
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Physiol. , 08 August 2016
Sec. Systems Biology Archive
Volume 7 - 2016 | https://doi.org/10.3389/fphys.2016.00342
This article is part of the Research Topic Systems Biology and the challenge of deciphering the metabolic mechanisms underlying cancer View all 11 articles
Cancer represents a complex disease originated from alterations in several genes leading to disturbances in important signaling pathways in tumor biology, favoring heterogeneity that promotes adaptability and pharmacological resistance of tumor cells. Metabolic reprogramming has emerged as an important hallmark of cancer characterized by the presence of aerobic glycolysis, increased glutaminolysis and fatty acid biosynthesis, as well as an altered mitochondrial energy production. The metabolic switches that support energetic requirements of cancer cells are closely related to either activation of oncogenes or down-modulation of tumor-suppressor genes, finally leading to dysregulation of cell proliferation, metastasis and drug resistance signals. Non-coding RNAs (ncRNAs) have emerged as one important kind of molecules that can regulate altered genes contributing, to the establishment of metabolic reprogramming. Moreover, diverse metabolic signals can regulate ncRNA expression and activity at genetic, transcriptional, or epigenetic levels. The regulatory landscape of ncRNAs may provide a new approach for understanding and treatment of different types of malignancies. In this review we discuss the regulatory role exerted by ncRNAs on metabolic enzymes and pathways involved in glucose, lipid, and amino acid metabolism. We also review how metabolic stress conditions and tumoral microenvironment influence ncRNA expression and activity. Furthermore, we comment on the therapeutic potential of metabolism-related ncRNAs in cancer.
Deregulation of cellular energetics has been pointed out as one of the emerging hallmarks of cancer, both during early cellular transformation and as a driving phenotype of several tumorigenic programs (Kroemer and Pouyssegur, 2008; Munoz-Pinedo et al., 2012). Under physiological conditions, cells maintain regulated and complex metabolic homeostasis by diverse signaling pathways that function as energetic sensors. Metabolic sensors act under a network of cooperative signaling cascades, not only to fulfill the energetic requirements of the cells, but also to influence cellular pathways like cell growth, proliferation, and death (Dumortier et al., 2013). In contrast, cancer cells loose this regulated homeostasis in several ways, including alterations in intrinsic and extrinsic molecular mechanisms that govern cellular metabolism, in order to provide the basic metabolic requirements of tumoral cells, such as quick biosynthesis of ATP, accelerated biosynthesis of macromolecules, and maintenance of optimal redox status (Cairns et al., 2011). To satisfy their metabolic needs, cancer cells also present changes in energetic pathways such as elevated glucose uptake, aerobic glycolysis and altered lipid and fatty-acid metabolism (Newsholme et al., 1985; Vander Heiden et al., 2009). This advantageous bioenergetic state is not only related to the metabolic requirements imposed by the higher biological activity of the tumoral cells, it can also promote a proliferative phenotype and facilitate cell survival and movement under adverse conditions like hypoxia or glucose and nutrient deprivation, becoming a major player in the development and evolution of a tumor (Jones and Thompson, 2009).
This metabolic shift, known as metabolic reprogramming, has been correlated with the activity of oncogenes and loss of tumor suppressor molecules (Esquela-Kerscher and Slack, 2006). Furthermore, once a tumor has developed and reached a certain volume, it becomes difficult to maintain optimal oxygen levels in its cells, creating a hypoxic environment. This also promotes a metabolic reprograming which includes an elevated glycolytic rate, preferentially through oxidative phosphorylation and suppression of gluconeogenesis, creating complex glucose-lactate fluxes, as well as a pro-tumorigenic environment (Reyes et al., 2014).
Non-coding RNAs (ncRNAs), mainly, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), have been defined as important regulators of several metabolic pathways. miRNAs are small ncRNAs (between 19 and 22 nt), with an important role as post-transcriptional regulators (Bartel, 2009). LncRNAs are transcripts from 200 nt to 100 kilobases (kb) lacking an open reading frame and without evident protein-coding function (Rinn and Chang, 2012). Both of them participate in many physiological processes through the modulation of gene expression at the epigenetic, transcriptional, and post-transcriptional levels (Figure 1).
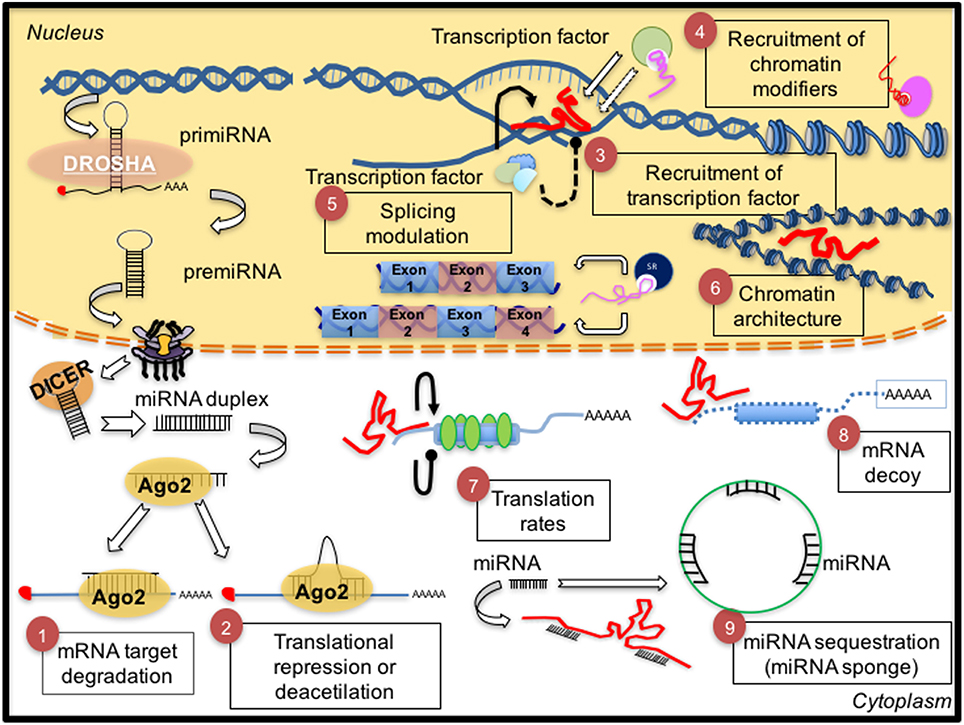
Figure 1. Biological and mechanical overview of non-coding RNAs. (1, 2) Biogenesis of microRNAs and their main mechanisms of action. The pri-miRNA is transcribed by pol II polymerase and digested by the RNase DROSHA originating a pre-miRNA (70 nt), which is exported to the cytoplasm by exportin 5. Then, another RNase, Dicer, digests the pre-miRNA to generate a mature duplex miRNA (~22 nt). One strand of this duplex is then incorporated in the miRISC complex (Ago2-microRNA) to target mRNA by perfect complementarity producing transcript degradation (1) or an imperfect one promoting translation repression (2). Conversely, (right side), general functions of lncRNAs are described. (3) Recruitment of transcription factors to promote transcription of target genes or (4) recruitment of chromatin modifiers and thus (6) promoting remodeling of the chromatin architecture. Other functions of lncRNAs are (5) control of alternative splicing of mRNA, and (7) control of translation rates favoring or inhibiting polysome loading to mRNAs, (8) acting as a decoy to preclude access of regulatory proteins to DNA. (9) The interaction between microRNAs and endogenous competent RNAs (ceRNAs) is a redundant system to regulate mRNA expression by lncRNAs-microRNAs; this mechanism is known as sponge function by lncRNAs. Thus, microRNA sequestration by lncRNA prevents microRNA functions on its target.
ncRNAs can actively regulate energetic signaling by targeting key metabolic transporters and enzymes, or by directly or indirectly controlling the expression of tumor suppressors or oncogenes (Iorio and Croce, 2012). Analysis of the correlation between oncogenic programs, metabolic reprograming and aberrant ncRNA expression has highlighted the crucial role of these metabolic aspects in initiation, promotion, and progression of cancer (Arora et al., 2015).
Several lines of evidence suggest that ncRNA plays an important role in the establishment of metabolic reprogramming in cancer cells, as well as the feedback regulation between alterations in energetic signaling and ncRNA expression or activity. In this review, we will discuss the evidence that describes the roles of ncRNAs as modulators of cancer metabolism and as molecules which contribute to the establishment of a diversity of mechanisms that govern the heterogeneity and plasticity of the energetic metabolism of cancer cells.
One of the most significant changes induced by cancer metabolic reprogramming involves the bypass of oxidative phosphorylation (Tricarboxylic Acid cycle) to a non-oxidative pathway lead by aerobic glycolysis and lactate production, in order to satisfy the energetic demands of the tumor cells (Vander Heiden et al., 2009). One of the better characterized metabolic phenotypes present in tumor cells is the Warburg effect, which gives preference to ATP generation through glycolysis, even under normal oxygen concentrations, over ATP synthesis through the electron transport chain in the mitochondria (Warburg, 1956; Gatenby and Gillies, 2004; Kim and Dang, 2006). Consequently, most of the glucose in the cell is converted to lactate, rather than being metabolized through the Krebs cycle (Warburg, 1956; Semenza et al., 2001; Gatenby and Gillies, 2004). Although the energetic balance established by glycolysis is less efficient (lower quantity of ATP generated per unit of glucose) than oxidative phosphorylation, it is quicker. However, oxidative phosphorylation is not completely abolished and still functions at a low level (Figure 2A). Therefore, this abnormal and accelerated metabolism meets the energetic needs for cellular functions and construction of biological blocks (fatty acids, lipids, nucleotides, and proteins) for cancer cells (Zheng, 2012).
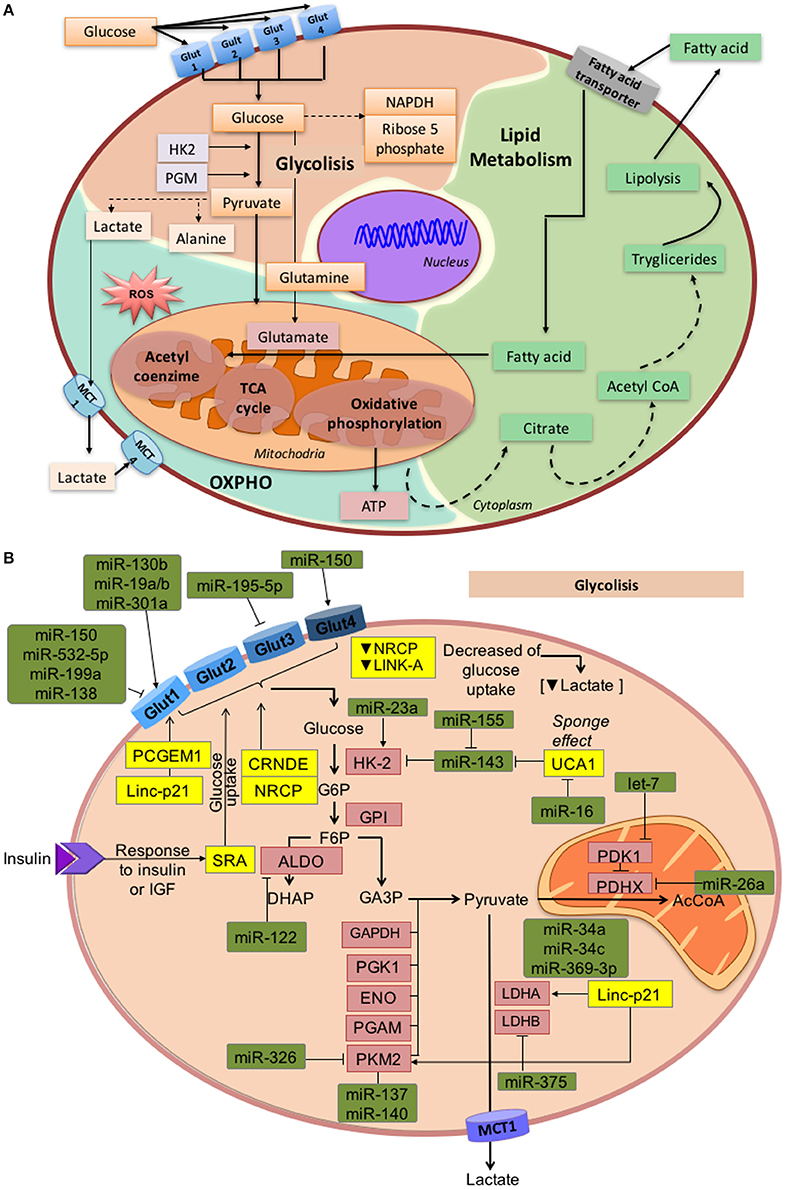
Figure 2. Overview of glycolysis, OXPHO, and lipid metabolism. (A) This figure describes the connections between metabolic sub-products that take part in different metabolic process, as well as enzymes and substrates that maintain the normal metabolic environment. Glycolysis occurs in the cytosol when D-glucose is internalized into the cell through the membrane transporters GLUT1, 2, 3, and 4. Through a system of coupled enzymatic reactions, D-glucose is converted into pyruvate, which enters into the TCA cycle, and OXPHO. When the amount of oxygen is limited, pyruvate is converted into lactate. Conversely, in the mitochondria, the TCA cycle is coupled to OXPHO which represents the largest source of metabolic energy. Pyruvate is oxidized and converted into acetyl coenzyme A, which enters the TCA cycle that generates reducing molecules (NADH and FADH2) to produce ATP by oxidative phosphorylation. Finally, fatty acids can be converted into acetyl coenzyme A by ß-oxidation to then generate energy through the TCA cycle and OXPHO (B). Glycolysis regulation by miRNAs and lncRNAs under oncogenic conditions. Expression of the GLUT transporter family is regulated by ncRNAs, thus altering the internalization rate of glucose. Other molecules are under ncRNAs regulation pathways, such as hexokinase-2 enzyme, which mediates the transformation of glucose to glucose 6-phosphate, PKM2 enzyme involved in pyruvate synthesis, LDHB and LDHA enzymes that convert pyruvate into lactate, and PDHK, responsible for the synthesis of Acetyl coenzyme A from pyruvate.
The first step in energy metabolism is the entry of glucose into the cells through glucose transporters (GLUTs). Until now, 14 isoforms of GLUTs have been identified, of which GLUT1, 2, 3, and 4 are well-characterized and expressed in different tissues, some of them in a specific manner (Thorens and Mueckler, 2010). ncRNAs actively regulate the intracellular glucose levels by modulating gene expression of glucose transporters. For instance, GLUT1 is targeted by miR-340, which is up-regulated in oral squamous cell carcinoma (Xu et al., 2016). In renal cell tumors, miR-199a, miR-138, miR-150, and miR-532-5p down-regulate GLUT1 expression, whereas miR-130b, miR-19a/b, and miR-301a increase GLUT-1 (Chow et al., 2010). Additionally, loss of miR-1291 enhances the development of renal tumors through targeting GLUT1 (Yamasaki et al., 2013). In prostate tumors, the PCGEM1 lncRNA promotes the expression of GLUT1. Similarly, lncRNA-p21 expression is related to HIF-1α and its responsive genes, such as GLUT1, promoting its expression in diverse cancer cell lines (Yang et al., 2014). In bladder cancer, down-modulation of miR-195-5p allows the expression of GLUT3 (Fei et al., 2012; Peschiaroli et al., 2013). Additionally, reduced levels of miR-150 negatively regulate GLUT4 expression in pancreatic cancer cells (Srivastava et al., 2011). Such alterations in the expression of ncRNAs and their effect over GLUT expression, represent possible mechanisms through which tumors may bypass regulatory energetic checkpoints by promoting glycolysis, as well as other oncogenic pathways like proliferation, migration, and invasion (Figure 2B).
ncRNAs can also affect the patterns and mechanisms of glucose uptake and glucose/lactate fluxes in cancer cells, promoting aggressive behavior through the establishment of a glycolytic phenotype. The CRNDE (Colorectal Neoplasia Differentially Expressed) lncRNA responds to insulin-like growth factors (IGF) promoting glucose uptake in colorectal cancer (Ellis et al., 2014). Furthermore, the over-expression of the ceruloplasmin lncRNA (NRCP) in ovarian and breast cancer cells, along with the LINK-A lncRNA in triple negative breast cancer, promotes glucose uptake, favoring lactate production and consequently, enhancing tumor progression (Rupaimoole et al., 2015; Lin et al., 2016). In breast tumors, ncRNAs can also function as modifiers of the tumor microenvironment. Under metastatic conditions, tumor cells secret vesicles that carry high levels of miR-122 to non-tumor cells, repressing glucose uptake in the normal cells and facilitating metastasis by increasing nutrient availability for the cancer cells (Fong et al., 2015; Figure 2B).
After glucose uptake, numerous enzymes take part in the catabolism of trioses, pyruvate, and finally lactate. Regulation of glycolytic enzymes by ncRNAs further increases this biological complexity. Hexokinases (HK) catalyze ATP-dependent phosphorylation of glucose to glucose-6-phosphate (Robey and Hay, 2006). Interestingly, HK2 is overexpressed in various tumors and contributes to the establishment of aerobic glycolysis (Mathupala et al., 2009; Vander Heiden et al., 2011). In lung, colon, prostate and head, and neck squamous cell cancers, loss of miR-143 allows HK2 expression (Fang et al., 2012; Peschiaroli et al., 2013). Similarly, miR-143 locus is deleted in other malignancies (Volinia et al., 2010), and has also been found down-regulated in cervical tumors (Michael et al., 2003; Lui et al., 2007). In bladder cancer cells, miR-155 repress miR-143, allowing up-regulation of HK2 (Jiang et al., 2012). Moreover, the up-regulation of hipoxia factors suppresses the expression of miR-199a-5p and promotes glycolysis in liver cancer, since the miRNA normally interferes with the expression of HK2 (Guo et al., 2015). The Urothelial Cancer-Associated 1 lncRNA (UCA1) modulates HK2 by activation of STAT3 through the repression of miR-143 (Li Z. et al., 2014). Another member of the hexokinases, HK1 is also regulated by miR-138 (Peschiaroli et al., 2013). Additionally, in colorectal cancer rosmarinic acid suppress miR-155 repressing the Warburg effect through the mechanism of inactivating the IL-6/STAT3 pathway (Xu et al., 2015).
Another important intermediate step in glycolysis is the conversion of fructose-1,6-bisphosphate to glyceraldehyde 3-phosphate by the aldose enzyme, which is a direct target of miR-122 in liver cells (Fabani and Gait, 2008).
Pyruvate kinase (PKM) regulates the final rate-limiting step of glycolysis, which catalyzes the generation of two molecules of pyruvate and two molecules of adenosine triphosphate (ATP; Mazurek, 2011). MiR-124, miR-137, and miR-340 regulate alternative splicing of the PKM gene in colorectal cancer. The switch from isoform PKM2 to PKM1 inhibits the glycolysis rate and promotes oxidative phosphorylation (Sun et al., 2012). PKM2 is also regulated by miR-326 which is down-modulated in glioblastoma cells (Kefas et al., 2010). Furthermore, pyruvate dehydrogenase kinase (PDHX) catalyzes the conversion of pyruvate to acetyl coenzyme A and is down-modulated by miR-26a in colorectal cancer, thus impairing mitochondrial metabolism (Chen et al., 2014). Let-7 is a microRNA that is commonly down-regulated in several cancer types. Since PDK1 is a physiological target of let-7, its down-regulation in tumors facilitates aerobic glycolysis. Furthermore, PDK1 is critical for Lin28A/B-mediated cancer proliferation, establishing a precise mechanism by which Lin28/let-7 facilitates the Warburg effect to promote cancer progression (Ma et al., 2014; Figure 2B).
Under aerobic glycolysis conditions, oncogenic lesions convert pyruvate to lactate through lactate dehydrogenase (LDH) to fulfill their energetic needs (Hatziapostolou et al., 2013). LDHB is a target of miR-375, which is down-regulated in esophageal squamous cell and maxillary sinus squamous cell carcinomas (Isozaki et al., 2012; Kinoshita et al., 2012). Another important enzyme is the LDHA, frequently overexpressed in tumor cells, and targeted by miR-34a, miR-34c, miR-369-3p, miR-374a, and miR-4524a/b, that are down-modulated in colorectal cancer tissues (Wang J. et al., 2015). Moreover, lncRNA-p21 positively modulates LDHA, Enolase 1, PDHX, Isozyme 4 (PDK4), Phosphoglycerate mutase (PGAM2), Glucose-6-Phosphate Isomerase (GPI), and Pyruvate Kinase (PKM2) in diverse tumors (Hung et al., 2014). The ability of cells to maintain optimal lactate fluxes depends on monocarboxylate transporters (MCTs). Specifically, MCT1 is targeted by miR-29a, miR-29b, and miR-124 in pancreatic cancer (Pullen et al., 2011). Additionally, let-7b, usually inhibited in tumors, has been shown to target basigin (BSG) which interacts with MCT1 (Fu et al., 2011; Figure 2B).
Cancer cells reprogram their metabolism, based on complex regulatory networks involving diverse oncogenic and tumor suppressor genes, including PI3K/Akt, Myc, hypoxia inducible factor (HIF), Ras, Src, p53, and PTEN that promote an increase glucose uptake and glycolysis (Dang et al., 2009; Luo and Semenza, 2011). Those genes are targets of ncRNAs regulation networks in cancer (Table 1).

Table 1. ncRNAs and their participation in cancer metabolic processes through oncogenic or tumor suppressor pathways.
Not only can the human genome-encoded ncRNAs modulate glucose metabolism in cancer cells. Kaposi's sarcoma-associated herpesvirus (KSHV), the etiological agent of Kaposi's sarcoma, has been shown to express microRNAs in its genome that collaborate to induce aerobic glycolysis in infected cells, mainly through the down-regulation of EGLN2 and HSPA9, which cooperate to form the glycolytic phenotype (Yogev et al., 2014).
Lipids constitute a mayor building block for organelles and cells to maintain cellular function and structure provide energy and orchestrate different cellular pathways. As part of lipid metabolism (anabolism and catabolism) a variety of biological intermediators are generated as second messengers (Huang and Freter, 2015), which manage multiple signaling pathways like cell growth, proliferation, differentiation, survival, apoptosis, inflammation, motility, and membrane homeostasis (Mattes, 2005; Krycer et al., 2010; Zechner et al., 2012). Alterations in lipid metabolism can affect cell function, promoting the establishment, and development of cancer (Santos and Schulze, 2012). In fact, lipid biosynthesis is limited to a subgroup of tissues and organs, including adipose, liver, and breast, but its reactivation or reprogramming is commonly observed in tumor cells (Menendez and Lupu, 2007; Abramson, 2011; Beloribi-Djefaflia et al., 2016). The activation or inhibition of lipid signaling pathways is aimed at fulfilling the cell energy requirements and responds to environmental changes. There are numerous enzymes regulating lipid metabolism in the cells and recently, diverse data show that expression of many of these enzymes are regulated by ncRNAs (Huang and Freter, 2015; Figure 3).
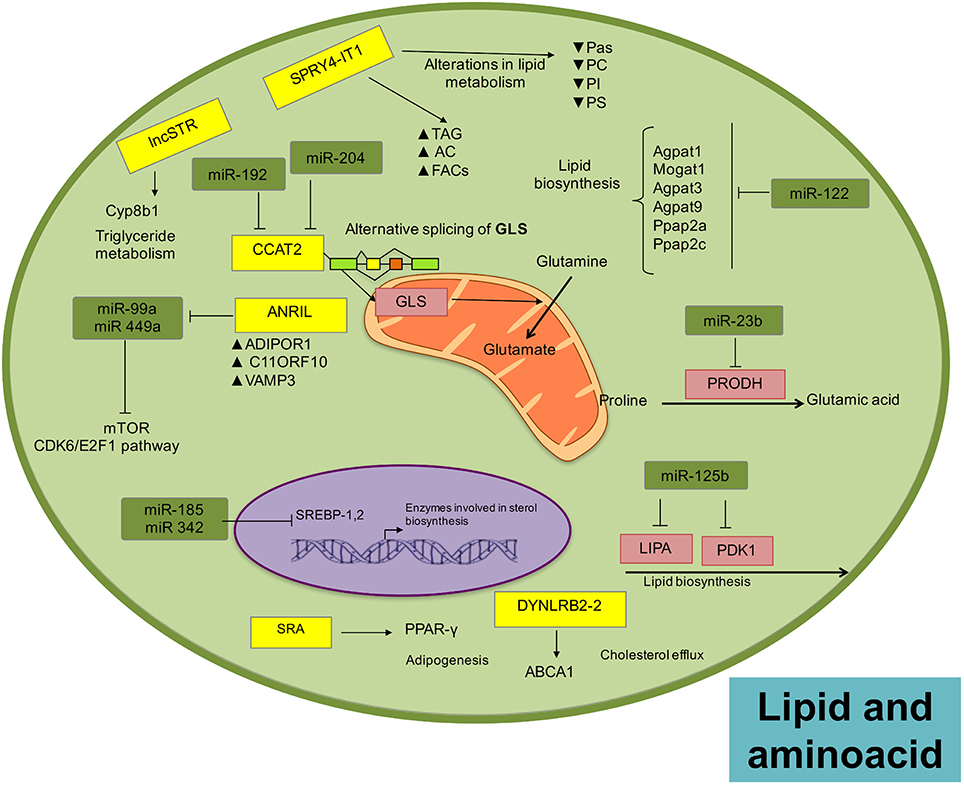
Figure 3. Landscape of lipid and amino acid metabolism regulated by ncRNAs in tumors. Micro and lncRNAs can regulate the metabolism of amino acids through the regulation of enzymes related with these metabolic pathways, favoring the disposition of amino acids as important sources of energy. There is also a fine regulatory loop between microRNAs and lncRNAs than can actively impact metabolic networks.
In prostate cancer cells, miR-185 and miR-342 control lipogenesis and cholesterol synthesis by down-modulating the expression of sterol regulatory element binding protein 1 and 2 (SREBP-1, SREBP-2), repressing their responsive genes, including fatty acid synthase (FASN) and 3-hydroxy-3-methylglutaryl CoA reductase (HMGCR; Li X. et al., 2013). In lymphocytic leukemias, metabolic enzymes related with lipid biosynthesis, such as lipase A (LIPA) and pyruvate dehydrogenase lipoamide kinase isozyme 1 (PDK1), are targets of miR-125b (Tili et al., 2012). Recently, miR-205 has been associated with lipid metabolism de-regulation in hepatocellular carcinoma, acting on acyl-CoA synthetase long-chain family member 1 (ACSL1), a lipid metabolism enzyme in liver (Liu et al., 2012; Cui M. et al., 2014). Additionally, the loss of miR-122, an abundant liver-specific miRNA, alters fat and cholesterol metabolism through modulation of genes involved in lipid synthesis, including Agpat1, Mogat1, Agpat3, Agpat9, Ppap2a, Ppap2c (Hsu et al., 2012; Tsai et al., 2012).
Over-expression of miR-27a in hepatitis C virus-infected liver cells vs. hepatitis B virus-infected cells has been recently described. MiR-27a targets the lipid synthetic transcription factor RXR and the lipid transporter ATP-binding cassette subfamily A member 1 in hepatocarcinoma. Moreover, miR-27a down-modulates the expression of several lipid metabolism-related genes (FASN, SREBP1, SREBP2, PPAR, PPAR, ApoA1, ApoB100, and ApoE3), some of which also participate in the production of infectious viral particles (Shirasaki et al., 2013).
The over-expression of HULC contributes to the malignant development of hepatocellular carcinoma by supporting abnormal lipid metabolism via activation of the acyl-CoA synthetase subunit ACSL1. This results in promotion of lipogenesis and the accumulation of intracellular triglycerides and cholesterol in experimental models. HULC induces methylation of the miR-9 promoter, regulating its expression and favoring alterations in lipid metabolism (Cui et al., 2015). LncRNA SPRY4-IT1 was first identified in adipose tissue (Ota et al., 2004) and was recently found up-regulated in melanoma (Khaitan et al., 2011). Expression of this lncRNA shows a strong correlation with lipid metabolism alterations, including the increase of acyl carnitine, fatty acyl chains, and triacylglycerol, as well as the down-modulation of phosphatidic acid, phosphatidylcholine, phosphatidylinositol, and phosphatidylserine. It is probable that the significant changes in lipid profiles are correlated with the oncogenic modulation of SPRY4-IT1 over the lipid phosphatase lipin 2, an enzyme that converts phosphatidate to diacylglycerol (Mazar et al., 2014).
The oncogene ANRIL is up-regulated in gastric, lung, hepatocellular, cervical, melanoma, ovarian, bladder cancer, among other tumors (Li Z. et al., 2016). Interestingly, ANRIL regulates genes involved in glucose and fatty acid metabolism (Bochenek et al., 2013), such as ADIPOR1. Furthermore, ANRIL can epigenetically regulate the expression of miRNAs in gastric cancer cells, particularly miR-99a/miR-449a, which target CDK6/E2F1 and mTOR pathways (Zhang et al., 2014), that regulate lipid metabolism and adipose tissue function (Cai et al., 2015).
The steroid receptor RNA activator gene is an unusual gene that expresses two different transcripts by alternative splicing of the first intron: (1) the lncRNA SRA and (2) the SRAP protein gene (Hube et al., 2006). SRA co-activates PPAR-gamma, inducing adipogenesis (Xu et al., 2010; Liu et al., 2014) and it may also regulate lipid metabolism (Marion-Letellier et al., 2015). Interestingly, the over-expression of SRA has been associated with poor prognosis in endometrial cancer (Smolle et al., 2015). The lncRNA-DYNLRB2-2 responds to oxidized-LDL to promote ABCA1-mediated cholesterol efflux (Hu et al., 2014). In prostate cancer, the ox-LDL/lncRNA-DYNLRB2-2 circuit might be involved in the promotion of proliferation, migration and invasion rates (Wan et al., 2015). Experiments in animal models showed that the lncLSTR (lncRNA-liver-specific triglyceride regulator), a liver-enriched lncRNA, physiologically contributes to triglyceride metabolism by enhancing Cyp8b1 expression, a molecule involved in bile acid synthesis. Furthermore, Cyp8b1 is down-modulated in primary hepatocytes in which lncLSTR is depleted, suggesting a regulatory activity over Cy8b1 as one of its downstream responsive genes (Li et al., 2015a).
Apart from other energetic sources, amino acids are important substrates that sustain mitochondrial metabolism and support the biosynthesis of proteins, lipids, and other macromolecules. Alterations in amino acid metabolism are also common in cancer cells (Figure 3).
Glutamine metabolism seems to have a critical role in cancer programs, and has been implicated in tumor formation and metastasis (DeBerardinis and Cheng, 2010), as well as being an important source of tumor energy (Li and Zhang, 2016). miRNAs have also been reported to regulate amino acid catabolism, for example, in kidney cancer miR-23b* regulates proline oxidase, which is the first enzyme involved in the conversion of proline to glutamic acid (Liu et al., 2012). Interestingly, the lncRNA CCAT2 participates in the alternative splicing of glutaminase (GLS), an enzyme that catalyzes the hydrolysis of glutamine to glutamate (Redis et al., 2016), where glutamate can be further deaminated to a-Ketoglutarate by glutamate dehydrogenase (GDH) and incorporated into the tricarboxylic acid cycle (Li and Zhang, 2016). Another lncRNA involved in glutamine metabolism is PCGEM1 an androgen-induced prostate specific lncRNA, which regulates expression of enzymes such as GLS, Glutathione Reductase (GSR), and type I gamma-glutamyltransferase (GGT1) in prostate tumors (Hung et al., 2014).
Redundant regulation by ncRNAs reveals that metabolic pathways in cancer are finely regulated, acting at different cellular levels. Consequently, understanding these processes will enable future development of anti-metabolite therapies to target specific energetic signals altered in oncogenic lesions.
The partial maintenance of mitochondrial function in glycolytic cells appears essential for cancer cell development. Thus, the tumor must balance the bioenergetic requirements to grow, proliferate, and survive within the energetic restrictions and metabolic pathways. Mitochondria are in fact, the main intracellular producers of reactive oxygen species (ROS) as part of adenosine triphosphate (ATP) production through oxidative phosphorylation (OXPHOS). This organelle is responsible for converting available nutrients into the fundamental blocks required for cell maintenance (Ahn and Metallo, 2015), such as fatty acids, cholesterol and proteins (Kamphorst et al., 2013). Therefore, mitochondrial alterations have been implicated in the etiology of many diseases including cancer. The metabolic reprogramming of the mitochondrial network in tumoral programs is achieved through several mechanisms, including ncRNAs transcribed both in the nuclear and in the mitochondrial genome (mtDNA). ncRNAs can actively regulate mitochondrial metabolism by controlling structural and functional mechanisms that respond to changes in energy requirements or environmental conditions (Benard et al., 2010; Figure 4).
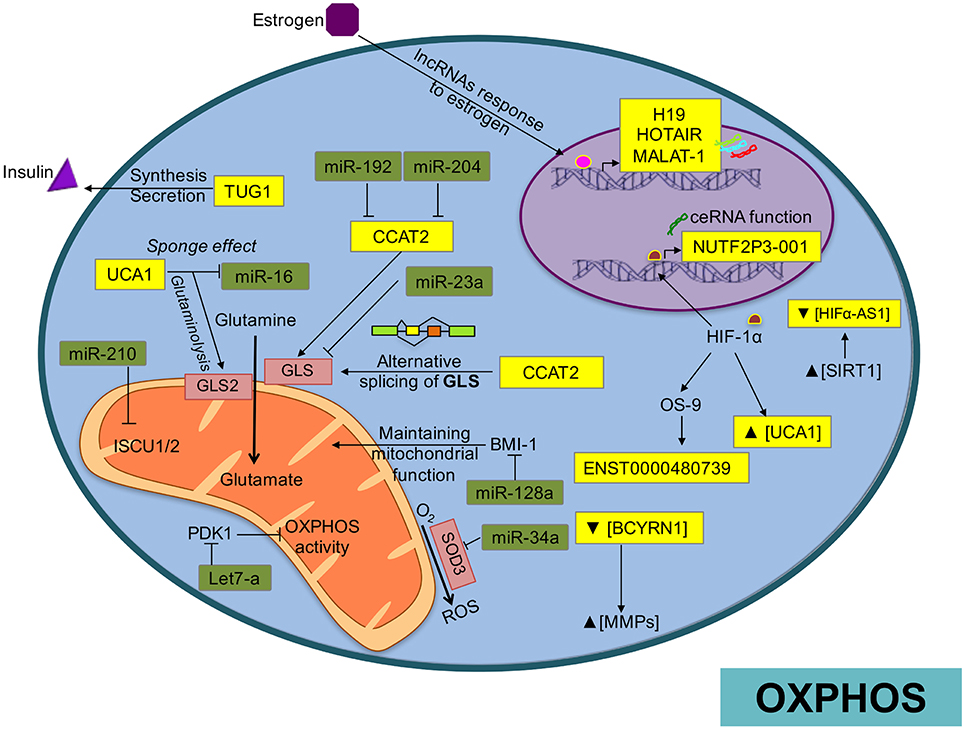
Figure 4. Overview of ncRNA regulatory network over OXPHO in cancer. In this picture we show how ncRNAs are involved in the regulation of OXPHOS, generation of ROS, or mediating alternative splicing of mitochondrial enzymes.
For example, miR-210 is up-regulated by hypoxia (Dang and Myers, 2015), and can block mitochondrial respiration through down-modulation of the electron transport chain (ETC) complexes (Huang and Zuo, 2014). Particularly, miR-210 targets ISCU1 and ISCU2, suppressing mitochondrial function and disrupting iron homeostasis in colon, breast, and esophageal cancer (Chen et al., 2010; Favaro et al., 2010). In breast cancer cells, miR-378* promotes a metabolic shift by inhibiting the expression of important regulators of energy metabolism such as estrogen-related receptor-γ and GA-binding protein transcription factor. This reduces the tricarboxylic acid cycle (TCA) rates, decreasing the dependency on OXPHOS, and increasing lactate production (Eichner et al., 2010). Similarly, in hepatocellular carcinomas miR-23a modulates a metabolic switch from OXPHOS to anaerobic glycolysis by targeting the glucose-6-phosphatase catalytic subunit (G6PC), which plays an important role in mitochondrial respiration (Wu et al., 1999; Wang et al., 2012). Likewise, overexpression of miR-125b in lymphocytic leukemia models represses many transcripts implicated in energetic and lipid metabolism including phosphatidylcholine transfer protein, lipase A, lysosomal acid, cholesterol esterase, glutathione synthetase, acyl-CoA synthetase short-chain family member 1, HK2, stearoyl-CoA desaturase 1, AKT2, and pyruvate dehydrogenase kinase 1 (PDK1; Tili et al., 2012).
Some of the most important by-products of the electron transport chain in the mitochondria are reactive oxygen species (mROS). Increased production of ROS can lead to activation of tumorigenic signaling and metabolic reprogramming. This tumorigenic signaling includes mechanisms to prevent imbalances in the production of mROS maintaining redox homeostasis (Sullivan and Chandel, 2014). Emerging evidence shows that control of ROS levels is mediated in part by ncRNAs. One of the first evidence was the cluster miR-17–92, overexpressed in small-cell lung cancer, which reduce DNA damage to a tolerable level and consequently lead to the accumulation of genetic instability (Ebi et al., 2009). miR-141 and miR-200a, contribute to ovarian tumorigenesis by targeting p38α and modulating oxidative stress response in mouse models (Mateescu et al., 2011). In addition, miR-21 and miR-34a promote tumor malignancy by the formation of ROS through the mediation of SOD3 and TNFα expression in cancer cells (Zhang et al., 2012). In medulloblastoma, miR-128a regulates ROS by specific inhibition of the Bmi-1 oncogene, which participates in maintaining mitochondrial function and redox homeostasis (Venkataraman et al., 2010). Let-7a promotes OXPHOS in breast cancer (Serguienko et al., 2015) and hepatocellular carcinoma by directly modulating PDK1, which as mentioned previously, is a negative regulator of OXPHOS activity (Ma et al., 2014). In bladder cancer, the lncRNA UCA1 participates in ROS formation and promotes mitochondrial glutaminolysis by its sponge effect on miR-16 (Li H. J. et al., 2015). SOD2, which has response elements for NF-κB, wipes out the superoxide anion radicals generated by OXPHOS and coverts them into hydrogen peroxide in cancer cells. Although it is know that the lncRNA Lethe prevents binding of NFκB to NFκB response elements resulting in the suppression of SOD2 (Rapicavoli et al., 2013), the impact of Lethe on energetic metabolism of cancer cells is poorly understood.
Apart from glucose, cancer cells exhibit increased glutamine intake and glutamine metabolism (glutaminolysis). The accelerated glutamine metabolism provides enough substrate to increase lipogenesis and nucleic acid biosynthesis, necessary for the proliferative phenotype of the cancer cells (Gao et al., 2009). Of particular importance, mitochondrial enzymes participate in the metabolism of glutamine and other metabolites (glutamate, proline, aspartate, and alanine) as part of the tumor programs (Dang, 2010). One of the major regulators of glutaminolysis is MYC. Along the same line the suppression of miR-23A/B by MYC enhances mitochondrial glutaminase expression and glutamine metabolism in prostate cancer (Gao et al., 2009). Additionally, the deregulation of the HOTTIP lncRNA by miR-192 and miR-204 can produce an abnormal glutaminolysis through positive regulation of GLS1 in hepatocellular tumors (Ge et al., 2015). Furthermore, the CCAT2 lncRNA modulates GSL alternative splicing through an allele-specific regulatory mechanism (Redis et al., 2016). Moreover, in bladder cancer cells the UCA1 lncRNA promotes glutamine metabolism through its sponge function over miR-16, allowing the expression of GLS2, enzyme that participates in the hydrolysis of glutamine to glutamate (Li H. J. et al., 2015).
The involvement of mitochondrial miRNAs (mitomiRs) and mitochondrial lncRNAs in regulating the OXPHOS system is of particular interest. These regulatory molecules have either a pro-oxidant or antioxidant effect (Bai et al., 2011; Aschrafi et al., 2012; Li P. et al., 2012). Therefore, mitochondrial ncRNAs may participate in the fine-tuning of the mitochondrial energy supply. A recent study identified 13 miRNAs significantly enriched in mitochondria of HeLa cells, which actively participate in cell cycle and apoptosis through regulation of mitochondrial activity (Bandiera et al., 2011; Demongeot et al., 2013). The lncRNAs encoded by mtDNA, ASncmtRNA-1/2, are down-regulated in cancer cells and take part in the mitochondrial reprograming of oncogenic pathways (Burzio et al., 2009). Biological activity of ASncmtRNAs results in survivin inhibition at the RNA level, probably mediated by microRNAs (Vidaurre et al., 2014). Survivin enhances the stability of oxidative phosphorylation complex II, which promotes cellular respiration (Rivadeneira et al., 2015).
Another type of non-coding RNA, the Plement-induced wimpy testis (PIWI)-interacting RNAs (piRNAs), have been recently recognized to be relevant in cancer metabolic reprogramming. piRNAs are small non-coding RNAs (26–31 nt) that form the piRNA-induced silencing complex (piRISC). The main function of piRNAs is to silence transposable elements (TEs) in the germ line, but also in cancer cells (Siomi et al., 2011), mainly through epigenetic regulation, genome re-arrangement, and stem cell self-renewal (Ross et al., 2014). piRNA expression has been detected in mitochondrial RNAs of HeLa cells, and are possibly implicated in diverse functions related to energetic homeostasis, bioenergetics and cell growth (Kwon et al., 2014).
Novel data suggest that the regulatory role of ncRNAs during carcinogenesis is not limited to cancer cells they are also implicated in the activation of the tumor stroma and in its transition into a cancer-associated microenvironment. In fact, tumor development involves a fine interplay between malignant and stromal cells. Secreted ncRNAs can serve as regulatory signals promoting cancer cell proliferation, migration, communication, and stromal modification, thereby enhancing an optimal microenvironment for oncogenesis (Soon and Kiaris, 2013). The tumor microenvironment presents a complex architecture, comprising fibroblasts, vascular endothelial cells, immune cells, adipocytes, and extracellular matrix, conforming the stromal tissue that surrounds and interacts tumor cells (Hanahan and Weinberg, 2011).
Importantly, cancer-associated fibroblasts (CAFs) can modify the metabolism of the adjacent cancer cells, as a consequence, its activity can promote tumor growth, invasion and angiogenesis (Franco et al., 2010). CAFs are originated from normal fibroblasts (NFs) that are in contact with tumor cells, receiving and sending signals to co-evolve with the tumor cells and support their biological requirements. Communication pathways between CAFs and neoplastic cells include ncRNA mediated signaling (Table 2; Erez et al., 2010).
Additionally, the metabolic status in cancer lesions is also balanced by different micro-environmental components. For instance, surrounding immune cells present active alternative pathways to overcome tumor energetic limitations. In particular, the metabolic switch in tumor cells promotes the presence of tumor-infiltrating lymphocytes (T cells) which is a crucial tumoral adaptation to dampen antitumor immunity (Molon et al., 2016; Zhao et al., 2016). Maintaining tumor metabolic homeostasis requires a balanced immune response, which is achieved by extracellular signals that can be induced or repressed by ncRNA activity (Table 2; Dumortier et al., 2013).
Another important component of the tumor microenvironment are the adipocytes, that are considered as an energy storage depot, as well as endocrine cells that produce hormones, growth factors, cytokines, and adipokines (Rajala and Scherer, 2003). Therefore, mature adipocytes influence tumor behavior through heterotypic signaling processes, providing fatty acids for rapid tumor growth, and also promoting homing, migration, and invasion of tumor cells (Nieman et al., 2011). ncRNAs can actively participate as important modulators of the lipid metabolism in tumors where adipocytes represent the major component of the tumoral microenvironment (Table 2).
Emerging data suggest a fine regulatory loop between the HIF system, microenvironment and tumor cells, governed by diverse regulatory molecules like ncRNAs. Given the fact that HIF target genes include many metabolism-induced genes, such as ncRNAS (Semenza, 2010; Masson and Ratcliffe, 2014), and both tumor and stromal hypoxia, along with deregulated metabolism, characterize aggressive cancer phenotypes, it is tempting to conclude that activation and regulation of HIF pathways by complex signaling processes is one of the most important causes for deregulated tumor metabolism (Höckel and Vaupel, 2001; Table 3). A more detailed overview about hypoxia and lncRNA is discussed in Chang et al. (2016).
Endogenous and exogenous hormone-signaling pathways serve as metabolic regulatory networks that control fuel and energy metabolism on both tumor and stromal cells, and connects nutrient availability with cell growth and proliferation. Currently, ncRNA modulation by hormones can reenforce hormone-signaling activity. For example, insulin, a major hormone in the homeostasis of energy and metabolism, has been implicated in the regulation of miRNA expression (Granjon et al., 2009). Additionally, the estrogen receptor activates autophagic fluxes as a response to metabolic damage, in part by regulating ncRNA expression (Bernales et al., 2007; Table 3).
When metabolic stress triggers energetic and nutritional changes in tumor cells, the metabolic stress responses collaborate to maintain homeostasis. Metabolic changes take place in reaction to stress in the tumor and stromal cells through the activation of several mechanisms, including autophagy, epithelial-mesenchymal transition (EMT), angiogenesis, and inflammation.
Autophagy is a catabolic process indispensable for the maintenance of cellular homeostasis. Alterations of autophagy are described in cancer and are due to alterations in the expression of various genes that promote or suppress it (Lozy and Karantza, 2012). Autophagic programs consist of the degradation of cellular organelles, cytoplasmic proteins and lipids, allowing recycling of the resulting catabolites for biosynthesis and energy metabolism, in order to satisfy nutrient, energy and hormonal demands of the tumor cells (Jing et al., 2015). The metabolic requirements of cancer cells are maintained, in part, by autophagy pathways present in tumor or stroma cells (Martinez-Outschoorn et al., 2011; Mathew and White, 2011). Considering the vast implications if ncRNAs in stress responses, their activity might contribute to the dynamics of autophagy during cancer progression (Leung and Sharp, 2010; Table 4).
Metabolism, mainly hypoxic conditions, can drive EMT through NF-κB, PI3K/Akt/mTOR, Notch, Wnt/β-catenin, and Hedgehog signaling pathways (Fan et al., 2013). EMT refers to a complex molecular and cellular program by which epithelial cells lose their epithelial attributes such as cell–cell adhesion, planar-basal polarity, and limited motility, but acquire mesenchymal features, including increased motility, invasiveness and development of escape routes for apoptosis (Polyak and Weinberg, 2009). Modulation of EMT pathways by ncRNAs has been described in several tumors (Table 4). Another important feature that characterizes the most advanced and aggressive tumors is angiogenesis; meaning the development of tumor neovasculature. This mechanism is crucial to satisfy nutrient and oxygen demands, as well as to provide routes for metabolic waste excretion (Carmeliet and Jain, 2000). To achieve this oncogenic hallmark, tumor cells induce pro-angiogenic factors or block anti-angiogenic signals, in part by modulating ncRNA expression profiles (Table 4). For a more detailed overview about ncRNAs implicated in EMT and angiogenesis refer to Wang W. et al. (2015) and Choudhry et al. (2016).
Finally, inflammation is considered as an oncogenic feature that allows the acquisition of carcinogenic capacities by the provision of biomolecules to the tumor and to the cells of the microenvironment, such as transcription factors which can enhance proliferative signaling, pro-angiogenic factors, invasion and metastasis (Hanahan and Weinberg, 2011; Table 4). A more detailed overview of ncRNAs implicated in inflammation is discussed in (O'connell et al., 2012).
The advent of novel knowledge and high throughput technologies, such as RNA-seq, Chip-seq, and metabolomic analysis, has allowed us to gain insight into the versatility of the mechanism that regulate metabolism and how the disturbance of specific factors, in particular ncRNAs, might impact the altered phenotypes of cancer cells. During the past years, we have gained important understanding about the biological activity of ncRNAs, although more research is needed to better understand the complex mechanisms that orchestrate tumor metabolism. Furthermore, pharmacological intervention of cell metabolism is emerging as a potential therapeutic strategy in some cancers (Ahn and Metallo, 2015) giving us the opportunity to explore new sources for biomarker discovery and development of new targeted drugs. The crucial role of ncRNAs in metabolism and associated mechanisms raises the possibility of developing ncRNA-targeted therapies. miRNA and lncRNAs mimics or inhibitors can be used to elevate or block the activity of metabolic-related genes to drive cancer initiation and/or progression programs. Figure 5 summarizes some of the actual and future therapeutic applications of metabolism-related ncRNAS in cancer treatment.
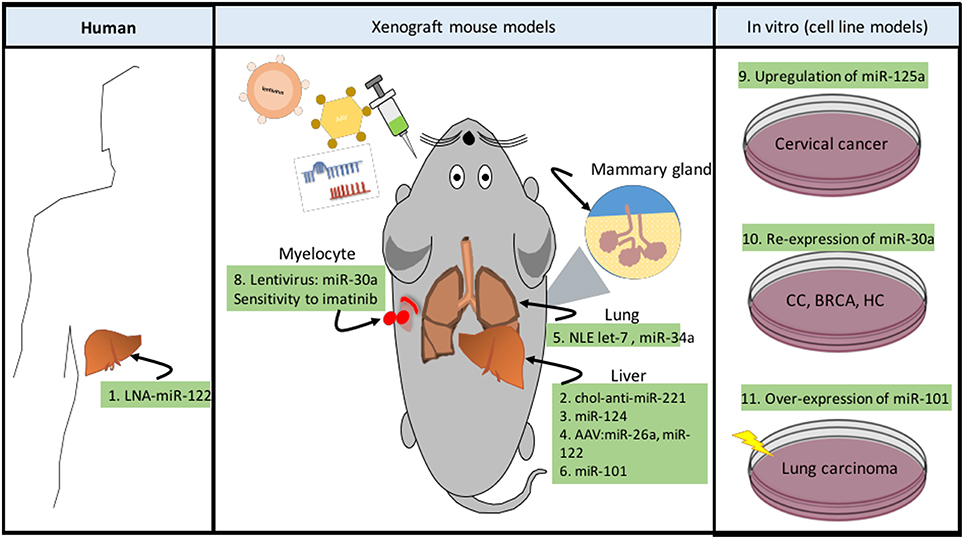
Figure 5. ncRNAs as novel therapeutic strategies in cancer metabolism. Targeting cancer metabolism represents a novel resource to develop anti-cancer therapies. Now a days, there are different techniques developed to specifically modulate metabolic pathways, some of them are dedicated to silencing (LNA) or re-expressing (miRNA mimic) ncRNA transcripts (Phan et al., 2014). These systems can be delivered by intratumoral, intraperitoneal, and intravenous injections, through systemic adenovirus-associated virus (AAV), or in complexes with neutral lipid emulsions (Drakaki et al., 2013). In addition to these technologies, cholesterol-modified miRNAs (chol-anti-miRs) exhibit improved pharmacokinetics and antitumor efficacy. Human (1) The development of hepatocellular carcinoma (HCC) in persons who are persistently infected with hepatitis C virus (HCV) is a growing problem. A phase II trial of the LNA anti-miR-122 is being carried out for treatment of HCV infection (Lindow and Kauppinen, 2012). Xenograft mouse models (2) chol-anti-miR-221 effectively suppresses liver tumor growth (Park et al., 2011). (3) Systemic administration of miR-124 suppresses liver cancer growth through suppression of the IL6/STAT3 inflammatory pathway (Hatziapostolou et al., 2011). (4) AAV delivery of miR-26a or miR-122 suppresses MYC-driven liver carcinogenesis without affecting normal hepatocytes (Kota et al., 2009; Hsu et al., 2012). (5) Neutral lipid emulsions (NLE) to deliver let-7 which targets RAS and MYC oncogenes, as well as miR-34, reduces tumor size in lung cancer (Trang et al., 2011). (6) miR-101 and miR-376b are miRNAs, which negatively regulate the autophagy pathway (Frankel et al., 2011; Korkmaz et al., 2012). Furthermore, overexpression of miR-101 suppressed tumor development and efficiently reduced tumor size in liver cancer (Su et al., 2009). (7) Over-expression of miR-101 can effectively reduce tamoxifen-induced autophagy and enhance the sensitivity of breast cancer cells to tamoxifen treatment (Frankel et al., 2011). (8) Recombinant lentivirus administration of miR-30a (inhibitor of autophagy by down-modulating BECN1), can enhance sensitivity to imatinib cytotoxicity in chronic myeloid leukemia, increasing tumor cell apoptosis (Yu et al., 2012). In vitro (cell line models). (9) Up-regulation of miR-125a in cervical cancer (CC) models sensitized to paclitaxel by down-regulating STAT3 (Fan et al., 2016). (10) Re-expression of miR-30a can sensitize tumor cells to cisplatin via mediating autophagy in HeLa, MCF-7 and HepG2 (Zou et al., 2012). (11) Over-expression of miR-101 sensitized human lung carcinoma cells to radiation treatment (Yan et al., 2010).
SR and AH coordinated the review process, SR, AH, FB, and AC performed the literature review, organized the information and wrote the paper. All authors read and approved the last version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Elizabeth Langley McCarron for her comments and critical review of the manuscript. FB and AC received a Ph.D., scholarship from the Mexican National Council of Science and Technology. This paper is part of the Ph.D., thesis project of FB and AC, students of the Biomedical Sciences Ph.D., Program of the National Autonomous University of Mexico.
Aakula, A., Leivonen, S. K., Hintsanen, P., Aittokallio, T., Ceder, Y., Børresen-Dale, A. L., et al. (2015). MicroRNA-135b regulates ERalpha, AR and HIF1AN and affects breast and prostate cancer cell growth. Mol. Oncol. 9, 1287–1300. doi: 10.1016/j.molonc.2015.03.001
Abramson, H. N. (2011). The lipogenesis pathway as a cancer target. J. Med. Chem. 54, 5615–5638. doi: 10.1021/jm2005805
Ahn, C. S., and Metallo, C. M. (2015). Mitochondria as biosynthetic factories for cancer proliferation. Cancer Metab. 3, 1. doi: 10.1186/s40170-015-0128-2
Arora, A., Singh, S., Bhatt, A. N., Pandey, S., Sandhir, R., and Dwarakanath, B. S. (2015). Interplay between metabolism and oncogenic process: role of microRNAs. Transl. Oncogenomics 7, 11–27. doi: 10.4137/TOG.S29652
Aschrafi, A., Kar, A. N., Natera-Naranjo, O., Macgibeny, M. A., Gioio, A. E., and Kaplan, B. B. (2012). MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell. Mol. Life Sci. 69, 4017–4027. doi: 10.1007/s00018-012-1064-8
Bai, X. Y., Ma, Y., Ding, R., Fu, B., Shi, S., and Chen, X. M. (2011). miR-335 and miR-34a Promote renal senescence by suppressing mitochondrial antioxidative enzymes. J. Am. Soc. Nephrol. 22, 1252–1261. doi: 10.1681/ASN.2010040367
Bandiera, S., Hanein, S., Lyonnet, S., and Henrion-Caude, A. (2011). Mitochondria as novel players of the cellular RNA interference. J. Biol. Chem. 286, le19. doi: 10.1074/jbc.L111.240259
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Beloribi-Djefaflia, S., Vasseur, S., and Guillaumond, F. (2016). Lipid metabolic reprogramming in cancer cells. Oncogenesis 5, e189. doi: 10.1038/oncsis.2015.49
Benard, G., Bellance, N., Jose, C., Melser, S., Nouette-Gaulain, K., and Rossignol, R. (2010). Multi-site control and regulation of mitochondrial energy production. Biochim. Biophys. Acta 1797, 698–709. doi: 10.1016/j.bbabio.2010.02.030
Bernales, S., Schuck, S., and Walter, P. (2007). ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy 3, 285–287. doi: 10.4161/auto.3930
Bhan, A., and Mandal, S. S. (2016). Estradiol-induced transcriptional regulation of long non-coding RNA, HOTAIR. Methods Mol. Biol. 1366, 395–412. doi: 10.1007/978-1-4939-3127-9_31
Bhattacharyya, S., Sul, K., Krukovets, I., Nestor, C., Li, J., and Adognravi, O. S. (2012). Novel tissue-specific mechanism of regulation of angiogenesis and cancer growth in response to hyperglycemia. J. Am. Heart Assoc. 1:e005967. doi: 10.1161/JAHA.112.005967
Blick, C., Ramachandran, A., Mccormick, R., Wigfield, S., Cranston, D., Catto, J., et al. (2015). Identification of a hypoxia-regulated miRNA signature in bladder cancer and a role for miR-145 in hypoxia-dependent apoptosis. Br. J. Cancer 113, 634–644. doi: 10.1038/bjc.2015.203
Bochenek, G., Häsler, R., El Mokhtari, N. E., König, I. R., Loos, B. G., Jepsen, S., et al. (2013). The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum. Mol. Genet. 22, 4516–4527. doi: 10.1093/hmg/ddt299
Burzio, V. A., Villota, C., Villegas, J., Landerer, E., Boccardo, E., Villa, L. L., et al. (2009). Expression of a family of noncoding mitochondrial RNAs distinguishes normal from cancer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 9430–9434. doi: 10.1073/pnas.0903086106
Cai, H., Dong, L. Q., and Liu, F. (2015). Recent advances in adipose mTOR signaling and function: therapeutic prospects. Trends Pharmacol. Sci. 37, 303–317. doi: 10.1016/j.tips.2015.11.011
Cairns, R. A., Harris, I. S., and Mak, T. W. (2011). Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95. doi: 10.1038/nrc2981
Carmeliet, P., and Jain, R. K. (2000). Angiogenesis in cancer and other diseases. Nature 407, 249–257. doi: 10.1038/35025220
Cha, S. T., Chen, P. S., Johansson, G., Chu, C. Y., Wang, M. Y., Jeng, Y. M., et al. (2010). MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res. 70, 2675–2685. doi: 10.1158/0008-5472.CAN-09-2448
Chai, S., Tong, M., Ng, K. Y., Kwan, P. S., Chan, Y. P., Fung, T. M., et al. (2014). Regulatory role of miR-142-3p on the functional hepatic cancer stem cell marker CD133. Oncotarget 5, 5725–5735. doi: 10.18632/oncotarget.2167
Chang, Y., Yan, W., He, X., Zhang, L., Li, C., Huang, H., et al. (2012). miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology 143, 177–187. doi: 10.1053/j.gastro.2012.04.009
Chang, Y. N., Zhang, K., Hu, Z. M., Qi, H. X., Shi, Z. M., Han, X. H., et al. (2016). Hypoxia-regulated lncRNAs in cancer. Gene 575, 1–8. doi: 10.1016/j.gene.2015.08.049
Chatterjee, A., Chattopadhyay, D., and Chakrabarti, G. (2014). miR-17-5p downregulation contributes to paclitaxel resistance of lung cancer cells through altering beclin1 expression. PLoS ONE 9:e95716. doi: 10.1371/journal.pone.0095716
Chen, B., Liu, Y., Jin, X., Lu, W., Liu, J., Xia, Z., et al. (2014). MicroRNA-26a regulates glucose metabolism by direct targeting PDHX in colorectal cancer cells. BMC Cancer 14:443. doi: 10.1186/1471-2407-14-443
Chen, Z., Li, Y., Zhang, H., Huang, P., and Luthra, R. (2010). Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 29, 4362–4368. doi: 10.1038/onc.2010.193
Chio, C. C., Lin, J. W., Cheng, H. A., Chiu, W. T., Wang, Y. H., Wang, J. J., et al. (2013). MicroRNA-210 targets antiapoptotic Bcl-2 expression and mediates hypoxia-induced apoptosis of neuroblastoma cells. Arch. Toxicol. 87, 459–468. doi: 10.1007/s00204-012-0965-5
Choudhry, H., Albukhari, A., Morotti, M., Haider, S., Moralli, D., Smythies, J., et al. (2015). Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 34, 4482–4490. doi: 10.1038/onc.2014.378
Choudhry, H., Harris, A. L., and Mcintyre, A. (2016). The tumour hypoxia induced non-coding transcriptome. Mol. Aspects Med. 47–48, 35–53. doi: 10.1016/j.mam.2016.01.003
Chow, T. F., Mankaruos, M., Scorilas, A., Youssef, Y., Girgis, A., Mossad, S., et al. (2010). The miR-17-92 cluster is over expressed in and has an oncogenic effect on renal cell carcinoma. J. Urol. 183, 743–751. doi: 10.1016/j.juro.2009.09.086
Comincini, S., Allavena, G., Palumbo, S., Morini, M., Durando, F., Angeletti, F., et al. (2013). microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol. Ther. 14, 574–586. doi: 10.4161/cbt.24597
Cui, H., Xie, N., Tan, Z., Banerjee, S., Thannickal, V. J., Abraham, E., et al. (2014). The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur. J. Immunol. 44, 2085–2095. doi: 10.1002/eji.201344126
Cui, M., Wang, Y., Sun, B., Xiao, Z., Ye, L., and Zhang, X. (2014). MiR-205 modulates abnormal lipid metabolism of hepatoma cells via targeting acyl-CoA synthetase long-chain family member 1 (ACSL1) mRNA. Biochem. Biophys. Res. Commun. 444, 270–275. doi: 10.1016/j.bbrc.2014.01.051
Cui, M., Xiao, Z., Wang, Y., Zheng, M., Song, T., Cai, X., et al. (2015). Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 75, 846–857. doi: 10.1158/0008-5472.CAN-14-1192
Dang, C. V. (2010). Glutaminolysis: supplying carbon or nitrogen or both for cancer cells? Cell Cycle 9, 3884–3886. doi: 10.4161/cc.9.19.13302
Dang, C. V., Le, A., and Gao, P. (2009). MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 15, 6479–6483. doi: 10.1158/1078-0432.CCR-09-0889
Dang, K., and Myers, K. A. (2015). The role of hypoxia-induced miR-210 in cancer progression. Int. J. Mol. Sci. 16, 6353–6372. doi: 10.3390/ijms16036353
Dávalos, A., Goedeke, L., Smibert, P., Ramírez, C. M., Warrier, N. P., Andreo, U., et al. (2011). miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 9232–9237. doi: 10.1073/pnas.1102281108
DeBerardinis, R. J., and Cheng, T. (2010). Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29, 313–324. doi: 10.1038/onc.2009.358
Demongeot, J., Hazgui, H., Bandiera, S., Cohen, O., and Henrion-Caude, A. (2013). MitomiRs, chloromiRs and modelling of the microRNA inhibition. Acta Biotheor. 61, 367–383. doi: 10.1007/s10441-013-9190-8
Doldi, V., Callari, M., Giannoni, E., D'aiuto, F., Maffezzini, M., Valdagni, R., et al. (2015). Integrated gene and miRNA expression analysis of prostate cancer associated fibroblasts supports a prominent role for interleukin-6 in fibroblast activation. Oncotarget 6, 31441–31460. doi: 10.18632/oncotarget.5056
Drakaki, A., Hatziapostolou, M., and Iliopoulos, D. (2013). Therapeutically targeting microRNAs in liver cancer. Curr. Pharm. Des. 19, 1180–1191. doi: 10.2174/138161213804805658
Dumortier, O., Hinault, C., and Van Obberghen, E. (2013). MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 18, 312–324. doi: 10.1016/j.cmet.2013.06.004
Ebi, H., Sato, T., Sugito, N., Hosono, Y., Yatabe, Y., Matsuyama, Y., et al. (2009). Counterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene 28, 3371–3379. doi: 10.1038/onc.2009.201
Eichner, L. J., Perry, M. C., Dufour, C. R., Bertos, N., Park, M., St-Pierre, J., et al. (2010). miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab. 12, 352–361. doi: 10.1016/j.cmet.2010.09.002
Ellis, B. C., Graham, L. D., and Molloy, P. L. (2014). CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim. Biophys. Acta 1843, 372–386. doi: 10.1016/j.bbamcr.2013.10.016
Elstrom, R. L., Bauer, D. E., Buzzai, M., Karnauskas, R., Harris, M. H., Plas, D. R., et al. (2004). Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899. doi: 10.1158/0008-5472.CAN-03-2904
Erez, N., Truitt, M., Olson, P., Arron, S. T., and Hanahan, D. (2010). Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell 17, 135–147. doi: 10.1016/j.ccr.2009.12.041
Esau, C., Kang, X., Peralta, E., Hanson, E., Marcusson, E. G., Ravichandran, L. V., et al. (2004). MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 279, 52361–52365. doi: 10.1074/jbc.C400438200
Esquela-Kerscher, A., and Slack, F. J. (2006). Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269. doi: 10.1038/nrc1840
Fabani, M. M., and Gait, M. J. (2008). miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA 14, 336–346. doi: 10.1261/rna.844108
Fan, Y. L., Zheng, M., Tang, Y. L., and Liang, X. H. (2013). A new perspective of vasculogenic mimicry: EMT and cancer stem cells (Review). Oncol. Lett. 6, 1174–1180. doi: 10.3892/ol.2013.1555
Fan, Z., Cui, H., Yu, H., Ji, Q., Kang, L., Han, B., et al. (2016). MiR-125a promotes paclitaxel sensitivity in cervical cancer through altering STAT3 expression. Oncogenesis 5, e197. doi: 10.1038/oncsis.2016.21
Fang, R., Xiao, T., Fang, Z., Sun, Y., Li, F., Gao, Y., et al. (2012). MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J. Biol. Chem. 287, 23227–23235. doi: 10.1074/jbc.M112.373084
Favaro, E., Ramachandran, A., Mccormick, R., Gee, H., Blancher, C., Crosby, M., et al. (2010). MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS ONE 5:e10345. doi: 10.1371/journal.pone.0010345
Fei, X., Qi, M., Wu, B., Song, Y., Wang, Y., and Li, T. (2012). MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 586, 392–397. doi: 10.1016/j.febslet.2012.01.006
Feng, R., Chen, X., Yu, Y., Su, L., Yu, B., Li, J., et al. (2010). miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 298, 50–63. doi: 10.1016/j.canlet.2010.06.004
Fish, J. E., Santoro, M. M., Morton, S. U., Yu, S., Yeh, R. F., Wythe, J. D., et al. (2008). miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 15, 272–284. doi: 10.1016/j.devcel.2008.07.008
Fong, M. Y., Zhou, W., Liu, L., Alontaga, A. Y., Chandra, M., Ashby, J., et al. (2015). Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 17, 183–194. doi: 10.1038/ncb3094
Fornari, F., Milazzo, M., Chieco, P., Negrini, M., Calin, G. A., Grazi, G. L., et al. (2010). MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 70, 5184–5193. doi: 10.1158/0008-5472.CAN-10-0145
Franco, O. E., Shaw, A. K., Strand, D. W., and Hayward, S. W. (2010). Cancer associated fibroblasts in cancer pathogenesis. Semin. Cell Dev. Biol. 21, 33–39. doi: 10.1016/j.semcdb.2009.10.010
Frankel, L. B., Wen, J., Lees, M., Høyer-Hansen, M., Farkas, T., Krogh, A., et al. (2011). microRNA-101 is a potent inhibitor of autophagy. EMBO J. 30, 4628–4641. doi: 10.1038/emboj.2011.331
Fu, T. Y., Chang, C. C., Lin, C. T., Lai, C. H., Peng, S. Y., Ko, Y. J., et al. (2011). Let-7b-mediated suppression of basigin expression and metastasis in mouse melanoma cells. Exp. Cell Res. 317, 445–451. doi: 10.1016/j.yexcr.2010.11.004
Gao, P., Tchernyshyov, I., Chang, T. C., Lee, Y. S., Kita, K., Ochi, T., et al. (2009). c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765. doi: 10.1038/nature07823
Gatenby, R. A., and Gillies, R. J. (2004). Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899. doi: 10.1038/nrc1478
Ge, Y., Yan, X., Jin, Y., Yang, X., Yu, X., Zhou, L., et al. (2015). fMiRNA-192 and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 11:e1005726. doi: 10.1371/journal.pgen.1005726
Ge, Y. Y., Shi, Q., Zheng, Z. Y., Gong, J., Zeng, C., Yang, J., et al. (2014). MicroRNA-100 promotes the autophagy of hepatocellular carcinoma cells by inhibiting the expression of mTOR and IGF-1R. Oncotarget 5, 6218–6228. doi: 10.18632/oncotarget.2189
Ghosh, G., Subramanian, I. V., Adhikari, N., Zhang, X., Joshi, H. P., Basi, D., et al. (2010). Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J. Clin. Invest. 120, 4141–4154. doi: 10.1172/JCI42980
Godlewski, J., Nowicki, M. O., Bronisz, A., Nuovo, G., Palatini, J., De Lay, M., et al. (2010). MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol. Cell 37, 620–632. doi: 10.1016/j.molcel.2010.02.018
Granjon, A., Gustin, M. P., Rieusset, J., Lefai, E., Meugnier, E., Güller, I., et al. (2009). The microRNA signature in response to insulin reveals its implication in the transcriptional action of insulin in human skeletal muscle and the role of a sterol regulatory element-binding protein-1c/myocyte enhancer factor 2C pathway. Diabetes 58, 2555–2564. doi: 10.2337/db09-0165
Gross, D. N., Van Den Heuvel, A. P., and Birnbaum, M. J. (2008). The role of FoxO in the regulation of metabolism. Oncogene 27, 2320–2336. doi: 10.1038/onc.2008.25
Gu, H., Liu, M., Ding, C., Wang, X., Wang, R., Wu, X., et al. (2016). Hypoxia-responsive miR-124 and miR-144 reduce hypoxia-induced autophagy and enhance radiosensitivity of prostate cancer cells via suppressing PIM1. Cancer Med. 5, 1174–1182. doi: 10.1002/cam4.664
Guo, C., Sah, J. F., Beard, L., Willson, J. K., Markowitz, S. D., and Guda, K. (2008). The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer 47, 939–946. doi: 10.1002/gcc.20596
Guo, W., Qiu, Z., Wang, Z., Wang, Q., Tan, N., Chen, T., et al. (2015). MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology 62, 1132–1144. doi: 10.1002/hep.27929
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Hatziapostolou, M., Polytarchou, C., Aggelidou, E., Drakaki, A., Poultsides, G. A., Jaeger, S. A., et al. (2011). An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 147, 1233–1247. doi: 10.1016/j.cell.2011.10.043
Hatziapostolou, M., Polytarchou, C., and Iliopoulos, D. (2013). miRNAs link metabolic reprogramming to oncogenesis. Trends Endocrinol. Metab. 24, 361–373. doi: 10.1016/j.tem.2013.03.002
He, J., Jing, Y., Li, W., Qian, X., Xu, Q., Li, F. S., et al. (2013). Roles and mechanism of miR-199a and miR-125b in tumor angiogenesis. PLoS ONE 8:e56647. doi: 10.1371/journal.pone.0056647
Höckel, M., and Vaupel, P. (2001). Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J. Natl. Cancer Inst. 93, 266–276. doi: 10.1093/jnci/93.4.266
Hsu, S. H., Wang, B., Kota, J., Yu, J., Costinean, S., Kutay, H., et al. (2012). Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Invest. 122, 2871–2883. doi: 10.1172/JCI63539
Hu, Y. W., Yang, J. Y., Ma, X., Chen, Z. P., Hu, Y. R., Zhao, J. Y., et al. (2014). A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. J. Lipid Res. 55, 681–697. doi: 10.1194/jlr.M044669
Huang, C., and Freter, C. (2015). Lipid metabolism, apoptosis and cancer therapy. Int. J. Mol. Sci. 16, 924–949. doi: 10.3390/ijms16010924
Huang, T. H., Wu, F., Loeb, G. B., Hsu, R., Heidersbach, A., Brincat, A., et al. (2009). Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J. Biol. Chem. 284, 18515–18524. doi: 10.1074/jbc.M109.006676
Huang, X., and Zuo, J. (2014). Emerging roles of miR-210 and other non-coding RNAs in the hypoxic response. Acta Biochim. Biophys. Sin. (Shanghai). 46, 220–232. doi: 10.1093/abbs/gmt141
Hube, F., Guo, J., Chooniedass-Kothari, S., Cooper, C., Hamedani, M. K., Dibrov, A. A., et al. (2006). Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 25, 418–428. doi: 10.1089/dna.2006.25.418
Hung, C. L., Wang, L. Y., Yu, Y. L., Chen, H. W., Srivastava, S., Petrovics, G., et al. (2014). A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. U.S.A. 111, 18697–18702. doi: 10.1073/pnas.1415669112
Iorio, M. V., and Croce, C. M. (2012). MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 4, 143–159. doi: 10.1002/emmm.201100209
Isobe, T., Hisamori, S., Hogan, D. J., Zabala, M., Hendrickson, D. G., Dalerba, P., et al. (2014). miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway. Elife 3:e01977. doi: 10.7554/eLife.01977
Isozaki, Y., Hoshino, I., Nohata, N., Kinoshita, T., Akutsu, Y., Hanari, N., et al. (2012). Identification of novel molecular targets regulated by tumor suppressive miR-375 induced by histone acetylation in esophageal squamous cell carcinoma. Int. J. Oncol. 41, 985–994. doi: 10.3892/ijo.2012.1537
Jalava, S. E., Urbanucci, A., Latonen, L., Waltering, K. K., Sahu, B., Jänne, O. A., et al. (2012). Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene 31, 4460–4471. doi: 10.1038/onc.2011.624
Jiang, S., Zhang, L. F., Zhang, H. W., Hu, S., Lu, M. H., Liang, S., et al. (2012). A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 31, 1985–1998. doi: 10.1038/emboj.2012.45
Jing, Z., Han, W., Sui, X., Xie, J., and Pan, H. (2015). Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 356, 332–338. doi: 10.1016/j.canlet.2014.09.039
Jones, R. G., and Thompson, C. B. (2009). Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 23, 537–548. doi: 10.1101/gad.1756509
Kamphorst, J. J., Cross, J. R., Fan, J., De Stanchina, E., Mathew, R., White, E. P., et al. (2013). Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. U.S.A. 110, 8882–8887. doi: 10.1073/pnas.1307237110
Kefas, B., Comeau, L., Erdle, N., Montgomery, E., Amos, S., and Purow, B. (2010). Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro Oncol. 12, 1102–1112. doi: 10.1093/neuonc/noq080
Khaitan, D., Dinger, M. E., Mazar, J., Crawford, J., Smith, M. A., Mattick, J. S., et al. (2011). The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 71, 3852–3862. doi: 10.1158/0008-5472.CAN-10-4460
Kim, H. R., Roe, J. S., Lee, J. E., Cho, E. J., and Youn, H. D. (2013). p53 regulates glucose metabolism by miR-34a. Biochem. Biophys. Res. Commun. 437, 225–231. doi: 10.1016/j.bbrc.2013.06.043
Kim, J. W., and Dang, C. V. (2006). Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 66, 8927–8930. doi: 10.1158/0008-5472.CAN-06-1501
Kinoshita, T., Nohata, N., Yoshino, H., Hanazawa, T., Kikkawa, N., Fujimura, L., et al. (2012). Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in maxillary sinus squamous cell carcinoma. Int. J. Oncol. 40, 185–193. doi: 10.3892/ijo.2011.1196
Korkmaz, G., Le Sage, C., Tekirdag, K. A., Agami, R., and Gozuacik, D. (2012). miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy 8, 165–176. doi: 10.4161/auto.8.2.18351
Kota, J., Chivukula, R. R., O'donnell, K. A., Wentzel, E. A., Montgomery, C. L., Hwang, H. W., et al. (2009). Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137, 1005–1017. doi: 10.1016/j.cell.2009.04.021
Kovaleva, V., Mora, R., Park, Y. J., Plass, C., Chiramel, A. I., Bartenschlager, R., et al. (2012). miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 72, 1763–1772. doi: 10.1158/0008-5472.CAN-11-3671
Kroemer, G., and Pouyssegur, J. (2008). Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell 13, 472–482. doi: 10.1016/j.ccr.2008.05.005
Krycer, J. R., Sharpe, L. J., Luu, W., and Brown, A. J. (2010). The Akt-SREBP nexus: cell signaling meets lipid metabolism. Trends Endocrinol. Metab. 21, 268–276. doi: 10.1016/j.tem.2010.01.001
Kumar, M., Lu, Z., Takwi, A. A., Chen, W., Callander, N. S., Ramos, K. S., et al. (2011). Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 30, 843–853. doi: 10.1038/onc.2010.457
Kwon, C., Tak, H., Rho, M., Chang, H. R., Kim, Y. H., Kim, K. T., et al. (2014). Detection of PIWI and piRNAs in the mitochondria of mammalian cancer cells. Biochem. Biophys. Res. Commun. 446, 218–223. doi: 10.1016/j.bbrc.2014.02.112
Lan, S. H., Wu, S. Y., Zuchini, R., Lin, X. Z., Su, I. J., Tsai, T. F., et al. (2014). Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology 59, 505–517. doi: 10.1002/hep.26659
Leung, A. K., and Sharp, P. A. (2010). MicroRNA functions in stress responses. Mol. Cell 40, 205–215. doi: 10.1016/j.molcel.2010.09.027
Li, B., Chen, P., Qu, J., Shi, L., Zhuang, W., Fu, J., et al. (2014). Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J. Biol. Chem. 289, 29365–29375. doi: 10.1074/jbc.M114.572693
Li, H. J., Li, X., Pang, H., Pan, J. J., Xie, X. J., and Chen, W. (2015). Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn. J. Clin. Oncol. 45, 1055–1063. doi: 10.1093/jjco/hyv132
Li, L., Li, C., Wang, S., Wang, Z., Jiang, J., Wang, W., et al. (2016). Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 76, 1770–1780. doi: 10.1158/0008-5472.can-15-1625
Li, L., Yuan, L., Luo, J., Gao, J., Guo, J., and Xie, X. (2013). MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin. Exp. Med. 13, 109–117. doi: 10.1007/s10238-012-0186-5
Li, P., Jiao, J., Gao, G., and Prabhakar, B. S. (2012). Control of mitochondrial activity by miRNAs. J. Cell. Biochem. 113, 1104–1110. doi: 10.1002/jcb.24004
Li, P., Ruan, X., Yang, L., Kiesewetter, K., Zhao, Y., Luo, H., et al. (2015a). A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 21, 455–467. doi: 10.1016/j.cmet.2015.02.004
Li, P., Shan, J. X., Chen, X. H., Zhang, D., Su, L. P., Huang, X. Y., et al. (2015b). Epigenetic silencing of microRNA-149 in cancer-associated fibroblasts mediates prostaglandin E2/interleukin-6 signaling in the tumor microenvironment. Cell Res. 25, 588–603. doi: 10.1038/cr.2015.51
Li, X., Chen, Y. T., Josson, S., Mukhopadhyay, N. K., Kim, J., Freeman, M. R., et al. (2013). MicroRNA-185 and 342 inhibit tumorigenicity and induce apoptosis through blockade of the SREBP metabolic pathway in prostate cancer cells. PLoS ONE 8:e70987. doi: 10.1371/journal.pone.0070987
Li, X., Deng, S. J., Zhu, S., Jin, Y., Cui, S. P., Chen, J. Y., et al. (2016). Hypoxia-induced lncRNA-NUTF2P3-001 contributes to tumorigenesis of pancreatic cancer by derepressing the miR-3923/KRAS pathway. Oncotarget 7, 6000–6014. doi: 10.18632/oncotarget.6830
Li, Z., Li, X., Wu, S., Xue, M., and Chen, W. (2014). Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 105, 951–955. doi: 10.1111/cas.12461
Li, Z., Yu, X., and Shen, J. (2016). ANRIL: a pivotal tumor suppressor long non-coding RNA in human cancers. Tumour Biol. 37, 5657–5661. doi: 10.1007/s13277-016-4808-5
Li, Z., and Zhang, H. (2016). Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. 73, 377–392. doi: 10.1007/s00018-015-2070-4
Liang, W. C., Fu, W. M., Wong, C. W., Wang, Y., Wang, W. M., Hu, G. X., et al. (2015). The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 6, 22513–22525. doi: 10.18632/oncotarget.4154
Lin, A., Li, C., Xing, Z., Hu, Q., Liang, K., Han, L., et al. (2016). The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat. Cell Biol. 18, 213–224. doi: 10.1038/ncb3295
Lin, C. W., Chang, Y. L., Chang, Y. C., Lin, J. C., Chen, C. C., Pan, S. H., et al. (2013). MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat. Commun. 4, 1877. doi: 10.1038/ncomms2876
Lindow, M., and Kauppinen, S. (2012). Discovering the first microRNA-targeted drug. J. Cell Biol. 199, 407–412. doi: 10.1083/jcb.201208082
Liu, S., Sheng, L., Miao, H., Saunders, T. L., Macdougald, O. A., Koenig, R. J., et al. (2014). SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J. Biol. Chem. 289, 13000–13009. doi: 10.1074/jbc.M114.564658
Liu, W., Le, A., Hancock, C., Lane, A. N., Dang, C. V., Fan, T. W., et al. (2012). Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc. Natl. Acad. Sci. U.S.A. 109, 8983–8988. doi: 10.1073/pnas.1203244109
Liu, X., Wang, C., Chen, Z., Jin, Y., Wang, Y., Kolokythas, A., et al. (2011). MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem. J. 440, 23–31. doi: 10.1042/BJ20111006
Lozy, F., and Karantza, V. (2012). Autophagy and cancer cell metabolism. Semin. Cell Dev. Biol. 23, 395–401. doi: 10.1016/j.semcdb.2012.01.005
Lu, K. H., Li, W., Liu, X. H., Sun, M., Zhang, M. L., Wu, W. Q., et al. (2013). Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 13:461. doi: 10.1186/1471-2407-13-461
Lui, W. O., Pourmand, N., Patterson, B. K., and Fire, A. (2007). Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 67, 6031–6043. doi: 10.1158/0008-5472.CAN-06-0561
Luo, W., and Semenza, G. L. (2011). Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget 2, 551–556. doi: 10.18632/oncotarget.299
Ma, X., Li, C., Sun, L., Huang, D., Li, T., He, X., et al. (2014). Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat. Commun. 5:5212. doi: 10.1038/ncomms6212
Marion-Letellier, R., Savoye, G., and Ghosh, S. (2015). Fatty acids, eicosanoids and PPAR gamma. Eur. J. Pharmacol. doi: 10.1016/j.ejphar.2015.11.004. [Epub ahead of print].
Martello, G., Rosato, A., Ferrari, F., Manfrin, A., Cordenonsi, M., Dupont, S., et al. (2010). A MicroRNA targeting dicer for metastasis control. Cell 141, 1195–1207. doi: 10.1016/j.cell.2010.05.017
Martinez-Outschoorn, U. E., Pavlides, S., Howell, A., Pestell, R. G., Tanowitz, H. B., Sotgia, F., et al. (2011). Stromal-epithelial metabolic coupling in cancer: integrating autophagy and metabolism in the tumor microenvironment. Int. J. Biochem. Cell Biol. 43, 1045–1051. doi: 10.1016/j.biocel.2011.01.023
Masson, N., and Ratcliffe, P. J. (2014). Hypoxia signaling pathways in cancer metabolism: the importance of co-selecting interconnected physiological pathways. Cancer Metab. 2:3. doi: 10.1186/2049-3002-2-3
Mateescu, B., Batista, L., Cardon, M., Gruosso, T., De Feraudy, Y., Mariani, O., et al. (2011). miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 17, 1627–1635. doi: 10.1038/nm.2512
Mathew, L. K., Skuli, N., Mucaj, V., Lee, S. S., Zinn, P. O., Sathyan, P., et al. (2014). miR-218 opposes a critical RTK-HIF pathway in mesenchymal glioblastoma. Proc. Natl. Acad. Sci. U.S.A. 111, 291–296. doi: 10.1073/pnas.1314341111
Mathew, R., and White, E. (2011). Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr. Opin. Genet. Dev. 21, 113–119. doi: 10.1016/j.gde.2010.12.008
Mathupala, S. P., Ko, Y. H., and Pedersen, P. L. (2009). Hexokinase-2 bound to mitochondria: cancer's stygian link to the “Warburg Effect” and a pivotal target for effective therapy. Semin. Cancer Biol. 19, 17–24. doi: 10.1016/j.semcancer.2008.11.006
Mattes, R. D. (2005). Fat taste and lipid metabolism in humans. Physiol. Behav. 86, 691–697. doi: 10.1016/j.physbeh.2005.08.058
Mazar, J., Zhao, W., Khalil, A. M., Lee, B., Shelley, J., Govindarajan, S. S., et al. (2014). The functional characterization of long noncoding RNA SPRY4-IT1 in human melanoma cells. Oncotarget 5, 8959–8969. doi: 10.18632/oncotarget.1863
Mazurek, S. (2011). Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int. J. Biochem. Cell Biol. 43, 969–980. doi: 10.1016/j.biocel.2010.02.005
Menendez, J. A., and Lupu, R. (2007). Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777. doi: 10.1038/nrc2222
Michael, M. Z., O'Connor, S. M., Van Holst Pellekaan, N. G., Young, G. P., and James, R. J. (2003). Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 1, 882–891.
Mikhaylova, O., Stratton, Y., Hall, D., Kellner, E., Ehmer, B., Drew, A. F., et al. (2012). VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell 21, 532–546. doi: 10.1016/j.ccr.2012.02.019
Mima, K., Nishihara, R., Nowak, J. A., Kim, S. A., Song, M., Inamura, K., et al. (2016). MicroRNA MIR21 and T cells in colorectal cancer. Cancer Immunol. Res. 4, 33–40. doi: 10.1158/2326-6066.CIR-15-0084
Molon, B., Calì, B., and Viola, A. (2016). T cells and cancer: how metabolism shapes immunity. Front. Immunol. 7:20. doi: 10.3389/fimmu.2016.00020
Muñoz-Pinedo, C., El Mjiyad, N., and Ricci, J. E. (2012). Cancer metabolism: current perspectives and future directions. Cell Death Dis. 3, e248. doi: 10.1038/cddis.2011.123
Nagel, R., Le Sage, C., Diosdado, B., Van Der Waal, M., Oude Vrielink, J. A., Bolijn, A., et al. (2008). Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 68, 5795–5802. doi: 10.1158/0008-5472.CAN-08-0951
Newsholme, E. A., Crabtree, B., and Ardawi, M. S. (1985). The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci. Rep. 5, 393–400. doi: 10.1007/BF01116556
Nieman, K. M., Kenny, H. A., Penicka, C. V., Ladanyi, A., Buell-Gutbrod, R., Zillhardt, M. R., et al. (2011). Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 17, 1498–1503. doi: 10.1038/nm.2492
O'connell, R. M., Rao, D. S., and Baltimore, D. (2012). microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 30, 295–312. doi: 10.1146/annurev-immunol-020711-075013
Ota, T., Suzuki, Y., Nishikawa, T., Otsuki, T., Sugiyama, T., Irie, R., et al. (2004). Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36, 40–45. doi: 10.1038/ng1285
Park, J. K., Kogure, T., Nuovo, G. J., Jiang, J., He, L., Kim, J. H., et al. (2011). miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 71, 7608–7616. doi: 10.1158/0008-5472.CAN-11-1144
Peschiaroli, A., Giacobbe, A., Formosa, A., Markert, E. K., Bongiorno-Borbone, L., Levine, A. J., et al. (2013). miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene 32, 797–802. doi: 10.1038/onc.2012.100
Phan, L. M., Yeung, S. C., and Lee, M. H. (2014). Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol. Med. 11, 1–19. doi: 10.7497/j.issn.2095-3941.2014.01.001
Polyak, K., and Weinberg, R. A. (2009). Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer 9, 265–273. doi: 10.1038/nrc2620
Pullen, T. J., Da Silva Xavier, G., Kelsey, G., and Rutter, G. A. (2011). miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol. Cell. Biol. 31, 3182–3194. doi: 10.1128/MCB.01433-10
Rajala, M. W., and Scherer, P. E. (2003). Minireview: The adipocyte–at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology 144, 3765–3773. doi: 10.1210/en.2003-0580
Ramón, L. A., Braza-Boïls, A., Gilabert-Estellés, J., Gilabert, J., España, F., Chirivella, M., et al. (2011). microRNAs expression in endometriosis and their relation to angiogenic factors. Hum. Reprod. 26, 1082–1090. doi: 10.1093/humrep/der025
Rapicavoli, N. A., Qu, K., Zhang, J., Mikhail, M., Laberge, R. M., and Chang, H. Y. (2013). A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2:e00762. doi: 10.7554/eLife.00762
Redis, R. S., Vela, L. E., Lu, W., Ferreira De Oliveira, J., Ivan, C., Rodriguez-Aguayo, C., et al. (2016). Allele-specific reprogramming of cancer metabolism by the long Non-coding RNA CCAT2. Mol. Cell. 61, 520–534. doi: 10.1016/j.molcel.2016.02.006
Reyes, R. K., Motiwala, T., and Jacob, S. T. (2014). Regulation of glucose metabolism in hepatocarcinogenesis by microRNAs. Gene Expr. 16, 85–92. doi: 10.3727/105221614X13919976902093
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi: 10.1146/annurev-biochem-051410-092902
Rivadeneira, D. B., Caino, M. C., Seo, J. H., Angelin, A., Wallace, D. C., Languino, L. R., et al. (2015). Survivin promotes oxidative phosphorylation, subcellular mitochondrial repositioning, and tumor cell invasion. Sci. Signal. 8, ra80. doi: 10.1126/scisignal.aab1624
Robey, R. B., and Hay, N. (2006). Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene 25, 4683–4696. doi: 10.1038/sj.onc.1209595
Ross, R. J., Weiner, M. M., and Lin, H. (2014). PIWI proteins and PIWI-interacting RNAs in the soma. Nature 505, 353–359. doi: 10.1038/nature12987
Rupaimoole, R., Lee, J., Haemmerle, M., Ling, H., Previs, R. A., Pradeep, S., et al. (2015). Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 13, 2395–2402. doi: 10.1016/j.celrep.2015.11.047
Ryu, H. S., Park, S. Y., Ma, D., Zhang, J., and Lee, W. (2011). The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS ONE 6:e17343. doi: 10.1371/annotation/2faafaa7-e359-4711-af5b-3597c705388d
Santos, C. R., and Schulze, A. (2012). Lipid metabolism in cancer. FEBS J. 279, 2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x
Semenza, G. L. (2010). HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 20, 51–56. doi: 10.1016/j.gde.2009.10.009
Semenza, G. L., Artemov, D., Bedi, A., Bhujwalla, Z., Chiles, K., Feldser, D., et al. (2001). The metabolism of tumours: 70 years later. Novartis Found. Symp. 240, 251–260; discussion 260–254. doi: 10.1002/0470868716.ch17
Seok, J. K., Lee, S. H., Kim, M. J., and Lee, Y. M. (2014). MicroRNA-382 induced by HIF-1alpha is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 42, 8062–8072. doi: 10.1093/nar/gku515
Serguienko, A., Grad, I., Wennerstrøm, A. B., Meza-Zepeda, L. A., Thiede, B., Stratford, E. W., et al. (2015). Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA. Oncotarget 6, 2451–2465. doi: 10.18632/oncotarget.3235
Shan, Y., Li, X., You, B., Shi, S., Zhang, Q., and You, Y. (2015). MicroRNA-338 inhibits migration and proliferation by targeting hypoxia-induced factor 1alpha in nasopharyngeal carcinoma. Oncol. Rep. 34, 1943–1952. doi: 10.3892/or.2015.4195
Shen, W. W., Zeng, Z., Zhu, W. X., and Fu, G. H. (2013). MiR-142-3p functions as a tumor suppressor by targeting CD133, ABCG2, and Lgr5 in colon cancer cells. J. Mol. Med. (Berl). 91, 989–1000. doi: 10.1007/s00109-013-1037-x
Shirasaki, T., Honda, M., Shimakami, T., Horii, R., Yamashita, T., Sakai, Y., et al. (2013). MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J. Virol. 87, 5270–5286. doi: 10.1128/JVI.03022-12
Siomi, M. C., Sato, K., Pezic, D., and Aravin, A. A. (2011). PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258. doi: 10.1038/nrm3089
Smolle, M. A., Bullock, M. D., Ling, H., Pichler, M., and Haybaeck, J. (2015). Long Non-Coding RNAs in Endometrial Carcinoma. Int. J. Mol. Sci. 16, 26463–26472. doi: 10.3390/ijms161125962
Soon, P., and Kiaris, H. (2013). MicroRNAs in the tumour microenvironment: big role for small players. Endocr. Relat. Cancer 20, R257–R267. doi: 10.1530/ERC-13-0119
Srivastava, S. K., Bhardwaj, A., Singh, S., Arora, S., Wang, B., Grizzle, W. E., et al. (2011). MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis 32, 1832–1839. doi: 10.1093/carcin/bgr223
Su, H., Yang, J. R., Xu, T., Huang, J., Xu, L., Yuan, Y., et al. (2009). MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 69, 1135–1142. doi: 10.1158/0008-5472.CAN-08-2886
Sullivan, L. B., and Chandel, N. S. (2014). Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2:17. doi: 10.1186/2049-3002-2-17
Sun, B., Li, J., Shao, D., Pan, Y., Chen, Y., Li, S., et al. (2015). Adipose tissue-secreted miR-27a promotes liver cancer by targeting FOXO1 in obese individuals. Onco. Targets Ther. 8, 735–744. doi: 10.2147/OTT.S80945
Sun, H., Wang, G., Peng, Y., Zeng, Y., Zhu, Q. N., Li, T. L., et al. (2015). H19 lncRNA mediates 17beta-estradiol-induced cell proliferation in MCF-7 breast cancer cells. Oncol. Rep. 33, 3045–3052. doi: 10.3892/or.2015.3899
Sun, Y., Oravecz-Wilson, K., Mathewson, N., Wang, Y., Mceachin, R., Liu, C., et al. (2015). Mature T cell responses are controlled by microRNA-142. J. Clin. Invest. 125, 2825–2840. doi: 10.1172/JCI78753
Sun, Y., Zhao, X., Zhou, Y., and Hu, Y. (2012). miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol. Rep. 28, 1346–1352. doi: 10.3892/or.2012.1958
Sun, Y. W., Chen, Y. F., Li, J., Huo, Y. M., Liu, D. J., Hua, R., et al. (2014). A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1alpha in pancreatic ductal adenocarcinoma. Br. J. Cancer 111, 2131–2141. doi: 10.1038/bjc.2014.520
Taguchi, A., Yanagisawa, K., Tanaka, M., Cao, K., Matsuyama, Y., Goto, H., et al. (2008). Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 68, 5540–5545. doi: 10.1158/0008-5472.CAN-07-6460
Thorens, B., and Mueckler, M. (2010). Glucose transporters in the 21st Century. Am. J. Physiol. Endocrinol. Metab. 298, E141–E145. doi: 10.1152/ajpendo.00712.2009
Tili, E., Michaille, J. J., Luo, Z., Volinia, S., Rassenti, L. Z., Kipps, T. J., et al. (2012). The down-regulation of miR-125b in chronic lymphocytic leukemias leads to metabolic adaptation of cells to a transformed state. Blood 120, 2631–2638. doi: 10.1182/blood-2012-03-415737
Tomasetti, M., Neuzil, J., and Dong, L. (2014). MicroRNAs as regulators of mitochondrial function: role in cancer suppression. Biochim. Biophys. Acta 1840, 1441–1453. doi: 10.1016/j.bbagen.2013.09.002
Trang, P., Wiggins, J. F., Daige, C. L., Cho, C., Omotola, M., Brown, D., et al. (2011). Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 19, 1116–1122. doi: 10.1038/mt.2011.48
Tsai, W. C., Hsu, S. D., Hsu, C. S., Lai, T. C., Chen, S. J., Shen, R., et al. (2012). MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 122, 2884–2897. doi: 10.1172/JCI63455
Vander Heiden, M. G., Cantley, L. C., and Thompson, C. B. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. doi: 10.1126/science.1160809
Vander Heiden, M. G., Lunt, S. Y., Dayton, T. L., Fiske, B. P., Israelsen, W. J., Mattaini, K. R., et al. (2011). Metabolic pathway alterations that support cell proliferation. Cold Spring Harb. Symp. Quant. Biol. 76, 325–334. doi: 10.1101/sqb.2012.76.010900
Venkataraman, S., Alimova, I., Fan, R., Harris, P., Foreman, N., and Vibhakar, R. (2010). MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS ONE 5:e10748. doi: 10.1371/journal.pone.0010748
Vidaurre, S., Fitzpatrick, C., Burzio, V. A., Briones, M., Villota, C., Villegas, J., et al. (2014). Down-regulation of the antisense mitochondrial non-coding RNAs (ncRNAs) is a unique vulnerability of cancer cells and a potential target for cancer therapy. J. Biol. Chem. 289, 27182–27198. doi: 10.1074/jbc.M114.558841
Volinia, S., Galasso, M., Costinean, S., Tagliavini, L., Gamberoni, G., Drusco, A., et al. (2010). Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 20, 589–599. doi: 10.1101/gr.098046.109
Volinia, S., Galasso, M., Sana, M. E., Wise, T. F., Palatini, J., Huebner, K., et al. (2012). Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc. Natl. Acad. Sci. U.S.A. 109, 3024–3029. doi: 10.1073/pnas.1200010109
Voorhoeve, P. M., Le Sage, C., Schrier, M., Gillis, A. J., Stoop, H., Nagel, R., et al. (2007). A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Adv. Exp. Med. Biol. 604, 17–46. doi: 10.1007/978-0-387-69116-9_2
Wan, F., Qin, X., Zhang, G., Lu, X., Zhu, Y., Zhang, H., et al. (2015). Oxidized low-density lipoprotein is associated with advanced-stage prostate cancer. Tumour Biol. 36, 3573–3582. doi: 10.1007/s13277-014-2994-6
Wang, B., Hsu, S. H., Frankel, W., Ghoshal, K., and Jacob, S. T. (2012). Stat3-mediated activation of microRNA-23a suppresses gluconeogenesis in hepatocellular carcinoma by down-regulating glucose-6-phosphatase and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha. Hepatology 56, 186–197. doi: 10.1002/hep.25632
Wang, B., Sun, F., Dong, N., Sun, Z., Diao, Y., Zheng, C., et al. (2014). MicroRNA-7 directly targets insulin-like growth factor 1 receptor to inhibit cellular growth and glucose metabolism in gliomas. Diagn. Pathol. 9, 211. doi: 10.1186/s13000-014-0211-y
Wang, J., Wang, H., Liu, A., Fang, C., Hao, J., and Wang, Z. (2015). Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget 6, 19456–19468. doi: 10.18632/oncotarget.3318
Wang, P., Zhang, J., Zhang, L., Zhu, Z., Fan, J., Chen, L., et al. (2013). MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology 145, 1133–1143. doi: 10.1053/j.gastro.2013.07.048
Wang, W., Zhang, E., and Lin, C. (2015). MicroRNAs in tumor angiogenesis. Life Sci. 136, 28–35. doi: 10.1016/j.lfs.2015.06.025
Wang, X., Sun, W., Shen, W., Xia, M., Chen, C., Xiang, D., et al. (2016). Long non-coding RNA DILC represses self-renewal of liver cancer stem cells via inhibiting autocrine IL-6/STAT3 axis. J. Hepatol. 64, 1283–1294. doi: 10.1016/j.jhep.2016.01.019
Wang, Y., Liu, X., Zhang, H., Sun, L., Zhou, Y., Jin, H., et al. (2014). Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting gamma-synuclein. Neoplasia 16, 1094–1106. doi: 10.1016/j.neo.2014.10.008
Warburg, O. (1956). On the origin of cancer cells. Science 123, 309–314. doi: 10.1126/science.123.3191.309
Wei, Z., Cui, L., Mei, Z., Liu, M., and Zhang, D. (2014). miR-181a mediates metabolic shift in colon cancer cells via the PTEN/AKT pathway. FEBS Lett. 588, 1773–1779. doi: 10.1016/j.febslet.2014.03.037
Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., et al. (1999). Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124. doi: 10.1016/S0092-8674(00)80611-X
Xiang, M., Birkbak, N. J., Vafaizadeh, V., Walker, S. R., Yeh, J. E., Liu, S., et al. (2014). STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-kappaB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci. Signal. 7, ra11. doi: 10.1126/scisignal.2004497
Xu, B., Gerin, I., Miao, H., Vu-Phan, D., Johnson, C. N., Xu, R., et al. (2010). Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS ONE 5:e14199. doi: 10.1371/journal.pone.0014199
Xu, P., Li, Y., Zhang, H., Li, M., and Zhu, H. (2016). MicroRNA-340 mediates metabolic shift in oral squamous cell carcinoma by targeting glucose transporter-1. J. Oral Maxillofac. Surg. 74, 844–850. doi: 10.1016/j.joms.2015.09.038
Xu, Y., Han, S., Lei, K., Chang, X., Wang, K., Li, Z., et al. (2015). Anti-Warburg effect of rosmarinic acid via miR-155 in colorectal carcinoma cells. Eur. J. Cancer Prev. doi: 10.1097/CEJ.0000000000000205. [Epub ahead of print].
Xue, M., Li, X., Li, Z., and Chen, W. (2014). Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1alpha-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biol. 35, 6901–6912. doi: 10.1007/s13277-014-1925-x
Yamakuchi, M., Lotterman, C. D., Bao, C., Hruban, R. H., Karim, B., Mendell, J. T., et al. (2010). P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 6334–6339. doi: 10.1073/pnas.0911082107
Yamakuchi, M., Yagi, S., Ito, T., and Lowenstein, C. J. (2011). MicroRNA-22 regulates hypoxia signaling in colon cancer cells. PLoS ONE 6:e20291. doi: 10.1371/journal.pone.0020291
Yamasaki, T., Seki, N., Yoshino, H., Itesako, T., Yamada, Y., Tatarano, S., et al. (2013). Tumor-suppressive microRNA-1291 directly regulates glucose transporter 1 in renal cell carcinoma. Cancer Sci. 104, 1411–1419. doi: 10.1111/cas.12240
Yan, D., Dong, X. D., Chen, X., Yao, S., Wang, L., Wang, J., et al. (2012). Role of microRNA-182 in posterior uveal melanoma: regulation of tumor development through MITF, BCL2 and cyclin D2. PLoS ONE 7:e40967. doi: 10.1371/journal.pone.0040967
Yan, D., Ng, W. L., Zhang, X., Wang, P., Zhang, Z., Mo, Y. Y., et al. (2010). Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS ONE 5:e11397. doi: 10.1371/journal.pone.0011397
Yang, F., Huo, X. S., Yuan, S. X., Zhang, L., Zhou, W. P., Wang, F., et al. (2013). Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell 49, 1083–1096. doi: 10.1016/j.molcel.2013.01.010
Yang, F., Zhang, H., Mei, Y., and Wu, M. (2014). Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol. Cell 53, 88–100. doi: 10.1016/j.molcel.2013.11.004
Yang, X., Cheng, Y., Li, P., Tao, J., Deng, X., Zhang, X., et al. (2015). A lentiviral sponge for miRNA-21 diminishes aerobic glycolysis in bladder cancer T24 cells via the PTEN/PI3K/AKT/mTOR axis. Tumour Biol. 36, 383–391. doi: 10.1007/s13277-014-2617-2
Ye, P., Liu, J., He, F., Xu, W., and Yao, K. (2014). Hypoxia-induced deregulation of miR-126 and its regulative effect on VEGF and MMP-9 expression. Int. J. Med. Sci. 11, 17–23. doi: 10.7150/ijms.7329
Ying, L., Huang, Y., Chen, H., Wang, Y., Xia, L., Chen, Y., et al. (2013). Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol. Biosyst. 9, 407–411. doi: 10.1039/c2mb25386k
Yogev, O., Lagos, D., Enver, T., and Boshoff, C. (2014). Kaposi's sarcoma herpesvirus microRNAs induce metabolic transformation of infected cells. PLoS Pathog. 10:e1004400. doi: 10.1371/journal.ppat.1004400
Yu, Y., Yang, L., Zhao, M., Zhu, S., Kang, R., Vernon, P., et al. (2012). Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia 26, 1752–1760. doi: 10.1038/leu.2012.65
Yuan, S. X., Yang, F., Yang, Y., Tao, Q. F., Zhang, J., Huang, G., et al. (2012). Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology 56, 2231–2241. doi: 10.1002/hep.25895
Zechner, R., Zimmermann, R., Eichmann, T. O., Kohlwein, S. D., Haemmerle, G., Lass, A., et al. (2012). FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15, 279–291. doi: 10.1016/j.cmet.2011.12.018
Zha, X., Sun, Q., and Zhang, H. (2011). mTOR upregulation of glycolytic enzymes promotes tumor development. Cell Cycle 10, 1015–1016. doi: 10.4161/cc.10.7.15063
Zhang, D., Wang, Y., Shi, Z., Liu, J., Sun, P., Hou, X., et al. (2015). Metabolic reprogramming of cancer-associated fibroblasts by IDH3alpha downregulation. Cell Rep. 10, 1335–1348. doi: 10.1016/j.celrep.2015.02.006
Zhang, E. B., Kong, R., Yin, D. D., You, L. H., Sun, M., Han, L., et al. (2014). Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget 5, 2276–2292. doi: 10.18632/oncotarget.1902
Zhang, X., Ng, W. L., Wang, P., Tian, L., Werner, E., Wang, H., et al. (2012). MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFalpha. Cancer Res. 72, 4707–4713. doi: 10.1158/0008-5472.CAN-12-0639
Zhao, E., Maj, T., Kryczek, I., Li, W., Wu, K., Zhao, L., et al. (2016). Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat. Immunol. 17, 95–103. doi: 10.1038/ni.3313
Zhao, Z., Chen, C., Liu, Y., and Wu, C. (2014). 17beta-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem. Biophys. Res. Commun. 445, 388–393. doi: 10.1016/j.bbrc.2014.02.006
Zheng, J. (2012). Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol. Lett. 4, 1151–1157. doi: 10.3892/ol.2012.928
Zheng, Y., Li, S., Ding, Y., Wang, Q., Luo, H., Shi, Q., et al. (2013). The role of miR-18a in gastric cancer angiogenesis. Hepatogastroenterology 60, 1809–1813.
Zhou, J., Xu, D., Xie, H., Tang, J., Liu, R., Li, J., et al. (2015). miR-33a functions as a tumor suppressor in melanoma by targeting HIF-1alpha. Cancer Biol. Ther. 16, 846–855. doi: 10.1080/15384047.2015.1030545
Zhou, S., Liu, L., Li, H., Eilers, G., Kuang, Y., Shi, S., et al. (2014). Multipoint targeting of the PI3K/mTOR pathway in mesothelioma. Br. J. Cancer 110, 2479–2488. doi: 10.1038/bjc.2014.220
Zhuang, J., Lu, Q., Shen, B., Huang, X., Shen, L., Zheng, X., et al. (2015). TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci. Rep. 5:11924. doi: 10.1038/srep11924
Keywords: metabolic reprogramming, cancer metabolism, ncRNA regulation, miRNAs
Citation: Beltrán-Anaya FO, Cedro-Tanda A, Hidalgo-Miranda A and Romero-Cordoba SL (2016) Insights into the Regulatory Role of Non-coding RNAs in Cancer Metabolism. Front. Physiol. 7:342. doi: 10.3389/fphys.2016.00342
Received: 25 May 2016; Accepted: 25 July 2016;
Published: 08 August 2016.
Edited by:
Firas H. Kobeissy, University of Florida, USAReviewed by:
Guanglong Jiang, Indiana University School of Medicine, USACopyright © 2016 Beltrán-Anaya, Cedro-Tanda, Hidalgo-Miranda and Romero-Cordoba. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alfredo Hidalgo-Miranda, YWhpZGFsZ29AaW5tZWdlbi5nb2IubXg=
Sandra L. Romero-Cordoba, c3JvbWVyb19jb3Jkb2JhQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.