- Cell and Molecular Biology, University of Pennsylvania, Philadelphia, PA, USA
The insulin/insulin-like growth factor signaling (IIS) pathway is well-established as a critical regulator of growth and metabolic homeostasis across the animal kingdom. Insulin-like peptides (ILPs), the functional analogs of mammalian insulin, were initially discovered in the silkmoth Bombyx mori and subsequently identified in many other insect species. Initial research focused on the role of insulin signaling in metabolism, cell proliferation, development, reproduction and aging. More recently however, increasing attention has been given to the role of insulin in the regulation of neuronal function and behavior. Here we review the role of insulin signaling in two specific insect behaviors: feeding and locomotion.
Introduction
In both vertebrates and invertebrates, insulin is a key metabolic hormone that modulates carbohydrate and lipid metabolism in response to an organisms' nutritional state. Dysregulation of insulin production, release, or downstream signaling leads to metabolic disease, including diabetes and obesity (Baker and Thummel, 2007; Hoffmann et al., 2013). The role of insulin in metabolic homeostasis as well as development, fertility and lifespan is well-established, whereas the function of insulin in the brain and behavior is not as well-understood (Britton et al., 2002; Ikeya et al., 2002; Rulifson et al., 2002; Broughton and Partridge, 2009). In mammals, insulin is known to act on the brain to modulate behaviors relating to reproduction, feeding, and memory (Gerozissis and Kyriaki, 2003). However, given the complexities inherent to the study of mammals, the use of simpler organisms, such as insects, provides an excellent opportunity to elucidate the role of insulin signaling in neuronal function and behavior (Teleman, 2010).
Insulin-like peptides (ILPs) were originally discovered in the silkmoth Bombyx mori and subsequently identified in migratory locusts, mosquitos, and scarab beetles among other insects (Lagueux et al., 1990; Riehle et al., 2006; Lavine et al., 2013). The study of insulin signaling in insects was significantly advanced by the identification of eight insulin-like genes in the genome of the fruit fly, Drosophila melanogaster (Brogiolo et al., 2001; Grönke et al., 2010; Colombani et al., 2012). In mammals, insulin and insulin-like growth factors signal through several different receptors whereas in Drosophila all ILPs signal through a single insulin receptor (InR) (Fernandez et al., 1995; Yamaguchi et al., 1995; Brogiolo et al., 2001). In the adult fly, three of these ILPs (2, 3, and 5), are expressed in the dorso-medial protocerebrum by two clusters of neurosecretory cells known as the Insulin-Producing Cells (IPCs). The IPCs are located in the pars intercerabalis, a brain region analogous to the mammalian hypothalamus, and their axons project to other regions in the brain and to the fly heart (Rulifson et al., 2002). Secreted ILPs can activate insulin signaling in the brain or head or enter the fly circulatory system to activate systemic insulin signaling in peripheral tissues. The regulation of ILP production and release has recently been reviewed by Nässel et al. (2013).
In Drosophila, the field of behavioral genetics began with the identification of the period gene as a critical regulator of rest:activity rhythms in the fly (Konopka and Benzer, 1971). Since then the fruit fly has been used extensively to identify genes and pathways underlying many complex behaviors. This review will focus primarily on Drosophila largely because of its simple and well-studied neuroanatomy as well as its genetically tractable nature, both of which facilitate the study of insulin's role in the brain and complex behavior.
The IPCs and Insulin Signaling Pathway Influence Feeding Behavior
Maintaining adequate energy stores is critical for animal survival and reproduction. As a result, neural and molecular mechanisms have evolved to modulate feeding behavior in response to an animal's internal physiological state as well as changes in food availability in the environment. Feeding begins with the motivation to eat, followed by a search for a suitable food source, which can be influenced by smell and taste among other factors. Feeding is terminated upon reaching a satiated state. In mammals it has long been known that insulin signaling suppresses food intake (Woods et al., 1979; Williams and Elmquist, 2012). Similarly, insulin has also been shown to play an important role at several stages of feeding behavior in insects, including Drosophila melanogaster. Studies in insects have also identified neuropeptides, many of which have mammalian orthologs, involved in the regulation of physiology and behavior, including feeding behavior (Nässel and Winther, 2010). Many of these other molecules and mechanisms regulating feeding behavior in fruit flies are reviewed by Itskov and Ribeiro (2013). Here we will focus solely only on instances in which insulin-signaling modulates Drosophila feeding behavior.
Hunger or deprivation due to limited food availability provokes animals to acquire food. Initiation of motivated foraging is crucial for survival so animals have developed mechanisms to ensure that this response occurs under appropriate conditions. The insulin signaling pathway links the fly's internal metabolic state with the initiation of feeding behavior. In larval IPCs, constitutive activation of a downstream effector of the InR, p70/S6 kinase (dS6K), reduces foraging motivation and food acquisition (Oldham and Hafen, 2003; Wu et al., 2005a). This attenuated feeding response may be mediated by increased ILP release since pan-neuronal overexpression of ilp2 or ilp4 also reduced motivated foraging (Wu et al., 2005a). Overall these data indicate that hunger normally downregulates S6K activity in the IPCs to reduce insulin release and drive deprived animals to search for and acquire food. Starvation-induced food acquisition is also modulated by insulin signaling in mushroom body neurons known as Kenyon cells (Zhao and Campos, 2012). Inhibition of insulin signaling in Kenyon cells throughout development reduces food intake following starvation. Interestingly, temporary suppression of synaptic transmission by these neurons also partially reduces food intake. Thus, insulin signaling in Kenyon cells during development may modulate the synaptic activity of these neurons to ultimately regulate food acquisition.
After assuming motivated foraging, animals must utilize their sense of smell in order to find an adequate food source. Starvation decreases the amount of time a fly takes to find food by heightening their sense of smell, or in more technical terms, by enhancing odor representation in neurons that process olfactory input, known as odorant receptor neurons (ORNs) (Root et al., 2011). Enhancement of odor representation is facilitated by the expression of short neuropeptide F (sNPF) and its receptor (sNPFR) in specific ORNs (Carlsson et al., 2010; Root et al., 2011). Both sNPF and its mammalian orthologue, Neuropeptide Y (NPY), are known to promote feeding behavior (Lee et al., 2004; Kageyama et al., 2012). Intriguingly, Root and colleagues found that insulin interacts with the sNPF pathway by acting as a satiety signal to decrease sNPFR expression in ORNs and in turn decrease motivated feeding (Root et al., 2011). Hence during starvation when insulin signaling is low, sNPFR levels are high to mediate increased ORN sensitivity to odors and to encourage food acquisition.
The gustatory system also plays a role in feeding behavior primarily by influencing food choice. When determining what to eat, animals must assess both the nutritional content and palatability of a food source. A hungry fly initially decides what to eat based on taste; choosing the sweeter option over the more calorie dense option. However, over time, this preference shifts toward the more calorie rich option (Stafford et al., 2012). This change in preference is at least partially mediated by insulin. Ilp2 and ilp3 mutants and decreased insulin signaling throughout the brain all show an increased initial preference for the more caloric food source instead of the sweeter option (Stafford et al., 2012). This suggests that insulin contributes to calorie sensing and food source preference by acting directly on neurons in feeding circuits.
In the absence of a preferred food source, animals will feed on less palatable food sources. Drosophila larvae prefer to feed on rich liquid food as opposed to solid food. This preference is partially mediated by insulin signaling in cells that promote food intake through signaling downstream of the receptor for NPF, a distinct peptide from sNPF but also a fly orthologue of NPY. Downregulating insulin signaling in NPFR neurons causes fed larvae to be hungry and also increases their consumption of the less preferred solid food (Wu et al., 2005b). Conversely, upregulation of the insulin pathway in NPFR neurons of deprived animals elicits attenuated feeding of solid food (Wu et al., 2005b). In states of starvation, animals adapt by undertaking risky behaviors including searching for food under less than optimal conditions and consuming normally aversive noxious food. Under unfavorable conditions like cold temperature, the NPF/IIS signaling cascade enables starving Drosophila larvae to adapt and continue searching for food in order to survive (Lingo et al., 2007). This pathway also regulates risk-sensitive food acquisition with respect to noxious or bitter compounds. Overexpression of NPFR as well as down regulation of insulin signaling in NPFR neurons both increase consumption of noxious or bitter compounds in non-deprived larvae (Wu et al., 2005b). These results indicate that the NPF and ILP signaling mediate foraging responses under adverse conditions presumably to promote survival.
After determining what to eat, animals must also determine how much they will eat. Under very poor nutrient conditions flies will compensate by eating more. Interestingly this change in feeding behavior does not occur in IPC ablated flies (Broughton et al., 2010). Similarly, inhibition of IPCs also results in attenuation of food intake under poor nutrient conditions (Cognigni et al., 2011). However, given that IPC ablation or inhibition presumably affects the expression levels of ILPs as well as other peptides expressed in these cells; alterations in feeding behavior may not simply be due to changes in insulin signaling. In addition to ILPs, the IPCs also express the cholecystokinin-like peptide, drosulfakinin (DSK) (Park et al., 2008; Söderberg et al., 2012). IPC specific reduction of dsk increases food consumption and alters the ability of the fly to discriminate between food choices. However, this effect is not independent of insulin because reduction of dsk also alters ilp transcript levels (Söderberg et al., 2012). Nonetheless, DSK likely modulates the amount of food a fly consumes by acting in conjunction with DILPs to convey fullness to the animal.
In addition to feeding behavior, Drosophila has also been used to investigate the mechanisms underlying physiological and behavioral responses to alcohol consumption (Devineni and Heberlein, 2013). The IPCs and insulin signaling in the brain are implicated in ethanol sensitivity and the behavioral response to this addictive drug. Inhibiting protein kinase A (PKA) activity in the IPCs increases ethanol sensitivity as does decreasing insulin receptor signaling suggesting that reduced PKA activity in IPCs results in decreased insulin production and/or release which ultimately causes increased ethanol sensitivity (Corl et al., 2005). On the other hand, exposure to ethanol can affect insulin signaling. Developmental ethanol exposure in the fly, as in many other organisms, has been shown to have detrimental effects. Flies that experienced developmental ethanol exposure are less viable, developmentally delayed, smaller in body size, and respond differently to alcohol as adults. These phenotypes are all due to reduced insulin signaling in exposed animals (McClure et al., 2011).
Some of the mechanisms involved in the regulation of feeding behavior discussed above have also been found to play a role in feeding behavior regulation in other insects not discussed here such as the African malaria mosquito Anopheles gambia and honey bees (Arsic and Guerin, 2008; Wang et al., 2010). Generally insulin conveys the organism's nutritional state, whether starved or fed, and accordingly drives feeding behavior in response to that state.
Effects of Insulin Signaling on Locomotor Activity
Many insects, including Drosophila, are crepuscular; they are primarily active at twilight or dawn and dusk. An internal clock entrained to the external environment largely through light and temperature cues, tightly regulates the timing of locomotor activity [Reviewed in Allada and Chung (2010)]. This well-conserved molecular oscillator is comprised of interlocked transcriptional/translational feedback loops, in which clock proteins negatively regulate their own transcription to create an endogenous rhythm of roughly 24 h [Reviewed by Zheng and Sehgal (2012)]. Briefly, in Drosophila, the core molecular oscillator consists of two transcription factors, CLOCK (CLK) and CYCLE (CYC), which drive the expression of the clock genes period (per) and timeless (tim). Eventually PER and TIM proteins re-enter the nucleus and inhibit the activity of CLK/CYC, thus preventing their own transcription. Ultimately the inhibition of CLK/CYC is relieved following the degradation of PER and TIM, allowing the cycle to begin again.
The anatomical and functional organization of the roughly 150 clock neurons in each hemisphere of the adult fly brain is well-established (Allada and Chung, 2010). Clock neurons located within the ventral lateral regions of the brain, known as the small ventral lateral neurons (sLNvs), are sufficient for driving locomotor rhythms under conditions where no external cues are present (constant darkness and temperature). The sLNvs produce the neuropeptide pigment dispersing factor (PDF) which rhythmically accumulates in the dorsal projections of the sLNvs. These axonal projections terminate near the PI, the aforementioned neuroendocrine center that encompasses the IPCs among other neurosecretory cells. The PI is implicated in the control of circadian rest:activity rhythms; however, neither the IPCs or the ILPs produced by these cells have been reported to directly regulate rest:activity rhythms (Helfrich-Forster et al., 2000). Nevertheless, there is data implicating insulin signaling in modulating the sensitivity of the circadian clock to oxidative stress, as measured in response to the redox cycling agent paraquat (Zheng et al., 2007). Activation of the insulin receptor initiates a signaling cascade that results in the retention of the forkhead transcription factor FOXO in the cytoplasm (Puig et al., 2003). Drosophila FOXO mutants and, by extension, perhaps elevated insulin signaling (which leads to the deactivation of FOXO), display increased sensitivity to oxidative stress. Thus, the metabolic state of the fly can influence the clock to affect behavioral rhythms.
In contrast to mammals, circadian regulation of the endocrine system has not been as extensively investigated in insects (Bloch et al., 2013). Neurohormones are excellent candidates for conveying temporal information from neuronal pacemakers to peripheral tissues because they are released from the brain and circulate in the blood allowing them to reach distal tissues and cells. ILPs are one such potential time-conveying neurohormone, but whether they are indeed regulated by the circadian system has yet to be definitively established in insects. In the blood feeding bug, Rhodnius prolixus, brains have been found to release ilps with a 24 h rhythm in vitro (Vafopoulou and Steel, 2002). Additionally, IPC projections in Rhodnius are described as closely associating with the axonal projections of PDF positive neurons in the circadian system, but whether these associations are indicative of functional synapses has not yet been determined (Vafopoulou and Steel, 2012). In Drosophila, microarray studies suggest the insulin signaling pathway may be regulated by the circadian system through the circadian expression of susi, a negative regulator of Phosphatidylinositol-3 kinase (PI3K) activity (Claridge-Chang et al., 2001; McDonald and Rosbash, 2001; Ueda et al., 2002; Wittwer et al., 2005). Additionally, slowpoke binding protein (SLOB) is expressed in a circadian manner in the IPCs and slob mutants have altered insulin-signaling, however a function for SLOB in behavior is unclear (Jaramillo et al., 2004; Sheldon et al., 2011).
In addition to being under circadian control, rest:activity behavior is also under the control of the homeostatic system which regulates rest duration and quality. Short sleep duration and poor sleep quality are associated with metabolic dysfunction (Spiegel et al., 2009). In Drosophila, octopamine, the insect equivalent of norepinephrine, promotes wakefulness by binding to octopamine receptors on the cell membranes of IPCs (Crocker and Sehgal, 2008; Crocker et al., 2010). Given that the major output of the IPCs is ILPs, we asked whether octopamine signaling in the IPCs promotes wakefulness by modulating the insulin signaling pathway. We found that this is not the case as increasing octopaminergic signaling in a ilp2-3 mutant background still results in decreased nighttime sleep similar to increasing octopaminergic signaling in a wildtype background (Erion et al., 2013). In addition, manipulations that either decreased or increased insulin signaling in the brain or fat body did not decrease sleep (Erion et al., 2013). These data do not rule out the possibility that sleep homeostasis is modulated by insulin signaling but does strongly suggest that it is not responsible for relaying the wake-promoting effects of octopamine.
Aspects of locomotor activity are sexually dimorphic in Drosophila. Males move more steadily compared to females as indicated by fewer activity/inactivity periods (or start/stop bouts), while the total distance traveled is similar for both sexes (Gatti et al., 2000; Belgacem and Martin, 2002). Restricted expression of the sex-determination transcription factor, transformer (tra), identified a subset of neurons in the PI, henceforth called feminizing cells (FCs), capable of feminizing the locomotor activity pattern of male flies (Gatti et al., 2000). The axonal projections of both FCs and IPCs terminate at the corpus cardiacum/corpus allatum (cc/ca) (Gatti et al., 2000; Ikeya et al., 2002). This gland synthesizes juvenile hormone (JH) which regulates many important processes in insects including metamorphosis, reproduction, and aging (Tu et al., 2005). JH and insulin have both been implicated in the sexual dimorphism of locomotor behavior. Feeding male flies an inhibitor of 3-Hydroxy-3-Methylgluaryl CoA Reductase (HMGCR), a key JH biosynthesis enzyme, feminizes their locomotor activity (Belgacem and Martin, 2002). HMGCR mutants and targeted reduction of HMGCR in the ca both abolish sexual dimorphism. Similarly, sexual dimorphism in locomotor activity is eliminated in insulin receptor mutants and flies with ablated IPCs. Interestingly, the insulin receptor is expressed in the ca and insulin receptor mutants have altered JH levels suggesting that there is a link between the insulin and JH pathways (Tatar et al., 2001; Belgacem and Martin, 2006). Furthermore, targeted reduction of the insulin receptor in the ca suppresses HMGCR expression in this tissue (Belgacem and Martin, 2007). Lastly, male mutants of takeout, a putative JH binding protein and known circadian output gene, also display feminization of their locomotor activity (Meunier et al., 2007). Thus, it appears that insulin, JH, and takeout may interact to regulate sexual dimorphism of locomotor behavior in flies; however their exact relationship remains unclear.
Conclusion
Given that behavior is so intricate, model organisms like Drosophila melanogaster as well as other insect species provide researchers with a framework to begin to teasing apart the many different pathways and molecules that contribute to behavior. In this review we have described the progress that has been made with respect to the regulation of feeding and locomotion by insulin, however many questions still remain (Figure 1). Future investigations will hopefully give us additional insight into the interactions between insulin and neural circuits responsible for regulating behavior.
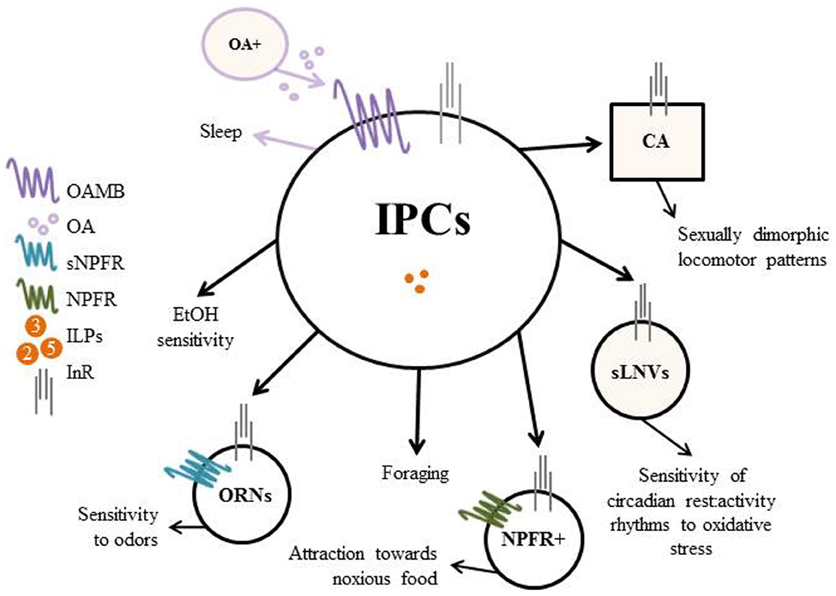
Figure 1. Regulation of behavior by the Insulin-Producing Cells (IPCs) in Drosophila. The IPCs modulate feeding and locomotor (shaded) behavior through the insulin pathway. Decreased insulin production/release from the brain IPCs results in decreased downstream insulin signaling and leads to increased ethanol (EtOH) sensitivity and motivated foraging. Decreased insulin signaling in sNPFR (short Neuropeptide F Receptor) expressing odor receptor neurons (ORNs) and in NPFR (Neuropeptide F Receptor) expressing neurons, increases the sensitivity of ORNs to odors and increases the attraction of flies towards normally aversive or noxious food sources, respectively. With respect to locomotion, octopaminergic neurons signal through the IPCs to promote wake; however, this effect is independent of insulin signaling. Insulin signaling in the corpus allatum (CA), a non-neuronal endocrine gland (indicated by square), drives sexual dimorphism of locomotor patterns. Lastly, insulin signaling in the circadian small ventrolateral neurons (sLNVs) modifies the sensitivity of rest: Lactivity rhythms to oxidative stress.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Allada, R., and Chung, B. Y. (2010). Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol. 72, 605–624. doi: 10.1146/annurev-physiol-021909-135815
Arsic, D., and Guerin, P. M. (2008). Nutrient content of diet affects the signaling activity of the insulin/target of rapamycin/p70 S6 kinase pathway in the African malaria mosquito Anopheles gambiae. J. Insect Physiol. 54, 1226–1235. doi: 10.1016/j.jinsphys.2008.06.003
Baker, K. D., and Thummel, C. S. (2007). Diabetic larvae and obese flies—emerging studies of metabolism in Drosophila. Cell Metab. 6, 257–266. doi: 10.1016/j.cmet.2007.09.002
Belgacem, Y. H., and Martin, J.-R. (2002). Neuroendocrine control of a sexually dimorphic behavior by a few neurons of the pars intercerebralis in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 99, 15154–15158. doi: 10.1073/pnas.232244199
Belgacem, Y. H., and Martin, J.-R. (2006). Disruption of insulin pathways alters trehalose level and abolishes sexual dimorphism in locomotor activity in Drosophila. J. Neurobiol. 66, 19–32. doi: 10.1002/neu.20193
Belgacem, Y. H., and Martin, J.-R. (2007). Hmgcr in the corpus allatum controls sexual dimorphism of locomotor activity and body size via the insulin pathway in Drosophila. PLoS ONE 2:e187. doi: 10.1371/journal.pone.0000187
Bloch, G., Hazan, E., and Rafaeli, A. (2013). Circadian rhythms and endocrine functions in adult insects. J. Insect Physiol. 59, 56–69. doi: 10.1016/j.jinsphys.2012.10.012
Britton, J. S., Lockwood, W. K., Li, L., Cohen, S. M., and Edgar, B. A. (2002). Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2, 239–249. doi: 10.1016/S1534-5807(02)00117-X
Brogiolo, W., Stocker, H., Ikeya, T., Rintelen, F., Fernandez, R., and Hafen, E. (2001). An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213–221. doi: 10.1016/S0960-9822(01)00068-9
Broughton, S., and Partridge, L. (2009). Insulin/IGF-like signalling, the central nervous system and aging. Biochem. J. 418, 1–12. doi: 10.1042/BJ20082102
Broughton, S. J., Slack, C., Alic, N., Metaxakis, A., Bass, T. M., Driege, Y., et al. (2010). DILP-producing median neurosecretory cells in the Drosophila brain mediate the response of lifespan to nutrition. Aging Cell 9, 336–346. doi: 10.1111/j.1474-9726.2010.00558.x
Carlsson, M. A., Diesner, M., Schachtner, J., and Nässel, D. R. (2010). Multiple neuropeptides in the Drosophila antennal lobe suggest complex modulatory circuits. J. Comp. Neurol. 518, 3359–3380. doi: 10.1002/cne.22405
Claridge-Chang, A., Wijnen, H., Naef, F., Boothroyd, C., Rajewsky, N., and Young, M. W. (2001). Circadian regulation of gene expression systems in the Drosophila head. Neuron 32, 657–671. doi: 10.1016/S0896-6273(01)00515-3
Cognigni, P., Bailey, A. P., and Miguel-Aliaga, I. (2011). Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 13, 92–104. doi: 10.1016/j.cmet.2010.12.010
Colombani, J., Andersen, D. S., and Léopold, P. (2012). Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336, 582–585. doi: 10.1126/science.1216689
Corl, A. B., Rodan, A. R., and Heberlein, U. (2005). Insulin signaling in the nervous system regulates ethanol intoxication in Drosophila melanogaster. Nat. Neurosci. 8, 18–19. doi: 10.1038/nn1363
Crocker, A., and Sehgal, A. (2008). Octopamine regulates sleep in drosophila through protein kinase A-dependent mechanisms. J. Neurosci. 28, 9377–9385. doi: 10.1523/JNEUROSCI.3072-08a.2008
Crocker, A., Shahidullah, M., Levitan, I. B., and Sehgal, A. (2010). Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron 65, 670–681. doi: 10.1016/j.neuron.2010.01.032
Devineni, A. V., and Heberlein, U. (2013). The evolution of Drosophila melanogaster as a model for alcohol research. Annu. Rev. Neurosci. 36, 121–138. doi: 10.1146/annurev-neuro-062012-170256
Erion, R., Diangelo, J. R., Crocker, A., and Sehgal, A. (2013). Interaction between sleep and metabolism in Drosophila with altered octopamine signaling. J. Biol. Chem. 287, 32406–32414. doi: 10.1074/jbc.M112.360875
Fernandez, R., Tabarini, D., Azpiazu, N., Frasch, M., and Schlessinger, J. (1995). The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 14, 3373–3384.
Gatti, S., Ferveur, J. F., and Martin, J. R. (2000). Genetic identification of neurons controlling a sexually dimorphic behaviour. Curr. Biol. 10, 667–670. doi: 10.1016/S0960-9822(00)00517-0
Gerozissis, K., and Kyriaki, G. (2003). Brain insulin: regulation, mechanisms of action and functions. Cell. Mol. Neurobiol. 23, 1–25. doi: 10.1023/A:1022598900246
Grönke, S., Clarke, D.-F., Broughton, S., Andrews, T. D., and Partridge, L. (2010). Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6:e1000857. doi: 10.1371/journal.pgen.1000857
Helfrich-Forster, C., Tauber, M., Park, J. H., Muhlig-Versen, M., Schneuwly, S., and Hofbauer, A. (2000). Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J. Neurosci. 20, 3339–3353.
Hoffmann, J., Romey, R., Fink, C., and Roeder, T. (2013). Drosophila as a model to study metabolic disorders. Adv. Biochem. Eng. Biotechnol. 1–21. doi: 10.1007/10_2013_196
Ikeya, T., Galic, M., Belawat, P., Nairz, K., and Hafen, E. (2002). Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12, 1293–1300. doi: 10.1016/S0960-9822(02)01043-6
Itskov, P. M., and Ribeiro, C. (2013). The dilemmas of the gourmet fly: the molecular and neuronal mechanisms of feeding and nutrient decision making in Drosophila. Front. Neurosci. 7:12. doi: 10.3389/fnins.2013.00012
Jaramillo, A. M., Zheng, X., Zhou, Y., Amado, D. A., Sheldon, A., Sehgal, A., et al. (2004). Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila. BMC Neurosci. 5:3. doi: 10.1186/1471-2202-5-3
Kageyama, H., Takenoya, F., Hirako, S., Wada, N., Kintaka, Y., Inoue, S., et al. (2012). Neuronal circuits involving neuropeptide Y in hypothalamic arcuate nucleus-mediated feeding regulation. Neuropeptides 46, 285–289. doi: 10.1016/j.npep.2012.09.007
Konopka, R. J., and Benzer, S. (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116. doi: 10.1073/pnas.68.9.2112
Lagueux, M., Lwoff, L., Meister, M., Goltzené, F., and Hoffmann, J. A. (1990). cDNAs from neurosecretory cells of brains of Locusta migratoria (Insecta, Orthoptera) encoding a novel member of the superfamily of insulins. Eur. J. Biochem. 187, 249–254. doi: 10.1111/j.1432-1033.1990.tb15302.x
Lavine, L. C., Hahn, L. L., Warren, I. A., Garczynski, S. F., Dworkin, I., and Emlen, D. J. (2013). Cloning and characterization of an mRNA encoding an insulin receptor from the horned scarab beetle Onthophagus nigriventris (Coleoptera: Scarabaeidae). Arch. Insect Biochem. Physiol. 82, 43–57. doi: 10.1002/arch.21072
Lee, K.-S., You, K.-H., Choo, J.-K., Han, Y.-M., and Yu, K. (2004). Drosophila short neuropeptide F regulates food intake and body size. J. Biol. Chem. 279, 50781–50789. doi: 10.1074/jbc.M407842200
Lingo, P. R., Zhao, Z., and Shen, P. (2007). Co-regulation of cold-resistant food acquisition by insulin- and neuropeptide Y-like systems in Drosophila melanogaster. Neuroscience 148, 371–374. doi: 10.1016/j.neuroscience.2007.06.010
McClure, K. D., French, R. L., and Heberlein, U. (2011). A Drosophila model for fetal alcohol syndrome disorders: role for the insulin pathway. Dis. Model. Mech. 4, 335–346. doi: 10.1242/dmm.006411
McDonald, M. J., and Rosbash, M. (2001). Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107, 567–578. doi: 10.1016/S0092-8674(01)00545-1
Meunier, N., Belgacem, Y. H., and Martin, J.-R. (2007). Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J. Exp. Biol. 210, 1424–1434. doi: 10.1242/jeb.02755
Nässel, D. R., Kubrak, O. I., Liu, Y., Luo, J., and Lushchak, O. V. (2013). Factors that regulate insulin producing cells and their output in Drosophila. Front. Physiol. 4:252. doi: 10.3389/fphys.2013.00252
Nässel, D. R., and Winther, A. M. E. (2010). Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92, 42–104. doi: 10.1016/j.pneurobio.2010.04.010
Oldham, S., and Hafen, E. (2003). Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13, 79–85. doi: 10.1016/S0962-8924(02)00042-9
Park, D., Veenstra, J. A., Park, J. H., and Taghert, P. H. (2008). Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS ONE 3:e1896. doi: 10.1371/journal.pone.0001896
Puig, O., Marr, M. T., Ruhf, M. L., and Tjian, R. (2003). Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006–2020. doi: 10.1101/gad.1098703
Riehle, M. A., Fan, Y., Cao, C., and Brown, M. R. (2006). Molecular characterization of insulin-like peptides in the yellow fever mosquito, Aedes aegypti: expression, cellular localization, and phylogeny. Peptides 27, 2547–2560. doi: 10.1016/j.peptides.2006.07.016
Root, C. M., Ko, K. I., Jafari, A., and Wang, J. W. (2011). Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 145, 133–144. doi: 10.1016/j.cell.2011.02.008
Rulifson, E. J., Kim, S. K., and Nusse, R. (2002). Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296, 1118–1120. doi: 10.1126/science.1070058
Sheldon, A. L., Zhang, J., Fei, H., and Levitan, I. B. (2011). SLOB, a SLOWPOKE channel binding protein, regulates insulin pathway signaling and metabolism in Drosophila. PLoS ONE 6:e23343. doi: 10.1371/journal.pone.0023343
Söderberg, J. A. E., Carlsson, M. A., and Nässel, D. R. (2012). Insulin-producing cells in the Drosophila brain also express satiety-inducing cholecystokinin-like peptide, drosulfakinin. Front. Endocrinol. (Lausanne). 3:109. doi: 10.3389/fendo.2012.00109
Spiegel, K., Tasali, E., Leproult, R., and Van Cauter, E. (2009). Effects of poor and short sleep on glucose metabolism and obesity risk. Nat. Rev. Endocrinol. 5, 253–261. doi: 10.1038/nrendo.2009.23
Stafford, J. W., Lynd, K. M., Jung, A. Y., and Gordon, M. D. (2012). Integration of taste and calorie sensing in Drosophila. J. Neurosci. 32, 14767–14774. doi: 10.1523/JNEUROSCI.1887-12.2012
Tatar, M., Kopelman, A., Epstein, D., Tu, M. P., Yin, C. M., and Garofalo, R. S. (2001). A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292, 107–110. doi: 10.1126/science.1057987
Teleman, A. A. (2010). Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 425, 13–26. doi: 10.1042/BJ20091181
Tu, M.-P., Yin, C.-M., and Tatar, M. (2005). Mutations in insulin signaling pathway alter juvenile hormone synthesis in Drosophila melanogaster. Gen. Comp. Endocrinol. 142, 347–356. doi: 10.1016/j.ygcen.2005.02.009
Ueda, H. R., Matsumoto, A., Kawamura, M., Iino, M., Tanimura, T., and Hashimoto, S. (2002). Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J. Biol. Chem. 277, 14048–14052. doi: 10.1074/jbc.C100765200
Vafopoulou, X., and Steel, C. G. H. (2002). Prothoracicotropic hormone of Rhodnius prolixus?: partial characterization and rhythmic release of neuropeptides related to Bombyx PTTH and bombyxin. Invertebr. Reprod. Dev. 42, 111–120. doi: 10.1080/07924259.2002.9652767
Vafopoulou, X., and Steel, C. G. H. (2012). Insulin-like and testis ecdysiotropin neuropeptides are regulated by the circadian timing system in the brain during larval-adult development in the insect Rhodnius prolixus (Hemiptera). Gen. Comp. Endocrinol. 179, 277–288. doi: 10.1016/j.ygcen.2012.08.018
Wang, Y., Mutti, N. S., Ihle, K. E., Siegel, A., Dolezal, A. G., Kaftanoglu, O., et al. (2010). Down-regulation of honey bee IRS gene biases behavior toward food rich in protein. PLoS Genet. 6:e1000896. doi: 10.1371/journal.pgen.1000896
Williams, K. W., and Elmquist, J. K. (2012). From neuroanatomy to behavior: central integration of peripheral signals regulating feeding behavior. Nat. Neurosci. 15, 1350–1355. doi: 10.1038/nn.3217
Wittwer, F., Jaquenoud, M., Brogiolo, W., Zarske, M., Wüstemann, P., Fernandez, R., et al. (2005). Susi, a negative regulator of Drosophila PI3-kinase. Dev. Cell 8, 817–827. doi: 10.1016/j.devcel.2005.04.002
Woods, S. C., Lotter, E. C., McKay, L. D., and Porte, D. (1979). Chronic intracerebroventricular infusion of insulin reduces food intake and body weight of baboons. Nature 282, 503–505. doi: 10.1038/282503a0
Wu, Q., Zhang, Y., Xu, J., and Shen, P. (2005a). Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 102, 13289–13294. doi: 10.1073/pnas.0501914102
Wu, Q., Zhao, Z., and Shen, P. (2005b). Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat. Neurosci. 8, 1350–1355. doi: 10.1038/nn1540
Yamaguchi, T., Fernandez, R., and Roth, R. A. (1995). Comparison of the signaling abilities of the Drosophila and human insulin receptors in mammalian cells. Biochemistry 34, 4962–4968. doi: 10.1021/bi00015a007
Zhao, X. L., and Campos, A. R. (2012). Insulin signalling in mushroom body neurons regulates feeding behaviour in Drosophila larvae. J. Exp. Biol. 215, 2696–2702. doi: 10.1242/jeb.066969
Zheng, X., and Sehgal, A. (2012). Speed control: cogs and gears that drive the circadian clock. Trends Neurosci. 35, 574–585. doi: 10.1016/j.tins.2012.05.007
Keywords: insulin signaling, Drosophila melanogaster, behavior, feeding, locomotion
Citation: Erion R and Sehgal A (2013) Regulation of insect behavior via the insulin-signaling pathway. Front. Physiol. 4:353. doi: 10.3389/fphys.2013.00353
Received: 03 October 2013; Accepted: 16 November 2013;
Published online: 03 December 2013.
Edited by:
Xanthe Vafopoulou, York University, CanadaReviewed by:
Graziano Fiorito, Stazione Zoologica Anton Dohrn, ItalyDavid B. Edelman, Bennington College, USA
Copyright © 2013 Erion and Sehgal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amita Sehgal, Cell and Molecular Biology, University of Pennsylvania, 3400 Civic Center Boulevard, Building 421, Philadelphia, PA 19104, USA e-mail:YW1pdGFAbWFpbC5tZWQudXBlbm4uZWR1
 Renske Erion
Renske Erion Amita Sehgal
Amita Sehgal