
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 10 January 2012
Sec. Aquatic Physiology
volume 2 - 2011 | https://doi.org/10.3389/fphys.2011.00107
This article is part of the Research Topic Aquaporins in fish - expression, localization and functional dynamics. View all 10 articles
The dogfish ortholog of aquaporin 4 (AQP4) was amplified from cDNA using degenerate PCR followed by cloning and sequencing. The complete coding region was then obtained using 5′ and 3′ RACE techniques. Alignment of the sequence with AQP4 amino acid sequences from other species showed that dogfish AQP4 has high levels (up to 65.3%) of homology with higher vertebrate sequences but lower levels of homology to Agnathan (38.2%) or teleost (57.5%) fish sequences. Northern blotting indicated that the dogfish mRNA was approximately 3.2 kb and was highly expressed in the rectal gland (a shark fluid secretory organ). Semi-quantitative PCR further indicates that AQP4 is ubiquitous, being expressed in all tissues measured but at low levels in certain tissues, where the level in liver > gill > intestine. Manipulation of the external environmental salinity of groups of dogfish showed that when fish were acclimated in stages to 120% seawater (SW) or 75% SW, there was no change in AQP4 mRNA expression in either rectal gland, kidney, or esophagus/cardiac stomach. Whereas quantitative PCR experiments using the RNA samples from the same experiment, showed a significant 63.1% lower abundance of gill AQP4 mRNA expression in 120% SW-acclimated dogfish. The function of dogfish AQP4 was also determined by measuring the effect of the AQP4 expression in Xenopus laevis oocytes. Dogfish AQP4 expressing-oocytes, exhibited significantly increased osmotic water permeability (Pf) compared to controls, and this was invariant with pH. Permeability was not significantly reduced by treatment of oocytes with mercury chloride, as is also the case with AQP4 in other species. Similarly AQP4 expressing-oocytes did not exhibit enhanced urea or glycerol permeability, which is also consistent with the water-selective property of AQP4 in other species.
Aquaporins (AQPs) are commonly known as cell membrane water channel proteins although various isoforms exist that exhibit other transport properties including having permeability’s for small solutes such as glycerol and urea. In mammals, there are 13 isoforms known (numbered 0–12), where AQPs 0, 1, 2, 4, 5 fall into a sub-group of water-selective channels and where AQP1 is the most ubiquitously expressed in mammalian tissues (Ishibashi et al., 2009; Zelanina, 2010). Most AQPs are reversibly inhibitable by mercury, although one exception to this is AQP4, whose original name was MIWC (mercury insensitive water channel; Hasegawa et al., 1994; Jung et al., 1994). Mammalian AQP4 is expressed in a number of tissues but is not as ubiquitous as AQP1 (Ishibashi et al., 2009), AQP4 is particularly abundant in the brain (Amiry-Moghaddam et al., 2010; Brian et al., 2010; Zelanina, 2010), but is also expressed in the retina (Goodyear et al., 2009; Hirrlinger et al., 2011), salivary gland (Delporte and Steinfeld, 2006), respiratory tract (Matsuzaki et al., 2009), heart and muscle (Butler et al., 2006; Wakayama, 2010), gastrointestinal tract (Ma and Verkman, 1999; Xu et al., 2009), and kidney (Nejsum, 2005). The mammalian AQP4 gene is also known to produce alternative splice forms (Crane et al. (2009); Strand et al., 2009; Fenton et al., 2010). Several of these variants produce proteins with a variable sized N-terminal region.
In lower vertebrates very little is known about AQPs. The sequence of AQP4 has been determined in both Pacific and Atlantic hagfish (Cutler, 2007a; Nishimoto et al., 2007) and information is also available on hagfish AQP9 (Cutler, 2006a), but very little information is available in other lower vertebrate taxonomic groups such as in the elasmobranches (Cutler et al., 2005; Cutler, 2006b; Cutler, 2007b). The studies presented in this article (and its companion article) were performed to begin to rectify this situation. However at the outset when little is known, basic information needs to be determined. This includes (1) showing which organs or tissues express AQP4 and at what level (2) determining the functional properties of expressed AQP4 protein to show functional similarities or difference to AQP4 in other species and (3) undertaking an initial investigation to see if AQP4 might be involved in body fluid homeostatic mechanisms by perturbing the fish’s osmotic environment to determine whether this effects the level of AQP4 expression.
Dogfish AQP4 was cloned and sequenced using well established methods (Cutler and Cramb, 2002a,b, 2008; Martinez et al., 2005) of degenerate PCR followed by 5′ and 3′ RACE. AQP4 degenerate primers were produced using an elasmobranch-targeted primer design with sequence information obtained from an aquaporin sequence cloned from the bull shark (Cutler et al., 2005). The sense primer was 5′-CAYRTIAGYGGI GCICAYRTIAAYCC-3′ and the antisense primer 5′-GGICCIACCCARTAIACCCART G-3′ the derived amino acid sequences of these primers are located at 117–125 and 257–264 of the alignment (as indicated) respectively (Figure 1.). Nucleotides using the base inosine (I) were incorporated into primers at positions of major sequence uncertainty. PCR conditions used with these primers varied but was essentially similar to: −96°C – 1 s, 55°C – 15 s, and 72°C – 30 s for 40 cycles. This amplifies an expected and actual cDNA fragment of 443 bp. cDNA for amplifications was made from dogfish total RNA from both rectal gland and kidney (extracted as in Cutler and Cramb, 2008), using Superscript III reverse transcriptase (Invitrogen) according to manufacturers instructions, with the exception that 65°C was used for RNA denaturation, as 1 μl of SUPERase•In thermostable RNase inhibitor was additionally added (Ambion). cDNA fragments were gel purified using a Gene clean kit (MP Biomedicals) and were cloned using a TOPO TA cloning kit for sequencing (Invitrogen). Sequencing reactions were performed by CUGI at Clemson University, S.C. 5′ and 3′ RACE primers were designed from the sequences obtained and RACE was performed using a Marathon RACE cDNA amplification kit (Clontech). Sequences were analyzed and aligned using Genejockey II software (Biosoft).
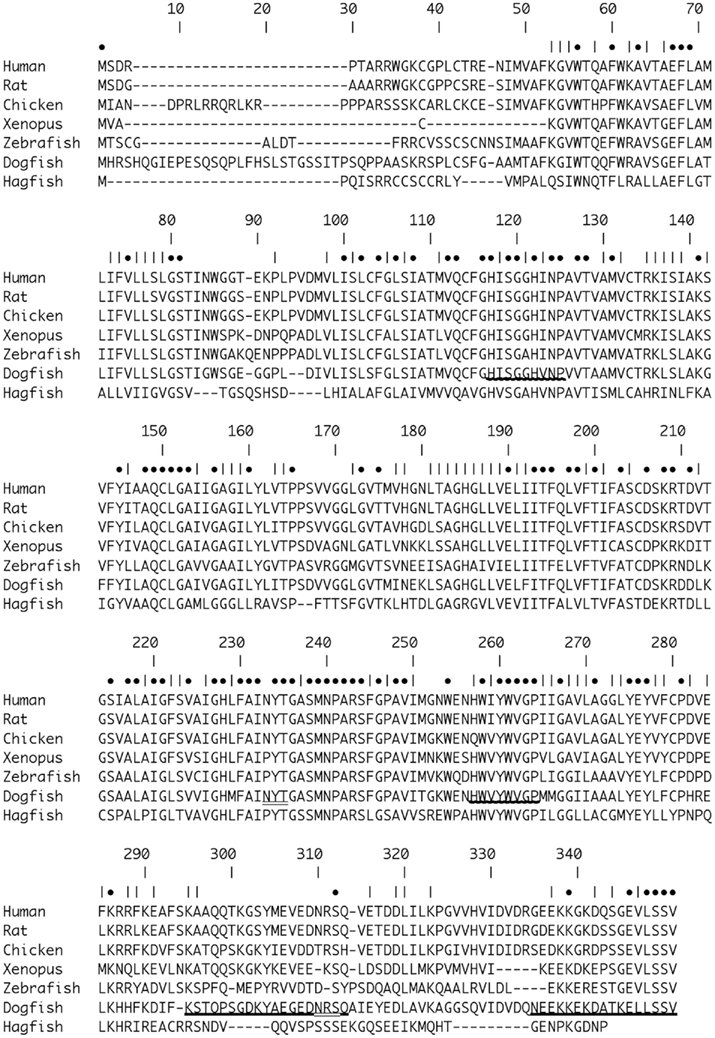
Figure 1. Alignment of Dogfish AQP4 [Accession number (Ac. No.) JF944824] amino acid sequence with AQP4 sequences from Human (Homo sapiens; Ac. No. NM_001650.4; 63.0%), Rat (Rattus norvegicus; Ac. No. AF144082; 62.5%), Chicken (Gallus gallus; Ac. No. NM_001004765; 65.3%), African Clawed Toad (Xenopus laevis; Ac. No. NM_001130949.1; 56.7%), Zebrafish (Danio rerio; NM_001003749; 57.5%), and Hagfish (Eptatretus burgeri; Ac. No. AB258403.1; 38.2%). Percentages in parentheses represent amino acid homologies. Numbers indicate position within the alignment. • Symbols indicate positions with identical amino acids. | Symbols indicate positions with chemically similar amino acids. Bold underline _ indicates the position of the peptide sequences used to raise the polyclonal antibodies. Wavy underline indicates the position of amino acids sequences used to make degenerate primers for initial AQP4 OCR amplifications. Double underline indicates the positions of putative N-glycosylation sites with the dogfish AQP4 sequence.
In order to determine whether AQP4 was directly involved in any kind of osmoregulatory or body fluid volume regulation processes in the shark, an attempt was made to disturb the shark regulatory system to try to cause changes in the expression of effector protein components. In other words, the external environmental salinity of the fish was manipulated to try to modulate the expression of aquaporin (and others) genes, such as AQP4. At the Mount Desert Island Biological Laboratory (MDIBL) at Salisbury Cove, in Maine, small (800–1500 g) adult mixed sex dogfish were (otherwise) randomly selected from a stock tank and were unfed during the experiment. All experimental animal protocols used were in compliance with IACUC regulations and had both GSU and MDIBL IACUC approval. The fish were placed in pairs in four-foot experimental tanks (approximately 1000 l). The acclimation of dogfish to dilute seawater (SW) in stages over 7 days was a protocol modified from Panabecker and Danzler (2005). A similar protocol to acclimate fish to 120% SW was then also devised. Two groups of sharks (six fish per group) were held in SW adjusted to either 75% salinity or 120% salinity in stages. The stages were 85% 3 days, 80% 2 days, and 75% 2 days or 110% 3 days, 115% 2 days, 120% 2 days. The acclimation of dogfish to dilute SW in stages over 7 days was a modified protocol from Panabecker and Danzler, 2005. A matching protocol to acclimate fish to 120% SW in stages was also then devised. Differing salinities were produced by the addition of de-chlorinated tap water, or sea salt (Instant Ocean) using a re-circulating system including a cooler to maintain temperature and biofilters. Control animals were kept in 100% salinity (normal) SW (around 31–32 ppt at MDIBL) for the same time period. Salinity was controlled using a model 85 dissolved oxygen, conductivity, salinity, and temperature meter (YSI). At the end of the 7 day experiment, fish were sacrificed and gill arches, kidney, rectal gland, esophagus (including the fundic stomach), and intestine were removed. Kidney and rectal gland were homogenized using a Polytron homogenizer (Kinematica). Gill epithelia were removed by scraping with a razor blade. Combined esophageal/cardiac stomach epithelia were removed by scraping with a microscope slide. The intestines were cleaned as much as possible and waste material removed. The intestinal epithelium was then also scraped using a microscope slide. Gill, esophageal/cardiac stomach, and intestinal epithelia were homogenized using a syringe and 16 gage needle. All tissues were homogenized in solution D for RNA extraction as previously outlined (Cutler and Cramb, 2008). Levels of AQP4 mRNA were then measured using Northern blots, performed using a 32P labeled AQP4 DNA probe (purified degenerate PCR fragment). Blots were quantified as previously, using electronic autoradiography via an Instant Imager (Canberra Packard). As the level of AQP4 expression in gill was too low to be detectable by this technique, quantitative PCR (QPCR) was instead employed using total RNA samples normalized utilizing rRNA levels measured with a gel documentation and analysis system (Syngene). This technique was used instead of the use of housekeeping genes as (1) few housekeeping are available for S. acanthias (2) In any case supposed housekeeping genes such as GAPDH (and others) can vary in different circumstances (De Jonge et al., 2007; McCurley and Callard, 2008) and (3) testing of housekeeping genes in any case requires comparison to rRNA levels. QPCR was performed using a MX 4000 QPCR machine and Brilliant II QPCR master mix (Stratagene). Statistical analysis of quantitative results was performed using Statview statistical analysis software (Abacus Concepts) using ANOVA and a Fisher post hoc test.
A full-length cDNA encompassing the entire AQP4 coding region was amplified from rectal gland cDNA using PCR employing highly accurate Phusion DNA polymerase (Finnzymes), the resulting DNA band was gel purified and cloned into a pXT7 expression vector, which contains a T7 promoter upstream of the multiple cloning site. Plasmid DNA for dogfish AQP4 was cut with restriction enzyme XbaI to linearize it and prevent run on of transcription. cRNA transcripts were produced using a mMessage mMachine kit (Ambion) utilizing T7 DNA-dependent RNA Polymerase. The cRNA produced was purified using phenol/chloroform extraction followed by isopropanol precipitation, dissolved in RNAse free water and quantified using a Biophotometer spectrophotometer (Eppendorf).
All experiments were done in accordance with IACUC approved protocols at Beth Israel Deaconess Medical Center. Xenopus laevis frogs (Harvard Institute of Medicine, Boston, MA, USA) were anesthetized in 1 l 0.5% (w/v) 3-aminobenzoic acid ethyl ester methanesulfonate salt (Tricaine) containing ice for 20 min. Oocytes were removed bilaterally from the abdominal cavity and the egg mass cut into small pieces and placed in calcium free ND96 buffer (in mM; 96 NaCl, 1 KCl, 1 MgCl2 5 Hepes, pH 7.5). Oocytes were then defolliculated in 2 mg/ml collagenase (Sigma-Aldrich), 0.2 mg/ml trypsin inhibitor (Sigma-Aldrich) in calcium free ND96 for 55 min with rotation on an Adams Nutator before washing three times with phosphate buffer [in mM; 100 K2HPO4, 0.1% (w/v) BSA, pH 6.5] and then allowing oocytes to incubate in phosphate buffer for 10 min at room temperature. Oocytes were transferred to calcium free ND96 and then to modified Barth’s solution [MBS; in mM; 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.82 MgSO4, 0.33 Ca(NO3)2, 0.41 CaCl2, 10 Hepes, pH 7.4, supplemented with 1% v/v penicillin/streptomycin] where they were maintained at 18°C. cRNA (10 ng) of saAQP4s was injected into oocytes using a Nanoject II Auto-Nanoliter Injector (Drummond Scientific Co.). Control oocytes were either injected with water alone or were uninjected. After 3 days incubation at 18°C, oocytes were tested for their ability to transport water, urea, or glycerol. Water transport kinetics were assessed at room temperature by quantitation of oocyte swelling after placement in hypotonic buffer (65% of normal MBS). Time lapse video microscopy was used to capture oocyte images every 10 s for 3 min. using an Olympus SZX7 binocular microscope equipped with a Scion CFW 1308C digital camera (1360 × 1024 pixel resolution).
Aquaporin activity was tested over a pH range of 6.6–8.6. For all experiments other than pH 7.4 oocytes were placed in MBS at the tested pH for 5 min and then were swelled in hypotonic MBS (65%) at the same pH. Studies investigating the inhibition of water permeability by mercury were performed at pH 7.4 in MBS containing 1 mM HgCl2.
The images were converted to black and white in ImageJ (Rasband, 1997–2011) using the Binary function and the cross-sectional pixel area was calculated with the Analyze Particle function. Data from ImageJ was exported to Microsoft Excel and areas from each image were normalized to 1.0 relative to a starting value. To calculate the permeability coefficient (Pf) the data were adjusted from area (A) to volume (V) by (A/Ao) = (V/Vo)3/2 (Zhang et al., 1990) then a second order polynomial equation was fit and the derivative of the equation was used to obtain the initial rate of swelling. Pf was calculated using the equation (Zhang et al., 1990):
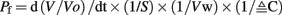
Where d(V/Vo)/dt is the rate from the curve fit, Vo is the initial volume of the oocyte, calculated as 5.2 × 10−4 cm3 based on a 1 mm diameter, S is the surface area of the oocyte (0.4 cm2 Zampighi et al., 1995), Vw is the molar volume of water (18 mol/cm3), and ΔC is the concentration difference of the applied hypo-osmotic solution in mol/cm3.
Solute fluxes were measured by isotopic uptake of [3H]glycerol and [14C]urea (American Radiolabeled Chemicals, St. Louis, MO, USA). Oocytes were incubated for varying times in MBS to which either 10 μCi/ml [14C]urea (55 mCi/mmol) and, or 10 μCi/ml [3H]glycerol (20–40 Ci/mmol), and then made to a final concentration of 2 mM with unlabelled urea or glycerol respectively. At the end of the incubation period for uptake, oocytes were washed six times with ice-cold buffer containing 5 mM solute. Individual oocytes were then placed in scintillation vials, had 300 μl 20% SDS added and vortex mixed for 10–15 s before addition of 4 ml Scintisafe scintillation cocktail. Vials were counted for 2 min in a Packard 1500 liquid scintillation analyzer.
A translation of the putative dogfish AQP4 cDNA sequence (Accession number JF944824) resulting from degenerate PCR and 5′ and 3′ RACE DNA fragments is shown in Figure 1. The combined cDNA produced, aligns with AQP4 sequences from other species. The homology of the dogfish amino acid sequence shares homology between 38.2 and 66.3%, ranging from hagfish (lowest) to chicken (highest; see Figure 1.) Interestingly, using the same alignment the homology of hagfish to human AQP4 was only 43.3%. The dogfish AQP4 sequence also shows two putative N-glycosylation sites at positions 233–235 and 310–312 of the amino acid alignment (Figure 1.) that are common to both human and rat AQP4, but that are absent in some other species.
The first priority following cloning experiments was to establish an expression profile for AQP4 in dogfish. This was performed in two different ways, initially using Northern blotting where a mRNA band of around 3.2 kb was detected, particularly in the rectal gland, but also with lower levels in the eye, esophagus/fundic stomach, skeletal muscle, stomach, brain, and kidney with no apparent signal from liver, gill, or intestine (Figure 2). Secondly this was followed up by a semi-quantitative PCR survey, to increase the sensitivity of detection (Figure 3). Similar results were obtained but bands were also detected with the following intensity, liver > gill > intestine. Although a band could be detected in intestine the level of expression was clearly extremely low.
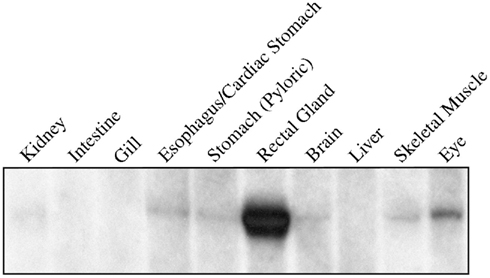
Figure 2. Northern blot of 5 μg of total RNA extracted from various tissues of the dogfish (S. acanthias) and probed with a 32P radioisotope-labeled cDNA fragment of the AQP4 gene. The probe hybridizes to a single band of 3.2 kbp.
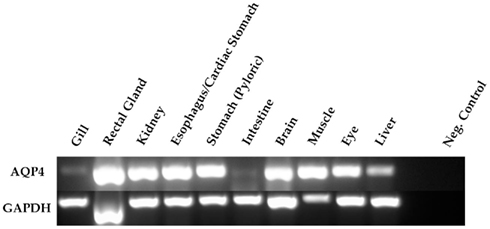
Figure 3. Semi-quantitative PCR amplification of dogfish AQP4 (274 bp) and GAPDH (271 bp) cDNA fragments. The primers used were also used for quantitative PCR and were designed across conserved intron–exon splice junctions of the respective genes to avoid genomic DNA amplification. 0.5 μl of cDNA template (made with the following microgram’s of total RNA; 1.15 gill; 0.67 rectal gland; 0.93 kidney; 0.88 esophagus/cardiac stomach; 0.99 stomach; 1.56 intestine; 0.41 brain; 0.21 muscle; 0.28 eye; 0.71 liver) was used in all reactions and GAPDH was used as a positive control. The negative (Neg.) control reactions were without cDNA.
In conjunction with the Northern blotting and semi-quantitative PCR, the level of mRNA expression measured in (100%) SW fish (Figure 4) showed higher levels of AQP4 mRNA expression in rectal gland, with lower levels in kidney and esophagus/cardiac stomach. Expression in gill and intestine could not be detected using this technique. The effect of adjusting the external environmental salinity of the fish made no significant difference to the level of AQP4 mRNA expression in rectal gland, kidney, or esophagus/cardiac stomach. In order to carry out a similar type of analysis on the gill (more sensitive) QPCR was employed to determine mRNA expression levels in the 75, 100, and 120% SW fish. In this case, the fish held in 120% SW showed a significant 63.1% lower level of AQP4 mRNA in comparison to the 100% SW control dogfish (Figure 5).
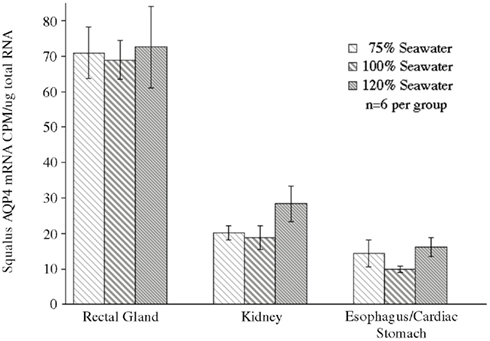
Figure 4. Analysis of a Northern blot experiment measuring the mRNA expression of aquaporin 4 (AQP4) in the tissue of dogfish acclimated to different external environmental salinities. No significant differences were seen in any tissue between any of the salinities.
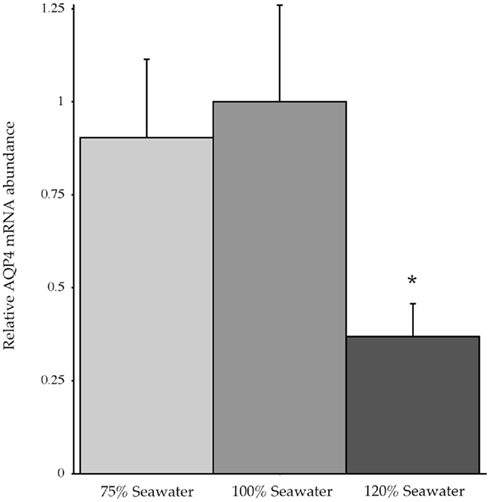
Figure 5. Relative dogfish AQP4 mRNA abundance in the gills of fish acclimated to 75, 100, or 120% seawater (SW). mRNA expression was determined using quantitative PCR, with primers designed at conserved exon–intron splice junctions within the gene sequence to avoid amplification of genomic DNA. n = 6 Fish per group. * = Statistically significant difference between 75 and 100% SW fish, where p < 0.05.
Experiments measuring the function of AQP4 expressed in Xenopus laevis oocytes, showed that water permeability (Pf) was significantly higher in oocytes expressing AQP4 than in controls (Figure 6). The level of oocyte permeability also did not vary across a range of pH values. The effect of mercury on permeability was also measured (Figure 7), and this did cause a small reduction in permeability but this was not statistically significant. Additionally, as some AQP isoforms also show a urea and glycerol permeability, the uptake of isotopic versions of these molecules was tested and no significantly higher level of permeability was found in AQP4 expressing-oocytes compared to controls (Figures 8 and 9).
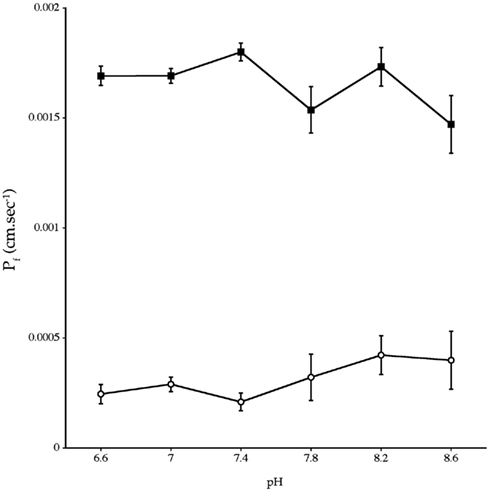
Figure 6. Osmotic water permeability (Pf) of Xenopus laevis oocytes micro-injected with dogfish AQP4 cRNA (-■-) or with H2O (-○-). The data are averages of four experiments where measurements were made at six different pH values with an average of eight oocytes per group, at each pH value, in each experiment.
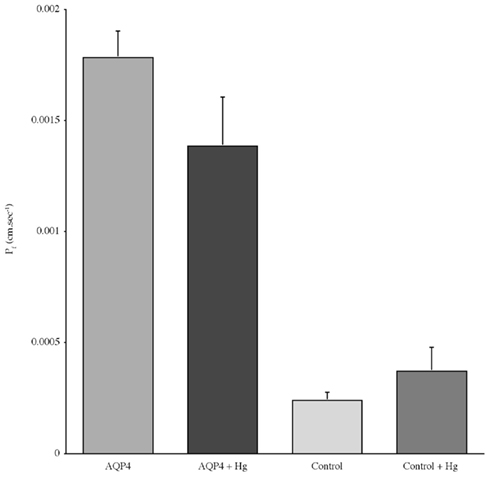
Figure 7. The effect of mercury chloride (Hg) on the osmotic water permeability (Pf) of Xenopus oocytes micro-injected with dogfish AQP4 cRNA or in with H2O (Control). Data represent averages of three experiments, with approximately eight oocytes per group in each. There was no statistically significant difference in Pf with or without mercury chloride.
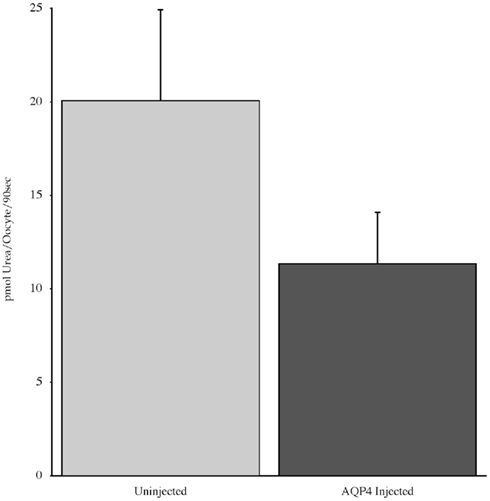
Figure 8. The uptake of C-labeled urea in Xenopus laevis oocytes micro-injected with dogfish AQP4 cRNA or uninjected. Results are averages from three experiments with approximately eight oocytes used per group.
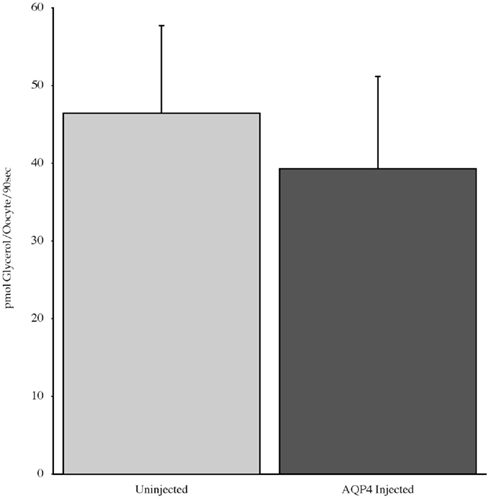
Figure 9. The uptake of C-labeled glycerol in Xenopus laevis oocytes micro-injected with dogfish AQP4 cRNA or uninjected. Results are averages from three experiments with approximately eight oocytes used per group.
This article documents the characteristics of an elasmobranch aquaporin for the first time. Despite the relatively ancient origin of elasmobranchs, in common with Agnathans such as the hagfish (elasmobranchs, around 528 million years old and Agnathans, around 564 million years old; Kumar and Hedges, 1998), dogfish AQP4 shares a much higher level of amino acid homology with mammalian AQP4 sequences than does the hagfish AQP4 sequence. As has been seen with other elasmobranch gene sequences, dogfish AQP4 has a higher level of homology (63%) to human AQP4 than the somewhat more recently developed teleost fish (450 million years old; Kumar and Hedges, 1998) AQP4 sequence from zebrafish (60.4%). This therefore may represent another example of an apparently slower rate of evolutionary change in cartilaginous fish in comparison to teleost fish.
Due to the relatively high level of amino acid homology of AQP4 sequences, it might be expected that dogfish AQP4 might be functionally similar to homologs in other species such as mammals. The oocyte functional expression studies here show that that was indeed the case in as much as, dogfish AQP4 expression produced significant oocyte water permeability irrespective of pH (other AQP isoforms can be affected by pH; Zelenina et al., 2003; MacIver et al., 2009), and it is permeability was insensitive to mercury (Hasegawa et al., 1994). The cysteine residue which is often present in most AQPs and is thought to be responsible for mercury-sensitive inhibition of AQPs is located at position 237 in the amino acid alignment (Figure 1; Yukutake et al., 2008). In AQP4 sequences from other species such as mammals, this residue is instead an alanine, hence explaining the mercury insensitivity of mammalian AQP4. Dogfish AQP4 also possesses an alanine residue at this position and this would therefore indicate that it is also likely to be insensitive to mercury. The Oocyte expression experiments also showed that dogfish AQP4 has no capacity to transport urea or glycerol in a similar way to mammalian AQP4 (Ishibashi et al., 2009).
In contrast to the tissue distribution of AQP4 mRNA expression in mammals, where there is significant expression in the GI tract but no expression in liver, in the dogfish there appears to be little (although still some) expression in intestine but definite expression seen in liver (Ishibashi et al., 2009). Dogfish AQP4 expression was seen at some level in every tissue tested and its expression is consequently more widespread than dogfish AQP1 (data not shown). Whereas, in mammals AQP1 is viewed as the ubiquitous aquaporin isoform (Zelanina et al., 2005; Ishibashi et al., 2009). There have been some suggestions that the intestine may play a role in osmoregulatory and/or body fluid homeostatic mechanisms in elasmobranch fish (Anderson et al. (2007)), but the low level of intestinal AQP4 mRNA expression in this study clearly suggests that this is unlikely to involve AQP4 at least in the dogfish.
Another aspect of interest is the presence of AQP4 expression in the rectal gland. Shark rectal gland has been a model for ion/fluid transport studies for decades, as the tissue is dedicated to regulated iso-osmotic fluid secretion. As many studies have concentrated on sodium and chloride ion transport over the years, it might be expected that the rectal gland function principally concerned the eradication of excess ions (Karnaky, 1997; Evans et al., 2004; Evans and Claiborne, 2008). However a series of nice studies by Solomon et al. (1984a,b, 1985) showed that ion secretion by the rectal gland was not stimulated when the animal was perfused with hypertonic shark ringer solution (they raised plasma salinity without changing body fluid volume), but was stimulated when body fluid volume was increased using isotonic shark ringer. This strongly suggests that the principle function of the rectal gland is actually to eradicate excess water but due to the passive nature of water transport, ions have to be transported to allow the water to follow by osmosis. Initial studies investigating osmotic water permeability of rectal gland cell membranes found little functional evidence to support the presence of AQPs (Zeidel et al., 2005).
However the fluid transport function of the rectal gland would suggest that this organ would likely contain AQPs. The Northern blot and PCR data in this study suggest that the rectal gland is likely to possess abundant aquaporin proteins but clearly further work needs to be done to determine what the role of AQP4 in the rectal gland is. A curious aspect of AQP4 rectal gland expression is that manipulation of the fish’s environmental salinity did not significantly alter AQP4 mRNA expression. It would be anticipated that placing the fish in a hyper-osmotic environment such as 120% SW would result in loss of water across the gills by osmosis and compel the fish’s regulatory system to retain both sodium chloride and urea to raise the internal osmotic concentration to match that of the external environment and to shut down rectal gland output. Placing the fish in a hypo-osmotic environment such as 75% SW, should have the opposite regulatory effect. Work in other Elasmobranch species shows that placing the fish in a dilute environment increases body fluid volume and consequently body mass (Anderson et al., 2007) as would be expected and this increases rectal gland secretion (Wong and Chan, 1977; Anderson et al. (2002)). The lack of change in AQP4 mRNA levels in either 75 or 120% SW suggests that either AQP4 is not involved directly in rectal gland fluid transport or that regulation occurs at another level (such as shutting down blood flow to the gland) eliminating any need to change AQP4 expression. The relatively high level of mRNA expression in the rectal gland might also indicate post-translational regulation of AQP4 protein production as seen with AQPs in other species (Ishibashi et al., 2009).
The change in AQP4 mRNA expression that was seen in the gill is also interesting. Its possible to hypothesize that in a hyper-osmotic environment such as 120% SW, there would likely tend to be a net outflow of water across the gills and if this produced osmotic problems (dehydration) for the fish, one possible response by the fish’s regulatory system would be to decrease branchial AQP4 expression to reduce gill permeability to slow down water loss. Dogfish tend to hold their body fluid osmotic concentration slightly above that of the external environment (Karnaky, 1997; Evans et al., 2004; Evans and Claiborne, 2008), which normally allows a small continual water influx across the gill, which presumably matches outflows through urine production and rectal/rectal gland fluid output. Again its possible to hypothesize that if gill osmotic water inflows were reversed this may ultimately cause problems for urine production, some minimal level of which is necessary to remove toxic metabolites, and this may explain the need to down-regulate branchial AQP4 mRNA expression, to reduce gill permeability. However, one complicating factor for that idea is that some elasmobranch species are known to drink in response to dehydration and this would reduce the need to adjust gill permeability (Anderson et al. (2007)). The other result from gill is also curious because a hypo-osmotic external environment such as 75% SW might have been expected to have the opposite effect of 120% SW (i.e., increased AQP4 mRNA expression in gill). However in that case, again it is possible to hypothesize that if there are additional water inflows across the gill, the fish can merely increase urine output and decrease urine concentration to mitigate the increased branchial influx. While it is possible to speculate as to the possible reasons for the level of branchial AQP4 mRNA expression in different environments, its clear further studies needs to be performed to elucidate the actual regulatory mechanisms at work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Christopher P. Cutler is in receipt of a grant from the National Science Foundation, NSF IOS 0844818.
Amiry-Moghaddam, M., Hoddevik, E. H., and Otterson, O. P. (2010). Aquaporins: multifarious roles in brain. Neuroscience 168, 859–861.
Anderson, W. G., Good, J. P., and Hazon, N. (2002). Changes in the secretion rate and vascular perfusion in the rectal gland of the European lesser spotted dogfish (Scyliorhinus canicula L.) in response to environmental and hormonal stimuli. J. Fish Biol. 60, 1580–1590.
Anderson, W. G., Taylor, J. R., Good, J. P., Hazon, N., and Grosell, M. (2007). Body fluid regulation in elesmobranch fish. Comp. Biochem. Physiol. 148, 3–13.
Brian, O. K., Tom, P., and Wang, D. (2010). Aquaporins: relevance to cerebrospinal fluid physiology and therapeutic potential in hydrocephalus. Cerebrospinal Fluid Res. 7, 15.
Butler, T. L., Au, C. G., Yang, B., Egan, J. R., Tan, Y. M., Hardeman, E. C., North, K. N., Verkman, A. S., and Winlaw, D. S. (2006). Cardiac aquaporin expression in humans rats and mice. Am. J. Physiol. 291, H705–H713.
Crane, J. M., Bennett, J. L., and Verkman, A. S. (2009). Live cell analysis of Aquaporin-4 M1/M23 interactions and regulated orthogonal array assembly in glial cells. J. Biol. Chem. 284, 35850–35860.
Cutler, C. P. (2006a). Cloning of an aquaporin 9 gene orthologue from the hagfish (Myxine glutinosa). Bull. Mt. Desert Isl. Biol. Lab. 45, 42–43.
Cutler, C. P. (2006b). Cloning of aquaporin 1e gene homologues in the dogfish shark (Squalus acanthias) and hagfish (Myxine glutinosa). Bull. Mt. Desert Isl. Biol. Lab. 45, 40–41.
Cutler, C. P. (2007a). Nucleotide and derived-amino acid sequence of a putative Atlantic hagfish (Myxine glutinosa) aquaporin 4 cDNA. Bull. Mt. Desert Isl. Biol. Lab. 46, 21–22.
Cutler, C. P. (2007b). Cloning and identification of four aquaporin genes in the dogfish shark (Squalus acanthias). Bull. Mt. Desert Isl. Biol. Lab. 46, 19–20.
Cutler, C. P., and Cramb, G. (2002a). Branchial expression of an aquaporin 3 (AQP-3) homologue is downregulated in the European eel (Anguilla anguilla) following seawater acclimation. J. Exp. Biol. 205, 2643–2651.
Cutler, C. P., and Cramb, G. (2002b). Two isoforms of the Na+/K+/2Cl-cotransporter (NKCC1) are expressed in the European eel (Anguilla anguilla). Biochim. Biophys. Acta 1566, 92–103.
Cutler, C. P., and Cramb, G. (2008). Differential expression of absorptive cation-chloride cotransporters in the intestinal and renal tissues of the European eel (Anguilla anguilla). Comp. Biochem. Physiol. 149, 63–73.
Cutler, C. P., Meischke, L., and Cramb, G. (2005). Evolutionary and comparative analysis of aquaporin water channel genes in fish. Bull. Mt. Desert Isl. Biol. Lab. 44, 55.
De Jonge, H. J. M., Fehrmann, R. S. N., de Bont, E. S. J. M., Hofstra, R. M. W., Gerbens, F., Kamps, W. A., de Vries, E. G. E., van der Zee, A. G. J., Meerman, G. J. T., and Elst, A. T. (2007). Evidence based selection of housekeeping genes. PLoS ONE 2, e898. doi:10.1371/journal.pone.0000898
Delporte, C., and Steinfeld, S. (2006). Distribution and roles of aquaporins in salivary glands. Biochim. Biophys. Acta 1758, 1061–1070.
Evans, D. H., Piermarini, P. M., and Choe, K. P. (2004). “Homeostasis: osmoregulation, pH regulation, and nitrogen excretion,” in Biology of Sharks and Their Relatives, eds J. C. Carrier, J. A. Musick, and M. R. Heithaus (Boca Raton: CRC Press), 247–268.
Evans, D. H., and Claiborne, J. B. (2008). “Osmotic and ionic regulation in fishes,” in Osmotic and Ionic Regulation: Cells and Animals, ed. D. H. Evans (Boca Raton: CRC Press), 295–366.
Fenton, R. A., Moeller, H. B., Zelenina, M., Snaebjornsson, M. T., Holen, T., and MacAulay, N. (2010). Differential water permeability and regulation of three aquaporin 4 isoforms. Cell. Mol. Life Sci. 67, 829–840.
Goodyear, M. J., Crewther, S. G., and Junghans, B. M. (2009). A role for aquaporin-4 in fluid regulation in the inner retina. Vis. Neurosci. 26, 159–165.
Hasegawa, H., Ma, T., Skach, W., Matthay, M. A., and Verkman, A. S. (1994). Molecular cloning of a mercurial-insensitive water channel expressed in selected water-transporting tissues. J. Biol. Chem. 269, 5497–5500.
Hirrlinger, P. G., Pannicke, T., Winkler, U., Claudepierre, T., Varshney, S., Schulze, C., Reichbach, A., Brunken, W. J., and Hirrlinger, J. (2011). Genetic deletion of laminin isoforms b2 and c3 induces a reduction in Kir4.1 and aquaporin-4 expression and function in the retina. PLoS ONE 6, e16106. doi:10.1371/journal.pone.0016106
Ishibashi, K., Hara, S., and Kondo, S. (2009). Aquaporin water channels in mammals. Clin. Exp. Nephrol. 13, 107–117.
Jung, J. S., Bhat, R. V., Preston, G. M., Guggino, W. B., Baraban, J. M., and Agre, P. (1994). Molecular Characterization of an aquaporin cDNA from brain: candidate osmoreceptor and regulator of water balance. Proc. Natl. Acad. Sci. U.S.A. 91, 13052–13056.
Karnaky, K. J. Jr. (1997). “Osmotic and ionic regulation,” in The Physiology of Fishes, ed. D. H. Evans (Boca Raton: CRC Press), 157–176.
Kumar, S., and Hedges, S. B. (1998). A molecular timescale for vertebrate evolution. Nature 392, 917–920.
Ma, T., and Verkman, A. S. (1999). Aquaporin water channels in gastrointestinal physiology. J. Physiol. 517, 317–326.
MacIver, B., Cutler, C. P., Yin, J., Hill, M. G., Zeidel, M. L., and Hill, W. G. (2009). Expression and functional characterization of four aquaporin water channels from the European eel (Anguilla anguilla). J. Exp. Biol. 212, 2856–2863.
Martinez, A.-S., Cutler, C. P., Wilson, G., Phillips, C., Hazon, N., and Cramb, G. (2005). Cloning and expression of three aquaporin homologues from the European eel (Anguilla anguilla), effects of seawater acclimation and cortisol treatment on renal expression. Biol. Cell 97, 615–627.
Matsuzaki, T., Hata, H., Ozawa, H., and Takata, K. (2009). Immunohistochemical localization of the aquaporins AQP1, AQP3, AQP4 and AQP5 in the mouse respiratory system. Acta Histochem. Cytochem. 42, 159–169.
McCurley, A. T., and Callard, G. V. (2008). Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 9, 102. doi:10.1186/1471-2199-9-102
Nejsum, L. N. (2005). The renal plumbing system: aquaporin water channels. Cell. Mol. Life Sci. 62, 1692–1706.
Nishimoto, G., Sasaki, G., Yaoita, E., Nameta, M., Li, H., Furuse, K., Fujinaka, H., Yoshida, Y., Mitsudome, A., and Yamamoto, T. (2007). Molecular characterization of water-selective AQP (EbAQP4) in hagfish: insight into ancestral origin of AQP4. Am. J. Physiol. 292, R644–R651.
Panabecker, T. L., and Danzler, W. H. (2005). Renal gene expression in Squalus acanthias following hyposmotic stress. Bull. Mt. Desert Isl. Biol. Lab. 44, 78–79.
Solomon, R., Taylor, M., Sheth, S., Silva, P., and Epstein, F. H. (1985). Primary role of volum expansion in stimulation of the rectal gland function. Am. J. Physiol. 248, R638–R640.
Solomon, R., Taylor, M., Stoff, J. S., Silva, P., and Epstein, F. H. (1984a). In vivo effect of volume expansion on rectal gland function. I. Humoral factors. Am. J. Physiol. 246, R63–R66.
Solomon, R., Taylor, M., Rosa, R., Silva, P., and Epstein, F. H. (1984b). In vivo effect of volume expansion on rectal gland function. II. Hemodynamic changes. Am. J. Physiol. 246, R67–R71.
Strand, L., Moe, S. E., Solbu, T. T., Vaadal, M., and Holen, T. (2009). Roles of aquapoin-4 isoforms and amino acids in square array assembly. Biochemistry 48, 5785–5793.
Wakayama, Y. (2010). Aquaporin expression in normal and pathological skeletal muscles: a brief review with focus on AQP4. J. Biomed. Biotechnol. 2010, 1–9.
Wong, T. M., and Chan, D. K. O. (1977). Physiological adjustments to dilution of the external medium in the lip-shark Hemiscyllium plagiosum (Bennett): II. Branchial, renal, and rectal gland function. J. Exp. Zool. 200, 85–96.
Xu, H., Zhang, W., Wei, W., Shen, L., and Wu, W. (2009). Differential expression of aquaporin-4 in human gastric normal and cancer tissues. Gastroenterol. Clin. Biol. 33, 72–76.
Yukutake, Y., Tsuji, S., Hirano, Y., Adachi, T., Takahashi, T., Fujihara, K., Agre, P., Yasui, M., and Suematsu, M. (2008). Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol. Cell 100, 355–363.
Zampighi, G. A., Kreman, M., Boorer, K. J., Loo, D. D., Bezanilla, F., Chandy, G., Hall, J. E., and Wright, E. M. (1995). A method for determining the unitary functional capacity of cloned channels and transporters expressed in Xenopus laevis oocytes. J. Membr. Biol. 148, 65–78.
Zeidel, J. D., Mathei, J. C., Campbell, J. D., Ruiz, W. G., Apodaca, G. L., Riordan, J., and Zeidel, M. L. (2005). Selective permeability barrier to urea in shark rectal gland. Am. J. Physiol. 289, F83–F89.
Zelanina, M., Zelenin, S., and Aperia, A. (2005). Water channels (aquaporins) and their role for postnatal adaptation. Pediatr. Res. 57, 47R–53R.
Zelenina, M., Bondar, A. A., Zelenin, S., and Aperia, A. (2003). Nickel and extracellular acidification inhibit the water permeability of human aquaporin-3 in lung epithelial cells. J. Biol. Chem. 278, 30037–30043.
Keywords: aquaporin 4, shark, dogfish, kidney, liver, rectal gland, gill, cardiac stomach
Citation: Cutler CP, MacIver B, Cramb G and Zeidel M (2012) Aquaporin 4 is a ubiquitously expressed isoform in the dogfish (Squalus acanthias) shark. Front. Physio. 2:107. doi: 10.3389/fphys.2011.00107
Received: 02 July 2011;
Accepted: 05 December 2011;
Published online: 10 January 2012.
Edited by:
Steffen Madsen, University of Southern Denmark, DenmarkReviewed by:
Martin Tresguerres, Scripps Institution of Oceanography UCSD, USACopyright: © 2012 Cutler, MacIver, Cramb and Zeidel. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Christopher P Cutler, Department of Biology, Georgia Southern University, 69 Georgia Avenue, Building 202, Statesboro, GA 30460-8042, USA. e-mail:Y2N1dGxlckBnZW9yZ2lhc291dGhlcm4uZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.