pathogen is localized in an specific organ of the host, as it is the case of M. refringens and the tissue is subsampled, or due to the presence of inhibitors that may be present in the sample and could affect the activity of the DNA polymerase.
Specificity is designed as the ability of an assay to amplify DNA only from the target agent. Sensitivity (or limit of detection) is defined as the smallest quantity of DNA that can be systematically detected. The highly sensitivity that makes PCR a very powerful technique for amplification of DNA can also be seen as a negative aspect because of inadvertent contamination by nucleic acids. Even very low levels of contamination can result in false-positive results. Consequently, qualification of the specificity and quantification of the sensitivity of a molecular assay is required and as the assay only detect nucleic acid not assuming a real infection, it needs to be validated against other techniques, such as histology or in situ hybridization, so that its reliability can be determined (Kleeman et al., 2002; Carnegie et al., 2003; Burreson and Ford, 2004; Burreson, 2008). This is specially important in the detection of the pathogen in new host species or in a new geographic location.
Available Molecular Tools for Non Exotic Molluscs Diseases Listed by EU
Detection of Marteilia refringens
Marteilia refringens is a protozoan parasite of the phylum Cercozoa and order Paramyxida (Cavalier-Smith and Chao, 2003; Feist et al., 2009) infecting the digestive system of several bivalve species and inducing physiological disorders and eventually death of the animal (Grizel et al., 1974; Alderman, 1979). In the case of paramyxeans, molecular studies have focused on the small-subunit ribosomal RNA gene (Le Roux et al., 1999; Berthe et al., 2000; Kleeman and Adlard, 2000; Itoh et al., 2003). However, closely related organisms, such as M. refringens and M. maurini, presently considered to be the same specie, (Le Roux et al., 2001) or M. refringens and M. sydneyi (Kleeman et al., 2002), have a high degree of sequence similarity, making the development of specific probes and primers more difficult. There has also been described PCRs focused on the ITS-1 region (Le Roux et al., 2001) and the IGS region of the parasite (López-Flores et al., 2004) although they are neither specie-specific as the newly described pathogen, Marteilia cochillia, is also amplified (Carrasco et al., 2012b).
At present time there is no a M. refingens specie-specific PCR available and an restriction analyse assay (RFLP) based on the PCR described by López-Flores et al. (2004) is needed to discriminate between Martelia cochillia and Martelia refringens (Carrasco et al., 2012b).
Detection of Bonamia ostreae and Bonamia exitiosa
Bonamia ostreae and B. exitiosa are Haplosporidian protozoan parasites (Carnegie and Cochennec-Laureau, 2004) infecting haemocytes of several oyster species and inducing physiological disorders and eventually death of the animal (Grizel, 1985; Dinamani et al., 1987; Cranfield et al., 2005).
In the case of Bonamia species several PCR assays have been developed targeting the 18S rDNA gene, (Carnegie et al., 2000; Cochennec et al., 2000; Abollo et al., 2008), or TaqMan® assays (Corbeil et al., 2006b; Marty et al., 2006). However, none of these methods are able to differentiate between Bonamia species. For this reason, differentiation between species has been achieved using PCR-restriction fragment length polymorphism (PCR-RFLP) assays (Hine et al., 2001; Cochennec-Laureau et al., 2003; Abollo et al., 2008), sequencing the products obtained by PCR assays or by means of light microscopic techniques, thus causing the diagnostic process to be more expensive and slower. Recently species- specific quantitative real time PCR assays for B. ostreae have finally been developed targeting a region of the actin-1 gene (Robert et al., 2009) and the 18S-ITS 1 rDNA region (Ramilo et al., 2013). Other approaches has also been reported to face the problem of distinguishing among species based on DNA sensors. DNA biosensors are analytical devices resulting from the integration between DNA sequence-specific probe with a signal transducer. Narcisi et al. (2011) developed six single strand DNA probes selected within different regions of B. ostreae and B. exitiosa with the aim to discriminate between both species. A post PCR nucleic acid work by comparing experimental data, from electrochemical genosensors, and bioinformatics data, derived from the simulation of the secondary structure folding and prediction of hybridization reaction, showed correspondence indicating the possibility to use the prediction method for ssDNA probes selection in the genosensors development.
Quantitative real time PCR is used to amplify and simultaneously detect and quantify a targeted DNA molecule following the cycling principle of basic PCR. It does not require post-PCR sample handling to visualize the result in an agarose gel, preventing potential PCR product carry-over contamination resulting much faster and higher throughput assays. The amplified DNA is detected as the reaction progresses in “real time” in contrast with the standard PCR where the product of the reaction is detected at the end. The data thus generated is analyzed by computer software to detect and quantify the targeted DNA in samples to determine the presence and abundance of a particular pathogen. Different quantitative real time PCRs have already been described to detect molluscs pathogens (Corbeil et al., 2006b; Marty et al., 2006; Robert et al., 2009; Martenot et al., 2010; Umeda and Yoshinaga, 2012; Carrasco et al., 2013; Ramilo et al., 2013). There have also been attempts to develop multiplex PCRs to detect the presence of different molluscs pathogens in the same reaction in order to facilitate their diagnosis (Penna et al., 2001; Xie et al., 2010, 2013; Umeda and Yoshinaga, 2012). Recently, Ramilo et al. (2013) has developed a specie-specific multiplex PCR for the diagnosis of B. ostreae and B. exitiosa simultaneously, using a cocktail of 3 primers. When there is coinfection of both species, there is competition for the forward primer between them that targets the same region of the 18S rDNA gene of both species, and for other reaction components frequently leading to detectable amplification of DNA of just one of the parasites, suggesting that the multiplex PCR assay should be improved.
Conclusion, Open Questions and Way Forward
Valid laboratory results are essential for diagnosis, surveillance, and trade. Although molecular tools are now moving from a developmental phase in specialized laboratories for research purposes to routine application, they still need formal validation. These diagnostic techniques are not standardized and differences exist in the quality of reagents, sample preparation, in controls, as well as in the interpretation of the results. The use of a “standardized” diagnostic tool for routine analysis should allow the implementation of a calibrated and controlled process in laboratories. It is recognized that for such purpose, studies conducted in parallel with the same isolates in several laboratories would be necessary. Balseiro et al. (2006) and Flannery et al. (2014), carried out respectively comparative studies of the different diagnostic techniques between different laboratories to detect B. ostreae, standing out the importance of the molecular tools. However, PCR used in those studies was not B. ostreae specie-specific.
The European Union Reference Laboratory (EURL) for mollusc diseases (IFREMER) is the reference institution in charge of the harmonization of the molluscs diagnostic techniques for the EU listed pathogens. To carry out this task, it has elaborated standard operating procedures in which protocols specifying the procedures from the tissue sample to the reactives used in pathogen diagnosis are summarized and organizes regularly interlaboratory comparison tests among all interested national reference laboratories in EU. In 2013 the EURL organized a qPCR interlaboratory comparison test for the detection of Bonamia sp., and the characterization of the species B. ostreae and B. exitiosa. Optimal results were obtained and at present time the multiplex qPCR designed by Ramilo et al. (2013) for the diagnosis of B. ostreae and B. exitiosa simultaneously is now recommended in the Commission Implementing Decision (EU) 2015/15543 as regard requirements of diagnostic tools. However, comparative studies among different laboratories of the different diagnostic techniques are still missing in order to validate these newly described molecular tools.
The development of the molecular tools has significantly improved the diagnosis of molluscs diseases due to their high specificity and sensitivity, higher reproducibility of the results and faster detection of the pathogen. In Table 2 it is summarized the recommended diagnostic methods for the diagnosis of molluscs diseases recommended by the OIE of the EU listed diseases by Council Directive 2006/88/EC4 taking into account the final purpouse of the diagnosis. Methods are classified depending if they are going to be used in a targeted surveillance program, as a presumptive diagnosis or as a confirmatory diagnosis. Methods used in targeted surveillance programs should not be time consuming and in most of the cases, the use of molecular tools is recommended.
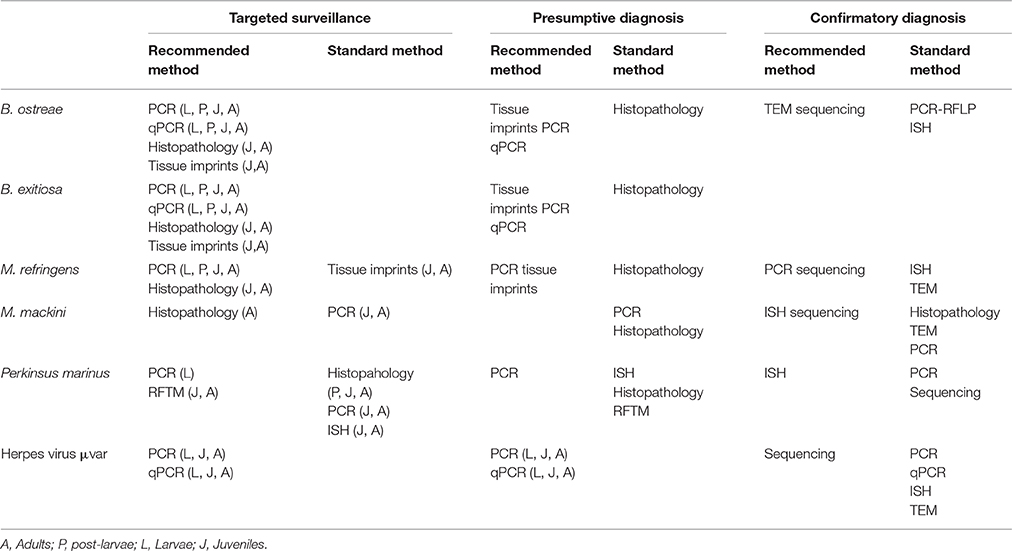
Table 2. Available diagnostic techniques recommended by O.I.E. (2016) for molluscs pathogens listed by EU.
Likewise, in the last few years, there have been greatly advances in the development of molecular tools with the use of new- generation sequencing technologies (NGS) and their ability to produce large volumes of data. The use of these technologies has drastically increased the number of molluscs genomic sequences in the databases.
Genomic research is basic to solve specific problems in bivalve aquaculture, including disease control. They can identify RNAs or proteins expressed in response to stress or pathogens and compensate the usual absence of clinical manifestations. All this new information will facilitate the study of bivalves and increase the success of molluscan aquaculture, facilitating the monitoring of the production, activation state of pathogens or immune status of the animals. Up to now, these methodologies have been applied to select key genes linked to resistence that could be used as markers for selection (Romero et al., 2012; Gomez-Chiarri et al., 2015), identify genes implicated in the immune response (Philipp et al., 2012; Moreira et al., 2012, 2014; Zhang et al., 2014; Gomez-Chiarri et al., 2015), or investigate biological processes (Clark et al., 2010; Joubert et al., 2010). Genomic advances happen in an exponential way. In the next incoming years the amount of available information will allow reserchers huge progresses regarding molecular biology of bivalve molluscs and pathogens.
Author Contributions
RA conducts the literature review and writing up of the paper and AF contributes to the revision of the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors want to thanks the Ministerio de Agricultura, Alimentacion y Medio Ambiente, to CSIC (project 201640E024) and to Ministerio de Economía y competitividad (project AGL2015-65705-R) for their financial support.
Footnotes
1. ^World Organisation for Animal Health. OIE. Manual of diagnostic test for aquatic animals. Chapter1.1.2. access online.
2. ^Commission regulation (EU) No. 175/2010 of 2 March 2010 implementing Council Directive 2006/88/EC as regards measures to control increased mortality in oysters of the species Crassostrea gigas in connection with the detection of Ostreid herpesvirus 1 μvar (OsHV-1 μvar).
3. ^Commission Implementing decision (EU) 2015/1554 of 11 September 2015, laying down rules for the application of Directive 2006/88/EC as regards requirements for surveillance and diagnostic methods.
4. ^Council Directive of 24 October 2006 on animal health requirements for aquaculture animals and products thereof and on the prevention and control of certain diseases in aquatic animals.
References
Abollo, E., Ramilo, A., Casas, S. M., Comesaña, P., Cao, A., Carballal, M. J., et al. (2008). First detection of the protozoan parasite Bonamia exitiosa (Haplosporidia) infecting flat oyster Ostrea edulis grown in European waters. Aquaculture 274, 201–207. doi: 10.1016/j.aquaculture.2007.11.037
Adlard, R. D., and Nolan, M. J. (2015). Elucidating the life cycle of Marteilia sydneyi, the aetiological agent of QX disease in the Sydney rock oyster (Sacccostrea glomerata). Int. J. Parasitol. 45, 419–426. doi: 10.1016/j.ijpara.2015.02.002
Anderson, T. J., McCault, T. F., Boulo, V., Robledo, J. A. F., and Lester, R. J. G. (1994). Light and electron immunohistochemical assays on paramyxean parasites. Aquat. Living Resour. 7, 47–52.
Andrews, J. D., and Ray, S. M. (1988). Epizootiology of the disease caused by the oyster pathogen Perkinsus marinus and its effects on the oyster industry. Am. Fish Soc. Spec. Publ. 18, 47–63.
Arzul, I., Chollet, B., Boyer, S., Bonnet, D., Gaillard, J., Baldi, Y., et al. (2014). Contribution to the understanding of the cycle of the protozoan parasite Marteilia refringens. Parasitology 141, 227–240. doi: 10.1017/S0031182013001418
Arzul, I., Magnabosco, C., Stone, D., Aranguren, R., Arcangeli, G., Deborah, C., et al. (2012). Distribution and variability of Bonamia exitiosa in flat oyster, Ostrea edulis, populations in Europe. Abstracts of Technical Papers, Presented at the 104th Annual Meeting, National Shellfisheries Association, Seattle, Washington, March 24–29, 2012. J. Shellfish Res. 31:231. doi: 10.2983/035.031.0124
Audemard, C., Le Roux, F., Barnaud, A., Collins, C., Sautour, B., Sauriau, P. G., et al. (2002). Needle in a haystack: involvement of the copepod Paracartia grani in the life cycle of the oyster pathogen Marteilia refringens. Parasitology 124, 315–323. doi: 10.1017/S0031182001001111
Azevedo, C., Corra, L., and Cachola, R. (1990). Fine structure of zoosporulation in Perkinsus atlanticus (Apicomplexa: Perkinsea). Parasitology 100, 351–358. doi: 10.1017/S0031182000078616
Balseiro, P., Fernandez-Conchas, R., Montes, J., Gómez-León, J., Novoa, B., and Figueras, A. (2006). Comparison of diagnosis techniques for the protozoan parasite Bonamia ostreae in flat oyster Ostrea edulis. Aquaculture 261, 1135–1143. doi: 10.1016/j.aquaculture.2006.05.014
Bannister, C. A., and Key, D. (1982). Bonamia a new threat to the native oyster fishery. Fish. Nat. MAFF Direct. Fish. Res. 71, 7.
Barber, J., and Davis, C. (1994). Prevalence of Bonamia ostreae in Ostrea edulis populations in Maine. J. Shellfish Res. 13, 298.
Batista, F. M., López-SanMartín, M., Grade, A., Navas, J. I., and Ruano, F. (2016). Detection of Bonamia exitiosa in the European flat oyster Ostrea edulis in southern Portugal. J. Fish Dis. 39, 607–611. doi: 10.1111/jfd.12396
Bayne, C. J. (2003). Origins and evolutionary relationships between the innate and adaptative arms of immune systems. Integr. Comp. Biol. 43, 293–299. doi: 10.1093/icb/43.2.293
Berthe, F. C. J. (2008). “New approaches to effective molluscs health management,” in Diseases in Asian Aquaculture VI. eds M. G. Bondad-Reantaso, C. V. Mohan, M. Crumlish, and R. P. Subasinghe (Manila: Fish Health section, Asian Fisheries Society), 343–352.
Berthe, F. C. J., Burreson, E., and Hine, M. (1999). Use of molecular tools for mollusc disease diagnosis. Bull. Eur. Assoc. Fish Pathology. 19, 277–278.
Berthe, F. C. J., and Hine, P. M. (2004). Bonamia exitiosa (Hine et al., 2001) is proposed instead of B. exitiosus as the valid name of Bonamia sp. infecting flat oysters Ostrea chilensis in New Zealand. Dis. Aquatic. Org. 57, 181.
Berthe, F. C. J., Le Roux, F., Peyretaillade, E., Peyret, P., Rodriguez, D., Gouy, M., et al. (2000). The existence of the phylum Paramyxea Desportes and Perkins, 1990 is validated by the phylogenetic analysis of the Marteilia refringens small subunit ribosomal RNA. J. Eukaryot. Microbiology. 47, 288–293. doi: 10.1111/j.1550-7408.2000.tb00049.x
Berthe, F. C. J., Roux, F., Adlard, R. D., and Figueras, A. (2004). Marteiliosis in molluscs: a review. Aquat. Living Resour. 17, 433–448. doi: 10.1051/alr:2004051
Boyer, S., Chollet, B., Bonnet, D., and Arzul, I. (2013). New evidence for the involvement of Paracartia grani (Copepoda, Calanoida) in the life cycle of Marteilia refringens (Paramyxea). Int. J. Parasitology. 43, 1089–1099. doi: 10.1016/j.ijpara.2013.07.008
Burreson, E. M. (2008). Misuse of PCR assay for diagnosis of mollusc protistan infections. Dis. Aquat. Org. 80, 81–83. doi: 10.3354/dao01925
Burreson, E. M., and Ford, S. (2004). A review of recent information on the Haplosporidia, with special reference to Haplosporidium nelsoni (MSX disease). Aquat. Living Resour. 17, 499–517. doi: 10.1051/alr:2004056
Carnegie, R. B., Barber, B. J., and Distel, D. L. (2003). Detection of the oyster parasite Bonamia ostreae by fluorescent in situ hibridization. Dis. Aquat. Org. 55, 247–252. doi: 10.3354/dao055247
Carnegie, R. B., Barber, B. J., Culloty, S. C., Figueras, A. J., and Distel, D. L. (2000). Development of a PCR assay for detection of the oyster pathogen Bonamia ostreae and support for its inclusion in the Haplosporidia. Dis. Aquat.Org. 42, 199–206. doi: 10.3354/dao042199
Carnegie, R. B., and Cochennec-Laureau, N. (2004). Microcell parasites of oysters: recent insights and future trends. Aquat. Living Resour. 17, 519–528. doi: 10.1051/alr:2004055
Carrasco, N., Andree, K., Lacuesta, B., Roque, A., Rodgers, C., and Furones, M. D. (2012b). Molecular characterizarion of the Marteilia parasite infecting the common edible cockle Cerastoderma edule in the Spanish Mediterranean coast. A new Marteilia species affecting bivalves in Europe? Aquaculture 324–325, 20–26. doi: 10.1016/j.aquaculture.2011.10.017
Carrasco, N., López-Flores, I., Alcaraz, M., Furones, M. D., Berthe, F. C., and Arzul, I. (2007). Dynamics of the parasite Marteilia refringens (Paramyxea) in Mytilus galloprovincialis and zooplankton populations in Alfacs Bay (Catalonia, Spain). Parasitology 134, 1541–1550. doi: 10.1017/S0031182007003009
Carrasco, N., Roozenburg, I., Voorbergen-laarman, M., Itoh, N., and Engelsma, M. Y. (2013). Development of a real-time pcr for detection of the oyster pathogen nocardia crassostreae based on its homogeneous 16s-23s rrna intergenic spacer region. J. Invertebr. Pathol. 114, 120–127. doi: 10.1016/j.jip.2013.07.002
Carrasco, N., Villalba, A., Andree, K. B., Engelsma, M. Y., Lacuesta, B., Ramilo, A., et al. (2012a). Bonamia exitiosa (Haplosporidia) observed infecting the European flat oyster Ostrea edulis cultured on the Spanish Mediterranean coast. J. Invertebr. Pathol. 110, 307–313. doi: 10.1016/j.jip.2012.03.015
Carriker, M. (1992). Introductions and transfers of molluscs: risk considerations and implications. J. Shellfish Res. 11, 507–510.
Cavalier-Smith, T., and Chao, E. E. (2003). Phylogeny and classification of phylum Cercozoa (Protozoa). Protist 154, 341–358. doi: 10.1078/143446103322454112
Clark, M. S., Thorne, M. A., Vieira, F. A., Cardoso, J. C., Power, D. M., and Peck, L. S. (2010). Insights into the shell deposition in the Antartic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genomics 11:362. doi: 10.1186/1471-2164-11-362
Cochennec, N., Le Roux, F., Berthe, F., and Gerard, A. (2000). Detection of Bonamia ostreae based on small subunit ribosomal probe. J. Invertebr. Pathol. 76, 26–32. doi: 10.1006/jipa.2000.4939
Cochennec-Laureau, N., Reece, K. S., Berthe, F. C. J., and Hine, P. M. (2003). Mikrocytos roughleyi taxonomic affiliation leads to the genus Bonamia (Haplosporidia). Dis. Aquat. Org. 54, 209–217. doi: 10.3354/dao054209
Corbeil, S., Arzul, I., Diggles, B., Heasman, M., Chollet, B., Berthe, F. C., et al. (2006b). Development of a TaqMan PCR assay for the detection of Bonamia species. Dis. Aquat. Org. 71, 75–80. doi: 10.3354/dao071075
Corbeil, S., Arzul, I., Robert, M., Berthe, F. C. J., Besnard-Cochennec, N., and Crane, M. S. J. (2006a). Molecular characterization of an Australian isolate of Bonamia exitiosa. Dis. Aquat. Org. 71, 81–85. doi: 10.3354/dao071081
Cranfield, H. J., Dunn, A., Doonan, I. J., and Michael, K. P. (2005). Bonamia exitiosa epizootic in Ostrea chilensis from Foveaux Strait, southern New Zealand between 1986 and 1992. ICES J. Mar. Sci. 62, 3–13. doi: 10.1016/j.icesjms.2004.06.021
Culloty, S. C., Cronin, M. A., and Mulcahy, M. F. (2004). Potential resistance of a number of populations of the oyster Ostrea edulis to the parasite Bonamia ostreae. Aquaculture 237, 41–58. doi: 10.1016/j.aquaculture.2004.04.007
Dinamani, P., Hine, P. M., and Jones, J. B. (1987). Occurrence and characteristics of the haemocyte parasite Bonamia sp. in the New Zealand dredge oyster Tiostrea lutaria. Dis. Aquat. Org. 3, 37–44. doi: 10.3354/dao003037
Dove, M. C., Nell, J. A., McOrrie, S., and O'Connor, W. A. (2013a). Assesment of QX and winter mortality disease resistance of mass selected Sydney rock oysters, Sacrostrea glomerata (Gould, 1850), in the Hawkesbury River and Merimbula Lake, NSW Australia. J. Shellfish Res. 32, 681–687. doi: 10.2983/035.032.0309
Dove, M. C., Nell, J. A., and O'Connor, W. A. (2013b). Evaluation of the progeny of the fourth generation Sydney rock oyster, Saccostrea glomerata (Gould, 1850) breeding lines for resistance to QX disease (Marteilia sydneyi) and winter mortality (Bonamia roughleyi). Aquaculture Res. 44, 1791–1800. doi: 10.1111/are.12012
FAO (2014). Global Aquaculture Production Volumen, and Value Statistics Database Updated to 2012. FAO Fisheries and Aquaculture Department.
Feist, S. W., Hine, P. M., Bateman, K. S., Grant, D. S., and Longshaw, M. (2009). Paramarteilia canceri sp. n. (Cercozoa) in the European edible crab (Cancer pagarus) with a proposal for the revision of the order Paramyxida Chatton, 1911. Folia Parasitol. 56, 73–85. doi: 10.14411/fp.2009.012
Flannery, G., Lynch, S. A., Longshaw, M., Stone, D., Martin, P., Ramilo, A., et al. (2014). Interlaboratory variability in screening for Bonamia ostreae, a protistan parasite of the European flat oyster Ostrea edulis. Dis. Aquat. Org. 110, 93–94. doi: 10.3354/dao02717
Ford, S. E., and Haskin, H. H. (1987). Infection and mortality patterns in strains of oyster Crassotrea virginica selected for resistance to the parasite Haplosporidium nelsoni MSX. J. Parasitol. 73, 386–376. doi: 10.2307/3282092
Friedman, C., and Perkins, F. O. (1994). Range extension of Bonamia ostreae to Maine, USA. J. Invertebr. Pathol. 64, 179–181. doi: 10.1016/S0022-2011(94)90075-2
Friedman, C. S., McDowell, T., Groff, J. M., Hollibaugh, J. T., Manzer, D., and Hedrick, R. P. (1989). Presence of Bonamia ostreae among populations of the European flat oyster, Ostrea edulis Linné, in California, USA. J. Shellfish Res. 8, 133–137.
Gaffney, P. M., and Bushek, D. (1996). Genetic aspects of disease resistance in oysters. J. Shellfish Res. 26, 877–885.
Gall, J. G., and Pardue, M. L. (1969). Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc. Natl. Acad. Sci. U.S.A. 63, 378–383. doi: 10.1073/pnas.63.2.378
Gomez-Chiarri, M., Guo, X., Tanguy, A., He, Y., and Proestou, D. (2015). The use os –omic tools in the study of diseases processes in marine bivalve mollusks. J. Invertebr. Pathol. 131, 137–154. doi: 10.1016/j.jip.2015.05.007
Grijalva-Chon, J. M., Castro-Longoria, R., Enriquez-Espinoza, T. L., Maeda-Martinez, A. N., and Mendoza-Cano, F. (2015). Molecular evidence of the protozoan parasite Marteilia refringens in Crassostrea gigas and Crassostrea corteziensis del Golfo de California. Lat. Am. J. Aquat. Res. 43, 776–780. doi: 10.3856/vol43-issue4-fulltext-16
Grizel, H. (1985). Etude des Recentes Epizooties de L'huître Plate (Ostrea edulis Linné) et leur Impact sur L'ostréiculture Bretonne. Thèse Doctorat es Sciences, Université des Sciences et Techniques du Languedoc, Montpellier.
Grizel, H., Bachére, E., Mialhe, E., and Tigé, G. (1986). “Solving parasite-related problems in cultured molluscs,” in Parasitology, ed M. J. Howell (Canberra, ACT: Australian Academy of Science), 301–308.
Grizel, H., Comps, M., Bonami, J. R., Cousserans, F., Duthoit, J. L., and Le Pennec, M. A. (1974). Recherche sur l'agent de la maladie de la glande digestive de Ostrea edulis Linne. Science et Pêche. Bull. Institut Pêches Maritimes 240, 7–29.
Grizel, H., and Héral, M. (1991). Introduction into France of the Japanese oyster (Crassotrea gigas). J. Conseil Int. Explor. 47, 399–403. doi: 10.1093/icesjms/47.3.399
Hand, R. E., Nell, J. A., Smith, I. R., and Maguire, G. R. (1998). Studies on triploid oysters in Australia. XI. Survival of diploid and triploid Sydney rock oysters (Saccostrea commercialis (Iredale and Rouhley)) through outbreaks of winter mortality caused by Mikrocytos roughleyi infestation. J. Shellfish Res. 17, 1129–1135.
Herrbach, B. (1971). Sur une affection parasitaire de la glande digestive de l'huître plate, Ostrea edulis Linné. Revue Travaux Institut Pêches Maritimes 35, 79–87.
Hervio, D., Bower, S. M., and Meyer, G. R. (1996). Detection, isolation, and experimental transmission of Mikrocytos mackini, a microcell parasite of Pacific oysters Crassotrea gigas (Thunberg). J. Invertebr. Pathol. 67, 72–79. doi: 10.1006/jipa.1996.0011
Hine, P. M., Cochennec-Laureau, N., and Berthe, F. C. J. (2001). Bonamia exitiosus n. sp. (Haplosporidia) infecting flat oysters Ostrea chilensis (Philippi) in New Zealand. Dis. Aquat. Org. 47, 63–72. doi: 10.3354/dao047063
Howard, D. W., Lewis, E. J., Keller, J., and Smith, C. S. (2004). Histological techniques for marine bivalve molluscs and crustaceans. NOAA Tech. Memo. NOS NCCOS 5, 218.
Hunt, P. W. (2011). Molecular diagnosis of infections and resistance in veterinary and human parasites. Vet. Parasitol. 180, 12–46. doi: 10.1016/j.vetpar.2011.05.027
Itoh, N., Oda, T., Yoshinaga, T., and Ogawa, K. (2003). Isolation and 18S ribosomal DNA gene sequences of Marteilioides chungmuensis (Paramyxea), an ovarian parasite of the Pacific oyster Crassostrea gigas. Dis. Aquat. Org. 54, 163–169. doi: 10.3354/dao054163
Joubert, C., Piquemal, D., Marie, B., Manchon, L., Pierrat, F., Zanella-Cléon, I., et al. (2010). Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: focus on biomineralization. BMC Genomics 11:613. doi: 10.1186/1471-2164-11-613
Kleeman, S. N., and Adlard, R. D. (2000). Molecular detection of Marteilia sydneyi, pathogen of Sydney rock oysters. Dis. Aquat. Org. 40, 137–146. doi: 10.3354/dao040137
Kleeman, S. N., Le Roux, F., Berthe, F. C. J., and Adlard, R. D. (2002). Specificity of PCR and in situ hybridization assays designed for detection of Marteilia sydneyi and M. refringens. Parasitology 125, 131–141. doi: 10.1017/s0031182002001919
Korringa, P. (1950). Investigations on Shell-Disease in Oyster Ostrea edulis L. Report V Meeting, ICES.
Lauckner, G. (1983). “Diseases of Mollusca: Bivalvia,” in Diseases of Marine Animals, Vol II, ed O. Kinne (Hamburg: Biologishe Anstalt Helgoland), 477–959.
Le Roux, F., Audemard, C., Barnaud, A., and Berthe, F. (1999). DNA probes as potential tools for the detection of Marteilia refringens. Mar. Biotechnol. 1, 588–597. doi: 10.1007/PL00011814
Le Roux, F., Lorenzo, G., Peyret, P., Audemard, C., Figueras, A., Vivarès, C., et al. (2001). Molecular evidence for the existence of two species of Marteilia in Europe. J. Eukaryot. Microbiol. 48, 449–454. doi: 10.1111/j.1550-7408.2001.tb00178.x
Longshaw, M., Feist, S. W., Matthews, R. A., and Figueras, A. (2001). Ultrastructural characterization of Marteilia species (Paramyxea) from Ostrea edulis, Mytilus edulis and Mytilus galloprovincialis in Europe. Dis. Aquat. Org. 44, 137–142. doi: 10.3354/dao044137
Longshaw, M., Stone, D. M., Wood, G., Green, M. J., and White, P. (2013). Detection of Bonamia exitiosa (Haplosporidia) in European flat oysters Ostrea edulis cultivated in mainland Britain. Dis. Aquat. Org. 106, 173–179. doi: 10.3354/dao02643
López-Flores, I., de la Herrán, R., Garrido-Ramos, M. A., Navas, J. I., Ruiz-Rejón, C., and Ruiz-Rejón, M. (2004). The molecular diagnosis of Marteilia refringens and differentiation between Marteilia strains infecting oysters and mussels based on the rDNA IGS sequence. Parasitology 129, 411–419. doi: 10.1017/S0031182004005827
López-Flores, I., Navas, J. I., Garrido, M. A., Ruiz, C., Ruiz, M., and de la Herrán, R. (2007a). “Utilización de marcadores moleculares para el diagnóstico e identificación taxonómica de protozoos parásitos en bivalvos,” in Genética y Acuicultura, eds P. Martinea and A. Figueras (Madrid: Fundación Observatorio Español de Acuicultura), 795–846.
López-Flores, I., Suárez-Santiago, V. N., Longet, D., Saulnier, D., Chollet, B., and Arzul, I. (2007b). Characterization of actin genes in Bonamia ostreae and their application to phylogeny of the Haplosporidia. Parasitology 134, 1941–1948. doi: 10.1017/S0031182007003307
Lupo, C., Osta Amigo, A., Mandard, Y. V., Peroz, C., and Renault, T. (2014). Improving early detection of exotic or emergent oyster diseases in France: identifying factors associated with shellfish farmer reporting behavior oy oyster mortality. Prev. Vet. Med. 116, 168–182. doi: 10.1016/j.prevetmed.2014.05.002
Lynch, S. A., Flannery, G., Hugh-Jones, T., Hugh-Jones, D., and Culloty, S. C. (2014). Thirty-year history of Irish (Rossmore) Ostrea edulis selectively bred for disease resistance to Bonamia ostreae. Dis. Aquat. Org. 110, 113–121. doi: 10.3354/dao02734
Macey, B. M., and Coyne, V. E. (2005). Improved growth rate and disease resistance in farmed Haliotis midae through probiotic treatment. Aquaculture 245, 249–261. doi: 10.1016/j.aquaculture.2004.11.031
Madsen, L. (2015). First Detection of Bonamia ostreae in Native Flat Oysters from the Limfjord in Denmark. Report from the Annual meeting of the National Reference Laboratories for Molluscs diseases, Saintes.
Madsen, L., Kamp, J., and Mellergaard, S. (2013). What can the Limfjord tell us about limiting factors for Bonamia ostreae in the northern Europe? Bull. Eur. Assoc. Fish Pathol. 33, 165–168.
Man, R., Burreson, E. M., and Baker, P. K. (1991). The decline of the Virginia oyster fishery in Chesapeake Bay: considerations for introduction of a non- endemic species, Crassostrea gigas (Thunberg, 1973). J. Shellfish Res. 10, 379–388.
Marteil, L. (1976). La conchyliculture française. 2. Biologie de l'huître et de la moule. Revue Travaux Institut Pêches Maritimes 40, 149–345.
Martenot, C., Oden, E., Travaille, E., Malas, J. P., and Houssin, M. (2010). Comparison of two real-time PCR methods for detection of ostreid herpesvirus 1 in the Pacific oyster Crassostrea gigas. J. Virol. Methods 70, 86–99. doi: 10.1016/j.jviromet.2010.09.003
Martin, A. G., Gérad, A., Cochennec, N., and Langlade, A. (1993). “Selecting flat oysters, Ostrea edulis, for survival against the parasite Bonamia ostreae: assessment of the resistance of a first selected generation,” in Production, Environment and Quality–Bourdeaux Aquaculture 92 (Spec. Publ. No. 18), eds G. Barnabé and P. Kestemont (Ghent: European Aquaculture Society), 547–554.
Marty, G., Bower, S., Clarke, K., Meyer, G., Lowe, G., Osborn, A., et al. (2006). Histopathology and a real-time PCR assay for detection of Bonamia ostreae in Ostrea edulis cultured in western Canada. Aquaculture 261, 33–42 doi: 10.1016/j.aquaculture.2006.07.024
Mialhe, E., Bachère, E., Boulo, V., and Cadoret, J. P. (1995). Strategy for research and international cooperation in marine invertebrate pathology, immunology and genetics. Aquaculture 132, 33–41. doi: 10.1016/0044-8486(94)00383-Y
Montes, J., Ferro-Soto, B., Conchas, R. F., and Guerra, A. (2003). Determining culture strategies in populations of the European flat oyster, Ostrea edulis, affected by bonamiosis. Aquaculture 220, 175–182. doi: 10.1016/S0044-8486(02)00628-2
Moreira, R., Balseiro, P., Romero, A., Dios, S., Posada, D., Novoa, B., et al. (2012). Gene expression analysis of clams Ruditapes philippinarum and Ruditapes decussatus following bacterial infection yields molecular insights into pathogen resistence and immunity. Dev. Comp. Immunol. 36, 140–149. doi: 10.1016/j.dci.2011.06.012
Moreira, R., Milan, M., Balseiro, P., Romero, A., Babbucci, M., Figueras, A., et al. (2014). Gene expression profile analysis of Manila clam (Ruditapes philippinarum) hemocytes after a Vibrio alginolyticus challenge using an immune-enriched oligo-microarray. BMC Genomics 15:267. doi: 10.1186/1471-2164-15-267
Narcisi, V., Arzul, I., Cargini, D., Mosca, F., Calzetta, A., Traversa, D., et al. (2010). Detection of Bonamia ostreae and Bonamia exitiosa (Haplosporidia) in Ostrea edulis from the Adriatic Sea (Italy). Dis. Aquat. Org. 89, 79–85. doi: 10.3354/dao02167
Narcisi, V., Mascini, M., Perez, G., Del Carlo, M., Tiscar, P. G., Yamanaka, H., et al. (2011). Electrochemical genosensors for the detection of Bonamia parasite. Selection of single-strand-DNA (ssDNA) probes by simulation of the secondary structure folding. Talanta 85, 1927–1932. doi: 10.1016/j.talanta.2011.07.019
Nell, J. A., and Perkins, B. (2006). Evaluation of the progeny of third-generation Sydney rock oyster Saccostrea glomerata (Gould 1850) breeding lines for resistance to QX disease Marteilia sydneyi and winter mortality Bonamia roughleyi. Aquat. Res. 37, 693–700. doi: 10.1111/j.1365-2109.2006.01482.x
Nell, J. A., Smith, I. R., and Mcphee, C. C. (2000). The Sydney rock oyster Saccostrea glomerata (Gould 1850) breeding programme: progress and goals. Aquat. Res. 31, 45–49. doi: 10.1046/j.1365-2109.2000.00387.x
O.I.E. World Organization for Animal Health (2016). “Section 2.4,” in Manual of Diagnostic Test for Aquatic Animals. Available online at: www.oie.int/en/international-standard-setting/aquatic-manual/access-online/
Ottaviani, E. (2011). Is the distinction between innate and adaptative immunity in invertebrates still as clear as thought? Ital. J. Zool. 78, 274–278. doi: 10.1080/11250003.2011.562247
Pande, A., Acosta, H., Brangenberg, N. A., and Keeling, S. E. (2015). Design of a detection survey for Ostreid herpesvirus-1 using hydrodynamic dispersion models to determine epidemiological units. Prev. Vet. Med. 119, 80–84. doi: 10.1016/j.prevetmed.2015.02.009
Penna, M. S., Khan, M., and French, R. A. (2001). Development of a multiplex PCR for the detection of Haplosporidium nelsoni, Haplosporidium costale and Perkinsus marinus in the eastern oyster (Crassostrea virginica, Gmelin 1971). Mol. Cell Probes 15, 385–390. doi: 10.1006/mcpr.2001.0386
Philipp, E. E., Kraemer, L., Melzner, F., Poustka, A. J., Thieme, S., Findeisen, U., et al. (2012). Massively parallel RNa sequencing identifies a complex immune gene repertoire in the lophotrochozoan Mytilus edulis. PLoS ONE 7:e33091. doi: 10.1371/journal.pone.0033091
Pichot, Y., Comps, M., Tigé, G., Grizel, H., and Rabouin, M. A. (1980). Recherches sur Bonamia ostreae gen. n., sp. n., parasite nouveau de l'huître plate Ostrea edulis L. Revue Travaux Institut Pêches Maritimes 43, 131–140.
Polanco, E., Montes, J., Outón, M. J., and Meléndez, M. I. (1984). Situation pathologique du stock d'huitres plates en Galice (Espagne) en relation avec Bonamia ostreae. Haliotis 14, 91–95.
Ramilo, A., Navas, J. I., Villalba, A., and Abollo, E. (2013). Species-specific diagnostic assay for Bonamia ostreae and B. exitiosa in European flat oyster Ostrea edulis: conventional, real-time and multiplex PCR. Dis. Aquat. Org. 104, 149–161. doi: 10.3354/dao02597
Robert, M., García, C., Chollet, B., Lopez-Flores, I., Ferrand, S., Francois, C., et al. (2009). Molecular detection and quantification of the protozoan Bonamia ostreae in the flat oyster, Ostrea edulis. Mol. Cell Probes 23, 264–271. doi: 10.1016/j.mcp.2009.06.002
Romero, A., Novoa, B., and Figueras, A. (2012). Genomics, immune studies and diseases in bivalve aquaculture. Int. Sci. J. 9, 110–121.
Samain, J. F., Dégremont, L., Soletchnik, P., Haure, J., Bédier, E., Roper, M., et al. (2007). Genetically based resistance to summer mortality in the Pacific oyster (Crassostrea gigas) and its relationship with physiological, immunological characteristics and infection processes. Aquaculture 268, 227–243. doi: 10.1016/j.aquaculture.2007.04.044
Segarra, A., Pépin, J. F., Arzul, I., Morga, B., Faury, N., and Renault, T. (2010). Detection and description of a particular Ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus Res. 153, 92–99. doi: 10.1016/j.virusres.2010.07.011
Shatkin, G., Shumway, S. E., and Hawes, R. (1997). Considerations regarding the possible introduction of the Pacific oyster (Crassotrea gigas) to the gulf of Maine: a review of global experience. J. Shellfish Res. 16, 463–477.
Umeda, K., and Yoshinaga, T. (2012). Development of real-time PCR assays for discrimination and quantification of two Perkinsus spp. in the Manila Clam Ruditapes phillipinarum. Dis. Aquat. Org. 99, 215–225. doi: 10.3354/dao02476
Van Banning, P. (1982). “Some aspects of the occurrence, importance and control of the oyster pathogen Bonamia ostreae in the Dutch oyster culture,” in Proceedings of the International Colloquium on Invertebrate Pathology XVth Annual Meeting, Society of Invertebrate Pathology, eds C. C Payne and H. D Burges (Brighton: University of Sussex), 261–265.
Van Banning, P. (1990). The life cycle of the oyster pathogen Bonamia ostreae with a presumptive phase in the ovarian tissue of the European flat oyster, Ostrea edulis. Aquaculture 84, 189–192. doi: 10.1016/0044-8486(90)90348-Q
Van Banning, P. (1991). Observations on bonamiosis in the stock of the European flat oyster, Ostrea edulis, in the Netherlands, with special reference to the recent developments in Lake Grevelingen. Aquaculture 93, 205–211. doi: 10.1016/0044-8486(91)90232-V
Van Hai, N., Buller, N., and Fotedar, R. (2009). The use of customised probiotics in the cultivation of western King prawns (Penaeus latisulcatus Kishinouve, 1986). Fish Shellfish Immunol. 27, 100–104. doi: 10.1016/j.fsi.2009.05.004
Villalba, A., Da Silva, P. M., and Fuentes, J. (2007). “La producción de estirpes de Ostrea edulis tolerantes a la bonamiosis como solución para el engorde de la ostra plana en Galicia,” in XII Congreso Nacional de Acuicultura, Centro de Investigacións Mariñas, Xunta de Galicia, Vilanova de Arousa, eds A. Cerviño, A. Guerra, and C. Pérez (Spain), 1499–1504.
Villalba, A., Iglesias, I., Ramilo, A., Darriba, S., Parada, J. M., No, E., et al. (2014). Cockle, Cerastoderma edule, fishery collapse in the Ria de Arousa (Galicia, NW Spain) associated with the protistan parasite Marteilia cochillia. Dis. Aquat. Org. 109, 55–80. doi: 10.3354/dao02723
Xie, Z. H., Xue, L. J., Pang, Y. S., Liu, J. B., Deng, X. W., and Xie, Z. Q. (2010). Development of a duplex real time PCR assay for detection of Perkinsus and Marteilia refringens in shellfish. Acta Hydrobiol. Sini. 34, 898–904. doi: 10.3724/SP.J.1035.2010.00898
Xie, Z., Xie, L., Fan, Q., Pang, Y., Deng, X., Xie, Z. Q., et al. (2013). A duplex quantitative real-time PCR assay for the detection of Haplosporidium and Perkinsus species in shellfish. Parasitol. Res. 112, 1597–1606. doi: 10.1007/s00436-013-3315-5
Xue, J., Xu, Y., Jin, L., Liu, G., Sun, Y., Li, S., et al. (2008). Effects of the traditional Chinese medicine on immune response in abalone, Haliotis discus hannai Ino. Fish Shellfish Immunol. 24, 752–758. doi: 10.1016/j.fsi.2008.02.005
Keywords: diagnostic techniques, quantitative real time PCR, diseases, molluscs, pathology, molecular tools
Citation: Aranguren R and Figueras A (2016) Moving from Histopathology to Molecular Tools in the Diagnosis of Molluscs Diseases of Concern under EU Legislation. Front. Physiol. 7:538. doi: 10.3389/fphys.2016.00538
Received: 23 March 2016; Accepted: 26 October 2016;
Published: 14 November 2016.
Edited by:
Luciana Mandrioli, University of Bologna, ItalyReviewed by:
Ana Roque, Institute for Research and Technology in Food and Agriculture, SpainPietro Giorgio Tiscar, University of Teramo, Italy
Copyright © 2016 Aranguren and Figueras. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonio Figueras, YW50b25pb2ZpZ3VlcmFzQGlpbS5jc2ljLmVz
 Raquel Aranguren
Raquel Aranguren Antonio Figueras
Antonio Figueras