- Department of Biological Sciences, University of Calgary, Calgary, AB, Canada
Demethylases play a pivitol role in numerous biological processes from covalent histone modification and DNA repair to specialized metabolism in plants and microorganisms. Enzymes that catalyze O- and N-demethylation include 2-oxoglutarate (2OG)/Fe(II)-dependent dioxygenases, cytochromes P450, Rieske-domain proteins and flavin adenine dinucleotide (FAD)-dependent oxidases. Proposed mechanisms for demethylation by 2OG/Fe(II)-dependent enzymes involve hydroxylation at the O- or N-linked methyl group followed by formaldehyde elimination. Members of this enzyme family catalyze a wide variety of reactions in diverse plant metabolic pathways. Recently, we showed that 2OG/Fe(II)-dependent dioxygenases catalyze the unique O-demethylation steps of morphine biosynthesis in opium poppy, which provides a rational basis for the widespread occurrence of demethylases in benzylisoquinoline alkaloid metabolism.
Introduction
Demethylation is a key aspect of many diverse biological processes including epigenetic regulation, DNA repair, toxin degradation, and the metabolism of bioactive metabolites. A number of enzyme families have been implicated in O- and N-demethylation reactions, including cytochromes P450 (P450), FAD-dependent amine oxidases, 2-oxoglutarate (2OG)/Fe(II)-dependent dioxygenases, and Rieske domain-containing proteins. Arguably, fewer demethylating enzymes have been characterized compared with methylating enzymes. For example, our recent isolation of thebaine 6-O- demethylase (T6ODM) and codeine O-demethylase (CODM) from opium poppy (Figure 1A) represent two of only a small number of plant enzymes capable of catalyzing an O-demethylation reaction (Hagel and Facchini, 2010). In contrast, myriad O-methyltransferases have been implicated in the structural elaboration of plant secondary metabolites such as phenylpropanoids, flavonoids, and alkaloids (Lam et al., 2007). Nevertheless, the discovery over the past decade of an impressive number of novel and functionally diverse N- and O-demethylases raises the possibility that dealkylating reactions are common. For example, landmark reports of histone demethylases have had a major impact in the field of chromatin remodeling despite the long-held assumption that histone methylation was irreversible (Mosammaparast and Shi, 2010). The possibility that the methylation status of a molecule is dynamic and reversible has not been seriously considered in the context of plant secondary metabolism. The discovery of novel enzymes such as T6ODM and CODM capable of demethylating thebaine and codeine (Hagel and Facchini, 2010) provides a rationale to re-evaluate many of the perceived metabolic schemes leading to approximately 2,500 benzylisoquinoline alkaloids (BIAs) reported in plants.
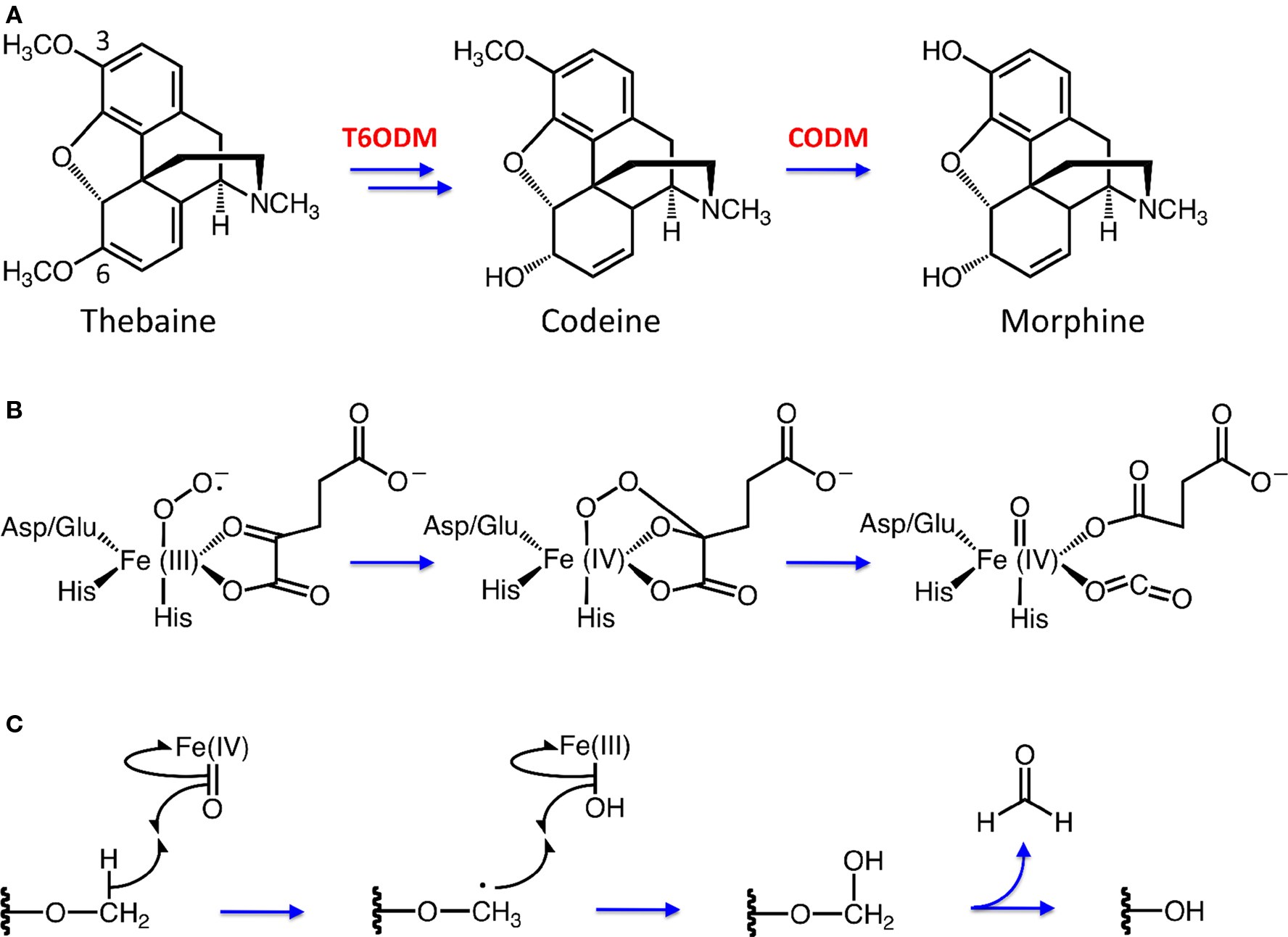
Figure 1. (A) Morphine biosynthesis in opium poppy requires two O-demethylation steps catalyzed by 2-oxoglutarate (2OG)/Fe(II)-dependent enzymes thebaine 6-O-demethylase (T6ODM) and codeine O-demethylase (CODM). (B) Formation of the iron-oxo intermediate, resulting in CO2 and succinate biproducts. (C) Demethylation by 2OG/Fe(II)-dependent dioxygenases proceeds through hydroxylation at the O-linked methyl group by an iron-oxo intermediate, followed with the release of formaldehyde.
In this article, we survey the known N- and O-demethylases found in nature and summarize the proposed biochemical mechanisms for these enzymes. Special attention is paid to 2OG/Fe(II)-dependent dioxygenases, a diverse family of proteins that are involved in numerous plant metabolic pathways. Finally, we provide a perspective on the potentially widespread significance of O-demethylases in BIA metabolism based on established roles for 2OG/Fe(II)-dependent enzymes in the O-demethylation of morphinan alkaloids.
Occurrence of O- and N-Demethylases
Specialized fungi and bacteria employ O-demethylases for catabolic processes such as the recycling of lignin and the degradation of environmental pollutants. The depolymerization of lignin by white-rot fungi results in the formation of monocyclic aromatics such as vanillate and syringate, which then undergo bacteria- mediated O-demethylation by either Rieske-type, [2Fe-2S] domain- containing proteins (Civolani et al., 2000; Providenti et al., 2006) or H4folate-dependent enzymes (Berman and Frazer, 1992; Abe et al., 2005). A Rieske-type oxygenase responsible for the O-demethylation of the widely used herbicide 2,3-dichloro-2-methoxybenzoic acid (dicamba) has been structurally characterized by X-ray crystallography (Dumitru et al., 2009). Additionally, microbes employ O-demethylating P450s for processes such as the biodegradation of 2,4,6-trichloroanisole (TCA) (Campoy et al., 2009), an environmental pollutant and contributing factor to wine off-odors (Silva Pereira et al., 2000).
Cytochromes P450 are important enzymes for O-demethylation in animals. Human P450s are capable of O-demethylating exogenous small molecules including a wide variety of drugs (Dowers and Jones, 2006; Wang et al., 2007, 2009) and environmental toxins (Le Gal et al., 2001). Unlike P450s involved in plant secondary metabolism, which generally display strict substrate stereo- and regiospecificities, many animal P450s show broad substrate acceptance profiles with single enzymes exhibiting multiple activities. For example, human CYP2D6 represents less than 2% of all hepatic P450s yet metabolizes approximately 25% of all clinically used drugs via, for example,O- and N-dealkylation, desulfuration, and hydroxylation of aromatic and aliphatic substrates (Wang et al., 2009). Among its diverse array of substrates are several plant alkaloids including codeine, the tetrahydroisoquinoline alkaloid emetine, the indole alkaloid ibogaine, and the pyrrolizidine alkaloids lasiocarpine and monocrotaline (Grobe et al., 2009; Wang et al., 2009). CYP2D6 exhibits O-demethylation activity toward ibogaine, a psychoactive metabolite derived from the iboga plant (Tabernanthe iboga) with potential use in the treatment of opiate and cocaine addictions (Obach et al., 1998). Emetine, which is derived from ipecac (Psychotria ipecacuanha) root and used both as an anti-protozoal agent and general emetic (Asano et al., 2001), undergoes CYP2D6-mediated O-demethylation to produce cephaeline. The capacity of CYP2D6 to act on alkaloid substrates likely contributes to the endogenous formation of morphine in humans (Boettcher et al., 2005). In vitro assays have indicated a role for this P450 in the phenol coupling of the simple BIA (R)-reticuline to form the promorphinan alkaloid salutaridine, a precursor of the pentacyclic morphinan alkaloids (Grobe et al., 2009). Furthermore, CYP2D6 produces morphine via the O-demethylation of codeine. Interestingly, CYP2D6 is regiospecific for the 3-O-linked methyl moiety of thebaine and codeine (Figure 1A). A mammalian P450 enzyme capable of the 6-O-demethylation of these compounds has not been reported. The plant cytochrome P450 CYP80G2 from Japanese goldthread (Coptis japonica) has also been reported to catalyze O-demethylation of alkaloid substrates (Ikezawa et al., 2008).
The isolation of T6ODM and CODM was achieved using a functional genomics approach that allowed us to avoid the assumption that the O-demethylations of thebaine and codeine are catalyzed by cytochromes P450 (Hagel and Facchini, 2010). A non-biased screen was used to identify genes differentially expressed in a morphine/codeine-free opium poppy mutant, in which the morphine pathway is blocked at thebaine (Figure 1A) (Hagel et al., 2008). The stem transcriptomes of this mutant and three morphine-accumulating varieties were independently compared using an opium poppy-specific microarray. Integration of all three pair-wise comparisons revealed a single cDNA, encoding a putative 2OG/Fe(II)-dependent dioxygenase, showing lower transcript levels in the mutant compared with the morphine-accumulating varieties. In vitro and in planta characterization identified the encoded enzyme as T6ODM, which specifically targets the 6-O-linked position for demethylation (Figure 1A). The gene encoding T6ODM is not expressed in the aerial organs of the morphine/codeine-free mutant. Sequence homology interrogation revealed a highly similar cDNA encoding CODM, which is specific for the 3-O-methyl group of thebaine and codeine (Figure 1A). T6ODM and CODM are the only known members of the 2OG/Fe(II)-dependent dioxygenase family capable of catalyzing O-demethylation reactions. The role of 2OG/Fe(II)-dependent dioxygenases in catalyzing the final steps of morphine biosynthesis in opium poppy was unexpected partly because upstream oxidative reactions, such as ring hydroxylation and C-C phenol coupling steps, are catalyzed by P450s. Additionally, prior to the discovery of T6ODM and CODM, 2OG/Fe(II)-dependent enzymes were not implicated in BIA metabolism. The recruitment of dioxygenases as O-demethylases is also a novel feature in the evolutionary diversification of plant natural products.
Several members of the 2OG/Fe(II)-dependent enzyme family are known to catalyze N-demethylation reactions. A well-studied example includes AlkB-type proteins, which catalyze the removal of N-linked methyl groups at positions 1 and 3 of purine and pyrimidine bases, respectively, in DNA and RNA (Yi et al., 2009). Other examples of N-demethylating enzymes that belong to the 2OG/Fe(II)-dependent protein family include Jumonji C (JmjC) domain-containing histone N-demethylases. JmjC-domains are found in more than 100 proteins in a broad spectrum of organisms from bacteria to eukaryotes and are variously capable of N-demethylating mono-, di- and trimethylated lysine residues of histone proteins (Mosammaparast and Shi, 2010). JmjC-domain proteins represent one of two types of enzymes known to N-demethylate histones. Proteins of the second type are FAD-dependent monoamine oxidases homologous to the histone demethylase LSD1. In plant alkaloid metabolism, the flavinylated oxidase berberine bridge enzyme (BBE) catalyzes N-demethylation of the alkaloid (S)-N-methylcoclaurine (Kutchan and Dittrich, 1995; Winkler et al., 2008). Myriad N-dealkylation reactions are also carried out by cytochromes P450, such as those involved in drug and xenobiotic metabolism in humans. For example, CYP2D6 is capable of N-demethylating a multitude of pharmaceuticals including the monoamine oxidase-B inhibitor selegiline, the antiretroviral ritonavir, and the antipsychotic clozapine (Wang et al., 2009). Certain plant P450s can catalyze the N-demethylation of small molecules, such as tobacco (Nicotiana tabacum) nicotine N-demethylase (CYP82E4) (Siminszky et al., 2005).
Proposed Demethylation Mechanisms: 2OG/Fe(II)-Dependent Dioxygenases
Several proposed demethylation mechanisms involve hydroxylation of O- or N-linked methyl groups prompting the departure of formaldehyde. 2-Oxoglutarate/Fe(II)-dependent dioxygenases such as AlkB homologues (Yi et al., 2009), JmjC-domain N-demethylases (Mosammaparast and Shi, 2010) and the morphinan alkaloid O-demethylases of opium poppy (Hagel and Facchini, 2010) employ molecular oxygen and 2OG cleavage to generate a reactive iron-oxo intermediate, which hydroxylates the primary substrate. Similarly, Rieske-type, non-heme oxygenases such as dicamba monooxygenase (DMO) are thought to activate molecular oxygen for the purpose of hydroxylating the O-linked methyl groups of monocyclic aromatic compounds (Dumitru et al., 2009). In contrast, FAD-dependent monoamine oxidases such as LSD1 first oxidize the amine substrate to form an iminium ion, which reacts with water to form a N,O-hemiacetal (Mosammaparast and Shi, 2010). However, the result is the same with the hemiacetal collapsing to form formaldehyde. A similar reaction mechanism has been proposed for the N-demethylation of (S)-N-methylcoclaurine by BBE (Kutchan and Dittrich, 1995).
Non-heme, iron-dependent oxygenases enact catalysis using a conserved 2-His + Asp/Glu facial triad, which acts to anchor iron at three coordination points (Purpero and Moran, 2007; Kovaleva and Lipscombe, 2008). The addition of molecular oxygen (O2) initially yields an Fe(III)-superoxo species that carries out nucleophilic attack on the 2OG keto group yielding a covalent linkage between the Fe(IV) center and 2OG (Figure 1B). Insertion of an oxygen atom into the C1-C2 bond of 2OG is accompanied by O-O-bond cleavage, and 2OG decomposes into a carbonate-succinate mixed anhydride bound to an Fe(IV)-oxo species (Purpero and Moran, 2007). Figure 1C briefly illustrates how this activated Fe(IV)-oxo species catalyzes the hydroxylation of an O-linked methyl group beginning with the abstraction of a hydrogen atom. This abstraction forms a substrate radical plus an Fe(III)-hydroxide species. Radical attack results in a hydroxylated substrate and restores iron to its Fe(II) state. Finally, the hydroxylated alkyl group departs as formaldehyde, completing O-demethylation of the substrate. We have proposed this mechanism for the O-demethylation of thebaine and codeine by T6ODM and CODM, respectively (Figure 1A) (Hagel and Facchini, 2010).
Physiological Roles for 2OG/Fe(II)-Dependent Dioxygenases
In plants, 2OG/Fe(II)-dependent enzymes hydroxylate a wide variety of substrates including proteins (Gorres and Raines, 2010), hormones (Yamaguchi, 2008), phenylpropanoids (Kai et al., 2008), flavonoids (Grotewold, 2006), alkaloids (Ziegler and Facchini, 2008; Hagel and Facchini, 2010), benzoxazinoids (Frey et al., 2009), and glucosinolates (Sønderby et al., 2010). Whereas most of these hydroxylations involve aliphatic substrates, 2OG/Fe(II)-dependent dioxygenases are also known to add hydroxyl functions to substituted aromatic rings. For example, the BX6 enzyme in grasses hydroxylates DIBOA-Glc (2,4-dihydroxy-2H-1,4-benzoxazin-3(4H)-one glucoside) in benzoxazinoid metabolism (Frey et al., 2009), a dioxygenase from Chrysosplenium americanum was shown to 6-hydroxylate partially methylated flavonols (Anzellotti and Ibrahim, 2000), and a 2OG/Fe(II)-dependent enzyme in Arabidopsis was recently shown to ortho-hydroxylate feruloyl-CoA to form scopoletin (Kai et al., 2008). Other reactions catalyzed by plant 2OG/Fe(II)-dependent enzymes include dehydration, O-demethylation, aldehyde oxidation, γ-ring formation, and the oxidative removal of methylsulfinyl groups (Hausinger, 2004; Hagel and Facchini, 2010; Sønderby et al., 2010). A mechanistically unique example is the ethylene-forming enzyme, 1-aminocyclopropane-1-carboxylate oxidase (ACCO). Although ACCO is related in terms of amino acid sequence to 2OG/Fe(II)-dependent dioxygenases it does not use 2OG, but instead requires bicarbonate to catalyze oxidation yielding ethylene, CO2 and hydrogen cyanide (Zhang et al., 2004). Conversely, 4-hydroxyphenylpyruvate dioxygenase (HPPD) is an example of an enzyme requiring 2OG and iron that likely evolved independently from other 2OG/Fe(II)-dependent enzymes (He and Moran, 2009). HPPD is often grouped together with other 2-oxoglutarate-dependent enzymes based on similar catalytic chemistries, although it is a structurally distinct enzyme.
A phylogenetic tree illustrates the relationships among various plant 2OG/Fe(II)-dependent oxygenases (Figure 2). High bootstrap support indicates a monophyletic clade containing the opium poppy O-demethylases T6ODM and CODM. We have recently found that the closely related enzyme, DIOX2, is capable of O-demethylating a variety of protoberberine alkaloids. The nearest-neighbor clade to opium poppy O-demethylases contains functionally characterized proteins with high sequence identity (i.e. > 50%) to T6ODM, CODM, and DIOX2. Phylogenetic analysis did not support monophylogeny between opium poppy dioxygenases and either desacetoxyvindoline 4-hydroxylase (D4H) or hyoscyamine 6β-hydroxylase(H6H) enzymes (Figure 2), which is consistent with the independent evolutionary origins of different alkaloid types (Ziegler and Facchini, 2008). D4H catalyzes the penultimate step in vindoline biosynthesis en route to the monoterpenoid indole alkaloid vinblastine in Catharanthus roseus, whereas H6H catalyzes the ultimate step in the formation of the tropane alkaloid scopolamine (Ziegler and Facchini, 2008). The substantial sequence identity (i.e. ∼40%) between a CjNCS from Coptis japonica, an enzyme reported to be involved in BIA metabolism (Minami et al., 2007) and opium poppy O-demethylases suggests a more recent ancestral link.
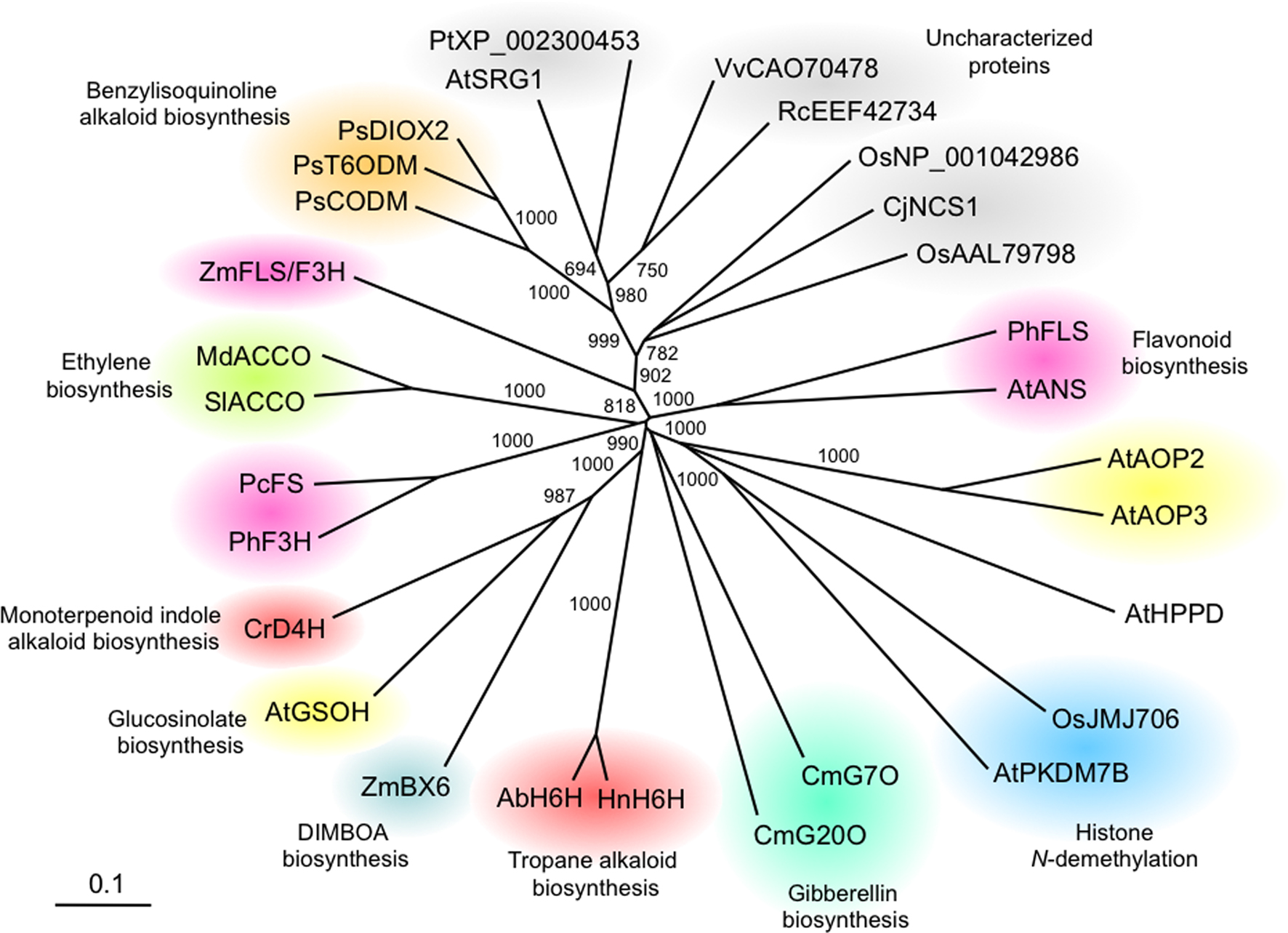
Figure 2. Unrooted neighbor-joining phylogenetic tree for selected plant 2-oxoglutarate (2OG)/Fe(II)-dependent dioxygenases. Bootstrap frequencies for each clade represent values relative to 1000 iterations. Phylogeny and amino acid alignments were performed using ClustalX, and phylogenetic data were displayed using TREEVIEW (Page, 1996). Protein functional categories are shown in color. Species and associated GenBank accession numbers are as follows: Arabidopsis thaliana anthocyanidin synthase AtANS (Q96323); Arabidopsis thaliana AtAOP2 (AAL14646); Arabidopsis thaliana AtAOP3 (AAL14647); Arabidopsis thaliana putative but-3-enyl hydroxylase AtGSOH (NP_180115); Arabidopsis thaliana H3K4 N-demethylase AtPKDM7B (AAQ65171); Arabidopsis thaliana 4-hydroxyphenylpyruvate dioxygenase At4HPPD (AAB58404); Arabidopsis thaliana senescence-related gene 1 AtSRG1 (NP_173145); Atropa belladonna hyoscyamine 6β-hydroxylaseAbH6H (BAA78340); Catharanthus roseus desacetoxyvindoline 4-hydroxylase CrD4H (AAC49827); Coptis japonica norcoclaurine synthase-1 CjNCS1(A2A1A0); Cucurbita maxima gibberellin 7-oxidase CmG7O (AAB64346); Cucurbita maxima gibberellin20-oxidase CmG20O (AAB64345); Hyoscyamus niger hyoscyamine 6β-hydroxylase HnH6H (AAA3338); Malus domestica ACC oxidase MdACCO (AAC36461); Oryza sativa H3K9 N-demethylase OsJMJ706 (NP_001065492); Oryza sativa putative dioxygenase (NP_001042986); Oryza sativa putative dioxygenase (AAL79798); Papaver somniferum thebaine 6-O-demethylase PsT6ODM (GQ500139); Papaver somniferum codeine O-demethylase PsCODM (GQ500141); Papaver somniferum PsDIOX2 (GQ500140); Petroselinum crispum flavone synthase I PcFS (AAP57393); Petunia hybrida flavanone 3-hydroxylase PhF3H (AAC49929); Petunia hybrida flavonol synthase PhFLS (CAA80264); Populus trichocarpa putative dioxygenase (XP_002300453); Ricinus communis putative dioxygenase (EEF42734); Solanum lycopersicum ACC oxidase SlACCO (P24157); Vitis vinifera VvDIOX-like (CAO70478); Zea mays DIBOA-Glc hydroxylase ZmBX6 (AAO65850); Zea mays flavonol synthase/flavanone 3-hydroxylase ZmFS/F3H (ACG44904).
Three 2OG/Fe(II)-dependent dioxygenases have been implicated in glucosinolate biosynthesis. Glucosinolates are largely found in the order Brassicales and are involved in plant resistance to insects and pathogens. In Arabidopsis, AOP2 and AOP3 are closely related proteins that convert methylsulfinylalkyl glucosinolates to alkenyl and hydroxyalkyl glucosinolates, respectively. In vivo evidence points to the involvement of a third enzyme, GSOH, in the hydroxylation of but-3-enyl to 2-hydroxy-but-3-enyl glucosinolate (Sønderby et al., 2010). Interestingly, phylogenetic analysis does not support a common origin between GSOH and AOP2/AOP3 enzymes (Figure 2), which appear to have been recruited independently for glucosinolate metabolism. A similar pattern is observed for dioxygenases involved in flavonoid metabolism, which appear as disparate clades containing one or two sequences (Figure 2). Overall, phylogenetic analysis indicates a low degree of conservation among plant 2OG/Fe(II)-dependent enzymes, suggesting that these enzymes represent a highly diversified class of proteins.
Possible Widespread Role of O-Demethylation in BIA Metabolism
The biosynthesis of BIAs begins with the formation of (S)-norcolaurine, which undergoes functionalization to yield the central intermediate (S)-reticuline. The formation of (S)-reticuline involves two O-methylation steps, including O-methylation at position 6 of the tetrahydroisoquinoline moiety catalyzed by norcoclaurine 6-O-methyltransferase (6OMT) and O-methylation at position 4’ of the benzyl ring catalyzed by 3’-hydroxy-N-methylcoclaurine 4’-O-methyltransferase (4’OMT) (Figure 3A). (S)-Reticuline can undergo diverse intramolecular coupling to yield a variety of backbone structures including the promorphinan, protoberberine, and aporphine alkaloids (Figure 3A). The protoberberine alkaloid (S)-scoulerine results from the oxidative cyclization of (S)-reticuline via a two-step mechanism involving the formation of a methylene iminium ion, followed by nucleophilic attack at the imine carbon by C-2’ to yield the C-8 berberine bridge (Kutchan and Dittrich, 1995; Winkler et al., 2008). This reaction is catalyzed by the flavinylated oxidase, BBE. Alternatively, (S)-reticuline can undergo configuration inversion at C-1 yielding (R)-reticuline, the substrate for para-ortho phenol coupling by salutaridine synthase (SalSyn) (Gesell et al., 2009). The promorphinan alkaloid salutaridine serves as the precursor for the pentacyclic morphinan alkaloids. Phenol coupling between C-8 and C-2’ or between C-8 and C-6’ of (S)-reticuline yields the aporphine alkaloids (S)-corytuberine or (S)-isoboldine, respectively. The former, ortho-ortho coupling is catalyzed by CYP80G2 in Coptis japonica (Ikezawa et al., 2008), whereas the latter, ortho-para coupling not yet been biochemically characterized in plants. Depending on the type of intramolecular rearrangement that occurs, the hydroxyl and methoxyl groups of (S)-reticuline acquire different positions on the resulting alkaloid. For example, the 3’-hydroxyl of (S)-reticuline becomes the 9-hydroxyl of (S)-scoulerine, the 4-hydroxyl of salutaridine, the 11-hydroxyl of (S)-corytuberine, and the 9-hydroxyl of (S)-isoboldine. Alkaloids derived from salutaridine, (S)-scoulerine, (S)-corytuberine or (S)-isoboldine that exhibit O-methylation patterns different from their precursors (Figure 3A) are shown in Figure 3B.
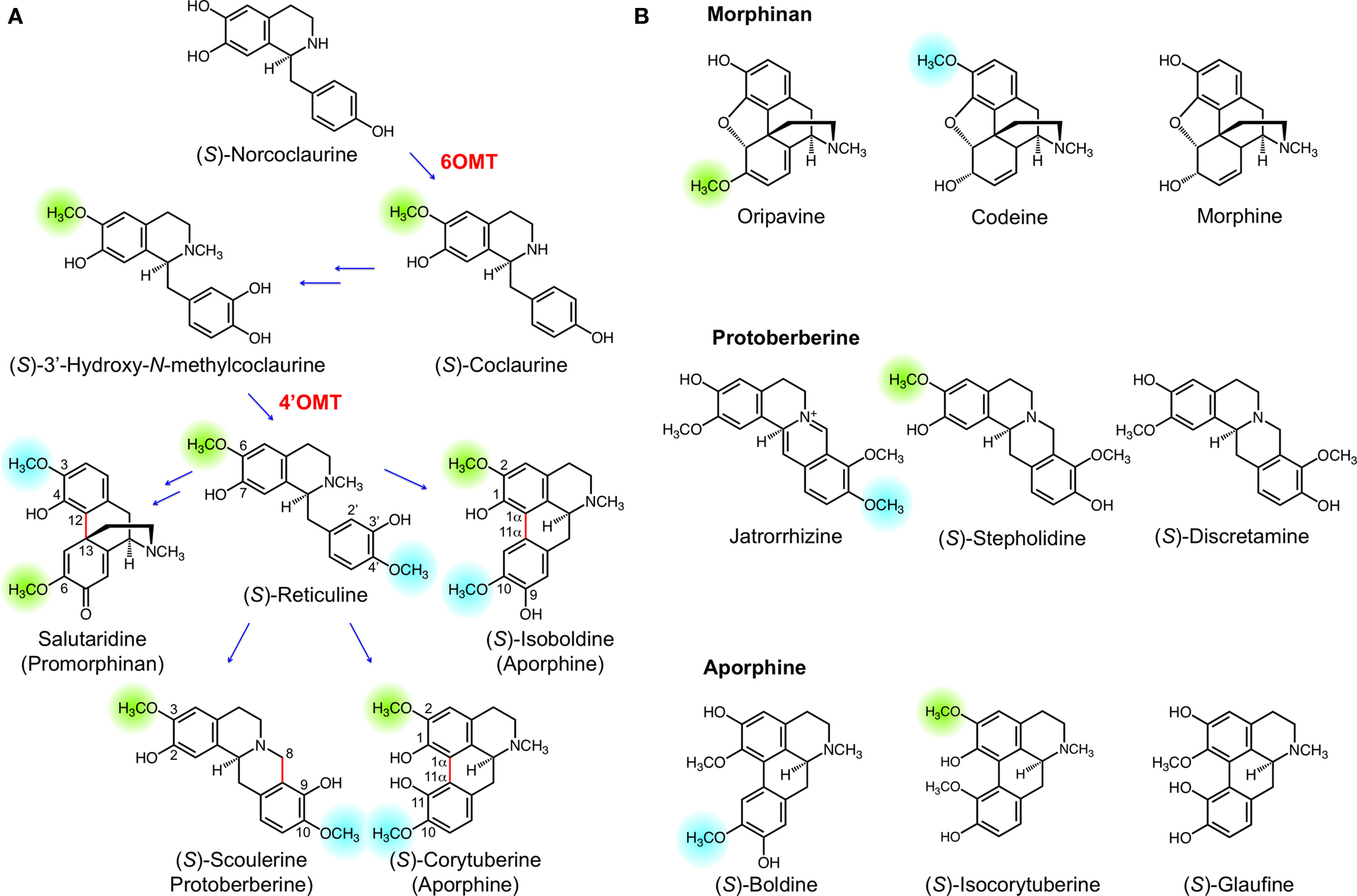
Figure 3. (A) Benzylisoquinoline alkaloid (BIA) biosynthesis begins with (S)-norcoclaurine, which acquires two O-methyl groups en route to the central intermediate (S)-reticuline. Intramolecular rearrangement of (S)-reticuline yields a variety of skeletal structures including promorphinan (e.g. salutaridine), protoberberine (e.g. (S)-scoulerine), and aprophine (e.g. (S)-corytuberine and (S)-isoboldine). (B) Examples of BIAs exhibiting different O-methylation patterns relative to the established or putative precursors shown in (A). Green and blue highlights indicate positions corresponding to the 6-O- and 4’-O-methyl moieties of (S)-reticuline. Enzymes indicated in red are norcoclaurine 6-O-methyltransferase (6OMT) and 3’-hydroxy-N-methylcoclaurine 4’-O-methyltransferase (4’OMT).
Comparing the O-methyl substitution patterns of oripavine, codeine and morphine with that of salutaridine reveals that O-demethylation has occurred (Figure 3B). For example, a 3-O-methyl (corresponding to the 4’-O-methyl of (S)-reticuline) is no longer present in oripavine. Conversely, codeine lacks a 6-O-methyl (corresponding to the 6-O-methyl of (S)-reticuline), while morphine lacks both O-methyl groups. These substitution patterns show that the O-methyl groups added by 6OMT and 4’OMT during (S)-reticuline formation are removed by O-demethylation, which is confirmed biochemically by our recent discovery of T6ODM and CODM (Hagel and Facchini, 2010).The O-demethylation of morphinan alkaloids by T6ODM and CODM provides a rational basis for hypothesizing the existence of other O-demethylases in BIA metabolism. Examples of protoberberine alkaloids exhibiting altered O-methyl substitution patterns relative to (S)-reticuline are also known to occur. For example, jatrorrhizine lacks a 3-O-methyl, (S)-stepholidine lacks a 10-O-methyl, and (S)-discretamine lacks both 3-O and 10-O-methyl groups (Figure 3B).Jatrorrhizine and (S)-stepholidine are found in several members of the order Ranunculales especially within Papaveraceae, Berberidaceae, and Menispermaceae families (Shulgin and Perry, 2002). (S)-Discretamine has been isolated from plants belonging to the Annonaceae family in the order Magnoliales. The biosynthesis of jatrorrhizine proceeds through (S)-scoulerine (Rueffer et al., 1983), which is also the likely intermediate in the formation of (S)-stepholidine and (S)-discretamine. The observed substitution pattern for jatrorrhizine was proposed to result from the oxidation of berberine (Rueffer et al., 1983), although it is also possible that 3-O-demethylation combined with 2-O- and 9-O-methylation leads to jatrorrhizine. In support of this hypothesis, opium poppy CODM was shown to efficiently 3-O-demethylate (S)-scoulerine (Hagel and Facchini, 2010).Our recent discovery of the protoberberine alkaloid 10-O-demethylase activity of DIOX2 further suggests the role of O-demethylation in the formation of BIAs such as (S)-stepholidine and (S)-discretamine.
O-Demethylation would appear to occur in other areas of BIA metabolism, such as aporphine alkaloid biosynthesis. For example, (S)-boldine, (S)-isocorytuberine and (S)-glaufine lack O-methyl groups corresponding to (S)-reticuline 6-O- and/or 4’-O-methyl groups (Figure 3). (S)-Isocorytuberine and (S)-glaufine have been isolated from Glaucium fimbrilligerum (Papaveraceae) (Karimova et al., 1979, 1980), and (S)-boldine is a major alkaloid of the boldo tree (Peumus boldus Molina, Monimiaceae) (O’Brien et al., 2006). Since the biosynthetic origin of these compounds has not yet been confirmed it cannot be ruled out that (S)-reticuline is not an intermediate in their biosyntheses. However, it is likely that at least some of these compounds are derived from (S)-reticuline through (S)-corytuberine or (S)-isoboldine. Biosynthesis through (S)-reticuline of (S)-boldine, (S)-isocorytuberine and (S)-glaufine could involve one or more O-demethylation steps, possibly by 2OG/Fe(II)-dependent dioxygenase enzymes. Additional work is needed to establish the biosynthetic networks connecting various specific BIAs. Nevertheless, sufficient evidence exists to strongly suggest a widespread involvement of O-demethylases in BIA metabolism.
Summary and Prospects
Enzymes that catalyze demethylation reactions belong to several different protein families including cytochromes P450, FAD-dependent oxidases, Rieske-domain proteins and others with non-heme irons centers. Of particular interest are 2OG/Fe(II)-dependent dioxygenases owing to their capacity to catalyze the O- and N-demethylation of various substrates, and because of their diverse roles in plant metabolic pathways. Our recent finding that these enzymes have the capacity for O-demethylation has expanded our view of the biosynthetic possibilities within the realm of plant alkaloid metabolism. Additional O-demethylases especially in, but not necessarily restricted to, the 2OG/Fe(II)-dependent dioxygenase family will likely be found to perform a central role in diversification of many BIA subcategories. Targeted metabolite and transcript profiling combined with the continued development of functional genomics tools such as virus-induced gene silencing will facilitate the discovery of novel demethylases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Peter J. Facchini holds the Canada Research Chair in Plant Metabolic Processes Biotechnology. Our work was funded by a Natural Sciences and Engineering Research Council of Canada Discovery Grant to Peter J. Facchini.
References
Abe, T., Masai, E., Miyauchi, K., Katayama, Y., and Fukuda, M. (2005). A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilus SYK-6. J. Bacteriol. 187, 2030–2037.
Anzellotti, D., and Ibrahim, R. K. (2000). Novel flavonol 2-oxoglutarate dependent dioxygenase: affinity purification, characterization, and kinetic properties. Arch. Biochem. Biophys. 382, 161–172.
Asano, T., Kushida, H., Sadakane, C., Ishihara, K., Wakui, Y., Yanagisawa, T., Kimura, M., Kamei, H., and Yoshida, T. (2001). Metabolism of ipecac alkaloids cephaeline and emetine by human hepatic microsomal cytochrome P450s, and their inhibitory effects on P450 enzyme activities. Biol. Pharm. Bull. 24, 678–682.
Berman, M. H., and Frazer, A. C. (1992). Importance of tetrahydrofolate and ATP in the anaerobic O-demethylation reaction for phenylmethylethers. Appl. Environ. Microbiol. 58, 925–931.
Boettcher, C., Fellermeier, M., Boettcher, C., Dräger, B., and Zenk, M. H. (2005). How human neuroblastoma cells make morphine. Proc. Natl. Acad. Sci. U.S.A. 102, 8495–8500.
Campoy, S., Àlvarez-Rodríguez, M. L., Recio, E., Rumbero, A., and Coque, J. J. R. (2009). Biodegradation of 2,4,6-TCA by the white-rot fungus Phelbia radiata is initiated by a phase I (O-demethylation)-phase II (O-conjugation) reactions system: implications for the chlorine cycle. Environ. Microbiol. 11, 99–110.
Civolani, C., Barghini, P., Roncetti, A. R., Ruzzi, M., and Schiesser, A. (2000). Bioconversion of ferulic acid into vanillic acid by means of a vanillate-negative mutant of Pseudomonas fluorescens strain BF13. Appl. Environ. Microbiol. 66, 2311–2317.
Dowers, T. S., and Jones, J. P. (2006). Kinetic isotope effects implicate a single oxidant for cytochrome P450-mediated O-dealkylation, N-oxygenation, and aromatic hydroxylation of 6- methoxyquinoline. Drug Metab. Dispos. 34, 1288–1290.
Dumitru, R., Jiang, W. Z., Weeks, D. P., and Wilson, M. A. (2009). Crystral structure of dicamba monooxygenase: a Rieske nonheme oxygenase that catalyzes oxidative demethylation. J. Mol. Biol. 392, 498–510.
Frey, M., Schullehner, K., Dick, R., Fiesselmann, A., and Gierl, A. (2009). Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70, 1645–1651.
Gesell, A., Rolf, M., Ziegler, J., Díaz Chávez, M. L., Huang, F. C., and Kutchan, T. M. (2009). CYP719B1 is salutaridine synthase, the C-C phenol-coupling enzyme of morphine biosynthesis in opium poppy. J. Biol. Chem. 284, 24432–24442.
Gorres, K. L., and Raines, R. T. (2010). Prolyl 4-hydroxylase. Crit. Rev. Biochem. Mol. Biol. 45, 106–124.
Grobe, N., Zhang, B., Fisinger, U., Kutchan, T. M., Zenk, M. H., and Guengerich, F. P. (2009). Mammalian cytochrome P450 enzymes catalyze the phenol-coupling step in endogenous morphine biosynthesis. J. Biol. Chem. 284, 24425–24431.
Grotewold, E. (2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57, 761–780.
Hagel, J. M., and Facchini, P. J. (2010). Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat. Chem. Biol. 6, 273–275.
Hagel, J. M., Weljie, A. M., Vogel, H. J., and Facchini, P. J. (2008). Quantitative 1H nuclear magnetic resonance metabolite profiling as a functional genomics platform to investigate alkaloid biosynthesis in opium poppy. Plant Physiol. 147, 1805–1821.
Hausinger, R. P. (2004). Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 39, 21–68.
He, P., and Moran, G. R. (2009). We two alone will sing: the two-substrate α-keto acid-dependent oxygenases. Curr. Opin. Chem. Biol. 13, 443–450.
Ikezawa, N., Iwasa, K., and Sato, F. (2008). Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C-C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 283, 8810–8821.
Kai, K., Mizutani, M., Kawamura, N., Yamamoto, R., Tamai, M., Yamaguchi, H., Sakata, K., and Shimizu, B. (2008). Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl-CoA by a 2-oxoglutarte-dependent dioxygenase in Arabidopsis thaliana. Plant J. 55, 989–999.
Karimova, S. U., Israilov, I. A., Yunusov, M. S., and Yunusov, S. Y. (1979). Structure of glaufine. Chem. Nat. Comp. 14, 699.
Karimova, S. U., Israilov, I. A., Yunusov, M. S., and Yunosov, S. Y. (1980). Alkaloids of Glaucium fimbrilligerum. Chem. Nat. Comp. 16, 177–180.
Kovaleva, E. G., and Lipscombe, J. D. (2008). Versatility of biological non-heme Fe(II) centers in oxygen activation reactions. Nat. Chem. Biol. 4, 186–193.
Kutchan, T. M., and Dittrich, H. (1995). Characterization and mechanism of the berberine bridge enzyme, a covalently flavinylated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J. Bioahem. 270, 24475–24481.
Lam, K. C., Ibrahim, R., Behdad, B., and Dayanandan, S. (2007). Structure, function and evolution of plant O-methyltransferases. Genome 50, 1001–1013.
Le Gal, A., Dréano, Y., Gervasi, P. G., and Berthou, F. (2001). Human cytochrome P450 2A6 is the major enzyme involved in the metabolism of three alkoxyethers used as oxyfuels. Toxicol. Lett. 124, 47–58.
Minami, H., Dubouzet, E., Iwasa, K., and Sato, F. (2007). Functional analysis of norcoclaurine synthase in Coptis japonica. J. Biol. Chem. 282, 6274–6282.
Mosammaparast, N., and Shi, Y. (2010). Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 78, 155–179.
Obach, R. S., Pablo, J., and Mash, D. C. (1998). Cytochrome P450 2D6 catalyzes the O-demethylation of the psychoactive alkaloid ibogaine to 12-hydroxyibogamine. Drug Metab. Dispos. 26, 764–768.
O’Brien, P., Carrasco-Pozo, C., and Speisky, H. (2006). Boldine and its antioxidant or health-promoting properties. Chem. Biol. Interact. 159, 1–17.
Page, R. D. (1996). TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358.
Providenti, M. A., O’Brien, J. M., Ruff, J., Cook, A. M., and Lambert, I. B. (2006). Metabolism of isovanillate, vanillate, and veratrate by Comamonas testosteroni strain BR6020. J. Bacteriol. 188, 3862–3869.
Purpero, V., and Moran, G. R. (2007). The diverse and pervasive chemistries of the α-keto acid dependent enzymes. J. Biol. Inorg. Chem. 12, 587–601.
Rueffer, M., Ekundayo, O., Nagakura, N., and Zenk, M. H. (1983). Biosynthesis of the protoberberine alkaloid jatrorrhizine. Tetrahedron Lett. 24, 2643–2644.
Shulgin, A. T., and Perry, W. E. (2002). The Simple Plant Isoquinolines. Berkeley, CA: Transform Press.
Silva Pereira, C., Figueiredo Marques, J. J., and San Romao, M. V. (2000). Cork taint in wine: scientific knowledge and public perception. A critical review. Crit. Rev. Microbiol. 26, 147–162.
Siminszky, B., Gavilano, L., Bowen, S. W., and Dewey, R. E. (2005). Conversion of nicotine to nornicotine in Nicotiana tabacum is mediated by CYP82E4, a cytochrome P450 monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 102, 14919–14924.
Sønderby, I. E., Geu-Flores, F., and Halkier, B. A. (2010). Biosynthesis of glucosinolates – gene discovery and beyond. Trends Plant Sci. 15, 283–290.
Wang, B., Yang, L. P., Zhang, X. Z., Huang, S. Q., Bartlam, M., and Zhou, S. F. (2009). New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab. Rev. 41, 573–643.
Wang, M. Z., Wu, J. Q., Bridges, A. S., Zeldin, D. C., Kornbluth, S., Tidwell, R. R., Hall, J. E., and Paine, M. F. (2007). Human enteric microsomal CYP4F enzymes O-demethylate the antiparasitic prodrug pafuramidine. Drug Metab. Dispos. 35, 2067–2075.
Winkler, A., Lyskowski, A., Riedl, S., Puhl, M., Kutchan, T. M., Macheroux, P., and Gruber, K. (2008). A concerted mechanism for berberine bridge enzyme. Nat. Chem. Biol. 4, 739–741.
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251.
Yi, C., Yang, C. G., and He, C. (2009). A non-heme iron-mediated chemical demethylation in DNA and RNA. Acc. Chem. Res. 42, 519–529.
Zhang, Z., Ren, J. S., Clifton, I. J., and Schofield, C. J. (2004). Crystal structure and mechanistic implications of 1-aminocyclopropane-1-carboxylic acid oxidase – the ethylene forming enzyme. Chem. Biol. 11, 1383–1394.
Keywords: O-demethylation, N-demethylation, 2-oxoglutarate/Fe(II)-dependent dioxygenase, benzylisoquinoline alkaloid biosynthesis
Citation: Hagel JM and Facchini PJ (2010) Biochemistry and occurrence of O-demethylation in plant metabolism. Front. Physio. 1:14. doi: 10.3389/fphys.2010.00014
Received: 30 April 2010;
Paper pending published: 17 May 2010;
Accepted: 05 June 2010;
Published online: 15 July 2010
Edited by:
Eleanore T. Wurtzel, City University of New York, USAReviewed by:
Sarah O’Connor, Massachussetts Institute of Technology, USAChristian Meyer, Institut National de la Recherche Agronomique, France
Copyright: © 2010 Hagel and Facchini. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Peter J. Facchini, Department of Biological Sciences, University of Calgary, 2500 University Drive N.W., Calgary, AB T2N 1N4, Canada. e-mail:cGZhY2NoaW5AdWNhbGdhcnkuY2E=

