
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Phys., 20 March 2024
Sec. Radiation Detectors and Imaging
Volume 12 - 2024 | https://doi.org/10.3389/fphy.2024.1388364
This article is part of the Research TopicMulti-Sensor Imaging and Fusion: Methods, Evaluations, and Applications, volume IIView all 16 articles
In the field of computer-assisted medical diagnosis, developing medical image segmentation models that are both accurate and capable of real-time operation under limited computational resources is crucial. Particularly for skin disease image segmentation, the construction of such lightweight models must balance computational cost and segmentation efficiency, especially in environments with limited computing power, memory, and storage. This study proposes a new lightweight network designed specifically for skin disease image segmentation, aimed at significantly reducing the number of parameters and floating-point operations while ensuring segmentation performance. The proposed ConvStem module, with full-dimensional attention, learns complementary attention weights across all four dimensions of the convolution kernel, effectively enhancing the recognition of irregularly shaped lesion areas, reducing the model’s parameter count and computational burden, thus promoting model lightweighting and performance improvement. The SCF Block reduces feature redundancy through spatial and channel feature fusion, significantly lowering parameter count while improving segmentation results. This paper validates the effectiveness and robustness of the proposed SCSONet on two public skin lesion segmentation datasets, demonstrating its low computational resource requirements. https://github.com/Haoyu1Chen/SCSONet.
In 2024, it is projected that around 99,700 cases of in situ melanoma will be diagnosed, with an estimated 13,120 deaths from skin cancer, of which melanoma accounts for 99% [1]. Early detection of melanoma can often lead to cure through simple outpatient surgery, as opposed to late-stage diagnosis significantly reducing survival rates from over 99%–32%. Early detection is thus crucial for improving survival chances [2].
Dermatologists typically use dermatoscopy, an intuitive method for skin lesion examination, which relies on experienced doctors manually inspecting images [3]. However, this method can be less accurate for inexperienced dermatologists [4].
Traditional image segmentation methods, such as threshold-based [5], edge-based [6], and clustering-based [7] approaches, have played a role but are often time-consuming and error-prone, with limited effectiveness on complex datasets. In contrast, deep learning enhances accuracy and adaptability in image segmentation, making skin disease diagnosis more efficient and widespread.
Over the years, with the enhancement of computing capabilities and advancements in deep learning technologies, segmentation methods based on convolutional neural networks have seen significant performance improvements [8]. Fully Convolutional Networks (FCN) were developed as pioneers for semantic segmentation [9]. The introduction of the U-Net network marked a major breakthrough in medical image segmentation [10]. Following that, the integration of Transformer technology through Vision Transformer (ViT) further enhanced the capabilities in medical image segmentation [11]. These advanced network technologies continue to push the performance and accuracy of medical image segmentation, providing more efficient and widespread technical support for the diagnosis of skin diseases.
Previous work on enhancing the performance of the U-Net network has tended to introduce more complex modules. However, in the field of medical image segmentation, the importance of model lightweighting is self-evident. In the modern medical field, especially in the application of medical image analysis, the importance of lightweight models is becoming increasingly prominent. These models can run efficiently on devices with limited memory and processing capabilities, and they show great potential in mobile healthcare and rapid response scenarios. Forn make high-qua instance, in emergencies, they can be used to quickly diagnose a patient’s condition, saving valuable treatment time. Moreover, these models are particularly valuable in remote areas because they cality medical diagnostic services more widespread and accessible, representing a significant advancement for typically resource-poor regions.Furthermore, the economic benefits of lightweight models cannot be overlooked. They can reduce the investment in hardware and operations for medical institutions, bringing cost benefits to medical systems around the world, especially in developing countries. By lowering medical costs, lightweight models provide more equal opportunities for medical services to a broader population, thereby helping to address socio-economic inequalities. In summary, the development of lightweight medical image segmentation models is not only a manifestation of technological progress but also an important part of social responsibility and commitment, aiming to improve the health level of all humanity by popularizing high-quality medical services.
To address the need for lightweight models, solutions like MobileNets [12–14] and transformer-based lightweight models such as MobileViT [15] have been developed for real-time image classification and segmentation of 2D images. Inception-ResNet optimizes inception modules and residual networks to enhance image feature extraction and detail restoration [16]. Additionally, in medical image segmentation, MISegNet [17] offers a powerful yet lightweight network for real-time segmentation of multimodal medical images. The UNeXt [18] model, combining UNet and MLP technologies, reduces parameters and computational load while maintaining high performance. MALUNet, through channel reduction and multiple attention mechanisms, shows superior performance in skin lesion segmentation, maintaining compactness and efficiency [19].
While existing lightweight medical image segmentation models have made progress in reducing computational resource consumption, they often overlook the issues of spatial and channel redundancy. Previous research has shown that there is considerable redundancy in both the spatial and channel dimensions of deep neural network feature maps. This redundancy can lead to insufficient extraction of key edge features in lesion areas, affecting the model’s performance and segmentation accuracy. Moreover, the presence of redundancy leads to wasteful use of computational resources. Therefore, addressing spatial and channel redundancy is crucial for enhancing the segmentation performance of lightweight medical image models.
In this study, we designed a U-shaped network architecture, the core of which is the Spatial-Channel Fusion Block (SCF Block). In addition, by incorporating ConvStem at the initial stage of feature extraction, we combined the stability of traditional convolution with the dynamic adaptability of Omni-dimensional Dynamic Convolution (ODConv) [20]. Additionally, our network introduces Channel Attention Bridge Block (CAB) and Spatial Attention Bridge Block (SAB) through skip connections, effectively achieving fusion of multi-level and multi-scale information. The core SCF Block, based on Spatial and Channel Reconstruction Convolution (SCConv) [21], significantly reduces feature redundancy through spatial-channel feature fusion technology, incorporating the Efficient Multi-Scale Attention Module (EMA) [22] and Partial Convolution (Pconv) [23] to establish short and long-range dependencies and enhance feature extraction capabilities. This ensures a substantial improvement in SCConv’s segmentation performance while reducing the parameter count and computational cost.In summary, our contributions are threefold:
• The Spatial-Channel Fusion Block (SCF Block) introduced aims to apply SCConv in the medical image segmentation field, reducing redundancy in the spatial and channel dimensions of feature mappings as well as in dense model parameters. It enhances the model’s ability to extract key edge features in lesion areas, significantly reducing parameter count and computational cost while ensuring segmentation accuracy.
• We introduced a unique lightweight feature extractor, ConvStem, that employs the ODConv convolution mechanism. By learning four different types of attention in parallel across the four core spatial dimensions, this mechanism not only enhances the model’s efficiency in capturing features but also significantly reduces the additional number of parameters. ConvStem merges the stability of traditional convolution with the dynamic adaptability of enhanced convolution structures, ensuring the model remains lightweight while effectively capturing a richer array of local features and details.
• We present SCSONet, a model characterized by innovative lightweight design and efficient feature extraction mechanisms. It significantly reduces the model’s parameters while maintaining segmentation accuracy. This approach not only streamlines the computational demands but also enhances the model’s applicability in real-world scenarios where resources are limited, ensuring both high performance and efficiency in medical image segmentation tasks.
In the evolution of medical image segmentation, Convolutional Neural Networks (CNN) have played a pivotal role. The introduction of Fully Convolutional Neural Networks (FCN) laid the foundation for precise segmentation and identification of target areas in images. UNet, with its encoder-decoder structure and efficient skip connections, significantly advanced medical image segmentation. Following UNet, architectures like 3D U-Net [24], V-Net [25], and U-Net++ [26] improved segmentation performance through enhanced convolution processes and connections. SF-Net [27] is an innovative multi-task framework that boosts tumor segmentation precision by fusing multimodal features and employing an uncertainty-based method for adaptive loss weight adjustment.TDGraphDTA Through multi-scale information interaction and graph optimization techniquesthe method enhances the accuracy of predictions and the interpretability of the model [28].BTSFDS-EI-MMRI [29] develops an advanced technique utilizing the Swin Transformer and CNNs for enhanced MRI image analysis, focusing on integrating semantic and edge features for improved accuracy.X-Net [30] combines CNNs and Transformers for improved medical image segmentation, employing a dual architecture for enhanced feature extraction and accuracy on small datasets.GSOMMIF-AL [31] introduces an adversarial approach for enhancing glioma segmentation from multi-modal MR images, emphasizing image fusion for better segmentation outcomes.MISMFIF [32] presents a cloud-enhanced medical image segmentation technique, integrating Transformers and CNNs for robust feature extraction and employing an interactive module for improved accuracy, showcasing cloud computing’s scalability and performance advantages.ASTCMSeg [33] presents a 3-D self-training framework for segmenting medical images across different modalities without paired data, focusing on anatomical consistency and a novel frequency domain approach for improved accuracy.ViT-UperNet [34], a hybrid model leveraging vision transformers and a unified-perceptual-parsing network, excels in medical image segmentation by combining self-attention with multi-scale feature fusion, significantly improving accuracy on cardiac MRI images.
As models grow in scale and complexity, so do their computational and storage costs, limiting their practical application in resource-constrained settings. This highlights the need for optimizing model efficiency without compromising performance, ensuring that advanced medical image segmentation technologies can be effectively deployed in diverse environments, particularly where computational resources are scarce.
To address these challenges, researchers are focusing on the design of lightweight segmentation networks for efficient visual processing. Innovations such as MobileNets with depthwise (DW) and pointwise (PW) convolutions, grouped convolutions from AlexNet [35], ODConv with multidimensional attention, PConv focusing on reducing redundant computation, and SCConv reducing feature map redundancy, are paving the way for more resource-efficient and practical models in medical image segmentation, especially in scenarios with limited computational and memory resources.
Recently, UNeXt, based on Multi-Layer Perceptrons (MLP) and UNet, has become a more suitable solution for practical applications in medical image segmentation due to its significant reduction in parameter count. MALUNet, as a lightweight medical image segmentation model incorporating various attention mechanisms, is better suited for clinical settings. However, despite these advancements, lightweight models still have performance gaps compared to larger models, with room for improvement in parameter efficiency and GFlops. Additionally, these methods have not fully addressed the redundancy in spatial and channel dimensions during the feature extraction process.
Given these considerations, this paper introduces an innovative lightweight UNet segmentation model based on the Spatial-Channel Fusion Block (SCF Blcok). This model effectively addresses spatial and channel redundancy issues by fusing multi-level, multi-scale information in skip connection paths, simultaneously enhancing segmentation accuracy and efficiency. Its innovation lies in its ability to deliver efficient, accurate segmentation results while maintaining low computational complexity, making it an ideal choice for practical applications and the mobile health domain. This approach offers a more efficient, practical solution for medical image analysis, also providing new directions for future developments in medical image segmentation technology.
The proposed skin lesion segmentation framework is shown in Figure 1, which consists of ConvStem, SCF Block, and SCAB modules. ConvStem enhances flexibility and reduces parameters through dynamic adjustment of convolution kernels via omni-dimensional attention, moving beyond static application. The SCF Block, comprising SCConv for spatial-channel reconstruction, PConv, and EMA for establishing dependencies and enhancing feature extraction, reduces spatial and channel redundancy, refining feature representation. SCAB, with CAB and SAB, improves multi-level and multi-scale information fusion, reducing feature loss during downsampling.
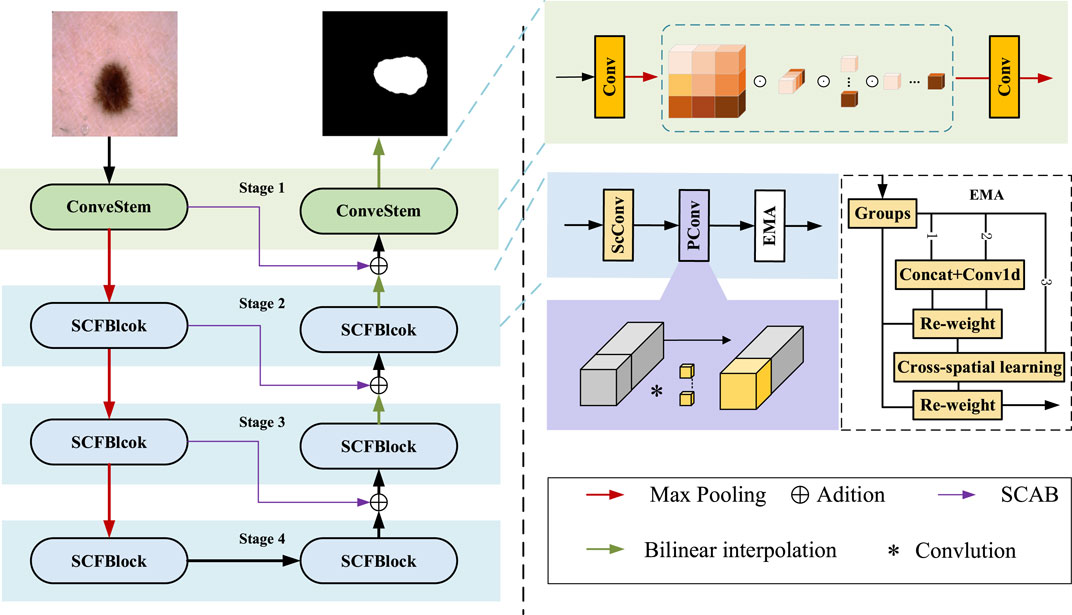
Figure 1. The framework of the proposed skin lsion segmentation method. It llustrates the framework for segmenting skin diseases, primarily consisting of three modules: the ConvStem module, the SCF Block, and the SCAB module for skip connections.
Lesion areas in medical images often present irregular shapes and vary greatly across different images, making accurate identification and segmentation a highly challenging task. To enhance the performance of medical image segmentation, particularly in capturing fine-grained and shape-sensitive local details, this study introduces the ConvStem module, as shown in the top right corner of Figure 1.
Traditional Convolutional Neural Networks are limited in simulating complex and irregular shape changes due to the fixed geometric structure of their basic modules. To overcome this limitation, the ConvStem module employs ODConv at the initial feature extraction stage, an innovative convolutional method with Omni-Dimensional attention mechanisms. ODConv combines the stability of traditional convolution with the flexibility of dynamic convolution, enabling the model to more effectively capture and process shape-aware features of irregularly shaped lesion areas in medical images. Through this design, the ConvStem module significantly enhances the network’s ability to recognize complex lesion morphologies in medical images, thereby providing richer and more accurate primary feature information for subsequent network layers’ feature learning and fusion.
The input image, denoted as
where
This application within ConvStem allows the convolution kernels to adjust dynamically to different inputs, moving away from a static, singular approach. This increases the model’s flexibility, reduces the number of parameters and computational burden, aiding in model lightweighting while boosting performance. Standard convolution captures basic features efficiently, while ODConv’s dynamic adjustment provides a deeper understanding and extraction for specific features, crucial for complex skin disease image analysis. Combining these convolutions, ConvStem outputs feature mappings that finely reflect shapes and local details, enabling our proposed SCSONet to produce more detailed lesion segmentation results, showcasing rich, multi-faceted feature information. After passing through ConvStem, the input F0 produces the outputs F1, F2 and F3, as described by the following Eq. 2.
F1, F2 and F3 are each connected to the decoder through the SCAB, which includes a Channel Attention Bridge Block (CAB) and a Spatial Attention Bridge Block (SAB).
The SAB uses max and average pooling operations at each stage to establish short and long-range dependencies and enhance feature extraction capability. After these operations, feature maps with channel C, height H, and width W are concatenated into feature maps with two channels, while height and width remain unchanged. Dilated convolution and the sigmoid function are then applied to obtain spatial attention maps for each stage. Finally, these are element-wise multiplied with the initial images of the stage, and the residuals are summed, restoring the original channel count for each stage.
The CAB is primarily designed to fuse features across different channel orders to better integrate information. The internal workings of this module can be represented by the following Eq. 3:
Pi is the feature map obtained at stage input. Is the total number of stages, FCi is the fully connected layer at stage, and σ is the sigmoid function.
The two bridge attention modules can fuse the multi-stage and multi-scale features of Stages 1–3 (Including the output from the subsequent SCF Block) to generate the attention maps in the spatial and channel dimension. And then, we add features obtained by bridge attention modules with features of the decoder part to reduce the feature semantic difference between the encoder and decoder while alleviating the information loss caused by the sampling process.
Although existing lightweight medical image segmentation models have made progress in reducing computational resource consumption, they often overlook issues of spatial and channel redundancy. To address these problems, the SCF Block was designed to significantly optimize the feature fusion process, particularly in reducing feature map redundancy across spatial and channel dimensions. By integrating the spatial-channel feature fusion technique of SCConv, the SCF module innovatively reduces feature redundancy, while the introduction of EMA and PConv enhances the model’s ability to capture short and long-range dependencies, further improving the efficiency and accuracy of feature extraction. This method, which focuses on feature fusion, not only reduces computational costs and model parameters but also greatly enhances the quality and precision of the segmentation results while maintaining the model’s lightweight stature. The SCF Block for Stage 2 can be represented as follows Eq. 4:
As shown in Figure 3. SCConv initially obtains spatially refined features Xw through SRU operations, and then acquires channel-refined features Y using CRU operations.
The Spatial Reconstruction Unit (SRU) reconstructs redundant features based on weights to suppress redundancy in the spatial dimension and enhance feature representation. The formula for calculating weights is as follows Eq. 5:
The formula for reconstruct is as follows Eq. 6:
The Channel Reconstruction Unit (CRU) employs a Split − Transform − and − Fuse strategy to reduce redundancy in the channel dimension, as well as computational and storage costs. After splitting, the spatially optimized feature Xw is divided into upper Xup and lower Xlow parts. In the Transform stage, Xup undergoes efficient convolution operations (i.e., GWC and PWC), and the outputs are aggregated to form a combined representative feature map Y1. The upper layer transformation stage can be represented as follows Eq. 7:
Xlow is input into a lower transformation stage where a cost-effective 1 × 1 PWC operation is applied to generate a feature map Y2 with shallow hidden details, complementing the rich feature extractor.
Global spatial information s1 and s2 are then collected through global average pooling, and channel soft attention operations produce feature importance vectors β1 and β2. These vectors guide the fusion of upper-layer features Y1 and lower-layer features Y2, generating refined features Y. The specific formula is as follows Eq. 8:
After passing through Partial Convolution (PConv), conventional convolution is applied to only a portion of the input channels for spatial feature extraction, with the remaining channels left unchanged, as shown on the right side of Figure 1. For continuous or regular memory access, the first or last continuous cp channels are computed as a representation of the entire feature map. Therefore, the Floating Point Operations (FLOPs) of a PConv are significantly reduced, as indicated by the Eq. 9:
With a typical partial ratio
To address the potential loss of important features due to SCConv’s spatial information compression, we introduced an Efficient Multi-Scale Attention (EMA) mechanism.
As shown on the right side of Figure 1, EMA reshapes part of the channel dimensions into batch dimensions, avoiding dimensionality reduction through standard convolution. This approach allows for different strategies in parallel subnetworks to maximally preserve multi-scale features of pathological sections. Moreover, EMA fuses output feature maps of two parallel subnets using a cross-space learning method, ensuring areas with potential targets in the final output feature map have higher feature weights.
By adjusting channel dimensions and applying multi-scale attention, EMA compensates for SCConv’s limitations in feature extraction, enhancing the capture of key features, optimizing feature representation, and improving the accuracy of medical image segmentation, effectively addressing SCConv’s limitations in handling fine-grained features.
The SCF Block represents a significant advancement in medical image segmentation, offering a robust solution for reducing redundancy while enhancing feature representation through spatial-channel fusion. By integrating SCConv, EMA, and PConv, it addresses the critical need for efficient, high-performance segmentation in medical imaging. This module’s innovative approach to capturing fine-grained details and dependencies not only improves segmentation accuracy but also ensures the model’s lightweight nature, making it an ideal choice for applications where computational resources are limited.
In this study, each image in the dataset is associated with a corresponding binary mask. Skin lesion segmentation is treated as a pixel-level binary classification task, distinguishing skin lesions from the background. The combination of Binary Cross-Entropy (BCE) loss and Dice similarity coefficient loss is used as the loss function to optimize network parameters, effectively addressing the challenges of skin lesion segmentation by balancing pixel-wise accuracy and overlap between the predicted and ground truth masks.
The loss function is BceDice loss, which can be expressed by the Eq. 10:
Where N is the total number of samples, yi is the real label,pi is the prediction. |X| and |Y| represent ground truth and X ∩ Y prediction, respectively. α1 and α2 refer to the weight of two loss functions. In this paper, both weights are taken as one by default.
The segmentation tasks were conducted on the ISIC2017 [36] and ISIC2018 datasets.Figure 4 showcasing a portion of the ISIC2017 and ISIC2018 datasets. The International Skin Imaging Collaboration (ISIC) dataset is a widely used open dataset in dermatological research. These datasets aim to facilitate computer-assisted dermatology diagnosis and research by providing a large collection of skin lesion images and related clinical metadata, supporting the development and validation of segmentation algorithms.
The ISIC2017 and ISIC2018 datasets contain 2,150 and 2,694 dermoscopic images with segmentation mask labels, respectively. For experimental purposes, the datasets were randomly split into training and testing sets at a 7:3 ratio. Specifically, the ISIC2017 dataset was divided into 1,500 images for training and 650 for testing, while the ISIC2018 dataset was divided into 1,886 images for training and 808 for testing. Comparative experiments were conducted on both ISIC2017 and ISIC2018, with ablation studies performed on ISIC2018.
All experiments were implemented in the PyTorch framework and conducted on an NVIDIA GeForce RTX 3070 Ti Laptop GPU with 8 GB of memory. Based on experience, all images were normalized and resized to 256 × 256, with data augmentation techniques including vertical flip, horizontal flip, and random rotation applied. The loss function used was the BceDice loss, represented by Eq. 10. AdamW was used as the optimizer with an initial learning rate of 0.001, employing a cosine annealing scheduler for learning rate adjustment, with a maximum of 50 iterations, a minimum learning rate of 0.00001, training epochs set to 300, and a batch size of 8.
Five metrics including Mean Intersection over Union (mIoU) and Dice similarity score (DSC), Eq. 11 are used to measure segmentation performances. In addition, Params is utilized to indicate the number of parameters, and the unit is Million (M). The computational complexity is calculated regarding the number of floating point operators (GFLOPs). Note that the parameters and GFLOPs of models are measured with 256 × 256 input size.
Where TP, FP, FN, TN represent true positive, false positive, false negative, and true negative.
In comparative experiments, the proposed SCSONet demonstrated significant advantages over advanced models like EGEUNet [37], showcasing its lightweight nature with fewer parameters and GFLOPs. Notably, SCSONet achieved the lowest GFLOPs among skin disease segmentation methods, at only 0.056, highlighting its efficiency. Figure 5 emphasized SCSONet’s reduced computational demand, making it an ideal choice for resource-constrained environments while maintaining high segmentation performance.
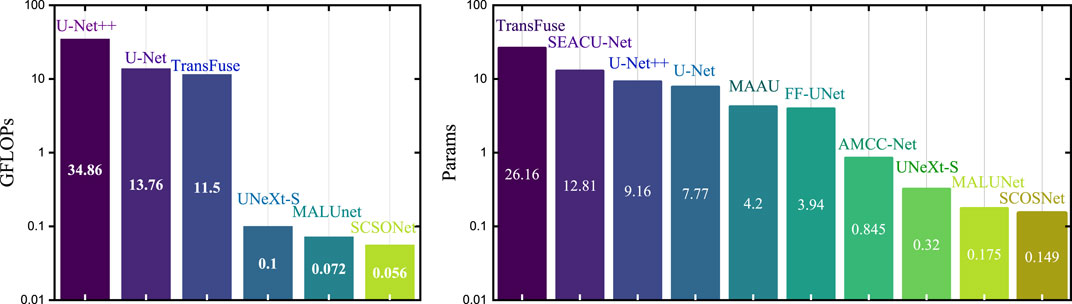
Figure 5. Histogram visualization with the Y-axis set as a logarithmic scale comparison with other methods on parameters and FLOPs.
Table 1 showcase SCSONet’s performance against other methods on the ISIC2017 and ISIC2018 datasets, illustrating its state-of-the-art overall performance. Specifically, compared to larger U-net models, SCSONet not only achieved superior performance but also significantly reduced parameters and GFLOPs by 451× and 1,224×, respectively. It outperformed other lightweight models by increasing mIoU by 7.56% over QGD-Net, with fewer parameters. Surpassing EGEUNet, it demonstrated better results in mIoU and DCS while reducing GFLOPs by 22.2%, able to train within 0.6 GB of VRAM. Its effectiveness is showcased in Figures 5, 6.
The qualitative comparison results, as shown in Figure 7, involve randomly selected test samples for qualitative evaluation. It is observed that SCSONet effectively differentiates between skin lesion areas and normal skin, achieving more accurate target area localization and boundary prediction compared to other models, which show issues with over-segmentation and under-segmentation. These comparisons demonstrate SCSONet’s effectiveness in skin lesion segmentation.
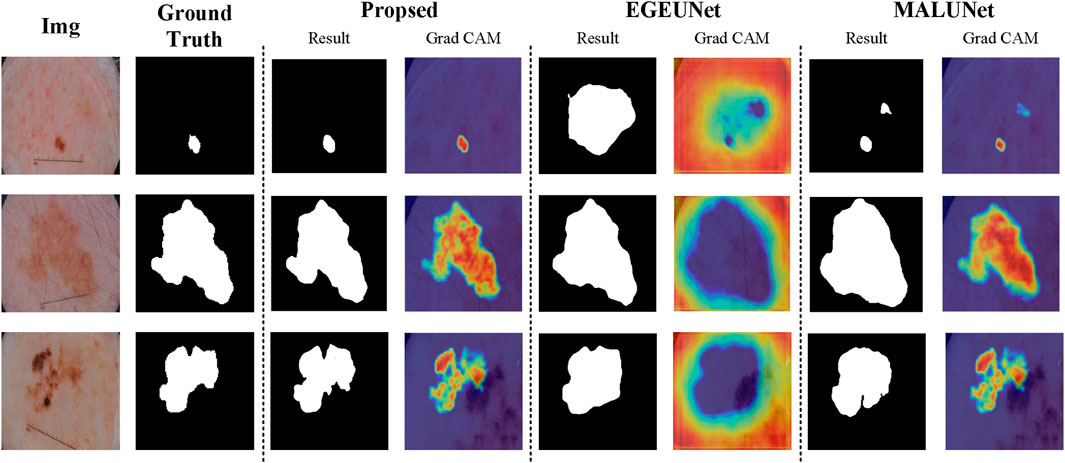
Figure 7. Comparison of segmentation results from different models on the ISIC2018 dataset and Grad CAM visualization (utilizing heatmaps to visualize the network prediction process.
As shown in Table 2, ablation studies were conducted to assess the effectiveness of each module within the proposed method. MALUNet served as the base model. Initially, ablation on the ConvStem module showed significant improvements in mIoU and DSC with notable reductions in parameters and GFLOPs, by replacing the first three convolutional layers in the base with ConvStem. Subsequently, replacing the base model’s last three layers with three SCF Blocks similarly resulted in performance enhancement and reductions in parameters and GFLOPs. The ablation study meticulously demonstrates the significant contributions of key modules within SCSONet—ConvStem and SCF—towards enhancing medical image segmentation performance. The ConvStem module, by incorporating Omni-Dimensional Dynamic Convolution (ODConv), significantly enhances the model’s capability to recognize the shapes of irregular lesion areas, substantially improving the efficiency of primary feature extraction. Meanwhile, the SCF module effectively reduces feature map redundancy through spatial-channel feature fusion technology, further enhancing the model’s segmentation precision and efficiency.Experimental results indicate that the inclusion of each module positively impacts model performance, particularly when used in combination, leading to optimal performance in terms of mIoU and DSC, while also achieving a significant reduction in the number of parameters and computational costs. These findings not only validate the effectiveness of the ConvStem and SCF modules in medical image segmentation tasks but also highlight the potential application of our lightweight network architecture in resource-constrained environments.Finally, for clearer visual comparison, experimental results are shown in Figure 8.
In Table 3, we conduct micro ablations on SCF Block.Further ablation studies within the SCF Block compared the effects of PConv, DepthwiseSeparableConv, and Dilated convolution. The results highlighted PConv’s significant contribution to SCConv’s performance enhancement, also confirming the role of EMA within the SCF Block for improving the segmentation capabilities of the network.
SCSONet stands out as the first lightweight model to reduce GFlops to around 0.056 while maintaining exceptional segmentation performance. Its effectiveness is showcased in Figures 5, 6, which clearly present experimental results and segmentation outcomes, respectively. Demonstrating robust performance on two public datasets, SCSONet’s primary clinical application is to assist in diagnosis, helping doctors quickly delineate focal areas or enabling non-specialists to diagnose diseases rapidly. Deploying this model in hospitals for semantic segmentation on small datasets can achieve higher segmentation accuracy.
In the field of medical image processing and analysis, hospitals often rely on high-performance GPUs and large computational devices, requiring substantial computational resources. However, for under-resourced or remote medical facilities, limited computational resources pose a significant barrier to implementing advanced medical image analysis. This gap hinders the widespread adoption and application of advanced medical imaging technologies, especially in regions that need them most. And also, for rapid lesion detection and diagnosis in the field or emergency situations, a model that can be easily integrated into mobile devices is equally necessary.To address this challenge, this paper proposes SCSONet, an innovative lightweight network architecture comprising ConvStem, SCF Block, and skip connections, aimed at bridging this gap by enabling efficient, high-quality medical image analysis with lower computational demands.
The ConvStem module with full-dimensional attention effectively enhances the recognition of irregularly shaped lesion areas while reducing the model’s parameter count and computational load, facilitating model lightweighting and performance improvement. The SCF Block, through spatial and channel feature fusion, efficiently reduces feature redundancy, significantly lowering parameter count while improving segmentation results. It addresses the challenges of resource-intensive traditional segmentation methods and high hardware requirements, offering an efficient solution for skin disease image segmentation tasks.
This study demonstrates the superior performance of the SCSONet model through optimization of parameters and floating-point operations (FLOPs), showcasing its strong generalizability and adaptability compared to other advanced models, while significantly reducing network parameters and computational costs. SCSONet achieves competitive segmentation performance with only 0.149 M parameters and 0.056GFLOPs, making it, to our knowledge, the first model to operate under such low computational load. Notably, SCSONet’s lightweight design allows it to be trained with just 0.6 GB of VRAM, a breakthrough feature that not only reduces the dependence on high-performance computing resources but also offers a new solution for medical image segmentation tasks in resource-limited environments. This design focus underscores the innovativeness and practical application value of our model, particularly in advancing mobile health technology and remote medical services.
While SCSONet exhibits a notable reduction in parameters and computational efficiency, it still has a gap compared to EGEUnet in terms of parameter quantity. Additionally, the limited datasets used for experiments and the model’s generalizability are areas for further inquiry. Additionally, during multiple training sessions, there were occasional instances of lower accuracy. This indicates that the model may not consistently achieve the expected high precision under certain specific datasets or training conditions, suggesting a sensitivity to training data or a deficiency in the optimization strategy under specific conditions. Although these instances are rare, they must be taken seriously as they could affect the model’s reliability and robustness in practical applications.
Future research should focus on extending the lightweight architecture to additional semantic segmentation tasks, alongside a thorough examination of its integration with hardware devices for enhanced performance. Investigating advanced training techniques and structural adjustments to the model will be crucial for augmenting its adaptability and consistency across diverse training scenarios. The ultimate objective is to refine segmentation efficiency without compromising accuracy, thereby rendering the model more effective for assisted diagnostics within medical image analysis. This approach aims to strike a balance between computational efficiency and diagnostic precision, facilitating broader application in real-world clinical settings.
Publicly available datasets were analyzed in this study. This data can be found here: https://challenge.isic-archive.com/data/#2017.
HC: Writing–review and editing, Writing–original draft, Software, Methodology, Conceptualization. ZL: Writing–review and editing, Writing–original draft, Software. XH: Writing–review and editing, Writing–original draft, Conceptualization. ZP: Writing–review and editing, Writing–original draft, Validation. YD: Writing–review and editing, Writing–original draft. LT: Formal Analysis, Writing–review and editing, Writing–original draft, Investigation, Conceptualization. LY: Writing–review and editing, Writing–original draft, Investigation, Data curation, Conceptualization.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Development Fund of Key Laboratory of Chongqing University Cancer Hospital (cquchkfjj005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA: A Cancer J Clinicians (2024) 74:12–49. doi:10.3322/caac.21820
2. Wolf AM, Oeffinger KC, Shih TY-C, Walter LC, Church TR, Fontham ET, et al. Screening for lung cancer: 2023 guideline update from the american cancer society. CA: A Cancer J Clinicians (2023) 74:50–81. doi:10.3322/caac.21811
3. Carli P, De Giorgi V, Soyer H, Stante M, Mannone F, Giannotti B. Dermatoscopy in the diagnosis of pigmented skin lesions: a new semiology for the dermatologist. J Eur Acad Dermatol Venereol (2000) 14:353–69. doi:10.1046/j.1468-3083.2000.00122.x
4. Dinnes J, Deeks JJ, Chuchu N, di Ruffano LF, Matin RN, Thomson DR, et al. Dermoscopy, with and without visual inspection, for diagnosing melanoma in adults. Cochrane Database Syst Rev (2018) 12. doi:10.1002/14651858.cd011902.pub2
5. Zhu S, Gao R. A novel generalized gradient vector flow snake model using minimal surface and component-normalized method for medical image segmentation. Biomed Signal Process Control (2016) 26:1–10. doi:10.1016/j.bspc.2015.12.004
6. Gupta D, Anand RS. A hybrid edge-based segmentation approach for ultrasound medical images. Biomed Signal Process Control (2017) 31:116–26. doi:10.1016/j.bspc.2016.06.012
7. Fraz MM, Jahangir W, Zahid S, Hamayun MM, Barman SA. Multiscale segmentation of exudates in retinal images using contextual cues and ensemble classification. Biomed Signal Process Control (2017) 35:50–62. doi:10.1016/j.bspc.2017.02.012
8. Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical image analysis using convolutional neural networks: a review. J Med Syst (2018) 42:226–13. doi:10.1007/s10916-018-1088-1
9. Shelhamer E, Long J, Darrell T, et al. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell (2017) 39:640–51. doi:10.1109/tpami.2016.2572683
10. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference Proceedings, Part III 18; October 5-9, 2015; Munich, Germany. Springer (2015). p. 234–41.
11. Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An image is worth 16x16 words: transformers for image recognition at scale (2020). Available at: https://arxiv.org/abs/2010.11929. (Accessed December 17, 2023).
12. Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, et al. Mobilenets: efficient convolutional neural networks for mobile vision applications (2017). Available at: https://arxiv.org/abs/1704.04861. (Accessed November 4, 2023).
13. Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C. Mobilenetv2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition; June 18 2018 to June 23 2018; Salt Lake City, UT, USA (2018). p. 4510–20.
14. Koonce B, Koonce B. Mobilenetv3. In: Convolutional neural networks with swift for tensorflow: image recognition and dataset categorization (2021). p. 125–44.
15. Mehta S, Rastegari M. Mobilevit: light-weight, general-purpose, and mobile-friendly vision transformer (2021). Available at: https://arxiv.org/abs/2110.02178. (Accessed January 3, 2024).
16. Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI conference on artificial intelligence. vol. 31; 4-9 February 2017; San Francisco, California, USA (2017).
17. Singh VK, Kalafi EY, Wang S, Benjamin A, Asideu M, Kumar V, et al. Prior wavelet knowledge for multi-modal medical image segmentation using a lightweight neural network with attention guided features. Expert Syst Appl (2022) 209:118166. doi:10.1016/j.eswa.2022.118166
18. Valanarasu JMJ, Patel VM. Unext: mlp-based rapid medical image segmentation network. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; September 18-22, 2022; Singapore. Springer (2022). p. 23–33.
19. Ruan J, Xiang S, Xie M, Liu T, Fu Y. Malunet: a multi-attention and light-weight unet for skin lesion segmentation. In: 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (IEEE); 6-8 December 2022; Las Vegas, Nevada, USA (2022). p. 1150–6.
20. Li C, Zhou A, Yao A. Omni-dimensional dynamic convolution (2022). Available at: https://arxiv.org/abs/2209.07947. (Accessed November 2, 2023).
21. Li J, Wen Y, He L. Scconv: spatial and channel reconstruction convolution for feature redundancy. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; June 18 2022 to June 24 2022; New Orleans, LA, USA (2023). p. 6153–62.
22. Ouyang D, He S, Zhang G, Luo M, Guo H, Zhan J, et al. Efficient multi-scale attention module with cross-spatial learning. In: ICASSP 2023-2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (IEEE); June 4 to June 9, 2023; Rhodes Island, Greece (2023). p. 1–5.
23. Chen J, Kao S-h., He H, Zhuo W, Wen S, Lee C-H, et al. Run, don’t walk: chasing higher flops for faster neural networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; June 17 2023 to June 24 2023; Vancouver, BC, Canada (2023). p. 12021–31.
24. Ahmed SF, Rahman FS, Tabassum T, Bhuiyan MTI. 3d u-net: fully convolutional neural network for automatic brain tumor segmentation. In: 2019 22nd International Conference on Computer and Information Technology (ICCIT) (IEEE); 18-20 December 2019; Dhaka, Bangladesh (2019). p. 1–6.
25. Milletari F, Navab N, Ahmadi S-A. V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV) (Ieee); 25-28 October 2016; Stanford, California, USA (2016). p. 565–71.
26. Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J. Unet++: a nested u-net architecture for medical image segmentation. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018 Proceedings 4; September 20, 2018; Granada, Spain. Springer (2018). p. 3–11.
27. Liu Y, Mu F, Shi Y, Chen X. Sf-net: a multi-task model for brain tumor segmentation in multimodal mri via image fusion. IEEE Signal Process. Lett (2022) 29:1799–803. doi:10.1109/LSP.2022.3198594
28. Zhu Z, Yao Z, Zheng X, Qi G, Li Y, Mazur N, et al. Drug–target affinity prediction method based on multi-scale information interaction and graph optimization. Comput Biol Med (2023) 167:107621. doi:10.1016/j.compbiomed.2023.107621
29. Zhu Z, He X, Qi G, Li Y, Cong B, Liu Y. Brain tumor segmentation based on the fusion of deep semantics and edge information in multimodal mri. Inf Fusion (2023) 91:376–87. doi:10.1016/j.inffus.2022.10.022
30. Li Y, Wang Z, Yin L, Zhu Z, Qi G, Liu Y. X-net: a dual encoding–decoding method in medical image segmentation. Vis Comp (2021) 1–11.
31. Liu Y, Shi Y, Mu F, Cheng J, Chen X. Glioma segmentation-oriented multi-modal mr image fusion with adversarial learning. IEEE/CAA J Automatica Sinica (2022) 9:1528–31. doi:10.1109/JAS.2022.105770
32. He X, Qi G, Zhu Z, Li Y, Cong B, Bai L. Medical image segmentation method based on multi-feature interaction and fusion over cloud computing. Simulation Model Pract Theor (2023) 126:102769. doi:10.1016/j.simpat.2023.102769
33. Zhuang Y, Liu H, Song E, Xu X, Liao Y, Ye G, et al. A 3-d anatomy-guided self-training segmentation framework for unpaired cross-modality medical image segmentation. IEEE Trans Radiat Plasma Med Sci (2024) 8:33–52. doi:10.1109/TRPMS.2023.3332619
34. Ruiping Y, Kun L, Shaohua X, Jian Y, Zhen Z. Vit-upernet: a hybrid vision transformer with unified-perceptual-parsing network for medical image segmentation. Complex Intell Syst (2024) 1–13. doi:10.1007/s40747-024-01359-6
35. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process Syst (2012) 25.
36. Berseth M. Isic 2017-skin lesion analysis towards melanoma detection (2017). Available at: https://arxiv.org/abs/1703.00523. (Accessed January 9, 2024).
37. Ruan J, Xie M, Gao J, Liu T, Fu Y. Ege-unet: an efficient group enhanced unet for skin lesion segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; October 8-12, 2023; Vancouver, BC, Canada. Springer (2023). p. 481–90.
38. Zhang Y, Liu H, Hu Q. Transfuse: fusing transformers and cnns for medical image segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference Proceedings, Part I 24; September 27–October 1, 2021; Strasbourg, France. Springer (2021). p. 14–24.
39. Le PT, Pham B-T, Chang C-C, Hsu Y-C, Tai T-C, Li Y-H, et al. Anti-aliasing attention u-net model for skin lesion segmentation. Diagnostics (2023) 13:1460. doi:10.3390/diagnostics13081460
40. Dayananda C, Yamanakkanavar N, Nguyen T, Lee B. Amcc-net: an asymmetric multi-cross convolution for skin lesion segmentation on dermoscopic images. Eng Appl Artif Intelligence (2023) 122:106154. doi:10.1016/j.engappai.2023.106154
41. Jiang X, Jiang J, Wang B, Yu J, Wang J. Seacu-net: attentive convlstm u-net with squeeze-and-excitation layer for skin lesion segmentation. Comp Methods Programs Biomed (2022) 225:107076. doi:10.1016/j.cmpb.2022.107076
42. Wu H, Chen S, Chen G, Wang W, Lei B, Wen Z. Fat-net: feature adaptive transformers for automated skin lesion segmentation. Med image Anal (2022) 76:102327. doi:10.1016/j.media.2021.102327
43. Fan R, Wang Z, Zhu Q. Egfnet: efficient guided feature fusion network for skin cancer lesion segmentation. In: 2022 the 6th International Conference on Innovation in Artificial Intelligence (ICIAI); October 26-28, 2024; Tipasa, ALGERIA (2022). p. 95–9.
44. Zhao C, Lv W, Zhang X, Yu Z, Wang S. Mms-net: multi-level multi-scale feature extraction network for medical image segmentation. Biomed Signal Process Control (2023) 86:105330. doi:10.1016/j.bspc.2023.105330
45. Wang J, Huang G, Zhong G, Yuan X, Pun C-M, Deng J. Qgd-net: a lightweight model utilizing pixels of affinity in feature layer for dermoscopic lesion segmentation. IEEE J Biomed Health Inform (2023) 27:5982–93. doi:10.1109/jbhi.2023.3320953
46. Ma Q, Mao K, Wang G, Xu L, Zhao Y. Lcaunet: a skin lesion segmentation network with enhanced edge and body fusion (2023). Available at: https://arxiv.org/pdf/2305.00837.pdf. (Accessed October 28, 2023).
Keywords: light-weight model, medical image segmentation, attention mechanism, mobile health, skin lesion segmentation
Citation: Chen H, Li Z, Huang X, Peng Z, Deng Y, Tang L and Yin L (2024) SCSONet: spatial-channel synergistic optimization net for skin lesion segmentation. Front. Phys. 12:1388364. doi: 10.3389/fphy.2024.1388364
Received: 19 February 2024; Accepted: 06 March 2024;
Published: 20 March 2024.
Edited by:
Guanqiu Qi, Buffalo State College, United StatesReviewed by:
Yafei Zhang, Kunming University of Science and Technology, ChinaCopyright © 2024 Chen, Li, Huang, Peng, Deng, Tang and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Yin, eWwxQGNxdS5lZHUuY24=; Li Tang, dGFuZ2xpX3dodUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.