- Structural Computational Biology Group, Structural Biology and BioComputing Programme, Spanish National Cancer Research Centre, Madrid, Spain
Protein kinases play a crucial role in a plethora of significant physiological functions and a number of mutations in this superfamily have been reported in the literature to disrupt protein structure and/or function. Computational and experimental research aims to discover the mechanistic connection between mutations in protein kinases and disease with the final aim of predicting the consequences of mutations on protein function and the subsequent phenotypic alterations. In this article, we will review the possibilities and limitations of current computational methods for the prediction of the pathogenicity of mutations in the protein kinase superfamily. In particular we will focus on the problem of benchmarking the predictions with independent gold standard datasets. We will propose a pipeline for the curation of mutations automatically extracted from the literature. Since many of these mutations are not included in the databases that are commonly used to train the computational methods to predict the pathogenicity of protein kinase mutations we propose them to build a valuable gold standard dataset in the benchmarking of a number of these predictors. Finally, we will discuss how text mining approaches constitute a powerful tool for the interpretation of the consequences of mutations in the context of disease genome analysis with particular focus on cancer.
The Human Kinome
Protein kinases are a family of enzymes that catalyze the transfer of a phosphate from ATP to a serine, threonine, or tyrosine hydroxyl group in the target protein. Phosphorylation often implies enzyme activation or inhibition, alteration of interaction surfaces, and conformational changes, among the most common consequences. It is due to the importance of the processes regulated, that protein kinases generally do not act alone but rather, they form part of a finely tuned signaling cascade that is strictly controlled spatiotemporally. Therefore, protein kinases are metaphorically referred to as the metabolic switches of the cell.
Protein kinases are one of the most ubiquitous families of signaling molecules in the human cell. The total number of genes encoding kinases has been a matter of discussion in the last decade and, for instance, in Wang (1998) estimated between 1000 and 2000 different human kinase genes. With the completion of the human genome, the current estimate is that 518 genes encode protein kinases, corresponding to more than 2% of the total number of genes in the human genome (Manning et al., 2002b).
All members of the superfamily share a characteristic domain – the protein kinase domain – that confers them the ability to phosphorylate other proteins. Empirical studies suggest that the residues conforming the ATP binding site tend to be conserved and that phosphotransfer is carried out by a shared set of amino acids (Schee and Bourne, 2005; Knight et al., 2007; López et al., 2007; Kinnings and Jackson, 2009; Tanramluk et al., 2009).
In spite of these similarities, experiments in yeast models (Manning et al., 2002a; Ubersax et al., 2003) suggest that although protein kinases individually present a remarkable substrate specificity, the superfamily as a whole is very promiscuous, phosphorylating a wide range of protein substrates. This observation, may be attributed to the different domain architectures present in the protein kinase superfamily. In addition to the aforementioned protein kinase domain committed to the general function of phosphorylation, a number of modular domains are combined to, for example, confer substrate specificity, to tightly control the activity of the enzyme or anchor the kinase to the membrane (Finn et al., 2010).
These differences in terms of functionality and domain architecture can be used to classify members of the protein kinase superfamily into different categories. Indeed, there are several different classifications of kinases from the main model organisms: yeast (Hunter and Plowman, 1997), worm (Manning, 2005), fruit fly (Manning et al., 2002a), and mouse (Caenepeel et al., 2004). The reference classification in humans is KinBase (Manning et al., 2002b; Miranda-Saavedra and Barton, 2007), which has also been incorporated into UniProt (Bairoch et al., 2005), albeit with minor modifications.
Mutations in the Protein Kinase Superfamily
Due to their important regulatory function, a number of mutations in protein kinases have been associated with different human diseases (Shchemelinin et al., 2006), including cancer. For example, Greenman et al. (2007) carried out the first large scale study of the variation of 518 human kinases in 210 samples of cancer tissues and cell-lines. Moreover, other high-throughput studies (Sjöblom et al., 2006; Wood et al., 2007) also yielding interesting information about the role that variation of human protein kinases plays in cancer. For a detailed review, refer to Baudot et al. (2009).
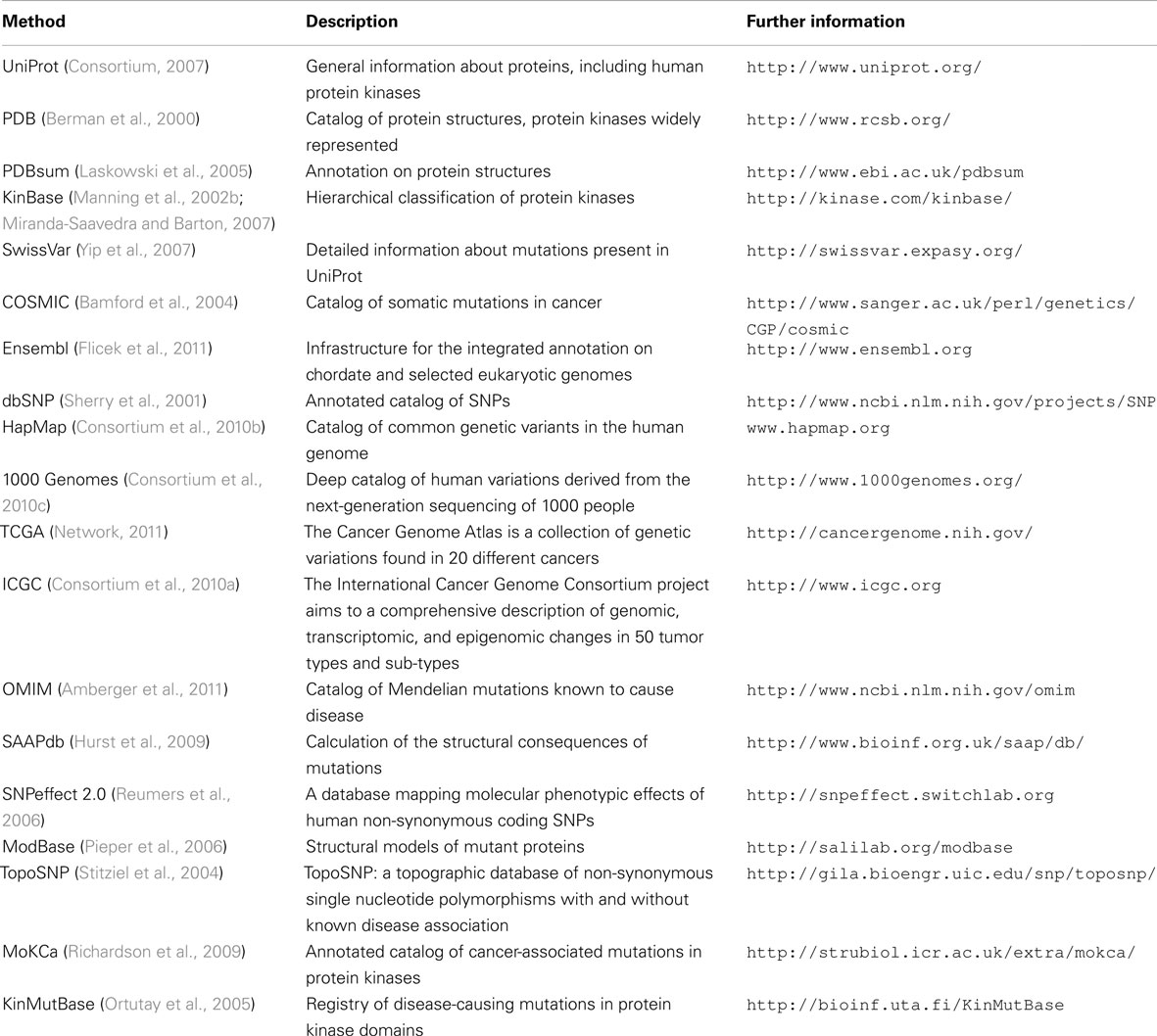
The results from these high-throughput resequencing projects is often available through research publications. However, in order to make the information more easily accessible, several efforts are devoted to compile, store, annotate, and characterize mutations, including mutations in the protein kinase superfamily. Some examples are UniProt (Yip et al., 2008), COSMIC (Bamford et al., 2004), SAAPdb (Hurst et al., 2009), MoKCa (Richardson et al., 2009), and KinMutBase (Ortutay et al., 2005). Together they constitute a powerful resource to understand disease association and the functional/structural properties of the mutations that affect human protein kinases.
Unfortunately, database curators are not able to store and annotate the vast amount of information provided by large-scale variation studies at the same pace it is generated. Mainly, because the process generally involves the manual inspection and curation of specific variation studies, which requires considerable resources. As a consequence, although growing in number, the mutations totally characterized, and well-understood only represent a small fraction of all the human variome.
Methods to Predict Pathogenic Mutations
In the section Mutations in the protein kinase superfamily we mentioned that high-throughput resequencing screenings represent a powerful set of techniques to discover large numbers of mutations. Of these, only a small fraction are causally implicated in disease onset and therefore, separating the wheat from the chaff is still a major challenge (Baudot et al., 2009). For a small subset of the new mutations discovered, experimental information is available regarding the relationship between the mutation and disease, and for a smaller number of cases the underlying biochemical mechanism is known. Little information is available for the remaining mutations. The requirement of a lot of investment, both in terms of time and money, means that it is not feasible to experimentally test the association of all these mutations to disease, and to characterize their functional effects. Nevertheless, this problem is very amenable to in silico predictors.
Cline and Karchin (2011) wisely summarized the two different approaches as follows: “A bench biologist interested in whether a mutation of interest impacts the transcription of a gene might perform site-directed mutagenesis on genomic DNA, transfect mutated DNA into cell culture, and use readouts of the gene’s transcriptional activity to measure changes with respect to wild type. In contrast, a bioinformatics approach typically involves computational analysis of the DNA sequence surrounding the mutation, possibly supplemented with information from published bench experiments.”
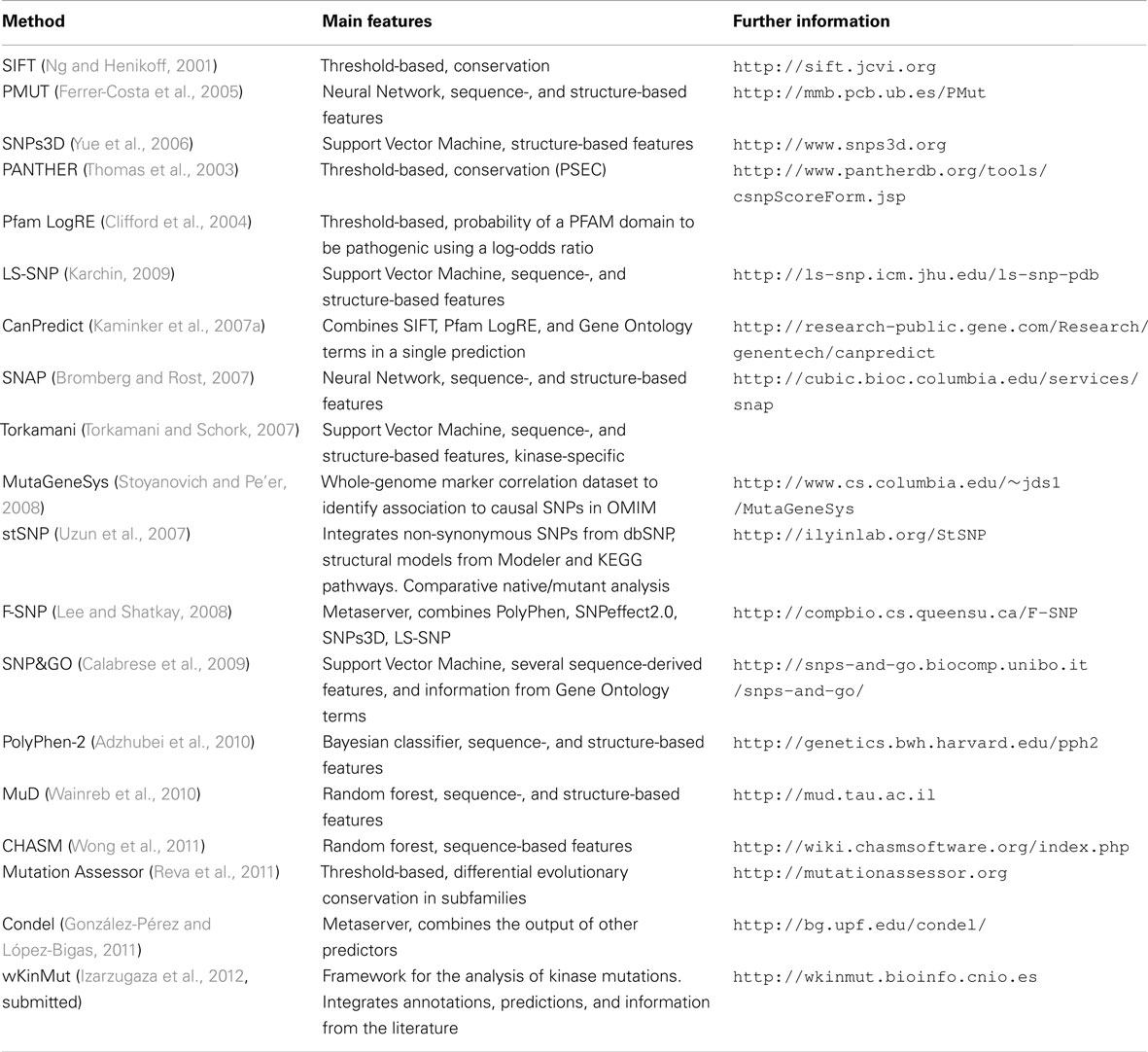
This is just one example of the very different methods available to predict in silico the probability of a newly discovered mutation being implicated in disease. Different approaches have been developed in the last decade (Table 1) and several detailed reviews on this subject have been published (Baudot et al., 2009; Karchin, 2009; Cline and Karchin, 2011).
These methodologies can be classified according to their underlying principles: Some methods make use of several features to identify relevant positions in a given protein, and hence, rules are derived to predict the pathogenicity of mutations. Another group of implementations assumes that evolutionarily conserved protein residues are important for protein structure, folding, and function, whereby mutations in these residues are considered deleterious (Ng and Henikoff, 2001). Variations on this principle lead to methods that predict deleterious mutations by assessing the changes in evolutionarily conserved PFAM motifs (Clifford et al., 2004). Furthermore, a group of methodologies use protein structures to characterize substitutions that significantly destabilize the folded state. A growing number of systems integrate prior knowledge in the form of both sequence-based and structure-based features from a set of mutations (for which their characterization as pathogenic or neutral exists) to train an automatic machine learning system. Once trained, the system can infer the pathogenicity of new mutations automatically. Different machine learning methods can be implemented depending on their individual needs. Among them, probably the most popular ones are: rule-based systems (Wang and Moult, 2001; Ramensky et al., 2002; Reva et al., 2011), decision trees (Krishnan and Westhead, 2003), random forests (Kaminker et al., 2007b; Wainreb et al., 2010), neural networks (Ferrer-Costa et al., 2002; Bromberg and Rost, 2007), Bayesian methods (Adzhubei et al., 2010), and SVMs (Karchin et al., 2005; Yue et al., 2005; Torkamani and Schork, 2007; Calabrese et al., 2009; Wainreb et al., 2010). In addition, some meta approaches have been implemented recently (Lee and Shatkay, 2008), for instance, Condel (González-Pérez and López-Bigas, 2011) integrates five of the most widely employed computational tools for sorting missense single nucleotide variations.
Methods also differ in the nature of the protein properties used to determine the pathogenicity of new mutations. Some of the predictors require sequence-oriented features that are easily applicable to any polymorphism. Recurrent examples of this category are: amino acid type, sequence conservation, domain type, functional annotations, post-translational modifications, and so on. A second set of predictors calculate features that require a protein structure. Common examples to illustrate these are: secondary structure, solvent accessibility, flexibility, etc. The major drawback of these methodologies is that although they may increase the accuracy, the need for either an experimentally solved or a precisely modeled protein structure implies a loss of coverage. The number of features and their combinations is infinite. Moreover, features can also either be general or apply only to a defined subset of proteins, as is the membership to a kinase group (Torkamani and Schork, 2007; Izarzugaza et al., 2012).
Benchmarking Prediction Methods
In the previous section we discussed the differences between the various methods, both in terms of implementation and prediction features. Equally important are the differences found in the composition of the datasets used to train the methods. This is particularly relevant in the case of machine learning approaches. Machine learning approaches are developed in two independent consecutive steps: during the initial development phase, the developers aim to optimize the combination of features, internal parameters, and prediction algorithms to obtain a trained classifier. In a later phase, blind tests are conducted to evaluate the performance simulating a more realistic scenario. Consequently, three separate datasets are needed: (i) a training dataset to allow the classifier to learn, (ii) a validation dataset to optimize the selection of parameters, and (iii) an evaluation dataset to conduct blind tests to assess the expected performance of the classifier.
Consequently, the datasets used highly influence the overall performance of the prediction and, if not pondered cautiously might become a source of evaluation errors. Probably, the most common of them being overtraining as a result from the evaluation of the methodologies with mutations that have also been considered in the training dataset. In other words, if a predictor were evaluated using a test set whose correct answers the method had previously been provided with, this may yield unfair over-estimation of the prediction capability. An extension of this problem, especially if the features considered predict at the protein level, is that mutations occurring in the same protein or closely related homologs should not span two different datasets.
The selection of a benchmark dataset that is fair and does not lead to artifacts is not a trivial task (Care et al., 2007) and clean datasets that were not used in the development of any of the methods are required. Following a similar approach to those in the detection of bio-entities from the literature (BioCreative), protein structure (CASP), and protein interaction prediction (CAPRI), a successful recent example is CAGI1. In summary, CAGI is intended to assess a battery of computational methods for predicting the phenotypic impacts of genome variation. Participants are provided a number of different sets of genetic variants and are expected to make predictions of resulting, molecular, cellular, or organismal phenotype. These predictions are later on evaluated by independent assessors against experimental characterizations.
Although CAGI constitutes an undoubtedly powerful tool to provide insights on the performance of state-of-the-art methodologies, the major drawback is that provided datasets are gathered from very specialized projects, and consequently are seldom universally applicably to all methodologies, which consequently, limits the benchmark. An example of the previous would be the intrinsic limitation to predict mutations outside the protein kinase superfamily for kinase-specific methodologies.
Complementary to the CAGI experiment, current text mining methodologies enable the generation of clean sets of experimentally validated mutation mentions from the literature. Those mutations that were not recorded in the databases used to provide the training and evaluation datasets are of special interest. Here we propose a pipeline for the curation of mutations automatically extracted from the literature and their use as a gold standard in the benchmarking of pathogenicity predictors. We will describe this approach thoroughly in the following sections.
Mining Kinase Mutations from the Literature
Previously, we discussed how the efforts of database curators to store and annotate mutations (Table 2) can hardly keep the pace of the vast amount of information generated by current large-scale variation studies. To bridge this growing gap, automatic extraction of entities and their relationships from the existing literature can be applied. This includes text mining techniques such as regular expressions, pattern recognition, and natural language processing, among others. Indeed, these approaches have been successfully applied to other fields of research, for instance for the automatic extraction of protein–protein interactions (Blaschke and Valencia, 2001; Krallinger et al., 2008c) and in the annotation of genes and proteins (Krallinger et al., 2008a, 2010). Despite the success of these methods, it must be born in mind that this technology does not aim to replace manual curation and validation. Rather, text mining approaches are better understood as systematic tools to assist the efforts of human curators by helping them to find information, prioritize documents, and highlight potentially relevant items (Krallinger et al., 2008a,b; Leitner et al., 2010).
Here we will use our recently published pipeline for extracting mutation mentions in protein kinases from the literature, SNP2L (Krallinger et al., 2009), as an example of a typical text mining workflow. The pipeline (Figure 1) integrates article retrieval, detection of mutations, and proteins in the corresponding article, correct mutation-protein association and, finally, validation of the results. To the best of our knowledge there is currently no pipeline similar to the one presented here. Two main aspects make our pipeline unique. First, our system is specifically designed to extract mutations occurring in the protein kinase superfamily. Second, we perform an additional filtering step to ensure the quality of the extracted mutations as we will disclose in the following sections.
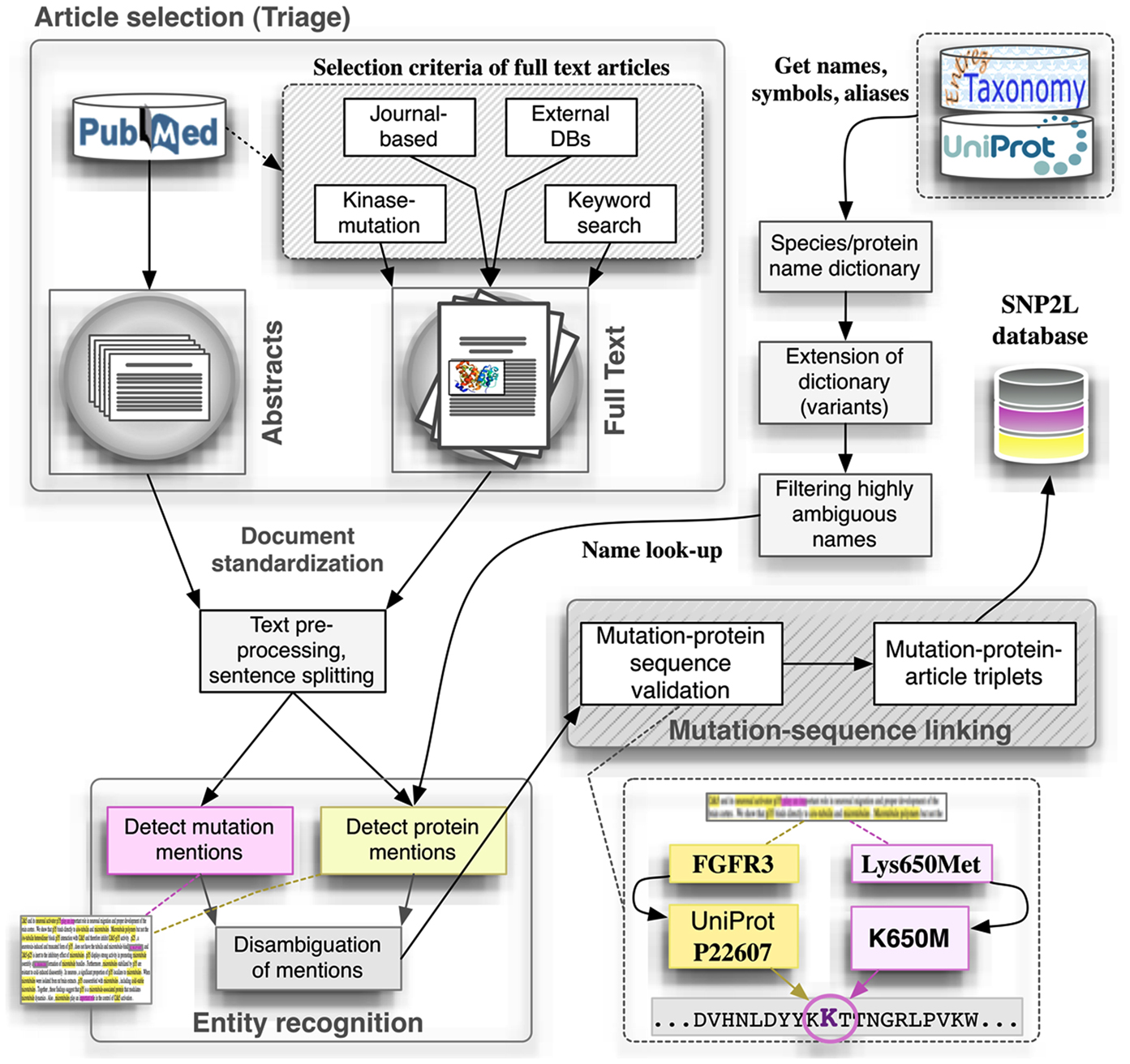
Figure 1. SNP2L Pipeline as an example of a typical automatic method to extract mutation mentions from the literature. The pipeline integrates article retrieval, detection of mutations and proteins in the corresponding article, correct mutation-protein association and, finally, validation of the results.
Article Selection (Triage): Constructing a Text Mining Corpus
Following a common approach in text mining, we tested SNP2L with two different datasets: One constituted by the whole collection of PubMed abstracts and the other by a collection of either manually or automatically selected full-text articles. In order to construct the corpus, full-text articles were automatically downloaded using an in-house retrieval system (Krallinger et al., 2008a) prioritized under three different criteria:
1. Relevance of the abstract: information contained in the corresponding abstracts such as the mention of mutations, mention of human kinases, and a combination of keywords (including “human kinase mutation”).
2. A priori relevance of the full-text articles: extracting all references in PubMed for human kinases contained in multiple databases (e.g., SwissProt, MINT, and IntAct).
3. Relevance of the journal: based on analyzing a fraction of mutation-mentioning abstracts of each journal and prioritizing a set of journals (and thus their articles) to retrieve their full-text articles. This set consisted of the following journals: American Journal of Human Genetics, European Journal of Human Genetics, Human Genetics, Human Mutation, and Human Molecular Genetics.
Before proceeding to the next step, all articles should be split in sentences using a sentence boundary detection system (Krallinger et al., 2008a).
Entity Recognition: Mutations and Protein Kinases
The consistent nomenclature used to describe mutations in the literature makes these entities especially amenable to this type of approach and accordingly, a growing number of such methods have been described in the literature over the years. A summary of several of these literature mining tools to extract information on mutations is presented in Table 3. In the example discussed here, we used MutationFinder (Caporaso et al., 2007) for the initial extraction of single aminoacid substitutions. MutationFinder constitutes a valuable tool to detect the mention of mutations in a given set of manuscripts and it relies on language expressions used to describe mutation events. MutationFinder is very competitive for recall and precision when compared to other strategies (Yip et al., 2007), and it has been evaluated using a manually generated gold standard collection of abstracts.
After recognizing all the mutations mentioned in the text, we attempted to identify all human protein kinases co-mentioned with them in the same document. Existing systems that try to link mentions of genes and proteins to database identifiers generally rely on approaches that compare the names appearing in the text to gene names or aliases contained in database records. The actual task of determining the exact database record for a gene/protein mention is commonly referred to as gene mention grounding or normalization, and has been evaluated in the second BioCreative community challenge, illustrating that dictionary look-up approaches can obtain competitive results for this purpose (Morgan et al., 2008).
Following this line, we constructed a lexicon specifically for human protein kinases, derived from gene and protein symbols, names, and aliases contained in the UniProt database (see Figure 1, Get names, symbols, and aliases). Because this gene/protein lexicon did not capture all representative typographical variants of a given name, we used a rule-based approach and heuristics for generating typographical variants for the kinase lexicon entries. With this respect, the alternative use of hyphens, capitalization (upper-case and capitalized names), and different word order variants were captured. The gene/protein lexicon was filtered to eliminate highly ambiguous names through comparison with a stop word list and by, after an initial look-up step, checking manually potential outlier names that show a very high mention frequency. The extended and pruned human kinase lexicon was then used for the detection of corresponding mentions in our document collections containing mutation mentions. As a given name can correspond to different records (ambiguity), both at the level of human genes as well as in case of genes from different species sharing the same name, we calculated for each article, two different scores reflecting (a) the contextual similarity of the article to the reference (UniProt) protein record and (b) the overall association of the article to human species terms from the total set of tagged species terms. A conceivable alternative would be to simply apply very strict protein-organism co-mention criteria based on relative textual distances, which is rather problematic in case of human proteins were often the organism source is not explicitly stated.
Mutation-Sequence Linking
The next step is to link mutation mentions with their corresponding human kinases. This step would be trivial if a single protein was mentioned per article, however, for most of the articles this is not the case and more than one protein is mentioned per article. A reasonable solution would be to check the existence of the amino acid at the specified position for each mutation mention-protein combination. In addition to this basic sequence look-up validation method additional mutation mapping strategies could be implemented. They should consider errors resulting from the wrong detection of the directionality of the extracted mutation mention (using the wild type as mutant residue and vice versa) and inconsistencies and alternative sequence counting between the article and the kinase sequence. For example:
– Sliding window algorithms that look for relative positions of mutations (pattern) rather than exact position co-occurrences. With this approach, mutation mentions would be scanned looking for positions relative to the starting one attending to the distance between all the mutations in the same abstract. The strength of this approach is that it is able to deal with alternative sequence coordinates. There are many examples in the literature: Mutations F175P, R178L, and Y530L in the proto-oncogene tyrosine-protein kinase Src, are mentioned in the considered article (PMID 2108315) as F172P, R175L, and Y527F respectively. Since the probability of finding simple patterns by chance can be high in some trivial cases, it is reasonable to consider only those cases where a minimum number of mutated positions (3 in our example) could be detected.
– Bidirectional mutation to sequence position mapping. Either the wild type or the mutant residue of an extracted mutation mention might be accepted in the corresponding sequence position.
– Pro-peptides and mature protein mutation mapping. In order to allow alternative residue counting due to the presence of a signal peptide, a displacement equal to the length of the corresponding signal peptide might be allowed.
– Methionine cleavage: the mutation mapping might be carried out taking into consideration the possibility of neglecting the N-terminal methionine.
Using the Literature to Generate a Benchmark Dataset
The main focus of this article has been the construction of a gold standard dataset to benchmark prediction methods. Following this thread of reasoning, mutations already present in common databases are discarded, while new ones form the benchmark dataset. This procedure will ensure a dataset that enables fair comparison and is less prone to over-estimation of the classifiers’ performance as we discussed previously in the Benchmarking prediction methods section.
In spite of constituting a powerful tool for the extraction of knowledge from the literature, text mining approaches to recover kinase mutations still have some limitations in terms of recall and a number mutations escape detection by even the most accurate state-of-the-art algorithms. Among the challenging aspects in this respect are the detection of mutations that are described in additional materials or contained in tables and figures. This is because they can not easily be converted efficiently to plain text. Another key issue is the appropriate detection of the kinase mentions, which can be referred to through a range of different typographical variations and aliases, of which text mining approaches can only cover some. To this issue one also needs to add the underlying limitations in terms of recall of the mutation extraction process (Caporaso et al., 2007) and inconsistencies of sequence descriptions in reference databases as compared to those examined in scientific articles.
Using the Literature to Understand the Consequences of Mutation
From a parallel perspective, text mining approaches can be used to enhance our understanding of both new and existing mutations. Text mining approaches output mutations extracted from the literature along with all their contextual information. Pointers to the relevant literature are provided, these include: experimental conditions, organism, or population sub-types, information regarding observed phenotypes including association to disease, or in a best case scenario, the underlying biochemical mechanisms.
This information can help to interpret the consequences of mutations and is often complementary to the valuable clues provided by the methods to predict the pathogenicity of mutations. Indeed, the emerging trend in the field is to integrate information from diverse sources (Lee and Shatkay, 2008; González-Pérez and López-Bigas, 2011), as we have done recently with the development of wKinMut2 to help in the interpretation of mutations in the protein kinase superfamily.
In addition to the predictions of pathogenicity directly from our in-house classifier (Izarzugaza et al., 2012) and the values of the features used in the classification, wKinMut combines information from different external sources to help in the interpretation of the prediction. These include the results from other classifiers focusing on different aspects of mutation pathogenicity (SIFT; Ng and Henikoff, 2001; MutationAssessor; Reva et al., 2011), the representation of the mutation in the context of its three-dimensional structure and records of the mutation in other databases such as SAAPdb (Hurst et al., 2009), UniProt (Yip et al., 2007), COSMIC (Bamford et al., 2004), and KinMutBase (Ortutay et al., 2005). Two text mining resources complement the framework: iHop (Hoffmann and Valencia, 2005) a literature mining system to extract gene–gene and protein–protein interactions and SNP2L (Krallinger et al., 2009) whose capabilities to detect mutation mentions from the literature have been described thoroughly here.
In summary, wKinMut can be useful to predict the pathogenicity of novel mutations and to interpret the biochemical mechanisms leading to pathogenicity and it can be applied to the interpretation of genomes from cancer patients.
Overview and Summary
Current research aims to discover the mechanistic connection between mutations and disease. We focused on the protein kinase superfamily due to the enormous wealth of mentions in the literature associating different diseases, including cancer, with mutations in members of this superfamily.
In this article we have reviewed the different possibilities and limitations of state-of-the-art computational methods for the prediction of the pathogenicity of mutations and we have discussed the difficulties that arise to benchmark and evaluate the performance of the classifiers. We have proposed our recently published pipeline, SNP2L, for the automatic extraction and curation of mentions in the literature to collect a gold standard dataset that might be used in the benchmarking of the different predictors. Finally, we have introduced wKinMut as an example the integration of text mining with prediction methodologies to help in the interpretation of the consequences of mutations in the context of disease genome analysis with particular focus on cancer. We think that such applications might be of interest in the interpretation of patient genomes in the emerging field of personalized/stratified medicine in, hopefully, a near future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P., Kondrashov, A. S., and Sunyaev, S. R. (2010). A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249.
Amberger, J., Bocchini, C., and Hamosh, A. (2011). A new face and new challenges for online mendelian inheritance in man (omim(®)). Hum. Mutat. 32, 564–567.
Bairoch, A., Apweiler, R., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., Martin, M. J., Natale, D. A., O’Donovan, C., Redaschi, N., and Yeh, L.-S. L. (2005). The universal protein resource (UniProt). Nucleic Acids Res. 33, D154–D159.
Baker, C. J. O., and Rene, W. (2006). Mutation mining – a prospector’s tale. J. Inform. Syst. Front. 8, 47–57.
Bamford, S., Dawson, E., Forbes, S., Clements, J., Pettett, R., Dogan, A., Flanagan, A., Teague, J., Futreal, P. A., Stratton, M. R., and Wooster, R. (2004). The cosmic (catalogue of somatic mutations in cancer) database and website. Br. J. Cancer 91, 355–358.
Baudot, A., Real, F., Izarzugaza, J., and Valencia, A. (2009). From cancer genomes to cancer models: bridging the gaps. EMBO Rep. 10, 359–366.
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., and Bourne, P. E. (2000). The protein data bank. Nucleic Acids Res. 28, 235–242.
Blaschke, C., and Valencia, A. (2001). The potential use of suiseki as a protein interaction discovery tool. Genome Inform. 12, 123–134.
Bromberg, Y., and Rost, B. (2007). Snap: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 35, 3823–3835.
Caenepeel, S., Charydczak, G., Sudarsanam, S., Hunter, T., and Manning, G. (2004). The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc. Natl. Acad. Sci. U.S.A. 101, 11707–11712.
Calabrese, R., Capriotti, E., Fariselli, P., Martelli, P. L., and Casadio, R. (2009). Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 30, 1237–1244.
Caporaso, J. G., Baumgartner, W. A., Randolph, D. A., Cohen, K. B., and Hunter, L. (2007). MutationFinder: a high-performance system for extracting point mutation mentions from text. Bioinformatics 23, 1862–1865.
Care, M. A., Needham, C. J., Bulpitt, A. J., and Westhead, D. R. (2007). Deleterious SNP prediction: be mindful of your training data! Bioinformatics 23, 664–672.
Clifford, R. J., Edmonson, M. N., Nguyen, C., and Buetow, K. H. (2004). Large-scale analysis of non-synonymous coding region single nucleotide polymorphisms. Bioinformatics 20, 1006–1014.
Cline, M., and Karchin, R. (2011). Using bioinformatics to predict the functional impact of SNVs. Bioinformatics 27, 441–448.
Consortium, I. C. G., Hudson, T. J., Anderson, W., Artez, A., Barker, A. D., Bell, C., Bernabé, R. R., Bhan, M. K., Calvo, F., Eerola, I., Gerhard, D. S., Guttmacher, A., Guyer, M., Hemsley, F. M., Jennings, J. L., Kerr, D., Klatt, P., Kolar, P., Kusada, J., Lane, D. P., Laplace, F., Youyong, L., Nettekoven, G., Ozenberger, B., Peterson, J., Rao, T. S., Remacle, J., Schafer, A. J., Shibata, T., Stratton, M. R., Vockley, J. G., Watanabe, K., Yang, H., Yuen, M. M. F., Knoppers, B. M., Bobrow, M., Cambon-Thomsen, A., Dressler, L. G., Dyke, S. O. M., Joly, Y., Kato, K., Kennedy, K. L., Nicolás, P., Parker, M. J., Rial-Sebbag, E., Romeo-Casabona, C. M., Shaw, K. M., Wallace, S., Wiesner, G. L., Zeps, N., Lichter, P., Biankin, A. V., Chabannon, C., Chin, L., Clément, B., de Alava, E., Degos, F., Ferguson, M. L., Geary, P., Hayes, D. N., Hudson, T. J., Johns, A. L., Kasprzyk, A., Nakagawa, H., Penny, R., Piris, M. A., Sarin, R., Scarpa, A., Shibata, T., van de Vijver, M., Futreal, P. A., Aburatani, H., Bayés, M., Botwell, D. D., Campbell, P. J., Estivill, X., Gerhard, D. S., Grimmond, S. M., Gut, I., Hirst, M., López-Otín, C., Majumder, P., Marra, M., McPherson, J. D., Nakagawa, H., Ning, Z., Puente, X. S., Ruan, Y., Shibata, T., Stratton, M. R., Stunnenberg, H. G., Swerdlow, H., Velculescu, V. E., Wilson, R. K., Xue, H. H., Yang, L., Spellman, P. T., Bader, G. D., Boutros, P. C., Campbell, P. J., Flicek, P., Getz, G., Guigó, R., Guo, G., Haussler, D., Heath, S., Hubbard, T. J., Jiang, T., Jones, S. M., Li, Q., López-Bigas, N., Luo, R., Muthuswamy, L., Ouellette, B. F., Pearson, J. V., Puente, X. S., Quesada, V., Raphael, B. J., Sander, C., Shibata, T., Speed, T. P., Stein, L. D., Stuart, J. M., Teague, J. W., Totoki, Y., Tsunoda, T., Valencia, A., Wheeler, D. A., Wu, H., Zhao, S., Zhou, G., Stein, L. D., Guigó, R., Hubbard, T. J., Joly, Y., Jones, S. M., Kasprzyk, A., Lathrop, M., López-Bigas, N., Ouellette, B. F., Spellman, P. T., Teague, J. W., Thomas, G., Valencia, A., Yoshida, T., Kennedy, K. L., Axton, M., Dyke, S. O., Futreal, P. A., Gerhard, D. S., Gunter, C., Guyer, M., Hudson, T. J., McPherson, J. D., Miller, L. J., Ozenberger, B., Shaw, K. M., Kasprzyk, A., Stein, L. D., Zhang, J., Haider, S. A., Wang, J., Yung, C. K., Cros, A., Liang, Y., Gnaneshan, S., Guberman, J., Hsu, J., Bobrow, M., Chalmers, D. R., Hasel, K. W., Joly, Y., Kaan, T. S., Kennedy, K. L., Knoppers, B. M., Lowrance, W. W., Masui, T., Nicolás, P., Rial-Sebbag, E., Rodriguez, L. L., Vergely, C., Yoshida, T., Grimmond, S. M., Biankin, A. V., Bowtell, D. D., Cloonan, N., deFazio, A., Eshleman, J. R., Etemadmoghadam, D., Gardiner, B. B., Kench, J. G., Scarpa, A., Sutherland, R. L., Tempero, M. A., Waddell, N. J., Wilson, P. J., McPherson, J. D., Gallinger, S., Tsao, M. S., Shaw, P. A., Petersen, G. M., Mukhopadhyay, D., Chin, L., DePinho, R. A., Thayer, S., Muthuswamy, L., Shazand, K., Beck, T., Sam, M., Timms, L., Ballin, V., Lu, Y., Ji, J., Zhang, X., Chen, F., Hu, X., Zhou, G., Yang, Q., Tian, G., Zhang, L., Xing, X., Li, X., Zhu, Z., Yu, Y., Yu, J., Yang, H., Lathrop, M., Tost, J., Brennan, P., Holcatova, I., Zaridze, D., Brazma, A., Egevard, L., Prokhortchouk, E., Banks, R. E., Uhlén, M., Cambon-Thomsen, A., Viksna, J., Ponten, F., Skryabin, K., Stratton, M. R., Futreal, P. A., Birney, E., Borg, A., Børresen-Dale, A. L., Caldas, C., Foekens, J. A., Martin, S., Reis-Filho, J. S., Richardson, A. L., Sotiriou, C., Stunnenberg, H. G., Thoms, G., van de Vijver, M., van’t Veer, L., Calvo, F., Birnbaum, D., Blanche, H., Boucher, P., Boyault, S., Chabannon, C., Gut, I., Masson-Jacquemier, J. D., Lathrop, M., Pauporté, I., Pivot, X., Vincent-Salomon, A., Tabone, E., Theillet, C., Thomas, G., Tost, J., Treilleux, I., Calvo, F., Bioulac-Sage, P., Clément, B., Decaens, T., Degos, F., Franco, D., Gut, I., Gut, M., Heath, S., Lathrop, M., Samuel, D., Thomas, G., Zucman-Rossi, J., Lichter, P., Eils, R., Brors, B., Korbel, J. O., Korshunov, A., Landgraf, P., Lehrach, H., Pfister, S., Radlwimmer, B., Reifenberger, G., Taylor, M. D., von Kalle, C., Majumder, P. P., Sarin, R., Rao, T. S., Bhan, M. K., Scarpa, A., Pederzoli, P., Lawlor, R. A., Delledonne, M., Bardelli, A., Biankin, A. V., Grimmond, S. M., Gress, T., Klimstra, D., Zamboni, G., Shibata, T., Nakamura, Y., Nakagawa, H., Kusada, J., Tsunoda, T., Miyano, S., Aburatani, H., Kato, K., Fujimoto, A., Yoshida, T., Campo, E., López-Otín, C., Estivill, X., Guigó, R., de Sanjosé, S., Piris, M. A., Montserrat, E., González-Díaz, M., Puente, X. S., Jares, P., Valencia, A., Himmelbauer, H., Quesada, V., Bea, S., Stratton, M. R., Futreal, P. A., Campbell, P. J., Vincent-Salomon, A., Richardson, A. L., Reis-Filho, J. S., van de Vijver, M., Thomas, G., Masson-Jacquemier, J. D., Aparicio, S., Borg, A., Børresen-Dale, A. L., Caldas, C., Foekens, J. A., Stunnenberg, H. G., van’t Veer, L., Easton, D. F., Spellman, P. T., Martin, S., Barker, A. D., Chin, L., Collins, F. S., Compton, C. C., Ferguson, M. L., Gerhard, D. S., Getz, G., Gunter, C., Guttmacher, A., Guyer, M., Hayes, D. N., Lander, E. S., Ozenberger, B., Penny, R., Peterson, J., Sander, C., Shaw, K. M., Speed, T. P., Spellman, P. T., Vockley, J. G., Wheeler, D. A., Wilson, R. K., Hudson, T. J., Chin, L., Knoppers, B. M., Lander, E. S., Lichter, P., Stein, L. D., Stratton, M. R., Anderson, W., Barker, A. D., Bell, C., Bobrow, M., Burke, W., Collins, F. S., Compton, C. C., DePinho, R. A., Easton, D. F., Futreal, P. A., Gerhard, D. S., Green, A. R., Guyer, M., Hamilton, S. R., Hubbard, T. J., Kallioniemi, O. P., Kennedy, K. L., Ley, T. J., Liu, E. T., Lu, Y., Majumder, P., Marra, M., Ozenberger, B., Peterson, J., Schafer, A. J., Spellman, P. T., Stunnenberg, H. G., Wainwright, B. J., Wilson, R. K., and Yang, H. (2010a). International network of cancer genome projects. Nature 464, 993–998.
Consortium, I. H., Altshuler, D. M., Gibbs, R. A., Peltonen, L., Altshuler, D. M., Gibbs, R. A., Peltonen, L., Dermitzakis, E., Schaffner, S. F., Yu, F., Peltonen, L., Dermitzakis, E., Bonnen, P. E., Altshuler, D. M., Gibbs, R. A., de Bakker, P. I. W., Deloukas, P., Gabriel, S. B., Gwilliam, R., Hunt, S., Inouye, M., Jia, X., Palotie, A., Parkin, M., Whittaker, P., Yu, F., Chang, K., Hawes, A., Lewis, L. R., Ren, Y., Wheeler, D., Gibbs, R. A., Muzny, D. M., Barnes, C., Darvishi, K., Hurles, M., Korn, J. M., Kristiansson, K., Lee, C., McCarrol, S. A., Nemesh, J., Dermitzakis, E., Keinan, A., Montgomery, S. B., Pollack, S., Price, A. L., Soranzo, N., Bonnen, P. E., Gibbs, R. A., Gonzaga-Jauregui, C., Keinan, A., Price, A. L., Yu, F., Anttila, V., Brodeur, W., Daly, M. J., Leslie, S., McVean, G., Moutsianas, L., Nguyen, H., Schaffner, S. F., Zhang, Q., Ghori, M. J. R., McGinnis, R., McLaren, W., Pollack, S., Price, A. L., Schaffner, S. F., Takeuchi, F., Grossman, S. R., Shlyakhter, I., Hostetter, E. B., Sabeti, P. C., Adebamowo, C. A., Foster, M. W., Gordon, D. R., Licinio, J., Manca, M. C., Marshall, P. A., Matsuda, I., Ngare, D., Wang, V. O., Reddy, D., Rotimi, C. N., Royal, C. D., Sharp, R. R., Zeng, C., Brooks, L. D., and McEwen, J. E. (2010b). Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58.
Consortium, G. P., Durbin, R. M., Abecasis, G. R., Altshuler, D. L., Auton, A., Brooks, L. D., Durbin, R. M., Gibbs, R. A., Hurles, M. E., and McVean, G. A. (2010c). A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073.
Erdogmus, M., and Sezerman, O. U. (2007). Application of automatic mutation-gene pair extraction to diseases. J. Bioinform. Comput. Biol. 5, 1261–1275.
Ferrer-Costa, C., Gelpí, J. L., Zamakola, L., Parraga, I., de la Cruz, X., and Orozco, M. (2005). PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics 21, 3176–3178.
Ferrer-Costa, C., Orozco, M., and de la Cruz, X. (2002). Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J. Mol. Biol. 315, 771–786.
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., Gavin, O. L., Gunasekaran, P., Ceric, G., Forslund, K., Holm, L., Sonnhammer, E. L. L., Eddy, S. R., and Bateman, A. (2010). The pfam protein families database. Nucleic Acids Res. 38, D211–D222.
Flicek, P., Amode, M. R., Barrell, D., Beal, K., Brent, S., Chen, Y., Clapham, P., Coates, G., Fairley, S., Fitzgerald, S., Gordon, L., Hendrix, M., Hourlier, T., Johnson, N., Kähäri, A., Keefe, D., Keenan, S., Kinsella, R., Kokocinski, F., Kulesha, E., Larsson, P., Longden, I., McLaren, W., Overduin, B., Pritchard, B., Riat, H. S., Rios, D., Ritchie, G. R. S., Ruffier, M., Schuster, M., Sobral, D., Spudich, G., Tang, Y. A., Trevanion, S., Vandrovcova, J., Vilella, A. J., White, S., Wilder, S. P., Zadissa, A., Zamora, J., Aken, B. L., Birney, E., Cunningham, F., Dunham, I., Durbin, R., Fernández-Suarez, X. M., Herrero, J., Hubbard, T. J. P., Parker, A., Proctor, G., Vogel, J., and Searle, S. M. J. (2011). Ensembl 2011. Nucleic Acids Res. 39, D800–D806.
Furlong, L. I., Dach, H., Hofmann-Apitius, M., and Sanz, F. (2008). Osirisv1.2: a named entity recognition system for sequence variants of genes in biomedical literature. BMC Bioinformatics 9, 84. doi:10.1186/1471-2105-9-84
González-Pérez, A., and López-Bigas, N. (2011). Improving the assessment of the outcome of nonsynonymous snvs with a consensus deleteriousness score, condel. Am. J. Hum. Genet. 88, 440–449.
Greenman, C., Stephens, P., Smith, R., Dalgliesh, G. L., Hunter, C., Bignell, G., Davies, H., Teague, J., Butler, A., Stevens, C., Edkins, S., O’Meara, S., Vastrik, I., Schmidt, E. E., Avis, T., Barthorpe, S., Bhamra, G., Buck, G., Choudhury, B., Clements, J., Cole, J., Dicks, E., Forbes, S., Gray, K., Halliday, K., Harrison, R., Hills, K., Hinton, J., Jenkinson, A., Jones, D., Menzies, A., Mironenko, T., Perry, J., Raine, K., Richardson, D., Shepherd, R., Small, A., Tofts, C., Varian, J., Webb, T., West, S., Widaa, S., Yates, A., Cahill, D. P., Louis, D. N., Goldstraw, P., Nicholson, A. G., Brasseur, F., Looijenga, L., Weber, B. L., Chiew, Y.-E., DeFazio, A., Greaves, M. F., Green, A. R., Campbell, P., Birney, E., Easton, D. F., Chenevix-Trench, G., Tan, M.-H., Khoo, S. K., Teh, B. T., Yuen, S. T., Leung, S. Y., Wooster, R., Futreal, P. A., and Stratton, M. R. (2007). Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158.
Hoffmann, R., and Valencia, A. (2005). Implementing the ihop concept for navigation of biomedical literature. Bioinformatics 21(Suppl. 2), ii252–ii258.
Horn, F., Lau, A. L., and Cohen, F. E. (2004). Automated extraction of mutation data from the literature: application of MuteXt to g protein-coupled receptors and nuclear hormone receptors. Bioinformatics 20, 557–568.
Hunter, T., and Plowman, G. D. (1997). The protein kinases of budding yeast: six score and more. Trends Biochem. Sci. 22, 18–22.
Hurst, J., McMillan, L., Porter, C., Allen, J., Fakorede, A., and Martin, A. (2009). The SAAPdb web resource: a large-scale structural analysis of mutant proteins. Hum. Mutat. 30, 616–624.
Izarzugaza, J. M., Pozo, A., Vazquez, M., and Valencia, A. (2012). Prioritization of pathogenic mutations in the protein kinase superfamily. BMC Genomics 13(Suppl. 4), S3. doi:10.1186/1471-2164-13-S4-S3
Kaminker, J. S., Zhang, Y., Watanabe, C., and Zhang, Z. (2007a). CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res. 35, W595–W598.
Kaminker, J. S., Zhang, Y., Waugh, A., Haverty, P. M., Peters, B., Sebisanovic, D., Stinson, J., Forrest, W. F., Bazan, J. F., Seshagiri, S., and Zhang, Z. (2007b). Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res. 67, 465–473.
Karchin, R. (2009). Next generation tools for the annotation of human SNPs. Brief Bioinform. 10, 35–52.
Karchin, R., Diekhans, M., Kelly, L., Thomas, D. J., Pieper, U., Eswar, N., Haussler, D., and Sali, A. (2005). LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics 21, 2814–2820.
Kinnings, S. L., and Jackson, R. M. (2009). Binding site similarity analysis for the functional classification of the protein kinase family. J. Chem. Inf. Model 49, 318–329.
Knight, J. D. R., Qian, B., Baker, D., and Kothary, R. (2007). Conservation, variability and the modeling of active protein kinases. PLoS ONE 2, e982. doi:10.1371/journal.pone.0000982
Krallinger, M., Izarzugaza, J. M. G., Rodriguez-Penagos, C., and Valencia, A. (2009). Extraction of human kinase mutations from literature, databases and genotyping studies. BMC Bioinformatics 10(Suppl. 8), S1. doi:10.1186/1471-2105-10-S8-S1
Krallinger, M., Leitner, F., Rodriguez-Penagos, C., and Valencia, A. (2008a). Overview of the protein-protein interaction annotation extraction task of BioCreative ii. Genome Biol. 9(Suppl. 2), S4.
Krallinger, M., Morgan, A., Smith, L., Leitner, F., Tanabe, L., Wilbur, J., Hirschman, L., and Valencia, A. (2008b). Evaluation of text-mining systems for biology: overview of the Second BioCreative community challenge. Genome Biol. 9(Suppl. 2), S1.
Krallinger, M., Valencia, A., and Hirschman, L. (2008c). Linking genes to literature: text mining, information extraction, and retrieval applications for biology. Genome Biol. 9(Suppl. 2), S8.
Krallinger, M., Leitner, F., and Valencia, A. (2010). Analysis of biological processes and diseases using text mining approaches. Methods Mol. Biol. 593, 341–382.
Krishnan, V. G., and Westhead, D. R. (2003). A comparative study of machine-learning methods to predict the effects of single nucleotide polymorphisms on protein function. Bioinformatics 19, 2199–2209.
Laskowski, R. A., Chistyakov, V. V., and Thornton, J. M. (2005). PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 33, D266–D268.
Lee, L. C., Horn, F., and Cohen, F. E. (2007). Automatic extraction of protein point mutations using a graph bigram association. PLoS Comput. Biol. 3, e16.
Lee, P. H., and Shatkay, H. (2008). F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 36, D820–D824.
Leitner, F., Chatr-aryamontri, A., Mardis, S. A., Ceol, A., Krallinger, M., Licata, L., Hirschman, L., Cesareni, G., and Valencia, A. (2010). The FEBS Letters/BioCreative ii.5 experiment: making biological information accessible. Nat. Biotechnol. 28, 897–899.
López, G., Valencia, A., and Tress, M. L. (2007). FireDB – a database of functionally important residues from proteins of known structure. Nucleic Acids Res. 35, D219–D223.
Manning, G., Plowman, G. D., Hunter, T., and Sudarsanam, S. (2002a). Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27, 514–520.
Manning, G., Whyte, D. B., Martinez, R., Hunter, T., and Sudarsanam, S. (2002b). The protein kinase complement of the human genome. Science 298, 1912–1934.
McDonald, R. T., Winters, R. S., Mandel, M., Jin, Y., White, P. S., and Pereira, F. (2004). An entity tagger for recognizing acquired genomic variations in cancer literature. Bioinformatics 20, 3249–3251.
Miranda-Saavedra, D., and Barton, G. J. (2007). Classification and functional annotation of eukaryotic protein kinases. Proteins 68, 893–914.
Morgan, A. A., Lu, Z., Wang, X., Cohen, A. M., Fluck, J., Ruch, P., Divoli, A., Fundel, K., Leaman, R., Hakenberg, J., Sun, C., hui Liu, H., Torres, R., Krauthammer, M., Lau, W. W., Liu, H., Hsu, C.-N., Schuemie, M., Cohen, K. B., and Hirschman, L. (2008). Overview of BioCreative ii gene normalization. Genome Biol. 9(Suppl. 2), S3.
Ng, P. C., and Henikoff, S. (2001). Predicting deleterious amino acid substitutions. Genome Res. 11, 863–874.
Ortutay, C., Väliaho, J., Stenberg, K., and Vihinen, M. (2005). KinMutBase: a registry of disease-causing mutations in protein kinase domains. Hum. Mutat. 25, 435–442.
Pieper, U., Eswar, N., Davis, F. P., Braberg, H., Madhusudhan, M. S., Rossi, A., Marti-Renom, M., Karchin, R., Webb, B. M., Eramian, D., Shen, M.-Y., Kelly, L., Melo, F., and Sali, A. (2006). MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 34, D291–D295.
Ramensky, V., Bork, P., and Sunyaev, S. (2002). Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30, 3894–3900.
Rebholz-Schuhmann, D., Marcel, S., Albert, S., Tolle, R., Casari, G., and Kirsch, H. (2004). Automatic extraction of mutations from Medline and cross-validation with OMIM. Nucleic Acids Res. 32, 135–142.
Reumers, J., Maurer-Stroh, S., Schymkowitz, J., and Rousseau, F. (2006). SNPeffect v2.0: a new step in investigating the molecular phenotypic effects of human non-synonymous SNPs. Bioinformatics 22, 2183–2185.
Reva, B., Antipin, Y., and Sander, C. (2011). Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118.
Richardson, C. J., Gao, Q., Mitsopoulous, C., Zvelebil, M., Pearl, L. H., and Pearl, F. M. G. (2009). MoKCa database–mutations of kinases in cancer. Nucleic Acids Res. 37, D824–D831.
Schee, E. D., and Bourne, P. E. (2005). Structural evolution of the protein kinase-like superfamily. PLoS Comput. Biol. 1, e49. doi:10.1371/journal.pcbi.0010049
Shchemelinin, I., Sefc, L., and Necas, E. (2006). Protein kinases, their function and implication in cancer and other diseases. Folia Biol. (Praha) 52, 81–100.
Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M., and Sirotkin, K. (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311.
Sjöblom, T., Jones, S., Wood, L. D., Parsons, D. W., Lin, J., Barber, T. D., Mandelker, D., Leary, R. J., Ptak, J., Silliman, N., Szabo, S., Buckhaults, P., Farrell, C., Meeh, P., Markowitz, S. D., Willis, J., Dawson, D., Willson, J. K. V., Gazdar, A. F., Hartigan, J., Wu, L., Liu, C., Parmigiani, G., Park, B. H., Bachman, K. E., Papadopoulos, N., Vogelstein, B., Kinzler, K. W., and Velculescu, V. E. (2006). The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274.
Stitziel, N. O., Binkowski, T. A., Tseng, Y. Y., Kasif, S., and Liang, J. (2004). topoSNP: a topographic database of non-synonymous single nucleotide polymorphisms with and without known disease association. Nucleic Acids Res. 32, D520–D522.
Stoyanovich, J., and Pe’er, I. (2008). MutaGeneSys: estimating individual disease susceptibility based on genome-wide SNP array data. Bioinformatics 24, 440–442.
Tanramluk, D., Schreyer, A., Pitt, W. R., and Blundell, T. L. (2009). On the origins of enzyme inhibitor selectivity and promiscuity: a case study of protein kinase binding to staurosporine. Chem. Biol. Drug Des. 74, 16–24.
Thomas, P. D., Campbell, M. J., Kejariwal, A., Mi, H., Karlak, B., Daverman, R., Diemer, K., Muruganujan, A., and Narechania, A. (2003). Panther: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141.
Torkamani, A., and Schork, N. J. (2007). Accurate prediction of deleterious protein kinase polymorphisms. Bioinformatics 23, 2918–2925.
Ubersax, J. A., Woodbury, E. L., Quang, P. N., Paraz, M., Blethrow, J. D., Shah, K., Shokat, K. M., and Morgan, D. O. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859–864.
Uzun, A., Leslin, C. M., Abyzov, A., and Ilyin, V. (2007). Structure SNP (StSNP): a web server for mapping and modeling nsSNPs on protein structures with linkage to metabolic pathways. Nucleic Acids Res. 35, W384–W392.
Wainreb, G., Ashkenazy, H., Bromberg, Y., Starovolsky-Shitrit, A., Haliloglu, T., Ruppin, E., Avraham, K. B., Rost, B., and Ben-Tal, N. (2010). Mud: an interactive web server for the prediction of non-neutral substitutions using protein structural data. Nucleic Acids Res. 38(Suppl.), W523–W528.
Wong, W. C., Kim, D., Carter, H., Diekhans, M., Ryan, M. C., and Karchin, R. (2011). CHASM and SNVBox: toolkit for detecting biologically important single nucleotide mutations in cancer. Bioinformatics 27, 2147–2148.
Wood, L. D., Parsons, D. W., Jones, S., Lin, J., Sjöblom, T., Leary, R. J., Shen, D., Boca, S. M., Barber, T. D., Ptak, J., Silliman, N., Szabo, S., Dezso, Z., Ustyanksky, V., Nikolskaya, T., Nikolsky, Y., Karchin, R., Wilson, P. A., Kaminker, J. S., Zhang, Z., Croshaw, R., Willis, J., Dawson, D., Shipitsin, M., Willson, J. K. V., Sukumar, S., Polyak, K., Park, B. H., Pethiyagoda, C. L., Pant, P. V. K., Ballinger, D. G., Sparks, A. B., Hartigan, J., Smith, D. R., Suh, E., Papadopoulos, N., Buckhaults, P., Markowitz, S. D., Parmigiani, G., Kinzler, K. W., Velculescu, V. E., and Vogelstein, B. (2007). The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113.
Yip, Y. L., Famiglietti, M., Gos, A., Duek, P. D., David, F. P. A., Gateau, A., and Bairoch, A. (2008). Annotating single amino acid polymorphisms in the UniProt/Swiss-Prot knowledgebase. Hum. Mutat. 29, 361–366.
Yip, Y. L., Lachenal, N., Pillet, V., and Veuthey, A.-L. (2007). Retrieving mutation-specific information for human proteins in UniProt/Swiss-Prot knowledgebase. J. Bioinform. Comput. Biol. 5, 1215–1231.
Yue, P., Li, Z., and Moult, J. (2005). Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 353, 459–473.
Keywords: disease, kinase, literature mining, mutation, pathogenicity prediction, protein kinase, text mining, variation
Citation: Izarzugaza JMG, Krallinger M and Valencia A (2012) Interpretation of the consequences of mutations in protein kinases: combined use of bioinformatics and text mining. Front. Physio. 3:323. doi: 10.3389/fphys.2012.00323
Received: 23 May 2012; Paper pending published: 05 June 2012;
Accepted: 23 July 2012; Published online: 22 August 2012.
Edited by:
Matteo Barberis, Humboldt University Berlin, Germany; Max Planck Institute for Molecular Genetics, Berlin, GermanyReviewed by:
Jonathan Christopher Fuller, Heidelberg Institute for Theoretical Studies, GermanyXavier De La Cruz Montserrat, Vall d’Hebron Research Institute, Spain
Copyright: © 2012 Izarzugaza, Krallinger and Valencia. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Jose M. G. Izarzugaza and Alfonso Valencia, Structural Computational Biology Group, Structural Biology and BioComputing Programme, Spanish National Cancer Research Centre, C/Melchor Fernandez Almagro, 3, 28029 Madrid, Spain. e-mail:am1nb256YWxlekBjbmlvLmVz;YXZhbGVuY2lhQGNuaW8uZXM=