- 1School of Artificial Intelligence, Chongqing University of Education, Chongqing, China
- 2School of Pharmacy (School of Traditional Chinese Medicine), Chongqing Medical and Pharmaceutical College, Chongqing, China
Ultrasound imaging has a history of several decades. With its non-invasive, low-cost advantages, this technology has been widely used in medicine and there have been many significant breakthroughs in ultrasound imaging. Even so, there are still some drawbacks. Therefore, some novel image reconstruction and image analysis algorithms have been proposed to solve these problems. Although these new solutions have some effects, many of them introduce some other side effects, such as high computational complexity in beamforming. At the same time, the usage requirements of medical ultrasound equipment are relatively high, and it is not very user-friendly for inexperienced beginners. As artificial intelligence technology advances, some researchers have initiated efforts to deploy deep learning to address challenges in ultrasound imaging, such as reducing computational complexity in adaptive beamforming and aiding novices in image acquisition. In this survey, we are about to explore the application of deep learning in medical ultrasound imaging, spanning from image reconstruction to clinical diagnosis.
1 Introduction
1.1 Brief introduction to medical imaging
Medical imaging relies on various physical phenomena to visualize human body tissues, internally and externally, through non-invasive or invasive techniques. Key modalities such as computed tomography (CT), magnetic resonance imaging (MRI), X-ray radiography, ultrasound, and digital pathology generate essential healthcare data, constituting around 90% of medical information [1]. Consequently, medical imaging plays a vital role in clinical assessment and healthcare interventions. Deep learning, as the cornerstone technology propelling the ongoing artificial intelligence (AI) revolution, exhibits significant potential in medical imaging. It spans from image reconstruction to comprehensive image analysis[2–8]. The integration of deep learning with medical imaging has spurred advancements, with the potential to reshape clinical practices and healthcare delivery. Empirical evidence has proven that deep learning algorithms exhibit performance comparable to that of medical professionals in diagnosing various medical conditions from imaging data [9]. At the same time, many applications of deep learning in clinics have emerged [10–16]. Consequently, there is a discernible trend towards certifying software applications for clinical utilization [17].
1.2 Literature reviews of deep learning in ultrasound beamforming
The development of medical ultrasound has a history of 80–90 years now. Medical ultrasound began as an investigative technology around the end of World War II [18]. With advancements in electronics, this technology improved. Ultrasound is being continually refined for better resolution, more portable devices, and more automated systems that can aid even in remote diagnostics. The most recent advancement in medical ultrasound is the incorporation of AI to help diagnosis.
The application of AI in medical ultrasound is long-standing [19–25]. With the explosion of deep learning, its application in medicine has become even more widespread. The medical ultrasound system mainly includes image reconstruction and image analysis, both of which have seen extensive applications of deep learning [26]. Deep learning has brought a revolutionary change in ultrasound beamforming, significantly enhancing image quality and improving computational efficiency. Ultrasound beamforming is a process of combining signals from multiple ultrasound elements to construct a focused image. Traditional methods rely heavily on user intervention and predefined parameters, which may limit the image quality and accuracy. Deep learning, on the other hand, uses neural network models to learn and generalize from examples. In the context of ultrasound beamforming, deep learning methods can learn to extract relevant features from raw ultrasound data and form a high-quality image without needing explicit instructions or predefined parameters. The process is relatively autonomous and adaptable. In training phase, a deep learning model is trained with a large amount of data (usually raw Radio Frequency (RF) data) which includes both inputs (ultrasound signals) and outputs (desired images). The model learns to identify patterns in the data and how to predict the output from given inputs. Once trained, the model can be used with new input data to predict the corresponding output images. The advantage is that this prediction process is usually faster than traditional beamforming methods as it bypasses the need for complex signal processing. Deep learning models such as Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) have been successfully used in ultrasound beamforming. They have shown promising results in enhancing image resolution, reducing speckle noise, improving contrast, and even performing advanced tasks like tissue characterization and acoustic aberration correction. Around 2017, applications of deep learning in beamforming began to appear in publications [27,28], and the interest in this area has been increasing ever since. In plane wave imaging, if only one plane wave is emitted, a very high frame rate can be achieved, but this will lead to poor image quality. Therefore, to improve image quality, a method called coherent plane wave compounding (CPWC) [29] has been proposed to solve this problem. However, using this method usually requires the emission of plane waves at multiple angles, which leads to a reduction in frame rate. Gasse et al. [27] propose a method using CNNs that allows for the acquisition of high-quality images even with the emission of only three plane waves. Luchies and Byram [28,30] discuss how to use deep neural networks (DNNs) to suppress off-axis scattering. The study is based on operations in the frequency domain through short-time Fourier transform. There are also some studies on bypassing beamforming [31–36]. The principal concept involves utilizing advanced deep learning methodologies to directly reconstruct images or conduct image segmentation from raw RF data. Deep learning has also been used to reduce artifacts in multi-line acquisition (MLA) and multi-line transmission (MLT) [37,38, 39]. Luijten et al. [35,40] investigate how deep learning can be applied to the adaptive beamforming process, addressing the computational challenges and aiming to produce better ultrasound images. Wiacek et al. [35,42] explore the use of DNNs to estimate normalized cross-correlation as a function of spatial lag. This estimation is specifically for coherence-based beamforming, such as short-lag spatial coherence (SLSC) beamforming [44]. Using sub-sampled RF data to reconstruct images can increase the frame rate, but the image quality will decrease. Some researchers [31,45–47] propose using deep learning to address this issue. More research is focused on the application of deep learning in plane wave imaging [48–56]. Some studies [57–59] discuss the training schemes. In addition, the ultrasound community also organized a challenge to encourage researchers to engage in deep learning research [60,61].
1.3 Overview of deep learning in clinical application of ultrasound
Deep learning plays a significant role in ultrasound clinical applications as it enhances the efficiency and accuracy of diagnosis, reducing human errors and paving the way for more sophisticated applications. Deep learning models can be trained to automatically detect and segment lesions in ultrasound images. This reduces the workload for radiologists and increases accuracy, as human interpretation can be subjective and variable. They can also be trained to classify diseases based on ultrasound images. Deep learning helps build more detailed 3D and 4D imaging from 2D ultrasound images, providing a more comprehensive picture of the patient’s condition. Deep learning algorithms can be used to predict clinical outcomes or progression of a disease based on ultrasound imaging data. From Ref. [19–25], it can be seen that the application of AI in medical ultrasound analysis predates that of beamforming. Medical ultrasound analysis mainly includes segmentation, classification, registration, and localization [62,63]. The integration of deep learning with ultrasound image analysis has spurred advancements, with the potential to reshape clinical practices. Breast cancer is a disease that seriously threatens people’s health [64]. The application of deep learning in breast ultrasound can effectively assist radiologists or clinicians in diagnosis. Becker et al. [65] are attempting to use a deep learning software (DLS) to classify breast cancer from ultrasound images. Xu et al. [66] focus on segmenting breast ultrasound images into functional tissues using CNNs. This segmentation aids in tumor localization, breast density measurement, and treatment response assessment, crucial for breast cancer diagnosis. Qian et al. [67] discuss a deep-learning system designed to predict Breast Imaging Reporting and Data System (BI-RADS) scores for breast cancer using multimodal breast-ultrasound images. Chen et al. [68] introduce a novel deep learning model for breast cancer diagnosis using contrast-enhanced ultrasound (CEUS) videos. Jabeen et al. [69] present a novel framework for classifying breast cancer from ultrasound images. The method employs deep learning and optimizes feature selection and fusion for enhanced classification accuracy. Raza et al. [70] propose a deep learning framework, DeepBreastCancerNet, designed for the detection and classification of breast cancer from ultrasound images. Deep learning is also widely applied in cardiac ultrasound. Degel et al. [71] discuss a novel approach to segment the left atrium in 3D echocardiography images using CNNs. Leclerc et al. [72] evaluate encoder-decoder deep CNN methods for assessing 2D echocardiographic images. The study introduces the Cardiac Acquisitions for Multi-structure Ultrasound Segmentation (CAMUS) dataset, the largest publicly available and fully annotated dataset for echocardiographic assessment, featuring images from 500 patients. Ghorbani et al. [73] investigate the application of deep learning models, particularly CNNs, to interpret echocardiograms. Narang et al. [74] explore the efficacy of a deep learning algorithm in assisting novice operators to obtain diagnostic-quality transthoracic echocardiograms. Ultrasonography is also a primary diagnostic method for thyroid diseases [75]. Thus, some studies on the assistance of deep learning in the diagnosis of thyroid diseases [76–81] emerged. There are also many applications of deep learning in prostate cancer detection [82–84] and prostate segmentation [85–91]. In ultrasound fetal imaging, deep learning also plays an increasingly important role[92–97]. In addition, there is a constant emergence of deep learning research in ultrasound brain imaging [98–103].
1.4 Other review articles on deep learning in ultrasound imaging
There are already some review articles about deep learning in medical ultrasound imaging. van Sloun et al. [104] presents an inclusive examination of the potential and application of deep learning strategies in ultrasound systems, spanning from the front end to more complex applications. In another article [105], they specifically discussed deep learning in beamforming. They introduce the potential role that deep learning can play in beamforming, as well as some of the existing achievements of deep learning in beamforming, and also look forward to new opportunities. Ref. [106] discusses the shortcomings of traditional signal processing methods in ultrasound imaging. The paper suggests a blend of model-based signal processing methods with machine learning approaches, stating that probability theory can seamlessly bridge the gap between conventional strategies and modern machine/deep learning approaches. However, these articles mainly focus on the principles of ultrasound imaging and do not discuss clinical applications. There are also some articles that provide reviews from the perspective of image analysis and clinical practices. Reference [62,63] discuss deep learning in medical ultrasound analysis from multiple perspectives. Afrin et al. [107] discuss the application of deep learning in different ultrasound methods for breast cancer management - from diagnosis to prognosis. Reference [108] presents an in-depth analysis of the application of AI in echocardiography interpretation. Khachnaoui et al. [109] discuss the role of ultrasound imaging in diagnosing thyroid lesions. In this review, our aim is to introduce the application of deep learning in medical ultrasound from the perspective of image reconstruction to clinical applications. The content seems to be quite broad, we aim to provide a comprehensive perspective on the application of deep learning in medical ultrasound and introduce the potential role of deep learning in ultrasound imaging from a system perspective.
2 Overview of medical ultrasound system
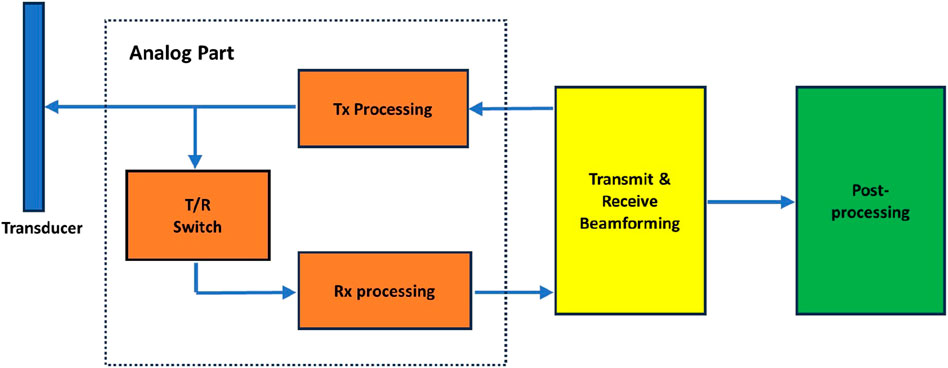
A medical ultrasound system consists of various interconnected modules, each of which is further segmented into numerous smaller components. Figure 1 illustrates a simplified block diagram of a medical ultrasound system. The entire signal processing pipeline of an ultrasound system is relatively complex, with even more detailed subdivisions for each module. For those interested in a deeper exploration, please refer to [110]. Here, we are only providing readers with a high-level overview, and a more in-depth introduction to the modules we are interested in will be covered subsequently. The dashed box in Figure 1 represents the analog signal processing module, which is not within the scope of discussion in this survey. We will focus on the discussion of the transmit and receive beamforming and introduce the post-processing as well. The transmit and receive beamforming are actually two distinct parts; however, in Figure 1, we categorize both under beamforming. In subsequent discussions, we will address these two parts separately. The categorization of post-processing here may be overly broad. In fact, after beamforming, there is a series of intermediate processing steps before the final post-processing. However, in this context, we refer to all these processing steps collectively as post-processing.
2.1 Transmit processing
In ultrasound systems, transmit beamforming is a technique that involves controlling the timing of excitation of multiple transducer elements to produce a directional beam or focus the beam within a specific area. By precisely adjusting the phase and amplitude of each element, an ultrasound beam with a specific direction and focal depth can be formed [111].
It can be seen from Figure 2, which illustrates the process of transmit beamforming, that the distance from each element on the transducer to the focal point is different. To ensure the beam ultimately focus at the point, it is necessary to control the emission timing of each element. It is noteworthy that in the typical transmission focusing, only a subset of elements are involved. In plane wave imaging, all elements on the transducer need to transmit, and the direction of the plane wave is controlled by adjusting the transmission timing of each element.
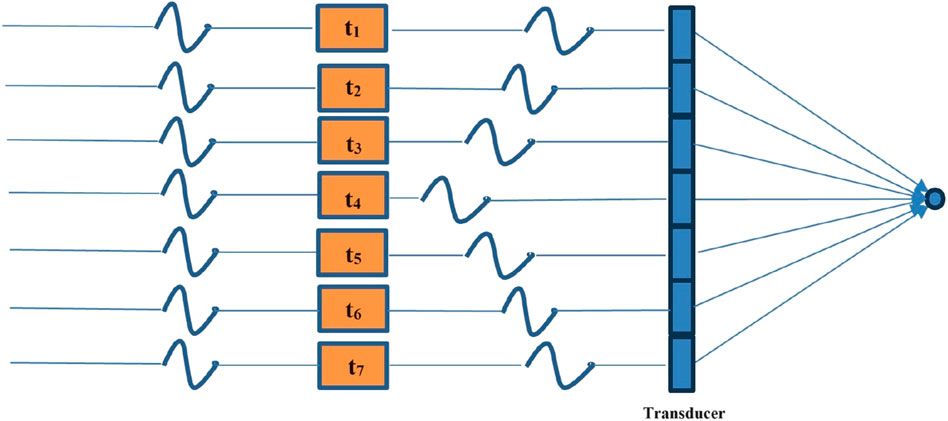
Figure 2. Schematic of transmit beamforming. By controlling the emission time of each element on the transducer, the waves emitted by each element can ultimately be focused on one point.
2.2 Receive processing
The receive beamforming is a crucial signal processing technique used to construct a high quality image from the echoes returning from the scanned tissue or organs in ultrasound imaging. When an ultrasound probe emits high-frequency sound waves, they travel through the body, echolocate off structures, and are then reflected back to the receiver. The reflected echoes are captured by multiple transducer elements arranged in an array on the probe. The schematic is illustrated in Figure 3.
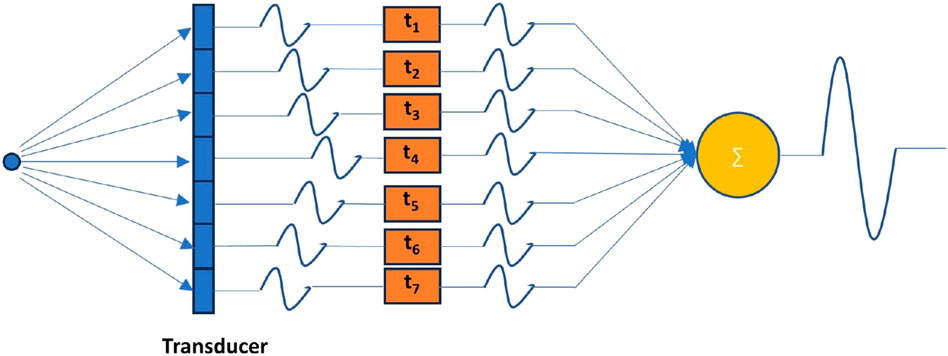
Figure 3. Schematic of receive beamforming. In order to align the echo received by each transducer element that is reflected from a certain point, it is necessary to properly delay the signal received by each element.
Receive beamforming involves combining the signals received by each of these elements in an intelligent way to construct a coherent and high-resolution representation of the scanned region. The most fundamental beamforming technique is the delay-and-sum (DAS) [112] method where the received signals from different transducer elements are delayed relative to each other to account for the different times of flight from the reflecting structure. They are then summed together, enhancing the signal from a specific direction or focal point while attenuating the signals from other directions. While the transmitted beam can be focused at a certain depth, receive beamforming allows dynamic focusing at various depths on receive. The delays are continuously adjusted as the echoes return from different depths, effectively focusing the beam at multiple depths in real-time. The apodization process involves weighting the received signals before they are summed, reducing side lobes and improving the lateral resolution. Advanced beamforming techniques use adaptive methods like Minimum Variance (MV) [113] to improve the image quality further by adapting to the signal environment, hence reducing the impact of off-axis scattering and noise. The result of receive beamforming is a narrow, well-defined beam that can accurately locate and display the internal structures of the body, thus providing detailed images for diagnosis. Advances in digital signal processing and hardware technology have significantly improved beamforming techniques, making them more sophisticated and effective.
2.3 Post processing
Ultrasound imaging can be roughly divided into pre-processing and post-processing [114]. Beamforming, as a key part of pre-processing, plays an important role in imaging quality, but post-processing is also an indispensable step. The post-processing is a research field that involves applying several steps after the channel data are mapped to the image domain via beamforming. These steps include further image processing to improve B-mode image quality, such as contrast, resolution, despeckling. It also involves spatiotemporal processing to suppress tissue clutter and to estimate motion. For 2D or 3D ultrasound data, post-processing is crucial for automatic analysis and/or quantitative measurements [115]. For instance, the recovery of quantitative volume parameters is a unique way of making objective, reproducible, and operator-independent diagnoses.
Medical ultrasound image analysis involves the use of diagnostic techniques, primarily those leveraging ultrasound, to create an image of internal body structures like blood vessels, joints, muscles, tendons, and internal organs. These images can then be used to measure certain characteristics such as distances and velocities. Medical ultrasound image analysis has extensive applications in various medical fields, including fetal, cardiac, trans-rectal, and intra-vascular examinations.
Common practices in the analysis of medical ultrasound images often encompass techniques such as segmentation and classification. Segmentation separates different types of organs and structures in the image, especially for regions of interest. Segmentation often uses edge detection, region growing, thresholding techniques, and more advanced techniques such as cascade classifiers, random forests, deep learning, etc. Classification is also a key part of image analysis. It classifies the images into normal images and abnormal images based on the previously extracted features, or further, performs disease classification. Common classification methods include neural networks, K-nearest neighbors (K-NN), decision trees, Support Vector Machines (SVM), etc. In recent years, deep learning-based classification models, such as CNNs, have been widely used, and with their powerful performance and accuracy, are extensively applied in the field of medical image analysis.
3 Deep learning in medical ultrasound imaging
We will discuss the application of deep learning in medical ultrasound imaging from several perspectives. First is the improvement of different beamforming techniques via deep learning, followed by a discussion on clinical application, and then the analysis of the application of deep learning in portable ultrasound devices and training schemes. Finally, we will briefly introduce the CNNs and transformer.
3.1 Image reconstruction
3.1.1 Bypass beamforming
Beamforming plays a crucial role in enhancing image quality. DAS algorithm, as a classic beamforming technique, is widely used in ultrasound imaging systems. Despite its operational simplicity and ease of implementation, this method also presents certain limitations and drawbacks. DAS beamforming generates relatively high side lobes and grating lobes, which are unwanted beam directions that may capture reflected signals from non-target areas, reducing image contrast and resolution. To suppress the side lobes, a common method is to use apodization, which is the application of weighting windows.
Usually, beamforming synthesizes the signals received by an array of elements to form a directional response or beam pattern, but this process can be computationally intensive. Deep learning approaches can potentially learn to perform the beamforming operation more efficiently, leading to faster image reconstruction without compromising quality. Simson et al. [31] address the challenge of reconstructing high-quality ultrasound images from sub-sampled raw data. Traditional beamforming methods, although adept at generating high-resolution images, impose considerable computational demands and their efficacy diminishes when dealing with sub-sampled data. To overcome this issue, the authors propose “DeepFormer,” an end-to-end, deep learning-based method designed to reconstruct high-quality ultrasound images in real-time, using sub-sampled raw data. Traditional beamforming algorithms often ignore the information between scan lines. Yet, a fully convolutional neural network (FCNN) is capable of capturing this information and utilizing it effectively; thus, enabling cross-scan line interpolation in sub-sampled data. As shown in Eq. 1 [31], the loss function used in DeepFormer is a combination of ℓ1 loss and Structural Similarity Imaging Metric (SSIM) [116].
Their results, which were tested on an in vivo dataset of some participants, indicate that DeepFormer is a promising approach for enhancing ultrasound image quality while also providing the speed necessary for clinical use. In addition, Nair et al. [32–35], introduced a concept with the objective of achieving high frame rates for automated imaging tasks over an extended field of view using single plane wave transmissions. They address the typical challenge of suboptimal image quality produced by single plane wave insonification and propose the use of DNNs to directly extract information from raw RF data to generate both an image and a segmentation map simultaneously. Unlike traditional beamforming, which generally only reconstructs images, they have utilized deep learning to achieve both image reconstruction and segmentation at the same time. They employed FCNN, the entire network includes an encoder and two decoders, one for image reconstruction and the other for image segmentation. As shown in Eq. 2 [35], the loss function of the entire network also adopts a combination of ℓ1 loss and Dice similarity coefficient (DSC) loss.
The DSC loss is used to measure the overlap between the predicted and true segmentation masks during the training of their DNN. Specifically, the DSC loss is utilized to quantify the similarity between the predicted DNN segmentation and the true segmentation. The DSC is calculated as a function of the overlap between these two segmentations, with a value of one indicating perfect overlap and 0 indicating no overlap. The DSC loss complements the mean absolute error loss by focusing on the segmentation performance of the network. While the mean absolute error provides a pixel-wise comparison between the predicted and reference images, the DSC loss offers a more holistic measure of the segmentation quality, especially important in medical imaging where the precise localization of structures is vital. This dual-loss approach enables the network to learn both the image reconstruction and segmentation tasks effectively, ensuring that the network parameters are optimized to generate accurate segmentations alongside the reconstructed images. The comparison between classic beamforming and this method is shown in Figure 4.
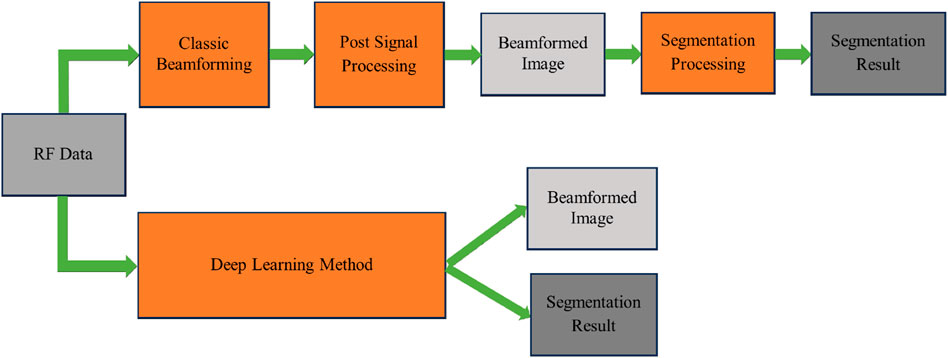
Figure 4. A comparison between DAS beamforming (top) and a deep learning method that bypasses the beamforming process [35] (bottom).
3.1.2 Adaptive beamforming
Traditional beamforming typically uses a fixed, predetermined set of weights applied to the received signals from each transducer element. These weights are usually uniform (DAS) or they use simple apodization (windowing) techniques. The resolution is generally limited by the fixed nature of the weights. The main lobe width does not adapt to different signal scenarios, which can lead to a less focused image. Traditional approaches may exhibit relatively higher side lobes, inducing higher levels of interference and clutter within the image. However, these methods are simpler to implement and faster in terms of computation, which makes them suitable for many real-time imaging applications.
The MV beamforming uses an adaptive approach to determine the weights applied to the signals. It calculates the weights that minimize the variance of the noise and interference, essentially optimizing the signal-to-noise ratio. The adaption of weights allows to generate a much narrower main lobe in the beam pattern, which translates to higher spatial resolution and better ability to distinguish between closely spaced scatterers. As a result of the narrower main lobe and suppressed side lobes, MV can provide significantly improved image resolution and contrast. It allows for clearer delineation of structures within the body, especially beneficial when visualizing small or closely spaced scatterers. The MV algorithm uses the data from the transducer elements to estimate the covariance matrix of the received signals. As shown in Eq. 3 [113], the weights are derived to minimize the output variance while maintaining the gain in the direction of the signal of interest.
where Rx is the covariance matrix of the received signals and a is a steering vector of ones. This optimization process, typically solved as a constrained minimization problem, is more complex than applying fixed weights as in traditional beamforming methods. This process involves calculating the correlations between the signals received at each pair of array elements. As the number of elements increases, the size of this matrix grows quadratically, thus increasing the computational burden. To compute the weights that will minimize the variance of the noise and interference, the MV algorithm requires the inversion of the covariance matrix. The inversion of a matrix is considered a process that requires significant computational resources, especially as the size of the matrix grows with the number of transducer elements. The algorithm must dynamically adapt and recalculate the weights for each focal point in real-time as the transducer moves and steers its beam. This continuous adaptation requires the algorithm to perform the above computations for each new set of received signals, which is computationally demanding.
Luijten et al. [35,40] examine the applicability of deep learning to augment the adaptive beamforming process, addressing the computational challenges and aiming to produce better ultrasound images. They develop a neural network architecture, termed Adaptive Beamforming by deep LEarning (ABLE), which can adaptively calculate apodization weights for image reconstruction from received RF data. This method aims to improve ultrasound image quality by efficiently mimicking adaptive beamforming methods without the high computational burden. The ABLE network consists of fully connected layers and employs an encoder-decoder structure to create a compact representation of the data, aiding in noise suppression and signal representation. The training of ABLE is performed using a specialized loss function designed to promote similarity between the target and the produced images while also encouraging unity gain in the apodization weights. The study demonstrates ABLE’s effectiveness on two different ultrasound imaging modalities: plane wave imaging with a linear array and synthetic aperture imaging with a circular array. Moreover, ABLE’s computational efficiency, as assessed by the number of required floating-point operations, is significantly lower than that of Eigen-Based Minimum Variance (EBMV) beamforming, highlighting its potential for real-time imaging applications. In the training strategy, the network employs a total loss function composed of an image loss and an apodization-weight penalty. The image loss is designed to promote similarity between the target image and the one produced by ABLE, while the weight penalty encourages the network to learn weights that facilitate a distortionless response in the beamforming process. This penalty is inspired by MV beamforming principles, which aim to minimize output power while ensuring a distortionless response in the desired direction. By incorporating this constraint, the network is guided to learn apodization weights that not only aim to reconstruct high-quality ultrasound images but also adhere to a fundamental beamforming criterion, ensuring the network’s predictions align with the physical beamforming process. The comparison between MV and ABLE is shown in Figure 5.
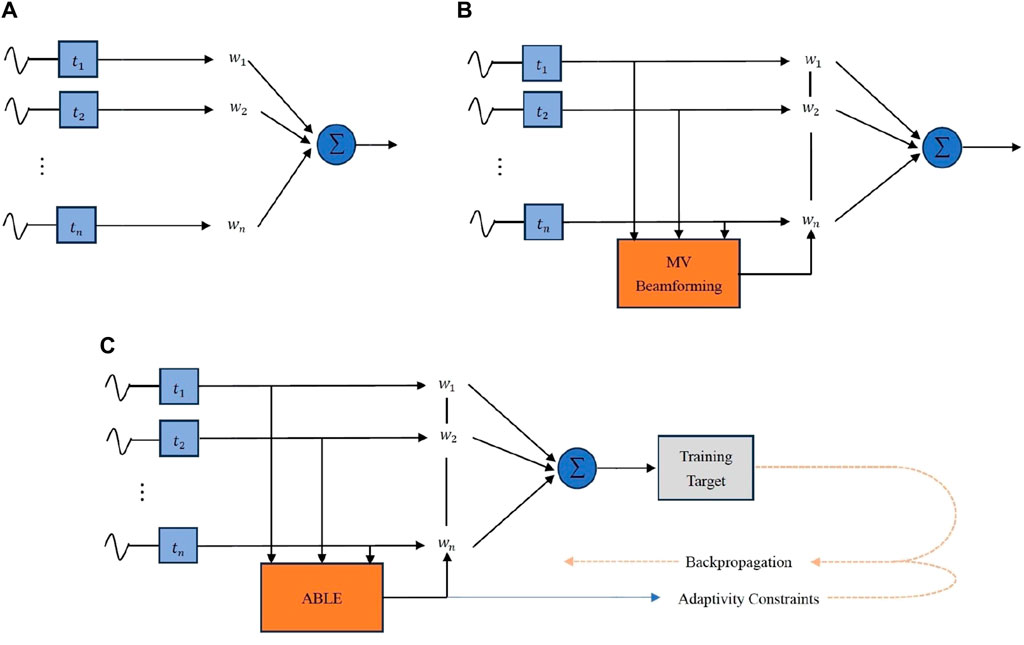
Figure 5. A comparison among (A)DAS, (B)MV and (C)ABLE [41]. The weights in DAS are typically pre-set fixed values, while the weights in MV and ABLE are adaptive. ABLE can be seen as an alternative form of MV. They both adaptively estimate weights through the received signals. The calculation of weights in MV requires a large amount of computation, while ABLE reduces the computational complexity.
3.1.3 Spatial coherence-based beamforming
Spatial coherence-based beamforming is a sophisticated method employed in ultrasound imaging that focuses on analyzing the spatial coherence of received echo signals to form diagnostic images. It improves image clarity by emphasizing echoes that show consistent phase or time delays across neighboring transducer elements, which indicates they are coming from a real reflector-like tissue structure, rather than random noise or scattering. By harnessing this spatial coherence, the beamformer can more effectively differentiate between signal and noise, leading to images with better resolution and contrast.
Typically, the DAS algorithm only utilizes one attribute, the signal strength, while spatial coherence reflects the similarity of signals [117]. Therefore, this is another property that can be used to enhance image quality. There are many studies based on spatial coherence, such as coherence factor (CF) [118], generalized coherence factor (GCF) [119], and phase coherence factor (PCF) [120]. Lediju et al. [44] have proposed a spatial coherence-based method named short-lag spatial coherence (SLSC). This method leverages the coherence of echoes that occur at short lags. The objective of this method is to overcome the limitations of traditional ultrasound imaging, caused by factors such as acoustic clutter, speckle noise, and phase aberration. SLSC images demonstrate improved visualization when compared to matched B-mode images by addressing these issues. By applying the SLSC imaging, the researchers aim to enhance ultrasound image quality and diagnostic accuracy, benefiting the field of medical imaging. The spatial coherence is calculated by Eq. 4 [44],
where xi is the aligned signal received by the ith element, si represents the sample index along the axial direction. In addition, N denotes the receive aperture, and m indicates the lag. From this equation, it can be seen that its computational complexity is relatively high. Wiacek et al. [35,42] have proposed a deep learning approach named CohereNet to estimate the normalized cross correlation as a function of lag. This network can be used to replace the SLSC beamforming. They delve into the potential of FCNNs as “universal approximators” that could learn any function. In CohereNet, a 7 × 64 input is adopted, which means the axial kernel chooses seven samples in the axial direction, while the aperture size is 64. The output is the spatial correlation at different lag distances. The network structure consists of an input layer, three fully connected layers using rectified linear unit (ReLU) as the activation function, followed by a fully connected layer using hyperbolic tangent (tanh) as the activation function, and an average pool output layer. In essence, CohereNet aims to utilize the capability of DNNs to enhance the beamforming process, thereby improving image quality and computational efficiency. As described in [43], the CohereNet is faster than SLSC, and this network also has high generality. The Figure 6 illustrates the comparison between DAS and CohereNet.
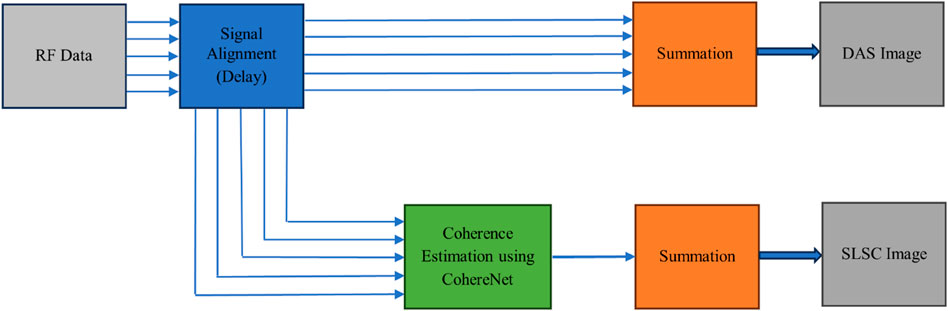
Figure 6. A comparison between DAS beamforming (top) and CohereNet [43] (bottom). The DAS algorithm obtains the final result by aligning the received signals and then weighting and summing them up. On the other hand, SLSC achieves the final result through calculating the spatial coherence. CohereNet reduces the computational complexity of SLSC.
3.2 Deep learning in clinical applications
Ultrasound imaging, due to its non-invasive characteristic and real-time imaging capabilities, has seen extensive use across various medical domains. This section delves into the clinical applications of deep learning, including breast imaging, cardiology, prostate imaging, fetal, thyroid, and brain.
3.2.1 Breast imaging
Breast ultrasound imaging is commonly utilized to detect potential breast diseases [121]. Although it falls short in identifying microcalcifications compared to X-ray mammography, it is instrumental in distinguishing benign masses like cysts and fibroadenomas from malignant ones. With the development of AI, especially the advent of deep learning technologies, it has also promoted the evolution of ultrasound breast imaging. Some open-source datasets, such as Breast Ultrasound Images Dataset (BUSI) [122], have also promoted the widespread application of deep learning. As depicted in Figure 7, the images within the BUSI dataset are classified into three distinct categories: normal, benign, and malignant.
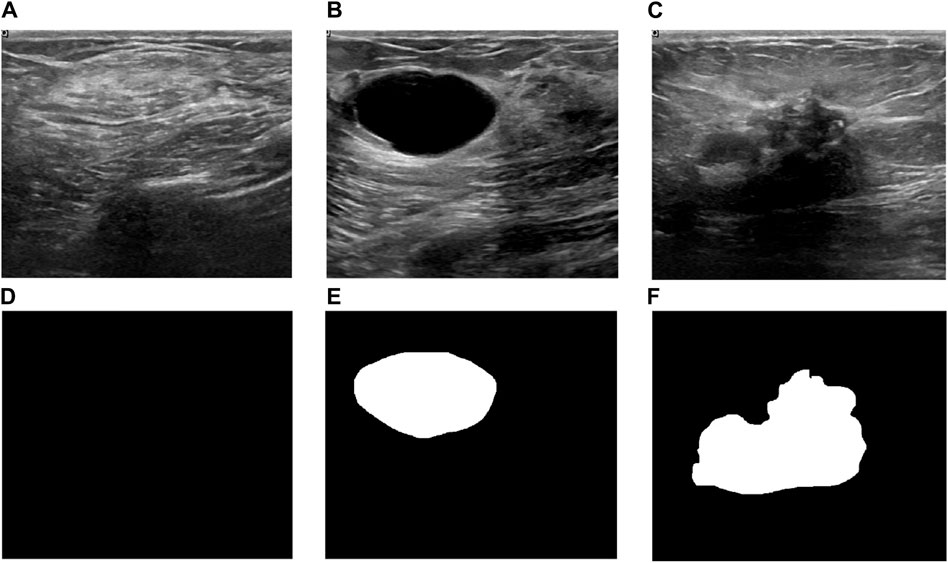
Figure 7. Three examples from BUSI dataset[122]. (A) A normal image and (D) its corresponding mask, (B) a benign image and (E) its corresponding mask, (C)a malignant image and (F) its corresponding mask.
In 2018, Becker et al.[65] reported using generic deep learning software (DLS) for the classification of breast cancer in ultrasound images. The study aimed to evaluate the effectiveness of a DLS in classifying breast cancer using ultrasound images and compare its performance against human readers with varying levels of breast imaging experience. They used Receiver Operating Characteristic (ROC) to assess the accuracy of diagnostic results. The DLS achieved diagnostic accuracy comparable to radiologists and performed better than a medical student with no prior experience. Although they did not discuss the technical details of deep learning, the study demonstrated that deep learning software could achieve high diagnostic accuracy in classifying breast cancer using ultrasound images, even with a limited number of training cases. The fast evaluation speed of the software supports the feasibility of real-time image analysis during ultrasound examinations. This indirectly illustrates the potential of deep learning in improving diagnostic processes. Xu et al. [66] develop a CNN based method for the automatic segmentation of breast ultrasound images into four major tissues: skin, fibroglandular tissue, mass, and fatty tissue, to aid in tumor localization, breast density measurement, and assessment of treatment response. They designed two CNN architectures CNN-I and CNN-II. CNN-I is an 8-layer CNN for pixel-centric patch classification. CNN-II is a smaller CNN to combine the outputs of three CNN-Is, each trained on orthogonal image planes, to provide comprehensive evaluation. CNNs were trained using the Adam optimization algorithm, and dropout methods were applied to prevent overfitting. The proposed method achieved high quantitative metrics for segmentation. Accuracy, Precision, Recall, and F1-measure all exceeded 80%. Jaccard similarity index (JSI) for mass segmentation reached 85.1%, outperforming previous methods. The proposed method provided better segmentation visualization and quantitative evaluation compared to previous studies. The automated segmentation method can offer objective references for radiologists, aiding in breast cancer diagnosis and breast density assessments. Qian et al. [67] have proposed a deep learning system to assess the breast cancer risk. The system was trained on a large dataset from two hospitals, encompassing 10,815 ultrasound images of 721 biopsy-confirmed lesions, and then prospectively tested on an additional 912 images of 152 lesions. The deep-learning system, when applied to bimodal (B-mode and color Doppler images) and multimodal (including elastography) images, achieved high accuracy in predicting BI-RADS scores. The system’s predictions align with radiologists’ assessments, demonstrating its potential utility in clinical settings. It could facilitate the adoption of ultrasound in breast cancer screening, particularly beneficial for women with dense breasts where mammography is less effective. This research underscores the potential of deep learning in enhancing breast ultrasound’s diagnostic power, offering a tool that aligns with current BI-RADS standards and supports radiologists in decision-making processes. Chen et al. [68] introduce a deep learning model for breast cancer diagnosis. They leverage the domain knowledge of radiologists, particularly their diagnostic patterns when viewing CEUS videos, to enhance the model’s diagnostic accuracy. The model integrates a 3D CNN with a domain-knowledge-guided temporal attention module (DKG-TAM) and a domain-knowledge-guided channel attention module (DKG-CAM). These modules are designed to mimic the attention patterns of radiologists, focusing on specific time slots in contrast-enhanced ultrasound (CEUS) videos and incorporating relevant features from both CEUS and traditional ultrasound images. The study utilizes a Breast-CEUS dataset comprising 221 cases, which includes CEUS videos and corresponding images, making it one of the largest datasets of its kind. Reference [69] addresses the challenge of breast cancer, the second leading cause of death among women worldwide. It highlights the importance of early detection through automated systems due to the time-consuming nature of manual diagnosis. The study introduces a new framework leveraging deep learning and feature fusion for classifying breast cancer using ultrasound images. The proposed framework comprises five main steps: data augmentation, model selection, feature extraction, feature optimization, feature fusion and classification. Operations like horizontal flip, vertical flip, and 90-degree rotation were applied to enhance the original dataset’s size and diversity. The pre-trained DarkNet-53 model was modified and trained using transfer learning techniques. Features were extracted from the global average pooling layer of the modified model. Two improved optimization algorithms, reformed differential evolution (RDE) and reformed gray wolf (RGW), were used to select the best features. The optimized features were fused using a probability-based approach and classified using machine learning algorithms. The study concludes that the proposed framework significantly improves the accuracy and efficiency of breast cancer classification from ultrasound images. It highlights the potential of the method to provide reliable support for radiologists, enhancing early detection and treatment planning. Rzaz et al. [70] present DeepBreastCancerNet, a new deep learning model designed for the detection and classification of breast cancer using ultrasound images. This model addresses the challenges of manual breast cancer detection, which is often time-consuming and prone to inaccuracies. The proposed DeepBreastCancerNet framework includes 24 layers, consisting of six convolutional layers, nine inception modules, and one fully connected layer. It employs both clipped ReLU and leaky ReLU activation functions, batch normalization, and cross-channel normalization to enhance model performance. Images were augmented through random translations and rotations to enhance the dataset’s size and diversity, thereby reducing overfitting. The architecture starts with a convolutional layer followed by max pooling, batch normalization, and leaky ReLU activation. Inception modules are used for extracting multi-scale features. The model ends with a global average pooling layer and a fully connected layer for classification. The proposed model achieved a classification accuracy of 99.35%, outperforming several state-of-the-art deep learning models. On a binary classification dataset, the model achieved an accuracy of 99.63%. The DeepBreastCancerNet model outperformed other pre-trained models like AlexNet, ResNet, and GoogLeNet in terms of accuracy, precision, recall, and F1-score. Ablation studies confirmed the importance of using both leaky ReLU and clipped ReLU activation functions and global average pooling for optimal performance.
3.2.2 Cardiology
In cardiology, echocardiography, particularly through ultrasound imaging of the heart, represents a pivotal area in medical ultrasound research, with abundant literature focusing on automated methods for segmenting and tracking the heart’s left ventricle - a crucial component evaluated in heart disease diagnosis. Echocardiography is a test that uses high-frequency sound waves to make pictures of your heart. It can show the size, shape, movement, pumping strength, valves, blood flow and other features of the heart. The quality of echocardiographic images can be influenced by multiple factors such as patient’s body habitus, lung disease, or surgical dressings, which can make interpretation difficult. Interpreting the results of an echocardiography exam requires significant expertise and experience. Sometimes not all views of the heart can be visualized adequately, which may limit the amount of information obtained from the test. Deep learning techniques can solve these problems to a certain extent.
Ref.[71] addressed the challenge of segmenting the left atrium (LA) in 3D ultrasound images using CNNs. The proposed method aims to automate this process, which is traditionally time-consuming and dependent on the observer. The introduction of shape priors and adversarial learning into the CNN framework enhances the accuracy and adaptability of the segmentation across different ultrasound devices. The framework integrates three existing methods: 3D Fully Convolutional Segmentation Network (V-Net), Anatomic Constraint via Autoencoder Network and Domain Adaptation with Adversarial Networks. The V-Net processes 3D image volumes and creates segmentation masks. Shape priors are incorporated through an autoencoder network trained on ground truth segmentation masks. This ensures that the segmentation masks adhere to anatomically plausible shapes. Domain adaptation is achieved by training a classifier to identify the data source, aiming to make the feature maps domain invariant. The combined approach of using shape priors and adversarial learning in CNNs significantly improves the segmentation of the left atrium in 3D ultrasound images. This method not only boosts accuracy but also ensures the generalizability of the model across different devices, making it a promising tool for clinical use.
2D echocardiographic image analysis is crucial in clinical settings for diagnosing cardiac morphology and function. Manual and semi-automatic annotations are still common due to the lack of accuracy and reproducibility of fully automatic methods. Challenges in segmentation arise from poor contrast, brightness inhomogeneities, speckle patterns, and anatomical variability. Leclerc et al.[72] evaluate the performance of state-of-the-art encoder-decoder deep CNNs for segmenting cardiac structures in 2D echocardiographic images and estimating clinical indices using the CAMUS dataset. CAMUS is the largest publicly available and fully annotated dataset for echocardiographic assessment, containing data from 500 patients. The CAMUS dataset enables comprehensive evaluation of deep learning methods for echocardiographic image analysis. Encoder-decoder networks, especially U-Net, demonstrate strong potential for accurate and reproducible cardiac segmentation, paving the way for fully automatic analysis in clinical practice. The study confirms that encoder-decoder networks, particularly U-Net, provide highly accurate segmentation results for 2D echocardiographic images. However, achieving inter-observer variability remains challenging, and more sophisticated architectures did not significantly outperform simpler U-Net designs. The findings suggest that further improvements in deep learning methods and larger annotated datasets are essential for advancing fully automatic cardiac image analysis.
Ghorbani et al. [73] have developed a deep learning model named EchoNet to interpret the echocardiograms. This model could identify local cardiac structures, estimate cardiac function, and predict systemic phenotypes like age, sex, weight, and height with significant accuracy. EchoNet is able to accurately predict various clinical parameters, such as ejection fraction and volumes, crucial for diagnosing and managing heart conditions. It also demonstrated the potential to predict systemic phenotypes that are not directly observable in echocardiogram images. By automating echocardiogram interpretation, such AI models could streamline clinical workflows, provide preliminary interpretations in regions lacking specialized cardiologists, and offer insights into phenotypes challenging for human evaluation. The research emphasized the potential of deep learning to enhance echocardiogram analysis, offering a step toward more automated, accurate, and comprehensive cardiovascular imaging diagnostics. Due to the lack of experience among novices, Narang et al. [74] proposed the use of deep learning techniques to assist them. The deep learning algorithm provides real-time guidance to novices, enabling them to capture essential cardiac views without prior experience in ultrasonography. The study involved eight nurses without prior echocardiography experience who used the AI guidance to perform echocardiographic scans on 240 patients. These scans were then compared with those obtained by experienced sonographers. The primary outcome was the ability of the AI-assisted novices to acquire echocardiographic images of sufficient quality to assess left and right ventricular size and function, as well as the presence of pericardial effusion. Results indicated that the novice-operated, AI-assisted echocardiograms were of diagnostic quality in a high percentage of cases, closely aligning with the quality of scans performed by experienced sonographers. The study suggests that AI-guided echocardiogram acquisition can potentially expand the accessibility of echocardiographic diagnostics to settings where expert sonographers are unavailable, thereby enhancing patient care in diverse clinical environments.
3.2.3 Thyroid
The thyroid gland, located in the neck and comprising two interconnected lobes, plays a critical role in hormone secretion, impacting protein synthesis, metabolic rate, and calcium homeostasis. These hormones are particularly influential in children’s growth and development. Despite its small size, the thyroid is susceptible to various disorders, such as hyperthyroidism, hypothyroidism, and nodule formation. Diagnosing these conditions involves a range of techniques, including blood tests for hormone levels, ultrasound imaging for gland volume and nodule detection, and fine-needle aspiration (FNA) for definitive tissue analysis. FNA, the most invasive of these methods, is being increasingly circumvented by leveraging ultrasound imaging with advanced deep learning and computer-aided diagnosis (CAD) systems to enhance diagnostic accuracy and nodule characterization.
Wang et al. [79] introduce a deep learning method for diagnosing thyroid nodules using multiple ultrasound images from an examination. The study proposes an architecture that includes three networks, addressing the challenge of using multiple views from an ultrasound examination for a comprehensive diagnosis. The research involves a dataset with 7803 images from 1046 examinations, employing various ultrasound equipment. The dataset is annotated at the examination level, categorizing examinations into malignant and benign based on ultrasound reports and pathological records. The method integrates features from multiple images using an attention-based feature aggregation network, aiming to reflect the diagnostic process of sonographers who consider multiple image views. The model demonstrated high diagnostic performance on the dataset, showcasing the potential of deep learning in enhancing the accuracy and objectivity of thyroid nodule diagnosis in ultrasound imaging. The attention-based network assigns weights to different images within an examination, focusing on those with significant features, which aligns with clinical practices where sonographers prioritize certain image views. Peng et al. [77] have developed a deep learning AI model called ThyNet. This model was designed to differentiate between malignant tumors and benign thyroid nodules, aiming to enhance radiologists’ diagnostic performance and reduce unnecessary FNAs. ThyNet was developed using 18,049 images from 8,339 patients across two hospitals and tested on 4,305 images from 2,775 patients across seven hospitals. The model’s performance was initially compared with 12 radiologists, and then a ThyNet-assisted diagnostic strategy was developed and tested in real-world clinical settings. The AI model, ThyNet, demonstrated superior diagnostic performance compared to individual radiologists, with an area under the receiver operating characteristic curve (AUROC) of 0.922, significantly higher than the radiologists’ AUROC of 0.839. When radiologists were assisted by ThyNet, their diagnostic performance improved significantly. The pooled AUROC increased from 0.837 to 0.875 with ThyNet assistance for image reviews and from 0.862 to 0.873 in a clinical setting involving image and video reviews. The ThyNet-assisted strategy significantly decreased the percentage of unnecessary FNAs from 61.9% to 35.2%, while also reducing the rate of missed malignancies from 18.9% to 17.0%.
3.2.4 Prostate
Prostate cancer ranks as the most frequently diagnosed malignancy among adult and elderly men, with early detection and intervention being crucial for reducing mortality rates. Transrectal ultrasound (TRUS) imaging, in conjunction with prostate-specific antigen (PSA) testing and digital rectal examination (DRE), plays a pivotal role in the diagnosis of prostate cancer. The delineation of prostate volumes and boundaries is critical for the accurate diagnosis, treatment, and follow-up of this cancer [123]. Typically, the delineation process involves outlining prostate boundaries on transverse parallel 2-D slices along its length, leading to the development of various (semi-)automatic methods for detecting these boundaries. In the diagnosis of prostate diseases, deep learning techniques provide some additional insights.
Azizi et al. [83] present a deep learning approach using Recurrent Neural Networks (RNNs), particularly Long Short-Term Memory (LSTM) networks, for prostate cancer detection through Temporal Enhanced Ultrasound (TeUS). The study aimed to leverage the temporal information inherent in TeUS to distinguish between malignant and benign tissue in the prostate. The authors utilized RNNs to model the temporal variations in ultrasound backscatter signals, demonstrating that LSTM networks outperformed other models in identifying cancerous tissues. The study analyzed data from 255 prostate biopsy cores from 157 patients. LSTM networks achieved an area under the curve (AUC) of 0.96, with sensitivity, specificity, and accuracy rates of 0.76, 0.98, and 0.93, respectively, highlighting the potential of RNNs in medical imaging analysis. The study also introduced algorithms for analyzing LSTM networks to understand the temporal features relevant for prostate cancer detection. This analysis revealed that significant discriminative features could be captured within the first half of the TeUS sequence, suggesting a potential reduction in data acquisition time for clinical applications. The research suggests that deep learning models, particularly LSTM-based RNNs, can significantly enhance prostate cancer detection using ultrasound imaging, offering a promising tool for improving diagnostic accuracy and potentially guiding biopsy procedures. Karimi et al. [89] introduces a method for the automatic segmentation of the prostate clinical target volume (CTV) in TRUS images, which is crucial for brachytherapy treatment planning. The method employs CNNs, specifically an ensemble of CNNs, to improve segmentation accuracy, particularly for challenging images with weak landmarks or strong artifacts. The method uses adaptive sampling to focus the training process on difficult-to-segment images and an ensemble of CNNs to estimate segmentation uncertainty, improving robustness and accuracy. For segmentations with high uncertainty, a statistical shape model (SSM) is used to refine the segmentation, utilizing prior knowledge about the expected shape of the prostate. The method achieved a Dice score of 93.9% ± 3.5% and a Hausdorff distance of 2.7 ± 2.3 mm, outperforming several other methods and demonstrating its effectiveness in reducing the likelihood of large segmentation errors. This study highlights the potential of deep learning and ensemble methods to enhance the accuracy and reliability of medical image segmentation, particularly in applications like prostate cancer treatment where precision is crucial.
3.2.5 Fetal
Ultrasonography is a pivotal technology in prenatal diagnosis, renowned for its safety for both the mother and fetus. This research area encompasses numerous subfields, often employing segmentation and classification techniques akin to those used in adult diagnostics but adapted for the smaller scale of fetal organs. This miniaturization introduces diagnostic challenges due to less pronounced signs of abnormalities. Furthermore, ultrasound imaging must penetrate maternal tissue and the placenta to reach the fetus, potentially introducing noise, exacerbated by the movements of both mother and fetus, emphasizing the need for enhanced automated diagnostic methods. Especially in underdeveloped areas with a shortage of medical personnel, such automatic diagnostic methods can provide tremendous help.
Van den Heuvel et al. [94] present a study where a system is developed to estimate the fetal head circumference (HC) from ultrasound data obtained using an obstetric sweep protocol (OSP). This protocol can be taught within a day to any healthcare worker without prior knowledge of ultrasound. The study aims to make ultrasound imaging more accessible in developing countries by eliminating the need for a trained sonographer to acquire and interpret images. The system uses two FCNNs. The first network detects frames containing the fetal head from the OSP data, and the second network measures the HC from these frames. The HC measurements are then used to estimate gestational age (GA) using the curve of Hadlock. The study, conducted on data from 183 pregnant women in Ethiopia, found that the system could automatically estimate GA with a reasonable level of accuracy, indicating its potential application in maternal care in resource-constrained settings. Pu et al. [96] developed an automatic fetal ultrasound standard plane recognition (FUSPR) model. This model is designed to operate in an Industrial Internet of Things (IIoT) environment and leverages deep learning to identify standard planes in fetal ultrasound imagery. The research introduces a distributed platform for processing ultrasound data using IIoT and high-performance computing (HPC) technology. The FUSPR model integrates a CNN and an RNN to learn spatial and temporal features of ultrasound video streams, aiming to improve the accuracy and robustness of fetal plane recognition. The system’s goal is to aid in gestational age assessment and fetal weight estimation by accurately identifying and analyzing key anatomical structures in ultrasound video frames. The study demonstrates that the FUSPR model significantly outperforms baseline models in recognizing four standard fetal planes from over 1000 ultrasound videos. The use of deep learning within the IIoT framework presents a promising approach to enhancing the efficiency and reliability of fetal ultrasound analysis, particularly in resource-constrained environments. A study by Xu et al.[124] introduced a novel segmentation framework incorporating vector self-attention layers (VSAL) and context aggregation loss (CAL) to address the challenges of fetal ultrasound image segmentation. The VSAL module allows for simultaneous spatial and channel attention, capturing both global and local contextual information. The CAL component further enhances the model’s ability to differentiate between similar-looking structures by considering both inter-class and intra-class dependencies. On the multi-target Fetal Apical Four-chamber dataset and one-target Fetal Head dataset, the proposed framework outperformed several state-of-the-art CNN-based, U-net[125], methods in terms of pixel accuracy (PA), dice coefficient (DCS), Hausdorff distance (HD) metrics, demonstrating its potential for improving fetal ultrasound image segmentation accuracy. The study showcases the effectiveness of self-attention techniques in enhancing the accuracy and reliability of fetal ultrasound image segmentation, offering a promising tool for improving prenatal diagnostics and care.
3.2.6 Brain
The brain, a pivotal organ in the nervous system, epitomizes complexity within the human body, orchestrating the functions of voluntary organs and muscles. Despite its critical role, the full extent of its operations remains partially elusive, prompting ongoing research to decipher its mechanisms. Notably, the brain undergoes a phenomenon termed “brain shift,” a potential deformation during surgical procedures that could impact surgical outcomes. Ultrasound technology, particularly when integrated with magnetic resonance (MR) imaging data, serves as a crucial aid in neurosurgical contexts. This integration is instrumental in addressing the challenges posed by brain shift and enhancing intraoperative navigation and decision-making.
Milletari et al.[98] discuss a deep learning approach using CNNs combined with a Hough voting strategy for segmenting deep brain regions in MRI and ultrasound images. The study showcases the use of this method for fully automatic localization and segmentation of anatomical regions of interest, utilizing the features produced by CNNs for robust, multi-region, and modality-flexible segmentation. The method is particularly designed to adapt to different imaging modalities, showing effectiveness in MRI and transcranial ultrasound volumes. It demonstrates the potential of CNNs in medical image analysis, particularly in the challenging context of brain imaging, where accurate segmentation of anatomical structures is critical. The study systematically explores the performance of various CNN architectures across different scenarios, offering insights into the effective application of deep learning techniques in medical imaging. Reference [99] presents a method for segmenting brain tumors during surgery using 3D intraoperative ultrasound (iUS) images. The technique employs a tumor model derived from preoperative magnetic resonance (MR) data for local MR-iUS registration, aiming to enhance the visualization of brain tumor contours in iUS. This multi-step process defines a region of interest based on the patient-specific tumor model, extracts hyperechogenic structures from this region in both modalities, and performs registration using gradient values and rigid and affine transformations to align the tumor model with the 3D-iUS data. The method’s effectiveness was assessed on a dataset of 33 patients, showing promising results in terms of computational time and accuracy, indicating its potential utility in supporting neurosurgeons during brain tumor resections. Di Ianni and Airan [102] introduce a deep learning-based image reconstruction method for functional ultrasound (fUS) imaging of the brain. The method significantly reduces the amount of data required for imaging while maintaining image quality, using a CNN to reconstruct power Doppler images from sparsely sampled ultrasound data. The approach enables high-quality fUS imaging of brain activity in rodents, with potential applications in various settings where dedicated ultrasound hardware is not available, thereby broadening the accessibility and utility of fUS imaging technology.
3.3 Deep learning in portable ultrasound system
Due to its portability and low cost, handheld ultrasound devices have great application prospects in areas such as emergencies, point-of-care, sports fields, and outdoors. At the same time, it is also suitable for assisting doctors in diagnosing diseases in remote and medically undeveloped areas. Portable ultrasound diagnostic devices appeared in the 1980s. Initially, they were mainly used to scan the bladder to measure the volume [126–129]. Compared to the common invasive method of catheterization through a urinary catheter, the bladder scanner does not cause any harm to the patient. Until now, the development of bladder scanners has been a direction in the advancement of portable ultrasound devices [130–132]. However, besides this field, portable ultrasound devices have many other applications, such as Color Doppler imaging [133], blood flow imaging [134], echocardiography [135], skin imaging [136] and so on. During the outbreak of COVID-19, portable ultrasound devices also played a positive role in assisting diagnosis [137–139].
Indeed, the compact size of portable ultrasound devices does present significant challenges for both hardware design and the development of imaging algorithms [140,141]. Despite its portable advantages, these challenges need to be meticulously addressed to ensure the efficient performance and accuracy of the device. With the advancement of semiconductor technology, technologies such as Field Programmable Gate Arrays (FPGAs) [142] and Application Specific Integrated Circuits (ASICs) [143] have been successively applied to portable ultrasound devices to overcome some of the challenges in hardware design. From the perspective of algorithm design, beamforming technology based on compressed sensing [144] has extensive research in portable ultrasound [145–148].
These methods have promoted the development of portable ultrasound devices, and with the advancement of artificial intelligence technology, the corresponding technologies have also brought new development directions for portable ultrasound devices. Zhou et al. [149] proposed to apply Generative Adversarial Network (GAN) to enhance the image quality of handheld ultrasound devices. They introduce a novel approach using a two-stage GAN to enhance image quality. The proposed two-stage GAN framework incorporates a U-Net network and a GAN to reconstruct high-quality ultrasound images from low-quality ones. The method focuses on reconstructing tissue structure details and speckles of the ultrasound images, essential for accurate diagnostics. The paper presents a comprehensive loss function combining texture, structure, and perceptual features to guide the GAN training effectively. The simulated, phantom and clinical data are used to demonstrate the method’s efficacy, showing significant improvements in image quality compared to original low-quality images and other algorithms. In addition, Soleimani et al. [103] developed a lightweight and portable ultrasound computed tomography (USCT) system for noninvasive imaging of the human head with high resolution. The study aims to compare the effectiveness of a deep neural network combining CNN and long short-term memory (LSTM) layers against traditional deterministic methods in creating tomographic images of the human head. The research shows that the proposed neural network is more effective in dealing with noisy and synthetic data compared to deterministic methods, which often require additional filtering to improve image quality. The findings suggest that the CNN + LSTM model is more versatile and generalizable, making it a superior choice for medical ultrasound tomography applications. The study contributes to the advancement of USCT by demonstrating the potential of deep learning approaches in improving the accuracy and reliability of noninvasive brain imaging techniques.
3.4 Training scheme
Vienneau et al. [57] discuss the training methods for DNNs in the context of ultrasound imaging. They address the issues with traditional ℓp norm loss functions when training DNNs, where lower loss values do not necessarily translate to improved image quality. Ref. [57] presents an effort to better align the optimization objective with the relevant image quality metrics. The authors suggest that their novel training scheme can potentially increase the maximum achievable image quality for ultrasound beamforming using DNNs. Luchies and Byram [58] investigate practical considerations of training DNN beamformers for ultrasound imaging. They discuss the use of combinations of multiple point target responses for training DNNs, as opposed to single point target responses. It also examines the impact of various hyperparameter settings on the quality of ultrasound images in simulated scans. The study demonstrates that DNN beamforming exhibits robustness when confronted with electronic noise, and it points out that mean squared error (MSE) validation loss is not a reliable predictor for image quality. Goudarzi and Rivaz [52] used real photographic images as the ground-truth echogenicity map in their simulations to provide the network with a diverse range of textures, contrasts, and object geometries during the training phase. This approach not only enhances the variety in the training dataset, which is crucial for preventing overfitting but also aligns the simulation settings more closely with the real experimental imaging settings of in vivo test data, thus minimizing unwanted domain shifts between training and test datasets.
3.5 Transformer/attention mechanism and CNN
CNNs have been the backbone of medical image analysis for years. It can be seen from our previous review that a larger number of architectures are based on the CNNs.They excel in extracting local features through convolutional layers, pooling, and activation functions. Networks such as U-Net[125] and its variants [150–153] have been particularly successful in medical image segmentation tasks due to their encoder-decoder architectures, which capture detailed spatial hierarchies. However, CNNs face limitations in modeling global context and long-range dependencies. This shortfall can lead to suboptimal performance in tasks where the relationship between distant regions in the image is crucial. In ultrasound imaging, this limitation manifests in difficulties handling speckle noise and artifacts, which require broader contextual understanding to be effectively mitigated. On the other hand, the advent of Transformer models and their self-attention mechanisms [154] has introduced new opportunities for enhancing ultrasound image analysis. The integration of U-net with transformer has also become a new direction for current research [155–159]. This section delves into the application of Transformers and attention mechanisms in medical imaging focusing on ultrasound, comparing their performance with traditional CNNs.
Transformers, originally designed for natural language processing, utilize a self-attention mechanism that allows the model to weigh the importance of different input elements dynamically [154]. This capability is particularly beneficial for medical image analysis [124,156,160–167], where different regions of an image may hold varying levels of significance for accurate diagnosis. The self-attention mechanism operates by creating attention scores between all pairs of input elements, which in the context of images, correspond to pixels or features. These scores determine how much attention each element should receive from the others. This global consideration enables the Transformer to capture long-range dependencies and contextual information that CNNs might miss due to their localized receptive fields [154]. In the realm of medical imaging, the Transformer models have been adapted to handle the unique challenges posed by this modality. For instance, TransUNet [156] architecture integrates CNNs and Transformers into a unified framework, where CNNs are employed to extract initial feature maps from medical images, and Transformers encode these features into tokenized patches to capture global context. This hybrid approach enables the model to retain detailed spatial information while benefiting from the global attention provided by Transformers; the GPA-TUNet[162] model integrates Group Parallel Axial Attention (GPA) with Transformers to enhance both local and global feature extraction. This hybrid approach leverages the strengths of Transformers in capturing long-range dependencies and the efficiency of GPA in highlighting local information. Another segmentation method specific to ultrasound images is the integration of a Vector Self-Attention Layer (VSAL) [124], which performs long-range spatial and channel-wise reasoning simultaneously. VSAL is designed to maintain translational equivariance and accommodate multi-scale inputs, which are critical for handling the variability in ultrasound images. This layer can be seamlessly integrated into existing CNN architectures, enhancing their performance by adding the benefits of self-attention. Studies [124] have shown that Transformer-based models significantly improve the accuracy of ultrasound image segmentation tasks. For example, in the segmentation of fetal ultrasound images, models incorporating VSAL and context aggregation loss (CAL) demonstrated superior performance compared to traditional CNNs.
The adaptive multimodal attention mechanism [160] is another advanced approach used in deep learning models to improve the generation of descriptive and coherent medical image reports. Yang et al. propose a novel framework for generating high-quality medical reports from ultrasound images using an adaptive multimodal attention network (AMAnet). This framework addresses the challenges of tedious and time-consuming manual report writing by leveraging deep learning techniques to automate the process. The core innovation of AMAnet lies in its adaptive multimodal attention mechanism, which integrates three key components: spatial attention, semantic attention, and a sentinel gate. The spatial attention mechanism focuses on the relevant regions of the ultrasound images, ensuring that the model captures essential visual details. Meanwhile, the semantic attention mechanism predicts crucial local properties, such as boundary conditions and tumor morphology, by using a multi-label classification network. These predicted properties are then used as semantic features to enhance the report generation process. The sentinel gate is a pivotal element in the AMAnet framework, designed to dynamically control the attention level on visual features and language model memories. This gate allows the model to decide whether to focus on current visual features or rely on the learned knowledge stored in the Long Short-Term Memory (LSTM) when generating the next word in the report. This adaptive mechanism is particularly beneficial in handling fixed phrases commonly found in medical reports, ensuring that the model can generate coherent and contextually appropriate text. The incorporation of semantic features and the adaptive attention mechanism contribute to the model’s superior performance, highlighting its potential for practical clinical applications. In practical terms, consider a scenario where the model is generating a report for an ultrasound image showing a tumor. The spatial attention mechanism might focus on the region where the tumor is located. The semantic attention mechanism will consider properties such as “irregular morphology” and “unclear boundary” predicted by the multi-label classification network. The sentinel gate will dynamically balance between these features and the language model’s internal memory to generate a sentence like “The ultrasound image shows an irregularly shaped tumor with unclear boundaries.” This adaptive attention mechanism ensures that the model generates accurate and contextually appropriate reports, enhancing its utility in clinical settings, which CNNs alone might struggle to achieve.
Chi et al. [168] propose a unified framework that combines the 2D and 3D Transformer-UNets into a single end-to-end network. This novel method enhances the segmentation of thyroid glands in ultrasound sequences, addressing several key limitations of existing deep learning models. The proposed Hybrid Transformer UNet (H-TUNet) integrates both intra-frame and inter-frame features through a combination of 2D and 3D Transformer UNets, significantly improving segmentation accuracy and efficiency. The framework is designed to exploit both the detailed intra-frame features and the broader inter-frame contextual information, resulting in a more accurate and robust segmentation of the thyroid gland in ultrasound images. The proposed method outperforms state-of-the-art CNN-based models, such as 3D UNet, in terms of segmentation accuracy, demonstrating the effectiveness of hybrid Transformer-2D-3D models in ultrasound image analysis. Wang et al.[169] presents a groundbreaking method for enhancing the safety and efficiency of robot-assisted prostate biopsy through advanced force sensing techniques. This method addresses the limitations of existing VFS techniques, particularly in accurately sensing the interaction force between surgical tools and prostate tissue. The core innovation of TransVFS is the spatio-temporal local–global transformer architecture. This model captures both local image details and global dependencies simultaneously, which is crucial for accurately estimating prostate deformations and the resulting forces during biopsy. The architecture includes efficient local–global attention modules that reduce the computational burden associated with processing 4D spatio-temporal data. This makes the method suitable for real-time force-sensing applications in clinical settings. The proposed method was extensively validated through experiments on prostate phantoms and beagle dogs. The results demonstrated that TransVFS outperforms state-of-the-art VFS methods and other spatio-temporal transformer models in terms of force estimation accuracy. Specifically, TransVFS provided significantly lower mean absolute errors in force estimation compared to the most competitive model, ResNet3dGRU. The paper highlights the practical benefits of TransVFS in improving the safety and efficacy of robot-assisted prostate biopsies. By providing accurate real-time force feedback, TransVFS can help reduce the risk of tissue damage and improve the precision of biopsy procedures, thereby enhancing patient outcomes. Ahmadi et al.[170] integrate a spatio-temporal architecture that combines anatomical features and the motion of the aortic valve to accurately classify AS severity. The Temporal Deformable Attention (TDA) mechanism is specifically designed to capture small local motions and spatial changes across frames, which are critical for assessing AS severity. The model incorporates a temporal coherent loss function to enforce sensitivity to small motions in spatially similar frames without explicit aortic valve localization labels. This loss helps the model maintain consistency in frame-level embeddings, enhancing its ability to detect subtle changes in the aortic valve’s movement. An innovative attention layer is introduced to aggregate disease severity likelihoods over a sequence of echocardiographic frames, focusing on the most clinically informative frames. This temporal localization mechanism enables the model to identify and prioritize frames that are critical for accurate AS diagnosis. The model was tested on both private and public datasets, demonstrating state-of-the-art accuracy in AS detection and severity classification. On the private dataset, the model achieved 95.2% accuracy in AS detection and 78.1% in severity classification. On the public TMED-2 dataset, the model achieved 91.5% accuracy in AS detection and 83.8% in severity classification. By reducing the reliance on Doppler measurements and enabling automated AS severity assessment from two-dimensional echocardiographic data, the proposed framework facilitates broader access to AS screening. This is particularly valuable in clinical settings with limited access to expert cardiologists and specialized Doppler imaging equipment.
Transformers address the limitations of CNNs by incorporating self-attention mechanisms that consider the entire input sequence (or image) simultaneously. This allows for a more comprehensive understanding of the image, capturing both local and global features effectively. Transformers can capture long-range dependencies and relationships across the entire image, which is essential for accurately interpreting ultrasound images that may contain complex structures and subtle differences. By dynamically adjusting the attention weights, Transformers can focus on the most relevant parts of the image, enhancing feature extraction and reducing the impact of irrelevant or noisy regions. Recent methods further develop the advantages via Integration with CNNs: Hybrid models, such as GPA-TUNet[162], combine the strengths of CNNs and Transformers, using CNN layers for initial feature extraction and Transformers for global context modeling. This integration leads to superior performance in segmentation tasks, particularly for images with large axial spans. Adaptive Attention Mechanisms: Models like AMAnet[160] incorporate adaptive attention mechanisms that dynamically control the focus on visual features and language model memories. This enables the model to generate coherent and contextually appropriate reports, enhancing its utility in clinical settings. Efficient Spatio-Temporal Processing: Methods like TransVFS[169,170] introduce factorized spatio-temporal processing strategies that significantly reduce computational complexity, making them suitable for real-time force-sensing applications in clinical settings. These advanced techniques demonstrate the potential of Transformers and attention mechanisms in enhancing the accuracy and reliability of medical ultrasound image analysis, offering a promising solution for improving diagnostic outcomes and patient care.
The integration of Transformer models and attention mechanisms into ultrasound image analysis represents a significant advancement over traditional CNN-based approaches. The ability of Transformers to capture long-range dependencies and model global context enhances the accuracy and reliability of medical image segmentation tasks. As research continues, these models [160,162,169,170] are likely to play an increasingly vital role in improving diagnostic accuracy and patient outcomes in medical imaging.
4 Discussion
With the rapid development of deep learning, its range of applications has also expanded into more fields. In this paper, we summarize the applications of deep learning in medical ultrasound imaging, focusing on its promoting effect on beamforming algorithms and clinical applications. We compared the classic beamforming algorithm and its corresponding deep learning alternatives. For both adaptive beamforming and SLSC beamforming algorithms, the use of deep learning can reduce computational complexity and enhance efficiency. Deep learning can enhance beamforming algorithms in medical ultrasound imaging in several ways. Data-Driven Optimization: Deep learning models can be trained on large datasets of ultrasound images to learn optimal beamforming parameters for different imaging conditions. This can result in better image quality compared to traditional beamforming techniques that use preset parameters. Feature Extraction: Neural networks, especially CNNs, are highly efficient at automatically extracting relevant features from ultrasound data. These features can then be employed to improve the spatial and contrast resolution of the images. Reducing Artifacts: Deep learning can help identify and reduce artifacts in ultrasound images, such as speckle noise, which can interfere with the clarity of the images and the diagnosis. Speeding Up Processing Time: Deep learning can significantly reduce the computational time required for beamforming, making real-time imaging more feasible and efficient. Advanced Reconstruction Techniques: Through the use of deep learning, more advanced beamforming algorithms, such as synthetic aperture and plane wave imaging, can be optimized for better resolution and frame rates. In summary, deep learning can play a crucial role in the advancement of beamforming algorithms by enhancing image quality, reducing noise, and improving the overall efficiency of medical ultrasound imaging.
In the section on “clinical applications”, we reviewed the application of deep learning in some clinical scenarios. The specific applications of deep learning in medical image analysis include the following aspects. Image registration and orientation: Deep learning can align the spatial orientation and adjust the pixel intensity of multiple images from different sources, times, directions, or modalities to increase the effective sample size and reduce non-biological differences. Tissue segmentation: Deep learning technology can achieve precise segmentation of target structures in medical images, which helps to improve the speed and accuracy of medical image analysis. Disease prediction and diagnosis: Deep learning can assist doctors in diagnosing various diseases, including tumors, inflammations, injuries, etc. For example, it has been successfully used in the diagnosis of many diseases such as lung cancer and breast cancer. Medical image feature learning: Intelligent calculations of medical imaging based on deep learning can automatically learn excellent feature expressions from large sample data.
Deep learning, as an advanced machine learning technique, has significant potential in improving the performance of beamforming algorithms in medical ultrasound. Deep learning may have a positive impact on beamforming algorithms in medical ultrasound in the future. Deep learning can improve the quality of ultrasound images by denoising, enhancing edges and contrast, and reconstructing details more finely. Accelerating the beamforming computational process through deep learning models could significantly reduce the time required to acquire high-quality ultrasound images. Deep learning models can optimize beamforming algorithms based on different patient characteristics and scanning conditions to achieve more personalized imaging. Deep learning models can increase the dynamic range of images, making it possible to display both high and low signal areas in the same image, and enhance resolution. It can also identify and reduce artifacts in ultrasound imaging, such as sidelobe contamination and Doppler artifacts. Beamforming algorithms integrated with deep learning can assist in real-time detection of lesions and measurement of biomarkers, providing more diagnostic information. Deep learning can be used for rapid reconstruction of three-dimensional and four-dimensional data, providing clinicians with a more comprehensive view. Deep learning can help perform more accurate tissue quantitative analysis, such as the measurement of tissue stiffness, which is particularly important for certain diagnoses. By continuously learning from clinical data, deep learning models can improve their performance over time, enhancing the accuracy and reliability of beamforming technology. Deep learning can be used to automatically determine the optimal beamforming parameters, simplifying clinical operations and reducing the workload of physicians.
The integration of AI in portable ultrasound devices with remote servers is also helpful. AI algorithms can analyze ultrasound images in real-time, helping to identify patterns, anomalies, or specific conditions. This can assist healthcare professionals in making more accurate and faster diagnoses. AI can enable remote monitoring of patients, analyzing ultrasound data transmitted to the remote server and alerting healthcare professionals to any concerning changes or findings that require immediate attention. AI can generate preliminary reports based on the ultrasound data, highlighting key findings and suggesting possible diagnoses. This can expedite the review process by healthcare professionals. AI also can help in organizing and managing vast amounts of ultrasound data, making it easier for healthcare professionals to access and retrieve patient information when needed.
Meanwhile, based on the Segment Anything (SA) project [171], Kirillov et al. developed a new segmentation model (SAM). The SAM demonstrates impressive zero-shot performance across various tasks, often matching or exceeding fully supervised methods. This indicates the model’s generalizability and potential applicability to a wide range of segmentation challenges. Numerous studies have adopted the SAM in the medical image segmentation [172,173]. With the continuous development of large language models (LLMs), AI technology based on these models can also be applied to medicine [174–178]. Based on these studies, it can be seen that LLMs can play a significant role in medical ultrasound imaging. It can be used to generate preliminary reports of ultrasound imaging by analyzing the textual descriptions provided by the sonographer or the data obtained from the ultrasound device. The reports will not only save time but also reduce the workload of radiologists. LLMs can generate descriptive annotations for the images based on the features identified through image processing techniques. By working through complex medical language and jargon, LLMs can translate these into more patient-friendly language. This helps patients better understand their medical condition and the significance of their ultrasound results. LLMs can be utilized in creating interactive training material for medical students and professionals. This can assist them in learning the nomenclature, understanding complex medical conditions, and being updated with the latest medical research associated with ultrasound imaging. LLMs can assist in data collection, research conduction, and generating insights from large bodies of medical texts or research papers, offering valuable contributions to the field of medical ultrasound imaging. LLMs can also be integrated with AI and machine learning algorithms aimed at identifying and diagnosing diseases from ultrasound imagery. The LLM can then provide detailed explanations or feedback based on the AI’s findings in a way that is understandable for the healthcare provider.
5 Conclusion
In conclusion, the future application of deep learning in medical ultrasound imaging is multifaceted. It can not only enhance image quality and diagnostic efficiency but also promote the development of personalized medicine and precision medicine. With the increasing availability of computational resources and the continuous improvement of algorithms, we can expect deep learning to play an increasingly important role in ultrasound imaging technology.
Author contributions
KS: Funding acquisition, Supervision, Writing–original draft, Writing–review and editing. JF: Conceptualization, Writing–original draft, Writing–review and editing. DC: Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation Project of CQ CSTC (Grant No. cstc2021jcyj-msxmX0305), by the Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJQN202102804), by the Research Program of Chongqing University of Education (Grant No. KY201925C), by the Chongqing Electronics Engineering Technology Research Center for Interactive Learning, and by the Chongqing Big Data Engineering Laboratory for Children.
Acknowledgments
We would like to thank the editors and the reviewers for their helpful remarks.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Beutel J. Handbook of medical imaging, 3. Bellingham, Washington, United States: Spie Press (2000).
2. Shen D, Wu G, Suk H-I. Deep learning in medical image analysis. Annu Rev Biomed Eng (2017) 19:221–48. doi:10.1146/annurev-bioeng-071516-044442
3. Cheplygina V, de Bruijne M, Pluim JP. Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med image Anal (2019) 54:280–96. doi:10.1016/j.media.2019.03.009
4. Wang G, Ye JC, De Man B. Deep learning for tomographic image reconstruction. Nat machine intelligence (2020) 2:737–48. doi:10.1038/s42256-020-00273-z
5. Ben Yedder H, Cardoen B, Hamarneh G. Deep learning for biomedical image reconstruction: a survey. Artif intelligence Rev (2021) 54:215–51. doi:10.1007/s10462-020-09861-2
6. Zhou SK, Greenspan H, Davatzikos C, Duncan JS, Van Ginneken B, Madabhushi A, et al. A review of deep learning in medical imaging: imaging traits, technology trends, case studies with progress highlights, and future promises. Proc IEEE (2021) 109:820–38. doi:10.1109/jproc.2021.3054390
7. Koetzier LR, Mastrodicasa D, Szczykutowicz TP, van der Werf NR, Wang AS, Sandfort V, et al. Deep learning image reconstruction for ct: technical principles and clinical prospects. Radiology (2023) 306:e221257. doi:10.1148/radiol.221257
8. Kiryu S, Akai H, Yasaka K, Tajima T, Kunimatsu A, Yoshioka N, et al. Clinical impact of deep learning reconstruction in mri. Radiographics (2023) 43:e220133. doi:10.1148/rg.220133
9. Liu X, Faes L, Kale AU, Wagner SK, Fu DJ, Bruynseels A, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. The lancet digital health (2019) 1:e271–97. doi:10.1016/s2589-7500(19)30123-2
10. Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, et al. Deep learning: a primer for radiologists. Radiographics (2017) 37:2113–31. doi:10.1148/rg.2017170077
11. Bizopoulos P, Koutsouris D. Deep learning in cardiology. IEEE Rev Biomed Eng (2018) 12:168–93. doi:10.1109/rbme.2018.2885714
12. Buda M, Wildman-Tobriner B, Hoang JK, Thayer D, Tessler FN, Middleton WD, et al. Management of thyroid nodules seen on us images: deep learning may match performance of radiologists. Radiology (2019) 292:695–701. doi:10.1148/radiol.2019181343
13. Murtaza G, Shuib L, Abdul Wahab AW, Mujtaba G, Mujtaba G, Nweke HF, et al. Deep learning-based breast cancer classification through medical imaging modalities: state of the art and research challenges. Artif Intelligence Rev (2020) 53:1655–720. doi:10.1007/s10462-019-09716-5
14. Van der Laak J, Litjens G, Ciompi F. Deep learning in histopathology: the path to the clinic. Nat Med (2021) 27:775–84. doi:10.1038/s41591-021-01343-4
15. Gul S, Khan MS, Bibi A, Khandakar A, Ayari MA, Chowdhury ME. Deep learning techniques for liver and liver tumor segmentation: a review. Comput Biol Med (2022) 147:105620. doi:10.1016/j.compbiomed.2022.105620
16. Zhu Z, Sun M, Qi G, Li Y, Gao X, Liu Y. Sparse dynamic volume transunet with multi-level edge fusion for brain tumor segmentation. Comput Biol Med (2024) 172:108284. doi:10.1016/j.compbiomed.2024.108284
17. Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med (2019) 25:44–56. doi:10.1038/s41591-018-0300-7
19. Buchan I, Covvey HD, Rakowski H. An artificial intelligence approach to automatic left ventricular border detection in 2-d echocardiography. In: Proceedings of the annual symposium on computer application in medical care (American medical informatics association) (1985). p. 691.
20. Goldberg V, Manduca A, Ewert DL, Gisvold JJ, Greenleaf JF. Improvement in specificity of ultrasonography for diagnosis of breast tumors by means of artificial intelligence. Med Phys (1992) 19:1475–81. doi:10.1118/1.596804
21. Han W, Birkeland R. Artificial intelligence as an approach to improve ultrasonic log scanning. Acoust Imaging (1993) 201–8. doi:10.1007/978-1-4615-2958-3_27
22. Buller D, Buller A, Innocent PR, Pawlak W. Determining and classifying the region of interest in ultrasonic images of the breast using neural networks. Artif Intelligence Med (1996) 8:53–66. doi:10.1016/0933-3657(95)00020-8
23. Wu K, Chen X, Ding M. Deep learning based classification of focal liver lesions with contrast-enhanced ultrasound. Optik (2014) 125:4057–63. doi:10.1016/j.ijleo.2014.01.114
24. Azizi S, Imani F, Zhuang B, Tahmasebi A, Kwak JT, Xu S, et al. Ultrasound-based detection of prostate cancer using automatic feature selection with deep belief networks. In: Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, october 5-9, 2015, proceedings, Part II 18. Springer (2015). p. 70–7.
25. Shi J, Zhou S, Liu X, Zhang Q, Lu M, Wang T. Stacked deep polynomial network based representation learning for tumor classification with small ultrasound image dataset. Neurocomputing (2016) 194:87–94. doi:10.1016/j.neucom.2016.01.074
26. Mischi M, Bell MAL, Van Sloun RJ, Eldar YC. Deep learning in medical ultrasound—from image formation to image analysis. IEEE Trans Ultrason Ferroelectrics, Frequency Control (2020) 67:2477–80. doi:10.1109/tuffc.2020.3026598
27. Gasse M, Millioz F, Roux E, Garcia D, Liebgott H, Friboulet D. High-quality plane wave compounding using convolutional neural networks. IEEE Trans Ultrason ferroelectrics, frequency Control (2017) 64:1637–9. doi:10.1109/tuffc.2017.2736890
28. Luchies A, Byram B. Deep neural networks for ultrasound beamforming. In: 2017 IEEE international ultrasonics symposium (IUS). IEEE (2017). p. 1–4. doi:10.1109/ULTSYM.2017.8092159
29. Montaldo G, Tanter M, Bercoff J, Benech N, Fink M. Coherent plane-wave compounding for very high frame rate ultrasonography and transient elastography. IEEE Trans Ultrason ferroelectrics, frequency Control (2009) 56:489–506. doi:10.1109/tuffc.2009.1067
30. Luchies AC, Byram BC. Deep neural networks for ultrasound beamforming. IEEE Trans Med Imaging (2018) 37:2010–21. doi:10.1109/tmi.2018.2809641
31. Simson W, Paschali M, Navab N, Zahnd G. Deep learning beamforming for sub-sampled ultrasound data. In: 2018 IEEE international ultrasonics symposium (IUS). IEEE (2018). p. 1–4.
32. Nair AA, Gubbi MR, Tran TD, Reiter A, Bell MAL. A fully convolutional neural network for beamforming ultrasound images. In: 2018 IEEE international ultrasonics symposium (IUS). IEEE (2018). p. 1–4.
33. Nair AA, Tran TD, Reiter A, Bell MAL. A deep learning based alternative to beamforming ultrasound images. In: 2018 IEEE International conference on acoustics, speech and signal processing (ICASSP). IEEE (2018). p. 3359–63.
34. Nair AA, Tran TD, Reiter A, Bell MAL. One-step deep learning approach to ultrasound image formation and image segmentation with a fully convolutional neural network. In: 2019 IEEE international ultrasonics symposium (IUS). IEEE (2019). p. 1481–4.
35. Nair AA, Washington KN, Tran TD, Reiter A, Bell MAL. Deep learning to obtain simultaneous image and segmentation outputs from a single input of raw ultrasound channel data. IEEE Trans Ultrason ferroelectrics, frequency Control (2020) 67:2493–509. doi:10.1109/tuffc.2020.2993779
36. Strohm H, Rothlübbers S, Eickel K, Günther M. Deep learning-based reconstruction of ultrasound images from raw channel data. Int J Comp Assist Radiol Surg (2020) 15:1487–90. doi:10.1007/s11548-020-02197-w
37. Senouf O, Vedula S, Zurakhov G, Bronstein A, Zibulevsky M, Michailovich O, et al. High frame-rate cardiac ultrasound imaging with deep learning. In: Medical image computing and computer assisted intervention–MICCAI 2018: 21st international conference, granada, Spain, september 16-20, 2018, proceedings, Part I. Springer (2018). p. 126–34.
38. Vedula S, Senouf O, Zurakhov G, Bronstein A, Zibulevsky M, Michailovich O, et al. High quality ultrasonic multi-line transmission through deep learning. In: Machine learning for medical image reconstruction: first international workshop, MLMIR 2018, held in conjunction with MICCAI 2018, granada, Spain, september 16, 2018, proceedings 1. Springer (2018). p. 147–55.
39. Vedula S, Senouf O, Zurakhov G, Bronstein A, Michailovich O, Zibulevsky M. Learning beamforming in ultrasound imaging (2018). arXiv preprint arXiv:1812.08043.
40. Luijten B, Cohen R, De Bruijn FJ, Schmeitz HA, Mischi M, Eldar YC, et al. Deep learning for fast adaptive beamforming. In: ICASSP 2019-2019 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE (2019). p. 1333–7.
41. Luijten B, Cohen R, De Bruijn FJ, Schmeitz HA, Mischi M, Eldar YC, et al. Adaptive ultrasound beamforming using deep learning. IEEE Trans Med Imaging (2020) 39:3967–78. doi:10.1109/tmi.2020.3008537
42. Wiacek A, Gonzalez E, Dehak N, Bell MAL. Coherenet: a deep learning approach to coherence-based beamforming. In: 2019 IEEE international ultrasonics symposium (IUS). IEEE (2019). p. 287–90.
43. Wiacek A, González E, Bell MAL. Coherenet: a deep learning architecture for ultrasound spatial correlation estimation and coherence-based beamforming. IEEE Trans Ultrason Ferroelectrics, Frequency Control (2020) 67:2574–83. doi:10.1109/tuffc.2020.2982848
44. Lediju MA, Trahey GE, Byram BC, Dahl JJ. Short-lag spatial coherence of backscattered echoes: imaging characteristics. IEEE Trans Ultrason ferroelectrics, frequency Control (2011) 58:1377–88. doi:10.1109/tuffc.2011.1957
45. Yoon YH, Khan S, Huh J, Ye JC. Efficient b-mode ultrasound image reconstruction from sub-sampled rf data using deep learning. IEEE Trans Med Imaging (2018) 38:325–36. doi:10.1109/tmi.2018.2864821
46. Mamistvalov A, Eldar YC. Compressed fourier-domain convolutional beamforming for sub-nyquist ultrasound imaging. IEEE Trans Ultrason Ferroelectrics, Frequency Control (2021) 69:489–99. doi:10.1109/tuffc.2021.3123079
47. Mamistvalov A, Amar A, Kessler N, Eldar YC. Deep-learning based adaptive ultrasound imaging from sub-nyquist channel data. IEEE Trans Ultrason Ferroelectrics, Frequency Control (2022) 69:1638–48. doi:10.1109/tuffc.2022.3160859
48. Qi Y, Guo Y, Wang Y. Image quality enhancement using a deep neural network for plane wave medical ultrasound imaging. IEEE Trans Ultrason Ferroelectrics, Frequency Control (2020) 68:926–34. doi:10.1109/tuffc.2020.3023154
49. Chen Y, Liu J, Luo X, Luo J. A self-supervised deep learning approach for high frame rate plane wave beamforming with two-way dynamic focusing. In: 2021 IEEE international ultrasonics symposium (IUS). IEEE (2021). p. 1–4.
50. Lu J-Y, Lee P-Y, Huang C-C. Improving image quality for single-angle plane wave ultrasound imaging with convolutional neural network beamformer. IEEE Trans Ultrason Ferroelectrics, Frequency Control (2022) 69:1326–36. doi:10.1109/tuffc.2022.3152689
51. Nguon LS, Seo J, Seo K, Han Y, Park S. Reconstruction for plane-wave ultrasound imaging using modified u-net-based beamformer. Comput Med Imaging Graphics (2022) 98:102073. doi:10.1016/j.compmedimag.2022.102073
52. Goudarzi S, Rivaz H. Deep reconstruction of high-quality ultrasound images from raw plane-wave data: a simulation and in vivo study. Ultrasonics (2022) 125:106778. doi:10.1016/j.ultras.2022.106778
53. Wasih M, Almekkawy M. A robust deep neural network approach for ultrafast ultrasound imaging using single angle plane wave. In: 2022 IEEE international ultrasonics symposium (IUS). IEEE (2022). p. 1–4.
54. Seoni S, Salvi M, Matrone G, Meiburger KM. Ultrasound image beamforming optimization using a generative adversarial network. In: 2022 IEEE international ultrasonics symposium (IUS). IEEE (2022). p. 1–4.
55. Gao J, Xu L, Zou Q, Zhang B, Wang D, Wan M. A progressively dual reconstruction network for plane wave beamforming with both paired and unpaired training data. Ultrasonics (2023) 127:106833. doi:10.1016/j.ultras.2022.106833
56. Mor E, Bar-Hillel A. A unified deep network for beamforming and speckle reduction in plane wave imaging: a simulation study. Ultrasonics (2020) 103:106069. doi:10.1016/j.ultras.2020.106069
57. Vienneau E, Luchies A, Byram B. An improved training scheme for deep neural network ultrasound beamforming. In: 2019 IEEE international ultrasonics symposium (IUS). IEEE (2019). p. 568–70.
58. Luchies AC, Byram BC. Training improvements for ultrasound beamforming with deep neural networks. Phys Med Biol (2019) 64:045018. doi:10.1088/1361-6560/aafd50
59. Tierney J, Luchies A, Berger M, Byram B. Image quality-based regularization for deep network ultrasound beamforming. In: 2020 IEEE international ultrasonics symposium (IUS). IEEE (2020). p. 1–3.
60. Bell MAL, Huang J, Hyun D, Eldar YC, Van Sloun R, Mischi M. Challenge on ultrasound beamforming with deep learning (cubdl). In: 2020 IEEE international ultrasonics symposium (IUS). IEEE (2020). p. 1–5.
61. Hyun D, Wiacek A, Goudarzi S, Rothlübbers S, Asif A, Eickel K, et al. Deep learning for ultrasound image formation: cubdl evaluation framework and open datasets. IEEE Trans Ultrason ferroelectrics, frequency Control (2021) 68:3466–83. doi:10.1109/tuffc.2021.3094849
62. Liu S, Wang Y, Yang X, Lei B, Liu L, Li SX, et al. Deep learning in medical ultrasound analysis: a review. Engineering (2019) 5:261–75. doi:10.1016/j.eng.2018.11.020
63. Wang Y, Ge X, Ma H, Qi S, Zhang G, Yao Y. Deep learning in medical ultrasound image analysis: a review. IEEE Access (2021) 9:54310–24. doi:10.1109/access.2021.3071301
64. Fujioka T, Mori M, Kubota K, Oyama J, Yamaga E, Yashima Y, et al. The utility of deep learning in breast ultrasonic imaging: a review. Diagnostics (2020) 10:1055. doi:10.3390/diagnostics10121055
65. Becker AS, Mueller M, Stoffel E, Marcon M, Ghafoor S, Boss A. Classification of breast cancer in ultrasound imaging using a generic deep learning analysis software: a pilot study. Br J Radiol (2018) 91:20170576. doi:10.1259/bjr.20170576
66. Xu Y, Wang Y, Yuan J, Cheng Q, Wang X, Carson PL. Medical breast ultrasound image segmentation by machine learning. Ultrasonics (2019) 91:1–9. doi:10.1016/j.ultras.2018.07.006
67. Qian X, Pei J, Zheng H, Xie X, Yan L, Zhang H, et al. Prospective assessment of breast cancer risk from multimodal multiview ultrasound images via clinically applicable deep learning. Nat Biomed Eng (2021) 5:522–32. doi:10.1038/s41551-021-00711-2
68. Chen C, Wang Y, Niu J, Liu X, Li Q, Gong X. Domain knowledge powered deep learning for breast cancer diagnosis based on contrast-enhanced ultrasound videos. IEEE Trans Med Imaging (2021) 40:2439–51. doi:10.1109/tmi.2021.3078370
69. Jabeen K, Khan MA, Alhaisoni M, Tariq U, Zhang Y-D, Hamza A, et al. Breast cancer classification from ultrasound images using probability-based optimal deep learning feature fusion. Sensors (2022) 22:807. doi:10.3390/s22030807
70. Raza A, Ullah N, Khan JA, Assam M, Guzzo A, Aljuaid H. Deepbreastcancernet: a novel deep learning model for breast cancer detection using ultrasound images. Appl Sci (2023) 13:2082. doi:10.3390/app13042082
71. Degel MA, Navab N, Albarqouni S. Domain and geometry agnostic cnns for left atrium segmentation in 3d ultrasound. In: Medical image computing and computer assisted intervention–MICCAI 2018: 21st international conference, granada, Spain, september 16-20, 2018, proceedings, Part IV 11. Springer (2018). p. 630–7.
72. Leclerc S, Smistad E, Pedrosa J, Østvik A, Cervenansky F, Espinosa F, et al. Deep learning for segmentation using an open large-scale dataset in 2d echocardiography. IEEE Trans Med Imaging (2019) 38:2198–210. doi:10.1109/tmi.2019.2900516
73. Ghorbani A, Ouyang D, Abid A, He B, Chen JH, Harrington RA, et al. Deep learning interpretation of echocardiograms. NPJ digital Med (2020) 3:10. doi:10.1038/s41746-019-0216-8
74. Narang A, Bae R, Hong H, Thomas Y, Surette S, Cadieu C, et al. Utility of a deep-learning algorithm to guide novices to acquire echocardiograms for limited diagnostic use. JAMA Cardiol (2021) 6:624–32. doi:10.1001/jamacardio.2021.0185
75. Ha EJ, Baek JH. Applications of machine learning and deep learning to thyroid imaging: where do we stand? Ultrasonography (2021) 40:23–9. doi:10.14366/usg.20068
76. Park VY, Han K, Seong YK, Park MH, Kim E-K, Moon HJ, et al. Diagnosis of thyroid nodules: performance of a deep learning convolutional neural network model vs. radiologists. Scientific Rep (2019) 9:17843. doi:10.1038/s41598-019-54434-1
77. Peng S, Liu Y, Lv W, Liu L, Zhou Q, Yang H, et al. Deep learning-based artificial intelligence model to assist thyroid nodule diagnosis and management: a multicentre diagnostic study. The Lancet Digital Health (2021) 3:e250–9. doi:10.1016/s2589-7500(21)00041-8
78. Buda M, Wildman-Tobriner B, Castor K, Hoang JK, Mazurowski MA. Deep learning-based segmentation of nodules in thyroid ultrasound: improving performance by utilizing markers present in the images. Ultrasound Med Biol (2020) 46:415–21. doi:10.1016/j.ultrasmedbio.2019.10.003
79. Wang L, Zhang L, Zhu M, Qi X, Yi Z. Automatic diagnosis for thyroid nodules in ultrasound images by deep neural networks. Med image Anal (2020) 61:101665. doi:10.1016/j.media.2020.101665
80. Zhao Z, Yang C, Wang Q, Zhang H, Shi L, Zhang Z. A deep learning-based method for detecting and classifying the ultrasound images of suspicious thyroid nodules. Med Phys (2021) 48:7959–70. doi:10.1002/mp.15319
81. Zhu Y-C, AlZoubi A, Jassim S, Jiang Q, Zhang Y, Wang Y-B, et al. A generic deep learning framework to classify thyroid and breast lesions in ultrasound images. Ultrasonics (2021) 110:106300. doi:10.1016/j.ultras.2020.106300
82. Feng Y, Yang F, Zhou X, Guo Y, Tang F, Ren F, et al. A deep learning approach for targeted contrast-enhanced ultrasound based prostate cancer detection. IEEE/ACM Trans Comput Biol Bioinformatics (2018) 16:1794–801. doi:10.1109/tcbb.2018.2835444
83. Azizi S, Bayat S, Yan P, Tahmasebi A, Kwak JT, Xu S, et al. Deep recurrent neural networks for prostate cancer detection: analysis of temporal enhanced ultrasound. IEEE Trans Med Imaging (2018) 37:2695–703. doi:10.1109/tmi.2018.2849959
84. Hassan MR, Islam MF, Uddin MZ, Ghoshal G, Hassan MM, Huda S, et al. Prostate cancer classification from ultrasound and mri images using deep learning based explainable artificial intelligence. Future Generation Comp Syst (2022) 127:462–72. doi:10.1016/j.future.2021.09.030
85. Wang Y, Deng Z, Hu X, Zhu L, Yang X, Xu X, et al. Deep attentional features for prostate segmentation in ultrasound. In: Medical image computing and computer assisted intervention–MICCAI 2018: 21st international conference, granada, Spain, september 16-20, 2018, proceedings, Part IV 11. Springer (2018). p. 523–30.
86. Anas EMA, Mousavi P, Abolmaesumi P. A deep learning approach for real time prostate segmentation in freehand ultrasound guided biopsy. Med image Anal (2018) 48:107–16. doi:10.1016/j.media.2018.05.010
87. Lei Y, Tian S, He X, Wang T, Wang B, Patel P, et al. Ultrasound prostate segmentation based on multidirectional deeply supervised v-net. Med Phys (2019) 46:3194–206. doi:10.1002/mp.13577
88. Wang Y, Dou H, Hu X, Zhu L, Yang X, Xu M, et al. Deep attentive features for prostate segmentation in 3d transrectal ultrasound. IEEE Trans Med Imaging (2019) 38:2768–78. doi:10.1109/tmi.2019.2913184
89. Karimi D, Zeng Q, Mathur P, Avinash A, Mahdavi S, Spadinger I, et al. Accurate and robust deep learning-based segmentation of the prostate clinical target volume in ultrasound images. Med image Anal (2019) 57:186–96. doi:10.1016/j.media.2019.07.005
90. Orlando N, Gillies DJ, Gyacskov I, Romagnoli C, D’Souza D, Fenster A. Automatic prostate segmentation using deep learning on clinically diverse 3d transrectal ultrasound images. Med Phys (2020) 47:2413–26. doi:10.1002/mp.14134
91. Orlando N, Gyacskov I, Gillies DJ, Guo F, Romagnoli C, D’Souza D, et al. Effect of dataset size, image quality, and image type on deep learning-based automatic prostate segmentation in 3d ultrasound. Phys Med Biol (2022) 67:074002. doi:10.1088/1361-6560/ac5a93
92. Fiorentino MC, Villani FP, Di Cosmo M, Frontoni E, Moccia S. A review on deep-learning algorithms for fetal ultrasound-image analysis. Med image Anal (2023) 83:102629. doi:10.1016/j.media.2022.102629
93. Sobhaninia Z, Rafiei S, Emami A, Karimi N, Najarian K, Samavi S, et al. Fetal ultrasound image segmentation for measuring biometric parameters using multi-task deep learning. In: 2019 41st annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE (2019). p. 6545–8.
94. van den Heuvel TL, Petros H, Santini S, de Korte CL, van Ginneken B. Automated fetal head detection and circumference estimation from free-hand ultrasound sweeps using deep learning in resource-limited countries. Ultrasound Med Biol (2019) 45:773–85. doi:10.1016/j.ultrasmedbio.2018.09.015
95. Xie H, Wang N, He M, Zhang L, Cai H, Xian J, et al. Using deep-learning algorithms to classify fetal brain ultrasound images as normal or abnormal. Ultrasound Obstet Gynecol (2020) 56:579–87. doi:10.1002/uog.21967
96. Pu B, Li K, Li S, Zhu N. Automatic fetal ultrasound standard plane recognition based on deep learning and iiot. IEEE Trans Ind Inform (2021) 17:7771–80. doi:10.1109/tii.2021.3069470
97. Komatsu M, Sakai A, Komatsu R, Matsuoka R, Yasutomi S, Shozu K, et al. Detection of cardiac structural abnormalities in fetal ultrasound videos using deep learning. Appl Sci (2021) 11:371. doi:10.3390/app11010371
98. Milletari F, Ahmadi S-A, Kroll C, Plate A, Rozanski V, Maiostre J, et al. Hough-cnn: deep learning for segmentation of deep brain regions in mri and ultrasound. Computer Vis Image Understanding (2017) 164:92–102. doi:10.1016/j.cviu.2017.04.002
99. Ilunga-Mbuyamba E, Avina-Cervantes JG, Lindner D, Arlt F, Ituna-Yudonago JF, Chalopin C. Patient-specific model-based segmentation of brain tumors in 3d intraoperative ultrasound images. Int J Comput Assist Radiol Surg (2018) 13:331–42. doi:10.1007/s11548-018-1703-0
100. Xie B, Lei T, Wang N, Cai H, Xian J, He M, et al. Computer-aided diagnosis for fetal brain ultrasound images using deep convolutional neural networks. Int J Comp Assist Radiol Surg (2020) 15:1303–12. doi:10.1007/s11548-020-02182-3
101. Hesse LS, Aliasi M, Moser F, Haak MC, Xie W, Jenkinson M, et al. Subcortical segmentation of the fetal brain in 3d ultrasound using deep learning. NeuroImage (2022) 254:119117. doi:10.1016/j.neuroimage.2022.119117
102. Di Ianni T, Airan RD. Deep-fus: a deep learning platform for functional ultrasound imaging of the brain using sparse data. IEEE Trans Med Imaging (2022) 41:1813–25. doi:10.1109/tmi.2022.3148728
103. Soleimani M, Rymarczyk T, Kłosowski G. Ultrasound brain tomography: comparison of deep learning and deterministic methods. IEEE Trans Instrumentation Meas (2023) 73:1–12. doi:10.1109/tim.2023.3330229
104. Van Sloun RJ, Cohen R, Eldar YC. Deep learning in ultrasound imaging. Proc IEEE (2019) 108:11–29. doi:10.1109/jproc.2019.2932116
105. van Sloun RJ, Ye JC, Eldar YC. 1 deep learning for ultrasound beamforming (2021). arXiv preprint arXiv:2109.11431.
106. Luijten B, Chennakeshava N, Eldar YC, Mischi M, van Sloun RJ. Ultrasound signal processing: from models to deep learning. Ultrasound Med Biol (2023) 49:677–98. doi:10.1016/j.ultrasmedbio.2022.11.003
107. Afrin H, Larson NB, Fatemi M, Alizad A. Deep learning in different ultrasound methods for breast cancer, from diagnosis to prognosis: current trends, challenges, and an analysis. Cancers (2023) 15:3139. doi:10.3390/cancers15123139
108. Akkus Z, Aly YH, Attia IZ, Lopez-Jimenez F, Arruda-Olson AM, Pellikka PA, et al. Artificial intelligence (ai)-empowered echocardiography interpretation: a state-of-the-art review. J Clin Med (2021) 10:1391. doi:10.3390/jcm10071391
109. Khachnaoui H, Guetari R, Khlifa N. A review on deep learning in thyroid ultrasound computer-assisted diagnosis systems. In: 2018 IEEE international conference on image processing, applications and systems (IPAS). IEEE (2018). p. 291–7.
110. Ali M, Magee D, Dasgupta U. Signal processing overview of ultrasound systems for medical imaging. Texas: SPRAB12, Texas Instruments (2008). p. 55.
111. Thomenius KE. Evolution of ultrasound beamformers. In: 1996 IEEE ultrasonics symposium. Proceedings (IEEE), 2. IEEE (1996). p. 1615–22. doi:10.1109/ultsym.1996.584398
112. Perrot V, Polichetti M, Varray F, Garcia D. So you think you can das? a viewpoint on delay-and-sum beamforming. Ultrasonics (2021) 111:106309. doi:10.1016/j.ultras.2020.106309
113. Synnevag JF, Austeng A, Holm S. Adaptive beamforming applied to medical ultrasound imaging. IEEE Trans Ultrason ferroelectrics, frequency Control (2007) 54:1606–13. doi:10.1109/tuffc.2007.431
114. Ortiz SHC, Chiu T, Fox MD. Ultrasound image enhancement: a review. Biomed Signal Process Control (2012) 7:419–28. doi:10.1016/j.bspc.2012.02.002
115. Basset O, Cachard C. Ultrasound image post-processing–application to segmentation. In: Physics for medical imaging applications. Springer (2007). p. 227–39.
116. Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans image Process (2004) 13:600–12. doi:10.1109/tip.2003.819861
117. Long J, Trahey G, Bottenus N. Spatial coherence in medical ultrasound: a review. Ultrasound Med Biol (2022) 48:975–96. doi:10.1016/j.ultrasmedbio.2022.01.009
118. Hollman K, Rigby K, O’donnell M. Coherence factor of speckle from a multi-row probe. In: 1999 IEEE ultrasonics symposium. Proceedings. International symposium (cat. No. 99CH37027), 2. IEEE (1999). p. 1257–60. doi:10.1109/ultsym.1999.849225
119. Li P-C, Li M-L. Adaptive imaging using the generalized coherence factor. IEEE Trans Ultrason ferroelectrics, frequency Control (2003) 50:128–41. doi:10.1109/tuffc.2003.1182117
120. Camacho J, Parrilla M, Fritsch C. Phase coherence imaging. IEEE Trans Ultrason ferroelectrics, frequency Control (2009) 56:958–74. doi:10.1109/tuffc.2009.1128
121. Fornage BD. Ultrasound of the breast. Ultrasound Q (1993) 11:1–40. doi:10.1097/00013644-199300000-00001
122. Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Dataset of breast ultrasound images. Data in brief (2020) 28:104863. doi:10.1016/j.dib.2019.104863
123. Shao F, Ling KV, Ng WS, Wu RY. Prostate boundary detection from ultrasonographic images. J Ultrasound Med (2003) 22:605–23. doi:10.7863/jum.2003.22.6.605
124. Xu L, Gao S, Shi L, Wei B, Liu X, Zhang J, et al. Exploiting vector attention and context prior for ultrasound image segmentation. Neurocomputing (2021) 454:461–73. doi:10.1016/j.neucom.2021.05.033
125. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, October 5-9, 2015, proceedings, part III 18. Springer (2015). p. 234–41.
126. Ravichandran G, Fellows G. The accuracy of a hand-held real time ultrasound scanner for estimating bladder volume. Br J Urol (1983) 55:25–7. doi:10.1111/j.1464-410x.1983.tb07073.x
127. Ireton RC, Krieger JN, Cardenas DD, Williams-Burden B, Kelly E, Souci T, et al. Bladder volume determination using a dedicated, portable ultrasound scanner. J Urol (1990) 143:909–11. doi:10.1016/s0022-5347(17)40133-9
128. Coombes GM, Millard RJ. The accuracy of portable ultrasound scanning in the measurement of residual urine volume. J Urol (1994) 152:2083–5. doi:10.1016/s0022-5347(17)32314-5
129. Teng C-H, Huang Y-H, Kuo B-J, Bih L-I. Application of portable ultrasound scanners in the measurement of post-void residual urine. J Nurs Res (2005) 13:216–24. doi:10.1097/01.jnr.0000387543.68383.a0
130. Luo H, Jin F, Yang D, Wang Y, Li C, Guo M, et al. Interfractional variation in bladder volume and its impact on cervical cancer radiotherapy: clinical significance of portable bladder scanner. Med Phys (2016) 43:4412–9. doi:10.1118/1.4954206
131. Zhao L, Liao L, Gao L, Gao Y, Chen G, Cong H, et al. Effects of bladder shape on accuracy of measurement of bladder volume using portable ultrasound scanner and development of correction method. Neurourology and Urodynamics (2019) 38:653–9. doi:10.1002/nau.23883
132. Ohira S, Komiyama R, Kanayama N, Sakai K, Hirata T, Yoshikata K, et al. Improvement in bladder volume reproducibility using a-mode portable ultrasound bladder scanner in moderate-hypofractionated volumetric modulated arc therapy for prostate cancer patients. J Appl Clin Med Phys (2022) 23:e13546. doi:10.1002/acm2.13546
133. Jeong E, Bae S, Park M, Jung W, Kang J, Song T-K. Color Doppler imaging on a smartphone-based portable us system: preliminary study. In: 2015 IEEE international ultrasonics symposium (IUS). IEEE (2015). p. 1–4.
134. Di Ianni T, Hoyos CAV, Ewertsen C, Kjeldsen TK, Mosegaard J, Nielsen MB, et al. A vector flow imaging method for portable ultrasound using synthetic aperture sequential beamforming. IEEE Trans Ultrason Ferroelectrics, Frequency Control (2017) 64:1655–65. doi:10.1109/tuffc.2017.2742599
135. Jafari MH, Girgis H, Van Woudenberg N, Moulson N, Luong C, Fung A, et al. Cardiac point-of-care to cart-based ultrasound translation using constrained cyclegan. Int J Comput Assist Radiol Surg (2020) 15:877–86. doi:10.1007/s11548-020-02141-y
136. Seviaryn F, Malyarenko E, Schreiner G, Seviaryna I, Maev RG. Handheld high-resolution ultrasonic scanner for quantitative assessment of skin conditions. In: 2019 IEEE international ultrasonics symposium (IUS). IEEE (2019). p. 2380–2.
137. Qian F, Zhou X, Zhou J, Liu Z, Nie Q. A valuable and affordable handheld ultrasound in combating covid-19. Crit Care (2020) 24:334–2. doi:10.1186/s13054-020-03064-5
138. Bennett D, De Vita E, Mezzasalma F, Lanzarone N, Cameli P, Bianchi F, et al. Portable pocket-sized ultrasound scanner for the evaluation of lung involvement in coronavirus disease 2019 patients. Ultrasound Med Biol (2021) 47:19–24. doi:10.1016/j.ultrasmedbio.2020.09.014
139. Aminlari A, Quenzer F, Hayden S, Stone J, Murchison C, Campbell C. A case of covid-19 diagnosed at home with portable ultrasound and confirmed with home serology test. J Emerg Med (2021) 60:399–401. doi:10.1016/j.jemermed.2020.10.022
140. Baran JM, Webster JG. Design of low-cost portable ultrasound systems. In: 2009 annual international conference of the IEEE engineering in medicine and biology society. IEEE (2009). p. 792–5.
141. Xu X, Venkataraman H, Oswal S, Bartolome E, Vasanth K. Challenges and considerations of analog front-ends design for portable ultrasound systems. In: 2010 IEEE international ultrasonics symposium (IEEE). IEEE (2010). p. 310–3.
142. Kim G-D, Yoon C, Kye S-B, Lee Y, Kang J, Yoo Y, et al. A single fpga-based portable ultrasound imaging system for point-of-care applications. IEEE Trans Ultrason ferroelectrics, frequency Control (2012) 59:1386–94. doi:10.1109/tuffc.2012.2339
143. Kang J, Yoon C, Lee J, Kye S-B, Lee Y, Chang JH, et al. A system-on-chip solution for point-of-care ultrasound imaging systems: architecture and asic implementation. IEEE Trans Biomed circuits Syst (2015) 10:412–23. doi:10.1109/tbcas.2015.2431272
144. Donoho DL. Compressed sensing. IEEE Trans Inf Theor (2006) 52:1289–306. doi:10.1109/tit.2006.871582
145. Zhou J, Hoyos S, Sadler BM. Asynchronous compressed beamformer for portable diagnostic ultrasound systems. IEEE Trans Ultrason Ferroelectrics, Frequency Control (2014) 61:1791–801. doi:10.1109/tuffc.2014.006384
146. Shin B, Jeon S, Ryu J, Kwon HJ. Compressed sensing for elastography in portable ultrasound. Ultrason Imaging (2017) 39:393–413. doi:10.1177/0161734617716938
147. George SS, Mitrovic J, Anand A, Ignjatovic Z. Low-complexity compressive beamforming for portable ultrasound imaging. In: 2017 IEEE international ultrasonics symposium (IUS). IEEE (2017). p. 1–4.
148. Mitrovic J, La Pietra L, Ignjatovic Z. Portable ultrasound through compressive beamforming with improved contrast. In: 2018 IEEE international ultrasonics symposium (IUS). IEEE (2018). p. 1–4.
149. Zhou Z, Wang Y, Guo Y, Qi Y, Yu J. Image quality improvement of hand-held ultrasound devices with a two-stage generative adversarial network. IEEE Trans Biomed Eng (2019) 67:298–311. doi:10.1109/tbme.2019.2912986
150. Zhou Z, Siddiquee MR, Tajbakhsh N, Liang J (2018). Unet++: a nested u-net architecture for medical image segmentation. In: Deep learn med image anal multimodal learn clin decis support (2018), deep learning in medical image analysis and multimodal learning for clinical decision support: 4th international workshop, DLMIA 2018, and 8th international workshop, ML-CDS 2018, held in conjunction with MICCAI 2018. Granada, Spain: Springer, Cham 11045, 3–11. doi:10.1007/978-3-030-00889-5_1
151. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3d u-net: learning dense volumetric segmentation from sparse annotation. In: Medical image computing and computer-assisted intervention–MICCAI 2016: 19th international conference, athens, Greece, october 17-21, 2016, proceedings, Part II 19. Springer (2016). p. 424–32.
152. Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, et al. Attention u-net: learning where to look for the pancreas (2018). arXiv preprint arXiv:1804.03999.
153. Ibtehaz N, Rahman MS. Multiresunet: rethinking the u-net architecture for multimodal biomedical image segmentation. Neural networks (2020) 121:74–87. doi:10.1016/j.neunet.2019.08.025
154. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Adv Neural Inf Process Syst (2017) 30. doi:10.5555/3295222.3295349
155. Gao Y, Zhou M, Metaxas DN. Utnet: a hybrid transformer architecture for medical image segmentation. In: Medical image computing and computer assisted intervention–MICCAI 2021: 24th international conference, strasbourg, France, september 27–october 1, 2021, proceedings, Part III 24. Springer (2021). p. 61–71.
156. Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, et al. Transunet: Transformers make strong encoders for medical image segmentation (2021). arXiv preprint arXiv:2102.04306.
157. Peiris H, Hayat M, Chen Z, Egan G, Harandi M. A robust volumetric transformer for accurate 3d tumor segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer (2022). p. 162–72.
158. Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, et al. Swin-unet: unet-like pure transformer for medical image segmentation. In: European conference on computer vision. Springer (2022). p. 205–18.
159. Shi L, Gao T, Zhang Z, Zhang J. Stm-unet: an efficient u-shaped architecture based on swin transformer and multiscale mlp for medical image segmentation. In: GLOBECOM 2023-2023 IEEE global communications conference (IEEE). IEEE (2023). p. 2003–8.
160. Yang S, Niu J, Wu J, Wang Y, Liu X, Li Q. Automatic ultrasound image report generation with adaptive multimodal attention mechanism. Neurocomputing (2021) 427:40–9. doi:10.1016/j.neucom.2020.09.084
161. Liang J, Yang X, Huang Y, Li H, He S, Hu X, et al. Sketch guided and progressive growing gan for realistic and editable ultrasound image synthesis. Med image Anal (2022) 79:102461. doi:10.1016/j.media.2022.102461
162. Li C, Wang L, Li Y. Transformer and group parallel axial attention co-encoder for medical image segmentation. Scientific Rep (2022) 12:16117. doi:10.1038/s41598-022-20440-z
163. Xu S, Quan H. Ect-nas: searching efficient cnn-transformers architecture for medical image segmentation. In: 2021 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE (2021). p. 1601–4.
164. Zhou H-Y, Guo J, Zhang Y, Yu L, Wang L, Yu Y. nnformer: interleaved transformer for volumetric segmentation (2021). arXiv preprint arXiv:2109.03201.
165. Liu D, Gao Y, Zhangli Q, Han L, He X, Xia Z, et al. Transfusion: multi-view divergent fusion for medical image segmentation with transformers. In: International conference on medical image computing and computer-assisted intervention. Springer (2022). p. 485–95.
166. Zhu Z, He X, Qi G, Li Y, Cong B, Liu Y. Brain tumor segmentation based on the fusion of deep semantics and edge information in multimodal mri. Inf Fusion (2023) 91:376–87. doi:10.1016/j.inffus.2022.10.022
167. Liu Y, Yu C, Cheng J, Wang ZJ, Chen X. Mm-net: a mixformer-based multi-scale network for anatomical and functional image fusion. IEEE Trans Image Process a Publ IEEE Signal Process Soc (2024) 33:2197–212. doi:10.1109/tip.2024.3374072
168. Chi J, Li Z, Sun Z, Yu X, Wang H. Hybrid transformer unet for thyroid segmentation from ultrasound scans. Comput Biol Med (2023) 153:106453. doi:10.1016/j.compbiomed.2022.106453
169. Wang Y, Ye Z, Wen M, Liang H, Zhang X. Transvfs: a spatio-temporal local-global transformer for vision-based force sensing during ultrasound-guided prostate biopsy. Med Image Anal (2024) 94:103130. doi:10.1016/j.media.2024.103130
170. Ahmadi N, Tsang M, Gu A, Tsang T, Abolmaesumi P. Transformer-based spatio-temporal analysis for classification of aortic stenosis severity from echocardiography cine series. IEEE Trans Med Imaging (2024) 43:366–76. doi:10.1109/tmi.2023.3305384
171. Kirillov A, Mintun E, Ravi N, Mao H, Rolland C, Gustafson L, et al. Segment anything. In: Proceedings of the IEEE/CVF international conference on computer vision. IEEE (2023). p. 4015–26.
172. Lin X, Xiang Y, Zhang L, Yang X, Yan Z, Yu L. Samus: adapting segment anything model for clinically-friendly and generalizable ultrasound image segmentation (2023). arXiv preprint arXiv:2309.06824.
173. Ma J, He Y, Li F, Han L, You C, Wang B. Segment anything in medical images. Nat Commun (2024) 15:654. doi:10.1038/s41467-024-44824-z
174. Singhal K, Azizi S, Tu T, Mahdavi SS, Wei J, Chung HW, et al. Large language models encode clinical knowledge. Nature (2023) 620:172–80. doi:10.1038/s41586-023-06291-2
175. Thirunavukarasu AJ, Ting DSJ, Elangovan K, Gutierrez L, Tan TF, Ting DSW. Large language models in medicine. Nat Med (2023) 29:1930–40. doi:10.1038/s41591-023-02448-8
176. Singhal K, Tu T, Gottweis J, Sayres R, Wulczyn E, Hou L, et al. Towards expert-level medical question answering with large language models (2023). arXiv preprint arXiv:2305.09617.
177. Wu S-H, Tong W-J, Li M-D, Hu H-T, Lu X-Z, Huang Z-R, et al. Collaborative enhancement of consistency and accuracy in us diagnosis of thyroid nodules using large language models. Radiology (2024) 310:e232255. doi:10.1148/radiol.232255
Keywords: medical ultrasound imaging, deep learning, ultrasound beamforming, medical image analysis, clinical diagnosis
Citation: Song K, Feng J and Chen D (2024) A survey on deep learning in medical ultrasound imaging. Front. Phys. 12:1398393. doi: 10.3389/fphy.2024.1398393
Received: 09 March 2024; Accepted: 03 June 2024;
Published: 01 July 2024.
Edited by:
Guanqiu Qi, Buffalo State College, United StatesReviewed by:
Wei Wang, Chengdu University of Information Technology, ChinaCongyao Zhagn, Sichuan University, China
Yang Xu, Chongqing University of Posts and Telecommunications, China
Aleksandar Vakanski, University of Idaho, United States
Copyright © 2024 Song, Feng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Feng, ZmpmbG9hdGluZzg5QDEyNi5jb20=
 Ke Song
Ke Song Jing Feng
Jing Feng Duo Chen
Duo Chen