- National Center of Neurology and Psychiatry, National Institute of Neuroscience, Kodaira, Tokyo, Japan
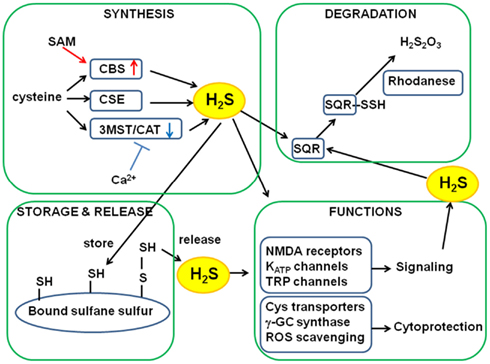
More than 15 years have passed since hydrogen sulfide (H2S) emerged as a biological signaling molecule – initially, in the nervous and vascular systems through the modulation of N-methyl d-aspartate (NMDA) receptors and the activation of transmembrane receptor potential (TRP)- and ATP-dependent K+ (KATP)-channels (Abe and Kimura, 1996; Hosoki et al., 1997; Dello Russo et al., 2000; Zhao et al., 2001; Teague et al., 2002; Nagai et al., 2004; Streng et al., 2008), and later, in nearly every organ system (Predmore and Lefer, 2010). H2S was found to play a role as a cytoprotectant in the nervous system (Kimura and Kimura, 2004; Whiteman et al., 2004); this finding led to the discovery of the cardioprotective effect of H2S (Elrod et al., 2007) (Figure 1). Previous studies have also described that H2S plays several roles: as a regulator of insulin release, inflammation, and angiogenesis, and as an oxygen (O2) sensor (Li et al., 2005; Yang et al., 2005; Kaneko et al., 2006; Olson et al., 2006; Zanardo et al., 2006; Cai et al., 2007; Papapetropoulos et al., 2009).
Previous studies report measurement of endogenous concentrations of sulfide by methods involving high concentrations of acids; therefore, contamination by free H2S released from acid-labile sulfur resulted in an overestimate of the free H2S levels (50–160 μM; Goodwin et al., 1989; Warenycia et al., 1989; Savage and Gould, 1990). A sulfur/silver electrode has frequently been used for the measurement of sulfide concentrations in biological samples. The electrode measures the level of S2−, and a pKa value of 13.9 results in the replacement of cysteine sulfide groups in proteins with hydroxyl groups, thereby releasing H2S from proteins. Because tissue and blood samples contain abundant proteins, this method estimates erroneously high concentrations of sulfide (Whitfield et al., 2008). Recently, the basal or steady state endogenous concentrations of H2S have been re-evaluated using methods that avoid release of contaminant H2S from proteins; these methods give concentration estimates of 20 nM to a few micromolar in tissue and blood samples (Furne et al., 2008; Ishigami et al., 2009; Wintner et al., 2010).
It is necessary to determine the active state concentrations of H2S. At least three factors influence H2S concentration: (1) rate of H2S production, (2) rate of H2S metabolism, and (3) storage of H2S as bound sulfane sulfur and its associated release.
Rate of H2S Production
H2S production by three enzymes has been studied extensively; these enzymes are cystathionine β-synthase (CBS), cystathionine γ-lyase (CSE), and 3-mercaptopyruvate sulfurtransferase (3MST; Stipanuk and Beck, 1982; Chiku et al., 2009; Shibuya et al., 2009a,b; Singh et al., 2009) (Figure 1). CBS and CSE metabolize cysteine and/or homocysteine to release H2S, whereas 3MST produces H2S from 3-mercaptopyruvate (3MP), which is produced by the action of cysteine aminotransferase (CAT) on cysteine and α-ketoglutarate (Cooper, 1983; Shibuya et al., 2009a,b). 3MST requires cofactors to reduce a persulfide intermediate generated between a cysteine residue of 3MST and a sulfide provided by 3MP. We recently found that thioredoxin and dihydrolipoic acid (DHLA) are endogenous reducing cofactors that facilitate H2S release from 3MST (Mikami et al., 2011a). We also found that the 3MST/CAT pathway is regulated by Ca2+ (Mikami et al., 2011b; Mikami and Kimura, 2012) and that the activity of CBS is enhanced by S-adenosyl methionine (SAM; Abe and Kimura, 1996). Thus, the physiological stimuli that alter the intracellular levels of Ca2+ and SAM should be investigated in order to clarify the regulation of H2S production.
Rate of H2S Metabolism
H2S is metabolized in the mitochondria, via the sulfide oxidation pathway (Hildebrandt and Grieshaber, 2008). The first step is catalyzed by a membrane-bound sulfide:quinone oxidoreductase (SQR), which oxidizes H2S to persulfide. SQRs were initially identified in invertebrates, and their mammalian counterpart SQRs were later discovered (Theissen et al., 2003; Theissen and Martin, 2008). In the presence of O2 and water, sulfur dioxygenase oxidizes persulfide to sulfite, which is combined with another persulfide molecule by sulfur transferase rhodanese in order to producing thiosulfate (Figure 1). Because H2S consumption by a dioxygenase is high in the presence of O2 (Furne et al., 2008), high H2S production is offset by rapid H2S clearance under aerobic conditions, accounting for very low basal levels of H2S. Small deviations in the rates of H2S production and clearance may lead to rapid and several-fold changes in the H2S levels (Vitvitsky et al., 2012).
Storage of H2S as Bound Sulfane Sulfur and Its Associated Release
In addition to enzymatic regulation of H2S levels, H2S may be stored in proteins as bound sulfane sulfur that is divalent sulfur bound mostly to sulfur of cysteine residues. (Ishigami et al., 2009) (Figure 1). Exogenously applied H2S is absorbed and stored as bound sulfane sulfur, and the rate of absorption varies according to tissue types. Cells expressing 3MST and CAT have higher levels of bound sulfane sulfur than control cells (Shibuya et al., 2009a,b). In contrast, cells expressing 3MST mutants – that lack the ability to produce H2S – retain control-like levels of bound sulfane sulfur. Therefore, H2S produced by enzymes is stored in cells as bound sulfane sulfur. Bound sulfane sulfur releases H2S under reducing conditions. In the presence of major cellular reducing substances, glutathione, cysteine, and dihydrolipoic acid (DHLA) at their physiologic concentrations, H2S is released from lysates of cultured neurons and astrocytes at pH 8.0–8.4 (Ishigami et al., 2009; Mikami et al., 2011a). When neurons are excited, sodium ions enter and potassium ions exit from cells, resulting in high potassium concentrations in the extracellular environment, which depolarizes the membrane of surrounding astrocytes. To recover from depolarization, cotransporters are activated in astrocytes (Brookes and Turner, 1994). The entrance of causes alkalinization. Approximately 10% of the astrocytes shifted their intracellular pH to 8.4, which can induce bound sulfane sulfur to release H2S (Ishigami et al., 2009).
The concentrations of H2S can vary locally in restricted areas of cells, while the reported values are those averaged over whole tissues, using tissue homogenates or blood samples (Furne et al., 2008; Ishigami et al., 2009; Wintner et al., 2010). Such local variations in H2S concentrations must be determined. Recently developed H2S-sensitive fluorescence probes are able to estimate local H2S levels in live cells (Lippert et al., 2011; Liu et al., 2011; Peng et al., 2011; Qian et al., 2011; Sasakura et al., 2011). However, the probes bind to H2S irreversibly; the detectable limit for H2S is approximately 5–30 μM, i.e., values higher than the basal or steady state concentrations; and the time to reach maximum sensitivity is between 3 min and 1 h. The development of probes able to detect rapid changes in H2S concentrations is awaited.
The metabolic balance between the production and clearance of H2S has a considerable effect on its endogenous concentrations. In addition, because of bound sulfane sulfur, which releases and absorbs H2S, the H2S concentrations change more rapidly and extensively. The discrepancy between the endogenous basal concentrations and the effective-concentrations of exogenously applied H2S has been discussed frequently. Some effects of H2S may be elicited by its entry into cells through the plasma membrane. However, the rate of permeation and how freely and rapidly H2S diffuses within the cytosol have not been determined. We also need to clarify the H2S concentrations reached when cells are stimulated and the associated mechanism.
In conclusion, the basal or steady state levels of H2S have been re-evaluated and found to be much lower than those previously reported. There is a difference between the steady state- and the effective-concentrations of H2S. Therefore, it is urgent to determine the local concentrations of H2S achieved when cells are stimulated.
Acknowledgments
This work was supported by a grant from National Institute of Neuroscience and KAKENHI (23659089) from Grant-in-Aid for Challenging Exploratory Research to Hideo Kimura.
References
Abe, K., and Kimura, H. (1996). The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 16, 1066–1071.
Brookes, N., and Turner, R. J. (1994). K+-induced alkalinization in mouse cerebral astrocytes mediatged by reversal of elecgtrogenic Na+-HCO3- cotransport. Am. J. Physiol. 267, C1633–C1640.
Cai, W. J., Wang, M. J., Moore, P. K., Jin, H. M., Yao, T., and Zhu, Y. C. (2007). The novel proangiogenic effect of hydrogen sulfide is dependent on Akt phosphorylation. Cardiovasc. Res. 76, 29–40.
Chiku, T., Padovani, D., Zhu, W., Singh, S., Vitvitsky, V., and Banerjee, R. (2009). H2S biogenesis by human cystathionine γ-lyase leads to the novel sulfur metabolites lanthionine and homolanthionine and is responsive to the grade of hyperhomocysteinemia. J. Biol. Chem. 284, 11601–11612.
Cooper, A. J. L. (1983). Biochemistry of sulfur-containing amino acids. Annu. Rev. Biochem. 52, 187–222.
Dello Russo, C., Tringali, G., Ragazzoni, E., Maggiano, N., Menini, E., Vairano, M., Preziosi, P., and Navarra, P. (2000) Evidence that hydrogen sulphide can modulate hypothalamo-pituitary-adrenal axis function: in vitro and in vivo studies in the rat. J. Neuroendocrinol. 12, 225–233.
Elrod, J. W., Calvert, J. W., Morrison, J., Doeller, J. E., Kraus, D. W., Tao, L., Jiao, X., Scalia, R., Kiss, L., Szabo, C., et al. (2007). Hydrogen sulfide attenuates myocardial ischemia-reperfusion injury by preservation of mitochondrial function. Proc. Natl. Acad. Sci. U.S.A. 104, 15560–15565.
Furne, J., Saeed, A., and Levitt, M. D. (2008). Whole tissue hydrogen sulfide concentrations are orders of magnitude lower than presently accepted values. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R1479–R1498.
Goodwin, L. R., Francom, D., Dieken, F. P., Taylor, J. D., Warenycia, M. W., Reiffenstein, R. J., and Dowling, G. (1989). Determination of sulfide in brain tissue by gas dialysis/ion chromatography: postmortem studies and two case reports. J. Anal. Toxicol. 13, 105–109.
Hildebrandt, T. M., and Grieshaber, M. K. (2008). Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS. J. 275, 3352–3361.
Hosoki, R., Matsuki, N., and Kimura, H. (1997). The possible role of hydrogen sulfide as an endogenous smooth muscle relaxant in synergy with nitric oxide. Biochem. Biophys. Res. Commun. 237, 527–531.
Ishigami, M., Hiraki, K., Umemura, K., Ogasawara, Y., Ishii, K., and Kimura, H. (2009). A source of hydrogen sulfide and a mechanism of its release in the brain. Antioxid. Redox Signal. 11, 205–214.
Kaneko, Y., Kimura, Y., Kimura, H., and Niki, I. (2006). L-cysteine inhibits insulin release from the pancreatic beta-cell: possible novel gasotransmitter. Diabetes 55, 1391–1397.
Kimura, Y., and Kimura, H. (2004). Hydrogen sulfide protects neurons from oxidative stress. FASEB J. 18, 1165–1167.
Li, L., Bhatia, M., Zhu, Y. Z., Zhu, Y. C., Ramnath, R. D., Wang, Z. J., Anuar, F. B., Whiteman, M., Salto-Tellez, M., and Moore, P. K. (2005). Hydrogen sulfide is a novel mediator of lipopolysaccharide-induced inflammation in the mouse. FASEB J. 19, 1196–1198.
Lippert, A. R., New, E. J., and Chang, C. J. (2011). Reaction based fluorescent probes for selective imaging of hydrogen sulfide in living cells. J. Am. Chem. Soc. 133, 10078–10080.
Liu, C., Pan, J., Li, S., Zhao, Y., Wu, L. Y., Berkman, C. E., Whorton, A. R., and Xian, M. (2011). Capture and visualization of hydrogen sulfide by a fluorescent probe. Angew. Chem. Int. Ed. Engl. 50, 10327–10329.
Mikami, Y., and Kimura, H. (2012). A mechanism of retinal protection from light-induced degeneration by hydrogen sulfide. Commun. Integr. Biol. 5, 1–3.
Mikami, Y., Shibuya, N., Kimura, Y., Nagahara, N., Ogasawara, Y., and Kimura, H. (2011a). Thioredoxin and dihydrolipoic acid are required for 3-mercaptopyruvate sulfurtransferase to produce hydrogen sulfide. Biochem. J. 439, 479–485.
Mikami, Y., Shibuya, N., Kimura, Y., Nagahara, N., Yamada, M., and Kimura, H. (2011b). Hydrogen sulfide protects the retina from light-induced degeneration by the modulation of Ca2+ influx. J. Biol. Chem. 286, 39379–39386.
Nagai, Y., Tsugane, M., Oka, J., and Kimura, H. (2004). Hydrogen sulfide induces calcium waves in astrocytes. FASEB J. 18, 557–559.
Olson, K. R., Dombkowski, R. A., Russell, M. J., Doellman, M. M., Head, S. K., Whitfield, N. L., and Madden, J. A. (2006). Hydrogen sulfide as an oxygen sensor/transducer in vertebrate hypoxic vasoconstriction and hypoxic vasodilation. J. Exp. Biol. 209, 4011–4023.
Papapetropoulos, A., Pyriochou, A., Altaany, Z., Yang, G., Marazioti, A., Zhou, Z., Jeschke, M. G., Branski, L. K., Herndon, D. N., Wang, R., and Szabo, C. (2009). Hydrogen sulfide is an endogenous stimulator of angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 21972–21977.
Peng, H., Cheng, Y., Dai, C., King, A. L., Predmore, B. L., Lefer, D. J., and Wang, B. (2011). A fluorescent probe for fast and quantitaqtive detection of hydrogen sulfide in blood. Angew. Chem. Int. Ed. Engl. 50, 9672–9675.
Predmore, B. L., and Lefer, D. J. (2010). Development of hydrogen sulfide-based therapeutics for cardiovascular disease. J. Cardiovasc. Transl. Res. 3, 487–498.
Qian, Y., Karpus, J., KIabil, O., Zhang, S.-Y., Zhu, H. -L., Banerjee, R., Zhao, J., and He, C. (2011). Selective fluorescent probes for live-cell monitoring of sulphide. Nat. Commun. 2, 495.
Sasakura, K., Hanaoka, K., Shibuya, N., Mikami, Y., Kimura, Y., Komatsu, T., Ueno, T., Terai, T., Kimura, H., and Nagano, T. (2011). Development of a highly selective fluorescence probe for hydrogen sulfide. J. Am. Chem. Soc. 133, 18003–18005.
Savage, J. C., and Gould, D. H. (1990). Determination of sulfide in brain tissue and rumen fluid by ion-interaction reversed-phase high-performance liquid chromatography. J. Chromatogr. 526, 540–545.
Shibuya, N., Mikami, Y., Kimura, Y., Nagahara, N., and Kimura, H. (2009a). Vascular endothelium exresses 3-mercaptopyruvate sulfurtransferase and produces hydrogen sulfide. J. Biochem. 146, 623–626.
Shibuya, N., Tanaka, M., Yoshida, M., Ogasawara, Y., Togawa, T., Ishii, K., and Kimura, H. (2009b). 3-Mercaptopyruvate sulfurtransferease produces hydrogen sulfide and bound sulfane sulfur in the brain. Antioxid. Redox Signal. 11, 703–714.
Singh, S., Padovani, D., Leslie, R. A., Chiku, T., and Banerjee, R. (2009). Relative contributions of cystathionine – synthase and – cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J. Biol. Chem. 284, 22457–22466.
Stipanuk, M. H., and Beck, P. W. (1982). Characterization of the enzymic capacity for cysteine desulphhydration in liver and kidney of the rat. Biochem. J. 206, 267–277.
Streng, T., Axelsson, H. E., Hedlund, P., Andersson, D. A., Jordt, S. E., Bevan, S., Andersson, K. E., Hogestatt, E. D., and Zygmunt, P. M. (2008). Distribution and function of the hydrogen sulfide-sensitive TRPA1 ion channel in rat urinary bladder. Eur. Urol. 53, 391–399.
Teague, B., Asiedu, S., and Moore, P. K. (2002). The smooth muscle relaxant effect of hydrogen sulphide in vitro: evidence for a physiological role to control intestinal contractility. Br. J. Pharmacol. 137, 139–145.
Theissen, U., Hoffmeister, M., Grieshaber, M., and Martin, W. (2003). Single eubacterial origin of eukaryotic sulfide:quinine oxidoreductase, a mitochondrial enzyme conserved from the early evolution of eukaryotes during anoxic and sulfidic times. Mol. Biol. Evol. 20, 1564–1574.
Theissen, U., and Martin, W. (2008). Sulfide:quinine oxidoreductase (SQR) from the lugworm Arenicola marina shows cyanide- and thioredoxin-dependent activity. FEBS J. 275, 1131–1139.
Vitvitsky, V., Kabil, O., and Banerjee, R. (2012). High turnover rates for hydrogen sulfide allow for rapid regulation of its tissue concentrations. Antioxid. Redox Signal. (in press).
Warenycia, M. W., Goodwin, L. R., Benishin, C. G., Reiffenstein, R. J., Grancom, D. M., Taylor, J. D., and Dieken, F. P. (1989). Acute hydrogen sulfide poisoning. Demonstration of selective uptake of sulfide by the brainstem by measurement of brain sulfide levels. Biochem. Pharmacol. 38, 973–981.
Whiteman, M., Armstrong, J. S., Chu, S. H., Jia-Ling, S., Wong, B. S., Cheung, N. S., Halliwell, B., and Moore, P. K. (2004). The novel neuromodulator hydrogen sulfide: an endogenous peroxynitrite ‘scavenger’? J. Neurochem. 90, 765–768.
Whitfield, N. L., Dreimier, E. L., Verdial, F. C., Skovgaard, N., and Olson, K. R. (2008). A reappraisal of H2S/sulfide concentration in vertebrate blood and its potential significance in ischemic preconditioning and vascular signaling. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1930–R1937.
Wintner, E. A., Deckwerth, T. L., Langston, W., Bengtsson, A., Leviten, D., Hill, P., Insko, M. A., Dumpit, R., VandenEkart, E., Toombs, C. F., and Szabo, C. (2010). A monobromobimane-based assay to measure the pharmacokinetic profile of reactive sulphide species in blood. Br. J. Pharmacol. 160, 941–957.
Yang, W., Yang, G., Jia, X., Wu, L., and Wang, R. (2005). Activation of KATP channels by H2S in rat insulin-secreting cells and the underlying mechanisms. J. Physiol. 569, 519–531.
Zanardo, R. C., Brancaleone, V., Distrutti, E., Fiorucci, S., Cirino, G., and Wallace, J. L. (2006). Hydrogen sulfide is an endogenous modulator of leukocyte-mediated inflammation. FASEB J. 20, 2118–2120.
Citation: Kimura H (2012) Metabolic turnover of hydrogen sulfide. Front. Physio. 3:101. doi: 10.3389/fphys.2012.00101
Received: 22 March 2012; Accepted: 30 March 2012;
Published online: 18 April 2012.
Copyright: © 2012 Kimura. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence:a2ltdXJhQG5jbnAuZ28uanA=