- Department of Mechanical Engineering, Whitacre College of Engineering, Texas Tech University, Lubbock, TX, United States
Current research strategies in the treatment of biofilm infections have focused on dispersal, in which bacteria are made to vacate the extracellular polymeric substance (EPS) surrounding them and return to a planktonic state where antimicrobial treatments are more effective. Glycoside hydrolases (GHs), which cleave bonds in EPS polysaccharides, have been shown to promote dispersal in Pseudomonas aeruginosa biofilms. The dispersal mechanism is possibly due to GHs’ ability to directly release bacteria from the EPS, disrupt EPS’ ability to regulate the environment, or reduce overall mechanical stability. In this work, passive microrheology is used to examine the relevance of the last mechanism by exploring the effects of three GHs (α-amylase, cellulase, and xylanase) known to disperse P. aeruginosa on local biofilm viscoelasticity. Compared to control studies in wild-type strains, it is found that treatment with all three GHs results in statistically relatively less elastic and stiffer biofilms, indicating that changes to mechanical stability may be a factor in effective dispersal. Both cellulase and xylanase were observed to have the greatest impact in creating a less stiff and elastic biofilm; these GHs have been observed to be effective at dispersal in the published results. Each GH was further tested on biofilms grown with strains that produced EPS missing specific polysaccharide components. Cellulase specifically targeted Psl, which forms the major structural and mechanical backbone of the EPS, explaining its efficacy in dispersal. However, xylanase did not appear to exhibit any affinity to any polysaccharide within the EPS based on the microrheology results. Overall, these results suggest that the local microrheology of the biofilms is impacted by GHs and that may be one of the factors that is causing the ability of these therapeutics to enhance dispersal.
Introduction
A biofilm is a hydrated mass of the self-secreted extracellular polymeric substance (EPS) encasing bacteria. The biofilm provides a favorable environment for the bacteria, enabling resistance to chemical/mechanical stresses and increasing survivability in a range of conditions [1–6]. Biofilms formed in medical settings have been found to strengthen virulence factors and increase negative patient outcomes of the associated bacteria. For instance, Pseudomonas aeruginosa biofilms enable the bacteria to evade host immune system response and antimicrobial agents [7,8], causing infections of millions of patients a year with a significant negative clinical impact [9–11].
Because of this, substantial efforts have been made to research strategies to treat and remove P. aeruginosa biofilms from wounds and clinical implants. Antimicrobial therapeutics alone are not enough to remove biofilms because bacteria can recolonize the biofilm causing reinfection. Current therapeutic approaches use a combination of physical removal via debridement/irrigation, followed by local treatment with high doses of antimicrobial therapeutics, which is aided by deeper access to the bacteria provided by the physical attack [12–18].
New treatment strategies have focused on causing biofilm dispersal. Dispersion is the process by which sessile bacteria in the biofilm become planktonic, escaping the biofilm and colonizing a new biofilm. In the planktonic state, the bacteria are more vulnerable to treatment, therefore making dispersal an attractive means of enhancing efficacy of antimicrobial therapeutics. Dispersal-based treatments cause active dispersion, in which bacteria respond to external stimuli, which can include molecules that target EPS, chemical triggers, quorum sensing, or other mechanisms [19–21]. Targeting the polysaccharides in the EPS that form the primary matrix of the biofilm has been identified particularly as an attractive means of causing dispersal due to their myriad roles in maintaining the environment of the biofilm, which, if interrupted, could cause active dispersal [22,23].
To attack polysaccharides, glycoside hydrolases (GHs) have been utilized, which hydrolyze specific glycosidic linkages within polysaccharides, cleaving polysaccharides into distinct units. Several GHs have been shown to effectively disperse P. aeruginosa biofilms of the PAO1 strain by “weakening” the EPS matrix [24–27]. For instance, both
One question of interest is what does “weakening” of the matrix mean: changes to viscoelasticity, reduction of surface attachment, reduced cell–cell adhesion, or changes to properties such as porosity. There is clear evidence of change to surface adhesion being one such property based on the aforementioned results [26]. However, polysaccharide EPS components are known to affect all of these properties, and manipulating any of them could be considered “weakening” the matrix of the biofilm.
P. aeruginosa EPS includes three polysaccharides. Alginate is composed of nonrepetitive monomers of
The weakening is likely a combination of various changes to the mechanical properties of the EPS that help induce dispersion, and there is some indication already of the importance of surface attachment. However, the impact on biofilm viscoelasticity as a potential mechanism is important to consider because the reduction of viscoelasticity would also indicate that the GHs have potential for not only increasing dispersal but also aiding debridement, which is still required to avoid recolonization.
There are a few previous studies to examine the effects of GHs on P. aeruginosa biofilm viscoelasticity. Studying the bulk rheology of alginate-dominant mucoid P. aeruginosa biofilms, Kovach et al.[45] found that alginate targeting alginate-lyase impacted viscoelasticity. However,
In this study, the effect of GHs known to be effective dispersal agents of P. aeruginosa biofilms is studied on PAO1 strain biofilms using microrheology with the aim of understanding whether GH’s potential reduction in local viscoelasticity on a baseline strain may be representative of generally observed dispersal efficacy.
Materials and methods
Microchannel fabrication
Microfluidic channels (Figure 1) were fabricated using soft lithography methods [48]. A high-resolution transparency mask (CAD/Art Services, Inc.) was placed with emulsion side down directly onto a 0.2-mm thick-layer of SU-8 (SU-8 2000, Microchem) spun (Laurell Spincoater) onto silicon wafers. SU-8 molds were patterned by exposure to UV light with a 380 nm filter (2000-EC series, Dymax). Polydimethylsiloxane, PDMS, (Sylgard 184, Dow Corning) was poured over the molds, followed by degassing and crosslinking. Channels were made by removing PDMS from the mold and punching 1-mm access ports. Channel bottoms, where PDMS was spun onto a glass slide, were bonded to channels using air plasma (Plasma Cleaner, Harrick Plasma) and then left overnight in an oven at 80°C and stored at 20°C until further use.
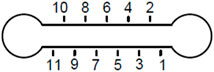
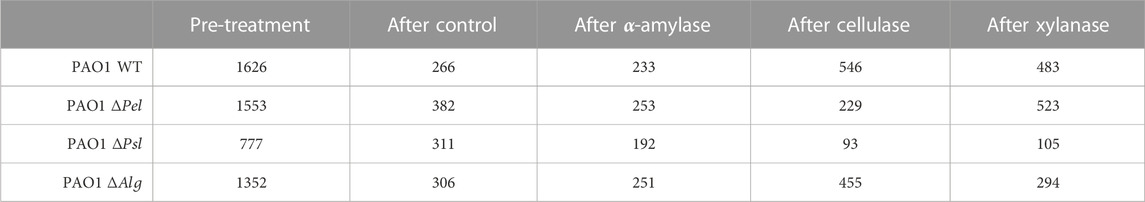
FIGURE 1. Microchannel schematic. Cross section of the channel is 1 × 6 mm2. Circular geometry on the sides has 1 mm radius each.
Bacteria growth medium preparation
Luria–Bertani (LB powder, Fisher Scientific, Catalog# BP1426-2) liquid broth solution was prepared by magnet-stirring LB powder in distilled water (5 g of powder in 200 mL water). This fresh LB broth solution was then autoclaved for sterilization.
Probe particle preparation
Probe particles used in the study were negatively charged, red-fluorescent (580 nm/605 nm), carboxylate-modified polystyrene microspheres (1
In a separate 20-mL vial, 1 mL of glycerol was vortex-mixed with 9 mL of freshly prepared LB broth to prepare a glycerol/LB solution. The small amount of added glycerol in the solution makes particles suspended in the solution for longer time, thereby helping particles to incorporate into the biofilm matrix as it forms. Then, 50
Bacterial strains and growth conditions
Four P. aeruginosa strains based on the widely used laboratory strain PAO1 were used in this work: wild-type (WT), strain lacking the EPS component Pel (∆pel), strain lacking the EPS component Psl (∆psl), and a strain lacking the EPS component alginate (∆alg).
Frozen bacterial stocks were stored at −20°C. Prior to culturing, 10 mL of freshly prepared sterile LB liquid broth was added to a 100-mL Erlenmeyer flask. Using an inoculating loop, a small amount of frozen bacterial stock was added to the flask. Afterward, the capped flask was incubated using a rotary shaker (Southwest Mini IncuShaker SH1000) at 200 rpm and 37°C for 24 h. Then, 100
The final culture solution was then inoculated onto the microchannels using pipette tips. After inoculation, the inlets and outlets were sealed to prevent evaporation. These channels were kept inside the incubator at 37°C under static conditions to allow for biofilm growth.
The final particle solution was syringe-pumped (at 30
After 48 h of inoculation, the biofilms were characterized via microrheology.
Treatment with glycoside hydrolases
Bacterial α-amylase (source Bacillus subtilis, 02100447; MP Biomedicals, LLC), fungal cellulase (source A. niger, 02150583; MP Biomedicals, LLC), and xylanase (source A. oryzae; Cat# X2753, Lot# SLBC5352V, Sigma-Aldrich) were utilized in experiments to treat the biofilms. First, 10% (w/v) of α-amylase/cellulase was prepared by vortex-mixing 0.1 gm of the lyophilized powder in 1 mL of 1x phosphate-buffered saline (PBS). Then, 500 units/mL xylanase was prepared by vortex-mixing 0.178 gm of the lyophilized powder in 1 mL of 1x PBS. These doses were chosen to be consistent with previously published results that saw them as effective for dispersal of biofilms [26,27].
To treat the biofilms after they were allowed to mature for 48 h, a small injection of GH solutions or a PBS control was added to the microchannels. Before treatments to the biofilms, all enzymes were activated by keeping at 37°C for 30 min. Approximately 7.5
Passive particle-tracking microrheology
Particle-tracking microrheology allows probing of biofilm viscoelasticity at the microscale of the biofilm microenvironments without affecting their microstructure[49]. This technique tracks the Brownian motion of particles embedded in a specimen. Each individual particle’s displacement reflects the material strain arising from the stress caused by the particle. By looking at the mean square displacement,
which ranges from 0 to 1 and represents the relative viscoelasticity of the solution. α = 1 represents purely viscous diffusion, whereas α = 0 is an elastic solid of the surrounding material [50]. Therefore, α is a measure of relative elasticity, with lower values indicating more elastic-like behavior. Additionally, we examine creep compliance, J, which is expressed as follows:
where a = particle radius, kB = Boltzmann constant, and T = temperature. This is a measure of inverse stiffness, with larger values meaning less stiff materials. It should be noted that we can calculate these values for individual particles and take MSD for an individual particle’s lag times or take the ensemble average MSD across many particles.
Epifluorescent microscopy was employed to visualize fluorescent particles using a Nikon Eclipse Ti-E microscope (×50 magnification). A SPECTRA X Light Engine was used to illuminate fluorescent particles. Particle tracks were captured using a Pco.edge 4.2 LT sCMOS camera at 40 frames per second for 25 s, allowing 1000 frames for each instance of imaging. Particle locations and tracks from image sequences were found using the Fiji installation of ImageJ with the plugin TrackMate [51,52]. This plugin finds particle centroids using a Laplacian of Gaussian filter, which allows subpixel localization, and generates 2D tracks from positions utilizing a simple linear assignment algorithm. Particle-tracking data were then imported to the MATLAB routine msdanalyzer [53] for the measurement of the MSD curves of individual particles and creating ensemble averages. Linear fits were made to log–log plots of MSD vs. lag time to find α to only lowest 4% of lag times on each MSD curve to the higher statistical significance of these data [54].
There are several ways to track particles; however, in this work, video microscopy is used. Two types of errors can occur when particle positions are tracked this way: static and dynamic. Static errors are inherent errors in measuring particle position due to factors such as fluctuations in light intensity, vibrations, and camera resolution limitations. Dynamic errors occur due to particle movement during exposure resulting in position inaccuracy [55–57]. Estimation of a dynamic error is difficult especially for heterogeneous mediums like biofilms, but the best practice to minimize the dynamic error is to choose low exposure times [57,58]. In our experiments, exposure time of ∼10% of the frame rate was used. To estimate the static error, a probe particle is typically fixed in a strong gel, and the MSD is measured [56]. Using the experimental setup described in the following paragraph, probe particles were fixed in a PDMS gel that was crosslinked overnight. An MSD floor for measurements from these particles was found to be ∼0.003
For a particular system, particle tracking occurred at two locations within a microchannel where separate biofilms were observed to have grown. These locations were found by looking for the fluorescence signature of the bacteria. Due to the channel design (Figure 1), it was possible to index locations and gather data within the same biofilm before and after treatments. Biofilms were grown in four different microchannels; this results in four biological replicates and two technical replicates for each experiment. The number of particle tracks for each condition varies, and the overall number of traces is reported in Table 1.
Data analysis
Distributions of
An unpaired Mann–Whitney (non-parametric) test, also supported in GraphPad Prism, was conducted to observe the statistical differences in the rheological parameters due to control or GH treatments. The null hypothesis was tested for equivalence in the median ranks of distributions. Statistical significance is demarcated in figures by * for p ≤ 0.05, ** for p ≤ 0.01, *** for p ≤ 0.001, and **** for p ≤ 0.0001.
Results
Raw data
Figure 2 presents typical experimental results from one location in a microfluidic channel. The data are shown for lag times up to 1 s, although tracking data were taken for 25 s. At early lag times, data of each individual particle are linear due to the higher statistical significance. Some data show more non-linearity/curvature at longer lag times, which is to be expected from the longer lag time data having relatively fewer data points to calculate the MSD despite tracking continued up to 25 total seconds.
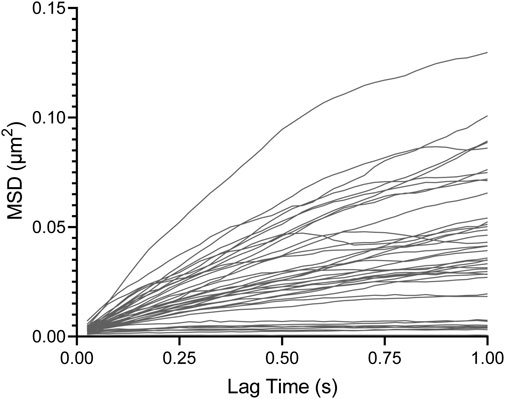
FIGURE 2. Individual particle MSD vs. lag time from WT biofilm sample post-treatment with cellulase.
Effects of GHs on wild-type viscoelasticity
Microrheology results of PAO1 WT biofilms are analyzed first to observe the effects of GHs in the presence of all three basic polysaccharides and to evaluate whether the GHs cause dispersal effect viscoelasticity as well for the baseline, widely studied PAO1 strain. This strain is a well-known producer of Pel and Psl and, although not a mucoid strain, also produces alginate in its EPS [26,27,45] and is often used as a baseline for comparison in many studies.
We first examine the results of the PBS control-treated biofilm. Figure 3 shows that the WT biofilm becomes statistically relatively more elastic and stiffer 2.5 h after the PBS control treatment based on decreasing median α and J. Furthermore, based on the narrowing of the distribution in the box–whisker plot, biofilms become less heterogeneous. These are consistent with the published results [35].
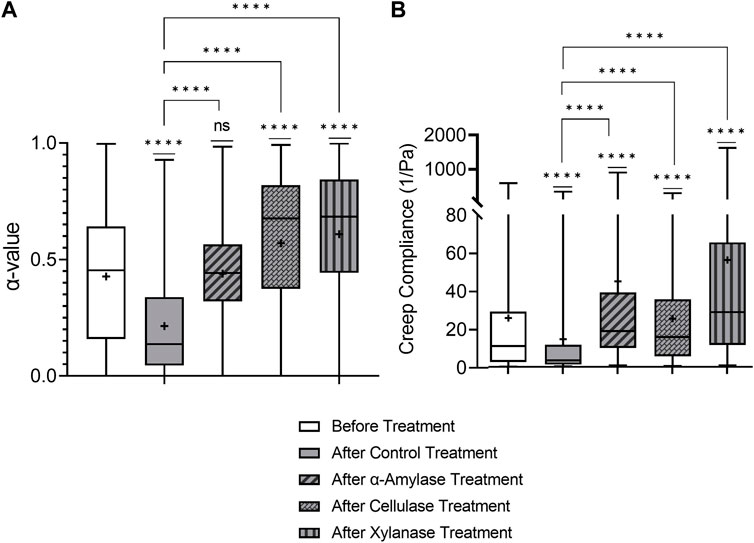
FIGURE 3. Effects on relative viscoelasticity, α, and creep compliance at lag time t = 1 s of PAO1 wild-type biofilms after 48 h and subjected to 2.5 h of PBS control and GH treatments.
All three GH-treated biofilms impacted the viscoelastic properties of PAO1 biofilms at a local level (Figure 3). All GH-treated biofilms exhibited increased median α and J values compared to the PBS control-treated biofilms and were also statistically less elastic and stiff. Additionally, the cellulase- and xylanase-treated biofilms were also statistically weaker than untreated biofilms in terms of median α and J values. Furthermore, all GH-treated biofilms are qualitatively more heterogeneous than the PBS control-treated biofilm.
Unlike previously mentioned bulk experiments on mucoid strains [45], all three GHs impacted the viscoelastic properties of PAO1 biofilms in microrheology; this is likely due to this strain (PAO1) expressing all three polysaccharides in P. aeruginosa biofilms. Importantly, these results indicate that one mechanism of efficacy of GHs as dispersal agents seems correlated with their ability to make biofilms less elastic and stiff based on the aforementioned observations from Figure 3, which shows the efficacy of all three known GHs as dispersal agents in weakening biofilm viscoelasticity.
Furthermore, these results indicate that similar to observations with dispersal experiments, the phenotypic expression of EPS components is quite important to the impact of GHs on biofilms. The three GHs chosen should each attack specific linkages within one or multiple polysaccharides present in the P. aeruginosa EPS; without the appropriate EPS component to act on, GH is ineffective. In this work, due to the strain expressing all three polysaccharides found in P. aeruginosa biofilms, each GH impacted viscoelasticity.
Interestingly, these impacts were not as initially expected, based on the current understanding of EPS impact on viscoelasticity. To better understand exactly how GHs impact viscoelasticity through interaction with EPS polysaccharides, we examined the effects of each GH on biofilms made from strains that were missing major EPS polysaccharide components, which we outline in the following sections.
Amylase and EPS interaction
α-Amylases cleave α-1,4 glycosidic bonds in amylose [59]; similar bonds are found in Pel in P. aeruginosa EPS [41], which is associated with less elastic and stiff biofilms [32,35,40]. Therefore, treatment with
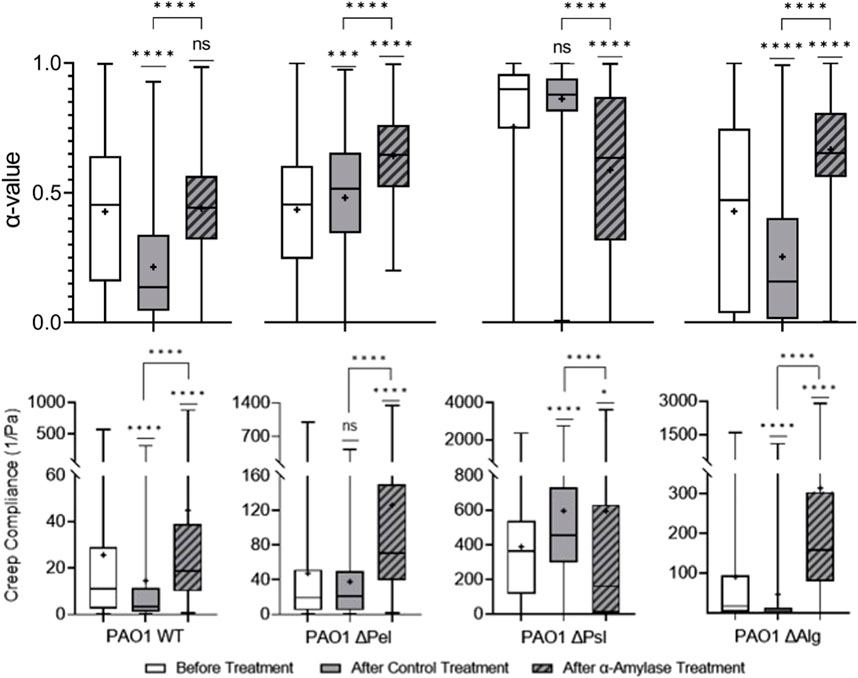
FIGURE 4. Effects of
The Δpel biofilm is both less elastic and stiff compared to the PBS control-treated biofilm. Theoretically,
The Δpsl strain
Finally, in Δalg (and the WT), both Psl and Pel were present, but the α-amylase treatment resulted in biofilms that were statistically less elastic and stiff compared with PBS control-treated biofilms. This is counterintuitive because typical results indicate that films with Psl that lack Pel should be more elastic and stiffer than biofilms with Pel. [32,35,40]. Why cleaving bonds in Pel, which should reduce its impact, has the reverse effect is unclear. Perhaps in this case, cleaving Pel rather than it not existing or being less alters the microstructure in a different manner, which explains the resultant less elastic film.
These results seem to indicate that the α-amylase’s interaction with EPS polysaccharides goes beyond cleaving Pel. Given the similarity between the α-amylase-treated Δpsl in this work and the ΔpelΔpsl in the previous work [35], it appears that the α-amylase is not interacting with alginate. Furthermore, given weakening of all strains that incorporate Psl, it appears that the α-amylase is also cleaving some bonds in Psl and Pel, which may explain its overall utility as a dispersal agent.
Cellulase and EPS interaction
Cellulase cleaves β-1,4 glycosidic bonds found in cellulose; [60] similar bonds are also found in Psl, the major structural component of the biofilm, and alginate, which is important to yield properties of biofilm, in P. aeruginosa EPS [28–30]. In previous studies, it was found that using bulk rheology on alginate-dominant biofilms, cellulase did not impact viscoelasticity [45]. Figure 3 shows that treatment with cellulase made PAO1 biofilms statistically less elastic and stiff compared with PBS control-treated biofilms based on increasing α and J values. Furthermore, the cellulase-treated film had greater heterogeneity.
Cellulase effects were consistent across the biofilms produced by all EPS knockout strains, making them relatively less elastic and stiff (Figure 5). Given that cellulase was expected to target both alginate and Psl, which were present in every strain studied, it is unsurprising that it was effective in impacting every strain.
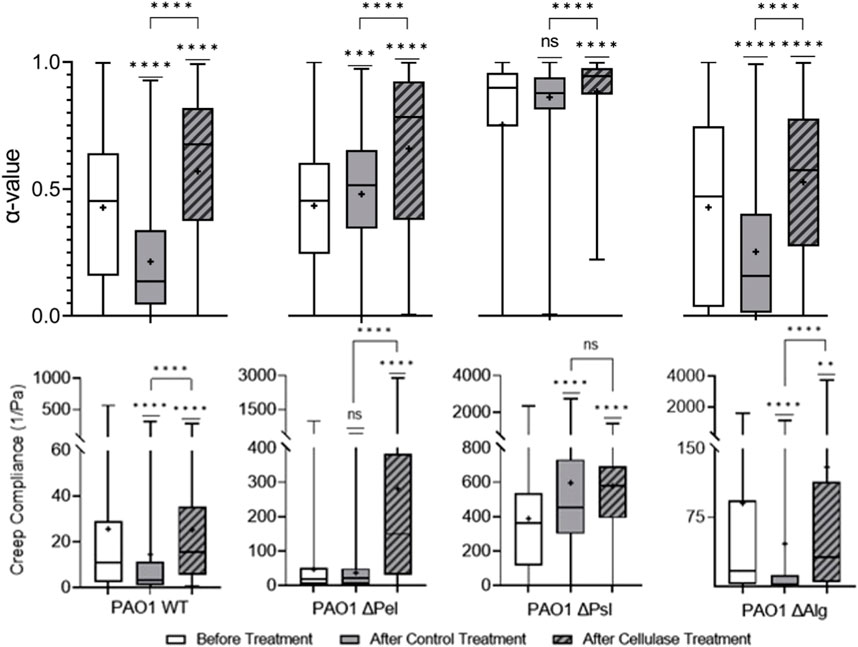
FIGURE 5. Effects of cellulase on relative viscoelasticity, α, and creep compliance at a lag time of t = 1 s on PAO1 biofilms.
Viscoelasticity changes were quite similar for both WT and
Looking at the cellulase-treated Δpsl biofilm, we note that the biofilm is statistically different in terms of the distribution of α compared to the control. However, there is a significant overlap in the distributions, and the means are in fact quite close to one another. Furthermore, the compliances are in fact statistically identical and similar in value. Examining these results, it would appear that the cellulase has minimal impact on the biofilm when Psl is not present.
Looking at the cellulase-treated Δalg biofilm, we see that relative viscosity and compliance both increase in statistically significant ways. In this film, the target of the cellulase is Psl, which as previously stated is the major mechanical component of most biofilms, and when cleaved by the cellulase, the remaining polysaccharide is Pel, and hence we expect a much less elastic and stiff film.
Given these results from the comparison of Δpsl and Δalg strains, it appears that cellulase primarily impact Psl, and, hence, can be seen to primarily weaken the biofilm due to its impact on that polysaccharide.
Xylanase and EPS interaction
Xylanase cleaves β-1,4 glycosidic bonds in xylose, [61] indicating that it should impact Psl, the major structural component of the biofilm, and alginate, which is important to yield properties of biofilm, in P. aeruginosa EPS [28–30]. Figure 3 shows that xylanase treatment made PAO1 biofilms statistically relatively less elastic and stiff compared to PBS control-treated biofilms based on increasing α and J values. It was assumed that this would be primarily due to the interaction with Psl, same as the cellulase.
Starting with the Δpel biofilm, the treatment results in a more elastic and stiffer biofilm. In this case without Pel, assuming that the xylanase has successfully cleaved bonds within Psl and alginate, the resultant biofilm would have very little complete microstructure left, and we would expect a decrease in overall elasticity and stiffness. However, the opposite is observed.
The Δpsl biofilm after xylanase treatment is statistically identical to the untreated biofilm in terms of both relative elasticity and compliance. Furthermore, it is statistically very similar to the PBS control-treated biofilms. This would appear to indicate that the xylanase is primarily interacting with the Psl and not alginate.
The Δalg biofilm, although statistically different in distribution, is slightly more elastic than the PBS control-treated biofilm and slightly less stiff in a statistically significant way. This would indicate very little interaction with the Psl.
It is unclear which EPS component the xylanase is impacting in the biofilm. From the WT results, xylanase clearly impacts viscoelasticity. From the Δpel, the increased elasticity and decreased compliance are counterintuitive to interactions with Psl or alginate. Δpsl results indicate a lack of interaction with alginate, and Δalg results indicate a lack of interaction with Psl. The results of the EPS knockout strains contribute little to clarify how the xylanase interacts with the EPS and further muddy the situation. Figure 6 exhibits that the results are significantly different and much harder to interpret.
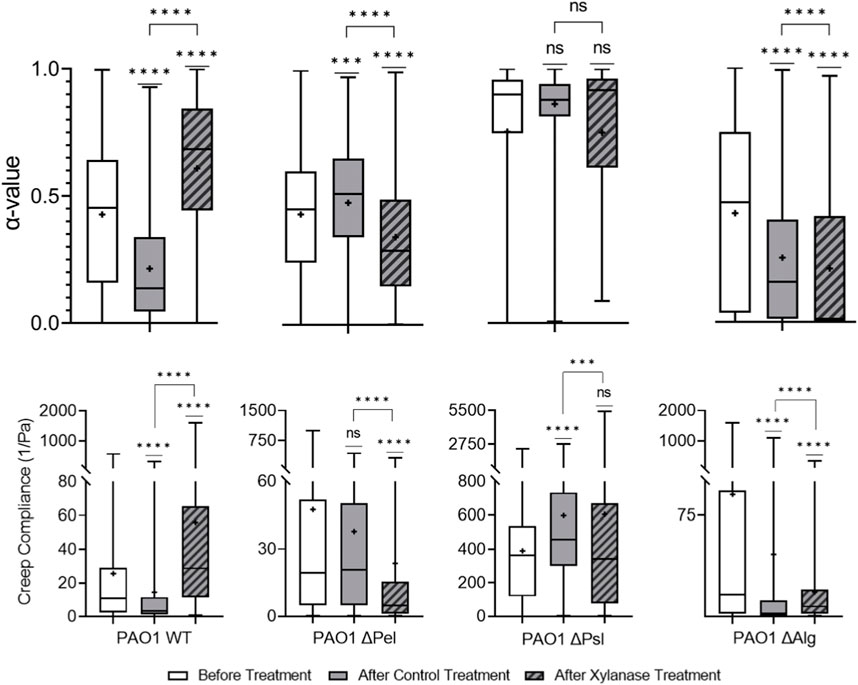
FIGURE 6. Effects of xylanase on relative viscoelasticity, α, and creep compliance at a lag time of t = 1 s on PAO1 biofilms.
Discussion
The impact of GHs on the viscoelasticity of PAO1 biofilms with all EPS polysaccharides present appears to confirm some correlation between the “weakening” of biofilms and a reduction in relative elasticity and increase in compliance of biofilms. Therefore, we can say with some confidence that biofilm reduction in viscoelasticity is correlated with active dispersal caused by these GHs as previously observed. Importantly, this effect seems to only occur when the appropriate EPS polysaccharide that the GH is assumed to impact is present. However, the actual interactions between the GHs and EPS are not as simple as is assumed based on the known glycosidic linkage they attack.
Reviewing the impact of microrheology on EPS knockout strains, we obtain interesting results from amylase impacting films without Pel and xylanase having difficult to interpret interactions with specific EPS components. It has been previously described that xylanase improves cell wall dewatering properties in plants, which might impact EPS water retention and overall biofilm viscoelasticity [62]. However, we might expect such effects to be consistent across all strains if the impact was indeed related to cell walls. It appears possible that both amylase and xylanase are having interactions with unexpected EPS polysaccharides, affecting non-polysaccharide EPS components, or the manner in which the microstructure is altered due to the specific bonds cleaved by the GHs is creating a more complicated microstructure than our tests here can discern without a thorough examination of microstructural changes which are extremely difficult to accomplish. In any case, it is clear that the interactions between the GHs and EPS are more complicated than it would appear which is predicted by the simple understanding of what bonds the GHs impact within specific EPS polysaccharides.
This is further evidenced by the difference between the cellulase and xylanase, which, despite both targeting the same polysaccharides, have different impacts on the WT results (Figure 3). Xylanase seemed to have a larger impact on J than cellulase. It was assumed that similar results on the EPS knockout strains would be observed for the xylanase compared to the cellulase. However, cellulase clearly targeted Psl, whereas xylanase had much more complicated EPS interactions. It is possible that the differences, both in the WT response and in the EPS knockout strains/EPS interactions, here reflect the ability of different GHs to diffuse through the biofilm. Indeed, one of the signatures of biofilms is the creation of microenvironments with altered mechanical, electrical, and chemical signatures that can effectively block different types of cells and antimicrobials. Although the xylanase and cellulase target the same bonds, we may be observing the effects of microenvironments blocking diffusion of the two molecules differently and that may explain the anomalous results between the two.
Conclusion
Examining the results of the microrheology studies in this work, we attempted to evaluate whether the effects of three well-known GH dispersal agents could be attributed to their ability to create less elastic and stiff biofilms at a local level. The results of all three GHs on the WT strain of PAO1 clearly indicate that all GHs create less elastic and stiff biofilms after typical treatments that have been shown to cause effective dispersal in previous studies. These results suggest that the cleaving of the bonds of the various polysaccharides in the EPS creates a less elastic and stiff environment, which is likely a cause for the efficacy of these GHs as dispersal agents.
Cellulase acted as expected and showed particularly strong efficacy in impacting Psl, which is typically attributed to be the most important polysaccharide in the microstructural integrity and mechanical properties of the EPS. The noted efficacy of cellulase in several studies as a dispersal agent seems to correspond to this work identification of its efficacy at affecting Psl polysaccharides within the EPS based on the microrheology results [26,27,63].
α−Amylase, known to attack Pel, was observed to not only clearly impact Pel based on the microrheology results but also impact biofilms without Pel, indicating that it was also potentially interacting with Psl. Given Pel’s lack of importance in biofilm elasticity, it is unsurprising that the α-amylase was observed to impact Psl, which may likely help explain its efficacy.
Finally, xylanase results suggest no consistent interaction with any particular polysaccharide and provide no clear indication of why/how it lowers the viscoelasticity of the WT strains. Although it clearly impacts the viscoelasticity of the WT, the mechanism remains unclear based on these results. The results indicate that the interactions between GHs and the EPS are more complicated than predicted by the specific bonds the GHs are known to interact with, which should be addressed in future studies to better understand what these interactions actually may be.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
BB conducted all experimental work, converted particle-tracking data into rheological data, aided in data interpretation, aided in background research, and aided in writing. GC created initial research plan, aided in data interpretation, aided in background research, and aided in writing. All authors contributed to the article and approved the submitted version.
Acknowledgments
GC gratefully acknowledge support from DOE/NNSA MSIPP Growing STEMS (grant number DOE NA0003988) and Dr. Betsy Snell, Program Manager. The authors would like to thank Dr. Kendra Rumbaugh and her student Rebecca Gabrilska from TTUHSC for their help throughout the work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Jefferson KK. What drives bacteria to produce a biofilm? Fems Microbiol Lett (2004) 236:163–73. doi:10.1111/j.1574-6968.2004.tb09643.x
2. Peterson BW, He Y, Ren YJ, Zerdoum A, Libera MR, Sharma PK, et al. Viscoelasticity of biofilms and their recalcitrance to mechanical and chemical challenges. Fems Microbiol Rev (2015) 39:234–45. doi:10.1093/femsre/fuu008
3. Gloag ES, Fabbri S, Wozniak DJ, Stoodley P. Biofilm mechanics: Implications in infection and survival. Biofilm (2020) 2:100017. doi:10.1016/j.bioflm.2019.100017
4. Muthami JM, Fernandez-Garcia L, Tomas M, Wood TK. What is the fate of the biofilm matrix? Environ Microbiol (2022) 24:4495–9. doi:10.1111/1462-2920.16097
5. Parrilli E, Tutino ML, Marino G. Biofilm as an adaptation strategy to extreme conditions. Rendiconti Lincei-Scienze Fisiche E Naturali (2022) 33:527–36. doi:10.1007/s12210-022-01083-8
6. Sauer K, Stoodley P, Goeres DM, Hall-Stoodley L, Burmolle M, Stewart PS, et al. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat Rev Microbiol (2022) 20:608–20. doi:10.1038/s41579-022-00767-0
7. Bodey GP, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis (1983) 5:279–313. doi:10.1093/clinids/5.2.279
8. Benthall G, Touzel RE, Hind CK, Titball RW, Sutton JM, Thomas RJ, et al. Evaluation of antibiotic efficacy against infections caused by planktonic or biofilm cultures of Pseudomonas aeruginosa and Klebsiella pneumoniae in Galleria mellonella. Int J Antimicrob Agents (2015) 46:538–45. doi:10.1016/j.ijantimicag.2015.07.014
9. Biswas K, Taylor MW, Turner SJ. Successional development of biofilms in moving bed biofilm reactor (MBBR) systems treating municipal wastewater. Appl Microbiol Biotechnol (2014) 98:1429–40. doi:10.1007/s00253-013-5082-8
10. Armbruster CR, Wolter DJ, Mishra M, Hayden HS, Radey MC, Merrihew G, et al. Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. MBio (2016) 7:e00538-16. doi:10.1128/mbio.00538-16
11. Munita JM, Aitken SL, Miller WR, Perez F, Rosa R, Shimose LA, et al. Multicenter evaluation of ceftolozane/tazobactam for serious infections caused by carbapenem-resistant Pseudomonas aeruginosa. Clin Infect Dis (2017) 65:158–61. doi:10.1093/cid/cix014
12. Hunter P. The mob response - the importance of biofilm research for combating chronic diseases and tackling contamination. Embo Rep (2008) 9:314–7. doi:10.1038/embor.2008.43
13. Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat Rev Microbiol (2017) 15:740–55. doi:10.1038/nrmicro.2017.99
14. Vuotto C, Donelli G. Novel treatment strategies for biofilm-based infections. Drugs (2019) 79:1635–55. doi:10.1007/s40265-019-01184-z
15. Jiang Y, Geng MX, Bai LP. Targeting biofilms therapy: Current research strategies and development hurdles. Microorganisms (2020) 8:1222. doi:10.3390/microorganisms8081222
16. Zhang KY, Li X, Yu C, Wang Y. Promising therapeutic strategies against microbial biofilm challenges. Front Cell Infect Microbiol (2020) 10:359. doi:10.3389/fcimb.2020.00359
17. Martin I, Waters V, Grasemann H. Approaches to targeting bacterial biofilms in cystic fibrosis airways. Int J Mol Sci (2021) 22:2155. doi:10.3390/ijms22042155
18. Su YJ, Yrastorza JT, Matis M, Cusick J, Zhao S, Wang GS, et al. Biofilms: Formation, research models, potential targets, and methods for prevention and treatment. Adv Sci (2022) 9:e2203291. doi:10.1002/advs.202203291
19. Mcdougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: Mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol (2012) 10:39–50. doi:10.1038/nrmicro2695
20. Verderosa AD, Totsika M, Fairfull-Smith KE. Bacterial biofilm eradication agents: A current review. Front Chem (2019) 7:824. doi:10.3389/fchem.2019.00824
21. Rumbaugh KP, Sauer K. Biofilm dispersion. Nat Rev Microbiol (2020) 18:571–86. doi:10.1038/s41579-020-0385-0
22. Fleming D, Rumbaugh KP. Approaches to dispersing medical biofilms. Microorganisms (2017) 5:15. doi:10.3390/microorganisms5020015
23. Pinto RM, Soares FA, Reis S, Nunes C, Van Dijck P. Innovative strategies toward the disassembly of the EPS matrix in bacterial biofilms. Front Microbiol (2020) 11:952. doi:10.3389/fmicb.2020.00952
24. Fleming D, Chahin L, Rumbaugh K. Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob Agents Chemother (2017) 61:e01998-16. doi:10.1128/AAC.01998-16
25. Fleming D, Rumbaugh K. The consequences of biofilm dispersal on the host. Scientific Rep (2018) 8:10738. doi:10.1038/s41598-018-29121-2
26. Redman WK, Welch GS, Rumbaugh KP. Differential efficacy of glycoside hydrolases to disperse biofilms. Front Cell Infect Microbiol (2020) 10:379. doi:10.3389/fcimb.2020.00379
27. Redman WK, Welch GS, Williams AC, Damron AJ, Northcut WO, Rumbaugh KP. Efficacy and safety of biofilm dispersal by glycoside hydrolases in wounds. Biofilm (2021) 3:100061. doi:10.1016/j.bioflm.2021.100061
28. Franklin MJ, Nivens DE, Weadge JT, Howell PL. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol (2011) 2:167. doi:10.3389/fmicb.2011.00167
29. Ghafoor A, Hay ID, Rehm BHA. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol (2011) 77:5238–46. doi:10.1128/aem.00637-11
30. Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. Fems Microbiol Rev (2012) 36:893–916. doi:10.1111/j.1574-6976.2011.00322.x
31. Wloka M, Rehage H, Flemming HC, Wingender J. Structure and rheological behaviour of the extracellular polymeric substance network of mucoid Pseudomonas aeruginosa biofilms. Biofilms (2005) 2:275–83. doi:10.1017/s1479050506002031
32. Kovach K, Davis-Fields M, Irie Y, Jain K, Doorwar S, Vuong K, et al. Evolutionary adaptations of biofilms infecting cystic fibrosis lungs promote mechanical toughness by adjusting polysaccharide production. Biofilms and Microbiomes (2017) 3:1. doi:10.1038/s41522-016-0007-9
33. Murgia X, Kany AM, Herr C, Ho D-K, De Rossi C, Bals R, et al. Micro-rheological properties of lung homogenates correlate with infection severity in a mouse model of Pseudomonas aeruginosa lung infection. Scientific Rep (2020) 10:16502. doi:10.1038/s41598-020-73459-5
34. Wells M, Schneider R, Bhattarai B, Currie H, Chavez B, Christopher G, et al. Perspective: the viscoelastic properties of biofilm infections and mechanical interactions with phagocytic immune cells. Front Cell Infect Microbiol (2023) 13:1102199. doi:10.3389/fcimb.2023.1102199
35. Rahman MU, Flemming D, Sinha I, Rumbaugh KP, Gordon V, Christopher G. Effect of Collagen and EPS components on the viscoelasticity of Pseudomonas aeruginosa biofilms. Soft Matter (2021) 17:6225–37. doi:10.1039/d1sm00463h
36. Byrd MS, Sadovskaya I, Vinogradov E, Lu HP, Sprinkle AB, Richardson SH, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol (2009) 73:622–38. doi:10.1111/j.1365-2958.2009.06795.x
37. Ma LM, Conover M, Lu HP, Parsek MR, Bayles K, Wozniak DJ. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. Plos Pathog (2009) 5:e1000354. doi:10.1371/journal.ppat.1000354
38. Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol (2012) 14:1913–28. doi:10.1111/j.1462-2920.2011.02657.x
39. Irie Y, Roberts AEL, Kragh KN, Gordon VD, Hutchison J, Allen RJ, et al. The Pseudomonas aeruginosa PSL polysaccharide is a social but noncheatable trait in biofilms. Mbio (2017) 8:e00374-17. doi:10.1128/mbio.00374-17
40. Chew SC, Kundukad B, Seviour T, Van Der Maarel JRC, Yang L, Rice SA, et al. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. mBio (2014) 5:e01536–14. doi:10.1128/mbio.01536-14
41. Le Mauff F, Razvi E, Reichhardt C, Sivarajah P, Parsek MR, Howell PL, et al. The Pel polysaccharide is predominantly composed of a dimeric repeat of α-1,4 linked galactosamine and N-acetylgalactosamine. Commun Biol (2022) 5:502. doi:10.1038/s42003-022-03453-2
42. Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci (2015) 112:11353–8. doi:10.1073/pnas.1503058112
43. Limoli DH, Jones CJ, Wozniak DJ. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr (2015) 3. doi:10.1128/microbiolspec.mb-0011-2014
44. Jennings LK, Dreifus JE, Reichhardt C, Storek KM, Secor PR, Wozniak DJ, et al. Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cel Rep (2021) 34:108782. doi:10.1016/j.celrep.2021.108782
45. Kovach KN, Fleming D, Wells MJ, Rumbaugh KP, Gordon VD. Specific disruption of established Pseudomonas aeruginosa biofilms using polymer-attacking enzymes. Langmuir (2020) 36:1585–95. doi:10.1021/acs.langmuir.9b02188
46. Hart JW, Waigh TA, Lu JR, Roberts IS. Microrheology and spatial heterogeneity of Staphylococcus aureus biofilms modulated by hydrodynamic shear and biofilm-degrading enzymes. Langmuir (2019) 35:3553–61. doi:10.1021/acs.langmuir.8b04252
47. Powell LC, Abdulkarim M, Stokniene J, Yang QE, Walsh TR, Hill KE, et al. Quantifying the effects of antibiotic treatment on the extracellular polymer network of antimicrobial resistant and sensitive biofilms using multiple particle tracking. Npj Biofilms and Microbiomes (2021) 7:13. doi:10.1038/s41522-020-00172-6
48. Mcdonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJ, et al. Fabrication of microfluidic systems in poly (dimethylsiloxane). ELECTROPHORESIS (2000) 21:27–40. doi:10.1002/(sici)1522-2683(20000101)21:1<27::aid-elps27>3.0.co;2-c
49. Bilings N, Birjiniuk A, Samad T, Doyle P, Ribbeck K. Material properties of biofilms–key methods for understanding permeability and mechanics. Rep Prog Phys (2016) 78:33. doi:10.1088/0034-4885/78/3/036601
50. Savin T, Doyle PS. Statistical and sampling issues when using multiple particle tracking. Phys Rev E (2007) 76:021501. doi:10.1103/physreve.76.021501
51. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods (2012) 9:676–82. doi:10.1038/nmeth.2019
52. Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, et al. TrackMate: An open and extensible platform for single-particle tracking. Methods (2017) 115:80–90. doi:10.1016/j.ymeth.2016.09.016
53. Tarantino N, Tinevez J-Y, Crowell EF, Boisson B, Henriques R, Mhlanga M, et al. TNF and IL-1 exhibit distinct ubiquitin requirements for inducing NEMO–IKK supramolecular structures. J Cel Biol (2014) 204:231–45. doi:10.1083/jcb.201307172
54. Michalet X. Mean square displacement analysis of single-particle trajectories with localization error: brownian motion in an isotropic medium. Phys Rev E (2010) 82:041914. doi:10.1103/physreve.82.041914
55. Rogers SS, Van Der Walle C, Waigh TA. Microrheology of bacterial biofilms in vitro: Staphylococcus aureus and Pseudomonas aeruginosa. Langmuir (2008) 24:13549–55. doi:10.1021/la802442d
57. Joyner K, Yang S, Duncan GA. Microrheology for biomaterial design. Apl Bioeng (2020) 4:041508. doi:10.1063/5.0013707
58. Kowalczyk A, Oelschlaeger C, Willenbacher N. Tracking errors in 2D multiple particle tracking microrheology. Meas Sci Tech (2015) 26:015302. doi:10.1088/0957-0233/26/1/015302
59. Sivaramakrishnan S, Gangadharan D, Nampoothiri KM, Soccol CR, Pandey A. alpha-amylases from microbial sources - an overview on recent developments. Food Tech Biotechnol (2006) 44:173–84.
60. Ejaz U, Sohail M, Ghanemi A. Cellulases: from bioactivity to a variety of industrial applications. Biomimetics (2021) 6:44. doi:10.3390/biomimetics6030044
61. Bhardwaj N, Kumar B, Verma P. A detailed overview of xylanases: an emerging biomolecule for current and future prospective. Bioresources and Bioprocessing (2019) 6:40. doi:10.1186/s40643-019-0276-2
62. Torres CE, Negro C, Fuente E, Blanco A. Enzymatic approaches in paper industry for pulp refining and biofilm control. Appl Microbiol Biotechnol (2012) 96:327–44. doi:10.1007/s00253-012-4345-0
Keywords: biofilm, microrheology, glycoside hydrolases, dispersal, Pseudomonas aeruginosa
Citation: Bhattarai B and Christopher GF (2023) Examining the role of glycoside hydrolases in local rheology of Pseudomonas aeruginosa biofilms. Front. Phys. 11:1239632. doi: 10.3389/fphy.2023.1239632
Received: 13 June 2023; Accepted: 21 August 2023;
Published: 07 September 2023.
Edited by:
Kelly Schultz, Lehigh University, United StatesReviewed by:
Angela Brown, Lehigh University, United StatesJing Yan, Yale University, United States
Jan Vermant, ETH Zürich, Switzerland
Copyright © 2023 Bhattarai and Christopher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gordon F. Christopher, R29yZG9uLmNocmlzdG9waGVyQHR0dS5lZHU=
 Bikash Bhattarai
Bikash Bhattarai Gordon F. Christopher
Gordon F. Christopher