- 1Department of Neurosurgery, The First Affiliated Hospital of Fujian Medical University, Fujian Key Lab of Individualized Cancer Immunotherapy, Fuzhou, China
- 2Department of Pathology, the First Affiliated Hospital of Fujian Medical University, Fuzhou, China
- 3The School of Medical Technology and Engineering, Fujian Medical University, Fuzhou, China
- 4Key Laboratory of OptoElectronic Science and Technology for Medicine of Ministry of Education, Fujian Provincial Key Laboratory of Photonics Technology, Fujian Normal University, Fuzhou, China
- 5Department of Pathology, The Second Affiliated Hospital, Fujian Medical University, Quanzhou, China
Accurate intraoperative identification of gliomas is of utmost importance. This task often remains a challenge for the pathologist and neurosurgeon because of the absence of full intraoperative microstructure feature details of the tumor. Here, multiphoton microscopy (MPM), based on second harmonic generation (SHG) and two-photon excited fluorescence (TPEF), is applied for label-free detecting the microstructure feature differences between normal brain tissue, low-grade, and high-grade gliomas. MPM can not only capture the difference of their qualitative microstructure features such as increased cellularity, nuclear atypia, microvascular proliferation, and necrosis that are significant for diagnosing and grading of glioma, but also visualize some additional features such as collagen deposition that cannot be seen by conventional methods. In addition, automated image analysis algorithms are developed to automatically and accurately calculate the quantitative diagnostic features: collagen content, the number and area of nuclei to further quantitatively analyze the microstructure features difference of collagen deposition, cellularity, and nuclear atypia between normal brain tissue, low-grade, and high-grade gliomas. With the development of two-photon fiberscope, combined MPM and image processing techniques may become an imaging tool for assisting intraoperatively diagnosing and grading gliomas.
Introduction
The goal of glioma surgery is “maximum safe resection” and the procedure is an invasive one. It is therefore important to determine before and during surgery whether the lesion is a glioma and whether the glioma is a high-grade or low-grade. Current techniques of intraoperative histopathological diagnosis including frozen sectioning and cytological preparations performed neuropathologist and skilled technicians. However, these techniques have limitations. First, in order to minimize damage to normal brain tissue, the size of tissue samples available for intraoperative neuropathological analysis is often small, which may affect diagnostic accuracy, especially when considering the heterogeneous nature of brain tumors [1]. Second, some brain tumors have histopathological features that are rarely seen and may be misclassified [2]. Third, the typical processing of intraoperative tissue samples, such as fixation and embedding, not only time-consume, but also leads to the appearance of artefacts and the loss of some biological components, which may affect the subjectivity of interpretation during subsequent diagnosis and delay surgery [3]. Four, in traditional intraoperative pathology, the pathologist often uses a light microscope to view the lesions on the pathology slides, which is relatively inefficient [4]. With the current shortage of pathologists and the increasing demand for screening biopsies for various diseases, the traditional method of reading further increases the burden and responsibility of pathologists [5]. Furthermore, and most importantly, this labelled pathological findings lack direct guidance for preoperative or intraoperative assessment, and do not simultaneously reflect the broader signal in biological tissue. Therefore, there is an urgent need for a new intraoperative imaging technique which can label-free provide pathological-images of the tissue microstructure feature.
Multiphoton microscopy (MPM), based on two photon excitation fluorescence (TPEF) and second-harmonic generation (SHG), represents a promising new adjunctive technology in the in vivo and real-time diagnosis of tissue [6, 7]. In comparison to conventional histopathology methods, MPM technique allows for the real-time simultaneous excitation of multiple endogenous signals in different components of biological tissue without the need for additional staining. Besides, combined with custom-developed image processing algorithms, it more reliably and rapidly shows the morphological characteristics and offer quantitative information of unstained tissues. Many previous reports have demonstrated that MPM enables tissue adjunctive diagnosis at the cellular level, for instance, skin aging [8], liver fibrosis [9], basal cell carcinoma [10], ovarian cancer [11], gastric cancer [12], breast cancer [13], colonic adenoma [14].
Previous studies also support the hypothesis that brain tumors and normal brain tissue can be well discerned based on label-free MPM imaging [15–22]. Gliomas were shown to have a specific morphochemistry in the TPEF images, with cell proliferation bearing enlarged nuclei. In unlabelled multiphoton images, several diagnostic features of some brain tumors in unstained multiphoton images have been determined and may be used as tumor markers [17]. Cytological and architectural features required for pathological tumor typing and grading in 55 lesions of the central nervous system can be retrieved by label-free multiphoton microscopy [21]. Moreover, a study of 382 tumor patients indicated that the combined analysis of texture parameters of the CARS and TPEF signal proved to be most suited for the discrimination of nontumor brain versus brain tumors leading to a correct rate of 96% [22].
In the present study, we aim to provide the basis for clinical translation of label-free multiphoton imaging as an intraoperative tool for gliomas delineation. Therefore, we imaged fresh unstained human normal brain tissue, low-grade, and high-grade gliomas using the combination of TPEF and SHG. Several qualitative microstructure features differences between normal brain tissue, low-grade, and high-grade gliomas, such as collagen deposition [23, 24], increased cellularity, nuclear atypia, microvascular proliferation, and necrosis were visualized. To describe the degree of collagen deposition, increased cellularity and nuclear atypia, some automated image analysis algorithms based on image segmentation technology were developed to automatically calculate the quantitative diagnostic features: the collagen content, the number, and area of nuclei.
Materials and Methods
Sample Preparation
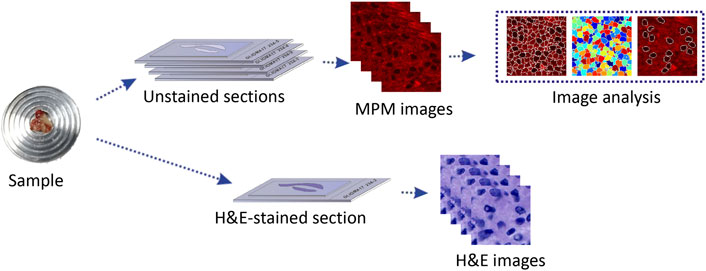
From 2014 to 2018, 20 glioma patients (10 high-grade gliomas, 10 low-grade gliomas) and 5 traumatic brain injury patients from the Department of Neurosurgery at the First Affiliated Hospital of Fujian Medical University were invited to join in the research. The research was approved by Fujian Medical University Clinical Research Screening Committee for Studies Involving Human Subjects. All of the subjects signed a written consent prior to the research. During neurosurgical procedures, the structurally normal tissue and glioma specimens were taken from the same patient to carry out the conventional diagnostic procedure. After resection, the specimens were delivered to the pathology department within 30 min. To verify the feasibility of MPM in tissue detail imaging, each specimen was divided into two parts. One portion of the fresh tissue was processed as five serial frozen slices by cryostat microtome. The middle slice with thickness of nearly 5 μm was stained with H&E staining for histological comparison with MPM outcomes. The remaining 4 slices with thickness of nearly 20 μm were sandwiched between microscope slide and a cover slip for MPM imaging (Figure 1). The other part of the fresh tissue is left in the pathology department to be processed into paraffin blocks for subsequent histopathological examinations (including standard hematoxylin and eosin staining, specific staining, and immunohistochemistry et) and molecular pathology examinations for the final diagnosis of glioma. Gliomas were classified and graded according to the criteria outlined by the 2016 World Health Organization. The pathological diagnoses were reaffirmed independently by two experienced neuropathologists.
Multiphoton Microscopic Imaging System
The MPM imaging system used in this study was previously mentioned in detail [25]. Briefly, this system is composed of a Zeiss LSM 510 system (Zeiss, Jena, Germany) and a Ti:sapphire femtosecond laser (Mira 900-F, Coherent Inc., Santa Clara, CA, United States), which can be tuned in the range of 700–980 nm. In this study, the laser power was set as 5 mw and excitation wavelength at 810 nm. 63×Plan-Apochromat oil immersion objective allows to focus the excitation beam and collect the backward signals. META detector is made up of an optimized 32-channel photomultiplier tube (PMT) array and high-quality reflective grating to detect the emission signals in the backward direction. In this system, 8 independent channels are included, of which individual channel(s) can be selected to capture emission signals, covering wavelength from 377 to 716 nm. And then images can be obtained. In this research, 2 independent channels were chosen to detect TPEF and SHG signals from the specimens. One channel detected TPEF signals, covering wavelength from 430 to 716 nm; the other channel detected SHG signals, covering wavelength from 387 to 419 nm. In order to strengthen the contrast of TPEF/SHG images, TPEF images were shown in red and SHG images were shown in green. All images were obtained at 2.56 μs per pixel and had a 12-bit pixel depth. The image acquisition time was 1.97 s/frame with a resolution of 512 pixels × 512 pixels.
Image Analysis Methods
Analysis of Collagen Density
MPM images were segmented and analyzed through custom-developed software in MATLAB Image Processing Toolbox (The MathWorks, Inc.). The patients enrolled in this study were split into training/test datasets at a ratio of 2:1. We collected 300 MPM images from the fresh ex vivo brain tissues without staining, obtaining 120, 120, and 60 images of diagnosed low-grade glioma, high-grade glioma and normal brain tissues, respectively. In our experiment, collagen content, number, and area of nuclei were automatically calculated by our self-written program. Collagen density was calculated as the ratio of SHG pixels to all pixels in every image. In order to calculate collagen content, MPM images were analyzed through the following algorithm: First, the SHG image was converted to grayscale image and then a Gaussian filter (size = 5, sigma = 1) was used to reduce noise and enhance the collagen details of image; Next, the enhanced results were carried out by Otsu threshold segmentation and mathematical morphological processing; Finally, the segmentation template was obtained, and collagen ratio was calculated according to the occupied area of the collagen. The flowchart of the algorithm is shown in Figure 2A.
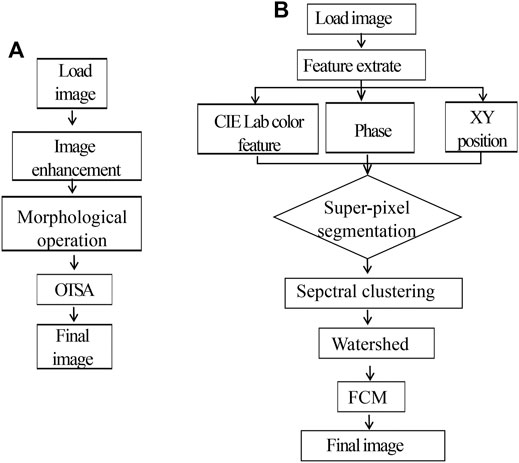
FIGURE 2. (A) Flowchart of the proposed collagen density segmentation. (B) Flowchart of the proposed nuclear density and size segmentation.
Analysis of Nucleus Density and Size
The segmentation approach based on watershed was used to depict cell boundaries and automatically calculate the number and area of nuclei. The flowchart of this algorithm is shown in Figure 2B. Detailed analysis steps were as following: first, the original MPM image in RGB color space was changed to CIE Lab space; then six-dimensional characteristic quantity was extracted, including image color space LAB color feature, phase, and XY position. Next, distance measurement was performed for the extracted six-dimensional eigenvectors, image pixels were clustered by similarity matrix spectrum, and segmentation was carried out with the marked watershed algorithm method. In the CIE Lab color space, the darkest one is selected with Fuzzy C-Means algorithm (FCM) as the final segmentation result; meanwhile, number and area of nuclei can be automatically calculated based on the segmentation results.
Results
Multiphoton Microscopic Imaging of Normal Human Brain Tissue
Figure 3 showed representative TPEF/SHG images and the corresponding H&E images of the normal brain tissue. Figures 3A–D showed the large-scale MPM images and the corresponding H&E stained images of the normal brain tissue (white matter), demonstrating the capability of MPM to displayed large-area histoarchitectural features. In the large-scale images, major components of the brain, such as extracellular matrix, cells, and blood vessels emitted strong multiphoton signals, which could be explicitly identified in multiphoton images. Figures 3E–L showed the selected regions at higher magnifications (yellow and white box in Figure 3C). In the magnified images, more details of cells and blood vessels were revealed. MPM showed that nonfluorescent cell nuclei were marked as dark spot while the tightly condensed matrix emitted strong TPEF signals was color-coded in red, highlighting the distinguishable individual cells. Those cells (yellow arrowheads) of round shape and uniform size were found to be evenly distributed in the normal brain tissue. A blood vessel (cyan arrowhead) could be identified owing to its tubular structure defined by elastin and collagen. The identified blood vessel was marked in yellow because collagen in the wall emitted strong SHG signals while elastin produced comparable TPEF signals. Except for blood vessel wall, SHG signals were not detected, indicating that the stroma in the normal brain tissue had limited collagen content. These results were consistent with what have been previously reported [23]. These characteristics can also be seen in the corresponding H&E-image. However, collagen could not be displayed in H&E images as clearly as this in MPM images.
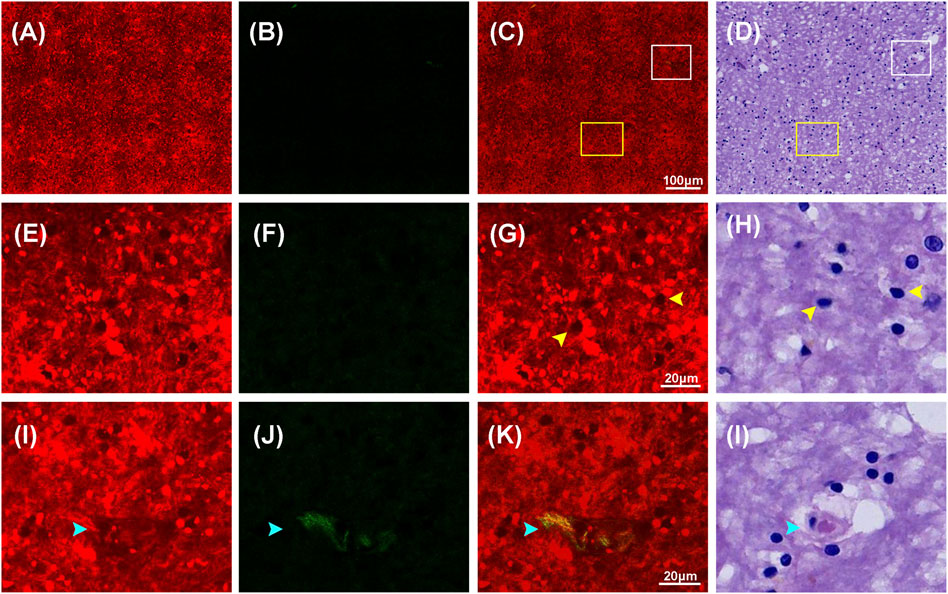
FIGURE 3. Representative MPM images and the corresponding images of H&E stained normal brain (white matter) tissue. (A–D) TPEF, SHG, TPEF/SHG overlaid images, and corresponding image of H&E stained normal brain tissue. (E–H) The magnified TPEF, SHG, TPEF/SHG overlaid images, and corresponding H&E image of the selected area (yellow square box) in Figure 3C. (I–L) The magnified TPEF, SHG, TPEF/SHG overlaid, and corresponding H&E image of the selected area (white square box) in Figure 3C. Yellow arrowheads: cells in normal brain tissue; cyan arrowhead: blood vessel in normal brain tissue.
Multiphoton Microscopic Imaging of Low-Grade Glioma
The structural differences of low-grade gliomas were featured as variably increased cellularity and slight nuclear atypia, which also helped to distinguish between normal and abnormal tissue. Figure 4 displays MPM images and the corresponding H&E images of low-grade gliomas. Compared with the normal brain tissue, low-grade gliomas showed moderate architectural and cytological abnormalities in MPM images. As it could be seen in the large-scale images in Figures 4A–D, cell density in low-grade gliomas was greatly increased compared to normal brain tissue. Figures 4E–L showed higher magnification images of selected areas from Figure 4C. In Figures 4E–H, MPM images could clearly reveal morphological details of tumor cells (yellow arrowheads) in low-grade gliomas. The tumor cells were of medium size with slightly enlarged nuclear, distinct cell borders, and round, oval or short fusiform nuclei. However, nuclear atypia was not obvious as it was only seen in a fraction of cells. In addition, the morphology of extracellular matrix was moderately changed. A few of collagen fibers were found to be irregularly scattered in the red extracellular matrix background. More collagen details were presented in Figures 4I–L. Through SHG signals, collagen fibres (pink arrowheads) with reticular morphology were shown in green. A few of collagen fibres were shown in yellow, meaning they concurrently generated comparable SHG and TPEF signals, indicating that they might have a distinct photochemical composition. These details were readily correlated with those in the corresponding H&E images. However, for these collagen differences between normal brain tissue and low-grade gliomas could also not be displayed clearly in H&E images as those in MPM images. The research findings showed that the microstructure features of low-grade gliomas could be well characterized by MPM technology.
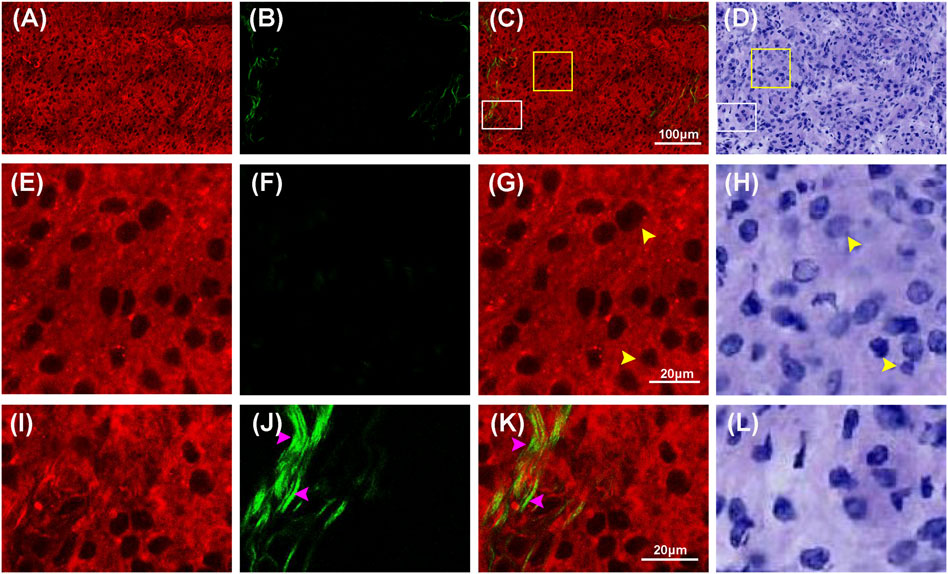
FIGURE 4. Representative MPM images and the corresponding images of H&E stained low-grade glioma. (A–D) TPEF, SHG, TPEF/SHG overlaid images, and corresponding image of H&E stained low-grade glioma. (E–H) The magnified TPEF, SHG, TPEF/SHG overlaid images, and corresponding H&E image of the selected area (yellow square box) in Figure 4C. (I–L) The magnified TPEF, SHG, TPEF/SHG overlaid images, and corresponding H&E image of the selected area (white square box) in Figure 4C. Yellow arrowheads: cells in low-grade glioma; pink arrowhead: collagen in low-grade glioma.
Multiphoton Microscopic Imaging of High-Grade Glioma
Compared to normal tissue and low-grade gliomas, high-grade gliomas have features such as variably increased cellularity, obvious nuclear atypia, microvascular proliferation, intense vascular proliferation, and necrosis. Figures 5, 6 showed representative TPEF/SHG images and the corresponding H&E images of the central tumor region and necrotic area of high-grade glioma (glioblastoma), respectively. Unlike normal brain tissue and low-grade gliomas, high-grade gliomas showed more severe architectural and cytological abnormalities in MPM images. Figures 5A–D clearly revealed the increased cellularity, obvious nuclear atypia, and intense vascular proliferation in the high-grade glioma tissue (tumor central region). In the magnified images (Figures 5E–L), the entire area of high-grade gliomas was almost filled with tumor cells that had indistinct cell borders and were unevenly distributed. The nuclei had larger areas and more severe nuclear pleomorphism. Intense vascular proliferation, seen as important histopathological hallmark of high-grade glioma, was easily detected via SHG signals that were emitted by collagen in the vessel walls. Blood vessels (cyan arrowheads) of different sizes and shapes were found to be scattered in the extracellular matrix. Compared with low-grade gliomas, more SHG signals were detected as denser collagen deposition (pink arrowheads) was notably presented in high-grade gliomas. Microvascular proliferation and necrosis, seen as two fundamental features of glioblastoma and two major predictors of the aggressive diffuse astrocytomas [26–28], were well characterized in Figure 6. Compared with its surrounding tumor cells, the tumor necrotic had been identified owing to their “pseudopalisading” pattern (Figures 6A,E) and higher TPEF signal intensity (yellow arrowheads). Near the necrotic core, the reactive hyperplasia of vascular endothelial cells in the blood vessel wall was visualized in the glomerulus-like arrangement (Figure 6K); emitted TPEF signals in the endothelial cells (cyan arrowheads) were weaker than those of the surrounding tumor cells. Thus, microvascular proliferation could be readily recognized in MPM images. These morphological characteristics of high-grade gliomas were also seen in the corresponding H&E images. Again, same as low-grade gliomas, collagen deposition was excluded since it could not well display in H&E images. These results indicated that characteristics of high-grade gliomas could be well captured by MPM. To some extent, MPM had a better effect than H&E-stained technology.
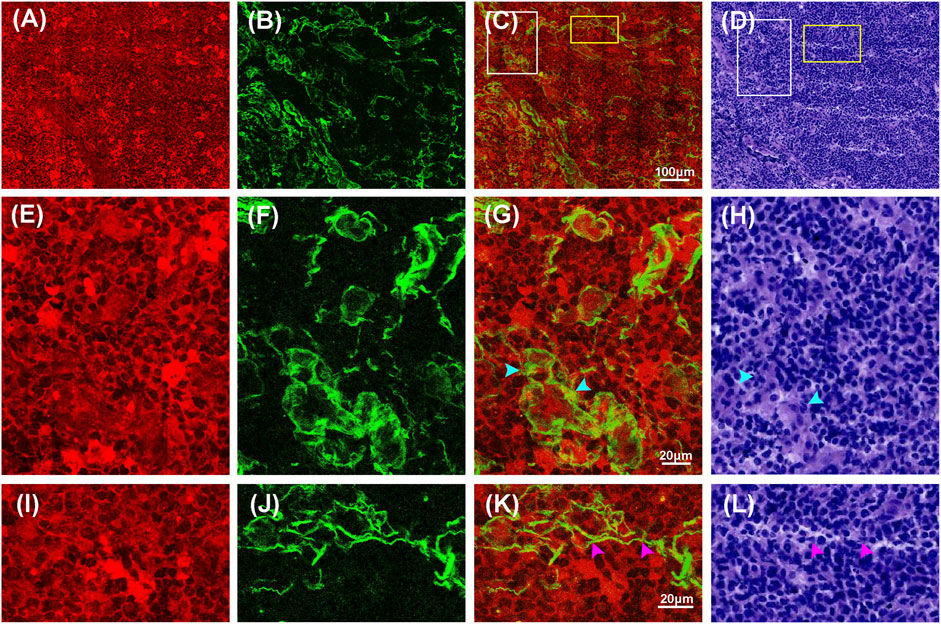
FIGURE 5. Representative MPM images and the corresponding H&E stained images of high-grade glioma (tumor central region). (A–D) TPEF, SHG, TPEF/SHG overlaid and corresponding H&E image of high-grade glioma. (E–H) The magnified TPEF, SHG, TPEF/SHG overlaid images, and corresponding H&E image of the selected area (white square box) in Figure 5C. (I–L) The magnified TPEF, SHG, TPEF/SHG overlaid images, and corresponding H&E image of the selected area (yellow square box) in Figure 5C. Cyan arrowheads: blood vessels in high-grade glioma; pink arrowhead: collagen in high-grade glioma.
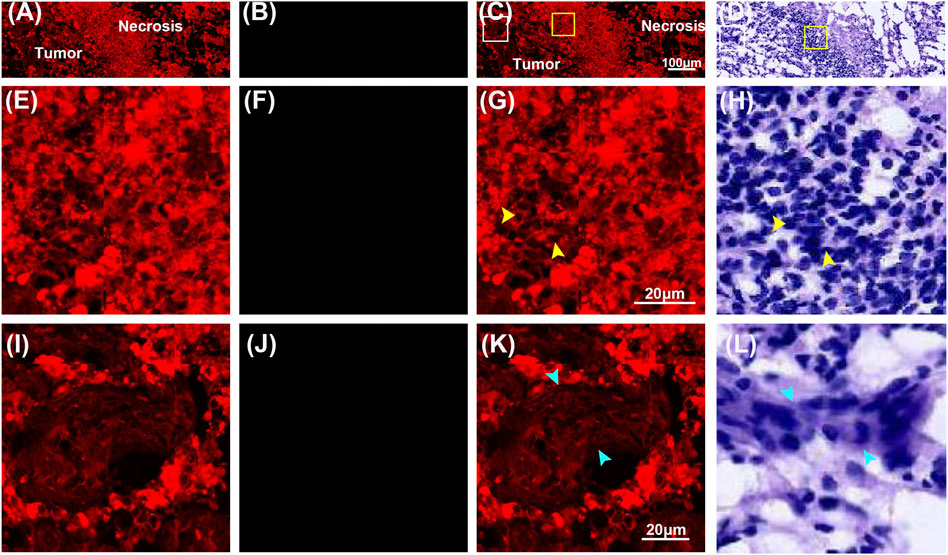
FIGURE 6. Representative MPM images and the corresponding images of H&E stained high-grade glioma (necrotic area). (A–D) TPEF, SHG, TPEF/SHG overlaid images, and the corresponding H&E image of high-grade glioma. (E–H) The magnified TPEF, SHG, TPEF/SHG overlaid images, and corresponding H&E image of the selected area (white square box) in Figure 6C. (I–L) The magnified TPEF, SHG, TPEF/SHG overlaid images, and corresponding H&E image of the selected area (yellow square box) in Figure 6C. Yellow arrowhead: tumor cells; cyan arrowheads: endothelial cells.
Automatic Identification of Glioma
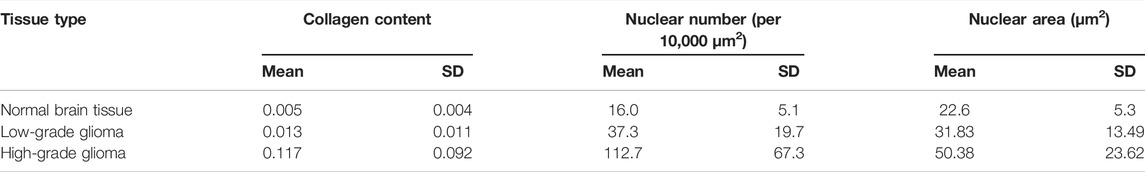
In order to further characterize the morphological differences between low-grade and high-grade gliomas, quantitative analysis was conducted. The collagen content, number and area of nuclei were automatically calculated through the proposed image analysis method. Figures 7A–D showed the representative collagen segmentation images. The collagen enhancement segmentation image (Figure 7B), perivascular collagen position segmentation image (Figure 7C), and final segmentation image (Figure 7D) can all be well correlated with original SHG images (Figure 7A) at the subcellular level. More segmentation details could be seen in the magnified images from Figures 7E–H. Results of comparing collagen density between normal brain tissue, low-grade gliomas, and high-grade gliomas using this collagen analysis method were shown in Figure 7I; Table 1, respectively. Quantitative analysis results showed that the collagen density of high-grade gliomas (0.157 ± 0.023) was significantly higher than that of low-grade gliomas (0.013 ± 0.005) and normal brain tissue (0.005 ± 0.001). The more severe the malignant tumor was, the higher the contents of collagen could be. These results were consistent with the other research findings [23].
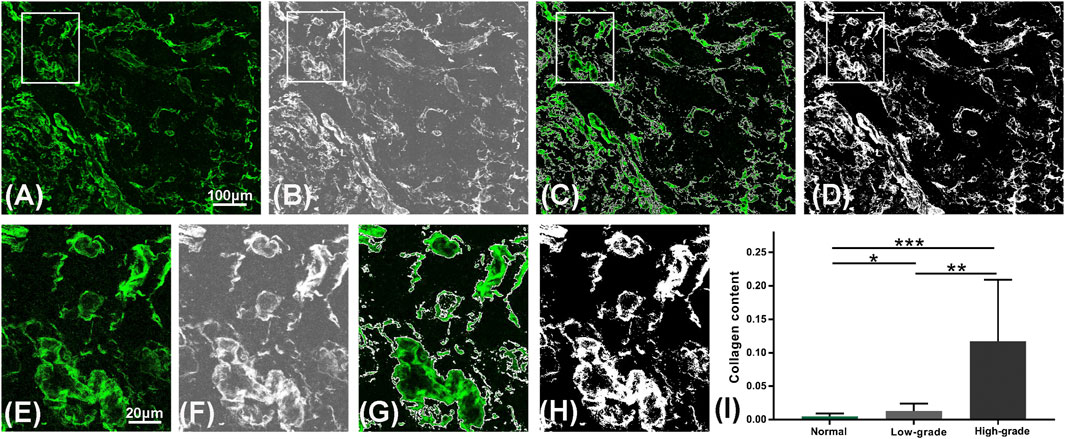
FIGURE 7. Quantitative analysis of collagen. (A–D) Representative segmentation results of the automatic analysis algorithm: an original SHG image in a high-grade glioma (A), the result after image enhancement (B), the segmentation result of collagen position (C) and the final segmentation result of collagen content (D) are displayed from left to right. (E–H) Magnified region marked in (A–D). (I) Collagen content variations in the normal brain tissue, low-grade and high-grade gliomas using the algorithm.
Figure 8 showed the processing results of nuclear segmentation. The number of nuclei, and the mean and standard deviation of the nuclear area could be automatically calculated. Figures 8A–E illustrated the representative nuclear segmentation results. As it was shown in Figure 8A, the original MPM image indicated low SNR as the cell boundaries were not clear and difficult to be precisely positioned by human eyes. The segmentation result after spectral clustering was shown in Figure 8B. In Figure 8C, image noises were reduced while cell boundaries were enhanced after watershed segmentation. Then the average gray-scale based on the above segmentation results was applied to correctly depict cells from the complicated background. The final segmentation image was shown in Figure 8D. In this image, cells can be automatically positioned and cell boundaries can be correctly depicted from the complicated background. In this way, cell number and nuclear area could be automatically quantified. MPM images (Figure 8A) well matched with segmentation results (Figure 8D) at the subcellular level. The comparison results of cell density, mean and standard deviation of nuclear parameters between normal brain tissue, low-grade gliomas, and high-grade gliomas using this analysis method were shown in Figures 8E,F and Table 1, respectively. Quantitative analysis results revealed that compared to low-grade glioma, nuclear number was sharply increased and presented a larger standard deviation in high-grade gliomas. In summary, MPM and image analysis method provided quantitative assessment of morphological changes of low-grade and high-grade glioma tissues. Above results showed that collagen density, cell density, and mean and standard deviation of nuclear area are representative features to different normal brain tissue, low-grade, and high-grade gliomas. Comprehensive analysis of them could rapidly offer additional assessment metrics to pathologists and surgeons. Thus, MPM combined with image analysis method has the potential to establish an objective standard about grading of gliomas.

FIGURE 8. Quantitative analysis of number and area of nuclei. (A–E) Representative nuclear segmentation results of the automated analysis algorithm: an original MPM image in a low-grade glioma (A), the result after super-pixel segmentation and spectral clustering (B), the result after watershed segmentation (C), and the final segmentation results (D) are displayed from left to right. (E) Number of nuclei of normal brain tissue, low-grade, and high-grade gliomas quantified using this algorithm. (F) Area of nuclei of normal brain, low-grade, and high-grade gliomas quantified using this algorithm.
Discussion
MPM, based on intrinsic TPEF and SHG signals, shows label-free images by allowing the nonlinear excitation of different intrinsic biomacromolecules, and obtaining the subcellular resolution from unstained and fresh tissue [29–31]. In this study, we used label-free MPM to obtain the microstructures of human normal, low-grade and high-grade brain tissues. Our results show that the morphology, content and morphology of the main components including cells, blood vessels and collagen in these three tissues have apparently different properties. Based on these differences, several diagnostic features including increased cellularity, nuclear atypia, intense vascular proliferation, microvascular proliferation, and necrosis can be extracted to distinguish between normal, low-grade and high-grade brain tissues, Meanwhile, collagen deposition, an additional feature that cannot be seen by the conventional methods, is clearly observed in MPM images. Compared to classic traditional intraoperative diagnosis techniques, MPM can affords real-time imaging of brain tissues without destructive procedures and staining. In addition, all images were acquired at a rate of 2.56 µs per pixel. Therefore, multichannel detection, combined with increased accuracy and speed, makes MPM highly suitable for characterizing morphologic features of cell, blood vessel and collagen at subcellular resolution.
Automated image analysis has become a powerful tool for solving biomedical problems in macroscopic image [32]. In this study, several image analysis algorithms were developed to calculate the determination of collagen location, collagen content, nuclei area, nuclei number and nuclei area variance in this study. Relay on this, the extent of collagen deposition, nuclei size, cellularity and nuclear atypia can be rapidly and quantitatively analysed. Thus, our research may be taken as an alternative and adjunctive approach to the diagnosis and grading of brain tumors. While compared with pathologist’s examination and traditional MPM technique, our proposed method can quickly display image characteristics and reduce amount of manual observation. Recent research studies indicate that MPM combined with image analysis method has major advantages in the feasibility of clinical translation [33].
Regarding the applicability of the proposed method in clinical settings, some limitations remain to be addressed. The number of patients enrolled in this study was relatively small. More data of patients should be evaluated. Besides, current label-free MPM images cannot get the depth in micrometer, the high-resolution images are often acquired by ex vivo histopathologic section or in vivo intraoperative biopsy. Fortunately, the advances of two-photon microendoscope and fiberscope have provided insights and feasibility in clinical practice [34–39]. The miniaturized two-photon fiberscope has enabled high-resolution optical recording of neural activity at the cellular level from freely moving animals in their natural behaviors [39]. Then small-diameter (350 um) gradient refractive index (GRIN) lens with side-view design could be inserted to solid tissues [37–39]. Under the fixation condition, the high-quality MPM images in many centimeters could be obtained from the object plane. From a recent clinical resection case of glioblastoma, multiphoton tomography showed its ability to rapidly identify the native glioma tissues without tissue staining or processing [40]. Thus, the proposed method in this study combined with MPM endomicroscopy could be taken as a powerful diagnosis tool for the auxiliary diagnosis and grading of gliomas in the operating room to offer real-time histology-like images and perform automated image analysis for higher diagnostic accuracy of neuropathology.
In the future, our research will focus on enrolling more patients, the intraoperative in vivo imaging through MPM microendoscopes and the optimization of algorithms for quantitative diagnosis. As probes and optical fiber are miniaturized and optimized, it is anticipated that our combined method may be used for auxiliary diagnosis and grading of gliomas in the operating room to offer real-time histology-like images and perform automated image analysis for higher diagnostic accuracy of neuropathology.
Conclusion
In summary, we demonstrated the capability of label-free MPM to identify differences of microstructure features between normal brain, low-grade and high-grade gliomas, revealing key microstructure features including collagen deposition, increased cellularity, nuclear atypia, microvascular proliferation, and necrosis were visualized. Furthermore, the combination of label-free MPM and image processing provided a supplementary methodology for quantitatively analysis microstructure features differences. Therefore, our presented method provides insights about both morphological and quantitative information of the gliomas tissues, which has the great potential for becoming a valid intraoperative imaging tool for assisting in diagnosing and grading of gliomas.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Fujian Medical University Clinical Research Screening Committee for Studies Involving Human Subjects. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
ZW, YL, and YX contributed to sample acquisition and subject recruitment; XW, SC contributed to sample processing and evaluation; JC, DK contributed to experiment design, plan and data analysis. ZW, NL, and NF wrote the initial version of the manuscript and all authors contributed to the revisions.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 62105063); The Natural science foundation of Fujian province (2018J01851, 2021J01811); and Special Funds of the Central Government Guiding Local Science and Technology Development (2017L3009); Fujian Provincial Health Technology Project (2020GGA054).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Hua Lin in the First Affiliated Hospital of Fujian Medical University for preparation of samples used in the study.
References
1. Lauko A, Lo A, Ahluwalia MS, Lathia JD. Cancer Cell Heterogeneity & Plasticity in Glioblastoma and Brain Tumors. Semin Cancer Biol (2021):1–14. doi:10.1016/j.semcancer.2021.02.014
2. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a Summary. Acta Neuropathol (2016) 131:803–20. doi:10.1007/s00401-016-1545-1
3. Uematsu Y, Owai Y, Okita R, Tanaka Y, Itakura T. The Usefulness and Problem of Intraoperative Rapid Diagnosis in Surgical Neuropathology. Brain Tumor Pathol (2007) 24:47–52. doi:10.1007/s10014-007-0219-z
4. Somerset HL, Kleinschmidt-DeMasters BK. Approach to the Intraoperative Consultation for Neurosurgical Specimens. Adv Anat Pathol (2011) 18:446–9. doi:10.1097/pap.0b013e3182169934
5. Malatesta M. Histological and Histochemical Methods - Theory and Practice. Eur J Histochem (2016) 60:60. doi:10.4081/ejh.2016.2639
6. Squirrell JM, Wokosin DL, White JG, Bavister BD. Long-term Two-Photon Fluorescence Imaging of Mammalian Embryos without Compromising Viability. Nat Biotechnol (1999) 17:763–7. doi:10.1038/11698
7. Centonze VE, White JG. Multiphoton Excitation Provides Optical Sections from Deeper within Scattering Specimens Than Confocal Imaging. Biophys J (1998) 75:2015–24. doi:10.1016/S0006-3495(98)77643-X
8. Cicchi R, Kapsokalyvas D, De Giorgi V, Maio V, Van Wiechen A, Massi D, et al. Scoring of Collagen Organization in Healthy and Diseased Human Dermis by Multiphoton Microscopy. J Biophoton (2010) 3:34–43. doi:10.1002/jbio.200910062
9. Tai DCS, Tan N, Xu S, Kang CH, Chia SM, Cheng CL, et al. Fibro-C-Index: Comprehensive, Morphology-Based Quantification of Liver Fibrosis Using Second Harmonic Generation and Two-Photon Microscopy. J Biomed Opt (2009) 14:044013. doi:10.1117/1.3183811
10. Balu M, Zachary CB, Harris RM, Krasieva TB, König K, Tromberg BJ, et al. In Vivo multiphoton Microscopy of Basal Cell Carcinoma. JAMA Dermatol (2015) 151:1068–74. doi:10.1001/jamadermatol.2015.0453
11. Kirkpatrick ND, Brewer MA, Utzinger U. Endogenous Optical Biomarkers of Ovarian Cancer Evaluated with Multiphoton Microscopy. Cancer Epidemiol Biomarkers Prev (2007) 16:2048–57. doi:10.1158/1055-9965.EPI-07-0009
12. Chen J, Zhuo S, Chen G, Yan J, Yang H, Liu N, et al. Establishing Diagnostic Features for Identifying the Mucosa and Submucosa of normal and Cancerous Gastric Tissues by Multiphoton Microscopy. Gastrointest Endosc (2011) 73:802–7. doi:10.1016/j.gie.2010.12.016
13. Wu X, Chen G, Lu J, Zhu W, Qiu J, Chen J, et al. Label-free Detection of Breast Masses Using Multiphoton Microscopy. PLoS One (2013) 8:e65933. doi:10.1371/journal.pone.0065933
14. Zhuo S, Yan J, Chen G, Chen J, Liu Y, Lu J, et al. Label-free Monitoring of Colonic Cancer Progression Using Multiphoton Microscopy. Biomed Opt Express (2011) 2:615–9. doi:10.1364/BOE.2.000615
15. Kantelhardt SR, Leppert J, Kantelhardt JW, Reusche E, Hüttmann G, Giese A. Multi-photon Excitation Fluorescence Microscopy of Brain-Tumour Tissue and Analysis of Cell Density. Acta Neurochir (Wien) (2009) 151:253–62. doi:10.1007/s00701-009-0188-6
16. Kantelhardt SR, Leppert J, Krajewski J, Petkus N, Reusche E, Tronnier VM, et al. Imaging of Brain and Brain Tumor Specimens by Time-Resolved Multiphoton Excitation Microscopy Ex Vivo1. Neuro Oncol (2007) 9:103–12. doi:10.1215/15228517-2006-034
17. Leppert J, Krajewski J, Kantelhardt SR, Schlaffer S, Petkus N, Reusche E, et al. Multiphoton Excitation of Autofluorescence for Microscopy of Glioma Tissue. Neurosurgery (2006) 58:759–67. doi:10.1227/01.NEU.0000204885.45644.22
18. Galli R, Uckermann O, Tamosaityte S, Geiger K, Schackert G, Steiner G, et al. CARS and Non-linear Microscopy Imaging of Brain Tumors. In: European Conference on Biomedical Optics, May 12, 2013 (2013). 89970E. doi:10.1117/12.2031729
19. Uckermann O, Galli R, Mackenroth L, Geiger K, Steiner G, Koch E, et al. Optical Biochemical Imaging: Potential New Applications in Neuro-Oncology. Eur Assoc Neuro Oncol Mag (2014) 4:20–6.
20. Jiang L, Wang X, Wu Z, Du H, Wang S, Li L, et al. Label-free Imaging of Brain and Brain Tumor Specimens with Combined Two-Photon Excited Fluorescence and Second Harmonic Generation Microscopy. Laser Phys Lett (2017) 14:105401. doi:10.1088/1612-202x/aa7c9a
21. Romeike BFM, Meyer T, Reichart R, Kalff R, Petersen I, Dietzek B, et al. Coherent Anti-stokes Raman Scattering and Two Photon Excited Fluorescence for Neurosurgery. Clin Neurol Neurosurg (2015) 131:42–6. doi:10.1016/j.clineuro.2015.01.022
22. Uckermann O, Galli R, Mark G, Meinhardt M, Koch E, Schackert G, et al. Label-free Multiphoton Imaging Allows Brain Tumor Recognition Based on Texture Analysis-A Study of 382 Tumor Patients. Neuro-Oncology Adv (2020) 2:2. doi:10.1093/noajnl/vdaa035
23. Payne LS, Huang PH. The Pathobiology of Collagens in Glioma. Mol Cancer Res (2013) 11:1129–40. doi:10.1158/1541-7786.MCR-13-0236
24. Huijbers IJ, Iravani M, Popov S, Robertson D, Al-Sarraj S, Jones C, et al. A Role for Fibrillar Collagen Deposition and the Collagen Internalization Receptor Endo180 in Glioma Invasion. PLoS ONE (2010) 5:e9808. doi:10.1371/journal.pone.0009808
25. Zhuo S, Chen J, Luo T, Zou D, Zhao J. Multimode Nonlinear Optical Imaging of the Dermis in Ex Vivo Human Skin Based on the Combination of Multichannel Mode and Lambda Mode. Opt Express (2006) 14:7810–20. doi:10.1364/oe.14.007810
26. Raza SM, Lang FF, Aggarwal BB, Fuller GN, Wildrick DM, Sawaya R. Necrosis and Glioblastoma: a Friend or a Foe? A Review and a Hypothesis. Neurosurgery (2002) 51:2–13. doi:10.1097/00006123-200207000-00002
27. Burger PC, Green SB. Patient Age, Histologic Features, and Length of Survival in Patients with Glioblastoma Multiforme. Cancer (1987) 59:1617–25. doi:10.1002/1097-0142(19870501)59:9<1617::aid-cncr2820590916>3.0.co;2-x
28. Homma T, Fukushima T, Vaccarella S, Yonekawa Y, Di Patre PL, Franceschi S, et al. Correlation Among Pathology, Genotype, and Patient Outcomes in Glioblastoma. J Neuropathol Exp Neurol (2006) 65:846–54. doi:10.1097/01.jnen.0000235118.75182.94
29. Denk W, Strickler JH, Webb WW. Two-Photon Laser Scanning Fluorescence Microscopy. Science (1990) 248:73–6. doi:10.1126/science.2321027
30. Zipfel WR, Williams RM, Webb WW. Nonlinear Magic: Multiphoton Microscopy in the Biosciences. Nat Biotechnol (2003) 21:1369–77. doi:10.1038/nbt899
31. Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, Webb WW. Live Tissue Intrinsic Emission Microscopy Using Multiphoton-Excited Native Fluorescence and Second Harmonic Generation. Proc Natl Acad Sci U.S.A (2003) 100:7075–80. doi:10.1073/pnas.0832308100
32. Ljosa V, Carpenter AE. Introduction to the Quantitative Analysis of Two-Dimensional Fluorescence Microscopy Images for Cell-Based Screening. Plos Comput Biol (2009) 5:e1000603. doi:10.1371/journal.pcbi.1000603
33. Streets AM, Li A, Chen T, Huang Y. Imaging without Fluorescence: Nonlinear Optical Microscopy for Quantitative Cellular Imaging. Anal Chem (2014) 86:8506–13. doi:10.1021/ac5013706
34. Helmchen F, Denk W, Kerr JND. Miniaturization of Two-Photon Microscopy for Imaging in Freely Moving Animals. Cold Spring Harb Protoc (2013) 2013:904–13. doi:10.1101/pdb.top078147
35. Piyawattanametha W, Cocker ED, Burns LD, Barretto RPJ, Jung JC, Ra H, et al. In Vivo brain Imaging Using a Portable 29 G Two-Photon Microscope Based on a Microelectromechanical Systems Scanning Mirror. Opt Lett (2009) 34:2309–11. doi:10.1364/ol.34.002309
36. Engelbrecht CJ, Johnston RS, Seibel EJ, Helmchen F. Ultra-compact Fiber-Optic Two-Photon Microscope for Functional Fluorescence Imaging In Vivo. Opt Express (2008) 16:5556–64. doi:10.1364/oe.16.005556
37. Jung JC, Mehta AD, Aksay E, Stepnoski R, Schnitzer MJ. In Vivo Mammalian Brain Imaging Using One- and Two-Photon Fluorescence Microendoscopy. J Neurophysiol (2004) 92:3121–33. doi:10.1152/jn.00234.2004
38. Levene MJ, Dombeck DA, Kasischke KA, Molloy RP, Webb WW. In Vivo multiphoton Microscopy of Deep Brain Tissue. J Neurophysiol (2004) 91:1908–12. doi:10.1152/jn.01007.2003
39. Murray TA, Levene MJ. Singlet Gradient index Lens for Deep In Vivo Multiphoton Microscopy. J Biomed Opt (2012) 17:021106. doi:10.1117/1.JBO.17.2.021106
Keywords: multiphoton microscopy, harmonic generation, two-photon excited fluorescence, high-grade gliomas, low-grade gliomas, image analysis
Citation: Wu Z, Wang X, Fang N, Lin Y, Zheng L, Xue Y, Cai S, Chen J, Lin N and Kang D (2022) Automatic and Label-Free Analysis of the Microstructure Feature Differences Between Normal Brain Tissue, Low-Grade, and High-Grade Gliomas Using the Combination of Multiphoton Microscopy and Image Analysis. Front. Phys. 10:865455. doi: 10.3389/fphy.2022.865455
Received: 10 February 2022; Accepted: 22 March 2022;
Published: 17 May 2022.
Edited by:
Yufei Ma, Harbin Institute of Technology, ChinaReviewed by:
Chandra Sekhar Yelleswarapu, University of Massachusetts Boston, United StatesXiaojun Yu, Northwestern Polytechnical University, China
Copyright © 2022 Wu, Wang, Fang, Lin, Zheng, Xue, Cai, Chen, Lin and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ni Lin, c3N5dG54QDEyNi5jb20=; Dezhi Kang, a2R6OTk5ODhAdmlwLnNpbmEuY29t
†These authors have contributed equally to this work
 Zanyi Wu1†
Zanyi Wu1† Xingfu Wang
Xingfu Wang Yuanxiang Lin
Yuanxiang Lin Jianxin Chen
Jianxin Chen Dezhi Kang
Dezhi Kang