- 1National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2National Cancer Center/National Clinical Research Center for Cancer/Hebei Cancer Hospital, Chinese Academy of Medical Sciences, Langfang, China
- 3Zhejiang Cancer Hospital, University of Chinese Academy of Sciences, Hangzhou, China
- 4State Key Laboratory of Oncology in South China, United Laboratory of Frontier Radiotherapy Technology of Sun Yat-sen University, Chinese Academy of Sciences Ion Medical Technology Co., Ltd., Sun Yat-sen University Cancer Center, Guangzhou, China
Purpose: Convolutional neural networks (CNNs) offer a promising approach to automating organ segmentation in radiotherapy. However, variations of segmentation protocols made by different medical centers may induce a well-trained CNN model in one center and may not perform well in other centers. In this study, we proposed a transfer learning method to improve the performance of deep-learning based segmentation models among different medical centers using nasopharyngeal cancer (NPC) data.
Methods: The NPC data included 300 cases (S_Train) from one institution (the source center) and 60 cases from another (the target center), divided into a training set of 50 cases (T_Train) and a test set of 10 target cases (T_Test). A ResNet CNN architecture was developed with 103 layers. We first trained Model_S and Model_T from scratch with the datasets S_Train and T_train, respectively. Transfer learning was then used to train Model_ST by fine-tuning the last 10 layers of Model_S with images from T_Train. We also investigated the effect of the numbers of re-trained layers on the performance. The performance of each model was evaluated using the dice similarity coefficient, and it was used as the evaluation metrics. We compared the dice similarity coefficient value using the three different models (Model_S, Model_T, and Model_ST).
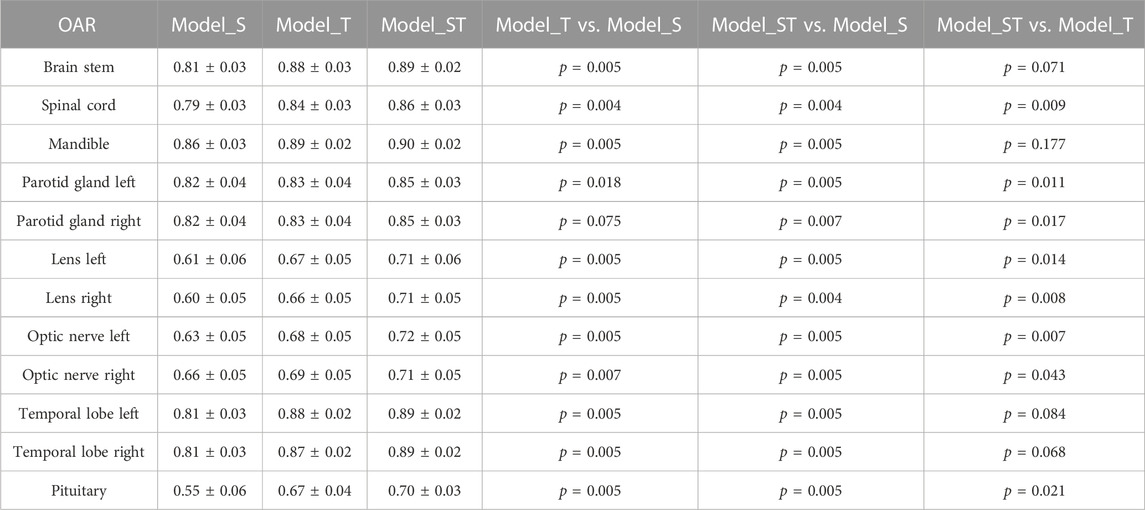
Results: When Model_S, Model_T, and Model_ST were applied to the T_Test dataset, the transfer learning (Model_ST) had the best performance. Compared with Model_S, the p-values of all organs at risk were less than 0.05. Compared with Model_T, the p-values of most organs at risk were less than 0.05, but there was no significant statistical difference in Model_ST for the brain stem (p = 0.071), mandible (p = 0.177), left temporal lobes (p = 0.084), and right temporal lobes (p = 0.068). Although there was no statistical difference for these organs, the mean accuracy of Model_ST was higher than that of Model_T. The proposed transfer learning can reduce the training time by up to 33%.
Conclusion: Transfer learning can improve organ segmentation for NPC by adapting a previously trained CNN model to a new image domain, reducing the training time and saving physicians from labeling a large number of contours.
1 Introduction
Segmentation of the organs at risk (OARs) plays a crucial role in modern radiation therapy. However, manual segmentation is time consuming. Over recent decades, a need of robust tools has emerged for automatic segmentation. The convolutional neural network (CNN) method is a type of supervised deep learning methods, which has demonstrated outstanding performance in various tasks, including organ and tumor segmentation for several disease sites [1–6]. There have been various notable developments in the use of deep learning methods for organ segmentation [7–10]. These methods have obviously improved the precision of automatic segmentation, and the performances of CNN models may achieve the level of experienced physicians for most OARs’ segmentation [11, 12].
Although most radiotherapy centers have made their own segmentation protocol according to the published report (e.g., ICRU and QUANTAC), there may still be some inter- and intra-observer variation in manual delineations [13, 14]. Some inconsistency in interpreting the boundary of OARs may occur among different centers, mainly because of differences in the expertise level and preferences for prescription and treatment planning. Due to this reason, the well-trained CNN model using the data from one medical center tends not to generalize the segmentation well to cases from medical centers compared to the one that supplied the training dataset [15, 16]. The specific deep learning models may need to be established for each medical center.
In general, training a deep learning model for image processing often requires a sufficiently large training dataset to tune millions of free parameters. For segmentation tasks in radiotherapy, the training data include not only a great number of images but also the corresponding manual contours. When retraining a CNN model for a new domain, expert labeling requires to be performed manually by the physician. This is very time consuming and not always possible.
When sufficiently large, standard datasets are not available for training the CNN, and the training data may not encompass a wide spectrum of the population; as a result, the performance of the trained CNN model may not be robust. Robustness can be improved by exploiting a technique known as transfer learning [17–19]. Xu et al. [20] applied the pre-trained model from ImageNet to segment brain 3D MR images. Zheng [21] applied shape learned from a different modality to improve the segmentation accuracy on the target modality. Van Opbroek et al. [22] presented four transfer classifiers to deal with inductive transfer learning for MRI segmentation. The team [23] also performed image weighting with kernel learning to reduce differences between the training and test data.
Some details of protocol for contouring each OAR are not exactly the same across medical centers; a well-trained CNN model from one center may not perform well for another center. Transfer learning might be a potential tool in radiotherapy for OAR segmentation across different centers which has not been adequately reported. This study aimed to train specific and accurate organ segmentation models using the NPC data for one center (the target institution) with transfer learning using only a small amount of training data based on a CNN model trained with a large dataset from another center (the source institution).
2 Materials and methods
2.1 Patient datasets
A retrospective study was conducted using the datasets from two centers, one was the source institution (our institution) and the other was the target institution. All the patients from the two institutes included in this study were diagnosed with nasopharyngeal carcinoma (NPC) who have received intensity-modulated radiation therapy (IMRT) or volumetric modulated arc therapy (VMAT). The datasets included 300 cases from the source institution and 60 cases from the target institution. The anonymized DICOM files including CT images and OARs’ contours were collected.
The simulation CT scans from the source institution were acquired on a SOMATOM Definition AS 40 (Siemens Healthcare, Forchheim, Germany) or Brilliance CT Big Bore (Philips Healthcare, Best, the Netherlands) system with contrast enhancement. Acquisition parameters were as follows: voltage: 120 kV; exposure: 270 mAs (Siemens) or 240 mAs (Philips); pitch: 1 (Siemens) or 0.938 (Philips); reconstruction filter: B31s kernel (Siemens) or B kernel (Philips); matrix size: 512 × 512; pixel size: 0.96–1.27 mm; and slice thickness: 3.0 mm. The CT images from the target institution were acquired on a LightSpeed RT (GE Medical Systems, Waukesha, WI, United States) or Brilliance CT Big Bore (Philips Healthcare, Best, the Netherlands) system with contrast enhancement. Acquisition parameters were as follows: voltage: 120 kV; exposure: 190 mAs (GE) or 200 mAs (Philips); pitch: 1.5 (GE) or 0.813–0.938 (Philips); reconstruction filter: STANDARD kernel (GE) or UB kernel (Philips); matrix size: 512 × 512; pixel size: 0.98–1.19 mm; and slice thickness: 3.0 mm. All the enrolled cases were real clinical cases.
In total, 12 OARs were involved in this study, including the brain stem, spinal cord, mandible, pituitary, bilateral parotid gland, bilateral lens, bilateral optic nerve, and bilateral temporal lobe. Each institution used the contours drawn according to its own protocol as the ground truth. For each institution, the contours of each OAR were first drawn by its own physician in charge and then checked and confirmed by its own radiotherapy expert group for quality control.
2.2 CNN architecture for transfer learning
In this study, we developed a CNN architecture for transfer learning with two steps.
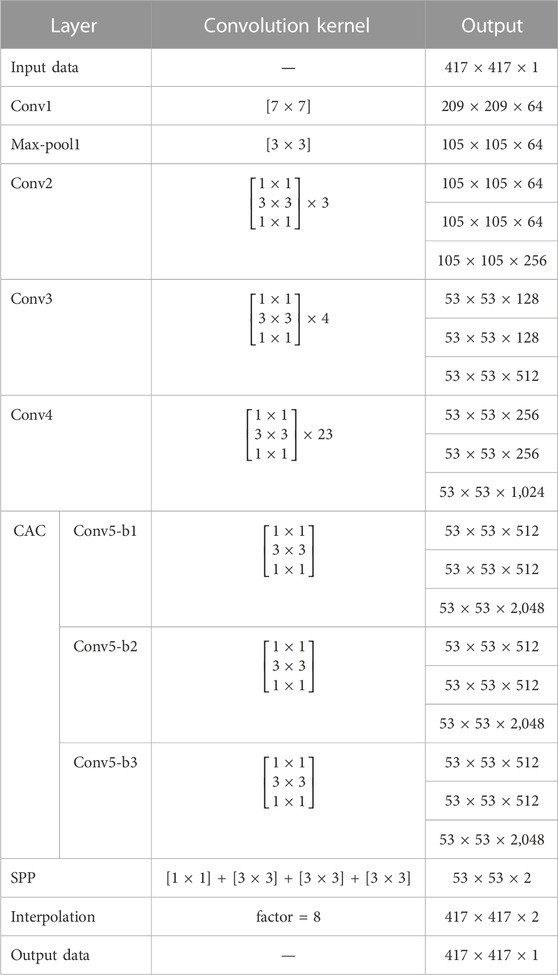
The first step was to develop a CNN model using the dataset from the source institution. There are many types of deep neural networks available for medical segmentation. The CNN we used was modified based on the ResNet-101 CNN [24, 25] with 103 convolution layers. The network architecture is illustrated in Figure 1, and details of the architecture are given in Table 1. The input to the CNN was the dataset of CT images, and its output was the corresponding segmentation probability maps for the organ. It has multiple deeper bottleneck architectures (DBAs), each of which consisted of three convolutional layers of 1 × 1, 3 × 3, and 1 × 1 and a connection. There were 3, 4, and 23 DBAs in Conv2, Conv3, and Conv4, respectively.

FIGURE 1. Architecture of the segmentation network for transfer learning. The input to the CNN was the dataset of CT images from the source institution, and its output was the corresponding segmentation probability maps for the organ. For transfer learning, the layers that were re-trained are shown in green and those not re-trained are shown in gray.
The second step was to fine-tune the existed CNN model (from the source institute) using a small dataset from the target center only by re-training the deeper layers. The parameters of Conv1 to Conv4 were locked, and transfer learning was implemented by re-training the last 10 layers (Conv5-b1, Conv5-b2, Conv5-b3, and SPP) using images and labels from the target center.
2.3 Setup of training
The model training and testing were implemented using the Caffe platform [26] on a NVIDIA TITAN XP GPU with an Intel® Core i7 processor (3.4 GHz). Backpropagation with the stochastic gradient descent (SGD) algorithm implementation in Caffe was used for parameter optimization. The learning rate policy was set to “poly” with a base learning rate of 0.00025 and power of 0.9. The batch size, momentum, and weight decay were set to 1, 0.9, and 0.0005, respectively. The maximum number of iterations was 200 K. The original images were 512 × 512. To avoid over-fitting, we applied general methods for data augmentation, including random scaling between 0.5 and 1.5 (scaling factors: 0.5, 0.75, 1, 1.25, and 1.5), random rotation (between −10° and 10°), and random cropping (crop size: 417 × 417). In detail, the original images were randomly resized with a factor of 0.5, 0.75, 1, 1.25, and 1.5 and then randomly rotated between −10° and 10°. The crop size was selected mainly based on the size of the image. Since the cropped images would be fed into the network, it is necessary to unify their size. Finally, all the training images were cropped to 417 × 417.
2.4 Experiments
First, we trained and validated an original CNN model (Model_S) with the 300 cases in the source center dataset for 12 OARs separately. Second, the 60 cases from the target center were randomly divided into a training set of 50 cases (T_Train) and a test set of 10 cases (T_Test). We then used the 50 cases (T_Train) in the target center dataset to train another CNN model (Model_T) from scratch for 12 OARs separately. Third, to build the transfer learning model (Model_ST), we transferred the learned weights from Model_S to the new model, locked most of the shallower layers, and fine-tuned the remaining deeper layers with the T_Train training dataset. Finally, the remaining 10 cases of the target center dataset (T_Test) were used as the test set for evaluation.
The dice similarity coefficient (DSC) [27] was used as the evaluation metrics. We compared the DSC value using the three different models (Model_S, Model_T, and Model_ST). Data were analyzed using SPSS version 24. The Wilcoxon signed-rank test for the paired samples non-parametric test was performed. A p-value of <0.05 was considered to be statistically significant. The training times for each of the models and configurations were recorded to compare the efficiency of the transfer learning.
3 Results
3.1 Evaluation of segmentation accuracy with transfer learning
For each organ, Model_S and Model_T were trained with the original network from starch, while Model_ST was trained with the proposed transfer learning. The comparisons of Model_S, Model_T, and Model_ST on the testing sets (T_Test) are shown in Table 2. Model_S, trained with the images from the source center dataset, performed worst (DSC = 0.74 ± 0.12) on the 10 T_Test cases from the target center. Model_T, trained with the T_Train training images from the target center dataset, had higher DSC (DSC = 0.78 ± 0.10) than Model_S when applied to T_Test. The p-values were less than 0.05 in all organs except the right parotid gland (p = 0.075). The transfer learning model Model_ST, fine-tuned using T_Train, had the best performance (DSC = 0.81 ± 0.09) when applied to T_Test. Compared with Model_S, the p-values of all OARs were less than 0.05. Compared with Model_T, the p values of most OARs were less than 0.05, but there was no significant statistical difference in Model_ST for the brain stem (p = 0.071), mandible (p = 0.177), left temporal lobes (p = 0.084), and right temporal lobes (p = 0.068). Although there was no statistical difference for these organs, the mean accuracy of Model_ST was higher than that of Model_T. The average DSC for all the OARs was increased by 0.03 with Model_ST compared to that with Model_T.
Figure 2 visualizes some typical examples of the contours segmented with Model_T and the proposed Model_ST. By comparison, it can be found that Model_ST had better performance in general.
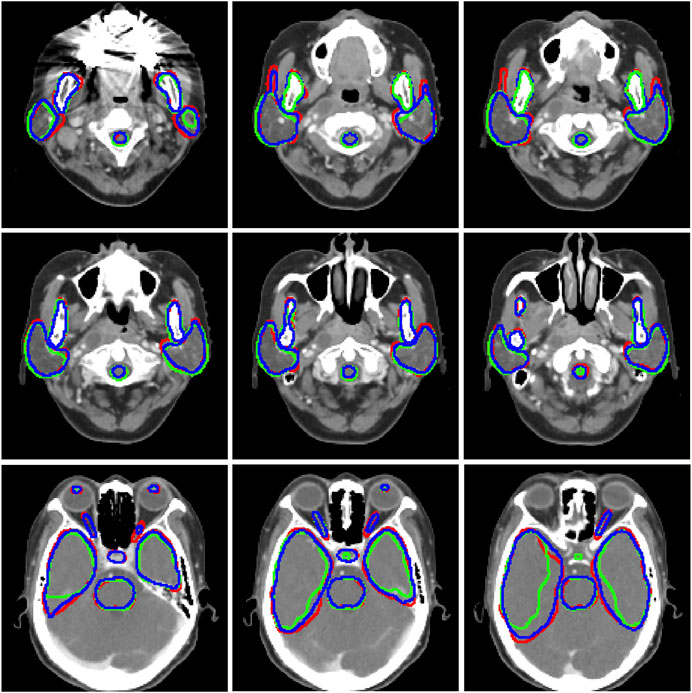
FIGURE 2. Segmentation results for tested cases. Red, ground truth; green, Model_T; and b, the proposed Model_ST.
3.2 Contouring and training time
The time for contouring one slice was 0.15 s for all the models used in this study because the same architecture was used to train each model. However, the training time varied significantly with the number of parameters to tune. We used 200 K iterations to train each model. It took ∼100 h to train Model_T with the original network, while the training time was reduced by 33% for the transfer learning.
4 Discussion
Deep learning networks are increasingly used in radiotherapy to delineate OARs, achieving promising results. However, when a model built in one medical center is applied in another, its performance tends to be poorer. This prevents a single model from being used across multiple medical centers. The results of this study showed that the proposed transfer learning method could improve the automatic segmentation of OARs by adapting a previously trained CNN model to a new domain. In addition, transfer learning could save up to 33% of the training time with the same training iterations.
For a task such as OAR segmentation, using data from the target center to fine-tune just the last few layers of a model trained using source center datasets could potentially yield a good performance. In addition, it was difficult to re-train all the free parameters of the network because of the limited amount of training data. For the proposed transfer learning, a large number of samples (300 cases) have been used to tune the parameters of the network, and then, the shallower layers were locked. The smaller number of parameters to be fine-tuned avoided over-fitting and improved the robustness of the segmentation model.
Training a CNN segmentation model usually needs large-scale image data with labels. The important findings of this study are, therefore, of great relevance for clinical practice. Transfer learning can improve organ segmentation to a high level with only a small amount of training data, which can reduce the need for time-consuming human interventions and improve the efficiency of model training.
There are several limitations in this study that need to be addressed in the future. First, the transfer learning in this study was based on the ResNet network. This may be a limitation because there are many other possible networks and OARs that were not considered. The network we chose has previously been demonstrated to be state of the art. Second, due to the limitation of data security and privacy, there were only 60 cases from the target domain. The amount of training set could not be too small, so we randomly selected only 10 from 60 cases as the testing set to verify the proposed method. Although the results show that it has significant improvement in all 10 testing cases, the small size of the testing set is another limitation of this study. Third, we demonstrate the power of the developed method only using the NPC data from two centers, while including more centers and more tumor sites to test our method would be more convincing. Finally, different scanners, scanning parameters, protocols, physicians, or contouring platforms may affect the performance of model on cases from different medical centers. It will be further explored to see which factor is most important in future research.
5 Conclusion
In this study, a transfer learning method was established to train specific organ segmentation models for one center using only a small amount of data based on a CNN model trained with a large dataset from another center. The findings demonstrated that the established method can improve predictive accuracy by adapting a previously trained deep learning model to a new image domain. This approach could save the training times and reduce the need for physicians to label a large number of contours. These findings suggest an approach for training segmentation models across medical centers.
Data availability statement
The datasets generated and/or analyzed during the current study are not publicly available due to data security requirement of our hospital. Requests to access the datasets should be directed to ZGFpX2ppYW5yb25nQGNpY2Ftcy5hYy5jbg==.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Cancer Hospital, Chinese Academy of Medical Sciences. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
All authors discussed and conceived the study design. XC wrote the programs, performed data analysis, and drafted the manuscript. JZu, YY, and JZg helped to collect the data. XC and JZu analyzed and interpreted the patients’ data. KM, JY, MC, and JD guided the study and participated in discussions and the preparation of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Beijing Natural Science Foundation (7222149), CAMS Innovation Fund for Medical Sciences (2021-I2M-C&T-A-016), National Natural Science Foundation of China (12005302 and 11875320), the Beijing Nova Program (Z201100006820058), and Beijing Hope Run Special Fund of Cancer Foundation of China (LC2021A15).
Conflict of interest
MC was employed by Chinese Academy of Sciences Ion Medical Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CAC, cascaded atrous convolution; CNN, convolutional neural networks; CT, computed tomography; DSC, dice similarity coefficient; OARs, organs at risk; and SPP, module and a spatial pyramid pooling.
References
1. Ibragimov B, Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys (2017) 44:547–57. doi:10.1002/mp.12045
2. Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys (2017) 44:6377–89. doi:10.1002/mp.12602
3. Cardenas CE, McCarroll RE, Court LE, Elgohari BA, Elhalawani H, Fuller CD, et al. Deep learning algorithm for auto-delineation of high-risk oropharyngeal clinical target volumes with built-in dice similarity coefficient parameter optimization function. Int J Radiat Oncol Biol Phys (2018) 101:468–78. doi:10.1016/j.ijrobp.2018.01.114
4. Tong N, Gou S, Yang S, Ruan D, Sheng K. Fully automatic multi-organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med Phys (2018) 45:4558–67. doi:10.1002/mp.13147
5. Zhu W, Huang Y, Zeng L, Chen X, Liu Y, Qian Z, et al. AnatomyNet: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med Phys (2019) 46:576–89. doi:10.1002/mp.13300
6. Wang Y, Zhou Y, Shen W, Park S, Fishman EK, Yuille AL. Abdominal multi-organ segmentation with organ-attention networks and statistical fusion. Med Image Anal (2019) 55:88–102. doi:10.1016/j.media.2019.04.005
7. Jin D, Guo D, Ho TY, Harrison AP, Xiao J, Tseng CK, et al. Accurate esophageal gross tumor volume segmentation in pet/ct using two-stream chained 3d deep network fusion (2019). Available at: https://arxiv.org/pdf/1909.01524.pdf.
8. Men K, Geng H, Cheng C, Zhong H, Huang M, Fan Y, et al. Technical Note: More accurate and efficient segmentation of organs-at-risk in radiotherapy with convolutional neural networks cascades. Med Phys (2019) 46:286–92. doi:10.1002/mp.13296
9. Gao Y, Huang R, Chen M, Wang Z, Deng J, Chen Y, et al. FocusNet: Imbalanced large and small organ segmentation with an end-to-end deep neural network for head and neck CT images. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; October 13–17, 2019; Shenzhen, China (2019).
10. Cardenas CE, Anderson BM, Aristophanous M, Yang J, Rhee DJ, McCarroll RE, et al. Auto-delineation of oropharyngeal clinical target volumes using 3D convolutional neural networks. Phys Med Biol (2018) 63:215026. doi:10.1088/1361-6560/aae8a9
11. Lustberg T, van Soest J, Gooding M, Peressutti D, Aljabar P, van der Stoep J, et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol (2018) 126:312–7. doi:10.1016/j.radonc.2017.11.012
12. Nikolov S, Blackwell S, Mendes R, Fauw JD, Ronneberger O. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy (2018). ArXiv.
13. Samaneh K, Anjali B, Dan N, Sarah MG, Raquibul H, Jiang S, et al. Segmentation of the prostate and organs at risk in male pelvic CT images using deep learning (2019). ArXiv.
14. Steenbakkers RJ, Duppen JC, Fitton I, Deurloo KE, Zijp L, Uitterhoeve AL, et al. Observer variation in target volume delineation of lung cancer related to radiation oncologist-computer interaction: A 'Big brother' evaluation. Radiother Oncol (2005) 77:182–90. doi:10.1016/j.radonc.2005.09.017
15. AlBadawy EA, Saha A, Mazurowski MA. Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing. Med Phys (2018) 45:1150–8. doi:10.1002/mp.12752
16. Perone CS, Ballester P, Barros RC, Cohen-Adad J. Unsupervised domain adaptation for medical imaging segmentation with self-ensembling. Neuroimage (2019) 194:1–11. doi:10.1016/j.neuroimage.2019.03.026
17. Shao L, Zhu F, Li X. Transfer learning for visual categorization: A survey. IEEE Trans Neural Netw Learn Syst (2015) 26:1019–34. doi:10.1109/tnnls.2014.2330900
19. Ravishankar H, Sudhakar P, Venkataramani R, Thiruvenkadam S, Vaidya V. Understanding the mechanisms of deep transfer learning for medical images (2017). Available at: https://arxiv.org/pdf/1704.06040v1.pdf.
20. Xu Y, Géraud T, Bloch I. From neonatal to adult brain MR image segmentation in a few seconds using 3D-like fully convolutional network and transfer learning. In: IEEE International Conference on Image Processing (ICIP); 2017; Beijing, China (2017). p. 4417–4421. doi:10.1109/ICIP.2017.8297117
21. Zheng Y. Cross-modality medical image detection and segmentation by transfer learning of shapel priors. In: IEEE 12th International Symposium on Biomedical Imaging (ISBI); 2015; Brooklyn, NY (2015). p. 424–427. doi:10.1109/ISBI.2015.7163902
22. van Opbroek A, Ikram MA, Vernooij MW, de Bruijne M. Transfer learning improves supervised image segmentation across imaging protocols. IEEE Trans Med Imaging (2015) 34:1018–30. doi:10.1109/tmi.2014.2366792
23. Van Opbroek A, Achterberg HC, Vernooij MW, De Bruijne M. Transfer learning for image segmentation by combining image weighting and kernel learning. IEEE Trans Med Imaging (2019) 38:213–24. doi:10.1109/tmi.2018.2859478
24. Men K, Boimel P, Janopaul-Naylor J, Zhong H, Huang M, Geng H, et al. Cascaded atrous convolution and spatial pyramid pooling for more accurate tumor target segmentation for rectal cancer radiotherapy. Phys Med Biol (2018) 63:185016. doi:10.1088/1361-6560/aada6c
25. He K, Zhang X, Ren S, Sun J. IEEE conference on computer vision & pattern recognition (2016). (unpublished).
26. Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, et al. Caffe: Convolutional architecture for fast feature embedding (2014). p. 1408. ArXiv.
Keywords: radiotherapy, automatic segmentation, transfer learning, deep learning, small training samples
Citation: Chen X, Zhu J, Yang Y, Zhang J, Men K, Yi J, Chen M and Dai J (2023) Investigating transfer learning to improve the deep-learning-based segmentation of organs at risk among different medical centers for nasopharyngeal carcinoma. Front. Phys. 11:1147900. doi: 10.3389/fphy.2023.1147900
Received: 19 January 2023; Accepted: 27 June 2023;
Published: 12 July 2023.
Edited by:
Yinqiang Zheng, The University of Tokyo, JapanReviewed by:
Wazir Muhammad, Florida Atlantic University, United StatesHua Wang, Victoria University, Australia
Copyright © 2023 Chen, Zhu, Yang, Zhang, Men, Yi, Chen and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianrong Dai, ZGFpX2ppYW5yb25nQGNpY2Ftcy5hYy5jbg==; Ming Chen, Y2hlbm1pbmdAc3lzdWNjLm9yZy5jbg==
 Xinyuan Chen
Xinyuan Chen Ji Zhu
Ji Zhu Yiwei Yang
Yiwei Yang Jie Zhang3
Jie Zhang3 Kuo Men
Kuo Men Junlin Yi
Junlin Yi Ming Chen
Ming Chen Jianrong Dai
Jianrong Dai