- 1School of Physical Science and Technology, Ningbo University, Ningbo, China
- 2State Key Laboratory of Surface Physics, Department of Physics, Multiscale Research Institute of Complex Systems, Fudan University, Shanghai, China
Improved single-molecule methods can largely increase our understanding of underlying molecular mechanism during cellular signal transduction. In contrast to conventional bulk methods, monitoring molecules one at a time can circumvent averaging effects and acquire unique information. With single-molecule techniques, quantitative characterizations can be achieved at microscopic level, especially for biochemical systems with strong heterogeneity. Here we review four fundamental single-molecule techniques including total internal reflection fluorescence imaging, single-molecule fluorescence recovery after photobleaching, single-molecule Förster resonance energy transfer, and fluorescence correlation/cross-correlation spectroscopy. These techniques are frequently employed in quantitatively investigating the molecular translocation, protein-protein interactions, aggregations, and conformational dynamics involved in the signal transduction both in vitro and in vivo. We also summarized the basic principles and implementations of these single-molecule techniques, as well as the conjunct applications extending the single-molecule measurements to multiple dimensions.
Introduction
Signal transduction governing cellular activities is a complex system of communication. It plays key roles in the ability of cells to perceive environmental cues and correctly response to the microenvironment (pH, temperature, pressure, ions, etc.), which is the basis of development, tissue repair, immunity, and other necessary life activities [1–4]. The execution of signal transduction requires the coordination of many biomolecules, the processes including but not limited to molecular diffusion, conformational changes, inter- and intra-molecular interactions. Monitoring individual behavior of each molecule and resolving the relationship between molecules involving in signal transduction in situ can help us illuminate the mechanism of signaling pathways.
Protein-protein interactions (PPIs) are at the core of signaling pathway studies. Currently, bulk biochemical measurements, such as Western Blot [5] or immunofluorescence [6], are still the gold standards for PPI investigations. However, these techniques suffer from disadvantages such as limited spatial resolution, false positive, and lack of time-resolved information. With the advent of single-molecule detection (SMD) techniques [7–9], it is now routine to image and track conformational changes, dynamics, and interactions of biomolecules at single-molecular level. Compared with conventional bulk methods, monitoring one molecule at a time can avoid averaging effects which may smear out the dynamical characteristic of individual molecules [10]. Especially for the highly complex and dynamic signal transduction processes in living cells, real-time analysis on physiological and kinetic characteristics of biomolecules at single-molecule level can further our understanding of the regulation mechanisms of life activities [11, 12].
SMD methods can be classified into two categories: fluorescence-based methods and force-based methods. The former includes fluorescence imaging, single-molecule tracking [13], single-molecule fluorescence resonance energy transfer (smFRET) [14], etc.; the latter includes optical tweezers [15], magnetic tweezers [16, 17], atomic force microscopy (AFM) [18], etc. In this review, we will focus on the fluorescence-based single-molecule techniques. Among which, total internal reflection fluorescence (TIRF) microscopy, fluorescence spectroscopy techniques, fluorescence recovery after photobleaching (FRAP) [19], fluorescence resonance energy transfer (FRET) [20], fluorescence correlation spectroscopy (FCS) [21], are especially suitable for cellular signal transduction studies. We will briefly summarize the basic principles, short-comings, and major biological applications of SMD methods for the quantitative investigation of biological signal transduction mechanisms.
TIRF
Total internal reflection fluorescence (TIRF) microscopy has a long history in cell biological applications [22]. The basic principle of TIRF is to utilize the evanescent wave appears at the interface of optically rarer medium (aqueous buffer) and optically denser medium (glass slide) when the excitation beam is incident at an angle greater than the critical angle. Owing to the non-propagating nature of evanescent wave, the illumination volume would be contained within a thin layer about half the wavelength (∼200 nm) around the glass slide interface. This can effectively reject the background fluorescence contributed by the contaminations in the buffer or other native pigments within the live cell to achieve better signal-to-noise level for single-molecule detection. Nowadays, TIRF microscopy is one of the most basic approaches for single-molecule observation in laboratories.
TIRF microscope (TIRFM [23, 24]) is relatively low cost and easy to set up in the laboratory. Depending on the nature of specimen, TIRFM can be set up in an objective style (easier to set up) or prism style (lower background, but requires thin samples), which illuminate from the bottom or from the top, respectively.
Despite the simple principle behind TIRF, it can do far more than merely imaging. To study signaling pathways, one is usually interested in the interaction partner, reaction stoichiometry, or even rate constants. These can be acquired when you design your TIRF experiment properly [25]. We will illustrate this with the single-molecule study on plant hormone brassinosteroid (BR) signaling pathway.
Song et al. [26] monitored the signaling transduction between BES1, BIN2, and 14-3-3 of the Arabidopsis BR pathway with TIRF microscopy. To study the interactions between several proteins, they used a TIRF setup accommodating multiple-color excitations as shown in Figure 1A [27]. For a single fluorophore, the photo-bleaching profile will appear as a step-wise trace in the time vs intensity trajectory (Figure 1B). Therefore, by counting the number of steps, we can determine the number of labeled molecules in the observation location. Based on this, Song et al. have found that BIN2 will bind stably on BES1 until the arrival of ATP to phosphorylate BES1 before dissociation (Figure 1C). 14-3-3 dimers would come afterwards to interact with phosphorylated BES1 (Figure 1B).
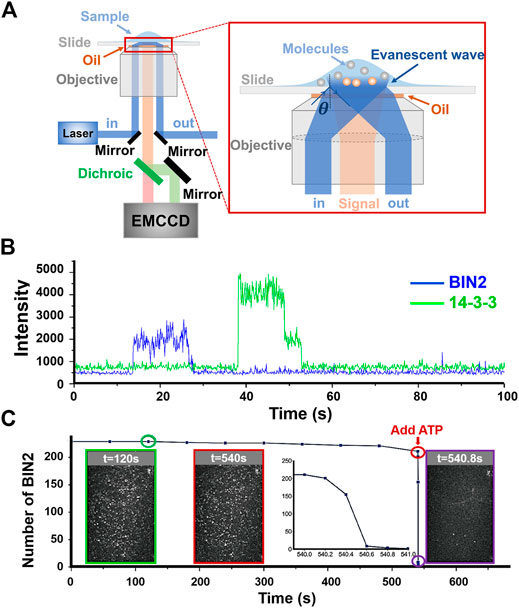
FIGURE 1. Signal transduction of the Arabidopsis BR pathway monitored with TIRF microscopy. (A) Schematic of multi-colored mirror-based TIRFM [27]. In mirror-based TIRFM, the excitation laser (blue) will be reflected by the mirror and then refracted by the objective lens, finally incident on the sample surface at a total reflection angle. The fluorescent signal (orange) will be collected by the objective and divided into two channels (green: donor; red: acceptor) by a dichroic before being detected by EMCCD. (B) Time-dependent fluorescent intensity traces tracking the interactions between BIN2 and 14-3-3. These two traces are collected by a multi-colored TIRFM design shown in A. The blue plateau represents the binding duration of BIN2, and green steps (with two photobleaching steps) means the arrival of 14-3-3 dimer. (C) Number changes of BIN2 after ATP arriving. Three intensity images show the distribution i.e. density of BIN2 binding stably on BES1at different times. The numbers of BIN2 have been counted and plotted in the inset. The TIRF images are the screen shots showing the number of bound BIN2 at time equals 120 (shown in green), 540 (red), and 540.8 (purple) sec, respectively. ATP was added at t = 540 s. Panel (C) is adapted from Ref. [26].
With properly proposed kinetic model, a series of single-molecule interaction assays at different conditions, i.e., reactant concentrations or other related parameters, can derive the rate constants for critical steps in the interaction reaction. A rather simple example of estimating the binding affinity with fluorophore bleaching effect can be found in Ref. [25]. For the case of BIN2 phosphorylating BES1, a mean rate constant is measured to be 2.3 ± 1.4 s−1. When the experiment is done in vitro, the chemical environment can be precisely controlled. For gaseous molecules that are vital for plant signaling pathways, such as ethylene or oxygen, an air-tight flow cell can provide tunable amount of soluble gases for the experiments. In Ref. [26], the researchers have found that BIN2 and BES1 interaction is activated by oxygen while trying to scavenge oxygen to prolong fluorescent probes’ photo-bleaching lifetime. This surprising finding may indicate a role of BIN2 in the stress response mechanism of plants.
Besides in vitro inquisitions described above, TIRFM is inherently suitable for in vivo observations especially for biological events happening around the cell membrane. For plant cells with a thick cell wall, a variation of TIRF named Variable-Angle TIRF (VA-TIRF) can be implemented [28, 29]. In fact, one of the first in vivo single molecule detections was done with TIRFM. Sako et al. [30] investigated the early signaling events of epidermal growth factor receptor (EGFR) on living A431 carcinoma cells. Instead of labeling the EGFR, Sako et al. designed the experiment with organic dye labeled epidermal growth factor (EGF). From TIRFM, they observed the binding event of Cy3-labeled EGF and EGFR. Two activated EGFR forming dimers were captured on the camera and an influx of Ca2+ were reported by calcium indicator, Fluo-3. When the ligand EGFs were labeled both with Cy3 and Cy5, single-molecule FRET can be detected when a pair of EGFRs bounds with Cy3-EGF and Cy5-EGF formed a dimer. Single-molecule FRET can provide distance measurements around molecular length scale, which we will discuss in further details in the following sections. Single-molecule fluorescence spectroscopy allows dynamics measurement on a fluidic substrate such as live cell-membrane or synthetic lipid bilayers. Srinivasan et al. [31] utilized a nano-disc [32], which is an artificial supported lipid bilayer, to confine a full-length EGFR monomer. Using smFRET, they reveal a conformational change upon EGF binding which transduces across the membrane.
TIRF microscopy can be readily augmented into a super-resolution imaging platform. In fact, techniques like PALM/STORM are usually performed on TIRF microscopes [33, 34]. In order to adapt for live cells with denser fluorophores, techniques like SOFI [35] or SRRF [36] can also be conducted on TIRFM. Recently, Sankaran et al. combined FCS [37], SRRF, and TIRF to study EGFR in a multi-parametric manner [38]. They use mApple labeled EGFR expressed in Chinese hamster ovary (CHO)-K1 cell, which is free of native EGFR. Some mApple-EGFRs are uniformly distributed on the cell surface and others are found in clusters. FCS analysis indicates that the diffusion constant and oligomeric states are distinctive for these two kinds of EGFRs. By observing mApple-EGFR and eGFP labelled Lifeact [39] simultaneously, they do not see any correlation between EGFR diffusion and cytoskeletal structures. The integration of different single-molecule techniques provides powerful means to extract critical information from signaling molecules. We will discuss this further in the FCS section.
FRAP
Fluorescence recovery after photobleaching (FRAP) is a microscopy technique for measuring the diffusion and reaction properties of fluorescently labeled molecules based on bleaching and recovery of the fluorescent signals. It has be widely applied to monitor the location, mobility, interaction, and translation process of molecules on both in vitro and in vivo systems [40–42]. To perform FRAP, one would first densely saturate the observation field of view with labeled specimens, bleach a certain spot with intense laser illumination, and then closely monitor the process of other unbleached molecules diffusing back into the spot. Basically, the translation diffusion coefficient of the labeled molecules can be fitted from the time-dependent fluorescence intensity recovery curves generated by the molecules moving back into the focal volume of the laser beam. There are three common FRAP methods: conventional FRAP, multi-photon FRAP (MPFRAP) [43, 44], and FRAP with spatial Fourier analysis (SFA-FRAP) [45, 46]. With the development of super-resolution imaging and local-illumination technology, single-molecule FRAP (smFRAP) was newly developed and enables the researchers to measure the dynamics, spatial locations, and relative concentrations of proteins on single-molecule level. FRAP has been widely applied to study various membrane protein dynamics on the lipid bilayer. Particularly, the technique has been combined with two-photon microscopy to restrict the photobleaching area and provide a better spatiotemporal resolution [47]. By combining single-point illumination and single-molecule fluorescence recovery after photobleaching (smFRAP), Mudumbi KC et al. [48] demonstrated a single-point single-molecule FRAP microscopy technique that enables determination of distribution and translocation rates for nuclear envelope transmembrane proteins (NETs) in vivo with a spatial resolution of less than 10-nm in real-time.
The nuclear envelope (NE) consists of the outer nuclear membrane (ONM) and the inner nuclear membrane (INM) (Figure 2A). NE transmembrane proteins (NETs) are embedded in either the ONM or the INM, playing crucial roles in both nuclear structure and functions. In order to fully understand the functional mechanisms of NETs undergoing various signaling transduction processes, quantitative determination of the spatial locations of NETs along the NE and translocation rates between the two membranes is essential.
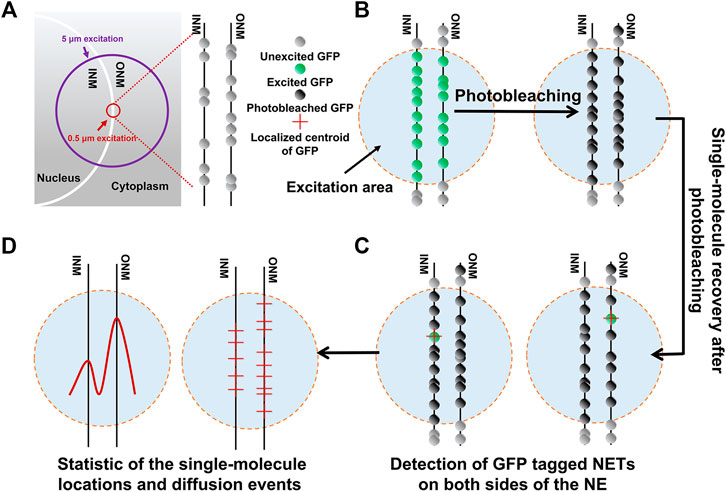
FIGURE 2. Study on NET distribution along the NE by single-point smFRAP. (A) Schematic of the GFP-tagged NET along the NE visualized in HeLa cell transfected system. The bright half circles represent the NE. The purple circle indicates the illumination area usually used in bulk FRAP experiments and the red circle indicates single-point illumination area limited to 0.5 μm used on single molecule level. (B) Photobleaching of both GFP-fused INM and ONM by excitation laser at a high power. (C) Single-molecule photorecovery of the unbleached NETs diffused into the laser excitation area again, as well as those that diffuse into the area are photobleached. (D) Locations and diffusion events of NETs along the ONM and INM of NET are compiled. Adapted from Ref. [48].
In Mudumbi KC et al., the single-point illumination by a high numerical aperture microscope objective was realized to generate a diffraction-limited illumination volume. First, the GFP-tagged NETs were quickly photobleached in the illumination area (Figure 2B). Then, the individual fluorescent GFP-NETs diffused into this photobleached area from outside the regions, the molecules were imaged with a regulated on-off laser excitation mode (Figure 2C). Finally, the two-dimensional super-resolution images showing all detected locations of GFP-NETs were reconstructed (Figure 2D). Through measuring the diffusion coefficients and the immobilized fractions of these NETs, the in vivo translocation rates and concentrations of NETs along the ONM and INM can be determined.
smFRET
Fluorescence resonance energy transfer (FRET) is widely applied to study PPIs both in vitro and in vivo [49]. It is one kind of dipole-dipole interactions between a pair of fluorophores occurring when the excitation spectrum of the acceptor overlaps with the emission spectrum of the donor. When donor and acceptor are in close proximity to each other, donor will transfer energy to acceptor resulting in acceptor emission. The energy transfer efficiency is related to the distance between donor and acceptor. The typical FRET distance between dipole-dipole centers is from 2 to 10 nm [50, 51].
In FRET experiments, the distance between two fluorophores is described by the FRET efficiency (E) as in the formula:
Here, R is the actual distance between two fluorophores. R0 is Förster radius defined by the nature of chosen FRET pair. When R equals to R0, the FRET efficiency is 0.5.
FRET efficiency can be experimentally measured as the fluorescence intensity ratio between the FRET pairs or as the fluorescence lifetimes of the donor with/without FRET. When detecting in vivo PPIs using FRET, we shall choose a FRET donor-acceptor pair with overlapping spectrums away from the emissions of cellular native pigments. For protein conformational changes measurement, one should choose a FRET pair with an R0 close to expected distances for best sensitivity. The FRET pair (donor and acceptor) must be carefully selected for the best experimental results.
Ha et al. [52] were the first to apply FRET to a single-molecule protein. Since then, rapid developments in microscopy and advanced fluorescent dyes allow researchers conduct single molecule FRET experiments as a routine in the laboratory. Besides TIRFM introduced in the previous section, confocal-based fluorescence lifetime imaging microscopes (FLIM) are also frequently used [53–55]. FLIM can detect smFRET events through both the intensities or lifetimes changes of fluorescence. Fluorescence intensity is more easily affected by the microenvironments around the fluorophore than fluorescence lifetime. Therefore, FLIM is better suited for smFRET detection in living cell with complex environment [56].
For a complete understanding of signaling pathways, it is desirable to have a real-time access to the activities of signal receptors and downstream signaling events, not only in vitro, but also in biologically relevant in vivo systems. Single-molecule FRET techniques are capable of realizing such detections owing to its characteristics of high sensitivity to small distance changes and time-scales matching the typical proteins dynamics and interactions.
G protein-coupled receptors (GPCRs) constitute the largest membrane superfamily involving in fundamental physiological processes of neurotransmission, immune reaction, behavioral regulation, and responses to external environments [57, 58]. As the cellular plasma membrane protein, GPCRs can transduce extracellular signals into the interior of the cell membrane, where downstream effector proteins are recruited and activated. In pharmacology, GPCRs signaling has been studied extensively, since GPCRs are the targets for as much as one-third of all therapeutic drugs today. Therefore, understanding how GPCRs works in signal transduction pathways is a significant and popular research area.
To study helix movements during GPCR activation, Gether et al. [59] labeled purified 2-adrenergic receptors (β2AR) with fluorophores to see if fluorescence signals would be influenced by receptor ligands. Their studies directly monitored conformational changes in a G-protein-coupled receptor and confirmed the notion of agonist-induced relative movements of helices 6 and 3. Using a series of β2ARs with a limited number of cysteines available for fluorophores labeling, they observed agonist-induced changes in fluorescence for labeled transmembrane helix 3 and helix 6. Using different ligands, the partial agonists caused only partial changes in the receptor fluorescence (for review, see Gether et al. [60]; Bissantz et al. [61]). In the first single-molecule conformational heterogeneity study of β2AR by Peleg et al. [62], the observation times were limited to a few milliseconds per molecule because the experiments were performed on diffusing molecules. In 2017, Lamichhane et al. [63] reconstituted labeled β2AR in nanodiscs, tethered them to a surface and monitored the structural changes of the G protein binding domain at single molecule level by TIRFM, which prolonged the observation time of individual molecules in the native-like environment of phospholipid nanodiscs, and uncovered the spontaneous transitions between two distinct inactive and active-like conformational states of G proteins.
Compared to in vitro experiments, in vivo smFRET assays have serval limitations due to the basic principle behind the technique: it is a spectrally dependent technique. It could be hard to distinguish fluorescent protein signals from autofluorescence background if the expression levels of the proteins are very low [64]. In addition, an external laser source may induce cell autofluorescence and the photobleaching of the donor and acceptor, which might interfere with the signal and lead to a serious problem when interactions are measured [65, 66]. In fact, with the development of super-resolution image and synthesis of new fluorescent probes, the signal to noise ratio have been improved, overcoming the interferences from autofluorescence.
Besides, GPCRs are known to form stable functional homodimers or heterodimers [67, 68], but the role of oligomeric status of class A and B receptors is debatable. The stoichiometry of the dimers versus higher-order oligomers is yet to be determined.
In 2021, Asher WB et al [69]. reported generally applicable method for using smFRET to detect and track transmembrane proteins diffusing within the plasma membrane of mammalian cells. They obtained the evidence for existences of receptor monomers, density-dependent dimers, and constitutive dimers, respectively. They also combine smFRET with FRAP to track individual complexes at high receptor density to address that dimerization is a rare event at low receptor concentrations. To perform the specific labeling of cell surface receptors, a self-labeling SNAPfast tag (SNAPf) was used to bind fluorophores covalently. They generated expression constructs that encode amino-terminally SNAPf-tagged (Sf)-mGluR2, which forms covalent disulfide-bonded receptor dimers, and showed that the receptor was functional. For single-molecule imaging, the CHO cell lines were used to express Sf-mGluR2 stably. The membrane-impermeant self-healing Lumidyne 555p and 655 dyes (Figure 3A) were chosen as the donor and acceptor fluorophores, respectively, because of their photostability as well as low levels of nonspecific labeling. In the smFRET-FRAP approach, active donor- and acceptor-labeled receptors in the TIRF-illumination field are selectively photobleached in TIRF mode (Figure 3B), producing an analysis region within the plasma membrane defined by the TIRF field. Unbleached acceptor- and donor-labeled receptors subsequently diffuse from the apical membrane outside the TIRF field into the analysis region, so that single molecules can be resolved and imaged under normal single-molecule TIRF imaging conditions (Figure 3C).
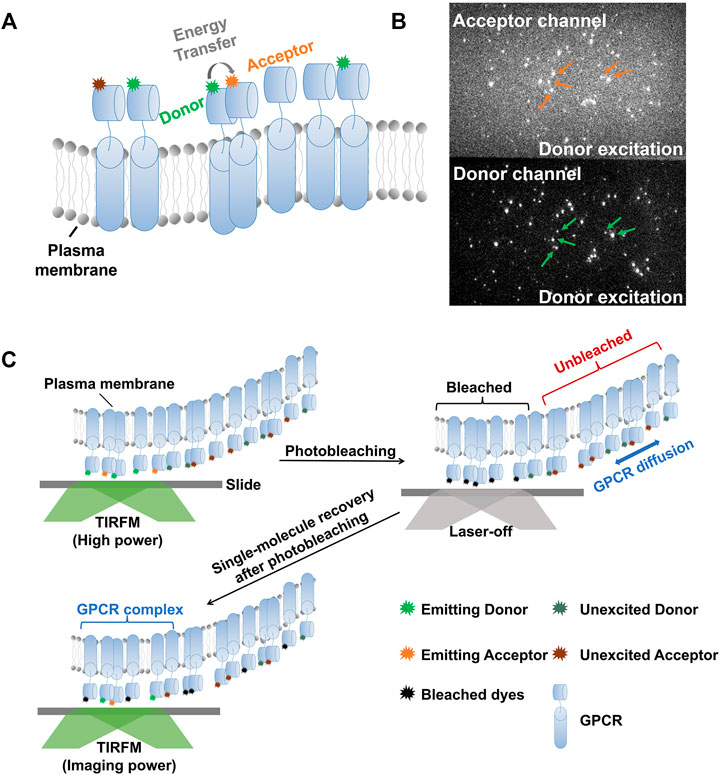
FIGURE 3. Schematics of smFRET combined with FRAP to study the dimerization of membrane proteins. (A) Label strategy for smFRET experiment to study dimerization of membrane proteins. When the membrane proteins (use GPCR as example, denoted by light bule cylinders) labeled with different fluorophores diffuse very close to each other, the acceptor will emit photons (orange stars) with energy transferred from donor (green star) upon donor excitation conditions, or else, the acceptor remain silent (dark orange stars). (B) Representative image of smFRET. The orange and green arrows denote the positions where both photons emitted by donor and acceptor can be collected, indicating FRET events. (C) Schematic of the smFRET-FRAP approach. Firstly, the active donor- and acceptor-labeled proteins in the TIRF-illumination field are selectively photobleached with high laser power. Both the donor and acceptor lasers are turn-on to make sure the thorough photobleaching. Then, unbleached acceptor- and donor-labeled receptors subsequently diffuse into the TIRF-illumination field from outside. Turn on the imaging laser i.e. donor excitation laser at lower power than bleaching one, and the dimerization can be monitored by FRET at single-molecule level.
According to the selection of FRET pairs, FRET can be classified into heteroFRET and homoFRET. HeteroFRET, i.e., conventional FRET described above, uses donor and acceptor labeled with different color fluorophores. Since the emission spectra of donor and acceptor overlap, quantification of molecules is hindered. Special procedures are required to avoid crosstalk and correction for bleed-through of the donor fluorescence into the acceptor channel is needed. On the contrary, homoFRET requires only a single fluorophore moiety for labelling. The labelling strategy is much easier to achieve, and the emission and excitation spectra overlap can be readily analyzed. However, traditional fluorescence intensity and lifetime analyses are not suitable for homoFRET. HomoFRET efficiency is detected via fluorescence anisotropy instead of fluorescence intensities.
HomoFRET has been widely used in complex in vivo experiments [70]. For example, Cameron W.D. et al. [71] studied NADPH-dependent antioxidant pathways in scavenging hydrogen peroxide (H2O2) produced by oxidative phosphorylation. To measure NADPH/NADP+ redox states, they explored genetically encoded sensors based on steady-state fluorescence anisotropy due to homoFRET between homologous fluorescent proteins, and created an Apollo sensor for NADP + called Apollo-NADP+. Using this sensor, they studied pancreatic beta cells responding to oxidative stress and demonstrated that NADPH is significantly depleted before H2O2 accumulation.
FCS/FCCS
Fluorescence correlation/cross-correlation spectroscopy (FCS/FCCS) is a powerful method for detecting diffusion, rotation, intersystem crossing, conformational changes, or other random effects when the fluorescent molecules diffuse through the observation volume based on correlating the fluctuations of the fluorescence intensity (Figure 4A) [72]. For strong signal of intensity fluctuations, the excitation volume should be limited to femtoliter level, making sure only a few molecules can be excited when diffusing through the excitation volume. Although it is not strictly single-molecule measurements, the detection usually reaches single-molecule level. FCS/FCCS is capable of detecting the fast dynamics of proteins, although the results are highly dependent on the data analysis models, which are usually multi-parametric. In FCS/FCCS analysis, the correlation of temporal fluctuations can be evaluated by
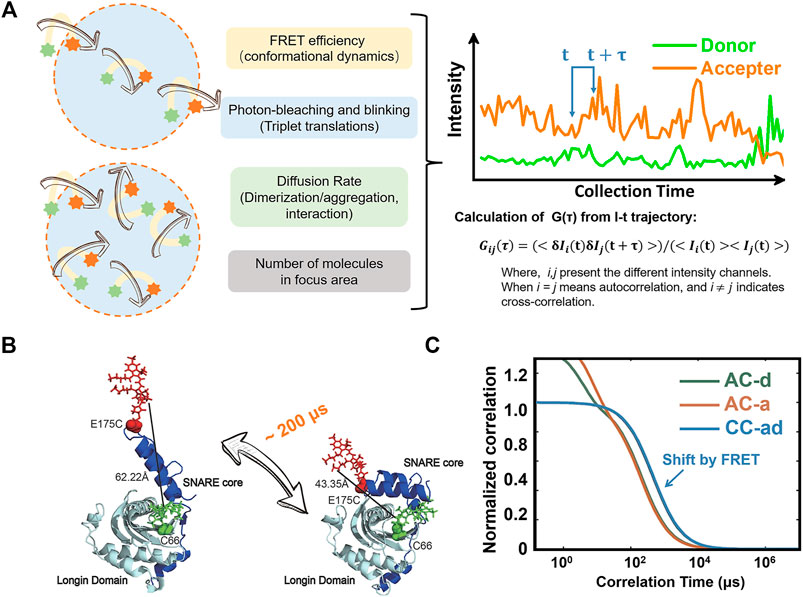
FIGURE 4. Sub-millisecond dynamics disclosed by smFRET-FCCS method. (A) Principle of FRET-FCCS [75, 76]. The time-dependent fluorescent intensity trajectories will fluctuate due to the labeled molecules diffusing in and out of the focus (light blue area in orange circle), the change of molecule numbers in focus area, photobleaching, blinking of fluorophores, and dynamics of proteins. Every course mentioned here can be described by a relevant model. (B) Experiment design of smFRET detection on Ykt6. (C) Auto- and cross- correlation traces in smFRET-FCCS experiment. The shift between auto- and cross- correlation traces denote that the conformational changes occurred during the diffussion through the focus areas. (AC-d: auto correlation trace of donor channel; AC-a: auto correlation trace of acceptor channel; CC-ad: cross- correlation trace of acceptor and donor channels). Panel (B,C) are adapted from Ref. [73].
For example, the interaction of lipids and proteins plays an important role in plasma membrane bioactivities and signaling transduction, much can be learned from their diffusion characteristics appropriately determined by FCS/FCCS method. Ykt6 plays a critical role in the membrane-trafficking process especially in brain neurons. Previous studies gave a potential model that an auto-inhibited conformation of single-lipidated Ykt6 existed, and a stable complex formed after the lipid dodecylphosphocholine (DPC) binding. However, the further details on alternative conformations or even the dynamics of Ykt6 protein was difficult to be resolved by traditional methods because of the small-but-fast conformational changes of rYkt6 upon DPC binding. In 2017, Dai Y. et al. [73] used the smFRET combined with FCS/FCCS to study the conformational state distributions and dynamics of Ykt6. In this work, the Ykt6 proteins were labeled at SNARE core and longin domain respectively (Figure 4B). Firstly, two discrete conformations of Ykt6 between the longin domain and the SNARE core were determined by smFRET under apo or saturated DPC conditions. And then, the dynamics between the longin domain and SNARE core with the increasing DPC was measured by FCCS (Figure 4C). Notably, owing to the parameters used to describe the static conformational information of Ykt6 (such as FRET efficiency, the population of different states etc.), which had already determined by smFRET experiment independently, the numbers of fitting parameters were reduced. Making the fitting of FCCS more robust. Finally, the protein dynamics on the microsecond time scale was measured quantitatively. The details on experiment performance and data analysis can be found in a previous protocol paper [74].
FCS/FCCS was also used to reveal the spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. In 2014, Honigmann A. et al. [77] used the beam-scanning STED-FCS to reveal the transient molecular interaction hotspots for a fluorescent sphingolipid analogue in the plasma membrane of live mammalian PtK2 cells. This is a method combining the technique of stimulated emission depletion (STED [78, 79]) super-resolution imaging with FCS and realizing on live cells. The interaction sites are smaller than 80 nm in diameter, and lipids are transiently trapped for several milliseconds in these areas. In their work, more homogenous diffusion of fluorescent phospholipid and cholesterol analogues with improved phase-partitioning properties were discovered. This phenomenon was independent of the preference for liquid-ordered or disordered membrane environments. Besides, compared with the single-point STED-FCS, the beam-scanning STED-FCS approach is capable of high sampling speed and can realize the detection of molecules at different positions and diffusing rates simultaneously.
Conclusion and Perspectives
In this review, we focused on the applications of single-molecule techniques in exploring various signaling pathways both in vitro and in vivo. The single-molecule techniques discussed here includes TIRF, FRAP, FRET, and FCS. Depending on the method used, the detection specialty includes but not limited to the conformational dynamics, oligomerization, PPIs, chemical modifications, and rates for each critical step.
In practice, in order to obtain more details during signal transduction, different single-molecule techniques are often used in combination. For example, smFRET assays, FCS, and FCCS can also be used to sensitively evaluate the dynamics and interactions of biomolecules. Combined with FRET, it can detect intramolecular conformational dynamics of proteins in a wide range of time scales [80, 81]. Benefiting from smFRET and FRET-FCCS methods, sub-milliseconds (∼200 μs) conformational dynamics measurements of the N-ethylmaleimide sensitive factor attachment protein receptors (SNARE) Ykt6 have been measured quantitatively, and the blocking effect after interacting with lipids has also been monitored by Dai et al. [73]. In addition, TIRF, SRRF, and FCS have been combined to study the localization and dynamics of EGFR [38]; smFRET and FRAP were combined to study the diffusive behavior of mGluR2 [69]; super-resolution STED and FCS were combined to study sphingolipid dynamics on live cell membranes [71], etc.
Besides, single-molecule manipulation techniques, such as AFM, are often applied to study on biomolecular interactions, mechanical property, and biochemical reaction kinetics with pulling force. However, various crucial details, such as the stoichiometry of active molecular interaction complex, the existence of functional internal conformations, as well as the activities of deformed states are not directly accessible via force measurements [82, 83]. To overcome these limitations, coupling with AFM, Lu’s group used smFRET to study the conformational change of 6-hydroxymethyl-7,8-dihydropterin protein kinase with specific coordination under mechanical pulling force [83].
From the examples illustrated above, we have seen a trend of combining several single-molecule fluorescent spectroscopy techniques to make the study of signal transduction more powerful and versatile. Table 1 lists some of the mainstream fluorescent single molecule techniques, together with their suitable applications and advantages. Through the combination of one or more techniques, it will be possible to monitor several parameters of different properties simultaneously and to further identify and characterize the signal transduction mechanisms under a precisely controlled experimental environment or during developmentally regulated events in living cells.
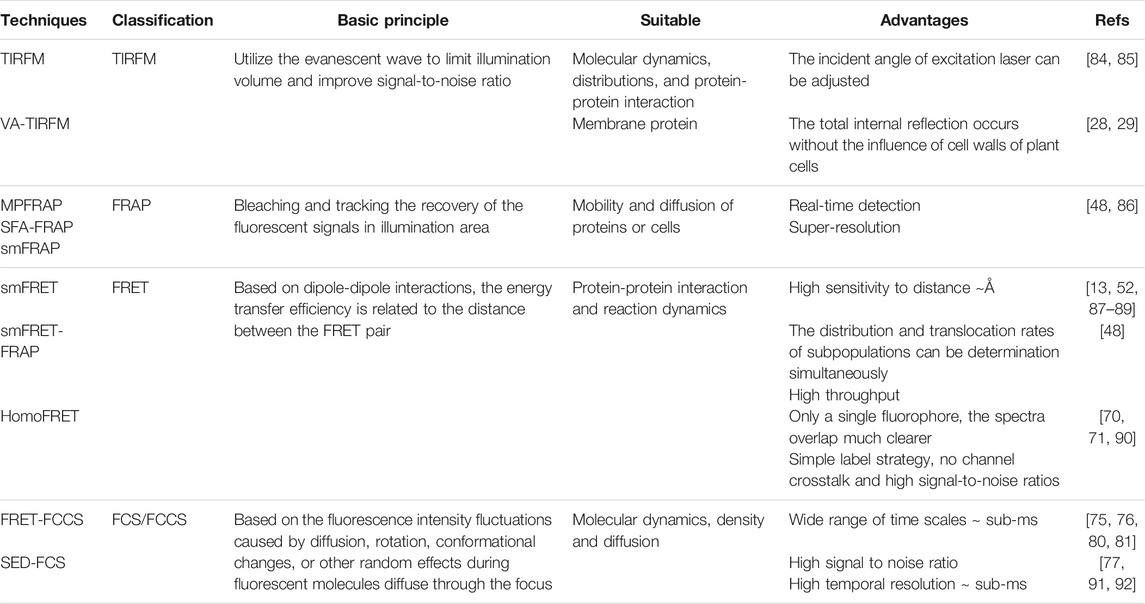
TABLE 1. Classification and advantages of different fluorescent single-molecule detection technique.
Author Contributions
PL and Y-WT conceived the manuscript. PL, TC and Y-WT wrote the drafts of manuscript and made the figures and table. PL and TC, edited the article. LC and Y-WT shared useful ideas and skills for writing.
Funding
The project is supported by the funding from National Natural Science Foundation of China (NSFC) (Grant numbers: No. 12174070, 21773039 and 12074341). Y-WT is also supported by Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX01), SCI & TECH Project (No. 20ZR1405800), and ZJLab.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank all the scientists whose results we discussed in this review and apologize to those whose works we omitted due to space limitations. We thank all members of the Y-WT lab for their inspiring input, helpful discussions, and continuous support. Thanks to Song Song and Yawei Dai again for sharing the data and figures obtained during working in Y-WT Lab.
References
1. Yang L, Xie X, Tu Z, Fu J, Xu D, Zhou Y. The Signal Pathways and Treatment of Cytokine Storm in COVID-19. Sig Transduct Target Ther (2021) 6:255. doi:10.1038/s41392-021-00679-0
2. Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell (2012) 149:274–93. doi:10.1016/j.cell.2012.03.017
3. Chen X, Ding Y, Yang Y, Song C, Wang B, Yang S, et al. Protein Kinases in Plant Responses to Drought, Salt, and Cold Stress. J Integr Plant Biol (2021) 63:53–78. doi:10.1111/jipb.13061
4. Castro B, Citterico M, Kimura S, Stevens DM, Wrzaczek M, Coaker G. Stress-induced Reactive Oxygen Species Compartmentalization, Perception and Signalling. Nat Plants (2021) 7:403–12. doi:10.1038/s41477-021-00887-0
5. Hossenlopp P, Seurin D, Segovia-Quinson B, Hardouin S, Binoux M. Analysis of Serum Insulin-like Growth Factor Binding Proteins Using Western Blotting: Use of the Method for Titration of the Binding Proteins and Competitive Binding Studies. Anal Biochem (1986) 154:138–43. doi:10.1016/0003-2697(86)90507-5
6. Poulter LW, Chilosi M, Seymour GJ, Hobbs S, Janossy G. Immunofluorescence Membrane Staining and Cytochemistry, Applied in Combination for Analysing Cell Interactions In Situ. In: JM Polak, and S Van Noorden, editors. Immunocytochemistry. Butterworth-Heinemann (1983). p. 233–48. doi:10.1016/B978-0-7236-0669-7.50020-9
7. Trautman JK, Macklin JJ, Brus LE, Betzig E. Near-field Spectroscopy of Single Molecules at Room Temperature. Nature (1994) 369:40–2. doi:10.1038/369040a0
8. Trautman JK, Macklin JJ. Time-resolved Spectroscopy of Single Molecules Using Near-Field and Far-Field Optics. Chem Phys (1996) 205:221–9. doi:10.1016/0301-0104(95)00391-6
9. Xie XS, Trautman JK. Optical Studies of Single Molecules at Room Temperature. Annu Rev Phys Chem (1998) 49:441–80. doi:10.1146/annurev.physchem.49.1.441
10. Kapanidis AN, Strick T. Biology, One Molecule at a Time. Trends Biochem Sci (2009) 34:234–43. doi:10.1016/j.tibs.2009.01.008
11. Iino R, Iida T, Nakamura A, Saita E-i., You H, Sako Y. Single-molecule Imaging and Manipulation of Biomolecular Machines and Systems. Biochim Biophys Acta (Bba) - Gen Subjects (2018) 1862:241–52. doi:10.1016/j.bbagen.2017.08.008
12. Okamoto K, Hiroshima M, Sako Y. Single-molecule Fluorescence-Based Analysis of Protein Conformation, Interaction, and Oligomerization in Cellular Systems. Biophys Rev (2018) 10:317–26. doi:10.1007/s12551-017-0366-3
13. Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing Gene Expression in Live Cells, One Protein Molecule at a Time. Science (2006) 311:1600–3. doi:10.1126/science.1119623
14. Ha T. Single-Molecule Fluorescence Resonance Energy Transfer. Methods (2001) 25:78–86. doi:10.1006/meth.2001.1217
15. Schafer DA, Gelles J, Sheetz MP, Landick R. Transcription by Single Molecules of RNA Polymerase Observed by Light Microscopy. Nature (1991) 352:444–8. doi:10.1038/352444a0
16. Lipfert J, Wiggin M, Kerssemakers JW, Pedaci F, Dekker NH. Freely Orbiting Magnetic Tweezers to Directly Monitor Changes in the Twist of Nucleic Acids. Nat Commun (2011) 2(1):439–10. doi:10.1038/ncomms1450
17. Neuman KC, Nagy A. Single-molecule Force Spectroscopy: Optical Tweezers, Magnetic Tweezers and Atomic Force Microscopy. Nat Methods (2008) 5(6):491–505. doi:10.1038/nmeth.1218
18. Zhang X, Wojcikiewicz EP, Moy VT. Dynamic Adhesion of T Lymphocytes to Endothelial Cells Revealed by Atomic Force Microscopy. Exp Biol Med (Maywood) (2006) 231:1306–12. doi:10.1177/153537020623100804
19. Thompson NL, Burghardt TP, Axelrod D. Measuring Surface Dynamics of Biomolecules by Total Internal Reflection Fluorescence with Photobleaching Recovery or Correlation Spectroscopy. Biophysical J (1981) 33:435–54. doi:10.1016/S0006-3495(81)84905-3
20. Clegg RM, Murchie AI, Zechel A, Lilley DM. Observing the Helical Geometry of Double-Stranded DNA in Solution by Fluorescence Resonance Energy Transfer. Proc Natl Acad Sci (1993) 90:2994–8. doi:10.1073/pnas.90.7.2994
21. Knox SL, Steinauer A, Alpha-Cobb G, Trexler A, Rhoades E, Schepartz A. Quantification of Protein Delivery in Live Cells Using Fluorescence Correlation Spectroscopy. In: DM Chenoweth, editor. Methods in Enzymology Chemical Tools for Imaging, Manipulating, and Tracking Biological Systems: Diverse Chemical, Optical and Bioorthogonal Methods. Academic Press (2020). p. 477–505. doi:10.1016/bs.mie.2020.05.007
22. Mattheyses AL, Simon SM, Rappoport JZ. Imaging with Total Internal Reflection Fluorescence Microscopy for the Cell Biologist. J Cel Sci (2010) 123:3621–8. doi:10.1242/jcs.056218
23. Axelrod D. Cell-substrate Contacts Illuminated by Total Internal Reflection Fluorescence. J Cel Biol (1981) 89:141–5. doi:10.1083/jcb.89.1.141
24. Axelrod D. [1] Total Internal Reflection Fluorescence Microscopy in Cell Biology. Methods Enzymol (2003) 361:1–33. doi:10.1016/s0076-6879(03)61003-7
25. Song S, Chang J, Ma C, Tan Y-W. Single-Molecule Fluorescence Methods to Study Plant Hormone Signal Transduction Pathways. Front Plant Sci (2017) 8:1888. doi:10.3389/fpls.2017.01888
26. Song S, Wang H, Sun M, Tang J, Zheng B, Wang X, et al. Reactive Oxygen Species‐mediated BIN 2 Activity Revealed by Single‐molecule Analysis. New Phytol (2019) 223:692–704. doi:10.1111/nph.15669
27. Friedman LJ, Chung J, Gelles J. Viewing Dynamic Assembly of Molecular Complexes by Multi-Wavelength Single-Molecule Fluorescence. Biophysical J (2006) 91:1023–31. doi:10.1529/biophysj.106.084004
28. Rohrbach A. Observing Secretory Granules with a Multiangle Evanescent Wave Microscope. Biophysical J (2000) 78:2641–54. doi:10.1016/S0006-3495(00)76808-1
29. Wan Y, Ash WM, Fan L, Hao H, Kim MK, Lin J. Variable-angle Total Internal Reflection Fluorescence Microscopy of Intact Cells of Arabidopsis thaliana. Plant Methods (2011) 7:27. doi:10.1186/1746-4811-7-27
30. Sako Y, Minoghchi S, Yanagida T. Single-molecule Imaging of EGFR Signalling on the Surface of Living Cells. Nat Cel Biol (2000) 2:168–72. doi:10.1038/35004044
31. Srinivasan S, Regmi R, Lin X, Dreyer CA, Chen X, Quinn SD, et al. Ligand-induced Transmembrane Conformational Coupling in Monomeric EGFR. bioRxiv (2021). doi:10.1101/2021.10.28.466294
32. Denisov IG, Sligar SG. Nanodiscs in Membrane Biochemistry and Biophysics. Chem Rev (2017) 117:4669–713. doi:10.1021/acs.chemrev.6b00690
33. Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, et al. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science (2006) 313:1642–5. doi:10.1126/science.1127344
34. Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nat Methods (2006) 3:793–6. doi:10.1038/nmeth929
35. Dertinger T, Colyer R, Iyer G, Weiss S, Enderlein J. Fast, Background-free, 3D Super-resolution Optical Fluctuation Imaging (SOFI). Pnas (2009) 106:22287–92. doi:10.1073/pnas.0907866106
36. Gustafsson N, Culley S, Ashdown G, Owen DM, Pereira PM, Henriques R. Fast Live-Cell Conventional Fluorophore Nanoscopy with ImageJ through Super-resolution Radial Fluctuations. Nat Commun (2016) 7:12471. doi:10.1038/ncomms12471
37. Schwille P, Korlach J, Webb WW. Fluorescence Correlation Spectroscopy with Single-Molecule Sensitivity on Cell and Model Membranes. Cytometry (1999) 36:176–82. doi:10.1002/(sici)1097-0320(19990701)36:3<176::aid-cyto5>3.0.co;2-f
38. Sankaran J, Balasubramanian H, Tang WH, Ng XW, Röllin A, Wohland T. Simultaneous Spatiotemporal Super-resolution and Multi-Parametric Fluorescence Microscopy. Nat Commun (2021) 12:1748. doi:10.1038/s41467-021-22002-9
39. Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, et al. Lifeact: a Versatile Marker to Visualize F-Actin. Nat Methods (2008) 5:605–7. doi:10.1038/nmeth.1220
40. Meyvis TKL, De Smedt SC, Van Oostveldt P, Demeester J. Fluorescence Recovery after Photobleaching: A Versatile Tool for Mobility and Interaction Measurements in Pharmaceutical Research. Pharm Res (1999) 16:1153–62. doi:10.1023/A:1011924909138
41. Rayan G, Guet J-E, Taulier N, Pincet F, Urbach W. Recent Applications of Fluorescence Recovery after Photobleaching (FRAP) to Membrane Bio-Macromolecules. Sensors (2010) 10:5927–48. doi:10.3390/s100605927
42. Houtsmuller AB. Fluorescence Recovery after Photobleaching: Application to Nuclear Proteins. In: J Rietdorf, editor. Microscopy Techniques. Berlin: Springer-Verlag Berlin (2005). p. 177–99. doi:10.1007/b102214
43. Brown EB, Wu ES, Zipfel W, Webb WW. Measurement of Molecular Diffusion in Solution by Multiphoton Fluorescence Photobleaching Recovery. Biophysical J (1999) 77:2837–49. doi:10.1016/S0006-3495(99)77115-8
44. Sullivan KD, Majewska AK, Brown EB. Single- and Two-Photon Fluorescence Recovery after Photobleaching. Cold Spring Harb Protoc (2015) 2015:pdb.top083519. doi:10.1101/pdb.top083519
45. Berk DA, Yuan F, Leunig M, Jain RK. Fluorescence Photobleaching with Spatial Fourier Analysis: Measurement of Diffusion in Light-Scattering media. Biophysical J (1993) 65:2428–36. doi:10.1016/S0006-3495(93)81326-2
46. Tsay TT, Jacobson KA. Spatial Fourier Analysis of Video Photobleaching Measurements. Principles and Optimization. Biophysical J (1991) 60:360–8. doi:10.1016/S0006-3495(91)82061-6
47. Coscoy S, Waharte F, Gautreau A, Martin M, Louvard D, Mangeat P, et al. Molecular Analysis of Microscopic Ezrin Dynamics by Two-Photon FRAP. Proc Natl Acad Sci (2002) 99:12813–8. doi:10.1073/pnas.192084599
48. Mudumbi KC, Schirmer EC, Yang W. Single-point Single-Molecule FRAP Distinguishes Inner and Outer Nuclear Membrane Protein Distribution. Nat Commun (2016) 7:12562. doi:10.1038/ncomms12562
49. Periasamy A, Day RN. Visualizing Protein Interactions in Living Cells Using Digitized GFP Imaging and FRET Microscopy. Methods Cel Biol (1999) 58:293–314. doi:10.1016/s0091-679x(08)61962-7
50. Förster T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann Phys (1948) 437:55–75. doi:10.1002/andp.19484370105
51. Förster T. Quantentheorie und chemische Bindung. Angew Chem (1949) 61:144–9. doi:10.1002/ange.19490610411
52. Ha T, Ting AY, Liang J, Caldwell WB, Deniz AA, Chemla DS, et al. Single-molecule Fluorescence Spectroscopy of Enzyme Conformational Dynamics and Cleavage Mechanism. Proc Natl Acad Sci (1999) 96:893–8. doi:10.1073/pnas.96.3.893
53. Brelje TC, Scharp DW, Sorenson RL. Three-dimensional Imaging of Intact Isolated Islets of Langerhans with Confocal Microscopy. Diabetes (1989) 38:808–14. doi:10.2337/diab.38.6.808
54. Becker W, Bergmann A, Biskup C, Kelbauskas L, Zimmer T, Klocker N, et al. High Resolution TCSPC Lifetime Imaging. High Resolution TCSPC Lifetime Imaging. in, eds. A Periasamy, and PTC So (San Jose, CA), 2003, 175. doi:10.1117/12.472866
55. Becker W, Bergmann A, Biskup C, Zimmer T, Kloecker N, Benndorf K. Multiwavelength TCSPC Lifetime Imaging. In: Multiphoton Microscopy in the Biomedical Sciences II. San Jose, CA: SPIE (2002). p. 79–84. doi:10.1117/12.470679
56. Datta R, Heaster TM, Sharick JT, Gillette AA, Skala MC. Fluorescence Lifetime Imaging Microscopy: Fundamentals and Advances in Instrumentation, Analysis, and Applications. J Biomed Opt (2020) 25:1. doi:10.1117/1.JBO.25.7.071203
57. Lefkowitz RJ. A Brief History of G-Protein Coupled Receptors (Nobel Lecture). Angew Chem Int Ed (2013) 52:6366–78. doi:10.1002/anie.201301924
58. Lagerström MC, Schiöth HB. Structural Diversity of G Protein-Coupled Receptors and Significance for Drug Discovery. Nat Rev Drug Discov (2008) 7:339–57. doi:10.1038/nrd2518
59. Gether U, Kobilka BK. G Protein-Coupled Receptors. J Biol Chem (1998) 273:17979–82. doi:10.1074/jbc.273.29.17979
60. Gether U. Uncovering Molecular Mechanisms Involved in Activation of G Protein-Coupled Receptors. Endocr Rev (2000) 21:90–113. doi:10.1210/edrv.21.1.0390
61. Bissantz C. Conformational Changes of G Protein‐Coupled Receptors during Their Activation by Agonist Binding. J Receptors Signal Transduction (2003) 23:123–53. doi:10.1081/rrs-120025192
62. Peleg G, Ghanouni P, Kobilka BK, Zare RN. Single-molecule Spectroscopy of the 2 Adrenergic Receptor: Observation of Conformational Substates in a Membrane Protein. Proc Natl Acad Sci (2001) 98:8469–74. doi:10.1073/pnas.151239698
63. Lamichhane R, Liu J, Pauszek III R, Millar D. Fluorophore Labeling, Nanodisc Reconstitution and Single-Molecule Observation of a G Protein-Coupled Receptor. Bio-protocol (2017) 7. doi:10.21769/bioprotoc.2332
64. Ishikawa-Ankerhold HC, Ankerhold R, Drummen GPC. Advanced Fluorescence Microscopy Techniques-FRAP, FLIP, FLAP, FRET and FLIM. Molecules (2012) 17:4047–132. doi:10.3390/molecules17044047
65. Ciruela F. Fluorescence-based Methods in the Study of Protein-Protein Interactions in Living Cells. Curr Opin Biotechnol (2008) 19:338–43. doi:10.1016/j.copbio.2008.06.003
66. De Los Santos C, Chang C-W, Mycek M-A, Cardullo RAFRAP, FLIMFRET:FRAP, FLIMFRET. Detection and Analysis of Cellular Dynamics on a Molecular Scale Using Fluorescence Microscopy. Mol Reprod Dev (2015) 82:587–604. doi:10.1002/mrd.22501
67. White JH, Wise A, Main MJ, Green A, Fraser NJ, Disney GH, et al. Heterodimerization Is Required for the Formation of a Functional GABAB Receptor. Nature (1998) 396:679–82. doi:10.1038/25354
68. Maurel D, Comps-Agrar L, Brock C, Rives M-L, Bourrier E, Ayoub MA, et al. Cell-surface Protein-Protein Interaction Analysis with Time-Resolved FRET and Snap-Tag Technologies: Application to GPCR Oligomerization. Nat Methods (2008) 5:561–7. doi:10.1038/nmeth.1213
69. Asher WB, Geggier P, Holsey MD, Gilmore GT, Pati AK, Meszaros J, et al. Single-molecule FRET Imaging of GPCR Dimers in Living Cells. Nat Methods (2021) 18:397–405. doi:10.1038/s41592-021-01081-y
70. Snell N, Rao V, Seckinger K, Liang J, Leser J, Mancini A, et al. Homotransfer FRET Reporters for Live Cell Imaging. Biosensors (2018) 8(4):89. doi:10.3390/bios8040089
71. Cameron WD, Bui CV, Hutchinson A, Loppnau P, Gräslund S, Rocheleau JV. Apollo-NADP+: a Spectrally Tunable Family of Genetically Encoded Sensors for NADP+. Nat Methods (2016) 13(4):352–8. doi:10.1038/nmeth.3764
72. Yu L, Lei Y, Ma Y, Liu M, Zheng J, Dan D, et al. A Comprehensive Review of Fluorescence Correlation Spectroscopy. Front Phys (2021) 9:110. doi:10.3389/fphy.2021.644450
73. Dai Y, Seeger M, Weng J, Song S, Wang W, Tan Y-W. Lipid Regulated Intramolecular Conformational Dynamics of SNARE-Protein Ykt6. Sci Rep (2016) 6:30282. doi:10.1038/srep30282
74. Li P, Dai Y, Seeger M, Tan Y-W. Quantifying Intramolecular Protein Conformational Dynamics under Lipid Interaction Using smFRET and FCCS. Methods Mol Biol (2019) 1860:345–59. doi:10.1007/978-1-4939-8760-3_23
75. Price ES, DeVore MS, Johnson CK. Detecting Intramolecular Dynamics and Multiple Förster Resonance Energy Transfer States by Fluorescence Correlation Spectroscopy. J Phys Chem B (2010) 114:5895–902. doi:10.1021/jp912125z
76. Price ES, Aleksiejew M, Johnson CK. FRET-FCS Detection of Intralobe Dynamics in Calmodulin. J Phys Chem B (2011) 115:9320–6. doi:10.1021/jp203743m
77. Honigmann A, Mueller V, Ta H, Schoenle A, Sezgin E, Hell SW, et al. Scanning STED-FCS Reveals Spatiotemporal Heterogeneity of Lipid Interaction in the Plasma Membrane of Living Cells. Nat Commun (2014) 5:5412. doi:10.1038/ncomms6412
78. Willig KI, Harke B, Medda R, Hell SW. STED Microscopy with Continuous Wave Beams. Nat Methods (2007) 4(11):915–8. doi:10.1038/nmeth1108
79. Sharma R, Singh M, Sharma R. Recent Advances in STED and RESOLFT Super-resolution Imaging Techniques. Spectrochimica Acta A: Mol Biomol Spectrosc (2020) 231:117715. doi:10.1016/j.saa.2019.117715
80. Eid JS. Two-photon Dual Channel Fluctuation Correlation Spectroscopy: Theory and Application. Ann Arbor, Ml: University of Illinois at Urbana-Champaign (2002). Available at: http://hdl.handle.net/2142/85426 Accessed: January 10, 2022.
81. Margittai M, Widengren J, Schweinberger E, Schroder GF, Felekyan S, Haustein E, et al. Single-molecule Fluorescence Resonance Energy Transfer Reveals a Dynamic Equilibrium between Closed and Open Conformations of Syntaxin 1. Proc Natl Acad Sci (2003) 100:15516–21. doi:10.1073/pnas.2331232100
82. Lu HP. Sizing up Single-Molecule Enzymatic Conformational Dynamics. Chem Soc Rev (2014) 43:1118–43. doi:10.1039/c3cs60191a
83. He Y, Lu M, Cao J, Lu HP. Manipulating Protein Conformations by Single-Molecule AFM-FRET Nanoscopy. ACS Nano (2012) 6:1221–9. doi:10.1021/nn2038669
84. Tokunaga M, Kitamura K, Saito K, Iwane AH, Yanagida T. Single Molecule Imaging of Fluorophores and Enzymatic Reactions Achieved by Objective-type Total Internal Reflection Fluorescence Microscopy. Biochem Biophysical Res Commun (1997) 235:47–53. doi:10.1006/bbrc.1997.6732
85. Reck-Peterson SL, Derr ND, Stuurman N. Imaging Single Molecules Using Total Internal Reflection Fluorescence Microscopy (TIRFM). Cold Spring Harb Protoc (2010) 2010:pdb.top73. doi:10.1101/pdb.top73
86. Tingey M, Li Y, Yang W. Protocol for Single-Molecule Fluorescence Recovery after Photobleaching Microscopy to Analyze the Dynamics and Spatial Locations of Nuclear Transmembrane Proteins in Live Cells. STAR Protoc (2021) 2:100490. doi:10.1016/j.xpro.2021.100490
87. McKinney SA, Joo C, Ha T. Analysis of Single-Molecule FRET Trajectories Using Hidden Markov Modeling. Biophysical J (2006) 91:1941–51. doi:10.1529/biophysj.106.082487
88. Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive Shuttling of a Motor Protein on DNA. Nature (2005) 437:1321–5. doi:10.1038/nature04049
89. Roy R, Hohng S, Ha T. A Practical Guide to Single-Molecule FRET. Nat Methods (2008) 5:507–16. doi:10.1038/NMETH.1208
90. Bastiaens PI, van Hoek A, Benen JA, Brochon JC, Visser AJ. Conformational Dynamics and Intersubunit Energy Transfer in Wild-type and Mutant Lipoamide Dehydrogenase from Azotobacter Vinelandii. A Multidimensional Time-Resolved Polarized Fluorescence Study. Biophysical J (1992) 63:839–53. doi:10.1016/S0006-3495(92)81659-4
91. Mueller VJ. Nanoscale Studies of Membrane Dynamics via STED- Fluorescence Correlation Spectroscopy (2012). doi:10.11588/heidok.00014199
92. Bianchini P, Cardarelli F, Luca MD, Diaspro A, Bizzarri R. Nanoscale Protein Diffusion by STED-Based Pair Correlation Analysis. PLoS One (2014) 9:e99619. doi:10.1371/journal.pone.0099619
Glossary
AFM atomic force microscopy
ATP adenosine-triphosphate
BR brassinosteroid
BES1 bri1-ems-suppressor 1
BIN2 brassinosteroid insensitive 2
Cy3 cyanine 3
Cy5 cyanine 5
Cy3-EGF cyanine 3-labeled epidermal growth factor
Cys cysteine
CHO chinese hamster ovary
Cy5-EGF cyanine 5-labeled epidermal growth factor
DNA deoxyribonucleic acid
DPC dodecylphosphocholine
EGFR epidermal growth factor receptor
EGF epidermal growth factor
ER endoplasmic reticulum
EMCCD electron multiplying charge coupled device
FLIM fluorescence lifetime imaging microscope
FRAP fluorescence recovery after photobleaching
FCS fluorescence correlation spectroscopy
FRET fluorescence resonance energy transfer
FRET-FCCS fluorescence resonance energy transfer combine with fluorescence correlation spectroscopy
FCCS fluorescence cross-correlation spectroscopy
GPCR G protein-coupled receptor
GFP green fluorescent protein
GFP-NET green fluorescent protein-labeled nuclear envelope transmembrane protein
INM inner nuclear membrane
NPC nuclear pore complex
NET nuclear envelope transmembrane protein
NE nuclear envelope
ONM outer nuclear membrane
PPI protein-protein interaction
TIRF total internal reflection fluorescence
TIRFM total internal reflection fluorescence microscopy
VA-TIRF variable-angle total internal reflection fluorescence
smFRET single-molecule fluorescence resonance energy transfer
PALM photoactivated localization microscopy
STORM stochastic optical reconstruction microscopy
sm single molecule
SMD single-molecule detection
SOFI super-resolution optical fluctuation imaging
SRRF super-resolution radial fluctuations
mApple-EGFR mApple labeled epidermal growth factor receptor
MPFRAP multi-photon fluorescence recovery after photobleaching
SFA-FRAP fluorescence recovery after photobleaching with spatial fourier analysis
smFRAP single-molecule fluorescence recovery after photobleaching
R0 the Förster distance between the donor and acceptor fluorophore at which FRET efficiency is 0.5
β2AR 2-adrenergic receptors
SNAPf SNAP fast tag
Sf SNAPf-tagged
mGluR2 metabotropic glutamate receptors 2
Sf-mGluR2 SNAPf-tagged metabotropic glutamate receptors 2
smFRET-FRAP single-molecule fluorescence resonance energy transfer combine with fluorescence recovery after photobleaching
SNARE soluble NSF attachment protein receptor
STED-FCS stimulated emission depletion combine with fluorescence correlation spectroscopy
Keywords: single-molecule, signal transduction, TIRF, single-molecule FRET, FRAP, FCS, protein-protein interaction
Citation: Li P, Chen T, Chen L and Tan Y-W (2022) Signal Transduction Mechanisms Quantitatively Observed One Molecule at a Time. Front. Phys. 10:855417. doi: 10.3389/fphy.2022.855417
Received: 15 January 2022; Accepted: 07 February 2022;
Published: 25 February 2022.
Edited by:
Mingxi Yao, Southern University of Science and Technology, ChinaReviewed by:
Guo Fu, Shanghai Institute of Optics and Fine Mechanics (CAS), ChinaQin Peng, Shenzhen Bay Laboratory, China
Copyright © 2022 Li, Chen, Chen and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-Wen Tan, eXd0YW5AZnVkYW4uZWR1LmNu
 Pei Li
Pei Li Ting Chen
Ting Chen Liang Chen
Liang Chen Yan-Wen Tan
Yan-Wen Tan