- 1International College, Chongqing University of Posts and Telecommunications, Chongqing, China
- 2Viterbi School of Engineering, University of Southern California, Los Angeles, CA, United States
To address the trade-off between segmentation performance and model lightweighting in computer-aided skin lesion segmentation, this paper proposes a lightweight network architecture, Multi-Conv Attention Network (MCAN). The network consists of two key modules: ISDConv (Inception-Split Depth Convolution) and AEAM (Adaptive Enhanced Attention Module). ISDConv reduces computational complexity by decomposing large kernel depthwise convolutions into smaller kernel convolutions and unit mappings. The AEAM module leverages dimensional decoupling, lightweight multi-semantic guidance, and semantic discrepancy alleviation to facilitate the synergy between channel attention and spatial attention, further exploiting redundancy in the spatial and channel feature maps. With these improvements, the proposed method achieves a balance between segmentation performance and computational efficiency. Experimental results demonstrate that MCAN achieves state-of-the-art performance on mainstream skin lesion segmentation datasets, validating its effectiveness.
Introduction
Melanoma, a highly malignant skin tumor, causes a significant number of deaths worldwide each year. Its incidence and mortality rates vary significantly depending on the region, the level of early diagnosis awareness, and the accessibility of primary care [1]. Early detection of melanoma is crucial for improving patient survival rates. However, due to the diversity and complexity of melanoma’s appearance, its accurate diagnosis often relies on the experience and expertise of doctors, which somewhat limits the efficiency and accuracy of early diagnosis.
In melanoma diagnosis, image segmentation is a key step that precisely separates the lesion area from healthy skin, helping doctors identify the lesion’s boundaries and assist in accurate diagnosis and treatment. Traditional segmentation methods rely heavily on complex preprocessing and manual feature extraction, making it difficult to handle the complexity of melanoma images. With the emergence of high-quality datasets, data-driven deep learning methods have rapidly gained popularity. Zhang et al. [2] proposed a novel framework that integrates multiple experts to jointly learn representations from diverse MRI modalities, effectively enhancing segmentation performance. Similarly, Li et al. [3] addressed challenges in brain tumor segmentation caused by missing modalities by utilizing a deformation-aware learning framework that reconstructs missing information, resulting in more reliable and accurate segmentation even in incomplete datasets. Among them, attention mechanisms, as an effective way to integrate local and global features, help the model focus on the lesion areas. Dong et al. [4] enhanced the capability to capture feature information by dynamically allocating attention weights across channel and spatial dimensions, addressing the complex features, blurry boundaries, and noise interference in skin lesion segmentation. Similarly, the GL-CSAM module designed by Sun et al. [5] aims to capture global contextual information, enhancing the model’s ability to perceive global features. However, they did not fully explore feature fusion between different convolutional layers. To address this issue, Qiu et al. [6] introduced a multi-level attention fusion mechanism that progressively extracts lesion boundary information using contextual information from different levels, alleviating the problem of blurry boundaries. Qi et al. [7] and Liu et al. [8] introduced single attention mechanisms to integrate contextual features, specifically designed for stroke lesion segmentation. The combination of standalone self-attention modules with convolutional layers has shown limited effectiveness in enhancing the model’s non-local feature modeling capabilities. To address this limitation, Yang et al. [9] introduced a multi-attention mechanism (spatial and reverse attention). Spatial attention is used to improve the extraction of useful features, while reverse attention enhances the network’s segmentation performance by applying reverse attention operations on skip connections, enabling more accurate analysis and localization of small lesion targets. Liu et al. [10] and Zhu et al. [11] enhanced the precision and detail of tumor segmentation by fusing information from multiple MRI modes such as T1, T2, and FLAIR. Zhu et al. [12] embedded a feature fusion module based on attention mechanism in the model structure to optimize the expression and integration of multi-modal features to improve segmentation accuracy. Liu et al. [13] examined the effectiveness of traditional objective evaluation indicators in the evaluation of image fusion results and proposed a statiscy-based framework to compensate for the shortcomings of existing indicators. These methods have improved the segmentation task to varying degrees at different stages, achieving commendable results. However, their network designs do not fully consider how to effectively utilize spatial information, and they lack dedicated mechanisms to enhance and preserve spatial information. These shortcomings may result in suboptimal performance when handling spatial correlations.
Moreover, it is worth noting that while introducing high-quality attention mechanisms, the parameter count of the model increases, potentially compromising the real-time performance during deployment. Although high-quality attention mechanisms can enhance model performance, they are often accompanied by an increase in parameter count, which can negatively impact the real-time performance of model deployment [14]. In response to such problems, most researchers have based their efforts on the potential of deep separable convolution to improve model efficiency and effectiveness. Zhou et al. [15] constructs expansion layers using depthwise separable convolutions to efficiently extract multi-scale features with low computational overhead, enhancing the feature representation capability. Liu et al. [16], Ma et al. [17], and Feng et al. [18] adopted a similar approach by integrating depthwise separable convolution layers into the encoder. However, they often struggle to achieve precise detailed description while maintaining low computational overhead. Ruan et al. [19] combined MLP to extract global feature information, followed by feature extraction using depthwise separable convolutions (DWConv). This effectively preserved significant features in the brain feature map while filtering out less relevant features. However, the lightweight processing of complex features remains limited. Similarly, Lei et al. [20] combined depthwise separable convolutions with bilinear interpolation to adjust the size of high-level features, making them match low-level features. However, this approach faces performance bottlenecks when further reducing the computational burden. Chen et al. [21] incorporated the advantages of asymmetric convolutions based on depthwise separable convolutions and designed an ultralight convolution module, further achieving the decoupling of spatial and channel dimensions. Existing methods still have limitations in lightweight design. Although different encoder designs effectively reduce computational load and ensure efficient feature extraction, they still lack precision in representing the blurry edges of skin lesions.
To address the contradiction between segmentation performance and lightweight design, this paper proposes a lightweight segmentation method. It aims to more accurately capture and segment the lesion area by leveraging channel and spatial redundancy, without increasing additional computational load. Specifically, the core of the segmentation framework is the Inception-Split ISDConv. Additionally, at the bridging layer stage, we introduce the AEAM, which combines the collaborative effects of spatial and channel attention with the feature calibration capabilities of the squeeze-and-excitation network. AEAM utilizes multi-scale depth-shared 1D convolutions to capture multi-semantic spatial information for each feature channel. It effectively integrates global contextual dependencies and multi-semantic spaces, while calculating channel similarity and contributions under the guidance of compressed spatial knowledge, thereby alleviating semantic differences in the spatial structure. Additionally, we introduce dynamic convolution in the encoder. Dynamic convolution dynamically aggregates multiple parallel convolution kernels based on input-relevant attention mechanisms. Assembling multiple convolution kernels is not only computationally efficient but also enhances representational capability due to the smaller size of the kernels.
The contributions of this paper can be summarized in the following three aspects:
1. In this study, a novel lightweight segmentation network named Multi-Conv Attention Network (MCAN) is proposed. It performs channel and spatial weighting on the spatial and channel redundancies in the feature map without increasing additional computational load, achieving an effect of information complementarity.
2. To address the unclear edges in skin lesions, this paper proposes ISDConv. This module performs multi-scale feature extraction using depthwise separable convolutions, multi-scale convolution kernels, and spatial and channel reconstruction convolutions. It reduces computational complexity and the number of parameters, thereby improving the model’s feature representation capability while maintaining efficient feature extraction.
3. To address the insufficient utilization of redundancies in the spatial and channel feature maps, this paper proposes the Adaptive Enhanced Attention Module (AEAM). Through dimension decoupling, lightweight multi-semantic guidance, and semantic discrepancy mitigation, AEAM achieves the collaborative effect between channel and spatial attention, enabling the model to capture and segment the lesion areas more accurately.
Related works
Attention mechanism
In the field of natural images, Li et al. [22] used a dual attention fusion module to effectively combine features from images from different sources, thereby enhancing the model’s ability to focus on important regions. The attention mechanism can enhance the extraction of key features in infrared and visible images, making the fused images clearer and retaining more meaningful details [23]. In medical image segmentation, the attention mechanism is primarily used to guide the model’s focus on the lesion areas in the image, assigning different weights to each pixel or feature, enhancing task-relevant features, and suppressing irrelevant background information. Huang et al. [24] prior convolutional attention mechanism that dynamically allocates attention weights across both channel and spatial dimensions. Shaker et al. [25] used a pair of mutually dependent branches based on spatial and channel attention to effectively learn discriminative features, improving the quality of segmentation masks. Fu et al. [26] used a Transformer-based spatial and channel attention module to extract global complementary information across different layers of the U-Net, which helps in learning detailed features at different scales. To address hair interference in dermoscopic images, Xiong et al. [27] proposed a multi-scale channel attention mechanism that enhances feature information and boundary awareness. Song et al. [28] argued that current popular attention mechanisms focus too much on external image features and lack research on latent features. They introduced an external-latent attention mechanism, using an entropy quantization method to summarize the distribution of latent contextual information. Similarly, Huang et al. [29] used Bi-Level Routing Attention in deep networks to discard irrelevant key-value pairs, achieving content-aware sparse attention for dispersed semantic information.
Network lightweighting
While pursuing high performance, researchers have also begun to focus on the lightweight and efficiency of medical image segmentation networks. Network structure design is one of the most popular approaches for lightweight optimization. Ma et al. [17] simplified the structure, reduced the number of parameters, and optimized the convolution operations, achieving a significant reduction in computational complexity and model size while maintaining segmentation accuracy. This enables the model to perform excellently even in resource-constrained environments, making it suitable for applications such as mobile healthcare and telemedicine. The UcUNet [30] network achieves lightweight and precise medical image segmentation by designing an efficient large-kernel U-shaped convolution module. This network leverages large-kernel convolutions to expand the receptive field while integrating depthwise separable convolutions to reduce the computational cost, thereby maintaining high segmentation accuracy with efficient computation. Liu et al. [16] combines the lightweight characteristics of HarDNet with multi-attention mechanisms, enhancing the network’s ability to capture key features and achieving more precise medical image segmentation. Sun et al. [31] introduces a contextual residual network, effectively integrating contextual information into the U-shaped network, enhancing the global understanding and stability of the segmentation. Nisa and Ismail [32] employs a dual-path structure with a ResNet encoder, combining ResNet’s feature extraction capabilities with U-Net’s segmentation advantages, offering an alternative effective solution for medical image segmentation. Zhao et al. [33] proposed a four-layer feature calibration branch based on an attention mechanism. The downsampling layer reduces the resolution of rectal cancer CT image feature maps to half of the original size, followed by pointwise convolution to enable interactions between channels. This method effectively expands the receptive field of subsequent convolutional layers and optimizes computational efficiency by reducing the cost of calculating spatial attention. Model compression, as another approach to simplifying network structures, removes structural redundancy while maintaining performance, making it more suitable for various applications in medical image analysis. Wang et al. [34] designed a sophisticated teacher network to learn multi-scale features, guiding a more lightweight student network to improve segmentation accuracy. Experiments showed that this method effectively acquires detailed morphological features of the brain from the teacher network. Hajabdollahi et al. [35] proposed a channel pruning algorithm for medical image segmentation tasks, which selects color channels during image processing and allows training of the target structure directly on the pre-selected key channels. However, these studies did not address how to utilize the redundancy effectively.
Based on the above research findings, this paper proposes a lightweight segmentation model that emphasizes spatial and channel features. This model improves segmentation accuracy and efficiency without increasing additional computational costs, providing a new and efficient solution for the medical imaging field.
Methods
The overall framework of MCA-Net
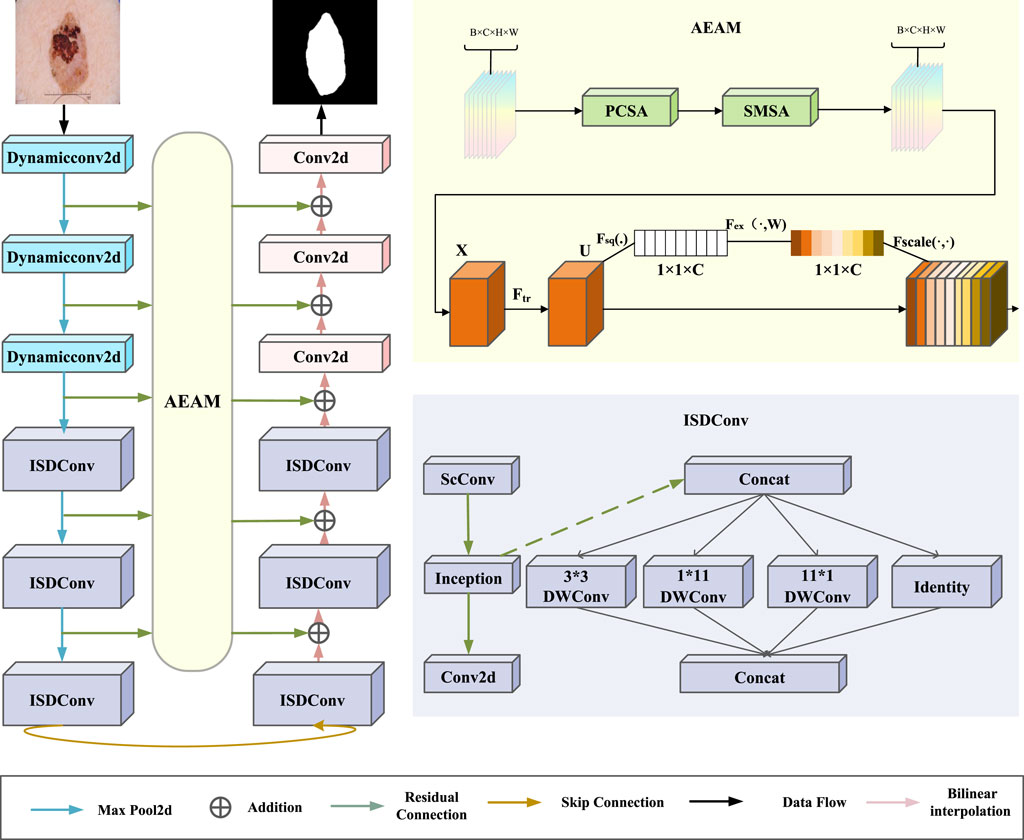
As illustrated in Figure 1, the proposed model framework consists primarily of the ISDConv module, the AEAM module, and dynamic convolution. The ISDConv module is composed of three parts: ScConv, Inception convolution, and standard convolution. By incorporating depthwise separable convolutions and group convolutions, ISDConv facilitates the model’s understanding of multi-scale information within images, thereby enhancing its ability to detect and classify objects of varying sizes.
The AEAM module operates in two stages: SEattention and SCSA. SEattention enhances the network’s representational capacity by explicitly modeling the interdependencies between convolutional feature channels. SCSA, in turn, is divided into two components: SMSA and PCSA. SMSA integrates multi-semantic information and employs a progressive compression strategy to inject discriminative spatial priors into the channel self-attention mechanism of PCSA, effectively guiding channel recalibration. Within PCSA, robust feature interaction based on a self-attention mechanism further mitigates the multi-semantic information discrepancy among sub-features in SMSA.
Inception-Split depth convolution
As shown in Figure 1, ISDConv consists of a ScConv, an Inception Convolution, and a standard Conv2d layer. The Inception Convolution achieves lightweight performance by efficiently decomposing a large kernel depthwise convolution into four parallel branches along the channel dimension. These branches consist of a small square kernel, two orthogonal large kernels, and an identity mapping. The use of a small square kernel reduces computational complexity, while the orthogonal large kernels capture different spatial information at varying scales. The identity mapping helps preserve the original input features, further enhancing the efficiency of the network. Additionally, this architecture incorporates 1 × 1 convolutions for dimensionality reduction before applying computationally expensive operations, minimizing the computational burden while preserving the model’s ability to learn rich, multi-scale features. These four branches not only achieve higher computational efficiency than the large kernel depthwise convolution but also maintain a large receptive field, enabling the model to capture spatial context effectively for improved performance.
One of the branches employs a
where,
where
Adaptive Enhanced Attention Module
This paper introduces the AEAM attention module, designed to achieve synergy between channel attention and spatial attention through dimensional decoupling, lightweight multi-semantic guidance, and semantic discrepancy mitigation. As shown in Figure 2, the AEAM module consists of two main components: SEattention and SCSA.
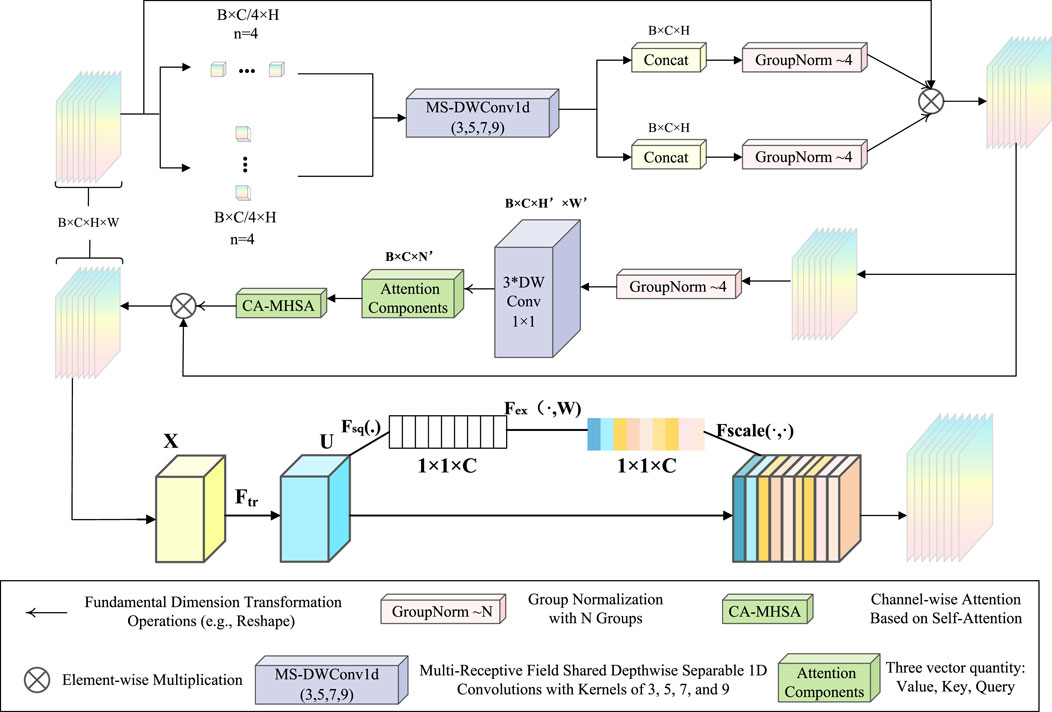
Figure 2. The overall framework of AEAM. SCSA uses multi-semantic spatial information to guide the learning of channel-wise self-attention.
The SCSA module is composed of two sequentially linked components: Shared Multi-Semantic Spatial Attention (SMSA) and Progressive Channel Self-Attention (PCSA). SMSA employs multi-scale, depth-sharing one-dimensional convolutions to extract spatial information at different semantic levels from four independent sub-features. This approach enables the efficient integration of diverse spatial semantics across sub-features. After SMSA modulates the feature maps, the resulting features are passed to PCSA. This component combines a progressive compression strategy with a channel-specific self-attention mechanism (CSA) to refine the feature representation further.
In this paper, a given input
The ablation formula is shown in Equation 4:
Where
The structure of the SE block is illustrated in the lower part of Figure 2. For any given transformation
The loss function
In this study, each image in the dataset is associated with a corresponding binary mask. Skin lesion segmentation is treated as a pixel-level binary classification task, distinguishing the skin lesions from the background. The combination of Binary Cross-Entropy (BCE) loss and the Dice Similarity Coefficient (DSC) loss is used as the loss function to optimize the network parameters. This approach effectively addresses the challenge of skin lesion segmentation by balancing pixel accuracy and overlap between the predicted and ground truth masks.
The loss function, referred to as the BceDice loss, can be expressed as Equation 5:
where
Experiment
Datasets
The ISIC (International Skin Imaging Collaboration) datasets are benchmark datasets widely used in medical image analysis, particularly for dermoscopic image segmentation, classification, and automated skin cancer detection. These datasets feature high-resolution dermoscopic images with comprehensive annotations, including lesion boundaries, diagnostic labels, and metadata. Covering a diverse range of skin conditions, they are designed to support tasks such as lesion segmentation, feature extraction, and disease classification. Notably, the ISIC2017 and ISIC2018 datasets have been instrumental in advancing research on melanoma detection and other skin diseases through the annual ISIC Challenges. Our research is specifically conducted on the ISIC2017 and ISIC2018 datasets. Figure 3 are some sample images from the ISIC2017 and ISIC2018 datasets.
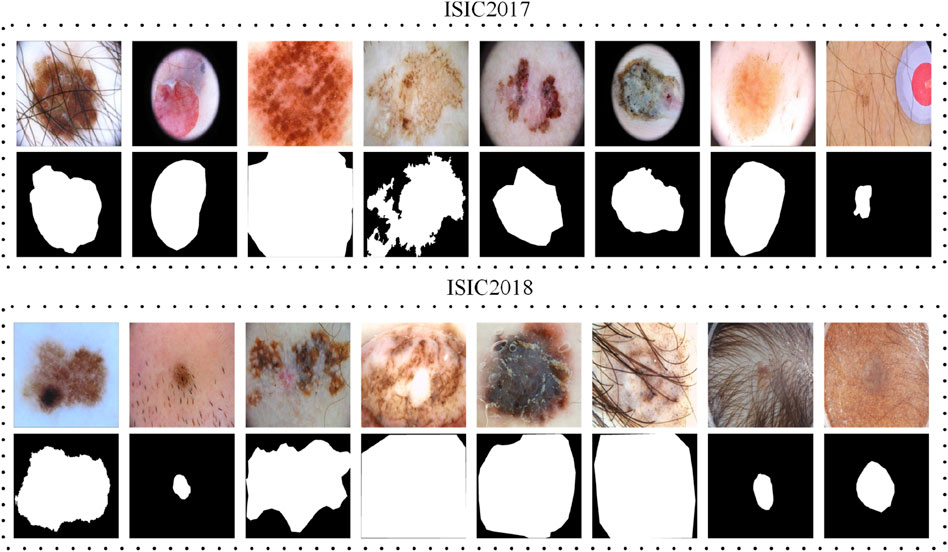
Figure 3. Examples of original images and their ground truth annotations from the ISIC2017 and ISIC2018 datasets.
Experiment details
All experiments were implemented using the PyTorch framework and performed on a laptop equipped with an NVIDIA GeForce RTX 3080 Ti GPU with 8 GB of memory. Based on established practices, all images were normalized and resized to 256
where
Evaluation metrics
In this study, segmentation performance is assessed using the mean Intersection over Union (mIoU), Dice Similarity Coefficient (DSC), and Accuracy (Acc), as defined in Equation 7. Additionally, the number of parameters is represented by Params, measured in millions (M), and computational complexity is quantified in GFLOPs. It is important to note that both Params and GFLOPs are calculated based on an input size of
Where, TP, FP, FN, and TN represent True Positives, False Positives, False Negatives, and True Negatives, respectively.
Segmentation result analysis
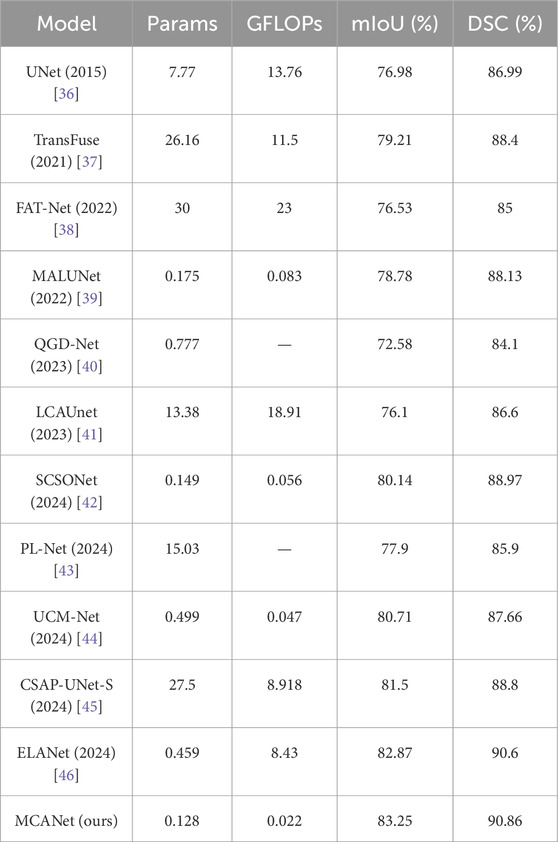
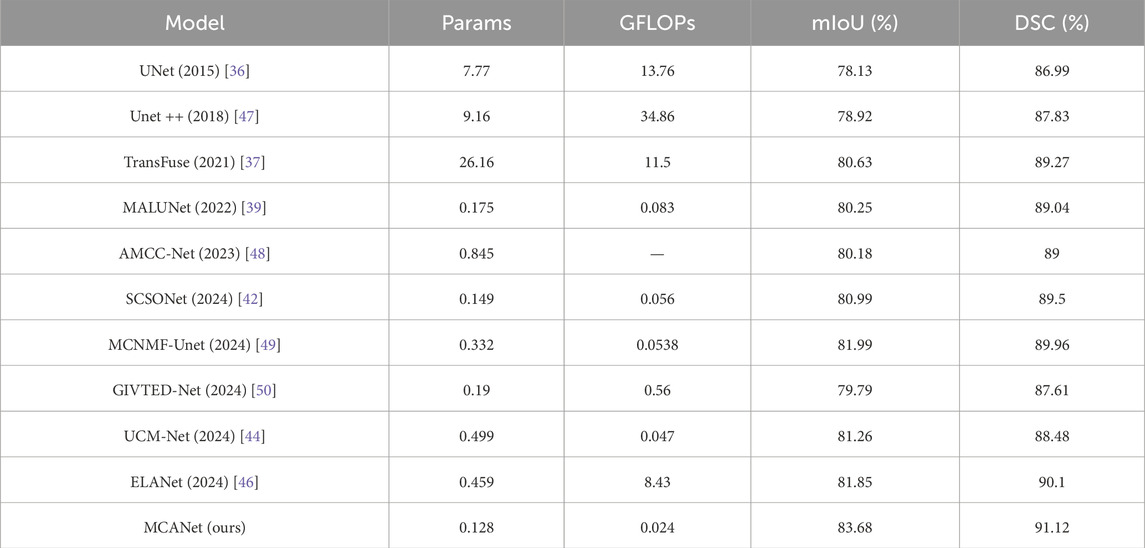
In this section, we conducted comparative experiments on melanoma segmentation using the ISIC2017 and ISIC2018 skin lesion segmentation datasets and evaluated the test results. The evaluation metrics include DSC, mIoU, params, and GFLOPs. The results are presented in Tables 1, 2, where we perform a comprehensive comparison of the proposed model with the following methods: UNet [36], Transfuse [37], FATNet [38], MALUNet [39], QGD-Net [40], LCA-UNet [41], SCSONet [42], PL-Net [43], UCM-Net [44], CSAP-UNet-S [45], and ELA-Net [46].
In addition, bar charts are utilized in this study to visually illustrate the performance of different models on various metrics, providing a clearer comparison between our method and others. Specifically, for the comparison of lightweight metrics, only models designed with lightweight objectives were selected, with the results presented in Figures 4, 5. The experimental results indicate that MCANet outperforms all other methods in both data sets in terms of DSC and mIoU metrics. Notably, MCANet achieves Dice scores exceeding 0.9 on the ISIC datasets, significantly outperforming all comparison models and demonstrating its superior segmentation performance.
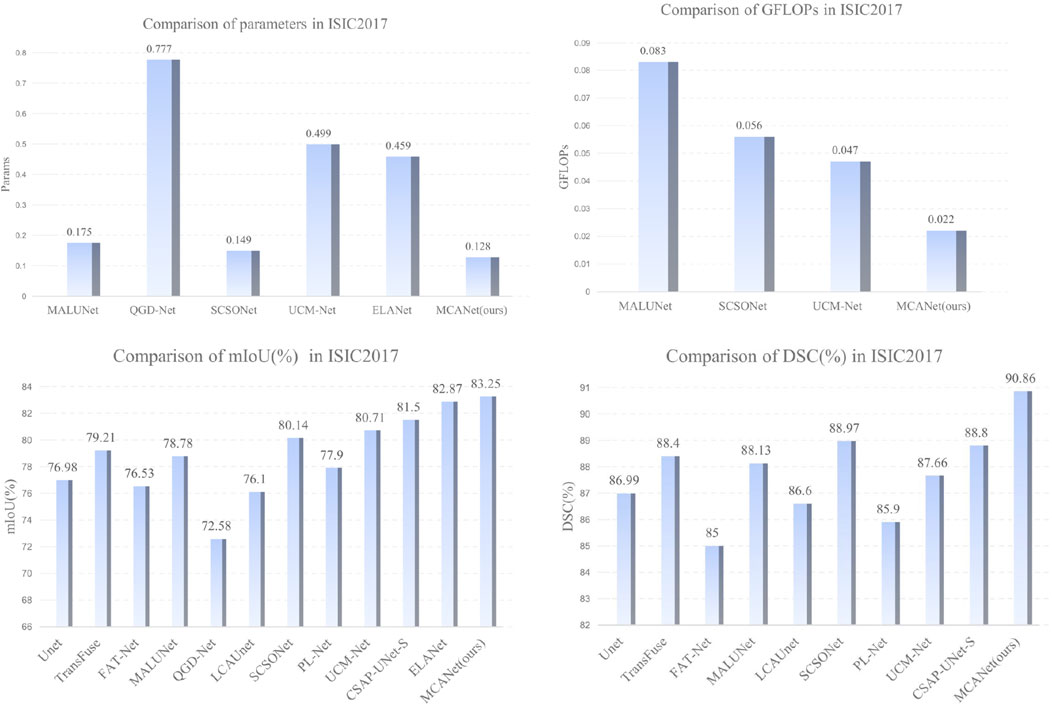
Figure 4. Performance comparison of different models across various metrics on the ISIC2017 dataset.
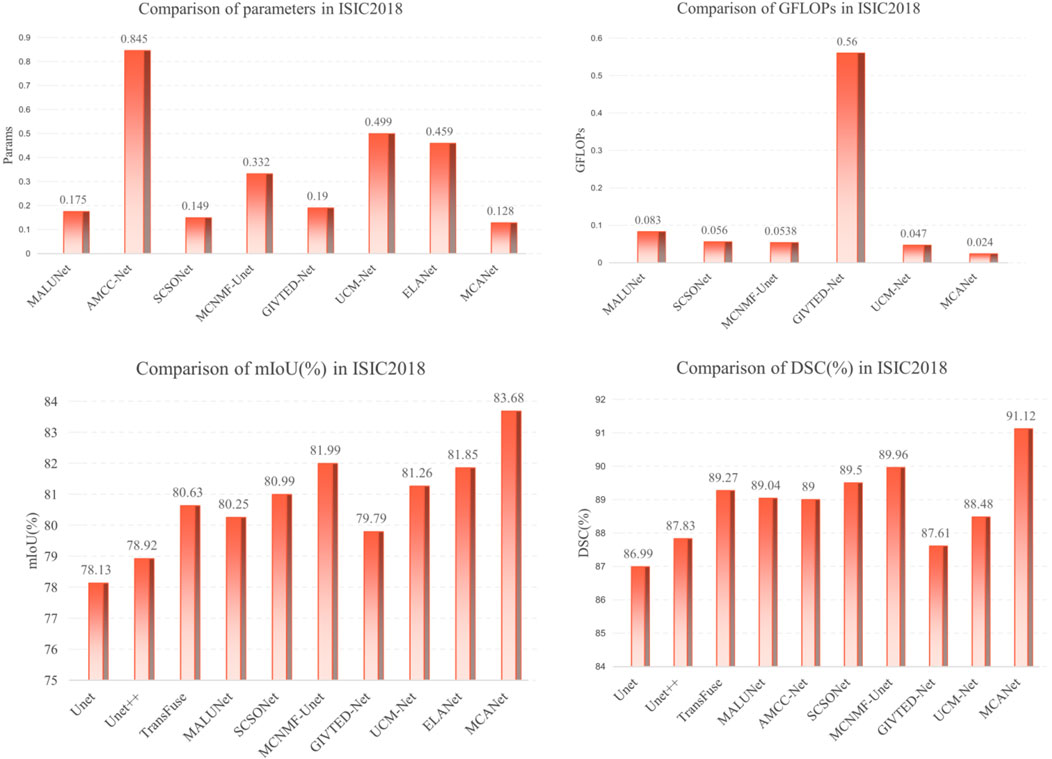
Figure 5. Performance comparison of different models across various metrics on the ISIC2018 dataset.
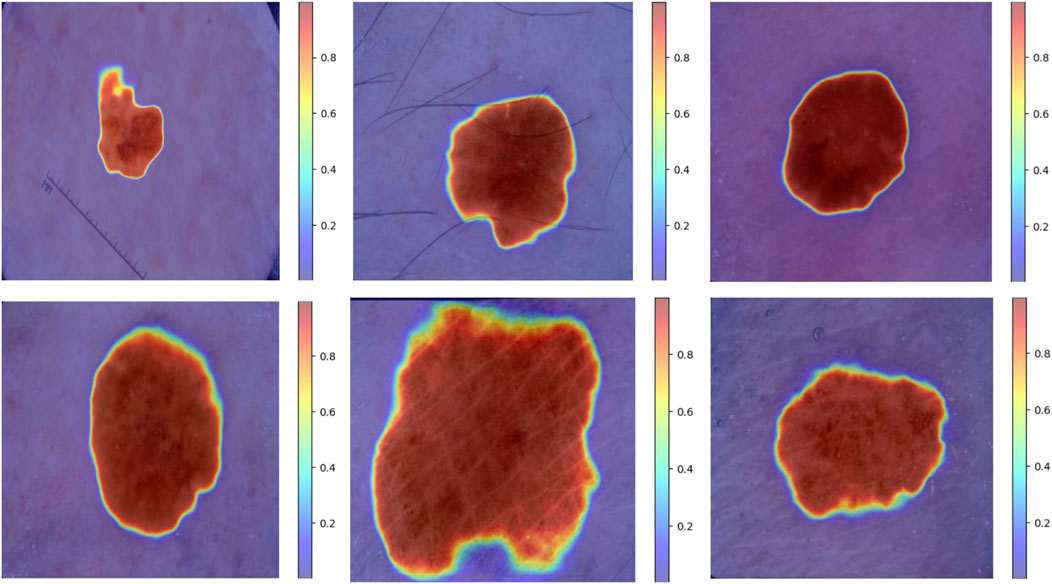
Furthermore, to further validate the segmentation performance of the model, we present the visual segmentation results on the ISIC dataset, as shown in Figures 6, 7. Although there are some differences between the MCANet segmentation results and mask images, MCANet outperforms other models in capturing detailed information from medical images, giving it a significant advantage in accurately segmenting the areas of the injury. Specifically, Figure 8 shows that MCANet can more accurately capture the target location in segmentation tasks involving smaller lesions, with finer and more precise segmentation of the lesion boundaries.However, our study also has some limitations. First, although MCANet demonstrates impressive performance on the ISIC datasets, its generalizability to other medical imaging datasets remains to be fully explored. In addition, while the model is lightweight in design, further optimization is required to meet the strict deployment constraints of resource-constrained devices, such as smartphones or embedded systems. Another limitation lies in the annotation quality of the datasets used, as potential noise in the segmentation masks may influence the model’s learning process. Finally, despite MCANet’s ability to capture detailed features, there are still some challenges in handling highly irregular or extremely small lesions, which may require more advanced attention mechanisms.To address these issues, future research will focus on several directions. First, extending the evaluation to additional datasets with diverse imaging modalities can help assess the robustness and versatility of MCANet. Second, incorporating techniques such as knowledge distillation or pruning could further improve the model’s efficiency for deployment in real-time scenarios. Third, exploring semi-supervised or unsupervised learning methods may reduce dependency on high-quality annotations, enabling better performance even with noisy labels. Finally, integrating advanced multi-scale feature extraction modules could enhance the model’s ability to handle challenging segmentation tasks involving complex lesion patterns.

Figure 6. Visual comparison of segmentation results of MCANet and other methods on the ISIC2017 dataset.

Figure 7. Visual comparison of segmentation results of MCANet and other methods on the ISIC2018 dataset.
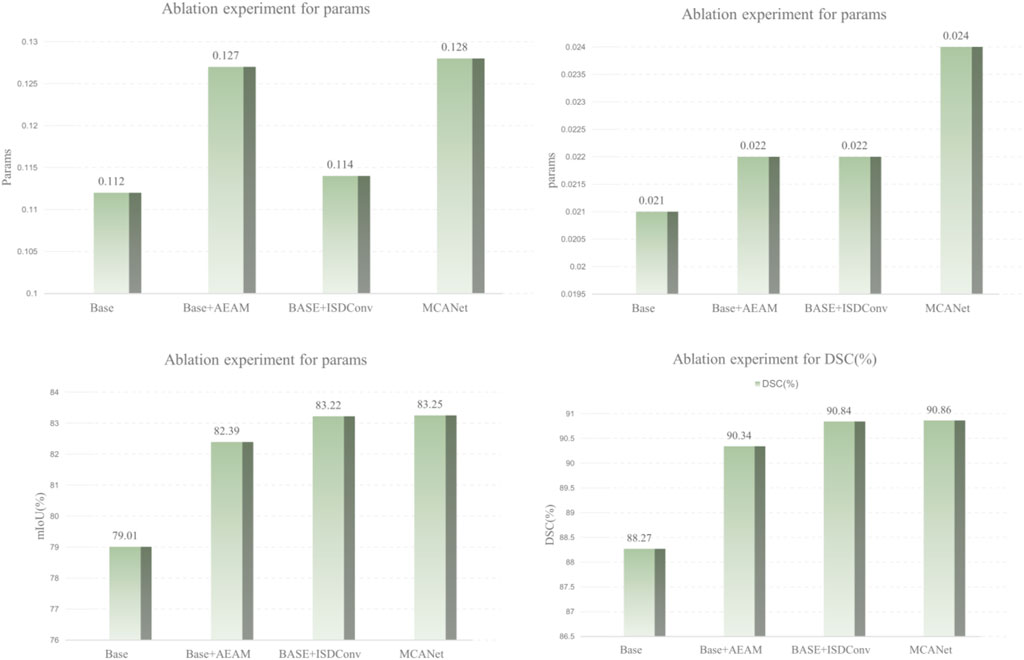
Ablation study on module effectiveness
To evaluate the contribution of each module in MCANet, we designed and conducted a series of ablation studies, with the results summarized in Table 3. Using the SCSONet baseline model as a reference, we performed comparative experiments with different combinations of the proposed modules on the ISIC dataset. Furthermore, to provide a clearer visualization of the impact of each module on segmentation performance, we used bar charts to illustrate variations in key metrics, such as DSC and mIoU, as shown in Figure 9.In the ablation study, “Base + AEAM” represents the integration of the proposed AEAM module into the baseline model, “Base + ISDConv” denotes the addition of the ISDConv module to the baseline, and “MCANet” refers to the complete network architecture proposed in this study. From Table 3 and the bar chart, it can be observed that integrating the proposed modules into the baseline model not only results in negligible increases in parameter count and computational complexity but also leads to significant improvements in segmentation performance. Specifically, as the modules are progressively added, the segmentation performance steadily improves, with the key metrics DSC and mIoU ultimately reaching 0.9086 and 0.8325, representing increases of
Moreover, the experimental results demonstrate that the proposed modules collaborate effectively, with the addition of individual modules not causing any degradation in overall performance but instead continuously improving segmentation accuracy. Additionally, our module design is highly adaptable, allowing for seamless integration into other network architectures without requiring significant modifications to the original structure. For instance, incorporating the AEAM or ISDConv modules into other networks results in varying degrees of performance improvement, validating the generalizability and practicality of the proposed modules.
In summary, the results of the ablation studies and their visual analysis demonstrate the significant contributions of the proposed modules to the model’s performance. These improvements not only enhance the segmentation capability of MCANet but also highlight the academic significance and practical applicability of our work in the field of medical image segmentation.
Conclusion
Medical image analysis typically requires significant computational resources, which directly impact diagnostic speed and accuracy. Advanced methods like deep learning are resource-intensive, making them difficult to implement in resource-constrained environments. To address this, we propose MCAN, a novel lightweight network architecture featuring ISDConv, AEAM, and dynamic convolution. Our model reduces computational costs while maintaining performance, achieving competitive segmentation with 0.128M parameters and 0.022 GFLOPs. However, due to the limited dataset, the model’s generalization ability requires further investigation.
Future research can focus on several key areas. Firstly, further optimization of lightweight techniques and attention mechanisms is needed, especially for specific types of medical images. For example, improving the prediction accuracy and robustness of melanoma images across different skin types is an important direction. Additionally, due to the limited dataset size in this study, further validation of the model’s generalization ability is required. Future work should aim to expand the dataset with more representative clinical data to assess the model’s performance in real-world clinical environments, particularly in resource-constrained settings such as mobile medical devices or low-resource hospitals. Finally, our method could be extended to multi-modal tasks, such as integrated diagnosis using CT and MRI, with a focus on improving the model’s fusion capability while maintaining computational efficiency.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
ZL: Conceptualization, Data curation, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. HW: Conceptualization, Formal Analysis, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. HC: Data curation, Investigation, Methodology, Project administration, Resources, Supervision, Writing–review and editing. CL: Conceptualization, Formal Analysis, Resources, Software, Writing–review and editing. AY: Data curation, Formal Analysis, Funding acquisition, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was sponsored by the Special key project of Chongqing technology innovation and application development (CSTB2024TIAD-STX0023, CSTB2024TIAD-STX0030, CSTB2024TIAD-STX0037), Science and Technology Research Program of Chongqing Municipal Education (KJQN202400618) and “Unveiling and Leading” Project by the Chongqing Municipal Bureau of Industry and Information Technology (2022-37).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Schadendorf D, Van Akkooi AC, Berking C, Griewank KG, Gutzmer R, Hauschild A, et al. Melanoma. The Lancet (2018) 392:971–84. doi:10.1016/s0140-6736(18)31559-9
2. Zhang Y, Li Z, Li H, Tao D. Prototype-driven and multi-expert integrated multi-modal mr brain tumor image segmentation. IEEE Trans Instrumentation Meas (2024) 74:1–14. doi:10.1109/tim.2024.3500067
3. Li Z, Zhang Y, Li H, Chai Y, Yang Y. Deformation-aware and reconstruction-driven multimodal representation learning for brain tumor segmentation with missing modalities. Biomed Signal Process Control (2024) 91:106012. doi:10.1016/j.bspc.2024.106012
4. Dong Z, Li J, Hua Z. Transformer-based multi-attention hybrid networks for skin lesion segmentation. Expert Syst Appl (2024) 244:123016. doi:10.1016/j.eswa.2023.123016
5. Sun Y, Dai D, Zhang Q, Wang Y, Xu S, Lian C. Msca-net: multi-scale contextual attention network for skin lesion segmentation. Pattern Recognition (2023) 139:109524. doi:10.1016/j.patcog.2023.109524
6. Qiu S, Li C, Feng Y, Zuo S, Liang H, Xu A. Gfanet: gated fusion attention network for skin lesion segmentation. Comput Biol Med (2023) 155:106462. doi:10.1016/j.compbiomed.2022.106462
7. Qi K, Yang H, Li C, Liu Z, Wang M, Liu Q, et al. X-net: brain stroke lesion segmentation based on depthwise separable convolution and long-range dependencies. In: Proceedings, Part III 22nd International Conference Medical Image Computing and Computer Assisted Intervention–MICCAI 2019; October 13–17, 2019; Shenzhen, China. Springer (2019) p. 247–55.
8. Liu X, Yang H, Qi K, Dong P, Liu Q, Liu X, et al. Msdf-net: multi-scale deep fusion network for stroke lesion segmentation. IEEE Access (2019) 7:178486–95. doi:10.1109/access.2019.2958384
9. Yang L, Fan C, Lin H, Qiu Y. Rema-net: an efficient multi-attention convolutional neural network for rapid skin lesion segmentation. Comput Biol Med (2023) 159:106952. doi:10.1016/j.compbiomed.2023.106952
10. Liu Y, Shi Y, Mu F, Cheng J, Chen X. Glioma segmentation-oriented multi-modal mr image fusion with adversarial learning. IEEE/CAA J Automatica Sinica (2022) 9:1528–31. doi:10.1109/jas.2022.105770
11. Zhu Z, Wang Z, Qi G, Mazur N, Yang P, Liu Y. Brain tumor segmentation in mri with multi-modality spatial information enhancement and boundary shape correction. Pattern Recognition (2024) 153:110553. doi:10.1016/j.patcog.2024.110553
12. Zhu Z, He X, Qi G, Li Y, Cong B, Liu Y. Brain tumor segmentation based on the fusion of deep semantics and edge information in multimodal mri. Inf Fusion (2023) 91:376–87. doi:10.1016/j.inffus.2022.10.022
13. Liu Y, Qi Z, Cheng J, Chen X. Rethinking the effectiveness of objective evaluation metrics in multi-focus image fusion: a statistic-based approach. IEEE Trans Pattern Anal Machine Intelligence (2024) 46:5806–19. doi:10.1109/tpami.2024.3367905
14. Khan TM, Naqvi SS, Meijering E. Esdmr-net: a lightweight network with expand-squeeze and dual multiscale residual connections for medical image segmentation. Eng Appl Artif Intelligence (2024) 133:107995. doi:10.1016/j.engappai.2024.107995
15. Zhou Y, Kang X, Ren F, Lu H, Nakagawa S, Shan X. A multi-attention and depthwise separable convolution network for medical image segmentation. Neurocomputing (2024) 564:126970. doi:10.1016/j.neucom.2023.126970
16. Liu T, Liu H, Yang B, Zhang Z. LDCNet: limb direction cues-aware network for flexible HPE in industrial behavioral biometrics systems. IEEE Trans Ind Inform (2023) 20:8068–78. doi:10.1109/tii.2023.3266366
17. Ma T, Wang K, Hu F. Lmu-net: lightweight u-shaped network for medical image segmentation. Med and Biol Eng and Comput (2024) 62:61–70. doi:10.1007/s11517-023-02908-w
18. Feng L, Wu K, Pei Z, Weng T, Han Q, Meng L, et al. Mlu-net: a multi-level lightweight u-net for medical image segmentation integrating frequency representation and mlp-based methods. IEEE Access (2024) 12:20734–51. doi:10.1109/access.2024.3360889
19. Ruan J, Xie M, Gao J, Liu T, Fu Y. Ege-unet: an efficient group enhanced unet for skin lesion segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer (2023) p. 481–90.
20. Lei T, Sun R, Du X, Fu H, Zhang C, Nandi AK. Sgu-net: shape-guided ultralight network for abdominal image segmentation. IEEE J Biomed Health Inform (2023) 27:1431–42. doi:10.1109/jbhi.2023.3238183
21. Chen Y, Dai X, Liu M, Chen D, Yuan L, Liu Z. Dynamic convolution: attention over convolution kernels. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (2020) p. 11030–9.
22. Li H, Cen Y, Liu Y, Chen X, Yu Z. Different input resolutions and arbitrary output resolution: a meta learning-based deep framework for infrared and visible image fusion. IEEE Trans Image Process (2021) 30:4070–83. doi:10.1109/tip.2021.3069339
23. Li H, Liu J, Zhang Y, Liu Y. A deep learning framework for infrared and visible image fusion without strict registration. Int J Comp Vis (2024) 132:1625–44. doi:10.1007/s11263-023-01948-x
24. Huang H, Chen Z, Zou Y, Lu M, Chen C, Song Y, et al. Channel prior convolutional attention for medical image segmentation. Comput Biol Med (2024) 178:108784. doi:10.1016/j.compbiomed.2024.108784
25. Shaker AM, Maaz M, Rasheed H, Khan S, Yang MH, Khan FS. Unetr++: delving into efficient and accurate 3d medical image segmentation. IEEE Trans Med Imaging (2024). doi:10.1109/TMI.2024.3398728
26. Fu Y, Liu J, Shi J. Tsca-net: Transformer based spatial-channel attention segmentation network for medical images. Comput Biol Med (2024) 170:107938. doi:10.1016/j.compbiomed.2024.107938
27. Xiong J, Tang M, Zong L, Li L, Hu J, Bian D, et al. Ina-net: an integrated noise-adaptive attention neural network for enhanced medical image segmentation. Expert Syst Appl (2024) 258:125078. doi:10.1016/j.eswa.2024.125078
28. Song E, Zhan B, Liu H. Combining external-latent attention for medical image segmentation. Neural Networks (2024) 170:468–77. doi:10.1016/j.neunet.2023.10.046
29. Huang Z, Cheng S, Wang L. Medical image segmentation based on dynamic positioning and region-aware attention. Pattern Recognition (2024) 151:110375. doi:10.1016/j.patcog.2024.110375
30. Yang S, Zhang X, Chen Y, Jiang Y, Feng Q, Pu L, et al. Ucunet: a lightweight and precise medical image segmentation network based on efficient large kernel u-shaped convolutional module design. Knowledge-Based Syst (2023) 278:110868. doi:10.1016/j.knosys.2023.110868
31. Sun Q, Dai M, Lan Z, Cai F, Wei L, Yang C, et al. Ucr-net: U-shaped context residual network for medical image segmentation. Comput Biol Med (2022) 151:106203. doi:10.1016/j.compbiomed.2022.106203
32. Nisa SQ, Ismail AR. Dual u-net with resnet encoder for segmentation of medical images. Int J Adv Comp Sci Appl (2022) 13. doi:10.14569/ijacsa.2022.0131265
33. Zhao Q, Zhong L, Xiao J, Zhang J, Chen Y, Liao W, et al. Efficient multi-organ segmentation from 3d abdominal ct images with lightweight network and knowledge distillation. IEEE Trans Med Imaging (2023) 42:2513–23. doi:10.1109/tmi.2023.3262680
34. Wang H, Zhang D, Song Y, Liu S, Wang Y, Feng D, et al. Segmenting neuronal structure in 3d optical microscope images via knowledge distillation with teacher-student network. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019). IEEE (2019) p. 228–31.
35. Hajabdollahi M, Esfandiarpoor R, Khadivi P, Soroushmehr SMR, Karimi N, Samavi S. Simplification of neural networks for skin lesion image segmentation using color channel pruning. Comput Med Imaging Graphics (2020) 82:101729. doi:10.1016/j.compmedimag.2020.101729
36. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: proceedings, part III 18th international conference Medical image computing and computer-assisted intervention–MICCAI 2015; October 5-9, 2015; Munich, Germany. Springer (2015) p. 234–41.
37. Zhang Y, Liu H, Hu Q. Transfuse: fusing transformers and cnns for medical image segmentation. In: proceedings, Part I 24th international conference Medical image computing and computer assisted intervention–MICCAI 2021; September 27–October 1, 2021; Strasbourg, France. Springer (2021) p. 14–24.
38. Wu H, Chen S, Chen G, Wang W, Lei B, Wen Z. Fat-net: feature adaptive transformers for automated skin lesion segmentation. Med image Anal (2022) 76:102327. doi:10.1016/j.media.2021.102327
39. Ruan J, Xiang S, Xie M, Liu T, Fu Y. Malunet: a multi-attention and light-weight unet for skin lesion segmentation. In: 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE (2022) p. 1150–6.
40. Wang J, Huang G, Zhong G, Yuan X, Pun CM, Deng J. Qgd-net: a lightweight model utilizing pixels of affinity in feature layer for dermoscopic lesion segmentation. IEEE J Biomed Health Inform (2023) 27:5982–93. doi:10.1109/jbhi.2023.3320953
41. Zhang Q, Bai R, Peng B, Wang Z, Liu Y. Fft pattern recognition of crystal hrtem image with deep learning. Micron (2023) 166:103402. doi:10.1016/j.micron.2022.103402
42. Chen H, Li Z, Huang X, Peng Z, Deng Y, Tang L, et al. Scsonet: spatial-channel synergistic optimization net for skin lesion segmentation. Front Phys (2024) 12:1388364. doi:10.3389/fphy.2024.1388364
43. Cheng J, Gao C, Lu H, Ming Z, Yang Y, Zhu M. Pl-net: progressive learning network for medical image segmentation (2021) arXiv preprint arXiv:2110.14484.
44. Weng S, Zhu T, Zhang T, Zhang C. Ucm-net: a u-net-like tampered-region-related framework for copy-move forgery detection. IEEE Trans Multimedia (2023) 26:750–63. doi:10.1109/tmm.2023.3270629
45. Fan X, Zhou J, Jiang X, Xin M, Hou L. Csap-unet: convolution and self-attention paralleling network for medical image segmentation with edge enhancement. Comput Biol Med (2024) 172:108265. doi:10.1016/j.compbiomed.2024.108265
46. Nie T, Zhao Y, Yao S. Ela-net: an efficient lightweight attention network for skin lesion segmentation. Sensors (2024) 24:4302. doi:10.3390/s24134302
47. Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J. Unet++: a nested u-net architecture for medical image segmentation. In: Proceedings 4 Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018; September 20, 2018; Granada, Spain. Springer (2018) p. 3–11.
48. Dayananda C, Yamanakkanavar N, Nguyen T, Lee B. Amcc-net: an asymmetric multi-cross convolution for skin lesion segmentation on dermoscopic images. Eng Appl Artif Intelligence (2023) 122:106154. doi:10.1016/j.engappai.2023.106154
49. Yuan L, Song J, Fan Y. Mcnmf-unet: a mixture conv-mlp network with multi-scale features fusion unet for medical image segmentation. PeerJ Comp Sci (2024) 10:e1798. doi:10.7717/peerj-cs.1798
Keywords: medical image segmentation, lightweight, melanoma, attention mechanism, Inception
Citation: Li Z, Wang H, Chen H, Lin C and Yan A (2024) Multi-Conv attention network for skin lesion image segmentation. Front. Phys. 12:1532638. doi: 10.3389/fphy.2024.1532638
Received: 22 November 2024; Accepted: 05 December 2024;
Published: 20 December 2024.
Edited by:
Bo Xiao, Imperial College London, United KingdomReviewed by:
Gang Hu, Buffalo State College, United StatesYafei Zhang, Kunming University of Science and Technology, China
Yimin Chen, University of Massachusetts Lowell, United States
Copyright © 2024 Li, Wang, Chen, Lin and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aochen Yan, YW9jaGVueWFAdXNjLmVkdQ==
 Zexin Li
Zexin Li Hanchen Wang1
Hanchen Wang1 Haoyu Chen
Haoyu Chen Chenxin Lin
Chenxin Lin Aochen Yan
Aochen Yan